Abstract
Background
Resistance to oxaliplatin (L-OHP) is a major barrier in the treatment of colorectal cancer (CRC). Autophagy is the main cause of L-OHP tolerance in CRC cells.
Method
The human colon cancer cell lines HCT116 and SW480 were treated with L-OHP to obtain the drug-resistant cell lines HCT116/L-OHP and SW480/L-OHP, respectively. To probe the relationship between autophagy and L-OHP tolerance of growth factor independent 1 (Gfi-1) and high-mobility group protein 1 (HMGB1) in CRC cells, gene knockout or overexpression was performed, and Western blotting was used to determine the levels of drug tolerance interrelated proteins. Transwell and CCK-8 assays were employed to analyze the proliferation of cancer cells. Immunofluorescence detection of LC3 reflected autophagy levels. Finally, the relationship between Gfi-1 and HMGB1 was detected by chromatin immunoprecipitation (ChIP).
Result
Compared to normal CRC cells, L-OHP-tolerant CRC cells exhibited greater autophagy (8.2 times greater in HCT116/L-OHP cells and 7.4 times greater in SW480/L-OHP cells). In addition, we detected low levels of Gfi-1 (0.6-fold for HCT116/L-OHP cells and 0.4-fold for SW480/L-OHP cells), and OE-Gfi-1 decreased HMGB1 levels (0.6-fold for HCT116/L-OHP + OE-Gfi-1 cells and 0.5-fold for SW480/L-OHP + OE-Gfi-1 cells). The inhibition of Gfi-1 further enhanced cell viability (1.7 times in HCT116+sh-Gfi-1 cells and 1.2 times in SW480+sh-Gfi-1 cells) and invasion (1.8 times in HCT116+sh-Gfi-1 cells and 2.1 times in SW480+sh-Gfi-1 cells) in CRC cells, thus promoting oxaliplatin resistance in these cells. The autophagy inhibitor 3-MA reversed the above effects. Furthermore, we noted that Gfi-1 can restrain HMGB1 expression by binding to its promoter (0.5 times in HCT116+OE-Gfi-1 cells and 0.5 times in SW480+OE-Gfi-1 cells). The inhibitory influence of 3-MA on HMGB1 reversed the influence of Gfi-1 on autophagy and malignant progression in CRC cells.
Conclusion
Our study suggested that Gfi-1 inhibited HMGB1 to reduce CRC autophagy levels, increasing CRC sensitivity to L-OHP.
Keywords: Oxaliplatin, Colorectal cancer, Autophagy, Growth factor independence 1, HMGB1
The abnormal increase in autophagy levels in L-OHP-resistant CRC cells is due to the promotion of HMGB1 expression by Gfi-1 inhibition, which leads to oxaliplatin resistance in CRC cells. Gfi-1 can inhibit autophagy through HMGB1 and facilitate the susceptibility of CRC/L-OHP to L-OHP.
1. Introduction
Colorectal cancer (CRC) is the third most common cancer and a main health issue responsible for cancer-related deaths [1,2]. Chemotherapy is still one of the main nonsurgical tumor control methods for patients with unresectable CRC. Oxaliplatin (L-OHP) is a small-molecule platinum compound and a commonly used anticancer drug for CRC [3]. However, related side effects and chemical sensitivity are the main reasons for treatment failure [4]. Therefore, it is essential to further understand the mechanism underlying L-OHP tolerance in CRC to improve the survival rate and prognosis of CRC patients.
Autophagy is an accommodative catabolic process that plays a pivotal role in sustaining the homeostasis of the internal environment of cells and plays a complex and critical role in cancer. Basic autophagy can restrain tumor progression by sustaining normal cellular homeostasis, while revitalized autophagy can help tumor cells overcome stress and survive, thereby facilitating tumor development [5]. Autophagy is also a pivotal mechanism of drug tolerance. Many studies have confirmed that L-OHP can restore autophagy in CRC cells in many ways. For example, a new protein encoded by the exosome CircATG4B induced CRC resistance to oxaliplatin by promoting autophagy [6]. Candida tropicalis could downregulate tumor cell-intrinsic PD-1 expression by enhancing tumor cell autophagy levels to promote CRC tumor growth and chemotherapeutic resistance to oxaliplatin [7]. miR-135b-5p upregulation in CRC could induce protective autophagy and promote oxaliplatin resistance [8]. Inhibition of protective autophagy may provide a new treatment strategy for cancer drug development that may help to prevent issues with disease progression and overcome drug resistance in CRC [9]. Therefore, it is necessary to investigate the relationship between L-OHP resistance and autophagy in CRC.
Research has shown that growth factor independence 1 (Gfi-1) plays major roles in tumorigenesis and cancer progression, including in chronic myeloid leukemia tumors [10], bone marrow cancer [11], and lung cancer [12]. The level of Gfi-1 is associated with drug resistance in tumors [[13], [14], [15]]. In addition, research has shown that Gfi-1 plays a role as a transcription factor in cancer development by regulating target genes, such as alpha 1-antitrypsin (AAT), alpha 1-antichymotrypsin (ACT) [16], HMG20B [17], and HMGB1 [18]. However, whether Gfi-1 participates in L-OHP tolerance in CRC by regulating autophagy remains unclear.
Increasing evidence has shown that high-mobility group protein 1 (HMGB1) plays a pivotal role in the development of cancer through multiple physiological and pathological processes (including cell metabolism, inflammation and cell death) [19]. Importantly, HMGB1 is considered a key regulatory factor for autophagy [20,21]. The regulation of HMGB1-mediated autophagy has been the focus of research on drug resistance in various cancers, including glioblastoma [22], non-small cell lung cancer [23] and CRC [24]. In addition, there is evidence suggesting that HMGB1-mediated autophagy contributes to CRC cell sensitivity to L-OHP [25], indicating that HMGB1-mediated autophagy may be a potential target for overcoming L-OHP resistance.
