Abstract
Extracellular vesicles (EVs), including exosomes, are increasingly recognized as potent mediators of intercellular communication due to their capacity to transport a diverse array of bioactive molecules. They assume vital roles in a wide range of physiological and pathological processes and hold significant promise as emerging disease biomarkers, therapeutic agents, and carriers for drug delivery. Exosomes encompass specific groups of membrane proteins, lipids, nucleic acids, cytosolic proteins, and other signaling molecules within their interior. These cargo molecules dictate targeting specificity and functional roles upon reaching recipient cells. Despite our growing understanding of the significance of exosomes in diverse biological processes, the molecular mechanisms governing the selective sorting and packaging of cargo within exosomes have not been fully elucidated. In this review, we summarize current insights into the molecular mechanisms that regulate the sorting of various molecules into exosomes, the resulting biological functions, and potential clinical applications, with a particular emphasis on their relevance in cancer and other diseases. A comprehensive understanding of the loading processes and mechanisms involved in exosome cargo sorting is essential for uncovering the physiological and pathological roles of exosomes, identifying therapeutic targets, and advancing the clinical development of exosome-based therapeutics.
Subject terms: Cell signalling, Cancer microenvironment
Exosome cargo sorting: insights into cellular communication and disease
Extracellular vesicles (EVs)—tiny structures produced by cells that are important for cell-to-cell communication—contain various molecules, a process known as cargo sorting. However, how these molecules are selected and packaged into EVs is not fully understood. This research by Lee, Shin, and Chae provides a detailed analysis of our current knowledge of cargo sorting in exosome biogenesis. They reviewed the literature on the types of molecules in exosome cargo, the factors and machinery controlling cargo selection and packaging, and the role of cargo sorting in diseases. They found that understanding exosome cargo sorting is vital for managing diseases and developing exosome-based treatments. This could help design exosomes that can more effectively deliver therapeutic substances to specific cells or tissues, improving the effectiveness and accuracy of these treatments in clinical use.
This summary was initially drafted using artificial intelligence, then revised and fact-checked by the author.
Introduction
Overview of extracellular vesicles (EVs)
EVs represent a diverse class of cell-derived membranous structures that play pivotal roles in regulating intercellular communication. These vesicles are involved in transmitting bioactive molecules and modulating various physiological and pathological processes. There are several subtypes of EVs, including exosomes, which were first characterized through electron microscopy. Initially, these EVs were considered waste bags responsible for expelling unwanted cellular products outside of the cell. Notably, transferrin was the first cargo discovered within these EVs1.
However, beginning in the early 2000s, the advent of advanced analytical methods, such as mass spectrometry and next-generation sequencing, revealed that EVs contain a diverse array of biomolecules, including proteins, lipids, metabolites, and nucleic acids, including various types of DNA and RNA2,3. This revelation reshaped our understanding of exosomes and highlighted the significance of EVs in intercellular communication beyond mere cellular waste removal. Furthermore, research on EVs has rapidly expanded as their potential for diagnostic and therapeutic applications in various diseases, including neurodegenerative disorders, cardiovascular dysfunction, metabolic diseases, and immune-related conditions, including cancer, became evident4.
EVs are secreted into the extracellular space and taken up by target cells, thereby modulating their phenotype. In contrast to the conventional ligand‒receptor-mediated communication system, EV-mediated signaling involves the direct transfer of diverse cargo into recipient cells, thereby enabling the direct regulation of cellular functions at multiple levels, including the genetic, signaling pathway, and functional levels. As innate vesicles, exosomes shield their cargo from enzymatic degradation and exhibit reduced immunogenicity within the circulatory system, making them ideal for transporting biomaterials5,6. Due to their efficacy in delivering biomaterials, EVs are highly promising candidates for a wide array of biomedical applications, including drug delivery, diagnostics, and therapeutic interventions4.
Exosome biogenesis is a complex, highly regulated process that involves several sequential stages, from the initial formation of early endosomes to the eventual release of fully mature exosomes into the extracellular environment. Among these stages, the selective sorting of cargo molecules into nascent exosome vesicles has emerged as a critical determinant of exosome functionality, diversity, and specificity7. In this comprehensive review, we aim to provide an in-depth overview of our current understanding of the cargo sorting mechanisms involved in exosome biogenesis. We explore the various classes of molecules that constitute exosome cargo, the regulatory factors and machinery that govern cargo selection and packaging, and the implications of cargo sorting in disease contexts. Understanding how exosomes manage cargo sorting is a crucial objective for disease management and a foundational step in advancing sophisticated exosome-based therapies7,8. This approach facilitates the design of exosomes that are better equipped to transport therapeutic payloads to specific cells or tissues, thereby enhancing the efficacy and precision of EV treatments in clinical applications.
Classification of EVs
EVs are primarily classified into three major groups based on their size and cellular origin, although EV classification is continuously evolving7. Within this classification, exosomes constitute the smallest EVs, typically measuring between 30 and 150 nm in diameter9. They originate from the endocytic pathway, specifically from late endosomes known as multivesicular bodies (MVBs), where exosome formation begins. Subsequent fusion of MVBs with the plasma membrane releases their internal vesicles into the extracellular space, resulting in exosomes10,11.
Exosomes can encapsulate a wide array of bioactive molecules, including microRNAs (miRNAs), proteins, and lipids. The composition of exosomal cargo is subject to regulation by various signaling pathways and external cellular conditions. Moreover, the composition is dynamic and capable of changing in response to different stimuli and cellular signals. Therefore, exosomes are considered one of the most important forms of EVs in intercellular signaling; exosomes play pivotal roles in transmitting bioactive molecules and responding to the everchanging cellular environment.
In addition to exosomes, EVs include microvesicles (MVs) and apoptotic bodies. MVs, which are larger than exosomes and range from 100–1000 nm in diameter, result from outward budding and fission of the cell’s plasma membrane. Their cargo predominantly includes cytosolic contents such as proteins, nucleic acids, lipids, and metabolites. However, the absence of specific markers for distinguishing MVs from exosomes remains a challenge3,12.
Apoptotic bodies, which make up the largest of the three EV groups, typically measure between 1,000 and 5000 nm. They emerge during apoptosis when cells fragment into smaller units and enveloped by membrane-bound vesicles. These vesicles contain DNA fragments, histones, chromatin remnants, cytosolic fractions, and degraded proteins. Typically, apoptotic bodies are cleared by phagocytic cells7,12.
Other types of EVs with potentially specialized functions have also been described. Large oncosomes (1 μm) and ARRDC1-mediated microvesicles (ARMMs) arise from outward protrusions of the plasma membrane that are excised and shed into the extracellular space. Recently, migrasomes derived from retraction fibers have been described and characterized13. Additionally, two nonvesicular extracellular nanoparticles (NVEPs), exomeres and supermeres, have been shown to be enriched in many cargoes previously associated with EVs. In this review, we focused on exosomes, the most studied type of EV.
Distinct characteristics of exosomes and the composition of exosomes
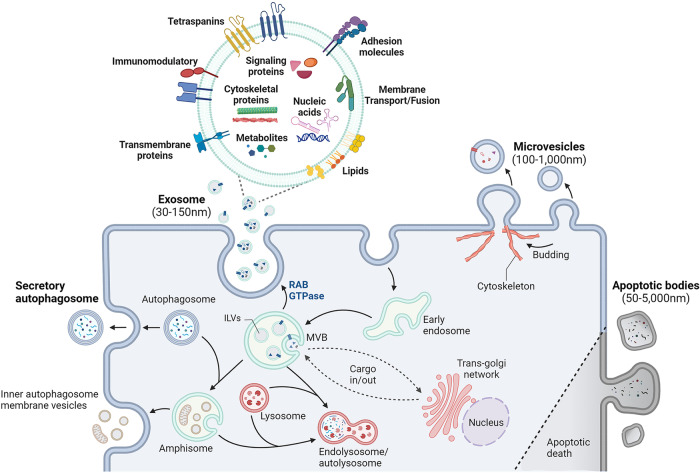
Exosomes have garnered significant attention due to their unique characteristics that distinguish them from other EVs (Fig. 1). The lipid bilayer membrane surrounding exosomes is composed of phosphatidylserine, phosphatidylcholine, sphingomyelin, ceramides, and cholesterol and plays a crucial role in maintaining exosome structural integrity and function4. Exosomes are rich in various forms of nucleic acids, including mRNAs, miRNAs, ribosomal RNAs (rRNAs), long noncoding RNAs (lncRNAs), transfer RNAs (tRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and p-element-induced wimpy testes (piwi)-interacting RNAs (piRNAs)14. Exosomes can also include double-stranded or single-stranded DNA (dsDNA and ssDNA, respectively) and mitochondrial DNA (mtDNA)15,16.
Fig. 1. Mechanisms of extracellular vesicle (EV) biogenesis and EV components.
EV biogenesis occurs through multiple pathways, including through multivesicular bodies (MVBs) within endosomes, which leads to the secretion of exosomes, and through the plasma membrane, which results in microvesicle (MV) generation. After endocytosis, early endosomes undergo maturation in MVBs, during which they form intraluminal vesicles (ILVs) through inward membrane budding. MVBs either fuse with lysosomes for degradation or dock at the cell periphery for exosome secretion, which is facilitated by the RAB GTPases and SNARE complexes. Other types of EVs include apoptotic bodies, which are released during apoptosis via membrane budding; migrasomes, which originate from retraction fibers and contain internal vesicles; and vesicles, which are formed from amphisomes and result from the fusion of outer autophagosome membranes with late-endosomes. Exosomes carry diverse macromolecules, including signaling proteins, transcriptional regulators, various RNA species, DNA, and lipids.
The primary membrane-bound and cytosolic proteins incorporated in exosomes include those involved in membrane transport or fusion (Rab GTPases and annexins), proteins associated with exosome biogenesis (the ESCRT complex, ALIX, and TSG101), heat shock proteins (HSP70 and HSP90), integrins, members of the tetraspanin family (CD63, CD81, and CD82), and cytoskeletal proteins3. Numerous antigen and receptor proteins are expressed on the surface of exosomes. These surface proteins interact with receptors on recipient cells to initiate intracellular signaling pathways. Additionally, exosomes also feature glycoproteins and glycolipids on their surface, along with signaling molecules such as cytokines, growth factors, small molecules, and metabolites7.
