Abstract
We have analyzed 20 randomly amplified polymorphic DNA (RAPD) primers against 36 Streptomyces strains, including 17 taxonomically undefined strains, 25 nonstreptomycete actinomycetes, and 12 outgroups consisting of gram-positive and -negative species. Most of the primers were useful in identifying unique DNA polymorphisms of all strains tested. We have used RAPD techniques to develop a genus-specific probe, one not necessarily targeting the ribosomal gene, for Streptomyces, and a strain-specific probe for the biological control agent Streptomyces lydicus WYEC108. In the course of these investigations, small-scale DNA isolations were also developed for efficiently isolating actinomycete DNA. Various modifications of isolation procedures for soil DNA were compared, and the reliability and specificity of the RAPD methodology were tested by specifically detecting the S. lydicus WYEC108 in DNA isolated from soil.
The biotechnologically important Streptomyces genus contains the largest number of species within the order Actinomycetales. Taxonomic status and phylogenetic analysis of Streptomyces have been based on a polyphasic approach (6, 35), including description and analysis of pigmentation, morphology, biochemical, and physiological properties (39, 40). More recently, molecular-biological techniques have been utilized for refining or extending classifications, especially those techniques targeting 16S rRNA genomic regions (6).
Developing taxon and species-specific probes and primers is important for taxonomic characterizations, phylogenetic analysis, screening for bioactive compounds, and basic ecological research (34). Genomic fingerprinting assays using randomly amplified polymorphic DNA (RAPD) are excellent methodologies for differentiating and tracking specific genetic elements within a complex genome or genomes. These methods were originally developed to identify genetic polymorphisms in plant (16, 27, 28), fungal (10), and prokaryotic genomes (12) and are fast and sensitive means for identifying small differences between similar complex genomes. RAPD methodology has been used for differentiation and tracking of specific strains within the actinomycetes, including Renibacterium (11), Mycobacterium (14, 15, 23), Corynebacterium (19), and Nocardia (20) spp. A limited number of taxon- and strain-specific primers and probes have been developed for and within the genus Streptomyces, targeting variable regions within the 16S rRNA gene; these regions are referred to as α, β, and γ (24, 25, 32, 36). A Streptomyces-specific probe targeting nucleotide position 929 (E. coli numbering) was shown to be specific for Streptomyces strains and species within the order Planctomycetales (32). We have analyzed 20 RAPD primers against 36 Streptomyces strains, including 17 taxonomically undefined strains (4), 25 nonstreptomycete actinomycetes, and 12 outgroups consisting of gram-positive and -negative species. Most of the primers were useful in identifying unique DNA polymorphisms of all strains tested. We have used RAPD techniques to develop a genus-specific probe, one not necessarily targeting the ribosomal gene, for Streptomyces. In the course of these investigations, small-scale DNA isolations were also developed for efficiently isolating actinomycete DNA in response to the large number of DNA extractions required. These methods were compared using RAPD primers to determine the template effectiveness for PCR and RAPD reproducibility. The usefulness of RAPD methodology for generating taxon-specific probes was tested by developing a strain-specific probe for the biological control agent, Streptomyces lydicus WYEC108 (4, 42). The probe was found to be specific for S. lydicus in DNA extracted from pure cultures of Streptomyces, non-Streptomyces actinomycetes, and outgroup bacteria. Further, community-extracted DNA from soil inoculated with S. lydicus and control soil shows the practicality and specificity of the probe.
MATERIALS AND METHODS
Bacteria and media.
A list of the bacterial strains used to investigate the RAPD fingerprint profiles and their respective growth media is presented in Table 1. The actinomycete strains were selected based on a previous work defining a probe homologous to streptomycete peroxidases (21). A cross-section of families are represented within the actinomycete order. Bacteria were grown on media as suggested by the culture suppliers.
TABLE 1.
Organisms investigated in this study
| Species | Family or group | Source or referencea | Mediumb |
|---|---|---|---|
| Actinomycete bacteria | |||
| Streptomyces albus | Streptomycetaceae | ATCC 3004T | YEME |
| Streptomyces ambofaciens | Streptomycetaceae | ELC | YEME |
| Streptomyces antibioticus | Streptomycetaceae | ATCC 8663T | YEME |
| Streptomyces badius | Streptomycetaceae | DC | YEME |
| Streptomyces coelicolor | Streptomycetaceae | ATCC 10147 | YEME |
| Streptomyces erythraeus | Streptomycetaceae | ATCC 11635 | YEME |
| Streptomyces griseifuscus | Streptomycetaceae | ELC | YEME |
| Streptomyces hygroscopicus YCED 9 | Streptomycetaceae | DC | CYD |
| Streptomyces lividans 1326 | Streptomycetaceae | ELC | YEME |
| Streptomyces lividans TK23 | Streptomycetaceae | DAH | YEME |
| Streptomyces lividans TK64 | Streptomycetaceae | DAH | YEME |
| Streptomyces lividans TK64.1 | Streptomycetaceae | DAH | YEME |
| Streptomyces lydicus WYEC108 | Streptomycetaceae | DC | CYD |
| Streptomyces olivaceus | Streptomycetaceae | ATCC 3335T | YEME |
| Streptomyces plicatus | Streptomycetaceae | ATCC 25483T | CYD |
| Streptomyces rimosus | Streptomycetaceae | ATCC 10970T | YEME |
| Streptomyces rochei A4 | Streptomycetaceae | MP | YEME |
| Streptomyces violaceus | Streptomycetaceae | ATCC 13734 | YEME |
| Streptomyces viridosporus T7A | Streptomycetaceae | DC | YEME |
| Streptomyces strain WYE 19 | Streptomycetaceae | DC | CYD |
| Streptomyces strain WYE 27 | Streptomycetaceae | DC | CYD |
| Streptomyces strain WYE 28 | Streptomycetaceae | DC | CYD |
| Streptomyces strain WYE 41 | Streptomycetaceae | DC | CYD |
| Streptomyces strain WYE 48 | Streptomycetaceae | DC | CYD |
| Streptomyces strain WYE 79 | Streptomycetaceae | DC | CYD |
| Streptomyces strain WYE 95 | Streptomycetaceae | DC | CYD |
| Streptomyces strain WYE 98 | Streptomycetaceae | DC | CYD |
| Streptomyces strain WYEC 102 | Streptomycetaceae | DC | CYD |
| Streptomyces strain WYEC 107 | Streptomycetaceae | DC | CYD |
| Streptomyces strain YCED 20 | Streptomycetaceae | DC | CYD |
| Streptomyces strain YCED 33 | Streptomycetaceae | DC | CYD |
| Streptomyces strain YCED 36 | Streptomycetaceae | DC | CYD |
| Streptomyces strain YCED 47 | Streptomycetaceae | DC | CYD |
| Streptomyces strain YCED 57 | Streptomycetaceae | DC | CYD |
| Streptomyces strain YCED 64 | Streptomycetaceae | DC | CYD |
| Streptomyces strain YCED 93 | Streptomycetaceae | DC | CYD |
| Actinoplanes garbadinensis | Micromonosporaceae | ATCC 31049T | NZA |
| Actinoplanes spp. | Micromonosporaceae | PIC | NZA |
| Arthrobacter globiformis | Micrococcaceae | ATCC 607 | TSB |
