Abstract
The recently proposed reorganization of the order Chlamydiales and description of new taxa are broadening our perception of this once narrowly defined taxon. We have recovered four strains of gram-negative cocci endosymbiotic in Acanthamoeba spp., representing 5% of the Acanthamoeba sp. isolates examined, which displayed developmental life cycles typical of members of the Chlamydiales. One of these endosymbiont strains was found stably infecting an amoebic isolate recovered from a case of amoebic keratitis in North America, with three others found in acanthamoebae recovered from environmental sources in North America (two isolates) and Europe (one isolate). Analyses of nearly full-length 16S rRNA gene sequences of these isolates by neighbor joining, parsimony, and distance matrix methods revealed their clustering with other members of the Chlamydiales but in a lineage separate from those of the genera Chlamydia, Chlamydophila, Simkania, and Waddlia (sequence similarities, <88%) and including the recently described species Parachlamydia acanthamoebae (sequence similarities, 91.2 to 93.1%). With sequence similarities to each other of 91.4 to 99.4%, these four isolates of intra-amoebal endosymbionts may represent three distinct species and, perhaps, new genera within the recently proposed family Parachlamydiaceae. Fluorescently labeled oligonucleotide probes targeted to 16S rRNA signature regions were able to readily differentiate two groups of intra-amoebal endosymbionts which corresponded to two phylogenetic lineages. These results reveal significant phylogenetic diversity occurring among the Chlamydiales in nontraditional host species and supports the existence of a large environmental reservoir of related species. Considering that all described species of Chlamydiales are known to be pathogenic, further investigation of intra-amoebal parachlamydiae as disease-producing agents is warranted.
All members of the order Chlamydiales are recognized pathogens of mammals, marsupials, or birds. The ability to produce respiratory disease, among other clinical presentations, is a feature of most species within the order and is especially characteristic of infections produced by Chlamydophila pneumoniae, Chlamydophila psittaci, certain serovars of Chlamydia trachomatis, and the recently described species Simkania negevensis (20, 21, 23). The recent finding of evidence for seroconversion to antigen of Parachlamydia acanthamoebae, an intra-amoebal Chlamydia-like bacterium, in a small number of humans experiencing community-acquired pneumonia, along with the findings of novel Parachlamydia-related 16S rRNA sequences in respiratory specimens, peripheral blood, and aortic tissue, suggests that this and related species of protozoal endosymbionts may also be of clinical significance, warranting further investigation (2, 5, 30).
Previously, we have reported on the common occurrence of uncultured bacterial endosymbionts in protozoa of the genus Acanthamoeba (13). While 20% of axenically growing Acanthamoeba isolates recovered from clinical and environmental sources were found to be host to gram-negative rod endosymbionts, 5% were host to gram-negative coccus endosymbionts; none could be cultured by standard microbiological techniques. Phylogenetic analyses of the gram-negative rod endosymbionts to date have included two lineages of alpha-Proteobacteria: one containing Rickettsiales-affiliated isolates and the other containing isolates related to the Paramecium caudatum symbiont Caedibacter caryophilus (14, 18). Subsequent ultrastructural studies of the coccoid endosymbionts demonstrated the presence of a developmental cycle suggestive of a relationship to members of the Chlamydiales (R. Gautom, R. Herwig, and T. R. Fritsche, Abstr. 96th Gen. Meet. Am. Soc. Microbiol., abstr. R-29, p. 474).
In this paper, we present further morphologic and phylogenetic analyses of four isolates of Chlamydiales endosymbionts found naturally infecting Acanthamoeba sp. trophozoites, three of which were recovered from environmental amoebic isolates originating in North America (two isolates) and Europe (one isolate), and one which was from amoebae infecting the corneal tissues of a patient in North America. Because these bacterial isolates could not be cultivated by standard microbiological techniques, we applied the culture-independent ribosomal RNA (rRNA) approach to determine phylogenetic relatedness to each other and to other strains for which sequence data are available. Fluorescence in situ hybridization (FISH) with oligonucleotide probes designed to target ribosomal signature regions was used to verify the origin of the retrieved sequences and further assist with characterization of these endosymbionts. The finding of a potentially large environmental reservoir of intracellular Chlamydia-like organisms has implications for the evolution of Chlamydiales and their preadaptation and ultimate recruitment to higher animals.
(Portions of this work were presented as an abstract at the 96th General Meeting of the American Society for Microbiology in New Orleans, La., 1996.)
MATERIALS AND METHODS
Isolation and maintenance of Acanthamoeba strains.
The techniques used for recovery and maintenance of acanthamoebae from clinical and environmental sources have been described in detail elsewhere (13, 39). Four isolates of Acanthamoeba spp. were included in this study and were recovered from infected corneal tissues (UWC22), soil samples from western Washington State (UWE1 and UWE25), and municipal sewage sludge from Munich, Germany (TUME1). All isolates were found to have gram-negative cocci occurring as endosymbionts within the cytoplasm which could be readily demonstrated using Giemsa, Hemacolor (Harleco, Gibstown, N.Y.), and other appropriate bacterial stains. The general phenotypic characteristics of three of these endosymbiont strains (UWE1, UWE25, and UWC22) have been described previously (13).
