Abstract
Aberrant accumulation of protein misfolding can cause aggregation and fibrillation and is one of the primary characteristic features of neurodegenerative diseases. Because they are disordered, misfolded, and aggregated proteins pose a significant setback in drug designing. The structural study of intermediate steps in these kinds of aggregated proteins will allow us to determine the conformational changes as well as the probable pathways encompassing various neurodegenerative disorders. The analysis of protein aggregates involved in neurodegenerative diseases relies on a diverse toolkit of biophysical techniques, encompassing both morphological and non-morphological methods. Additionally, Thioflavin T (ThT) assays and Circular Dichroism (CD) spectroscopy facilitate investigations into aggregation kinetics and secondary structure alterations. The collective application of these biophysical techniques empowers researchers to comprehensively unravel the intricate nature of protein aggregates associated with neurodegeneration. Furthermore, the topics covered in this review have summed up a handful of well-established techniques used for the structural analysis of protein aggregation. This multifaceted approach advances our fundamental understanding of the underlying mechanisms driving neurodegenerative diseases and informs potential therapeutic strategies.
Keywords: Neurodegenerative diseases, Protein aggregation, Structural analysis
1. Introduction
Aggregation is a major stumbling block in the yield and functionality of many protein products, including therapeutic proteins as well as pharmacological compounds. This eventually hampers their rapid commercialization and affects almost all stages of production viz., expression, purification, packaging, and distribution. Thereby, protein aggregation needs to be carefully controlled to ensure satisfactory product quality, according to the WHO Guidelines for the Management of Protein Aggregation in Pharmaceuticals. Therefore, a deeper comprehension to understand the mechanisms underlying protein aggregation, at the fundamental level, in order to optimize the primary sequence or the operative parameters of the concerned protein, is required [1].
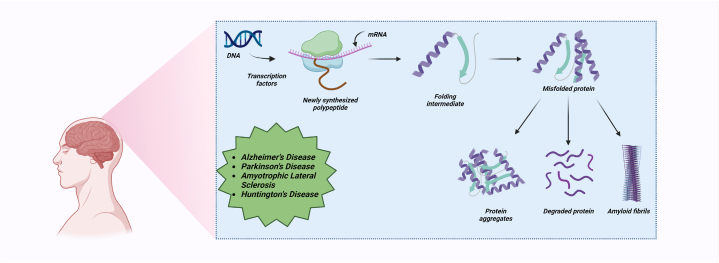
Neurological diseases are the primary contributor to both physical and cognitive impairment on a global scale, with a current prevalence rate of roughly 15% among the global population [2]. The total number of patients has seen a significant increase over the course of the last three decades [3]. Recent research suggests that the global prevalence of epilepsy exceeds 50 million individuals, whereas dementia affects around 47.5 million individuals [4]. Moreover, according to the findings of the Global Burden of Disease research, it has been reported that in the year 2019, there was a significant occurrence of major neurological diseases, resulting in a total of 10.06 million deaths and 349.22 million DALYs (disability-adjusted life years) [5]. These protein aggregates are typically composed of β-sheet conformation resulting in amyloid misfolding that is scattered throughout the brain. Protein aggregates are produced either because of the misfolding of a particular protein or the random misfolding of endogenic proteins that act as seeds for aggregation (Fig. 1). The key factors responsible for the formation of proteinaceous aggregates include hydrophobicity, low net charge, as well as stability [6]. We further advocate illustrating the conformation i.e., size or shape of the aggregate, as well as potential structural alterations to the protein species so as to get a better understanding of the aggregation of proteins.
Fig. 1.
Schematic representation of the fate of proteins involved in protein aggregation.
The protein folding mechanism includes diverse categories of interaction like van der Waals interaction, H-bonding, hydrophobic interactions, and electrostatic interactions, that are essential for maintaining the structural integrity of proteins. Furthermore, research suggests that H-bonds play a fundamental role in both protein folding and unfolding as well [[7], [8], [9]]. Despite the fact that cells have mechanisms to govern the integrity of their proteins, proteins frequently misfold as a result of dominant-negative mutations, trafficking errors, the loss of binding partners, errors in post-translational regulation, ROS damage, and fluctuations in environmental conditions like (pH, temperature, protein concentration etc.) [10,11]. The natively folded/unfolded monomeric protein type slowly transforms into a partially folded intermediate and then converts to soluble oligomers and protofibrils. Eventually, these oligomers form beta-rich fibrils with typical morphology of amyloid as observed under an electron microscope (Fig. 2) [12].So, for proper cell functioning, protein folding into its correct native state is essential as misfolding can lead to aggregation further resulting in many types of diseases [10,13].
Fig. 2.
Schematic description of the aggregation of amyloids [12].
2. Techniques employed for the analysis of aggregates
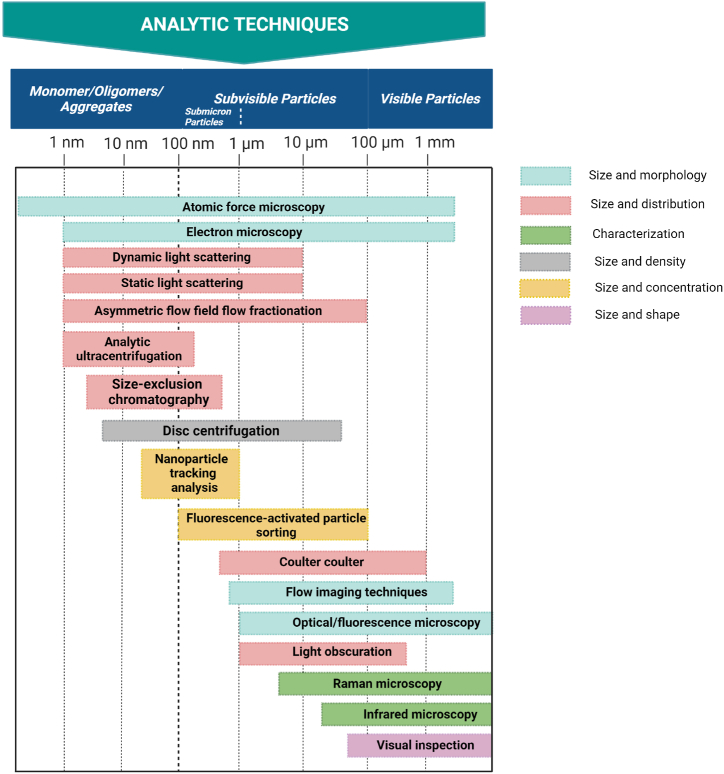
There are two types of particles that can be used to classify insoluble aggregates: the first one being the subvisible particles (SVPs), and the other one, visible particles (VPs). Methods such as spectroscopy, flow cytometry, coulter counter, fluorescence microscopy, as well as data acquisition, have all been utilized on a consistent basis in the process of characterizing these particles (Table 1& 2). In this context, the term "aggregation" pertains to soluble aggregates, whilst the term "particulates" is intended to refer to both subvisible particles (100 nm–100 m) and macroscopic insoluble aggregates that emerge as visible particles (>100 m) (Fig. 3). The European Immunogenicity Platform (EIP) has incorporated the Protein Characterization Subcommittee (EIP-PCS) to converse about the existing type of methodologies for analyzing protein aggregates [14]. Seeing that aggregation is a very complicated process a single technique is not sufficient to describe the complete assembly process (see Table 2).
Table 1.
Investigative strategies used to contemplate protein structure, folding, and aggregation.
| TECHNIQUES | PRINCIPLE | SIZES | OBSERVABLE |
|---|---|---|---|
| Atomic force microscopy | Topographical scanning | 0.01 nm | Molecular resolution |
| Size exclusion chromatography | Separation through porous matrix based on size. | 5–50 nm | hydrodynamic scattering |
| Analytical ultracentrifugation | Sedimentation rate in response to centrifugal force. | 1–100 nm | Molecular weight and confirmation |
| Field flow fractionation | Separation by flow retention based on the diffusion coefficient | 1–1000 nm | Hydrodynamic diameter |
| Dynamic light scattering | The fluctuation of scattered light signals | 0.5–10 μm | Hydrodynamic diameter |
| Mass spectroscopy | Detection of mass/charge ratio of ionized molecule | Atomic resolution Dalton | Mass/charge ratio |
| Optical microscopy | Visualization of protein particles | 1 μm-mm | Size and morphology |
| Electron microscopy | Visualization of protein particles and detection of chemicals at high resolution | nm-mm | Size and morphology |
Fig. 3.
An overview of an analytical technique that can be used for analysis of aggregates, subvisible (SVP) and visible particles (VPS). Insoluble aggregates can be categorized as subvisible particles (SVP) and visible particles (VPS).
Table 2.
| TECHNIQUES | DESCRIPTION | ADVANTAGES | DISADVANTAGES |
|---|---|---|---|
| UV–Vis Absorption Spectroscopy | Measures the absorbance of light by molecules, detecting changes in absorbance due to protein aggregation | Rapid and widely accessible. | Limited to detecting changes in absorbance only. |
| Fluorescence Spectroscopy | Measures the emission of fluorescence light by fluorophores within proteins, detecting changes in fluorescence due to aggregation. | Sensitive and applicable to a wide range of proteins. | Requires suitable fluorescent probes. |
| Absorbance Spectroscopy (e.g., FTIR) | Measures the absorbance of infrared or other electromagnetic radiation by molecular vibrations, revealing changes in protein structure associated with aggregation. | Provides structural information about aggregates. | Requires expertise in spectral interpretation. |
| Circular Dichroism (CD) Spectroscopy | Measures changes in protein secondary and tertiary structure caused by aggregation, providing insights into conformational changes. | Provides information on protein conformation. | Cannot directly visualize aggregates. |
| Thioflavin T (ThT) Fluorescence Assay | Detects amyloid-like fibril formation in proteins by binding to β-sheet structures, indicating amyloid aggregation. | Highly specific for amyloid aggregates. | Limited to certain types of protein aggregates. |
| Fluorescence Resonance Energy Transfer (FRET) | Measures energy transfer between fluorophores attached to proteins, detecting changes in FRET signal due to aggregation. | Provides insights into proximity and interactions. | Requires suitable labeling and expertise. |
| Nuclear Magnetic Resonance (NMR) Spectroscopy | Probes changes in protein conformation and dynamics associated with aggregation, offering high structural resolution. | High structural resolution. | Limited to small to moderately sized proteins. |
Moreover, for analyzing protein aggregates, even though there is a long list of techniques, we have made an effort to be as comprehensive as possible and to explain the most popular analytical approaches. Various analytical techniques can be used to investigate protein aggregates within the micrometer (μm) range and (nm) range. The most esteemed of these methods include.
2.1. Direct non-morphological methods
2.1.1. Absorption spectroscopy
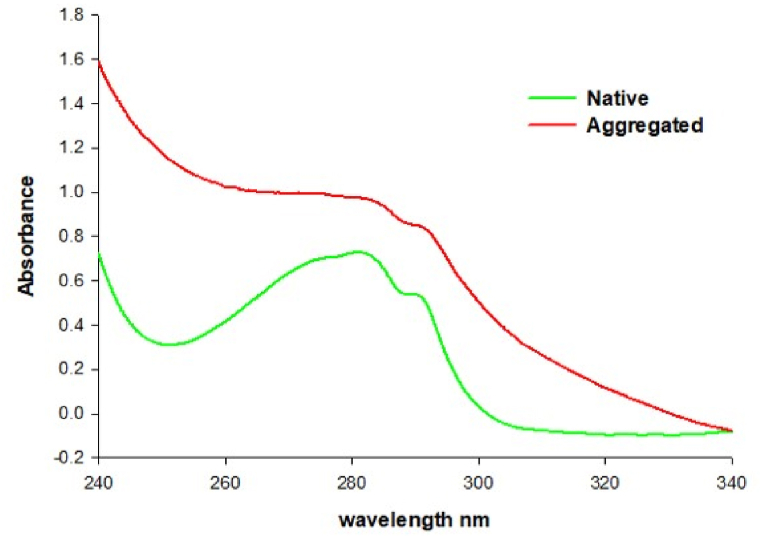
Absorption spectroscopy is amongst the most prevalent and commonly used technique for estimating the concentration of protein in a solution and detecting alterations in the tertiary structure of a protein [17]. Tryptophan (W), tyrosine (Y), and phenylalanine (F) are the three aromatic amino acids, found in a protein, detects a well-demarcated UV absorption peak in the region of 240–320 nm [18]. The principle of the instrument is based on the absorption of light by protein at particular wavelengths, and by using precise formulas, the quantity of light that is absorbed can be determined by measuring the imitated light after passing through the solution, aiding in the quantitative estimation of protein. The Monochromatic light is set at a range or specified wavelength; however, scattering has been noted in our study on aggregates [7]. Different absorption spectra are obtained for a native protein and the aggregated one (Fig. 4).
Fig. 4.
UV absorption spectra of native and aggregated protein.