| Arthrobacter globiformis | Micrococcaceae | ATCC 8010T | TSB |
| Arthrobacter ramosus | Micrococcaceae | ATCC 13727T | TSB |
| Brevibacterium linens | Brevibacteriaceae | ATCC 9172T | NB |
| Corynebacterium xerosis | Corynebacteriaceae | ATCC 373T | AB |
| Couchioplanes caeruleus | Micromonosporaceae | ATCC 33937 | NZA |
| Dactylosporangium fulvum | Micromonosporaceae | ATCC 43301T | YEME |
| Dactylosporangium roseum | Micromonosporaceae | ATCC 31744 | YEME |
| Frankia spp. | Frankiaceae | ATCC 33029 | FM |
| Frankia spp. | Frankiaceae | ATCC 33255 | FM |
| Gordonia corallina | Gordoniaceae | ATCC 31338 | AB |
| Microbispora aerata | Streptosporangiaceae | ATCC 15448T | AB |
| Micrococcus luteus | Micrococcaceae | ATCC 4698T | LB |
| Micromonospora citra | Micromonosporaceae | ATCC 35571T | AB |
| Micromonospora globosa | Micromonosporaceae | ATCC 31465 | AB |
| Micromonospora spp. | Micromonosporaceae | DC | CYD |
| Mycobacterium lacticola | Mycobacteriaceae | ATCC 14468 | NB |
| Mycobacterium smegmatis | Mycobacteriaceae | ATCC 14468 | NB |
| Nocardioides simplex | Nocardioidaceae | ATCC 15799 | TSB |
| Rhodococcus opacus | Nocardiaceae | ATCC 51881T | NB |
| Streptosporangium longisporum | Streptosporangiaceae | ATCC 25212T | AB |
| Terrabacter tumescens | Intrasporangiaceae | ATCC 21109 | TSB |
| Thermomonospora mesophila | Thermomonosporaceae | ATCC 27303T | AB |
| Nonactinomycete bacteria | |||
| Ralstonia eutrophia JMP134 | Burkholderia | DON | NB |
| Bacillus subtilis | Bacillus or Staphylococcus | ATCC 19659 | NB |
| Clostridium bifermentans KMR-1 | Clostridiaceae | RC | BHI |
| Enterobacter cloaceae 96-37 | Enterobacteriaceae | ATCC 43560 | YEG |
| Escherichia coli JM101 | Enterobacteriaceae | YAN | LB |
| Escherichia coli | Enterobacteriaceae | ATCC 11303 | LB |
| Flavobacterium spp. | Flavobacteriaceae | RC | GLU |
| Burkholderia cepacia DLC 62 | Burkholderia | SND | NB |
| Pseudomonas putida mt-2 | Pseudomonas | ATCC 33015 | NB |
| Pseudomonas stutzerii KC | Pseudomonas | CRI | ACE |
| Salmonella enterica serovar Typhimurium TA1538 | Enterobacteriaceae | WAT | NB |
| Staphylococcus aureus | Bacillus or Staphylococcus | ATCC 6538 | NB |
The superscript “T” designates type strains. PIC, Presque Isle Cultures; ICPB, International Cultures of Plant Bacteria, University of California at Davis; ATCC, American Type Culture Collection; ELC, Eli Lilly & Company; DAH, D. A. Hopwood, John Innes Institute; SND, soil associated with North Dakota lignite coal; MP, M. Pasti (27a); DC, D. Crawford, University of Idaho; RC, R. Crawford, University of Idaho; WAT, M. Watanabe et al. (38a); DON, R. H. Don and J. M. Pemberton (4b); YAN, C. Yanisch-Perron et al. (40a); CRI, C. S. Criddle et al. (4a).
YEME, yeast extract-malt extract (yeast extract, 4 g; malt extract, 10 g; water to 1 liter); CYD, casein-yeast extract-dextrose (casein [acid hydrolysate], 5 g; yeast extract, 5 g; glucose, 10 g; water to 1 liter); NZA, N-Z amine with soluble starch and glucose; TSB, tryptic soy broth (casein hydrolysate, 17 g; soy meal hydrolysate, 3 g; NaCl, 5 g; K2HPO4, 2.5 g; glucose, 2.5 g; water to 1 liter); NB, nutrient broth (nutrient broth powder [Difco], 8 g; water to 1 liter); AB, Actinomyces broth (actinomyces broth powder [Difco]); YEG, yeast extract-glycerol (yeast extract, 1 g; casein hydrolysate, 2 g; K2HPO4, 1.4 g; KH2PO4, 0.6 g; MgSO4, 0.5 g; K2SO4, 1 g; glycerol, 20 ml; water to 800 ml); LB, L broth (tryptone, 10 g; yeast extract, 5 g; NaCl, 0.5 g; water to 1 liter); GLU, glutamate medium l-Na-glutamate, 4 g; K2HPO4 · 3H2O, 0.82 g; KH2PO4, 0.19 g; NaNO3, 0.5 g; MgSO4 · 7H2O, 0.1 g; water to 1 liter); ACE, acetate medium [Na-acetate, 3 g; KH2PO4, 2 g; K2HPO4, 3.5 g; (NH4)2SO4, 1 g; MgSO4 · 7H2O, 0.5 g; NaNO3, 2 g; Ca(NO3)2 (0.15M), 1 ml]; BHI, brain heart infusion broth (Difco; 37 g, water to 1 liter); FM, Frankia medium (ATCC medium 1060).
Template DNA isolations.
Nonactinomycete DNA was isolated by genomic isolation methods (2). Actinomycetes were grown in liquid culture at 28°C to late exponential phase. Genomic DNA was obtained from cultures by the methods of Rao et al. (29) or 5′→3′ (5 Prime→3 Prime. The procedures described by Marmur (22) and Heath et al. (13) were modified in order to process small volume samples or for more effective lysis of actinomycete spores or mycelia. Culture volumes of 50 ml or greater were processed essentially as described by Rao et al. (29).
Small-scale volumes were processed in 2-ml microcentrifuge tubes with appropriate modifications. The method of Heath et al. (13) was modified for 1-ml culture volumes. It was important to test cells at periodic intervals during the lysozyme procedure by dividing the cell mixture into 10-μl aliquots and treating them with sodium dodecyl sulfate (SDS). Cells were incubated in the lysozyme mixture until substantial clearing was observed with the SDS-treated test aliquot. At this point, the entire tube contents were processed.
The method of Marmur (22) was altered in order to process 1-ml culture volumes. Mixture constituents were as described, with the following modifications. Culture, 1 ml, was centrifuged and resuspended in 1 ml of TE (Tris-HCl, 50 mM; EDTA, 20 mM; pH 8.0). Lysis solution, 0.38 ml, was added, followed by 0.40 ml of sodium perchlorate solution. Phenol-chloroform was added to fill the 2-ml centrifuge tube, and the culture was extracted. The aqueous upper phase was transferred into another tube and extracted with chloroform-isoamyl alcohol. Then, 2 ml of 95% ethanol was added to the aqueous phase, and the DNA was spooled out, washed in 80% ethanol, and air dried. The DNA was resuspended in 0.1× SSC (15 mM sodium chloride, 15 mM sodium citrate; pH 7.0). RNase was added to a final concentration of 1 mg ml−1. The mixture was extracted once again with chloroform-isoamyl alcohol and centrifuged; the aqueous phase was transferred to another tube, and SSC was added (1×, final concentration). The DNA was precipitated with 90% ethanol, centrifuged, washed in 70% ethanol, centrifuged again, and air dried. The DNA was then dissolved in 500 μl of TE. Some samples (listed in Fig. 1) were processed with an initial hot-phenol treatment according to the method of Beyazova and Lechevalier (3) prior to this DNA isolation procedure.
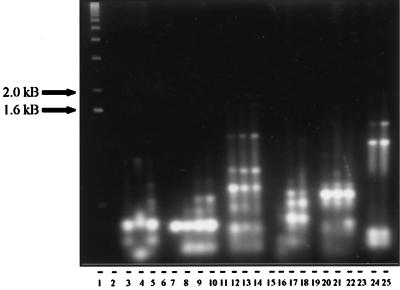
FIG. 1.
Comparison of RAPD profiles of miniprep DNA extraction methods. Isolates were prepared using modifications of the Marmur (M), 5′→3′, Heath (H), Beyazova pretreatment (B), or large-scale Rao (R) methods. Lane 1, 1-kb ladder; lane 2, S. badius (M); lane 3, S. badius (5′→3′); lane 4, S. badius (R); lane 5, S. badius (H); lane 6, empty; lane 7, S. viridosporus T7A (M); lane 8, S. viridosporus T7A (5′→3′); lane 9, S. viridosporus T7A (R); lane 10, S. viridosporus T7A (H); lane 11, empty; lane 12, WYE 27 (M); lane 13, WYE 27 (5′→3′); lane 14, WYE 27 (R); lane 15, empty; lane 16, YCED 5 (M); lane 17, YCED 5 (H); lane 18, YCED 5 (H); lane 19, empty; lane 20, WYEC108 (B); lane 21, WYEC108 (H); lane 22, WYEC108 (R); lane 23, empty; lane 24, WYE 49 (B); lane 25, WYE 49 (B).
Primers.
