Abstract
Recent progress in molecular microbial ecology has revealed that traditional culturing methods fail to represent the scope of microbial diversity in nature, since only a small proportion of viable microorganisms in a sample are recovered by culturing techniques. To develop methods to investigate the full extent of microbial diversity, we used a bacterial artificial chromosome (BAC) vector to construct libraries of genomic DNA isolated directly from soil (termed metagenomic libraries). To date, we have constructed two such libraries, which contain more than 1 Gbp of DNA. Phylogenetic analysis of 16S rRNA gene sequences recovered from one of the libraries indicates that the BAC libraries contain DNA from a wide diversity of microbial phyla, including sequences from diverse taxa such as the low-G+C, gram-positive Acidobacterium, Cytophagales, and Proteobacteria. Initial screening of the libraries in Escherichia coli identified several clones that express heterologous genes from the inserts, confirming that the BAC vector can be used to maintain, express, and analyze environmental DNA. The phenotypes expressed by these clones include antibacterial, lipase, amylase, nuclease, and hemolytic activities. Metagenomic libraries are a powerful tool for exploring soil microbial diversity, providing access to the genetic information of uncultured soil microorganisms. Such libraries will be the basis of new initiatives to conduct genomic studies that link phylogenetic and functional information about the microbiota of environments dominated by microorganisms that are refractory to cultivation.
The biosphere is dominated by microorganisms (32), yet most microbes in nature have not been studied. Traditional methods for culturing microorganisms limit analysis to those that grow under laboratory conditions (14, 25). The recent surge of research in molecular microbial ecology provides compelling evidence for the existence of many novel types of microorganisms in the environment in numbers and varieties that dwarf those of the comparatively few microorganisms amenable to laboratory cultivation (7, 13, 31). Corroboration comes from estimates of DNA complexity and the discovery of many unique 16S rRNA gene sequences from numerous environmental sources (8, 10, 28). Collectively, the genomes of the total microbiota found in nature, which we termed the metagenome (11), contain vastly more genetic information than is contained in the culturable subset. Given the profound utility and importance of microorganisms to all biological systems, methods are needed to access the wealth of information within the metagenome.
Cloning large fragments of DNA isolated directly from microbes in natural environments provides a method to access soil metagenomic DNA. Previously, we investigated the use of the bacterial artificial chromosome (BAC) vector to express Bacillus cereus genomic DNA (20). The advantage of BAC vectors is that they maintain very large DNA inserts (greater than 100 kb) stably in Escherichia coli (23), facilitating the cloning of large fragments of DNA. Our results demonstrated that expression of heterologous DNA from B. cereus in an E. coli BAC system was detectable at a reasonable frequency (20), validating the idea that the low-copy BAC vector (one to two per cell) (23) could be used to express foreign DNA from foreign promoters in E. coli.
Here we describe the construction and initial screening of two BAC libraries made with DNA isolated directly from soil. We found detectable levels of several biochemical activities from BAC library clones. Sequence analysis of selected BAC plasmids encoding such activities and of 16S rRNA genes in one of the libraries confirms the novelty of the genomic information cloned in our libraries. The results show that DNA extracted directly from soil is a valuable source of new genetic information and is accessible by using BAC libraries. Our results demonstrate that both traditional and functional genomics of uncultured microorganisms can be carried out by this approach and that screening of metagenome libraries for activities or gene sequences can provide a basis for conducting genomic analyses of uncultured microorganisms.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strain DH10B and the BAC vector pBeloBAC11 were provided by H. Shizuya (15). Bacillus subtilis strain BR151(pPL608) is strain 1E32 (lys-3 metB10 trpC2) from the Bacillus Genetic Stock Center, Ohio State University. λ-TnphoA was used as described before (20).
DNA extraction from soil.
Soil was collected from the West Madison Agricultural Research Station in Madison, Wis. (4). Soil (5 g) was suspended in 10 ml of buffer (34), and incubated at 60°C for 2 h with occasional gentle shaking. Soil was sieved to remove particulates larger than 1 cm, and no plant roots were visible in the sample. The suspension was extracted with an equal volume of chloroform, and the DNA in the supernatant was precipitated with isopropanol (34). Precipitated DNA was dissolved in 500 μl of water and electrophoresed in a low-melting-temperature agarose gel (SEAPlaque; FMC Bioproducts). After electrophoresis at 100 V for 1 h, a strip from each side of the gel was cut off and stained to localize DNA. DNA-containing regions were cut from the rest of the gel (unstained) and stored overnight in 0.5× TE (Tris-EDTA) plus polyamines as described before (URL http://informa.bio.caltech.edu/idx_www_tree.html). In numerous experiments, this method produced DNA of appropriate size and cleanliness for subsequent cloning.