The reduction of light produced by an absorption spectrophotometer has a significant contribution to the capacity of the instrument to detect reflectance [19]. If the protein of interest contains aromatic residues and the absorbance is monitored over a broad range (at least 240–350 nm), then it is possible to obtain an Ultraviolet light absorption spectrum with resolution up to 0.01 nm using spectrophotometers that contain diode arrays provided that the protein of interest contains aromatic residues. This range of values is accurate enough to make it possible for the resultant analysis to separate the peaks of the spectrum into individual components [18]. At this subsequent step, the energy of photons is used up by the electrons of the atoms that make up the matter. The reduced light that passes through the sample is analyzed via optical methods. Spectroscopy techniques are applied in accordance with the energy of absorbed photons, which may correspond to ultraviolet, visible, or infrared absorption [20,21]. Absorption spectroscopy is a widely employed analytical method that offers both advantages and disadvantages. On the positive side, it provides quantitative insights into the concentration of substances in a sample, making it invaluable for determining the presence and number of various analytes. Its high sensitivity allows for the detection of even trace amounts of compounds, making it suitable for applications ranging from environmental monitoring to pharmaceutical quality control. Its versatility spans across different types of analytes, from gases to liquids and solids. Moreover, absorption spectroscopy is often non-destructive, preserving the integrity of precious or irreplaceable samples. Additionally, its high selectivity, stemming from the uniqueness of absorption spectra for each molecule or compound, aids in the identification and characterization of complex mixtures. On the flip side, absorption spectroscopy has limitations such as its inability to provide detailed structural information, susceptibility to sample interference, and the need for complex instrumentation for certain applications. Researchers must weigh these pros and cons when selecting absorption spectroscopy as their analytical tool of choice.
2.1.2. Intrinsic fluorescence spectroscopy
Because of its resilience, adequate sensitivity, and no invasiveness, fluorescence spectroscopy has emerged as an important technique in biochemical research [22]. It operates through the integration of a UV light supply and a remarkable UV optical device in a precision microscope equipped with a highly sensitive spectrophotometer. Tryptophan, an amino acid found in proteins, has intrinsic fluorescence properties due to its aromatic structure. When exposed to UV light, tryptophan molecules absorb photons and transition to an excited state, emitting fluorescence in the 300–350 nm range. This natural fluorescence is used in intrinsic fluorescence spectroscopy to understand structural and environmental changes within proteins, particularly when they are in an aggregated state. Furthermore, tryptophan fluorescence quenching is a crucial parameter for understanding the microenvironment within protein aggregates. The degree of quenching can be influenced by factors such as interactions between tryptophan residues and nearby molecules or the aggregation process itself. Quenching studies help researchers assess the compactness and structural changes within protein aggregates [23]. Micro-spectroscopy provides additional information on the optic topographies as well as the structural properties of the protein. The spectroscopic data of the protein's intrinsic fluorescence can be collected via excitation of protein at 280 nm and emission at 300 nm in order to capture photons to the nearby IR-region with the help of a spectrophotometer. Moreover, it is useful in analyzing aggregation in proteins containing aromatic amino acids particularly tryptophan residues (Fig. 5). Furthermore, this technique is sensitive enough to detect early stages of protein aggregation, even before visible aggregates form. This can be crucial for understanding the kinetics of aggregation and identifying potential aggregation-prone regions of the protein.
Fig. 5.
Intrinsic Fluorescence spectra of protein in native and aggregated state.
Furthermore, in order to specifically excite the protein's tryptophan (Trp) residues for intrinsic spectrophotometric analysis, the sample is stimulated at a wavelength of 280 nm. The tryptophan spectrum of fluorescence is much dependent on its surrounding conditions. Generally, a blue shift and red shift detect the folding and unfolding patterns of proteins in polar to non-polar and vice versa, reducing solvent polarity [22]. Trp is the most prevalent of the three fluorescence amino acid residues that are found in proteins. It is found in cytoplasmic proteins at a concentration of around 1 M and in membrane proteins at values of about 3 M, in an insoluble state. Because of the poor absorptivity of Phe as well as its low quantum yield, the consequence of this amino acid on the intrinsic fluorescence of proteins is almost negligible [24]. Despite the fact that tyrosine (Tyr) has a comparable optical absorption to that of Trp [25,26]. Likewise, the energy emitted by Tyr is presumed to be quenched, either due to transferring photons to Tyr or perhaps through interaction of Trp with the peptide chain of the protein [10]. Moreover, tryptophan seems to have a sensitivity to collisional quenching that is unmatched by any other amino acid. This peculiar sensitivity is likely caused by the tendency of excited-state indole to transfer electrons. Tryptophan may be inhibited either by quenchers that are introduced to the protein from the outside or by adjacent groups within the protein itself. The utilization of emission spectrum, anisotropy, and quenched tryptophan residues in proteins is used to investigate the structures and functions of proteins that have been the primary objective of many studies and publications [27,28].
Intrinsic fluorescence can also be used to study the binding interactions between proteins and potential aggregation inhibitors or chaperone molecules. Changes in fluorescence can indicate alterations in the binding affinity or mode of interaction [29]. Anisotropy measurements in intrinsic fluorescence spectroscopy investigate the rotational mobility of tryptophan residues within aggregated proteins [30]. Increased anisotropy may indicate reduced rotational motion due to aggregation. The emission spectrum of tryptophan fluorescence in aggregated proteins serves as a critical indicator of the aggregate's structural properties and the microenvironment surrounding tryptophan residues. Shifts or alterations in the emission spectrum can reveal changes in the local structure, solvent exposure, or polarity within the aggregate, often associated with protein aggregation processes. Nevertheless, the most effective method for separating proteins from salt solution is visualization via intrinsic fluorescence [31]. It offers several advantages and disadvantages. On the positive side, intrinsic fluorescence spectroscopy provides valuable insights into the structural and conformational properties of molecules and proteins. This technique leverages the natural fluorescence exhibited by certain biomolecules, such as aromatic amino acids (tryptophan, tyrosine, and phenylalanine) and coenzymes (NADH and FAD), making it particularly useful in biochemistry and biophysics. It can reveal information about the microenvironment of these fluorophores, including changes in protein folding, binding interactions, and conformational alterations. This intrinsic property eliminates the need for chemical modifications or labels, preserving the sample's native state and reducing potential artifacts. Furthermore, intrinsic fluorescence spectroscopy is non-destructive and can be performed in real-time, enabling dynamic studies of biological processes. However, intrinsic fluorescence spectroscopy also comes with certain limitations. One major drawback is its lack of specificity. While it can provide valuable structural information, it may not distinguish between different molecules with similar fluorophores, limiting its applicability in complex mixtures. Additionally, the technique's sensitivity can be influenced by environmental factors, such as pH, temperature, and ionic strength, which may impact the accuracy and reproducibility of measurements. The instrumentation required for fluorescence spectroscopy can be relatively complex and costly, and sample turbidity or impurities can interfere with measurements. Intrinsic fluorescence spectroscopy is most effective when combined with complementary techniques, such as circular dichroism or mass spectrometry, to obtain a more comprehensive understanding of molecular structure and behavior.
2.1.3. Fourier transform infrared spectroscopy (FTIR)
The use of Fourier transform infrared spectroscopy (FTIR) and micro-spectroscopy has significantly enhanced our understanding of the underlying processes involved in protein misfolding and aggregation in several neurodegenerative disorders, such as Parkinson's disease, Alzheimer's disease, Huntington's disease, amyotrophic lateral sclerosis, and prion diseases. The Carbon–Oxygen (double bond), Carbon–Nitrogen (triple bond) stretching vibration, and Nitrogen–Hydrogen bending, correlate with the FTIR Amide-I (approx. 1650 cm−1) and Amide-II (approx. 1540 cm−1) bands [32]. The amide-I bands correspond to alpha-helices, beta-sheets, turns, and unordered conformations, while aggregated proteins have a frequency of roughly ∼1620 to 1625 cm−1 owing to the hydrophobic environment [33]. Nilsson et al. have provided a wealth of detailed documentation about the interpretation of FTIR data [34]. The analysis of the primary sequence of a peptide is the first and foremost stage in the process of deciphering an FTIR spectrum. The monomeric form of sequences that are abundant in glutamine (Q) and asparagine (N) is inherently disordered, and these sequences have the potential to aggregate forming amyloids [35]. Amyloids are fibrillar protein aggregates abundant in beta-sheet structures that could self-propagate through a conformational change in the chain of reactions. Because of the high concentration of the aforementioned amino acids, the IR vibrations of the amyloid fibrils overlap with those of the amide I band. Further, the curve fitting is done, and the probable secondary structural changes associated with protein aggregation are determined using the second derivative [1].
Furthermore, the investigation of protein misfolding and aggregation via the use of Fourier-transform infrared spectroscopy (FTIR) extends beyond in vitro experimentation, as it may also be applied to intracellular and tissue-based analyses. Protein aggregation studies may be challenging due to the tiny size of protein aggregates, which initially exist as oligomers in the nanoscale range but can grow to enormous sizes in the micromolar range. The spectral differences are minimal, posing a challenge that necessitates the acquisition of high signal spectra in relation to the background. The synchrotron, equipped with an infrared source, offers a high level of brightness, making it well-suited for addressing the aforementioned challenge. Thereby, this technique has been utilized to investigate the process of aggregation in diseased tissues of individuals afflicted with Scrapie's disease, Huntington's disease (HD) [36] Alzheimer's disease (AD) [33], and Parkinson's disease (PD) [37]]. Nevertheless, recent advancements in established methodologies, such as in vivo imaging technology, have the potential to enhance our understanding of protein aggregation processes and treatment approaches. Fourier-transform infrared spectroscopy (FTIR) is a powerful analytical technique that offers numerous advantages while having some limitations. One of its primary advantages is its ability to provide valuable insights into the chemical composition and molecular structure of a wide range of substances. FTIR works by measuring the absorption and transmission of infrared light as it interacts with molecules, allowing for the identification of functional groups, chemical bonds, and even quantitative analysis of components within a sample. In recent times, advancements in scattering-type near-field scanning optical microscopy (s-SNOM) have facilitated the capture of infrared (IR) spectral data at scales below a micron [38]. The underlying principle of this imaging approach involves utilizing an atomic force microscope (AFM) tip as an optical antenna, or 'near-field probe,' to concentrate incident electromagnetic radiation into regions smaller than the wavelength of the incoming light [39]. Through the integration of a wideband mid-infrared (MIR) laser and a spectroscopic interferometer into an s-SNOM setup, researchers have successfully demonstrated nanoFTIR, achieving IR nano-spectroscopy with a resolution of 20 nm [40]. Researchers have harnessed this technique to explore both morphological features and the unique protein signatures associated with these aggregates. By examining the spatial distribution of protein aggregates at the nanoscale, scientists can gain insights into their shape, size, and arrangement. This information is crucial for understanding the underlying mechanisms of aggregation [41]. Moreover, it allows researchers to visualize the topography of protein aggregates, revealing details such as fibril structures, amorphous clusters, and their interactions with surrounding molecules. Each protein has a unique infrared absorption spectrum associated with its chemical bonds. When proteins aggregate, their spectral characteristics change. Nanoscale infrared spectroscopy enables the identification of specific vibrational modes related to protein secondary structures (such as α-helices, β-sheets, and random coils) [42]; thereby, plays a pivotal role in characterizing structural heterogeneity within individual amyloid fibrils [43] and prefibrillar aggregates [44]. Through the analysis of aggregate secondary structure, nanoIR offers insights into morphological characteristics and distinctive spectral signatures, specifically enabling differentiation between the β-rich core and α-rich regions within aggregates [44].
However, FTIR has limitations, including the potential for interference from water vapor and other atmospheric gases, which may require specialized sample handling or instrument configurations. Additionally, it may not provide detailed structural information, necessitating complementary techniques for a more comprehensive analysis. Nevertheless, FTIR remains a valuable tool for its versatility and ability to reveal crucial information about the chemical nature of substances.
2.1.4. Circular dichroism (CD)
Circular Dichroism is a prominent and effective technique for monitoring changes in the secondary and tertiary structure of proteins, such as aggregation, unfolding caused by heat or chemical, as well as different types of interactions associated with it. The use of this technology offers several advantages, such as the capacity to examine the structural attributes of proteins in a liquid medium, accurately simulating physiological conditions, using a little protein amount, and achieving prompt outcomes. Additionally, the data acquisition process is expedited. CD spectroscopy has been used to assess intricate processes like collagen fibrillogenesis [45].
The term "circular dichroism" refers to the discrepancy in absorption between left- and right-handed circularly polarised light, which may vary depending on the wavelength. CD measures the intra- or intermolecular asymmetry, also known as helicity, inside the molecular structure [46]. This property allows CD to effectively identify samples that are randomly orientated. CD signals may arise from many chemical systems with varying levels of complexity, resulting in distinct bands that have diverse physical sources. In the fundamental scenario, CD develops from inherent asymmetry or the asymmetric disruption of a molecule [47].
CD is used to evaluate the 2D structure of a protein, dynamics of protein folding and misfolding, and binding characteristics of proteins, in a very short amount of time. When the amide bond of the polypeptide chain is in alignment with the protein backbone, the light falling on the molecule causes the photons to undergo a transformation or split into several transitions due to “excitation”. In the context of complex molecules or tiny aggregates, CD often arises from the influence of short-range, excitonic interactions among chromophores. Chiral molecules such as DNA agglomerates, compacted chromatins, and proteins, exhibit very pronounced circular dichroism (CD) signals characterized by non-conservative, atypical band shapes and irregular graphs beyond the absorbance region. Systems that are organized hierarchically, such as thylakoid membranes or lamellar clusters of LHC II [48], exhibit the presence of three distinct forms of signals that are overlaid onto one another [46]. As a result, numerous structural components exhibit distinctive CD spectra. Proteins containing antiparallel β-sheets (β-helices) exhibit negative bands at a 218 nm wavelength and positive bands at 195 nm wavelength. Circular Dichroism is also used for screening conformational changes caused by temperature, mutation, radiation, denaturants, or binding interactions in addition to providing an estimation of the unknown structure of the proteins [24]. The far-UV CD within the range of 190 nm–250 nm gives valuable conformational data of amyloidal proteins utilizing the algorithm of g-factor calculation, a verifiable factor that is influenced by merely molecular interactions rather than the path length of light [27].