Exosomal cargoes located on the external membrane surface can directly engage with recipient cells to initiate signaling events or facilitate specific targeting. Conversely, internal cargoes are released into recipient cells upon exosome uptake, thereby influencing cellular functions at the genetic, protein, or lipid level. The amalgamation of these external and internal cargoes renders exosomes highly versatile vehicles for intercellular communication and the transfer of biological information. Consequently, the selective sorting and packaging of specific cargo into exosomes constitute a fundamental aspect of their biogenesis and function. Understanding the intricacies of cargo sorting mechanisms in exosome formation is essential for deciphering the physiological significance of exosomes and holds great promise for advancing diagnostic and therapeutic strategies across a wide spectrum of diseases.
Exosome biogenesis and selective cargo sorting processes
In recent years, considerable progress has been made in unraveling the mechanisms underlying the formation and secretion of exosomes. In conventional membrane trafficking pathways, cargoes destined for specific organelles typically recruit the machinery necessary for their own sorting and transport17. Generally, the recruited trafficking machinery isolates cargoes on patches of the membrane, reshapes the surrounding membrane into vesicular structures, and ultimately detaches vesicles from the source membrane11. Exosome cargoes appear to follow a comparable fundamental process.
Exosome biogenesis commences with the formation of multiple intraluminal vesicles (ILVs) within endocytic compartments known as multivesicular endosomes or MVBs18 (Fig. 1). ILVs emerge through the inward budding of the endosome’s limiting membrane and encapsulate cargoes destined for exosomes. These ILVs are enriched with specific cargoes and interact with trafficking effectors (Table 1) on endosomal and plasma membrane patches. These unique interactions lead to membrane bending and scission processes that give rise to exosomes. Subsequently, MVBs traverse the plasma membrane, where they fuse and release ILVs into the extracellular space as exosomes. In contrast, plasma membrane-derived MVs are generated via direct budding of the plasma membrane into the extracellular space19. In this section, we describe the current understanding of the basic mechanisms of exosome cargo sorting.
Table 1.
Components that regulate cargo loading in exosome biogenesis.
| Component | Cargo | Refs. |
|---|---|---|
| ESCRT-dependent pathways (classical) | ||
| HRS | Ub modifications | 21 |
| PD-L1 | 53 | |
| TSG101 | CD63 | 23 |
| MHC-II | ||
| FMRP | miR-155 | 74 |
| ESCRT-dependent pathways (variation) | ||
| Syntenin | CD63 | 22 |
| Syndecan1 | ||
| Syndecan1 | β Integrin | 24,44 |
| Fibronectin | ||
| ALIX | Syntenin | 22 |
| PAR1 | 52 | |
| PD-L1 | 53 | |
| YBX1 | miR-133 | 72 |
| ESCRT-independent pathways | ||
| CD9 | CD10 | 39 |
| β-Catenin | ||
| CD63 | PMEL17 | 38 |
| LMP1 | 29 | |
| CD81 | RAC | 39 |
| CD82 | β-Catenin | 39 |
| Ezrin | ||
| Others (unknown) | ||
| hnRNPA1 |
miR-27a-3p miR-27b-3p |
69 |
| miR-92a-3p | ||
| miR-221-3p | ||
| miR-21 | ||
| hnRNPA2B1 | miR-198 | 70 |
| miR-601 | ||
| miR-451 | ||
| miR-575 | ||
| miR-125a-3p | ||
| miR-887 | ||
| AFAP1-AS1 IncRNA | 71 | |
| AGAP2-AS1 IncRNA | ||
| SYNCRIP (hnRNP-Q or NSAP1) | miR-3470a | 67 |
| miR-194-2-3p | ||
| La protein | miR-126 | 14 |
| miR-145 | ||
| miR-486 | ||
| miR-122 | ||
| miR-142 | ||
ESCRT-mediated exosome biogenesis
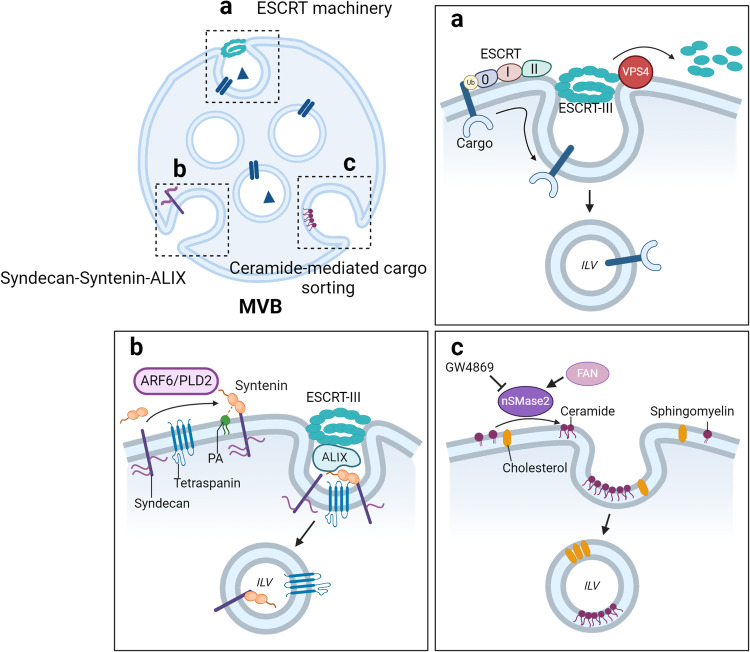
The ESCRT pathway was one of the first pathways associated with exosome biogenesis and involves the coordinated action of all four ESCRT complexes (ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III) in conjunction with disassembly and deubiquitylating enzymes present on the endosome membrane. This process also involves the ESCRT accessory protein VPS420 (Fig. 2a).
Fig. 2. Exosome biogenesis mechanisms and cargo sorting pathways.
Multiple molecular mechanisms of intraluminal vesicle (ILV) generation in multivesicular bodies (MVBs) have been revealed. a The classical ESCRT-dependent pathway involves the recognition of ubiquitinated proteins in the endosomal membrane by the ESCRT-0, -I, -II, and -III subcomplexes. The ATPase VPS4 cooperates in a stepwise manner to mediate ILV formation. b In the syndecan-syntenin-ALIX pathway, membrane budding and cargo clustering can occur independently of the early ESCRT machinery, with VPS4 required for the scission step. c Ceramide, which is generated from sphingomyelin by nSMase2, plays a key role in the ESCRT-independent pathway of ILV biogenesis. Ceramide can form lipid raft microdomains and trigger the conversion of ILVs into MVBs. nSMase2 is activated by FAN and can be pharmacologically inhibited by small molecules such as GW4869. The cargoes sorted through this pathway include flotillin, cholesterol, and tetraspanins, which are localized to lipid rafts.
The ESCRT-0, -I, and -II complexes are equipped with ubiquitin-binding domains that allow them to capture ubiquitinated cargo21, such as epidermal growth factor receptor (EGFR) and ligand‒receptor signaling complexes. Moreover, ESCRT-I, -II, and -III play crucial roles in remodeling the membrane for ILV binding. To complete exosome biogenesis, ESCRT-II induces the formation of ESCRT-III filaments, which facilitate the severing of the nascent exosome neck from the endosome membrane17. ESCRT-III is thought to be directed to the vesicle bud neck either by sensing negative membrane curvature or by promoting membrane bending to facilitate the separation of ILVs from the endosome membrane. However, the specific roles of ESCRT-III are still under investigation.
The AAA ATPase VPS4 interacts with ESCRT-III to facilitate the final stages of ILV formation by promoting membrane scission, resulting in the leakage of ILVs into the MVB lumens. Importantly, the deletion of multiple ESCRT protein subunits or VPS4 can significantly impact exosome biogenesis, leading to alterations in exosome number, size, and protein composition to varying extents22,23. The roles of ESCRT proteins in ILV biogenesis are conserved, as evidenced by studies in budding yeast, where deletion of ESCRT proteins results in prevacuolar, endosomal compartments that lack ILVs21,24.
Variations in the ESCRT pathway
Exosomes carrying different cargoes exhibit distinct requirements for ESCRT proteins during biogenesis, likely reflecting differences in how cargoes are recruited into the ESCRT pathway. Defining these requirements can be challenging in mammalian cells due to the presence of multiple ESCRT protein isoforms with partially redundant functions and the pleiotropic effects of ESCRT deletion23. However, recent insights have shed light on such variations, as exemplified by the syndecan-syntenin-ALIX pathway22 (Fig. 2b).
In MCF7 breast cancer cells, syntenin, a cytoplasmic adapter protein, recruits ALIX to MVBs, where its interaction with ESCRT-III induces ILV formation22. Regulation of syndecan-syntenin-ALIX-mediated exosome biogenesis involves activation of the oncogenic tyrosine kinase SRC. SRC phosphorylates syndecan1, syntenin, and ALIX, thereby stimulating exosome biogenesis25. The recruitment of syntenin to endosomes may occur through the activation of phospholipase D 2 (PLD2)26 by the GTPase ADP-ribosylation factor 6 (ARF6). Subsequently, PLD2 generates phosphatidic acid at the MVB-limiting membrane, thus facilitating syntenin binding. Alternatively, phosphatidic acid generation at endosomes can result from PLD1 activation by Ral GTPases, which further promotes exosome biogenesis27. The localization of ALIX to MVBs is also driven by its association with the late endosome-specific lipid lysobisphosphatidic acid (LBPA), which supports ESCRT-III-dependent ILV formation and exosome production in HeLa cells28. Importantly, the syndecan-syntenin-ALIX pathway can be utilized by the Epstein–Barr virus to load exosomes with latent membrane protein 1 (LMP1), the major Epstein–Barr virus oncoprotein29.
