Abstract
Immense volumes of radioactive wastes, which were generated during nuclear weapons production, were disposed of directly in the ground during the Cold War, a period when national security priorities often surmounted concerns over the environment. The bacterium Deinococcus radiodurans is the most radiation-resistant organism known and is currently being engineered for remediation of the toxic metal and organic components of these environmental wastes. Understanding the biotic potential of D. radiodurans and its global physiological integrity in nutritionally restricted radioactive environments is important in development of this organism for in situ bioremediation. We have previously shown that D. radiodurans can grow on rich medium in the presence of continuous radiation (6,000 rads/h) without lethality. In this study we developed a chemically defined minimal medium that can be used to analyze growth of this organism in the presence and in the absence of continuous radiation; whereas cell growth was not affected in the absence of radiation, cells did not grow and were killed in the presence of continuous radiation. Under nutrient-limiting conditions, DNA repair was found to be limited by the metabolic capabilities of D. radiodurans and not by any nutritionally induced defect in genetic repair. The results of our growth studies and analysis of the complete D. radiodurans genomic sequence support the hypothesis that there are several defects in D. radiodurans global metabolic regulation that limit carbon, nitrogen, and DNA metabolism. We identified key nutritional constituents that restore growth of D. radiodurans in nutritionally limiting radioactive environments.
Most of the wastes generated during global nuclear weapons production between 1945 and 1986 were discharged into the ground and are now contaminating the subsurface at thousands of sites (16, 27; http://www.em.doe.gov/bemr96). In 1992, the United States Department of Energy (DOE) surveyed 91 of ∼3,000 contaminated sites at 18 research facilities in the United States and reported that millions of cubic meters of such wastes contain mixtures of radionuclides (e.g., 235U), heavy metals (e.g., Hg and Pb), and toxic organic compounds (e.g., toluene) (27). It is estimated that one-third of the DOE waste sites are radioactive, and radiation levels are as high as 10 mCi/liter (27).
Among the technologies currently being developed (19) for treatment of these environmental wastes are bioremediation strategies in which the extremely radiation-resistant organism Deinococcus radiodurans R1 is used (3). This bacterium has been the subject of whole-genome optical mapping (15) and sequencing (33) and has recently been engineered to express metal-remediating and organic compound-degrading genes (2, 14).
D. radiodurans R1 is a nonpathogenic, desiccation-resistant (22, 23), solvent-tolerant (14) soil bacterium that can survive acute (short) exposure to an ionizing irradiation dose of 1.5 Mrads, a dose that induces 150 to 200 DNA double-strand breaks per chromosome (7, 15). Also, it has been shown that this bacterium can grow in the presence of chronic (continuous) gamma irradiation (14); this is a process that requires simultaneous semiconservative DNA replication and homologous recombination (24). The molecular mechanisms that underlie the extreme radiation resistance phenotype have been the subject of several investigations (8–11, 17, 23). However, the role of the metabolic repertoire and physiological state of D. radiodurans at the time of irradiation has been characterized far less. Little is known about this relationship other than that (i) exponentially growing D. radiodurans cells are more sensitive to radiation than stationary-phase cells are (22, 31), (ii) increasing the concentration of Mn2+ in cells decreases genomic redundancy along with radiation resistance (6), and (iii) freezing or desiccating D. radiodurans substantially increases its radiation resistance (7, 18, 26).
In radioactive environments that do not kill an organism but rather limit or prevent metabolism, genetic damage accumulates, and survival depends on repairing and preventing the accumulation of irreversible (lethal) genetic damage (7–10). Genetic recovery of D. radiodurans after such DNA damage occurs is heavily dependent on energy metabolism and protein synthesis (22), and we have recently demonstrated the remarkable ability of D. radiodurans to grow in the presence of 6,000 rads/h under nutrient-rich conditions with no effect on its viability, growth rate, or ability to express cloned genes (14). By comparison, Escherichia coli is very quickly killed in such environments (14).
The nutrient conditions at DOE radioactive waste sites are poor, and the effect of such nutrient conditions on the growth and survival of D. radiodurans was not known previously. Using a defined synthetic minimal medium, we examined the effect of nutrient conditions on the ability of D. radiodurans to survive acute or chronic exposure to radiation. We found that while the metabolic state of cells had little effect on cell survival following treatment with acute ionizing radiation, nutrient conditions had a profound effect on the survival and growth of D. radiodurans during chronic exposure to irradiation. Under nutrient-limiting conditions during chronic irradiation, DNA repair was found to be limited by the metabolic capabilities of the organism and not by any nutritionally induced defect in genetic repair. The results of our analyses support the hypothesis that global RNA synthesis is suppressed in D. radiodurans grown in nutrient-poor environments and the hypothesis that there are several defects in its metabolic pathways.
MATERIALS AND METHODS
Growth of cells.
