Abstract
Extremophiles and their products have been a major focus of research interest for over 40 years. Through this period, studies of these organisms have contributed hugely to many aspects of the fundamental and applied sciences, and to wider and more philosophical issues such as the origins of life and astrobiology. Our understanding of the cellular adaptations to extreme conditions (such as acid, temperature, pressure and more), of the mechanisms underpinning the stability of macromolecules, and of the subtleties, complexities and limits of fundamental biochemical processes has been informed by research on extremophiles. Extremophiles have also contributed numerous products and processes to the many fields of biotechnology, from diagnostics to bioremediation. Yet, after 40 years of dedicated research, there remains much to be discovered in this field. Fortunately, extremophiles remain an active and vibrant area of research. In the third decade of the twenty-first century, with decreasing global resources and a steadily increasing human population, the world’s attention has turned with increasing urgency to issues of sustainability. These global concerns were encapsulated and formalized by the United Nations with the adoption of the 2030 Agenda for Sustainable Development and the presentation of the seventeen Sustainable Development Goals (SDGs) in 2015. In the run-up to 2030, we consider the contributions that extremophiles have made, and will in the future make, to the SDGs.
Keywords: Bioeconomy, Bioenergy, Biomining, Bioproducts, Extremophiles, Gene discovery, Global Sustainability Goals, Metagenomics, Sustainable Development Goals, Thermophiles
Introduction
Extremophiles and their products have contributed much to areas such as diagnostics and bioprocessing (Coker 2016). The most successful commercial examples of extremophilic products are the DNA manipulating enzymes (Taq polymerase, Vent® polymerase), and numerous other extremophilic enzymes that play important roles in commercial drug biosynthesis, household products and large-scale biotransformations (Antranikian and Egorova 2007). Extremophilic organisms also continue to yield novel metabolic pathways, enzymes and control elements (e.g., Bräsen et al. 2014; Weisse et al. 2016; Saitsev et al. 2018; Johnsen et al. 2019, 2020, 2023; Stracke et al. 2020; Kuprat et al. 2021; Lewis et al. 2021).
In a world where research fields rise and fall, it is perhaps surprising that extremophile research remains a highly active and exciting topic. The continued interest in extremophile research has many causes. The concept that very high levels of evolutionary novelty, such as in the hyperthermophilic archaea, will be associated with molecular and functional novelty is borne out by recent metagenomic studies of hot pool and deep-sea hydrothermal vent microbiomes (Strazzulli et al. 2017; Iacono et al. 2020; Reichart et al. 2021) and metaviromes (Dávila-Ramos et al. 2019), and by the discovery of completely unique viruses in hydrothermal systems (Prangishvilli et al. 2017). Similarly, the notion that studies of organisms living at the outer edges of the ‘biological envelope’ can yield unique insights into the mechanisms of structural and functional adaptation and survival strategies has been supported by decades of research (see, for example, Gerday and Glansdorff 2007; Schmerling et al. 2022). Extremophiles remain one of the dominant themes in the field of astrobiology and the search for life on other planetary bodies (Thombre et al. 2020).
The global biotechnology industry has also retained an ongoing interest in extremophiles and their products (Krüger et al. 2018; Straub et al. 2018; Pfeiffer et al. 2021). The lesson of the past 30 years, since the heyday of Recombinant Biocatalysis Inc. and Diversa Corpn. (DeSantis et al. 2002), is that the road to commercial success is often driven top-down (from industry needs) rather than bottom-up (from scientific discoveries). Nonetheless, achieving true progress towards completely novel products and innovations requires a solid scientific basis—and often novel scientific discoveries that may be exploited, such as gene editing using the CRISPR Cas 9 system. CRISPR loci have been described in halophiles by Francisco Mojica (Mojica et al. 1993), who later proposed a role for these sequences in microbial immunity (Mojica et al. 2005). Proof of the continued interest in extremophiles as sources of academic and biotechnological novelty is evident in the fact that extremophile research is still well-funded (e.g., in EU-programmes such as Horizon 2020).
It is necessary to add a caveat at this point. While extremophiles are already used in many fields of biotechnology and, as discussed below, hold great potential for future exploitation, these organisms are not without their practical limitations. The stringent optimal growth conditions for some extremophiles, and the costs associated with their growth at large (multi-kilolitre) scales, together impose significant challenges for industrial application.
However, the aim of this review is to look forward, not back; to speculate on the future of extremophiles in a changing world where issues of climate change, sustainability, constraints on energy, food systems, healthy soil and clean water supplies and global health challenges are all increasingly immediate and concerning. We also aim to highlight conceptual and practical gaps in our technical and knowledge base that currently inhibit our ability to fully exploit the resources available in the world of extremophiles (and other biological groups).
In less than 20 years since the commercialization of Next Generation nucleic acid sequencing platforms, the global research community is deluged in DNA sequence data. At the time of writing, many hundreds of publicly-available extremophilic metagenomic DNA sequence datasets, representing many Terabase-pairs of sequence data) are accessible in NCBI and other platforms. These datasets represent a resource of enormous scale: tens of thousands of complete (and incomplete) biosynthetic pathways and many millions of protein coding genes, a significant proportion of which (20–40%; e.g., Chen et al. 2021) cannot currently be annotated and are therefore of totally unknown function (Salzberg 2019). This sequence resource will continue to grow, possibly at an exponential rate. While this represents an ever-increasing resource, the gap between digitalized and experimental data will grow proportionally.
We note, as have others, that the rate of sequence data generation generally exceeds the rate of development of systems and processes capable of manipulating, interpreting, understanding and exploiting this sequence resource. The general issues of ‘data overload’ have been discussed in some detail elsewhere (Baker 2010).
Gene discovery
The latest technologies available for the identification of genes (and/or gene products) in metagenomic DNA are broadly divided into functional metagenomic methods (most commonly function-based screening of metagenomic plasmid, phage, fosmid, cosmid or BAC/YAC expression libraries (Uchiyama and Miyazaki 2009; Iacono et al. 2022)), and gene-mining from assembled metagenome sequence data (Kenshole et al. 2021). Both approaches have substantial limitations.
In the former, expression screening is restricted to a very limited range of expression hosts, almost none of which are extremophiles (Uchiyama and Miyazaki 2009; Tripathi and Srivastava 2019). Issues such as the constraints of variable codon-usage, promoter sequence variations, the complexities of in vivo gene expression and domain-specific post-translational modification all limit the efficiency of expression screening of extremophile metagenomic DNA extracts. Even where screening is successful, scale-up of expression is largely constrained to a few highly engineered non-extremophilic expression hosts (Tripathi and Srivastava 2019). Expression screening is also limited by assay development: it is notable that two decades of gene mining metagenomic expression screening (often termed functional metagenomics) has largely exploited readily available chromogenic hydrolase substrates.
Some of the current gaps in the technology are obvious: ‘plug-and-play’ laboratory expression hosts for the major groups of extremophiles (e.g., thermophilic bacteria and archaea, psychrophiles, acidophiles, halophiles), where intracellular expression is highly impacted by the extracellular environment (temperature, pH, salt), are only available for a limited range of organisms. Probably the first thermophilic organism to be genetically manipulated was Thermus thermophiles (Oshima 1995), and this organism has now been engineered as a thermophilic host for the over-expression of proteins that cannot be expressed in mesophilic systems (De Rose et al. 2021a). However, the very high G + C content of the DNA of this organism generally means that genes to be expressed in this host system have to be synthesized.
The constant development of novel genetic tools for extremophilic archaea (Leigh et al. 2011; Atomi et al. 2012; Farkas et al. 2013; Pfeifer et al. 2021) and bacteria (Averhoff et al. 2021) is particularly promising. Examples include genetic methods to manipulate thermophiles Pyrococcus furiosus and Thermococcus spp. (Lipscomb et al. 2011; Sato et al. 2005; Thiel et al. 2014; Birien et al. 2018) and Sulfolobus (Wagner et al. 2012; Quehenberger et al. 2017) as well as halophilic (Peck et al. 2000; Bitan-Banin et al. 2003; Allers et al. 2004) and methanogenic (Susanti et al. 2019) archaea. Within the extremophilic bacteria, it is now possible to over-produce oxygen-sensitive proteins; e.g., in the anaerobe Thermoanaerobacter kivui (Basen et al. 2018; Katsyv et al. 2021a), and a two-enzyme cascade from Methanothermus fervidus has been used successfully to produce the extremolyte cyclic di-phosphoglycerate, a small molecule that can stabilize proteins and DNA and has commercial application in the healthcare and cosmetic industries (de Rose et al. 2021b).
Thermus thermophilus is a particularly suitable candidate as a production platform for thermostable proteins, given its very high natural transformation frequencies (Cava et al. 2009; Averhoff et al. 2021). Six inducible promoters, the Parg, PdnaK, Pscs-mdh Pnar, Psip, and PpilA4 promoters, have been characterized and two systems for efficient protein over-production have been reported (Moreno et al 2003, 2005; Fujino et al. 2020; Kirchner and Averhof 2023). However, it has to be noted that the Pnar promoter is only induced under anaerobic conditions in the presence of nitrate and the Psip is only induced by addition of 10 mM silica, which leads to growth inhibition (Moreno et al. 2005; Fujino et al. 2020).
It has been proposed that a viable alternative for the development of cell-based systems for the in vitro overexpression and production of extremophilic proteins is the use of active learning in the optimization of the physico-chemical and biochemical process conditions (Borkowski et al. 2020).
While these developments provide some insights into the current and future potential of extremophilic protein production platforms, there remains considerable room for improvement. To the authors’ knowledge, no extremophilic organism has ever been engineered for very high-level (multi-g per L) protein production, a critical requirement for economic viability of any commercial commodity enzyme. However, an L-aminoacylase from Thermococcus litoralis, over-expressed in Escherichia coli or Pseudomonas systems at room temperature, is being used for large scale L-amino acid and amino acid analogue production at 50°C by Dr Reddys/Chirotech (Toogood et al. 2002). The concurrent use of a racemase enzyme capable of converting the unused D-enantiomer into the L-enantiomer gives full kinetic resolution (100% conversion) production.
Metagenomic gene mining
The scope of gene mining using functional metagenomics is hugely constrained by the limited range of assay methods that can be readily adapted to high throughput screening (Markel et al. 2020). The development of new assay systems requires a strongly interdisciplinary approach; particularly the close collaboration between biochemists and synthetic organic chemists. Nevertheless, some very significant advances in metagenomic screening technology have been introduced over the past two decades. High throughput robotic liquid handling systems, coupled with advanced microfluidics technology (e.g., de Boer et al. 2005; Strutt et al. 2023) have made it possible to screen very large recombinant expression libraries in practical timescales.
It is argued that the future of ‘gene mining’ lies in silico: identification of novel genes in assembled metagenomic sequence datasets using bioinformatics tools (Koonin et al. 2021). The strengths of this approach are based around the scalability of the process, the speed and capacity of modern computational platforms and the increasing role of Artificial Intelligence (AI) systems (e.g., Saldívar-González et al. 2022). The release of AlphaFold, a Google DeepMind AI-based program, has dramatically increased the accuracy of protein structure prediction (Kryshtafovych et al. 2021), although the algorithm is only reliable for proteins that have related structures available in the protein PDB database. It has been reported that the two enzymes responsible for the biosynthesis of the extremolyte cyclic di-phosphoglycerate, 2PGK and cDPGS (for which no similar protein structures have been solved), give inaccurate AlphaFold predictions (De Rose et al. 2023).