Library construction.
Two BAC libraries were constructed, designated SL1 and SL2. For constructing SL1, the isolated gel zone containing DNA was digested with Gelase (Epicentre, Madison, Wis.). Preparation of the vector pBeloBAC11, digestion of the insert DNA with HindIII, ligation, and transformation were done according to the above-cited URL and to Rondon et al. (20). White colonies were picked onto plates gridded to be compatible with 96-well microtiter plates. The library was replicated into duplicate sets of 96-well microtiter plates with freezing medium and stored at −80°C (33).
The protocol modifications used in construction of SL2 are as follows. Vector preparation was as described above but included a ligation step followed by gel purification to remove any self-ligated product. The pBeloBAC11 vector was subsequently electroeluted from the gel slice and dialyzed against 1× TE. Approximately 100 μg of metagenomic DNA was run on a preparative pulsed-field gel (Bio-Rad CHEFMapper; 0.5 s switch time, 9 V/cm, 0.5× TBE, 120° included angle, 5 h), and DNA greater than 50 kb was isolated, electroeluted, and dialyzed against 1× TE. Following HindIII digestion, insert DNA was loaded onto a second preparative gel and size-selected to retain DNA of 40 kb or larger. Ligation, transformation, and storage steps were performed as described above.
PCR amplification, cloning, and sequencing of 16S rRNA gene sequences from SL1.
Pools of 48 BAC clones were prepared, and BAC DNA was isolated from pooled cultures as described before (20). Details of the PCR protocol designed to amplify 16S rRNA gene sequences in the presence of contaminating E. coli DNA will be presented elsewhere (M. R. Liles, J. Handelsman, and R. M. Goodman, unpublished data). PCR amplification of 16S rRNA genes (50-μl reaction volume) used 100 ng of pooled BAC DNA, a 200 μM concentration of the four deoxynucleoside triphosphates, 2.5 U of Taq polymerase (Promega, Madison, Wis.), and 200 μM eubacterium-specific primers in conjunction with terminally modified E. coli-specific competitive oligonucleotides to preferentially amplify nonhost 16S rRNA genes. Reactions were performed in a Robocycler 96 (Stratagene, Inc., La Jolla, Calif.), using 1 min of denaturation at 94°C, then 40 cycles of 30 s at 94°C, 90 s at 58°C, and 150 s at 72°C, followed by 5 min of extension at 72°C. The presence of non-E. coli 16S rRNA product was determined by restriction digestion of PCR products with multiple enzymes, including AluI, HaeIII, and HinfI. Full-length PCR products containing unique restriction fragments were subsequently reamplified under the same PCR conditions, and the resulting product was cloned into the TA cloning vector pGEM-T (Promega). Sequence information from cloned 16S rRNA genes was obtained by using the T7 and SP6 primers in BigDye sequencing reactions and analyzed with an ABI model 377 automated sequencer at the University of Wisconsin-Madison Biotechnology Center. The resulting sequence was compared with the nonredundant sequence database at the National Center for Biotechnology Information (NCBI) using BLAST (1).
Library screening.