In this study, we investigated the mechanism and role of Gfi-1-mediated autophagy in oxaliplatin resistance in CRC through HMGB1 and provided a novel perspective on the potential mechanism of Gfi-1 in oxaliplatin sensitivity.
2. Materials and methods
2.1. Cell culture
HCT116 (ATCC, CCL-247) and SW480 (ATCC, CCL-228) cells were obtained from the American Type Culture Collection (ATCC) in the United States and passaged in the laboratory for fewer than 6 months after resuscitation. The median lethal concentrations (IC50s) of oxaliplatin (Sigma, USA) in the human colon cancer cell lines SW480 and HCT116 were measured, and the initial concentration of L-OHP needed to induce the generation of drug-resistant subclones was determined based on the IC50. The half maximal inhibitory concentration (IC50) of L-OHP was detected for the cell lines treated with different concentrations of L-OHP (0, 0.01, 0.05, 0.1, 0.2, 0.5, and 1 μmol/L) for 24 h. Cell viability was determined by a CCK8 assay, and an inhibition curve was generated. The drug concentration at a 50 % inhibition rate was calculated.
The drug-resistant cell line was constructed based on previous reference [26,27]. The cells in the logarithmic growth phase were cultured in 10 % FBS, 1 % penicillin/streptomycin and L-15 medium supplemented with 0.25 μmol/L oxaliplatin, after which the cells were allowed to grow and passed through three generations stably to reach the next concentration (0.5 μmol/L). By repeatedly increasing the drug concentration, the cells were allowed to grow stably for 2–3 months at a drug concentration of 2.0 μmol/L. This was the drug-resistant cell line CRC/L-OHP to be constructed. In subsequent experiments, the cells were cultured in RPMI 1640 medium supplemented with 3 μmol/L oxaliplatin. Cell culture was carried out in a 37 °C environment containing 5 % CO2 for proliferation. The autophagy inhibitor 3-MA (1 mM, Sigma, USA) was used in this study.
2.2. Cell transfection
Cell transfection was performed based on previous reference [28]. In a 6-well plate, when the cells grew to 90 % confluence, they were transfected with 1 μg of plasmids (HMGB1, Gli1, sh-Gli1, and the corresponding negative control plasmids) according to the instructions for the Lipofectamine 3000 reagent (Invitrogen, USA). Then, the cells were incubated at 37 °C and cultured in 5 % CO2 for 48 h, after which the transfection efficiency was measured for subsequent experiments.
2.3. Immunofluorescence detection of LC3
Methodology technique was performed based on previous reference [28]. The cells were digested with trypsin and centrifuged (1200 rpm, 5 min), and the supernatant was discarded. The cells were resuspended in culture substrate and treated with 2 × 105 cells/mL in a special Petri dish. The cells in each group were treated according to different experimental requirements. After the culture was completed, the supernatant was removed with a pipette, and the excess medium was removed with PBS. Anti-LC3 (ab192890, Abcam) was added for 2 h, and after washing with PBS, the corresponding fluorescent secondary antibody was incubated for 1 h. DAPI (ab104139, Abcam) was used for nuclear staining. Finally, the cells were fixed at room temperature with paraformaldehyde for 15 min and washed 3 times with PBS. The fluorescence intensity of the cells was observed on an inverted fluorescence microscope (Lecia, Germany).
2.4. CCK-8 determination of cell viability
Cell viability was performed based on previous reference [28]. Cells (4 × 103 cells) were inoculated into a 96-well plate and treated with the designated treatments or doses of L-OHP for 24 h. After incubation, 20 μL of CCK (Solarbio, China) solution was added, the absorbance was measured at 450 nm according to the manufacturer's specifications, and the cell viability was calculated.
2.5. Transwell assay
Transwell was performed based on previous reference [28]. Cells in the logarithmic growth phase were washed with PBS. The cells were then suspended in serum-free culture substrate, and the density was adjusted to 5 × 105/mL. Culture substrate containing 10 % FBS was added to the lower chamber of a 24-well plate, and the Transwell chamber was sealed with matrix glue. A 100 μL cell suspension was added to the upper chamber and placed in an incubator for 24 h of cultivation. The chamber and culture substrate were removed, and the cells were gently removed with a cotton swab. Then, the cells were fixed with 4 % paraformaldehyde in a new 24-well plate for 20–30 min. The fixative was discarded, and the cells were stained with 0.1–0.2 % crystal violet for 5–10 min and washed with PBS 3 times to remove crystal violet that was not bound to the cells. Three fields of view were selected under a high-power microscope for observation and counting after suitable air drying.
2.6. Western blot
Western blot was performed based on previous reference [28]. Total proteins were extracted with RIPA lysis buffer, the concentration was measured by BCA, and the proteins were separated by Tris-glycine/SDS gel electrophoresis. The isolated proteins were transferred to PVDF membranes and blocked with 5 % nonfat milk powder. The membranes were then incubated overnight at 4 °C with the following primary antibodies: anti-ABCG2 (ab207732, 1/1000), anti-P-gp (ab261736, 1/500), anti-MRP2 (ab172630, 1/2000), anti-GST-pi (ab138491, 1/2000), anti-GAPDH (ab8245, 1/1000), and anti-GAPDH (ab8245, 1/1000). After the membranes were washed 3 times with TBST, they were then incubated with secondary goat anti-rabbit IgG antibodies (1:1000) for 1 h at room temperature. ECL chemiluminescence was used for development, a chemiluminescence instrument was used for exposure and observation, and ImageJ was used for protein band analysis.
