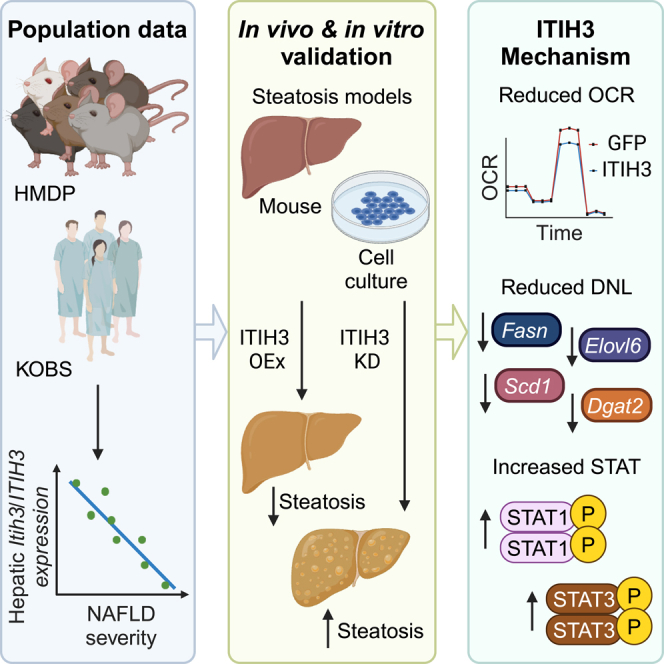
Summary
Recent studies demonstrate that liver secretory proteins, also known as hepatokines, regulate normal development, obesity, and simple steatosis to non-alcoholic steatohepatitis (NASH) progression. Using a panel of ∼100 diverse inbred strains of mice and a cohort of bariatric surgery patients, we found that one such hepatokine, inter-trypsin inhibitor heavy chain 3 (ITIH3), was progressively lower in severe non-alcoholic fatty liver disease (NAFLD) disease states highlighting an inverse relationship between Itih3/ITIH3 expression and NAFLD severity. Follow-up animal and cell culture models demonstrated that hepatic ITIH3 overexpression lowered liver triglyceride and lipid droplet accumulation, respectively. Conversely, ITIH3 knockdown in mice increased the liver triglyceride in two independent NAFLD models. Mechanistically, ITIH3 reduced mitochondrial respiration and this, in turn, reduced liver triglycerides, via downregulated de novo lipogenesis. This was accompanied by increased STAT1 signaling and Stat3 expression, both of which are known to protect against NAFLD/NASH. Our findings indicate hepatokine ITIH3 as a potential biomarker and/or treatment for NAFLD.
Subject areas: Biochemistry, Biological sciences, Cell biology, Physiology
Graphical abstract
Highlights
-
•
In both mice and humans, hepatokine ITIH3 is inversely related to NAFLD severity
-
•
ITIH3 overexpression rescues both mice and hepatocytes from steatosis
-
•
ITIH3 reduced mitochondrial respiration and downregulated de novo lipogenesis
-
•
ITIH3 increased STAT1 signaling and Stat3 expression
Biochemistry; Biological sciences; Cell biology; Physiology
Introduction
The most common cause of chronic liver disease on a global scale is non-alcoholic fatty liver disease (NAFLD).1,2,3,4,5,6 NAFLD is an umbrella term ranging from simple steatosis (fat accumulation in hepatocytes) to complex non-alcoholic steatohepatitis (NASH), fibrosis and cirrhosis, eventually leading to hepatocellular carcinoma (HCC).1,2,3 An important feature of NAFLD is their differential prevalence and disease phenotypes between males and females.2,7,8,9,10,11,12 Given that obesity and insulin resistance are strongly associated with NAFLD,2,6,13,14 the prevalence of NAFLD is rising concurrently with the obesity epidemic, creating a significant current and future healthcare challenge. Despite this, factors governing the progression of the disease or prospective drug targets are yet unknown. To this end, we used an integrative multiomics approach using a well-characterized mouse population, the hybrid mouse diversity panel (HMDP),15 and identified a liver secreted protein, ITIH3 as a potential candidate protein involved in NAFLD pathogenesis.
Secretory proteins, cytokines and hormones have paracrine or endocrine functions on neighboring cells or other tissues, maintaining systemic nutrient and energy homeostasis. Hence, the abundance and structure of these chemical messengers change in response to the normal/disease conditions. Several proteins circulating in the blood are synthesized by the liver and are altered during the different stages of liver pathologies. Recent works demonstrated that genes encoding secretory proteins are abundantly expressed in livers of people with type 2 diabetes.16 Furthermore, genes encoding fibrinogenic factors, angiogenic factors, and redox-associated factors are reported to regulate the pathophysiology of type 2 diabetes.17,18 Therefore, the metabolic disturbance in the liver often regulates the whole-body energy metabolism. Indeed, several liver-derived secretory proteins or hepatokines affect the metabolism of peripheral organs.19,20,21
Inter-α-trypsin inhibitors (ITI) are plasma protease inhibitors that are assembled from light and heavy chain precursor proteins. ITI genes are transcribed in the liver as light chain and heavy chain polypeptides.22 A single light chain with a combination of different heavy chains, such as ITIH1, ITIH2, ITIH3, ITIH4, and ITIH5, forms a mixture of complex proteins. So far, the difference in their rearrangements results in different functional polypeptides namely, urinary trypsin inhibitor (UTI), inter-α-trypsin inhibitors (ITI), pre-α-inhibitor (PαI), or inter-α-trypsin inhibitor family heavy chain related protein (IHRP).23 ITI proteins act as hyaluronic acid binding proteins,24 anti-inflammatory agents,25 extracellular matrix stabilizers23 and inhibitors of tumor cell invasion.26 These liver secreted protease inhibitors play important roles during normal development, tissue remodeling,27,28 cancer,29 insulin resistance,30 and HCC.31 Previous studies have demonstrated the importance of hepatokines in hepatic steatosis, but there is not much progress made toward the identification and/or functional importance of ITI proteins in hepatic steatosis or NAFLD. In addition, comparative analysis of mouse liver and plasma proteome revealed a strong correlation between both in pathology.32 Taken together, it is imperative to further our understanding on hepatokines in the context of liver pathology to diagnose, treat and prevent hepatic diseases.
Our current focus is to investigate the role of hepatokine ITIH3 on NAFLD pathogenesis. To this end, we used both mouse and human populations and show that ITIH3 is strongly and negatively associated with the disease progression suggesting a protective role against NAFLD/NASH development. Using both loss- and gain-of-function strategies in mouse and cell culture experiments, we further confirmed that ITIH3 negatively regulates liver lipid accumulation. Furthermore, we identified ITIH3 to protect against NAFLD by regulating both mitochondrial metabolism and de novo lipogenesis (DNL).
Results
ITIH3 is a potential candidate gene for protection against NAFLD
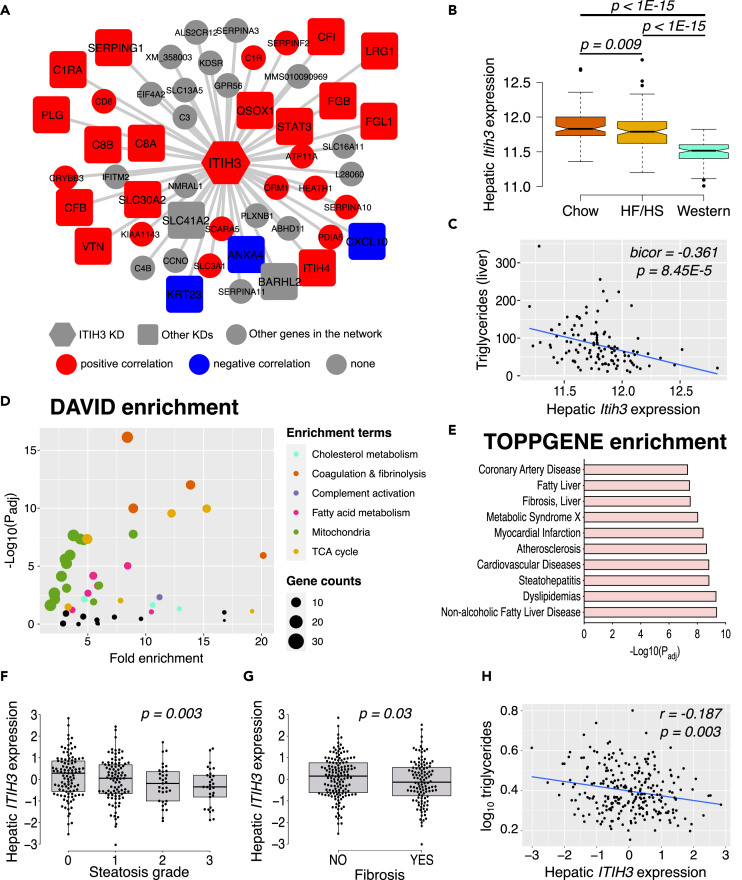
We had earlier used an integrative multiomics approach using transcriptomic data from approximately 100 different mouse strains called HMDP and identified several “key driver” genes underlying hepatic TG accumulation.15 Using similar approaches, here we show that ITIH3 is a potential “key driver” gene, that is strongly correlated with other inflammatory and coagulation factors (Figure 1A). Particularly, we found STAT3, a transcription factor widely known to ameliorate steatosis and liver fibrosis33,34,35 and LRG1, a secreted hepatokine that was reported to inhibit M1 polarization of hepatic macrophages to alleviate NASH,36 to be positively correlated; whereas CXCL10, a pro-inflammatory cytokine reported to increase steatosis and NASH37 and KRT23, a reported biomarker for NASH and progression to HCC,38 to be negatively correlated with hepatic Itih3 expression. Additionally, we found another ITI family gene namely, ITIH4, to be positively correlated with hepatic Itih3 expression. Follow-up investigations on livers isolated from HMDP strains maintained on chow (healthy), high fat-high sucrose HF/HS (steatosis)39 and western (NASH/fibrosis)40 diets revealed that as NAFLD progresses, liver Itih3 expression significantly decreases (Figure 1B). Focusing on the HMDP strains maintained on HF/HS diet revealed us that liver Itih3 expression was negatively correlated with liver triglyceride (TG) levels (Figure 1C). Furthermore, immunoblot analyses of C57BL/6J mice revealed that ITIH3 protein levels in the liver decreased with diet-induced NAFLD/NASH progression (Figure S1). Taken together, we reasoned that loss in hepatic Itih3 expression worsens the NAFLD conditions.