D. radiodurans R1 was grown on a nutrient-rich medium, TGY (1% Bacto Tryptone [Difco], 0.5% yeast extract, 0.1% glucose), or on minimal medium (Table 1) in the absence or in the presence of chronic irradiation at a rate of 6,000 rads/h (137Cs Gammacell 40 irradiation unit; Atomic Energy of Canada Limited) at 22°C, as described previously (14). To facilitate growth, liquid minimal medium was inoculated with cells (104 to 105 cells/ml) that had been pregrown on solid minimal medium. All chemicals were obtained from Sigma Chemical Co.; Bacto Agar and Noble agar were obtained from Difco. In liquid cultures, cell density was determined at 600 nm by using a Beckman spectrophotometer. For acute high-level exposure to radiation, stationary-phase cultures were irradiated without a change of broth on ice at a rate of 1.33 Mrads/h (model 109 60Co Gammacell irradiation unit; J. L. Shepard and Associates). Following irradiation, cell viability was determined by performing a plate assay as described previously (7).
TABLE 1.
Minimal nutrient requirements for growth of D. radiodurans in the absence and in the presence of gamma irradiation (6,000 rads/h)a
| Class | Component | Concn necessary for growth
|
|
|---|---|---|---|
| (I) Without radiation | (II) With radiation | ||
| Buffer | Potassium phosphate buffer (pH 7.5 to 8.0) | 20 mM | 20 mM |
| Salts | Magnesium chloride tetrahydrate | 0.2 mM | 0.2 mM |
| Calcium chloride dihydrate | 0.1 mM | 0.1 mM | |
| Manganese(II) acetate tetrahydrate | 5.0 μM | 5.0 μM | |
| Ammonium molybdate tetrahydrate | 5.0 μM | 5.0 μM | |
| Ferrous sulfate heptahydrate | 5.0 μM | 5.0 μM | |
| Amino acids | l-Histidine | 25 μg/ml | 200 μg/ml |
| l-Cysteine | 30 μg/ml | 30 μg/ml | |
| l-Glutamine | 500 μg/ml | ||
| l-Alanine | 500 μg/ml | ||
| l-Arginine | 800 μg/ml | ||
| l-Asparagine | 800 μg/ml | ||
| Glycine | 300 μg/ml | ||
| l-Leucine | 500 μg/ml | ||
| l-Lysine | 300 μg/ml | ||
| l-Methionine | 100 μg/ml | ||
| l-Proline | 370 μg/ml | ||
| l-Serine | 300 μg/ml | ||
| l-Threonine | 200 μg/ml | ||
| l-Tryptophan | 200 μg/ml | ||
| l-Tyrosine | 200 μg/ml | ||
| l-Valine | 200 μg/ml | ||
| Vitaminsb | Nicotinic acid | 1.0 μg/ml | 1.0 μg/ml |
| Carbon source | 2 mg/ml | 2 mg/ml | |
Basal salt medium was autoclaved and then supplemented with sterile preparations of salts, amino acids, and nicotinic acid at the concentrations indicated. For solid medium, 1.5% (wt/vol) Noble agar was added before basal salt medium was autoclaved.
Replacing nicotinic acid with Basal Medium Eagle Vitamin Solution (Gibco BRL) improved growth slightly. For growth in liquid minimal medium, the cells used for inoculation were pregrown on solid minimal medium.
rel Gene function assay.
rel function was assayed by using the procedures described by Cashel (4). All eubacteria that have been tested so far are capable of forming guanine nucleotide analogs of GDP and GTP that have a pyrophosphate group esterified to the 3′-hydroxyl of the ribose moiety; these analogs are designated ppGpp and pppGpp, respectively. The RNA control locus relA encodes (p)ppGpp synthetase and sometimes forms a hybrid locus containing spoT, which encodes 3′-pyrophosphohydrolase (e.g., in Bacillus subtilis [32]). RelA is synthesized in bacteria in response to amino acid starvation and indirectly reduces protein synthesis by repressing stable RNA synthesis when the concentrations of amino acids cannot keep up with the demand during protein biosynthesis. RelA-induced RNA suppression has pleiotropic effects, including reductions in DNA replication, transcription, translation, and growth. Together with SpoT, RelA participates in integrating carbon metabolism and nitrogen metabolism (5). RelA activity in D. radiodurans cells was determined as follows. Cells were grown on solid TGY for 60 h or on solid minimal (fructose) medium (Table 1) for 170 h. Approximately 108 cells were suspended in phosphate-free labeling medium (PFLM) (0.1 M MOPS [morpholinepropanesulfonic acid], 0.2% dextrose, 100 μCi of carrier-free [32P]orthophosphoric acid per ml) that contained or did not contain 1 mg of serine hydroxamate per ml (serine hydroxamate blocks translation and is used to induce relA-spoT activity). The authenticities of (p)ppGpp and pppGpp spots were established by using Escherichia coli wild-type strain CF1648 and a mutant E. coli strain (CF1652) lacking relA (20) as controls. A cell suspension (25 μl) was mixed with an equal volume of 13 M formic acid, placed on dry ice, and freeze-thawed twice. A 4-μl portion of the mixture was then spotted onto a cellulose-polyethyleneimine thin-layer chromatography (TLC) plate (Fisher Scientific), and the TLC plate was developed with 1.5 M KH2PO4 (pH 3.4). When the solvent front reached 15 cm, the plate was air dried and exposed overnight to X-ray film.
CSLM.