Electron microscopy.
Amoebic isolates containing endosymbionts were examined by electron microscopy using a variation of published methods (16). Briefly, aliquots of amoebae in broth were fixed with 2% glutaraldehyde in 0.1 M cacodylate. Fixed amoebae were then pelleted in agar and embedded. Thin sections were stained with uranyl acetate and lead citrate and examined with a Phillips CM-10 electron microscope.
DNA isolation and PCR amplification of nearly full-length 16S rDNA.
UWC22, UWE1, and UWE25 amoebae in log-phase growth were pelleted, washed three times with cold (4°C) sterile distilled water, and freeze-thawed three times followed by aspiration and expulsion through a 24-gauge needle. The cell slurry was resuspended in 5 ml of cold (4°C) sterile physiologic (0.15 M) saline and gently centrifuged (5 min at 120 × g). The supernatant was filtered through a 5-μm syringe filter, and the bacteria were pelleted by centrifugation (10 min at 3,000 × g). Extraction of bacterial DNA was performed by standard procedures (34).
Amplification of ribosomal gene sequences of these three isolates was performed with 1 μg of extracted DNA using fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and rP2 (5′-ACGGCTACCTTGTTACGACTT3′) broad-range eubacterial primers (40) along with standard Gen Amp reagents (Perkin-Elmer, Norwalk, Conn.) according to the manufacturer's recommendations. Thermal cycling consisted of 35 cycles of denaturation at 94°C for 1.5 min, annealing at 42°C for 1 min, and elongation at 72°C for 4 min; cycling was completed with a final elongation step of 20 min.
TUME1 amoebae were harvested from axenic cultures, washed twice with double-distilled water, and resuspended in 500 μl of UNSET lysis buffer (8 M urea, 0.15 M NaCl, 2% sodium dodecyl sulfate [SDS], 0.001 M EDTA, 0.1 M Tris-HCl [pH 7.5]) at 60°C for 5 min (19). Lysates were extracted twice with phenol-chloroform, and DNA was precipitated with 2 volumes of absolute ethanol.
Oligonucleotide primers targeting 16S ribosomal DNA (rDNA) signature regions that are conserved within the Chlamydiales were used for PCR to obtain nearly full-length bacterial 16S rRNA gene fragments of TUME1 (32). Forward and reverse primer sequences were 5′-CGGATCCTGAGAATTTGATC-3′ (Escherichia coli 16S rDNA positions −2 to 18) and 5′-TGTCGACAAAGGAGGTGATCCA-3′ (E. coli 16S rDNA positions 1554 to 1537), respectively. Amplification reactions were performed in a reaction volume of 50 μl in a thermal capillary cycler with reaction mixtures, including a 20 mM MgCl2 reaction buffer, prepared as recommended by the manufacturer (Idaho Technology, Idaho Falls, Idaho) with Taq DNA polymerase (Promega, Madison, Wis.). Thermal cycling consisted of an initial denaturation step at 94°C for 30 s, followed by 30 cycles of denaturation at 94°C for 20 s, annealing at 50°C for 15 s, and elongation at 72°C for 30 s, with a final elongation step at 72°C for 1 min. Positive controls containing purified DNA from E. coli were included along with negative controls (no DNA added).
The presence and size of all amplification products were determined by agarose gel electrophoresis and ethidium bromide staining.
Cloning and sequence analysis.
Amplified DNA from UWC22, UWE1, and UWE25 was purified by electrophoresis in low-melting-point agarose and ligated into the cloning vector Bluescript II (Stratagene, La Jolla, Calif.), while amplified DNA from TUME1 was ligated directly into the cloning vector pCR2.1 (Invitrogen, Carlsbad, Calif.), with subsequent transformation of E. coli by each vector. The nucleotide sequences of the cloned DNA fragments were determined by automated dideoxynucleotide methods with the Taq Dye Deoxy Terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) for UWC22, UWE1, and UWE25 and the Thermo Sequenase cycle sequencing kit (Amersham Life Science, Little Chalfont, England) for TUME1.
Phylogenetic analysis.
Obtained sequences were added to the 16S rRNA sequence database maintained at the Technische Universität München (encompassing about 16,000 published and unpublished homologous small-subunit rRNA primary structures). Alignment of the new sequences was performed using the program package ARB and its automated alignment tool (O. Strunk and W. Ludwig, www.biol.chemie.tu-muenchen.de/pub/ARB/), with refinement of positioning by visual inspection and by secondary-structure analysis. The ARB parsimony, distance matrix, and maximum-likelihood treeing methods, combined with and without use of filters which exclude highly variable regions, were applied to different data sets.
Oligonucleotide probes and FISH.