SL1 was replicated onto Q-trays (Genetix, Christchurch, Mass.) made for the Q-BOT (Genetix), each containing 250 ml of Luria-Bertani (LB) agar plus 10 μg of chloramphenicol (Cm) per ml using a 96-pin array. Individual screens were carried out on Q-trays containing assay-specific agar medium with Cm. Following replication onto the Q-trays, colonies were incubated for 3 days at 30°C before scoring phenotypes or performing overlays. For detecting β-lactamase activity, plates were overlaid with top agar containing 0.01% nitrocefin (Becton Dickinson Microbiological Systems, Cockeysville, Md.). A red precipitate surrounding the colony indicates activity. For detecting cellulase activity, plates were overlaid with top agar containing 0.1% Ostazin brilliant red hydroxyethyl cellulose (Sigma, St. Louis, Mo.), dissolved in sterile water. A yellow halo around the colony indicates cellulase activity. For detecting protease, keratinase, chitinase, and lipase activity, the library was replicated to plates containing LB agar plus 1% commercial nonfat dry milk, 0.5% keratin powder (ICN Biomedical, Los Angeles, Calif.), 0.5% chitin powder (Fluka, Buchs, Switzerland), or 3% Bacto Lipid (Difco, Detroit, Mich.), respectively, and scored after 3 days for the presence of a clear halo. Esterase activity was detected on LB plates containing 1% Tween 20 (Sigma) by monitoring formation of a powdery halo surrounding the colonies. Amylase activity was detected on Bacto Starch agar plates by flooding plates with Bacto Stabilized Gram iodine (Difco) after 3 days of growth; active colonies were surrounded by a bright orange halo. DNase activity was tested on Bacto DNase methyl green agar (Difco); positive colonies were surrounded by a bright orange halo. Siderophore activity was tested with standard CAS medium (21) containing an additional 0.1 mM FeSO4 and monitoring for production of an orange halo. In this medium, the background activity of strain DH10B is suppressed, but siderophore overproduction can still be detected. In all cases, DH10B/pBeloBAC11 was used as a negative control, and either control strains or purified enzymes were used as positive controls.
Screening of SL2 for hemolytic activity was done by overlaying 5 ml of blood agar on plates, followed by 2 days of incubation at 28°C. Blood agar contained LB-Cm medium with 7% (vol/vol) defibrinated sheep blood (Hemostat Laboratories, Dixon, Calif.).
All clones that were positive in an initial screen were retested at least once. The BAC plasmid was isolated and introduced by transformation into a fresh background, and the phenotype was rechecked to determine whether the cloned DNA was responsible for the phenotype.
Antibacterial screening.
Colonies were grown for 2 days at 37°C and overlaid with 5 ml of LB soft agar containing 0.5 ml of B. subtilis BR151(pPL608) grown in LB-Cm to an optical density at 600 nm of 0.2. Plates were then incubated overnight at 37°C and scored for activity by looking for a zone of inhibition in the B. subtilis lawn.
Transposon mutagenesis.
Mutagenesis of BAC clones with TnphoA (19) and sequencing from transposon ends were done as described before (20).
Sequencing clone SL1-36C7.
Clone SL1-36C7 was sheared by sonication for 5 s at 80% power using an ultrasonic homogenizer 4/10 series with microtip (Cole-Parmer, Chicago, Ill.). The ends of the DNA were blunted with T4 DNA polymerase (New England BioLabs, Beverly, Mass.). Fragments were ligated to the vector pCR-BLUNT (Invitrogen, Carlsbad, Calif.) according to the manufacturer's protocol and transformed into E. coli TOP10 cells. Transformants were plated onto LB agar containing 100 μg of kanamycin per ml. Colonies were picked into 96-deep-well plates (Marsh Biomedical Products, Inc., Rochester, N.Y.) and grown at 37°C for 16 h. Cultures were pelleted, and DNA was isolated using a Qiagen Biorobot 9600. DNA sequencing reactions were performed using Applied Biosystems dye terminator chemistry and analyzed on ABI 377 machines. The sequence data generated provided sixfold coverage of the DNA.
Sequence manipulation and alignment of clone SL1-36C7.
The DNA sequence was assembled using the program Sequencher on a Macintosh G3 personal computer. Open reading frame (ORF) analysis was performed using EditSeq, in which ORFs greater than 100 bp were identified. Putative ORFs were translated and queried using BLAST against the NCBI nonredundant protein database. Additional annotation was obtained using PSI-BLAST (1). The sequence of the ORF responsible for antibacterial activity has GenBank accession no. AF246145.
Subcloning the antibacterial ORF from SL1-36C7.
The ORF was amplified using primers 5′-CATATGTCTTTCATGAAACGGTTTTTCTGT-3′, encoding an NdeI site at the 5′ end, and 5′-CTCGAGCCTCGTAGAGTTGGGTTTGCC-3′, encoding an XhoI site at the 3′ end, using the original BAC SL1-36C7 clone as a template. Amplified DNA was ligated to NdeI- and XhoI-digested pET22b (Novagen, Madison, Wis.). The resulting construct encoded the antibacterial ORF with a hexahistidine tag on the 3′ end. The plasmid was transformed into E. coli strain BL21(DE3) (Novagen). Transformants were tested for antibacterial activity by overlaying with B. subtilis as described above.
