Abstract
The rep-PCR DNA fingerprint technique, which uses repetitive intergenic DNA sequences, was investigated as a way to differentiate between human and animal sources of fecal pollution. BOX and REP primers were used to generate DNA fingerprints from Escherichia coli strains isolated from human and animal sources (geese, ducks, cows, pigs, chickens, and sheep). Our initial studies revealed that the DNA fingerprints obtained with the BOX primer were more effective for grouping E. coli strains than the DNA fingerprints obtained with REP primers. The BOX primer DNA fingerprints of 154 E. coli isolates were analyzed by using the Jaccard band-matching algorithm. Jackknife analysis of the resulting similarity coefficients revealed that 100% of the chicken and cow isolates and between 78 and 90% of the human, goose, duck, pig, and sheep isolates were assigned to the correct source groups. A dendrogram constructed by using Jaccard similarity coefficients almost completely separated the human isolates from the nonhuman isolates. Multivariate analysis of variance, a form of discriminant analysis, successfully differentiated the isolates and placed them in the appropriate source groups. Taken together, our results indicate that rep-PCR performed with the BOX A1R primer may be a useful and effective tool for rapidly determining sources of fecal pollution.
Despite the fact that elevated levels of Escherichia coli are correlated with increased risk of several diseases, fecal contamination of water is a widespread problem in the United States (30). A 1996 report to Congress stated that 47% of the river miles assessed in Minnesota could not be used for swimming due to high levels of fecal coliform bacteria. For some rivers the problem is pervasive; the fecal coliform counts for more than 90% of the Minnesota River and its tributaries are consistently elevated.
Determining the source of fecal pollution is necessary to develop effective control strategies. The possible sources of fecal contamination include surface runoff from manure-treated agricultural land or farm animal feedlots, failing or inadequate septic systems, sewer overflow, and wildlife. Contamination of water with fecal coliform bacteria of human origin may signal the presence of other potential human pathogens, such as Salmonella spp., Shigella spp., hepatitis A virus, and Norwalk group viruses (8, 17). Farm animals may also harbor human pathogens, including the potentially fatal organism E. coli O157:H7. Poultry are a primary reservoir of Salmonella spp, as are swine, which may also carry Shigella spp. (14).
A number of analytical methods for differentiating between human and nonhuman sources of fecal pollution have been evaluated. These methods include determining percentages and identities of fecal streptococci (3, 4, 23), determining differences in RNA coliphage distribution (6, 18), pulsed-field gel electrophoresis (11, 26), and determining whether Bacteroides fragilis HSP40 phages are present (29). However, a completely satisfactory technique has not been found yet. Historically, the ratio of fecal coliforms to fecal streptococci has been used (5, 7), but it has been shown that this method is unreliable (21).
There have been several reports of the use of antibiotic resistance profiles to determine sources of E. coli. It has been found that isolates obtained from humans, chickens, and dairy cows have higher resistance indices than strains obtained from wild animals (14). A larger percentage of water isolates from urban areas compared to isolates from rural areas exhibit resistance to antibiotics, presumably because human isolates are present (12). More recently, Parveen et al. (19) reported that human E. coli isolates clustered near isolates obtained from sewage treatment plant effluents and that isolates from animal feces were more similar to nonpoint source isolates.
In two recent studies workers have demonstrated that antibiotic resistance profiles of fecal streptococci can be used to differentiate between human and animal sources of fecal pollution. In one study, more than 10,000 fecal streptococcal isolates were obtained from 236 samples of human sewage and septage, cattle feces, poultry feces, and pristine waters (33). The average rates of correct classification into one of four possible groups (human, cattle, poultry, and wild) ranged from 64 to 78%. More recently, Hagedorn et al. (9) validated this method by using 13 antibiotics and more than 7,000 isolates from 147 samples obtained from humans, dairy cattle, beef cattle, chickens, deer, and waterfowl. Correct classification into one of the six groups described above was 87%.
More modern methods have also been evaluated to determine whether they can be used to differentiate between sources of fecal contamination of water. Parveen et al. (20) performed a ribotype analysis of E. coli isolates obtained from a bay in Florida. When their database was used, 67 and 100% of the isolates from human and animal feces, respectively, were correctly classified as members of human or nonhuman source groups. Ribotyping has also been used to determine the sources of E. coli contaminating Little Soos Creek in Washington state (25).