Oligonucleotide probes Bn9658 (a probe described previously [2]), specific for P. acanthamoebae and targeting E. coli positions 658 to 675) and C22658 (designed from sequence data derived in this study from the endosymbiont infecting Acanthamoeba sp. isolate UWC22) were used to differentiate between two groups of endosymbionts (Table 1). Probe C22658 is designated S-*-ParaC-0658-a-A-18 according to the standard proposed by Alm et al. (1). Both probes were synthesized and directly labeled with the hydrophilic sulfoindocyanine dye Cy3 or Cy5 (Interactiva, Ulm, Germany).
TABLE 1.
Overall sequence similarities for retrieved 16S rRNA sequences of endosymbionts of Acanthamoeba sp. strains UWE1, UWE25, UWC22, and TUME1 and representative members of the Chlamydiales
| Organism | % Sequence similarity to:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chlamydophila abortus B577 | Chlamydophila psittaci 6BC | Chlamydophila felis FP Baker | Chlamydophila caviae GPIC | Chlamydophila pecorum E58 | Chlamydia trachomatis HAR-13 | Chlamydia suis S45 | Chlamydia muridarum MoPn | Chlamydophila pneumoniae TW-183 | Parachlamydia acanthamoebae Bn9 | Simkania negevensis Z | Endosymbiont UWE1 | Endosymbiont UWE25 | Endosymbiont UWC22 | Endosymbiont TUME1 | |
| Chlamydophila abortus B577 | |||||||||||||||
| Chlamydophila psittaci 6BC | 99.7 | ||||||||||||||
| Chlamydophila felis FP Baker | 98.0 | 98.4 | |||||||||||||
| Chlamydophila caviae GPIC | 98.9 | 98.9 | 97.9 | ||||||||||||
| Chlamydophila pecorum E58 | 96.2 | 96.5 | 95.9 | 96.1 | |||||||||||
| Chlamydia trachomatis HAR-13 | 95.0 | 95.2 | 94.8 | 95.3 | 95.1 | ||||||||||
| Chlamydia suis S45 | 94.3 | 94.7 | 94.4 | 94.5 | 94.5 | 97.3 | |||||||||
| Chlamydia muridarum MoPn | 95.6 | 95.7 | 95.5 | 95.7 | 95.5 | 98.4 | 97.7 | ||||||||
| Chlamydophila pneumoniae TW-183 | 95.8 | 96.2 | 95.1 | 95.3 | 95.8 | 93.9 | 93.5 | 94.6 | |||||||
| Parachlamydia acanthamoebae Bn9 | 86.2 | 86.7 | 86.6 | 86.9 | 86.4 | 86.2 | 87.2 | 87.0 | 87.0 | ||||||
| Simkania negevensis Z | 83.5 | 83.8 | 83.9 | 83.9 | 83.6 | 84.2 | 84.4 | 84.2 | 83.7 | 88.2 | |||||
| Endosymbiont UWE1 | 85.8 | 85.5 | 84.8 | 85.4 | 84.4 | 84.3 | 85.4 | 85.0 | 85.6 | 93.1 | 85.7 | ||||
| Endosymbiont UWE25 | 86.2 | 86.0 | 85.6 | 86.0 | 85.6 | 85.4 | 85.9 | 86.1 | 86.3 | 92.5 | 85.4 | 93.0 | |||
| Endosymbiont UWC22 | 86.8 | 86.7 | 86.2 | 86.7 | 86.2 | 85.7 | 86.4 | 86.4 | 86.1 | 91.2 | 85.5 | 92.9 | 91.9 | ||
| Endosymbiont TUME1 | 86.8 | 87.2 | 86.8 | 87.2 | 86.8 | 86.3 | 87.0 | 87.0 | 86.6 | 91.2 | 85.9 | 92.3 | 91.4 | 99.4 | |
| Waddlia chondrophila WSU-85-1044 | 84.4 | 84.6 | 84.4 | 84.6 | 84.2 | 84.7 | 84.8 | 84.9 | 84.4 | 87.2 | 84.4 | 87.1 | 87.9 | 87.0 | 87.1 |
For in situ hybridization studies, infected amoebic isolates UWC22, UWE25, and TUME1 were washed with Page's saline (39), and living cells were allowed to adhere to six-well slides for 3 h, followed by a single 10-s dip in sterile distilled water, a single 10-s dip in 80% ethanol for fixation and dehydration, and air drying. Amoebae were hybridized at 46°C with the fluorescently labeled probes according to published methods (35), but without SDS in the hybridization and washing buffers. Optimal hybridization stringency conditions were determined for each probe, including formamide concentrations used in the hybridization buffer (25). Positive and negative controls were carried out with probes Eub338 (targeting most members of the domain Bacteria) and Bet42a (specific for the beta subgroup of proteobacteria) (25). Slides were examined with a confocal laser scanning microscope (LSM 510; Carl Zeiss, Oberkochen, Germany) equipped with two HeNe lasers (543 and 633 nm) and optical sectioning capabilities. Image analysis processing was performed with the standard software package delivered with the instrument (version 1.5).
Nucleotide sequence accession numbers.