Protein expression and purification.
The recombinant antibacterial gene was expressed by growing cells in 4xYT medium (32 g of tryptone, 20 g of yeast extract, and 5 g of NaCl per liter) with Cm and inducing with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) or by letting the cells leak overnight without IPTG (9). The tagged protein was purified using Talon metal affinity resin (Clontech).
RESULTS
Construction of metagenomic BAC libraries.
To access genomic information from as large a pool of soil microbes as possible, including those that are not readily culturable, we developed methods to extract and clone large DNA fragments from soil. We chose soil from a site near Madison, Wis., that had been characterized previously and found to contain a diverse community of bacteria and archaea (4, 8, 10). We used this DNA to construct metagenomic libraries in pBeloBAC11. We prepared two libraries, designated SL1 and SL2.
Molecular characteristics of SL1 and SL2.
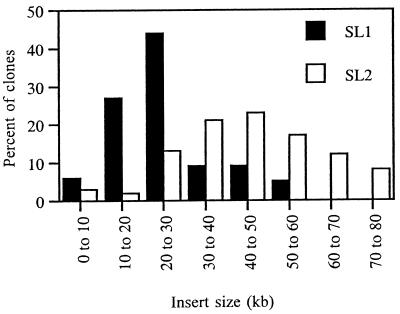
SL1 is a prototype metagenome library and consists of 3,648 clones arrayed in 38 96-well microtiter plates. We examined approximately 2% (n = 81) of the clones for inserts; 97% contained insert DNA, with an average insert size of 27 kb (Fig. 1). We estimated that there is approximately 100 Mbp of DNA contained in SL1. Based on restriction digest analysis, the clones fell into two classes: those with NotI sites within the insert, and those with no internal NotI sites. Given that the recognition sequence of NotI is 5′-GCGGCCGC-3′, this suggests that the library contains DNA that varies in GC content. As extraction and cloning steps may contribute to biases in the DNA represented in the library, it is important to monitor indicators of diversity.
FIG. 1.
Representation of insert size range in the soil metagenome libraries. Clones within a range of 10 kb are grouped together. For SL1, n = 81; for SL2, n = 132.
We constructed a second soil library (SL2), implementing several improvements to the method used for SL1. SL2 contains 24,576 clones, with an average insert size of 44.5 kb, in which greater than 60% of the inserts are larger than 40 kb, based on analysis of 132 clones (0.5% of total) (Fig. 1). We estimated that SL2 contains 1,000 Mbp of DNA; given an average of 1 kb per gene, SL2 might contain one million genes. These statistics demonstrate that improvements to our original method resulted in a library that contains considerably more metagenomic DNA with a larger average insert size.
Phylogenetic analysis of SL1.
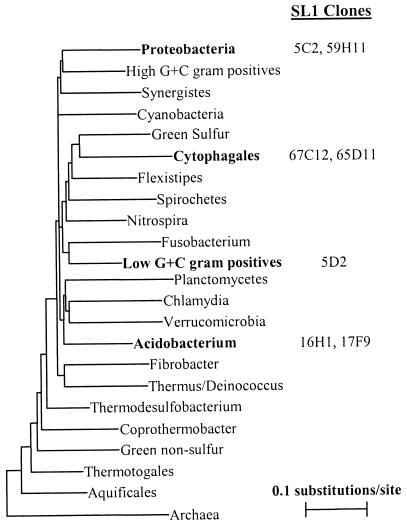
To begin to link physiological function and phylogenetic analysis of uncultured microorganisms, we developed a method to amplify 16S rRNA gene sequences from BAC plasmid preparations from pooled cultures in the presence of contaminating E. coli genomic DNA. Once a positive was identified from a given pool of 48 BAC clones, the clones from that pool were examined individually to identify the clones carrying a 16S rRNA gene. This suggested that the sequence was encoded on a BAC clone and not the result of a contaminant in the PCR process. Final confirmation of the 16S rRNA gene sequence will come from sequence analysis of the individual BAC clones. We recovered seven sequences from SL1, listed in Table 1, that fall into four different bacterial phyla (Fig. 2). These data revealed that our methods for DNA extraction and cloning successfully recover DNA from widely diverse prokaryotes, including gram-positive bacteria.
TABLE 1.
