Abstract
A rapid increase in antimicrobial resistant bacterial infections around the world is causing a global health crisis. The Gram-negative bacterium Acinetobacter baumannii is categorized as a Priority 1 pathogen for research and development of new antimicrobials by the World Health Organization due to its numerous intrinsic antibiotic resistance mechanisms and ability to quickly acquire new resistance determinants. Specialized phage enzymes, called depolymerases, degrade the bacterial capsule polysaccharide layer and show therapeutic potential by sensitizing the bacterium to phages, select antibiotics, and serum killing. The functional domains responsible for the capsule degradation activity are often found in the tail fibers of select A. baumannii phages. To further explore the functional domains associated with depolymerase activity, tail-associated proteins of 71 sequenced and fully characterized phages were identified from published literature and analyzed for functional domains using InterProScan. Multisequence alignments and phylogenetic analyses were conducted on the domain groups and assessed in the context of noted halo formation or depolymerase characterization. Proteins derived from phages noted to have halo formation or a functional depolymerase, but no functional domain hits, were modeled with AlphaFold2 Multimer, and compared to other protein models using the DALI server. The domains associated with depolymerase function were pectin lyase-like (SSF51126), tailspike binding (cd20481), (Trans)glycosidases (SSF51445), and potentially SGNH hydrolases. These findings expand our knowledge on phage depolymerases, enabling researchers to better exploit these enzymes for therapeutic use in combating the antimicrobial resistance crisis.
Keywords: Acinetobacter, bacteriophage, protein domains, depolymerase, pectin lyase, tail spike, antimicrobial resistance
1 Introduction
Antimicrobial resistance (AMR) is a major threat to human health, with watch lists created for several bacteria due to their high levels of resistance. A global delay in action against AMR, coupled with the COVID-19 pandemic, has further exacerbated the AMR crisis. The Centers for Disease Control and Prevention (CDC) published a special report in 2022 detailing a 15% increase in resistant nosocomial infections from 2019 to 2020. Of particular concern is the significant increase in carbapenem-resistant Acinetobacter baumannii infections, with a 78% increase between 2019 and 2020 (COVID-19: U.S. Impact on Antimicrobial Resistance Special Report 2022, 2022). Due to multidrug-resistant outbreaks involving A. baumannii, the World Health Organization classified it as a number one priority pathogen that is urgently in need of alternative treatment options (World Health Organization, 2021).
Novel treatment options are desperately needed to address the critical lack of therapeutics against A. baumannii infections. One treatment approach is the use of its evolutionary predator–bacteriophages (phages). Phages, and their biological products such as endolysins and depolymerases, are attracting renewed interest as therapeutic options due to their high specificity, potential synergy with antibiotics, unique mode of action, safety, natural abundance, and modification potential (Melo et al., 2020). The narrow host range of phages, as well as the requirement of the phages to successfully enter the cell and propagate to produce progeny phages, complicates the use of phages in therapy. Recent advancements in engineering host receptor binding proteins to expand a phage's host range have shown great success, though it does not eliminate the requirement of the phage to successfully propagate in the cell (Dams et al., 2019; Yehl et al., 2019).
The use of specialized phage enzymes, such as depolymerases or endolysins, instead of active phages could be a potential solution. There are two main groups of phage-associated enzymes responsible for the degradation of carbohydrate-containing polymers: hydrolases (EC 3) and lyases (EC 4) (Latka et al., 2017). Hydrolases target the peptidoglycan, capsular polysaccharides, or the O-antigen side chains of lipopolysaccharides by catalyzing the cleavage of O-glycosidic bonds using water. Lyases utilize β-elimination to introduce a double bond between the C4 and C5 of the non-reducing uronic acid after cleavage of the glycosidic bond between a monosaccharide and the C4 of uronic acid (Sutherland, 1999). These phage depolymerases are found as integral components of the virion structure or as soluble proteins that diffuse out following the host cell lysis (Drulis-Kawa et al., 2015). Phage depolymerases are most often identified within tail fibers or tail spikes and generally form as homotrimeric complexes.
Many groups have investigated phage-encoded depolymerases and endolysins for therapeutic use (Liu et al., 2019a; Kim et al., 2020; Khan et al., 2021; Abdelkader et al., 2022; Chen et al., 2022; Drobiazko et al., 2022). The potential therapeutic benefit of depolymerases is the removal of the capsule layer from the infecting bacterium, which exposes the bacterium to the innate immune system and thus activates serum killing. The potential use of depolymerases for therapy against A. baumannii has been studied and demonstrated in mouse models with a promising therapeutic effect (Liu et al., 2019a; Oliveira et al., 2019; Wang et al., 2020). In this study, we analyzed the tail fibers of published Acinetobacter phages and identified different functional domains present. These data will enable further research and development on these exciting enzymes for therapeutic use.
2 Methods
2.1 Analysis of Acinetobacter phage genomes for tail fibers
A total of 114 Acinetobacter phages were identified in the literature as of 22 July 2021. Phage genomes were downloaded from the NCBI database using the accession numbers presented in each article. Information on the presence or absence of halo formation was collected for each phage. All coding sequences (CDS) in the morphogenesis module of the phage genomes were translated and analyzed using the InterProScan (Jones et al., 2014; Blum et al., 2021) plugin for Geneious Prime 2021.2.2 to identify any functional domains present within the tail fibers. InterProScan ran using the following applications: conserved domains database (CDD), Gene3D, high-quality automated and manual annotation of proteins (HAMAP), protein analysis through evolutionary relationships (PANTHER), Pfam-A, PRINTS, PROSITE profiles, simple modular architecture research tool (SMART), and SUPERFAMILY (Jones et al., 2014; Blum et al., 2021).
The CDD is composed of curated protein domain and protein family models that are searched using reverse position-specific BLAST to match protein sequences with domain and family models (Yang et al., 2020). Gene3D is a comprehensive database of protein domain assignments for sequences from major sequence databases including Ensembl, UniProt, and RefSeq, where domains are directly mapped from structures in the class, architecture, topology, homology (CATH) database or predicted with a library of representative profile hidden Markov models (HMMs) derived from CATH superfamilies (Cuff et al., 2011; Lees et al., 2012). HAMAP is an automatic annotation pipeline that uses a collection of family profiles and manually curated signatures to determine protein family membership of a query protein sequence (Pedruzzi et al., 2015). PANTHER is used to classify sequences into evolutionary groupings (protein class, family, subfamily) using phylogenetic trees, and functional groupings with gene ontology terms and pathways (Mi et al., 2021). The Pfam-A database is a comprehensive collection of protein families, where each family is represented by a curated set of multiple sequence alignments and HMMs (Mistry et al., 2021). The PRINTS database is comprised of a collection of protein family ‘fingerprints' or a group of conserved motifs that provide distinctive signatures for particular protein families and structural/functional domains (Attwood et al., 2012). PROSITE profiles is a database of protein families and domains with specific signatures on constant and variable properties of proteins that can enable the formation of hypotheses about the protein's function (Sigrist et al., 2012). The SMART database is used for the identification and annotation of protein domains and the analysis of protein domain architectures using manually curated models (Letunic et al., 2021). SUPERFAMILY is a database composed of HMMs of structural protein domains that have an evolutionary relationship (Gough et al., 2001).
Functional domain groups were formed for comparison using the identified InterProScan hit results. Multisequence protein alignments were generated using Clustal Omega v.1.2.3 (Sievers and Higgins, 2018). Phylogenetic trees were built from the resulting protein sequence alignments using Randomized Axelerated Maximum Likelihood (RAxML)v.8.2.11 with the following settings: protein model GAMMA BLOSUM62, rapid bootstrapping and search for best-scoring ML tree with 100 bootstrap replicates, and parsimony random seed of 1 (Stamatakis, 2014).
Further investigation into the tertiary structure of specific proteins was completed using AlphaFold v.2.3.1 (Jumper et al., 2021) using the multimer_v3 models. The proteins were modeled as homotrimers as this is the most common oligomeric state phage depolymerases adopt (Latka et al., 2017). The models were visually inspected to ensure good structural integrity in the fold. Only the model with the highest confidence according to AlphaFold was analyzed further. The predictions were run on the Digital Research Alliance of Canada superclusters and required 300 h of A100 GPU computing time. The models were submitted to the DALI server (Holm, 2022) to compare the predicted structures to pre-existing ones. The models were made accessible in the PDB format in the Supplementary material. In addition, all abbreviations are provided in Supplementary Table 1.
3 Results and discussion
3.1 Characteristics of the phages used in the analysis
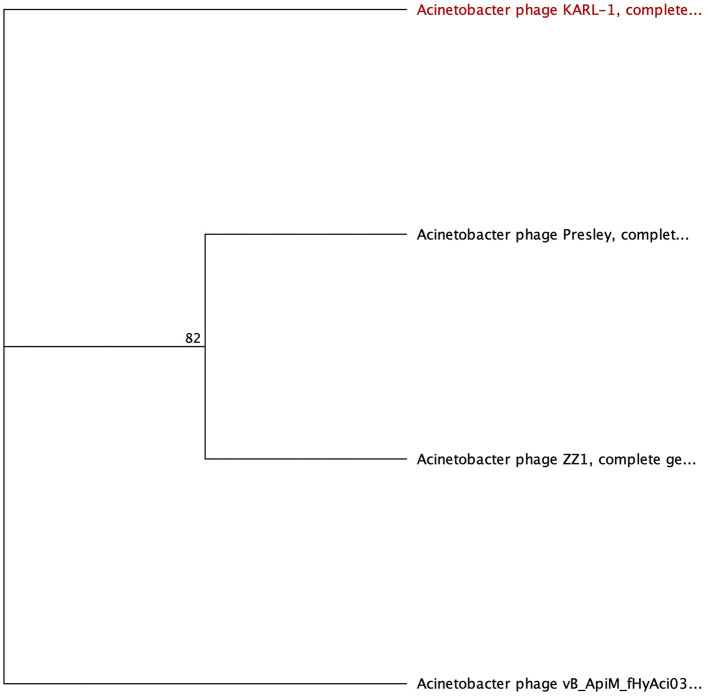
Of the 114 phages documented, a total of 43 phages had no genomic data or had poor genome assemblies and were excluded from the analysis; thus, 71 characterized and sequenced phages were identified for further study (Table 1). Based on the literature review of the viruses documented at the time, the genomes of 31 myoviruses, 33 podoviruses, and 7 siphoviruses were downloaded from the NCBI database and 94 tail fibers were identified and translated. The majority of phages (69%) were predicted to encode one tail fiber, while 20 phages (28%) encoded two tail fibers, and two phages (3%, B9 and fHyAci03) encoded three tail fibers (Table 1). Furthermore, the presence of halo formation around the phage plaques, or expression of a depolymerase, is noted in Table 1 under “Halo or depolymerase”. In total, 43 phages were identified as having halo formation or a functional depolymerase was expressed. Only three phages were explicitly described as lacking halo formation (KARL-1, TAC1, and Loki).
Table 1.