A list of PCR primers and sequences is given in Table 2. Primers were chosen based on previous success in generating nonambiguous RAPD profiles (1, 7, 8) or primer sets comprising high-GC nucleotides synthesized for RAPD procedures (Genosys Biotechnologies). The eubacterial consensus sequence primer, CLF-4, targets positions 324 to 343 (E. coli numbering) within the 16S ribosomal DNA (rDNA) gene sequence. It was developed from homology matches to 47 16S rDNA sequences from Clostridium, Pseudomonas, Streptomyces, and Flavobacterium strains. The Streptomyces-specific primer was developed to target positions 1661 to 1678 (Streptomyces ambofaciens rrnD, accession no. M27245) homologous to 22 16S rDNA Streptomyces sequences. PCR utilizing primers CLF-4 in combination with the Streptomyces specific primer amplifies the bacterial 16S rDNA fragment at positions 324 to 966 (E. coli numbering) or positions 1054 to 1678 (S. ambofaciens rrnD gene cluster). Primers were optimized using Primer Design (PC-Gene).
TABLE 2.
PCR primers
| Primer | Sequence (5′ to 3′) | Source or reference |
|---|---|---|
| 1247 | AAGAGCCCGT | Akopyanz et al. 1 |
| 1253 | GTTTCCGCCC | Akopyanz et al. 1 |
| 1281 | AACGCGCAAC | Akopyanz et al. 1 |
| 1283 | GCGATCCCCA | Akopyanz et al. 1 |
| 1290 | GTGGATGCGA | Akopyanz et al. 1 |
| D8635 | GAGCGGCCAAAGGGAGCAGAC | Akopyanz et al. 1 |
| D9355 | CCGGATCCGTGATGCGGTGCG | Akopyanz et al. 1 |
| NP-2 | CGGGGGACTGTTGGGCGCCATCT | Fani et al. 7 |
| A-1 | GAGGCCCTTC | Fritsch et al. |
| 70-31 | GCCCCTCTTG | Genosys Biotech |
| 70-32 | GGACCGACTG | Genosys Biotech |
| 70-33 | CTGTCGGCTC | Genosys Biotech |
| 70-34 | GGACCGCTAG | Genosys Biotech |
| 70-35 | CATGTCCGCC | Genosys Biotech |
| 70-36 | GCACGTGAGG | Genosys Biotech |
| 70-37 | CTATCGCCGC | Genosys Biotech |
| 70-38 | GAGAGGGAGG | Genosys Biotech |
| 70-39 | CCGGGGTTAC | Genosys Biotech |
| 70-40 | CGCAGACCTC | Genosys Biotech |
| 10-40 | CGCAGACCTC | Genosys Biotech |
| CLF-4 | AGACACGGCCCAGACTCCTA | PC-Gene database |
| Streptomyces specific | GCGTCGAATTAAGCCACA | Wellington et al. 39 |
PCR amplifications.
Amplifications using RAPD primers were performed on a PTC-100 thermal cycler (MJ Research). RAPD amplification reaction mixtures consisted of 1 μl of template DNA (100 ng), 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.1% (wt/vol) Triton X-100, 0.1 mM deoxynucleotide triphosphates, 1.5 mM MgCl2, 200 nM primer, and 1.5 U of Taq polymerase (Promega). Amplification was performed with an initial denaturation step of 4 min at 95°C and then 40 cycles of 40-s denaturation at 94°C, 45 s at 38°C for primer annealing, and 1.5 min at 72°C for primer extension. A 5-min extension and cooling to 4°C completed the reaction sequence.
Specific amplifications of 16S rDNA of Streptomyces mixtures were modified to 10 ng of template DNA, 50 μM deoxynucleotide triphosphates, and 50 nM CLF-4 and Streptomyces specific primers. Two sets of thermal cycler conditions were performed and compared for product specificity and signal intensity. The first set of conditions consisted of an initial DNA denaturation for 4 min at 95°C, followed by 30 cycles of 40-s denaturation at 94°C, annealing for 45 s at 60°C, and a 1.5-min extension at 72°C. A final extension step of 5 min at 72°C was performed. In order to decrease nonactinomycete DNA amplification, the annealing temperature was increased to 69, 71, and optimally to 73°C. “Touchdown” PCR (5) was employed in the second set of conditions. This method takes advantage of the fact that the PCR reaction begins at or above the expected annealing temperature. The annealing temperature is incrementally decreased every cycle to a final “touchdown” annealing temperature. In this manner, the correctly matched primer to template combination has an advantage (due to the exponential nature of PCR) over misprimed combinations. Initial denaturation was performed at 94°C for 5 min, followed by 20 cycles of 30-s denaturation, annealing at 65°C for 40 s, with a decrease in annealing temperature of 0.5°C/cycle, and then a final extension at 72°C. This was followed by 10 cycles at the touchdown annealing temperature of 55°C, followed by a final 3-min extension at 72°C and a 4°C hold temperature. In order to decrease the nonactinomycete signal, touchdown annealing temperatures were modified to 71 to 69°C (Δ0.1°C/cycle), 73 to 71°C, and 75 to 73°C.
Direct DNA extraction from soil. (i) Freeze-thaw DNA extraction method.
Soil samples used for DNA isolations were collected from the rhizosphere of potato tubers at a field site (University of Idaho Agricultural Experiment Station, Parma) where research investigating the potential for use of biological control by Streptomyces lydicus WYEC108 to prevent fungal potato diseases was accomplished. Depending upon the plot, the soil had been unamended or seeded with WYEC108 spores or mycelia. A modification of the methods of Tsai and Olson (38) was employed. Soil (0.5 g) was mixed with 1.0 ml of 50 mM Tris–20 mM EDTA. Samples were vortexed and then centrifuged (12,000 × g) for 10 min at 4°C; the supernatant was then aspirated. The pellet was resuspended in 1 ml of TE containing 1 mg of lysozyme per ml and incubated in a 37°C water bath with agitation for 1 h. A 10% (wt/vol) solution of SDS (150 μl) was added, followed by 250 μl of 5 M NaCl. Three cycles of freezing in a −70°C dry ice-ethanol bath and thawing in a 65°C water bath (3 min at each temperature) were conducted to lyse the microbial cells in the soil. The efficiency of this method was found to be >99%, as determined by dilution plate counts (no colonies detectable on the 10−1 dilution plate at an initial concentration of 3 × 109 CFU ml−1). After the freeze-thaw cycles, the slurry was centrifuged, and the supernatant was then carefully removed and treated with RNase (200 μg ml−1, final concentration) for 15 min at 37°C. This was followed by a proteinase K treatment (50 μg ml−1, final concentration) for 30 min at 37°C. The mixture was extracted with 500 μl of chloroform-isoamyl alcohol (24:1), inverted several times, and centrifuged. The supernatant was removed and reextracted until very little to no proteinaceous interface remained. The aqueous phase was extracted with 1/4 to 1/3 volume of water-saturated ethyl ether. The ether was then aspirated, and the residual material was driven off at 60°C. DNA was precipitated with 0.6 volume of −20°C isopropanol for 1 h at −20°C. The crude DNA pellet was obtained by centrifugation at 12,000 × g for 10 min at 4°C. The supernatant was aspirated and the pellet gently washed with 70% ethanol (−20°C). Ethanol was removed by aspiration after centrifugation and drying. The pellet was then suspended in 25 μl of TE (10/1, pH 8.0).
DNA was gel purified by electrophoresis on low-melting-point (LMP) agarose (1.5% [wt/vol] Ultrapure; Bethesda Research Laboratories) containing TAE buffer (40 mM Tris-acetate, 1 mM EDTA). Polyvinylpyrrolidine-40K (PVP), at a 2% (wt/vol) final concentration, was incorporated into the gel to eliminate electrophoretic comigration of humic acid phenolics with the nucleic acids (41). Electrophoresis was performed overnight in PVP-LMP agarose at low voltage (5 V cm−1). The DNA separated in the PVP-LMP agarose gel was visualized by ethidium bromide staining. DNA of approximately 25 kDa was excised from a gel slice of approximately 250 mg of agarose. DNA was purified from the agarose gel with the Qiaex gel extraction kit (Qiagen).
(ii) Bead-mill homogenization DNA extraction.
DNA was also extracted from soil by bead-mill homogenization as described by More et al. (26). Subsequent gel purification was performed as previously described.
(iii) Gene-Releaser DNA extraction.
