Abstract
The sequence of the gene iaaL of Pseudomonas savastanoi EW2009 was used to design primers for PCR amplification. The iaaL-derived primers directed the amplification of a 454-bp fragment from genomic DNA isolated from 70 strains of P. savastanoi, whereas genomic DNA from 93 non-P. savastanoi isolates did not yield this amplified product. A previous bacterial enrichment in the semiselective liquid medium PVF-1 improved the PCR sensitivity level, allowing detection of 10 to 100 CFU/ml of plant extract. P. savastanoi was detected by the developed enrichment-PCR method in knots from different varieties of inoculated and naturally infected olive trees. Moreover, P. savastanoi was detected in symptomless stem tissues from naturally infected olive plants. This enrichment-PCR method is more sensitive and less cumbersome than the conventional isolation methods for detection of P. savastanoi.
Pseudomonas savastanoi and its pathovars savastanoi, fraxini, and nerii incite a disease of olive (Olea europaea L.), ash (Fraxinus excelsior L.), other Oleaceae plants and oleander (Nerium oleander L.) that is characterized by tumorous outgrowths (4, 19). This development of knots is dependent on bacterial production of the phytohormone indoleacetic acid (IAA) and cytokinins (3, 7, 16, 18). P. savastanoi can conjugate IAA with lysine to form 3-indoleacetyl-ɛ-l-lysine (IAA-lysine) (6). The two enzymes involved in IAA biosynthesis are tryptophan monooxygenase, which converts tryptophan to indoleacetamide, and indoleacetamide hydrolase, which catalyzes the conversion of indoleacetamide to IAA (12). The enzyme involved in the conversion of IAA to IAA-lysine is (indole-3-acetyl)-l-lysine synthethase (5). The genes for tryptophan monooxygenase (iaaM), indoleacetamide hydrolase (iaaH), and IAA-lysine synthethase (iaaL) reside on the 52-kb plasmid pIAA1 in the oleander P. savastanoi strain EW2009, and they have been sequenced (3, 5, 14, 20). Sequence analysis revealed that iaaM and iaaH have significant similarity with homologous genes of other plant-associated bacteria (13, 20). In contrast, to date, no nucleotide homologies have been found with the iaaL gene.
Detection of P. savastanoi is currently based on bacterial isolation followed by pathogenicity tests and biochemical or serological techniques (2, 8, 17, 21). These conventional methods are time-consuming and expensive, requiring bacterial isolation. We used the published sequence of iaaL (14) to design specific primers for amplification of this gene. We report here the development of a new sensitive and specific detection method for P. savastanoi based on amplification of iaaL after a bacterial enrichment. The developed enrichment-PCR assay can be applied to specifically detect low levels of P. savastanoi in inoculated and naturally infected plants.
Specificity of the PCR assay.
Seventy strains of P. savastanoi isolated from olive, oleander, ash, and jasmine (Jasminus officinalis L.) plants from different countries, 23 outgroup strains, and 70 saprophytic isolates from olive plants were used to test for the specificity of the primers designed (Table 1). All strains were routinely grown in King's medium B (9). The bacterial DNA preparation method was based on the protocol described by Llop et al. (10). The primers designed for iaaL amplification were as follows: primer IAALF, 5′-GGCACCAGCGGCAACATCAA-3′; primer IAALR, 5′-CGCCCTCGGAACTGCCATAC-3′. PCR reactions were performed by combining the following reagents in a reaction mix: 10× Taq buffer (GIBCO-BRL), 1.5 mM MgCl2, 5% formamide, 0.2 mM concentrations of each deoxynucleoside triphosphate (Pharmacia LKB), 0.6 μM concentrations of each primer, and 1.5 U of Taq DNA polymerase (GIBCO-BRL) per reaction. Then, 5 μl of the DNA extraction from bacterial cultures (or plant samples) was added. Samples were amplified through 1 cycle of 94°C (5 min), followed by 35 cycles of 94°C (30 s), 62°C (30 s), and 72°C (30 s) and then 1 cycle of 72°C for 5 min in a 9600 Perkin-Elmer thermocycler. Negative controls with uninfected olive plants were included in every DNA extraction series. Furthermore, to help detect carryover contamination, duplicate samples and two sets of two negative controls each with sterile purified water were routinely included in every reaction. One set was loaded just after reaction mix preparation, and the other one was added just after loading the samples. Amplification was also conducted in a separate laboratory. Amplified products (5 μl) were separated by electrophoresis (100 V) on a 1.5% agarose gel or digested with the restriction enzyme HaeIII (GIBCO-BRL) at 37°C for 2 h and separated by electrophoresis (100 V) on a 2% agarose gel. Electrophoresis gels were stained with ethidium bromide.