Acinetobacter baumannii bacteriophages and their tail fiber proteins used in the analysis.
| Name | Accession | Length (bp) | Protein IDs | Halo or depolymerase | Type | References |
|---|---|---|---|---|---|---|
| Ab 121 | MT623546 | 102499 | QMP18972.1 | N/A | Myovirus | Wu et al., 2021 |
| AB1 | HM368260 | 45159 | ADO14448.1, ADO14447.1 | Yes | Myovirus | Li et al., 2012 |
| AbTJ | MK340941 | 42670 | QAU04146.1 | Yes | Myovirus | Xu et al., 2020 |
| IME-AB2 | JX976549 | 43665 | AFV51555.1, AFV51556.1 | N/A | Myovirus | Peng et al., 2014 |
| SH-Ab 15599 | MH517022 | 143204 | AXF41547.1 | Yes | Myovirus | Hua et al., 2018 |
| AbP2 | MF346584 | 45373 | ASJ78889.1, ASJ78888.1 | Yes | Myovirus | Yang et al., 2019 |
| Abp9 | MN166083 | 44820 | QEA11049.1 | Yes | Myovirus | Jiang et al., 2020 |
| TaPaz | MZ043613 | 93703 | QVW53859.1, QVW53860.1 | Yes | Myovirus | Shchurova et al., 2021 |
| AbTZA1 | NC_049445 | 168223 | YP_009882247.1 | N/A | Myovirus | Nir-Paz et al., 2019 |
| AM24 | KY000079 | 97177 | APD20249.1 | Yes | Myovirus | Popova et al., 2019 |
| AP22 | HE806280 | 46387 | CCH57762.1, CCH57761.1 | Yes | Myovirus | Popova et al., 2012 |
| BS46 | MN276049 | 94068 | QEP53229.1 | Yes | Myovirus | Popova et al., 2020b |
| DMU1 | MT992243 | 43482 | QOI69765.1 | N/A | Siphovirus | Pehde et al., 2021 |
| YMC-13-01-C62 (C62) | KJ817802 | 44844 | AID17959.1, AID17960.1 | N/A | Myovirus | Jeon et al., 2016b |
| YMC11/12/R1215 (R1215) | KP861231 | 44866 | AJT61417.1, AJT61416.1 | N/A | Myovirus | Jeon et al., 2016a |
| YMC11/12/R2315 (R2315) | KP861229 | 44846 | AJT61314.1, AJT61315.1 | N/A | Myovirus | Jeon et al., 2016a |
| KARL-1 | MH713599 | 166560 | YP_009881534.1, YP_009881544.1 | No | Myovirus | Jansen et al., 2018 |
| YMC13/03/R2096 (R2096) | KM672662 | 98170 | AIW02800.1, AIW02768.1 | N/A | Myovirus | Jeon et al., 2019 |
| Ab124 | MT633129 | 40471 | QMP19165.1 | N/A | Podovirus | Wu et al., 2021 |
| AB3 | KC311669 | 31185 | AGC35305.1 | Yes | Podovirus | Zhang et al., 2015 |
| Presley | KF669658 | 77792 | AGY48147.1 | N/A | Podovirus | Farmer et al., 2013 |
| SH-Ab 15497 | MG674163 | 43420 | AUG85465.1 | N/A | Siphovirus | Hua et al., 2018 |
| Abp1 | JX658790 | 42185 | AFV51022.1 | Yes | Podovirus | Huang et al., 2013 |
| Fri1 | KR149290 | 41805 | AKQ06854.1 | Yes | Podovirus | Knirel et al., 2020 |
| TAC1 | MK170160 | 101770 | AZF88430.1 | No | Myovirus | Asif et al., 2020 |
| IME200 | KT804908 | 41243 | ALJ97635.1 | Yes | Podovirus | Liu et al., 2019b |
| vB_AbaM_Acibel004 (Acibel004) | KJ473422 | 99730 | AHY26763.1, AHY26738.1 | N/A | Myovirus | Merabishvili et al., 2014 |
| vB_AbaM_B09_Aci01-1 (Aci01-1) | MH800198 | 103628 | AYD85523.1 | N/A | Myovirus | Essoh et al., 2019 |
| vB_AbaM_B09_Aci02-2 (Aci02-2) | MH800199 | 104354 | AYD85686.1 | N/A | Myovirus | Essoh et al., 2019 |
| vB_AbaM_B09_Aci05 (Aci05) | MH746814 | 102789 | AYD82311.1 | N/A | Myovirus | Essoh et al., 2019 |
| vB_AbaM_B9 (B9) | MH133207 | 93641 | AWD93202.1, AWD93211.1, AWD93192.1 | Yes | Myovirus | Oliveira et al., 2018 |
| vB_AbaM_IME285 (IME285) | MH853786 | 45063 | AYP68900.1 | Yes | Myovirus | Wang et al., 2020 |
| vB_AbaM_ME3 (ME3) | KU935715 | 234900 | AND75265.1, AND75267.1 | N/A | Myovirus | Buttimer et al., 2016 |
| vB_AbaM_PhT2 (PhT2) | MN864865 | 166330 | QHJ75775.1 | N/A | Myovirus | Kitti et al., 2014 |
| vB_AbaP_46-62_Aci07 (Aci07) | MH800200 | 42330 | AYD85862.1 | Yes | Podovirus | Essoh et al., 2019 |
| Petty | KF669656 | 40739 | AGY48011.1 | Yes | Podovirus | Hernandez-Morales et al., 2018 |
| phiAB1 | HQ186308 | 41526 | ADQ12745.1 | N/A | Podovirus | Li et al., 2012 |
| phiAB6 | KT339321 | 40570 | ALA12264.1 | Yes | Podovirus | Lai et al., 2016 |
| vB_AbaP_APK32 (APK32) | MK257722 | 41142 | AZU99395.1 | Yes | Podovirus | Popova et al., 2020a |
| SH-Ab 15519 | KY082667 | 40493 | APD19440.1 | Yes | Podovirus | Hua et al., 2018 |
| vB_AbaP_Acibel007 (Acibel007) | KJ473423 | 42654 | AHY26817.1 | N/A | Podovirus | Merabishvili et al., 2014 |
| vB_AbaP_APK48 (APK48) | MN294712 | 41105 | QFG06960.1 | Yes | Podovirus | Popova et al., 2020a |
| vB_AbaP_APK116 (APK116) | MN807295 | 41765 | QHS01530.1 | Yes | Podovirus | Popova et al., 2020a |
| vB_AbaP_APK2 (APK2) | MK257719 | 41476 | AZU99242.1 | Yes | Podovirus | Popova et al., 2020a |
| vB_AbaP_APK37 (APK37) | MK257723 | 41981 | AZU99445.1 | Yes | Podovirus | Popova et al., 2020a |
| vB_AbaP_APK44 (APK44) | MN604238 | 41461 | QGK90444.1 | Yes | Podovirus | Popova et al., 2020a |
| vB_AbaP_AS12 (AS12) | KY268295 | 41402 | APW79830.1 | Yes | Podovirus | Popova et al., 2017 |
| vB_AbaP_APK87 (APK87) | MN604239 | 42402 | QGK90498.1 | Yes | Podovirus | Popova et al., 2020a |
| vB_AbaP_APK89 (APK89) | MN651570 | 41198 | QGK90394.1 | Yes | Podovirus | Popova et al., 2020a |
| vB_AbaP_APK93 (APK93) | MK257721 | 41668 | AZU99342.1 | Yes | Podovirus | Popova et al., 2020a |
| vB_AbaP_B5 (B5) | MF033349 | 41608 | ASN73455.2 | Yes | Podovirus | Oliveira et al., 2017 |
| vB_AbaP_AS11 (AS11) | KY268296 | 41642 | AQN32697.1 | Yes | Podovirus | Popova et al., 2017 |
| vB_AbaP_PD-6A3 (PD-6A3) | NC_028684 | 41563 | YP_009190472.1 | Yes | Podovirus | Wu et al., 2019 |
| vB_AbaP_PD-AB9 (PD-AB9) | KT388103 | 40938 | ALM01895.1 | N/A | Podovirus | Wu et al., 2019 |
| vB_AbaP_B09_Aci08 (Aci08) | MH763831 | 42067 | AYD82867.1 | Yes | Podovirus | Essoh et al., 2019 |
| vB_AbaS_TRS1 (TRS1) | KX268652 | 40749 | ANT40742.1, ANT40741.1 | N/A | Siphovirus | Turner et al., 2016 |
| vB_ApiM_fHyAci03 (fHyAci03) | MH460829 | 165975 | AXF40726.1, AXF40736.1, AXF40808.1 | N/A | Myovirus | Pulkkinen et al., 2019 |
| vB_ApiP_P1 (P1) | MF033350 | 41208 | ASN73504.1 | Yes | Podovirus | Oliveira et al., 2017 |
| vB_AbaP_B1 (B1) | MF033347 | 40879 | ASN73353.1 | Yes | Podovirus | Oliveira et al., 2017 |
| WCHABP1 | KY829116 | 45888 | ARQ94727.1, ARQ94726.1 | Yes | Myovirus | Zhou et al., 2018 |
| WCHABP12 | KY670595 | 45415 | ARB06756.1, ARB06757.1 | Yes | Myovirus | Zhou et al., 2018 |
| vB_AbaP_B3 (B3) | MF033348 | 40598 | ASN73401.1 | Yes | Podovirus | Oliveira et al., 2017 |
| vB_AbaP_D2 (D2) | MH042230 | 39964 | AVP40472.1 | Yes | Podovirus | Yuan et al., 2021 |
| vB_AbaP_PMK34 (PMK34) | MN433707 | 41847 | QGF20174.1 | Yes | Podovirus | Abdelkader et al., 2020 |
| vB_ApiP_P2 (P2) | MF033351 | 41514 | ASN73558.1 | Yes | Podovirus | Oliveira et al., 2017 |
| ZZ1 | HQ698922 | 166687 | AEJ90215.1, AEJ90224.1 | N/A | Myovirus | Jin et al., 2012 |
| fEg-Aba01 | MT344103 | 33779 | QJT69876.1, QJT69875.1 | Yes | Siphovirus | Badawy et al., 2020 |
| fLi-Aba02 | MT344104 | 35093 | QJT69930.1, QJT69929.1 | Yes | Siphovirus | Badawy et al., 2020 |
| fLi-Aba03 | MT344105 | 34931 | QJT69984.1, QJT69983.1 | Yes | Siphovirus | Badawy et al., 2020 |
| phiAC-1 | JX560521 | 43216 | AFU62318.1 | N/A | Myovirus | Kim et al., 2012 |
| Loki | LN890663 | 41308 | CUS06481.1 | No | Siphovirus | Turner et al., 2017 |
3.2 Domains of the phage tail fibers
Investigation into each functional domain identified was conducted, and information on the presence of a halo around the plaques produced by each phage, or expression of a functional depolymerase, was used. Six major functional domains were identified in 65 tail fibers: lysozyme, G3DSA:2.60.40.3940 (immunoglobulin-like), galactose-binding domain-like, pectin lyase-like (PLD), SGNH hydrolase, and phage_tailspike_middle. Less common domains, such as Concanavalin A-like lectins/glucanases, (trans)glycosidases, and peptidase_S74_CIMCD, were found in 13 tail fibers, and 16 tail fibers had no protein domain hits (Figure 1). The functional domains associated with halo formation, a common identifiable characteristic of bacteriophage depolymerases, were found to be pectin lyase-like and phage_tailspike_middle. Other domains potentially associated with depolymerase activity were (trans)glycosidases and SGNH hydrolases.
Figure 1.
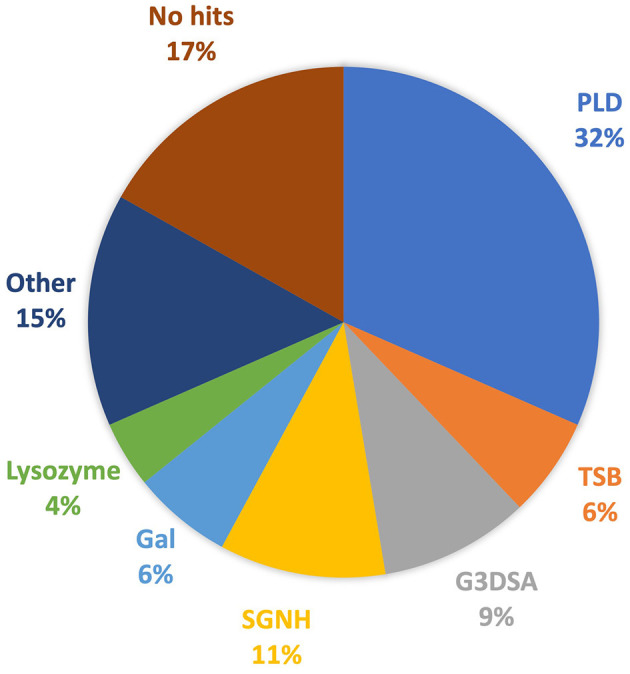
Protein domains identified in A. baumannii tail fibers analyzed with InterProScan: pectin lyase-like domain (PLD; blue), tailspike binding protein (TSB; orange), G3DSA:2.60.40.3940 (G3DSA; gray), SGNH domain (yellow), galactose-binding domain (Gal; light blue), lysozyme (green), other (navy), and no hits (brown).
3.2.1 Pectin lyase-like domains
Polysaccharide lyases, including pectin and pectate lyases, are enzymes that cleave (1,4)-glycosidic bonds through a β-elimination mechanism (Sutherland, 1999). The predominant functional domain identified in the analyzed tail fibers is the pectin lyase-like domain (PLD), which is present in 30 tail fibers. This domain is exclusive to Ackermannviridae (1), Autographiviridae (22), and myoviruses (seven) (Table 2), which are associated with depolymerase activity studied in various recombinantly expressed proteins (Liu et al., 2019b; Oliveira et al., 2019; Popova et al., 2020a; Abdelkader et al., 2022).
Table 2.