An alternative branch of the ESCRT pathway involves the accessory ESCRT-III proteins CHMP1, CHMP5, and increased sodium tolerance protein 1 (IST1). These proteins play pivotal roles in orchestrating the creation of a distinct subgroup of exosomes within cells subjected to conditions of glutamine deprivation and/or inhibition of the mTOR-Akt pathway30. These exosomes originate within recycling endosomes marked by RAB11, in contract to conventional RAB5- or RAB7-positive endosomes. These investigations shed light on the role of accessory ESCRT-III proteins in the generation of stress-induced exosomes.
Furthermore, GPR143, an atypical G-protein coupled receptor, has emerged as a regulator of ESCRT-dependent exosome production. GPR143 recruits HRS (ESCRT-0) to endosomes to modulate interactions with cargo proteins. This orchestration leads to altered protein sorting in ILVs and modifications to the exosomal proteome composition. Notably, the expression of GPR143 facilitates the secretion of oncogenic exosomes that contain integrins that promote cell motility and cancer metastasis31.
ESCRT-independent exosome biogenesis
Ongoing research has revealed that the formation of MVBs and ILVs is not solely contingent upon the ESCRT complex. Rather, emerging research has demonstrated the existence of ESCRT-independent mechanisms that govern exosome generation.
Lipids in exosome biogenesis
Exosomes are rich in cholesterol, phosphatidylcholine, phosphatidylserine, sphingomyelin, and ceramide, each of which contributes distinct functions in terms of exosome biogenesis and uptake, and these lipids have a consequential impact on recipient cells32. Ceramide assumes a prominent role in exosome biogenesis33. Recent reports indicate that certain cargo molecules can undergo ESCRT-independent sorting into ILVs through a lipid-dependent pathway (Fig. 2c). For example, proteolipid protein (PLP), an abundant membrane protein within the central nervous system34, exhibits a proclivity for endosomal sorting in oligodendrocytes, a propensity retained even in the absence of essential ESCRT components, such as TSG101, ALIX, or HRS. This persistence of sorting competence is observed despite interventions such as small interfering RNA (siRNA)-mediated depletion or the expression of dominant-negative VPS433.
In addition to PLP, the enzyme neutral sphingomyelinase 2 (nSMase 2), which is responsible for the generation of ceramide from sphingomyelin within endosomes, serves as a promoter of ILV and exosome biogenesis. The functional involvement of ceramide within MVBs may be facilitated by diverse ancillary pathways that further amplify exosome biogenesis. The autophagy-related protein microtubule-associated protein 1 A/1B-light chain 3 (LC3) plays a role in recruiting factors associated with nSMase (FAN), an activator of nSMase, to endosome membranes. This recruitment facilitates ceramide-mediated ILV formation35. Furthermore, the activation of RAB31 augments both exosome production and the packaging of EGFR into exosomes derived from cancer cells36. Importantly, it is proposed that this augmentation occurs through the ceramide-dependent pathway of ILV production; these results underscore the potential significance of ILV in the context of cancer cell exosome biogenesis.
Tetraspanins in exosome biogenesis
Tetraspanins, integral membrane surface proteins frequently enriched in small EVs, prominently engage in interactions with integrins and associated proteins at the cell membrane, thus orchestrating the formation of highly organized tetraspanin-enriched microdomains (TEMs)37. The ‘classic’ tetraspanins that have been identified as exosomal cargoes, including CD63, CD81, and CD9, not only contribute to exosome biogenesis but also function as molecular guides for the sorting of other TEM-associated proteins into exosomes (Table 1). For example, the melanocyte-specific glycoprotein PMEL readily undergoes sorting into ILVs upon interacting with CD6338. Similarly, the membrane metalloproteinase CD10 engages with CD9 to facilitate its entry into ILVs. Additionally, CD9 and CD82 interact with E-cadherin to promote exosomal secretion of β-catenin. Nevertheless, efforts aimed at precisely elucidating the intricate mechanisms by which tetraspanins contribute to exosome biogenesis have yielded conflicting results.
Depending on the specific tetraspanin under consideration, its impact on exosome biogenesis may manifest redundantly; thus, specific tetraspanins may have minimal consequences for this process39. Alternatively, disrupting TEMs through the depletion of one tetraspanin may yield altered localization and/or protein‒protein interactions among other tetraspanins, ultimately leading to an increase in exosome biogenesis. Thus, inhibition of exosome biogenesis at another membrane site, such as the endosome membrane, may consequently promote exosome biogenesis at one membrane location, such as the plasma membrane. This phenomenon collectively culminates in an overall increase in exosome abundance40. Consequently, caution is needed when assessing the classification of exosomes solely based on the presence or absence of any given tetraspanin within a heterogeneous mixture of small EVs.
Cargo sorting in exosome biogenesis
Protein composition and enrichment in exosomes
Exosomes serve as versatile carriers for a diverse array of biomolecules, including transmembrane proteins, membrane-associated proteins, and soluble proteins sequestered within their lumens. Proteomic analyses have revealed the rich and intricate protein content of exosomes, thus highlighting their potential roles in intercellular communication and disease diagnosis.
Proteomic characterizations of EVs derived from cellular sources, including cancer cell lines, have revealed distinct protein profiles, often indicative of the cellular origins of these vesicles41,42. In a separate study, 497 EV preparations sourced from cell lines, tissue explants, and both murine and human plasma were evaluated, revealing commonalities in the protein signatures of these EVs43. These investigations have led to the identification of common proteins, some of which are associated with exosome biogenesis and tend to serve as consistent exosome markers. Furthermore, a subset of the proteome demonstrates cell-type specificity, rendering them promising candidates for disease diagnosis and patient prognosis.
Notably, the mechanisms responsible for sorting and packaging proteins into exosomes remain to be fully elucidated. However, a significant portion of these proteins are cell-surface receptors, suggesting that they may originate from processes such as plasma membrane budding or shedding. Additionally, proteomic analyses have played a pivotal role in characterizing disease stages across various cancer types, thus underscoring the potential of selective exosomal protein packaging as a means to predict disease progression, assess therapeutic responses, and gauge therapeutic outcomes.
Tetraspanins and other membrane proteins that are sorted in exosomes
Exosomes are highly enriched in tetraspanin family members, including CD81, CD63, CD9, CD82, and CD3739. While these tetraspanins lack intrinsic enzyme-linked receptors or catalytic activity, they play crucial roles in facilitating the sorting of protein cargoes, particularly tetraspanin-interacting proteins, such as integrins44, ICAM-145, IGFS-846, major histocompatibility complex class II proteins47, and syndecan22.
The surfaces of exosomes harbor various other transmembrane proteins with scaffolding functions, including flotillin 1 and 248, IL-6R49, EGFR, T-cell receptor50, chimeric antigen receptor51, GPCR receptors52, PD-L153, TGFB54, and ADAM proteases55. In addition to transmembrane proteins, the surface of exosomes is also rich in many membrane-interacting proteins, especially proteins with glycosylphosphatidylinositol (GIP) anchors, such as proteoglycans, glypican-156, DAF, and MAC-IP, which further augment the protein landscape of the exosome surface.
A variety of proteins, including small GTPases that contribute to exosome biogenesis and are attached via posttranslational prenylation, have been found on the inner leaflet of exosome membranes. Additionally, proteins myristoylated at the N-terminus, such as BASP-1 and Src signaling kinases, are among those that undergo sorting into exosomes. Similarly, N-terminal myristoylation is used for loading lentiviral gag into viruses or exosomes. Other posttranslational modifications, such as ubiquitination, SUMOylation, and phosphorylation, have also been implicated in cargo sorting57.
The abundant proteins within exosomes include molecular chaperones that interact with or are part of ESCRT complexes, including ALIX, TSG101, and syntenin. In addition to biogenesis-related proteins, exosomes are enriched in molecular chaperones such as HSP70, HSP90, and HSP20.
Exosomes as RNA carriers
Despite the well-established process of cargo selection for transmembrane receptors through ubiquitylation and recognition by ESCRT components in exosome biogenesis, the mechanisms that govern the incorporation of cytoplasmic cargoes, including RNAs and RNA-binding proteins (RBPs), into exosomes have not been fully elucidated. Multiple reports using next-generation sequencing and microarray technologies to characterize RNA content in exosomes sourced from cell cultures, tissues, or biological fluids have revealed the enrichment of specific RNAs. These included mRNA fragments (≤1 kb in length), miRNAs, snRNAs, tRNA fragments, snoRNAs, mitochondrial RNAs (mtRNAs), piRNAs, vault RNAs (vtRNAs), and Y RNAs. Circular RNAs (circRNAs), rRNA fragments, and long non-coding RNAs (lncRNAs) have also been identified in exosomes58,59.
For the protein cargo, the current literature indicates that the RNA composition of exosomes varies depending on the cellular context. Emerging evidence suggests that RBPs play a pivotal role in orchestrating the selective sorting of various small RNAs, including miRNAs, tRNAs, Y RNAs, vault RNAs, and others, into exosomes. RBPs are primarily localized to sites of exosome biogenesis and function as adaptors between RNA cargo and exosome biogenesis machinery14,60 (Table 1). Generally, sorting mechanisms are classified as active RNA-loading processes61. In these processes, specific regulatory mechanisms are involved in actively selecting and incorporating certain RNAs into exosomes. Passive loading, on the other hand, is primarily driven by the intracellular concentration of a specific RNA, and the presence of a specific RNA in exosomes is largely dependent on the abundance of that RNA within the source cell62.
A distinctive category of active RNA loading is selective loading, which involves the specific sorting of particular RNA species in exosomes. This selective sorting has been demonstrated for miRNAs that possess specific motifs containing EXOmotifs, with CGGGAG being the strongest example. Conversely, miRNAs with CELL motifs, such as AGAAC, CAGU, or AUUA, tend to be retained within source cells63,64. Moreover, specific motifs have been characterized for several RBPs, including hnRNPA2B1 (GGAG, AGG, or UAG), hnRNPK (AsQGnA), SYNCRPI (GGCU), and FMRP (AAUGC). These motifs contribute to the selective incorporation of associated RNAs into exosomes, often regardless of the overall RNA composition of the source cells65,66. Additionally, 40 miRNA seed sequences were identified as motifs enriched in lncRNAs associated with prostate cancer exosomes. The selection of these miRNAs is governed by cell type-specific preferences, where specific GC-rich miRNA sequence elements are recognized by RBPs.