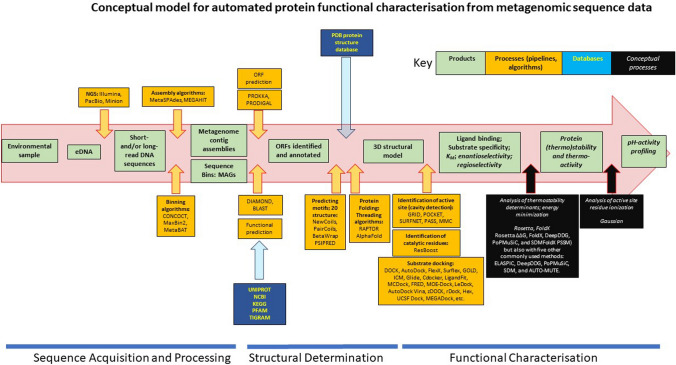
A very wide array of bioinformatic tools is available for processing nucleic acid and protein sequence data along the pipeline from raw sequence to accurate prediction of a folded and functional protein (Fig. 1) (https://bio.tools/), and new algorithms and pipelines are released almost daily. Some, such as AlphaFold, have moved the field forward in quantum leaps (Jones and Thornton 2022). However, some fundamental gaps remain. Most of the tools for structural and functional prediction are homology-based, providing little useful information for the ‘dark sequence’ fraction of genomes, which can be as high as 50% in particularly novel extremophiles such as the Asgard archaea (MacLeod et al. 2019). Bioinformatic tools for prediction of subtle inter-molecular interactions, such as oligomerization, protein–ligand binding and enzyme–substrate docking generally lack sufficient resolution, and the computational capacity to quantitate some protein properties, such as enzyme catalytic rates as well as Intrinsically Disordered Regions (Necci et al. 2021), are not currently available. Furthermore, the many bioinformatic tools that do exist to process steps along the sequence-function pipeline (Fig. 1) are generally not coordinated or interlinked, and there are some fundamental discontinuities in data-exchange. For example, major protein functional databases such as BRENDA (https://www.brenda-enzymes.org/) are currently not coordinated with many processing algorithms, and new data on protein function may often not flow back into the sequence-function pipeline.
Fig. 1.
Conceptual model for automated protein functional characterisation from metagenomic sequence
We freely acknowledge that the advances in science will resolve these issues in time, but argue that the development of coordinated bioinformatic systems and processes designed to accurately extract functional data on gene products from sequence data should be an immediate priority.
Nevertheless, there remains enormous scope (and need) for the development of new approaches to the discovery of new genes and processes, particularly those that come from the unknown (dark sequence) fraction of metagenomic data. Recent advances in correlative analysis of transcriptomes, proteomes and metabolomes in complex communities and enrichment cultures (i.e., meta-transcriptomics, meta-proteomics, meta-metabolomics) or in single cells open new horizons for bioprospecting. For example, the development of novel technologies such as the combination of (meta)genomics and activity-based protein profiling (ABPP: Cravatt et al. 2008; Fang et al. 2021) makes it possible to target specific active enzyme classes in vivo, for subsequent quantification and identification by downstream LC–MS/MS. This approach has been successfully used for the identification of extremozymes in pure cultures (e.g., Sulfolobus spp., Haloferax volcanii and Thermococcus sp.: Zweerink et al. 2017) as well as for the direct profiling of extremophilic microbial community samples in their native environment (environmental ABPP, eABPP: Ninck et al. 2022).
Extremophiles and the sustainable development goals
In a changing world, the United Nations has defined a list of goals that aim to transform our world. (17 sustainable development goals; SDG). The SDGs are a call to action to end poverty and inequality, ensure the sustainability of the processes underlying our societies and economies, and protect the biosphere as the foundation of these objectives. We postulate that basic and applied research in extremophiles can strongly contribute to achieving these goals. In this review, we focus on specific SDGs where we believe extremophiles have made and will continue to make a significant contribution (Fig. 2).
Fig. 2.
Contributions of extremophiles to the UN Sustainable Development Goals
SDG3: good health and well-being
This SDG incorporates many elements relating to communicable and non-communicable human diseases, for most of which extremophilic organisms and their products have no obvious contribution. However, several extremophilic enzymes have played critically important roles in diagnostics and disease research. The most obvious and well-known is Taq DNA polymerase, originally sourced from the extreme thermophile Thermus aquaticus (Innis et al. 1988). This enzyme has a projected market value in 2023 of around $US0.35 billion (https://www.futuremarketinsights.com/reports/dna-polymerase-market). The DNA polymerase market is dominated by Taq polymerase and various engineered variants, but with contributions from other hyperthermophilic functional homologues. Interestingly, the process (PCR) and its core enzyme, which until recently were known only in the research and diagnostic fields, have now become a matter of public knowledge, thanks to the globally important process of Covid-19 PCR testing.
Haloarchaeal carotenoids, in particular bacterioruberin, are especially exciting as they might represent the first case of extremolytes with a demonstrated therapeutic effect in vitro. Bacterioruberin, a rare C50 carotenoid synthesized by most haloarchaea has been shown to exhibit the highest antioxidant capacity of all known carotenoids, including ß-carotene (Mandelli et al. 2012), and were recently shown in in vitro studies to have a cytotoxic effect on human breast cancer lines (Giani et al. 2021, 2023). A novel C50 carotenoid were also recently identified from Natrialba sp. M6, and was also shown to exert significantly higher anticancer effects than standard anticancer drugs, as well the ability to suppress the replication of the hepatitis viruses HCV and HBV (Hegazy et al. 2020).
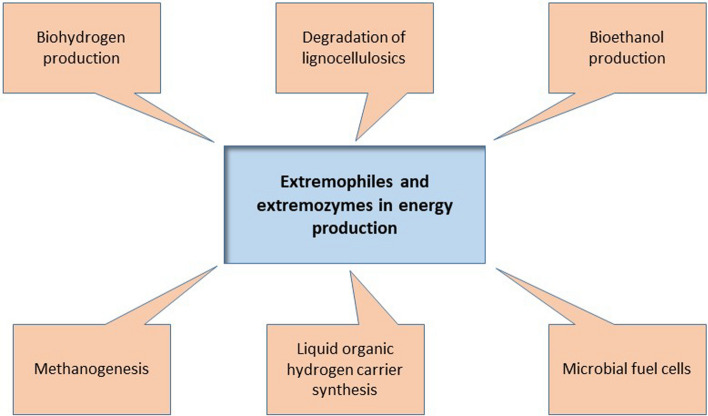
SDG7: Affordable and clean energy is goal number 7 as defined by the United Nations, and arguably the one where extremophiles may make the greatest contribution (summarized in Fig. 3). Currently, the world’s economy is based on the combustion of fossil fuels but concerns about anthropogenic climate change and global warming have led to many initiatives, including those exploiting extremophiles, to replace fossil-fuel based energy carriers. The biological production of fuels from different feedstocks has been experimentally addressed for some decades; early studies focused on sugars as feedstock but this approach competes with human nutrition (the food versus fuel discussion (Naik et al. 2010)). Therefore, sustainable production of fuels and other products from biomass by fermentation should rely on plant-based feedstocks that are not used for human nutrition, such as wood and its constituents (lignocellulose). The use of thermophilic enzymes for degradation of lignocellulosic substrates to simple sugars, for subsequent fermentation to generate value-added products, has been a focus of research and development for more than three decades (e.g., Blumer-Schuette et al. 2014; Botha et al. 2018; Singh et al. 2021).
Fig. 3.
Extremophiles and SDG7 (Affordable and Clean Energy)
Numerous thermophilic bacteria and fungi are known to degrade cellulose and lignocellulose, yielding ethanol and hydrogen (Andlar et al. 2018; Chukwuma et al. 2021). High temperatures facilitate the degradation of the complex feedstocks and, indeed, growth rates on crystalline cellulose in thermophiles are generally superior to those of mesophiles (Blumer-Schuette et al. 2015), one of the reasons that the US DOE-funded BioEnergy Science Center has focused on biomass conversion in two model thermophilic lignocellulolytic bacteria, Clostridium thermocellum and Caldicellulosiruptor bescii (Gilna et al. 2017). Both bacteria have been shown to be highly cellulolytic, efficiently degrading high loads of unpretreated lignocellulose (Basen et al. 2014a; Straub et al. 2019; Kubis et al. 2022), but they differ in their mechanism of plant biomass deconstruction. While C. thermocellum harbors the outer cell wall-attached cellulosome, a huge multienzyme apparatus harboring Carbohydrate Active enZymes (CAZymes) such as glycosyl hydrolases and pectate lyases (Artzi et al. 2017), Caldicellulosiruptor sp. mostly use secreted multidomain CAZymes for biomass deconstruction (Yang et al. 2009; Kataeva et al. 2013) next to surface-attached glycosyl hydrolases (Conway et al. 2016). Both organisms harbor arsenals of thermostable CAZymes for putative industrial applications. As an example, the multidomain endo/exo-cellulase CelA from Caldicellulosiruptor bescii outperformed the commercial (fungal) cellulase blend CTec2 on crystalline cellulose (Brunecky et al. 2017). Caldicellulosiruptor species are known to achieve high hydrogen yields from sugars, close to the maximum of 4 mols H2/mol C6 unit (the Thauer limit) (Van de Werken et al. 2008). This yield may even be improved in the future by co-culture experiments that oxidize the acetate/ethanol produced by fermentation to H2 + CO2. Thermodynamically, this is challenging, but recent advances in understanding production of molecular hydrogen from high potential electrons by the process called electron bifurcation (Müller et al. 2018; Peters et al. 2016; Buckel and Thauer 2018) will lead to further improvement.
Production of fuels such as ethanol from biomass or biomass-derived sugars by (hyper)thermophiles has been studied extensively and is still an area of very productive research (Olson et al. 2015). Genetic tools have been established for several cellulolytic microbes (C. bescii and C. thermocellum: Guss et al. 2012; Chung et al. 2013; Lipscomb et al. 2016), and used to improve ethanol yields in both organisms (Chung et al. 2014; Argyros et al. 2011; Olson et al. 2015 and references therein). However, this remains challenging as a potentially commercial process as they are not native ethanol producers (C. bescii) and produce ethanol as one of many products (C. thermocellum). Conversely, the related thermophilic genera, Thermoanaerobacterium and Thermoanaerobacter, naturally produce ethanol at comparably high yields (Wiegel et al. 1981), which have been further improved by deleting the production of other fermentation products such as lactate and acetate (Argyros et al. 2011; Olson et al. 2015). The understanding of ethanol production in these species may serve as a template for improvement of ethanol production in C. thermocellum or C. bescii or in other thermophilic anaerobes. One key enzyme may be the bifunctional aldehyde/alcohol dehydrogenase AdhE (Lo et al. 2015; Hitscher et al. 2021), which is essential for ethanol production from acetyl-Coenzyme A in some fermentative organisms. Fermentative microbes, however, preferentially produce acetate from acetyl-CoA as a means of generating additional ATP: this represents an energetic barrier to ethanol production in such organisms. Moreover, reduced ferredoxin (Fdred) is often produced during pyruvate oxidation, and Fdred is not utilized by AdhE. A viable alternative is the use of a membrane-bound enzyme complex that oxidizes Fdred and reduces NAD+ at the same time; the energy-conserving Rnf-complex (Biegel et al. 2010). The use of genetic engineering to overproduce this complex may be a route to higher ethanol yields (Williams-Rhaesa et al. 2018). Other thermophiles contain an enzyme, aldehyde-ferredoxin oxidoreductase (AOR), which reduces acetate to acetaldehyde (Mukund and Adams 1991), where acetate may then be converted to ethanol by an alcohol dehydrogenase. Some metabolic engineering of AOR-containing pathways has already been performed: for example, in the hyperthermophilic archaeon P. furiosus (Basen et al. 2014b) and in Caldicellulosiruptor sp. (Rubinstein et al. 2020). AOR not only oxidizes acetate but also higher carbon chain acids when provided to the bacteria in the medium (Hitschler et al. 2021; Nissen and Basen 2019); thus, fuels with higher energy density such as butanol or even hexanol can be produced. It is possible that this conceptual process will become a reality in the near future, using extremophiles in the production of long chain alcohols (kerosene). Recently, production of ethanol was achieved at 95°C by an engineered pathway in P. furiosus, using CO as a substrate (Lipscomb et al. 2023).