Soil samples, 0.5 g, were washed three times in 100 mM phosphate buffer (pH 8.0) (25). After final centrifugation, the pellet was resuspended in 1 ml of TE (10/0.1, pH 8.0). The slurry was treated in an ultrasonic water bath for 10 min. Then, 20 μl of Gene-Releaser beads (BioVentures) were added to 100 μl of the soil slurry, which was then heated in a microwave oven for 7 min at 700 W. Samples were then gel purified or amplified directly.
Southern hybridization.
Specific gel-eluted DNA fragments, 25 ng, were labeled with [α-32P]dCTP by random priming (Gibco BRL). These fragments were used as probes to detect homologous sequences from PCR-generated DNA fragments amplified with the identical primer used to generate the probe. In this manner, the RAPD fingerprint profiles from each organism could be scored as homologous or nonhomologous to the probe, thus testing whether the probe was unique to one organism or toward specific groups of bacteria. DNA was transferred from the agarose gel to nylon membranes (Hybond-N; Amersham Life Science) by vacuum blotting. Blotted membranes were dried under vacuum at 80°C. Prehybridization and hybridization were carried out at 65°C, unless otherwise noted, as described by Sambrook et al. (30). Next, 10 ml of hybridization solution with labeled probe and membrane were incubated overnight. Low-stringency (2× SSC–0.5% SDS, room temperature) and high-stringency (0.1× SSC–0.5% SDS, 65 to 75°C) washes were performed. If required, the probe was stripped from the membrane in 0.1× SSC–0.5% SDS twice for 20 min at 95°C, and the membrane was reprobed with a different fragment.
Nucleotide sequence accession number.
The 0.3-kDa probe has been sequenced and has been assigned GenBank number AF239669. It is a unique sequence, one not highly homologous to any sequences currently in the databases.
RESULTS
DNA isolation procedures.
Table 3 illustrates the range in DNA concentration and purity obtained from different procedures with various actinomycete strains.
TABLE 3.
DNA isolation comparisonsa
| Strain | Procedure | A260/A280 | DNA concn (μg/μl) |
|---|---|---|---|
| S. badius 252 | Marmur | 0.89 | 0.246 |
| 5′→3′ | 1.20 | 0.717 | |
| Rao | 1.83 | 1.68 | |
| Heath | 1.87 | 2.98 | |
| S. viridosporus T7A | Marmur | 1.12 | 0.342 |
| 5′→3′ | 1.48 | 0.951 | |
| Rao | 1.92 | 5.40 | |
| Heath | 1.84 | 0.500 | |
| Streptomyces sp. strain WYE 27 | Marmur | 1.28 | 1.077 |
| 5′→3′ | 1.15 | 0.426 | |
| Rao | 1.90 | 4.14 | |
| Heath | 1.54 | 3.71 | |
| Streptomyces sp. strain YCED 5 | Marmur | 1.09 | 0.291 |
| 5′→3′ | 1.01 | 0.327 | |
| Heath | 1.90 | 0.97 |
Cell pellets were normalized on all small-scale DNA-extracted samples to 0.2-ml centrifuged pellet volume.
A260/A280 ratios show that Marmur (22) and 5′→3′ (5 Prime→3 Prime) procedures provided DNA contaminated with phenol or protein (means of 1.09 and 1.23, respectively). The small-scale procedure of Heath et al. (13) produced DNA of high purity (mean A260/A280 value of 1.80). DNA purified using the large-scale procedure of Rao et al. (29) was used as a standard for comparative purposes. The relatively large culture volumes (250 ml), lengthy processing times, and large waste volumes make this method less desirable when processing large numbers of samples. The ease of extraction and DNA purity were significantly enhanced by hot phenol extraction (3) of the intact cells prior to the Marmur procedure (data not shown).
Though two of the small-scale DNA extraction procedures produced a final product contaminated with phenol or protein, the RAPD fingerprint profiles were consistent within strains (Fig. 1). The only preparations in which no amplification was observed were with the lowest A260/A280 ratios (Fig. 1, lanes 2 and 16). It is apparent that the use of RAPD methodology is a rigorous technique applicable to a diversity of samples and DNA isolation methods.
RAPD fingerprint profiles.
RAPD fingerprint profiles were generated using 20 primers on genomic DNA from Streptomyces strains, non-Streptomyces actinomycetes, and outgroup bacteria in order to determine whether this methodology might be useful in developing genus-specific probes (in particular, Streptomyces-specific probes) within the order Actinomycetales. Profiles of most interest were those that produced clearly distinguishable major products of between 0.2 and 1.2 kDa. PCR-generated DNA fragments were subsequently labeled and tested for genus specificity. Promising fragments were chosen based on size homology to other Streptomyces products and absence of homology to outgroup or non-Streptomyces actinomycetes PCR products. Probes were developed following gel purification of major, well-resolved RAPD PCR fragments of the correct size. Hybridization of the fragment to RAPD fingerprint blots determined whether the potential probe showed sequence homology toward other Streptomyces strains and the extent of this homology within the genus.
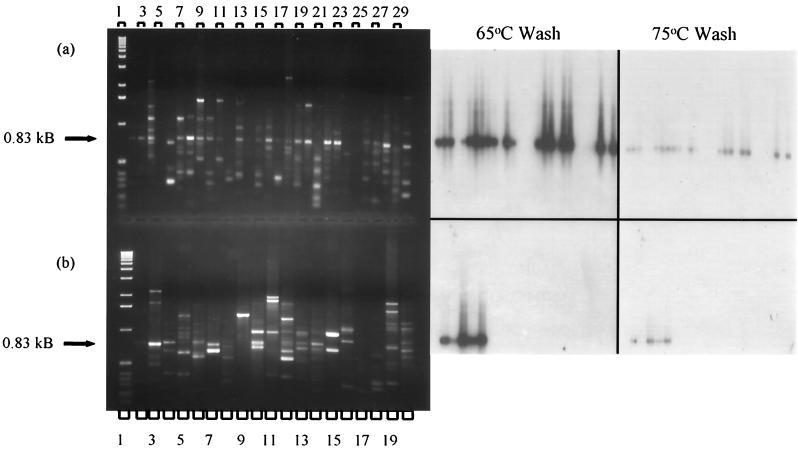
A primer 1253-generated 0.8-kDa RAPD product from S. lividans 1326 was gel purified, labeled, and probed against primer 1253-amplified DNA from actinomycetes and outgroup strains (Fig. 2). High-stringency washes of the blotted DNA showed that the probe was homologous to most, but not all, Streptomyces DNA, and otherwise only to the Arthrobacter or Nocardiodes simplex (basonym, Arthrobacter) products (Fig. 2).
FIG. 2.
RAPD fingerprint profiles and Southern (65 and 75°C wash) of Streptomyces, suprageneric, and outgroup DNA primed with 1253. (a) RAPD fingerprint profiles of Streptomyces spp. Lane 1, 1-kb ladder; lane 2, Streptomyces lividans 1326; lane 3, Streptomyces lividans TK23; lane 4, Streptomyces antibioticus; lane 5, Streptomyces albus; lane 6, Streptomyces coelicolor; lane 7, Streptomyces rimosus; lane 8, Streptomyces ambofaciens; lane 9, Streptomyces violaceus; lane 10, Streptomyces griseifuscus; lane 11, Streptomyces olivaceus; lane 12, Streptomyces badius 252; lane 13, Streptomyces lividans TK64; lane 14, Streptomyces lividans TK64.1; lane 15, Streptomyces viridosporus T7A; lane 16, Streptomyces plicatus; lane 17, Streptomyces erythraeus; lane 18, Streptomyces rochei A4; lane 19, Streptomyces strain YCED 20; lane 20, Streptomyces strain YCED 36; lane 21, Streptomyces strain YCED 64; lane 22, Streptomyces strain WYE 28; lane 23, Streptomyces strain WYE 79; lane 24, Streptomyces strain WYE 27; lane 25, Streptomyces strain WYE 98; lane 26, Streptomyces lydicus WYEC108; lane 27, Streptomyces hygroscopicus YCED 9; lane 28, Streptomyces strain YCED 33; lane 29, Streptomyces strain WYEC102; lane 30, Streptomyces strain WYEC107. (b) RAPD fingerprint profiles of suprageneric (lanes 3 to 15) and outgroup (lanes 16 to 20) strains. Lane 1, 1-kb ladder; lane 2, Streptomyces lividans 1326, positive control; lane 3, Arthrobacter ramosus; lane 4, Nocardioides simplex; lane 5, Arthrobacter globiformis 607; lane 6, Arthrobacter globiformis 8010; lane 7, Brevibacterium linens; lane 8, Micrococcus luteus; lane 9, Gordonia corallina; lane 10, Rhodococcus opacus; lane 11, Corynebacterium xerosis; lane 12, Dactylosporangium roseum; lane 13, Streptosporangium longisporum; lane 14, Microbispora aerata; lane 15, Frankia sp. strain 33255; lane 16, Burkholderia cepacia DLC 62; lane 17, Bacillus subtilis; lane 18, Pseudomonas putida mt-2; lane 19, Flavobacterium spp.; lane 20, Ralstonia eutropha JMP134. (Photograph, righthand side) Southern blot tests of homology with RAPD PCR products. The probe is a gel-purified 0.8-kb RAPD fragment generated from Streptomyces lividans 1326.