Bacterial cells were harvested, washed with 0.1 M Tris-HCl–0.01 M EDTA buffer (pH 8.0), fixed in 77% ethanol (0°C), and stained with acridine orange. The stained preparations were visualized with a Bio-Rad model MRC-600 confocal scanning laser microscope (CSLM) interfaced with a Zeiss Axiovert microscope, as well as a Merdian model ULTIMA ACAS 570 CSLM; ×100 immersion objectives were used. Images were reproduced by using a New Codonics model NP1600 Postscript printer. Acridine orange-stained double-stranded nucleic acid forms a complex that has an absorption maximum at wavelengths between 450 and 490 nm; this complex fluoresces green and is used to localize DNA with a 520-nm barrier filter. An acridine orange-stained single-stranded nucleic acid complex has an absorption maximum at wavelengths between 510 and 560 nm; this complex fluoresces red and is used to localize RNA with a 590-nm barrier filter (12).
Nucleic acid manipulation.
Total nucleic acid and DNA were prepared and electrophoresis was performed as described previously (7–10, 29).
RESULTS
Development of a minimal medium suitable for analysis of D. radiodurans growth.
In the only previous description of a D. radiodurans minimal medium that was used to assess growth, Shapiro et al. described a synthetic medium that contained very high concentrations (30 to 600 μg/ml each) of 17 amino acids, which resulted in a medium containing more than 5 mg of amino acids per ml (28). In addition, this minimal medium included a large variety of minerals and vitamins that we have shown to be unnecessary for D. radiodurans growth. The excessive concentrations of nonessential nutrients made this medium neither minimal nor useful for our growth studies. In fact, D. radiodurans can grow in this medium in the absence of typical Embden-Meyerhof-Parnas (EMP) pathway substrates (e.g., fructose, glucose, and maltose). We, therefore, tried to develop a synthetic medium that is truly minimal, highly characterized, and suitable for testing the metabolic capabilities of D. radiodurans, as guided by our analysis of the genomic sequence.
To develop a synthetic minimal medium, we systematically tested many combinations of different amounts of carbohydrates, amino acids, salts, and vitamins in both liquid media and solid media (prepared with Noble agar). By a process of elimination, we identified minimal nutrient constituents and the concentrations of these nutrients necessary for luxuriant growth (Table 1). The synthetic medium which we developed for D. radiodurans is distinct from the media described by other workers (25, 28) in that it is much simpler and growth of D. radiodurans in the medium is completely dependent on a nonamino carbon source (e.g., fructose) (Fig. 1). In addition to a metabolizable carbon source, growth of D. radiodurans is dependent on exogenous amino acids and a vitamin; sulfur-rich amino acids together with nicotinic acid were particularly effective at supporting growth. However, we found that there was not a specific amino acid combination that was necessary since many different combinations of amino acids supported growth. A factor that strongly influenced the extent of growth was the total amino acid concentration in the growth medium (Fig. 2), not the composition of the amino acid pool. We found that carbon sources supported luxuriant to slow growth in the following order: fructose > pyruvate > lactate > glucose > oxaloacetate > acetate > glycerol (Fig. 1). There are numerous examples of free-living bacteria that exhibit absolute specificity for sugar metabolism (e.g., Arthrobacter strains can utilize fructose but not glucose [30]). Surprisingly, the tricarboxylic acid cycle intermediates fumarate, citrate, malate, and succinate did not support growth (Fig. 1).
FIG. 1.
Growth of D. radiodurans in liquid minimal media (Table 1, composition I) containing different carbon sources at a concentration of 2 mg/ml. An optical density at 600 nm (OD600) of 1.0 was equivalent to ∼1 × 108 CFU/ml. Cells were pregrown on solid minimal (fructose) medium before they were inoculated into liquid minimal medium.
FIG. 2.
Relationship between amino acid concentration and growth of D. radiodurans in liquid minimal medium. The nutrient conditions were the conditions described in Table 1 except for the amino acid composition. Fructose was the carbon source. The amino acid composition was as follows: glutamine, 25%; cysteine, 18%; and a mixture containing tyrosine, tryptophan, and phenylalanine and buffered with 5% glycine, 10%. Cultures were inoculated with 5 × 106 CFU/ml by using cells that were pregrown on solid minimal (fructose) medium (Table 1). Optical densities at 600 nm (OD600) were determined 96 h after inoculation.
Sensitivity of D. radiodurans grown in synthetic medium to chronic irradiation and acute irradiation.
