Abstract
A binary classification of macrophage activation as inflammatory or resolving does not capture the diversity of macrophage states observed in tissues. However, framing macrophage activation as a continuous spectrum of states over-looks the intracellular and extracellular networks that regulate and coordinate macrophage responses. Here, we suggest that the systems biology concept of network motifs, which incorporate rules of local molecular interactions, is useful for reframing macrophage activation. Because network motifs can be used to regulate distinct biological functions, they offer a simplified unit that can be compared across organismal, tissue, and disease contexts. Moreover, defining macrophage states as combinations of functional modules regulated by network motifs offers a framework to ultimately predict and target macrophage responses arising in complex environments.
Evolution of macrophage classification: from binary to a continuum of states
Macrophages are innate immune cells that perform a wide range of functions, from fighting pathogens and tumors, to assisting in wound healing, to maintaining tissue homeostasis [1]. To accomplish this, macrophages tailor their functional responses to meet the demands of their current microenvironment. Establishing a framework for defining macrophage activation states in specific biological contexts is helpful to understand macrophage biology and to identify points of dysregulation in disease. Of note, some key macrophage functions directly conflict with other functions (e.g., secreting proinflammatory versus anti-inflammatory cytokines), which is interesting as it suggests that a simplified framework might be sufficient to explain such activation states, despite the diversity of their functional tasks.
The canonical example of a simplified framework is the M1–M2 paradigm of mammalian macrophage activation (Figure 1A, Key figure). The M1 program was originally defined in murine macrophages based on the increased microbicidal activity induced by IFN-γ (see Glossary) [2]. M1 stimuli have expanded to include the bacterial endotoxin lipopolysaccharide (LPS) and are grouped by their ability to induce aspects of a proinflammatory response. In contrast, the M2 program was defined based on increased endocytosis in response to IL-4 [3]. Typical M2 stimuli have expanded to include IL-13 and IL-10, among others, and are generally associated with wound healing and helminth responses [4,5]. Although a useful framework, particularly for in vitro research, broader application of the M1–M2 classification has obscured the connection to the macrophage functions it originally described.
Moreover, a binary classification does not reflect the complexity of macrophage states observed in vivo. To address the shortcomings of the M1–M2 categories, alternate frameworks have been proposed, such as defining macrophage states as a blend of their homeostatic functional activities encompassing host defense, wound healing, and immune regulation [6]. To standardize nomenclature across diverse experimental scenarios, a recommendation was made to define the initial state of the macrophage, the stimulation cues, and the markers used to classify the response [5]. Using these criteria, a systems-level transcriptome-based analysis of human monocyte-derived macrophages (MDMs) in 299 different contexts that varied the initial macrophage state and the combinations of stimuli, concluded that macrophages exhibit at least nine distinct macrophage activation programs, thus extending the M1–M2 model to include a spectrum of states [7]. Several additional systems-level studies using transcriptomic, proteomic, and epigenomic approaches in both mice and humans added detail to the spectrum of macrophage states observed in vitro [8,9].
Around this time, single-cell RNA sequencing (scRNA-seq) was developed and rapidly adopted by immunologists. With scRNA-seq, it is possible to take snapshots of individual macrophages isolated directly from in vivo environments (e.g., normal tissue, tumors, wound beds, and infected lungs). These snapshots confirmed extensive variability between macrophages in tissues, where cues were highly multiplexed and varied in space and time. Binary M1–M2 classification was increasingly rejected in favor of a spectrum of states, with a focus on defining transcriptional signatures to identify macrophage subsets within complex tissues that could be compared across biological contexts [10–13]. However, while transcriptional signatures are useful, they do not contribute to an updated framework for defining macrophage states based on functional responses, motivating a different approach.
Key figure
Two views of mammalian macrophage activation
Figure 1.