16S rRNA gene sequences obtained from SL1
| SL1 clone | Insert size (kb) | Phylogenetic group | % Identity to closest relative (no. of bp used) | GenBank accession no. |
|---|---|---|---|---|
| 5C2 | 23 | Proteobacteria | 100, Caulobacter sp. strain FWC21 (1,437) | AF245033 |
| 5D2 | 36 | Low G+C, gram positive | 94, Paenibacillus kobensis (1,119) | AF245034 |
| 16H1 | 50 | Acidobacterium | 96, clone RB41 (1,437) | AF245035 |
| 17F9 | 30 | Acidobacterium | 99, clone 32-21 (1,543) | AF245036 |
| 59H11 | 76 | Proteobacteria | 96, Porphyrobacter sp. (1,462) | AF245037 |
| 65D11 | 27 | Cytophagales | 90, Haliscomenobacter hydrossis (920) | AF245038 |
| 67C12 | 37 | Cytophagales | 89, Hymenobacter roseosalivarius (905) | AF245039 |
FIG. 2.
Phylogenetic placement of 16S rRNA sequences from SL1. Neighbor-joining analysis on 75 sequences from the represented phyla were used to construct the tree.
Interestingly, the acidobacteria sequences that we recovered were more similar to 16S rRNA gene sequences identified by culture-independent methods than to 16S rRNA gene sequences from cultured species in this phylum (Table 1). These clones offer the opportunity to further investigate the biology of these organisms, which are perhaps refractory to cultivation, by sequence analysis of the entire BAC insert and by functional analysis of the genes encoded therein.
Screening SL1 and SL2 for biological activity.
To begin to investigate the functional diversity of the metagenomic DNA captured in the BAC libraries and to identify clones expressing metagenomic DNA, we carried out initial screens of SL1 for gene expression on solid medium. We found clones expressing DNase (one clone), antibacterial (one clone), lipase (two clones), and amylase (eight clones) activities. Clones expressing cellulase, chitinase, esterase, keratinase, protease, or hemolytic activity or siderophore production were not found. In all cases, BACs were isolated from the putative positive clones and retransformed into DH10B; the resultant transformants were retested to confirm that the activity was encoded on the BAC insert. These results show that SL1 contains heterologous DNA sequences that can be expressed in E. coli at detectable levels. The fact that 4 of the 12 activities screened were identified in SL1 suggests that this method can be used successfully to extract and identify useful genetic information from environmental DNA. In the only screen of SL2, for hemolytic activity, we identified 29 active clones. Further screening of SL2 is expected to yield other interesting activities.
Characterization of active clones from SL1. (i) DNase-producing clone.
The DNase-producing clone SL1-11G4 contains an insert of approximately 25 kb in size, as estimated by restriction digestion followed by agarose gel analysis (not shown). One transposon insertion that abolished activity was located in a potential ORF with homology to a family of single-strand-specific nucleases typified by S1 nuclease from Aspergillus oryzae (17) and including sequences of plant (e.g., Hemerocallis, Hordeum, and Zinnia), fungal (Aspergillus and Penicillium), protozoal (Leishmania), and bacterial (Mesorhizobium) origin (3, 6, 27). Extended sequence analysis of the region (not shown) identified a complete ORF belonging to this family. The predicted amino acid sequence of the protein from SL1-11G4 was most similar to the nucleotidase from Leishmania donovani (6), with a similarity score of 10−14 (1). Residues important for activity that are conserved in other members are also conserved in the SL1-11G4 sequence (not shown) (3). We have not yet sequenced other regions of SL1-11G4 to determine the likely origin of this DNA fragment.
(ii) Antibacterial clone.
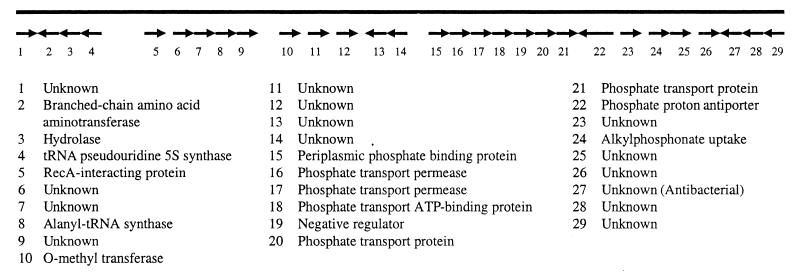
One clone (SL1-36C7) produced an activity that was inhibitory to B. subtilis, weakly active against Staphylococcus aureus, and not active against E. coli. The insert DNA of SL1-36C7 was completely sequenced (Fig. 3); the fragment appears to be of bacterial origin, given the homology of potential genes on the insert to genes of known function. Notable in the clone is the presence of a gene cluster with similarity to the phosphate transport cluster (pstCAB-phoU) of E. coli (30). This demonstrates the potential for BAC clones to contain complete, intact operons.