TABLE 1.
Bacterial strains used
| Strain(s)a (no. of isolates) | Origin | Host | Amplification with iaaL primers | Source |
|---|---|---|---|---|
| Pseudomonas savastanoi | ||||
| 1613-1, 1624-a, 1613-4, 1624-b, 1628, 1629, 1637-b, 1637-c, 1649, 1651-a, 1651-b, 1651-c, 1657-2, 1657-8, 1663, 1664-1, 1664-6, 1668, 1669, 1670, 78/55, 1862-3 | Spain | Olive | + | This study |
| CFBP 1670, NCPPB 1479 | Yugoslavia | Olive | + | |
| NCPPB 2780, NCPPB 3335, CFBP 2073 | France | Olive | + | |
| NCPPB 2327, NCPPB 1506 | Italy | Olive | + | |
| PBa206, PBa223, PBa221, PBa230, PBa225, PVFi1, PVFi2, ITM101, ITM317 | Italy | Olive | + | N. Iacobellis |
| NCPPB 1342, NCPPB 1344 | United States | Olive | + | |
| ITM301, ITM302 | United States | Olive | + | N. Iacobellis |
| CFBP 2074 | Algeria | Olive | + | |
| CFBP 71 | Tunisia | Olive | + | |
| NCPPB 64 | Portugal | Olive | + | |
| BPIC 264, BPIC346 | Greece | Olive | + | P. G. Psallidas |
| 1610, 1611, 910-1, 910-4, 1889-2, 1889-3, 1889-4, 1889-5, 1889-6, 1889-7 | Spain | Oleander | + | This study |
| ITM519, ITM601, PBa204 | Oleander | + | N. Iacobellis | |
| NCPPB 640 | Yugoslavia | Oleander | + | |
| CFBP 2088 | Algeria | Oleander | + | |
| BPIC 344, BPIC 345 | Greece | Oleander | + | P. G. Psallidas |
| NCPPB 1464 | United Kingdom | Ash | + | |
| F2, F3 | Ash | + | N. Iacobellis | |
| NCPPB 3474 | France | Ash | + | |
| BPIC 464, BPIC 465 | Greece | Jasmine | + | P. G. Psallidas |
| Outgroup speciesb (23) | − | IVIAc | ||
| Microbiota from olive plants | ||||
| Fluorescent pseudomonads (9) | Spain | Olive | − | This study |
| Pseudomonas putida (1) | Spain | Olive | +d | This study |
| Pantoea agglomerans (10) | Spain | Olive | − | This study |
| Unidentified isolates (52) | Spain | Olive | − | This study |
CFBP, Collection Française de Bactéries Phytopathogènes, Angers, France; NCPPB, National Collection of Plant Pathogenic Bacteria, York, England; BPIC, Benaki Phytopathological Institute Collections, Athens, Greece.
Agrobacterium tumefaciens (1 strain); Clavibacter michiganensis subsp. michiganensis (1 strain); Erwinia amylovora (1 strain); Pectobacterium carotovorum (2 strains); Pectobacterium chrysanthemi (1 strain); Pseudomonas chlororaphis (1 strain); Pseudomonas marginalis (1 strain); Pseudomonas syringae pv. syringae (12 strains); Rhodococcus facians (1 strain) Serratia fonticola (1 strain); Xanthomonas hortorum pv. pelargonii (1 strain).
IVIA, bacterial collection from the Instituto Valenciano de Investigaciones Agrarias, Moncada, Valencia, Spain.
DNA from this isolate yielded a ca. 440-bp fragment. After HaeIII digestion, the amplified DNA yielded 310- and 130-bp fragments that were perfectly distinguishable from those of the iaaL gene of P. savastanoi.
A 454-bp product was obtained from all P. savastanoi strains, showing that iaaL sequences were present in strains from different countries and isolated from four hosts (Table 1). Two fragments of 279 and 175 bp were generated from all amplified fragments after HaeIII digestion, a result in agreement with previously published sequence data (14). Genomic DNA from outgroup species did not produce any discrete bands upon amplification. Similarly, we found no amplification of the 454-bp fragment with genomic DNA from isolates of olive microbiota (Table 1). Only the DNA from one saprophytic isolate identified by biochemical tests as Pseudomonas putida gave a smaller product of ca. 440 bp upon amplification. However, it was perfectly distinguishable from that of P. savastanoi because after HaeIII digestion we did not find the corresponding pattern of P. savastanoi iaaL (Table 1). Formamide (5%) was required in the amplification cocktail to eliminate nonspecific bands.