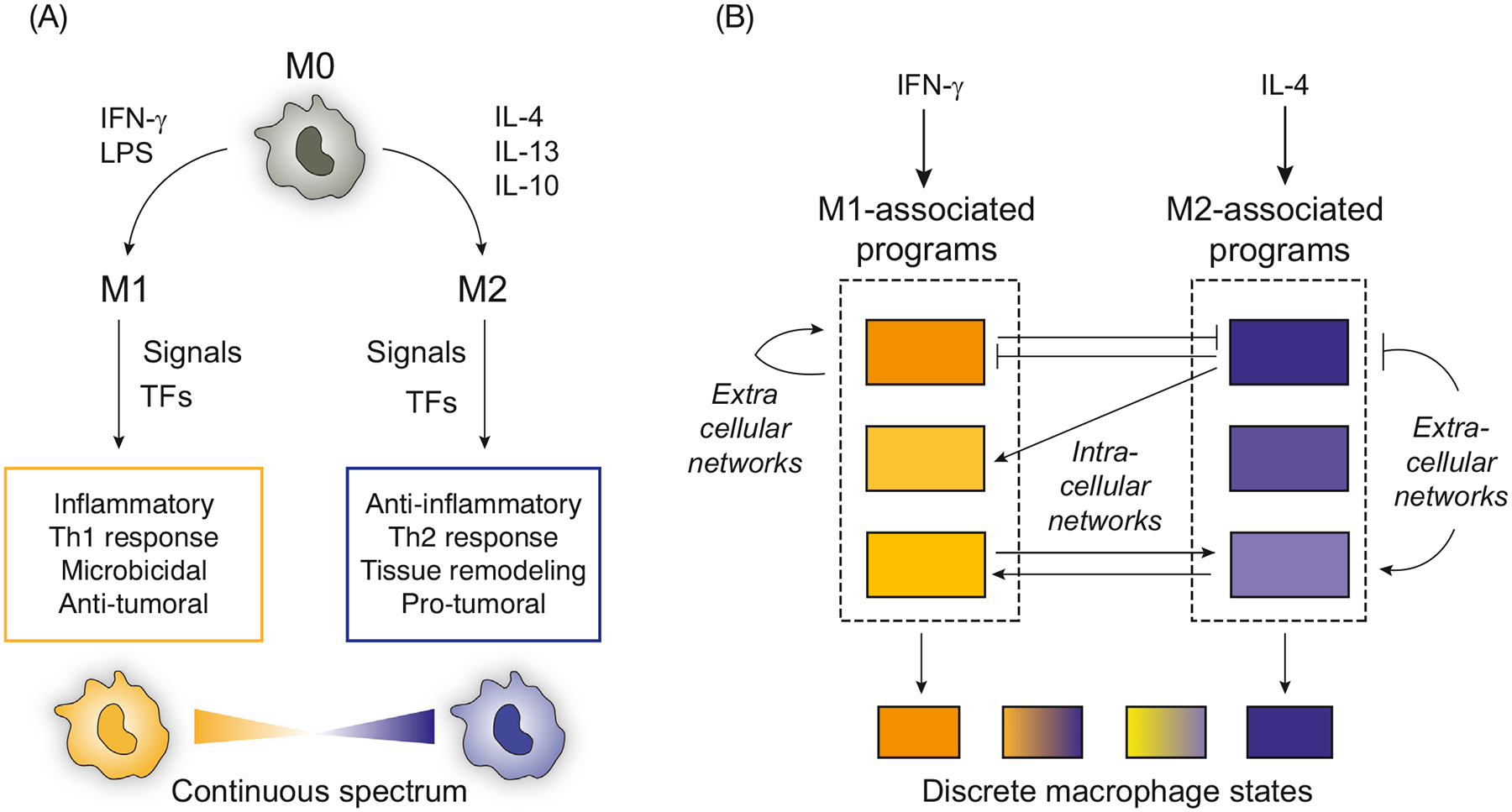
(A) A traditional framework for macrophage activation defines M1 and M2 at extreme ends of a spectrum of macrophage states. (B) By contrast, a systems biology view can reframe macrophage activation in response to defined stimuli as a set of interacting intracellular and extracellular network motifs that regulate distinct functions. Macrophage states might be defined as combinatorial sets of functions that can more flexibly describe diversity in different contexts. Abbreviations: LPS, lipopolysaccharide; TF, transcription factor.