2.7. Electrophoretic mobility shift assay (EMSA)
EMSA was performed based on previous reference [29]. EMSAs were performed using the Lightshift Chemiluminescent EMSA kit (Pierce, Rockford IL). HMGB1 was extracted from HCT116 cells. Gfi-1-specific binding oligonucleotides of the HMGB1 promoter were labeled with biotin at the 3′ end using a DNA biotin labeling kit (Pierce). The extract was incubated with an excess of biotin-labeled ds-DNA probe. The binding reaction was carried out at room temperature for 30 min in binding buffer supplemented with 0.1 μg of the nonspecific competitor poly-dIdC (Pierce). A monoclonal HMGB1 antibody (1 μL/reaction) was used in the HMGB1 supershift experiments. The complexes were resolved on a 5 % polyacrylamide gel, transferred to a nylon membrane and visualized with a chemiluminescent nucleic acid detection module kit (Pierce).
2.8. Luciferase assay
Luciferase assay was performed based on previous reference [18]. HCT116 cells were transfected with 1 μg of Gfi-1-pcDNA3.1 and 0.3 μg of HMGB1 reporter plasmids using Lipofectamine 3000 following the manufacturer's instructions (Invitrogen). The pRL-SV40 luciferase reporter was used as an internal control. After incubation for 24 h, the luciferase activity of the cell lysis supernatant was determined using a dual-luciferase reporter assay system according to the manufacturer's instructions.
2.9. Chromatin immunoprecipitation (ChIP)
As mentioned earlier [18], ChIP experiments were conducted, and the purified DNA was quantified by qPCR using a ChIP reagent kit (Millipore). Briefly, CRC cells were crosslinked with fresh 1 % formaldehyde and lysed in SDS lysis buffer containing 1 % protease inhibitor. The harvested cell lysate was subjected to ultrasound treatment to cleave the cross-linked DNA into fragments with a growth rate of 200–1000 base pairs. Three micrograms of anti-Gfi-1 antibody or control IgG was used for immunoprecipitation of the protein/DNA complexes. After elution from the antibody, the complexes were dissociated in a solution containing 5 mol/L sodium chloride. ChIP specimens were analyzed via RT‒qPCR using HMGB1 promoter-specific primers. The primer sequences are listed in Table 1.
Table 1.
Primers for detecting Gfi-1 located in the promoter region of HMGB1.
| Site | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| HMGB1 | GAAGCAGTACACGTCGTA | AGAAAACCGTTTGATTTG |
2.10. Statistics and analysis
The analysis results are expressed as the mean ± SD. GraphPad Prism 8.0 was used for all the statistical analyses. All experiments were performed in at least 3 parallel groups and repeated 3 times. ANOVA and t tests were used for analysis. p < 0.05 was considered to indicate statistical significance.
3. Results
3.1. Inhibiting autophagy can increase the sensitivity of CRC cells to L-OHP
Autophagy is interrelated with L-OHP tolerance. To probe the influence of autophagy on oxaliplatin resistance in CRC, we used 3-MA (an autophagy inhibitor) to investigate the relationship between autophagy and L-OHP tolerance in CRC. Compared with normal CRC cells (HCT116 and SW480 cells), CRC/L-OHP cells (HCT116/L-OHP and SW480/L-OHP cells) exhibited higher levels of LC3 expression (Fig. 1A). Transmission electron microscopy revealed that L-OHP tolerance promoted the formation of autophagosomes in the L-OHP-treated group but not in the CRC group (Fig. 1B), indicating that autophagy was activated in the CRC/L-OHP group. Moreover, compared with the levels in CRC/L-OHP cells, the addition of 3-MA inhibited the activity and invasion of CRC cells (Fig. 1C and D). Moreover, the addition of 3-MA inhibited the levels of the drug tolerance-related proteins ABCG2, P-gp, MRP2, and GST-pi in CRC/L-OHP cells (Fig. 1E). Compared with those in the normal CRC cell group, L-OHP tolerance promoted the expression of LC3 II/I and Beclin1 and inhibited the expression of p62, and these effects were reversed after the addition of 3-MA (Fig. 1F). These results suggest that autophagy promotes the insensitivity of CRC cells to L-OHP. Reactivation of autophagy can help tumor cells overcome stress and increase drug sensitivity, thereby facilitating tumor development [5].
Fig. 1.
Restraining autophagy can increase the sensitivity of CRC/L-OHP cells to L-OHP (n = 3)
A: Immunofluorescence assay for the LC3 level in CRC (scale = 100 μm, T test); B: Observation of autophagosome formation using transmission electron microscopy (scale bar = 1 μm); C: CCK-8 assay for checking cell viability (T test); D: Transwell assay for checking cell invasion (T test); E: Western blot analysis for measuring the levels of the drug tolerance-related proteins ABCG2, P-gp, MRP2, and GST-pi (two-way ANOVA, Uncropped images of gels were supplemented in Fig. S1); F: Western blot analysis for measuring the levels of LC3, Beclin1, and p62 (two-way ANOVA, Uncropped images of gels were supplemented in Fig. S2). **P < 0.01, ***P < 0.001 vs. the CRC group. #P < 0.05, ##P < 0.01, ###P < 0.001, vs. the CRC/L-OHP group.
3.2. Gfi-1 can promote the sensitivity of drug-fast CRC cells to L-OHP
In previous studies, Gfi-1 has been linked to drug resistance [13,18]. Therefore, we explored the influence of Gfi-1 on L-OHP tolerance in CRC by knocking down or overexpressing Gfi-1. Western blot analysis showed a low level of Gfi-1 in CRC/L-OHP cells (Fig. 2A), and protein immunoblotting revealed the successful transfection of Gfi-1 in CRC/L-OHP cells (Fig. 2B). In drug-resistant CRC cells, overexpression of Gfi-1 decreased cell viability and weakened invasion ability (Fig. 2C and D). Overexpressing Gfi-1 decreased the levels of the drug tolerance-interrelated proteins ABCG2, P-gp, MRP2, and GST-pi in CRC cells (Fig. 2E). The transfection efficiency analysis showed that sh-Gfi1-2 had better interference efficiency than sh-Gfi1-1; therefore, sh-Gfi1-2 was used in the following experiments (Fig. 2F). Moreover, compared with that of normal CRC cells, knocking down Gfi-1 resulted in increased viability and invasion of CRC cells (Fig. 2G and H). In addition, knocking down Gfi-1 increased the levels of ABCG2, P-gp, MRP2, and GST-pi (Fig. 2I). The results indicated that while overexpression of Gfi-1 could sensitize CRC/L-OHP cells to oxaliplatin, downregulation of Gfi-1 could desensitize CRC cells to oxaliplatin.