In this paper, we describe the use of the rep-PCR DNA fingerprinting technique to differentiate E. coli strains obtained from known animal and human sources. In rep-PCR DNA fingerprinting, PCR amplification of the DNA between adjacent repetitive extragenic elements is used to obtain strain-specific DNA fingerprints which can be easily analyzed with pattern recognition computer software. The rep-PCR technique was chosen because this technique is simple, can differentiate between closely related strains of bacteria, and can be used for high-throughput studies (32). Previously, rep-PCR has been used successfully to classify and differentiate among strains of E. coli (15), Rhizobium meliloti (2), Bradyrhizobium japonicum (10), Streptomyces spp. (24), Xanthomonas spp. (1), and several other bacteria (31).
MATERIALS AND METHODS
E. coli sources and isolation.
Table 1 lists the sources of the isolates used in this study, the number of isolates obtained from each source, the numbers of individuals sampled, and locations of the sources. The human isolates were obtained from rectal swabs obtained from students enrolled in a microbiology lab class at the University of Minnesota. Duck fecal samples were obtained during a duck-banding study in northern Minnesota, and rectal swabs of Canada geese were taken from animals at a wildlife management area. Chicken, pig, sheep, and cow fecal samples were collected at the 1999 Minnesota State Fair; samples were collected within 1 day of the arrival of animals at the fair. Except for the chicken samples, all of which were animals from the same farm, manure samples were gathered from animals from farms throughout the state of Minnesota. Rectal swabs and fecal matter were stored on ice and streaked within 12 h of collection onto mFC agar (Difco, Detroit, Mich.). After overnight incubation at 44.5°C, blue colonies were streaked onto the surfaces of MacConkey agar (Difco) plates and transferred onto ChromAgar ECC (Chromagar Microbiology, Paris, France). E. coli isolates form blue colonies on ChromAgar ECC, which differentiates them from other coliform and gram-negative bacteria, which form red and colorless colonies, respectively. After overnight incubation at 37°C, pink colonies that were obtained from the MacConkey plates and were also positive for E. coli on ChromAgar ECC plates were used to inoculate citrate agar, EC broth supplemented with 4-methylumbelliferyl-d-glucuronide (Difco) and 1% tryptone (Difco), and methyl red—Voges-Proskauer (Difco) broth. Isolates that did not grow on citrate agar, were positive for gas production and fluorescence on EC broth containing 4-methylumbelliferyl-d-glucuronide, produced indole from tryptophan, and produced acidic end products when they were grown in methyl red–Voges-Proskauer broth were designated E. coli isolates and used for subsequent studies. Approximately 26 isolates were obtained from each animal source, and 52 isolates were obtained from humans. Up to three isolates were obtained from a single individual.
TABLE 1.
E. coli isolates used in this study
| Animal source | No. of isolates | No. of individuals sampled | Location in Minnesota |
|---|---|---|---|
| Human | 52 (29)a | 21 (14) | Minneapolis |
| Duck | 26 (23) | 10 (10) | Roseau Riverb |
| Geese | 26 (21) | 9 (8) | Lac Qui Parleb |
| Chicken | 26 (20) | 13 (10) | Owatonna |
| Pig | 26 (21) | 10 (9) | Statewide |
| Sheep | 26 (19) | 11 (10) | Statewide |
| Cow | 26 (21) | 13 (12) | Statewide |
| Total | 208 (154) | 87 (73) |
The numbers in parentheses are the numbers of isolates used for statistical analysis of BOX-derived PCR fingerprints.
Wildlife management areas.