InterProScan hit table of the 30 tail fibers containing a pectin lyase-like domain.
| Sequence | Name | Type | InterPro ID | Minimum | Maximum | Length |
|---|---|---|---|---|---|---|
| AB1; gp76 | Pectin_lyas_fold | Gene3D | IPR012334 | 155 | 503 | 349 |
| Beta_helix | Pfam | IPR039448 | 262 | 426 | 165 | |
| pbh1 | SMART | IPR006626 | 368 | 648 | 156 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 201 | 535 | 335 | |
| phiAC-1; gp69 | Pectin lyase-like | SUPERFAMILY | IPR011050 | 153 | 422 | 270 |
| Abp1; gp47 | pyocin_knob | CDD | 671 | 761 | 91 | |
| Pectin_lyas_fold | Gene3D | IPR012334 | 283 | 624 | 342 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 291 | 615 | 325 | |
| IME-AB2; gp71 | pyocin_knob | CDD | 599 | 685 | 87 | |
| Pectin_lyas_fold | Gene3D | IPR012334 | 207 | 547 | 341 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 214 | 544 | 331 | |
| AB3; gp4 | pbh1 | SMART | IPR006626 | 420 | 683 | 92 |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 277 | 566 | 290 | |
| Petty; gp39 | pyocin_knob | CDD | 703 | 775 | 73 | |
| Pectin_lyas_fold | Gene3D | IPR012334 | 329 | 586 | 258 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 333 | 604 | 272 | |
| Acibel007; gp46 | Pectin_lyas_fold | Gene3D | IPR012334 | 186 | 474 | 289 |
| YMC-13-01-C62; gp45 | Pectin lyase-like | SUPERFAMILY | IPR011050 | 234 | 545 | 312 |
| YMC13/03/R2096; gp34 | Pectin_lyas_fold | Gene3D | IPR012334 | 347 | 814 | 468 |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 376 | 660 | 285 | |
| YMC11/12/R2315; gp83 | Pectin lyase-like | SUPERFAMILY | IPR011050 | 234 | 545 | 312 |
| YMC11/12/R1215; gp21 | Pectin lyase-like | SUPERFAMILY | IPR011050 | 234 | 545 | 312 |
| Fri1; gp49 | Pectin_lyas_fold | Gene3D | IPR012334 | 292 | 765 | 474 |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 292 | 623 | 332 | |
| phiAB6; gp40 | Pectin_lyas_fold | Gene3D | IPR012334 | 192 | 387 | 196 |
| Pectate_lyase_3 | Pfam | IPR024535 | 225 | 433 | 209 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 221 | 495 | 275 | |
| IME200; gp48 | Pectin_lyas_fold | Gene3D | IPR012334 | 155 | 493 | 339 |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 171 | 469 | 299 | |
| SH-Ab 15519; gp45 | Pectin_lyas_fold | Gene3D | IPR012334 | 142 | 488 | 347 |
| Pectate_lyase_3 | Pfam | IPR024535 | 176 | 383 | 208 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 171 | 468 | 298 | |
| AS11; gp45 | Pectin_lyas_fold | Gene3D | IPR012334 | 291 | 767 | 477 |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 292 | 621 | 330 | |
| B1; gp45 | phage_tailspike_middle | CDD | 173 | 581 | 409 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 328 | 564 | 237 | |
| B3; gp42 | Pectin_lyas_fold | Gene3D | IPR012334 | 203 | 561 | 359 |
| Pectate_lyase_3 | Pfam | IPR024535 | 225 | 433 | 209 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 222 | 557 | 336 | |
| P2; gp48 | Pectin_lyas_fold | Gene3D | IPR012334 | 230 | 541 | 312 |
| Beta_helix | Pfam | IPR039448 | 298 | 462 | 165 | |
| pbh1 | SMART | IPR006626 | 377 | 684 | 180 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 231 | 567 | 337 | |
| D2; gp2 | Pectin_lyas_fold | Gene3D | IPR012334 | 203 | 569 | 367 |
| Pectate_lyase_3 | Pfam | IPR024535 | 225 | 432 | 208 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 221 | 556 | 336 | |
| SH-Ab 15599; gp196 | Pectin_lyas_fold | Gene3D | IPR012334 | 302 | 666 | 365 |
| Pectate_lyase_3 | Pfam | IPR024535 | 322 | 425 | 104 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 316 | 656 | 341 | |
| Aci08; gp46 | Pectin_lyas_fold | Gene3D | IPR012334 | 229 | 506 | 278 |
| pbh1 | SMART | IPR006626 | 336 | 428 | 86 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 328 | 571 | 244 | |
| APK2; gp43 | Pectin_lyas_fold | Gene3D | IPR012334 | 155 | 493 | 339 |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 171 | 469 | 299 | |
| APK93; gp43 | Pectin_lyas_fold | Gene3D | IPR012334 | 155 | 493 | 339 |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 171 | 469 | 299 | |
| APK37; gp44 | Pectin_lyas_fold | Gene3D | IPR012334 | 225 | 586 | 362 |
| Pectate_lyase_3 | Pfam | IPR024535 | 238 | 364 | 127 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 233 | 542 | 310 | |
| PMK34; gp45 | Pectin_lyas_fold | Gene3D | IPR012334 | 203 | 570 | 368 |
| Pectate_lyase_3 | Pfam | IPR024535 | 225 | 433 | 209 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 221 | 557 | 337 | |
| APK44; gp44 | Pectin lyase-like | SUPERFAMILY | IPR011050 | 204 | 490 | 287 |
| APK87; gp48 | Pectin lyase-like | SUPERFAMILY | IPR011050 | 234 | 526 | 293 |
| APK89; gp46 | Pectin_lyas_fold | Gene3D | IPR012334 | 206 | 612 | 407 |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 260 | 566 | 307 | |
| Ab124; gp46 | Pectin_lyas_fold | Gene3D | IPR012334 | 140 | 492 | 353 |
| Pectate_lyase_3 | Pfam | IPR024535 | 176 | 383 | 208 | |
| Pectin lyase-like | SUPERFAMILY | IPR011050 | 171 | 468 | 298 |
Tail fibers containing PLD exhibit diversity, with an average pairwise identity of 19.2% and lengths ranging from 693 to 921 amino acids (AAs). Among these tail fibers, 23 phages with a PLD domain were explicitly documented to exhibit halo formation around their plaques (Table 1). Although the majority of tail fibers share a common PLD SUPERFAMILY (SSF51126), phage Acibel007 (gp46) deviates with only a Gene3D hit (2.160.20.10; Pectin_lyas_fold) (Table 2). Some variations involve Pfam, SMART, or CDD hits, with 10 tail fibers overlapping a Pfam hit, eight featuring Pectate_lyase_3 (PF12708), and two labeled as Beta_helix (PF13229) for myovirus phage AB1 (gp76) and Autographiviridae phage P2 (gp48). Additionally, four tail fibers contain a SMART PbH1 (SM00710) hit, corresponding to parallel beta-helix repeats in pectate lyases and rhamnogalacturonase A.
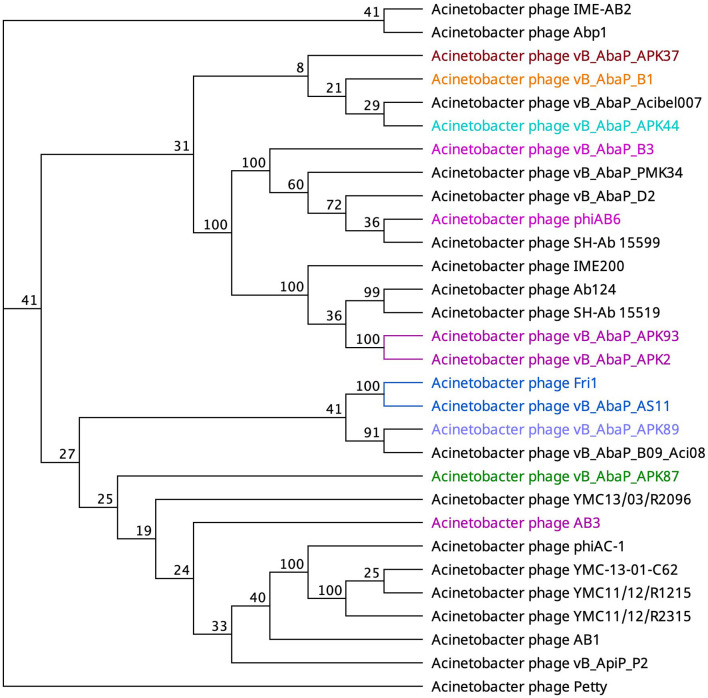
An alternate domain architecture features a PLD SUPERFAMILY (SSF51126) overlapping a Gene3D (2.160.20.10) and a CDD pyocin_knob hit at the CTD (cd19958) (Table 2). This layout is present in the tail fibers of two podoviruses (Abp1 gp47 and IME-AB2 gp71) and one myovirus (Petty gp39) (Table 2). Phage proteins sharing this domain range from 21.9% to 29.6% identity (Supplementary Table 2). Finally, Autographiviridae phage B1 gp45 encoded the only tail fiber with both a PLD SUPERFAMILY hit and a phage_tailspike_middle hit from the CDD database (Table 2). A phylogenetic tree illustrates that tail fiber proteins do not group based on capsule targets but mainly on viral morphology (Figure 2). Generally, the myoviruses group together except for SH-Ab 155599 and IME-AB2, which group with podoviruses.
Figure 2.
Unrooted phylogenetic tree of PLD containing tail fiber proteins with bootstrap support percent at branch nodes. The colors of tips correspond to documented capsule targets of the depolymerases: blue, K19; pink, K2; teal, K44; orange, K9; green, K87; purple, K89; and burgundy, K37.
3.2.2 Phage tailspike middle
The tail fibers of six phages feature a hit to the phage_tailspike_middle domain from the CDD (Table 3). This model characterizes the middle beta-helical domain of Acinetobacter bacteriophage tail spike proteins, encompassing a distinct N-terminal domain unrelated to the beta-helical substructure but implicated in virion binding. The C-terminal domain, highly variable, is suggested to play a role in receptor binding. Among these phages, AM24, IME285, BS46, and WCHABP12 (Tables 1, 3) exhibit halo formation around their plaques, with recombinantly expressed tail spike proteins from AM24 (gp50), BS46 (gp47), and IME285 (AYP68900.1) that are confirmed as functional depolymerases (Popova et al., 2019; Knirel et al., 2020; Wang et al., 2020).
Table 3.
InterProScan hit table of the six tail fibers containing a phage_tailspike_middle domain.
| Sequence name | Name | Type | Min (AA) | Max (AA) | Length (AA) |
|---|---|---|---|---|---|
| AM24; gp50 | phage_tailspike_middle | CDD | 266 | 669 | 404 |
| WCHABP12; gp16 | phage_tailspike_middle | CDD | 150 | 553 | 404 |
| B1; gp45 | phage_tailspike_middle | CDD | 173 | 581 | 409 |
| Pectin lyase-like | SUPERFAMILY | 328 | 564 | 237 | |
| B5; gp47 | phage_tailspike_middle | CDD | 179 | 581 | 403 |
| IME285; gp35 | phage_tailspike_middle | CDD | 150 | 553 | 404 |
| BS46; gp47 | phage_tailspike_middle | CDD | 243 | 646 | 404 |
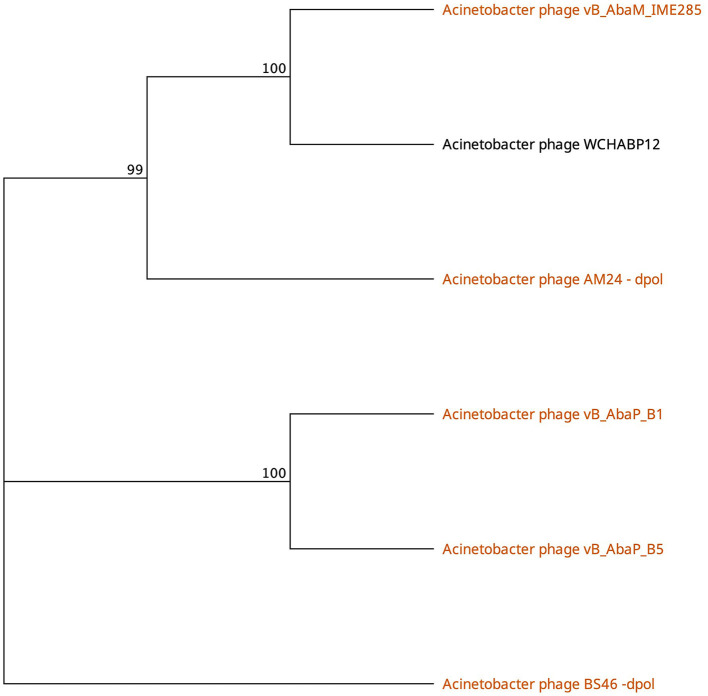
Phages featuring tail fibers with the phage_tailspike_middle domain are confined to myoviruses and Autographiviridae lineages. The six identified proteins, ranging from 732 to 848 amino acids, share a pairwise identity of 60.1% (Supplementary Table 3). These proteins cluster by taxonomy, with a high % identity observed within myovirus-derived tail fibers (WCHABP12, gp16; IME285, AYP68900.1; BS46, gp47; and AM24, gp50) at 72.4–97.4% (Figure 3, Supplementary Table 2). Autographiviridae proteins B1 (gp45) and B5 (gp47) share 40.7% identity (Figure 3, Supplementary Table 3). Intriguingly, B5′s tail fiber exhibits greater similarity to myovirus phage tail fibers, with % identity ranging from 69.1% to 73.2%. The most diverse tail fiber in this group belongs to B1, featuring a SUPERFAMILY pectin lyase-like hit (SSF51126) overlapping the CDD domain (Figure 3, Table 3). This 761-AA-long tail fiber shares 22.5–40.7% identity with other tail fibers in this group.