The recovered 16S rDNA sequences have been deposited in GenBank under accession numbers AF083616 (endosymbiont of Acanthamoeba sp. UWC22), AF083614 (endosymbiont of Acanthamoeba sp. UWE1), AF083615 (endosymbiont of Acanthamoeba sp. UWE25), and AF098330 (endosymbiont of Acanthamoeba sp. TUME1).
RESULTS
Morphologic analyses.
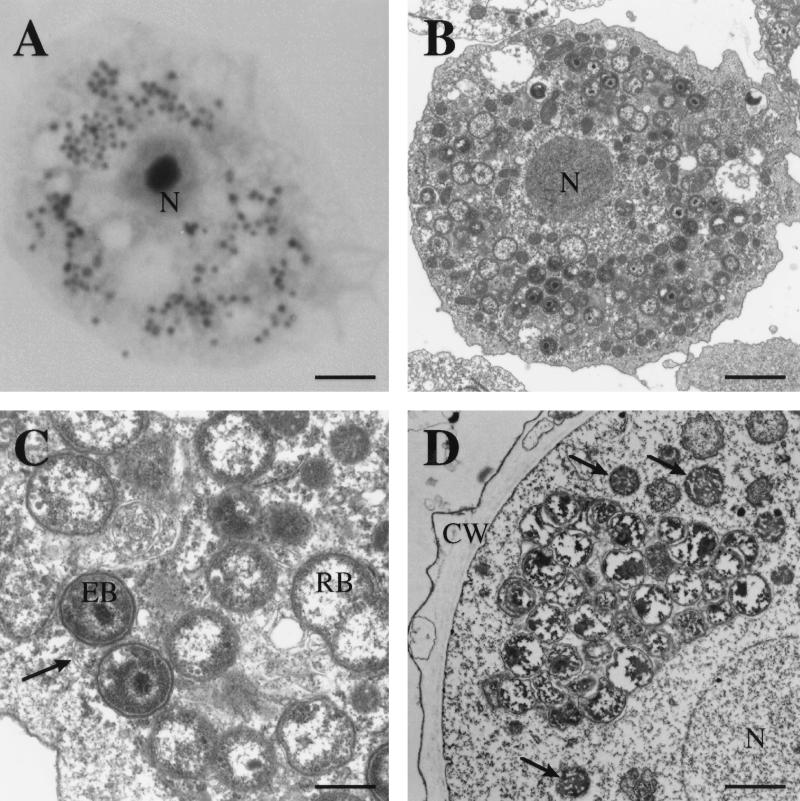
As seen by light microscopy, the organisms are coccoid in appearance and stain gram negative. Use of Giemsa and related stains revealed that they are dispersed throughout the cytoplasm and do not form discrete inclusions, although small clusters and morulae are occasionally noted (Fig. 1A). By electron microscopy, large numbers of cocci are seen in each amoebic trophozoite and display a developmental life cycle typical of Chlamydiales, consisting of smaller electron-dense forms (elementary bodies) and larger dividing forms (reticulate bodies) (Fig. 1B and C). Rather than growing as discrete intravacuolar inclusions, the bacteria are found dispersed throughout the cytoplasm but appear to be surrounded by vacuolar membranes which insinuate themselves around each bacterial cell (Fig. 1C). Some bacteria are found in food vacuoles but appear to be in various stages of disintegration; these forms may have been nonviable bacteria released from other amoebae previously and phagocytosed. When present in amoebic cysts, the bacteria appear to form inclusions and lack evidence of a developmental cycle, suggesting that they are in a resting state (Fig. 1D).
FIG. 1.
(A) Acanthamoeba sp. trophozoite (strain UWE25) infected with coccoid bacterial endosymbionts as seen using Hemacolor stain. N, nucleus. Bar, 7 μm. (B) Low-power electron micrograph of an Acanthamoeba trophozoite; note numerous bacteria scattered throughout the cytoplasm in various stages of differentiation. Bar, 1 μm. (C) High-power view of intracellular bacteria; note developmental stages (elementary bodies [EB] and reticulate bodies [RB]) typical of Chlamydiales and the presence of vacuolar membranes (arrow) between bacteria and surrounding each bacterium. Bar, 1 μm. (D) Chlamydia-like bacterial inclusion seen in a cyst of Acanthamoeba. The bacteria appear to be inactive, with an absence of developmental stages. CW, cyst wall. The arrows indicate mitochondria. Bar, 1 μm.
Comparative sequence results and phylogenetic analysis.