Given the presence of more than 4,000 identified RBPs in the human genome at present67 (a list of RBPs is available in the online database RBPbase v0.2.0 Alpha, https://rbpbase.shiny.embl.de) and given that RBPs constitute 25% of the EV protein content, RBPs are undoubtedly key players in the active sorting of RNA into EVs. Among these RBPs, hnRNPA2B1 has been extensively reported to be a pivotal factor in regulating the loading of numerous miRNAs68,69 (Table 1). Although the exact mechanism through which hnRNPA2B1 engages with the exosome biogenesis machinery has not been determined, its association with intracellular ceramide-rich MVB structures suggests a potential link to the nSMase2-ceramide pathway in exosome biogenesis. Intriguingly, the role of hnRNPA2B1 in facilitating colorectal cancer liver metastasis and bladder cancer lymphatic metastasis is supported by its regulation of exosomal sorting of tumor cell miRNAs and lncRNAs such as AFAP1-AS1, AGAP2-AS1, H19, and LNMAT2, respectively70,71 (Table 1). Notably, studies involving RBP YBX1 have indicated its potential role in the sorting of a variety of small RNAs (miRNAs, tRNAs, Y RNAs, and vault RNAs) into exosomes, as demonstrated by the coprecipitation of YBX1 with RNAs and the subsequent reduction in the levels of these associated RNAs following YBX1 deletion72. Similarly, the RBP La directly binds miRNAs and preferentially directs them into high-density vesicles while excluding low-density counterparts.
RNA sorting into exosomes extends beyond the binding of specific RNA sequence motifs and encompasses various RNA or RBP modifications. For example, SUMOylation has been implicated in the loading of miRNAs into exosomes, with hnRNPAB1 being the key mediator of this process70. Phosphorylation has also been linked to exosome cargo packaging, as exemplified by exosomal 5’pppRNA during latent EBV infection73. Additionally, LC3 conjugation has been associated with the loading of small noncoding RNA species, including snoRNAs and miRNAs, within the framework of the secretory autophagy pathway. Similarly, LC3 conjugation has been described for hnRNPK- and SAFB-mediated loading of these small ncRNA species.
Although the importance of RBPs in mediating RNA sorting into exosomes is evident, the specific mechanisms by which these RBPs interact with the exosome biogenesis machinery are unclear. Notably, FMRP is recruited to MVBs through interactions with the RAB-interacting lysosomal protein and HRS. This intriguing connection links FMRP-mediated miRNA sorting with the ESCRT-mediated process of exosome biogenesis74.
Moreover, membrane contact sites between the ER and MVBs have emerged as crucial locations for the generation of RNA-enriched exosomes. A recent study identified integral membrane proteins, namely, the ER tether protein VAP-A and the ceramide transport protein CERT, as crucial components in the biogenesis of both large and small EVs loaded with RNA and RBPs75. Given the role of the ER as a structural scaffold for various RNA granules, the regulation of RBP-RNA complexes localized to the ER could play a significant role in driving the formation of RNA-containing exosomes. These findings illuminate the multifaceted landscape of RNA sorting into exosomes and highlight the intricate interplay between RBPs, RNA modifications, and the exosome biogenesis machinery in shaping the cargo profile of these versatile vesicles.
Exosomes as vehicles for DNA and mtDNA transport
While RNA has been extensively investigated as a cargo molecule in exosomes, considerably less research has been dedicated to understanding the origin and clinical significance of DNA in these vesicles. Various DNA species, including genomic dsDNA76, dsDNA-binding histone proteins77, ssDNA, mitochondrial DNA (mtRNA)78, and viral DNA, are associated with exosomes. The presence of DNA within exosomes has generated significant interest, particularly in the context of liquid biopsy biomarker analysis79,80. Interestingly, recent studies employing advanced imaging and biochemical analysis have revealed the presence of DNA both on the surface and within exosomes. However, the mechanisms orchestrating the integration of DNA into exosomes have not been fully elucidated.
Traditionally, the majority of EV-containing DNA was thought to originate from apoptotic bodies. However, ongoing investigations have revealed a more intricate scenario in which nonapoptotic EVs have emerged as carriers of DNA. Notably, a substantial portion of DNA is found within large EVs, including both apoptotic and nonapoptotic subtypes. This phenomenon is evident in studies comparing large EVs and exosomes from prostate cancer cells and patient plasma80. Intriguingly, large nonapoptotic EVs seem to accumulate more DNA, especially in cancer cells with nuclear shape instability, suggesting that these cells play a role in encapsulating and exporting cytosolic DNA that is generated due to genomic instability in rapidly dividing cancer cells.
Furthermore, recent research has suggested that DNA secretion via exosomes exerts a crucial cytoprotective function by alleviating cellular stress associated with the accumulation of deleterious cytoplasmic DNA and micronuclei. Consequently, therapy-induced DNA damage in cancer cells may contribute to exosome-mediated DNA packaging. However, the mechanisms governing DNA recruitment into exosomes have not been elucidated. Understanding the process of DNA entry into MVBs remains a challenge. Nevertheless, some reports have successfully demonstrated the presence of DNA within exosomes16. Importantly, senescence and DNA damage have been linked to increased DNA levels in MVBs and exosomes, thus offering a potential explanation for the prevalence of DNA in exosomes, particularly in the context of exosomes derived from cancer cells.
This evolving field holds great promise for shedding light on the role of exosomes as carriers of DNA, with potential implications for diagnostic and therapeutic applications. However, further research is needed to unravel the intricate mechanisms underlying DNA sorting into exosomes and to fully understand the clinical relevance of DNA cargo within these versatile vesicles.
Exosomes as lipid carriers
Lipids constitute another crucial class of macromolecules that are packaged into exosomes. These vesicles are particularly rich in various lipids, including cholesterol, phosphatidylcholine, phosphatidylserine, sphingomyelin, and ceramide. These lipids play diverse and essential roles in exosome biogenesis, uptake, and the functional impact of exosomes on recipient cells.
In the context of cancer, in an extensive, large-scale lipidomic analysis, researchers quantified >200 lipid species in exosomes derived from PC3 prostate cancer cells. This analysis revealed that certain lipids, including cholesterol, sphingomyelin, and glycosphingolipids such as ceramides and phosphatidylserine, were more abundant in exosomes than in other phospholipids and were generally less abundant in exosomes81. Furthermore, exosomes isolated from the urine of prostate cancer patients exhibit higher levels of specific lipids, notably ceramides, than urine-derived exosomes from healthy patients. This observation suggests the potential utility of these lipids as fluid-based biomarkers82. However, a separate study reported reduced ceramide levels in urine exosomes from stage 2 benign prostate hyperplasia patients compared to urine exosomes from stage 3 prostate cancer patients, thus complicating the potential use of ceramides as biomarkers. Additionally, exosomes from colorectal cancer, glioblastoma, and hepatocellular carcinoma cells also demonstrated cholesterol, sphingomyelin, and phosphatidylserine enrichment in contrast to their parent cells83,84. These findings highlight commonalities in the lipid content of exosomes across different cancer types. Further research aimed at elucidating the mechanisms underlying lipid packaging in exosomes and uncovering the functional roles of these lipid components holds great promise for enhancing the potential of these components as biomarkers and therapeutic tools.
Therapeutic insights into small-molecule compounds that target exosome biogenesis and secretion in cancer
Exosomes facilitate fundamental cell-to-cell communication, a process that is conserved across all life forms. These minuscule vesicles play pivotal roles in a wide range of (patho-)physiological processes, including maintaining cellular balance, transmitting infections, driving cancer progression, and contributing to cardiovascular diseases. Consequently, strategies aimed at blocking the release or biogenesis of exosomes or regulating exosomal cargo hold promise for slowing the progression of diseases, particularly cancers85,86.
The role of exosomes in systemic aspects of human disease, particularly cancer, has garnered significant attention. Exosomes are present in all body fluids, and their cargo signature can be used to predict cancer type, stage, and therapeutic response43,87. However, the importance of tumor-derived exosomes in shaping the tumor microenvironment homeostasis should not be underestimated. Experimental studies have consistently shown that inhibiting exosome production in cancer and stromal cells reduces cancer growth and metastasis88. This finding underscores the critical role of exosome secretion in cancer development.
Exosomes influence all aspects of cancer progression, including carcinogenesis, host-microbiota interactions, metastasis, immune responses, drug resistance, and the formation of premetastatic niches in distant organs89. Various factors, such as cellular stress (e.g., hypoxia, nutrient deprivation, and oxidative stress) and the presence of oncogenes, such as Src, EGFRvIII, and KRAS, can impact the composition of exosomal cargo25,90,91. For example, the presence of Src in cells increases the release of exosomes containing proinflammatory proteins, while the presence of KRAS leads to an increase in the number of exosomes containing proteins and miRNAs involved in cell growth and survival. Modulating signaling in exosome biogenesis and secretion pathways can affect exosome contents, suggesting that regulating exosomal composition is a promising avenue for developing new cancer therapies.
However, research into the intricate mechanisms governing the sorting of exosomal cargo is in its nascent stages, and specific inhibitors have not yet been identified. The formation of exosomes is also intricately intertwined in this process. Here, we introduce the various small molecules known to be involved in exosome formation, including 33 inhibitors that were assessed based on the pathways of exosome biogenesis or secretion that they primarily target. These inhibitors play a crucial role in regulating exosome secretion and have shown promise in the development of new cancer therapies. Table 2 summarizes these pathways and their corresponding inhibitors.
Table 2.