Fig. 2.
Gfi-1 can increase the sensitivity of CRC-resistant cells to L-OHP (n = 3)
A: Western blot for detecting Gfi-1 expression (T test, Uncropped images of gels were supplemented in Fig. S3); B: Western blot for detecting Gfi-1 overexpression efficiency (one-way ANOVA, Uncropped images of gels were supplemented in Fig. S4); C: CCK-8 for detecting cell viability (T test); D: Transwell for detecting cell invasion (T test); E: Measurement of the levels of ABCG2, P-gp, MRP2, and GST-pi (GAPDH was used as an internal reference protein, two-way ANOVA, Uncropped images of gels were supplemented in Fig. S5); F: Western blot for detecting the knockdown efficiency of Gfi-1 (one-way ANOVA, Uncropped images of gels were supplemented in Fig. S6); G: CCK-8 for detecting cell viability (T test); H: Transwell for detecting cell invasion (T test); I: Measurement of the levels of ABCG2, P-gp, MRP2, and GST-pi (two-way ANOVA, Uncropped images of gels were supplemented in Fig. S7). *P < 0.05, **P < 0.01, ***P < 0.001 vs. the CRC or CRC/L-OHP group.
3.3. Gfi-1 inhibits autophagy by regulating HMGB1 expression
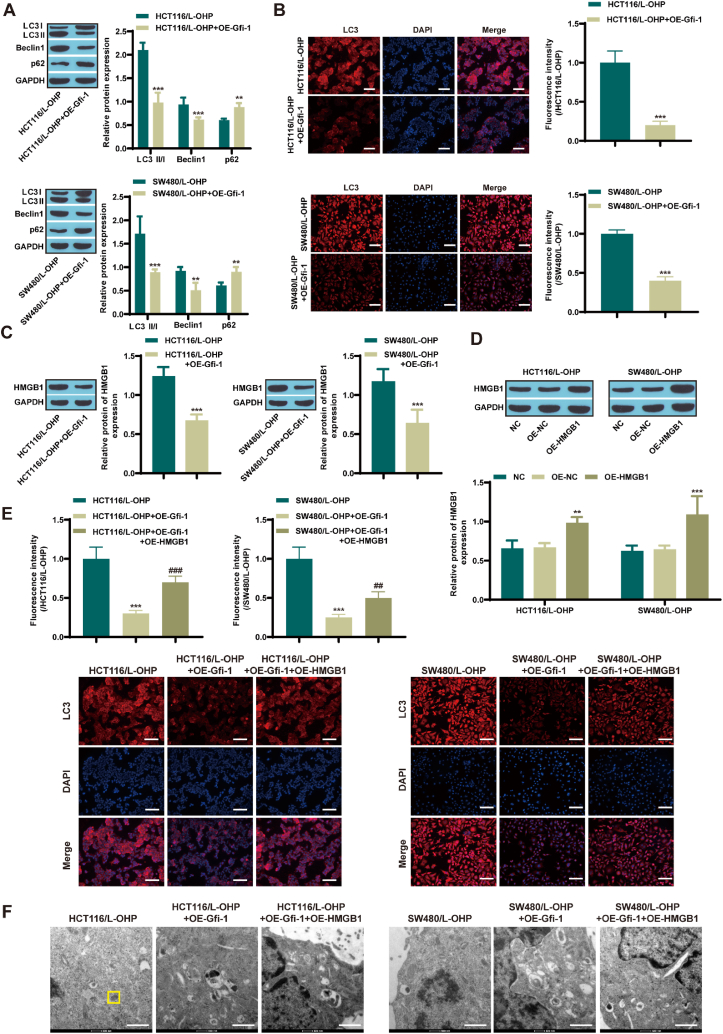
Research on HMGB1-mediated autophagy has focused on drug resistance in various cancers [[22], [23], [24]]. Next, we investigated the relationships among Gfi-1, HMGB1, and autophagy in CRC cells. Western blot analysis revealed that Gfi-1 overexpression increased autophagy in resistant CRC cells (Fig. 3A). Immunofluorescence also showed the same results (Fig. 3B). In addition, overexpression of Gfi-1 reduced HMGB1 levels in HCT116/L-OHP and SW480/L-OHP cells (Fig. 3C). Next, we overexpressed HMGB1 in CRC/L-OHP cells, and Western blot analysis confirmed the successful transfection of HMGB1 (Fig. 3D). Compared with CRC/L-OHP, overexpressing Gfi-1 resulted in a reduction in LC3 expression levels, while further overexpression of HMGB1 restored LC3 levels (Fig. 3E). Transmission electron microscopy revealed that, compared with that in the CRC/L-OHP group, the formation of autophagosomes was inhibited in the Gfi-1-overexpressing group, and the formation of autophagosomes was promoted in the HMGB1-overexpressing group (Fig. 3F). The above results indicate that Gfi-1 restrains CRC cell autophagy and that HMGB1 facilitates CRC cell autophagy, which is interrelated with L-OHP resistance.
Fig. 3.