CD spectroscopy has resulted in significant developments in the realm of protein aggregation studies. Vibrational Circular Dichroism (VCD) is another advancement of Circular Dichroism (CD) that serves as a valuable tool for investigating the chirality shown by amyloid fibrils [49]. The use of VCD may also be employed for the elucidation of the structure of huge macromolecules. The development of amyloid fibrils is often associated with increased vibrational circular dichroism (VCD) spectra and a downward shift of the amide band. The study conducted by Measey et al. demonstrates that the observed increase in VCD intensity may be mostly attributed to interactions between different sheets, rather than interactions within the same sheet [50].
CD allows examination of amyloid-like structure in real-time, using a limited number of samples and within short duration. This methodology enables the concurrent examination of turbidity as well as scattering of light amid the process of thermal-induced folding and unfolding, hence simplifying investigations into protein aggregation [51]. The use of circular dichroism (CD) theory in conjunction with Vacuum-ultraviolet circular dichroism (VUVCD) spectroscopy has been employed in the investigation of the intermolecular architectures of fundamental regions of β2-microglobulin (β2m) found in amyloid fibrils. The research findings indicate that both the backbone and side chain components have an impact on the structural characteristics of amyloid fibrils [52].
Synchrotron radiation circular dichroism (SRCD) spectroscopy facilitates protein folding investigations via small-angle X-ray scattering and IR spectroscopy. SRCD offers advantages over standard CD, notably a high signal-to-noise ratio, quick data retrieval, time-resolved assessments, and high-throughput screening rates [53]. A SRCD spectroscopy examinations on α-synuclein revealed that lower concentrations of trehalose suppresses aggregation and promotes correct unfolding [54].
Protein misfolding and fibrillation incited by high temperatures often lead to the development of an amyloid/fibril-like structure. This fibrillar structure arrangement shall undoubtedly be examined in real-time with fewer test measures in a very brief timeframe, utilizing CD. Since both the turbidity and the light dissipated at 90° during the thermal folding/unfolding transition can be investigated simultaneously using this technique, it is well suited for research on fibrillation and aggregation of proteins (Fig. 6) [51]. CD spectroscopy's prevalence in aggregation research has grown with a diverse range of advancements and breakthroughs throughout the course of its history.
Fig. 6.
Circular dichroism measurements of aggregated protein showing prevalent beta sheet.
2.1.5. Nuclear magnetic resonance
The use of nuclear magnetic resonance (NMR) in the investigation of aggregation and formulation design has shown to be of great importance across several disciplines. This includes the examination of protein aggregation processes, the analysis of amyloid fibril aggregates, and the advancement of therapeutic proteins. The use of heteronuclear spin relaxation rates has been employed in the determination of weak association constants for the transitory production of oligomers of bovine -low molecular weight -protein tyrosine phosphatase (BPTP) under circumstances of equilibrium [55]. Nevertheless, the detection of soluble oligomers in irreversible aggregation mechanisms poses a greater challenge of detection by NMR due to the formation of transient pseudo-equilibrium intermediates.
Considerable research efforts using nuclear magnetic resonance (NMR) spectroscopy have been directed towards the investigation of amyloid fibrils which exhibit substantial involvement in the pathogenesis of several neurodegenerative disorders, including Alzheimer's, Parkinson's, and other diseases. The process of α-synuclein (α-Syn) fibril production, which is involved in the development of Parkinson's disease, has been investigated via the use of Paramagnetic Relaxation Enhancement (PRE) Nuclear Magnetic Resonance (NMR) spectroscopy. This technique enables the visualization and characterization of the many interactions occurring among the structurally heterogeneous and disordered monomers involved in this process. The PRE-NMR technique entails the attachment of a nitroxide spin-label to a specific area of a protein. In its oxidized form, this spin-label exhibits paramagnetic properties, which enhances relaxation during the 1H–15 N heteronuclear single quantum coherence (HSQC) experiment [56]. Nuclear magnetic resonance (NMR) characteristics, namely the longitudinal and the transverse relaxation rates, (R1) and (R2), respectively, are of significant importance in the investigation of protein aggregation phenomena.
The targeted binding of the labeled paramagnetic spin to proteins increases the transverse relaxation rates of neighboring nuclei atoms that may be employed for the identification of spatial constraints [[57], [58], [59]].The interchange dynamics between polydisperse, NMR-invisible (or "dark") protofibrils and amyloid (A) monomers were studied using an innovative technique dubbed dark-state exchange saturation transfer (DEST). This structural and kinetic investigation of protofibril formation was very important because the accumulation of toxic, soluble clumping forms of Amyloid-beta produces bigger assemblies that lead to the development of Alzheimer's disease [60].
Under situations of stress, it is possible for the human immunoglobulin k-IV light chain variable domain (LEN) to form amyloids. The identification of residues experiencing slow millisecond movements was achieved by the use of CPMG relaxation NMR studies [61,62], whereas multidimensional solution NMR investigations were conducted at physiological and acidic pH levels. The study found that flexible residues at the dimer interface might cause partially misfolded conformers, potentially forming stable tertiary and quaternary structures that can prevent aggregation [63,64].
Nuclear magnetic resonance (NMR) spectroscopy takes into account the structural characteristics of therapeutic proteins, although fails to account for the existence of a solvent, often water. The solvent is a crucial factor in modulating protein dynamics. The proposal introduces a novel technique known as water proton nuclear magnetic resonance (NMR) for the purpose of quantifying protein aggregation by using the transverse relaxation (T2) time of water protons [64]. In another study, a combined methodology was used that included DOSY-NMR (diffusion-ordered spectroscopy nuclear magnetic resonance) and DLS (dynamic light scattering) to assess the diffusion coefficients and particle size distributions of five insulin medication formulations that are commercially available. The scientists disclosed that DLS exhibited superior efficacy in the detection of bigger aggregates compared to DOSY-NMR as a result of its heightened sensitivity towards high molecular weight entities. On the other hand, it was observed that DOSY-NMR is more suited for elucidating the behaviour of excipients in the formulation. The results presented in this work do not decrease the potential of Nuclear Magnetic Resonance (NMR) as a valuable tool for investigating aggregates. Instead, they underscore the need to carefully choose the proper NMR approach, taking into account existing knowledge about the system under investigation [60,65].
An additional method used for investigating supramolecular systems is solid-state nuclear magnetic resonance (ssNMR). This technique facilitates the acquisition of structural and dynamic insights into intricate biological systems, particularly in the context of protein aggregation and fibrils, such as those prevalent in amyloid formations [[66], [67], [68], [69]].
In summary, nuclear magnetic resonance (NMR) plays a significant role in facilitating the elucidation of novel mechanisms to combat aggregation, which serves as the fundamental cause of several neurodegenerative disorders [70,71].
2.1.6. Fluorescence resonance energy transfer (FRET)
FRET is a useful approach for examining the kinetic and structural characteristics of protein aggregates and their interactions. FRET occurs when two fluorophores, a donor and an acceptor, are within a certain range of distances (typically a few to ∼10 nm). A light source excites the donor fluorophore, which then transfers its energy to the acceptor fluorophore and causes the acceptor to emit fluorescence. FRET can detect changes at a nanoscale scale because the efficiency of energy transfer is inversely related to the sixth power of the distance between the donor and acceptor. Distance Measurements, Conformational Changes, Aggregation Monitoring, Cellular Imaging, and Protein-Protein Interactions are some of the uses of FRET in protein aggregates [72,73]. Real-time and dynamic insights into protein aggregation formation, conformational changes, and interactions are provided by FRET and other fluorescence-based approaches. They may be used with other approaches to provide a thorough knowledge of aggregate structures and behavior. They provide useful information for comprehending the mechanics of aggregation. One significant advantage of FRET is its ability to provide real-time, quantitative information about molecular interactions and structural dynamics within living cells and biological macromolecules. It is commonly employed in cell biology, structural biology, and drug discovery to investigate protein-protein interactions, DNA-protein binding, and conformational changes in biomolecules [73].
However, FRET also has limitations. It requires the use of fluorescent labels, which may affect the native behavior of the molecules under study. Careful selection of fluorophores and controls is essential to minimize artifacts. FRET is highly dependent on the fluorophore's photophysical properties, and photobleaching or phototoxicity can be issues in long-term experiments. Additionally, FRET data interpretation can be complex and requires precise calibration and control experiments to ensure accuracy.
2.1.7. Super-resolution microscopy
Super-resolution microscopy techniques, such as stimulated emission depletion (STED) microscopy and structured illumination microscopy (SIM), have transformed the field of cellular imaging by exceeding the diffraction limit of conventional light microscopy [74,75]. Utilizing the phenomena of stimulated emission, STED microscopy is a super-resolution method that enhances spatial resolution. The point spread function is narrowed using STED microscopy, which concentrates a laser beam to eliminate fluorescence emission from the outside areas of the excitation spot. It enables better clarity and resolution for the visualization of protein aggregates, exposing previously hidden features. This method is particularly helpful for examining how proteins are arranged and organized inside aggregates as well as for describing the shape and size of the aggregates [76,77]. STED microscopy can shed light on the interactions and spatial arrangements of various proteins within aggregates. One of the primary advantages of super-resolution microscopy is its ability to surpass the classical diffraction limit of light microscopy, enabling the observation of structures as small as a few nanometers. Techniques like STED (stimulated emission depletion), SIM (structured illumination), and PALM/STORM (photoactivated localization microscopy/stochastic optical reconstruction microscopy) have ushered in a new era of imaging by revealing fine subcellular structures, individual molecules, and their interactions [78]. Super-resolution microscopy is invaluable in cell biology and neuroscience for studying cellular organelles, protein distributions, and neuronal synapses [79,80]. Super-resolution microscopy is non-invasive and can be used with living cells and tissues, allowing dynamic, real-time observations of biological processes. Moreover, it provides detailed spatial information, enabling scientists to gain a deeper understanding of biological mechanisms at the nanoscale. However, super-resolution microscopy also has limitations. It typically requires specialized equipment and expertise, making it less accessible than conventional microscopy techniques. Sample preparation can be complex, and fluorescent labeling may interfere with biological processes. Furthermore, the extended acquisition times required for super-resolution imaging may introduce photobleaching and phototoxicity issues, especially when imaging live cells [81].
SIM is another super-resolution method which increases resolution by employing structured illumination patterns to record high-frequency data that is above the diffraction threshold [82]. To extract finer information from the sample, the light pattern is rotated and shifted. SIM is a good tool for cellular context-specific protein aggregation visualization [83]. It can offer details on how aggregates are distributed throughout cells and how they interact with cellular frameworks. Understanding the location of various aggregate morphologies in cellular compartments and being able to discriminate between them is made easier by SIM's capacity to record structural information with enhanced resolution.
2.1.8. Optical tweezers
Optical tweezers are a revolutionary tool for detecting and characterizing protein aggregation [84]. These laser-based trapping devices enable precise manipulation and measurement of individual proteins or aggregates, providing insights into their mechanical properties and interactions. By measuring the forces required to unfold or dissociate proteins within aggregates, they can reveal the stability and structural changes associated with aggregation. Moreover, they can be combined with advanced microscopy techniques to enable real-time monitoring of aggregation processes at the single-molecule level [85]. This level of sensitivity and control is crucial for understanding the kinetics [86], energetics, and molecular forces driving protein aggregation pathways.
Optical tweezers hold promise in advancing our understanding of protein aggregation and facilitating the development of targeted interventions and therapeutics for protein misfolding diseases [87]. They can be used to exert controlled forces on individual proteins or aggregates, providing insights into their unfolding and mechanical stability. By measuring force-induced conformational changes, researchers gain valuable information about the structural integrity of aggregated proteins.
A recent study by Zaltron et al. revealed that Optical tweezers capture real-time dynamics during protein aggregation processes [88]. Moreover, optical tweezers can be combined with advanced microscopy techniques to enable real-time, single-molecule observations of aggregation processes. This level of sensitivity enables the precise tracking of aggregation kinetics, the study of intermediate states, and the exploration of forces involved in aggregation-related events [89]. In summary, optical tweezers offer a powerful and specific means to explore the mechanical and dynamic aspects of protein aggregation, providing crucial insights for understanding protein misfolding diseases and developing targeted therapeutic strategies.
2.1.9. Fluorescence recovery after photobleaching (FRAP)
FRAP is a microscopy-based method for examining the dynamics and mobility of biomolecules, particularly protein aggregates, inside of live cells. This technique involves bleaching a specific area of a cell, typically with a high-intensity laser, which renders fluorescently tagged molecules, including protein aggregates, non-fluorescent. Their fluorescence eventually returns when these non-fluorescent molecules drift into and out of the bleached area. Understanding the mobility and cycling of protein aggregates within cellular compartments can be gained from the rate of fluorescence recovery.