Figure 1.
ITIH3 is negatively associated with NAFLD/NASH in both mice and humans
(A) Overlay of HMDP bicorrelation directionality on ITIH3 key driver (KD) network from our previous study.15 ITIH3 is represented in hexagon shape, other key driver genes are represented in square shapes, and the rest of the network genes are represented in circle shapes. Red represents positive and blue represents negative correlation with liver Itih3 expression from HMDP strains maintained on HF/HS diet (n = 113 HMDP strains).
(B) Liver Itih3 expression from HMDP strains maintained on chow (n = 96 HMDP strains) or HF/HS (n = 113 HMDP strains) or western (n = 102 HMDP strains) diet.
(C) Correlation plot between hepatic Itih3 expression and triglyceride in HMDP strains maintained on HF/HS diet (n = 113 HMDP strains).
(D) DAVID pathway and (E) ToppGene disease enrichment analyses of highly correlated HMDP liver genes (listed in Table S1) with liver Itih3 expression (bicor > |±0.3|; p < 1E-05). Hepatic ITIH3 expression from KOBS cohort (n = 262) grouped by (F) steatosis grade and (G) Fibrosis.
(H) Correlation plot between hepatic ITIH3 expression and triglycerides. Data are presented as median and interquartile range (boxplots). p values were calculated by (A and C) bicor; (B) one-factor ANOVA corrected by post-hoc “Holm-Sidak’s” multiple comparisons test; (F and G) ANCOVA corrected for age, BMI, and sex; (H) partial correlation adjusting for age, BMI, and sex. HMDP, hybrid mouse diversity panel; bicor, biweight midcorrelation; BMI, body mass index.
To further explore the gene networks and pathways associated with hepatic Itih3 expression, we computed Itih3 correlated hepatic genes (listed in Table S1) from HMDP strains maintained on HF/HS diet (bicor > |±0.3|; p < 1E-05). Follow up enrichment analysis by DAVID41 and ToppGene42 results in identification of NAFLD-related metabolic pathways such as cholesterol metabolism, mitochondria, fatty acid metabolism and TCA cycle (Figure 1D); and NAFLD-related disease terms such as fatty liver, liver cirrhosis, steatohepatitis, dyslipidemia, and NAFLD (Figure 1E). Finally, to understand the clinical relevance of hepatic ITIH3 expression in human NAFLD/NASH, we used the RNA-seq data from a total of 262 liver biopsies from the Kuopio Obesity Surgery (KOBS) cohort. We found a negative correlation between hepatic ITIH3 expression and NAFLD/NASH-related phenotypes such as steatosis grade (Figure 1F), fibrosis stage (Figure 1G; Figure S2) and triglyceride levels (Figure 1H). Taken together, since our population studies in both mice and human revealed that hepatic Itih3/ITIH3 expression was negatively correlated with NAFLD/NASH phenotypes and associated with mitochondria and metabolism gene networks, we hypothesized that hepatic ITIH3 protects against NAFLD and were further interested to investigate the ITIH3’s role in NAFLD.
ITIH3 attenuates hepatic steatosis both in vivo and in vitro
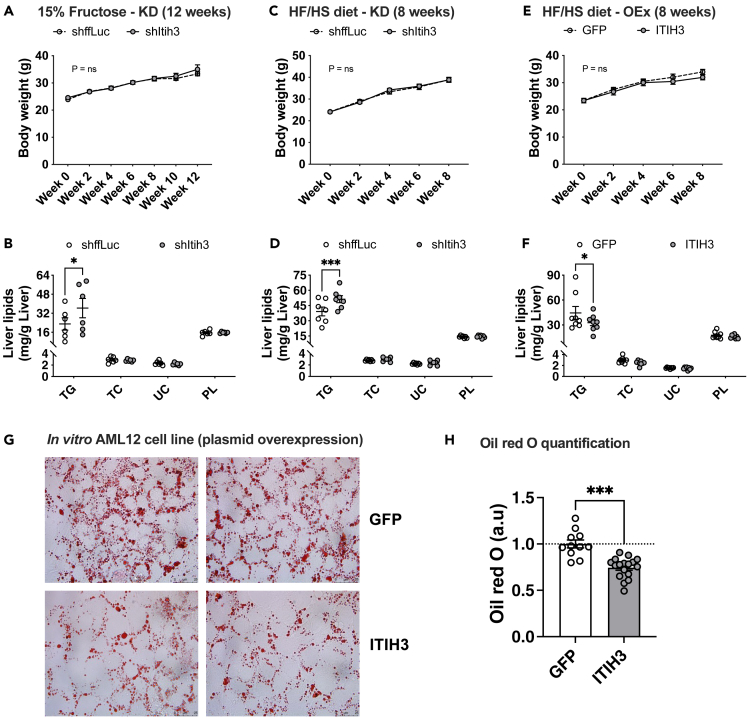
To functionally validate the protective role of ITIH3 against liver steatosis, we performed fructose water or HF/HS diet induced steatosis models in 8-week-old male C57BL/6J mice using both silencing and/or overexpression strategies via AAV8 vectors. For ITIH3 knockdown studies, liver specific AAV vectors harboring shRNA against ffLuc (control) or Itih3 was used; for overexpression studies, liver specific AAV vectors harboring cDNAs of GFP (control) or ITIH3 was used. The validity of these strategies is shown in Figure S3. All these animals were subjected to either 15% fructose water for 12 weeks or HF/HS diet for 8 weeks. We noted that both ITIH3 knockdown and overexpression did not alter the body weights in both steatosis models (Figures 2A, 2C, and 2E). We next analyzed the hepatic lipid profiles in the presence or absence of ITIH3. We noticed that knocking down ITIH3 significantly increased the intrahepatic triglyceride content in both steatosis models (Figures 2B and 2D) without altering total cholesterol (TC), unesterified cholesterol (UC), and phospholipids (PL). To further confirm this, we observed a significant reduction in the liver TG content with no changes in TC, UC, and PL in ITIH3 overexpression mice (Figure 2F). Furthermore, H&E staining demonstrated that ITIH3 overexpression significantly lowered liver lipid droplet accumulation (Figure S4). However, we did not observe any significant differences with liver weights, plasma TG or TC levels (Figure S5). We also found no significant differences in fasting plasma glucose, insulin, or homeostatic model assessment for insulin resistance (HOMA-IR) (Figure S6).
Figure 2.
ITIH3 lowers liver triglyceride accumulation both in vivo and in vitro
Comparisons of body weight measurements and hepatic lipid levels from (A and B) 15% fructose in drinking water or (C and D) HF/HS fed ITIH3 silencing (KD) mice or (E and F) HF/HS fed ITIH3 overexpressing (OEx) mice, respectively. Oil Red O staining of AML12 cells overexpressing GFP or ITIH3 with (G) representative images and (H) quantification. Data are presented as mean ± SEM (n = 6–8 mice per group; n = 2 independent experiments for AML12 cells). p values were calculated by (A, C, and E) repeated measures two-factor ANOVA; (B, D, and F) two-factor ANOVA corrected by post-hoc “Holm-Sidak’s” multiple comparisons test; (H) t test. ∗p < 0.05; ∗∗∗p < 0.001. TG, triglyceride; TC, total cholesterol; UC, unesterified cholesterol; PL, phospholipids.
We next explored ITIH3’s role in in vitro cell culture models by overexpressing Itih3 plasmid in oleic acid treated AML12 liver cells. Oil red O staining was performed to analyze the lipid content. We report that ITIH3 overexpression results in reduction of total lipid droplet content in comparison with GFP overexpression as measured by Oil red O staining (Figures 2G and 2H). We did not perform any in vitro knockdown studies as AML12 cells inherently had very low ITIH3 expression. Taken together, we conclude that ITIH3 confers hepatoprotection against steatosis both in vivo and in vitro.