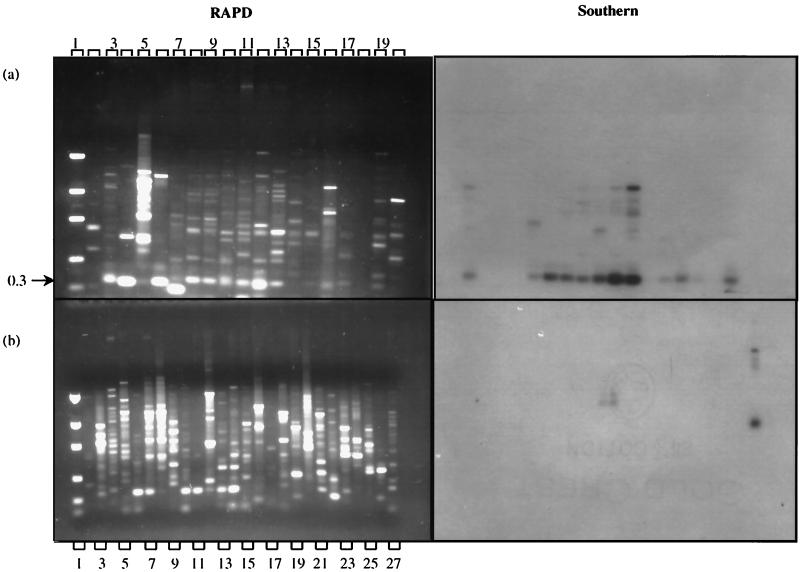
A 0.3-kDa fragment from S. badius 252 amplified with primer 70-34 was labeled and hybridized to 70-34-amplified DNA (Fig. 3). The probe was homologous to most 0.3-kDa DNA fragments from the RAPD blot but to none of the tested non-Streptomyces actinomycetes or outgroups (Southern, Fig. 3). Weak hybridization signals were detected from Gordonia corallina and Rhodococcus opaca DNA, but at a higher molecular mass than that of the 0.3-kDa product, thus making these fragments easily distinguishable. These weaker signals disappeared upon washes carried out at 75°C, while the Streptomyces signals were retained. This probe satisfied the criteria of Streptomyces specificity on those strains tested and might be useful as a Streptomyces-specific probe.
FIG. 3.
RAPD fingerprint profiles and Southern blots of Streptomyces, suprageneric, and outgroup DNA primed with 70-34. (a) RAPD fingerprint profiles of Streptomyces spp. Lane 1, DNA mass marker ladder; lane 2, no DNA control; lane 3, Streptomyces lividans 1326; lane 4, Streptomyces antibioticus; lane 5, Streptomyces albus; lane 6, Streptomyces coelicolor; lane 7, Streptomyces rimosus; lane 8, Streptomyces ambofaciens; lane 9, Streptomyces ambofaciens; lane 10, Streptomyces violaceus; lane 11, Streptomyces griseifuscus; lane 12, Streptomyces olivaceus; lane 13, Streptomyces badius 252; lane 14, Streptomyces viridosporus T7A; lane 15, Streptomyces rochei A4; lane 16, Streptomyces lydicus WYEC108; lane 17, Streptomyces hygroscopicus YCED 9; lane 18, Streptomyces hygroscopicus YCED 9; lane 19, Streptomyces plicatus; lane 20, Streptomyces erythraeus. (b) RAPD fingerprint profiles of suprageneric (lanes 3 to 15) and outgroup (lanes 16 to 26) strains. Lane 1, DNA mass marker ladder; lane 2, no DNA control; lane 3, Arthrobacter ramosus; lane 4, Nocardioides simplex; lane 5, Brevibacterium linens; lane 6, Micrococcus luteus; lane 7, Dactylosporangium fulvum; lane 8, Dactylosporangium roseum; lane 9, Corynebacterium xerosis; lane 10, Streptosporangium longisporum; lane 11, Microbispora aerata; lane 12, Frankia sp. strain 33255; lane 13, Gordonia corallina; lane 14, Rhodococcus opacus; lane 15, Thermomonospora mesophila; lane 16, Burkholderia cepacia DLC 62; lane 17, Bacillus subtilis; lane 18, Staphylococcus aureus; lane 19, Flavobacterium spp.; lane 20, Escherichia coli JM101; lane 21, Pseudomonas putida mt-2; lane 22, Ralstonia eutropha JMP134; lane 23, Enterobacter cloacae 96-3; lane 24, Salmonella enterica serovar Typhimurium strain TA 1538; lane 25, Pseudomonas stutzerii KC; lane 26, Clostridium bifermentans KMR-1; lane 27, Streptomyces badius 252, positive control. (Righthand side panels). Southern blot tests of homology with RAPD PCR products. The probe is a gel-purified 0.3-kb RAPD fragment generated from Streptomyces badius 252.
Development of S. lydicus WYEC108-specific probe.
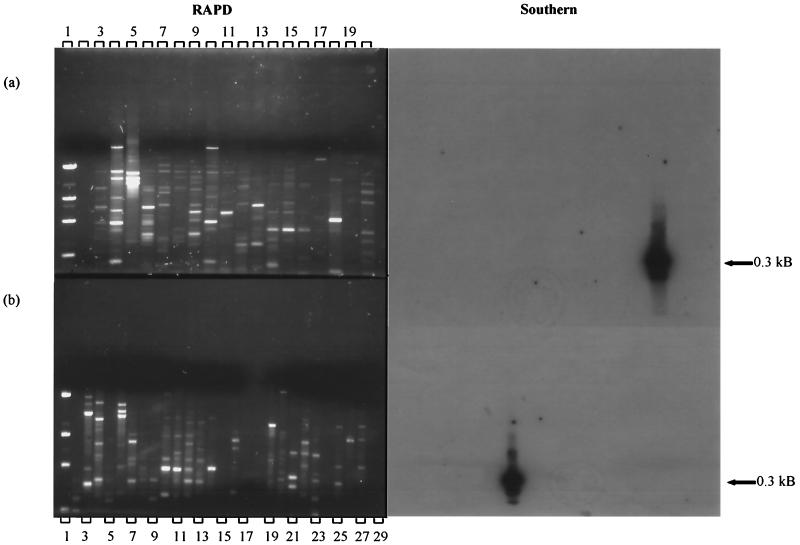
In order to determine whether RAPD methodology might be useful in producing a probe specific for WYEC108, PCR amplifications were analyzed for unique DNA fragments. DNA fragments, 0.3 and 0.8 kDa, amplified by primer set 70-40 (Fig. 4a, lane 18) appeared to be specific to WYEC108. This was verified by labeling the isolated fragment with 32P, blotting, and hybridizing it to the genomic DNA from a number of Streptomyces strains, nonstreptomycete actinomycetes, and outgroups. It is apparent that each probe's specificity is to WYEC108 (Southern, Fig. 4a, lane 18; Fig. 4b, lane 14).
FIG. 4.
