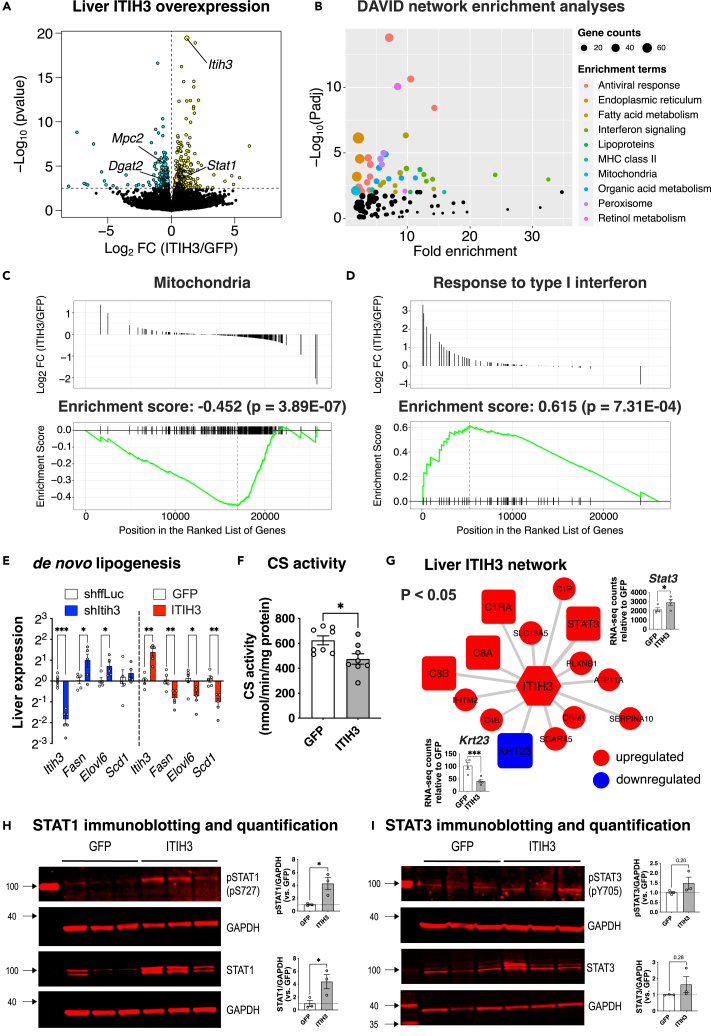
Figure 4.
ITIH3 protects against steatosis via downregulating mitochondria and de novo lipogenesis while upregulating STAT1 signaling
(A) Global genome-wide transcriptomics of livers extracted from GFP or ITIH3 overexpressing mice. Turquoise and yellow represent down-regulated and up-regulated genes, respectively.
(B) DAVID pathway enrichment analyses of 480 DEGs (listed in Table S2). Gene set enrichment analysis (GSEA) of ranked DEGs revealed a significant enrichment of (C) Mitochondria and (D) Response to type I interferon. Follow-up (E) qPCR analyses of Itih3 and liver DNL genes such as Fasn, Elovl6, and Scd1 in ITIH3 knockdown and overexpression mice and their respective controls and (F) citrate synthase (CS) activity measured in GFP or ITIH3 overexpression mice.
(G) Overlay of DEGs on ITIH3 network from Figure 1A. Blue represents genes going down and red represents genes going up in ITIH3 overexpressing mice (p < 0.05). Inset shows RNA counts of Stat3 and Krt23 in ITIH3 overexpressing mice. Follow-up immunoblot analyses and their respective quantification of liver proteins such as (H) pSTAT1 and STAT1, (I) pSTAT3 and STAT3 in GFP or ITIH3 overexpressing mice. GAPDH was used as a loading control. Data are presented as mean ± SEM (n = 4 mice for transcriptomics, n = 5–6 mice for qPCR, n = 8 for CS activity and n = 3 mice for immunoblot analyses per group). p values were calculated by (A and G) Wald test; (E) multiple t tests corrected by post-hoc “Benjamini, Krieger, Yekutieli” FDR approach for multiple comparisons test; (F, H, and I) t test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.