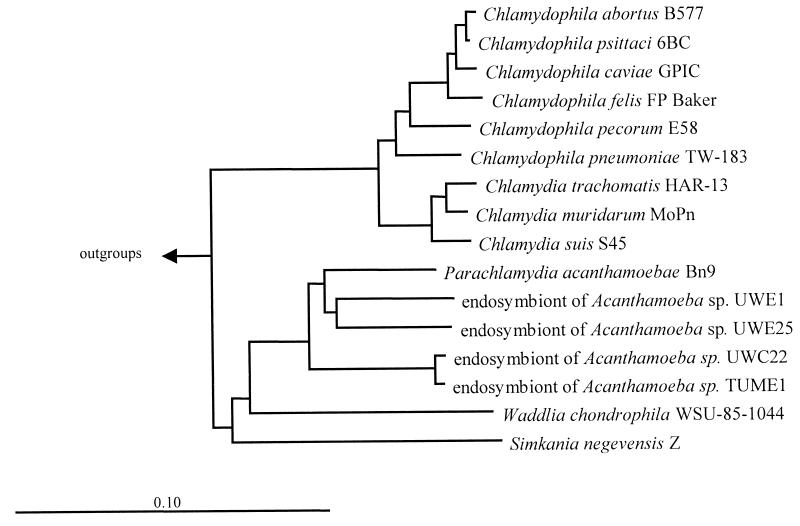
Nearly full-length 16S rDNA sequences from the four endosymbiont strains were amplified, cloned, and sequenced. Comparative sequence analysis using parsimony, distance matrix, and maximum-likelihood treeing methods revealed that the four clustered unequivocally with other members of the Chlamydiales but formed a lineage with P. acanthamoebae and, more distantly, S. negevensis and Waddlia chondrophila that is distinct from Chlamydia and Chlamydophila spp. (Fig. 2). Further analysis of tree topology revealed the presence of two lineages of intra-amoebal endosymbionts: one contains the closely related isolates UWC22 and TUME1 (99.1% sequence similarity), and a second includes UWE1, UWE25, and P. acanthamoebae (sequence similarities of 91.5 to 93.2%). All four isolates warrant inclusion, along with P. acanthamoebae, in the recently proposed family Parachlamydiaceae and may represent up to three new genera and species (Table 1) (11, 36). With sequence similarities of 83.5 to 88.2% and 84.2 to 87.9% to all other Chlamydiales, S. negevensis and W. chondrophila, respectively, would appear to reside in separate families, as has been proposed (11).
FIG. 2.
Neighbor-joining dendrogram showing phylogenetic relationships of the coccoid endosymbionts of Acanthamoeba sp. isolates UWC22, TUME1, UWE1, and UWE25 to other members of the Chlamydiales and outgroups (bar represents estimated evolutionary distance).
FISH analysis.
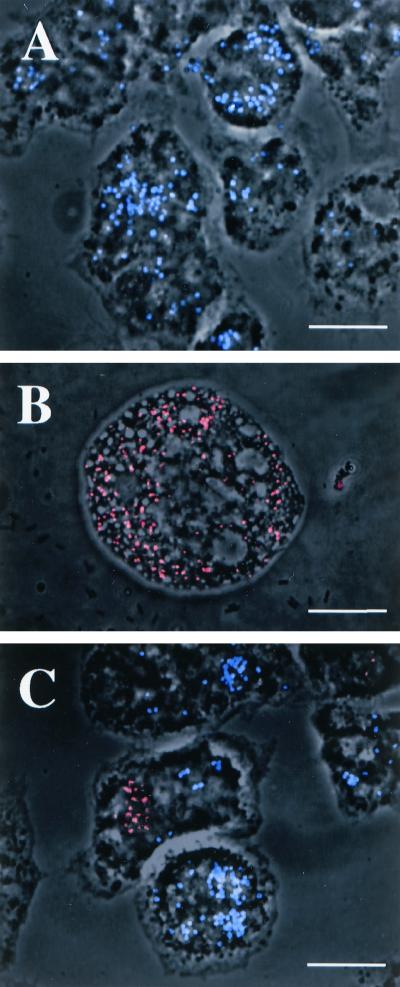
The oligonucleotide probe Bn9658, specific for P. acanthamoebae, fully matched the corresponding sequence in endosymbionts from Acanthamoeba sp. strains UWE1 and UWE25, whereas probe C22658 was designed to recognize the endosymbionts present in Acanthamoeba sp. strains UWC22 and TUME1 (Fig. 3). These probes displayed two mismatches between each other and at least three mismatches to other members of the Chlamydiales and other bacteria.
FIG. 3.
Difference alignment of the 16S rRNA region of intra-amoebal Chlamydiales targeted by probes Bn9658 and C22658 with their corresponding E. coli positions; other bacterial species showing 2 to 4 nucleotide mismatches are also noted. Dots represent nucleotide matches.
The probe Bn9658 gave strong signals when applied to the endosymbiont of Acanthamoeba sp. strain UWE25 and no signal when applied to the endosymbionts of strains UWC22 and TUME1, whereas probe C22658 gave strong signals when applied to the endosymbionts of UWC22 and TUME1 and no signal when applied to the endosymbionts of UWE25 (Fig. 4). Both probes performed optimally at a formamide concentration of 30% when applied separately, determined on the basis of a formamide dilution series (25). Because the two probes act as competitors for the same hybridization site, differentiation of the two groups of endosymbionts in mixed cultures using both probes simultaneously was demonstrated at a formamide concentration of 10% in the hybridization buffer and a hybridization temperature of 46°C. The optimal formamide concentration for the horseradish peroxidase-labeled probe Bn9658 had been described previously as 40% at a hybridization temperature of 35°C (2). Horseradish peroxidase cannot be used at the higher temperature, requiring a higher concentration of formamide to achieve the same hybridization stringency.