RAPD fingerprint profiles and Southern blots of Streptomyces, suprageneric, and outgroup DNA primed with 70-40. (a) RAPD fingerprint profiles of Streptomyces spp. Lane 1, DNA mass marker ladder; lane 2, no DNA control; lane 3, Streptomyces lividans 1326; lane 4, Streptomyces antibioticus; lane 5, Streptomyces albus; lane 6, Streptomyces coelicolor; lane 7, Streptomyces rimosus; lane 8, Streptomyces ambofaciens; lane 9, Streptomyces ambofaciens; lane 10, Streptomyces violaceus; lane 11, Streptomyces griseifuscus; lane 12, Streptomyces olivaceus; lane 13, Streptomyces badius 252; lane 14, Streptomyces viridosporus T7A; lane 15, Streptomyces plicatus; lane 16, Streptomyces erythraeus; lane 17, Streptomyces rochei A4; lane 18, Streptomyces lydicus WYEC108; lane 19, Streptomyces hygroscopicus YCED 9; lane 20, Streptomyces hygroscopicus YCED 9. (b) Suprageneric (lanes 3 to 13) and outgroup (lanes 19 to 29) strains. Lane 1, DNA mass marker ladder; lane 2, no DNA control; lane 3, Arthrobacter ramosus; lane 4, Brevibacterium linens; lane 5, Micrococcus luteus; lane 6, Dactylosporangium fulvum; lane 7, Corynebacterium xerosis; lane 8, Streptosporangium longisporum; lane 9, Microbispora aerata; lane 10, Frankia sp. strain 33255; lane 11, Gordonia corallina; lane 12, Rhodococcus opacus; lane 13, Thermomonospora mesophila; lane 14, Streptomyces lydicus WYEC108; lane 15, Streptomyces viridosporus T7A; lane 16, Streptomyces rimosus; lane 17, sterile soil sample inoculated with S. lydicus WYEC108; lane 18, unautoclaved soil sample inoculated with S. lydicus WYEC108; lane 19, Burkholderia cepacia DLC 62; lane 20, Pseudomonas stutzerii KC; lane 21, Staphylococcus aureus; lane 22, Flavobacterium spp.; lane 23, Escherichia coli JM101; lane 24, Pseudomonas putida mt-2; lane 25, Ralstonia eutropha JMP134; lane 26, Enterobacter cloacae 96-3; lane 27, Salmonella enterica serovar Typhimurium TA 1538; lane 28, Clostridium bifermentans KMR-1; lane 29, Bacillus subtilis. (Righthand side panels) Southern blot tests of homology with RAPD PCR products. The probe is a gel-purified 0.3-kb RAPD fragment generated from Streptomyces lydicus WYEC108.
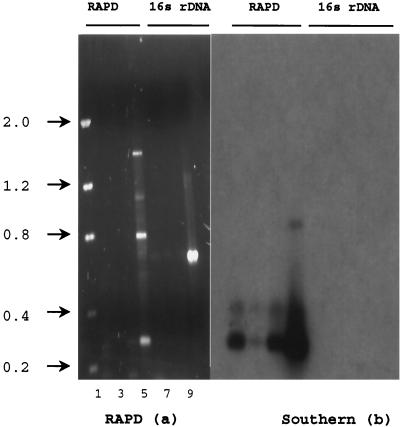
The 0.3-kDa RAPD fragment hybridized to DNA extracted from sterilized field soil (Fig. 5b). A total of 50 mg (wet weight) of WYEC108 spores was added to 0.5 g of sterile soil in 4.5 ml of 100 mM phosphate buffer (pH 8.0). Serial dilutions were performed, and 100 μl from each dilution tube was subjected to DNA purification using Gene-Releaser and gel purification. The 0.3-kDa probe hybridized to all samples containing S. lydicus WYEC108 spores amplified with primer 70-40 but to none of the other DNA fragments (Fig. 5b).
FIG. 5.
(a) RAPD fingerprint profile (a) generated with primer 70-40 (lanes 2 to 5) and 16S rDNA primers that were streptomycetes specific and universal CLF-4 (lanes 6 to 10). (b) Southern blot using 0.3-kb S. lydicus WYEC108-specific probe. Lane 1, DNA mass ladder; lane 2, DNA extracted from WYEC108 spores (10 ng of DNA); lane 3, DNA extracted from WYEC108 spores in soil (10 ng); lane 4, 1:100 dilution of WYEC108 spore-amended soil (5 ng); lane 5, positive control template DNA from WYEC108. Lanes 6 to 9 show samples identical to those of lanes 2 to 5 but amplified with 16S rDNA primers. Lane 10, no DNA control.
It is probable that the DNA extracted from soil contained some substances inhibitory to PCR, since a 10−3 dilution of WYEC108 spores in soil possessed a more intense PCR signal than the 10−1 dilution (Fig. 5a, lanes 3 and 4). Dilution plate counts and DNA Dipstick strips (Invitrogen) verified that DNA was extracted from 2.7 × 105 CFU (10−3 dilution), and the 10−1 spore-amended soil contained twice as much DNA as the 10−3 spore-amended soil (10 versus 5 ng μl−1). Dot blots of WYEC108 spores serially diluted into sterile soil allowed amplification and positive hybridization signal detection of as few as 10 cells using the 0.3-kDa probe (data not shown). Therefore, it seems probable that the signal intensity differences were due to inhibition of the PCR reaction and not to PCR or probe sensitivity. In order to test the 16S rDNA primer's ability to amplify soil-extracted DNA, the Streptomyces-specific primer and CF-1 were used to amplify DNA extracted from WYEC108. The only sample to elicit a PCR product of the correct length, 624 nucleotides, was the nonsoil positive control template DNA (Fig. 5a, lane 9). The 0.3-kDa probe did not hybridize to the 16S rDNA product (Fig. 5b, lanes 6 to 9) nor to another biological control agent, S. hygroscopicus YCED 9 DNA amplified with RAPD primer 70-32 or the Streptomyces-specific 16S rDNA primer pair (data not shown).
In order to further test the specificity of the probe toward WYEC108 and to determine if the probe could be used to identify WYEC108 from DNA extracted from environmental samples, soil samples from WYEC108-inoculated potato rhizospheres were collected and analyzed by RAPD fingerprinting. Soil samples were collected from three WYEC108-inoculated treatments and an uninoculated control. Each sample was homogeneously mixed, air dried, and sieved (425-μm [pore size] mesh). Then, 0.5 g of soil from each treatment was processed for DNA isolations by freeze-thaw and bead-mill homogenization. Samples were electrophoresed and gel purified. Positive controls were provided by WYEC108 spore-inoculated sterilized or native field soil. The purified samples were amplified with primer 70-40.
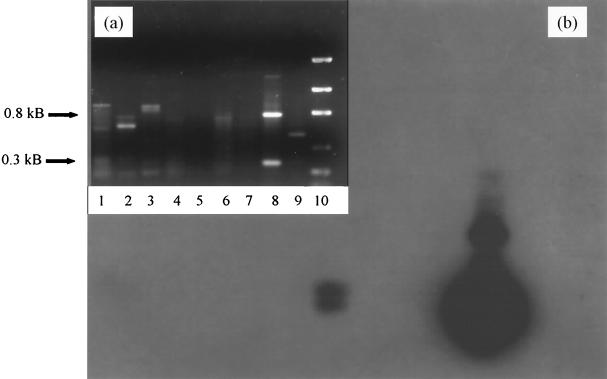
PCR products generated from primer 70-40 are shown in Fig. 6. The 0.8- and 0.3-kDa products from the positive control are clearly visible (lane 8). The Southern blot (Fig. 6b) shows an intense signal at 0.3 kDa and a cross-reacting, less-intense 0.8-kDa band. The only other positive signal is observed from the WYEC108 spore-amended native soil which had been extracted with the bead-mill methodology (lane 4). Sterilized soil inoculated with the same quantity of WYEC108 spores and processed similarly did not elicit a hybridization signal (lane 3). This phenomenon was previously observed from other autoclaved samples using this soil. The PCR-inhibiting humic acid-derived phenolics will normally decrease in concentration upon dilution, and successful amplification reactions will result, but the autoclaved soil into which WYEC108 spores were inoculated did not amplify at identical dilutions. It is known that autoclaved soil has modified physical-chemical characteristics, and these characteristics or other byproducts of the autoclaving process might be responsible for the difficulty in amplifying these samples. This sample was also subjected to the freeze-thaw DNA extraction procedure. No signal was apparent from these samples, even though the spore concentration and soil type were similar (Fig. 6a, lanes 5, 6, and 7).
FIG. 6.