When less is more: from a continuum of states to a combination of functional modules
While the binary M1–M2 dichotomy is oversimplified, a continuum of macrophage states over-looks the underlying regulatory networks revealed in decades of experimental studies. Because macrophages continually sense and respond to varying cues in their microenvironment, the near-continuous spectrum of states observed in tissues might result from their frequent transitions between states (an artifact of their own plasticity). Rather, the signaling, transcriptional, and metabolic programs that regulate macrophages may in fact place limits on the number of states they occupy. By considering which macrophage states emerge from these underlying regulatory networks, it might be possible to generate a new framework to describe macrophage activation.
Systems biology combines ‘omics approaches, which provides a list of the parts (e.g., genes, proteins, and even cells), with theory and modeling to predict how those parts will interact to determine responses. In a systems biology framework, biological functions emerge from modules of biological components; the boundaries of these modules are defined by the degree to which the functional module is separable from other functional modules [14,15]. Modules for each task often follow regulatory principles referred to as network motifs, which can explain how cells use continuous molecular interactions to switch between functional responses [16]. Sigmoidal signal response curves, mutual activation and inhibition, and positive and negative feedback are network motifs that enable cells to turn functional modules ‘off’ and ‘on’.
We propose that defining macrophage activation states as a combination of functional modules (e.g., secretion, microbial killing, and phagocytosis) that are regulated by underlying network motifs offers a potentially useful framework to compare macrophages across biological contexts. As a first step, we highlight examples of how network motifs enable switch-like transitions, combinatorial activity, and stability in macrophage functions even in complex environments (Figure 1B). As with the original M1 and M2 classifications, most of these examples were worked out in murine bone marrow-derived macrophages (BMDMs), but the network motif framework provides a means of comparing these signal–response relationships across organisms and disease contexts.
Switch-like activation and positive feedback can regulate a proinflammatory macrophage state
To defend against pathogens, macrophages trigger an inflammatory state, but too much inflammation may cause tissue damage. To carefully regulate these responses, murine BMDMs exhibit switch-like activation of the inflammatory program in response to stimulation of the pattern-recognition receptor toll-like receptor 4 (TLR4) with increasing doses of Kdo2-lipid A (KLA), the essential component of LPS in most gram-negative bacteria [17]. This means that at low doses of KLA (e.g., doses in the range of homeostatic bacterial titers), macrophages remain in a resting state, but once a critical dose threshold is reached, macrophages abruptly activate inflammatory functions. This switch-like response is due to ultrasensitivity in the mitogen-activated protein kinase (MAPK) signaling networks downstream of TLR4, a three-kinase motif that is conserved across species, enabling abrupt changes in phosphorylation in response to small changes in input [18]. Consistent with this observation, human MDMs showed a conserved ultrasensitivity of TLR4-induced tumor-necrosis factor alpha (TNF) production and the TLR4 ligand threshold for TNF production varied little across individuals [17]. Overall, this mechanism illustrates how network motifs can encode functional states with near-discontinuous transitions such that macrophages can be classified as ‘on’ or ‘off’ with respect to TLR4-mediated proinflammatory functions.
Positive feedback is another common network motif used to stabilize two divergent functional states, as has been observed in proinflammatory activation in macrophages. In a murine macrophage cell line, TLR4-activated transcription factor (TF) NF-kB has positively regulated itself to further stabilize the proinflammatory state [19]. In a human macrophage cell line, when paracrine signaling from the TLR4-activated inflammatory cytokine TNF-a was blocked, overall proinflammatory secretion decreased, demonstrating extracellular positive feedback [19,20]. Such observations are consistent with the network theory that switch-like responses and positive feedback can combine to create discontinuous, bistable states in biological systems [21].