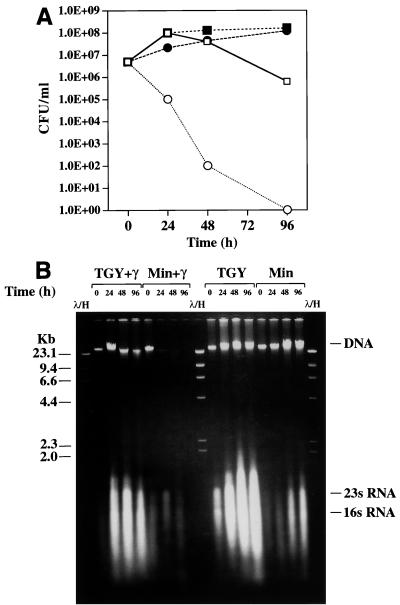
To investigate the effect of the nutritional state of D. radiodurans on the extreme radiation phenotype of this organism, cells were exposed to continuous gamma irradiation in a 137Cs irradiator (6,000 rads/h) under different growth conditions. Control cultures were incubated in the absence of irradiation at the same temperature. When cells were grown on rich medium (TGY), growth was not affected by continuous exposure to 6,000 rads/h compared to growth on TGY in the absence of irradiation. By contrast, growth on the synthetic medium (Table 1, composition I) was eliminated by chronic exposure to 6,000 rads/h. To determine if chronic irradiation under minimal conditions was bactericidal or bacteriostatic, a series of inoculated minimal medium plates were exposed to 0.1, 0.2, 0.3, 0.4, or 0.5 Mrad in the irradiator. Following exposure, the plates were incubated in the absence of irradiation in order to monitor survival. We found that a dose of 0.3 Mrad was lethal to cells; this contrasts with the ability of D. radiodurans to survive 1.7 Mrads of acute irradiation without lethality if cells are allowed to grow and recover in rich medium (7–10). We also examined cell viability and the DNA repair capabilities of chronically irradiated D. radiodurans cells incubated in either liquid TGY or minimal medium (Fig. 3). Following incubation in the 137Cs irradiator, cells were collected at intervals for up to 96 h. Some of the cells were plated to examine survival (Fig. 3A), and the remainder were frozen until total DNA was prepared and examined to determine whether degradation occurred (Fig. 3B). Rapid degradation of DNA occurred in cells incubated in minimal medium, whereas there was little evidence of DNA degradation in cells incubated in TGY. Some DNA degradation and a loss of viability in cells incubated in TGY were observed with the 48- and 96-h samples, and we believe that this was due to depletion of metabolizable nutrients and the inevitable accumulation of DNA damage in slowly replicating or nonreplicating cells. In contrast, the viability of cells that were incubated in minimal medium and irradiated decreased very substantially almost immediately. After 24 h of irradiation (144,000 rads) (Fig. 3B), the DNA was highly degraded, and all of the cells were dead by 96 h (576,000 rads) (Fig. 3).
FIG. 3.
Effect of nutrient conditions on the viability and DNA content of D. radiodurans exposed to chronic gamma irradiation in liquid culture. Cells were irradiated at a rate of 6,000 rads/h (144,000 rads/day) at 23°C. Both irradiated and control cultures were diluted to a concentration 5 × 106 CFU/ml at the start of the experiment. (A) Survival curves. Symbols: ■, control, TGY, no irradiation; □, TGY, gamma irradiation; ●, control, minimal (fructose) medium (Table 1, composition I), no irradiation; ○, minimal (fructose) medium (Table 1, composition I), gamma irradiation. (B) Total DNA was prepared from cells obtained at each of the time points shown in panel A. Each lane contained DNA from ∼3 × 106 cells, as determined by hemocytometer counting (7). TGY+γ, cells that were grown in TGY and received gamma irradiation; Min+γ, cells that were grown in minimal (fructose) medium (Table 1, composition I) and received gamma irradiation; TGY and Min, controls incubated in the absence of irradiation. Lanes λ/H contained lambda phage DNA cut with HindIII. DNA sizes (in kilobases) are indicated on the left. The gel migration positions of DNA and rRNA are indicated on the right. Gel electrophoresis was performed with a 0.66% agarose gel for 17 h at 45 V.
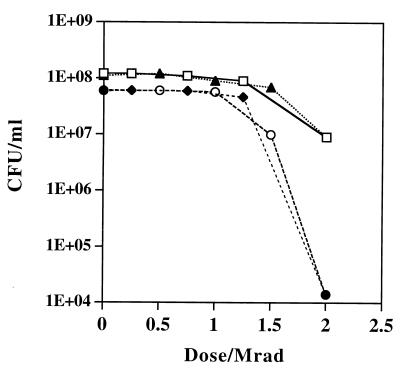
To rule out the possibility that cells grown in minimal medium (Table 1, composition I) were inherently sensitive to irradiation and the possibility that certain nutrition-dependent repair factors were not present in our synthetic medium, TGY- and minimal medium-grown D. radiodurans cells were tested to determine their ability to survive during extremely high doses of acute gamma irradiation. Figure 4 shows that irrespective of the preirradiation growth medium or the postirradiation recovery substrate, the levels of resistance of D. radiodurans were very similar up to a dose of 1.2 Mrads; at doses greater than 1.2 Mrads, minimal medium-grown cells exhibited enhanced sensitivity.
FIG. 4.
Effect of growth substrate and recovery substrate on survival of D. radiodurans following acute gamma irradiation. Cells were grown to the early stationary phase and irradiated on ice at a rate of 1.33 Mrads/h. Symbols: □, cells pregrown in liquid TGY, irradiated, and plated onto solid TGY; ▴, cells pregrown in liquid TGY, irradiated, and plated onto solid minimal (fructose) medium (Table 1, composition I); ○, cells pregrown in liquid minimal (fructose) medium, irradiated, and plated onto solid TGY; ⧫, cells pregrown in liquid minimal (fructose) medium, irradiated, and plated onto solid minimal (fructose) medium.