(a) RAPD profile of DNA isolated from soil and amplified with primer 70-40. (b) Southern blot probed with the S. lydicus WYEC108 0.3-kb fragment. (a) RAPD fingerprint profile. Lane 1, control field soil; lane 2, control field soil from a different location of the plot; lane 3, bead-mill DNA extraction procedure of 10−2 dilution (initial inoculum concentration, 3 × 109 CFU g−1); lane 4, bead-mill DNA extraction procedure of 10−2 dilution of nonsterilized field soil inoculated with WYEC108 spores; lane 5, DNA extracted by the freeze-thaw method from 10−3 dilution of sterile inoculated soil; lane 6, DNA extracted by the freeze-thaw method from a 10−2 dilution of nonsterile inoculated soil; lane 7, DNA extracted by the freeze-thaw method from a 10−7 dilution of nonsterile inoculated soil; lane 8, WYEC108-positive control template; lane 9, no DNA control; lane 10, DNA mass ladder.
In order to test the specificity of the 0.3-kDa probe to WYEC108, soil from the control plot (WYEC108 uninoculated) was processed for DNA extraction. The fingerprint profile and hybridization signal were negative (Fig. 6a and b, lanes 1 and 2). This, in conjunction with the RAPD fingerprint and hybridization tests on archived actinomycete DNA templates, indicates that the probe generated by the RAPD procedure is specific for WYEC108.
DISCUSSION
Total genomic DNA isolation methods were modified in order to process numerous samples quickly and efficiently in microcentrifuge tubes. The growth characteristics and ease of DNA extraction varies widely not only between different actinomycete strains but even for the same strain grown on different media. For example, S. badius 252 produces a heavy polysaccharide residue upon DNA extraction growing in rich media, requiring several phenol-chloroform extractions. The residue is significantly less from DNA extracted in minimal-medium-grown cells, requiring a single organic extraction. While DNA was extracted from cells cultured in liquid media, further investigation showed that small-scale DNA isolations can also be performed from agar-grown mycelia and spores (17). Dramatic differences in yield and purity were observed dependent upon the method used. The Marmur (22) modifications and 5′→3′ methods generally produced DNA of low purity, although pretreatment with hot phenol was found to increase DNA purity significantly (data not shown) as has been observed previously (3).
The modified Heath method (13) provided DNA of high purity and the greatest recovery of any of the small-scale isolation procedures investigated (average yield of 1.77 μg μl−1, i.e., three times greater than with the other methods).
RAPD fingerprint profiles also appear to be highly reproducible between Taq polymerases. Major RAPD fragments were consistently amplified regardless of the source of the commercial enzyme. It is known that DNA polymerases derived from a particular species (e.g., Thermus aquaticus) produced identical amplification products of major bands (31). In some cases, minor bands would qualitatively and quantitatively differ, and band intensity was not necessarily an indicator of template concentration (18). We have also found that major products are reproducible and can reliably be used as DNA fingerprints and as probes. Throughout our experimentation, unless minor products could be reproducibly amplified, they were treated as artifacts.
Primer 1253 amplified a unique conserved DNA fragment shown to be specific for Streptomyces and Arthrobacter. Arthrobacter globiformis has been used to root actinomycete phylogenetic trees (32, 33). Similarity matrices based on partial sequencing of 16S rRNAs (9) do not show a close relationship of Arthrobacter to Streptomyces relative to other suprageneric groups. Sequence analysis of this probe might be useful in determining what genetic elements these two somewhat disparate phylogenetic taxa have in common. The probe, in conjunction with the Streptomyces probe and the actinomycetes probe mentioned earlier, might be useful in microbial ecology research as a tool for quantifying these groups in the environment over time or in soil after ecological disturbances. Primer 70-34 amplifies a fragment, subsequently used as a probe, specific for Streptomyces spp. Sequence analysis of the probe and modification of the nucleotide composition might make the probe generally homologous to more Streptomyces strains. It might be useful as a tool to further refine the taxonomic structure of Streptomyces, possibly as a clade-specific probe.
In the course of investigating genus-specific DNA probes, it was observed that one such probe generated from primer 70-32 hybridized only to S. griseofuscus. A more feasible approach for the RAPD methodology became apparent, i.e., to use RAPD DNA as a method to generate strain-specific probes by screening unique fragments from the test strain. RAPD primers were screened in this manner using a Streptomyces biological control strain, S. lydicus WYEC108 (4, 42).
A strain-specific probe unique to S. lydicus WYEC108 genome was developed. While semiselective plating shows persistence of this organism in the rhizoplane and rhizosphere of a number of crops (unpublished data), the difficulty in precisely quantifying and interpreting morphologically similar forms from environmental samples remains a challenge. The use of a strain-specific probe would verify and assist in confirming, quantifying, and localizing WYEC108 in the rhizoplane and rhizosphere. Primer 70-40 allowed amplification of two DNA fragments unique to WYEC108. The 0.3-kDa fragment was isolated, labeled, and probed against WYEC108, streptomycete, actinomycete, and outgroup DNA. Its sequence uniqueness was verified by the fact that this probe hybridized only to S. lydicus WYEC108 DNA amplified with primer 70-40.
The RAPD methodology is especially powerful for analyzing complex genomes in that it combines the sensitivity of PCR (we have been able to amplify DNA from as few as 10 spores) with the specificity of oligonucleotide probe hybridization. A one-nucleotide mismatch in a 20-mer can be detected (36). S. lydicus WYEC108 DNA was successfully extracted and amplified from soil. The efficacy of the RAPD method was shown in that the probe hybridized to WYEC108 soil-extracted DNA and to none of the control soils. The specificity of the WYEC108 probe was shown considering the fact that 1 g of soil may contain at least 4,000 various genomes (37).
Positive hybridization signals were detected from DNA isolated by bead-mill homogenization from nonsterile WYEC108-inoculated soil. Bead-mill extraction was found to be superior to SDS–freeze-thaw treatments. None of the freeze-thaw DNA from WYEC108-spiked soil samples hybridized to the WYEC108-specific probe, while the DNA from the bead-mill homogenized samples did. More et al. (26) found the DNA extraction efficiency to be twice that of the freeze-thaw procedure. For instance, the freeze-thaw procedure demonstrated that 94% of Bacillus endospores survived the freeze-thaw procedure but only 2% remained viable after bead-mill homogenization. They also showed that lysis of total cells was less efficient than cells that could be cultured. It is possible that the freeze-thaw procedure, while reducing the viable counts >99%, might leave a majority of cells nonculturable, though they still retained their DNA. Even if the extraction efficiency were 99.9% at 109 cells g−1, the DNA of 106 cells g−1 would remain nonextracted. The problem of chimera formation due to PCR amplification of small DNA fragments (18) was avoided by gel purification of only higher-molecular-mass (>20-kDa) DNA during the purification of soil-extracted DNA.
The methods used in this study are generally applicable for family, genus, clade, or strain differentiation in screening for bioactive principles, taxonomic characterization, phylogenetic analysis, or community profile characterization of bacterial group to strain fluctuations within the community.
ACKNOWLEDGMENTS
We thank David Knaebel for his thoughtful insight and helpful conversations.