Gfi-1 restrains autophagy by regulating HMGB1 expression (n = 3)
A: Western blot analysis of the levels of LC3, Beclin1, and p62 (two-way ANOVA, Uncropped images of gels were supplemented in Fig. S8); B: Immunofluorescence analysis of the LC3 level in CRC tissue (scale = 100 μm, t-test); C: Western blot analysis of HMGB1 expression (t-test, Uncropped images of gels were supplemented in Fig. S9); D: Western blot analysis of HMGB1 overexpression efficiency (one-way ANOVA, Uncropped images of gels were supplemented in Fig. S10); E: Immunofluorescence analysis of the LC3 level in CRC tissue (scale = 100 μm, one-way ANOVA); F: Observation of autophagosome formation via transmission electron microscopy (scale bar = 1 μm). **P < 0.01, ***P < 0.001 vs. the CRC/L-OHP group. ##P < 0.01, ###P < 0.001, vs. the CRC/L-OHP + OE-Gfi-1 group.
3.4. Gfi-1 binds to the HMGB1 promoter and inhibits its expression
First, the start site of HMGB1 was determined through NCBI (https://www.ncbi.nlm.nih.gov/), and the binding site between Gfi-1 and the HMGB1 promoter was determined through JASPAR (https://jaspar.genereg.net/) (Fig. 4A). EMSAs revealed that Gif-1 could directly interact with the HMGB1 promoter region (Fig. 4B). ChIP‒qPCR detection showed that Gfi-1 bound to the site of the HMGB1 promoter region (Fig. 4C). Transfection with pGL3-HMGB1-luc alone resulted in lower luciferase activity than cotransfection with Gfi-1-pcDNA3.1 (Fig. 4D). We also observed that the expression of HMGB1 mRNA prominently decreased after Gfi-1 was overexpressed, as measured by RT‒qPCR (Fig. 4E). These data confirm that Gfi-1 directly restrains the transcription of HMGB1 by binding to the promoter region of HMGB1, as reported by Xian et al. [18].
Fig. 4.
Gfi-1 binds to the HMGB1 promoter and restrains its expression (n = 3)
A: Gfi-1 and HMGB1 promoter-binding site; B: The binding between Gif-1 and HMGB1 was determined by EMSA (Uncropped images of gels were supplemented in Fig. S11); C: HMGB1 expression was detected by chromatin immunoprecipitation (T test); D: Luciferase activities were measured (T test); E: RT‒qPCR for determining the level of HMGB1 expression (T test). **P < 0.01, ***P < 0.001 vs. the IgG or CRC group.
3.5. Gfi-1 inhibits autophagy through HMGB1 and promotes the sensitivity of CRC cells/L-OHP to L-OHP
The above results showed that Gfi-1 and HMGB1 are associated with autophagy and drug tolerance in CRC cells and that Gfi-1 binds to the HMGB1 promoter. We further validated that Gfi-1 restrains autophagy through HMGB1 and promotes the susceptibility of CRC cells/L-OHP to L-OHP by overexpressing Gfi-1 and HMGB1. Compared with the NC group, the Gfi-1-overexpressing group exhibited reduced cell viability and decreased invasion of drug-fast CRC cells. The overexpression of HMGB1 increased the viability and invasion of drug-resistant CRC cells, and the autophagy inhibitor 3-MA reversed these effects (Fig. 5A and B). Similarly, Western blot results showed that overexpressing Gfi-1 decreased the levels of the drug tolerance-interrelated proteins ABCG2, P-gp, MRP2, and GST-pi in drug-resistant CRC cells, while overexpression of HMGB1 increased the levels of ABCG2, P-gp, MRP2, and GST-pi in drug-fast CRC cells. The autophagy inhibitor 3-MA inhibited the expression of these proteins (Fig. 5C). Compared with those in the NC group, Gfi-1 overexpression inhibited the expression of LC3 II/I and Beclin1 and promoted the expression of p62. HMGB1 overexpression promoted the expression of LC3 II/I and Beclin1 and inhibited the expression of p62 in drug-resistant CRC cells, and the autophagy suppressor 3-MA reversed these effects (Fig. 5D).
Fig. 5.
Gfi-1 can promote the sensitivity of CRC-resistant cells to L-OHP (n = 3)
A: CCK-8 for checking cell viability (one-way ANOVA); B: Transwell assay for checking cell invasion (one-way ANOVA); C: Detection of the levels of ABCG2, P-gp, MRP2, and GST-pi (GAPDH was used as an internal reference protein, two-way ANOVA, Uncropped images of gels were supplemented in Fig. S12); D: Western blot for measuring the levels of LC3, Beclin1, and p62 (two-way ANOVA, Uncropped images of gels were supplemented in Fig. S13). P *<0.05, P * *<0.01, P * * *<0.001, vs. the NC group; ##P < 0.01, ###P < 0.001, vs. the OE-Gfi-1 group; ^P < 0.05, ^^P < 0.01, ^^^P < 0.001, vs. the OE-Gfi-1+OE-HMGB1 group.
4. Discussion
Most patients with early colorectal cancer can be treated with surgery, but the prognosis of patients with advanced colorectal cancer is poor. Oxaliplatin is widely used as a first-line treatment for CRC; however, L-OHP tolerance is the main barrier to treatment, and the response in such cases is not satisfactory [30]. Although significant progress has been made in recent years, the molecular mechanism underlying CRC tolerance to L-OHP is still entirely unclear [30,31]. Hence, there is an urgent need to find valid biomarkers and targets for CRC diagnosis and therapy.