Biofuel production from biomass is always accompanied by the production of carbon dioxide, and although there is no net production of carbon dioxide in the process it is desirable to limit further increases in atmospheric CO2 levels. CO2 produced by processes under high temperatures can be removed by hot acetogenesis. Acetogens such as Moorella thermoacetica or Thermoanaerobacter kivui fix CO2 by the Wood-Ljungdahl pathway to acetyl-CoA that is further reduced to acetate (Basen and Müller 2017; Rosenbaum and Müller 2023). Electrons for the reduction of CO2 can be derived from oxidation of molecular hydrogen, from methanol (or other methyl-group containing substrates) or carbon monoxide. Even electrons derived from an anode can be used (electrosynthesis). Although the major pathways, the enzymes and genes involved are known (Katsyv et al. 2021b), there are still gaps: for example, what is the role of the two different energy-converting hydrogenases (Schölmerich and Müller 2019) and are there more, cytochrome-containing, energy conservation mechanisms (Rosenbaum and Müller 2021)? The latter is of special importance since acetogens are energy limited and can only produce low energy products such as acetate or some ethanol. Higher carbon chain compounds are only produced by a few species and only on very minor amounts. Metabolic engineering of (thermophilic) acetogens is well on its way but production of value-added compounds is restricted thermodynamically. One of the challenges of the future is to overcome these energetic barriers (Katsyv and Müller 2020).
CO2 can also be reduced to methane by methanogenesis, in a pathway similar to acetogenesis, carried out by thermophilic methanogens (Thauer et al. 2008). Electrons can be derived from molecular hydrogen but also electrochemically and the company Elektrochaea (https://www.electrochaea.com) is leading the technological development of commercial biomethanization, or CO2-based biological CH4 production (CO2-BMP), a process with potential as chemical energy storage of excess electricity (Bernacchi and Herwig 2016; Bernacchi et al. 2016). Recent physiological screening of 80 cultivated methanogenic archaea yielded a number of high-performance strains for future development of high temperature biomethanization (Mauerhofer et al. 2021). Biomethanation of carbon monoxide has also been achieved by using a synthetic hyperthermophilic archaeal consortia (Zipperle et al. 2021).
Thermophilic bacteria such as the anaerobe Thermovibrio ammonificans, which contain a highly thermostable α-carbonic anhydrase, are being developed for CO2 capture (James et al. 2014). This enzyme has been immobilized in industrial-scale bioreactors to remove CO2 from the environment (CO2 solutions, Quebec, Canada; www.co2solutions.com).
Molecular hydrogen has been considered as an attractive, alternative energy carrier (Rosen and Koohi-Fayegh 2016), as has ‘hythane’, a mixture of hydrogen and methane (Ghosh and Kar 2022). Hydrogen can be produced in various ways. If generated by water splitting using renewable energy sources (i.e., solar-, wind-, hydro- or geothermal-power) there is no net CO2 production in the production process. However, such processes are not yet operating at a scale large enough to impact fossil fuel usage. High temperature processing may improve production efficiencies. The thermophilic archaeon Thermococcus onnurineus and the thermophilic bacterium Thermoanaerobacter kivui have the highest-ever reported rates of hydrogen production from formic acid (Müller 2019; Burger et al. 2022). While the former uses a membrane-bound enzyme for formate oxidation (Kim et al. 2010; Bae et al. 2015), the latter has a soluble enzyme: hydrogen-dependent CO2 reductase (Schuchmann and Müller 2015; Dietrich et al. 2022). A current limitation is the low cell density these organisms achieve in bioreactors, offering considerable scope for future improvement. An interesting aspect is that carbon monoxide, a component of synthesis gas (a waste product from steel milling) can be oxidized to molecular hydrogen (and CO2) at high temperatures (Kim et al. 2014; Weghoff and Müller 2016).
One of the biggest challenges in the hydrogen industry is storage and transport of this highly explosive gas. One possibility is to bind molecular hydrogen to create a liquid, organic hydrogen carrier (LOHC), such as formate. Formate is produced from H2 + CO2 by chemical catalysis but also by the hydrogen-dependent CO2 reductase (HDCR; Müller 2019). The HDCR from the thermophile T. kivui has the highest-ever reported rates for CO2 reduction with molecular hydrogen to formate (Schwarz et al. 2018; Schwarz and Müller 2020). LOHC formate can be shipped over long distances and reoxidized at the target location to hydrogen (and CO2). Recently, a biobattery has been developed in which cells produce formate from hydrogen and carbon dioxide during the day and then reverse the reaction at night to generate molecular hydrogen (Schwartz et al. 2022).
SDG9: industry, innovation and infrastructure
Establishment of advanced extremophilic protein expression and metabolic engineering platform
Although extremophiles and their extremozymes continue to attract attention for commercial and industrial applications, some taxa, particularly extremophilic archaea, are still under-exploited compared to their mesophilic counterparts. The different lifestyles of extremophiles, with their molecular and physiological adaptations to the biotic limits of temperature, pH and salt, often match the harsh process conditions required for industrial processes such as lignocellulosic biomass conversion. However, the ability to engineer most extremophiles for industrial use remains limited. For a limited number of model extremophilic organisms such as Sulfolobus acidocaldarius, Saccharolobus solfataricus, Pyrococcus furiosus, Thermococcus kodakarensis, Thermus thermophilus, Haloferax volcanii, Halobacterium salinarum, Methanocaldococcus jannaschii and Methanothermobacter thermautotrophicus, advanced genetic systems have been developed to facilitate protein expression, gene deletion and metabolic engineering (e.g., Schocke et al. 2019). However, one major challenge for future commercial applications is the need to generate very high cell densities in large-scale bioreactors. Some advances have been reported: for example, process engineering developments for S. acidocaldarius culture medium optimization have yielding up to 35 g dry cell weight per litre fermentation broth (OD650 50–60: Quehenberger et al. 2019). This compares well with wet cell yields of > 100 g.L−1 for some bacterial and yeast fermentations (Shay et al. 1987) although industrial fermentations achieve very much higher biomass yields.
Sulfolobus cell biomass has been used for production of archaeal liposomes (archaeosomes), which can serve as delivery systems for drugs such as vaccines, proteins, peptides and nucleic acids or for trehalose production (Rastädter et al. 2020). However, further process engineering developments are critical prerequisites to establishing extremophiles as metabolic engineering platforms and cell factories.
SDG12: responsible consumption and production
Currently, the world’s fossil-fuel based economy is essentially linear, where the production of raw materials, the manufacturing of goods and their subsequent consumption patterns are frequently unsustainable, energy-inefficient processes which produce high volumes of waste and greenhouse gasses. For the transition to a circular economy, there is an urgent requirement for novel products that can be readily recycled back into the value chain or degraded without harmful effects for the environment.
Haloarchaea are a particularly interesting group of extremophiles that produce a rich repertoire of products with potential or actual uses in medicine (e.g., haloarchaeal bacteriorhodopsins in bioelectronics: reviewed by Pfeifer et al. 2021), or in material production (e.g., polyhydroxyalkanoates (PHA)). PHAs are a group of diverse bio-polyesters produced by microorganisms as carbon polyester storage compounds, and can be developed as biodegradable and sustainably sourced alternatives to fossil fuel-based plastics. Alongside the widely exploited mesophilic bacterial producers, haloarchaea have been developed as tools for PHA production, as they offer certain advantages over bacterial producers (Dietrich et al. 2017; Pfeifer et al. 2021). In an effort to connect CO2 fixation (and thereby removal) to production of added-value compounds Methanococcus maripaludis, a mesophilic autotrophic methanogenic archaeon, was recently engineered to produce PHAs by diverting the flow of acetyl-CoA to various biosynthetic pathways, paving the way for similar attempts with extremophiles (Thevasundaram et al. 2022). While not yet developed to industrial scale, this technology is considered to have considerable future promise in replacing fossil fuel-based plastics.
The establishment of safe and sustainable food systems has emerged as one of the most important focus points in the international agenda (e.g. Horizon Europe Cluster 6, SDGs 2, 12, 13). In the search for sustainable alternative protein sources suitable for human and animal diets, microbial protein is emerging as one of many promising alternatives (Matassa et al. 2016; Bajic et al. 2023).
Autotrophic, hydrogenotrophic thermophilic methanogens excrete all 20 proteinogenic amino acids into culture media, with the amino acid mixture composition varying depending on growth conditions (Taubner et al. 2023). The physiological reason for this remains unexplored, but the trait has been observed in syntrophic interactions involving methanogens in various ecosystems, and is therefore probably of ecophysiological significance (e.g. Imachi et al. 2020). A recently established startup company, Arkeon GmbH (https://arkeon.bio/), is developing a one-step fermentation process to upscale the production of amino acids from the genus Methanothermobacter as a sustainable protein source for the food industry (Turrell 2023). The added value of this technology is that the process can use CO2 from industrial sources, while the CH4 output could be used as a biofuel.
Another example with relevance to SDG12 (and SDG3) would be in the use of thermophilic sugar isomerases for the synthesis of new nutraceuticals (De Rose et al. 2021a), as sugar-free products which could combat chronic ‘lifestyle’ diseases such as diabetes, obesity, hyperlipidemia, and hypertension (all linked to high intakes of sugar and fatty foods). A thermostable D-Lyxose isomerase enzyme from a hyperthermophilic Thermofilum sp. shows activity up to 95°C. This enzyme is capable of producing D-mannose and L-ribose, both of which have applications in the food, cosmetic and pharmaceutical industries.
Extremophilic enzymes, the robust structures of which are well suited to the demands of synthetic industrial processes, play important roles in sustainable chemical synthesis. In particular, the unique stereoselectivity and enantiospecificity of some of these enzymes finds application in the synthesis of new drug intermediates (Littlechild 2015a, b). Enzymes that have evolved under different evolutionary pressures often show novel structural features, changes in active site tunnels, increased substrate promiscuity or different substrate stereo-specificities from related homologues that have previously been identified. Sequence-based screening of thermophilic metagenomes (Wohlgemuth et al. 2018) has yielded novel limonene and α/β class epoxide hydrolases; enzymes of special interest to the pharmaceutical industries (Ferrandi, et al. 2015, 2018).
The aminotransferases, which are capable of introducing chiral amines, are another important group of enzymes for the production of pharmaceuticals. A range of stereoselective aminotransferases, from the thermophilic archaea Sulfolobus sulfotaricus (Sayer et al. 2012), Geoglobus acetivorans and Archaeoglobus fulgidus (Isupov et al. 2019), have been developed as part of the enzyme toolkit for simplifying chemical synthesis pathways. For example, recombinant gluconate dehydratase from the hyperthermophilic crenarchaeon Thermoproteus tenax has been used for the production of chiral 2-keto-3 deoxy gluconate (KDG: Matsubara et al. 2014). The resource-efficient catalyst preparation (two precipitations), in combination with a one-step biocatalytic process, substitutes for a ten-step chemical synthesis and results in stereochemically pure KDG without side-product formation (90% yield).