This work was supported by U.S. Department of Agriculture National Research Initiative Competitive Grant 96-35500-3189 and by the Idaho Agriculture Experiment Station.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Dresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 2nd ed. New York, N.Y: Green Publishing Associates/John Wiley & Sons; 1992. [Google Scholar]
- 3.Beyazova M, Lechevalier M P. Taxonomic utility of restriction endonuclease fingerprinting of large DNA fragments from Streptomyces strains. Int J Syst Bacteriol. 1993;43:674–682. [Google Scholar]
- 4.Crawford D L, Lynch J M, Whipps J M, Ousley M A. Isolation and characterization of actinomycete antagonists of a fungal root pathogen. Appl Environ Microbiol. 1993;59:3899–3905. doi: 10.1128/aem.59.11.3899-3905.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Criddle C S, DeWitt J T, Grbic-Galic D, McCarty P L. Transformation of carbon tetrachloride by Pseudomonas sp. strain KC under denitrification conditions. Appl Environ Microbiol. 1990;56:3240–3246. doi: 10.1128/aem.56.11.3240-3246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4b.Don R H, Pemberton J M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981;145:681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embley T M, Stackebrandt E. The molecular phylogeny and systematics of the actinomycetes. Annu Rev Microbiol. 1994;48:257–289. doi: 10.1146/annurev.mi.48.100194.001353. [DOI] [PubMed] [Google Scholar]
- 7.Fani R, Damiani G, Serio D C, Gallori E, Grifoni A, Bazzicalupo M. Use of random amplified polymorphic DNA (RAPD) for generating specific DNA probes for microorganisms. Mol Ecol. 1993;2:243–250. doi: 10.1111/j.1365-294x.1993.tb00014.x. [DOI] [PubMed] [Google Scholar]
- 8.Fritsch P, Hanson M A, Spore C D, Pack P E, Rieseberg L H. Constancy of RAPD primer amplification strength among distantly related taxa of flowering plants. Plant Mol Biol Reporter. 1993;11:10–20. [Google Scholar]
- 9.Goodfellow M. Suprageneric classification of actinomycetes. In: Williams S T, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 4. Baltimore, Md: The Williams & Wilkins Co.; 1989. pp. 2333–2339. [Google Scholar]
- 10.Grajal-Martin M J, Simon C J, Muehlbauer F. Use of random amplified polymorphic DNA (RAPD) to characterize race 2 of Fusarium oxysporum f. sp. pisi. Phytopathology. 1993;83:612–614. [Google Scholar]
- 11.Grayson T H, Atienzar F A, Gilpin M L. Molecular diversity of Renibacterium salmoninarum isolates determined by randomly amplified polymorphic DNA analysis. Appl Environ Microbiol. 2000;66:435. doi: 10.1128/aem.66.1.435-438.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadrys H, Balick M, Schierwater B. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol Ecol. 1992;1:55–63. doi: 10.1111/j.1365-294x.1992.tb00155.x. [DOI] [PubMed] [Google Scholar]
- 13.Heath L S, Sloan G L, Heath H E. A simple and generally applicable procedure for releasing DNA from bacterial cells. Appl Environ Microbiol. 1986;51:1138–1140. doi: 10.1128/aem.51.5.1138-1140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harn H-J, Shen K L, Lee J H. Evidence of transmission of Mycobacterium tuberculosis by random amplified polymorphic DNA (RAPD) fingerprinting in Taipei City, Taiwan. J Clin Pathol. 1997;50:505. doi: 10.1136/jcp.50.6.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kauppinen J, Mantyjarvi R, Katila M L. Random amplified polymorphic DNA genotyping of Mycobacterium malmoense. J Clin Pathol. 1994;32:1827. doi: 10.1128/jcm.32.7.1827-1829.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein-Lankhorst R M, Vermunt A, Weide R, Liharska T, Zabel P. Isolation of molecular markers for tomato (L. esculentum) using random amplified polymorphic DNA (RAPD) Theor Appl Genet. 1991;83:108–114. doi: 10.1007/BF00229232. [DOI] [PubMed] [Google Scholar]
- 17.Kutchma A, Roberts M A, Knaebel D B, Crawford D L. Small-scale isolation of genomic DNA from Streptomyces mycelia and spores. BioTechniques. 1998;24:452–456. doi: 10.2144/98243st05. [DOI] [PubMed] [Google Scholar]
- 18.Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 19.Linton C J, Jalal H, Millar M R. Rapid discrimination of Mycobacterium tuberculosis strains by random amplified polymorphic DNA analysis. J Clin Microbiol. 1994;32:2169. doi: 10.1128/jcm.32.9.2169-2174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louie L, Louie M, Simor A E. Investigation of a pseudo-outbreak of Nocardia asteroides infection by pulsed-field gel electrophoresis and randomly amplified polymorphic DNA PCR. J Clin Microbiol. 1997;35:1582. doi: 10.1128/jcm.35.6.1582-1584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahadevan B. Enzymological and genetic characterization of chitinase in Streptomyces lydicus WYEC108, an antifungal biocontrol agent. M.S. thesis. Moscow: University of Idaho; 1992. [Google Scholar]
- 22.Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 23.Matsiota-Bernard P, Waser S, Legakis N J. Rapid discrimination of Mycobacterium avium strains from AIDS patients by randomly amplified polymorphic DNA analysis. J Clin Microbiol. 1997;35:1585. doi: 10.1128/jcm.35.6.1585-1588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehling A, Wehmeier U F, Pipersberg W. Application of random amplified polymorphic DNA (RAPD) assays in identifying conserved regions of actinomycete genomes. FEMS Microb Lett. 1995;128:119–126. doi: 10.1111/j.1574-6968.1995.tb07510.x. [DOI] [PubMed] [Google Scholar]
- 25.Mehling A, Wehmeier U F, Piepersberg W. Nucleotide sequences of streptomycete 16S ribosomal DNA: towards a specific identification system for streptomycetes using PCR. Microbiology. 1995;141:2139–2147. doi: 10.1099/13500872-141-9-2139. [DOI] [PubMed] [Google Scholar]
- 26.More M I, Herrick J B, Silva M C, Ghiorse W C, Madsen E L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol. 1994;60:1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munthali M, Ford-Lloyd B V, Newbury H J. The random amplification of polymorphic DNA for fingerprinting plants. PCR Methods Appl. 1992;2:274–276. doi: 10.1101/gr.1.4.274. [DOI] [PubMed] [Google Scholar]
- 27a.Pasti M B, Pometto III A L, Nuti N P, Crawford D L. Lignin-degrading ability of actinomycetes isolated from termite (Termitidae) gut. Appl Environ Microbiol. 1990;56:2213–2218. doi: 10.1128/aem.56.7.2213-2218.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rafalski J A, Tingey S V, Williams J G K. RAPD markers—a new technology for genetic mapping and plant breeding. AgBiotech News Inf. 1991;3:645–648. [Google Scholar]
- 29.Rao R N, Richardson M A, Kushtoss S A. Cosmid shuttle vectors for cloning and analysis of Streptomyces DNA. Methods Enzymol. 1987;153:166–198. doi: 10.1016/0076-6879(87)53053-1. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Schierwater B, Ender A. Different thermostable DNA polymerases may amplify different RAPD products. Nucleic Acids Res. 1993;21:4647–4648. doi: 10.1093/nar/21.19.4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stackebrandt E. Aspects on the evolution and phylogeny of the actinomycetes. Actinomycetology. 1991;5:56–63. [Google Scholar]
- 33.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 34.Stackebrandt E, Liesack W, Webb R, Witt D. Towards a molecular identification of Streptomyces species in pure culture and in environmental samples. Actinomycetology. 1991;5:38–44. [Google Scholar]
- 35.Stackebrandt E, Liesack W, Witt D. Ribosomal RNA and rDNA sequence analyses. Gene. 1992;115:255–260. doi: 10.1016/0378-1119(92)90567-9. [DOI] [PubMed] [Google Scholar]
- 36.Stackebrandt E, Witt D, Kemmerling C, Kroppenstedt R, Liesack W. Designation of streptomycete 16S and 23S rRNA-based target regions for oligonucleotide probes. Appl Environ Microbiol. 1991;57:1468–1477. doi: 10.1128/aem.57.5.1468-1477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torsvik V, Goksoyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai Y, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Watanabe M, Ishidate M, Jr, Nohmi T. Nucleotide sequence of Salmonella typhimurium nitroreductase gene. Nucleic Acids Res. 1990;18:1059. doi: 10.1093/nar/18.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wellington E M, Stackebrandt E, Sanders D, Wolstrop J, Jorgensen N O. Taxonomic status of Kitasatosporia, and proposed unification with Streptomyces on the basis of phenotype and 16S rRNA analysis and emendation of Streptomyces Waksman and Henrici 1943, 339AL. Int J Syst Bacteriol. 1992;42:156–160. doi: 10.1099/00207713-42-1-156. [DOI] [PubMed] [Google Scholar]
- 40.Williams S T, Goodfellow M, Alderson G, Wellington E M H, Sneath P H A, Sackin M J. Numerical classification of Streptomyces and related genera. J Gen Microbiol. 1983;129:1743–1813. doi: 10.1099/00221287-129-6-1743. [DOI] [PubMed] [Google Scholar]
- 40a.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC 19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 41.Young C C, Burghoff R L, Keim L G, Minak-Bernero V, Lute J R, Hinton S M. Polyvinylpyrrolidone-agarose gel electrophoresis purification of polymerase chain reaction-amplifiable DNA from soil. Appl Environ Microbiol. 1993;59:1972–1974. doi: 10.1128/aem.59.6.1972-1974.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan W M, Crawford D L. Characterization of Streptomyces lydicus WYEC 108 as a potential biocontrol agent against fungal root and seed rots. Appl Environ Microbiol. 1995;61:3119–3128. doi: 10.1128/aem.61.8.3119-3128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]