FIG. 4.
FISH reactions demonstrating the application of specific oligonucleotide probes to the detection of intracellular Chlamydia-like endosymbionts of Acanthamoeba spp. (A) Positive reaction of the Cy-5 (blue)-labeled probe Bn9658 with the endosymbionts of UWE25 amoebae. (B) Positive reaction of the Cy-3 (red)-labeled probe C22658 with the endosymbionts of UWC22 amoebae. (C) Positive reactions with probes Bn9658 (blue) and C22658 (red) and their abilities to simultaneously differentiate between the UWC22 and UWE25 endosymbionts following a 3-h cocultivation of infected amoebic trophozoites. In some cases bacteria released from one amoebic isolate are seen to have been taken up by the other amoebic isolate during cocultivation. Bars, 15 μm.
DISCUSSION
Phylogenetically, members of the Chlamydiales comprise a unique bacterial assemblage which is highly divergent from other groups within the domain Bacteria in the 16S rDNA-based universal tree. Among the Bacteria, rDNA sequence analysis has revealed their closest relatives to be members of the Planctomycetales, a group of free-living, mostly aquatic bacteria which also display developmental cycles (10).
While little is known about the evolution of traditional Chlamydiales species, the recent description of the Parachlamydiaceae occurring as endosymbionts in protozoa, specifically Acanthamoeba spp., has measurably broadened the possible evolutionary origins of the group (2, 11). Of the six isolates of Parachlamydiaceae described in the literature for which partial or nearly full-length 16S rDNA sequencing has been performed, including the four presented here, sequence dissimilarities suggest the existence of up to four genera and species; two of the isolates studied previously appear to be highly (>99% sequence similarity) related (2, 5). The finding of additional related endosymbionts in these and other protozoa would therefore not be surprising, given the species diversity and geographic range noted to date, and suggests that a large environmental reservoir for them may exist. We conclude that the phylogenetic diversity of the Chlamydiales is extensive but is underrepresented in the literature as a result of bias towards the study of species recognized as being medically and economically important.
Considering that all described species of Chlamydiales are known to be pathogenic and capable of producing respiratory tract disease, among other clinical presentations, analysis of the intra-amoebal forms for pathogenic potential should be stressed as well. Recently, seroepidemiologic evidence has been presented that “Hall's coccus,” an endosymbiont of acanthamoebae which appears to be identical to P. acanthamoebae, may be responsible for some cases of community-acquired pneumonia (5).
Other evidence comes from the amplification of Chlamydiales 16S rDNA fragments (216 to 224 bp) in respiratory tract specimens from humans with pulmonary disease, an aortic specimen, and peripheral blood mononuclear cells using broad-range primers designed to detect any member of the order (29). While sequences characteristic of Chlamydophila pneumoniae (three specimens) and Chlamydia trachomatis (one specimen) were recovered from the 42 respiratory specimens examined, four sequences (two respiratory, one blood, and one aortic tissue) were also detected which group in the second major Chlamydiales lineage, which includes the Simkaniaceae and Parachlamydiaceae. Because S. negevensis (Fig. 2) has been reported to cause both community-acquired pneumonia in adults and acute bronchiolitis in infants (20, 23), further analysis of the Parachlamydiaceae as disease-producing agents should be undertaken.
Acanthamoebae and other free-living protozoa are uniquely positioned ecologically to support the dissemination of environmental respiratory pathogens. Able to colonize water supply, cooling, and humidification systems, they also serve as amplification vehicles and possibly reservoirs for Mycobacterium avium and a variety of Legionella spp.; even C. pneumoniae has been shown to survive and replicate within acanthamoebae (8, 9, 12, 27, 29, 33, 37). Such intraprotozoal multiplication followed by aerosolization and inhalation of bacterium-laden vesicles has been proposed as a mechanism to explain the epidemiology of legionellosis and the apparent lack of case-to-case spread (4, 6). The ubiquity of acanthamoebae in the environment, the presence of a resistant cyst stage, and their ability to support growth of a variety of intracellular pathogens make them prime suspects in the epidemiology of respiratory disease caused by other organisms as well.
Many free-living protozoa mimic the role of professional phagocytes in their abilities to ingest and destroy large numbers of bacteria. Such bacterium-protozoan interactions have undoubtedly provided selective pressure resulting in the emergence of environmental species capable of escaping intracellular destruction, which are known to include Legionella spp., Burkholderia pickettii, Listeria monocytogenes, Vibrio cholerae, and M. avium, among others (3, 7, 24, 26, 37, 38). The adaptation of legionellae and M. avium to an intracellular existence in free-living protozoa may also have been a driving force in the evolution of virulence mechanisms which permit their survival within pulmonary macrophages (3, 22, 37).