Previous reports have shown that autophagy, as a phylactic mechanism, exposes cancer cells to various anticancer drugs [32]. Song et al. reported that hypoxia-induced autophagy is interrelated to chemotherapy resistance in hepatocellular carcinoma cells [33]. Ahn et al. suggested that restraint of autophagy can enhance resistance to Src family tyrosine kinase (SFK) inhibitors [34]. In addition, studies have shown that autophagy mediated by miR-637 promotes the tolerance of CRC to L-OHP [35]. P-glycoprotein (P-gp), multidrug resistance associated protein 1 (MRP1/ABCC1), MRP2, and cassette efflux transporter G2 (ABCG2) are considered the main factors that induce multidrug resistance [36]. It has been reported that some GSTs are interrelated with the mechanism underlying the multidrug tolerance of tumor cells to anticancer drugs. Glutathione S-transferases (GSTs) are a family of multimolecular enzymes with multiple drug detoxification functions [37]. Here, we found that L-OHP-resistant CRC cells exhibited high levels of autophagy, and the expression levels of the tolerance-related proteins ABCG2, P-gp, MRP2, and GST-pi increased. 3-MA can reduce the autophagy level of CRC cells and restrain CRC progression. It is unclear how the expression levels of ABCG2, P-gp, MRP2, and GST pi in CRC/L-OHP are decreased upon treatment with 3-MA. This will be explored and validated in future research.
Independent growth factor 1 (Gfi-1) is a pivotal transcription factor in cancer evolution. Cai et al. reported that Gfi-1 facilitates the proliferation of cervical cancer cells [38]. Lee et al. suggested that Gfi-1 regulates the development of acute myeloid leukemia by restraining SOCS1 [39]. Here, we found that Gfi-1 facilitates an increase in L-OHP sensitivity in CRC cells, while inhibiting Gfi-1 expression can desensitize CRC cells to L-OHP. This finding indicates that Gfi-1 may be a potential target for increasing the sensitivity of CRC cells to L-OHP. Previous studies have demonstrated that Gfi-1 is involved in macrophage-mediated gemcitabine tolerance in pancreatic ductal adenocarcinoma [18]. Moreover, Gfi-1 levels are intimately interrelated with panobinostat tolerance in acute myeloid leukemia [13].
Gfi-1 can regulate the expression of multifarious targets, including HMGB1 [18]. HMGB1 is a nucleoprotein that contributes to tumorigenesis [40]. In addition, the role of HMGB1 in autophagy is well characterized. Li et al. demonstrated that autophagy-based unconventional secretion of HMGB1 in glioblastoma promotes chemosensitivity to temozolomide [41]. Guo et al. suggested that HMGB1-mediated autophagy is critical to the vulnerability of multiple myeloma cells [42]. Notably, HMGB1-mediated autophagy is crucial for the drug tolerance of many cancers [[22], [23], [24]]. Here, we observed that Gfi-1 restrains HMGB1 expression in CRC by binding to the promoter of HMGB1, as reported by Xian et al. [18]. The autophagy suppressor 3-MA can reverse the inhibitory influence of Gfi-1 on the sensitivity of CRC cells to L-OHP and reduce the autophagy level of CRC/L-OHP cells.
In this work, we first confirmed that autophagy plays a pivotal role in the chemosensitivity of L-OHP-resistant CRC, as reported by Pan [6]. Elevated autophagy levels were observed in L-OHP-resistant CRC cells. Importantly, our study further demonstrated that the expression of the transcription inhibitor Gfi-1 was decreased in CRC/L-OHP cells and was crucial for autophagy regulation in L-OHP-resistant CRC cells. Inhibiting Gfi-1 further enhanced the autophagy and drug resistance phenotype of L-OHP-resistant CRC cells. Notably, Gfi-1 could restrain HMGB1 expression by binding to its promoter in CRC cells. As an important autophagy regulatory factor, HMGB1 is involved in Gfi-1-mediated autophagy and CRC cell resistance to L-OHP. Similar to the results of Liu et al.'s previous study [25], HMGB1 mediated autophagy and L-OHP tolerance in CRC cells. Nevertheless, the pharmacological effects of Gfi1 need to be verified in vivo, which will greatly improve the findings of this study. Nanostructured drug delivery systems with modifiable chemical properties have been identified as promising candidates for reversing drug resistance in CRC [43]. In addition, tumor-associated fibroblast-derived exosomes have also been found to be associated with drug resistance [44]. In the future, we will focus on the study of other mechanisms of oxaliplatin resistance, and animal experiments will be performed for further verification of potential mechanisms.
5. Conclusion
The abnormal increase in autophagy levels in CRC cells is due to the promotion of HMGB1 expression by Gfi-1 inhibition, which leads to oxaliplatin resistance. Gfi-1 can inhibit autophagy through HMGB1 and facilitate the susceptibility of CRC/L-OHP cells to L-OHP. Gfi-1 may become a target of L-OHP tolerance in CRC cells, providing new ideas and strategies for clinical cancer chemotherapy.
Funding
This study was approved by Yunnan Provincial Department of Science and Technology and Kunming Medical University Basic Research - Key Funded Project (202301AY070001-016).
Data availability
The data will be made available upon request.