It is now accepted that using enzymes in chemical synthesis is an economically viable route for producing a variety of new pharmaceuticals (Alcántara et al. 2022) and extremophile enzymes will play an important role in this achievement.
SDGs 14, 15: life below water, life on land
These goals address the conservation, restoration and sustainable use of marine and terrestrial ecosystems. Currently, a significant proportion of global marine, freshwater and terrestrial ecosystems are polluted by industrial and urban wastes (e.g., petroleum, mining waste, agricultural runoff and hazardous chemicals). Bioremediation is a particularly attractive option for reducing the negative impacts of pollution, due to its sustainable and cost-effectiveness nature. The tolerance of extremophiles to environmental extremes (including high concentrations of heavy metals or xenobiotics) makes them particularly suited for some bioremediation processes. A few examples of advances in this area of research are discussed here, but for thorough reviews we direct the readers elsewhere (Krzmarzick et al. 2018; Kaushik et al. 2021).
Restoration of sites contaminated with crude oil or petroleum necessitates the degradation of a variety of aliphatic, branched and aromatic hydrocarbon compounds as well as other organic compounds. Pathways for activation and degradation have been found among the members of Halobacteriota, Sulfolobales, while methanogenic archaea often perform the final part of the process (Shukla and Singh 2020; Park and Park 2018). Excitingly, novel lineages performing anaerobic oxidation of alkanes are continuously being discovered among bacteria and archaea (termed ANME, anaerobic methane-oxidizing archaea, and ANKA, anaerobic multicarbon alkane-degrading archaea), with the latter using phylogenetically and functionally divergent variants of the methyl-coenzyme M reductase (MCR), the key enzyme of methanogenesis operating in reverse (Wegener et al. 2022).
Extremophiles encode multiple survival strategies against heavy metal toxicity in their environment, which can be harnessed for bioremediation of heavy metal and radionuclide contaminated sites (Marques 2018; Krzmarzick et al. 2018; Kaushik et al. 2021). These include bioadsorption to their cell surface or extracellular polymeric substances produced by them and biotransformation through enzymatic oxidation or reduction and subsequent precipitation or accumulation of organic or mineral metal complexes. Most of these strategies are found in thermophilic and acidophilic archaea and bacteria, and in Haloarchaea. Genetically tractable organisms such as members of the genus Deinococcus have successfully been engineered for enhanced radiation and heavy metal resistance and detoxification (Brim et al. 2000, 2003). As an alternative strategy, native microbial communities in contaminated sites have also been harnessed for natural bioremediation, assisted by biostimulation or bioaugmentation. This has been extensively researched in the case of acid mine drainage (Rambabu et al. 2020).
Extremophiles have also the potential to play a part in addressing the major driver of climate change; rising atmospheric CO2 concentrations. Natural mineralization of CO2 into mineral carbonates is a very slow process, where the rate limiting step is the hydration of dissolved CO2 to form HCO3− (bicarbonate). This reaction can be performed biologically by carbonic anhydrases (reviewed in Bose and Satyanarayana 2017; de Oliveira Maciel et al. 2022; Steger et al. 2022), widespread bidirectional enzymes that maintain the carbonate equilibrium in both prokaryotes and eukaryotes, a crucial function for a number of physiological processes such as pH maintenance, ion transport and respiration. Biomineralization of CO2 at industrial combustion sites necessitates the use of thermo-alkali-stable carbonic anhydrases, as alkaline pH is needed for the reaction balance to shift towards bicarbonate. A number of enzymes with the desired properties have been characterized from archaea (Sulfurihydrogenibium azorense, S. yellowstonense YO3AOP1; De Luca et al. 2015) and bacteria (Bacillus halodurans), while metagenomics has revealed the presence of novel carbonic anhydrase variants in eDNA (environmental DNA) sequence data from extremophilic environments (reviewed in Steger et al. 2022). Lab-scale and pilot studies using these thermo-alkali-stable carbonic anhydrases have been shown to be effective in sequestering CO2 from flue gas (Capasso et al. 2012; Faridi and Satyanarayana 2016). While industrial scale processes are still in development, the construction of a CO2 capture facility utilizing CA was announced by the companies Saipem and Novozymes in 2021 (https://www.novozymes.com/en/news/novozymes-deliver-strategy-carbon-capture-collaboration-agreement).
Extremophiles as models for life on other planets
Studying extremophiles can provide valuable insights into the potential for life on other planets, as these organisms demonstrate the capacity to thrive in environments that were once considered inhospitable (Cavicchioli 2002). By studying how such organisms survive and reproduce in extreme environments, we can better define the boundaries of habitability and refine our criteria for identifying potential habitable zones on other planets. Further, extremophiles can help us identify potential biosignatures—signs of past or present life—on other planets. By understanding the types of molecules and metabolic processes that extremophiles use to survive, we can search for similar signatures in the atmospheres or surface materials of other planets. Hence, extremophiles provide a basis for astrobiologists to develop models and theories on the potential types of life that could exist on other planets.
As an example, the exposure of extremophiles to simulated extreme conditions of other planets (such as Martian temperatures or the high-pressure environments of some of Jupiter’s moons), can yield insights into whether similar organisms could potentially survive there (e.g. Taubner et al. 2015, 2018; Favreau et al. 2023). Mars, with its cold arid conditions but a history of surface liquid water, is a prime target for the search for evidence of past life or even extant life. The hypothetical existence of extremophiles on other planetary bodies and systems (e.g., the Venusian atmosphere, the vapour plumes of the Saturnian moon Enceladus, and the icy depths of Europa) has been discussed in some detail (e.g., Seckbach and Stan-Lotter 2020; Schultz et al. 2023).
Studies of extremophiles contribute to our understanding of the limits of life’s resilience. This knowledge is important in preventing contamination of other planets with Earth microbes and will provide a valid basis of the design of future planetary protection protocols.
Extremophile research can contribute to our understanding of the origins of life. The extreme environments of modern Earth may resemble the conditions on early Earth over the period when life is believed to have originated. By studying how these organisms survive in such conditions, we can infer how life might have emerged and evolved in the early stages of our planet’s history. In addition, extremophiles may provide clues to the synthesis of important molecules and structures such as nucleic acids, protein-based catalysts and lipid membranes, and provide clues for prebiotic chemistry.
Hydrothermal vents on the ocean floor are extreme environments where life thrives in the complete absence of sunlight, relying completely on chemosynthesis for energy acquisition. One of the emerging theories is that these environments are analogous to the conditions that might have existed during the early evolution of life. Further, the burst of metagenomic studies of extreme environments has refined our understanding of the genetic makeup and evolutionary relationships of organisms, as well as eukaryogenesis, the process that led to the creation of the first eukaryotic cells. Extremophiles are often found to be deeply rooted in the tree of life, whereas in recent years new clades of archaea were discovered that are rooted in the eukaryotic tree of life lending further support for the symbiogenesis theory for the creation of eukaryotic cells.
Conclusions
Over the past 50 years, extremophiles have contributed to many fields of biotechnology, spanning the full spectrum from basic research to large-scale industrial applications. The unique properties of these organisms and their constituents, driven by evolutionary processes and the properties of their environments, are frequently found to be well matched with the stringent requirements of the process or system in which they are used.
The future applications of extremophiles and extremophilic products, particularly in addressing the critical needs of the SDGs, will come, as they have in the past, from expanding the extremophilic ‘toolbox’ (Antranikian and Streit 2022) and from precise matching the properties of extremophilic ‘tools’ with process requirements (as expressed in the concept of the Ideal Biocatalyst: Burton et al. 2002).
We argue most strongly for the need for a continued emphasis on fundamental research in the field of extremophiles, since new tools to address the current and future challenges of society, industry and the environment inevitably come from fundamental understanding of the diversity, structure, function and properties of extremophiles and their constituents.
We also argue for a need to value the fundamental resources from which extremophiles are acquired; most especially a need to secure and protect specialized and/or limited extremophilic environments which are under threat from destructive commercial exploitation (e.g., mining Li-rich hypersaline lakes for the electronic industry, and tapping hydrothermal sources for energy production). Such habitats contain substantially untapped genetic resources (e.g., Antranikian et al. 2017) which, with future developments in computational power and sophistication (Kruger et al. 2020), will be increasingly accessible in the search for usable and valuable bioproducts.
Acknowledgements
The authors dedicate this article to Professor Mosè Rossi, one of the founding fathers of the field of extremophile research, and a leader in the microbiology and enzymology of thermophiles.
