Abstract
Epithelial ovarian cancer (EOC) is associated with a dismal prognosis due to development of resistance to chemotherapy and metastasis in the peritoneal cavity and distant organs. In order to identify new targets and treatment modalities we searched the literature for up- and and down-regulated circRNAs with efficacy in preclinical EOC-related in vivo systems. Our search yielded circRNAs falling into the following categories: cisplatin and paclitaxel resistance, transmembrane receptors, secreted factors, transcription factors, RNA splicing and processing factors, RAS pathway-related components, proteolysis and cell-cycle regulation, signaling-related proteins, and circRNAs regulating proteins in additional categories. These findings can be potentially translated by validation and manipulation of the corresponding targets, inhibition of circRNAs with antisense oligonucleotides (ASO), small interfering RNAs (siRNA) or small hairpin RNA (shRNA) or by reconstituting their activity.
Keywords: Inhibition and reconstitution of circ RNA, small hairpin RNA (shRNA), small interfering RNA (siRNA), target validation, xenograft models, review
The incidence of ovarian cancer (OC) in the USA is 22,000 cases per year and 14,000 deaths, and 29,000 deaths in Europe (1). The vast majority are epithelial ovarian cancers (EOC). This type of tumor is typically treated with a combination of surgery and chemotherapy, which often includes cisplatin (DDP) and paclitaxel (PTX). An exciting development was the treatment of homologous recombination-deficient ovarian tumors with breast cancer antigen (BRCA) mutations or deletions with poly-ADP-ribose polymerase (PARP) inhibitors (2). Three PARP inhibitors (olaparib, rucaparib and niraparib) have been approved for the treatment of OCs in different clinical settings. In addition, anti-angiogenic therapy with Bevacizumab was approved (3). Nevertheless, recurrence due to resistance mechanisms is commonly observed and significantly impairs the therapeutic benefit of treatment (4). Epithelial ovarian cancer (EOC) gains a growth advantage due to interactions with fat cells, mesothelial cells, and cancer-associated fibroblasts. These interactions enhance the activation of pathways that support its growth (5-7). Also, its dissemination in ascites fluid and formation of nodules in the peritoneum are a unique mechanism of metastasis hampering therapeutic intervention (8,9). Immunotherapy with checkpoint inhibitors such as Pembrolizumab has shown limited success (10). Diverse mechanisms of immune escape in a tumor-location specific manner have been observed (11). Taken together, there is a tremendous medical need for identification of new targets and modalities for treatment of OC. Therefore, we searched the literature for circular RNAs (circRNAs) that are up- or down-regulated in EOC and upon their modulation exhibit efficacy in preclinical in vivo models of EOC.
Circular RNA in Cancer
circRNA can be generated during mRNA processing by intra-molecular mRNA pairing followed by circularization, RNA-binding protein-mediated and lariat-driven circularization (12). Physiologically, circRNAs are involved in a plethora of cellular functions (13). In cancer, circRNAs can exert oncogenic as well as tumor-suppressive functions modulating proliferation, angiogenesis, migration, and metastasis (14-17). In addition, circRNAs can act as scaffolds or sponges for proteins, regulate transcription, interact with RNAs, modulate translation and, in some cases, they can encode proteins (18). Another important issue is their impact on regulation of inter-cell interactions and orchestration of the tumor micro-environment through their exosome-mediated transmission (19). Recently, circRNAs have been engineered for protein production (20). The function of circRNAs in EOC as biomarkers, prognostic markers and as targets for therapy have been summarized in several recent reviews (21-23). Here, we focus on circRNAs which exhibit in vivo activity in EOC-related preclinical models. The identified targets can be either inhibited or reconstituted with small molecules or antibody-derived entities and the corresponding circRNAs can be reconstituted or inhibited with ASO, siRNA or shRNA or by intervention with clustered regularly interspaced short palindromic repeats/CRISPR associated enzymes (CRISPR-CAS) (24,25).
CircRNAs Deregulated in EOC
Circular RNAs and cisplatin (DDP) resistance.
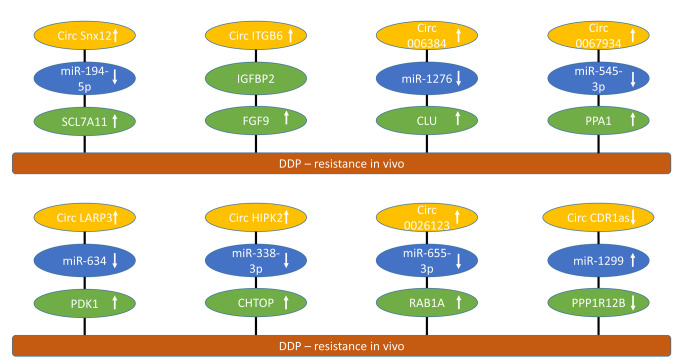
Circ sorting nexin 12 (Snx12) up-regulates solute carrier family 7 member 11 (SCL7A11). circSnx12 (Figure 1) was up-regulated in EOC tissues and cells resistant to DDP (26). circSnx12 confers DDP-resistance in SKOV3 and A2780 EOC cells by inducing ferroptosis resistance (26). miR-194-5p was sponged by circSnx12 resulting in up-regulation of SCL7A11. The SLC superfamily includes 458 proteins, which transport a variety of substances across cell membranes (27). Knockdown of circSnx12 rendered DDP-resistant cells sensitive to DDP in vitro (SKOV3 and A2780) and in vivo (SKOV3) after subcutaneous (s.c.) implantation into nude mice (26). Transmembrane protein SLC7A11 is a key component of the glutamate antiporter system which is involved in glutathione (GSH) synthesis and resistance to ferroptosis (28). Inhibition of ferroptosis is a common phenomenon in EOC. Ferroptosis is associated with lipid peroxidation, iron accumulation and GSH deprivation (29). DDP resistance is a major challenge in the treatment of EOC (30,31).
Figure 1. circRNAs involved in cisplatin resistance of epithelial ovarian cancer with efficacy in preclinical in vivo models. circRNAs mediating cisplatin resistance are shown. Upward arrows indicate up-regulation, and downward arrows indicate down-regulation. CHTOP: Chromatin target of CHTOP; circCDRas: circ cerebellar degenerated-related protein 1 antisense; HIPK2: circ homeodomain containing protein kinase 2; circITGB6: circ integrin β6; circLAPR3: circ lysophosphatic acid receptor 3; circSNX12: circ sorting nexin 12; CLU: clusterin; DDP: cisplatin; FGF9: fibroblast growth factor 9; miR: micro RNA; DNAJB6: DNA homolog subfamily B, member 6; KLF4: Krueppel-like factor 4; PDK1: pyruvate dehydrogenase kinase 1; PPA-1: inorganic pyrophosphatase 1; PPP1R12B: protein phosphatase 1 regulatory subunit 12B; RAB1A: ras-related protein RAB1A; SCL7A11: solute carrier family 7, member 11.
Circ integrin β6 (ITGB6) stabilizes fibroblast growth factor 9 (FGF9). Elevated levels of circITGB6 (Figure 1) correlated with DDP chemoresistance and poor prognosis in EOC patients (32). circITGB6 induced chemoresistance of ID8 OC cells in nude mice after intraperitoneal injection in a tumor micro-environment (TME)-dependent manner (32). circITGB6 induced macrophage polarization towards the M2 phenotype (32). circITGB6 interacted with insulin-growth factor binding protein 2 (IGFBP2) directly, forming a ternary complex with FGF9. M2 macrophages have been shown to play a role in the progression and chemoresistance of EOC (33). Macrophage-based evaluation of therapy options for EOC are under investigation (34). It has been shown that FGF9 promotes DDP resistance in colorectal cancer patients by activation of wingless-related integration site (WNT)/β catenin signaling (35). In lung cancer, interaction between FGF9 and fibroblast growth factor receptor 1 (FGFR1) promotes proliferation, epithelial-mesenchymal transition (EMT), M2 polarization and liver metastasis (36).
Circ0063804 up-regulates clusterin (CLU). High expression of circ006384 (Figure 1) indicated poor prognosis in EOC patients (37). In OVCAR3 cells, silencing of circ006384 inhibited proliferation and DDP-resistance and induced apoptosis (37). circ006384 sponged miR-1276 and up-regulated CLU. circ006384 silencing inhibited growth of OVCAR3 xenografts after s.c. implantation into nude mice (37). CLU is a chaperone involved in the protein folding pathway with oncogenic and cytoprotective effects similar to heat-shock proteins (38-40). Custirsen (OGX-011), a second-generation antisense inhibitor of CLU has been evaluated in prostate cancer, hepatocellular carcinoma and lung cancer patients, but did not reach the expected clinical endpoints (41,42).
Circ0067934 up-regulates inorganic pyrophosphatase PPA1. circ0067934 (Figure 1) was found to be over-expressed in EOC samples and was associated with advanced tumor stages and lymph node metastasis (43). Down-regulation of circ0067934 reduced DDP resistance of cell line A2780/DDP in vitro and in xenografts in nude mice (43). circ0067934 sponged miR-545-3p and up-regulated PPA1. Up-regulation of PPA1 also enhanced proliferation, invasion and reduced pro-apoptotic JUN-kinase (JNK) signaling in A2780 cells (43). It was shown that the role of PPA1 in energy metabolism is to promote progression of malignant tumors (44). PPA1-mediated dephosphorylation was found to positively regulate the activity of yes-associated protein 2 (YAP2), promoting survival and DDP-resistance of EOC cells (45). YAP2, which is an effector of the HIPPO pathway, functions as an EOC oncogene (46). Dephosphorylated YAP was shown to be involved in the pathogenesis of EOC (47).
Circ lysophosphatic acid receptor 3 (circLARP3) up-regulates pyruvate dehydrogenase kinase1 (PDK1). circLARP3 (Figure 1) was up-regulated in DDP-resistant EOC tissues and cell lines (48). Knockdown of circLARP3 enhanced DDP-sensitivity of SKOV3/DDP and A2780/DDP cells in vitro and of SKOV3/DDP xenografts after s.c. injection into nude mice (48). circLARP3 sponged miR-634 and up-regulated PDK1. The latter was shown to be involved in promotion of proliferation, migration, glycolysis, and inhibition of apoptosis of EOC cell lines. It was demonstrated that PDK1 plays an oncogenic and prognostic role in progression and metastasis of EOC (49). Perturbation of PDK1 modulates growth, angiogenesis, and metabolic pathways in EOC xenografts (50). It was also found that PDK1 can contribute to DDP-resistance in EOC through activation of the epidermal growth factor receptor (EGFR) pathway (51).
Circ homeodomain-interacting protein kinase 2 (circHIPK2) up-regulates chromatin target of PRMT1 (CHTOP). circHIPK2 (Figure 1) was up-regulated in EOC patients and associated with DDP-resistance (52). In SKOV3/DDP and A2789/DDP EOC cells, knockdown of circHIPK2 inhibited proliferation, migration, invasion, cell-cycle progression, increased apoptosis, reversed DDP resistance in vitro and restrained tumor growth and DDP-resistance in A2780/DDP xenografts after s.c. implantation into nude mice. circHIPK2 sponged miR-338-3p and up-regulated CHTOP. The latter functions in the regulation of gene transcription and mRNA transport (53) and is involved in the chemoresistance of EOC (54). CHTOP binds to arginine methyltransferases PRMT1 and PRMT5 promoting methylation of arginine residues (54). Its knockdown inhibited stemness, invasion and metastasis and enhanced DDP-induced apoptosis in EOC cells (55).
circ0026123 up-regulates Ras-related protein RAB1A. The expression of circ0026123 (Figure 1) was found to be elevated in DDP-resistant EOC tumors and cells (56). Down-regulation of circ0026123 inhibited EOC cell growth, angiogenesis, migration, and invasion and interfered with DDP resistance (56). circ0026123 inhibition decreased EOC growth in nude mice and enhanced cytotoxicity of DDP (56). miR-655-3p was sponged by circ0026123 resulting in up-regulation of RAB1A. It was shown previously that miR-655-3p inhibits proliferation and migration of EOC cells by targeting RAB1A (57). RAB GTPases are coordinators of vesicle trafficking in normal and cancer cells (58,59). They can act as oncogenes as well as tumor suppressors. RAB-mediated membrane delivery can shift normal cell behavior towards malignancy (59). In EOC, deregulation of GTPases of the RAS superfamily is commonly observed (60).
Circ cerebellar degenerated-related protein 1 antisense (circCDR1as) down-regulates protein phosphatase 1 regulatory subunit 12B (PPP1R12B). Down-regulation of circCDR1as in EOC was associated with DDP- resistance (61). circCDR1 (Figure 1) suppressed DDP-resistance in SKOV/DDP and HO8910/DDP cells in vitro and inhibited DDP-chemoresistance in nude mice after s.c. administration. circCDR1as sponged miR-1299, up-regulated PPP1R12B and inhibited AKT/mammalian target of rapamycin (mTOR) signaling (61). PPP1R12B contains a terminal leucine zipper and is involved in dimerization and protein-protein interaction (62). In colon cancer, PPP1R12B promotes proliferation, migration, inhibits apoptosis and up-regulates phosphatidyl-inosite-3-kinase (PI3K)/AKT signaling (63,64).
CircRNAs and Paclitaxel (PTX) Resistance
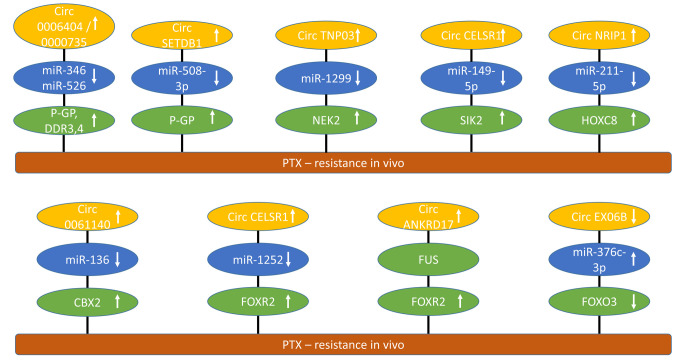
Circ0006404 and circ0000735 up-regulate P-glycoprotein (P-GP). circ0006404 and circ0000735 (Figure 2) were suppressed in PTX-treated SKOV3 cells and regulated response to PTX in vitro and in xenograft models (65). PTX resistance is a major problem in the treatment of EOC (66,67). Both sponged miR-346 and miR-526, resulting in up-regulation of Dickkopf 3 and 4 (DKK3,4) and P-GP. DKK proteins are antagonists of WNT signaling through interactions with LDL receptor related protein 5 and 6 (LRP5 and LRP6) (68). P-GP, also known as ABC subfamily B member 1 (ABCC1) and multidrug resistance protein 1 (MDR1), is an ATP-binding efflux protein, which is involved in resistance against several chemotherapeutic agents (69). It is a member of a family of ABC transporters of more than 40 proteins, which in addition to P-GP includes multi-drug resistance protein (MRP) and breast cancer resistance protein (BCRP) (70).
Figure 2. circRNAs involved in paclitaxel resistance of epithelial ovarian cancer with efficacy in preclinical in vivo models. CircRNAs mediating paclitaxel resistance are shown. Up-ward arrows indicate up-regulation, downward arrows correlate with down-regulation. CBX2: Chromobox 2; circANKRD17: circ ankyrin repeat domain 17; circCELSR1: circ cadherin EGF LAG receptor seven-pass G-type receptor 1; EXO6B: circ exocyst complex component 6B; circNRIP1: circ nuclear receptor interacting protein 1; circSETDB1: circ bifurcated lysine methyltransferase 1; circTNPO3: circ transportin 3; FOXO3: forkhead box protein O3; FOXR2: forkhead box 2; HOXC8: homeobox C8; miR: micro RNA; NEK2: never in mitosis (NIMA) related kinase 2; P-GP: P-glycoprotein; PTX: paclitaxel; SIK2: salt-induced kinase 2.
CircSET domain bifurcated histone lysine methyltransferase 1 (circSETDB1) up-regulates P-glycoprotein (P-GP). circSETDB1 (Figure 2) was highly expressed in PTX-resistant EOC (71). Knockdown of SETDB1 inhibited proliferation, half-maximal inhibitory concentration of PTX, cell-cycle progression and induced apoptosis in PTX-resistant EOC cells. In nude mice, knockdown of circSETDB1 enhanced PTX sensitivity of xenografts. circSETDB1 sponged miR-508-3p and up-regulated P-GP (72).
Circ transportin 3 (circTNPO3) up-regulates never in mitosis (NIMA) related kinase 2 (NEK2). circTNPO3 (Figure 2) was found to be over-expressed in EOC and associated with PTX resistance (73). Downregulation of circTNPO3 rendered PTX resistant cell lines derived from SKOV3 and Hey1-8 cells more sensitive to PTX. In nude mice, knockdown of circTNPO3 enhanced the anti-tumor effect of PTX in EOC xenografts. circTNPO3 sponged miR-1299 and up-regulated NEK2. The latter is involved in mitotic processes, such as centrosome duplication and separation, microtubule stabilization, kinetochore attachment and spindle assembly checkpoint (74). NEK2 expression correlated with rapid relapse and poor outcome in several types of cancer and its therapeutic potential is under investigation (75). In EOC, NEK2 is over-expressed, promotes proliferation and migration and is associated with drug resistance (76,77).
Circ Cadherin EGF LAG seven-pass G-type receptor 1 (circCELSR1) up-regulates salt-induced kinase 2 (SIK2). circCELSR1 (Figure 2) was up-regulated in PTX-resistant EOC and cell lines (78). In PTX-resistant EOC cell lines, knockdown of circCELSR1 enhanced PTX-sensitivity as well as cell apoptosis and repressed cell viability, colony formation and cell-cycle progression. circCELSR1 silencing impeded PTX-resistance in EOC xenografts in nude mice. SIK2 was up-regulated by sponging of miR-149-5p by circCELSR1. Three SIKs have been identified, which loosely resemble AMP-activated protein kinase (AMPK) (79). They can promote oncogenic as well as tumor-suppressive functions (80). In EOC, SIK2 functions as a centrosome kinase, which is required for bipolar mitotic spindle cell formation (81). SIK2 has been shown to promote EOC motility by phosphorylation of myosin-light chain kinase (MYLK) (82). It also enhances synthesis of fatty acids and cholesterol and promotes tumor growth through PI3K/AKT signaling in EOC (83). Structure-based design of selective SIK inhibitors is an ongoing effort (84).