Furthermore, this technique is used for the purpose of quantifying the size distributions of nanomaterials present in biological fluids [90]. The procedure entails positioning a specimen onto a confocal laser scanning microscope, where fluorescently tagged molecules or nanoparticles are subjected to photobleaching within a region of one micron in size, achieved by applying a high-intensity excitation pulse. The pace at which the fluorescence inside the bleach region returns to its original level is directly related to the rate at which the fluorescent species diffuses [90]. The analysis and interpretation of FRAP data have mostly focused on a singular average diffusion coefficient. The first development of a Fluorescence Recovery After Photobleaching (FRAP) model for quantifying continuous distributions of diffusion coefficients was achieved by Verkman and Periasamy [91]. However, this model had limitations in its ability to distinguish between species exhibiting distinct diffusion coefficients. The study conducted by Hauser et al. shown that the inclusion of spatial information may greatly improve the ability to distinguish between two diffusing components in fluorescence recovery after photobleaching (FRAP) analysis [92]. Xiong et al. proposed an enhanced methodology for this technique namely, continuous diffusion coefficient distributions (cFRAP), which can easily detect comparable size distributions [90]. The approach exhibits flexibility by enabling the adjustment of the recovery time to align with the sample rate of the used microscope, hence optimizing the process for a certain diffusion coefficient. The researchers have successfully shown that the use of cFRAP-sizing methodology allows for precise assessment of protein aggregation levels in undiluted blood serum [90]. The findings conducted by Xiong et al. provides evidence that cFRAP has a high level of proficiency in the analysis of protein aggregates within blood sample after dilution [90].
FRAP (Fluorescence Recovery After Photobleaching) and optical tweezers are undoubtedly more modern techniques used to investigate the structural analysis of protein aggregation, particularly in the context of cellular and subcellular dynamics.
2.2. Indirect/non-morphological methods
Another, one of the broadly employed methods for monitoring protein aggregation is Indirect fluorometry. Indirect fluorimetry can be a useful method for determining protein aggregation when dealing with proteins that may not naturally fluoresce but can be labeled with fluorescent markers or probes. The aforementioned procedure involves using a fluorescent dye, such as ThT (ThioflavinT) [93], which is highly specific for visualizing cross beta-sheets that are further responsible for producing amyloid fibrils as well as amorphous aggregates in proteins. As ThT has the property to interact with amyloids, it shows heavy fluorescence when excited at 450 nm within the range of 480 nm–490 nm. Therefore, indirect fluorimetry is considered the most widely used technique to keep track of amyloidogenic protein aggregates [94]. Furthermore, the histology dye, Congo red (CR) is often employed for the identification of beta-amyloids. This dye's specificity is its affinity for binding protein fibrils that increase the beta-sheet conformation [34]. Unpredictably, findings from current research reveal that the dye still has the capacity to prevent protein misfolding and aggregation [95]. A distinctive change in the absorbance maximum from 490 to 540 nm and green birefringence through polarised light is observed by CR [94]. Congo red (CR) dye exhibits a unique change in absorbance maximum from 490 to 540 nm and green birefringence through polarized light. Initially, it absorbs light most strongly at 490 nm. However, as it binds to beta-amyloid fibrils, this peak shifts to a higher wavelength, specifically around 540 nm. This indicates the dye has bound to the target structure. When Congo red binds to these structures, it can exhibit birefringence, splitting incident light into two different rays with different velocities and orientations. This green birefringence pattern, observed under a polarizing microscope, is a distinctive visual indicator of the presence of amyloid fibrils or related structures.
Other dyes, such as Nile Red, include lipophilic properties that enable them to potentially interact with hydrophobic regions of protein aggregates, thereby intensifying the fluorescence signal. Sutter and coauthors documented the utilization of the fluorescent dye Nile Red as a sensitive probe for detecting substantial denatured aggregates [96]. Furthermore, the compound 8-Anilino-1-naphthalenesulfonic acid (ANS) is used as a hydrophobic probe in order to selectively attach to hydrophobic regions on proteins [97,98]. These hydrophobic areas are often seen during the first stages of protein aggregation. When the substance comes into contact with these hydrophobic regions, there is an increase in its fluorescence intensity. Similarly, Bis-ANS (4,4′-Dianilino-1,1′-binaphthyl-5,5′-disulfonic acid), with its penchant for binding to exposed hydrophobic domains on proteins, furnishes insights into the nascent stages of protein aggregation, offering valuable mechanistic insights into aggregate nucleation.
Amytracker dyes are specialized fluorescent probes used for detecting and studying amyloid fibrils [99]and protein aggregates, particularly in neurodegenerative diseases. These dyes offer high specificity for amyloid structures, emit fluorescence in multiple spectral variants for multiplex imaging, and enable real-time monitoring of aggregate formation. They are invaluable for visualizing aggregate distribution, compatible with various techniques, and aid in drug discovery for targeting protein aggregation. Amytracker dyes provide quantitative insights into aggregate concentration and offer versatile tools for researching complex protein misfolding processes.
The fluorescent dye 2-(4′-methylaminophenyl) benzothiazole, is often used in academic research to investigate protein aggregation. Its utility is particularly notable in the examination of amyloid fibrils and other aggregates that include a high content of β-sheets. This dye is classified as an amyloid-specific probe, implying that it has a distinct affinity for binding to amyloid aggregates and has the potential to generate fluorescence upon interaction [100]. Moreover, Methylene Blue, esteemed for its pleiotropic binding properties, has been enlisted for its proclivity to associate with amyloid aggregates and additional proteinaceous structures, affording opportunities for in-depth exploration within the domain of protein aggregation research [101]. In conclusion, researchers may study protein aggregation and structural changes using a wide range of fluorescent dyes. These dyes empower scientists to delve deeper into the molecular intricacies of diseases and gain insights that may ultimately lead to novel therapeutic strategies.2.3. Morphological methods.
The direct as well as indirect methods cannot differentiate amongst the aggregated species morphologically. The morphological method is used to analyze protein aggregation that can differentiate among the morphological characteristics of the aggregated species. Turbidity is one of the optical kinetics methods by which aggregation can be determined virtually [102,103]. The turbidity method also shows some significant disadvantages like weak sensitivity as well as an inability to differentiate between the various forms of aggregates [93].
2.2.1. Dynamic light scattering (DLS)
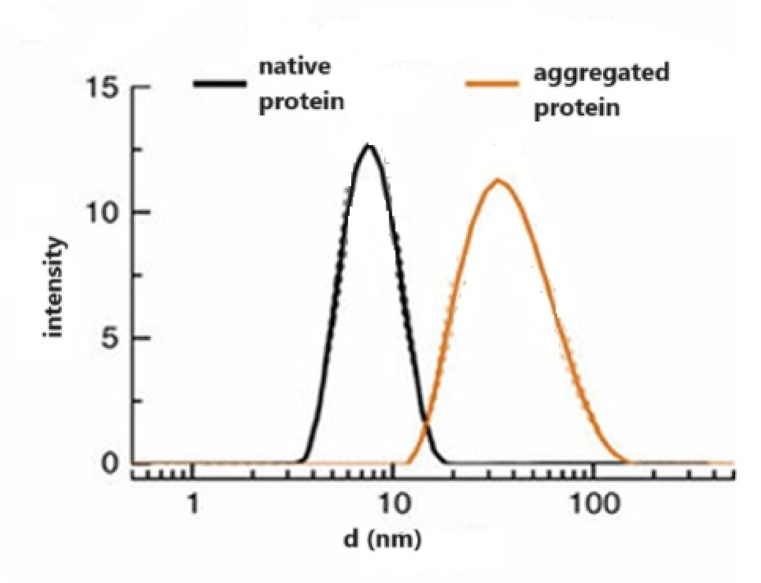
Dynamic Light Scattering (DLS), also termed as Photon Correlation Spectroscopy (PCS) permits the precise measurement of particle sizes ranging from 1 nm to several microns in due time [104]. It is a prevalent technique used in both colloidal sciences and protein characterization studies (Fig. 7) [105]. The technique is based on the analysis of scattered light variations that occur due to the Brownian motion of particles [106]. When a monochromatic beam of light strikes a particle in Brownian motion, it causes a Doppler shift resulting in change in the wavelength of the spherical particle (predominantly, the incident light is near-IR at 830 nm or red light at 633 nm) [105]. In addition, this procedure is one of the most often used techniques to regulate the particle size within a range of 0.001 mm to several microns, which is challenging to attain with other techniques [105]. In biology labs, DLS has been used on a consistent basis for a variety of purposes, including the detection of aggregate particles in complex molecular solutions, determination of the sizes of peptides, nucleic acids, and polysaccharides, and monitoring of ligand-ligand interactions [107].
Fig. 7.
Particle size distribution of aggregates. DLS data of aggregating species taken at different time points and represented as color traces [90]. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
DLS offers several advantages and has its share of limitations. One of the primary advantages of DLS is its non-invasive nature. It allows for the analysis of nanoparticles or macromolecules without the need for any labels or dyes, preserving the sample's native properties. DLS is also a fast technique, capable of providing results within minutes, making it suitable for real-time monitoring of particle size and dynamics. Furthermore, DLS has a broad detection range, enabling the measurement of particles in the sub-nanometer to several micrometer range, making it versatile for a wide range of applications.
However, DLS is not without its limitations. It is highly sensitive to sample quality and purity, as impurities or contaminants can lead to inaccurate results. Additionally, DLS provides a volume-weighted size distribution, which means larger particles contribute more to the signal, potentially masking the presence of smaller particles. The technique is also sensitive to polydispersity, which can complicate data interpretation. Furthermore, DLS requires a dilute sample, which may not be feasible for high-concentration or complex samples. Finally, DLS is primarily a qualitative technique, and while it can provide size information, it does not offer detailed structural or morphological insights into particles or molecules.
In summary, dynamic light scattering is a valuable tool for quickly assessing the size distribution and diffusion properties of particles or molecules in a solution. Its non-invasive, label-free nature and broad detection range make it widely applicable, but researchers must be aware of its sensitivity to impurities, polydispersity, and its inability to provide detailed structural information when interpreting results. Careful sample preparation and complementary techniques may be necessary to address these limitations and obtain a comprehensive understanding of the sample under investigation.
2.2.2. Mass spectroscopy
Mass spectrometry (MS) has become a prominent technique in the field of protein aggregation research due to its high sensitivity. It provides valuable information about the initiation of amorphous aggregation and, more specifically, the formation of amyloid fibrils. This includes the characterization of monomers and oligomers, the study of aggregation mechanisms, and the screening of inhibitors [108]. Furthermore, this technique is used for the investigation of proteins, peptides and oligomers [109]. It is based on the principle that determines the proportion of mass of the charged particles with respect to their individual charges. Mass spectrometry-based methodologies play an essential part in finding and characterizing protein using high-mass detector that allows ion recognition up to 1,000,000 m/z [110]. With this technique, it is possible to identify substantial protein molecules with a molecular mass of approximately 300,000 g/mol or more. It has become a particularly helpful instrument for the analysis of proteins as well as peptides because of the innovation of "soft" ionisation technologies, most notably electrospray (ESI) ionisation and matrix-assisted laser desorption (MALDI) ionisation. "Soft" ionisation is a term used in mass spectrometry to describe ionisation techniques that produce ions with minimal fragmentation or damage to the analyte molecules. A deliberate process of buffer selection is carried out so as to avoid unwanted concentrations of ions that might hamper matrix formation as well as desorption and ionisation [93]. Soft ionisation techniques are particularly valuable in the analysis of proteins and peptides because they allow for the generation of intact molecular ions, which can provide valuable information about the molecular weight and structure of these biomolecules. This technique offers numerous advantages and some limitations, which are important to consider in a scientific context.
One of the primary advantages of mass spectrometry is its exceptional sensitivity and specificity. MS can detect and quantify trace levels of molecules, making it invaluable in fields like proteomics, metabolomics, and environmental monitoring. It provides detailed structural information by measuring the mass-to-charge ratio (m/z) of ions, allowing for the identification of compounds, including their isotopic composition and fragmentation patterns.
Another advantage is its wide applicability. Mass spectrometry can analyze a diverse range of analytes, from small molecules to large biomolecules like proteins and nucleic acids. This versatility allows scientists to study complex mixtures and elucidate the composition of unknown samples.
MS is also highly quantitative, making it suitable for precise concentration measurements. It can be used for isotope ratio analysis, quantifying the abundance of specific isotopes, which is crucial in radiometric dating and stable isotope analysis.
However, mass spectrometry has limitations. One challenge is sample preparation, as certain samples may require extensive cleanup or derivatization to be compatible with MS analysis. Matrix effects can also impact accuracy, especially in complex samples. Additionally, high-resolution mass spectrometers and expert data analysis are often needed for comprehensive results, which can be costly and require specialized training.
2.2.3. Hydrogen-deuterium exchange mass spectrometry (HDX-MS)
Hydrogen/Deuterium exchange (HDX) is an intricate mass spectroscopic application for conformational observation of proteins. H/D exchange is used to analyze thermally exhilarated structural changes in protein [111]. The information regarding conformational and dynamic systems is acquired by comparing the level of deuterium uptake in diverse proteins [112]. A flexible method for examining the structural dynamics and solvent accessibility of proteins, including those seen in protein aggregates, is hydrogen-deuterium exchange mass spectrometry (HDX-MS). Proteins are subjected to a solution of deuterated (heavy) water during HDX-MS, causing hydrogen atoms to exchange positions with deuterium atoms. Protein structure and solvent accessibility are a couple of the variables that affect how quickly hydrogen and deuterium are exchanged. The protein is quickly quenched and broken down into peptides after a certain exchange duration. Mass spectrometry is then used to calculate the deuteration-related mass differences. Applications of HDX-MS to the study of protein aggregates include Quantitative Analysis, Interactions and Binding Sites, Structural Dynamics, Stability and Folding, and Aggregation-Related Conformational Changes.