Mutual inhibition in intracellular networks allows macrophages to respond to complex environments
Macrophages in tissues often encounter conflicting cues and therefore require network motifs that can coordinate functional responses in these complex environments. Mutual inhibition in biochemical networks is a common network motif for generating distinct and stable responses to two, often opposing, inputs. Consistent with this, in murine BMDMs, co-stimulation with IFN-γ and IL-4 mutually inhibited some of the changes induced by each cytokine alone, as measured by RNA-seq and chromatin immunoprecipitation followed by sequencing (ChIP-seq) [22]. This mechanism is orchestrated, in part, by cross-regulation of TFs. For instance, IFN-γ-stimulated genes (ISGs) regulated by TFs STAT1 and IRF1 are resistant to IL-4 inhibition; however, some IFN-γ-target genes contain binding sites for auxiliary TFs, such as AP-1, that are inhibited by IL-4 [22]. This means that macrophages exposed to both cues would be expected to simultaneously exhibit some IFN-γ and IL-4 markers, while others would be significantly reduced. Understanding the co-stimulated IFN-γ and IL-4 macrophage state as a combination of mutually inhibited versus uninhibited functional modules is more precise than simply classifying macrophages as a mixed M1–M2 state.
Inspired by this study in macrophage populations, we used scRNA-seq to explore how individual murine BMDMs responded to co-stimulation with LPS + IFN-γ and IL-4. As expected, opposing stimuli led to both unaffected and mutually exclusive gene expression (defined as having an odds ratio < 1, P < 0.05) [23]. LPS + IFN-γ-stimulated Il12b and Il6 were not coexpressed with IL-4-stimulated Arg1 and Chil3 in individual macrophages, suggesting that these functional modules were subject to mutual inhibition. This pattern was also observed between the TFs Nfkbiz and Klf4, which positively regulate Il6 and Il12b, and Arg1 and Chil3, respectively, indicating that mutual inhibition might be enforced via TFs, as found in another study that indicated that KLF4 negatively regulates Il12b and Il6 [24]. Importantly, BMDMs in a single-cell secretion microwell assay exhibited mutually exclusive secretion of IL-12p40 and IL-6 versus Chi3l3 at 48 h post-stimulation with LPS + IFN-γ and IL-4, suggesting that mutual inhibition could produce a distinct combination of functional subsets of macrophages in response to opposing cues [23].
Extracellular circuits and negative feedback stabilize macrophage functions in homeostasis and disease
Regulatory network motifs can also be established via extracellular paracrine signals between macrophages and other cell types. An elegant example is the circuit established between macrophages and fibroblasts, two cell types that exist in most mammalian tissues [25]. When cocultured in vitro, murine BMDMs promote fibroblast proliferation via platelet-derived growth factor-β (PDGF-β) and, in turn, embryonic fibroblasts induce the proliferation of macrophages via the growth factor macrophage-colony stimulating factor 1 (CSF1) [25]. Mathematical modeling and experimental validation demonstrated that negative feedback makes the circuit robust to perturbations: CSF1 receptor internalization maintains stability in the macrophage cell number [26], while fibroblasts are limited by the carrying capacity of the growth environment.
The overall stability of the circuit relied on cell–cell contacts between macrophages and other cell types that can be identified in a complex tissue. The relevance of such simplified two-cell circuit motifs was explored in a recent study that computationally mapped cell–cell interactions from human breast cancer scRNA-seq data to reveal a hierarchical network of repeating two-cell circuit motifs, structured similarly to the interactions described earlier. Specifically, tumor-associated macrophages and cancer-associated fibroblasts exhibited the strongest interaction score [27]. Of note, when the two-cell circuit was reconstructed in vitro with primary murine fibroblasts and BMDMs, it recapitulated the hierarchy of in vivo interactions, which is interesting because it provides evidence that functions emerging from small circuit motifs can be preserved and mapped from human in vivo, to murine in vitro contexts.
From network motifs to predictive models of macrophage activation states
The earlier examples illustrate how identifying network motifs in macrophage responses help to define functional modules that can be compared across organisms, tissues, and diseases. Although connecting these motifs and modules into larger networks that define macrophage states across varied contexts is not straightforward, we argue that computational modeling is the most promising approach and network motif patterns provide structure in these larger models. For example, a large-scale, logic-based differential equation computational model was developed to predict macrophage activation in response to single and pairwise stimulation of nine cytokine inputs [28]. The model, which was calibrated and validated using published datasets and transcriptional data from murine macrophages, showed that combinations of conflicting stimuli, including IL-4 with LPS and IFN-γ, produced mutual inhibition of several signaling pathways [28]. In another example, a mass action-based mechanistic model calibrated against human and mouse experimental data from the literature also predicted IFN-γ and IL-4 antagonism that resulted in a spectrum of macrophage responses [29].