Circ nuclear receptor interacting protein 1 (circNRIP1) up-regulates homeobox C8 (HOXC8). circNRIP1 (Figure 2) was highly expressed in PTX-resistant EOC and cell lines (85). Silencing of circNRIP1 in PTX-resistant SKOV3 and A2780 cells repressed PTX-resistance in vitro and of SKOV3/PTX xenografts in nude mice after s.c. implantation. circNRIP1 sponged miR-211-5p and up-regulated HOXC8. The latter belongs to a family of at least 39 HOX genes in mammals, which act as transcription factors and contribute to the hallmarks of cancer (86,87). HOX genes play an important role in serous OC (88). HOXC8 is associated with poor prognosis in EOC (89).
Circ0061140 up-regulates chromobox 2 (CBX2). circ0061140 (Figure 2) was up-regulated in PTX-resistant EOC tissues and cells such as SKOV3 and HeyA8 cells (90). In nude mice, knockdown of circ0061140 inhibited tumor growth and PTX resistance of SKOV3 xenografts after s.c. implantation. circ0061140 sponged miR-136 and up-regulated CBX2. The latter is part of the polycomb group complex (PcG), which silences genes by remodeling of chromatin and is involved in renewal of cancer stem cells (91-93). CBX2 has prognostic value and therapeutic potential for EOC (94). CBX2 has been identified as a driver of anoikis escape and dissemination of EOC (94).
Circ cadherin EGF LAG seven pass G-type receptor 1 (CELSR1) up-regulates forkhead box 2 (FOXR2). circCELSR1 (Figure 2) was highly expressed in EOC and correlated with PTX resistance (95). Silencing of circCELSR1 enhanced PTX-induced cytotoxicity in SKOV3/PTX and HeyA-8/PTX cells. circCELSR1 knockdown increased the anti-tumor effect of PTX in SKOV3/PTX xenografts after s.c. implantation into nude mice. mir-1252 was sponged by circCELSR1 resulting in up-regulation of FOXR2. The latter is part of the forkhead family of transcription factors, which comprises 43 members (96) and exhibits oncogenic function (97). FOXR2 promotes metastasis and growth of EOC by stimulating angiogenesis and activating hedgehog signaling (98). FOXR2 also is involved in DDP resistance of EOC (99).
Circ ankyrin repeat domain protein 17 (circANKRD17) stabilizes FOXR2. circANKRD17 (Figure 2) was highly expressed in EOC and correlated with PTX resistance (100). Knockdown of circANKRD17 decreased PTX-resistance, decreased cell viability and induced apoptosis in vitro and in EOC xenografts. circANKRD17 interacted with fused in sarcoma (FUS), which bound to FOXR2, resulting in its stabilization. FUS, also known as heterogeneous nuclear ribonucleoprotein (hnRNP), represents a subunit of a complex involved in maturation of pre-mRNA with a C-terminus, which binds to RNA (101,102).
Circ exocyst complex component 6B (circEXOC6B) is down-regulated and leads to inhibition of forkhead box protein O3 (FOXO3). circEXOC6B (Figure 2) was down-regulated in EOC samples (103). circEXOC6B suppressed proliferation and motility and decreased PTX resistance in EOC cells in vitro and in xenografts in nude mice. circEXOC6B sponged miR-376c-3p leading to down-regulation of FOXO3. The latter is a member of the FOXO transcription factors, which play a role in response to antitumor drugs (104,105). FOXO3 is expressed in ovarian tissues and acts as an apoptosis initiator in granulosa cells in chickens (106). FOXO3 has been shown to inhibit proliferation, invasiveness, and the tumorigenic potential of cancer cells (107).
CircRNAs Targeting Transmembrane Receptors and Secreted Proteins
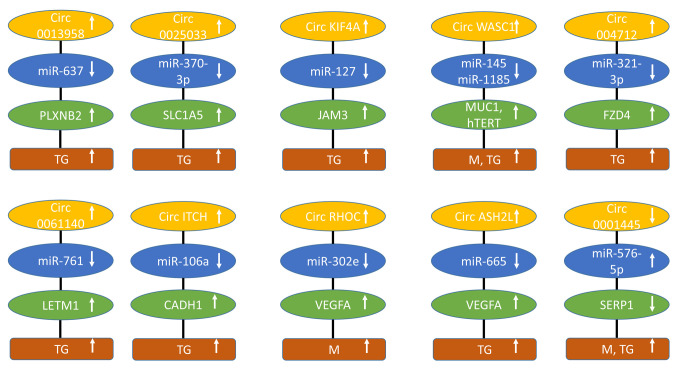
Circ0013958 up-regulates PlexinB2 (PLXNB2). circ0013958 (Figure 3) was up-regulated in EOC tissues and cells (108). Its knockdown inhibited proliferation, invasion and promoted apoptosis of SKOV3 and CAOV3 OC cells in vitro. Xenograft growth of SKOV3 cells was hampered by knockdown of circ0013958 in nude mice after s.c. implantation. circ0013958 sponged miR-637 and up-regulated PLXNB2. The latter is member of a family of nine transmembrane receptors, which interact with at least 20 members of semaphorins as ligands (109,110). Plexins B family members are involved in neural development, axon growth guidance, as well as the modulation of the cytoskeletal and adhesive machinery. In cancer, they have been shown to be involved in adhesion, proliferation, motility, invasion, and angiogenesis (110,111). PLXNB2 binds to semaphorins 4C and 4D, activates AKT and extracellular signal-regulated kinase 1,2 (ERK1/2) signaling and promotes proliferation of EOC cells (112,113).
Figure 3. circRNAs regulating transmembrane receptors and secreted factors of epithelial ovarian cancer with efficacy in preclinical in vivo models. Upward arrows indicate up-regulation, downward arrows correlate with down-regulation. circASHL2: circ histone lysine methyltransferase 2L; circITCH: circ Itchy homolog; circKLF4: circ kinesin family member 4A; circRHOC: circ RAS homology family member C; circWHSC1: circ Wolf- Hirschhorn syndrome candidate 1; CADH1: cadherin 1; FZD4: frizzled 4; hTERT: human telomerase reverse transcriptase; JAM3: junctional adhesion molecule 3; LETM1: leucine zipper and EF-hand transmembrane protein 1; M: metastasis; miR: micro RNA; MUC1: mucin 1; PLXNB2: plexin B2; SLC1A5: solute carrier 1A5; SFRP1: secreted frizzled related protein 1; TG: tumor growth.
Circ0025033 up-regulates solute carrier 1A5 (SLC1A5). circ0025033 (Figure 3) was up-regulated in EOC and positively regulated viability, DNA synthesis in SKOV3 and A2780 EOC cells (114). Knockdown of circ0025033 inhibited growth of A2780 EOC xenografts after s.c. implantation into nude mice. In addition, it acted as a mediator of angiogenesis. It sponged miR-370-3p and up-regulated SLC1A5, also known as alanine, serine, cysteine transporter 2 (ASCT2), which is also involved in glutamine transport and plays a role in proliferation, migration, invasion, angiogenesis, and apoptosis of cancer cells and is associated with a poor prognosis (115,116). SLC1A5 acts as a promoter of EOC (117).
Circ kinesin family member 4A (circKIF4A) up-regulates junctional adhesion molecule 3 (JAM3). circKIF4A (Figure 3) was highly expressed in EOC tissues (118). Its knockdown inhibited proliferation and invasion of SKOV3 and CAO3 EOC cells in vitro. Intra-tumoral injected si-circ KIF4A inhibited growth of SKOV3 xenografts in nude mice and i.v. injected si-circ KIF4A decreased lung metastases of tail-vein injected SKOV3 cells. circKIF4A acted as a sponge for miR-127 and up-regulated JAM3. JAMs are adhesion molecules of the immuno-globulin superfamily, which are connected to the cytoskeleton by interaction with PDZ-domain containing scaffold proteins recruiting and assembling signaling complexes (119,120). JAMs are involved in angiogenesis and can exert pro- and anti-tumoral functions (121,122). In a murine model of EOC, JAM3 has been shown to play a key role in the development of tumors (123). High expression of JAM-A is associated with poor survival in patients with EOC (124).
Circ Wolf-Hirschhorn syndrome candidate 1 (circWHSC1) up-regulates mucin1 (MUC1) and human telomerase reverse transcriptase (hTERT). circWHSC1 (Figure 3) was found to be over-expressed in EOC tissues (125). It promoted viability, invasion, and migration of OVCAR3 and CAOV3 EOC cell lines. Exosomal circWHSC1 increased s.c. growth of CAOV3 xenografts after i.p. injection and peritoneal dissemination into nude mice. circWHSC1 sponged miR-45 and -1185 and up-regulated MUC1 and hTERT. MUC1, a transmembrane glycoprotein, promotes metastasis and progression in EOC (126) and regulates AKT signaling by up-regulation of EGFR expression in OC cells (127). hTERT is involved in proliferation and metastasis and inhibits apoptosis of EOC cells (128).
Circ004712 up-regulates frizzled 4 (FZD4). circ004712 (Figure 3) was aberrantly over-expressed in EOC tissues and cells (129). Down-regulation of circ004712 inhibits proliferation, attenuated migration and invasion and stimulated apoptosis in SKOV3 and OVCAR3 EOC cells in vitro. Knockdown of circ004712 blocked growth of SKOV3 xenografts after s.c. injection into nude mice. circ004712 sponged miR-331-3p resulting in up-regulation of FZD4. The latter plays a role in WNT signaling in EOC (130).
Circ0061140 up-regulates leucine zipper and EF-hand transmembrane protein 1 (LETM1). circ0061140 (Figure 3) was notably up-regulated in EOC and cell lines (131). Knockdown of circ0061140 suppressed proliferation, migration, invasion and angiogenesis-promoting activity and triggered apoptosis in EOC cell lines in vitro. Knockdown of circ0061140 inhibited growth of EOC-related xenografts in nude mice. circ0061140 sponged miR-761 and up-regulated LETM1. The latter is located in the inner mitochondrial membrane, acts as protein Ca-exchange protein and is required for mitochondrial homeostasis and cellular viability (132,133). In EOC, LETM1 has been identified as a promoter of tumorigenesis and metastasis (134).
Circ E3 ubiquitin ligase itchy homolog (circITCH) up-regulates cadherin-1 (CADH1). circITCH (Figure 3) was down-regulated in EOC. It inhibited proliferation, invasion, glycolysis and promoted apoptosis of A2780 and OVCAR3 EOC cells (135). circITCH suppressed proliferation, invasion, and glycolysis of A2780 and OVCAR3 OC cells in vitro. Over-expression of circITCH repressed the growth of SKOV3 xenografts after s.c. implantation into nude mice. circITCH sponged miR-106a resulting in increased levels of miR-106a and reduced levels of CDH1 (135). Expression of CDH1 in EOC is a controversial issue because increased levels (136,137) as well as decreased levels of CDH1 have been associated with progression of EOC (138-141).
CircRAS homology family member C (circRHOC) up-regulates vascular endothelial growth factor A (VEGFA). circRHOC (Figure 3) was over-expressed in EOC (142). In A2780 and CAOV3 EOC cells, circRHOC increased viability, migration, and invasion. Down-regulation of circ RHOC attenuated dissemination of A2780 EOC cells after i.p. injection into nude mice. circRHOC functioned as a sponge for miR-302e and up-regulated VEGFA.
Circ histone lysine methyltransferase 2L (circ ASH2L) up-regulates VEGFA. circASH2L (Figure 3) was up-regulated in EOC tissues and cell lines (143). circASH2L enhanced proliferation and invasion in SKOV3 EOC cells by sponging miR-665 and up-regulating VEGFA. Knockdown of circASH2L in SKOV3 cells inhibited growth, angiogenesis, and lymphangiogenesis after s.c. implantation into nude mice. VEGFA also up-regulated kinase domain insert receptor (KDR), also known as VEGFR2 (144) and Fms related receptor tyrosine kinase 4 (FLT4), which binds VEGFC and VEGFD (145). Angiogenesis plays an important role in generation and progression of EOC (146). Therefore, anti-angiogenic approaches play an important role in the treatment of EOC (147). Bevacizumab, which neutralizes VEGFA has been approved for treatment of EOC in various clinical scenarios (148).
Circ0001445 up-regulates secreted frizzled-related protein 1 (SFRP1). Expression of circ0001445 (Figure 3) was decreased in EOC (149). It blocked proliferation and migration and promoted apoptosis in SKOV3 and HO8910 EOC cells. Over-expression of circ0001445 in HO8910 EOC cells inhibited tumorigenesis and i.p. metastasis in nude mice. circ0001445 sponged miR-576-5p and up-regulated SFRP1. The latter is a member of a family of five proteins, which bind to WNT ligands or to frizzled receptors with the consequence of interfering with WNT signaling (150,151). In EOC, it was shown that SFRP1 inhibits proliferation, migration, and invasion, and WNT/β-catenin signaling (152). Methylation-associated silencing of SFRP1 was observed in high-grade serous ovarian carcinoma (153).
CircRNAs Targeting Transcription Factors
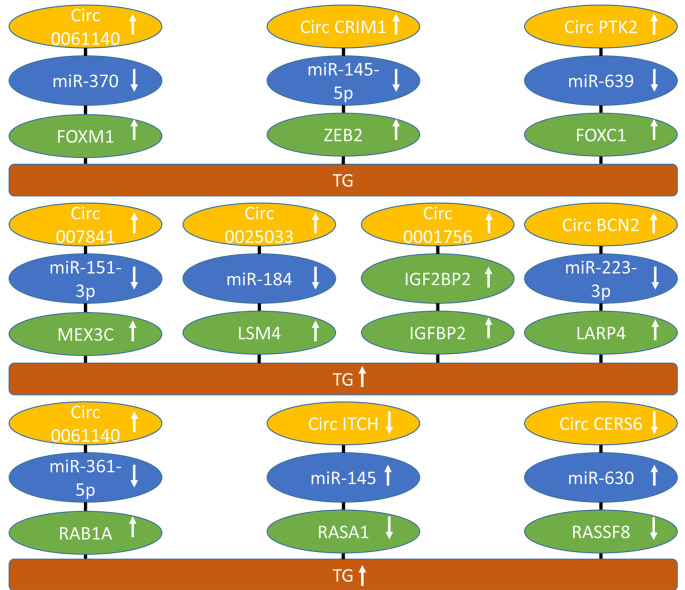
Circ0061140 up-regulates forkhead box M1 (FOXM1). circ0061140 (Figure 4) was up-regulated in EOC and cell lines (154). Knockdown of circ0061140 suppressed proliferation and invasion of A2780 and SKOV3 OC cell lines and inhibited growth of SKOV3 xenografts after s.c. implantation into nude mice. circ0061140 sponged miR-370 and up-regulated transcription factor FOXM1. The latter is frequently over-expressed in cancer (155). In EOC, FOXM1 is involved in proliferation, invasion and metastasis, genomic instability, cancer stem cell (CSC) maintenance, cellular metabolism, and chemo-resistance (156). Peritoneal spread and migration is mediated by expression of FOXM1 (157,158). Small molecules targeting the FOXM1 DNA binding domain exhibit antitumor activity in EOC (159,160).
Figure 4. circRNAs regulating transcription factors, RNA binding and splicing factors and RAS pathway-related factors with activity in preclinical in vivo models of epithelial ovarian cancer. Upward arrows indicate up-regulation, downward arrows correlate with down-regulation. CHD1L: Chromosomal helicase DNA binding protein; circBCN2: circ basonuclin 2; circCERS6: circ ceramide synthase 6; circCRIM1: circ cysteine rid motor neuron protein 1; circITCH: Itchy homolog ubiquitin protein ligase; circPTK2: circ protein tyrosine kinase 2; FOXC1: forkhead box C1; FOXM1: forkhead box M1; IGF2BP2: insulin growth factor binding protein 2; LARP4: La ribonucleoprotein 4; LSM4: Sm-like protein 4; MEX3C: Mex-3 RNA binding family member C; miR: micro RNA; RAB1A: RAS-related protein RAB1A; RASA1: RAS p21 protein activator 1; RASSF8: RASassociated domain family nuclear 8; TG: tumor growth; ZEB2: zinc finger E-box homeo box 2.
Circ cysteine-rich motor neuron protein 1 (circCRIM1) up-regulates CRIM1 and zinc finger E-box binding homeobox 2 (ZEB2). circCRIM1 (Figure 4) was over-expressed in EOC (161). Its knockdown inhibited cell viability, migration, invasion and promoted apoptosis in CAOV3 and OVCAR3 EOC cells. circCRIM increased growth of CAOV3 xenografts after s.c. implantation into nude mice. circCRIM1 sponged miR-145-5p resulting in up-regulation of CRIM1 and, in addition, it sponged miR-383-5p leading to up-regulation of ZEB2. CRIM1 regulates processing of bone morphogenetic protein (BMP) preprotein into mature protein and delivery of BMPs to the cell surface (162). It stimulates capillary formation, angiogenesis and EMT (162,163). ZEB2, together with other transcription factors, such as SLUG, SNAIL, TWIST and ZEB1 acts as a mediator EMT (164). ZEB2 is highly expressed in EOC and correlates with poor prognosis (165). In EOC, ZEB2 has been shown to act as a repressor of E-cadherin (166).
Circular RNA protein tyrosine kinase 2 (circPTK2) up-regulates chromosomal helicase DNA binding protein CHD1L and forkhead-box C1 (FOXC1). circPTK2 (Figure 4) was over-expressed in EOC and positively affected migration, invasion, angiogenesis and EMT in SKOV3 and OVCAR3 cells (167). OVCAR3 cells transfected with circPTK2 showed increased tumor growth after s.c. implantation into nude mice. circPTK2 expression was positively correlated with CDH1L mRNA and interacted with miR-639 resulting in up-regulation of FOXC1. CDH1L acts as an oncogene promoting unleashed proliferation, inhibition of apoptosis by binding to apoptotic nuclear receptor protein NUR77 and activation of AKT signaling (168). CDH1L is over-expressed in EOC and promotes its invasiveness and metastasis by regulation of methionyl aminopeptidase 2 (169,170). The forkhead family of transcription factors plays an important role in ovarian function (171). FOXC1 is involved in cancer cell proliferation, migration, angiogenesis, and cancer stem cell maintenance and is a potential target for therapy (172).