While HDX-MS is a strong tool, it does have certain drawbacks, such as possible difficulties in data interpretation and the requirement for careful experimental design. The method may not be able to catch minute details at the atomic level, but it does offer information on the general behavior of protein aggregates. However, it advances our knowledge of the processes that lead to protein aggregation and its effects on numerous disorders.
2.2.4. Atomic force microscopy (AFM)
AFM can capture a 3D morphology of biological specimens having a resolution of a few nanometers, has emerged among the most robust and adaptable single-molecule methodologies in recent years [113]. This technique is immensely helpful in analyzing amyloid fibrils' association process that provides vital information for understanding the procedure of aggregation because of fibrillation [114]. Using time-lapse imaging, atomic force microscopy (AFM) might be used to capture the process by which biological macromolecules associate with one another. The development of AFM has a crucial influence on research into improperly folded proteins that do not adopt or stay in their natural functional & conformational states, which might be because of aggregation [113]. Furthermore, AFM has also been used to visualize protein-protein interactions as a function of pH. According to the results of the AFM, the diameters of the cross-section of amyloid species fluctuate between the nanometer and micrometer scales. Oligomeric species may be measured as spheroid or toroid-particles with a typical diameter ranging from 1 to 15 nm, and they can also take the form of extended protofilaments often measuring hundreds of nanometers in length as well as a diameter (cross-sectional) of 1–2 nm. The appearance of protofibrils is often that of stretched out, linear or curvilinear aggregates, with an average height in the range of 1–5 nm and lengths between hundreds of nanometers [113]. This technique has been employed to analyze the natural configuration of amyloid fibrils and their different morphological forms [115]. Amyloid fibers are distinguished by their well-ordered, stretched, and relatively uniform fibrillar configurations while being composed of a wide variety of different polypeptides. Topological characteristics may be discerned with a resolution of a tenth or a hundredth of a nanometer or less. It is now possible, as a result of observations made using time-space AFM, to analyze both the shape as well as the formation of aggregates of Aβ [24]. AFM is a versatile tool that can be used in various environments, including air, liquid, and even vacuum conditions, making it suitable for studying a wide range of samples and processes. Furthermore, AFM can be used for both imaging and force spectroscopy, providing information not only about surface morphology but also about mechanical properties and molecular interactions.
One of AFM's primary advantages is its ability to provide topographical information at the nanometer and atomic scales. It achieves this by scanning a sharp tip over the sample surface and measuring the interaction forces between the tip and the atoms or molecules on the surface. This high-resolution capability allows scientists to visualize and manipulate materials and biological samples at the nanoscale.
However, there are limitations to AFM. One significant limitation is the relatively slow scanning speed, which can make imaging large areas or fast dynamic processes time-consuming. Additionally, AFM imaging can be influenced by tip-sample interactions, and the choice of tip and imaging parameters requires careful optimization to obtain accurate and meaningful results. Moreover, AFM is primarily a surface technique, so it provides limited information about the three-dimensional structure of samples.
2.2.5. Size exclusion chromatography (SEC)
SEC is a method for separating compounds from one another based on the variations in the molecular sizes of the substances being analyzed [116]. This method operates on the presumption that monomer and oligomer extinction coefficients are identical. The extent of agglomeration of a sample may be determined by measuring its absorbance at the range of 280 nm or even lesser (214–220 nm) [14]. However, SEC is able to examine protein aggregates up to a certain size since bigger aggregates are being clarified out from the system by the frits or via column. As a result, huge amounts of samples are required and are thereby lost in aggregation during processing.
Further, SEC is similar to Gel-permeation chromatography (GPC), the distinction lies in the types of samples that may be analyzed using either technique and the leading use of SEC is in the investigation of bio-therapeutic originations. Moreover, this technique has been employed in the determination of the amounts of reversible self-assembled, amalgamated (non-reversible) aggregates that can be soluble or insoluble with an increased molecular weight that may impact the quality and the efficacy of the product [117]. While numerous methods have been discovered to evaluate the aggregation of proteins, the SEC technique is most commonly selected for regular and verified tests due to the fact that it is both quick and reliable. When double-checked, the data obtained via SEC may be depended on to provide correct findings, exactly as the results obtained when employing an orthogonal technique such as sedimentation velocity-analytical ultracentrifugation (SV-AUC) [118]. In analytical SEC, the columns are often rather tiny, and the amount of sample needed to produce a result range from only a few micrograms to tens of micrograms.
2.2.6. Analytical ultracentrifugation (AUC)
AUC, which is often performed in the sedimentation velocity (SV-AUC) mode, is a robust tool that can be applied to analyze and characterize the clumping/precipitating behavior of biomolecules, thereby formation of aggregates in the solution. It measures the size of the aggregate ranging from 1 to 100 nm. Amid the myriad of experimental and practical characteristics. The orientation and quality seem to be the primary causes of discrepancies in the measurement of tiny quantities of aggregates amidst the various practical and theoretical factors which affect the precision and accuracy of the AUC investigations. It is feasible to quantify aggregated species, although the disorganization of aggregate particles brought about during sample preparation, dissolution, or the background effect is only to a limited extent [119]. Therefore, the data analyzed on AUC can provide in-depth knowledge on aggregation while overcoming challenges associated with SEC. So, the AUC may be utilized as an independent technique to check the aggregate information provided by SEC as well as the features of biopharmaceuticals that correlate with it [120]. One of the primary advantages of AUC is its ability to provide detailed information about the hydrodynamic properties, molecular weight, shape, and interactions of macromolecules and particles in their native state. AUC is a non-invasive, label-free technique that can be applied to a wide range of biological molecules, from small proteins to large complexes, as well as synthetic nanoparticles. It offers high-resolution separation and analysis, making it valuable for characterizing complex mixtures and determining the stoichiometry of protein complexes.
AUC is versatile in terms of the type of information it can provide. It can be used to study sedimentation velocity, which yields information about size and shape, and sedimentation equilibrium, which provides insights into molecular weight and interactions. Additionally, AUC can be performed under various conditions, including different buffer compositions, temperatures, and concentrations, allowing researchers to investigate macromolecule behavior under physiologically relevant conditions.
However, there are limitations to AUC. One limitation is the relatively long data acquisition time, which can make it less suitable for studying fast dynamic processes. AUC also requires a relatively large sample volume compared to other analytical techniques, which may not be feasible for precious or limited samples. Furthermore, data analysis can be complex and may require specialized software and expertise.
2.2.7. Asymmetric flow field-flow fractionation
Asymmetrical flow field-flow fractionation, sometimes known as AF4 for short, is an analyzing technique that can separate distinct proteins with diameter sizes varying from a few nm range to mm range [121]. In AF4, the separation process takes place in a narrow flow channel. It works on the principle based on the sedimentation rate in response to centrifugal force [14]. Initially, a crossflow is produced that directs the laminar flow perpendicularly across the membrane. There are several types of membranes that can be utilized, such as cellulose triacetate (CTA), regenerated cellulose (RC), poly(ether sulfone) (PES), polypropylene (PP), polyamide (PA), polycarbonate (PC), or polyvinylidene difluoride (PVDF). These materials vary in terms of their thickness, surface properties, surface charge, smoothness, and mechanical and chemical stability [122]. Thereby, in order to minimize adsorption effects, it is essential that both the eluent and the sample be compatible with the membrane. Furthermore, the analytes are guided toward the membrane via this cross-route, without being diffused out due to the semi-permeability nature of the membrane. The tiny proteins would return promptly to the streamlines of laminar channel than the bigger ones due to their relatively high diffusion coefficient. As a result, smaller proteins would elute first from the channel before the bigger molecules [14].
2.2.8. X-ray diffraction
Investigations using X-ray diffraction (XRD) reveal the existence of a typical structure known as a "cross beta-sheet," wherein β-sheets move parallel to the longitudinal axis whereas the β-strands present in a single sheet are organized perpendicularly to one another. The distance between the two β-strands and the β-sheets is 4.8 Å and 10 Å, respectively. Furthermore, it is essential to take into account that the in vitro fibrillar morphology can vary depending on the environment of the solution such as the concentration of the protein, the pH of the solution, the temp, buffer components, and the involvement of particular additives [123]. X-ray crystallography: X-ray crystallography is a fundamental technique in structural biology that has been widely used to determine the atomic structures of proteins, including those within protein aggregates. A protein or protein aggregation is grown into well-ordered crystals, and these crystals are then exposed to X-rays. As X-rays reflect off the crystal lattice, a diffraction pattern is created that provides details on the arrangement of atoms inside the crystal. Protein aggregates can benefit from the use of X-ray crystallography in a variety of ways, including crystal growth, X-ray diffraction, phase determination, model building and refinement, validation, and insights into protein aggregates. X-ray crystallography has restrictions despite it's achievements. It involves getting high-quality crystals, which may be difficult for aggregates in particular. Furthermore, the method only captures static images of protein structures and might not fully capture the dynamics of protein aggregation. The resolution and applicability of X-ray crystallography to complex systems like protein aggregates have increased as a result of developments in X-ray sources, detectors, and computational techniques. X-ray crystallography is a useful tool for revealing atomic-level details about the structures of protein aggregates, even if it may not necessarily be the principal method for investigating these structures.
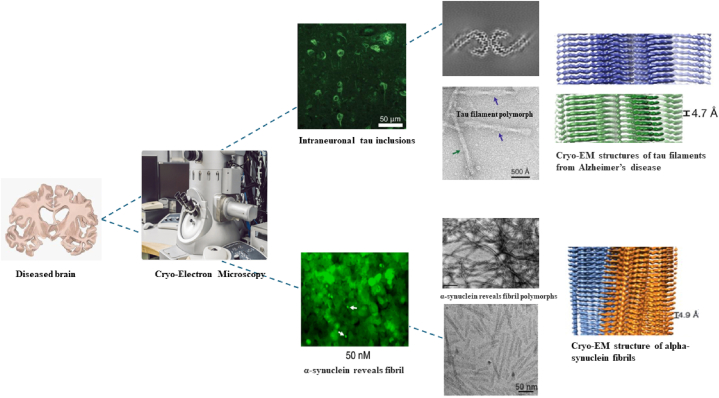
2.2.9. Cryo-electron microscopy (Cryo-EM)
Cryo-Electron Microscopy (Cryo-EM) is a powerful structural biology technique that allows scientists to visualize biological macromolecules, including proteins, at near-atomic resolution. Cryo-EM includes photographing specimens in their natural condition and retain their biological structures and interactions. By using fast freezing to immobilize biological samples in a thin layer of vitrified ice, the approach preserves the sample's original conformation and avoids the development of crystalline artifacts [124]. In order to scan the frozen samples, an electron microscope is used, which operates at extremely low temperatures, often about −196 °C (−320 °F). The structural characteristics of amyloid fibrils, such as protofilament structure, packing, and long-range helical twist, are known to exhibit considerable variability. The presence of variations in the fibril structures within the sample imposes limitations on the achievable resolution with most of the techniques. Nevertheless, the use of single particle cryo-electron microscopy (cryo-EM) enables the computational extraction of brief segments from the fibrils and their subsequent classification into subsets that exhibit structural homogeneity [125]. The field of cryo-electron microscopy (cryo-EM) has seen significant progress in capturing high-resolution structures of α-syn (α-synuclein), Aβ (amyloid-β), tau, and TDP-43 (transactive response element DNA-binding protein of 43 kDa) aggregates that are associated with various diseases [126], some ofthese are being depicted in Fig. 8. Recent studies utilizing cryo-electron microscopy (cryo-EM) have uncovered intriguing differences between ex-vivo α-synuclein filaments extracted from individuals affected by multiple system atrophy (MSA) and their counterparts formed in vivo [127]. These disparities strongly suggest the involvement of co-factors within the in vivo environment, which likely influence α-synuclein aggregation dynamics. Moreover, investigations employing cryo-EM in combination with solid-state nuclear magnetic resonance (NMR) spectroscopy have delved into α-synuclein filaments influenced by tau proteins. The findings reveal that tau-promoted α-synuclein filament structures share certain similarities with those observed in vitro. However, distinct conformational variations are evident in the N- and C-terminal regions, indicating a nuanced interplay between α-synuclein and tau in the context of protein aggregation [127,128]. These insights deepen our understanding of the intricate molecular mechanisms underlying neurodegenerative disorders like MSA, where α-synuclein and tau pathologies intersect. Such discoveries hold promise for the development of targeted therapeutic interventions aimed at modulating these complex interactions to mitigate disease progression and potentially pave the way for novel treatment strategies.
Fig. 8.
Schematic representation showing Cryo-EM micrographs of alpha-synuclein and tau fibrils revealing structural heterogeneity [[127], [128], [129], [130], [131]].
2.2.10. Small-angle X-ray scattering (SAXS)
Small-Angle X-ray Scattering (SAXS) is a powerful technique used to study the overall shape, size, and structural organization of macromolecules, including protein aggregates, in solution [132]. SAXS involves exposing a sample to X-rays and measuring the scattering pattern of the X-rays at low angles. This method enables the identification of important structural properties by providing useful information on the spatial distribution of electrons inside the sample. In the study of protein aggregates using SAXS, researchers collect scattering data, analyze it using techniques like Guinier analysis and Porod analysis, and use mathematical models to interpret and reconstruct the structure of the protein aggregates. This information can be crucial for understanding the behavior and properties of protein aggregates in various biological and biomedical contexts [133]. Studying protein aggregates, which can exhibit a variety of structural complexity, is a valuable application of SAXS since it is particularly well-suited for researching flexible or partly disordered systems. It is a solution-based method that enables the analysis of samples in almost natural environments, such as in various buffer solutions.