The minimal nutrient conditions required to support growth at a dose of 6,000 rads/h were determined by increasing the concentrations of the nutrients shown in Table 1. When cells were exposed to continuous irradiation, growth was restored only when high concentrations of amino acids were provided together with an EMP or Entner-Doudoroff carbon substrate (Table 1 and Fig. 1). Increasing the concentration of the carbon source or the concentrations of the non-amino acid supplements to values greater than the values shown in Table 1 had no effect on resistance. Similarly, in the absence of a carbon source, high concentrations of amino acids alone did not support growth in the irradiator. Table 1 (composition II) shows the minimal medium nutrients and the concentrations of these nutrients that supported luxuriant D. radiodurans growth at a dose of 6,000 rads/h.
Physiologic genomic analysis of D. radiodurans.
In the course of our annotation of the D. radiodurans genome (33), we found that most of the metabolic pathway genes that were key to our analysis were present. For example, the EMP and Entner-Doudoroff pathways were found to be intact, and D. radiodurans could grow on fructose, glucose, maltose, and mannose, as expected. However, we found three examples in which the primary biosynthetic pathways of amino acids were incomplete (Table 2). Furthermore, we found that D. radiodurans cannot utilize ammonia as a nitrogen source and that growth of this organism is entirely dependent on exogenous amino acids. We, therefore, carefully examined the genomic sequence for defects that could affect nitrogen assimilation. Generally, the key step in assimilating inorganic nitrogen into amino acids is the synthesis of glutamine and glutamate from ammonia catalyzed by glutamine synthetase (glnA). In D. radiodurans R1 there are two copies of glnA; glnA-1 (chromosomal position, 2049790) is disrupted by a frameshift mutation, while glnA-2 (chromosomal position, 447280) appears to be intact. The glutamate synthase subunit genes (gltB and gltD) also appear to be functional (chromosomal position of operon, 181526); GltB/D integrates carbon metabolism and nitrogen metabolism by synthesizing glutamine from glutamate. Nevertheless, we found that strain R1 could not use 2-oxoglutarate as a growth substrate in minimal medium supplemented with a variety of inorganic nitrogen sources (e.g., ammonium sulfate), suggesting that there may be a defect in assimilation of ammonia in the glutamine synthetase-glutamate synthase cycle.
TABLE 2.
Effectiveness of carbon sources as precursors for D. radiodurans growtha
| Carbon and energy source | Growthb | Biosynthetic pathwayc | Genome datad | Defect |
|---|---|---|---|---|
| Pyruvate | +++ | Alanine | Complete | |
| Valine | Complete | |||
| Leucine | Complete | |||
| Oxaloactate | ++ | Isoleucine | Complete | |
| Threonine | Complete | |||
| Lysine | Incomplete | dapABDF absent | ||
| Methionine | Complete | |||
| Aspartic acid | Complete | |||
| Phosphoglycerate | + | Serine | Incomplete | serCB absent |
| Glycine | Complete | |||
| Cysteine | Incomplete | cysEJDN absent |
Acetate and lactate are effective growth substrates, while the tricarboxylic acid cycle intermediates malate, succinate, fumarate, and citrate, are ineffective.
+++, excellent growth; ++, good growth; +, poor growth.
The pathways are the entry points for biosynthesis for the amino acids indicated from the carbon and energy sources listed.
For genomic comparisons, we used metabolic pathways present in E. coli. All of the genes necessary for histidine biosynthesis were present.
Correlation between amino acid-limited growth and relA activity.
RelA and SpoT are responsible for integrating carbon metabolism and nitrogen metabolism (5), and when regulation of these gene products is defective, the genes can have pleiotropic effects on cells that can include dependence on exogenous amino acids for growth and suppression of cellular RNA levels (5). We, therefore, examined rel function in D. radiodurans at both a genomic informatic level and an experimental level.
We found that D. radiodurans rel encodes a predicted protein that is most similar (58% identity) (1) to the RelA/SpoT protein [(p)ppGpp synthetase/3′-pyrophosphohydrolase] of Bacillus subtilis (32). We tested the functional integrity of the putative rel locus in D. radiodurans by monitoring the synthesis of ppGpp and pppGpp under amino acid deprivation conditions (Fig. 5A). Cells were shifted from TGY or minimal medium to labeled phosphate medium (PFLM) containing a single carbon and energy source with or without serine hydroxamate (Fig. 5B and C). Under these conditions, ppGpp and pppGpp were rapidly synthesized and expressed at levels comparable to the levels observed in E. coli (Fig. 5). These data support the hypothesis that rel function is normal in D. radiodurans and is induced under amino-acid-limiting conditions.
FIG. 5.