CircRNAs Up-regulating RNA Binding and Splicing Proteins
Circ0007841 up-regulates Mex-3 RNA binding family member C (MEX3C). circ0007841 (Figure 4) was highly expressed in EOC and promoted proliferation, invasion, and migration in SKOV3 and OVCAR3 EOC cells (173). Knockdown of circ0007841 in SKOV3 EOC cells suppressed tumor growth after s.c. implantation into nude mice. The underlying mechanism implicated sponging of miR-151-3p and up-regulation of RNA binding protein MEX3C. RNA binding proteins have been implicated in human cancer and other diseases by diverse mechanisms, such as mRNA splicing, polyadenylation, translation, regulation of its stability and subcellular localization (174,175). MEX3C contains two RNA binding domains and a RING-finger domain and constitutes an RNA binding ubiquitin ligase (176). MEX3C has been shown to promote lipid metabolism to support bladder tumorigenesis by activation of the JNK pathway (177). In addition, MEX3B has been shown to mediate decay of SOCS3 mRNA promoting Janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) signaling and facilitating hepatocellular carcinoma metastasis (178).
Circ0025033 up-regulates Sm-like protein 4 (LSM4). circ0025033 (Figure 4) was up-regulated in EOC and its knockdown blocked colony formation, migration, invasion, and glycolysis of EOC cells (179). In nude mice, circ0025033 knockdown inhibited growth of EOC xenografts after s.c. implantation. circ0025033 sponged miR-184 resulting in over-expression of ribonucleoprotein LMS4. The latter is a member of an RNA binding protein family with 14 members (180). LMS4 binds to the 3´UTR of small nuclear RNA U6snRNA (181,182).
Circ0001756 up-regulates insulin growth factor 2 mRNA binding protein (IGF2BP2). circ0001756 (Figure 4) was up-regulated in EOC and its knockdown in SKOV7 and A2780 EOC cells inhibited proliferation, invasion, EMT and tumor growth of SKOV3 xenografts after tail vein injection (183). circ0001756 was shown to bind and promote expression of IGF2BP2 to interact with RAS-associated protein 5A (RAB5A) and to activate EGFR/mitogen-activated protein kinase (MAPK) signaling (183). IGF2BP2 has been identified as a post-transcriptional driver and as an oncogene in human cancer (184,185). RAB5A mediates cell proliferation by APPL1 related epidermal growth factor signaling in EOC (186). IGF2BP2 has been shown to promote aggressiveness and stemness of EOC by ERB-B2 receptor tyrosine kinase 4(ERBB4)/PI3K/AKT signaling (187).
Circbasonuclin 2 (circBCN2) up-regulates La ribonucleoprotein 4 (LARP4). circBCN2 (Figure 4) was found to be down-regulated in EOC (188). Its over-expression decreased proliferation, migration, invasion of EOC cells in vitro and inhibited growth of EOC xenografts in nude mice. circBCN2 sponges miR-223-3p resulting in up-regulation of RNA binding protein LARP4. It is member of a family of proteins with a conserved 90 aa signature sequence, which are deregulated in cancer and can exert an oncogenic or tumor-suppressive function (189,190). It has been shown that LARP4 inhibits motility of EOC cells (191,192).
CircRNAs Targeting RAS Pathway-related Proteins
Circ0061140 up-regulates RAS-related protein 1A (RAB1A). circ0061140 (Figure 4) was up-regulated in EOC tissues and cells and its down-regulation impeded proliferation, migration, invasion, EMT and angiogenesis capability of EOC cells in vitro and in nude mice (193). It sponged miR-361-5p and up-regulated RAB1A. The latter is a small GTPase, which is involved in vesicular trafficking between the endoplasmic reticulum and the Golgi apparatus (194). RAB1A was shown to mediate proliferation and migration of colorectal cancer (195).
Circ itchy homolog E3 ubiquitin protein ligase (circITCH) up-regulates RAS p21 protein activator 1 (RASA1). circITCH (Figure 4) was down-regulated in EOC tissues and cell lines (196). It inhibited cell viability, motility, wound-healing, and invasion of EOC cell lines and tumor growth of EOC xenografts in nude mice. circITCH sponged miR-145 and up-regulated RASA1, which is also known as RAS-GTPase activating protein (RAS-GAP). RASA1 acts as a GAP with dual specificity that enhances and accelerates GTPase activity of RAS and ras-related protein (RAP) (197). RASA1 is composed of several domains, such as src homology regions 2 and 3 (SH2, SH3), a pleckstrin domain (PH), a phospho-lipid binding domain (C2) and a GTPase activating domain (GAP) (197). In cancer, RASA1 is involved in cell growth, proliferation, and apoptosis (197).
Circ ceramide synthase (circ CERS6) up-regulates RAS-association domain family member 8 (RASSF8). circ CERS6 (Figure 4) was markedly reduced in EOC tissues and cell lines (198). circCERS6 inhibited proliferation, migration, invasion and EMT of EOC cells. circCERS6 over-expression in EOC cells blocked tumor growth in nude mice. circCERS6 sponged miR-630 and up-regulated RASSF8. The latter belongs to a protein family of ten members (199). RASSF8 acts as a tumor suppressor by maintaining adherents junctions and plays a role in epithelial migration and suppression of WNT and nuclear factor ĸB (NFĸB) signaling (200). RASSF8 is in involved in regulation of the cell-cycle and apoptosis (201,202). RASSF8 has been shown to be involved in proliferation and migration of EOC (203).
circRNAs Targeting Ubiquitinylation and Cell-cycle Related Proteins
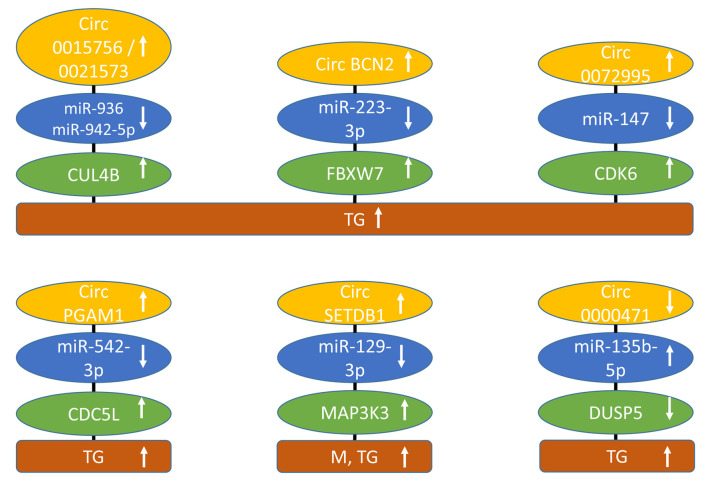
Circ0015756 and circ0021573 up-regulate cullin 4B (CUL4B). circ0015756 (Figure 5) was up-regulated in EOC tissues and cells (204). Knockdown of 0015756 inhibited proliferation, migration, and invasion in OC90 and SKOV3 EOC cells. Depletion of circ0015756 hindered growth of SKOV3 xenografts in nude mice. The underlying mechanism involved binding of miR-942-5p to circ0015756 and subsequent up-regulation of CUL4B (204). circ0021573 was also over-expressed in EOC tissues and its inhibition impeded cell proliferation, migration and invasion, and induced apoptosis of EOC cells in vitro and diminished growth of EOC-related xenografts in nude mice (205). circ0021573 sponged miR-936 and also up-regulated CUL4B. The latter is a scaffold protein that participates in the formation of the E3 ubiquitin ligase complex (206). CUL4B is over-expressed in a variety of tumors and can affect the cell-cycle, promoter methylation, histone deacetylation, and DNA damage and repair, resulting in context-dependent oncogenic and tumor-suppressive properties (207,208). It is well documented that ubiquitinylation can have pro-tumoral as well as tumor-suppressive effects such as degradation of p53 (209,210). CUL4B is up-regulated in EOC and correlates with poor prognosis and induces expression of cyclin-dependent kinase 2 (CDK2) and cyclin D (211).
Figure 5. circRNAs regulating ubiquitinylation and cell-cycle related factors of epithelial ovarian cancer with efficacy in preclinical in vivo models. Upward arrows indicate up-regulation, and downward arrows indicate down-regulation. CDC5L: Cell-division 5-like; CDK6: cyclin-dependent kinase 6; circBCN2: circ basonuclin 2; circPGAM1: circ phosphoglycerate mutase 1; circSETDB1: circ SET domain bifurcated lysine methyltransferase 1; Cul 4B: cullin 4B; DUSP5: dual specificity phosphatase 5; FBXW7: F box and WD repeat domain containing 7; M: metastasis; MAPK3K3: mitogen-related protein kinase kinase kinase 3; miR: micro-RNA; TG: tumor growth.
Circ basonuclin 2 (circBCN2) up-regulates F box and WD repeat domain containing 7 (FBXW7). circBCN2 (Figure 5) was down-regulated in EOC patients and decreased expression correlated with poor prognosis (212). It inhibited cell growth, proliferation, migration, invasion, and cell-cycle progression in SKOV3 and HO-8910 OC cells. circBCN2 sponged miR-223-3p and up-regulated FBXW7. Over-expression of circBCN2 in SKOV3 EOC cells suppressed lung metastasis after tail vein injection into nude mice. FBXW7, a member of the F-box protein family, is a component of the Skp, F-box containing complex (SCF). This complex, an E3 ubiquitin ligase, regulates substrate specificity, ubiquitinates proteins and triggers their degradation in the proteasome. It acts as a tumor suppressor by ubiquitinylation of substrates such as the transcription factors c-MYC and JUN and cyclin E and induces myeloid leukemia cell differentiation protein 1 (MCL1), receptor tyrosine kinase NOTCH1 and mTOR (213,214). FBXW7 is aberrantly expressed in several types of cancer (213,214). In EOC, FBW7 has been shown to suppress N6-methyladenosine binding protein YTHDF2 (215).
Circ0072995 up-regulates cyclin-dependent kinase 6 (CDK6). circ0072995 (Figure 5) was up-regulated in EOC tissues and cell lines (216). High expression of circ0072995 correlated with pathological grading. Its knockdown suppressed proliferation, migration, and induced apoptosis of HO8910 and A2780 EOC cancer cells. Down-regulation of circ0072995 inhibited tumor growth of HO8910 xenografts after s.c. implantation into nude mice. It sponged miR-147 resulting in up-regulation of CDK6. Targeting of CDK4 and CDK6, which are critical mediators of S-phase entry, has led to approved drugs for treatment of hormone receptor positive breast cancer (217). CDK6 also exerts kinase-independent functions as a chromatin-bound co-factor by inducing genes involved in angiogenesis, cell-cycle inhibition, stem cell activation and activation of the immune response (218). CDK6 has been shown to contribute to progression of EOC and to protect EOC cells from DDP-induced cell death (219,220). Furthermore, it has been shown that CDK4/6 inhibition promotes immune infiltration in EOC and synergizes with programmed death (PD)-blockade (221).
Circ phosphoglycerate mutase1 (circPGAM1) up-regulates cell division 5-like (CDC5L). circPGAM1 (Figure 5) was up-regulated in EOC tissues and its silencing resulted in inhibition of proliferation, migration, and invasion, and promoted apoptosis in CAOV3 and OVCAR3 EOC cells (222). In OVCAR3 and CAVOV3 xenografts, circPGAM1 stimulated tumor growth after s.c. implantation into nude mice. circPGAM1 sponged miR-542-3p and up-regulated CDC5L. The latter interacted with the promoter of pseudopodium enriched atypical kinase 1 (PEAK1) and increased its expression. PEAK1 activated ERK1/2 and JAK2 signaling pathways. CDC5L functions as a cell-cycle regulator at G2/M transition and is involved in pre-mRNA splicing and DNA damage repair (223). Depletion of CDC5L was shown to inhibit mitotic progression and mitotic catastrophe (224). PEAK1 has intrinsic tyrosine kinase activity and, in addition, exhibits scaffolding functions. It promotes proliferation and motility and is involved in regulation of actin cytoskeleton and focal adhesions and represents a potential target for anti-cancer drugs (225,226).
circRNAs Targeting Kinases and Phosphatases
CircSET domain bifurcated histone lysine methyltransferase 1 (circSETDB1) up-regulates mitogen-activated protein kinase kinase kinase 3 (MAP3K3). circSETDB1 (Figure 5) was up-regulated in EOC tissues and cell lines (227). Silencing of circSETDB1 repressed proliferation, invasion and migration, and induced apoptosis in A2780 and SKOV3 EOC cell lines. CircSETDB1 depletion inhibited tumor formation and metastases of i.p. injected SKOV3 cells in nude mice. circ SETB1 sponged miR-129-3p and up-regulated MAP3K3, a kinase, which activates ERK1/2, c-jun terminal kinase (JNK) and mitogen-activated protein kinase p38 (228). MAP3K3 over-expression was associated with poor survival in EOC patients (229). MAP3K3 promotes growth of EOC through activation of NFĸB signaling (230). MAP3K3 also confers resistance to apoptosis in EOC cells through stimulation of NFĸB (231).
Circ0000471 up-regulates dual-specificity phosphatase 5 (DUSP5). circ0000471 (Figure 5) was found down-regulated in EOC tissues and cell lines (232). Over-expression of circ0000471 blocked proliferation, migration and invasion, and triggered apoptosis in EOC cells. Its mechanism of action involves sponging of miR-135b-5p and down-regulating DUSP5. Over-expression of circ0000471 inhibited EOC-derived xenografts in nude mice. DUSP5 is a member of dual-specificity phosphatases, which inactivate different mitogen-activated kinases (MAPK), such as ERKs, JNK and p38 (233). DUSP5 has been shown to suppress EOC progression by inhibiting interleukin 33 (IL33) signaling (234).
circRNAs Targeting Components of Signaling Systems
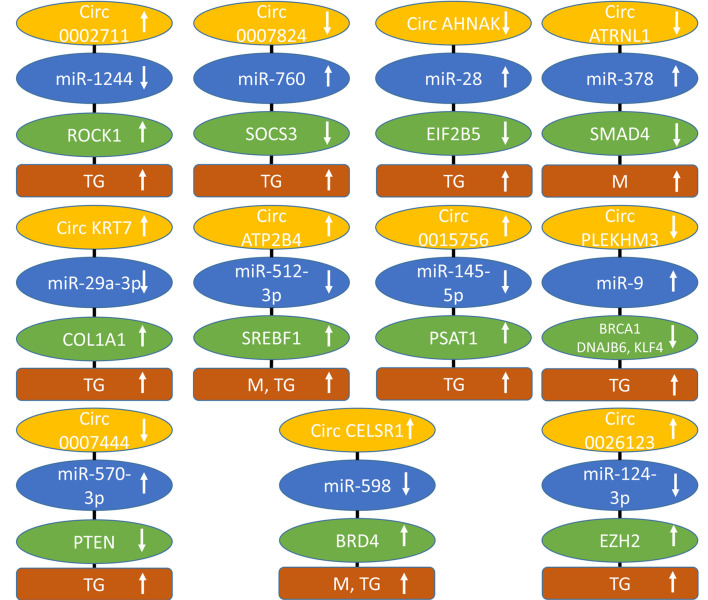
Circ0002711 up-regulates Rho-associated protein kinase 1 (ROCK1). The level of circ0002711 (Figure 6) was up-regulated in EOC tissues (235). Knockdown of circ0002711 inhibited cell proliferation and aerobic glycolysis in SKOV3 and OV90 EOC cells. Decreased expression of circ0002711 mediated inhibition of SKOV3 xenografts after s.c. implantation. circ0002711 sponged miR-1244 and up-regulated the Ser-Thr kinase ROCK1. The latter is a downstream effector of GTPases RHOA and RHOC and is frequently deregulated in cancer. It mediates cytoskeleton contractability, migration, survival, and proliferation of tumor cells (236,237). For cancer, a ROCK1 inhibitor has not yet being approved due the narrow therapeutic window, whereas four different ROCK1 inhibitors have been approved for other diseases (237). It has been shown that ROCK1 promotes proliferation, invasion, and migration of EOC cells (238,239).
Figure 6. circRNAs regulating signaling and factors of other categories of epithelial ovarian cancer with efficacy in preclinical in vivo models. Upward arrows indicate up-regulation, and downward arrows indicate down-regulation. BRCA1: Breast cancer related antigen 1; circATP2B4: circ ATPase plasma membrane Ca2+ transporter 4; circAHNAK: circ neuroblast differentiation associated gene AHNAK; circATRNL1: circ attractinlike 1; circKRT7: circ keratin 7; circPLEKM3: pleckstrin homology domain family member 3; COL1A1: collagen 1A1; DNAJB6; DNA J heat shock family (hsp40) member B6; EIF2B5: eucaryotic initiation factor 2B, subunit ε; EZH2: enhancer of zeste homolog 2; M: metastasis; miR: micro RNA; PSAT1: phosphoserine aminotransferase 1; PTEN: phosphatase and tensin homolog; ROCK1: RHO-associated protein kinase 1; SMAD4: decapentaplegic homolog 4; SOCS3: suppressor of cytokine signaling 3; SREBF1: sterol regulatory element binding factor 1; TG: tumor growth.
Circ0007874 up-regulates suppressor of cytokine signaling 3 (SOCS3). circ0007874 (Figure 5) was decreased in EOC cell lines (240). It suppressed growth and migration of A2780 and SKOV3 EOC cell lines. Over-expression of circ0007874 inhibited growth of A2780 EOC cells in nude mice after s.c. implantation. It sponged miR-760 and up-regulated SOCS3. The latter is a member of a family of eight proteins, , each characterized by varying N-termini, an SH2 domain that binds to proteins phosphorylated by tyrosine kinases, and a C-terminus responsible for recruiting E3 ligases and ubiquitin signaling molecules (241). SOCS3 is induced by cytokines which activate STAT3 via JAK2 and inhibits this pathway by a variety of mechanisms (242). SOCS3 suppresses progression of EOC by blockade of the JAK/STAT signaling pathway (243). Reduced expression of SOCS3 predicts poor outcome in EOC (244).
Circ neuroblast differentiation associated gene (circAHNAK) induces eukaryotic initiation factor 2B subunit ε (EIF2B5). circAHNAK (Figure 6) was decreased in patients with EOC (245). Expression of circAHNAK in EOC cells promoted apoptosis and inhibited cell proliferation, migration, invasion, EMT and JAK2/STAT signaling. circAHNAK retarded growth of EOC xenografts in nude mice. It sponged miR-28 and up-regulated EIF2B5, a component of EIF2B, which is composed of five subunits (246). The mechanism through which EIF2B suppresses JAK2/STAT signaling is not yet resolved. Low expression of EIF2B5 predicts a poor prognosis in EOC (247). Inactivation of EIF2B5 by promoter methylation mediated metastasis of EOC cell lines (248).