SAXS, on the other hand, provides less resolution than methods like X-ray crystallography or cryo-electron microscopy, providing data on general shapes as opposed to specific atomic structures [134].
SAXS aids in the comprehension of the size distribution, conformational changes, and interactions with other molecules in protein aggregates. A more complete picture of aggregate behavior and morphology may be obtained by integrating SAXS data with other structural approaches.
2.2.11. Cryo-electron tomography (Cryo-ET)
A sophisticated imaging method called cryo-electron tomography (Cryo-ET) is used to see the three-dimensional structures of biological specimens, such as protein aggregation, under conditions that are close to their natural ones. Researchers may reconstitute the three-dimensional architecture of macromolecules within their natural biological setting using a cryo-electron microscopy (cryo-EM) extension. Cryo-ET offers insights into protein aggregates spatial distribution, interactions with biological components, and effects on cellular structure, making it particularly useful for researching protein aggregates within cells, tissues, or other complex settings [126]. In order to fully comprehend the biological functions of protein aggregates and the mechanisms underlying their creation, it enables researchers to study protein aggregates in their natural environment.
The necessity for specialized equipment and powerful computational tools for data collecting and processing are only two examples of the technological difficulties that Cryo-ET faces [72]. Additionally, cryo-ET often has a lesser resolution than other high-resolution methods like single-particle cryo-EM or X-ray crystallography. However, it continues to be an effective tool for researching the structural organization of protein aggregates in intricate biological settings.
3. In vivo biophysical characterization of protein aggregates
In vivo characterization using biophysical tools is a crucial method in biological and biomedical research, allowing scientists to study the properties, behaviors, and responses of biological molecules, cells, tissues, and organisms in their natural environments. This non-invasive approach provides real-time insights into dynamic biological events, such as cellular signaling, protein interactions, and physiological responses, which are essential for understanding biological processes. In vivo biophysical tools include techniques like MRI, PET, fluorescence microscopy, and EPR spectroscopy, which help researchers gain a comprehensive understanding of complex biological systems. The biological relevance of in vivo studies is crucial, as they faithfully replicate physiological conditions, ensuring the results are biologically meaningful and reflective of living systems in their natural context. These techniques are widely used in disease research, drug development, neuroscience, biomechanics, and environmental monitoring, facilitating disease progression tracking, treatment efficacy assessment, drug candidate evaluation, and tissue mechanical properties investigation. However, ethical considerations, particularly concerning animal and human subjects, are essential.
VISTA, or vibrational imaging with synchronous two-photon excitation, is an advanced microscopy technique designed for high spatial-resolution imaging of label-free protein aggregates in living biological samples. This approach leverages Raman spectroscopy and two-photon microscopy principles to provide detailed information about protein aggregate distribution and composition without the need for fluorescent labels. VISTA offers several advantages, including label-free imaging, high spatial resolution, in vivo compatibility, and chemical specificity. However, it requires specialized instrumentation, may have lower signal intensity, and demands expertise in data analysis [135].
In conclusion, in vivo characterization using biophysical tools is a cornerstone of modern biological and medical research, providing valuable insights into living system dynamics and advancing our understanding of health, disease, and intervention effects.
4. Future applications of current methods
The future is filled with tremendous potential for enhancing our comprehension of protein aggregation's structural aspects. Our grasp of the intricate realm of protein aggregates is on the brink of transformation through the fusion of current methodologies and upcoming techniques. Other techniques, including Small-Angle X-ray Scattering (SAXS), offer a dynamic picture of aggregation processes as techniques like Cryo-Electron Microscopy (Cryo-EM) push the limits of resolution by showing atomic-scale features. Super-resolution microscopy and fluorescence-based methods shed light on aggregate interactions and their locations within cellular environments. The emergence of in-cell methodologies and advanced computer modeling opens the door to unraveling real-time dynamics and guiding drug development initiatives. This convergence of innovation presents a future where targeted therapies could hold the key to addressing aggregation-related diseases at their most fundamental levels. Imminent expansion in the structural analysis of protein aggregation shows significant potential for advancing our knowledge of the underlying molecular mechanisms behind aggregation-related disorders. Modern approaches and technological developments will provide revolutionary insights into the mechanics of protein aggregation and its effects on health and disease. These advancements have profound clinical implications, potentially leading to the development of more effective therapies, diagnostics, and preventive measures for diseases associated with protein aggregation, such as Alzheimer's, Parkinson's, or Huntington's disease. Furthermore, in the long run, these innovations poise to shape the long-term future of medicine and our understanding of fundamental biological processes, with far-reaching implications for healthcare and disease management.
5. Conclusion
Aggregates have a variety of sizes and other physicochemical characteristics, making them diverse. As a direct consequence of this, a single technique is not merely sufficient to adequately analyze all the properties of aggregate particles. The findings acquired with one approach are inextricably related to those procured with another since every technique measures a different prospect of the enormous characteristic of aggregates. Development in protein aggregation research is driven by numerous approaches, including the structural study of protein aggregation. Besides, using numerous biophysical studies, the structural properties of amyloid at the insoluble and solvable processes have been reviewed, however, further research on the intermediate stage of the aggregated proteins is still under consideration. Herein, a short description of protein aggregation's structural analysis strategies using various biophysical techniques has been discussed. Using Fluorescence spectroscopy, FTIR, NMR, and CD, structural modifications during protein aggregation can be understood which will help in developing therapeutics for many diseases induced by protein aggregation. Thus, along with other multidisciplinary methods, the insights obtained using these strategies will advance our level of awareness of protein aggregation and misfolding and related therapeutics for neurodegenerative diseases as well.
Funding
SB acknowledges ICMR-SRF Fellowship (45/28/2019-Bio/BMS). The authors thank for the grants from the Indian Council of Medical Research (ICMR) ISRM/12/(127)/2020. AAis thankful for the funds from the PMRF.
Data availability statement
There is no data required to be made available in the review article.
CRediT authorship contribution statement
Sania Bashir: Conceptualization, Writing – original draft. Ayesha Aiman: Conceptualization, Writing – original draft. Anis Ahmad Chaudhary: Writing – original draft, Writing – review & editing. Nashrah Khan: Writing – original draft. Ishfaq Ahmad Ahanger: Writing – original draft. Neha Sami: Writing – original draft. Eman Abdullah Almugri: Writing – original draft. Mohamed A.M. Ali: Writing – original draft. Mohammad Shahid: Writing – original draft. Asimul Islam: Conceptualization, Writing – original draft, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge the College of Medicine and Deanship of scientific research, Prince Sattam bin Abdul-Aziz University, Alkharj, KSA for their continuous support to carry out this research.
The authors extend their appreciation to the Deanship of Scientific Research at Imam Mohammad Ibn Saud Islamic University (IMSIU) for funding and supporting this work through Research Partnership Program no.RP-21-09-86.
References
- 1.Khan E., K Mishra S., Kumar A. Emerging methods for structural analysis of protein aggregation. Protein Pept. Lett. 2017;24:331–339. doi: 10.2174/0929866524666170206123150. [DOI] [PubMed] [Google Scholar]
- 2.Feigin V.L., Vos T., Nichols E., Owolabi M.O., Carroll W.M., Dichgans M., Deuschl G., Parmar P., Brainin M., Murray C. The global burden of neurological disorders: translating evidence into policy. Lancet Neurol. 2020;19:255–265. doi: 10.1016/S1474-4422(19)30411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Schependom J., D’haeseleer M. Advances in neurodegenerative diseases. 2023;12:1709. doi: 10.3390/jcm12051709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukherjee S., Ali S., Hashmi S., Jahan S. History. Applications of Stem Cells and Derived Exosomes in Neurodegenerative Disorders. Springer; 2023. Origin and types of neurological disorders; pp. 1–32. [Google Scholar]
- 5.Chen F., Wu Y., Chen X., Chen Y., Chen X., Wu Y., Wei P., Kang D., Ding C. Global, regional, and national burden and attributable risk factors of transport injuries: global Burden of Disease Study 1990–2019. Chinese Med J. 2023;136:1762–1764. doi: 10.1097/CM9.0000000000002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregersen N., Bross P., Vang S., Christensen J.H. Protein misfolding and human disease. Annu. Rev. Genom. Hum. Genet. 2006;7:103–124. doi: 10.1146/annurev.genom.7.080505.115737. [DOI] [PubMed] [Google Scholar]
- 7.Bashir S., Shamsi A., Ahmad F., Hassan M.I., Kamal M.A., Islam A. Biophysical elucidation of fibrillation inhibition by sugar osmolytes in α-lactalbumin: multispectroscopic and molecular docking approaches. ACS Omega. 2020;5:26871–26882. doi: 10.1021/acsomega.0c04062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolen D.W., Rose G.D. Structure and energetics of the hydrogen-bonded backbone in protein folding. Annu. Rev. Biochem. 2008;77:339–362. doi: 10.1146/annurev.biochem.77.061306.131357. [DOI] [PubMed] [Google Scholar]
- 9.Tanford C. Protein denaturation: Part C. Theoretical models for the mechanism of denaturation. Adv. Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- 10.Bashir S., Sami N., Bashir S., Ahmad F., Hassan M.I., Islam A. Management of insulin through Co-solute engineering: a therapeutic approach. Frontiers in Protein Structure, Function, and Dynamics. 2020:283–315. [Google Scholar]
- 11.Uversky V.N. The triple power of D (3): protein intrinsic disorder in degenerative diseases. Front. Biosci. 2014;19:181–258. doi: 10.2741/4204. [DOI] [PubMed] [Google Scholar]
- 12.Dinca V., Kasotakis E., Mourka A., Ranella A., Farsari M., Mitraki A., Fotakis C. Fabrication of amyloid peptide micro‐arrays using laser‐induced forward transfer and avidin‐biotin mediated assembly. Phys. Status Solidi C. 2008;5:3576–3579. [Google Scholar]
- 13.Yadav K., Yadav A., Vashistha P., Pandey V.P., Dwivedi U.N. Protein misfolding diseases and therapeutic approaches. Curr. Protein Pept. Sci. 2019;20:1226–1245. doi: 10.2174/1389203720666190610092840. [DOI] [PubMed] [Google Scholar]
- 14.Den Engelsman J., Garidel P., Smulders R., Koll H., Smith B., Bassarab S., Seidl A., Hainzl O., Jiskoot W. Strategies for the assessment of protein aggregates in pharmaceutical biotech product development. Pharmaceut. Res. 2011;28:920–933. doi: 10.1007/s11095-010-0297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Housmans J.A., Wu G., Schymkowitz J., Rousseau F. A guide to studying protein aggregation. FEBS J. 2023;290:554–583. doi: 10.1111/febs.16312. [DOI] [PubMed] [Google Scholar]
- 16.Pignataro M.F., Herrera M.G., Dodero V.I. Evaluation of peptide/protein self-assembly and aggregation by spectroscopic methods. Molecules. 2020;25:4854. doi: 10.3390/molecules25204854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nilapwar S.M., Nardelli M., Westerhoff H.V., Verma M. vol. 500. Elsevier; 2011. pp. 59–75. (Absorption Spectroscopy. In Methods in Enzymology). [DOI] [PubMed] [Google Scholar]
- 18.Raynal B., Lenormand P., Baron B., Hoos S., England P. Quality assessment and optimization of purified protein samples: why and how? Microb. Cell Factories. 2014;13:1–10. doi: 10.1186/s12934-014-0180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reule A. Errors in spectrophotometry and calibration procedures to avoid them. Journal of research of the National Bureau of Standards. Section A, Physics and chemistry. 1976;80:609. doi: 10.6028/jres.080A.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson K., Walker J. Cambridge university press; 2010. Principles and Techniques of Biochemistry and Molecular Biology. [Google Scholar]
- 21.Zuber, Agnieszka A., Klantsataya Elizaveta, Akash Bachhuka. Biosensing. Reference Module in Materials Science and Materials Engineering. Elsevier; 2019. pp. 105–126. [Google Scholar]