Figure 3.
Unrooted phylogenetic tree of Tailspike_middle_domain containing tail fiber proteins with bootstrap support percent at branch nodes. Orange tips indicate halo formation documented against K9 capsule types.
3.2.3 CATH/G3DSA 2.60.40.3940 domain
The CATH superfamily 2.60.40.3940 is characterized by a predominantly beta class (2), sandwich architecture (2.60), and immunoglobulin-like topology (2.60.40) within a novel homologous superfamily (2.60.40.3940). In the Obolenskvirus clade of Acinetobacter myophages, nine phages, namely, AP22 (gp53), AB1 (gp77), YMC-13-01-C62 (gp46), YMC11/12/R2315 (gp84), YMC11/12/R1215 (gp20), WCHABP12 (gp15), WCHABP1 (gp6), AbP2 (gp18), and Abp9 (gp49), feature the G3DSA 2.60.40.3940 domain in the CTD of one of their tail fiber proteins (Table 4). These proteins, encoded in the morphogenesis region and positioned upstream of other tail fiber proteins, vary from 258 to 283 amino acids, sharing an overall pairwise identity of 83.0% (Supplementary Table 4). An analysis of the multisequence alignment reveals high sequence conservation at the N-terminal domain (NTD), indicating its involvement in virion attachment. Conversely, the CTD, associated with the 2.60.40.3940 hit, displays a breakdown in sequence identity among some proteins, suggesting a potential role in host cell recognition. This role is supported by the crystal structure of Acinetobacter phage AP22 gp53 CTD (PDB: 4MTM), resembling a homotrimeric globular lectin-like protein with a slender midsection, akin to host-cell binding proteins of Escherichia coli phages T7 gp17 (Garcia-Doval and Van Raaij, 2012) and T4 gp12 (Van Raaij et al., 2001). Furthermore, AP22 gp53C binds to ethylene glycol and glycerol molecules, which are surrogates of an oligosaccharide backbone. Overall, this data suggests that tail fibers encoding a 2.60.40.3940 domain are involved in recognizing oligosaccharide moieties of Acinetobacter baumannii.
Table 4.
InterProScan hit table of the nine tail fibers containing a CATH/G3DSA 2.60.40.3940 domain.
| Sequence Name | Name | Type | Min (AA) | Max (AA) | Length (AA) |
|---|---|---|---|---|---|
| AP22; gp53 | G3DSA:2.60.40.3940 | Gene3D | 176 | 271 | 96 |
| AB1; gp77 | G3DSA:2.60.40.3940 | Gene3D | 184 | 283 | 100 |
| Agglutinin HPA-like | SUPERFAMILY | 192 | 282 | 91 | |
| C62; gp46 | G3DSA:2.60.40.3940 | Gene3D | 181 | 276 | 96 |
| R2315; gp84 | G3DSA:2.60.40.3940 | Gene3D | 182 | 276 | 95 |
| R1215; gp20 | G3DSA:2.60.40.3940 | Gene3D | 181 | 276 | 96 |
| WCHABP12; gp15 | G3DSA:2.60.40.3940 | Gene3D | 187 | 281 | 95 |
| WCHABP1; gp6 | G3DSA:2.60.40.3940 | Gene3D | 187 | 281 | 95 |
| AbP2; gp18 | G3DSA:2.60.40.3940 | Gene3D | 183 | 276 | 94 |
| Abp9; gp49 | G3DSA:2.60.40.3940 | Gene3D | 165 | 258 | 94 |
3.2.4 SGNH hydrolase domain
The SGNH hydrolase superfamily comprises 16 well-studied protein families with a conserved catalytic fold and mechanism (Anderson et al., 2022). These enzymes, named after their catalytic Ser, His, Gly, and Asn residues, function as esterases and lipases, playing vital roles in biomass conversion, pathogenesis, and cell signaling (Akoh et al., 2004; Anderson et al., 2022). The SGNH hydrolase domain was identified in the tail fibers of 10 phages: SH-Ab 15497, fHyAci03, KARL-1, PhT2, fEg-Aba01, fLi-Aba02, fLi-Aba03, DMU1, PD-6A3, and AbTZA1, with a pairwise identity of 33.7%. Four of these phages exhibited halo formation (PD-6A3, fEg-Aba01, fLi-Aba02, and fLi-Aba03) (Wu et al., 2019; Badawy et al., 2020) (Table 5, Supplementary Table 5). Based on the structural organization of the domains, the tail fibers can be grouped into two architectures. One group features two fibritin domains: one at the NTD and one directly upstream of the SGNH hydrolase domain (Table 5). This domain layout is restricted to four members of the subfamily Tevenvirinae: vB_ApiM_fHyAci03 (fHyAci03), KARL-1, vB_AbaM_PhT2 (PhT2), and AbTZA1, which tend to group together (Figure 4). Fibritin belongs to a class of chaperones that catalyze specific phage-assembly processes, promoting the assembly of the long tail fibers and their attachment to the tail baseplate (Tao et al., 1997). Furthermore, fibritin also serves as a sensing device, controlling the retraction of the long tail fibers in adverse environments to prevent infection (Tao et al., 1997). These four proteins with this domain layout have similar lengths, ranging from 621 to 626 AA and share between 55% and 97.6% AA identity (Table 5 and Supplementary Table 5).
Table 5.
InterProScan hit table of the 10 tail fibers containing an SGNH hydrolase domain.
| Sequence | Name | Type | Min (AA) | Max (AA) | Length (AA) |
|---|---|---|---|---|---|
| SH-Ab 15497; gp21 | SGNH hydrolase | Gene3D | 267 | 522 | 256 |
| SASA | Pfam | 358 | 519 | 162 | |
| SGNH hydrolase | SUPERFAMILY | 338 | 520 | 183 | |
| fHyAci03; gp175 | SGNH hydrolase | Gene3D | 463 | 615 | 153 |
| G3DSA:1.20.5.320 | Gene3D | 353 | 446 | 94 | |
| G3DSA:1.20.5.320 | Gene3D | 3 | 89 | 87 | |
| Fibritin_C | Pfam | 357 | 412 | 56 | |
| SGNH hydrolase | SUPERFAMILY | 472 | 606 | 135 | |
| Fibritin | SUPERFAMILY | 6 | 106 | 101 | |
| KARL-1; gp114 | SGNH hydrolase | Gene3D | 466 | 618 | 153 |
| G3DSA:1.20.5.320 | Gene3D | 356 | 449 | 94 | |
| G3DSA:1.20.5.320 | Gene3D | 3 | 89 | 87 | |
| Fibritin_C | Pfam | 360 | 415 | 56 | |
| SGNH hydrolase | SUPERFAMILY | 475 | 609 | 135 | |
| Fibritin | SUPERFAMILY | 6 | 106 | 101 | |
| PhT2; gp163 | SGNH hydrolase | Gene3D | 450 | 624 | 175 |
| G3DSA:1.20.5.320 | Gene3D | 357 | 449 | 93 | |
| G3DSA:1.20.5.320 | Gene3D | 2 | 89 | 88 | |
| Fibritin_C | Pfam | 362 | 416 | 55 | |
| SGNH hydrolase | SUPERFAMILY | 476 | 601 | 126 | |
| Fibritin | SUPERFAMILY | 4 | 106 | 103 | |
| fEg-Aba01; gp19 | SGNH hydrolase | Gene3D | 258 | 481 | 224 |
| SASA | Pfam | 315 | 477 | 163 | |
| SGNH hydrolase | SUPERFAMILY | 233 | 479 | 247 | |
| fLi-Aba02; gp21 | SGNH hydrolase | Gene3D | 258 | 481 | 224 |
| SASA | Pfam | 315 | 477 | 163 | |
| SGNH hydrolase | SUPERFAMILY | 233 | 479 | 247 | |
| fLi-Aba03; gp21 | SGNH hydrolase | Gene3D | 258 | 481 | 224 |
| SASA | Pfam | 315 | 477 | 163 | |
| SGNH hydrolase | SUPERFAMILY | 233 | 479 | 247 | |
| DMU1; gp20 | SGNH hydrolase | Gene3D | 267 | 521 | 255 |
| SASA | Pfam | 356 | 518 | 163 | |
| SGNH hydrolase | SUPERFAMILY | 337 | 519 | 183 | |
| PD-6A3; gp13 | SGNH hydrolase | Gene3D | 430 | 682 | 253 |
| SASA | Pfam | 522 | 664 | 143 | |
| SGNH hydrolase | SUPERFAMILY | 427 | 675 | 249 | |
| AbTZA1; gp180 | SGNH hydrolase | Gene3D | 450 | 624 | 175 |
| G3DSA:1.20.5.320 | Gene3D | 357 | 449 | 93 | |
| G3DSA:1.20.5.320 | Gene3D | 2 | 89 | 88 | |
| Fibritin_C | Pfam | 361 | 416 | 56 | |
| SGNH hydrolase | SUPERFAMILY | 476 | 601 | 126 | |
| Fibritin | SUPERFAMILY | 4 | 106 | 103 |
Figure 4.
Unrooted phylogenetic tree of G3DSA:2.60.40.3940 domain containing tail fiber proteins with bootstrap support percent at branch nodes. Green tips indicate halo formation documented in the literature.
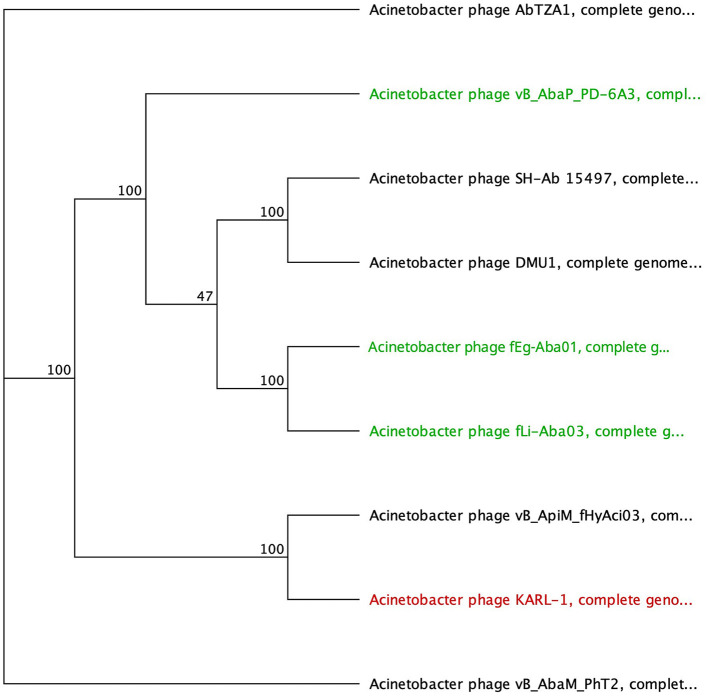
The second group comprises hypothetical proteins from six phages, featuring a lone SGNH domain at the CTD of the protein (Figure 4, Table 5). This domain structure is present in one Autographiviridae member, PD-6A3, and five siphoviruses (SH-Ab 15497, fEg-Aba01, fLi-Aba02, fLi-Aba03, DMU1), which group more closely together (Figure 4). PD-6A3 encodes an endolysin with activity against A. baumannii cells, which could potentially be responsible for the observed halo formation (Wu et al., 2019). In contrast, siphophages, fEg-Aba01, fLi-Aba02, and fLi-Aba03, encode two putative tail fiber proteins: one with an SGNH hydrolase domain and, in the case of fLi-Aba02 and fLi-Aba03, a Concanavalin A-like lectins/glucanases domain. The siphovirus proteins are very close in length, ranging from 620 to 660 AA, sharing 41 to 100% AA identity (Table 5, Supplementary Table 5). Phages with 100% AA identity are fEg-Aba01, fLi-Aba02, and fLi-Aba03. Comparatively, the podophage PD-6A3 tail fiber protein is significantly longer at 817 AA, although it shares approximately 41% identity with the five siphoviruses of the same layout (Figure 4, Table 5, Supplementary Table 5).