Funding
Open access funding provided by University of Pretoria.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alcántara AR, Domínguez de María P, Littlechild JA, Schürmann M, Sheldon RA, Wohlgemuth R. Biocatalysis as key to sustainable industrial chemistry. Chemsuschem. 2022;15:e202102709. doi: 10.1002/cssc.202102709. [DOI] [PubMed] [Google Scholar]
- Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andlar M, Rezić T, Marđetko N, Kracher D, Ludwig R, Šantek B. Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng Life Sci. 2018;18:768–778. doi: 10.1002/elsc.201800039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antranikian G, Egorova K. Extremophiles a unique resource of biocatalysts for industrial biotechnology. In: Gerday C, Glansdorff N, editors. Chapter 27 in Physiology and Biochemistry of Extremophiles. Wiley Publ; 2007. pp. 359–406. [Google Scholar]
- Antranikian G, Streit WR. Microorganisms harbor keys to a circular bioeconomy making them useful tools in fighting plastic pollution and rising CO2 levels. Extremophiles. 2022;26:10. doi: 10.1007/s00792-022-01261-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antranikian G, Suleiman M, Schäfers C, Adams MW, Bartolucci S, Blamey JM, Birkeland NK, Bonch-Osmolovskaya E, da Costa MS, Cowan D, Danson M. Diversity of bacteria and archaea from two shallow marine hydrothermal vents from Vulcano Island. Extremophiles. 2017;21:733–742. doi: 10.1007/s00792-017-0938-y. [DOI] [PubMed] [Google Scholar]
- Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, Olson DG, Foden JM, Miller BB, Lynd LR, Hogsett DA, Caiazza NC. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol. 2011;77:8288–8294. doi: 10.1128/AEM.00646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artzi L, Bayer EA, Morais S. Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat Rev Microbiol. 2017;15:83–95. doi: 10.1038/nrmicro.2016.164. [DOI] [PubMed] [Google Scholar]
- Atomi H, Imanaka T, Fukui T. Overview of the genetic tools in the Archaea. Front Microbiol. 2012;3:337. doi: 10.3389/fmicb.2012.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averhoff B, Kirchner L, Pfefferle K, Yaman D. Natural transformation in Gram-negative bacteria thriving in extreme environments: from genes and genomes to proteins, structures and regulation. Extremophiles. 2021;25:425–436. doi: 10.1007/s00792-021-01242-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SS, Lee HS, Jeon JH, Lee JH, Kang SG, Kim TW. Enhancing bio-hydrogen production from sodium formate by hyperthermophilic archaeon, Thermococcus onnurineus NA1. Bioproc Biosyst Eng. 2015;38:989–993. doi: 10.1007/s00449-014-1336-9. [DOI] [PubMed] [Google Scholar]
- Bajić B, Vučurović D, Vasić Đ, Jevtić-Mučibabić R, Dodić S. Biotechnological production of sustainable microbial proteins from agro-industrial residues and by-products. Foods. 2023;12:107. doi: 10.3390/foods12010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. Next-generation sequencing: adjusting to data overload. Nature Meth. 2010;7:495–499. doi: 10.1038/nmeth0710-495. [DOI] [Google Scholar]
- Basen M, Geiger I, Henke L, Müller V. A genetic system for the thermophilic acetogenic bacterium Thermoanaerobacter kivui. Appl Environ Microbiol. 2018;84:e02210–02217. doi: 10.1128/AEM.02210-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basen M, Rhaesa AM, Kataeva I, Prybol CJ, Scott IM, Poole FL, Adams MWW. Degradation of high loads of crystalline cellulose and of unpretreated plant biomass by the thermophilic bacterium Caldicellulosiruptor bescii. Bioresource Technol. 2014;152:384–392. doi: 10.1016/j.biortech.2013.11.024. [DOI] [PubMed] [Google Scholar]
- Basen M, Schut GJ, Nguyen DM, Lipscomb GL, Benn RA, Prybol CJ, Vaccaro BJ, Poole FL, Kelly RM, Adams MWW. Single gene insertion drives bioalcohol production by a thermophilic archaeon. Proc Natl Acad Sci USA. 2014;111:17618–17623. doi: 10.1073/pnas.1413789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basen M, Müller V. “Hot” acetogenesis. Extremophiles. 2017;21:15–26. doi: 10.1007/s00792-016-0873-3. [DOI] [PubMed] [Google Scholar]
- Bernacchi S, Herwig C. Challenges and solutions for development of gas limited bioprocesses illustrated by the biological methane production (BMP) process development. Curr Biochem Eng. 2016;3:165–176. doi: 10.2174/1570180813666160527114628. [DOI] [Google Scholar]
- Bernacchi S, Krajete A, Herwig C. Experimental workflow for developing a feed forward strategy to control biomass growth and exploit maximum specific methane productivity of Methanothermobacter marburgensis in a biological methane production process (BMPP) AIMS Microbiol. 2016;2:262–277. doi: 10.3934/microbiol.2016.3.262. [DOI] [Google Scholar]
- Biegel E, Müller V. Bacterial Na+-translocating ferredoxin: NAD+ oxidoreductase. Proc Natl Acad Sci USA. 2010;107:18138–18142. doi: 10.1073/pnas.1010318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birien T, Thiel A, Henneke G, Flament D, Moalic Y, Jebbar M. Development of an effective 6-methylpurine counterselection marker for genetic manipulation in Thermococcus barophilus. Genes. 2018;9:77. doi: 10.3390/genes9020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan-Banin G, Ortenberg R, Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J Bacteriol. 2003;185:772–778. doi: 10.1128/JB.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer-Schuette SE, Brown SD, Sander KB, Bayer EA, Kataeva I, Zurawski JV, Conway JM, Adams MWW, Kelly RM. Thermophilic lignocellulose deconstruction. FEMS Microbiol Rev. 2014;38:393–448. doi: 10.1111/1574-6976.12044. [DOI] [PubMed] [Google Scholar]
- Borkowski O, Koch M, Zettor A, Pandi A, Batista AC, Soudier P, Faulon JL. Large scale active-learning-guided exploration for in vitro protein production optimization. Nature Commun. 2020;11:1872. doi: 10.1038/s41467-020-15798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose H, Satyanarayana T. Microbial carbonic anhydrases in biomimetic carbon sequestration for mitigating global warming: prospects and perspectives. Front Microbiol. 2017;8:1615. doi: 10.3389/fmicb.2017.01615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha J, Mizrachi E, Myburg AA, Cowan DA. Carbohydrate active enzyme domains from extreme thermophiles: components of a modular toolbox for lignocellulose degradation. Extremophiles. 2018;22:1–2. doi: 10.1007/s00792-017-0974-7. [DOI] [PubMed] [Google Scholar]
- Bräsen C, Esser D, Rauch B, Siebers B. Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol Mol Biol Rev. 2014;78:176–197. doi: 10.1128/MMBR.00041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brim H, McFarlan SC, Fredrickson JK, Minton KW, Zhai M, Wackett LP, Daly MJ. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nature Biotechnol. 2000;18:85–90. doi: 10.1038/71986. [DOI] [PubMed] [Google Scholar]
- Brim H, Venkateswaran A, Kostandarithes HM, Fredrickson JK, Daly MJ. Engineering Deinococcus geothermalis for bioremediation of high-temperature radioactive waste environments. Appl Environ Microbiol. 2003;69:4575–4582. doi: 10.1128/AEM.69.8.4575-4582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunecky R, Donohoe BS, Yarbrough JM, Mittal A, Scott BR, Ding H, Taylor LE, Russell JF, Chung D, Westpheling J, Teter SA, Himmel ME, Bomble YJ. The multi-domain Caldicellulosiruptor bescii CelA cellulase excels at the hydrolysis of crystalline cellulose. Sci Rep. 2017;7:9622. doi: 10.1038/s41598-017-08985-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel W, Thauer RK. Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem Rev. 2018;118:3862–3886. doi: 10.1021/acs.chemrev.7b00707. [DOI] [PubMed] [Google Scholar]
- Burger Y, Schwarz FM, Müller V. Formate-driven H2 production by whole cells of Thermoanaerobacter kivui. Biotechnol Biofuels Bioprod. 2022;15:4. doi: 10.1186/s13068-022-02147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton SG, Cowan DA, Woodley JM. The search for the ideal biocatalyst. Nature Biotechnol. 2002;20:37–45. doi: 10.1038/nbt0102-37. [DOI] [PubMed] [Google Scholar]
- Capasso C, De Luca V, Carginale V, Caramuscio P, Cavalheiro C, Cannio R, Rossi M. Characterization and properties of a new thermoactive and thermostable carbonic anhydrase. Chem Eng Trans. 2012;27:271–276. [Google Scholar]
- Cava F, Hidalgo A, Berenguer J. Thermus thermophilus as biological model. Extremophiles. 2009;13:213–231. doi: 10.1007/s00792-009-0226-6. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R. Extremophiles and the search for extraterrestrial life. Astrobiology. 2002;2:281–292. doi: 10.1089/153110702762027862. [DOI] [PubMed] [Google Scholar]
- Chen C, Zhou Y, Fu H, Xiong X, Fang S, Jiang H, Wu J, Yang H, Gao J, Huang L. Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nature Commun. 2021;12:1106. doi: 10.1038/s41467-021-21295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukwuma OB, Rafatullah M, Tajarudin HA, Ismail N. A review on bacterial contribution to lignocellulose breakdown into useful bio-products. Int J Environ Res Public Health. 2021;18:6001. doi: 10.3390/ijerph18116001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D, Cha M, Guss AM, Westpheling J. Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc Natl Acad Sci USA. 2014;111:8931–8936. doi: 10.1073/pnas.1402210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung DW, Farkas J, Westpheling J. Overcoming restriction as a barrier to DNA transformation in Caldicellulosiruptor species results in efficient marker replacement. Biotechnol Biofuels. 2013;6:82. doi: 10.1186/1754-6834-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker JA. Extremophiles and biotechnology: current uses and prospects. F1000Res. 2016;5:f1000. doi: 10.12688/f1000research.7432.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JM, Pierce WS, Le JH, Harper GW, Wright JH, Tucker AL, Zurawski JV, Lee LL, Blumer-Schuette SE, Kelly RM. Multidomain, surface layer-associated glycoside hydrolases contribute to plant polysaccharide degradation by Caldicellulosiruptor species. J Biol Chem. 2016;291:6732–6747. doi: 10.1074/jbc.M115.707810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Ann Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- Dávila-Ramos S, Castelán-Sánchez HG, Martínez-Ávila L, Sánchez-Carbente MdR, Peralta R, Hernández-Mendoza A, Dobson ADW, Gonzalez RA, Pastor N, Batista-García RA. A review on viral metagenomics in extreme environments. Front Microbiol. 2019;10:3389. doi: 10.3389/fmicb.2019.02403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer AR, Bruyneel B, Krabbe JG, Lingeman H, Niessen WM, Irth H. A microfluidic-based enzymatic assay for bioactivity screening combined with capillary liquid chromatography and mass spectrometry. Lab Chip. 2005;5:1286–1292. doi: 10.1039/b506559c. [DOI] [PubMed] [Google Scholar]
- De Luca V, Vullo D, Scozzafava A, Carginale V, Rossi M, Supuran CT, Capasso C. An α-carbonic anhydrase from the thermophilic bacterium Sulphurihydrogenibium azorense is the fastest enzyme known for the CO2 hydration reaction. Bioorg Med Chem. 2013;21:1465–1469. doi: 10.1016/j.bmc.2012.09.047. [DOI] [PubMed] [Google Scholar]
- de Oliveira MA, Christakopoulos P, Rova U, Antonopoulou I. Carbonic anhydrase to boost CO2 sequestration: Improving carbon capture utilization and storage (CCUS) Chemosphere. 2022;299:134419. doi: 10.1016/j.chemosphere.2022.134419. [DOI] [PubMed] [Google Scholar]