FIG. 3.
Schematic diagram of sequence analysis of clone SL1-36C7. Putative ORFs are indicated by the arrows; below is a list of the ORFs with the protein to which each is most similar. The potential operon includes ORFs 15 to 21. The antibacterial ORF is number 27.
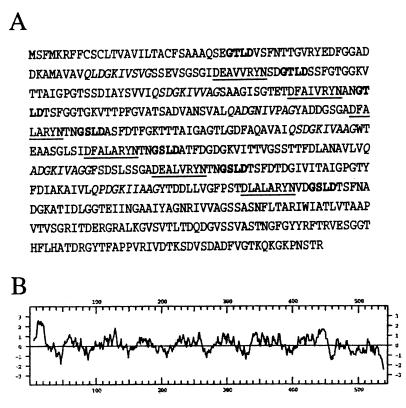
We identified the locus responsible for the antibacterial activity by insertional inactivation via transposon mutagenesis. A single candidate gene appeared to encode the activity. The nucleotide sequence of this gene showed no similarity to known sequences, suggesting that the gene encoded a protein of novel structure. The predicted protein contains a putative amino-terminal signal sequence and at least seven long sequence repeats (Fig. 4A). The hydrophobicity plot of the predicted amino acid sequence is characteristic of a membrane protein (Fig. 4B). The gene encoding this putative ORF was cloned individually into expression plasmid pET22b as a hexahistidine-tagged construct. When transformed into E. coli expression strain BL21(DE3), a new protein of approximately 55 kDa was produced (not shown). The subcloned gene conferred antibacterial activity on the host strain, confirming that this gene was sufficient to produce the inhibitory activity. However, the partially purified protein was not itself active.
FIG. 4.
(A) Predicted amino acid sequence of the antibacterial ORF. The predicted protein has 543 residues. Highly conserved sequences within the repeats are in bold, underlined, or italics to highlight the repeats. (B) Kyte-Doolittle hydrophobicity profile of the antibacterial ORF.
To investigate whether the antibacterial activity was due to a diffusable molecule, we fractionated cells and cell growth medium in an attempt to isolate the active substance. Antibacterial activity was consistently detected on undisturbed agar on which clone SL1-36C7 had been cultured. However, we were unable to extract or concentrate a clone-specific activity from liquid or agar cultures, using a variety of growth conditions, filtration steps, organic extractions, and pH variations. This suggests that the activity is not due to a diffusable small molecule; rather, some aspect of the protein itself, or its effect upon the host cell, is likely responsible for the inhibitory activity.
(iii) Amylase-producing, lipase-producing, and hemolytic clones.
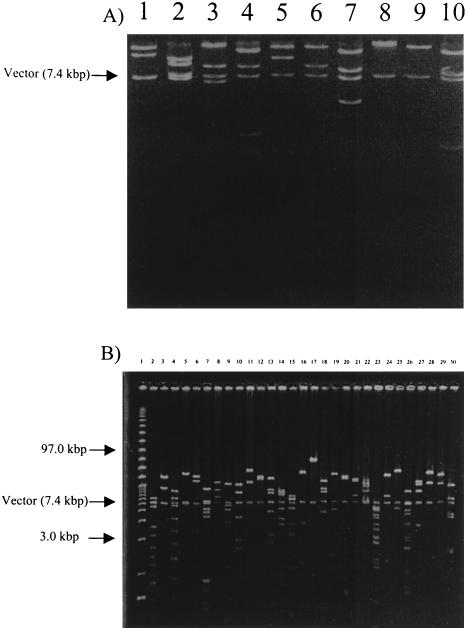
We identified eight clones that produce amylase activity and two lipase-producing clones from SL1 and 29 hemolytic clones from SL2. Restriction digest analysis of these clones (Fig. 5) suggests that they result from independent cloning events and are not the result of duplicate clones. The variety of restriction patterns demonstrates the molecular diversity of DNA cloned in the BAC libraries.