Since our data collection, the Acinetobacter podovirus Aristophanes was published. This phage does not produce a halo but encodes a tail spike SGNH hydrolase domain (gp41) (Timoshina et al., 2021). Functional study of this protein revealed a tail deacetylase causing O-acetylation of one of the K26 sugar residues which causes a slight decrease in turbidity of the host (Timoshina et al., 2021). This finding suggests that the other phages may also utilize this structural protein as a deacetylase. To get more information on the potential functions of the phages DMU1, PD-6A3, and SH Ab 15497, the proteins were modeled with AlphaFold Multimer as homotrimers, and the resulting models were submitted to the DALI server PDB search. The top hits are to a xyloglucan-active beta-galactosidase from Xanthomonas citri (7KMM) for all models, followed by a protein with an unknown function from Arabidopsis thaliana (2APJ) for DMU1 and PD-63A and a homo-dimer acetylxylan esterase from Clostridium acetobutylicum (1ZMB) for SH Ab 15497.
3.2.5 Galactose binding domain
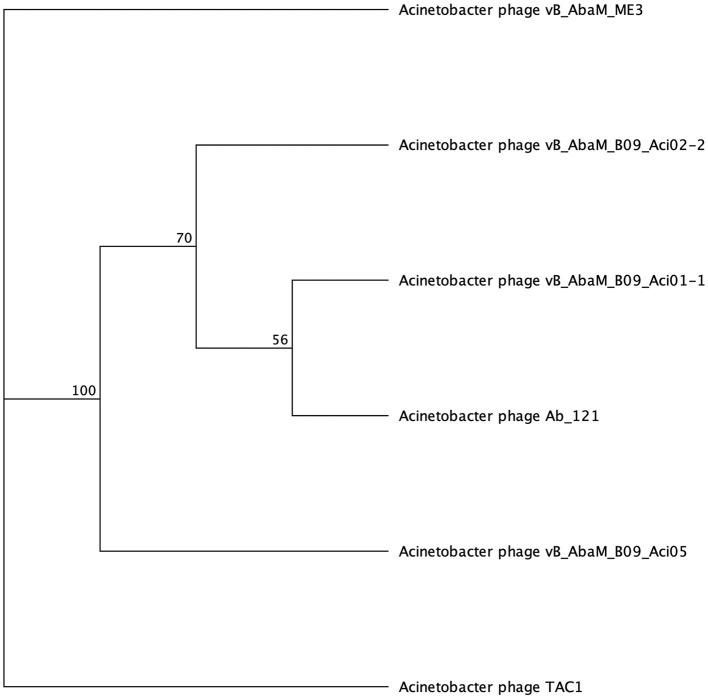
Galactose binding domains are present in several different protein families in eukaryotes and prokaryotes and bind to specific ligands, such as cell-surface-attached carbohydrates (Ito et al., 1991). The members of this domain exhibit a β-sandwich forming a jelly roll fold. In the tail fibers of six myophages (ME3, TAC1, Aci05, Aci02-2, Aci01-1, and Ab_121), a galactose-binding domain was identified (Figure 5, Table 6). Notably, literature reports indicate that five of these phages do not exhibit halo formation (Lee et al., 2011; Essoh et al., 2019; Asif et al., 2020). Tail fibers encoding this domain varied significantly in sequence length, spanning from 1,177 to 5,419 AA (Table 6), with diverse percent amino acid identity ranging from 12.9 to 98.8% (Supplementary Table 6). ME3, the first jumbo Acinetobacter phage identified, presented the most divergent tail fiber with an average % identity ranging from 12.4–13.2 % (Figure 5, Supplementary Table 6) (Buttimer et al., 2016). The tail fiber protein encoded by ME3 is 5,419 AA long and features multiple hits from Gene3D, SUPERFAMILY, Pfam, and PROSITE profiles (Figure 5, Table 5).
Figure 5.
Unrooted phylogenetic tree of SGNH hydrolase-encoding tail fiber proteins. Bootstrap support percent is listed at branch nodes. Green tips indicate halo formation in literature, red tips indicate lack of halo formation documented, and black tips indicate no data on halo formation available.
Table 6.
InterProScan hit table of the six tail fibers containing a galactose binding domain.
| Sequence name | Name | Type | Min (AA) | Max (AA) | Length (AA) |
|---|---|---|---|---|---|
| ME3; gp104 | G3DSA:2.60.120.260 | Gene3D | 4,468 | 4,628 | 161 |
| G3DSA:2.60.120.260 | Gene3D | 1,014 | 1,172 | 159 | |
| G3DSA:2.60.120.260 | Gene3D | 4,179 | 4,331 | 153 | |
| G3DSA:2.60.120.260 | Gene3D | 3,438 | 3,577 | 140 | |
| G3DSA:2.60.120.260 | Gene3D | 5,266 | 5,398 | 133 | |
| G3DSA:2.60.120.260 | Gene3D | 5,018 | 5,128 | 111 | |
| F5_F8_type_C | Pfam | 4,505 | 4,608 | 104 | |
| DUF1983 | Pfam | 4,359 | 4,410 | 52 | |
| FA58C_3 | PROSITE_PROFILES | 4,461 | 4,623 | 163 | |
| Galactose-binding domain-like | SUPERFAMILY | 4,178 | 4,331 | 154 | |
| Galactose-binding domain-like | SUPERFAMILY | 4,483 | 4,622 | 140 | |
| Galactose-binding domain-like | SUPERFAMILY | 5,264 | 5,397 | 134 | |
| Galactose-binding domain-like | SUPERFAMILY | 3,442 | 3,571 | 130 | |
| Aci05; gp30 | G3DSA:2.60.120.260 | Gene3D | 800 | 922 | 123 |
| Galactose-binding domain-like | SUPERFAMILY | 798 | 895 | 98 | |
| Aci01-1; gp31 | G3DSA:2.60.120.260 | Gene3D | 1,265 | 1,398 | 134 |
| G3DSA:2.60.120.260 | Gene3D | 800 | 922 | 123 | |
| A-type inclusion protein, putative-related | Panther | 424 | 1,488 | 1,065 | |
| Galactose-binding domain-like | SUPERFAMILY | 800 | 878 | 79 | |
| Aci02-2; gp32 | G3DSA:2.60.120.260 | Gene3D | 1,265 | 1,398 | 134 |
| G3DSA:2.60.120.260 | Gene3D | 800 | 922 | 123 | |
| A-type inclusion protein, putative-related | Panther | 423 | 1,485 | 1,063 | |
| Galactose-binding domain-like | SUPERFAMILY | 800 | 878 | 79 | |
| TAC1; gp42 | G3DSA:1.20.5.340 | Gene3D | 920 | 1,470 | 221 |
| G3DSA:2.60.120.260 | Gene3D | 777 | 919 | 143 | |
| G3DSA:1.10.287.1490 | Gene3D | 427 | 563 | 137 | |
| Myosin heavy chain - related | Panther | 184 | 1,020 | 837 | |
| MPN010-like | SUPERFAMILY | 1,408 | 1,460 | 53 | |
| Ab 121; gp12 | G3DSA:2.60.120.260 | Gene3D | 1,265 | 1,398 | 134 |
| G3DSA:2.60.120.260 | Gene3D | 800 | 922 | 123 | |
| A-type inclusion protein, putative-related | Panther | 91 | 1,436 | 1,346 | |
| Galactose-binding domain-like | SUPERFAMILY | 798 | 895 | 98 |
The second most divergent tail fiber belongs to TAC1, displaying an identity range of 13–42.5% (Figure 5, Supplementary Table 6), containing a myosin heavy chain Panther hit (PTHR18921) overlapping a Gene3D hit (1.10.287.1490, Phosphatidylinositol 3-kinase regulator activity). TAC1′s tail fiber also included two additional Gene3D hits corresponding to a galactose-binding domain (2.60.120.260) and a myosin heavy chain (1.20.5.340). Phages Ab_121, Aci01-1, and Aci02-2 encode a 2,211 AA tail fiber protein with an identity range of 96.8% and 98.9% (Supplementary Table 6). Aci01-1 and Aci02-2 have matching Panther (A-type inclusion protein), Gene3D (galactose binding domain), and SUPERFAMILY (galactose binding domain) hits spanning the length of the protein (Figure 5, Table 6). Phage Ab_121 tail fiber has the same Gene3D hits but differs in the location of the Panther and SUPERFAMILY hits. Phage Aci05 also has similar domain hits to phages Ab_121, Aci01-1, and Aci02-2, although it lacks a Panther and one of the Gene3D hits.
3.2.6 Lysozyme domain
Four phages were identified to encode a lysozyme domain in their tail fibers (Table 6). Among them, three are unclassified Tevenvirinae myoviruses (ZZ1, gp162; fHyAci03, gp165; and KARL-1, gp124) while the remaining phage is an unclassified podovirus (Presley, gp80). Notably, only the characterization article of KARL-1 discussed halo formation, which was not observed previously (Jansen et al., 2018). Presley shares 12% identity to the myovirus proteins (Supplementary Table 7).
The most highly annotated proteins with the lysozyme domain belong to Tevenvirinae, reflecting the extensive research on the classical coliphage T4 (Table 7). The sequence length of these proteins varies from 593 to 600 AA long with notable conservation in percent identity ranging from 83.4% and 99.7% (Supplementary Table 7). All three proteins hit needle_T4 from the HAMAP database (Figure 6). Furthermore, each phage has three distinct SUPERFAMILY database hits corresponding to gp5 N-terminal domain-like (SSF69255), lysozyme-like (SSF53955), and phage fiber proteins (SSF69349) (Figure 6, Table 7). Three Gene3D hits are present on all proteins (2.40.50.260, 1.10.530.40, and 3.10.450.190), as well as two Pfam hits (Gp5_OB, PF06714 and Phage_lysozyme, PF00959). The lysozyme functionality of these proteins is supported by a Panther hit (T4-type lysozyme 1-related, PTHR37406), CDD hit (T4-like_lys, cd00735), and a PRINTS hit (T4 lysozyme, PR00684).
Table 7.
InterProScan hit table of the four tail fibers containing a lysozyme domain.
| Sequence | Name | Type | Min (AA) | Max (AA) | Length (AA) |
|---|---|---|---|---|---|
| ZZ1; gp162 | T4-like_lys | CDD | 173 | 331 | 159 |
| Needle_t4 | HAMAP | 3 | 593 | 591 | |
| T4lysozyme | PRINTS | 175 | 332 | 121 | |
| T4-type lysozyme 1-related | Panther | 173 | 332 | 160 | |
| Gp5_OB | Pfam | 33 | 171 | 139 | |
| Phage_lysozyme | Pfam | 195 | 320 | 126 | |
| G3dsa:1.10.530.40 | Gene3D | 172 | 337 | 166 | |
| G3dsa:2.40.50.260 | Gene3D | 1 | 128 | 128 | |
| G3dsa:3.10.450.190 | Gene3D | 390 | 461 | 72 | |
| Phage fiber proteins | SUPERFAMILY | 391 | 585 | 195 | |
| Lysozyme-like | SUPERFAMILY | 145 | 337 | 193 | |
| Gp5 N-terminal domain-like | SUPERFAMILY | 6 | 128 | 123 | |
| Presley; gp80 | LT-like | CDD | 256 | 376 | 121 |
| G3dsa:1.10.530.10 | Gene3D | 242 | 387 | 146 | |
| Slt | Pfam | 273 | 353 | 81 | |
| Lysozyme-like | SUPERFAMILY | 256 | 373 | 118 | |
| fHyAci03; gp165 | T4-like_lys | CDD | 173 | 331 | 159 |
| Baseplate hub and tail lysozyme CDS | CDS | 1 | 594 | 594 | |
| Needle_t4 | HAMAP | 3 | 594 | 592 | |
| T4lysozyme | PRINTS | 175 | 332 | 121 | |
| T4-type lysozyme 1-related | Panther | 173 | 332 | 160 | |
| Gp5_OB | Pfam | 33 | 171 | 139 | |
| Phage_lysozyme | Pfam | 195 | 320 | 126 | |
| G3dsa:1.10.530.40 | Gene3D | 172 | 337 | 166 | |
| G3dsa:2.40.50.260 | Gene3D | 1 | 128 | 128 | |
| G3dsa:3.10.450.190 | Gene3D | 376 | 463 | 88 | |
| Lysozyme-like | SUPERFAMILY | 141 | 338 | 198 | |
| Phage fiber proteins | SUPERFAMILY | 393 | 586 | 194 | |
| Gp5 N-terminal domain-like | SUPERFAMILY | 6 | 128 | 123 | |
| KARL-1; gp124 | T4-like_lys | CDD | 173 | 331 | 159 |
| Needle_t4 | HAMAP | 3 | 594 | 592 | |
| T4lysozyme | PRINTS | 175 | 332 | 121 | |
| T4-type lysozyme 1-related | Panther | 173 | 332 | 160 | |
| Gp5_OB | Pfam | 33 | 171 | 139 | |
| Phage_lysozyme | Pfam | 195 | 320 | 126 | |
| G3DSA:1.10.530.40 | Gene3D | 172 | 337 | 166 | |
| G3DSA:2.40.50.260 | Gene3D | 1 | 128 | 128 | |
| G3DSA:3.10.450.190 | Gene3D | 376 | 463 | 88 | |
| Lysozyme-like | SUPERFAMILY | 141 | 338 | 198 | |
| Phage fiber proteins | SUPERFAMILY | 393 | 586 | 194 | |
| gp5 N-terminal domain-like | SUPERFAMILY | 6 | 128 | 123 |
Figure 6.