E. coli preparation and PCR conditions.
E. coli was prepared and PCR were performed essentially as described by Rademaker and de Bruijn (22), with a few minor modifications. Briefly, the E. coli isolates were grown for about 18 h in Luria-Bertani liquid medium (16), washed in 1 M NaCl, resuspended in sterile water, and frozen at −80°C until they were used. rep-PCR fingerprints were obtained by using primer BOX A1R (5′-CTACGGCAAGGCGACGCTGACG-3′) (31) or primers REP 1R (5′-IIIICGICGICATCIGGC-3′) and REP 2I (5′-ICGICTTATCIGGCCTAC-3′) (22, 28). PCR mixtures were prepared as described previously (22) by using 2-μl portions of whole-cell suspensions of each isolate as the templates. A control reaction mixture containing 2 μl of water instead of E. coli was also included in each set of PCR. Each PCR was performed with a model PTC 100 apparatus (MJ Research, Waltham, Mass.) by using the faster protocol specific for this thermocycler (22). The PCR performed with primer BOX A1R was initiated by incubating the reaction mixture at 95°C for 2 min, and this was followed by 30 cycles consisting of 94°C for 3 s, 92°C for 30 s, 50°C for 1 min, and 65°C for 8 min. The reaction was terminated with an extension step consisting of 65°C for 8 min. For PCR performed with primers REP 1R and REP 2I the annealing temperature was 40°C. Five microliters of 6× loading dye was added to each 25-μl PCR mixture, and 10 μl of each reaction mixture was separated on a 1.5% horizontal agarose gel. A 1-kb size ladder (0.5 μg/well; Life Technologies, Rockville, Md.) was loaded into the two terminal wells and in the middle of the gel. The gels were electrophoresed at 4°C for 18 h at 70 V and stained for 20 min with a solution containing 0.5 μg of ethidium bromide per ml. Gel images were captured with a FOTO/Analyst Archiver electronic documentation system (Fotodyne Inc., Hartland, Wis.).
Computer-assisted rep-PCR DNA fingerprint analysis.
Gel images were normalized, bands were identified and the data were statistically analyzed by using Bionumerics software (version 1.5; Applied Maths, Kortrijk, Belgium). Lanes that were blank because the PCR failed and lanes in which limited numbers of PCR products were produced were not included in the analysis. The positions of fragments (bands) on each gel were normalized by using the 1-kb ladder from 298 to 5,090 bp as an external reference standard. Normalization with the same set of external standards allowed us to compare multiple gels. Three or four bands that were common to most of the isolates on each gel were also used as internal reference standards. The external and internal standards corrected for smiling or other irregularities during electrophoresis. DNA fragments less than 300 bp long were not used in analyses because they tended to be indistinct. Fingerprint images were added to a database and compared by performing a statistical analysis.
Similarity coefficients were generated by the band-based method of Jaccard by using fuzzy logic and area-sensitive options. The Jaccard similarity coefficient for each pair of fingerprints was calculated by dividing the number of bands that occurred in both fingerprints by the total number of bands (common and unique) in both fingerprints. The fuzzy logic option allowed band matching values to gradually decrease with the distance between bands, and the area-sensitive option took into account differences in area between matching bands.
Statistical analysis was used to determine the relatedness of DNA fingerprints and to determine whether the isolates could be successfully assigned to the correct source groups. The DNA fingerprints were compared to each other by calculating Jaccard similarity coefficients. Similarity coefficients were first determined by using the BOX- and REP-derived fingerprints individually and then were determined by using a combined data set, designated the BOX-plus-REP fingerprints. Jackknife analysis was used to determine how accurately the similarity coefficients were able to predict the source group of each isolate. This was done as follows. The isolates were first manually assigned to the correct source groups, and then each isolate was individually removed from the database. The level of similarity of the removed isolate to the isolates remaining in each source group was determined, and an average group similarity coefficient was calculated from the individual similarity values for each group. The isolate that was removed was placed in the source group having the greatest average group similarity coefficient, and the percentage of isolates from each source group correctly assigned was then calculated.
A dendrogram was constructed by using Jaccard similarity coefficients. A binary band-matching character table was generated by using the BOX-derived PCR DNA fingerprint data, and this table was analyzed by multivariate analysis of variance (MANOVA), a form of discriminant analysis. MANOVA was done, accounting for the covariance structure.
RESULTS AND DISCUSSION
Assignment of isolates to source groups.