Circ attractin-like1 (circ ATRNL1) up-regulates mothers of decapentapledgic homolog 4 (SMAD4). circATRNL1 (Figure 6) was down-regulated in the tissues of EOC patients and suppressed proliferation, migration, invasion in SKOV3 and CAOV3 EOC cells and inhibited angiogenesis (249). In an i.p. xenograft model, circATRNL1 suppressed abdominal metastasis of SKOV3 and CAOV3 EOC cells. circATRNL1 bound miR-378 and up-regulated SMAD4. miR-378 activated AKT via targeting transcription factor SMAD4. SMADs transduce extracellular signaling to the nucleus and SMAD4 is activated by transforming growth factor β (TGFβ) and related ligands and induces cell-cycle arrest, apoptosis, and suppression of cancer development (250-252). It was independently demonstrated that SMAD4 inhibited proliferation of EOC cells (253).
circRNAs Targeting Additional Categories of Proteins
Circ keratin 7 (circ KRT7) up-regulates collagen 1A1 (COL1A1). circKRT7 (Figure 6) was highly expressed in EOC tissues (254). Down-regulation of circKRT7 suppressed proliferation and invasion of ES2 and SKOV3 EOC cells. Inhibition of circKRT7 decreased growth of ES2 xenografts in nude mice. circKRT7 sponged miR-29a-3p and up-regulated COLA1, a major component of type I collagen, which is the main structural component of the extracellular matrix. Collagen is composed of a triple helix with two identical α1 chains and an additional α2 chain that differs slightly in its chemical composition. The ECM plays an important role in cancer genesis and progression (255). COL1A1 was elevated in ascites from EOC patients in comparison to normal peritoneal fluids (256). COL1A1 was mainly derived from fibroblasts and bound to β1 integrin, activating AKT signaling and promoting migration and invasion (256). The extension of understanding of the functions of the extracellular matrix (ECM) will enable the design of new therapeutics for the treatment of cancer (257). Furthermore, it was shown that COL1A1 was involved in DDP resistance of EOC (258).
Circ ATPase plasma membrane Ca2+ transporter 4 (ATP2B4) up-regulates sterol regulatory element-binding factor 1 (SREBF1). circATP2B4 (Figure 6) was up-regulated in EOC tissue samples and positively correlated with disease progression (259). circATP2B4 targets EOC cells as well as tumor-associated macrophages. EOCs releasing circATP2B4 in extracellular vesicles (EV) enhance migration, invasion and EMT of SKOV3 and A2780 EOC cells. circATP2B4 was highly expressed in EVs from EOC cells and could be transferred into macrophages resulting in their polarization into protumoral M2 macrophages. SKOV3 cells mixed with conditional macrophages stimulated with EVs from A2780 cells exhibited significantly increased tumor weight and peritoneal metastases after i.p. injection into nude mice in comparison to the corresponding control group. It was shown that circATP2B4 sponges miR-532-3p and up-regulated SREBF1 resulting in activation of SREBF1/PI3K/AKT signaling and macrophage polarization. SREBF1 mediates lipogenesis, activates PI3K-AKT-mTOR signaling and suppresses ferroptosis (260). In EOC, SREBF1 is required for tumor growth (261) and targeting of lipogenesis is an important topic for treatment of EOC (262). A correlation between high level of M2 macrophages and low survival in EOC patients has been noted, therefore preclinical evaluation of macrophage-targeted therapies for EOC are under evaluation (263).
Circ0015756 up-regulates phosphoserine aminotransferase 1 (PSAT1). circ0015756 (Figure 6) was highly expressed in EOC tissues and cells (264). Knockdown of circ0015756 suppressed growth, migration, and invasion of EOC cell in vitro and impeded growth of EOC xenografts in nude mice. circ0015756 acted as a sponge for miR-145-5p and up-regulated PSAT1, which is involved in serine biosynthesis and functions as a pro-proliferative and pro-survival factor (265). In cancer, PSAT1 provides anabolic support of tumor cells, stimulating proliferation, survival, autophagy, migration, invasion and non-enzymatically regulates the IGF1 pathway and nuclear PKM2 to promote EMT and metastasis (266). In EOC, PSAT1 is markedly over-expressed and correlates with lymph node metastasis, distant metastasis, and presence of ascites (267). Its inhibition decreases growth of EOC cells, induces apoptosis and cell-cycle arrest (267). PSAT1 increases the proportion of GSH and reduced nicotinamide dinucleotide phosphate (NADPH) in EOC cells, increasing oxidative stress tolerance (267).
Circ pleckstrin homology domain family M member 3 (circPLEKHM3) up-regulates breast cancer antigen 1 (BRCA1). Down-regulation of circPLEKHM3 (Figure 6) was associated with a poor prognosis in patients with EOC (268). circPLEKM3 repressed proliferation, migration and EMT of A2780 and OV90 EOC cells in vitro and in vivo of corresponding xenografts after s.c. implantation. circPLEKHM3 was shown to bind to miR-9 and to suppress its activity. BRCA1, DNAJ heat shock family (Hsp40), member B6 (DNAJB6) and Krueppel-like transcription factor 4 (KLF4) were identified as targets of miR-9. It was shown that circPLEKHM3 inactivates AKT1 and WNT/β catenin signaling. BRCA1 is part of a complex that repairs double-strand breaks and regulates DNA repair, transcription, and cell-cycle regulation in response to DNA damage and is frequently inactivated in EOC (269,270). DNAJB6 interacts with hsp70 chaperones, inhibits WNT and NFĸB signaling and negatively regulates tumor growth and metastasis (271). The other target of circPLEKHM3, KLF4, has been shown to be down-regulated in EOC and its over-expression caused EOC proliferation, migration, and invasion by inhibiting TGFβ induced EMT (272).
Circ0007444 up-regulates phosphatase and tensin homolog (PTEN). Low expression of circ0007444 (Figure 6) predicted poor prognosis in patients with EOC (273). circ0007444 inhibited proliferation, migration, invasion and promoted apoptosis of SKOV3 and OVCAR3 EOC cells in vitro and suppressed growth of OVCAR3 xenografts after s.c. implantation into nude mice. circ0007444 sponged miR-570-3p and up-regulated PTEN. The latter acts as a tumor suppressor by dephosphorylating phosphatidylinositol-3,4,5 triphosphate and other phosphoinositides and inhibiting PI3K/AKT/mTOR signaling (274). PTEN also acts as a scaffold protein both in the nucleus and the cytoplasm through a phosphatase-independent function acting as a chromosomal stability controller (275). In EOC, PTEN is inactivated in 5-7% of the tumors (276).
Circ cadherin RGF LAG seven pass-G-type receptor 1 (circCELSR1) up-regulates bromodomain-containing protein 4 (BRD4). circCELSR1 (Figure 6) was over-expressed in EOC cancer cells (277). Its knockdown inhibited proliferation, migration and promoted apoptosis in SKOV3 and A2780 EOC cells. In nude mice, knockdown of circCELSR1 suppressed growth and metastasis of SKOV3 cells injected into the abdominal cavity of nude mice. BRD4 was up-regulated due to sponging of miR-598 by circCELSR1. BRD4 is a member of the bromodomain and extraterminal protein family (BET) with two tandem bromodomains, which can bind to acetylated histones and other proteins and activate oncogenic proteins by interaction with RNA polymerase II (278,279). Several BET inhibitors, which mimic the acetyl moiety occluding the acetyl binding site pocket unique to the BET family of proteins, have been designed (278,279). In EOC, cyclin E1 (CCNE1) and BRD4 amplification are associated with poor outcomes in patients (280). BRD4 amplification facilitates oncogenic gene expression in EOC and confers sensitivity to BET inhibitors (281). BRD4 inhibition has been shown to be synthetically lethal together with PARP inhibition through induction of homologous recombination deficiency (282). BET inhibition also has been shown to resensitize EOC cells to approved anticancer agents such as DDP and PARP inhibitors (283).
Circ0026123 up-regulates enhancer of zeste homolog 2 (EZH2). circ0026123 (Figure 6) was significantly increased in EOC in comparison to adjacent normal tissues (284). Silencing of circ0026123 inhibited proliferation, migration, and CSC differentiation markers in SKOV3 cells. Down-regulation of circ0026123 suppressed tumor growth of SKOV3 cells after s.c. implantation into nude mice. All of these effects are based on sponging miR-124-3p by circ0026123 and subsequent up-regulation of EZH2. The latter is the catalytic subunit of the polycomb repressive complex (PRC2), which mediates repression of target genes by trimethylation of Lys 27 in histone 3 (285). EZH2 is involved in cancer cell proliferation, invasion, metastasis, angiogenesis, and resistance to chemotherapy (286). In EOC, EZH2 is over-expressed, promotes proliferation, inhibits apoptosis, and enhances angiogenesis. Currently, multiple inhibitors targeting EZH2 are undergoing clinical trials (287,288).
Technical Issues
Searching the literature, we identified up- and down-regulated EOC-related circRNAs and their corresponding targets. Only circRNAs which exhibit activity in EOC-related xenograft models in nude mice were selected. The corresponding targets can be validated with respect to their priority for drug development by further in vitro and in vivo experiments. According to the target class identified, up-regulated targets can be inhibited with small molecules, antibody-derived moieties, or proteolysis-targeting chimeras (PROTACS) (289,290). Down-regulated targets can be up-regulated by gene transfer or small molecules (291). The latter are often associated with specificity issues and target deconvolution is necessary.
Up-regulated circRNAs can be inhibited with ASO, siRNA or shRNA and down-regulated circRNAs can be reconstituted by gene transfer (292,293). However, several technical hurdles have to be tackled. These issues are not discussed in further details in this review. Among the issues are pharmacokinetic and pharmacodynamic aspects of the corresponding agents, immunogenicity, specificity, and delivery (15-17). The optimization process depends on the specific approach and the clinical scenario to be pursued. Lipid derived nanoparticles (LNPs) are frequently used for delivery of corresponding inhibitors of circRNA or circRNAs for reconstitution experiments. They consist of variations of basically four compounds: a cationic or ionizable lipid, cholesterol, a helper lipid and a polyethylene glycol lipid (294). Conjugation of siRNA with N-acetyl galactosamine (GalNAc) resulted in liver transfer (295) and functionalization of siRNA with antibodies has enabled transfer to endothelial cells and tumor cells (296). Four siRNAs for different indications have been approved now including Patisiran for treatment of transerytrin-mediated amyloidosis as the first one (297).
Concluding Remarks
We identified up- and down-regulated circRNAs with efficacy in EOC-related preclinical in vivo systems. The vast majority act by sponging miRNAs. They can be assigned to the following categories according to the targets they regulate: DDP- and PTX-resistance, transmembrane receptors and secreted factors, transcription factors, RNA binding and processing factors, RAS pathway-related factors, ubiquitinylation- and cell-cycle proteins, signaling-related components, and targets that fit into additional categories.
Figure 1 and Figure 2 show that resistance to DDP and PTX can be mediated by a variety of targets, which complicates the treatment of chemo-resistance, one of the major problems of the treatment of EOC. PLXND2, SCL1A5, JAM3, FZD4 and SFRP1 and corresponding circRNAs were identified as potential targets for treatment of EOC (Figure 3). The role of transcription factors, RNA splicing and processing related proteins and components of the RAS-related signaling system as targets of circRNAs are outlined in Figure 4. The significance of ubiquitinylation, proteolysis and cell-cycle proteins is highlighted in Figure 5. PSAT-1 and SREBF-1 were identified as targets for interference with metabolism and lipogenesis of EOC (Figure 6). EZH2 and BRD4 have emerged as targets for interference with epigenetic mechanisms and induction of macrophage polarization as a strategy for treatment of EOC (Figure 6). Reconstitution of activity of tumor suppressor genes, such as BRCA1 and PTEN might be of therapeutic benefit in patients with EOC (Figure 6).
Since circRNAs exhibit a unique splice junction, elimination of circRNAs with the CRISPR-CAS technology might be a new mode of therapeutic intervention (24,25). Also, investigations on the representation of individual circRNAs will pave the way for identification of circRNAs with a dominant role in EOC. The role of defined circRNA in cancer-related networks, the issue of synthetic lethality of inhibition of circRNAs together with other targets should be examined in more detail. Demonstration of activity of circRNA modulating agents in patient-derived xenograft models (PDX) would increase the confidence for this approach with respect to clinical translation.
Conflicts of Interest
FB is and UHW was an employee of Roche.
Authors’ Contributions
FB and UHW contributed equally to all aspects of the paper.
References
- 1.Kurnit KC, Fleming GF, Lengyel E. Updates and new options in advanced epithelial ovarian cancer treatment. Obstet Gynecol. 2021;137(1):108–121. doi: 10.1097/AOG.0000000000004173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Y, Xu S, Cheng S, Yang J, Wang Y. Clinical application of PARP inhibitors in ovarian cancer: from molecular mechanisms to the current status. J Ovarian Res. 2023;16(1):6. doi: 10.1186/s13048-023-01094-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh N, Jayraj AS, Sarkar A, Mohan T, Shukla A, Ghatage P. Pharmacotherapeutic treatment options for recurrent epithelial ovarian cancer. Expert Opin Pharmacother. 2023;24(1):49–64. doi: 10.1080/14656566.2022.2112030. [DOI] [PubMed] [Google Scholar]
- 4.Veneziani AC, Scott C, Wakefield MJ, Tinker AV, Lheureux S. Fighting resistance: post-PARP inhibitor treatment strategies in ovarian cancer. Ther Adv Med Oncol. 2023;15:17588359231157644. doi: 10.1177/17588359231157644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee A, Bilecz AJ, Lengyel E. The adipocyte microenvironment and cancer. Cancer Metastasis Rev. 2022;41(3):575–587. doi: 10.1007/s10555-022-10059-x. [DOI] [PubMed] [Google Scholar]
- 6.Zheng A, Wei Y, Zhao Y, Zhang T, Ma X. The role of cancer-associated mesothelial cells in the progression and therapy of ovarian cancer. Front Immunol. 2022;13:1013506. doi: 10.3389/fimmu.2022.1013506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pogge von Strandmann E, Reinartz S, Wager U, Müller R. Tumor-host cell interactions in ovarian cancer: Pathways to therapy failure. Trends Cancer. 2017;3(2):137–148. doi: 10.1016/j.trecan.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Tan DS, Agarwal R, Kaye SB. Mechanisms of transcoelomic metastasis in ovarian cancer. Lancet Oncol. 2006;7(11):925–934. doi: 10.1016/S1470-2045(06)70939-1. [DOI] [PubMed] [Google Scholar]
- 9.Mei S, Chen X, Wang K, Chen Y. Tumor microenvironment in ovarian cancer peritoneal metastasis. Cancer Cell Int. 2023;23(1):11. doi: 10.1186/s12935-023-02854-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumitru A, Dobrica EC, Croitoru A, Cretoiu SM, Gaspar BS. Focus on PD-1/PD-L1 as a therapeutic target in ovarian cancer. Int J Mol Sci. 2022;23(20):12067. doi: 10.3390/ijms232012067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vázquez-García I, Uhlitz F, Ceglia N, Lim JLP, Wu M, Mohibullah N, Niyazov J, Ruiz AEB, Boehm KM, Bojilova V, Fong CJ, Funnell T, Grewal D, Havasov E, Leung S, Pasha A, Patel DM, Pourmaleki M, Rusk N, Shi H, Vanguri R, Williams MJ, Zhang AW, Broach V, Chi DS, Da Cruz Paula A, Gardner GJ, Kim SH, Lennon M, Long Roche K, Sonoda Y, Zivanovic O, Kundra R, Viale A, Derakhshan FN, Geneslaw L, Issa Bhaloo S, Maroldi A, Nunez R, Pareja F, Stylianou A, Vahdatinia M, Bykov Y, Grisham RN, Liu YL, Lakhman Y, Nikolovski I, Kelly D, Gao J, Schietinger A, Hollmann TJ, Bakhoum SF, Soslow RA, Ellenson LH, Abu-Rustum NR, Aghajanian C, Friedman CF, McPherson A, Weigelt B, Zamarin D, Shah SP. Ovarian cancer mutational processes drive site-specific immune evasion. Nature. 2022;612(7941):778–786. doi: 10.1038/s41586-022-05496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu Y, Chen Y, Agbede O, Eshaghi E, Peng C. Circular RNAs in epithelial ovarian cancer: from biomarkers to therapeutic targets. Cancers (Basel) 2022;14(22):5711. doi: 10.3390/cancers14225711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185(12):2016–2034. doi: 10.1016/j.cell.2022.04.021. [DOI] [PubMed] [Google Scholar]