Limitations of the proposed framework
One challenge in applying this framework is that the time scale over which macrophage functions change varies widely, from highly stable functions associated with tissue homeostasis to more transiently activated inflammatory functions associated with acute stress. These differences in time scales could obscure the mapping of functional modules to macrophage states. Another challenge is that macrophages in different contexts will respond differently to the same inputs, even if the underlying network motif is conserved. In other words, the history of the macrophage state can impact how new cues change the cell’s functional responses. One approach to address this is to integrate mechanistic models connecting macrophage signaling to responses with data-driven models of transcriptomic and proteomic data that define the microenvironmental context [30]. A complementary approach is to define markers of macrophage activation for particular stimuli and measure how these signatures change with microenvironmental context, thus acquiring information about the underlying macrophage functional state [31].
Concluding remarks
The broad M1–M2 classification has become disconnected from the biological networks that regulate macrophage activation and it does not precisely describe the diversity of macrophage subsets observed across healthy and diseased tissues. In this opinion piece, we highlighted examples of network motifs that regulate specific macrophage functions and are conserved across biological contexts. We argue that they may be a useful starting point for defining macrophage activation states. However, given the complexity of macrophages across biological contexts (see Outstanding questions), we envision that network motifs can serve as a bridge to the development of computational models connecting functional modules and which can be calibrated with systems-level measurements; these may be applied to predict how macrophage states change relative to new biological contexts.
Outstanding questions.
How stable are the functions that emerge from network motifs when cues are changing in space and time? Can some aspects of function remain stable while other are reversible?
How much do responses from macrophages differentiated from bone marrow or blood monocytes differ from those of tissue-resident macrophages?
If we rely on the M1-inflammatory versus M2-resolving model baseline as a reference, will this limit our ability to effectively define and predict macrophage states?
What is the relative importance of intrinsic versus extrinsic motifs in regulating macrophage functions in tissues?
Could defining network motifs mathematically and combining them in silico push forward the development of predictive models of innate immunity in complex environments?
Highlights.
Systems-level ‘omics approaches and single-cell analyses have expanded our knowledge of macrophage diversity across tissues, exposing the limits of the M1 ‘inflammatory’ versus M2 ‘resolving’ binary classification and suggesting a spectrum of states.
Both the binary and the spectrum framework are disconnected from the intracellular and extracellular networks that regulate macrophage functional responses.
Recent studies have demonstrated that macrophage functions are regulated by conserved molecular interactions called network motifs, suggesting that macrophage states could be effectively described as a combinatorial set of functions.
Evidence that network motifs encode conserved functional modules in macrophages could further motivate the use of larger computational models of regulatory networks to predict macrophages states across varied biological contexts.
Significance.
Macrophages perform diverse functions across tissues and must adapt to changing demands in complex environments. Here, the concept of biological network motifs is proposed to help define macrophage functional modules according to the molecular interactions that regulate them. These modules could provide building blocks to help describe the diversity of macrophage states observed in healthy and diseased tissues.
Acknowledgments
We thank the anonymous reviewers for useful advice in writing this opinion. Research in our laboratory is supported by grants from the National Institutes of Health (2R01GM123011 and 1U01CA238728) and the National Science Foundation (MCB2231765).
Glossary
- IFN-γ
major cytokine produced by type 1 CD4+ T helper cells (Th1 cells); usually associated with inflammation and cell-mediated immunity; canonical stimulus of the ‘M1-like’ macrophage response
- IL-4
major cytokine produced by type 2 CD4+ T helper cells (Th2 cells); usually associated with humoral immune responses; canonical stimulus of the ‘M2-like’ macrophage response
- Macrophage-colony stimulating factor 1 (CSF1)
or M-CSF; lineage-specific growth factor for macrophages
- Mutual inhibition
a network motif in which two species or biological pathways negatively regulate each other
- Negative feedback
a network motif in which activation of a reaction or biological process leads to inhibition of the same process
- Network motifs
local interactions that adhere to regulatory principles; they define modules that are the basic building blocks of complex networks
- Positive feedback
a network motif in which activation of a reaction or biological process further amplifies the same process
- Toll-like receptor 4 (TLR4)
receptor for the endotoxin (e.g., lipopolysaccharide or LPS) produced by gram-negative bacteria that mediates an antibacterial inflammatory response. It is considered a prototypical stimulus of the ‘M1-like’ macrophage response
- Ultrasensitivity
describes an output response that is more sensitive to stimulus change than the hyperbolic response associated with Michaelis-Menten enzyme kinetics
Footnotes
Declaration of interests
No interests are declared.