CRediT authorship contribution statement
Weijun Liu: Writing – original draft, Visualization, Methodology, Investigation, Conceptualization. Zhenyong Zhang: Methodology, Investigation, Formal analysis. Liju Zhang: Investigation, Data curation. Xiaoming Jiang: Methodology, Formal analysis. Changxian Chen: Writing – original draft. Xi Wu: Writing – original draft. Quan Zhao: Writing – review & editing, Supervision, Resources, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29859.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Baidoun F., Elshiwy K., Elkeraie Y., Merjaneh Z., Khoudari G., Sarmini M.T., et al. Colorectal cancer epidemiology: recent trends and impact on outcomes. Curr. Drug Targets. 2021;22(9):998–1009. doi: 10.2174/1389450121999201117115717. [DOI] [PubMed] [Google Scholar]
- 2.Biller L.H., Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. 2021;325(7):669–685. doi: 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 3.Lin J.F., Hu P.S., Wang Y.Y., Tan Y.T., Yu K., Liao K., et al. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal Transduct. Targeted Ther. 2022;7(1):54. doi: 10.1038/s41392-022-00889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li T., Hu P.S., Zuo Z., Lin J.F., Li X., Wu Q.N., et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer. 2019;18(1):112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X., He S., Ma B. Autophagy and autophagy-related proteins in cancer. Mol. Cancer. 2020;19(1):12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Z., Zheng J., Zhang J., Lin J., Lai J., Lyu Z., et al. A novel protein encoded by exosomal CircATG4B induces oxaliplatin resistance in colorectal cancer by promoting autophagy. Adv. Sci. 2022;9(35) doi: 10.1002/advs.202204513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu J., Chen Q., Bing Z., Shen S., Hou Y., Lv M., et al. C. tropicalis promotes CRC by down-regulating tumor cell-intrinsic PD-1 receptor via autophagy. J. Cancer. 2023;14(10):1794–1808. doi: 10.7150/jca.79664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Wang X., Zhang H., Deng T., Liu R., Liu Y., et al. The HSF1/miR-135b-5p axis induces protective autophagy to promote oxaliplatin resistance through the MUL1/ULK1 pathway in colorectal cancer. Oncogene. 2021;40(28):4695–4708. doi: 10.1038/s41388-021-01898-z. [DOI] [PubMed] [Google Scholar]
- 9.Usman R.M., Razzaq F., Akbar A., Farooqui A.A., Iftikhar A., Latif A., et al. Role and mechanism of autophagy-regulating factors in tumorigenesis and drug resistance. Asia Pac. J. Clin. Oncol. 2021;17(3):193–208. doi: 10.1111/ajco.13449. [DOI] [PubMed] [Google Scholar]
- 10.Salarpour F., Goudarzipour K., Mohammadi M.H., Ahmadzadeh A., Faraahi S., Allahbakhshian A., et al. Evaluation of growth factor independence 1 expression in patients with de novo acute myeloid leukemia. J. Cancer Res. Therapeut. 2020;16(1):23–27. doi: 10.4103/jcrt.JCRT_129_17. [DOI] [PubMed] [Google Scholar]
- 11.Khandanpour C., Eisfeld C., Nimmagadda S.C., Raab M.S., Weinhold N., Seckinger A., et al. Prevalence of the GFI1-36N SNP in multiple myeloma patients and its impact on the prognosis. Frontiers in oncology. 2021;11 doi: 10.3389/fonc.2021.757664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H., Lin Z., Nian Z., Zhang W., Liu W., Yan F., et al. Hematopoietic transcription factor GFI1 promotes anchorage independence by sustaining ERK activity in cancer cells. The Journal of clinical investigation. 2022;132(17) doi: 10.1172/jci149551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng B., Tang S., Zhe N., Ma D., Yu K., Wei D., et al. Low expression of GFI-1 Gene is associated with Panobinostat-resistance in acute myeloid leukemia through influencing the level of HO-1. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;100:509–520. doi: 10.1016/j.biopha.2018.02.039. [DOI] [PubMed] [Google Scholar]
- 14.Petrusca D.N., Mulcrone P.L., Macar D.A., Bishop R.T., Berdyshev E., Suvannasankha A., et al. GFI1-Dependent repression of SGPP1 increases multiple myeloma cell survival. Cancers. 2022;14(3) doi: 10.3390/cancers14030772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Arca D., Severi L., Ferrari S., Dozza L., Marverti G., Magni F., et al. Serum mass spectrometry proteomics and protein set identification in response to FOLFOX-4 in drug-resistant ovarian carcinoma. Cancers. 2023;15(2) doi: 10.3390/cancers15020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashour N., Angulo J.C., González-Corpas A., Orea M.J., Lobo M.V.T., Colomer R., et al. Epigenetic regulation of Gfi1 in endocrine-related cancers: a role regulating tumor growth. Int. J. Mol. Sci. 2020;21(13) doi: 10.3390/ijms21134687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maiques-Diaz A., Nicosia L., Basma N.J., Romero-Camarero I., Camera F., Spencer G.J., et al. HMG20B stabilizes association of LSD1 with GFI1 on chromatin to confer transcription repression and leukemia cell differentiation block. Oncogene. 2022;41(44):4841–4854. doi: 10.1038/s41388-022-02471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xian G., Zhao J., Qin C., Zhang Z., Lin Y., Su Z. Simvastatin attenuates macrophage-mediated gemcitabine resistance of pancreatic ductal adenocarcinoma by regulating the TGF-β1/Gfi-1 axis. Cancer letters. 2017;385:65–74. doi: 10.1016/j.canlet.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Wang S., Zhang Y. HMGB1 in inflammation and cancer. J. Hematol. Oncol. 2020;13(1):116. doi: 10.1186/s13045-020-00950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chai W., Ye F., Zeng L., Li Y., Yang L. HMGB1-mediated autophagy regulates sodium/iodide symporter protein degradation in thyroid cancer cells. Journal of experimental & clinical cancer research : CR. 2019;38(1):325. doi: 10.1186/s13046-019-1328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Zhou H., Zheng H., Zhou X., Shen G., Teng X., et al. Autophagy-based unconventional secretion of HMGB1 by keratinocytes plays a pivotal role in psoriatic skin inflammation. Autophagy. 