- 14.Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301. doi: 10.7150/ijms.28047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weidle UH, Birzele F. Triple-negative breast cancer: Identification of circRNAs with efficacy in preclinical in vivo models. Cancer Genomics Proteomics. 2023;20(2):117–131. doi: 10.21873/cgp.20368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weidle UH, Nopora A. Up-regulated circular RNAs in colorectal cancer: New entities for therapy and tools for identification of therapeutic targets. Cancer Genomics Proteomics. 2023;20(2):132–153. doi: 10.21873/cgp.20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weidle UH, Sela T, Brinkmann U, Niewoehner J. Circular RNAs with efficacy in preclinical in vitro and in vivo models of esophageal squamous cell carcinoma. Cancer Genomics Proteomics. 2022;19(3):283–298. doi: 10.21873/cgp.20320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206. doi: 10.1038/s41571-021-00585-y. [DOI] [PubMed] [Google Scholar]
- 19.Dai J, Su Y, Zhong S, Cong L, Liu B, Yang J, Tao Y, He Z, Chen C, Jiang Y. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. 2020;5(1):145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen R, Wang SK, Belk JA, Amaya L, Li Z, Cardenas A, Abe BT, Chen CK, Wender PA, Chang HY. Engineering circular RNA for enhanced protein production. Nat Biotechnol. 2023;41(2):262–272. doi: 10.1038/s41587-022-01393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu Y, Chen Y, Agbede O, Eshaghi E, Peng C. Circular RNAs in epithelial ovarian cancer: from biomarkers to therapeutic targets. Cancers (Basel) 2022;14(22):5711. doi: 10.3390/cancers14225711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Mei J, Wang H, Gu D, Ding J, Liu C. The emerging roles of circular RNAs in ovarian cancer. Cancer Cell Int. 2020;20:265. doi: 10.1186/s12935-020-01367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foruzandeh Z, Zeinali-Sehrig F, Nejati K, Rahmanpour D, Pashazadeh F, Seif F, Alivand MR. CircRNAs as potent biomarkers in ovarian cancer: a systematic scoping review. Cell Mol Biol Lett. 2021;26(1):41. doi: 10.1186/s11658-021-00284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Li X, Xue W, Zhang L, Yang LZ, Cao SM, Lei YN, Liu CX, Guo SK, Shan L, Wu M, Tao X, Zhang JL, Gao X, Zhang J, Wei J, Li J, Yang L, Chen LL. Screening for functional circular RNAs using the CRISPR–Cas13 system. Nat Methods. 2021;18(1):51–59. doi: 10.1038/s41592-020-01011-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Nguyen TM, Zhang XO, Wang L, Phan T, Clohessy JG, Pandolfi PP. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021;22(1):41. doi: 10.1186/s13059-021-02263-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin K, Zhang F, Wang H, Wang N, Qiu H, Jia X, Gong S, Zhang Z. circRNA circSnx12 confers Cisplatin chemoresistance to ovarian cancer by inhibiting ferroptosis through a miR-194-5p/SLC7A11 axis. BMB Rep. 2023;56(2):184–189. doi: 10.5483/BMBRep.2022-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pizzagalli MD, Bensimon A, Superti-Furga G. A guide to plasma membrane solute carrier proteins. FEBS J. 2021;288(9):2784–2835. doi: 10.1111/febs.15531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X, Li J, Kang R, Klionsky DJ, Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021;17(9):2054–2081. doi: 10.1080/15548627.2020.1810918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Qiu C, Hou M, Wang X, Huang C, Zou J, Liu T, Qu J. Ferroptosis in ovarian cancer: a novel therapeutic strategy. Front Oncol. 2021;11:665945. doi: 10.3389/fonc.2021.665945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, Xie HJ, Li YY, Wang X, Liu XX, Mai J. Molecular mechanisms of platinum based chemotherapy resistance in ovarian cancer (Review) Oncol Rep. 2022;47(4):82. doi: 10.3892/or.2022.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortiz M, Wabel E, Mitchell K, Horibata S. Mechanisms of chemotherapy resistance in ovarian cancer. Cancer Drug Resist. 2022;5(2):304–316. doi: 10.20517/cdr.2021.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Luo F, Jiang X, Zhang W, Xiang T, Pan Q, Cai L, Zhao J, Weng D, Li Y, Dai Y, Sun F, Yang C, Huang Y, Yang J, Tang Y, Han Y, He M, Zhang Y, Song L, Xia JC. CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype. J Immunother Cancer. 2022;10(3):e004029. doi: 10.1136/jitc-2021-004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowak M, Klink M. The role of tumor-associated macrophages in the progression and chemoresistance of ovarian cancer. Cells. 2020;9(5):1299. doi: 10.3390/cells9051299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An Y, Yang Q. Tumor-associated macrophage-targeted therapeutics in ovarian cancer. Intl Journal of Cancer. 2021;149(1):21–30. doi: 10.1002/ijc.33408. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z, Zhang Y, Qin X, Wang Y, Fu J. FGF9 promotes cisplatin resistance in colorectal cancer via regulation of Wnt/β-catenin signaling pathway. Exp Ther Med. 2020;19(3):1711–1718. doi: 10.3892/etm.2019.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang MM, Wu SZ, Yang SH, Wu CC, Wang CY, Huang BM. FGF9/FGFR1 promotes cell proliferation, epithelial-mesenchymal transition, M2 macrophage infiltration and liver metastasis of lung cancer. Transl Oncol. 2021;14(11):101208. doi: 10.1016/j.tranon.2021.101208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You J, Han Y, Qiao H, Han Y, Lu X, Lu Y, Wang X, Kai H, Zheng Y. Hsa_circ_0063804 enhances ovarian cancer cells proliferation and resistance to cisplatin by targeting miR-1276/CLU axis. Aging (Albany NY) 2022;14(11):4699–4713. doi: 10.18632/aging.203474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoter A, Naim HY. Heat shock proteins and ovarian cancer: important roles and therapeutic opportunities. Cancers (Basel) 2019;11(9):1389. doi: 10.3390/cancers11091389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poon S, Treweek TM, Wilson MR, Easterbrook-Smith SB, Carver JA. Clusterin is an extracellular chaperone that specifically interacts with slowly aggregating proteins on their off-folding pathway. FEBS Letters. 2002;513(2-3):259–266. doi: 10.1016/s0014-5793(02)02326-8. [DOI] [PubMed] [Google Scholar]
- 40.Wilson MR, Easterbrook-Smith SB. Clusterin is a secreted mammalian chaperone. Trends Biochem Sci. 2000;25(3):95–98. doi: 10.1016/s0968-0004(99)01534-0. [DOI] [PubMed] [Google Scholar]
- 41.Chi KN, Zoubeidi A, Gleave ME. Custirsen (OGX-011): a second-generation antisense inhibitor of clusterin for the treatment of cancer. Expert Opin Investig Drugs. 2008;17(12):1955–1962. doi: 10.1517/13543780802528609. [DOI] [PubMed] [Google Scholar]
- 42.Wilson MR, Zoubeidi A. Clusterin as a therapeutic target. Expert Opin Ther Targets. 2017;21(2):201–213. doi: 10.1080/14728222.2017.1267142. [DOI] [PubMed] [Google Scholar]
- 43.Yin Y, Li J, Rong J, Zhang B, Wang X, Han H. Circ_0067934 reduces JNK phosphorylation through a microRNA-545-3p/PPA1 axis to enhance tumorigenesis and cisplatin resistance in ovarian cancer. Immunopharmacol Immunotoxicol. 2022;44(2):261–274. doi: 10.1080/08923973.2022.2038193. [DOI] [PubMed] [Google Scholar]
- 44.Wang S, Wei J, Li S, Luo Y, Li Y, Wang X, Shen W, Luo D, Liu D. PPA1, an energy metabolism initiator, plays an important role in the progression of malignant tumors. Front Oncol. 2022;12:1012090. doi: 10.3389/fonc.2022.1012090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang P, Bai Y, Song B, Wang Y, Liu D, Lai Y, Bi X, Yuan Z. PP1A-mediated dephosphorylation positively regulates YAP2 activity. PLoS One. 2011;6(9):e24288. doi: 10.1371/journal.pone.0024288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall CA, Wang R, Miao J, Oliva E, Shen X, Wheeler T, Hilsenbeck SG, Orsulic S, Goode S. Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 2010;70(21):8517–8525. doi: 10.1158/0008-5472.CAN-10-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai H, Xu Y. The role of LPA and YAP signaling in long-term migration of human ovarian cancer cells. Cell Commun Signal. 2013;11(1):31. doi: 10.1186/1478-811X-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Yin Z, Wu Y, Zhan Q, Huang H, Fan J. Circular RNA lysophosphatidic acid receptor 3 (circ-LPAR3) enhances the cisplatin resistance of ovarian cancer. Bioengineered. 2022;13(2):3739–3750. doi: 10.1080/21655979.2022.2029109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao S, Shang W, Huang L, Xu R, Wu M, Wang F. The oncogenic and prognostic role of PDK1 in the progression and metastasis of ovarian cancer. J Cancer. 2021;12(3):630–643. doi: 10.7150/jca.47278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venturoli C, Piga I, Curtarello M, Verza M, Esposito G, Venuto S, Navaglia F, Grassi A, Indraccolo S. Genetic perturbation of pyruvate dehydrogenase kinase 1 modulates growth, angiogenesis and metabolic pathways in ovarian cancer xenografts. Cells. 2021;10(2):325. doi: 10.3390/cells10020325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang M, Cong Q, Zhang XY, Zhang MX, Lu YY, Xu CJ. Pyruvate dehydrogenase kinase 1 contributes to cisplatin resistance of ovarian cancer through EGFR activation. J Cell Physiol. 2019;234(5):6361–6370. doi: 10.1002/jcp.27369. [DOI] [PubMed] [Google Scholar]
- 52.Cao Y, Xie X, Li M, Gao Y. CircHIPK2 contributes to DDP resistance and malignant behaviors of DDP-resistant ovarian cancer cells both in vitro and in vivo through circHIPK2/miR-338-3p/CHTOP ceRNA pathway. Onco Targets Ther. 2021;14:3151–3165. doi: 10.2147/OTT.S291823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Izumikawa K, Ishikawa H, Simpson RJ, Takahashi N. Modulating the expression of Chtop, a versatile regulator of gene-specific transcription and mRNA export. RNA Biol. 2018;15(7):849–855. doi: 10.1080/15476286.2018.1465795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feng X, Bai X, Ni J, Wasinger VC, Beretov J, Zhu Y, Graham P, Li Y. CHTOP in chemoresistant epithelial ovarian cancer: a novel and potential therapeutic target. Front Oncol. 2019;9:557. doi: 10.3389/fonc.2019.00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng X, Li L, Wang L, Luo S, Bai X. Chromatin target of protein arginine methyltransferase regulates invasion, chemoresistance, and stemness in epithelial ovarian cancer. Biosci Rep. 2019;39(4):BSR20190016. doi: 10.1042/BSR20190016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei L, He W, Zhao H, Zhao P. Circ_0026123 promotes cisplatin resistance and progression of ovarian cancer by upregulating RAB1A through sequestering miR-543. Anticancer Drugs. 2022;33(10):1069–1080. doi: 10.1097/CAD.0000000000001373. [DOI] [PubMed] [Google Scholar]
- 57.Zha JF, Chen DX. MiR-655-3p inhibited proliferation and migration of ovarian cancer cells by targeting RAB1A. Eur Rev Med Pharmacol Sci. 2019;23(9):3627–3634. doi: 10.26355/eurrev_201905_17786. [DOI] [PubMed] [Google Scholar]
- 58.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 59.Tzeng HT, Wang YC. Rab-mediated vesicle trafficking in cancer. J Biomed Sci. 2016;23(1):70. doi: 10.1186/s12929-016-0287-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng KW, Agarwal R, Mills GB. Ras-Superfamily GTP-ases in Ovarian Cancer. Cancer Treat Res. 2009;149:229–240. doi: 10.1007/978-0-387-98094-2_11. [DOI] [PubMed] [Google Scholar]
- 61.Wu H, Zhao X, Wang J, Jiang X, Cheng Y, He Y, Sun L, Zhang G. Circular RNA CDR1as alleviates cisplatin-based chemoresistance by suppressing MiR-1299 in ovarian cancer. Front Genet. 2022;12:815448. doi: 10.3389/fgene.2021.815448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grassie ME, Moffat LD, Walsh MP, MacDonald JA. The myosin phosphatase targeting protein (MYPT) family: A regulated mechanism for achieving substrate specificity of the catalytic subunit of protein phosphatase type 1δ. Arch Biochem Biophys. 2011;510(2):147–159. doi: 10.1016/j.abb.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 63.Tan J, Lu T, Xu J, Hou Y, Chen Z, Zhou K, Ding Y, Jiang B, Zhu Y. MicroRNA-4463 facilitates the development of colon cancer by suppression of the expression of PPP1R12B. Clin Transl Oncol. 2022;24(6):1115–1123. doi: 10.1007/s12094-021-02752-0. [DOI] [PubMed] [Google Scholar]
- 64.Ding C, Tang W, Wu H, Fan X, Luo J, Feng J, Wen K, Wu G. The PEAK1-PPP1R12B axis inhibits tumor growth and metastasis by regulating Grb2/PI3K/Akt signalling in colorectal cancer. Cancer Lett. 2019;442:383–395. doi: 10.1016/j.canlet.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 65.Chen YY, Tai YC. Hsa_circ_0006404 and hsa_circ_0000735 regulated ovarian cancer response to docetaxel treatment via regulating p-GP expression. Biochem Genet. 2022;60(1):395–414. doi: 10.1007/s10528-021-10080-9. [DOI] [PubMed] [Google Scholar]
- 66.Colombo N, Parma G, Bocciolone L, Franchi D, Sideri M, Maggioni A. Medical therapy of advanced malignant epithelial tumours of the ovary. Forum (Genova) 2000;10(4):323–332. [PubMed] [Google Scholar]
- 67.Ortiz M, Wabel E, Mitchell K, Horibata S. Mechanisms of chemotherapy resistance in ovarian cancer. Cancer Drug Resist. 2022;5(2):304–316. doi: 10.20517/cdr.2021.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giralt I, Gallo-Oller G, Navarro N, Zarzosa P, Pons G, Magdaleno A, Segura MF, Sánchez de Toledo J, Moreno L, Gallego S, Roma J. Dickkopf proteins and their role in cancer: a family of Wnt antagonists with a dual role. Pharmaceuticals (Basel) 2021;14(8):810. doi: 10.3390/ph14080810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Engle K, Kumar G. Cancer multidrug-resistance reversal by ABCB1 inhibition: A recent update. Eur J Med Chem. 2022;239:114542. doi: 10.1016/j.ejmech.2022.114542. [DOI] [PubMed] [Google Scholar]
- 70.Liu X. ABC Family Transporters. Adv Exp Med Biol. 2019;1141:13–100. doi: 10.1007/978-981-13-7647-4_2. [DOI] [PubMed] [Google Scholar]
- 71.Huang C, Qin L, Chen S, Huang Q. CircSETDB1 contributes to paclitaxel resistance of ovarian cancer cells by sponging miR-508-3p and regulating ABCC1 expression. Anticancer Drugs. 2023;34(3):395–404. doi: 10.1097/CAD.0000000000001465. [DOI] [PubMed] [Google Scholar]
- 72.Waghray D, Zhang Q. Inhibit or evade multidrug resistance P-glycoprotein in cancer treatment. J Med Chem. 2018;61(12):5108–5121. doi: 10.1021/acs.jmedchem.7b01457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xia B, Zhao Z, Wu Y, Wang Y, Zhao Y, Wang J. Circular RNA circTNPO3 regulates paclitaxel resistance of ovarian cancer cells by miR-1299/NEK2 signaling pathway. Mol Ther Nucleic Acids. 2020;21:780–791. doi: 10.1016/j.omtn.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fang Y, Zhang X. Targeting NEK2 as a promising therapeutic approach for cancer treatment. Cell Cycle. 2016;15(7):895–907. doi: 10.1080/15384101.2016.1152430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kokuryo T, Yokoyama Y, Yamaguchi J, Tsunoda N, Ebata T, Nagino M. NEK2 is an effective target for cancer therapy with potential to induce regression of multiple human malignancies. Anticancer Res. 2019;39(5):2251–2258. doi: 10.21873/anticanres.13341. [DOI] [PubMed] [Google Scholar]
- 76.Liu XL, Liu HM, Han N, Li FH, Sun F, Fan DM, Xu Q. PCAT1 promotes the proliferative and migratory potentials of ovarian cancer via targeting NEK2. Eur Rev Med Pharmacol Sci. 2019;23(19):8239–8248. doi: 10.26355/eurrev_201910_19133. [DOI] [PubMed] [Google Scholar]
- 77.Liu X, Gao Y, Lu Y, Zhang J, Li L, Yin F. Upregulation of NEK2 is associated with drug resistance in ovarian cancer. Oncol Rep. 2014;31(2):745–754. doi: 10.3892/or.2013.2910. [DOI] [PubMed] [Google Scholar]
- 78.Wei S, Qi L, Wang L. Overexpression of circ_CELSR1 facilitates paclitaxel resistance of ovarian cancer by regulating miR-149-5p/SIK2 axis. Anticancer Drugs. 2021;32(5):496–507. doi: 10.1097/CAD.0000000000001058. [DOI] [PubMed] [Google Scholar]
- 79.Darling NJ, Cohen P. Nuts and bolts of the salt-inducible kinases (SIKs) Biochem J. 2021;478(7):1377–1397. doi: 10.1042/BCJ20200502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Du WQ, Zheng JN, Pei DS. The diverse oncogenic and tumor suppressor roles of salt-inducible kinase (SIK) in cancer. Expert Opin Ther Targets. 2016;20(4):477–485. doi: 10.1517/14728222.2016.1101452. [DOI] [PubMed] [Google Scholar]
- 81.Ahmed AA, Lu Z, Jennings NB, Etemadmoghadam D, Capalbo L, Jacamo RO, Barbosa-Morais N, Le XF, Australian Ovarian Cancer Study Group, Vivas-Mejia P, Lopez-Berestein G, Grandjean G, Bartholomeusz G, Liao W, Andreeff M, Bowtell D, Glover DM, Sood AK, Bast RC Jr. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell. 2010;18(2):109–121. doi: 10.1016/j.ccr.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi X, Yu X, Wang J, Bian S, Li Q, Fu F, Zou X, Zhang L, Bast RC Jr, Lu Z, Guo L, Chen Y, Zhou J. SIK2 promotes ovarian cancer cell motility and metastasis by phosphorylating MYLK. Mol Oncol. 2022;16(13):2558–2574. doi: 10.1002/1878-0261.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao J, Zhang X, Gao T, Wang S, Hou Y, Yuan P, Yang Y, Yang T, Xing J, Li J, Liu S. SIK2 enhances synthesis of fatty acid and cholesterol in ovarian cancer cells and tumor growth through PI3K/Akt signaling pathway. Cell Death Dis. 2020;11(1):25. doi: 10.1038/s41419-019-2221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tesch R, Rak M, Raab M, Berger LM, Kronenberger T, Joerger AC, Berger B, Abdi I, Hanke T, Poso A, Strebhardt K, Sanhaji M, Knapp S. Structure-based design of selective salt-inducible kinase inhibitors. J Med Chem. 2021;64(12):8142–8160. doi: 10.1021/acs.jmedchem.0c02144. [DOI] [PubMed] [Google Scholar]