Unrooted phylogenetic tree of lysozyme-encoding tail fiber proteins. Bootstrap support percent is listed at branch nodes. Red tips indicate a lack of halo formation documented, and black tips indicate no data on halo formation available.
3.2.7 Other domains
The analysis of the phage tail fibers also revealed nine unique functional domains found in three Autographiviridae, four myoviruses, and five siphovirus phages; of which, seven were reported to have halo formation: fEgAba01 (gp20), fLiAba02 (gp22), fLiAba03 (gp22), Aci07 (gp45), TaPaz (gp78), APK116 (gp43), and AS12 (gp42) (Table 8) (Popova et al., 2017, 2020a; Essoh et al., 2019; Badawy et al., 2020; Shchurova et al., 2021). Further analysis will be focused on the abovementioned phages due to their halo formation and in the case of AKP116, AS12, and TaPaz, the expression of a recombinant depolymerase enzyme. The six remaining phage tail fibers with unique domains belong to Acibel004 (gp123; 2201 AA), ME3 (gp106; 574 AA), TRS1 (gp30; 526 AA), fHyAci03 (gp247; 1259 AA), R2096 (gp66; 414 AA), and ZZ1 (gp171; 510 AA) (Table 8).
Table 8.
InterProScan hit table of the 11 tail fibers encoding rare domains.
| Sequence name | Name | Type | Min (AA) | Max (AA) | Length (AA) |
|---|---|---|---|---|---|
| YMC13/03/R2096; gp66 | Collagen | Pfam | 50 | 96 | 47 |
| ME3; gp106 | G3DSA:3.90.1340.10 | Gene3D | 418 | 482 | 65 |
| The receptor-binding domain of short tail fiber protein gp12 | SUPERFAMILY | 423 | 574 | 152 | |
| Collar | Pfam | 430 | 488 | 59 | |
| Phage_fiber_2 | Pfam | 244 | 280 | 37 | |
| TRS1; gp30 | G3DSA:2.10.10.20 | Gene3D | 75 | 119 | 45 |
| AS12; gp42 | Peptidase_S74_CIMCD | CDD | 788 | 896 | 109 |
| G3DSA:1.10.10.10 | Gene3D | 788 | 901 | 114 | |
| ICA | PROSITE_PROFILES | 788 | 901 | 114 | |
| Loki; gp20 | G3DSA:2.60.40.10 | Gene3D | 475 | 577 | 103 |
| FN3 | PROSITE_PROFILES | 481 | 578 | 98 | |
| Phage-tail_3 | Pfam | 265 | 411 | 147 | |
| Fibronectin type III | SUPERFAMILY | 483 | 653 | 171 | |
| fHyAci03; gp247 | pyocin_knob | CDD | 334 | 408 | 75 |
| Phage_T4_gp36 | Pfam | 828 | 933 | 106 | |
| Aci07; gp45 | (trans)glycosidases | SUPERFAMILY | 292 | 498 | 207 |
| APK116; gp43 | pyocin_knob | CDD | 645 | 730 | 86 |
| TaPaz; gp78 | Peptidase_S74_CIMCD | CDD | 749 | 875 | 127 |
| ICA | PROSITE_PROFILES | 749 | 878 | 130 | |
| Peptidase_S74 | Pfam | 749 | 807 | 59 | |
| fLiAba02; gp22 | G3DSA:2.60.120.200 | Gene3D | 82 | 256 | 175 |
| Concanavalin A-like lectins/glucanases | SUPERFAMILY | 80 | 246 | 167 | |
| fLiAba03; gp22 | G3DSA:2.60.120.200 | Gene3D | 82 | 256 | 175 |
| Concanavalin A-like lectins/glucanases | SUPERFAMILY | 80 | 246 | 167 |
The tail fiber proteins from the podovirus AS12 (gp42, 901 AA) and the myovirus TaPaz (gp78, 878 AA) have peptidase domain hits at their CTD (Table 8). The functional domains of these tail fibers are of special interest because they have already been characterized as depolymerases (Popova et al., 2017; Shchurova et al., 2021). A PROSITE profiles hit (PS51688) was identified, which is an intramolecular chaperone auto-processing (ICA) domain that can catalyze the trimerization-dependent auto-proteolysis using two conserved serine and lysine residues. This domain has been identified in bacteriophage-encoded endosialidases and tail spike and fiber proteins (Schwarzer et al., 2007). The protein domain responsible for endosialidase activity in the ICA domain-containing tail fibers is restricted to the NTD of the proteins (Schwarzer et al., 2007). The final shared database hit overlaps the PROSITE profiles hit and is from the CDD database (cd10144) to the Peptidase S74 protein family of known phage endosialidases (Table 8). TaPaz gp78 has a Pfam Peptidase S74 hit (PF13884), while AS12 differs with an overlapping Gene3D hit (1.10.10.10), which is found with winged helix DNA-binding proteins (Table 8).
Phages APK116 (gp43, 861 AA) and PhiAB1 (gp41, 882 AA) both contain a lone pyocin knob domain at the CTD from CDD (cd19958) (Table 8). This domain layout is similar to those of Friunavirus members discussed above, except it lacks a pectin-lyase domain hit. Phage APK116 was documented to have halo formation, and gp43 was recombinantly expressed and shown to function as a depolymerase (Popova et al., 2020a). APK116 gp43 and PhiAB1 gp41 were modeled with AlphaFold and the models were submitted to the DALI server (Figure 7). The top DALI models for these two proteins are Acinetobacter phage AS12 depolymerase gp42 (6EU4) (Table 8), followed by the E. coli CAB120 depolymerase tail spike protein (6W4Q) for phiAB1 and poly(beta-d-mannuronate) c5 epimerase from Azotobacter vinelandii (5LW3) for APK116.
Figure 7.
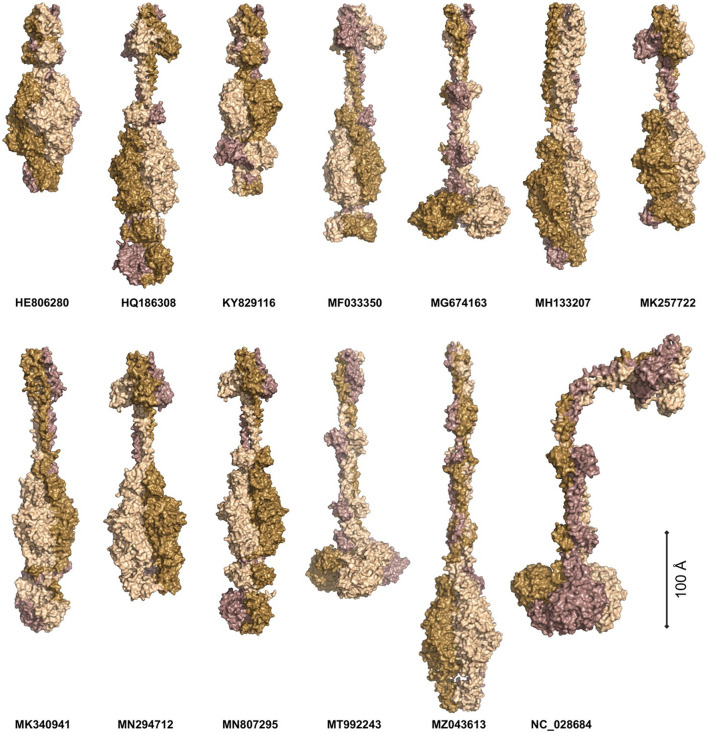
The models of Acinetobacter phage tail fiber proteins. The proteins were predicted as homotrimers with AlphaFold. The models were predicted with at least 90% confidence (AP22, HE806280, WCHABP1: KY829116), 80% (PhiAB1: HQ186308, P1: MF033350, B9: MH133207, APK32: MK257722, AbTj: MK340941, APK48: MN294712), 70% (SH-Ab 15497: MG674163, APK116: MN807295, DMU1: MT992243, TaPaz: MZ043613), and 60% (PD-6A3: NC_028684). The monomers are color-coded to highlight the symmetric nature of the complexes. The proteins are scaled proportionally to give an order of magnitude in size.
Two siphophages, fLiAba02 and fLiAba03 (gp22; identical proteins), were found to be the only phage tail fibers to have a Concanavalin A-like lectins/glucanases (CALG) SUPERFAMILY (SSF49899) domain hit (Table 8). Lectins are described as non-immune origin proteins possessing binding affinity toward glycoconjugates in a specific and reversible manner. As discussed above, these phages have an additional tail fiber encoding an SGNH hydrolase. To further investigate the function of the CALG domains, other characterized depolymerases were searched. Paenibacillus sp. 32352 is a soil-dwelling bacterium that produces the enzyme Pn3Pase, which degrades the capsular polysaccharide of Streptococcus pneumoniae serotype 3 (Pn3P) (Middleton et al., 2018; Wantuch et al., 2021). This protein encodes two SUPERFAMILY domain hits of interest: CALG (SSF49899; CTD) and (trans)glycosidases (SSF51445; NTD). Transglycosylases are a class of glycosyl hydrolase enzymes that can catalyze the transformation of one glycoside to another (Romero-Téllez et al., 2019). The functional domain responsible for the depolymerization of Pn3P was found to be the (trans)glycosidase domain, as knockouts of this region result in the loss of depolymerase activity compared to the loss of the CALG domain (Wantuch et al., 2021). The findings of the Pn3Pase mutation experiment can further be applied to phage Aci07 (gp45). As mentioned above, this phage has also documented halo formation. This is the only tail fiber with a hit to (trans)glycosidases from the SUPERFAMILY database (SSF51445) (Table 8). This is the same SUPERFAMILY domain as the Pn3Pase depolymerase discussed above, which suggests that gp45 of Aci07 may be a functional depolymerase.
3.2.8 Tail fibers without domain hits
Four phage tail fibers with no domain hits or information on halo formation will not be investigated further: ABP2 gp17, TRS1 gp29, Acibel004 gp148 and IME-AB2 gp72, and PD-AB9. Six phage tail fibers had no domain hits present, although their function as depolymerases was experimentally confirmed by researchers studying their recombinant proteins. These phages include AP22 (gp54), APK32 (gp46), APK48 (gp43), P1 (gp43), B9 (gp69), and TaPaz (gp79) (Oliveira et al., 2017, 2018; Knirel et al., 2020; Popova et al., 2020a; Shchurova et al., 2021). Furthermore, halo formation was detailed in the following phages but the protein responsible was not confirmed: AbTj (gp53) and WCHABP1 (gp5) (Zhou et al., 2018; Xu et al., 2020; Drobiazko et al., 2022). All the eight abovementioned proteins were modeled as homotrimers with AlphaFold Multimer and showed narrow midsections and larger globular regions (Figure 7).
The protein models were submitted to DALI PDB search to identify similar structural models (Figure 7, Supplementary Table 8) (Jumper et al., 2021; Holm, 2022). The top P1 DALI hit is to its own crystal structure (6E1R), followed by the putative pectin lyase gp18 (7CHU) of the Geobacillus virus E2 (Table 8). APK32 top DALI model is to Acinetobacter phage AS12 gp42 (6EU4), the characterized depolymerase discussed in the above section documenting rare domains (Table 8). The AP22 DALI hit is to its own structure (4Y9V), followed by hits to the O-specific polysaccharide lyases of Pseudomonas phages LKA1 (4RU4) and phi297 (4RU5) (Supplementary Table 8). The B9 and APK48 depolymerase models hit is to a poly(beta-d-mannuronate) c5 epimerase from Azotobacter vinelandii (5LW3 and 2PYH) and the putative pectin lyase gp18 (7CHU-A) of the Geobacillus virus E2. The TaPaz top hit is to the E. coli phage HK620 tail spike depolymerase (4XLA) and the poly(beta-d-mannuronate) c5 epimerase from Azotobacter vinelandii (5LW3). The top AbTj AlphaFold model DALI hit is to Acinetobacter phage phiAB6 tail spike depolymerase (5JSD), suggesting that this protein is responsible for the halo formation documented with the AbTj plaques. Similarly, the WCHABP1 depolymerase was modeled to a glycan biofilm modifying enzyme from Pantoea stewartii (6TGF), followed by a hit to the phiAB6 tail spike. These findings highlight a breakdown in the amino acid sequence of the proteins, but a conservation in structure, which has led to the missing functional domain hits of these proteins. The use of AlphaFold Multimer to model the proteins, and the DALI server for PDB search of the resulting models shows the power of these methods for investigating the function of tail fiber proteins lacking domain hits to investigate their potential functions.