(A) Production of pp(p)Gpp in D. radiodurans and E. coli. 32P-labeled ppGpp and ppGpp were detected by polyethyleneimine-cellulose chromatography (see Materials and Methods). Stationary-phase cells grown in either TGY or minimal medium (Table 1) were suspended in PFLM (see Materials and Methods) with or without the amino acid analogue serine hydroxamate. Lanes 1 and 2, D. radiodurans cells obtained from minimal (fructose) medium (Table 1, composition I) and incubated in PFLM containing serine hydroxamate (equivalent to lanes 4 and 5 in panel B); lanes 3 and 4, D. radiodurans cells obtained from minimal (fructose) medium (Table 1) and incubated in PFLM containing the 16 amino acids listed in Table 1 (final amino acid concentration, 50 μg/ml); lane 5, D. radiodurans cells obtained from TGY and incubated in PFLM containing serine hydroxamate; lane 6, E. coli (relA deleted) incubated in PFLM containing serine hydroxamate (control); lane 7, E. coli wild type incubated in PFLM containing serine hydroxamate (control). (B) Formation of ppGpp and pppGpp in D. radiodurans. Cells were treated as described above in the presence of serine hydroxamate. Lane 1, control, no cells; lanes 2 and 3, cells grown in TGY; lanes 4 and 5, cells grown in minimal (fructose) medium (Table 1). (C) Formation of ppGpp and pppGpp in D. radiodurans grown in minimal medium. Cells were treated as described above. Lane 1, cells treated in the absence of serine hydroxamate; lane 2, cells treated in the presence of serine hydroxamate.
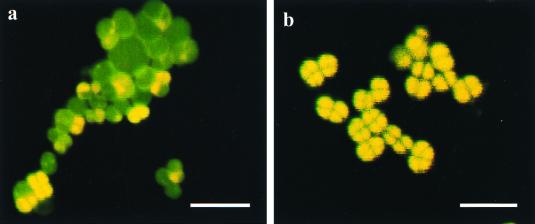
Consistent with induction of rel functions, we found that cells grown in minimal medium (Table 1, composition I) contained substantially reduced levels of cellular RNA, as determined by gel electrophoresis of the total nucleic acid (Fig. 3B). The finding that the cellular RNA content was decreased was verified directly by examining cells with a CSLM after acridine orange staining (Fig. 6). Differential staining of DNA and RNA showed that cells grown in minimal medium (Table 1, composition I) contained much lower levels of RNA than cells grown in nutrient-rich medium (TGY).
FIG. 6.
CSLM. Bacterial cells were stained with acridine orange. Acridine orange-stained single-stranded nucleic acids result in complexes that fluoresce red and were used to localize RNA. Acridine orange-stained double-stranded nucleic acid complexes fluoresce green and were used to localize DNA. Note that when DNA and RNA were both present, the cells were yellow. (a) Minimal (fructose) medium (Table 1, composition I). (b) TGY. Bars = 5 μm.
DISCUSSION
The prevailing nutrient conditions at DOE radioactive waste sites are very poor (19). We, therefore, developed a synthetic medium for D. radiodurans that is suitable for testing the relationship among the metabolic capabilities of this organism, the ambient nutrient conditions, and the radiation resistance of the organism. D. radiodurans growth is not affected by chronic exposure to 6,000 rads/h under nutrient-rich conditions (14). However, we found that cells are not able to grow and are killed in radioactive nutritionally restricted environments in which luxuriant growth occurs in the absence of irradiation. This phenotypic reversal from radiation resistance to radiation sensitivity is of great interest and concern since it brings into question the suitability of D. radiodurans as a bioremediation host at radioactive waste sites. Our data show that the resistance of cells grown in minimal medium to acute gamma irradiation (0 to 1.2 Mrads) is similar to the resistance of cells grown under nutrient-rich conditions (in TGY) (Fig. 4) and that the principal factor that limits survival and growth under chronically irradiated conditions (Fig. 3) is the ability of cells to utilize certain nutrients. In our study we identified key nutritional constituents that restore growth of D. radiodurans in radioactive nutrient-limiting environments (Table 1, composition II).
The results described above reinforce our previous assertion that DNA repair requires nutrient-rich conditions. In previous studies of D. radiodurans cells exposed to acute irradiation (0.5 to 2.0 Mrads), DNA repair required fresh TGY (7). It has been reported that in the absence of such conditions double-strand-break-induced DNA degradation is relentless and leads to cell death (7, 22). When DNA degradation and repair are considered in the context of D. radiodurans that is also burdened with de novo synthesis of cell components on minimal medium, the added metabolic demands of repairing 4 to 10 copies (13) of the genome (3.28 Mbp) during chronic irradiation are probably profound. Indeed, we found that degradation of DNA in chronically irradiated cells incubated in minimal medium is rapid and progressive (Fig. 3B). In the context of our growth experiments performed with the irradiator, therefore, we speculate that in minimal medium (Table 1, composition I) the rate of DNA repair and synthesis is overwhelmed by the rate of DNA degradation.
For growth under chronic irradiation conditions, phenotypic reversion from radiation sensitivity (in minimal medium) (Table 1, composition I) to radiation resistance could be induced by enriching the growth substrate. Specifically, we found that restoration of radiation resistance during growth was highly dependent on an exogenous abundant amino acid source (Table 1, composition II). Our genomic analysis showed that D. radiodurans contains most amino acid-biosynthetic pathways (Table 2). However, the primary biosynthetic pathways for cysteine, lysine, and serine are incomplete (Table 2); nevertheless, D. radiodurans can grow in the absence of these pathways, which supports the hypothesis that there are secondary biosynthetic pathways for these three amino acids in D. radiodurans. Generally, transamination reactions facilitate interconversion of many amino acids, as well as de novo synthesis from tricarboxylic acid cycle intermediates. Since D. radiodurans is not able to assimilate inorganic nitrogen, the existence of such transamination abilities is consistent with our finding that while D. radiodurans growth in minimal medium does not depend on specific amino acids, it does depend on a nonspecific exogenous amino acid source.