- 85.Li M, Cai J, Han X, Ren Y. Downregulation of circNRIP1 suppresses the paclitaxel resistance of ovarian cancer via regulating the miR-211-5p/HOXC8 axis. Cancer Manag Res. 2020;12:9159–9171. doi: 10.2147/CMAR.S268872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 87.Brotto DB, Siena ÁDD, de Barros II, Carvalho SDCES, Muys BR, Goedert L, Cardoso C, Plaça JR, Ramão A, Squire JA, Araujo LF, Silva WAD Jr. Contributions of HOX genes to cancer hallmarks: Enrichment pathway analysis and review. Tumour Biol. 2020;42(5):101042832091805. doi: 10.1177/1010428320918050. [DOI] [PubMed] [Google Scholar]
- 88.Idaikkadar P, Morgan R, Michael A. HOX genes in high grade ovarian cancer. Cancers (Basel) 2019;11(8):1107. doi: 10.3390/cancers11081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu S, Liu R, Su M, Wei Y, Yang S, He S, Wang X, Qiang F, Chen C, Zhao S, Qian L, Shao M, Mao G. Overexpression of HOXC8 is associated with poor prognosis in epithelial ovarian cancer. Reprod Sci. 2016;23(7):944–954. doi: 10.1177/1933719115625845. [DOI] [PubMed] [Google Scholar]
- 90.Zhu J, Luo JE, Chen Y, Wu Q. Circ_0061140 knockdown inhibits tumorigenesis and improves PTX sensitivity by regulating miR-136/CBX2 axis in ovarian cancer. J Ovarian Res. 2021;14(1):136. doi: 10.1186/s13048-021-00888-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan HL, Morey L. Emerging roles for polycomb-group proteins in stem cells and cancer. Trends Biochem Sci. 2019;44(8):688–700. doi: 10.1016/j.tibs.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 93.Hu K, Yao L, Xu Z, Yan Y, Li J. Prognostic value and therapeutic potential of CBX family members in ovarian cancer. Front Cell Dev Biol. 2022;10:832354. doi: 10.3389/fcell.2022.832354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wheeler LJ, Watson ZL, Qamar L, Yamamoto TM, Post MD, Berning AA, Spillman MA, Behbakht K, Bitler BG. CBX2 identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer. Oncogenesis. 2018;7(11):92. doi: 10.1038/s41389-018-0103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang S, Cheng J, Quan C, Wen H, Feng Z, Hu Q, Zhu J, Huang Y, Wu X. circCELSR1 (hsa_circ_0063809) contributes to paclitaxel resistance of ovarian cancer cells by regulating FOXR2 expression via miR-1252. Mol Ther Nucleic Acids. 2020;19:718–730. doi: 10.1016/j.omtn.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Katoh M, Katoh M. Human FOX gene family (Review) Int J Oncol. 2004;25(5):1495–1500. [PubMed] [Google Scholar]
- 97.Liu AP, Northcott PA. Pursuing FOXR2-driven oncogenesis. Cancer Res. 2022;82(17):2977–2979. doi: 10.1158/0008-5472.CAN-22-2259. [DOI] [PubMed] [Google Scholar]
- 98.Li B, Huang W, Cao N, Lou G. Forkhead-box R2 promotes metastasis and growth by stimulating angiogenesis and activating hedgehog signaling pathway in ovarian cancer. J Cell Biochem. 2018;119(9):7780–7789. doi: 10.1002/jcb.27148. [DOI] [PubMed] [Google Scholar]
- 99.Liu M, Peng J. FTX regulated miR-153-3p/FOXR2 to promote cisplatin resistance in ovarian cancer. Comput Math Methods Med. 2022;2022:2318170. doi: 10.1155/2022/2318170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100.Liang YX, Zhang LL, Yang L. circANKRD17(has_circ_0007883) confers paclitaxel resistance of ovarian cancer via interacting with FUS to stabilize FOXR2. Mol Cell Biochem. 2023;478(4):835–850. doi: 10.1007/s11010-022-04548-4. [DOI] [PubMed] [Google Scholar]
- 101.Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Höglund M, Forster A, Rabbitts TH, Ron D, Mandahl N, Mitelman F. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics. 1996;37(1):1–8. doi: 10.1006/geno.1996.0513. [DOI] [PubMed] [Google Scholar]
- 102.Gittings LM, Foti SC, Benson BC, Gami-Patel P, Isaacs AM, Lashley T. Heterogeneous nuclear ribonucleoproteins R and Q accumulate in pathological inclusions in FTLD-FUS. Acta Neuropathol Commun. 2019;7(1):18. doi: 10.1186/s40478-019-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zheng Y, Li Z, Yang S, Wang Y, Luan Z. CircEXOC6B suppresses the proliferation and motility and sensitizes ovarian cancer cells to paclitaxel through miR-376c-3p/FOXO3 axis. Cancer Biother Radiopharm. 2022;37(9):802–814. doi: 10.1089/cbr.2020.3739. [DOI] [PubMed] [Google Scholar]
- 104.Calissi G, Lam EW, Link W. Therapeutic strategies targeting FOXO transcription factors. Nat Rev Drug Discov. 2021;20(1):21–38. doi: 10.1038/s41573-020-0088-2. [DOI] [PubMed] [Google Scholar]
- 105.Beretta GL, Corno C, Zaffaroni N, Perego P. Role of FoxO proteins in cellular response to antitumor agents. Cancers (Basel) 2019;11(1):90. doi: 10.3390/cancers11010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cui C, Han S, Yin H, Luo B, Shen X, Yang F, Liu Z, Zhu Q, Li D, Wang Y. FOXO3 is expressed in ovarian tissues and acts as an apoptosis initiator in granulosa cells of chickens. Biomed Res Int. 2019;2019:6902906. doi: 10.1155/2019/6902906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Y, Ao X, Ding W, Ponnusamy M, Wu W, Hao X, Yu W, Wang Y, Li P, Wang J. Critical role of FOXO3a in carcinogenesis. Mol Cancer. 2018;17(1):104. doi: 10.1186/s12943-018-0856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liang Y, Meng K, Qiu R. Circular RNA Circ_0013958 functions as a tumor promoter in ovarian cancer by regulating miR-637/PLXNB2 axis. Front Genet. 2021;12:644451. doi: 10.3389/fgene.2021.644451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alto LT, Terman JR. Semaphorins and their signaling mechanisms. Methods Mol Biol. 2017;1493:1–25. doi: 10.1007/978-1-4939-6448-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Malik MF, Ye L, Jiang WG. The Plexin-B family and its role in cancer progression. Histol Histopathol. 2014;29(2):151–165. doi: 10.14670/HH-29.151. [DOI] [PubMed] [Google Scholar]
- 111.Bica C, Tirpe A, Nutu A, Ciocan C, Chira S, Gurzau ES, Braicu C, Berindan-Neagoe I. Emerging roles and mechanisms of semaphorins activity in cancer. Life Sci. 2023;318:121499. doi: 10.1016/j.lfs.2023.121499. [DOI] [PubMed] [Google Scholar]
- 112.Masuda K, Furuyama T, Takahara M, Fujioka S, Kurinami H, Inagaki S. Sema4D stimulates axonal outgrowth of embryonic DRG sensory neurones. Genes Cells. 2004;9(9):821–829. doi: 10.1111/j.1365-2443.2004.00766.x. [DOI] [PubMed] [Google Scholar]
- 113.Xiang G, Cheng Y. MiR-126-3p inhibits ovarian cancer proliferation and invasion via targeting PLXNB2. Reproductive Biology. 2018;18(3):218–224. doi: 10.1016/j.repbio.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 114.Ma H, Qu S, Zhai Y, Yang X. circ_0025033 promotes ovarian cancer development via regulating the hsa_miR-370-3p/SLC1A5 axis. Cell Mol Biol Lett. 2022;27(1):94. doi: 10.1186/s11658-022-00364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu Y, Zhao T, Li Z, Wang L, Yuan S, Sun L. The role of ASCT2 in cancer: A review. Eur J Pharmacol. 2018;837:81–87. doi: 10.1016/j.ejphar.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 116.Teixeira E, Silva C, Martel F. The role of the glutamine transporter ASCT2 in antineoplastic therapy. Cancer Chemother Pharmacol. 2021;87(4):447–464. doi: 10.1007/s00280-020-04218-6. [DOI] [PubMed] [Google Scholar]
- 117.Huang X, Luo Y, Li X. Circ_0072995 promotes ovarian cancer progression through regulating miR-122-5p/SLC1A5 axis. Biochem Genet. 2022;60(1):153–172. doi: 10.1007/s10528-021-10092-5. [DOI] [PubMed] [Google Scholar]
- 118.Sheng S, Hu Y, Yu F, Tong W, Wang S, Cai Y, Zhu J. circKIF4A sponges miR-127 to promote ovarian cancer progression. Aging (Albany NY) 2020;12(18):17921–17929. doi: 10.18632/aging.103389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hartmann C, Schwietzer YA, Otani T, Furuse M, Ebnet K. Physiological functions of junctional adhesion molecules (JAMs) in tight junctions. Biochim Biophys Acta Biomembr. 2020;1862(9):183299. doi: 10.1016/j.bbamem.2020.183299. [DOI] [PubMed] [Google Scholar]
- 120.Bazzoni G. The JAM family of junctional adhesion molecules. Curr Opin Cell Biol. 2003;15(5):525–530. doi: 10.1016/s0955-0674(03)00104-2. [DOI] [PubMed] [Google Scholar]
- 121.Naik TU, Naik MU, Naik UP. Junctional adhesion molecules in angiogenesis. Front Biosci. 2008;13(13):258. doi: 10.2741/2676. [DOI] [PubMed] [Google Scholar]
- 122.Lauko A, Mu Z, Gutmann DH, Naik UP, Lathia JD. Junctional adhesion molecules in cancer: a paradigm for the diverse functions of cell-cell interactions in tumor progression. Cancer Res. 2020;80(22):4878–4885. doi: 10.1158/0008-5472.CAN-20-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leinster DA, Colom B, Whiteford JR, Ennis DP, Lockley M, McNeish IA, Aurrand-Lions M, Chavakis T, Imhof BA, Balkwill FR, Nourshargh S. Endothelial cell junctional adhesion molecule C plays a key role in the development of tumors in a murine model of ovarian cancer. FASEB J. 2013;27(10):4244–4253. doi: 10.1096/fj.13-230441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ivana B, Emina M, Marijana M, Irena J, Zoran B, Radmila J. High expression of junctional adhesion molecule-A is associated with poor survival in patients with epithelial ovarian cancer. Int J Biol Markers. 2019;34(3):262–268. doi: 10.1177/1724600819850178. [DOI] [PubMed] [Google Scholar]
- 125.Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38(1):437. doi: 10.1186/s13046-019-1437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Deng J, Wang L, Chen H, Li L, Ma Y, Ni J, Li Y. The role of tumour-associated MUC1 in epithelial ovarian cancer metastasis and progression. Cancer Metastasis Rev. 2013;32(3-4):535–551. doi: 10.1007/s10555-013-9423-y. [DOI] [PubMed] [Google Scholar]
- 127.Ma Q, Song J, Wang S, He N. MUC1 regulates AKT signaling pathway by upregulating EGFR expression in ovarian cancer cells. Pathol Res Pract. 2021;224:153509. doi: 10.1016/j.prp.2021.153509. [DOI] [PubMed] [Google Scholar]
- 128.Wang QA, Liang X, Yang Y. Downregulation of hTERT contributes to ovarian cancer apoptosis and inhibits proliferation of ovarian cancer cells. Transl Cancer Res. 2020;9(3):1448–1454. doi: 10.21037/tcr.2020.01.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou X, Jiang J, Guo S. Hsa_circ_0004712 downregulation attenuates ovarian cancer malignant development by targeting the miR-331-3p/FZD4 pathway. J Ovarian Res. 2021;14(1):118. doi: 10.1186/s13048-021-00859-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ricken A, Lochhead P, Kontogiannea M, Farookhi R. Wnt signaling in the ovary: identification and compartmentalized expression of wnt-2, wnt-2b, and frizzled-4 mRNAs. Endocrinology. 2002;143(7):2741–2749. doi: 10.1210/endo.143.7.8908. [DOI] [PubMed] [Google Scholar]
- 131.Ma L, Liu W, Li M. Circ_0061140 contributes to ovarian cancer progression by targeting miR-761/LETM1 signaling. Biochem Genet. 2023;61(2):628–650. doi: 10.1007/s10528-022-10277-6. [DOI] [PubMed] [Google Scholar]
- 132.Natarajan GK, Mishra J, Camara AKS, Kwok WM. LETM1: A single entity with diverse impact on mitochondrial metabolism and cellular signaling. Front Physiol. 2021;12:637852. doi: 10.3389/fphys.2021.637852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li Y, Tran Q, Shrestha R, Piao L, Park S, Park J, Park J. LETM1 is required for mitochondrial homeostasis and cellular viability (Review) Mol Med Rep. 2019;19(5):3367–3375. doi: 10.3892/mmr.2019.10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang J, Ding W, Xu Y, Tao E, Mo M, Xu W, Cai X, Chen X, Yuan J, Wu X. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging (Albany NY) 2020;12(5):4558–4572. doi: 10.18632/aging.102911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lin C, Xu X, Yang Q, Liang L, Qiao S. Circular RNA ITCH suppresses proliferation, invasion, and glycolysis of ovarian cancer cells by up-regulating CDH1 via sponging miR-106a. Cancer Cell Int. 2020;20:336. doi: 10.1186/s12935-020-01420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dong LL, Liu L, Ma CH, Li JS, Du C, Xu S, Han LH, Li L, Wang XW. E-cadherin promotes proliferation of human ovarian cancer cells in vitro via activating MEK/ERK pathway. Acta Pharmacol Sin. 2012;33(6):817–822. doi: 10.1038/aps.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rodriguez FJ, Lewis-Tuffin LJ, Anastasiadis PZ. E-cadherin's dark side: possible role in tumor progression. Biochim Biophys Acta. 2012;1826(1):23–31. doi: 10.1016/j.bbcan.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sawada K, Mitra AK, Radjabi AR, Bhaskar V, Kistner EO, Tretiakova M, Jagadeeswaran S, Montag A, Becker A, Kenny HA, Peter ME, Ramakrishnan V, Yamada SD, Lengyel E. Loss of E-cadherin promotes ovarian cancer metastasis via alpha 5-integrin, which is a therapeutic target. Cancer Res. 2008;68(7):2329–2339. doi: 10.1158/0008-5472.CAN-07-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou B, Xu H, Xia M, Sun C, Li N, Guo E, Guo L, Shan W, Lu H, Wu Y, Li Y, Yang D, Weng D, Meng L, Hu J, Ma D, Chen G, Li K. Overexpressed miR-9 promotes tumor metastasis via targeting E-cadherin in serous ovarian cancer. Front Med. 2017;11(2):214–222. doi: 10.1007/s11684-017-0518-7. [DOI] [PubMed] [Google Scholar]
- 140.Sui X, Jiao YN, Yang LH, Liu J. MiR-9 accelerates epithelial-mesenchymal transition of ovarian cancer cells via inhibiting e-cadherin. Eur Rev Med Pharmacol Sci 23(3. 2019;Suppl):209–216. doi: 10.26355/eurrev_201908_18649. [DOI] [PubMed] [Google Scholar]
- 141.Bačić B, Haller H, Mrklić I, Košta V, Čarić A, Tomić S. Prognostic role of E-cadherin in patients with advanced serous ovarian cancer. Arch Gynecol Obstet. 2013;287(6):1219–1224. doi: 10.1007/s00404-012-2684-9. [DOI] [PubMed] [Google Scholar]
- 142.Wang LL, Zong ZH, Liu Y, Guan X, Chen S, Zhao Y. CircRhoC promotes tumorigenicity and progression in ovarian cancer by functioning as a miR-302e sponge to positively regulate VEGFA. J Cell Mol Med. 2019;23(12):8472–8481. doi: 10.1111/jcmm.14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen J, Li X, Yang L, Li M, Zhang Y, Zhang J. CircASH2L promotes ovarian cancer tumorigenesis, angiogenesis, and lymphangiogenesis by regulating the miR-665/VEGFA axis as a competing endogenous RNA. Front Cell Dev Biol. 2020;8:595585. doi: 10.3389/fcell.2020.595585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Melincovici CS, Boşca AB, Şuşman S, Mărginean M, Mihu C, Istrate M, Moldovan IM, Roman AL, Mihu CM. Vascular endothelial growth factor (VEGF) - key factor in normal and pathological angiogenesis. Rom J Morphol Embryol. 2018;59(2):455–467. [PubMed] [Google Scholar]
- 145.Wang J, Huang Y, Zhang J, Wei Y, Mahoud S, Bakheet AMH, Wang L, Zhou S, Tang J. Pathway-related molecules of VEGFC/D-VEGFR3/NRP2 axis in tumor lymphangiogenesis and lymphatic metastasis. Clin Chim Acta. 2016;461:165–171. doi: 10.1016/j.cca.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 146.Gómez-Raposo C, Mendiola M, Barriuso J, Casado E, Hardisson D, Redondo A. Angiogenesis and ovarian cancer. Clin Transl Oncol. 2009;11(9):564–571. doi: 10.1007/s12094-009-0406-y. [DOI] [PubMed] [Google Scholar]
- 147.Mei C, Gong W, Wang X, Lv Y, Zhang Y, Wu S, Zhu C. Anti-angiogenic therapy in ovarian cancer: Current understandings and prospects of precision medicine. Front Pharmacol. 2023;14:1147717. doi: 10.3389/fphar.2023.1147717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nakai H, Matsumura N. The roles and limitations of bevacizumab in the treatment of ovarian cancer. Int J Clin Oncol. 2022;27(7):1120–1126. doi: 10.1007/s10147-022-02169-x. [DOI] [PubMed] [Google Scholar]
- 149.Wu Y, Zhou J, Li Y, Shi X, Shen F, Chen M, Chen Y, Wang J. Hsa_circ_0001445 works as a cancer suppressor via miR-576-5p/SFRP1 axis regulation in ovarian cancer. Cancer Med. 2023;12(5):5736–5750. doi: 10.1002/cam4.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vincent KM, Postovit LM. Matricellular proteins in cancer: a focus on secreted Frizzled-related proteins. J Cell Commun Signal. 2018;12(1):103–112. doi: 10.1007/s12079-017-0398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Surana R, Sikka S, Cai W, Shin EM, Warrier SR, Tan HJG, Arfuso F, Fox SA, Dharmarajan AM, Kumar AP. Secreted frizzled related proteins: Implications in cancers. Biochim Biophys Acta. 2014;1845(1):53–65. doi: 10.1016/j.bbcan.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 152.Zhang H, Sun D, Qiu J, Yao L. SFRP1 inhibited the epithelial ovarian cancer through inhibiting Wnt/β-catenin signaling. Acta Biochimica Polonica. 2019;66(4):393–400. doi: 10.18388/abp.2019_2757. [DOI] [PubMed] [Google Scholar]