- De Rose SA, Kuprat T, Isupov MN, Reinhardt A, Schönheit P, Littlechild JA. Biochemical and structural characterisation of a novel D-Lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp. Front Bioeng Biotechnol. 2021;9:711487. doi: 10.3389/fbioe.2021.711487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rose SA, Finnigan W, Harmer NJ, Littlechild JA, the HotSolute consortium Production of the extremolyte cyclic 2,3-diphosphoglycerate using Thermus thermophilus as a whole-cell factory. Front Catal. 2021;1:803416. doi: 10.3389/fctls.2021.803416. [DOI] [Google Scholar]
- De Rose SA, Isupov M, Worthy H, Harmer N, Siebers B, Littlechild JA (2023) Structural characterisation of cyclic 2,3-diphosphoglycerate synthetase from Methanothermus fervidus. Front Microbiol (in press) [DOI] [PMC free article] [PubMed]
- DeSantis G, Zhu Z, Greenberg WA, Wong K, Chaplin J, Hanson SR, Farwell B, Nicholson LW, Rand CL, Weiner DP, Robertson DE, Burk MJ. An enzyme library approach to biocatalysis: development of nitrilases for enantioselective production of carboxylic acid derivatives. J Am Chem Soc. 2002;124:9024–9025. doi: 10.1021/ja0259842. [DOI] [PubMed] [Google Scholar]
- Dietrich HM, Righetto RD, Kumar A, Wietrzynski W, Trischler R, Schuller SK, Wagner J, Schwarz FM, Engel BD, Müller V, Schuller J. Membrane-anchored HDCR nanowires drive hydrogen-powered CO2 fixation. Nature. 2022;607:823–830. doi: 10.1038/s41586-022-04971-z. [DOI] [PubMed] [Google Scholar]
- Dietrich K, Dumont MJ, Del Rio LF, Orsat V. Producing PHAs in the bioeconomy - towards a sustainable bioplastic. Sustain Prod Consum. 2017;9:58–70. doi: 10.1016/j.spc.2016.09.001. [DOI] [Google Scholar]
- Fang H, Peng B, Ong SY, Wu Q, Li L, Yao SQ. Recent advances in activity-based probes (ABPs) and affinity-based probes (AfBPs) for profiling of enzymes. Chem Sci. 2021;12:8288–8310. doi: 10.1039/D1SC01359A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faridi S, Satyanarayana T. Novel alkalistable α-carbonic anhydrase from the polyextremophilic bacterium Bacillus halodurans: characteristics and applicability in flue gas CO2 sequestration. Environ Sci Pollut Res. 2016;23:15236–15249. doi: 10.1007/s11356-016-6642-0. [DOI] [PubMed] [Google Scholar]
- Farkas JA, Picking JW, Santangelo TJ. Genetic techniques for the Archaea. Annu Rev Genet. 2013;47:539–561. doi: 10.1146/annurev-genet-111212-133225. [DOI] [PubMed] [Google Scholar]
- Favreau C, Tribondeau A, Marugan M, Guyot F, Marie A, Puppo R, Dufour T, Huguet A, Zirah S, Kish A. Molecular acclimation of Halobacterium salinarum to halite brine inclusions. Front Microbiol. 2023;13:1075274. doi: 10.3389/fmicb.2022.1075274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandi EE, Sayer C, Isupov MN, Annovazzi C, Marchesi C, Iacobone G, Peng X, Bonch-Osmolovskaya E, Wohlgemuth R, Littlechild JA, Monti D. Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. FEBS J. 2015;282:2879–2894. doi: 10.1111/febs.13328. [DOI] [PubMed] [Google Scholar]
- Ferrandi EE, Sayer C, De Rose SA, Guazzelli E, Marchesi C, Saneei V, Isupov MN, Littlechild JA, Monti D. New thermophilic α/β Class epoxide hydrolases found in metagenomes from hot environments. Front Bioeng Biotechnol. 2018;6:144. doi: 10.3389/fbioe.2018.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino Y, Goda S, Suematsu Y, Doi K. Development of a new gene expression vector for Thermus thermophilus using a silica-inducible promoter. Microb Cell Fact. 2020;19:126. doi: 10.1186/s12934-020-01385-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerday C, Glansdorff N. Physiology and biochemistry of extremophiles. ASM Press; 2007. p. 429. [Google Scholar]
- Ghosh S, Kar D. Biohythane: a potential biofuel of the future. Appl Biochem Biotechnol. 2022;28:1–9. doi: 10.1007/s12010-022-04291-y. [DOI] [PubMed] [Google Scholar]
- Giani M, Montoyo-Pujol YG, Peiró G, Martínez-Espinosa RM. Halophilic carotenoids and breast cancer: from salt marshes to miomedicine. Mar Drugs. 2021;19:594. doi: 10.3390/md19110594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani M, Montoyo-Pujol YG, Peiró G, Martínez-Espinosa RM. Haloarchaeal carotenoids exert an in vitro antiproliferative effect on human breast cancer cell lines. Sci Rep. 2023;13:7148. doi: 10.1038/s41598-023-34419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilna P, Lynd LR, Mohnen D, Davis MF, Davison BH. Progress in understanding and overcoming biomass recalcitrance: a BioEnergy science center (BESC) perspective. Biotechnol Biofuels. 2017;10:285. doi: 10.1186/s13068-017-0971-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss AM, Olson DG, Caiazza NC, Lynd LR. Dcm methylation is detrimental to plasmid transformation in Clostridium thermocellum. Biotechnol Biofuels. 2012;5:30. doi: 10.1186/1754-6834-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy GE, Abu-Serie MM, Abo-Elela GM, Ghozlan H, Sabry SA, Soliman NA, Abdel-Fattah YR. In vitro dual (anticancer and antiviral) activity of the carotenoids produced by haloalkaliphilic archaeon Natrialba sp. M6. Sci Rep. 2020;10:5986. doi: 10.1038/s41598-020-62663-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitschler L, Nissen LS, Kuntz M, Basen M. Alcohol dehydrogenases AdhE and AdhB with broad substrate ranges are important enzymes for organic acid reduction in Thermoanaerobacter sp. strain X514. Biotechnol Biofuels. 2021;14:187. doi: 10.1186/s13068-021-02038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono R, Cobucci-Ponzano B, De Lise F, Curci N, Maurelli L, Moracci M, Strazzulli A. Spatial metagenomics of three geothermal sites in Pisciarelli hot spring focusing on the biochemical resources of the microbial consortia. Molecules. 2020;25:4023. doi: 10.3390/molecules25174023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono R, Strazzulli A, Giglio R, Bitetti F, Cobucci-Ponzano B, Moracci M. Valorization of biomasses from energy crops for the discovery of novel thermophilic glycoside hydrolases through metagenomic analysis. Int J Mol Sci. 2022;23:10505. doi: 10.3390/ijms231810505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, Matsui Y, Miyazaki M, Murata K, Saito Y, Sakai S, Song C, Tasumi E, Yamanaka Y, Yamaguchi T, Kamagata Y, Tamaki H, Takai K. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature. 2020;577:519–525. doi: 10.1038/s41586-019-1916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis NA, Myambo KB, Gelfand DH, Brows MA. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci USA. 1988;85:9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isupov MN, Boyko KM, Sutter J-M, James P, Sayer C, Schmidt M, Schönheit P, Nikolaeva AY, Stekhanova TN, Mardanov AV, Ravin NV, Bezsudnova EY, Popov VO, Littlechild JA. Thermostable branched-chain amino acid transaminases from the archaea Geoglobus acetivorans and Archaeoglobus fulgidus: biochemical and structural characterization. Front Bioeng Biotechnol. 2019;7:7. doi: 10.3389/fbioe.2019.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Isupov MN, Sayer C, Saneeii V, Berg S, Lioliou M, Kotlar HK, Littlechild JA. The structure of a tetrameric α-carbonic anhydrase from Thermovibrio ammonificans reveals a core formed around intermolecular disulfides that contribute to its thermostability. Acta Cryst. 2014;D70:2607–2618. doi: 10.1107/S1399004714016526. [DOI] [PubMed] [Google Scholar]
- Johnsen U, Ortjohann M, Reinhardt A, Turner JM, Stratton C, Weber KR, Sanchez KM, Maupin-Furlow J, Davies C, Schönheit P. Discovery of a novel transcriptional regulator of sugar catabolism in archaea. Mol Microbiol. 2023;120:224–240. doi: 10.1111/mmi.15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen U, Reinhardt A, Landan G, Tria F, Turner JM, Davies CP. New views on an old enzyme: allosteric regulation and evolution of archaeal pyruvate kinases. FEBS J. 2019;286:2471–2489. doi: 10.1111/febs.14837. [DOI] [PubMed] [Google Scholar]
- Johnsen U, Sutter JM, Reinhardt A, Pickl A, Wang R, Xiang H, Schönheit P. D-Ribose catabolism in archaea: discovery of a novel oxidative pathway in Haloarcula species. J Bacteriol. 2020;202:10–128. doi: 10.1128/JB.00608-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Thornton JM. The impact of AlphaFold2 one year on. Nature Meth. 2022;19:15–20. doi: 10.1038/s41592-021-01365-3. [DOI] [PubMed] [Google Scholar]
- Kataeva I, Foston MB, Yang SJ, Pattathil S, Biswal AK, Poole FL, Basen M, Rhaesa AM, Thomas TP, Azadi P, Olman V, Saffold TD, Mohler KE, Lewis DL, Doeppke C, Zeng YN, Tschaplinski TJ, York WS, Davis M, Mohnen D, Xu Y, Ragauskas AJ, Ding SY, Kelly RM, Hahn MG, Adams MWW. Carbohydrate and lignin are simultaneously solubilized from unpretreated switchgrass by microbial action at high temperature. Energy Environ Sci. 2013;6:2186–2195. doi: 10.1039/c3ee40932e. [DOI] [Google Scholar]
- Katsyv A, Jain S, Basen M, Müller V. Electron carriers involved in autotrophic and heterotrophic acetogenesis in the thermophilic bacterium Thermoanaerobacter kivui. Extremophiles. 2021;25:513–526. doi: 10.1007/s00792-021-01247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsyv A, Schoelmerich MC, Basen M, Müller V. The pyruvate:ferredoxin oxidoreductase of the thermophilic acetogen, Thermoanaerobacter Kivui. FEBS Open Bio. 2021;11:1332–1342. doi: 10.1002/2211-5463.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsyv A, Müller V. Overcoming energetic barriers in acetogenic C1 conversion. Front Bioeng Biotechnol. 2020;8:621166. doi: 10.3389/fbioe.2020.621166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Alatawi A, Djiwanti SR, Pande A, Skotti E, Soni V. Microbial rejuvenation of polluted environment. Springer; 2021. Potential of extremophiles for bioremediation; pp. 293–328. [Google Scholar]
- Kenshole E, Herisse M, Michael M, Pidot SJ. Natural product discovery through microbial genome mining. Curr Opin Chem Biol. 2021;60:47–54. doi: 10.1016/j.cbpa.2020.07.010. [DOI] [PubMed] [Google Scholar]
- Kim MS, Choi AR, Lee SH, Jung HC, Bae SS, Yang TJ, Jeon JH, Lim JK, Youn H, Kim TW, Lee HS, Kang SG. A novel CO-responsive transcriptional regulator and enhanced H2 production by an engineered Thermococcus onnurineus NA1 strain. Appl Environ Microbiol. 2014;81:1708–1714. doi: 10.1128/AEM.03019-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Lee HS, Kim ES, Bae SS, Lim JK, Matsumi R, Lebedinsky AV, Sokolova TG, Kozhevnikova DA, Cha SS, Kim SJ, Kwon KK, Imanaka T, Atomi H, Bonch-Osmolovskaya EA, Lee JH, Kang SG. Formate-driven growth coupled with H2 production. Nature. 2010;467:352–3525. doi: 10.1038/nature09375. [DOI] [PubMed] [Google Scholar]
- Kirchner L, Averhoff B. A temperature dependent pilin promoter for production of thermostable enzymes in Thermus thermophilus. Microb Cell Fact. 2023;22:187. doi: 10.1186/s12934-023-02192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Wolf YI. Evolution of microbial genomics: conceptual shifts over a quarter century. Trends Microbiol. 2021;29:582–592. doi: 10.1016/j.tim.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger A, Schäfers C, Schröder C, Antranikian G. Towards a sustainable biobased industry—highlighting the impact of extremophiles. New Biotechnol. 2018;40:144–153. doi: 10.1016/j.nbt.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Krüger A, Schäfers C, Busch P, Antranikian G. Digitalization in microbiology–paving the path to sustainable circular bioeconomy. New Biotechnol. 2020;59:88–96. doi: 10.1016/j.nbt.2020.06.004. [DOI] [PubMed] [Google Scholar]