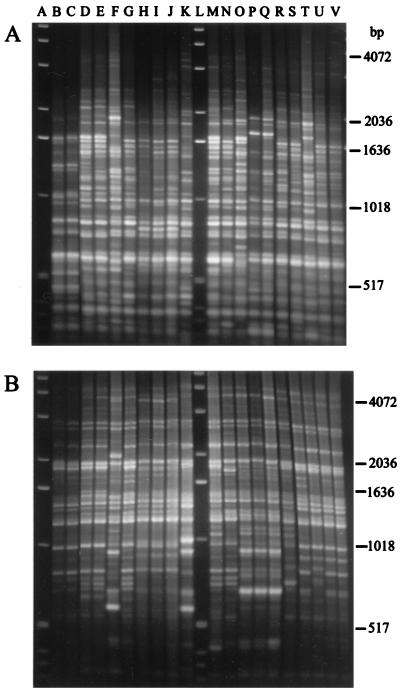
Figure 1A shows typical fingerprints for E. coli isolates generated by using rep-PCR performed with primer BOX A1R. Complex fingerprint patterns were obtained for all of the isolates studied. In general, the band patterns of isolates from different animal sources were very similar, and the data indicated that the isolates were closely related. While the fingerprint patterns for E. coli isolates obtained from the same animal were similar, they were not always identical. Approximately one-quarter of the bands were common to 80% or more of the isolates, and a few bands were shared by more than 90% of the isolates. Individual lanes generally contained from 25 to 30 PCR product bands, although almost 40 bands were obtained for some E. coli isolates. The sizes of the PCR products ranged from slightly less than 300 bp to about 4,500 bp. The initial studies were performed with both the BOX and REP primers. However, 25% fewer PCR products were usually present in the fingerprints generated with the REP primers than in the fingerprints obtained with the BOX primer (Fig. 1B). This was largely due to the scarcity of PCR products less than 750 bp long.
FIG. 1.
rep-PCR DNA fingerprint patterns of E. coli strains obtained from beef and dairy cows. (A) PCR DNA fingerprint patterns generated with primer BOX A1R. Lanes A and L contained an external standard, a 1-kb molecular weight ladder. (B) PCR DNA fingerprint patterns generated with primers REP 1R and REP 2I. Lanes A and L contained an external standard, a 1-kb molecular weight ladder. The E. coli strains used for the fingerprint analysis shown in panel B are identical to the strains used for the analysis shown in panel A, except that the strains in lanes O and T are reversed.
A total of 208 isolates were used as templates for PCR performed with the BOX and REP primers. These 208 isolates consisted of 26 isolates from each nonhuman animal source and 52 human isolates. Approximately 74% of the isolates (154 isolates) produced high-quality DNA fingerprints when primer BOX A1R was used. These isolates are listed in Table 1. Some of the isolates that were successfully used as templates when the BOX primer was used did not produce reliable fingerprints when the REP primers were used. In our initial studies, fingerprints were obtained for only 125 isolates with both the BOX primer and the REP primers.
Statistical analysis was used to verify that these 125 isolates were assigned to the correct source groups. Jaccard similarity coefficients were calculated for both the BOX-derived fingerprints and the REP-derived fingerprints individually and for a combined data set, the BOX-plus-REP fingerprints. The isolates were manually assigned to the correct groups, and a Jackknife analysis was performed. The Jackknife analysis was used to determine how accurately the similarity coefficients predicted the source groups. The results of this analysis are shown in Table 2. DNA fingerprints generated by using the REP primers were almost as useful as BOX-derived DNA fingerprints for correctly classifying human and sheep isolates; about 90% of the isolates belonging to both groups were correctly classified. However, REP-derived fingerprints lagged considerably behind BOX-derived fingerprints in the ability to effectively group the remaining isolates from the other animal sources (chickens, cows, ducks, geese, and pigs). There was no improvement in the grouping of strains when BOX-plus-REP DNA fingerprint data were used compared to BOX-derived fingerprints alone. Consequently, only BOX-derived DNA fingerprints were used in the remainder of our study. Previously, Lipman and coworkers (15) found that rep-PCR performed with REP primers was less reliable than PCR performed with enterobacterial repetitive intergenic consensus (ERIC) primers for differentiating among E. coli strains from cows with clinical mastitis. However, in their study, these authors generated only a limited number of PCR fragments with ERIC primers.
TABLE 2.
Percentages of isolates correctly assigned to source groups by using BOX-derived, REP-derived, and BOX-plus-REP PCR DNA fingerprintsa
| Data set | % of isolates correctly assigned to source groups for E. coli isolates from:
|
||||||
|---|---|---|---|---|---|---|---|
| Humans | Geese | Ducks | Sheep | Pigs | Chickens | Cows | |
| BOX | 94.7 | 89.5 | 80.0 | 93.3 | 93.8 | 100.0 | 100.0 |
| REP | 89.5 | 52.6 | 35.0 | 86.7 | 50.0 | 81.3 | 65.0 |
| BOX-plus-REP | 89.5 | 73.7 | 80.0 | 93.3 | 87.5 | 100.0 | 100.0 |
Based on average similarity data.