- 153.Kardum V, Karin V, Glibo M, Skrtic A, Martic TN, Ibisevic N, Skenderi F, Vranic S, Serman L. Methylation-associated silencing of SFRP1 gene in high-grade serous ovarian carcinomas. Ann Diagn Pathol. 2017;31:45–49. doi: 10.1016/j.anndiagpath.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 154.Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. 2018;13:55–63. doi: 10.1016/j.omtn.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 155.Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, Hu CJ, Bai JY. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal. 2018;16(1):57. doi: 10.1186/s12964-018-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu C, Barger CJ, Karpf AR. FOXM1: A multifunctional oncoprotein and emerging therapeutic target in ovarian cancer. Cancers (Basel) 2021;13(12):3065. doi: 10.3390/cancers13123065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Parashar D, Nair B, Geethadevi A, George J, Nair A, Tsaih SW, Kadamberi IP, Gopinadhan Nair GK, Lu Y, Ramchandran R, Uyar DS, Rader JS, Ram PT, Mills GB, Pradeep S, Chaluvally-Raghavan P. Peritoneal spread of ovarian cancer harbors therapeutic vulnerabilities regulated by FOXM1 and EGFR/ERBB2 signaling. Cancer Res. 2020;80(24):5554–5568. doi: 10.1158/0008-5472.CAN-19-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fang L, Zhang B, Zhu N. MiR-877 suppresses tumor metastasis via regulating FOXM1 in ovarian cancer. J BUON. 2021;26(1):229–234. [PubMed] [Google Scholar]
- 159.Zhang Z, Xue ST, Gao Y, Li Y, Zhou Z, Wang J, Li Z, Liu Z. Small molecule targeting FOXM1 DNA binding domain exhibits anti-tumor activity in ovarian cancer. Cell Death Discov. 2022;8(1):280. doi: 10.1038/s41420-022-01070-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhou ZY, Han XY, Sun LQ, Li SY, Xue ST, Li ZR. Structure-based virtual screening identified novel FOXM1 inhibitors as the lead compounds for ovarian cancer. Front Chem. 2022;10:1058256. doi: 10.3389/fchem.2022.1058256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Du Y, Liu X, Zhang S, Chen S, Guan X, Li Q, Chen X, Zhao Y. CircCRIM1 promotes ovarian cancer progression by working as ceRNAs of CRIM1 and targeting miR-383-5p/ZEB2 axis. Reprod Biol Endocrinol. 2021;19(1):176. doi: 10.1186/s12958-021-00857-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zeng H, Tang L. CRIM1, the antagonist of BMPs, is a potential risk factor of cancer. Curr Cancer Drug Targets. 2014;14(7):652–658. doi: 10.2174/1568009614666140725094125. [DOI] [PubMed] [Google Scholar]
- 163.Wu KZ, Zhang CD, Zhang C, Pei JP, Dai DQ. miR-665 suppresses the epithelial-mesenchymal transition and progression of gastric cancer by targeting CRIM1. Cancer Manag Res. 2020;12:3489–3501. doi: 10.2147/CMAR.S241795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Brabletz S, Schuhwerk H, Brabletz T, Stemmler MP. Dynamic EMT: a multi-tool for tumor progression. EMBO J. 2021;40(18):e108647. doi: 10.15252/embj.2021108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Prislei S, Martinelli E, Zannoni GF, Petrillo M, Filippetti F, Mariani M, Mozzetti S, Raspaglio G, Scambia G, Ferlini C. Role and prognostic significance of the epithelial-mesenchymal transition factor ZEB2 in ovarian cancer. Oncotarget. 2015;6(22):18966–18979. doi: 10.18632/oncotarget.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wu Q, Guo R, Lin M, Zhou B, Wang Y. MicroRNA-200a inhibits CD133/1+ ovarian cancer stem cells migration and invasion by targeting E-cadherin repressor ZEB2. Gynecol Oncol. 2011;122(1):149–154. doi: 10.1016/j.ygyno.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 167.Wu SG, Zhou P, Chen JX, Lei J, Hua L, Dong Y, Hu M, Lian CL, Yang LC, Zhou J. circ-PTK2 (hsa_circ_0008305) regulates the pathogenic processes of ovarian cancer via miR-639 and FOXC1 regulatory cascade. Cancer Cell Int. 2021;21(1):277. doi: 10.1186/s12935-021-01985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cheng W, Su Y, Xu F. CHD1L: a novel oncogene. Mol Cancer. 2013;12(1):170. doi: 10.1186/1476-4598-12-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.He WP, Zhou J, Cai MY, Xiao XS, Liao YJ, Kung HF, Guan XY, Xie D, Yang GF. CHD1L protein is overexpressed in human ovarian carcinomas and is a novel predictive biomarker for patients survival. BMC Cancer. 2012;12:437. doi: 10.1186/1471-2407-12-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.He WP, Guo YY, Yang GP, Lai HL, Sun TT, Zhang ZW, Ouyang LL, Zheng Y, Tian LM, Li XH, You ZS, Xie D, Yang GF. CHD1L promotes EOC cell invasiveness and metastasis via the regulation of METAP2. Int J Med Sci. 2020;17(15):2387–2395. doi: 10.7150/ijms.48615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Uhlenhaut NH, Treier M. Forkhead transcription factors in ovarian function. Reproduction. 2011;142(4):489–495. doi: 10.1530/REP-11-0092. [DOI] [PubMed] [Google Scholar]
- 172.Gilding LN, Somervaille TCP. The diverse consequences of FOXC1 deregulation in cancer. Cancers (Basel) 2019;11(2):184. doi: 10.3390/cancers11020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huang K, Liu D, Su C. Circ_0007841 accelerates ovarian cancer development through facilitating MEX3C expression by restraining miR-151-3p activity. Aging (Albany NY) 2021;13(8):12058–12066. doi: 10.18632/aging.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Qin H, Ni H, Liu Y, Yuan Y, Xi T, Li X, Zheng L. RNA-binding proteins in tumor progression. J Hematol Oncol. 2020;13(1):90. doi: 10.1186/s13045-020-00927-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gebauer F, Schwarzl T, Valcárcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22(3):185–198. doi: 10.1038/s41576-020-00302-y. [DOI] [PubMed] [Google Scholar]
- 176.Pereira B, Le Borgne M, Chartier NT, Billaud M, Almeida R. MEX-3 proteins: recent insights on novel post-transcriptional regulators. Trends Biochem Sci. 2013;38(10):477–479. doi: 10.1016/j.tibs.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 177.Chao H, Deng L, Xu F, Yu Z, Xu X, Huang J, Zeng T. MEX3C regulates lipid metabolism to promote bladder tumorigenesis through JNK pathway. Onco Targets Ther. 2019;12:3285–3294. doi: 10.2147/OTT.S199667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Xiao Y, Li Y, Shi D, Wang X, Dai S, Yang M, Kong L, Chen B, Huang X, Lin C, Liao W, Xu B, Chen X, Wang L, Chen X, Ouyang Y, Liu G, Li H, Song L. MEX3C-mediated decay of SOCS3 mRNA promotes JAK2/STAT3 signaling to facilitate metastasis in hepatocellular carcinoma. Cancer Res. 2022;82(22):4191–4205. doi: 10.1158/0008-5472.CAN-22-1203. [DOI] [PubMed] [Google Scholar]
- 179.Hou W, Zhang Y. Circ_0025033 promotes the progression of ovarian cancer by activating the expression of LSM4 via targeting miR-184. Pathol Res Pract. 2021;217:153275. doi: 10.1016/j.prp.2020.153275. [DOI] [PubMed] [Google Scholar]
- 180.Chen L, Lin YH, Liu GQ, Huang JE, Wei W, Yang ZH, Hu YM, Xie JH, Yu HZ. Clinical significance and potential role of LSM4 overexpression in hepatocellular carcinoma: an integrated analysis based on multiple databases. Front Genet. 2022;12:804916. doi: 10.3389/fgene.2021.804916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Séraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18(12):3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3'-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18(20):5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ji J, Li C, Wang J, Wang L, Huang H, Li Y, Fang J. Hsa_circ_0001756 promotes ovarian cancer progression through regulating IGF2BP2-mediated RAB5A expression and the EGFR/MAPK signaling pathway. Cell Cycle. 2022;21(7):685–696. doi: 10.1080/15384101.2021.2010166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Barghash A, Golob-Schwarzl N, Helms V, Haybaeck J, Kessler SM. Elevated expression of the IGF2 mRNA binding protein 2 (IGF2BP2/IMP2) is linked to short survival and metastasis in esophageal adenocarcinoma. Oncotarget. 2016;7(31):49743–49750. doi: 10.18632/oncotarget.10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression. Cell Mol Life Sci. 2013;70(15):2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhao Z, Liu XF, Wu HC, Zou SB, Wang JY, Ni PH, Chen XH, Fan QS. Rab5a overexpression promoting ovarian cancer cell proliferation may be associated with APPL1-related epidermal growth factor signaling pathway. Cancer Sci. 2010;101(6):1454–1462. doi: 10.1111/j.1349-7006.2010.01558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang S, Li Z, Zhu G, Hong L, Hu C, Wang K, Cui K, Hao C. RNA-binding protein IGF2BP2 enhances circ_0000745 abundancy and promotes aggressiveness and stemness of ovarian cancer cells via the microRNA-3187-3p/ERBB4/PI3K/AKT axis. J Ovarian Res. 2021;14(1):154. doi: 10.1186/s13048-021-00917-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lu M, Gong B, Wang Y, Li J. CircBNC2 affects epithelial ovarian cancer progression through the miR-223-3p/LARP4 axis. Anticancer Drugs. 2023;34(3):384–394. doi: 10.1097/CAD.0000000000001423. [DOI] [PubMed] [Google Scholar]
- 189.Stavraka C, Blagden S. The La-related proteins, a family with connections to cancer. Biomolecules. 2015;5(4):2701–2722. doi: 10.3390/biom5042701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Koso H, Yi H, Sheridan P, Miyano S, Ino Y, Todo T, Watanabe S. Identification of RNA-binding protein LARP4B as a tumor suppressor in glioma. Cancer Res. 2016;76(8):2254–2264. doi: 10.1158/0008-5472.CAN-15-2308. [DOI] [PubMed] [Google Scholar]
- 191.Egiz M, Usui T, Ishibashi M, Zhang X, Shigeta S, Toyoshima M, Kitatani K, Yaegashi N. La-related protein 4 as a suppressor for motility of ovarian cancer cells. Tohoku J Exp Med. 2019;247(1):59–67. doi: 10.1620/tjem.247.59. [DOI] [PubMed] [Google Scholar]
- 192.Lin W, Ye H, You K, Chen L. Up-regulation of circ_LARP4 suppresses cell proliferation and migration in ovarian cancer by regulating miR-513b-5p/LARP4 axis. Cancer Cell Int. 2020;20:5. doi: 10.1186/s12935-019-1071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhang Y, Di Q, Chen J, Chang M, Ma Y, Yu J. Circ_0061140 contributes to the malignant progression in ovarian cancer cells by mediating the RAB1A level through sponging miR-361-5p. Biochem Genet. 2022;60(6):1946–1962. doi: 10.1007/s10528-022-10200-z. [DOI] [PubMed] [Google Scholar]
- 194.Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91(1):119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Peng C, Li X, Ye Z, Wu W. Rab1A promotes cell proliferation and migration by upregulating Gli1 in colorectal cancer. Sci Rep. 2021;11(1):16243. doi: 10.1038/s41598-021-95798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hu J, Wang L, Chen J, Gao H, Zhao W, Huang Y, Jiang T, Zhou J, Chen Y. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem Biophys Res Commun. 2018;505(1):222–228. doi: 10.1016/j.bbrc.2018.09.060. [DOI] [PubMed] [Google Scholar]
- 197.Zhang Y, Li Y, Wang Q, Su B, Xu H, Sun Y, Sun P, Li R, Peng X, Cai J. Role of RASA1 in cancer: A review and update (Review) Oncol Rep. 2020;44(6):2386–2396. doi: 10.3892/or.2020.7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li X, Jiang X, Lu J, Lin Y, Jiang L, Li Y, Wan F, Wang C. CircCERS6 suppresses the development of epithelial ovarian cancer through mediating miR-630/RASSF8. Biochem Genet. 2022;60(6):2611–2629. doi: 10.1007/s10528-022-10227-2. [DOI] [PubMed] [Google Scholar]
- 199.Sherwood V, Recino A, Jeffries A, Ward A, Chalmers AD. The N-terminal RASSF family: a new group of Ras-association-domaincontaining proteins, with emerging links to cancer formation. Biochem J. 2010;425(2):303–311. doi: 10.1042/BJ20091318. [DOI] [PubMed] [Google Scholar]
- 200.Lock FE, Underhill-Day N, Dunwell T, Matallanas D, Cooper W, Hesson L, Recino A, Ward A, Pavlova T, Zabarovsky E, Grant MM, Maher ER, Chalmers AD, Kolch W, Latif F. The RASSF8 candidate tumor suppressor inhibits cell growth and regulates the Wnt and NF-ĸB signaling pathways. Oncogene. 2010;29(30):4307–4316. doi: 10.1038/onc.2010.192. [DOI] [PubMed] [Google Scholar]
- 201.van der Weyden L, Adams DJ. The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim Biophys Acta. 2007;1776(1):58–85. doi: 10.1016/j.bbcan.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Avruch J, Xavier R, Bardeesy N, Zhang XF, Praskova M, Zhou D, Xia F. Rassf family of tumor suppressor polypeptides. J Biol Chem. 2009;284(17):11001–11005. doi: 10.1074/jbc.R800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang L, Chen H, He F, Zhang S, Li A, Zhang A, Zhang A. MicroRNA-320a promotes epithelial ovarian cancer cell proliferation and invasion by targeting RASSF8. Front Oncol. 2021;11:581932. doi: 10.3389/fonc.2021.581932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Du Z, Wang L, Xia Y. Circ_0015756 promotes the progression of ovarian cancer by regulating miR-942-5p/CUL4B pathway. Cancer Cell Int. 2020;20(1):572. doi: 10.1186/s12935-020-01666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu L, Han B, Liu L, Cui H, Liu H, Jia R, Zhang X, Lu X. Circ_0021573 acts as a competing endogenous RNA to promote the malignant phenotypes of human ovarian cancer cells. Reprod Biol. 2023;23(1):100704. doi: 10.1016/j.repbio.2022.100704. [DOI] [PubMed] [Google Scholar]
- 206.Hannah J, Zhou P. Distinct and overlapping functions of the cullin E3 ligase scaffolding proteins CUL4A and CUL4B. Gene. 2015;573(1):33–45. doi: 10.1016/j.gene.2015.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34(11):562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Li Y, Wang X. The role of cullin4B in human cancers. Exp Hematol Oncol. 2017;6:17. doi: 10.1186/s40164-017-0077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mansour MA. Ubiquitination: Friend and foe in cancer. Int J Biochem Cell Biol. 2018;101:80–93. doi: 10.1016/j.biocel.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 210.Thirunavukarasou A, Singh P, Govindarajalu G, Bandi V, Baluchamy S. E3 ubiquitin ligase Cullin4B mediated polyubiquitination of p53 for its degradation. Mol Cell Biochem. 2014;390(1-2):93–100. doi: 10.1007/s11010-014-1960-3. [DOI] [PubMed] [Google Scholar]
- 211.Duan PJ, Zhao JH, Xie LL. Cul4B promotes the progression of ovarian cancer by upregulating the expression of CDK2 and CyclinD1. J Ovarian Res. 2020;13(1):76. doi: 10.1186/s13048-020-00677-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Liu T, Yuan L, Zou X. Circular RNA circ-BNC2 (hsa_circ_0008732) inhibits the progression of ovarian cancer through microRNA-223-3p/ FBXW7 axis. J Ovarian Res. 2022;15(1):95. doi: 10.1186/s13048-022-01025-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17(1):115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang L, Ye X, Liu Y, Wei W, Wang Z. Aberrant regulation of FBW7 in cancer. Oncotarget. 2014;5(8):2000–2015. doi: 10.18632/oncotarget.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Xu F, Li J, Ni M, Cheng J, Zhao H, Wang S, Zhou X, Wu X. FBW7 suppresses ovarian cancer development by targeting the N(6)-methyladenosine binding protein YTHDF2. Mol Cancer. 2021;20(1):45. doi: 10.1186/s12943-021-01340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ding J, Wang Q, Guo N, Wang H, Chen H, Ni G, Li P. CircRNA circ_0072995 promotes the progression of epithelial ovarian cancer by modulating miR-147a/CDK6 axis. Aging (Albany NY) 2020;12(17):17209–17223. doi: 10.18632/aging.103668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Goel S, Bergholz JS, Zhao JJ. Targeting CDK4 and CDK6 in cancer. Nat Rev Cancer. 2022;22(6):356–372. doi: 10.1038/s41568-022-00456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Nebenfuehr S, Kollmann K, Sexl V. The role of CDK6 in cancer. Int J Cancer. 2020;147(11):2988–2995. doi: 10.1002/ijc.33054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cao J, Zhang Y, Mu J, Yang D, Gu X, Zhang J. Exosomal miR-21-5p contributes to ovarian cancer progression by regulating CDK6. Hum Cell. 2021;34(4):1185–1196. doi: 10.1007/s13577-021-00522-2. [DOI] [PubMed] [Google Scholar]