ITIH3 protects against steatosis by lowering mitochondrial respiration
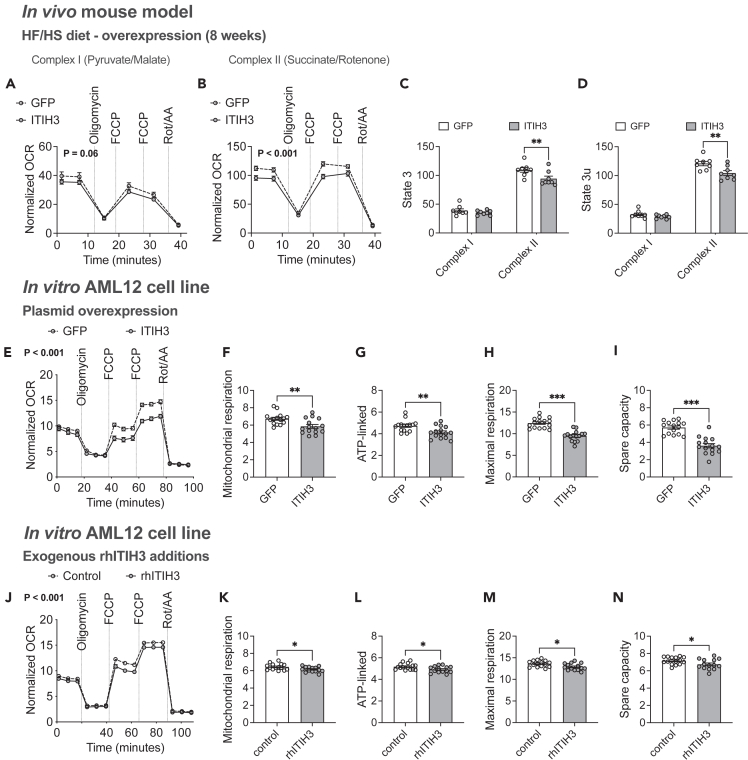
Several studies have demonstrated that mitochondrial respiration is elevated during hepatic steatosis due to high substrate availability and ATP demand resulting in hyperactive OXPHOS phenotypes.15,43 Chronic hyperactive OXPHOS phenotypes are maladaptive leading to oxidative damages. Therefore, lowering mitochondrial respiration is the key to rescue from steatosis as we have demonstrated earlier.15,44 Given that our DAVID gene enrichment analyses revealed hepatic Itih3 expression to be associated with mitochondria and TCA cycle (Figure 1D), we measured mitochondrial respiration capacities in both in vivo and in vitro conditions using XF Pro Seahorse bioanalyzer as described in the STAR methods section. First, we isolated liver mitochondria from HF/HS fed animals overexpressing either GFP or ITIH3. We observed that ITIH3 overexpression significantly reduced the mitochondrial respiration profile especially in the presence of complex II substrates (Figures 3A and 3B). Also, we noted that ITIH3 overexpression reduced complex II mediated mitochondrial respiration at both State 3 (ADP-stimulated) and State 3u (FCCP-stimulated) respiration (Figures 3C and 3D).
Figure 3.
ITIH3 lowers mitochondrial respiration both in vivo and in vitro
Respirometry traces of isolated liver mitochondria from GFP or ITIH3 overexpressing mice offered (A) pyruvate with malate (Complex I) or (B) succinate with rotenone (Complex II) and their respective (C) State 3 and (D) State 3u mitochondrial respiration. Respirometry traces of intact AML12 cells (E) overexpressing GFP or ITIH3, or (J) exogenously treated with control or rhITIH3 and their respective (F and K) mitochondrial (datapoint 15 subtracted from 3), (G and L) ATP-linked (datapoint 6 subtracted from 3), (H and M) maximal respiration (datapoint 15 subtracted from 12) and (I and N) spare capacity (datapoint 3 subtracted from 12), respectively. Data are presented as mean ± SEM (n = 8 mice per group; n = 14–15 replicates per group for AML12 cells). p values were calculated by (A, B, E and J) repeated measures two-factor ANOVA; (C and D) multiple t tests corrected by post-hoc ‘Holm-Sidak’s’ multiple comparisons test; (F–I and K–N) t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To further address the role of ITIH3 in vitro, AML12 liver cells overexpressing GFP or ITIH3 for 48 h were used to measure cellular respiration using XF Pro Seahorse bioanalyzer. We observed that ITIH3 overexpression significantly reduced the cellular respiration profile (Figure 3E). Furthermore, individual measurements revealed that ITIH3 overexpression reduced mitochondrial, ATP-linked, maximal, and spare capacity related respiration (Figures 3F–3I). Since, ITIH3 is a secreted hepatokine, we next treated the AML12 liver cells with recombinant human ITIH3 protein (rhITIH3) for 24 h and repeated our cellular bioenergetic experiments to corroborate these results. As expected, we observed significantly reduced cellular respiration profile and corresponding individual measurements only in the rhITIH3 treated cells (Figures 3J–3N). Altogether, we conclude that ITIH3 confers hepatoprotection against steatosis by lowering mitochondrial respiration.
ITIH3 protects against steatosis by downregulating mitochondria and de novo lipogenesis genes while upregulating Stat3 expression and STAT1 signaling
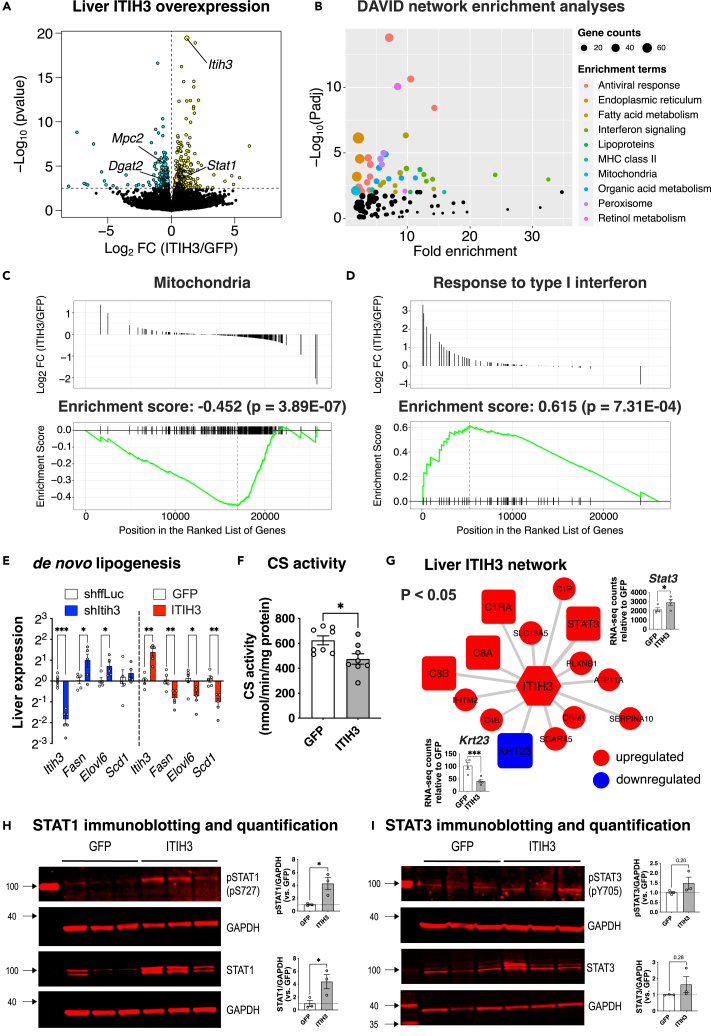
To further explore the mechanistic roles of ITIH3 in the observed metabolic alterations, we performed whole genome RNA sequencing on the extracted liver tissues from ITIH3 overexpression groups. We found 480 differentially expressed genes (DEGs) in ITIH3 overexpression animals (Figure 4A; Table S2). Key NAFLD/NASH genes that were significantly changed in ITIH3 overexpression groups include Dgat2, Mpc2, and Stat1. Notably, both DGAT245,46,47 and MPC248,49,50 are considered as promising drug targets against NAFLD/NASH and were both downregulated in ITIH3 overexpression mice. On the other hand, STAT1, which plays an essential role in response to interferons, was reported to inhibit both liver mitochondrial biogenesis51 and liver fibrosis52,53 and was found to be upregulated in ITIH3 overexpression. Follow-up network enrichment analyses of the 480 DEGs by DAVID41 revealed NAFLD-related metabolic pathways such as mitochondria, fatty acid metabolism, organic acid metabolism, and retinol metabolism (Figure 4B). Interestingly, interferon signaling pathways were also enriched. Furthermore, gene set enrichment analysis (GSEA) of ranked DEGs identified mitochondria as a downregulated gene set (Enrichment score: −0.452, p = 3.89E-07) and response to Type I interferon as an upregulated gene set (Enrichment score: 0.615, p = 7.31E-04) (Figures 4C and 4D).
Figure 4.
ITIH3 protects against steatosis via downregulating mitochondria and de novo lipogenesis while upregulating STAT1 signaling
(A) Global genome-wide transcriptomics of livers extracted from GFP or ITIH3 overexpressing mice. Turquoise and yellow represent down-regulated and up-regulated genes, respectively.
(B) DAVID pathway enrichment analyses of 480 DEGs (listed in Table S2). Gene set enrichment analysis (GSEA) of ranked DEGs revealed a significant enrichment of (C) Mitochondria and (D) Response to type I interferon. Follow-up (E) qPCR analyses of Itih3 and liver DNL genes such as Fasn, Elovl6, and Scd1 in ITIH3 knockdown and overexpression mice and their respective controls and (F) citrate synthase (CS) activity measured in GFP or ITIH3 overexpression mice.