- Kryshtafovych A, Schwede T, Topf M, Fidelis K, Moult J. Critical assessment of methods of protein structure prediction (CASP)-Round XIV. Proteins. 2021;89:1607–1617. doi: 10.1002/prot.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzmarzick MJ, Taylor DK, Fu X, McCutchan AL. Diversity and niche of archaea in bioremediation. Archaea. 2018;2018:3194108. doi: 10.1155/2018/3194108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubis MR, Holwerda EK, Lynd LR. Declining carbohydrate solubilization with increasing solids loading during fermentation of cellulosic feedstocks by Clostridium thermocellum: documentation and diagnostic tests. Biotechnol Biofuels Bioprod. 2022;15:12. doi: 10.1186/s13068-022-02110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuprat T, Ortjohann M, Johnsen J, Schönheit P. Glucose metabolism and acetate switch in archaea: the enzymes in Haloferax volcanii. J Bacteriol. 2021;203:10–128. doi: 10.1128/JB.00690-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh JA, Albers SV, Atomi H, Allers T. Model organisms for genetics in the domain Archaea: methanogens, halophiles Thermococcales and Sulfolobales. FEMS Microbiol Rev. 2011;35:577–608. doi: 10.1111/j.1574-6976.2011.00265.x. [DOI] [PubMed] [Google Scholar]
- Lewis AM, Recalde A, Bräsen C, Counts JA, Nussbaum P, Bost J, Schocke L, Shen L, Willard DJ, Quax TEF, Peeters E, Siebers A-V, Kelly RM. The biology of thermoacidophilic archaea from the order Sulfolobales. FEMS Microbiol Rev. 2021;17:45. doi: 10.1093/femsre/fuaa063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb GL, Conway JM, Blumer-Schuette SE, Kelly RM, Adams MWW. A highly thermostable kanamycin resistance marker expands the tool kit for genetic manipulation of Caldicellulosiruptor bescii. Appl Environ Microbiol. 2016;82:4421–4428. doi: 10.1128/AEM.00570-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb GL, Crowley AT, Nguyen DM, Keller MW, O’Quinn HC, Tanwee TN, Vailionis JL, Zhang K, Zhang Y, Kelly RM, Adams MW. Manipulating fermentation pathways in the hyperthermophilic archaeon Pyrococcus furiosus for ethanol production up to 95°C driven by carbon monoxide oxidation. Appl Environ Microbiol. 2023;10:e00012–23. doi: 10.1128/aem.00012-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb GL, Stirrett K, Schut GJ, Yang F, Jenney FE, Scott RA, Adams MWW, Westpheling J. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl Environ Microbiol. 2011;77:2232–2238. doi: 10.1128/AEM.02624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlechild JA. Enzymes from extreme environments and their industrial applications. Front Bioeng Biotechnol. 2015;3:161. doi: 10.3389/fbioe.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlechild JA. Archaeal enzymes and applications in industrial biocatalysts. Archaea. 2015;30:147671. doi: 10.1155/2015/147671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo J, Zheng TY, Hon S, Olson DG, Lynd LR. The bifunctional alcohol and aldehyde dehydrogenase dene, adhE, is necessary for ethanol production in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. J Bacteriol. 2015;197:1386–1393. doi: 10.1128/JB.02450-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod F, Kindler GS, Wong HL, Chen R, Burns B. Asgard archaea: diversity, function, and evolutionary implications in a range of microbiomes. AIMS Microbiol. 2019;30:48–61. doi: 10.3934/microbiol.2019.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelli F, Miranda VS, Rodrigues E, Mercadante AZ. Identification of carotenoids with high antioxidant capacity produced by extremophile microorganisms. World J Microbiol Biotechnol. 2012;28:1781–1790. doi: 10.1007/s11274-011-0993-y. [DOI] [PubMed] [Google Scholar]
- Markel U, Essani KD, Besirlioglu V, Schiffels J, Streit WR, Schwaneberg U. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem Soc Rev. 2020;49:233–262. doi: 10.1039/C8CS00981C. [DOI] [PubMed] [Google Scholar]
- Marques CR. Extremophilic microfactories: applications in metal and radionuclide bioremediation. Front Microbiol. 2018;9:1191. doi: 10.3389/fmicb.2018.01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassa S, Boon N, Pikaar I, Verstraete W. Microbial protein: future sustainable food supply route with low environmental footprint. Microb Biotechnol. 2016;9:568–575. doi: 10.1111/1751-7915.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Köhling R, Schönenberger B, Kouril T, Esser D, Bräsen C, Siebers B, Wohlgemuth R. One-step synthesis of 2-keto-3-deoxy-D-gluconate by biocatalytic dehydration of D-gluconate. J Biotechnol. 2014;191:69–77. doi: 10.1016/j.jbiotec.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Mauerhofer LM, Zwirtmayr S, Pappenreiter P, Bernacchi S, Seifert AH, Reischl B, Schmider T, Taubner RS, Paulik C, Rittmann SK. Hyperthermophilic methanogenic archaea act as high-pressure CH4 cell factories. Commun Biol. 2021;4:289. doi: 10.1038/s42003-021-01828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor CS, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Mojica FJM, Juez G, Rodriguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol. 1993;9:613–621. doi: 10.1111/j.1365-2958.1993.tb01721.x. [DOI] [PubMed] [Google Scholar]
- Moreno R, Haro A, Castellanos A, Berenguer J. High-level overproduction of His-tagged Tth DNA polymerase in Thermus thermophilus. Appl Environ Microbiol. 2005;71:591–593. doi: 10.1128/AEM.71.1.591-593.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno R, Zafra O, Cava F, Berenguer J. Development of a gene expression vector for Thermus thermophilus based on the promoter of the respiratory nitrate reductase. Plasmid. 2003;49:2–8. doi: 10.1016/S0147-619X(02)00146-4. [DOI] [PubMed] [Google Scholar]
- Mukund S, Adams MWW. The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an aldehyde ferredoxin oxidoreductase - evidence for its participation in a unique glycolytic pathway. J Biol Chem. 1991;266:14208–14216. doi: 10.1016/S0021-9258(18)98669-2. [DOI] [PubMed] [Google Scholar]
- Müller V. New horizons in acetogenic conversion of one-carbon substrates and biological hydrogen storage. Trends Biotechnol. 2019;37:1344–1354. doi: 10.1016/j.tibtech.2019.05.008. [DOI] [PubMed] [Google Scholar]
- Müller V, Chowdhury N, Basen M. Electron bifurcation: a long-hidden energy-coupling mechanism. Annu Rev Microbiol. 2018;72:331–353. doi: 10.1146/annurev-micro-090816-093440. [DOI] [PubMed] [Google Scholar]
- Naik SN, Goud VV, Rout PK, Dalai AK. Production of first and second generation biofuels: a comprehensive review. Renew Sust Energ Rev. 2010;14:578–597. doi: 10.1016/j.rser.2009.10.003. [DOI] [Google Scholar]
- Necci M, Piovesan D, Tosatto SC. Critical assessment of protein intrinsic disorder prediction. Nat Methods. 2021;18:472–481. doi: 10.1038/s41592-021-01117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninck S, Klaus T, Kochetkova TV, Esser SP, Sewald L, Kaschani F, Bräsen C, Probst AJ, Kublanov I, Siebers B, Kaiser M. Environmental activity-based protein profiling for function-driven enzyme discovery from natural communities. BioRxiv. 2022 doi: 10.1101/2022.11.11.516116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen LS, Basen M. The emerging role of aldehyde:ferredoxin oxidoreductases in microbially-catalyzed alcohol production. J Biotechnol. 2019;306:105–117. doi: 10.1016/j.jbiotec.2019.09.005. [DOI] [PubMed] [Google Scholar]
- Olson DG, Sparling R, Lynd LR. Ethanol production by engineered thermophiles. Curr Opin Biotechnol. 2015;33:130–141. doi: 10.1016/j.copbio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Oshima T. Genes and genetic manipulation in Thermus thermophilus. In: Sharp R, Williams R, editors. Thermus Species. Boston, MA, USA: Springer; 1995. pp. 185–205. [Google Scholar]
- Park C, Park W. Survival and energy producing strategies of alkane degraders under extreme conditions and their biotechnological potential. Front Microbiol. 2018;9:1081. doi: 10.3389/fmicb.2018.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck RF, Dassarma S, Krebs MP. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol Microbiol. 2000;35:667–676. doi: 10.1046/j.1365-2958.2000.01739.x. [DOI] [PubMed] [Google Scholar]
- Peters JW, Miller AF, Jones AK, King PW, Adams MWW. Electron bifurcation. Curr Opin Chem Biol. 2016;31:146–152. doi: 10.1016/j.cbpa.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Pfeifer K, Ergal I, Koller M, Basen M, Schuster B, Rittmann KMR. Archaea in biotechnology. Biotechnol Adv. 2021;47:107668. doi: 10.1016/j.biotechadv.2020.107668. [DOI] [PubMed] [Google Scholar]
- Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV, Krupovic M. The enigmatic archaeal virosphere. Nat Rev Microbiol. 2017;15:724–739. doi: 10.1038/nrmicro.2017.125. [DOI] [PubMed] [Google Scholar]
- Quehenberger J, Albersmeier A, Glatzel H, Hackl M, Kalinowski J, Spadiut O. A defined cultivation medium for Sulfolobus acidocaldarius. J Biotechnol. 2019;301:56–67. doi: 10.1016/j.jbiotec.2019.04.028. [DOI] [PubMed] [Google Scholar]
- Quehenberger J, Shen L, Albers SV, Siebers B, Spadiut O. Sulfolobus–a potential key organism in future biotechnology. Front Microbiol. 2017;12:2474. doi: 10.3389/fmicb.2017.02474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambabu K, Banat F, Pham QM, Ho SH, Ren NQ, Show PL. Biological remediation of acid mine drainage: review of past trends and current outlook. Environ Sci Ecotech. 2020;2:100024. doi: 10.1016/j.ese.2020.100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastädter K, Wurm DJ, Spadiut O, Quehenberger J. The cell membrane of Sulfolobus spp.-homeoviscous adaption and biotechnological applications. Int J Mol Sci. 2020;21:3935. doi: 10.3390/ijms21113935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichart NJ, Bowers RM, Woyke T, Hatzenpichler R. High potential for biomass-degrading enzymes revealed by hot spring metagenomics. Front Microbiol. 2021;12:668238. doi: 10.3389/fmicb.2021.668238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen MA, Koohi-Fayegh S. The prospects for hydrogen as an energy carrier: an overview of hydrogen energy and hydrogen energy systems. Energy Ecol Environ. 2016;1:10–29. doi: 10.1007/s40974-016-0005-z. [DOI] [Google Scholar]
- Rosenbaum FP, Müller V. Energy conservation under extreme energy limitation: the role of cytochromes and quinones in acetogenic bacteria. Extremophiles. 2021;25:413–424. doi: 10.1007/s00792-021-01241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum FP, Müller V. Moorella thermoacetica: a promising cytochrome- and quinone-containing acetogenic bacterium as a platform for a CO2-based bioeconomy. Green Carbon. 2023;1:2–13. doi: 10.1016/j.greenca.2023.06.002. [DOI] [Google Scholar]
- Rubinstein GM, Lipscomb GL, Williams-Rhaesa AM, Schut GJ, Kelly RM, Adams MWW. Engineering the cellulolytic extreme thermophile Caldicellulosiruptor bescii to reduce carboxylic acids to alcohols using plant biomass as the energy source. J Ind Microbiol Biotechnol. 2020;47:585–597. doi: 10.1007/s10295-020-02299-z. [DOI] [PubMed] [Google Scholar]
- Saitsev V, Johnsen U, Reher M, Ortjohann M, Taylor GL, Danson MJ, Schönheit P, Crennell SJ. Insights into the substrate specificity of archaeal entner-doudoroff aldolases: the structures of Picrophilus torridus 2-keto-3-deoxygluconate aldolase and Sulfolobus solfataricus 2-keto-3-deoxy-6-phosphogluconate aldolase in complex with 2-keto-3-deoxy-6-phosphogluconate. Biochemistry. 2018;57:3797–3806. doi: 10.1021/acs.biochem.8b00535. [DOI] [PubMed] [Google Scholar]