2021;17(2):529–552. doi: 10.1080/15548627.2020.1725381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao M., Zhang Y., Jiang Y., Wang K., Wang X., Zhou D., et al. YAP promotes autophagy and progression of gliomas via upregulating HMGB1. Journal of experimental & clinical cancer research : CR. 2021;40(1):99. doi: 10.1186/s13046-021-01897-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei T., Huang J., Xie F., Gu J., Cheng Z., Wang Z. HMGB1-mediated autophagy promotes gefitinib resistance in human non-small cell lung cancer. Acta biochimica et biophysica Sinica. 2022;54(4):fpage–lpage. doi: 10.3724/abbs.2022023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu F., Ai F.Y., Zhang D.C., Tian L., Yang Z.Y., Liu S.J. LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med. 2020;9(3):1079–1091. doi: 10.1002/cam4.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu W., Zhang Z., Zhang Y., Chen X., Guo S., Lei Y., et al. HMGB1-mediated autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin via MEK/ERK signaling pathway. Cancer Biol. Ther. 2015;16(4):511–517. doi: 10.1080/15384047.2015.1017691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C., Zhang Y., Lin S., Liu Y., Li W. Suppressing the KIF20A/NUAK1/Nrf2/GPX4 signaling pathway induces ferroptosis and enhances the sensitivity of colorectal cancer to oxaliplatin. Aging. 2021;13(10):13515–13534. doi: 10.18632/aging.202774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun W., Ge Y., Cui J., Yu Y., Liu B. Scutellarin resensitizes oxaliplatin-resistant colorectal cancer cells to oxaliplatin treatment through inhibition of PKM2. Molecular therapy oncolytics. 2021;21:87–97. doi: 10.1016/j.omto.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Z., Zhao Y., Mang Y., Zhu J., Yu L., Li L., et al. MiR-21-5p promotes sorafenib resistance and hepatocellular carcinoma progression by regulating SIRT7 ubiquitination through USP24. Life Sci. 2023;325 doi: 10.1016/j.lfs.2023.121773. [DOI] [PubMed] [Google Scholar]
- 29.Yu S., Yerges-Armstrong L.M., Chu Y., Zmuda J.M., Zhang Y. E2F1 effects on osteoblast differentiation and mineralization are mediated through up-regulation of frizzled-1. Bone. 2013;56(2):234–241. doi: 10.1016/j.bone.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Gan Y., Liu J., Li J., Zhou Z., Tian R., et al. Downregulation of MEIS1 mediated by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and oxaliplatin resistance in colorectal cancer. Signal Transduct. Targeted Ther. 2022;7(1):87. doi: 10.1038/s41392-022-00902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q., Deng T., Zhang H., Zuo D., Zhu Q., Bai M., et al. Adipocyte-derived exosomal MTTP suppresses ferroptosis and promotes chemoresistance in colorectal cancer. Adv. Sci. 2022;9(28) doi: 10.1002/advs.202203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doherty J., Baehrecke E.H. Life, death and autophagy. Nat. Cell Biol. 2018;20(10):1110–1117. doi: 10.1038/s41556-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song J., Qu Z., Guo X., Zhao Q., Zhao X., Gao L., et al. Hypoxia-induced autophagy contributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy. 2009;5(8):1131–1144. doi: 10.4161/auto.5.8.9996. [DOI] [PubMed] [Google Scholar]
- 34.Ahn J.H., Lee M. Suppression of autophagy sensitizes multidrug resistant cells towards Src tyrosine kinase specific inhibitor PP2. Cancer Lett. 2011;310(2):188–197. doi: 10.1016/j.canlet.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Li C., Liu X., Wang Y., Zhao R., Yang Y., et al. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48:277–288. doi: 10.1016/j.ebiom.2019.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar A., Jaitak V. Natural products as multidrug resistance modulators in cancer. Eur. J. Med. Chem. 2019;176:268–291. doi: 10.1016/j.ejmech.2019.05.027. [DOI] [PubMed] [Google Scholar]
- 37.Sato K., Tsuchida S., Tamai K. [Anti-cancer drug resistance and glutathione S-transferases] Gan to kagaku ryoho Cancer & chemotherapy. 1989;16(3 Pt 2):592–598. [PubMed] [Google Scholar]
- 38.Cai H., Zhang F., Li Z. Gfi-1 promotes proliferation of human cervical carcinoma via targeting of FBW7 ubiquitin ligase expression. Cancer Manag. Res. 2018;10:2849–2857. doi: 10.2147/cmar.s161130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee M., Kuo Y., Chou W., Hou H., Hsiao M., Tien H. Gfi-1 is the transcriptional repressor of SOCS1 in acute myeloid leukemia cells. J. Leukoc. Biol. 2014;95(1):105–115. doi: 10.1189/jlb.0912475. [DOI] [PubMed] [Google Scholar]
- 40.Khambu B., Huda N., Chen X., Antoine D., Li Y., Dai G., et al. HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers. The Journal of clinical investigation. 2018;128(6):2419–2435. doi: 10.1172/jci91814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z., Fu W.J., Chen X.Q., Wang S., Deng R.S., Tang X.P., et al. Autophagy-based unconventional secretion of HMGB1 in glioblastoma promotes chemosensitivity to temozolomide through macrophage M1-like polarization. Journal of experimental & clinical cancer research : CR. 2022;41(1):74. doi: 10.1186/s13046-022-02291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo X., He D., Zhang E., Chen J., Chen Q., Li Y., et al. HMGB1 knockdown increases MM cell vulnerability by regulating autophagy and DNA damage repair. Journal of experimental & clinical cancer research : CR. 2018;37(1):205. doi: 10.1186/s13046-018-0883-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei M., Liu K., Qu X., Wang K., Chen Q., Zhang Y., et al. Enzyme-catalyzed synthesis of selenium-doped manganese phosphate for synergistic therapy of drug-resistant colorectal cancer. J. Nanobiotechnol. 2023;21(1):72. doi: 10.1186/s12951-023-01819-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu X., Liu B., Wang L., Liu L., Zhao W., Liu C., et al. Loss of cancer-associated fibroblast-derived exosomal DACT3-AS1 promotes malignant transformation and ferroptosis-mediated oxaliplatin resistance in gastric cancer. Drug Resist. Updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2023;68 doi: 10.1016/j.drup.2023.100936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data will be made available upon request.