In eubacteria, global regulation of various metabolic processes, including transcription and protein synthesis, is known to be under stringent control of the relA and spoT loci (21); under amino-acid-limiting conditions relA is induced. It is known that in eubacteria relA activity exhibits global suppression of RNA synthesis and induction of amino acid synthesis (20). In D. radiodurans, in which three major biosynthetic pathways are disrupted, the relA function may be easily triggered. We, therefore, examined RNA suppression in D. radiodurans since the presence of such suppression not only would support relA function but also could help explain the inhibition of D. radiodurans growth under nutrient-limiting conditions during chronic irradiation. Growth of D. radiodurans under amino-acid-limiting conditions resulted in rel-associated functions; pp(p)Gpp- was induced (Fig. 5), and the cellular RNA content decreased substantially (Fig. 3B and 6). In most types of cells, about 80% of the total cellular RNA is rRNA (12). Thus, the rRNA content is a good indicator of the total cellular RNA content and the translational potential of a cell. Global suppression of RNA synthesis in D. radiodurans would probably inhibit metabolism, and this could explain the inability of cells exposed to radiation in minimal medium to survive and grow. If this occurs, these cells are not able to generate the levels of precursors required for cell division, as well as DNA repair, and, consequently, the cells become overwhelmed by accumulated genetic damage. However, there is no direct evidence that correlates relA function and radiation sensitivity in D. radiodurans grown in minimal medium.
Our analysis of the minimal nutrient requirements for growth at a dose of 6,000 rads/h showed that in addition to a carbon source that is effectively metabolized by D. radiodurans, cells need to have a rich source of amino acids. In order to bioremediate radioactive DOE waste sites with genetically engineered D. radiodurans designed for growth on aromatic compounds (15), biostimulation with amino acids will probably be necessary. While further genetic engineering could correct defects in D. radiodurans amino acid-biosynthetic pathways, the concept of including a rich source of amino acids in bioremediating protocols is reasonable given the abundance of inexpensive, amino acid-rich by-products of the dairy industry.
ACKNOWLEDGMENTS
This research was funded largely by grant FG07-97ER20293 from the Environmental Science Management Program, Office of Biological and Environmental Research, DOE. Some of this work was also supported by grants FG02-97ER62492 and DE-FG02-98ER62583 from the DOE and by grant 5R01-GM39933-09 from the National Institutes of Health.
We thank Michael Cashel at the National Institutes of Health for his participation and for advice concerning the pp(p)Gpp synthesis analysis.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brim H, McFarlan S C, Fredrickson J K, Minton K W, Zhai M, Wackett L P, Daly M J. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nature Biotechnol. 2000;18:85–90. doi: 10.1038/71986. [DOI] [PubMed] [Google Scholar]
- 3.Brooks B W, Murray R G E, Johnson J L, Stackebrandt E, Woese C R, Fox G E. Red-pigmented micro cocci: a basis for taxonomy. Int J Sys Bacteriol. 1980;30:627–646. [Google Scholar]
- 4.Cashel M. Detection of (p)ppGpp accumulation patterns in Escherichia coli mutants. Methods Mol Genet. 1994;3:341–356. [Google Scholar]
- 5.Cashel M, Rudd K E. The stringent response in Escherichia coli and Salmonella typhimurium. In: Neidhardt F C, et al., editors. Cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. pp. 1410–1438. [Google Scholar]
- 6.Chou F I, Tan S T. Manganese(II) induces cell division and increases in superoxide dismutase and catalase activities in an aging deinococcal culture. J Bacteriol. 1990;172:2029–2035. doi: 10.1128/jb.172.4.2029-2035.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daly M J, Ouyang L, Fuchs P, Minton K W. In vivo damage and recA-dependent repair of plasmid and chromosomal DNA in the radiation-resistant bacterium Deinococcus radiodurans. J Bacteriol. 1994;176:3508–3517. doi: 10.1128/jb.176.12.3508-3517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly M J, Ouyang L, Minton K W. Interplasmidic recombination following irradiation of the radioresistant bacterium Deinococcus radiodurans. J Bacteriol. 1994;176:7506–7515. doi: 10.1128/jb.176.24.7506-7515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly M J, Minton K W. Interchromosomal recombination in the extremely radioresistant bacterium Deinococcus radiodurans. J Bacteriol. 1995;177:5495–5505. doi: 10.1128/jb.177.19.5495-5505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly M J, Minton K W. An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J Bacteriol. 1996;178:4461–4471. doi: 10.1128/jb.178.15.4461-4471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dardalhon-Samsonoff M, Averbeck D. DNA-membrane complex restoration in Micrococcus radiodurans after X-irradiation: relation to repair, DNA synthesis and DNA degradation. Int J Radiat Biol. 1980;38:31–52. doi: 10.1080/09553008014550931. [DOI] [PubMed] [Google Scholar]