4 Conclusion
Overall, this investigation into phage tail fiber domains has provided valuable insights into the diversity and functional characteristics of these proteins within the Acinetobacter phage. The domains associated with the depolymerase function were found to be pectin lyase-like (SSF51126), tail spike binding (cd20481), (trans)glycosidases (SSF51445), and potentially SGNH hydrolase. Furthermore, phage tail fibers with confirmed, or potential, depolymerase activity, but no functional domain hits, were modeled with AlphaFold Multimer and searched against the PDB database with the DALI server and hit to templates of other known depolymerase proteins, highlighting the power of this approach while investigating novel tail fiber proteins lacking functional domains. Although this study enhances our understanding, it is essential to recognize its limitations and the dynamic nature of scientific knowledge. Future research endeavors should further explore the role of these domains in phage-host interactions. This exploration will ensure a comprehensive grasp of their implications for phage therapy and bacterial pathogenesis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
DP compiled raw data, performed analysis on the data, and wrote and edited the paper. WC edited the paper and acquired funding. FG analyzed the data and edited the paper. All authors contributed to the article and approved the submitted version.
Funding Statement
This study was partially supported by the National Research Council (NRC) Canada's Ideation Small Team Project (National Program Office) and Vaccines and Emerging Infection Research Initiative (Human Health Therapeutics Research Center).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1230997/full#supplementary-material
Abbreviations used throughout the article.
Heatmap resulting from a Clustal Omega multisequence alignment of PLD containing proteins.
Heatmap resulting from a Clustal Omega multisequence alignment of Tailspike domain containing proteins.
Heatmap resulting from a Clustal Omega multisequence alignment of G3DSA containing proteins.
Heatmap resulting from a Clustal Omega multisequence alignment of SGNH hydrolase domain containing proteins.
Heatmap resulting from a Clustal Omega multisequence alignment of Galactose-binding domain containing proteins.
Heatmap resulting from a Clustal Omega multisequence alignment of lysozyme domain containing proteins.
Top five DALI hits from PDB90 for AlphaFold-modeled proteins.
References
- Abdelkader K., Gutiérrez D., Grimon D., Ruas-Madiedo P., Lood C., Lavigne R., et al. (2020). Lysin LysMK34 of Acinetobacter baumannii bacteriophage PMK34 has a turgor pressure-dependent intrinsic antibacterial activity and reverts colistin resistance. Appl. Environ. Microbiol. 86. 10.1128/AEM.01311-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelkader K., Gutiérrez D., Latka A., Boeckaerts D., Drulis-Kawa Z., Criel B., et al. (2022). The specific capsule depolymerase of phage PMK34 sensitizes Acinetobacter baumannii to serum killing. Antibiotics 11, 677. 10.3390/antibiotics11050677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akoh C. C., Lee G.-C., Liaw Y.-C., Huang T.-H., Shaw J.-F. (2004). GDSL family of serine esterases/lipases. Prog. Lipid Res. 43, 534–552. 10.1016/j.plipres.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Anderson A. C., Stangherlin S., Pimentel K. N., Weadge J. T., Clarke A. J. (2022). The SGNH hydrolase family: a template for carbohydrate diversity. Glycobiology 32, 826–848. 10.1093/glycob/cwac045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M., Alvi I. A., Tabassum R., Rehman S. U. (2020). TAC1, an unclassified bacteriophage of the family Myoviridae infecting Acinetobacter baumannii with a large burst size and a short latent period. Arch. Virol. 165, 419–424. 10.1007/s00705-019-04483-8 [DOI] [PubMed] [Google Scholar]
- Attwood T. K., Coletta A., Muirhead G., Pavlopoulou A., Philippou P. B., Popov I., et al. (2012). The PRINTS database: a fine-grained protein sequence annotation and analysis resource–its status in 2012. Database (Oxford) 2012, bas019. 10.1093/database/bas019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badawy S., Pajunen M. I., Haiko J., Baka Z. A. M., Abou-Dobara M. I., El-Sayed A. K. A., et al. (2020). Identification and functional analysis of temperate siphoviridae bacteriophages of Acinetobacter baumannii. Viruses 12, 604. 10.3390/v12060604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M., Chang H.-Y., Chuguransky S., Grego T., Kandasaamy S., Mitchell A., et al. (2021). The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354. 10.1093/nar/gkaa977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttimer C., O'Sullivan L., Elbreki M., Neve H., McAuliffe O., Ross R. P., et al. (2016). Genome sequence of jumbo phage vB_AbaM_ME3 of Acinetobacter baumanni. Genome Announcements 4, 16. 10.1128/genomeA.00431-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu M., Zhang P., Xu M., Yuan W., Bian L., et al. (2022). Phage-derived depolymerase as an antibiotic adjuvant against multidrug-resistant Acinetobacter baumannii. Front. Microbiol. 13, 845500. 10.3389/fmicb.2022.845500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID-19: U.S. Impact on Antimicrobial Resistance Special Report 2022 (2022). National Center for Emerging and Zoonotic Infectious Diseases. Atlanta, GA: U.S. Department of Health and Human Services, CDC. 10.15620/cdc:117915 [DOI] [Google Scholar]
- Cuff A. L., Sillitoe I., Lewis T., Clegg A. B., Rentzsch R., Furnham N., et al. (2011). Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Res. 39, D420–D426. 10.1093/nar/gkq1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dams D., Brøndsted L., Drulis-Kawa Z., Briers Y. (2019). Engineering of receptor-binding proteins in bacteriophages and phage tail-like bacteriocins. Biochem. Soc. Trans. 47, 449–460. 10.1042/BST20180172 [DOI] [PubMed] [Google Scholar]
- Drobiazko A. Y., Kasimova A. A., Evseev P. V., Shneider M. M., Klimuk E. I., Shashkov A. S., et al. (2022). Capsule-targeting depolymerases derived from acinetobacter baumannii prophage regions. IJMS 23, 4971. 10.3390/ijms23094971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drulis-Kawa Z., Majkowska-Skrobek G., Maciejewska B. (2015). Bacteriophages and phage-derived proteins – application approaches. CMC 22, 1757–1773. 10.2174/0929867322666150209152851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essoh C., Vernadet J.-P., Vergnaud G., Coulibaly A., Kakou-N'Douba A., N'Guetta A. S.-P., et al. (2019). Complete genome sequences of five Acinetobacter baumannii phages from Abidjan, Côte d'Ivoire. Microbiol. Resour. Announc 8, 18. 10.1128/MRA.01358-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer N. G., Wood T. L., Chamakura K. R., Kuty Everett G. F. (2013). Complete genome of Acinetobacter baumannii N4-like podophage Presley. Genome Announc. 1:e00852–13. 10.1128/genomeA.00852-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Doval C., Van Raaij M. J. (2012). Structure of the receptor-binding carboxy-terminal domain of bacteriophage T7 tail fibers. Proc. Natl. Acad. Sci. U.S.A. 109, 9390–9395. 10.1073/pnas.1119719109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J., Karplus K., Hughey R., Chothia C. (2001). Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313, 903–919. 10.1006/jmbi.2001.5080 [DOI] [PubMed] [Google Scholar]
- Hernandez-Morales A. C., Lessor L. L., Wood T. L., Migl D., Mijalis E. M., Cahill J., et al. (2018). Genomic and biochemical characterization of acinetobacter podophage petty reveals a novel lysis mechanism and tail-associated depolymerase activity. J. Virol. 92. 10.1128/JVI.01064-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. (2022). Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–W215. 10.1093/nar/gkac387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Luo T., Yang Y., Dong D., Wang R., Wang Y., et al. (2018). Phage therapy as a promising new treatment for lung infection caused by carbapenem-resistant Acinetobacter baumannii in mice. Front. Microbiol. 8, 2659. 10.3389/fmicb.2017.02659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Le S., Peng Y., Zhao Y., Yin S., Zhang L., et al. (2013). Characterization and genome sequencing of phage Abp1, a New phiKMV-like virus infecting multidrug-resistant Acinetobacter baumannii. Curr. Microbiol. 66, 535–543. 10.1007/s00284-013-0308-7 [DOI] [PubMed] [Google Scholar]
- Ito N., Phillips S. E., Stevens C., Ogel Z. B., McPherson M. J., Keen J. N., et al. (1991). Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase. Nature 350, 87–90. 10.1038/350087a0 [DOI] [PubMed] [Google Scholar]
- Jansen M., Wahida A., Latz S., Krüttgen A., Häfner H., Buhl E. M., et al. (2018). Enhanced antibacterial effect of the novel T4-like bacteriophage KARL-1 in combination with antibiotics against multi-drug resistant Acinetobacter baumannii. Sci. Rep. 8, 14140. 10.1038/s41598-018-32344-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., D'Souza R., Pinto N., Ryu C. -M., Park J., Yong D., et al. (2016a). Characterization and complete genome sequence analysis of two Myoviral bacteriophages infecting clinical carbapenem-resistant Acinetobacter baumannii isolates. J. Appl. Microbiol. 121, 68–77. 10.1111/jam.13134 [DOI] [PubMed] [Google Scholar]
- Jeon J., Park J.-H., Yong D. (2019). Efficacy of bacteriophage treatment against carbapenem-resistant Acinetobacter baumannii in Galleria mellonella larvae and a mouse model of acute pneumonia. BMC Microbiol. 19, 70. 10.1186/s12866-019-1443-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., Ryu C. M., Lee J. Y., Park J. H., Yong D., Lee K. (2016b). In vivo application of bacteriophage as a potential therapeutic agent to control OXA-66-like carbapenemase-producing Acinetobacter baumannii strains belonging to sequence type 357. Appl. Environ. Microbiol. 82. 10.1128/AEM.00526-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Tan J., Hao Y., Wang Q., Yan X., Wang D., et al. (2020). Isolation and characterization of a novel myophage Abp9 against pandrug resistant Acinetobacater baumannii. Front. Microbiol. 11. 10.3389/fmicb.2020.506068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Li Z. J., Wang S. W., Wang S. M., Huang D. H., Li Y. H., et al. (2012). Isolation and characterization of ZZ1, a novel lytic phage that infects Acinetobacter baumannii clinical isolates. BMC Microbiol. 12. 10.1186/1471-2180-12-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F. M., Gondil V. S., Li C., Jiang M., Li J., Yu J., et al. (2021). A novel acinetobacter baumannii bacteriophage endolysin LysAB54 with high antibacterial activity against multiple gram-negative microbes. Front. Cell. Infect. Microbiol. 11, 637313. 10.3389/fcimb.2021.637313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Oh C., Choresca C. H., Shin S. P., Han J. E., Jun J. W., et al. (2012). Complete genome sequence of bacteriophage phiAC-1 infecting acinetobacter soli strain KZ-1. J. Virol. 86, 13131–13132. 10.1128/JVI.02454-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lee D.-W., Jin J.-S., Kim J. (2020). Antimicrobial activity of LysSS, a novel phage endolysin, against Acinetobacter baumannii and Pseudomonas aeruginosa. J. Global Antimicrob. Resist. 22, 32–39. 10.1016/j.jgar.2020.01.005 [DOI] [PubMed] [Google Scholar]
- Kitti T., Thummeepak R., Thanwisai A., Boonyodying K., Kunthalert D., Ritvirool P., et al. (2014). Characterization and detection of endolysin gene from three Acinetobacter baumannii bacteriophages isolated from sewage water. Indian J. Microbiol. 54, 383–388. 10.1007/s12088-014-0472-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirel Y. A., Shneider M. M., Popova A. V., Kasimova A. A., Senchenkova S. N., Shashkov A. S., et al. (2020). Mechanisms of Acinetobacter baumannii capsular polysaccharide cleavage by phage depolymerases. Biochemistry Moscow 85, 567–574. 10.1134/S0006297920050053 [DOI] [PubMed] [Google Scholar]
- Lai M.-J., Chang K.-C., Huang S.-W., Luo C.-H., Chiou P.-Y., Wu C.-C., et al. (2016). The tail associated protein of Acinetobacter baumannii Phage ΦAB6 is the host specificity determinant possessing exopolysaccharide depolymerase activity. PLoS ONE 11, e0153361. 10.1371/journal.pone.0153361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latka A., Maciejewska B., Majkowska-Skrobek G., Briers Y., Drulis-Kawa Z. (2017). Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 101, 3103–3119. 10.1007/s00253-017-8224-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-N., Tseng T.-T., Lin J.-W., Fu Y.-C., Weng S.-F., Tseng Y.-H. (2011). Lytic myophage Abp53 encodes several proteins similar to those encoded by host Acinetobacter baumannii and phage phiKO2. Appl. Environ. Microbiol. 77, 6755–6762. 10.1128/AEM.05116-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees J., Yeats C., Perkins J., Sillitoe I., Rentzsch R., Dessailly B. H., et al. (2012). Gene3D: a domain-based resource for comparative genomics, functional annotation and protein network analysis. Nucleic Acids Res. 40, D465–D471. 10.1093/nar/gkr1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Khedkar S., Bork P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. 10.1093/nar/gkaa937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Chen B., Song Z., Song Y., Yang Y., Ma P., et al. (2012). Bioinformatic analysis of the Acinetobacter baumannii phage AB1 genome. Gene 507, 125–134. 10.1016/j.gene.2012.07.029 [DOI] [PubMed] [Google Scholar]