(G) Overlay of DEGs on ITIH3 network from Figure 1A. Blue represents genes going down and red represents genes going up in ITIH3 overexpressing mice (p < 0.05). Inset shows RNA counts of Stat3 and Krt23 in ITIH3 overexpressing mice. Follow-up immunoblot analyses and their respective quantification of liver proteins such as (H) pSTAT1 and STAT1, (I) pSTAT3 and STAT3 in GFP or ITIH3 overexpressing mice. GAPDH was used as a loading control. Data are presented as mean ± SEM (n = 4 mice for transcriptomics, n = 5–6 mice for qPCR, n = 8 for CS activity and n = 3 mice for immunoblot analyses per group). p values were calculated by (A and G) Wald test; (E) multiple t tests corrected by post-hoc “Benjamini, Krieger, Yekutieli” FDR approach for multiple comparisons test; (F, H, and I) t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
It has been proposed that hepatic DNL is elevated in NAFLD, which is one of the central reasons for accumulation of hepatic lipids.54 Furthermore, since our bioenergetic analyses revealed that ITIH3 negatively affects mitochondrial respiration capacity (Figure 3), we reasoned that reduced mitochondrial capacity implies reduced TCA cycle flux and substrate (citrate) generation for DNL thus lowering it. To test this, we first investigated the expression of DNL genes in our in vivo liver samples. We observed that DNL-related genes such as Fasn, Elovl6, and Scd1 were significantly elevated in ITIH3 silencing compared to shffLuc control animals (Figure 4E). In contrast, ITIH3 overexpression lowered the DNL-related genes compared to GFP control animals (Figure 4E). Furthermore, we observed that ITIH3 overexpression lowered TCA cycle flux as measured by citrate synthase (CS) activity (Figure 4F). Overall, this further highlights the importance of ITIH3 in conferring hepatoprotection against steatosis via lowering DNL. Next, when we reanalyzed the ITIH3 network (Figure 1A), we found that 13 genes were positively regulated including Stat3 and complement factors and one gene (Krt23), a reported biomarker for NASH and HCC progression,38 was negatively regulated in ITIH3 overexpression mice (Figure 4G). Finally, western blot analyses demonstrated that ITIH3 overexpression increased both activated STAT1 (phosphorylated STAT1, pSTAT1) and total STAT1 protein levels in the liver (Figure 4H). We also found an increased trend of both activated STAT3 (phosphorylated STAT3, pSTAT3) and total STAT3 liver protein levels in ITIH3 overexpression mice (Figure 4I). Further investigation into whether ITIH3 regulated STAT3 via IL-6/JAK pathway revealed us that ITIH3 overexpression had no effect on the levels of Il1b, Il6, Socs3, Jak1 RNA, or JAK1 protein (Figure S7). Overall, ITIH3 overexpression studies demonstrated that hepatic ITIH3 negatively regulates DNL gene expression and mitochondria, while positively regulating Stat3 expression and STAT1 signaling.
Discussion
In this study, we demonstrate that hepatokine ITIH3 inversely regulates NAFLD pathology by providing multiple evidence ranging from population studies to in vitro and in vivo studies. Population studies using ∼100 HMDP strains maintained on different diet conditions and 262 human liver biopsies revealed that hepatic Itih3 expression was inversely associated with NAFLD progression and hepatic TG accumulation. ITIH3 loss- or gain-of-function studies in both animal and cell culture models revealed inverse relationship between ITIH3 and hepatic lipid content, mitochondrial respiration, and DNL. Altogether, our results reveal a hepatoprotective role of ITIH3 on NAFLD that is distinct from its known functions.
Inter-tissue crosstalk by endocrine factors or secretory proteins is conserved throughout evolution and is mediated by almost all organisms. As a result, many secretory proteins have been identified. Recent works utilizing “omics” technologies have enabled the identification of systemic regulators of whole-body homeostasis.55,56 Previously, cell or tissue specific secretome have been studied by using various methodological approaches such as proteomic analysis of cell culture medium or by examining the transcriptome data for the presence of signal peptides.57,58 Most importantly, inter-tissue communication was also addressed by analyzing plasma proteomics followed by candidate biomarker identification.59,60 A recent work from Seldin et al. report the importance of novel endocrine factors responsible for inter-tissue communication.61 Most notably, several secretory proteins based pharmacological agents such as GLP-1 receptor antagonists, FGF-19/21 analogs, and MOTS-c analogs are at different stages of clinical trials for NAFLD/NASH treatments.62 Here, we report a hitherto unknown role of a hepatokine ITIH3 in NAFLD protection.
Liver is the central secretory organ of the human body; it secretes majority of the blood proteins that have profound impact in the systemic homeostasis. During liver abnormalities, liver enzymes in the circulation are routinely measured to diagnose the liver diseases. Remarkably, plasma protein profiling of type 2 diabetes, NAFLD and NASH cohort patients resulted in the identification of proteins regulating blood coagulation or fibrinolysis factors, carrier proteins that play an important role in cholesterol metabolism and proteins involved in immune system regulation and inflammation.63 Several studies have also shown that hepatokines regulate inflammation, insulin resistance, type 2 diabetes, obesity, steatosis, NAFLD, and cancer.16,19,21,64,65 Thus, it was imperative to investigate the role of hepatokines that directly contribute to NAFLD. To this end, our integrative multiomics approach using ∼100 HMDP strains identified the hepatokine ITIH3 as a potential NAFLD candidate gene that was strongly associated with several complement and coagulation factors. Indeed, our DAVID and ToppGene enrichment analyses prioritized NAFLD related gene networks and pathways such as fatty acid metabolism, mitochondria, TCA cycle, dyslipidemia, metabolic syndrome, fatty liver, steatohepatitis, fibrosis, complement activation, coagulation, and fibrinolysis pathways with respect to hepatic Itih3 expression. Finally, our follow-up in vitro and in vivo validation studies verified the hepatoprotective role of ITIH3 against steatosis. All these data demonstrate that the hepatokine ITIH3 could be employed as a biomarker or potential treatment for severe NAFLD/NASH pathologies. Nevertheless, future investigations involving comparative proteome analyses of liver and plasma are warranted to determine what extrinsic vs. intrinsic metabolic changes are mediated by ITIH3.
Several studies have shown that mitochondrial dysfunction is causal in developing NAFLD and its progression.15,66 Despite the importance of hepatokines in NAFLD progression, little is known on the role of these proteins in mitochondrial function. Given that our DAVID enrichment prioritized the involvement of fatty acid metabolism, mitochondria, and TCA cycle with respect to ITIH3 and to further explore the mechanistic connections between ITIH3 and NAFLD, we focused on ITIH3’s role in mitochondrial function. During the initial stages of NAFLD (hepatic steatosis) there is an acute increase in the mitochondrial functions to combat the excessive nutrient availability and ATP demand resulting in a hyperactive OXPHOS phenotype.15,43 Over time, this will result in elevated ROS generation which may lead to reduced mitochondrial function, thus progressing to severe stages of NAFLD, such as NASH and fibrosis.43,66,67,68 Hyperactive OXPHOS will also lead to elevated TCA cycle flux resulting in augmented DNL thereby causing hepatic steatosis.69 Indeed, we had earlier shown that lowering mitochondrial function will reduce DNL and hepatic steatosis.15,44 In this study, both our in vitro and in vivo experiments confirmed that ITIH3 reduced the mitochondrial functions that were elevated during steatosis. Our findings thus emphasize ITIH3’s role as a regulator of mitochondrial function.
Several factors are known to contribute to the intrahepatic lipid accumulation during hepatic steatosis such as elevated hepatic free fatty acid uptake coming from the diet or adipose tissue lipolysis, augmented DNL, reduced fatty acid oxidation and reduced VLDL secretion. Nevertheless, a large number of studies have demonstrated the upregulation and the causal effects of DNL genes such as Fasn, Elovl6, and Scd1 during NAFLD.70,71,72 Furthermore, both animal and human studies have demonstrated that DNL is a distinguishing feature of NAFLD.54,73,74,75,76 Indeed, our in vivo studies demonstrated that ITIH3 knockdown increased DNL genes such as Fasn, Elovl6, and Scd1, while ITIH3 overexpression downregulated them. Similarly, ITIH3 overexpression reduced TCA cycle flux. Thus, our results corroborate with previous literature as well as highlight the importance of ITIH3 as a negative regulator of liver DNL.
Finally, we wanted to gain a clear mechanistic understanding of how the hepatokine ITIH3 exerts its hepatoprotective functions. Our network modeling analyses gave us a clue in which we observed that STAT3 was directly connected to ITIH3 and positively correlated with hepatic Itih3 expression. Accumulating evidence suggested that phosphorylated STAT3 translocate into the nucleus to activate the transcription of anti-steatosis and anti-fibrosis related genes.33,34,35,77 Additionally, we observed strong positive correlation between expression of ITIH3 and STAT3 in our human NAFLD cohort (bicor = 4.2, p = 7.1E-13). We therefore speculated that ITIH3 works via STAT3 to ameliorate NAFLD. Indeed, our global gene expression analyses revealed us that liver Stat3 expression was high in ITIH3 overexpressing mice but immunoblot analyses revealed only an increased trend. Surprisingly, we also observed that ITIH3 overexpression significantly upregulated liver STAT1 at both the RNA and protein levels, as well as activation (pSTAT1-pS727). Indeed, STAT1 has been validated by others to inhibit both liver mitochondrial biogenesis via negatively regulating Pgc1α51 and liver fibrosis via negatively regulating TGF-β signaling.52,53 Based on these findings, we suspect that ITIH3 functions through STAT1 and possibly STAT3 signaling, although further studies are required to confirm this.
In conclusion, our study demonstrated that the hepatokine ITIH3 plays a unique role in controlling hepatic lipid profile, mitochondrial metabolism, and DNL, all of which have implications for NAFLD treatment. We have also identified two potential pathways, STAT1 and STAT3, which may contribute to the observed hepatoprotection in ITIH3 overexpression.