- 220.Dall'Acqua A, Sonego M, Pellizzari I, Pellarin I, Canzonieri V, D'Andrea S, Benevol S, Sorio R, Giorda G, Califano D, Bagnoli M, Militello L, Mezzanzanica D, Chiappetta G, Armenia J, Belletti B, Schiappacassi M, Baldassarre G. CDK6 protects epithelial ovarian cancer from platinum-induced death via FOXO3 regulation. EMBO Mol Med. 2017;9(10):1415–1433. doi: 10.15252/emmm.201607012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhang QF, Li J, Jiang K, Wang R, Ge JL, Yang H, Liu SJ, Jia LT, Wang L, Chen BL. CDK4/6 inhibition promotes immune infiltration in ovarian cancer and synergizes with PD-1 blockade in a B cell-dependent manner. Theranostics. 2020;10(23):10619–10633. doi: 10.7150/thno.44871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhang C, Li Y, Zhao W, Liu G, Yang Q. Circ-PGAM1 promotes malignant progression of epithelial ovarian cancer through regulation of the miR-542-3p/CDC5L/PEAK1 pathway. Cancer Med. 2020;9(10):3500–3521. doi: 10.1002/cam4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang Z, Mao W, Wang L, Liu M, Zhang W, Wu Y, Zhang J, Mao S, Geng J, Yao X. Depletion of CDC5L inhibits bladder cancer tumorigenesis. J Cancer. 2020;11(2):353–363. doi: 10.7150/jca.32850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Mu R, Wang YB, Wu M, Yang Y, Song W, Li T, Zhang WN, Tan B, Li AL, Wang N, Xia Q, Gong WL, Wang CG, Zhou T, Guo N, Sang ZH, Li HY. Depletion of pre-mRNA splicing factor Cdc5L inhibits mitotic progression and triggers mitotic catastrophe. Cell Death Dis. 2014;5(3):e1151. doi: 10.1038/cddis.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wang Y, Kelber JA, Tran Cao HS, Cantin GT, Lin R, Wang W, Kaushal S, Bristow JM, Edgington TS, Hoffman RM, Bouvet M, Yates JR 3rd, Klemke RL. Pseudopodium-enriched atypical kinase 1 regulates the cytoskeleton and cancer progression [corrected] Proc Natl Acad Sci USA. 2010;107(24):10920–10925. doi: 10.1073/pnas.0914776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kelber JA, Klemke RL. PEAK1, a novel kinase target in the fight against cancer. Oncotarget. 2010;1(3):219–223. doi: 10.18632/oncotarget.100703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Li B, Zhang L. CircSETDB1 knockdown inhibits the malignant progression of serous ovarian cancer through miR-129-3p-dependent regulation of MAP3K3. J Ovarian Res. 2021;14(1):160. doi: 10.1186/s13048-021-00875-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298(5600):1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 229.Jia W, Dong Y, Tao L, Pang L, Ren Y, Liang W, Jiang J, Cheng G, Zhang WJ, Yuan X, Li F. MAP3K3 overexpression is associated with poor survival in ovarian carcinoma. Hum Pathol. 2016;50:162–169. doi: 10.1016/j.humpath.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 230.Zhang Y, Wang SS, Tao L, Pang LJ, Zou H, Liang WH, Liu Z, Guo SL, Jiang JF, Zhang WJ, Jia W, Li F. Overexpression of MAP3K3 promotes tumour growth through activation of the NF-ĸB signalling pathway in ovarian carcinoma. Sci Rep. 2019;9(1):8401. doi: 10.1038/s41598-019-44835-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 231.Samanta AK, Huang HJ, Bast RC Jr, Liao WS. Overexpression of MEKK3 confers resistance to apoptosis through activation of NFĸB. J Biol Chem. 2004;279(9):7576–7583. doi: 10.1074/jbc.M311659200. [DOI] [PubMed] [Google Scholar]
- 232.Yu S, Yu M, Chen J, Tang H, Gong W, Tan H. Circ_0000471 suppresses the progression of ovarian cancer through mediating mir-135b-5p/dusp5 axis. Am J Reprod Immunol. 2023;89(4):e13651. doi: 10.1111/aji.13651. [DOI] [PubMed] [Google Scholar]
- 233.Shen J, Zhang Y, Yu H, Shen B, Liang Y, Jin R, Liu X, Shi L, Cai X. Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med. 2016;5(8):2061–2068. doi: 10.1002/cam4.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wang L, Hu J, Qiu D, Gao H, Zhao W, Huang Y, Jiang T, Zhou J, Chen Y. Dual-specificity phosphatase 5 suppresses ovarian cancer progression by inhibiting IL-33 signaling. Am J Transl Res. 2019;11(2):844–854. [PMC free article] [PubMed] [Google Scholar]
- 235.Xie W, Liu LU, He C, Zhao M, Ni R, Zhang Z, Shui C. Circ_0002711 knockdown suppresses cell growth and aerobic glycolysis by modulating miR-1244/ROCK1 axis in ovarian cancer. J Biosci. 2021;46:21. [PubMed] [Google Scholar]
- 236.Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases. 2014;5:e29846. doi: 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Barcelo J, Samain R, Sanz-Moreno V. Preclinical to clinical utility of ROCK inhibitors in cancer. Trends Cancer. 2023;9(3):250–263. doi: 10.1016/j.trecan.2022.12.001. [DOI] [PubMed] [Google Scholar]
- 238.Pan L, Meng Q, Li H, Liang K, Li B. LINC00339 promotes cell proliferation, migration, and invasion of ovarian cancer cells via miR-148a-3p/ROCK1 axes. Biomed Pharmacother. 2019;120:109423. doi: 10.1016/j.biopha.2019.109423. [DOI] [PubMed] [Google Scholar]
- 239.Yang Q, Dong YJ. LncRNA SNHG20 promotes migration and invasion of ovarian cancer via modulating the microRNA-148a/ROCK1 axis. J Ovarian Res. 2021;14(1):168. doi: 10.1186/s13048-021-00889-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Li L, Yu P, Zhang P, Wu H, Chen Q, Li S, Wang Y. Upregulation of hsa_circ_0007874 suppresses the progression of ovarian cancer by regulating the miR-760/SOCS3 pathway. Cancer Med. 2020;9(7):2491–2499. doi: 10.1002/cam4.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sobah ML, Liongue C, Ward AC. SOCS proteins in immunity, inflammatory diseases, and immune-related cancer. Front Med (Lausanne) 2021;8:727987. doi: 10.3389/fmed.2021.727987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Dai L, Li Z, Tao Y, Liang W, Hu W, Zhou S, Fu X, Wang X. Emerging roles of suppressor of cytokine signaling 3 in human cancers. Biomed Pharmacother. 2021;144:112262. doi: 10.1016/j.biopha.2021.112262. [DOI] [PubMed] [Google Scholar]
- 243.Chakraborty R, Darido C, Alnakli A, Hu H, Vickery K. A review on the dual role of SOCS3 in cancer. Res Oncol. 2022;18(1):1–9. doi: 10.21608/resoncol.2022.114548.1159. [DOI] [Google Scholar]
- 244.Heinzelmann-Schwarz V, Gardiner-Garden M, Fink D, Sutherland R, O'Brien P, Hacker N. Reduced expression of SOCS3 predicts poor outcome in epithelial ovarian cancer. Geburtshilfe Frauenheilkd 66(S. 2005;1):P2. doi: 10.1055/s-2005-920800. [DOI] [Google Scholar]
- 245.He SL, Zhao X, Yi SJ. CircAHNAK upregulates EIF2B5 expression to inhibit the progression of ovarian cancer by modulating the JAK2/STAT3 signaling pathway. Carcinogenesis. 2022;43(10):941–955. doi: 10.1093/carcin/bgac053. [DOI] [PubMed] [Google Scholar]
- 246.Wortham NC, Proud CG. eIF2B: recent structural and functional insights into a key regulator of translation. Biochem Soc Trans. 2015;43(6):1234–1240. doi: 10.1042/BST20150164. [DOI] [PubMed] [Google Scholar]
- 247.Hou L, Jiao Y, Li Y, Luo Z, Zhang X, Pan G, Zhao Y, Yang Z, He M. Low EIF2B5 expression predicts poor prognosis in ovarian cancer. Medicine (Baltimore) 2020;99(5):e18666. doi: 10.1097/MD.0000000000018666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.He SL, Chen YL, Chen QH, Tian Q, Yi SJ. LncRNA KCNQ1OT1 promotes the metastasis of ovarian cancer by increasing the methylation of EIF2B5 promoter. Mol Med. 2022;28(1):112. doi: 10.1186/s10020-022-00521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wang J, Li Y, Zhou JH, Shen FR, Shi X, Chen YG. CircATRNL1 activates Smad4 signaling to inhibit angiogenesis and ovarian cancer metastasis via miR-378. Mol Oncol. 2021;15(4):1217–1233. doi: 10.1002/1878-0261.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhao M, Mishra L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14(2):111–123. doi: 10.7150/ijbs.23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 252.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zhou J, Zhang X, Li W, Chen Y. MicroRNA-145-5p regulates the proliferation of epithelial ovarian cancer cells via targeting SMAD4. J Ovarian Res. 2020;13(1):54. doi: 10.1186/s13048-020-00656-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.An Q, Liu T, Wang MY, Yang YJ, Zhang ZD, Lin ZJ, Yang B. circKRT7-miR-29a-3p-COL1A1 axis promotes ovarian cancer cell progression. Onco Targets Ther. 2020;13:8963–8976. doi: 10.2147/OTT.S259033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Cox TR. The matrix in cancer. Nat Rev Cancer. 2021;21(4):217–238. doi: 10.1038/s41568-020-00329-7. [DOI] [PubMed] [Google Scholar]
- 256.Li M, Wang J, Wang C, Xia L, Xu J, Xie X, Lu W. Microenvironment remodeled by tumor and stromal cells elevates fibroblast-derived COL1A1 and facilitates ovarian cancer metastasis. Exp Cell Res. 2020;394(1):112153. doi: 10.1016/j.yexcr.2020.112153. [DOI] [PubMed] [Google Scholar]
- 257.Xu S, Xu H, Wang W, Li S, Li H, Li T, Zhang W, Yu X, Liu L. The role of collagen in cancer: from bench to bedside. J Transl Med. 2019;17(1):309. doi: 10.1186/s12967-019-2058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Januchowski R, Świerczewska M, Sterzyńska K, Wojtowicz K, Nowicki M, Zabel M. Increased expression of several collagen genes is associated with drug resistance in ovarian cancer cell lines. J Cancer. 2016;7(10):1295–1310. doi: 10.7150/jca.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wang F, Niu Y, Chen K, Yuan X, Qin Y, Zheng F, Cui Z, Lu W, Wu Y, Xia D. Extracellular vesicle-packaged circATP2B4 mediates M2 macrophage polarization via miR-532-3p/SREBF1 axis to promote epithelial ovarian cancer metastasis. Cancer Immunol Res. 2023;11(2):199–216. doi: 10.1158/2326-6066.CIR-22-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Yi J, Zhu J, Wu J, Thompson CB, Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc Natl Acad Sci U S A. 2020;117(49):31189–31197. doi: 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Nie LY, Lu QT, Li WH, Yang N, Dongol S, Zhang X, Jiang J. Sterol regulatory element-binding protein 1 is required for ovarian tumor growth. Oncol Rep. 2013;30(3):1346–1354. doi: 10.3892/or.2013.2575. [DOI] [PubMed] [Google Scholar]
- 262.Chaudhry S, Thomas SN, Simmons GE Jr. Targeting lipid metabolism in the treatment of ovarian cancer. Oncotarget. 2022;13:768–783. doi: 10.18632/oncotarget.28241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.An Y, Yang Q. Tumor-associated macrophage-targeted therapeutics in ovarian cancer. Int J Cancer. 2021;149(1):21–30. doi: 10.1002/ijc.33408. [DOI] [PubMed] [Google Scholar]
- 264.Pan Y, Huang Q, Peng X, Yu S, Liu N. Circ_0015756 promotes ovarian cancer progression via the miR-145–5p/PSAT1 axis. Reprod. 2022;22(4):100702. doi: 10.1016/j.repbio.2022.100702. [DOI] [PubMed] [Google Scholar]
- 265.Baek JY, Jun DY, Taub D, Kim YH. Characterization of human phosphoserine aminotransferase involved in the phosphorylated pathway of L-serine biosynthesis. Biochem J 373(Pt. 2003;1):191–200. doi: 10.1042/BJ20030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Chen Y, Yang X, Li C. Phosphoserine aminotransferase 1: a metabolic enzyme target of cancers. Curr Cancer Drug Targets. 2023;23(3):171–186. doi: 10.2174/1568009622666220829105300. [DOI] [PubMed] [Google Scholar]
- 267.Zhang Y, Li J, Dong X, Meng D, Zhi X, Yuan L, Yao L. PSAT1 regulated oxidation-reduction balance affects the growth and prognosis of epithelial ovarian cancer. Onco Targets Ther. 2020;13:5443–5453. doi: 10.2147/OTT.S250066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J, Li X, Lu B, Cheng X, Liu P, Lu W, Lu Y. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer. 2019;18(1):144. doi: 10.1186/s12943-019-1080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 2004;95(11):866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Groelly FJ, Fawkes M, Dagg RA, Blackford AN, Tarsounas M. Targeting DNA damage response pathways in cancer. Nat Rev Cancer. 2023;23(2):78–94. doi: 10.1038/s41568-022-00535-5. [DOI] [PubMed] [Google Scholar]
- 271.Meng E, Shevde LA, Samant RS. Emerging roles and underlying molecular mechanisms of DNAJB6 in cancer. Oncotarget. 2016;7(33):53984–53996. doi: 10.18632/oncotarget.9803. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 272.Chen Z, Wang Y, Liu W, Zhao G, Lee S, Balogh A, Zou Y, Guo Y, Zhang Z, Gu W, Li C, Tigyi G, Yue J. Doxycycline inducible Krüppel-like factor 4 lentiviral vector mediates mesenchymal to epithelial transition in ovarian cancer cells. PLoS One. 2014;9(8):e105331. doi: 10.1371/journal.pone.0105331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Wu X, Liu D, Wang S, Liu J. Circ_0007444 inhibits the progression of ovarian cancer via mediating the miR-570-3p/PTEN axis. Onco Targets Ther. 2021;14:97–110. doi: 10.2147/OTT.S266186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100(4):387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 275.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19(9):547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 276.Nero C, Ciccarone F, Pietragalla A, Scambia G. PTEN and gynecological cancers. Cancers (Basel) 2019;11(10):1458. doi: 10.3390/cancers11101458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zeng XY, Yuan J, Wang C, Zeng D, Yong JH, Jiang XY, Lan H, Xiao SS. circCELSR1 facilitates ovarian cancer proliferation and metastasis by sponging miR-598 to activate BRD4 signals. Mol Med. 2020;26(1):70. doi: 10.1186/s10020-020-00194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Petersen S, Wilson AJ, Hirst J, Roby KF, Fadare O, Crispens MA, Beeghly-Fadiel A, Khabele D. CCNE1 and BRD4 co-amplification in high-grade serous ovarian cancer is associated with poor clinical outcomes. Gynecol Oncol. 2020;157(2):405–410. doi: 10.1016/j.ygyno.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Rhyasen GW, Yao Y, Zhang J, Dulak A, Castriotta L, Jacques K, Zhao W, Gharahdaghi F, Hattersley MM, Lyne PD, Clark E, Zinda M, Fawell SE, Mills GB, Chen H. BRD4 amplification facilitates an oncogenic gene expression program in high-grade serous ovarian cancer and confers sensitivity to BET inhibitors. PLoS One. 2018;13(7):e0200826. doi: 10.1371/journal.pone.0200826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Sun C, Yin J, Fang Y, Chen J, Jeong KJ, Chen X, Vellano CP, Ju Z, Zhao W, Zhang D, Lu Y, Meric-Bernstam F, Yap TA, Hattersley M, O'Connor MJ, Chen H, Fawell S, Lin SY, Peng G, Mills GB. BRD4 inhibition is synthetic lethal with PARP inhibitors through the induction of homologous recombination deficiency. Cancer Cell. 2018;33(3):401–416.e8. doi: 10.1016/j.ccell.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Andrikopoulou A, Liontos M, Koutsoukos K, Dimopoulos MA, Zagouri F. Clinical perspectives of BET inhibition in ovarian cancer. Cell Oncol (Dordr) 2021;44(2):237–249. doi: 10.1007/s13402-020-00578-6. [DOI] [PubMed] [Google Scholar]
- 284.Yang X, Wang J, Li H, Sun Y, Tong X. Downregulation of hsa_circ_0026123 suppresses ovarian cancer cell metastasis and proliferation through the miR 124 3p/EZH2 signaling pathway. Int J Mol Med. 2021;47(2):668–676. doi: 10.3892/ijmm.2020.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Duan R, Du W, Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol. 2020;13(1):104. doi: 10.1186/s13045-020-00937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Eich ML, Athar M, Ferguson JE 3rd, Varambally S. EZH2-targeted therapies in cancer: hype or a reality. Cancer Res. 2020;80(24):5449–5458. doi: 10.1158/0008-5472.CAN-20-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Hanaki S, Shimada M. Targeting EZH2 as cancer therapy. J Biochem. 2021;170(1):1–4. doi: 10.1093/jb/mvab007. [DOI] [PubMed] [Google Scholar]
- 288.Jones BA, Varambally S, Arend RC. Histone methyltransferase EZH2: a therapeutic target for ovarian cancer. Mol Cancer Ther. 2018;17(3):591–602. doi: 10.1158/1535-7163.MCT-17-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Li X, Pu W, Zheng Q, Ai M, Chen S, Peng Y. Proteolysis-targeting chimeras (PROTACs) in cancer therapy. Mol Cancer. 2022;21(1):99. doi: 10.1186/s12943-021-01434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Arora P, Singh M, Singh V, Bhatia S, Arora S. PROTACs in treatment of cancer: a review. Mini Rev Med Chem. 2021;21(16):2347–2360. doi: 10.2174/1389557521666210226150740. [DOI] [PubMed] [Google Scholar]
- 291.Wang X, Ma C, Rodríguez Labrada R, Qin Z, Xu T, He Z, Wei Y. Recent advances in lentiviral vectors for gene therapy. Sci China Life Sci. 2021;64(11):1842–1857. doi: 10.1007/s11427-021-1952-5. [DOI] [PubMed] [Google Scholar]
- 292.Lei B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301. doi: 10.7150/ijms.28047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wang H, Meng Q, Qian J, Li M, Gu C, Yang Y. Review: RNA-based diagnostic markers discovery and therapeutic targets development in cancer. Pharmacol Ther. 2022;234:108123. doi: 10.1016/j.pharmthera.2022.108123. [DOI] [PubMed] [Google Scholar]
- 294.Paunovska K, Loughrey D, Dahlman JE. Drug delivery systems for RNA therapeutics. Nat Rev Genet. 2022;23(5):265–280. doi: 10.1038/s41576-021-00439-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Adams D, Gonzalez-Duarte A, O'Riordan WD, Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL, Lin KP, Vita G, Attarian S, Planté-Bordeneuve V, Mezei MM, Campistol JM, Buades J, Brannagan TH 3rd, Kim BJ, Oh J, Parman Y, Sekijima Y, Hawkins PN, Solomon SD, Polydefkis M, Dyck PJ, Gandhi PJ, Goyal S, Chen J, Strahs AL, Nochur SV, Sweetser MT, Garg PP, Vaishnaw AK, Gollob JA, Suhr OB. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379(1):11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 296.Marques AC, Costa PC, Velho S, Amaral MH. Lipid nanoparticles functionalized with antibodies for anticancer drug therapy. Pharmaceutics. 2023;15(1):216. doi: 10.3390/pharmaceutics15010216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, Liang XJ. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5(1):101. doi: 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]