- 12.Darzynkiewicz Z. Simultaneous analysis of cellular RNA and DNA content. Methods Cell Biol. 1994;41:401–420. doi: 10.1016/s0091-679x(08)61731-8. [DOI] [PubMed] [Google Scholar]
- 13.Hansen M T. Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J Bacteriol. 1978;134:71–75. doi: 10.1128/jb.134.1.71-75.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange C C, Wackett L P, Minton K W, Daly M J. Engineering a recombinant Deinococcus radiodurans for organopollutant degradation in radioactive mixed waste environments. Nature Biotechnol. 1998;16:929–933. doi: 10.1038/nbt1098-929. [DOI] [PubMed] [Google Scholar]
- 15.Lin J, Qi R, Aston C, Jing J, Anantharaman T S, Mishra B, White O, Daly M J, Minton K W, Venter J C, Schwartz D C. Whole genome shotgun optical mapping of Deinococcus radiodurans using genomic DNA molecules. Science. 1999;285:1558–1561. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 16.Macilwain C. Science seeks weapons clean-up role. Nature. 1996;383:375–379. [Google Scholar]
- 17.Mattimore V, Udupa K S, Berne G A, Battista J R. Genetic characterization of forty ionizing radiation-sensitive strains of Deinococcus radiodurans: linkage information from transformation. J Bacteriol. 1995;177:5232–5237. doi: 10.1128/jb.177.18.5232-5237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattimore V, Battista J R. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol. 1996;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCullough J, Hazen T C, Benson S M, Blaine-Metting F, Palmisano A C. Bioremediation of metals and radionuclides. U.S. Germantown, Md: Department of Energy Office of Biological and Environmental Research; 1999. [Google Scholar]
- 20.Mechold U, Cashel M, Steiner K, Gentry D, Malke H. Functional analysis of relA/spoT gene homolog from Streptococcus equisimilis. J Bacteriol. 1996;178:1401–1411. doi: 10.1128/jb.178.5.1401-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mechold U, Malke H. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J Bacteriol. 1997;179:2658–2667. doi: 10.1128/jb.179.8.2658-2667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minton K W. DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol Microbiol. 1994;13:9–15. doi: 10.1111/j.1365-2958.1994.tb00397.x. [DOI] [PubMed] [Google Scholar]
- 23.Minton K W. Repair of ionizing-radiation damage in the radiation resistant bacterium Deinococcus radiodurans. Mutat Res DNA Repair. 1996;363:1–7. doi: 10.1016/0921-8777(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 24.Minton K W, Daly M J. A model for repair of radiation-induced DNA DSB's in the extreme radiophile Deinococcus radiodurans. Bioessays. 1995;17:457–464. doi: 10.1002/bies.950170514. [DOI] [PubMed] [Google Scholar]
- 25.Raj H D, Duryee F L, Deeney A M, Wang C H, Anderson A W, Elliker P R. Utilization of carbohydrates and amino acids by Micrococcus radiodurans. Can J Microbiol. 1960;6:289–298. doi: 10.1139/m60-033. [DOI] [PubMed] [Google Scholar]
- 26.Richmond R C, Sridhar R, Daly M J. Physicochemical survival pattern for the radiophile Deinococcus radiodurans: a polyextremophile model for life on Mars. SPIE. 1999;3755:210–222. [Google Scholar]
- 27.Riley R G, Zachara J M, Wobber F J. Chemical contaminants on DOE lands and selection of contaminant mixtures for subsurface science research. U.S. Washington, D.C.: Department of Energy Office of Energy Research Subsurface Science Program; 1992. [Google Scholar]
- 28.Shapiro A, DiLello D, Loudis M C, Keller D E, Hutner S H. Minimal requirements in defined media for improved growth of some radioresistant pink tetracocci. Appl Environ Microbiol. 1977;33:1129–1133. doi: 10.1128/aem.33.5.1129-1133.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith M D, Lennon E, McNeil L B, Minton K W. Duplication insertion of drug resistance determinants in the radioresistant bacterium Deinococcus radiodurans. J Bacteriol. 1988;170:2126–2135. doi: 10.1128/jb.170.5.2126-2135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobel M E, Kruwlwich K. Metabolism of d-fructose by Arthrobacter pyridinolis. J Bacteriol. 1973;113:907–913. doi: 10.1128/jb.113.2.907-913.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thornley M J. Radiation resistance among bacteria. J Appl Bacterial. 1963;26:334–345. [Google Scholar]
- 32.Wendrich T M, Marahiel M A. Cloning and characterization of a relA/SpoT homologue from Bacillus subtilis. Mol Microbiol. 1977;26:65–79. doi: 10.1046/j.1365-2958.1997.5511919.x. [DOI] [PubMed] [Google Scholar]
- 33.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, Moffet K S, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan J J, Lam P, McDonald L, Utterback T, Zalewski C, Makarova K S, Aravind L, Daly M J, Minton K W, Fleishmann R D, Ketchum K A, Nelson K E, Salzberg S, Venter J C, Fraser C M. Complete genome sequencing of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]