Potential exosome inhibitors and their targets for cancer treatment.
| Inhibitor | Targets | Pathways | Cancer cell types | Refs. |
|---|---|---|---|---|
| Exosome biogenesis | ||||
| SyntOFF | Syntenin | Syndecan-Syntenin-Alix | Breast cancer | 98 |
| Imipramine | Acid sphingomyelinase (aSMase) | aSMase | Prostate cancer | 93 |
| GW4869 | Neutral sphingomyelinase (nSMase) | nSMase-ceramide pathway | Breast cancer, prostate cancer, melanoma, glioma, and myeloma | 92,95 |
| Spiroepoxide | Neutral sphingomyelinase (nSMase) | nSMase-ceramide pathway | - | 92 |
| Simvastatin | HMG-CoA reductase | Cholesterol synthesis | - | 94 |
| Glibenclamide(Glyburide) | ATP-binding cassette transporter | Potassium channels block | Breast cancer, prostate cancer | 93 |
| Indomethacin | Cyclooxygenase I/II | ABCA3 and lipid transporter | - | 95 |
| Exosome release | ||||
| Y27632 | Rho-associated protein kinases (ROCK) | Cytoskeletal reorganization | Breast cancer | 95 |
| U0126 | MEK1/2 | Ras/Raf/ERK signaling | Prostate cancer | 95 |
| Manumycin A | Farnesyltransferase (FTase) | Ras/Raf/ERK1/2 signaling | Prostate cancer | 97 |
| Tipifarnib | Farnesyltransferase (FTase) and Rab | Ras/Raf/ERK1/2 signaling | Rhabdomyosarcoma, prostate cancer | 97 |
| CPPG | GRM3 | RAB27a and Alix | Multiple myeloma | 99 |
| LY341495 | GRM3 | RAB27a | Breast cancer | 99 |
| Sulfisoxazole | Endothelin receptor A | Multiple process | Breast cancer | 95,100 |
| Macitentan | Endothelin receptor A | Multiple process | Breast, colon and lung cancer | 101 |
- : no tested in cancer cells
Among these inhibitors are those that target sphingomyelinases, such as imipramine, GW4869, cambinol, and spiroepoxide. The latter three selectively inhibit neutral sphingomyelinase (nSMase or aSMase). Each of these inhibitors operates through distinct mechanisms, ultimately reducing exosome secretion by blocking ceramide-modulated inward budding of MVBs and the subsequent release of exosomes from MVBs92. Additionally, inhibitors such as glyburide (glibenclamide) target ATP-sensitive K+ channels, while others, such as indomethacin, target ATP-binding cassette transporters. These inhibitors regulate cellular cholesterol and phospholipid concentrations, ultimately inhibiting the release of MVs and exosomes93. Simvastatin, an HMG-CoA reductase inhibitor, prevents the synthesis of cholesterol and decreases the intracellular concentrations of exosome-associated proteins such as ALIX, CD63, and CD8194. Furthermore, proteins involved in cytoskeletal organization are key targets for various inhibitors, as they are essential for both exosome release and endocytic processes. Y27632, a competitive inhibitor of the ROCK family, which includes ROCK1 and ROCK2, plays a crucial role in this context. Y27632 competes with ATP for binding to the catalytic sites of ROCK1 and ROCK2, resulting in a decrease in MV and exosome-sized secretion. Additionally, inhibitors targeting protein kinases, such as Y27632, U0126, imatinib, and dasatinib, are effective at preventing the activation of ERK, which is necessary for microvesiculation95.
Several inhibitors target the ESCRT-dependent pathway of exosome production. These inhibitors act on Ras farnesyltransferase enzymes by inhibiting their activation and disrupting the Ras/Raf/ERK1/2 signaling pathway, which is crucial in the ESCRT-dependent pathway related to exosome biogenesis96,97. SyntOFF binds to the PDZ domain of syntenin, thereby disrupting the syndecan-syntenin-ALIX pathway98. This disruption results in reduced proliferation and metastasis in breast cancer patients and decreased exosome secretion. In multiple myeloma and bone marrow stromal cells, the glutamate antiporter system facilitates glutamate export and promotes exosome secretion by upregulating the expression of Rab27a, TSG101, ALIX, and VAMP7. This mechanism contributes to bortezomib resistance in multiple myeloma. Targeting GRM3 with the antagonist (RS) alpha-cyclopropyl-4-phosphonophenylglycine (CPPG) reduces exosome secretion and overcomes drug resistance in multiple myeloma99. Recently, endothelin A receptor (ETA) antagonists, such as sulfisoxazole (SFX) and macitentan (MAC), have emerged as new agents for inhibiting exosome secretion. SFX, which was originally an antibacterial drug, has been shown to inhibit components within or related to the ESCRT-dependent pathway, including ALIX and VPS4B, as well as some RAB proteins in breast cancer cell lines95,100. Another ETA antagonist, MAC, inhibits exosomal PD-L1 secretion101. This inhibition reduces the number of exosomes that bind to PD-1 on T cells, thereby enhancing antitumor immunity by revitalizing exhausted T cells. These findings suggest that targeting various steps of exosome biogenesis and secretion holds promise for the development of effective drugs.99,102
Exosome engineering for selective cargo sorting: active vs. passive processes
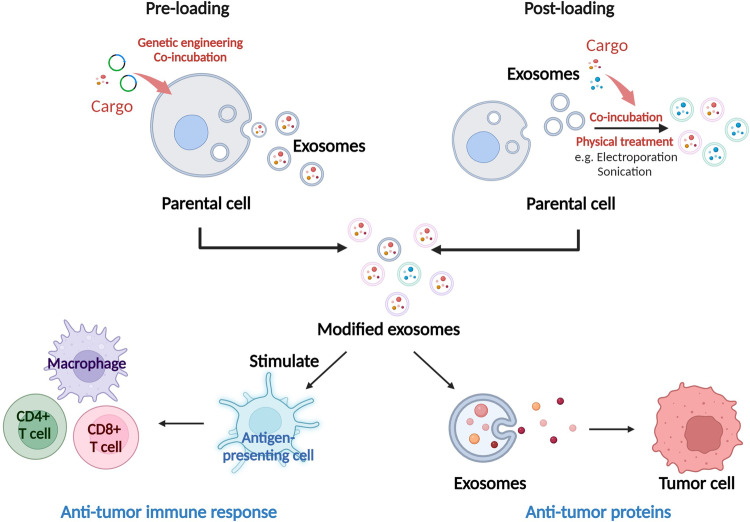
Exosomes are reservoirs of diverse biomolecular cargo, including lipids, proteins, and nucleic acids. Recent advancements in genetic tools, such as RNAi, CRISPR-Cas9, and recombinant DNA technology, coupled with enhanced exosome characterization techniques, have begun to unveil the intricate mechanisms underlying cargo sorting into exosomes (Fig. 2). Currently, research on exosome modification predominantly revolves around two methods: active and passive approaches103. Active strategies involve the integration of target substances during exosomal biogenesis, often through genetic cell modifications. Passive approaches involve the loading of a combination of exogenous substances after exosome secretion via techniques such as electroporation or chemical conjugation (Fig. 3). Key objectives in exosome bioengineering include accurate cargo loading and enhancing exosomal targeting.
Fig. 3. Exosome engineering strategies for loading cancer therapeutic cargo into exosomes.
Exosomes can be engineered to target internal and modified surface cargoes for cancer therapy. Cargo loading strategies are achieved by incubating therapeutic agents (e.g., pharmacological inhibitors, miRNAs, and recombinant proteins) directly with isolated exosomes (post-loading) or by exposing them to exosome-secreting donor cells, followed by the isolation of loaded exosomes (pre-loading). Post-loading methods require physical treatments to disrupt membrane integrity and allow the cargo to enter the interior of exosomes. Alternatively, exosome-producing cells with genetic expression constructs that encode therapeutic cargoes linked to an exosome sorting domain can be generated. This leads to sorting therapeutic cargo into exosomes. Modified exosomes containing tumor antigens can stimulate antigen-presenting cells and drive antitumor immune responses in the human body. Engineered exosomes can also directly release antitumor cargo to attack cancer cells.
The primary components of exosomal cargo typically include small RNAs and proteins. Exosomal cargo ‘selection’ or ‘sorting’ refers to processes that locally enrich cargo molecules during nascent exosome formation. Exosome selection and sorting mechanisms involve exosome biogenesis machinery that is responsible for capturing and aggregating cargoes. In active loading, parental cells undergo genetic engineering to overexpress a desired protein fused with an exosome scaffold, which is subsequently integrated into secreted vesicles during biogenesis104. Despite extensive exploration of endogenous scaffolds for exosomal cargo engineering, achieving versatile strategies for loading biotherapeutic cargo into exosomes remains a challenge due to the diversity of proteins that participate in exosome biogenesis and cargo loading. Importantly, the choice of cargo fusion partner profoundly impacts loading efficiency. It is essential to consider that fusion of a specific cargo with one exosome biogenesis regulator may result in suboptimal loading into exosomes, whereas fusion with another may yield superior engineering performance. Ideally, exosome engineering scaffolds should enable the efficient packaging of multiple copies of proteins without disrupting the exosome biogenesis pathway or the luminal content of exosomes. Therefore, various studies have systematically compared different exosome-sorting domains for endogenous engineering using the latest innovations in exosome characterization to determine the efficiency of these methods at the single-exosome level. In a recent study, 150 enhanced green fluorescent protein molecules were loaded per vesicle using TSPAN14 as the exosome scaffold105. Other identified exosome-sorting domains include the syntenin106, ARRDC1, BASP-1, syndecan-1, and HIV-I Nef proteins. To facilitate the post-encapsulation release of biotherapeutic cargo, advanced systems, such as light-induced dimerization systems107 and small-molecule-controlled protein associations108, have been developed. By leveraging these innovations in endogenous exosome engineering, exosomes have been bioengineered for delivering CRISPR/Cas9109, the IkBa superrepressor110, Cre recombinase111, and lysosomal enzymes (Fig. 3).