- Saldívar-González FI, Aldas-Bulos VD, Medina-Franco JL, Plisson F. Natural product drug discovery in the artificial intelligence era. Chem Sci. 2022;13:1526–1546. doi: 10.1039/D1SC04471K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL. Next-generation genome annotation: we still struggle to get it right. Genome Biol. 2019;20:92. doi: 10.1186/s13059-019-1715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Fukui T, Atomi H, Imanaka T. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl Environ Microbiol. 2005;71:3889–3899. doi: 10.1128/AEM.71.7.3889-3899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayer C, Bommer M, Isupov M, Ward J, Littlechild J. Crystal structure and substrate specificity of the thermophilic serine:pyruvate aminotransferase from Sulfolobus solfataricus. Acta Crystallogr D Biol Crystallogr. 2012;68:763–772. doi: 10.1107/S0907444912011274. [DOI] [PubMed] [Google Scholar]
- Schmerling C, Kouril T, Snoep J, Bräsen C, Siebers B. Enhanced underground metabolism challenges life at high temperature–metabolic thermoadaptation in hyperthermophilic archaea. Curr Opin Syst Biol. 2022;30:100423. doi: 10.1016/j.coisb.2022.100423. [DOI] [Google Scholar]
- Schocke L, Bräsen C, Siebers B. Thermoacidophilic Sulfolobus species as source for extremozymes and as novel archaeal platform organisms. Curr Opin Biotechnol. 2019;59:71–77. doi: 10.1016/j.copbio.2019.02.012. [DOI] [PubMed] [Google Scholar]
- Schölmerich MC, Müller V. Energy conservation by a hydrogenase-dependent chemiosmotic mechanism in an ancient metabolic pathway. Proc Natl Acad Sci USA. 2019;116:6329–6334. doi: 10.1073/pnas.1818580116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchmann K, Müller V. Direct and reversible hydrogenation of CO2 to formate by a bacterial carbon dioxide reductase. Science. 2013;342:1382–1385. doi: 10.1126/science.1244758. [DOI] [PubMed] [Google Scholar]
- Schultz J, dos Santos A, Patel N, Rosado AS. Life on the edge: bioprospecting extremophiles for astrobiology. J Indian Inst Sci. 2023;103:721–737. doi: 10.1007/s41745-023-00382-9. [DOI] [Google Scholar]
- Schwarz F, Schuchmann K, Müller V. Hydrogenation of CO2 at ambient pressure catalyzed by a highly active thermostable biocatalyst. Biotechnol Biofuels. 2018;11:237. doi: 10.1186/s13068-018-1236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F, Müller V. Whole-cell biocatalysis for hydrogen storage and syngas conversion to formate using a thermophilic acetogen. Biotechnol Biofuels. 2020;13:32. doi: 10.1186/s13068-020-1670-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz FM, Moon J, Oswald F, Müller V. Biological hydrogen storage and release through multiple cycles of bi-directional hydrogenation of CO2 to formic acid in a single process unit. Joule. 2022;6:1304–1319. doi: 10.1016/j.joule.2022.04.020. [DOI] [Google Scholar]
- Seckbach J, Stan-Lotter H. Extremophiles as astrobiological models. John Wiley & Sons; 2020. p. 385. [Google Scholar]
- Shay LK, Hunt HR, Wegner GH. High-productivity fermentation process for cultivating industrial microorganisms. J Ind Microbiol. 1987;2:79–85. doi: 10.1007/BF01569506. [DOI] [Google Scholar]
- Shukla AK, Singh AK. Exploitation of potential extremophiles for bioremediation of xenobiotics compounds: a biotechnological approach. Curr Genom. 2020;21:161–167. doi: 10.2174/1389202921999200422122253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Mathur AS, Gupta RP, Barrow CJ, Tuli DK, Puri M. Enzyme systems of thermophilic anaerobic bacteria for lignocellulosic biomass conversion. Int J Biol Macromol. 2021;168:572–590. doi: 10.1016/j.ijbiomac.2020.12.004. [DOI] [PubMed] [Google Scholar]
- Steger F, Reich J, Fuchs W, Rittmann SKR, Gübitz GM, Ribitsch D, Bochmann G. Comparison of carbonic anhydrases for CO2 sequestration. Int J Mol Sci. 2022;23:957. doi: 10.3390/ijms23020957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke C, Meyer BH, Hagemann A, Jo E, Lee A, Albers S-V, Cha J, Bräsen C, Siebers B. Salt stress response of Sulfolobus acidocaldarius involves complex trehalose metabolism utilizing a novel trehalose-6-phosphate synthase (TPS)/trehalose-6-phosphate phosphatase (TPP) pathway. Appl Environ Microbiol. 2020;86:e01565–e1620. doi: 10.1128/AEM.01565-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub CT, Counts JA, Nguyen DM, Wu CH, Zeldes BM, Crosby JR, Conway JM, Otten JK, Lipscomb GL, Schut GJ, Adams MW. Biotechnology of extremely thermophilic archaea. FEMS Microbiol Rev. 2018;42:543–578. doi: 10.1093/femsre/fuy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub CT, Khatibi PA, Wang JP, Conway JM, Williams-Rhaesa AM, Peszlen IM, Chiang VL, Adams MWW, Kelly RM. Quantitative fermentation of unpretreated transgenic poplar by Caldicellulosiruptor bescii. Nature Commun. 2019;10:3548. doi: 10.1038/s41467-019-11376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazzulli A, Iacono R, Giglio R, Moracci M, Cobucci-Ponzano B. Metagenomics of hyperthermophilic environments: Biodiversity and biotechnology. In: Chénard C, Lauro FM, editors. Chapter in Microbial Ecology of Extreme Environments. Springer Publ; 2017. pp. 103–135. [Google Scholar]
- Strutt R, Xiong B, Abegg FF, Dittrich PS (2023) Open microfluidics: droplet microarrays as next generation multiwell plates for high throughput screening. Lab on a Chip. https://pubs.rsc.org/en/content/articlelanding/2024/lc/d3lc01024d [DOI] [PMC free article] [PubMed]
- Susanti D, Frazier MC, Mukhopadhyay B. A genetic system for Methanocaldococcus jannaschii: an evolutionary deeply rooted hyperthermophilic methanarchaeon. Front Microbiol. 2019;10:1256. doi: 10.3389/fmicb.2019.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubner RS, Baumann LMF, Steiner M, Pfeifer K, Reischl B, Korynt K, Bauersachs T, Mähnert B, Clifford EL, Peckmann J, Schuster B, Birgel D, Rittmann SKR. Lipidomics and comparative metabolite excretion analysis of methanogenic archaea reveal organism-specific adaptations to varying temperatures and substrate concentrations. mSystems. 2023;8:e0115922. doi: 10.1128/msystems.01159-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubner RS, Pappenreiter P, Zwicker J, Smrzka D, Pruckner C, Kolar P, Bernacchi S, Seifert AH, Krajete A, Bach W, Peckmann J. Biological methane production under putative enceladus-like conditions. Nature Commun. 2018;9:748. doi: 10.1038/s41467-018-02876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubner R-S, Schleper C, Firneis MG, Rittmann SK-MR. Assessing the ecophysiology of methanogens in the context of recent astrobiological and planetological studies. Life. 2015;5:1652–1686. doi: 10.3390/life5041652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol. 2008;6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- Thevasundaram K, Gallagher JJ, Cherng F, Chang MC. Engineering nonphotosynthetic carbon fixation for production of bioplastics by methanogenic archaea. Proc Natl Acad Sci USA. 2022;119:e2118638119. doi: 10.1073/pnas.2118638119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel A, Michoud G, Moalic Y, Flament D, Jebbar M. Genetic manipulations of the hyperthermophilic piezophilic archaeon Thermococcus barophilus. Appl Environ Microbiol. 2014;80:2299–2306. doi: 10.1128/AEM.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thombre RS, Vaishampayan PA, Gomez F. Physiological and biotechnological aspects of extremophiles. Academic Press; 2020. Applications of extremophiles in astrobiology; pp. 89–104. [Google Scholar]
- Toogood HS, Hollingsworth EJ, Brown RC, Taylor IN, Taylor SJ, McCague R, Littlechild JA. A thermostable L-aminoacylase from Thermococcus litoralis: cloning, overexpression, characterization, and applications in biotransformations. Extremophiles. 2002;6:111–122. doi: 10.1007/s007920100230. [DOI] [PubMed] [Google Scholar]
- Tripathi NK, Shrivastava A. Recent developments in bioprocessing of recombinant proteins: expression hosts and process development. Front Bioeng Biotechnol. 2019;7:3389. doi: 10.3389/fbioe.2019.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrell C. From air to your plate: tech startups making food from atmospheric CO2. Nature Biotechnol. 2023;41:1362–1364. doi: 10.1038/s41587-023-01992-5. [DOI] [PubMed] [Google Scholar]
- Uchiyama T, Miyazaki K. Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr Opin Biotechnol. 2009;20:616–622. doi: 10.1016/j.copbio.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Van de Werken HJG, Verhaart MRA, Vanfossen AL, Willquist K, Lewis DL, Nichols JD, Goorissen HP, Mongodin EF, Nelson KE, Van Niel EWJ, Stams AJM, Ward DE, De Vos WM, Van Der Oost J, Kelly RM, Kengen SWM. Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl Environ Microbiol. 2008;74:6720–6729. doi: 10.1128/AEM.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Van Wolferen M, Wagner A, Lassak K, Meyer BH, Reimann J, Albers SV. Versatile genetic tool box for the crenarchaeote Sulfolobus acidocaldarius. Front Microbiol. 2012;3:214. doi: 10.3389/fmicb.2012.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener G, Laso-Pérez R, Orphan VJ, Boetius A. Anaerobic degradation of alkanes by marine archaea. Ann Rev Microbiol. 2022;76:553–577. doi: 10.1146/annurev-micro-111021-045911. [DOI] [PubMed] [Google Scholar]
- Weghoff MC, Müller V. CO metabolism in the thermophilic acetogen Thermoanaerobacter kivui. Appl Environ Microbiol. 2016;82:2312–2319. doi: 10.1128/AEM.00122-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiße R, Faust A, Schmidt M, Schönheit P, Scheidig AJ. Structure of NDP-forming Acetyl-CoA synthetase ACD1 reveals a large rearrangement for phosphoryl transfer. Proc Natl Acad Sci USA. 2016;113:E519–E528. doi: 10.1073/pnas.1518614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegel J, Ljungdahl LG. Thermoanaerobacter ethanolicus gen. nov., spec. nov., a new extreme thermophilic anaerobic bacterium. Arch Microbiol. 1981;128:343–348. doi: 10.1007/BF00405910. [DOI] [Google Scholar]
- Williams-Rhaesa AM, Rubinstein GM, Scott IM, Lipscomb GL, Poole FL, Kelly RM, Adams MWW. Engineering redox-balanced ethanol production in the cellulolytic and extremely thermophilic bacterium, Caldicellulosiruptor bescii. Metab Eng Commun. 2018;7:e00073. doi: 10.1016/j.mec.2018.e00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth R, Littlechild J, Monti D, Schnorr K, van Rossum T, Siebers B, Menzel P, Kublanov IV, Rike AG, Skretas G, Szabo Z, Peng X, Young MJ. Discovering novel hydrolases from hot environments. Biotechnol Adv. 2018;36:2077–2100. doi: 10.1016/j.biotechadv.2018.09.004. [DOI] [PubMed] [Google Scholar]
- Yang SJ, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, Davis M, Westpheling J, Adams MWW. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol. 2009;75:4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipperle A, Reischl B, Schmider T, Stadlbauer M, Kushkevych I, Pruckner C, Vítězová M, Rittmann SK. Biomethanation of carbon monoxide by hyperthermophilic artificial archaeal co-cultures. Fermentation. 2021;7:276. doi: 10.3390/fermentation7040276. [DOI] [Google Scholar]
- Zweerink S, Kallnik V, Ninck S, Nickel S, Verheyen J, Blum M, Wagner A, Feldmann I, Sickmann A, Albers S-V, Bräsen C, Kaschani F, Siebers B, Kaiser M. Activity based protein profiling as a robust method for enzyme identification and screening in extremophilic archaea. Nature Commun. 2017;8:15352. doi: 10.1038/ncomms15352. [DOI] [PMC free article] [PubMed] [Google Scholar]