FIG. 5.
(A) Restriction digest analysis of amylase and lipase clones from SL1 digested with HindIII. The arrow indicates the BAC vector. Lanes 1 to 8, amylase clones; lanes 9 and 10, lipase clones. (B) Restriction digests of hemolytic clones from SL2, digested with NotI. The arrow indicates the BAC vector. Lane 1, size markers.
DISCUSSION
The probable magnitude of the soil metagenome, encompassing the collective genomes of microbes in soil, requires large-scale approaches for analysis, and its inaccessibility via traditional methods demands approaches that are culture independent. Our results demonstrate the feasibility of cloning environmental DNA into BAC libraries maintained in E. coli, an approach that is both large scale and independent of culturing methods. This approach provides a route to study the phylogenetic, physical, and functional properties of the metagenome. We readily detected gene expression from foreign DNA cloned into the low-copy BAC vector in a small library (SL1) containing only 100 Mbp of DNA. We also constructed a much larger library (SL2) using improved methods, demonstrating that construction of metagenomic BAC libraries on a large scale is possible.
The direct cloning approach described here provides a route to expand the investigation of soil microbial diversity to the majority of microbes that may not be cultivatable by standard methods. The fact that the antibacterial and nuclease clones we identified were of novel sequence supports the prediction that BAC libraries of environmental DNA provide a source of novel genes.
We recovered several BAC clones containing 16S rRNA gene sequences (Table 1 and Fig. 2). Phylogenetic inference (including analysis of genes other than rRNA-encoding genes) is a crucial step in metagenome analysis, as BAC libraries can serve as a link between phylogeny of uncultured soil microbes and their physiological and genetic activities encoded on metagenomic fragments captured in BAC libraries.
Of particular interest to us is the identification of two clones in SL1 containing 16S rRNA genes from members of the recently described acidobacterium division. Sequences from this phylum are frequently recovered in soil diversity studies (16, 18), indicating the widespread and abundant nature of this group in soil, although only three cultured strains have been identified as belonging to this division. Identification of BAC clones containing DNA from this intriguing phylum will enable us to analyze further the biology of these microbes by sequence and functional analysis of these clones.
Other methods to extract functional sequences from environmental DNA have focused on PCR-based methods to access novel gene fragments (22) and on cloning small fragments into high-copy expression vectors or lambda phage cloning vectors (5, 12, 24). Complementary to this has been a hybridization approach to identify novel 16S rRNA gene sequences from large cloned fragments of environmental DNA, followed by sequence analysis to identify other genes on these clones (26). The BAC method is distinct from these in that it combines cloning large fragments of DNA for phylogenetic and genomic analysis with screening clones directly for functional gene expression, taking advantage of the unique properties of the BAC vector. This method complements existing approaches to exploit the genetic diversity of noncultured microbes. Since many soil microbes produce important secondary metabolites and other useful products, and genes required for secondary metabolite production along with the accessory resistance and regulatory genes are often clustered in one contiguous segment on the chromosome of the producer organism (29), BAC libraries offer a new source for natural-product discovery.
Most of the story of microbiology is based on the pure-culture technique. While leading to remarkable discoveries, this story is now seen to be incomplete in light of accumulating evidence that culturing provides poor access to many microorganisms in the environment (2, 25). Discovery of novel 16S rRNA gene sequences from environmental samples provides a window into a world of microbial diversity that is astonishing in its magnitude and breadth (13). The challenge we have begun to address here is to develop methods to move beyond cataloging 16S rRNA gene sequences toward an understanding of the physiology and functional roles of microbes in nature.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (grants 1 RO3 AI42786-01 to J.H. and R.M.G. and CA24487 to J.C.), National Research Service Award 1 F32 GM18871-01A1 to M.R.R., Cellular and Molecular Biology Training Grant GM07237 to S.F.B., and the McKnight Foundation and the University of Wisconsin-Madison (University-Industry Research Program and the College of Agricultural and Life Sciences).