While many species of facultative bacteria develop in and ultimately lyse their protozoal host, a variety of obligate endosymbionts of ciliates and amoebae are well known to stably infect their host, suggesting the occurrence of longer-term coevolution (15, 17, 31). The Parachlamydia-like endosymbionts described here also appear to be well adapted to their natural amoebic hosts and have been maintained in continuous cultivation for several years. The presence of these bacteria in amoebic cysts in an apparent nondividing resting stage (Fig. 1D) further supports the stable symbiotic nature of the relationship.
Considering that, until recently, all Chlamydiales were considered to be pathogenic, the finding of a potentially large environmental and geographically dispersed reservoir of related organisms which are adapted to stable intracellular growth in specific hosts provides an important link in the evolution of the order. The existence of a pool of protozoal symbionts stably adapted to the intracellular milieu may have provided the genetic material from which recruitment to vertebrates accidentally occurred, with the attendant appearance of pathogenic properties. Such recruitment would be facilitated by the frequent occurrence of these endosymbionts in Acanthamoeba spp. (and perhaps other protists), the ubiquity of acanthamoebae in the environment, and the ease with which these amoebae transiently colonize the respiratory tract of humans and animals (28).
ACKNOWLEDGMENTS
We are grateful for the assistance of Daniel E. Possin of the University of Washington for preparation of the electron micrographs.
This study was supported in part by Public Health Service grant F06 TW02279-01 to T.R.F. and by Deutsche Forschungsgemeinschaft grant WA 1027/2-1 to M.W. and K.-H.S.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R, Springer N, Schönhuber W, Ludwig W, Schmid E N, Müller K, Michel R. Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl Environ Microbiol. 1977;63:115–121. doi: 10.1128/aem.63.1.115-121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker J, Brown M R W. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology. 1994;140:1253–1259. doi: 10.1099/00221287-140-6-1253. [DOI] [PubMed] [Google Scholar]
- 4.Berk S G, Ting R S, Turner G W, Ashburn R J. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl Environ Microbiol. 1998;64:279–286. doi: 10.1128/aem.64.1.279-286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birtles R J, Rowbotham T J, Storey C, Marrie T J, Raoult D. Chlamydia-like obligate parasite of free-living amoebae. Lancet. 1997;349:925–926. doi: 10.1016/s0140-6736(05)62701-8. [DOI] [PubMed] [Google Scholar]
- 6.Brieland J K, Fantone J C, Remick D G, LeGendre M, McClain M, Engleberg N C. The role of Legionella pneumophila-infected Hartmanella vermiformis as an infectious particle in a murine model of Legionnaires' disease. Infect Immun. 1997;12:5330–5333. doi: 10.1128/iai.65.12.5330-5333.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirillo J D, Falkow S, Tompkins L S. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect Immun. 1994;62:3254–3261. doi: 10.1128/iai.62.8.3254-3261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirillo J D, Falkow S, Tompkins L S, Bermudez L E. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun. 1997;65:3759–3767. doi: 10.1128/iai.65.9.3759-3767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Essig A, Heinemann M, Simnacher U, Marre R. Infection of Acanthamoeba castellanii by Chlamydia pneumoniae. Appl Environ Microbiol. 1997;63:1396–1399. doi: 10.1128/aem.63.4.1396-1399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett K D E, Andersen A A. The ribosomal intergenic spacer and domain I of the 23S rRNA gene are phylogenetic markers for Chlamydia spp. Int J Syst Bacteriol. 1997;47:461–473. doi: 10.1099/00207713-47-2-461. [DOI] [PubMed] [Google Scholar]
- 11.Everett K D E, Bush R M, Andersen A A. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae with descriptions of five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49:415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 12.Fields B S. The molecular ecology of Legionella. Trends Microbiol. 1997;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 13.Fritsche T R, Gautom R K, Seyedirashti S, Bergeron D L, Lindquist T D. Occurrence of bacterial endosymbionts in Acanthamoeba spp. isolated from corneal and environmental specimens and contact lenses. J Clin Microbiol. 1993;31:1122–1126. doi: 10.1128/jcm.31.5.1122-1126.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritsche T R, Horn M, Seyedirashti S, Gautom R K, Schleifer K-H, Wagner M. In situ detection of novel bacterial endosymbionts of Acanthamoeba spp. phylogenetically related to members of the order Rickettsiales. Appl Environ Microbiol. 1999;65:206–212. doi: 10.1128/aem.65.1.206-212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautom R, Fritsche T R. Transmissibility of bacterial endosymbionts between isolates of Acanthamoeba spp. J Eukaryot Microbiol. 1995;42:452–456. doi: 10.1111/j.1550-7408.1995.tb05890.x. [DOI] [PubMed] [Google Scholar]