The entire BOX-derived DNA fingerprint data set generated with 154 isolates was analyzed by using Jaccard similarity coefficients and Jackknife analysis. The percentage of isolates assigned to each group was calculated. Table 3 shows that almost 83% of the isolates obtained from humans were assigned to the human group. In some instances, however, human isolates were misidentified as members of the goose, pig, and duck groups. This relationship was not reciprocal, as goose isolates were most often misidentified as chicken isolates (9.5%) and never were classified as members of the human group. Overall, our results show that when primer BOX A1R was used, the rep-PCR technique very successfully classified E. coli isolates in the correct source groups. All of the chicken and cow isolates and between 78 and 90% of the human, goose, duck, pig, and sheep isolates were correctly identified when this primer was used. Based on these results, we concluded that the source group of an unknown isolate can most likely be identified by comparison to the average similarity coefficients for the source groups. The percentages of isolates correctly classified based on maximum-similarity data were similar, although usually not identical, to the percentages of isolates correctly classified by using average similarity coefficients (data not shown). For example, while duck and goose isolates were classified slightly better when maximum-similarity values were used, average similarity data described groups of pig and cow isolates better. Nevertheless, our results indicate that Jaccard similarity coefficients may be useful for identifying sources of unknown environmental isolates. To do this, the DNA fingerprint of an unknown isolate can be directly compared to a library of BOX-derived DNA fingerprints of isolates from human and animal sources. After average group similarity coefficients are determined for the source groups, the unknown isolate can be placed in the source group with which it exhibits the highest level of similarity.
TABLE 3.
Assignment of isolates to animal source groups by using BOX PCR DNA fingerprints and Jackknife analysis
| Assigned group | % of E. coli isolates in assigned groupa:
|
||||||
|---|---|---|---|---|---|---|---|
| Human | Goose | Duck | Sheep | Pig | Chicken | Cow | |
| Human | 82.8 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Goose | 6.9 | 81.0 | 4.3 | 5.3 | 0.0 | 0.0 | 0.0 |
| Duck | 3.4 | 0.0 | 78.3 | 0.0 | 0.0 | 0.0 | 0.0 |
| Sheep | 0.0 | 4.8 | 8.7 | 89.5 | 4.8 | 0.0 | 0.0 |
| Pig | 6.9 | 0.0 | 4.3 | 5.3 | 81.0 | 0.0 | 0.0 |
| Chicken | 0.0 | 9.5 | 0.0 | 0.0 | 4.8 | 100 | 0.0 |
| Cow | 0.0 | 4.8 | 4.3 | 0.0 | 9.5 | 0.0 | 100 |
Values in boldface indicate percentages of isolates correctly assigned to source groups.
Dendrogram construction.
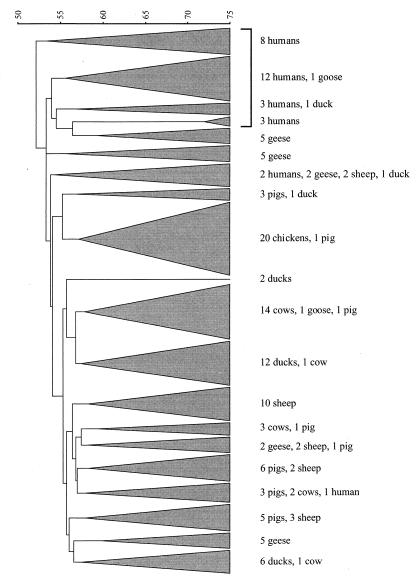
To determine the relatedness of strains, a dendrogram based on BOX-derived fingerprint data was constructed by using Jaccard similarity coefficients and the neighbor-joining clustering method (Fig. 2). As shown in Fig. 2, 26 of the 29 human isolates were grouped into four clusters at the top of the dendrogram. These clusters also included two waterfowl isolates. Other types of animal isolates also clustered together when this analysis was performed. All of the chicken isolates fell into a single cluster, as did the majority of the cow, duck, and sheep isolates. Our results indicate that while the dendrogram may have been useful for separating isolates into human and nonhuman source groups, the isolates were clearly closely related. The average distance between 18 of the 20 clusters, which accounted for more than 96% of the isolates, was less than 10%. Similarly, although Hagedorn et al. (9) were able to classify fecal streptococci isolates into groups (humans, dairy cattle, beef cattle, chickens, deer, and waterfowl) by using antibiotic resistance patterns, some overlap occurred between the human and nonhuman (chicken) clusters.