We thank Brian Manske for preparation of Fig. 2; Brad Borlee for help in storage and propagation of SL1; Doreen Gillespie, Jennifer Wendland, Andrea Borchardt, Amy Klimowicz, and Christian Reisenfeld for help with SL2; Dennis Holt for helpful discussions and support; and Doreen Gillespie for a critical reading of the manuscript.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffeer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoyagi S, Sugiyama M, Fukuda H. BEN1 and ZEN1 cDNAs encoding S1-type DNases that are associated with programmed cell death in plants. FEBS Lett. 1998;429:134–138. doi: 10.1016/s0014-5793(98)00563-8. [DOI] [PubMed] [Google Scholar]
- 4.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cottrell M T, Moore J A, Kirchman D L. Chitinases from uncultured marine microorganisms. Appl Environ Microbiol. 1999;65:2553–2557. doi: 10.1128/aem.65.6.2553-2557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debrabant A, Gottleib M, Dwyer D M. Isolation and characterization of the gene encoding the surface membrane 3′-nucleotidase/nuclease of Leishmania donovani. Mol Biochem Parasitol. 1995;71:51–63. doi: 10.1016/0166-6851(95)00035-y. [DOI] [PubMed] [Google Scholar]
- 7.Giovannoni S J, Britschgi T B, Meyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 8.Goodman R M, Bintrim S B, Handelsman J, Quirino B F, Rosas J C, Simon H M, Smith K P. A dirty look: soil microflora and rhizosphere biology. In: Flores H E, Lynch J P, Eissenstat D, editors. Radical biology: advances and perspectives on the function of plant roots. Rockville, Md: American Society of Plant Physiologists; 1997. pp. 219–231. [Google Scholar]
- 9.Grossman T H, Kawasaki E K, Punreddy S R, Osburne M S. Spontaneous cAMP-dependent derepression of gene expression in stationary phase plays a role in recombinant expression instability. Gene. 1998;209:95–103. doi: 10.1016/s0378-1119(98)00020-1. [DOI] [PubMed] [Google Scholar]
- 10.Handelsman J, Rondon M R, Bintrim S B, Goodman R M. The agroecosystem as a source of microbial metabolites. Dev Ind Microbiol. 1998;37:49–52. [Google Scholar]
- 11.Handelsman J, Rondon M R, Brady S F, Clardy J, Goodman R M. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 12.Henne A, Daniel R, Schmitz R A, Gottschalk G. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl Environ Microbiol. 1999;65:3901–3907. doi: 10.1128/aem.65.9.3901-3907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugenholtz P, Pace N R. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 1996;14:190–197. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 15.Kim U-J, Birren B B, Slepak T, Mancino V, Boysen C, Kang H-L, Simon M I, Shizuya H. Construction and characterization of a human bacterial artificial chromosome library. Genomics. 1996;34:213–218. doi: 10.1006/geno.1996.0268. [DOI] [PubMed] [Google Scholar]
- 16.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee R R, Kitamoto K, Yamada O, Kumagi C. Cloning, characterization and overproduction of nuclease S1 gene (nucS) from Aspergillus oryzae. Appl Microbiol Biotechnol. 1995;44:425–431. doi: 10.1007/BF00169939. [DOI] [PubMed] [Google Scholar]
- 18.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmenn A, Schleifer K H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Ecol. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 19.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rondon M R, Raffel S J, Goodman R M, Handelsman J. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc Natl Acad Sci USA. 1999;96:6451–6455. doi: 10.1073/pnas.96.11.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 22.Seow K-T, Meurer G, Gerlitz M, Wendt-Pienkowski E, Hutchinson C R, Davies J. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J Bacteriol. 1997;179:7360–7368. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Short J M. Recombinant approaches for accessing biodiversity. Nat Biotechnol. 1997;15:1322–1323. doi: 10.1038/nbt1297-1322. [DOI] [PubMed] [Google Scholar]
- 25.Staley J T, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 26.Stein J L, Marsh T L, Wu K Y, Shizuya H, DeLong E F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan J T, Ronson C W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torsvik V, Sorheim R, Goksøyr J. Total bacterial diversity in soil and sediment communities—a review. J Ind Microbiol. 1996;17:170–178. [Google Scholar]
- 29.Vining L C. Secondary metabolism, inventive evolution and biochemical diversity—a review. Gene. 1992;115:135–140. doi: 10.1016/0378-1119(92)90551-y. [DOI] [PubMed] [Google Scholar]
- 30.Wanner B L. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1357–1381. [Google Scholar]
- 31.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 32.Whitman W B, Coleman D C, Wiebe W J. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo S-S, Jiang J, Gill B S, Paterson A H, Wing R A. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 1994;22:4922–4931. doi: 10.1093/nar/22.23.4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]