Limitations of the study
Although our research addresses a potential mechanism of action for ITIH3 regarding mitochondrial metabolism and DNL via STAT1 and/or STAT3, the exact signaling sequences warrant additional investigation. Furthermore, although our hypothesis was formulated using mouse and human population, all validation studies were performed using mice or murine cell lines. Finally, our current study does not investigate the potential role of ITIH3 in mitigating hepatic steatosis in female sex.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit monoclonal ITIH3 | SinoBiological | Cat# 16138-R049 |
| rabbit monoclonal STAT1 | Cell Signaling | Cat# 14994; RRID: AB_2737027 |
| rabbit monoclonal Phospho-STAT1 (Ser727) | Cell Signaling | Cat# 8826; RRID: AB_2773718 |
| rabbit polyclonal STAT3 | Proteintech | Cat# 10253-2-AP; RRID: AB_2302876 |
| rabbit monoclonal Phospho-STAT3 (Tyr705) | Cell Signaling | Cat# 9145; RRID: AB_2491009 |
| rabbit monoclonal JAK1 | Cell Signaling | Cat# 3344; RRID: AB_2265054 |
| mouse monoclonal ACTIN | abcam | Cat# ab8226; RRID: AB_306371 |
| rabbit monoclonal GAPDH | Cell Signaling | Cat# 5174; RRID: AB_10622025 |
| Bacterial and virus strains | ||
| AAV8.TBG.PI.mItih3.rBG | Penn Vector Core facility | N/A |
| AAV8.TBG.PI.eGFP.rBG | Penn Vector Core facility | N/A |
| AAV8.TBG.PI.shltih3.rBG | Penn Vector Core facility | N/A |
| AAV8.TBG.PI.shFFluc.rBG | Penn Vector Core facility | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Human ITIH3 | SinoBiological | Cat# 16138-H08H |
| QIAzol Lysis Reagent | Qiagen | Cat# 79306 |
| Chloroform, HPLC grade | Thermo Fisher | Cat# C606-4 |
| Isopropanol, 99.5%, for molecular biology | Acros Organics | Cat# 32727-0010 |
| Oleic Acid-Albumin from bovine serum | Sigma-Aldrich | Cat# O3008-5ML |
| High fat/high sucrose (HF/HS) diet | Research Diets | Cat# D12266B |
| Critical commercial assays | ||
| miRNeasy Mini Kit | Qiagen | Cat# 217004 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher | Cat# 4368813 |
| PowerUp SYBR Green Master Mix | Thermo Fisher | Cat# A25778 |
| Serum Triglyceride Determination Kit | Sigma-Aldrich | Cat# TR0100-1KT |
| Cholesterol Quantitation Kit | Sigma-Aldrich | Cat# MAK043-1KT |
| Phospholipids C | Wako Diagnostics | Cat# 997-01801 |
| Glucose Liquid Reagent for Diagnostic Set | Thermo Fisher | Cat# SB-1070-125 |
| Mouse Ultrasensitive Insulin ELISA Jumbo | ALPCO | Cat# 80-INSMSU-E10 |
| Deposited data | ||
| Chow HMDP liver | GEO | GSE16780 |
| HF/HS HMDP liver | GEO | GSE64769 |
| Western HMDP liver | GEO | GSE66568 |
| Experimental models: Cell lines | ||
| AML12 | ATCC | Cat# CRL-2254; RRID: CVCL_0140 |
| Experimental models: Organisms/strains | ||
| C57BL6J, male, 8 weeks old | Jackson Laboratories | RRID: IMSR_JAX:000664 |
| Oligonucleotides | ||
| B2m-Forward: TACGTAACACAGTTCCACCCGCCTC | This paper | N/A |
| B2m-Reverse: GCAGGTTCAAATGAATCTTCAGAGCATC | This paper | N/A |
| Tbp-Forward: CAAACCCAGAATTGTTCTCCTT | This paper | N/A |
| Tbp-Reverse: ATGTGGTCTTCCTGAATCCCT | This paper | N/A |
| Itih3-Forward: TGCTCACAATGTTGTCACCAC | This paper | N/A |
| Itih3-Reverse: CTTGACCAAACCGGCTGTC | This paper | N/A |
| Fasn-Forward: TGCACCTCACAGGCATCAAT | This paper | N/A |
| Fasn-Reverse: GTCCCACTTGATGTGAGGGG | This paper | N/A |
| Elovl6-Forward: GAAAAGCAGTTCAACGAGAACG | This paper | N/A |
| Elovl6-Reverse: AGATGCCGACCACCAAAGATA | This paper | N/A |
| Scd1-Forward: TTCCCTCCTGCAAGCTCTAC | This paper | N/A |
| Scd1-Reverse: CAGAGCGCTGGTCATGTAGT | This paper | N/A |
| Software and algorithms | ||
| R software environment for analyses | https://www.r-project.org/ | N/A |
| DESeq2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | N/A |
| clusterProfiler | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html | N/A |
| DAVID | https://david.ncifcrf.gov/ | N/A |
| ToppGene | https://toppgene.cchmc.org/ | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Karthickeyan Chella Krishnan (chellakn@ucmail.uc.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All HMDP liver microarray raw data can be accessed at the Gene Expression Omnibus under the accession numbers: GSE16780, GSE64769 and GSE66568.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Animals
All mice were purchased from the Jackson Laboratory and bred at UCLA and University of Cincinnati according to approved IACUC protocols. Animal health was monitored daily by vivarium personnel and were maintained under standard housing condition with 12 h light/dark cycle. For steatosis models, 8-week-old male mice were fed a high fat/high sucrose (HF/HS) diet (Research Diets-D12266B) for 8 weeks or 15% w/v fructose in drinking water for 12 weeks.
Human population cohort
The study detail and its characteristics have been reported elsewhere78,79 and in Table S3. The study included a total of 262 participants of the Kuopio Obesity Surgery (KOBS) study who underwent the Roux-en-Y gastric bypass surgery at Kuopio University Hospital, and from whom both detailed liver histology and RNA sequencing data were available. Total RNA was extracted and purified using the miRNeasy Mini Kit (Qiagen). RNA sequencing libraries underwent 50-nucleotide long paired-end sequencing on Illumina HiSeq 2500 machine, followed by read alignment, normalization and differential expression analysis considering the technical and confounding factors (namely RIN, uniquely aligned reads %, 3’ bias, age, sex, and BMI) as described previously.79 The gene level count values were normalized using a trimmed-mean of M values converted to count per million using edgeR80 and inverse normal transformed. The study protocol was approved by the Ethics Committee of the Northern Savo Hospital District. It was performed in accordance with the Helsinki Declaration. Written informed consent was obtained from all the participants.
AAV expression system
Liver ITIH3 gene expression was modulated by AAV-mediated gene transfer, as described, and successfully applied in our previous studies.44,81,82 For ITIH3 overexpression, the cDNA of ITIH3 (NM_008407.2) was cloned into the AAV8 expression plasmid under a TBG promoter. For ITIH3 deficiency, the AAV8-TBG vector expressing shRNA sequences against ITIH3 (TTGGCAACAATCTGAATTATAA) was created as previously described.44,81 All AAV syntheses were performed on a fee-for-service basis at the University of Pennsylvania’s Penn Vector Core facility.
ITIH3 loss- and gain-of-function studies
Eight-week-old male C57BL/6J mice were intraperitoneally injected with respective AAVs (∼1 × 1012 genome copies diluted in 200 μL saline). We injected control mice with AAVs expressing shRNA against firefly luciferase gene (for knockdown) or GFP (for overexpression). After this, the mice were subjected to 8 weeks of HF/HS diet or 12 weeks of 15% w/v fructose in drinking water. Body weight and body composition were measured for every two weeks until euthanasia. On the last day of the experiment, mice were fasted for 4 h and animals were euthanized, and tissues were collected. For analyzing plasma lipids, retro-orbital blood was collected, and plasma was separated. Liver tissues were collected for weight, mitochondria isolation, lipid measurements, histology and RNA and protein isolation.
Method details
HMDP liver gene expression analysis
Snap frozen liver tissues of HMDP mice maintained on regular chow (healthy)83 or HF/HS (steatosis)39 or western diet (NASH/fibrosis)40 were used for RNA isolation. Global gene expression analysis was done by using Affymetrix HT_MG430A arrays, and filtering criteria for microarray data was done as previously described.84
Liver and plasma lipid analysis
Liver lipids were extracted as described,85 while plasma was directly used. Calorimetric assays from Sigma (triglyceride, total cholesterol and unesterified cholesterol) and Wako (phospholipids) were used to measure respective lipids according to the manufacturer’s instructions.
Isolation of liver mitochondria and bioenergetics
Mitochondria were isolated from liver tissue and respiration was measured as described.15,44 Briefly, mitochondria were obtained by dual centrifugation and resuspended in respiration buffer, while mitochondrial protein was estimated by BCA method. Seahorse XF Pro Analyzer (Agilent) was used to measure mitochondrial respiration. For Complex I respiration, 5 mM pyruvate (Complex I substrate), 0.5 mM malate and 4 mM ADP were used. For Complex II respiration, 5 mM succinate (Complex II substrate), 2μM rotenone (Complex I inhibitor) and 4 mM ADP were used. Then, oxygen consumption rates (OCR) were measured before and after the sequential injections of 2.5μM oligomycin, 4μM FCCP, and 1μM of rotenone/antimycin A. Measures were normalized by total protein.
Cellular bioenergetics
Cellular bioenergetics on AML12 (male) cells were performed as described.15,82 Briefly, GFP or ITIH3 transfected cells and control or exogenous rhITIH3 treated cells were plated on XF cell culture plate (10,000 cells per well) and OCR were measured before and after the sequential injections of 1.5μM oligomycin, 1μM FCCP, and 0.5μM of rotenone/antimycin A. Measures were normalized by cell number.