- 22.Ghisaidoobe A.B., Chung S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: a focus on Förster resonance energy transfer techniques. Int. J. Mol. Sci. 2014;15:22518–22538. doi: 10.3390/ijms151222518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yammine A., Gao J., Kwan A.H. Tryptophan fluorescence quenching assays for measuring protein-ligand binding affinities: principles and a practical guide. Bio-protocol. 2019;9:e3253. doi: 10.21769/BioProtoc.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 25.Barik S. The uniqueness of tryptophan in biology: properties, metabolism, interactions and localization in proteins. Int. J. Mol. Sci. 2020;21:8776. doi: 10.3390/ijms21228776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saunders M., Wishnia A., Kirkwood J.G. The nuclear magnetic resonance spectrum of ribonuclease1. J. Am. Chem. Soc. 1957;79:3289–3290. [Google Scholar]
- 27.Ladokhin A.S. Fluorescence spectroscopy in peptide and protein analysis. Encyclopedia of analytical chemistry. 2000:5762–5779. [Google Scholar]
- 28.Lakowicz J.R. Springer; 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 29.Sridharan R., Zuber J., Connelly S.M., Mathew E., Dumont M.E. Fluorescent approaches for understanding interactions of ligands with G protein coupled receptors. Biochim. Biophys. Acta Biomembr. 2014;1838:15–33. doi: 10.1016/j.bbamem.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain N., Bhattacharya M., Mukhopadhyay S. Chain collapse of an amyloidogenic intrinsically disordered protein. Biophys. J. 2011;101:1720–1729. doi: 10.1016/j.bpj.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole R.A., Hawe A., Jiskoot W., Braeckmans K. Analysis of Aggregates and Particles in Protein Pharmaceuticals. 2012. Fluorescence spectroscopy to characterize protein aggregates and particles; pp. 201–226. [Google Scholar]
- 32.Haris P.I., Severcan F. FTIR spectroscopic characterization of protein structure in aqueous and non-aqueous media. J. Mol. Catal. B Enzym. 1999;7:207–221. [Google Scholar]
- 33.Miller L.M., Wang Q., Telivala T.P., Smith R.J., Lanzirotti A., Miklossy J. Synchrotron-based infrared and X-ray imaging shows focalized accumulation of Cu and Zn co-localized with β-amyloid deposits in Alzheimer's disease. J. Struct. Biol. 2006;155:30–37. doi: 10.1016/j.jsb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson M.R. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Man V.H., Roland C., Sagui C. Amyloid properties of asparagine and glutamine in prion-like proteins. ACS Chem. Neurosci. 2016;7:576–587. doi: 10.1021/acschemneuro.5b00337. [DOI] [PubMed] [Google Scholar]
- 36.Bonda M., Perrin V., Vileno B., Runne H., Kretlow A., Forró L., Luthi-Carter R., Miller L.M., Jeney S. Synchrotron infrared microspectroscopy detecting the evolution of Huntington's disease neuropathology and suggesting unique correlates of dysfunction in white versus gray brain matter. Anal. Chem. 2011;83:7712–7720. doi: 10.1021/ac201102p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szczerbowska-Boruchowska M., Dumas P., Kastyak M.Z., Chwiej J., Lankosz M., Adamek D., Krygowska-Wajs A. Biomolecular investigation of human substantia nigra in Parkinson's disease by synchrotron radiation Fourier transform infrared microspectroscopy. Arch. Biochem. Biophys. 2007;459:241–248. doi: 10.1016/j.abb.2006.12.027. [DOI] [PubMed] [Google Scholar]
- 38.Taubner T., Hillenbrand R., Keilmann F. Nanoscale polymer recognition by spectral signature in scattering infrared near-field microscopy. Appl. Phys. Lett. 2004;85:5064–5066. [Google Scholar]
- 39.Dominguez G., Mcleod A., Gainsforth Z., Kelly P., Bechtel H.A., Keilmann F., Westphal A., Thiemens M., Basov D. Nanoscale infrared spectroscopy as a non-destructive probe of extraterrestrial samples. Nat. Commun. 2014;5:5445. doi: 10.1038/ncomms6445. [DOI] [PubMed] [Google Scholar]
- 40.Huth F., Govyadinov A., Amarie S., Nuansing W., Keilmann F., Hillenbrand R. Nano-FTIR absorption spectroscopy of molecular fingerprints at 20 nm spatial resolution. Nano Lett. 2012;12:3973–3978. doi: 10.1021/nl301159v. [DOI] [PubMed] [Google Scholar]
- 41.Ruggeri F.S., Šneideris T., Chia S., Vendruscolo M., Knowles T.P. Characterizing individual protein aggregates by infrared nanospectroscopy and atomic force microscopy. JoVE. 2019 doi: 10.3791/60108. [DOI] [PubMed] [Google Scholar]
- 42.Amenabar I., Poly S., Nuansing W., Hubrich E.H., Govyadinov A.A., Huth F., Krutokhvostov R., Zhang L., Knez M., Heberle J. Structural analysis and mapping of individual protein complexes by infrared nanospectroscopy. Nat. Commun. 2013;4:2890. doi: 10.1038/ncomms3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waeytens J., Van Hemelryck V., Deniset-Besseau A., Ruysschaert J.-M., Dazzi A., Raussens V. Characterization by nano-infrared spectroscopy of individual aggregated species of amyloid proteins. Molecules. 2020;25:2899. doi: 10.3390/molecules25122899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banerjee S., Holcombe B., Ringold S., Foes A., Naik T., Baghel D., Ghosh A. Nanoscale infrared spectroscopy identifies structural heterogeneity in individual amyloid fibrils and prefibrillar aggregates. J. Phys. Chem. B. 2022;126:5832–5841. doi: 10.1021/acs.jpcb.2c04797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drzewiecki K.E., Grisham D.R., Parmar A.S., Nanda V., Shreiber D.I. Circular dichroism spectroscopy of collagen fibrillogenesis: a new use for an old technique. Biophys. J. 2016;111:2377–2386. doi: 10.1016/j.bpj.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garab G., van Amerongen H. Linear dichroism and circular dichroism in photosynthesis research. Photosynth. Res. 2009;101:135–146. doi: 10.1007/s11120-009-9424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Holde K.E. 2006. Principles of Physical Biochemistry. [Google Scholar]
- 48.Simidjiev I., Barzda V., Mustárdy L., Garab G. Isolation of lamellar aggregates of the light-harvesting chlorophyll a/b protein complex of photosystem II with long-range chiral order and structural flexibility. Anal. Biochem. 1997;250:169–175. doi: 10.1006/abio.1997.2204. [DOI] [PubMed] [Google Scholar]
- 49.Kurouski D. Advances of vibrational circular dichroism (VCD) in bioanalytical chemistry. A review. Anal. Chim. Acta. 2017;990:54–66. doi: 10.1016/j.aca.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Measey T.J., Schweitzer-Stenner R. Vibrational circular dichroism as a probe of fibrillogenesis: the origin of the anomalous intensity enhancement of amyloid-like fibrils. J. Am. Chem. Soc. 2011;133:1066–1076. doi: 10.1021/ja1089827. [DOI] [PubMed] [Google Scholar]
- 51.Benjwal S., Verma S., Röhm K.H., Gursky O. Monitoring protein aggregation during thermal unfolding in circular dichroism experiments. Protein Sci. 2006;15:635–639. doi: 10.1110/ps.051917406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsuo K., Hiramatsu H., Gekko K., Namatame H., Taniguchi M., Woody R.W. Characterization of intermolecular structure of β2-microglobulin core fragments in amyloid fibrils by vacuum-ultraviolet circular dichroism spectroscopy and circular dichroism theory. J. Phys. Chem. B. 2014;118:2785–2795. doi: 10.1021/jp409630u. [DOI] [PubMed] [Google Scholar]
- 53.Wallace B.A. Protein characterisation by synchrotron radiation circular dichroism spectroscopy. Q. Rev. Biophys. 2009;42:317–370. doi: 10.1017/S003358351000003X. [DOI] [PubMed] [Google Scholar]
- 54.Ruzza P., Hussain R., Biondi B., Calderan A., Tessari I., Bubacco L., Siligardi G. Effects of trehalose on thermodynamic properties of alpha-synuclein revealed through synchrotron radiation circular dichroism. Biomolecules. 2015;5:724–734. doi: 10.3390/biom5020724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernadó P., Åkerud T., García de la Torre J., Akke M., Pons M. Combined use of NMR relaxation measurements and hydrodynamic calculations to study protein association. Evidence for tetramers of low molecular weight protein tyrosine phosphatase in solution. J. Am. Chem. Soc. 2003;125:916–923. doi: 10.1021/ja027836h. [DOI] [PubMed] [Google Scholar]
- 56.Wu K.-P., Baum J. Detection of transient interchain interactions in the intrinsically disordered protein α-synuclein by NMR paramagnetic relaxation enhancement. J. Am. Chem. Soc. 2010;132:5546–5547. doi: 10.1021/ja9105495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Battiste J.L., Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 58.Iwahara J., Schwieters C.D., Clore G.M. Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 59.Clore G.M., Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pandya A., Howard M.J., Zloh M., Dalby P.A. An evaluation of the potential of NMR spectroscopy and computational modelling methods to inform biopharmaceutical formulations. Pharmaceutics. 2018;10:165. doi: 10.3390/pharmaceutics10040165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carr H.Y., Purcell E.M. Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys. Rev. 1954;94:630. [Google Scholar]
- 62.Meiboom S., Gill D. Modified spin‐echo method for measuring nuclear relaxation times. Rev. Sci. Instrum. 1958;29:688–691. [Google Scholar]
- 63.Mukherjee S., Pondaven S.P., Jaroniec C.P. Conformational flexibility of a human immunoglobulin light chain variable domain by relaxation dispersion nuclear magnetic resonance spectroscopy: implications for protein misfolding and amyloid assembly. Biochemistry. 2011;50:5845–5857. doi: 10.1021/bi200410c. [DOI] [PubMed] [Google Scholar]
- 64.Feng Y., Taraban M.B., Yu Y.B. Water proton NMR—a sensitive probe for solute association. Chem. Commun. 2015;51:6804–6807. doi: 10.1039/c5cc00741k. [DOI] [PubMed] [Google Scholar]
- 65.Patil S.M., Keire D.A., Chen K. Comparison of NMR and dynamic light scattering for measuring diffusion coefficients of formulated insulin: implications for particle size distribution measurements in drug products. AAPS J. 2017;19:1760–1766. doi: 10.1208/s12248-017-0127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naito A., Kawamura I. Solid-state NMR as a method to reveal structure and membrane-interaction of amyloidogenic proteins and peptides. Biochim. Biophys. Acta Biomembr. 2007;1768:1900–1912. doi: 10.1016/j.bbamem.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 67.Weingarth M., Baldus M. Solid-state NMR-based approaches for supramolecular structure elucidation. Acc. Chem. Res. 2013;46:2037–2046. doi: 10.1021/ar300316e. [DOI] [PubMed] [Google Scholar]
- 68.Linser R. Solid-state NMR spectroscopic trends for supramolecular assemblies and protein aggregates. Solid State Nucl. Magn. Reson. 2017;87:45–53. doi: 10.1016/j.ssnmr.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 69.Duer M.J. John Wiley & Sons; 2008. Solid State NMR Spectroscopy: Principles and Applications. [Google Scholar]
- 70.Karamanos T.K., Jackson M.P., Calabrese A.N., Goodchild S.C., Cawood E.E., Thompson G.S., Kalverda A.P., Hewitt E.W., Radford S.E. Structural mapping of oligomeric intermediates in an amyloid assembly pathway. Elife. 2019;8 doi: 10.7554/eLife.46574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arar S., Haque M.A., Kayed R. Protein aggregation and neurodegenerative disease: structural outlook for the novel therapeutics. Proteins: Struct., Funct., Bioinf. 2023:1–16. doi: 10.1002/prot.26561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turk M., Baumeister W. The promise and the challenges of cryo‐electron tomography. FEBS Lett. 2020;594:3243–3261. doi: 10.1002/1873-3468.13948. [DOI] [PubMed] [Google Scholar]
- 73.Wan Q., Mouton S.N., Veenhoff L.M., Boersma A.J. A FRET-based method for monitoring structural transitions in protein self-organization. Cell reports methods. 2022;2 doi: 10.1016/j.crmeth.2022.100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valli J., Garcia-Burgos A., Rooney L.M., e Oliveira B.V.d.M., Duncan R.R., Rickman C. Seeing beyond the limit: a guide to choosing the right super-resolution microscopy technique. J. Biol. Chem. 2021:297. doi: 10.1016/j.jbc.2021.100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jing Y., Zhang C., Yu B., Lin D., Qu J. Super-resolution microscopy: shedding new light on in vivo imaging. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.746900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cosentino M., Canale C., Bianchini P., Diaspro A. AFM-STED correlative nanoscopy reveals a dark side in fluorescence microscopy imaging. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodrigues B.I.A. 2022. Assessment of Protein Aggregation Using Super-resolution Microscopy. [Google Scholar]
- 78.Coltharp C., Xiao J. Superresolution microscopy for microbiology. Cell Microbiol. 2012;14:1808–1818. doi: 10.1111/cmi.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fuhrmann M., Gockel N., Arizono M., Dembitskaya Y., Nägerl U.V., Pennacchietti F., Damenti M., Testa I., Willig K.I. Super-resolution microscopy opens new doors to life at the nanoscale. J. Neurosci. 2022;42:8488–8497. doi: 10.1523/JNEUROSCI.1125-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sigal Y.M., Zhou R., Zhuang X. Visualizing and discovering cellular structures with super-resolution microscopy. Science. 2018;361:880–887. doi: 10.1126/science.aau1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen R., Tang X., Zhao Y., Shen Z., Zhang M., Shen Y., Li T., Chung C.H.Y., Zhang L., Wang J. Single-frame deep-learning super-resolution microscopy for intracellular dynamics imaging. Nat. Commun. 2023;14:2854. doi: 10.1038/s41467-023-38452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feiner-Gracia N., Pujals S., Delcanale P., Albertazzi L. Elsevier; 2018. pp. 219–236. (Advanced Optical Microscopy Techniques for the Investigation of Cell-Nanoparticle Interactions. In Smart Nanoparticles for Biomedicine). [Google Scholar]