FIG. 2.
Dendrogram showing the relatedness of E. coli strains isolated from humans, geese, ducks, sheep, pigs, chickens, and cows as determined by a PCR DNA fingerprint analysis performed with primer BOX A1R. Relationships were determined by using Jaccard similarity coefficients and the neighbor-joining clustering method.
Clustering of isolates by MANOVA.
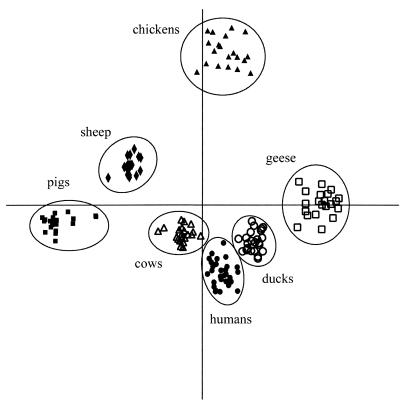
MANOVA, a clustering technique based on discriminant analysis, can be used to determine even small differentiating features in user-specified groups. In this study, isolates were first manually assigned to the correct groups, and a binary band-matching character table was generated by using the BOX-derived fingerprint data. This table was analyzed by MANOVA by using an option that accounted for covariance structure. Since seven groups were specified (humans, cows, pigs, chickens, sheep, ducks, and geese), a total of six discriminants were determined, and P values calculated. The P value was the probability that random subdivision of the groups would yield the same degree of discrimination. Figure 3 maps isolates based on the first two discriminants. The MANOVA successfully sorted the E. coli isolates into the correct source groups (the human, cow, sheep, duck, goose, chicken, and pig groups) with no overlaps (Fig. 3). The first, second, and third discriminants accounted for 33.0, 24.5, and 18.2%, respectively, of the discrimination. Together, the first three discriminants accounted for 75.7% of the variation, and the first five discriminants accounted for 93.8% of the variation. The P values for the first five discriminants were ≤0.002, indicating that the specified groups were valid. Together, these results indicate that the MANOVA of the BOX-derived PCR fingerprint data effectively clustered the human and animal isolates. Moreover, BOX-derived PCR fingerprint data may be very useful for determining the sources of unknown environmental E. coli isolates.
FIG. 3.
MANOVA of BOX-derived PCR DNA fingerprints from E. coli strains obtained from animal and human sources. Binary band-matching character tables were analyzed by MANOVA, accounting for the covariance structure. The E. coli isolates were obtained from humans (●), geese (□), ducks (○), sheep (⧫), pigs (■), chickens (▴), and cows (▵). The first discriminant is represented by the distance along the x axis, and the second discriminant is represented by the distance along the y axis.
In this study, we found that rep-PCR DNA fingerprint analysis was a useful tool for differentiating between E. coli isolates obtained from six species of animals and humans. The animal isolates included those from two types of waterfowl (geese and ducks) and common farm animals (cows, pigs, sheep, and chickens). Since genotypic analyses are less subject to environmental effects than phenotypic analyses are, we believe that rep-PCR may be a method of choice for differentiating and grouping E. coli isolates obtained from animals and humans. Other advantages of rep-PCR are its simplicity, accuracy, and speed, which are desirable for high-throughput analysis. While other genotypic analysis methods, such as ribotyping, have been examined to determine their ability to differentiate coliform bacteria (25), these methods tend to require extensive manipulation of DNA and the use of labeled gene probes. Because of this, these methods are not amenable to high-throughput analyses. In addition, in the rep-PCR analyses performed here, DNA fingerprints were generated by using whole cell suspensions, which eliminated the need for DNA purification.