- 16.Hall J, Voelz H. Bacterial endosymbionts of Acanthamoeba sp. J Parasitol. 1985;71:89–95. [PubMed] [Google Scholar]
- 17.Heckmann K, Görtz H-D. Prokaryotic symbionts of ciliates. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 3865–3890. [Google Scholar]
- 18.Horn M, Fritsche T R, Gautom R K, Schleifer K-H, Wagner M. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus. Environ Microbiol. 1999;1:357–367. doi: 10.1046/j.1462-2920.1999.00045.x. [DOI] [PubMed] [Google Scholar]
- 19.Hugo E R, Gast R J, Byers T J, Stewart V J. Purification of amoeba mtDNA using the UNSET procedure. In: Lee J J, Solldo A T, editors. Protocols in protozoology. Lawrence, Kans: Allen Press; 1992. pp. D7.1–D7.2. [Google Scholar]
- 20.Kahane S, Greenberg D, Friedman M G, Haikin H, Dagan R. High prevalence of “Simkania Z,” a novel Chlamydia-like bacterium, in infants with acute bronchiolitis. J Infect Dis. 1998;177:1425–1429. doi: 10.1086/517830. [DOI] [PubMed] [Google Scholar]
- 21.Kahane S, Metzer E, Friedman M G. Evidence that the novel microorganism “Z” belong to a new genus in the family Chlamydiaceae. FEMS Microbiol Lett. 1995;126:203–208. doi: 10.1111/j.1574-6968.1995.tb07417.x. [DOI] [PubMed] [Google Scholar]
- 22.King C H, Shotts E B, Wooley R E, Porter K G. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl Environ Microbiol. 1988;54:3023–3033. doi: 10.1128/aem.54.12.3023-3033.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman D, Kahane S, Lieberman D, Friedman M G. Pneumonia with serological evidence of acute infection with the Chlamydia-like microorganism “Z.”. Am J Respir Crit Care Med. 1997;156:578–582. doi: 10.1164/ajrccm.156.2.9608081. [DOI] [PubMed] [Google Scholar]
- 24.Ly T M, Muller H E. Ingested Listeria monocytogenes survive and multiply in protozoa. J Med Microbiol. 1990;33:51–54. doi: 10.1099/00222615-33-1-51. [DOI] [PubMed] [Google Scholar]
- 25.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligonucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 26.Michel R, Hauröder B. Isolation of an Acanthamoeba strain with intracellular Burkholderia pickettii infection. Zentbl Bakteriol Mikrobiol Hyg B. 1997;285:541–557. doi: 10.1016/s0934-8840(97)80116-8. [DOI] [PubMed] [Google Scholar]
- 27.Michel R, Müller K-D, Amann R, Schmid E N. Legionella-like slender rods multiplying within a strain of Acanthamoeba sp. isolated from drinking water. Parasitol Res. 1998;60:84–88. doi: 10.1007/s004360050362. [DOI] [PubMed] [Google Scholar]
- 28.Michel R, Rohl R, Schneider H. Isolation of free-living amoebae from nasal mucosa of healthy individuals. Zentralbl Bakteriol Mikrobiol Hyg B. 1982;176:155–159. [PubMed] [Google Scholar]
- 29.Newsome A L, Scott T M, Benson R F, Fields B F. Isolation of an amoeba naturally harboring a distinctive Legionella species. Appl Environ Microbiol. 1998;64:1688–1693. doi: 10.1128/aem.64.5.1688-1693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ossewarde J M, Meijer A. Molecular evidence for the existence of additional members of the order Chlamydiales. Microbiology. 1999;145:411–417. doi: 10.1099/13500872-145-2-411. [DOI] [PubMed] [Google Scholar]
- 31.Preer J R, Preer L B. Endosymbionts of protozoa. In: Krieg N R, editor. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 795–811. [Google Scholar]
- 32.Pudjiatmoko, Fukushi H, Ochial Y, Yamaguchi T, Hirai K. Phylogenetic analysis of the genus Chlamydia based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1997;47:425–431. doi: 10.1099/00207713-47-2-425. [DOI] [PubMed] [Google Scholar]
- 33.Rowbotham T J. Current view on the relationship between amoebae, legionellae, and man. Isr J Med Sci. 1986;22:678–689. [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Springer N, Amann R, Ludwig W. The design and application of ribosomal RNA-targeted, fluorescent oligonucleotide probes for the identification of endosymbionts in protozoa. In: Clapp J P, editor. Methods in molecular biology. vol. 50: species diagnostics protocols: PCR and other nucleic acid methods. Totowa, N.J: Humana Press, Inc.; 1996. pp. 133–144. [DOI] [PubMed] [Google Scholar]
- 36.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 37.Steinert M, Birkness K, White E, Fields B, Quinn F. Mycobacterium avium bacilli grow saprophytically in coculture with Acanthmaoeba polyphaga and survive within cyst walls. Appl Environ Microbiol. 1998;64:2256–2261. doi: 10.1128/aem.64.6.2256-2261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thom S, Warhurst D, Drasar B S. Association of Vibrio cholerae with fresh water amoebae. J Med Microbiol. 1992;36:303–306. doi: 10.1099/00222615-36-5-303. [DOI] [PubMed] [Google Scholar]
- 39.Visvesvara G S. Pathogenic and opportunistic free-living amoebae. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. pp. 1383–1390. [Google Scholar]
- 40.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]