- Liu Y., Leung S. S. Y., Guo Y., Zhao L., Jiang N., Mi L., et al. (2019a). The capsule depolymerase Dpo48 rescues Galleria mellonella and mice from Acinetobacter baumannii systemic infections. Front. Microbiol. 10, 545. 10.3389/fmicb.2019.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Mi Z., Mi L., Huang Y., Li P., Liu H., et al. (2019b). Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. PeerJ 2019, 6173. 10.7717/peerj.6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo L. D. R., Oliveira H., Pires D. P., Dabrowska K., Azeredo J. (2020). Phage therapy efficacy: a review of the last 10 years of preclinical studies. Crit. Rev. Microbiol. 46, 78–99. 10.1080/1040841X.2020.1729695 [DOI] [PubMed] [Google Scholar]
- Merabishvili M., Vandenheuvel D., Kropinski A. M., Mast J., De Vos D., Verbeken G., et al. (2014). Characterization of newly isolated lytic bacteriophages active against Acinetobacter baumannii. PLoS ONE 9, e104853. 10.1371/journal.pone.0104853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Ebert D., Muruganujan A., Mills C., Albou L.-P., Mushayamaha T., et al. (2021). PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49, D394–D403. 10.1093/nar/gkaa1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton D. R., Zhang X., Wantuch P. L., Ozdilek A., Liu X., LoPilato R., et al. (2018). Identification and characterization of the Streptococcus pneumoniae type 3 capsule-specific glycoside hydrolase of Paenibacillus species 32352. Glycobiology 28, 90–99. 10.1093/glycob/cwx097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G. A., Sonnhammer E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir-Paz R., Gelman D., Khouri A., Sisson B. M., Fackler J., Alkalay-Oren S., et al. (2019). Successful treatment of antibiotic-resistant, poly-microbial bone infection with bacteriophages and antibiotics combination. Clin. Infect. Dis. 69, 2015–2018. 10.1093/cid/ciz222 [DOI] [PubMed] [Google Scholar]
- Oliveira H., Costa A. R., Ferreira A., Konstantinides N., Santos S. B., Boon M., et al. (2018). Functional analysis and antivirulence properties of a new depolymerase from a myovirus that infects Acinetobacter baumannii capsule K45. J. Virol. 93, e01163. 10.1128/JVI.01163-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira H., Costa A. R., Konstantinides N., Ferreira A., Akturk E., Sillankorva S., et al. (2017). Ability of phages to infect Acinetobacter calcoaceticus-Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains: specific genomic pattern variation of phages. Environ. Microbiol. 19, 5060–5077. 10.1111/1462-2920.13970 [DOI] [PubMed] [Google Scholar]
- Oliveira H., Mendes A., Fraga A. G., Ferreira A., Pimenta A. I., Mil-Homens D., et al. (2019). K2 capsule depolymerase is highly stable, is refractory to resistance, and protects larvae and mice from Acinetobacter baumannii sepsis. Appl. Environ. Microbiol. 85, e00934–e00919. 10.1128/AEM.00934-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I., Rivoire C., Auchincloss A. H., Coudert E., Keller G., de Castro E., et al. (2015). HAMAP in 2015: updates to the protein family classification and annotation system. Nucleic Acids Res. 43, D1064–D1070. 10.1093/nar/gku1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehde B. M., Niewohner D., Keomanivong F. E., Carruthers M. D. (2021). Genome Sequence and characterization of Acinetobacter phage DMU1. PHAGE 2, 50–56. 10.1089/phage.2020.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng F., Mi Z., Huang Y., Yuan X., Niu W., Wang Y., et al. (2014). Characterization, sequencing and comparative genomic analysis of vB_AbaM-IME-AB2, a novel lytic bacteriophage that infects multidrug-resistant Acinetobacter baumannii clinical isolates. BMC Microbiol. 14, 181. 10.1186/1471-2180-14-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova A. V., Lavysh D. G., Klimuk E. I., Edelstein M. V., Bogun A. G., Shneider M. M., et al. (2017). Novel Fri1-like viruses infecting Acinetobacter baumannii—vB_AbaP_AS11 and vB_AbaP_AS12— characterization, comparative genomic analysis, and host-recognition strategy. Viruses 9, 188. 10.3390/v9070188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova A. V., Myakinina V. P., Platonov M. E., Volozhantsev N. V. (2012). Molecular genetic characterization of multidrug-resistant Acinetobacter baumannii strains and assessment of their sensitivity to phage AP22. Mol. Genet. Microbiol. Virol. 27, 154–159. 10.3103/S0891416812040064 [DOI] [PubMed] [Google Scholar]
- Popova A. V., Shneider M. M., Arbatsky N. P., Kasimova A. A., Senchenkova S. N., Shashkov A. S., et al. (2020a). Specific interaction of novel friunavirus phages encoding tailspike depolymerases with corresponding Acinetobacter baumannii capsular types. J. Virol. 95, 20. 10.1128/JVI.01714-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova A. V., Shneider M. M., Mikhailova Y. V., Shelenkov A. A., Shagin D. A., Edelstein M. V., et al. (2020b). Complete genome sequence of Acinetobacter baumannii phage BS46. Microbiol Resour Announc 9. 10.1128/MRA.00398-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova A. V., Shneider M. M., Myakinina V. P., Bannov V. A., Edelstein M. V., Rubalskii E. O., et al. (2019). Characterization of myophage AM24 infecting Acinetobacter baumannii of the K9 capsular type. Arch. Virol. 164, 1493–1497. 10.1007/s00705-019-04208-x [DOI] [PubMed] [Google Scholar]
- Pulkkinen E., Wicklund A., Oduor J. M. O., Skurnik M., Kiljunen S. (2019). Characterization of vB_ApiM_fHyAci03, a novel lytic bacteriophage that infects clinical Acinetobacter strains. Arch. Virol. 164, 2197–2199. 10.1007/s00705-019-04284-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Téllez S., Lluch J. M., González-Lafont À., Masgrau L. (2019). Comparing hydrolysis and transglycosylation reactions catalyzed by thermus thermophilus β-glycosidase. A combined MD and QM/MM study. Front. Chem. 7, 200. 10.3389/fchem.2019.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer D., Stummeyer K., Gerardy-Schahn R., Mühlenhoff M. (2007). Characterization of a novel intramolecular chaperone domain conserved in endosialidases and other bacteriophage tail spike and fiber proteins. J. Biol. Chem. 282, 2821–2831. 10.1074/jbc.M609543200 [DOI] [PubMed] [Google Scholar]
- Shchurova A. S., Shneider M. M., Arbatsky N. P., Shashkov A. S., Chizhov A. O., Skryabin Y. P., et al. (2021). Novel Acinetobacter baumannii myovirus TaPaz encoding two tailspike depolymerases: characterization and host-recognition strategy. Viruses 13, 978. 10.3390/v13060978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Higgins D. G. (2018). Clustal omega for making accurate alignments of many protein sequences: clustal omega for many protein sequences. Prot. Sci. 27, 135–145. 10.1002/pro.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. J. A., De Castro E., Cerutti L., Cuche B. A., Hulo N., Bridge A., et al. (2012). New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–D347. 10.1093/nar/gks1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland I. W. (1999). Polysaccharases for microbial exopolysaccharides. Carbohydr. Polym. 38, 319–328. 10.1016/S0144-8617(98)00114-3 [DOI] [Google Scholar]
- Tao Y., Strelkov S. V., Mesyanzhinov V. V., Rossmann M. G. (1997). Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure 5, 789–798. 10.1016/S0969-2126(97)00233-5 [DOI] [PubMed] [Google Scholar]
- Timoshina O., Yu, Shneider M. M., Evseev P. V., Shchurova A. S., Shelenkov A. A., et al. (2021). Novel Acinetobacter baumannii bacteriophage aristophanes encoding structural polysaccharide deacetylase. Viruses 13, 1688. 10.3390/v13091688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D., Wand M. E., Briers Y., Lavigne R., Sutton J. M., Reynolds D. M. (2017). Characterisation and genome sequence of the lytic Acinetobacter baumannii bacteriophage vB_AbaS_Loki. PLoS ONE 12, e0172303. 10.1371/journal.pone.0172303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D., Wand M. E., Sutton J. M., Centron D., Kropinski A. M., Reynolds D. M. (2016). Genome sequence of vB_AbaS_TRS1, a viable prophage isolated from Acinetobacter baumannii strain A118. Genome Announc. 4, 16. 10.1128/genomeA.01051-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raaij M. J., Schoehn G., Burda M. R., Miller S. (2001). Crystal structure of a heat and protease-stable part of the bacteriophage T4 short tail fibre. J. Mol. Biol. 314, 1137–1146. 10.1006/jmbi.2000.5204 [DOI] [PubMed] [Google Scholar]
- Wang C., Li P., Zhu Y., Huang Y., Gao M., Yuan X., et al. (2020). Identification of a novel acinetobacter baumannii phage-derived depolymerase and its therapeutic application in mice. Front. Microbiol. 11, 1407. 10.3389/fmicb.2020.01407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wantuch P. L., Jella S., Duke J. A., Mousa J. J., Henrissat B., Glushka J., et al. (2021). Characterization of the β-glucuronidase Pn3Pase as the founding member of glycoside hydrolase family GH169. Glycobiology 31, 266–274. 10.1093/glycob/cwaa070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2021). Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: 2021. Geneva: World Health Organization. Available online at: https://apps.who.int/iris/handle/10665/341666 (accessed October 18, 2021).
- Wu M., Hu K., Xie Y., Liu Y., Mu D., Guo H., et al. (2019). A novel phage PD-6A3, and its endolysin Ply6A3, with extended lytic activity against Acinetobacter baumannii. Front. Microbiol. 9, 3302. 10.3389/fmicb.2018.03302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Dai J., Guo M., Li J., Zhou X., Li F., et al. (2021). Pre-optimized phage therapy on secondary Acinetobacter baumannii infection in four critical COVID-19 patients. Emerg. Microbes Infect. 10, 612–618. 10.1080/22221751.2021.1902754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li X., Kang G., Bai L., Wang P., Huang H. (2020). Isolation and characterization of AbTJ, an Acinetobacter baumannii phage, and functional identification of its receptor-binding modules. Viruses 12, 205. 10.3390/v12020205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Derbyshire M. K., Yamashita R. A., Marchler-Bauer A. (2020). NCBI's conserved domain database and tools for protein domain analysis. CP Bioinformat. 69, e90. 10.1002/cpbi.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Liu X., Shi Y., Yin S., Shen W., Chen J., et al. (2019). Characterization and genome annotation of a newly detected bacteriophage infecting multidrug-resistant Acinetobacter baumannii. Arch. Virol. 164, 1527–1533. 10.1007/s00705-019-04213-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehl K., Lemire S., Yang A. C., Ando H., Mimee M., Torres M. D. T., et al. (2019). Engineering phage host-range and suppressing bacterial resistance through phage tail fiber mutagenesis. Cell 179, 459–469. 10.1016/j.cell.2019.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Li X., Wang L., Li G., Cong C., Li R., et al. (2021). The endolysin of the Acinetobacter baumannii phage vB_AbaP_D2 shows broad antibacterial activity. Microb. Biotechnol. 14, 403–418. 10.1111/1751-7915.13594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu X., Li X.-J. (2015). Bioinformatic analysis of phage AB3, a phiKMV-like virus infecting Acinetobacter baumannii. Genet. Mol. Res. 14, 190–198. 10.4238/2015.January.16.2 [DOI] [PubMed] [Google Scholar]
- Zhou W., Feng Y., Zong Z. (2018). Two new lytic bacteriophages of the myoviridae family against carbapenem-resistant Acinetobacter baumannii. Front. Microbiol. 9, 850. 10.3389/fmicb.2018.00850 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Abbreviations used throughout the article.
Heatmap resulting from a Clustal Omega multisequence alignment of PLD containing proteins.
Heatmap resulting from a Clustal Omega multisequence alignment of Tailspike domain containing proteins.
Heatmap resulting from a Clustal Omega multisequence alignment of G3DSA containing proteins.
Heatmap resulting from a Clustal Omega multisequence alignment of SGNH hydrolase domain containing proteins.
Heatmap resulting from a Clustal Omega multisequence alignment of Galactose-binding domain containing proteins.
Heatmap resulting from a Clustal Omega multisequence alignment of lysozyme domain containing proteins.
Top five DALI hits from PDB90 for AlphaFold-modeled proteins.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.