Previously, multiple antibiotic resistance profiles of E. coli isolates were used to differentiate between point sources and nonpoint sources (19). Parveen et al. showed that isolates from point sources were more diverse than isolates from nonpoint sources and that E. coli isolates from human and animal feces clustered with isolates from both point and nonpoint sources. Other researchers have demonstrated that antibiotic resistance analysis of fecal streptococci is useful for differentiating source groups (9, 33). However, grouping may be influenced by a strain's prior exposure to antibiotics. In addition, fecal streptococci also persist longer in the environment, which may limit their usefulness for determining sources of recent contamination (13, 27). We contend that since fecal coliforms, not fecal streptococci, are the most widely used indicators of water quality, it is important to be able to group E. coli isolates.
In conclusion, our results indicate that rep-PCR DNA fingerprinting performed with the BOX A1R primer is a promising method for determining the source groups of E. coli isolates and may prove to be useful for determining the sources of closely related E. coli strains obtained from environmental samples.
ACKNOWLEDGMENTS
This work was supported in part by funding from The Environmental and Natural Resources Trust Fund through The Legislative Commission on Minnesota Resources and by funding from the University of Minnesota Agricultural Experiment Station (to M.J.S.).
REFERENCES
- 1.Bouzar H, Jones J B, Stall R E, Louws F J, Schneider M, Rademaker J L W, de Bruijn F J, Jackson L E. Multiphasic analysis of xanthomonads causing bacterial spot disease on tomato and pepper in the Caribbean and Central America: evidence for common lineages within and between countries. Phytopathology. 1999;89:328–335. doi: 10.1094/PHYTO.1999.89.4.328. [DOI] [PubMed] [Google Scholar]
- 2.de Bruijn F J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devriese L A, Van De Kerckhove A, Kilpper-Bälz R, Schleifer K H. Characterization and identification of Enterococcus species isolated from the intestines of animals. Int J Syst Bacteriol. 1987;37:257–259. [Google Scholar]
- 4.Devriese L A, Pot B, Collins M D. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J Appl Bacteriol. 1993;75:399–408. doi: 10.1111/j.1365-2672.1993.tb02794.x. [DOI] [PubMed] [Google Scholar]
- 5.Feachem R. An improved role for faecal coliform to faecal streptococci ratios in the differentiation between human and nonhuman pollution sources. Water Res. 1975;9:689–690. [Google Scholar]
- 6.Furuse K, Ando A, Osawa S, Watanabe I. Distribution of ribonucleic acid coliphages in raw sewage from treatment plants in Japan. Appl Environ Microbiol. 1981;41:1139–1143. doi: 10.1128/aem.41.5.1139-1143.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geldreich E E, Kenner B A. Concepts of fecal streptococci in stream pollution. J Water Pollut Control Fed. 1969;41:R336–R352. [PubMed] [Google Scholar]
- 8.Guzewich J J, Morse D L. Sources of shellfish in outbreaks of probable viral gastroenteritis: implications for control. J Food Prot. 1986;49:389–394. doi: 10.4315/0362-028X-49.5.389. [DOI] [PubMed] [Google Scholar]
- 9.Hagedorn C, Robinson S L, Filtz J R, Grubbs S M, Angier T A, Reneau R B., Jr Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl Environ Microbiol. 1999;65:5522–5531. doi: 10.1128/aem.65.12.5522-5531.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Judd A K, Schneider M, Sadowsky M J, de Bruijn F J. Use of repetitive sequences and the polymerase chain reaction technique to classify genetically related Bradyrhizobium japonicum serocluster 123 strains. Appl Environ Microbiol. 1993;59:1702–1708. doi: 10.1128/aem.59.6.1702-1708.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kariuki S, Gilks C, Kimari J, Obanda A, Muyodi J, Waiyaki P, Hart C A. Genotype analysis of Escherichia coli strains isolated from children and chickens living in close contact. Appl Environ Microbiol. 1999;65:472–476. doi: 10.1128/aem.65.2.472-476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasper C W, Burgess J L, Knight I T, Colwell R R. Antibiotic resistance indexing of Escherichia coli to identify sources of fecal contamination in water. Can J Microbiol. 1990;36:891–894. doi: 10.1139/m90-154. [DOI] [PubMed] [Google Scholar]