- 83.Lumkwana D., Engelbrecht L., Loos B. Monitoring autophagy using super-resolution structured illumination and direct stochastic optical reconstruction microscopy. Methods in Cell Biology; Elsevier. 2021;165:139–152. doi: 10.1016/bs.mcb.2020.12.005. [DOI] [PubMed] [Google Scholar]
- 84.Lee K., Kinnunen M., Khokhlova M.D., Lyubin E.V., Priezzhev A.V., Meglinski I., Fedyanin A.A. Optical tweezers study of red blood cell aggregation and disaggregation in plasma and protein solutions. J. Biomed. Opt. 2016;21:35001. doi: 10.1117/1.JBO.21.3.035001. [DOI] [PubMed] [Google Scholar]
- 85.Lehmann K., Shayegan M., Blab G.A., Forde N.R. Optical tweezers approaches for probing multiscale protein mechanics and assembly. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.577314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bustamante C., Alexander L., Maciuba K., Kaiser C.M. Single-molecule studies of protein folding with optical tweezers. Annu. Rev. Biochem. 2020;89:443–470. doi: 10.1146/annurev-biochem-013118-111442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ritchie D.B., Woodside M.T. Probing the structural dynamics of proteins and nucleic acids with optical tweezers. Curr. Opin. Struct. Biol. 2015;34:43–51. doi: 10.1016/j.sbi.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zaltron A., Merano M., Mistura G., Sada C., Seno F. Optical tweezers in single-molecule experiments. The European Physical Journal Plus. 2020;135:896. [Google Scholar]
- 89.Maciuba K., Zhang F., Kaiser C.M. Facile tethering of stable and unstable proteins for optical tweezers experiments. Biophys. J. 2021;120:2691–2700. doi: 10.1016/j.bpj.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiong R., Vandenbroucke R.E., Broos K., Brans T., Van Wonterghem E., Libert C., Demeester J., De Smedt S.C., Braeckmans K. Sizing nanomaterials in bio-fluids by cFRAP enables protein aggregation measurements and diagnosis of bio-barrier permeability. Nat. Commun. 2016;7 doi: 10.1038/ncomms12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Periasamy N., Verkman A. Analysis of fluorophore diffusion by continuous distributions of diffusion coefficients: application to photobleaching measurements of multicomponent and anomalous diffusion. Biophys. J. 1998;75:557–567. doi: 10.1016/S0006-3495(98)77545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seiffert S., Oppermann W. Systematic evaluation of FRAP experiments performed in a confocal laser scanning microscope. J. Microsc. 2005;220:20–30. doi: 10.1111/j.1365-2818.2005.01512.x. [DOI] [PubMed] [Google Scholar]
- 93.Kumar V., Sami N., Kashav T., Islam A., Ahmad F., Hassan M.I. Protein aggregation and neurodegenerative diseases: from theory to therapy. Eur. J. Med. Chem. 2016;124:1105–1120. doi: 10.1016/j.ejmech.2016.07.054. [DOI] [PubMed] [Google Scholar]
- 94.Gregoire S., Irwin J., Kwon I. Techniques for monitoring protein misfolding and aggregation in vitro and in living cells. Kor. J. Chem. Eng. 2012;29:693–702. doi: 10.1007/s11814-012-0060-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Frid P., Anisimov S.V., Popovic N. Congo red and protein aggregation in neurodegenerative diseases. Brain Res. Rev. 2007;53:135–160. doi: 10.1016/j.brainresrev.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 96.Sutter M., Oliveira S., Sanders N.N., Lucas B., van Hoek A., Hink M.A., Visser A.J., De Smedt S.C., Hennink W.E., Jiskoot W. Sensitive spectroscopic detection of large and denatured protein aggregates in solution by use of the fluorescent dye Nile red. J. Fluoresc. 2007;17:181–192. doi: 10.1007/s10895-007-0156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Siraj S., Yameen D., Shamsi A., Khan F., Islam A., Haque M.M. Interaction of Thioflavin T (ThT) and 8-anilino-1-naphthalene sulfonic acid (ANS) with macromolecular crowding agents and their monomers: biophysical analysis using in vitro and computational approaches. J. Mol. Liq. 2023;374 [Google Scholar]
- 98.Singh K., Hussain I., Mishra V., Akhtar M.S. New insight on 8-anilino-1-naphthalene sulfonic acid interaction with TgFNR for hydrophobic exposure analysis. Int. J. Biol. Macromol. 2019;122:636–643. doi: 10.1016/j.ijbiomac.2018.10.208. [DOI] [PubMed] [Google Scholar]
- 99.Pretorius E., Page M.J., Hendricks L., Nkosi N.B., Benson S.R., Kell D.B. Both lipopolysaccharide and lipoteichoic acids potently induce anomalous fibrin amyloid formation: assessment with novel Amytracker™ stains. J. R. Soc. Interface. 2018;15 doi: 10.1098/rsif.2017.0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kitts C.C., Vanden Bout D.A. A spectroscopic study of 2-[4′-(dimethylamino) phenyl]-benzothiazole binding to insulin amyloid fibrils. J. Fluoresc. 2010;20:881–889. doi: 10.1007/s10895-010-0634-0. [DOI] [PubMed] [Google Scholar]
- 101.How S.-C., Cheng Y.-H., Lo C.-H., Lai J.-T., Lin T.-H., Bednarikova Z., Antosova A., Gazova Z., Wu J.W., Wang S.S.-S. Exploring the effects of methylene blue on amyloid fibrillogenesis of lysozyme. Int. J. Biol. Macromol. 2018;119:1059–1067. doi: 10.1016/j.ijbiomac.2018.08.038. [DOI] [PubMed] [Google Scholar]
- 102.Harper J.D., Lansbury P.T., Jr. Models of amyloid seeding in Alzheimer's disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 103.Mahler H.-C., Friess W., Grauschopf U., Kiese S. Protein aggregation: pathways, induction factors and analysis. J. Pharmaceut. Sci. 2009;98:2909–2934. doi: 10.1002/jps.21566. [DOI] [PubMed] [Google Scholar]
- 104.Falke S., Betzel C. Dynamic. Light scattering (DLS) principles, perspectives, applications to biological samples. Radiation in Bioanalysis: Spectroscopic Techniques and Theoretical Methods. 2019:173–193. [Google Scholar]
- 105.Wang C., Zhong X., Ruffner D.B., Stutt A., Philips L.A., Ward M.D., Grier D.G. Holographic characterization of protein aggregates. J. Pharmaceut. Sci. 2016;105:1074–1085. doi: 10.1016/j.xphs.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stetefeld J., McKenna S.A., Patel T.R. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophysical reviews. 2016;8:409–427. doi: 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lorber B., Fischer F., Bailly M., Roy H., Kern D. Protein analysis by dynamic light scattering: methods and techniques for students. Biochem. Mol. Biol. Educ. 2012;40:372–382. doi: 10.1002/bmb.20644. [DOI] [PubMed] [Google Scholar]
- 108.Pukala T.L. Mass spectrometric insights into protein aggregation. Essays Biochem. 2023;67:243–253. doi: 10.1042/EBC20220103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Turunen O., Jänis J., Fenel F., Leisola M. vol. 388. Elsevier; 2004. pp. 156–167. (Engineering the Thermotolerance and pH Optimum of Family 11 Xylanases by Site-Directed Mutagenesis. In Methods in Enzymology). [DOI] [PubMed] [Google Scholar]
- 110.Bronsoms S., Trejo S.A. Applications of mass spectrometry to the study of protein aggregation. Insoluble Proteins: Methods and Protocols. 2015:331–345. doi: 10.1007/978-1-4939-2205-5_19. [DOI] [PubMed] [Google Scholar]
- 111.Del Mar C., Greenbaum E.A., Mayne L., Englander S.W., Woods V.L., Jr. Structure and properties of α-synuclein and other amyloids determined at the amino acid level. Proc. Natl. Acad. Sci. USA. 2005;102:15477–15482. doi: 10.1073/pnas.0507405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oganesyan I., Lento C., Wilson D.J. Contemporary hydrogen deuterium exchange mass spectrometry. Methods. 2018;144:27–42. doi: 10.1016/j.ymeth.2018.04.023. [DOI] [PubMed] [Google Scholar]
- 113.Ruggeri F.S., Šneideris T., Vendruscolo M., Knowles T.P. Atomic force microscopy for single molecule characterisation of protein aggregation. Arch. Biochem. Biophys. 2019;664:134–148. doi: 10.1016/j.abb.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chiang Y.-L., Chang Y.-C., Chiang I.-C., Mak H.-M., Hwang I.-S., Shih Y.-L. Atomic force microscopy characterization of protein fibrils formed by the amyloidogenic region of the bacterial protein MinE on mica and a supported lipid bilayer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dong J., Mao G. Direct study of C12E5 aggregation on mica by atomic force microscopy imaging and force measurements. Langmuir. 2000;16:6641–6647. [Google Scholar]
- 116.Held D., Kilz P. Size-exclusion chromatography as a useful tool for the assessment of polymer quality and determination of macromolecular properties. Chemistry Teacher International. 2021;3:77–103. [Google Scholar]
- 117.Hong P., Koza S., Bouvier E.S. A review size-exclusion chromatography for the analysis of protein biotherapeutics and their aggregates. J. Liq. Chromatogr. Relat. Technol. 2012;35:2923–2950. doi: 10.1080/10826076.2012.743724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McAllister C., Karymov M.A., Kawano Y., Lushnikov A.Y., Mikheikin A., Uversky V.N., Lyubchenko Y.L. Protein interactions and misfolding analyzed by AFM force spectroscopy. J. Mol. Biol. 2005;354:1028–1042. doi: 10.1016/j.jmb.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 119.Pekar A., Sukumar M. Quantitation of aggregates in therapeutic proteins using sedimentation velocity analytical ultracentrifugation: practical considerations that affect precision and accuracy. Anal. Biochem. 2007;367:225–237. doi: 10.1016/j.ab.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 120.Berkowitz S.A. Role of analytical ultracentrifugation in assessing the aggregation of protein biopharmaceuticals. AAPS J. 2006;8:E590–E605. doi: 10.1208/aapsj080368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Drexel R., Sogne V., Dinkel M., Meier F., Klein T. Asymmetrical flow field-flow fractionation for sizing of gold nanoparticles in suspension. J. Vis. Exp. 2020;163 doi: 10.3791/61757. [DOI] [PubMed] [Google Scholar]
- 122.Wagner M., Holzschuh S., Traeger A., Fahr A., Schubert U.S. Asymmetric flow field-flow fractionation in the field of nanomedicine. Anal. Chem. 2014;86:5201–5210. doi: 10.1021/ac501664t. [DOI] [PubMed] [Google Scholar]
- 123.Chaturvedi S.K., Siddiqi M.K., Alam P., Khan R.H. Protein misfolding and aggregation: mechanism, factors and detection. Process Biochem. 2016;51:1183–1192. [Google Scholar]
- 124.Mitra A.K. Visualization of biological macromolecules at near-atomic resolution: cryo-electron microscopy comes of age. Acta Crystallogr. F: Structural Biology Communications. 2019;75:3–11. doi: 10.1107/S2053230X18015133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fitzpatrick A.W., Saibil H.R. Cryo-EM of amyloid fibrils and cellular aggregates. Curr. Opin. Struct. Biol. 2019;58:34–42. doi: 10.1016/j.sbi.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Creekmore B.C., Chang Y.-W., Lee E.B. The Cryo-EM effect: structural biology of neurodegenerative disease aggregates. J. Neuropathol. Exp. Neurol. 2021;80:514–529. doi: 10.1093/jnen/nlab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hojjatian A., Dasari A.K., Sengupta U., Taylor D., Daneshparvar N., Yeganeh F.A., Dillard L., Michael B., Griffin R.G., Borgnia M. Distinct cryo-EM structure of α-synuclein filaments derived by tau. bioRxiv. 2021;2020(2031) doi: 10.1016/j.bbrc.2021.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hojjatian A., Dasari A.K., Sengupta U., Taylor D., Daneshparvar N., Yeganeh F.A., Dillard L., Michael B., Griffin R.G., Borgnia M.J. Tau induces formation of α-synuclein filaments with distinct molecular conformations. Biochem. Biophys. Res. Commun. 2021;554:145–150. doi: 10.1016/j.bbrc.2021.03.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guerrero-Ferreira R., Taylor N.M., Mona D., Ringler P., Lauer M.E., Riek R., Britschgi M., Stahlberg H. Cryo-EM structure of alpha-synuclein fibrils. Elife. 2018;7 doi: 10.7554/eLife.36402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fitzpatrick A.W., Falcon B., He S., Murzin A.G., Murshudov G., Garringer H.J., Crowther R.A., Ghetti B., Goedert M., Scheres S.H. Cryo-EM structures of tau filaments from Alzheimer's disease. Nature. 2017;547:185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li B., Ge P., Murray K.A., Sheth P., Zhang M., Nair G., Sawaya M.R., Shin W.S., Boyer D.R., Ye S. Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat. Commun. 2018;9:3609. doi: 10.1038/s41467-018-05971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Da Vela S., Svergun D.I. Methods, development and applications of small-angle X-ray scattering to characterize biological macromolecules in solution. Current research in structural biology. 2020;2:164–170. doi: 10.1016/j.crstbi.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Barbosa L.R.S., Spinozzi F., Mariani P., Itri R. Small‐angle X‐ray scattering applied to proteins in solution. Proteins in Solution and at Interfaces: Methods and Applications in Biotechnology and Materials Science. 2013:49–72. [Google Scholar]
- 134.Doniach S., Lipfert J. vol. 469. Elsevier; 2009. pp. 237–251. (Use of Small Angle X-Ray Scattering (SAXS) to Characterize Conformational States of Functional RNAs. In Methods in Enzymology). [DOI] [PubMed] [Google Scholar]
- 135.Lin L.-E., Miao K., Qian C., Wei L. High spatial-resolution imaging of label-free in vivo protein aggregates by VISTA. Analyst. 2021;146:4135–4145. doi: 10.1039/d1an00060h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There is no data required to be made available in the review article.