Following the discovery of functional RNA transfer via exosomes112, substantial efforts have been directed toward creating bioengineered exosomes as nanosized biomimetic delivery agents for therapeutic nucleic acid cargo. Encapsulating RNA into exosomes has been approached through both brute-force overexpression and engineered targeting. Overexpressing Cre recombinase mRNA within donor cells results in its presence within exosomes, albeit generally with low efficiency. Notably, recent progress has been made with siRNA sequences incorporated into a Dicer-independent pre-miRNA stem–loop (premiR-451), which has demonstrated enhanced loading efficiency using miRNA sorting machinery113. For exosome loading of nucleic acid cargo, strategies similar to those discussed earlier for therapeutic proteins involving the fusion of a nucleic acid-binding protein with an exosome-sorting protein have been adopted. This methodology has successfully facilitated the loading of small RNAs114 as well as longer RNA species such as mRNAs115. Consequently, in experimental scenarios, careful management of co-delivered passively loaded proteins is crucial. In therapeutic applications, co-delivering mRNA, along with therapeutic proteins, can confer advantages. A recent approach elegantly combined endogenous and exogenous exosome loading strategies. The approach involved expressing a DNA aptamer within producer cells to sequester target mRNAs while efficiently loading this complex into exosomes via a CD9-zinc finger fusion protein. To release the mRNA from the bound DNA aptamer, purified mRNA/DNA aptamer-loaded exosomes were electroporated to encapsulate a Klenow fragment exonuclease. This modification enabled mRNA translation in recipient cells both in vitro and in vivo116.
Exosomes as promising delivery systems for anticancer drugs and future perspectives
Exosomes are natural vesicles secreted by various cells in the body and serve as essential carriers of information between cells. Exosomes are innate carriers of functional genetic information that exhibit low toxicity and widespread tissue distribution in vivo. They also possess more than 30 times greater cellular uptake than alternative delivery systems such as nanoparticles or liposomes117. Additionally, exosomes exhibit resistance to harsh environmental conditions, such as low blood pH, making them promising biocarriers for drugs, nucleic acids, and imaging agents in cancer therapy. The potential of exosomes as delivery systems for anticancer drugs, particularly compounds with low solubility and limited off-target delivery, is of significant interest.
Manipulating exosomes by inhibiting their biogenesis or modifying their surface membranes and loading them with cargo, such as proteins, nucleic acids, or drugs, has demonstrated remarkable potential in reducing tumor progression and metastasis. Notably, exosomes are at the forefront of clinical trials for diverse therapeutic applications; there are 118 ongoing trials as of 2022 (https://www.clinicaltrials.gov/). While preclinical studies have shown promising therapeutic efficacy, ongoing clinical trials are further evaluating the potential of these agents. Table 3 provides a comprehensive list of clinical trials utilizing exosomes as therapeutic tools. However, despite the immense promise of EVs for therapeutic and diagnostic purposes, several significant challenges must be addressed.
Table 3.
Overview of clinical trials using exosomes for cancer treatment.
| Sr. no. | Disease | Exosome source | Status | Remarks | Clinical trial identification | Ref. |
|---|---|---|---|---|---|---|
| 1 | Pancreatic cancer | Ultrasound-guided portal venous blood exosome | Recruiting | Safety of sampling portal venous blood, analyzing mRNA markers | NCT03821909 | 118 |
| 2 | Colon cancer | Curcumin conjugated with plant exosomes | Active, Phase I | Comparing exosome-loaded curcumin on immune modulation, phospholipid profile of normal and malignant patients | NCT01294072 | Not available |
| 3 | Sarcoma | Blood samples | Recruiting | Evaluation of cancer pathogenesis, progression, and treatment efficacy of exosomes | NCT03800121 | 119 |
| 4 | Prostate cancer | Urine exosomes | Completed | Validation of non-digital rectal examination (DRE) exosome gene expression test of prostate cancer in biopsy | NCT02702856 | 120 |
| 5 | Pancreatic cancer | Blood samples from patients | Active, not recruiting | Exosome purification for RNA sequencing and proteomics | NCT02393703 | 121 |
| 6 | Lung metastasis osteosarcoma | Blood samples | Recruiting | Identification of levels of circulating exosomal RNA with or without lung metastasis | NCT03108677 | 122 |
| 7 | Gallbladder carcinoma | Exosomal blood samples | Recruiting | Establishing a correlation between exosome biomarkers and gallbladder carcinoma | NCT03581435 | Not available |
| 8 | Stage IV pancreatic adenocarcinoma | Mesenchymal stromal cells-derived exosomes with KRAS G12D siRNA | Phase-I, not recruiting | Mesenchymal-derived exosomes with KRAS G12D in treating individuals with pancreatic cancer with KRAS G12D mutation | NCT03608631 | 5 |
| 9 | Pancreatic ductal adenocarcinoma | Portal vein blood | Completed | Test 3 CTC isolation methods and analyses for onco exosomes in pancreatic cell culture media by flow cytometry | NCT03032913 | 123 |
| 10 | Oral mucositis, head and neck cancer | Grape extract exosomes | Active, phase-I, not recruiting | The ability of plant exosomes to prevent oral mucositis in head and neck cancer | NCT01668849 | Not available |
| 11 | Non-small cell lung cancer(NSCLC) | Plasma exosomes | Not recruiting | New radiotherapy combined with immunotherapy | NCT02890849 | 53 |
| 12 | Non-small cell lung cancer(NSCLC) | Dendritic-derived exosomes | Completed Phase-2 | No induction of T cells monitored in patients | NCT01159288 | Not available |
| 13 | Thyroid cancer | Urine exosomal thyroglobulin and galectin3 | Active, not recruiting | Identifying urinary exosomal proteins (thyroglobulin and galectin 3) | NCT03488134 | 124 |
| 14 | Colon cancer | Blood samples | Recruiting | Novel ways of diagnosing and predicting the spread to other organs, such as liver | NCT03432806 | Not available |
| 15 | Prostate cancer | Urine samples | Active, not recruiting | Validated urine test to predict the incidence of high-grade prostate cancer in initial prostate biopsy | NCT03235687 | 121 |
| 16 | Triple-negative breast cancer | Serum exosomes | Phase-I, recruiting | Assessing response to pembrolizumab in the primary tumor, circulating lymphocytes | NCT02977468 | Not available |
| 17 | Thyroid cancer | Urine exosomes | Active, not recruiting | Evaluation of new therapeutic mechanisms and medications for poorly differentiated or anaplastic thyroid cancer | NCT02862470 | Not available |
| 18 | Lung cancer | Blood samples | Recruiting | Drug efficacy, surgical effect evaluation, recurrence monitoring, prognosis judgment, molecular differentiation by analyzing blood ctDNA | NCT03317080 | 125 |
Technical challenges in exosome-based therapies
There are various technical obstacles in developing exosome-based therapies, including isolation techniques, characterization methods, and the standardization of clinically suitable exosome preparations. While conventional low-throughput techniques such as ultracentrifugation are widely used for exosome isolation, emerging techniques such as tangential flow filtration and size exclusion chromatography offer the potential to handle larger volumes of exosomes. Scaling up production for clinical applications involving bioreactors and media supplements, such as fetal bovine serum containing exosomes presents additional complexities and underscores the critical importance of rigorous characterization. Standardization is paramount, particularly in defining exosome function, determining effective doses, and unraveling exosome mechanisms of action.
Confronting the inherent heterogeneity of exosomes and refining isolation protocols to select vesicles with optimal functional activity is essential. The development of potency assays capable of gauging the effectiveness of specific exosome populations is a priority and will facilitate the regulatory approval process. Concerns regarding the loading of exogenous cargo into exosomes, which could interfere with endogenous cargo and give rise to off-target effects, especially in complex conditions such as cancer, have been noted. Identifying the safest cell source for exosome isolation to mitigate immunogenicity and unwanted cargo is critical and will require the use of appropriate preclinical models and meticulous cargo selection.
Efforts to target exosomes to specific sites for therapeutic intervention necessitate a deeper understanding of the delivery mechanisms. Balancing natural processing with payload targeting to minimize off-target effects is a formidable hurdle. While engineering exosomes to increase tropism for specific cells or organs has shown promise, the production of therapeutically relevant quantities of exosomes remains challenging. Recent advancements involving the incorporation of nanobody fragments or RNA aptamers on exosome surfaces have shown some degree of targeting specificity.
Moreover, exosome-based biomarkers, whether based on proteins, nucleic acids, lipids, or glycans, hold immense potential for addressing diseases such as cancer and neurological conditions. However, the translation of these discoveries into clinical applications requires technological innovations, particularly in the development of high-throughput exosome isolation methods and standardized assays. Cross-validation of exosome disease biomarkers across different laboratories and sample cohorts will expedite the transition from the laboratory to clinical practice.
Concluding remarks
Gaining a deeper understanding of the intricate machinery governing cargo selection and packaging during exosome biogenesis holds great promise for advancing our comprehension of intercellular signaling within the human body. This research has the potential to shed light on fundamental questions, including why distinct cell types produce unique exosome profiles and how external environmental conditions and the cellular state influence exosome composition. Moreover, considering the involvement of exosomes in numerous disease contexts, these insights offer promise for the development of targeted therapeutic interventions that precisely diagnose and regulate disease-associated exosomes.
Furthermore, elucidating the cargo selection and packaging processes is pivotal for harnessing exosomes as a versatile platform for therapeutic intervention. Researchers can engineer exosomes to transport specific cargoes, such as therapeutic drugs or genetic material, and direct them to specific cells or tissues. This precision is paramount for achieving highly desirable therapeutic outcomes, as it minimizes off-target effects and enhances treatment efficacy. Continued investigations into exosome biology and engineering hold the potential to significantly impact health care and enhance our capacity to address various diseases.
Acknowledgements
This work was supported by National Research Foundation of Korea grants funded by the Ministry of Education (2018R1A6A1A03025810, 2022R1I1A1A01068234, and 2022R1I1A4053049) and by the Ministry of Science and ICT (RS-2023-00211744 and 2023R1A2C2005342). The figures were created with BioRender.com.