RNA isolation, Library Preparation, and sequencing
Flash-frozen liver samples were homogenized in QIAzol (Qiagen), and after chloroform phase separation, RNA was isolated according to the manufacturer’s protocol using miRNeasy columns (Qiagen). Libraries were prepared from these extracted liver RNA (Agilent Bioanalyzer RIN >7) using NEBNext Ultra II Directional RNA Library Prep kit (New England Biolabs) per the manufacturers’ instructions. The pooled libraries were sequenced with an Illumina NextSeq 2000 instrument (Illumina) by the Genomics, Epigenomics, and Sequencing Core (GESC) at the University of Cincinnati. Reads were aligned to the mouse genome mm10 using STAR aligner86 and quantified using the Bioconductor R packages as described in the RNA-seq workflow.87 Follow up enrichment analyses were done using DAVID41 and ToppGene.42 Gene set enrichment analysis (GSEA) was performed using the clusterProfiler R package.88
RNA isolation for qPCR
First, total RNA from frozen liver tissues or AML12 cells was extracted using TRIzol (Invitrogen). Next, first-strand complementary DNA (cDNA) was made using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) following manufacturer’s instructions. Finally, PowerUp SYBR Green Master Mix (Applied Biosystems) was used to measure relative normalized expression using the 2−ΔΔCt method.89 The geometric mean of B2m and Tbp was used for normalization as described.90 All qPCR primer sequences are listed in key resources table.
Immunoblotting analyses
Liver lysates were prepared in RIPA buffer (Teknova) and proteins were resolved in 4%–12% Bis-Tris gels (Invitrogen). Proteins were then transferred to polyvinylidene difluoride (PVDF) membrane (Thermo Scientific) and probed by using rabbit monoclonal ITIH3 (SinoBiological #16138-R049), rabbit monoclonal STAT1 (Cell Signaling #14994), rabbit monoclonal Phospho-STAT1 (Ser727) (Cell Signaling #8826), rabbit polyclonal STAT3 (Proteintech #10253-2-AP), rabbit monoclonal Phospho-STAT3 (Tyr705) (Cell Signaling #9145), rabbit monoclonal JAK1 (Cell Signaling #3344), mouse monoclonal ACTIN (abcam #ab8226) and rabbit monoclonal GAPDH (Cell Signaling #5174), and their respective secondary antibodies. Bands were quantified by using ImageJ.
Quantification and statistical analysis
Statistical analysis
Graphs and statistical analyses were performed using Prism v10.0.2 (GraphPad Software). Errors bars plotted on graphs are presented as the mean ± standard error of the mean (SEM) unless reported otherwise. The critical significance value (α) was set at 0.05, and if the p values were less than α, we reported that by rejecting the null hypothesis, the observed differences were statistically significant.
Acknowledgments
We thank Sarada Charugundla for liver lipid analyses; Anidya Soni, Meghan Curran, Dharshini Kumar, Zhiqiang Zhou, and Yonghong Meng for assistance in animal experiments. We acknowledge CSC–IT Center for Science, Finland, for computational resources. The graphical abstract was created with BioRender.com.
This work was supported by NIH R00DK120875 (K.C.K). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author contributions
K.C.K. conceived the study. K.C.K., N.K.T., and U.M. designed, performed experiments, or analyzed the data. I.S.R., and A.P.R. performed cell culture experiments. M.P. cloned and constructed the AAV vectors. D.K., P.P., and J.P. collected, analyzed, or prepared the figures for the KOBS cohort data. K.C.K. and N.KT. drafted the manuscript, and all authors read or revised the manuscript.
Declaration of interests
The authors declare no conflict of interests.
Published: April 10, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109709.
Supplemental information
References
- 1.Adams L.A., Lymp J.F., St Sauver J., Sanderson S.O., Lindor K.D., Feldstein A., Angulo P. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–121. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Browning J.D., Szczepaniak L.S., Dobbins R., Nuremberg P., Horton J.D., Cohen J.C., Grundy S.M., Hobbs H.H. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 3.Kopec K.L., Burns D. Nonalcoholic fatty liver disease: a review of the spectrum of disease, diagnosis, and therapy. Nutr. Clin. Pract. 2011;26:565–576. doi: 10.1177/0884533611419668. [DOI] [PubMed] [Google Scholar]
- 4.Clark J.M., Brancati F.L., Diehl A.M. Nonalcoholic fatty liver disease. Gastroenterology. 2002;122:1649–1657. doi: 10.1053/gast.2002.33573. [DOI] [PubMed] [Google Scholar]
- 5.Falck-Ytter Y., Younossi Z.M., Marchesini G., McCullough A.J. Clinical features and natural history of nonalcoholic steatosis syndromes. Semin. Liver Dis. 2001;21:17–26. doi: 10.1055/s-2001-12926. [DOI] [PubMed] [Google Scholar]
- 6.Marchesini G., Bugianesi E., Forlani G., Cerrelli F., Lenzi M., Manini R., Natale S., Vanni E., Villanova N., Melchionda N., Rizzetto M. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology. 2003;37:917–923. doi: 10.1053/jhep.2003.50161. [DOI] [PubMed] [Google Scholar]
- 7.Pan J.J., Fallon M.B. Gender and racial differences in nonalcoholic fatty liver disease. World J. Hepatol. 2014;6:274–283. doi: 10.4254/wjh.v6.i5.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruhl C.E., Everhart J.E. Determinants of the association of overweight with elevated serum alanine aminotransferase activity in the United States. Gastroenterology. 2003;124:71–79. doi: 10.1053/gast.2003.50004. [DOI] [PubMed] [Google Scholar]
- 9.Clark J.M., Brancati F.L., Diehl A.M. The prevalence and etiology of elevated aminotransferase levels in the United States. Am. J. Gastroenterol. 2003;98:960–967. doi: 10.1111/j.1572-0241.2003.07486.x. [DOI] [PubMed] [Google Scholar]
- 10.Ioannou G.N., Boyko E.J., Lee S.P. The prevalence and predictors of elevated serum aminotransferase activity in the United States in 1999-2002. Am. J. Gastroenterol. 2006;101:76–82. doi: 10.1111/j.1572-0241.2005.00341.x. [DOI] [PubMed] [Google Scholar]
- 11.Lazo M., Hernaez R., Eberhardt M.S., Bonekamp S., Kamel I., Guallar E., Koteish A., Brancati F.L., Clark J.M. Prevalence of nonalcoholic fatty liver disease in the United States: the Third National Health and Nutrition Examination Survey, 1988-1994. Am. J. Epidemiol. 2013;178:38–45. doi: 10.1093/aje/kws448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider A.L.C., Lazo M., Selvin E., Clark J.M. Racial differences in nonalcoholic fatty liver disease in the U.S. population. Obesity. 2014;22:292–299. doi: 10.1002/oby.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Alwis N.M.W., Day C.P. Non-alcoholic fatty liver disease: the mist gradually clears. J. Hepatol. 2008;48:S104–S112. doi: 10.1016/j.jhep.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 14.McCullough A.J. The clinical features, diagnosis and natural history of nonalcoholic fatty liver disease. Clin. Liver Dis. 2004;8:521–533. doi: 10.1016/j.cld.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Chella Krishnan K., Kurt Z., Barrere-Cain R., Sabir S., Das A., Floyd R., Vergnes L., Zhao Y., Che N., Charugundla S., et al. Integration of Multi-omics Data from Mouse Diversity Panel Highlights Mitochondrial Dysfunction in Non-alcoholic Fatty Liver Disease. Cell Syst. 2018;6:103–115.e7. doi: 10.1016/j.cels.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misu H., Takamura T., Matsuzawa N., Shimizu A., Ota T., Sakurai M., Ando H., Arai K., Yamashita T., Honda M., et al. Genes involved in oxidative phosphorylation are coordinately upregulated with fasting hyperglycaemia in livers of patients with type 2 diabetes. Diabetologia. 2007;50:268–277. doi: 10.1007/s00125-006-0489-8. [DOI] [PubMed] [Google Scholar]
- 17.Takamura T., Sakurai M., Ota T., Ando H., Honda M., Kaneko S. Genes for systemic vascular complications are differentially expressed in the livers of type 2 diabetic patients. Diabetologia. 2004;47:638–647. doi: 10.1007/s00125-004-1366-y. [DOI] [PubMed] [Google Scholar]
- 18.Takeshita Y., Takamura T., Hamaguchi E., Shimizu A., Ota T., Sakurai M., Kaneko S. Tumor necrosis factor-alpha-induced production of plasminogen activator inhibitor 1 and its regulation by pioglitazone and cerivastatin in a nonmalignant human hepatocyte cell line. Metabolism. 2006;55:1464–1472. doi: 10.1016/j.metabol.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Meex R.C., Hoy A.J., Morris A., Brown R.D., Lo J.C.Y., Burke M., Goode R.J.A., Kingwell B.A., Kraakman M.J., Febbraio M.A., et al. Fetuin B Is a Secreted Hepatocyte Factor Linking Steatosis to Impaired Glucose Metabolism. Cell Metabol. 2015;22:1078–1089. doi: 10.1016/j.cmet.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Vernia S., Cavanagh-Kyros J., Garcia-Haro L., Sabio G., Barrett T., Jung D.Y., Kim J.K., Xu J., Shulha H.P., Garber M., et al. The PPARalpha-FGF21 hormone axis contributes to metabolic regulation by the hepatic JNK signaling pathway. Cell Metabol. 2014;20:512–525. doi: 10.1016/j.cmet.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misu H., Takamura T., Takayama H., Hayashi H., Matsuzawa-Nagata N., Kurita S., Ishikura K., Ando H., Takeshita Y., Ota T., et al. A liver-derived secretory protein, selenoprotein P, causes insulin resistance. Cell Metabol. 2010;12:483–495. doi: 10.1016/j.cmet.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Bourguignon J., Vercaigne D., Sesboüé R., Martin J.P., Salier J.P. Inter-alpha-trypsin-inhibitor (ITI): two distinct mRNAs in baboon liver argue for a discrete synthesis of ITI and ITI derivatives. FEBS Lett. 1983;162:379–383. doi: 10.1016/0014-5793(83)80791-1. [DOI] [PubMed] [Google Scholar]