References
- 1.Murray PJ (2017) Macrophage polarization. Annu. Rev. Physiol 79, 541–566 [DOI] [PubMed] [Google Scholar]
- 2.Nathan CF et al. (1983) Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med 158, 670–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stein M et al. (1992) Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J. Exp. Med 176, 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez FO and Gordon S (2014) The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray PJ et al. (2014) Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosser DM and Edwards JP (2008) Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol 8, 958–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue J et al. (2014) Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong AJ et al. (2016) A stringent systems approach uncovers gene-specific mechanisms regulating inflammation. Cell 165, 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jha AK et al. (2015) Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 [DOI] [PubMed] [Google Scholar]
- 10.Ma RY et al. (2022) Macrophage diversity in cancer revisited in the era of single-cell omics. Trends Immunol. 43, 546–563 [DOI] [PubMed] [Google Scholar]
- 11.Zilionis R et al. (2019) Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dick SA et al. (2022) Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci. Immunol 7, eabf7777 [DOI] [PubMed] [Google Scholar]
- 13.Ghosh P et al. (2023) Machine learning identifies signatures of macrophage reactivity and tolerance that predict disease outcomes. EBioMedicine 94, 104719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartwell LH et al. (1999) From molecular to modular cell biology. Nature 402, C47–C52 [DOI] [PubMed] [Google Scholar]
- 15.Atay O and Skotheim JM (2014) Modularity and predictability in cell signaling and decision making. Mol. Biol. Cell 25, 3445–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyson JJ et al. (2003) Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol 15, 221–231 [DOI] [PubMed] [Google Scholar]
- 17.Gottschalk RA et al. (2016) Distinct NF-kappaB and MAPK activation thresholds uncouple steady-state microbe sensing from anti-pathogen inflammatory responses. Cell Syst. 2, 378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang CY and F JE, Jrerrell JE Jr (1996) Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. U. S. A 93, 10078–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung MH et al. (2014) Switching of the relative dominance between feedback mechanisms in lipopolysaccharide-induced NF-kappaB signaling. Sci. Signal 7, ra6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue Q et al. (2015) Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses to TLR4 stimulation. Sci. Signal 8, ra59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrell JE (1996) Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem. Sci 21, 460–466 [DOI] [PubMed] [Google Scholar]
- 22.Piccolo V et al. (2017) Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat. Immunol 18, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munoz-Rojas AR et al. (2021) Co-stimulation with opposing macrophage polarization cues leads to orthogonal secretion programs in individual cells. Nat. Commun 12, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao X et al. (2011) Kruppel-like factor 4 regulates macrophage polarization. J. Clin. Invest 121, 2736–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X et al. (2018) Circuit design features of a stable two-cell system. Cell 172, 744–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adler M et al. (2018) Endocytosis as a stabilizing mechanism for tissue homeostasis. Proc. Natl. Acad. Sci. U. S. A 115, E1926–E1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer S et al. (2023) The tumor microenvironment shows a hierarchy of cell-cell interactions dominated by fibroblasts. Nat. Commun 14, 5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X et al. (2021) Network analysis reveals a distinct axis of macrophage activation in response to conflicting inflammatory cues. J. Immunol 206, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao C et al. (2019) A mechanistic integrative computational model of macrophage polarization: Implications in human pathophysiology. PLoS Comput. Biol 15, e1007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gottschalk RA (2023) Signaling is the pathway to macrophage function. Trends Immunol. 44, 496–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheu KM et al. (2023) Quantifying stimulus-response specificity to probe the functional state of macrophages. Cell Syst. 14, 180–195 [DOI] [PMC free article] [PubMed] [Google Scholar]