- 13.Kibbey H J, Hagedorn C, McCoy E L. Use of fecal streptococci as indicators of pollution in soil. Appl Environ Microbiol. 1978;35:711–717. doi: 10.1128/aem.35.4.711-717.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krumperman P H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol. 1983;46:165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipman L J A, de Nijs A, Lam T J G M, Gaastra W. Identification of Escherichia coli strains from cows with clinical mastitis by serotyping and DNA polymorphism patterns with REP and ERIC primers. Vet Microbiol. 1995;43:13–19. doi: 10.1016/0378-1135(94)00070-d. [DOI] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 17.Orskov F, Orskov I. Enterobacteriaceae. In: Broude A I, editor. Medical microbiology and infectious diseases. Philadelphia, Pa: The W. B. Saunders Co.; 1981. pp. 340–352. [Google Scholar]
- 18.Osawa S, Furuse K, Wantanabe I. Distribution of ribonucleic acid coliphages in animals. Appl Environ Microbiol. 1981;41:164–168. doi: 10.1128/aem.41.1.164-168.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parveen S, Murphree R L, Edmiston L, Kasper C W, Portier K M, Tamplin M L. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl Environ Microbiol. 1997;63:2607–2612. doi: 10.1128/aem.63.7.2607-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parveen S, Portier K M, Robinson K, Edmiston L, Tamplin M L. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl Environ Microbiol. 1999;65:3142–3147. doi: 10.1128/aem.65.7.3142-3147.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pourcher A-M, Devriese L A, Hernandez J F, Delattre J M. Enumeration by a miniaturized method of Escherichia coli, Streptococcus bovis, and enterococci as indicators of the origin of faecal pollution of waters. J Appl Bacteriol. 1991;70:525–530. doi: 10.1111/j.1365-2672.1991.tb02752.x. [DOI] [PubMed] [Google Scholar]
- 22.Rademaker J L W, de Bruijn F J. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer-assisted pattern analysis. In: Caetano-Anollés G, Gresshoff P M, editors. DNA markers: protocols, applications, and overviews. J. New York, N.Y: Wiley and Sons; 1997. pp. 151–171. [Google Scholar]
- 23.Rutkowski A A, Sjogren R E. Streptococcal population profiles as indicators of water quality. Water Air Soil Pollut. 1987;34:273–284. [Google Scholar]
- 24.Sadowsky M J, Kinkel L L, Bowers J H, Schottel J L. Use of repetitive intergenic DNA sequences to classify pathogenic and disease-suppressive Streptomyces strains. Appl Environ Microbiol. 1996;62:3489–3493. doi: 10.1128/aem.62.9.3489-3493.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samadpour M, Chechowitz N. Little Soos Creek microbial source tracking: a survey. Seattle: University of Washington Department of Environmental Health; 1995. [Google Scholar]
- 26.Simmons G M, Jr, Herbein S A, James C M. Managing nonpoint fecal coliform sources to tidal inlets. Water Resour Update. 1995;100:64–74. [Google Scholar]
- 27.Sinton L W, Donnison A M, Hastie C M. Faecal streptococci as faecal pollution indicators: a review. II. Sanitary significance, survival, and use. N Z J Mar Freshwater Res. 1993;27:117–137. [Google Scholar]
- 28.Stern M J, Ames G F-L, Smith N H, Robinson E C, Higgins C F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984;37:1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- 29.Tartera C, Lucena F, Jofre J. Human origin of Bacteroides fragilis bacteriophages present in the environment. Appl Environ Microbiol. 1989;55:2696–2701. doi: 10.1128/aem.55.10.2696-2701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.United States Environmental Protection Agency. Ambient water quality criteria for bacteria—1986. Report EPA440/5-84-002. Washington, D.C.: United States Environmental Protection Agency; 1986. [Google Scholar]
- 31.Versalovic J, de Bruijn F J, Lupski J R. Repetitive sequence-based PCR (rep-PCR) DNA fingerprinting of bacterial genomes. In: de Bruijn F J, Lupski J R, Weinstock G M, editors. Bacterial genomes: physical structure and analysis. New York, N.Y: Chapman and Hall; 1998. pp. 437–454. [Google Scholar]
- 32.Versalovic J, Schneider M, de Bruijn F J, Lupski J R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 33.Wiggins B A, Andrews R W, Conway R A, Corr C L, Dobratz E J, Dougherty D P, Eppard J R, Knupp S R, Limjoco M C, Mettenburg J M, Rinehardt J M, Sonsino J, Torrijos R L, Zimmerman M E. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl Environ Microbiol. 1999;65:3483–3486. doi: 10.1128/aem.65.8.3483-3486.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]