- 23.Bost F., Diarra-Mehrpour M., Martin J.P. Inter-alpha-trypsin inhibitor proteoglycan family--a group of proteins binding and stabilizing the extracellular matrix. Eur. J. Biochem. 1998;252:339–346. doi: 10.1046/j.1432-1327.1998.2520339.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen L., Mao S.J., McLean L.R., Powers R.W., Larsen W.J. Proteins of the inter-alpha-trypsin inhibitor family stabilize the cumulus extracellular matrix through their direct binding with hyaluronic acid. J. Biol. Chem. 1994;269:28282–28287. [PubMed] [Google Scholar]
- 25.Fries E., Kaczmarczyk A. Inter-alpha-inhibitor, hyaluronan and inflammation. Acta Biochim. Pol. 2003;50:735–742. [PubMed] [Google Scholar]
- 26.Kobayashi H., Shinohara H., Ohi H., Sugimura M., Terao T., Fujie M. Urinary trypsin inhibitor (UTI) and fragments derived from UTI by limited proteolysis efficiently inhibit tumor cell invasion. Clin. Exp. Metastasis. 1994;12:117–128. doi: 10.1007/BF01753978. [DOI] [PubMed] [Google Scholar]
- 27.Schreitmüller T., Hochstrasser K., Reisinger P.W., Wachter E., Gebhard W. cDNA cloning of human inter-alpha-trypsin inhibitor discloses three different proteins. Biol. Chem. Hoppe Seyler. 1987;368:963–970. doi: 10.1515/bchm3.1987.368.2.963. [DOI] [PubMed] [Google Scholar]
- 28.Gomis-Rüth F.X., Maskos K., Betz M., Bergner A., Huber R., Suzuki K., Yoshida N., Nagase H., Brew K., Bourenkov G.P., et al. Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1. Nature. 1997;389:77–81. doi: 10.1038/37995. [DOI] [PubMed] [Google Scholar]
- 29.Ossowski L. Plasminogen activator dependent pathways inthe dissemination of human tumor cells in the chick embryo. Cell. 1988;52:321–328. doi: 10.1016/s0092-8674(88)80025-4. [DOI] [PubMed] [Google Scholar]
- 30.Kim T.H., Koo J.H., Heo M.J., Han C.Y., Kim Y.I., Park S.Y., Cho I.J., Lee C.H., Choi C.S., Lee J.W., et al. Overproduction of inter-alpha-trypsin inhibitor heavy chain 1 after loss of Galpha(13) in liver exacerbates systemic insulin resistance in mice. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aan4735. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura N., Hatano E., Iguchi K., Sato M., Kawaguchi H., Ohtsu I., Sakurai T., Aizawa N., Iijima H., Nishiguchi S., et al. Elevated levels of circulating ITIH4 are associated with hepatocellular carcinoma with nonalcoholic fatty liver disease: from pig model to human study. BMC Cancer. 2019;19:621. doi: 10.1186/s12885-019-5825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai K.K.Y., Kolippakkam D., Beretta L. Comprehensive and quantitative proteome profiling of the mouse liver and plasma. Hepatology. 2008;47:1043–1051. doi: 10.1002/hep.22123. [DOI] [PubMed] [Google Scholar]
- 33.Wang H., Lafdil F., Kong X., Gao B. Signal transducer and activator of transcription 3 in liver diseases: a novel therapeutic target. Int. J. Biol. Sci. 2011;7:536–550. doi: 10.7150/ijbs.7.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ki S.H., Park O., Zheng M., Morales-Ibanez O., Kolls J.K., Bataller R., Gao B. Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: role of signal transducer and activator of transcription 3. Hepatology. 2010;52:1291–1300. doi: 10.1002/hep.23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L., Zhang Y., Wang L., Fan F., Zhu L., Li Z., Ruan X., Huang H., Wang Z., Huang Z., et al. Amelioration of high fat diet induced liver lipogenesis and hepatic steatosis by interleukin-22. J. Hepatol. 2010;53:339–347. doi: 10.1016/j.jhep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Xu L., Han J., Yang Z.H.E., Yang Y., Chen J., Wu X., Hong Y.A.N., Wang Q.I. 2022. Lrg1 Inhibits the Activation of Hepatic Macrophages to Alleviate NAFLD by Enhancing TGF-Β1 Signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., Shen J., Man K., Chu E.S.H., Yau T.O., Sung J.C.Y., Go M.Y.Y., Deng J., Lu L., Wong V.W.S., et al. CXCL10 plays a key role as an inflammatory mediator and a non-invasive biomarker of non-alcoholic steatohepatitis. J. Hepatol. 2014;61:1365–1375. doi: 10.1016/j.jhep.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Starmann J., Fälth M., Spindelböck W., Lanz K.L., Lackner C., Zatloukal K., Trauner M., Sültmann H. Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hui S.T., Parks B.W., Org E., Norheim F., Che N., Pan C., Castellani L.W., Charugundla S., Dirks D.L., Psychogios N., et al. The genetic architecture of NAFLD among inbred strains of mice. Elife. 2015;4 doi: 10.7554/eLife.05607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hui S.T., Kurt Z., Tuominen I., Norheim F., C Davis R., Pan C., Dirks D.L., Magyar C.E., French S.W., Chella Krishnan K., et al. The Genetic Architecture of Diet-Induced Hepatic Fibrosis in Mice. Hepatology. 2018;68:2182–2196. doi: 10.1002/hep.30113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 42.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shum M., Ngo J., Shirihai O.S., Liesa M. Mitochondrial oxidative function in NAFLD: Friend or foe? Mol. Metabol. 2021;50 doi: 10.1016/j.molmet.2020.101134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chella Krishnan K., Floyd R.R., Sabir S., Jayasekera D.W., Leon-Mimila P.V., Jones A.E., Cortez A.A., Shravah V., Péterfy M., Stiles L., et al. Liver Pyruvate Kinase Promotes NAFLD/NASH in Both Mice and Humans in a Sex-Specific Manner. Cell. Mol. Gastroenterol. Hepatol. 2021;11:389–406. doi: 10.1016/j.jcmgh.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gluchowski N.L., Gabriel K.R., Chitraju C., Bronson R.T., Mejhert N., Boland S., Wang K., Lai Z.W., Farese R.V., Jr., Walther T.C. Hepatocyte Deletion of Triglyceride-Synthesis Enzyme Acyl CoA: Diacylglycerol Acyltransferase 2 Reduces Steatosis Without Increasing Inflammation or Fibrosis in Mice. Hepatology. 2019;70:1972–1985. doi: 10.1002/hep.30765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calle R.A., Amin N.B., Carvajal-Gonzalez S., Ross T.T., Bergman A., Aggarwal S., Crowley C., Rinaldi A., Mancuso J., Aggarwal N., et al. ACC inhibitor alone or co-administered with a DGAT2 inhibitor in patients with non-alcoholic fatty liver disease: two parallel, placebo-controlled, randomized phase 2a trials. Nat. Med. 2021;27:1836–1848. doi: 10.1038/s41591-021-01489-1. [DOI] [PubMed] [Google Scholar]
- 47.Yenilmez B., Wetoska N., Kelly M., Echeverria D., Min K., Lifshitz L., Alterman J.F., Hassler M.R., Hildebrand S., DiMarzio C., et al. An RNAi therapeutic targeting hepatic DGAT2 in a genetically obese mouse model of nonalcoholic steatohepatitis. Mol. Ther. 2022;30:1329–1342. doi: 10.1016/j.ymthe.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCommis K.S., Hodges W.T., Brunt E.M., Nalbantoglu I., McDonald W.G., Holley C., Fujiwara H., Schaffer J.E., Colca J.R., Finck B.N. Targeting the mitochondrial pyruvate carrier attenuates fibrosis in a mouse model of nonalcoholic steatohepatitis. Hepatology. 2017;65:1543–1556. doi: 10.1002/hep.29025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Habibi M., Ferguson D., Eichler S.J., Chan M.M., LaPoint A., Shew T.M., He M., Lutkewitte A.J., Schilling J.D., Cho K.Y., et al. Mitochondrial Pyruvate Carrier Inhibition Attenuates Hepatic Stellate Cell Activation and Liver Injury in a Mouse Model of Metabolic Dysfunction-associated Steatotic Liver Disease. bioRxiv. 2023 doi: 10.1101/2023.02.13.528384. Preprint at. [DOI] [Google Scholar]
- 50.McCommis K.S., Chen Z., Fu X., McDonald W.G., Colca J.R., Kletzien R.F., Burgess S.C., Finck B.N. Loss of Mitochondrial Pyruvate Carrier 2 in the Liver Leads to Defects in Gluconeogenesis and Compensation via Pyruvate-Alanine Cycling. Cell Metabol. 2015;22:682–694. doi: 10.1016/j.cmet.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sisler J.D., Morgan M., Raje V., Grande R.C., Derecka M., Meier J., Cantwell M., Szczepanek K., Korzun W.J., Lesnefsky E.J., et al. The Signal Transducer and Activator of Transcription 1 (STAT1) Inhibits Mitochondrial Biogenesis in Liver and Fatty Acid Oxidation in Adipocytes. PLoS One. 2015;10 doi: 10.1371/journal.pone.0144444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeong W.I., Park O., Radaeva S., Gao B. STAT1 inhibits liver fibrosis in mice by inhibiting stellate cell proliferation and stimulating NK cell cytotoxicity. Hepatology. 2006;44:1441–1451. doi: 10.1002/hep.21419. [DOI] [PubMed] [Google Scholar]