Author contributions
Conceptualization: Y.J.L. and Y.C.C.; writing-original draft preparation: Y.J.L. and Y.C.C.; review and editing: Y.J.L., K.J.S., and Y.C.C.; visualization (figures): K.J.S. and Y.J.L.; visualization (tables): Y.J.L.; project administration and funding acquisition: Y.J.L., K.J.S., and Y.C.C.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yu Jin Lee, Email: yjlee0627@unist.ac.kr.
Young Chan Chae, Email: ychae@unist.ac.kr.
References
- 1.Harding C, Heuser J, Stahl P. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: demonstration of a pathway for receptor shedding. Eur. J. Cell Biol. 1984;35:256–263. [PubMed] [Google Scholar]
- 2.Shurtleff MJ, Temoche-Diaz MM, Schekman R. Extracellular vesicles and cancer: caveat lector. Annu. Rev. Cancer Biol. 2018;2:395–411. doi: 10.1146/annurev-cancerbio-030617-050519. [DOI] [Google Scholar]
- 3.Jeppesen DK, et al. Reassessment of exosome composition. Cell. 2019;177:428–445 e418. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamerkar S, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendt, M. et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight3, e99263 (2018). [DOI] [PMC free article] [PubMed]
- 7.Dixson AC, Dawson TR, Di Vizio D, Weaver AM. Context-specific regulation of extracellular vesicle biogenesis and cargo selection. Nat. Rev. Mol. Cell Biol. 2023;24:454–476. doi: 10.1038/s41580-023-00576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li SP, Lin ZX, Jiang XY, Yu XY. Exosomal cargo-loading and synthetic exosome-mimics as potential therapeutic tools. Acta Pharmacol. Sin. 2018;39:542–551. doi: 10.1038/aps.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurung S, Perocheau D, Touramanidou L, Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021;19:47. doi: 10.1186/s12964-021-00730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 12.Buzas EI. The roles of extracellular vesicles in the immune system. Nat. Rev. Immunol. 2023;23:236–250. doi: 10.1038/s41577-022-00763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma L, et al. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 2015;25:24–38. doi: 10.1038/cr.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020;21:585–606. doi: 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sansone P, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl Acad. Sci. 2017;114:E9066–E9075. doi: 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokoi A, et al. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019;5:eaax8849. doi: 10.1126/sciadv.aax8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Gruenberg J. Life in the lumen: the multivesicular endosome. Traffic. 2020;21:76–93. doi: 10.1111/tra.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sedgwick AE, D’Souza-Schorey C. The biology of extracellular microvesicles. Traffic. 2018;19:319–327. doi: 10.1111/tra.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juan T, Furthauer M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin. Cell Dev. Biol. 2018;74:66–77. doi: 10.1016/j.semcdb.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 21.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 22.Baietti MF, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 23.Colombo M, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 24.Coonrod EM, Stevens TH. The yeast vps class E mutants: the beginning of the molecular genetic analysis of multivesicular body biogenesis. Mol. Biol. Cell. 2010;21:4057–4060. doi: 10.1091/mbc.e09-07-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imjeti NS, et al. Syntenin mediates SRC function in exosomal cell-to-cell communication. Proc. Natl Acad. Sci. USA. 2017;114:12495–12500. doi: 10.1073/pnas.1713433114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghossoub R, et al. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014;5:3477. doi: 10.1038/ncomms4477. [DOI] [PubMed] [Google Scholar]
- 27.Ghoroghi, S. et al. Ral GTPases promote breast cancer metastasis by controlling biogenesis and organ targeting of exosomes. Elife10, e61539 (2021). [DOI] [PMC free article] [PubMed]
- 28.Matsuo H, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 29.Nkosi, D. et al. Epstein-Barr virus LMP1 promotes syntenin-1- and Hrs-induced extracellular vesicle formation for its own secretion to increase cell proliferation and migration. mBio11, e00589-20 (2020). [DOI] [PMC free article] [PubMed]
- 30.Fan SJ, et al. Glutamine deprivation alters the origin and function of cancer cell exosomes. EMBO J. 2020;39:e103009. doi: 10.15252/embj.2019103009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YJ, et al. GPR143 controls ESCRT-dependent exosome biogenesis and promotes cancer metastasis. Dev. Cell. 2023;58:320–334 e328. doi: 10.1016/j.devcel.2023.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Donoso-Quezada J, Ayala-Mar S, Gonzalez-Valdez J. The role of lipids in exosome biology and intercellular communication: function, analytics and applications. Traffic. 2021;22:204–220. doi: 10.1111/tra.12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trajkovic K, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 34.Knapp PE. Proteolipid protein: is it more than just a structural component of myelin? Dev. Neurosci. 1996;18:297–308. doi: 10.1159/000111420. [DOI] [PubMed] [Google Scholar]
- 35.Leidal AM, et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 2020;22:187–199. doi: 10.1038/s41556-019-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei D, et al. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021;31:157–177. doi: 10.1038/s41422-020-00409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009;19:434–446. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 38.van Niel G, et al. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell. 2011;21:708–721. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andreu Z, Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fordjour FK, Guo C, Ai Y, Daaboul GG, Gould SJ. A shared, stochastic pathway mediates exosome protein budding along plasma and endosome membranes. J. Biol. Chem. 2022;298:102394. doi: 10.1016/j.jbc.2022.102394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hurwitz SN, et al. Proteomic profiling of NCI-60 extracellular vesicles uncovers common protein cargo and cancer type-specific biomarkers. Oncotarget. 2016;7:86999–87015. doi: 10.18632/oncotarget.13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kugeratski FG, et al. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat. Cell Biol. 2021;23:631–641. doi: 10.1038/s41556-021-00693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoshino A, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–1061.e1018. doi: 10.1016/j.cell.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rieu S, Geminard C, Rabesandratana H, Sainte-Marie J, Vidal M. Exosomes released during reticulocyte maturation bind to fibronectin via integrin alpha4beta1. Eur. J. Biochem. 2000;267:583–590. doi: 10.1046/j.1432-1327.2000.01036.x. [DOI] [PubMed] [Google Scholar]
- 45.Segura E, et al. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood. 2005;106:216–223. doi: 10.1182/blood-2005-01-0220. [DOI] [PubMed] [Google Scholar]
- 46.Liang Y, et al. Complex N-linked glycans serve as a determinant for exosome/microvesicle cargo recruitment. J. Biol. Chem. 2014;289:32526–32537. doi: 10.1074/jbc.M114.606269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Escola JM, et al. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 48.Phuyal S, Hessvik NP, Skotland T, Sandvig K, Llorente A. Regulation of exosome release by glycosphingolipids and flotillins. FEBS J. 2014;281:2214–2227. doi: 10.1111/febs.12775. [DOI] [PubMed] [Google Scholar]
- 49.Arnold, P. et al. Joint reconstituted signaling of the IL-6 receptor via extracellular vesicles. Cells9, 1307 (2020). [DOI] [PMC free article] [PubMed]
- 50.Blanchard N, et al. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J. Immunol. 2002;168:3235–3241. doi: 10.4049/jimmunol.168.7.3235. [DOI] [PubMed] [Google Scholar]
- 51.Fu W, et al. CAR exosomes derived from effector CAR-T cells have potent antitumour effects and low toxicity. Nat. Commun. 2019;10:4355. doi: 10.1038/s41467-019-12321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nager AR, et al. An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell. 2017;168:252–263.e214. doi: 10.1016/j.cell.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen G, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shelke GV, et al. Endosomal signalling via exosome surface TGFbeta-1. J. Extracell. Vesicles. 2019;8:1650458. doi: 10.1080/20013078.2019.1650458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keller MD, et al. Decoy exosomes provide protection against bacterial toxins. Nature. 2020;579:260–264. doi: 10.1038/s41586-020-2066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diamandis EP, Plebani M. Glypican-1 as a highly sensitive and specific pancreatic cancer biomarker. Clin. Chem. Lab. Med. 2016;54:e1–e2. doi: 10.1515/cclm-2015-0773. [DOI] [PubMed] [Google Scholar]
- 57.Carnino JM, Ni K, Jin Y. Post-translational modification regulates formation and cargo-loading of extracellular vesicles. Front Immunol. 2020;11:948. doi: 10.3389/fimmu.2020.00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sork H, et al. Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci. Rep. 2018;8:10813. doi: 10.1038/s41598-018-28485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim, K. M., Abdelmohsen, K., Mustapic, M., Kapogiannis, D. & Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA810.1002/wrna.1413 (2017). [DOI] [PMC free article] [PubMed]
- 60.Das S, et al. The extracellular RNA communication consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell. 2019;177:231–242. doi: 10.1016/j.cell.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fabbiano F, et al. RNA packaging into extracellular vesicles: an orchestra of RNA-binding proteins? J. Extracell. Vesicles. 2020;10:e12043. doi: 10.1002/jev2.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brinkman K, et al. Extracellular vesicles from plasma have higher tumour RNA fraction than platelets. J. Extracell. Vesicles. 2020;9:1741176. doi: 10.1080/20013078.2020.1741176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Batagov AO, Kurochkin IV. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3’-untranslated regions. Biol. Direct. 2013;8:12. doi: 10.1186/1745-6150-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garcia-Martin R, et al. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature. 2022;601:446–451. doi: 10.1038/s41586-021-04234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mittelbrunn M, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stoorvogel W. Functional transfer of microRNA by exosomes. Blood. 2012;119:646–648. doi: 10.1182/blood-2011-11-389478. [DOI] [PubMed] [Google Scholar]
- 67.Gebauer F, Schwarzl T, Valcarcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 2021;22:185–198. doi: 10.1038/s41576-020-00302-y. [DOI] [PubMed] [Google Scholar]
- 68.Zhang H, et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer. 2020;19:43. doi: 10.1186/s12943-020-01168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Y, et al. Heterogeneous nuclear ribonucleoprotein A1 loads batched tumor-promoting MicroRNAs into small extracellular vesicles with the assist of caveolin-1 in A549 cells. Front Cell Dev. Biol. 2021;9:687912. doi: 10.3389/fcell.2021.687912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Villarroya-Beltri C, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen C, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J. Clin. Invest. 2020;130:404–421. doi: 10.1172/JCI130892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shurtleff MJ, et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl Acad. Sci. USA. 2017;114:E8987–E8995. doi: 10.1073/pnas.1712108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baglio SR, et al. Sensing of latent EBV infection through exosomal transfer of 5’pppRNA. Proc. Natl. Acad. Sci. USA. 2016;113:E587–E596. doi: 10.1073/pnas.1518130113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wozniak, A. L. et al. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J. Cell. Biol.219, e201912074 (2020). [DOI] [PMC free article] [PubMed]
- 75.Barman, B., Dawson, T. R. & Weaver, A. M. ER membrane contact sites: key platforms for biogenesis of RNA-containing extracellular vesicles. Contact5, 25152564221121444 (2022). [DOI] [PMC free article] [PubMed]
- 76.Thakur BK, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takahashi A, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017;8:15287. doi: 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cai J, Wu G, Jose PA, Zeng C. Functional transferred DNA within extracellular vesicles. Exp. Cell Res. 2016;349:179–183. doi: 10.1016/j.yexcr.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 79.Hagey DW, et al. Extracellular vesicles are the primary source of blood-borne tumour-derived mutant KRAS DNA early in pancreatic cancer. J. Extracell. Vesicles. 2021;10:e12142. doi: 10.1002/jev2.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vagner T, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles. 2018;7:1505403. doi: 10.1080/20013078.2018.1505403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Llorente A, et al. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta. 2013;1831:1302–1309. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 82.Skotland T, et al. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer. 2017;70:122–132. doi: 10.1016/j.ejca.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 83.Lydic TA, et al. Rapid and comprehensive ‘shotgun’ lipidome profiling of colorectal cancer cell derived exosomes. Methods. 2015;87:83–95. doi: 10.1016/j.ymeth.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haraszti RA, et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles. 2016;5:32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J. Control. Release. 2015;219:278–294. doi: 10.1016/j.jconrel.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 86.Teng F, Fussenegger M. Shedding light on extracellular vesicle biogenesis and bioengineering. Adv. Sci. 2020;8:2003505. doi: 10.1002/advs.202003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shimada Y, et al. Serum-derived exosomal PD-L1 expression to predict anti-PD-1 response and in patients with non-small cell lung cancer. Sci. Rep. 2021;11:7830. doi: 10.1038/s41598-021-87575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsumoto A, et al. Accelerated growth of B16BL6 tumor in mice through efficient uptake of their own exosomes by B16BL6 cells. Cancer Sci. 2017;108:1803–1810. doi: 10.1111/cas.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lobb RJ, Lima LG, Moller A. Exosomes: key mediators of metastasis and pre-metastatic niche formation. Semin. Cell Dev. Biol. 2017;67:3–10. doi: 10.1016/j.semcdb.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 90.Choi D, et al. The impact of oncogenic EGFRvIII on the proteome of extracellular vesicles released from glioblastoma cells. Mol. Cell. Proteomics. 2018;17:1948–1964. doi: 10.1074/mcp.RA118.000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jafari R, Rahbarghazi R, Ahmadi M, Hassanpour M, Rezaie J. Hypoxic exosomes orchestrate tumorigenesis: molecular mechanisms and therapeutic implications. J. Transl. Med. 2020;18:474. doi: 10.1186/s12967-020-02662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jo, H., Shim, K. & Jeoung, D. Exosomes: diagnostic and therapeutic implications in cancer. Pharmaceutics15, 1465 (2023). [DOI] [PMC free article] [PubMed]
- 93.Kosgodage, U. S., Trindade, R. P., Thompson, P. R., Inal, J. M. & Lange, S. Chloramidine/bisindolylmaleimide-I-mediated inhibition of exosome and microvesicle release and enhanced efficacy of cancer chemotherapy. Int. J. Mol. Sci.18, 1007 (2017). [DOI] [PMC free article] [PubMed]
- 94.Kulshreshtha A, et al. Simvastatin mediates inhibition of exosome synthesis, localization and secretion via multicomponent interventions. Sci. Rep. 2019;9:16373. doi: 10.1038/s41598-019-52765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Catalano M, O’Driscoll L. Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. J. Extracell. Vesicles. 2020;9:1703244. doi: 10.1080/20013078.2019.1703244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hanson PI, Cashikar A. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 2012;28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- 97.Datta A, et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: a drug repurposing strategy for advanced cancer. Sci. Rep. 2018;8:8161. doi: 10.1038/s41598-018-26411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Henkels KM, Muppani NR, Gomez-Cambronero J. PLD-specific small-molecule inhibitors decrease tumor-associated macrophages and neutrophils infiltration in breast tumors and lung and liver metastases. PLoS One. 2016;11:e0166553. doi: 10.1371/journal.pone.0166553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang F, et al. System Xc(-) inhibition blocks bone marrow-multiple myeloma exosomal crosstalk, thereby countering bortezomib resistance. Cancer Lett. 2022;535:215649. doi: 10.1016/j.canlet.2022.215649. [DOI] [PubMed] [Google Scholar]
- 100.Im EJ, et al. Sulfisoxazole inhibits the secretion of small extracellular vesicles by targeting the endothelin receptor A. Nat. Commun. 2019;10:1387. doi: 10.1038/s41467-019-09387-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee CH, et al. Macitentan improves antitumor immune responses by inhibiting the secretion of tumor-derived extracellular vesicle PD-L1. Theranostics. 2022;12:1971–1987. doi: 10.7150/thno.68864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rabas, N. et al. PINK1 drives production of mtDNA-containing extracellular vesicles to promote invasiveness. J. Cell. Biol.220, e202006049 (2021). [DOI] [PMC free article] [PubMed]
- 103.Cully M. Exosome-based candidates move into the clinic. Nat. Rev. Drug Discov. 2021;20:6–7. doi: 10.1038/d41573-020-00220-y. [DOI] [PubMed] [Google Scholar]
- 104.Wiklander, O. P. B., Brennan, M. A., Lotvall, J., Breakefield, X. O. & El Andaloussi, S. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med.11, eaav8521 (2019). [DOI] [PMC free article] [PubMed]
- 105.Silva AM, et al. Quantification of protein cargo loading into engineered extracellular vesicles at single-vesicle and single-molecule resolution. J. Extracell. Vesicles. 2021;10:e12130. doi: 10.1002/jev2.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Corso G, et al. Systematic characterization of extracellular vesicle sorting domains and quantification at the single molecule—single vesicle level by fluorescence correlation spectroscopy and single particle imaging. J. Extracell. Vesicles. 2019;8:1663043. doi: 10.1080/20013078.2019.1663043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yim N, et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat. Commun. 2016;7:12277. doi: 10.1038/ncomms12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gee P, et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat. Commun. 2020;11:1334. doi: 10.1038/s41467-020-14957-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ye Y, et al. An engineered exosome for delivering sgRNA:Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater. Sci. 2020;8:2966–2976. doi: 10.1039/D0BM00427H. [DOI] [PubMed] [Google Scholar]
- 110.Choi H, et al. Exosome-based delivery of super-repressor IkappaBalpha relieves sepsis-associated organ damage and mortality. Sci. Adv. 2020;6:eaaz6980. doi: 10.1126/sciadv.aaz6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sterzenbach U, et al. Engineered exosomes as vehicles for biologically active proteins. Mol. Ther. 2017;25:1269–1278. doi: 10.1016/j.ymthe.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 113.Reshke R, et al. Reduction of the therapeutic dose of silencing RNA by packaging it in extracellular vesicles via a pre-microRNA backbone. Nat. Biomed. Eng. 2020;4:52–68. doi: 10.1038/s41551-019-0502-4. [DOI] [PubMed] [Google Scholar]
- 114.Li Z, et al. In vitro and in vivo RNA inhibition by CD9-HuR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 2019;19:19–28. doi: 10.1021/acs.nanolett.8b02689. [DOI] [PubMed] [Google Scholar]
- 115.Kojima R, et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 2018;9:1305. doi: 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang S, et al. Selective encapsulation of therapeutic mRNA in engineered extracellular vesicles by DNA aptamer. Nano Lett. 2021;21:8563–8570. doi: 10.1021/acs.nanolett.1c01817. [DOI] [PubMed] [Google Scholar]
- 117.Pan Y, et al. A simple and sensitive method for exosome detection based on steric hindrance-controlled signal amplification. Chem. Commun. (Camb.) 2020;56:13768–13771. doi: 10.1039/D0CC06113A. [DOI] [PubMed] [Google Scholar]
- 118.Yang J, et al. Plasma-derived exosomal ALIX as a novel biomarker for diagnosis and classification of pancreatic cancer. Front Oncol. 2021;11:628346. doi: 10.3389/fonc.2021.628346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chanteloup G, et al. Monitoring HSP70 exosomes in cancer patients’ follow up: a clinical prospective pilot study. J. Extracell. Vesicles. 2020;9:1766192. doi: 10.1080/20013078.2020.1766192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McKiernan J, et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2016;2:882–889. doi: 10.1001/jamaoncol.2016.0097. [DOI] [PubMed] [Google Scholar]
- 121.Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bao Q, et al. Extracellular vesicle RNA sequencing reveals dramatic transcriptomic alterations between metastatic and primary osteosarcoma in a liquid biopsy approach. Ann. Surg. Oncol. 2018;25:2642–2651. doi: 10.1245/s10434-018-6642-z. [DOI] [PubMed] [Google Scholar]
- 123.Buscail, E. et al. Liquid biopsy approach for pancreatic ductal adenocarcinoma. Cancers (Basel)11, 10.3390/cancers11060852 (2019). [DOI] [PMC free article] [PubMed]
- 124.Tang W, Huang C, Tang C, Xu J, Wang H. Galectin-3 may serve as a potential marker for diagnosis and prognosis in papillary thyroid carcinoma: a meta-analysis. Onco Targets Ther. 2016;9:455–460,. doi: 10.2147/OTT.S94514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen K, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC) Clin. Cancer Res. 2019;25:7058–7067. doi: 10.1158/1078-0432.CCR-19-1213. [DOI] [PubMed] [Google Scholar]