- 53.Martí-Rodrigo A., Alegre F., Moragrega Á.B., García-García F., Martí-Rodrigo P., Fernández-Iglesias A., Gracia-Sancho J., Apostolova N., Esplugues J.V., Blas-García A. Rilpivirine attenuates liver fibrosis through selective STAT1-mediated apoptosis in hepatic stellate cells. Gut. 2020;69:920–932. doi: 10.1136/gutjnl-2019-318372. [DOI] [PubMed] [Google Scholar]
- 54.Lambert J.E., Ramos–Roman M.A., Browning J.D., Parks E.J. Increased De Novo Lipogenesis Is a Distinct Characteristic of Individuals With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2014;146:726–735. doi: 10.1053/j.gastro.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gehlenborg N., O'Donoghue S.I., Baliga N.S., Goesmann A., Hibbs M.A., Kitano H., Kohlbacher O., Neuweger H., Schneider R., Tenenbaum D., Gavin A.C. Visualization of omics data for systems biology. Nat. Methods. 2010;7:S56–S68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 56.Civelek M., Lusis A.J. Systems genetics approaches to understand complex traits. Nat. Rev. Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hathout Y. Approaches to the study of the cell secretome. Expert Rev. Proteomics. 2007;4:239–248. doi: 10.1586/14789450.4.2.239. [DOI] [PubMed] [Google Scholar]
- 58.Alvarez-Llamas G., Szalowska E., de Vries M.P., Weening D., Landman K., Hoek A., Wolffenbuttel B.H.R., Roelofsen H., Vonk R.J. Characterization of the human visceral adipose tissue secretome. Mol. Cell. Proteomics. 2007;6:589–600. doi: 10.1074/mcp.M600265-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Makridakis M., Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J. Proteonomics. 2010;73:2291–2305. doi: 10.1016/j.jprot.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Lawlor K., Nazarian A., Lacomis L., Tempst P., Villanueva J. Pathway-based biomarker search by high-throughput proteomics profiling of secretomes. J. Proteome Res. 2009;8:1489–1503. doi: 10.1021/pr8008572. [DOI] [PubMed] [Google Scholar]
- 61.Seldin M.M., Koplev S., Rajbhandari P., Vergnes L., Rosenberg G.M., Meng Y., Pan C., Phuong T.M.N., Gharakhanian R., Che N., et al. A Strategy for Discovery of Endocrine Interactions with Application to Whole-Body Metabolism. Cell Metabol. 2018;27:1138–1155.e6. doi: 10.1016/j.cmet.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim K., Kim K.H. Targeting of Secretory Proteins as a Therapeutic Strategy for Treatment of Nonalcoholic Steatohepatitis (NASH) Int. J. Mol. Sci. 2020;21:2296. doi: 10.3390/ijms21072296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niu L., Geyer P.E., Wewer Albrechtsen N.J., Gluud L.L., Santos A., Doll S., Treit P.V., Holst J.J., Knop F.K., Vilsbøll T., et al. Plasma proteome profiling discovers novel proteins associated with non-alcoholic fatty liver disease. Mol. Syst. Biol. 2019;15 doi: 10.15252/msb.20188793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choi J.W., Wang X., Joo J.I., Kim D.H., Oh T.S., Choi D.K., Yun J.W. Plasma proteome analysis in diet-induced obesity-prone and obesity-resistant rats. Proteomics. 2010;10:4386–4400. doi: 10.1002/pmic.201000391. [DOI] [PubMed] [Google Scholar]
- 65.Oike Y., Akao M., Yasunaga K., Yamauchi T., Morisada T., Ito Y., Urano T., Kimura Y., Kubota Y., Maekawa H., et al. Angiopoietin-related growth factor antagonizes obesity and insulin resistance. Nat. Med. 2005;11:400–408. doi: 10.1038/nm1214. [DOI] [PubMed] [Google Scholar]
- 66.Pessayre D., Fromenty B. NASH: a mitochondrial disease. J. Hepatol. 2005;42:928–940. doi: 10.1016/j.jhep.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 67.Begriche K., Igoudjil A., Pessayre D., Fromenty B. Mitochondrial dysfunction in NASH: causes, consequences and possible means to prevent it. Mitochondrion. 2006;6:1–28. doi: 10.1016/j.mito.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 68.Begriche K., Massart J., Robin M.A., Bonnet F., Fromenty B. Mitochondrial adaptations and dysfunctions in nonalcoholic fatty liver disease. Hepatology. 2013;58:1497–1507. doi: 10.1002/hep.26226. [DOI] [PubMed] [Google Scholar]
- 69.Sunny N.E., Parks E.J., Browning J.D., Burgess S.C. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metabol. 2011;14:804–810. doi: 10.1016/j.cmet.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kotronen A., Seppänen-Laakso T., Westerbacka J., Kiviluoto T., Arola J., Ruskeepää A.L., Oresic M., Yki-Järvinen H. Hepatic stearoyl-CoA desaturase (SCD)-1 activity and diacylglycerol but not ceramide concentrations are increased in the nonalcoholic human fatty liver. Diabetes. 2009;58:203–208. doi: 10.2337/db08-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsuzaka T., Atsumi A., Matsumori R., Nie T., Shinozaki H., Suzuki-Kemuriyama N., Kuba M., Nakagawa Y., Ishii K., Shimada M., et al. Elovl6 promotes nonalcoholic steatohepatitis. Hepatology. 2012;56:2199–2208. doi: 10.1002/hep.25932. [DOI] [PubMed] [Google Scholar]
- 72.Dorn C., Riener M.O., Kirovski G., Saugspier M., Steib K., Weiss T.S., Gäbele E., Kristiansen G., Hartmann A., Hellerbrand C. Expression of fatty acid synthase in nonalcoholic fatty liver disease. Int. J. Clin. Exp. Pathol. 2010;3:505–514. [PMC free article] [PubMed] [Google Scholar]
- 73.Ferré P., Foufelle F. Hepatic steatosis: a role for de novo lipogenesis and the transcription factor SREBP-1c. Diabetes Obes. Metabol. 2010;12:83–92. doi: 10.1111/j.1463-1326.2010.01275.x. [DOI] [PubMed] [Google Scholar]
- 74.Belew G.D., Jones J.G. De novo lipogenesis in non-alcoholic fatty liver disease: Quantification with stable isotope tracers. Eur. J. Clin. Invest. 2022;52 doi: 10.1111/eci.13733. [DOI] [PubMed] [Google Scholar]
- 75.Knebel B., Fahlbusch P., Dille M., Wahlers N., Hartwig S., Jacob S., Kettel U., Schiller M., Herebian D., Koellmer C., et al. Fatty Liver Due to Increased de novo Lipogenesis: Alterations in the Hepatic Peroxisomal Proteome. Front. Cell Dev. Biol. 2019;7:248. doi: 10.3389/fcell.2019.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwarz J.-M., Clearfield M., Mulligan K. Conversion of Sugar to Fat: Is Hepatic de Novo Lipogenesis Leading to Metabolic Syndrome and Associated Chronic Diseases? J. Am. Osteopath. Assoc. 2017;117:520–527. doi: 10.7556/jaoa.2017.102. [DOI] [PubMed] [Google Scholar]
- 77.Zhao J., Qi Y.F., Yu Y.R. STAT3: A key regulator in liver fibrosis. Ann. Hepatol. 2021;21 doi: 10.1016/j.aohep.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 78.Benhammou J.N., Ko A., Alvarez M., Kaikkonen M.U., Rankin C., Garske K.M., Padua D., Bhagat Y., Kaminska D., Kärjä V., et al. Novel Lipid Long Intervening Noncoding RNA, Oligodendrocyte Maturation-Associated Long Intergenic Noncoding RNA, Regulates the Liver Steatosis Gene Stearoyl-Coenzyme A Desaturase As an Enhancer RNA. Hepatol. Commun. 2019;3:1356–1372. doi: 10.1002/hep4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Männistö V., Kaminska D., Käkelä P., Neuvonen M., Niemi M., Alvarez M., Pajukanta P., Romeo S., Nieuwdorp M., Groen A.K., Pihlajamäki J. Protein Phosphatase 1 Regulatory Subunit 3B Genotype at rs4240624 Has a Major Effect on Gallbladder Bile Composition. Hepatol. Commun. 2021;5:244–257. doi: 10.1002/hep4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chella Krishnan K., Sabir S., Shum M., Meng Y., Acín-Pérez R., Lang J.M., Floyd R.R., Vergnes L., Seldin M.M., Fuqua B.K., et al. Sex-specific metabolic functions of adipose Lipocalin-2. Mol. Metabol. 2019;30:30–47. doi: 10.1016/j.molmet.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chella Krishnan K., Vergnes L., Acín-Pérez R., Stiles L., Shum M., Ma L., Mouisel E., Pan C., Moore T.M., Péterfy M., et al. Sex-specific genetic regulation of adipose mitochondria and metabolic syndrome by Ndufv2. Nat. Metab. 2021;3:1552–1568. doi: 10.1038/s42255-021-00481-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bennett B.J., Farber C.R., Orozco L., Kang H.M., Ghazalpour A., Siemers N., Neubauer M., Neuhaus I., Yordanova R., Guan B., et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parks B.W., Sallam T., Mehrabian M., Psychogios N., Hui S.T., Norheim F., Castellani L.W., Rau C.D., Pan C., Phun J., et al. Genetic architecture of insulin resistance in the mouse. Cell Metabol. 2015;21:334–347. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 86.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Love M.I., Anders S., Kim V., Huber W. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000Res. 2015;4:1070. doi: 10.12688/f1000research.7035.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 90.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All HMDP liver microarray raw data can be accessed at the Gene Expression Omnibus under the accession numbers: GSE16780, GSE64769 and GSE66568.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.