Detection of P. savastanoi in plants.
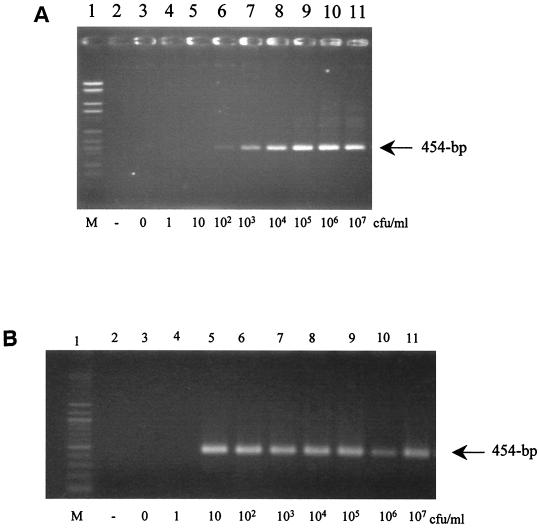
Olive plant samples (1 g of stem tissue from uninfected greenhouse-grown plants comminuted in 50 ml of sterile distilled water) were subsequently amended with known amounts of P. savastanoi 1628 from ca. 1 to 107 CFU/ml of the plant extract. Then, 0.5 ml of amended plant samples from each bacterial concentration was amplified after performing the DNA extraction described above or was added to 5 ml of the nonselective King's medium B (9) and the semiselective PVF-1 medium (17) for bacterial enrichments. Samples were incubated for 3 days at 26°C, and bacterial DNA extracts using 500 μl of each sample were then subjected to PCR amplification as described above. Three replicates of the experiment were performed. The detection limit was 102 to 103 CFU/ml of plant extract (Fig. 1A). A previous bacterial enrichment using King's medium B slightly improved the detection level, allowing detection of ca. 102 CFU/ml of plant extract. However, preenrichment in PVF-1 improved the detection level, allowing detection of ca. 10 to 100 CFU/ml of plant extract (Fig. 1B).
FIG. 1.
Sensitivity of the PCR assay in detecting the iaaL gene of P. savastanoi pv. savastanoi in amended plant samples without enrichment (A) or with PVF-1 enrichment (B). PCR products from amended plant samples were inoculated with P. savastanoi 1628. Lanes 1, molecular marker (M); lanes 2, PCR-negative control; lanes 3, noninoculated plant material; lanes 4 to 11, PCR-amplified products obtained from amended plant samples inoculated with increased concentrations of bacteria from 1 to 107 CFU/ml of plant extract.
We then evaluated the detection of P. savastanoi in knots from inoculated and naturally infected plants. The stems of 1-year-old olive plants were wounded and subsequently inoculated with 10 μl of a bacterial suspension (ca. 109 CFU/ml) of strain 1628 to develop knots. They were analyzed 8 months after inoculation. In addition, knots from olive, oleander, and Spanish broom (Retama sphaerocarpa [Boiss] L.) from naturally infected plants were also collected. From inoculated and naturally infected samples, 500 μl of comminuted knot tissues in sterile water (1:50 [wt/vol]) were subjected to the protocol described above for amended plant samples, and subsequently the amplified product was digested with HaeIII. Conventional bacterial isolations were performed from each sample before or after bacterial enrichments by plating serial dilutions on King's B (9) and PVF-1 (17) media. The identity of the putative P. savastanoi colonies was confirmed by biochemical tests (21) and by the developed PCR assay. In all of the knot samples analyzed from inoculated plants, we found the expected PCR product of 454 bp and also the corresponding restriction profile (Table 2). We were also able to isolate the inoculated P. savastanoi strain (Table 2). We compared the detection efficiency of the developed PCR assay with that of the isolation method in knot samples from naturally infected plants, either with a previous bacterial enrichment step or not. When analyzing 37 knot samples, P. savastanoi was detected by PCR in 21 of the 37 samples, without preenrichment (Table 2). One of these amplified products was double strand sequenced, and the obtained sequence was 100% similar to the previous iaaL reported sequence. A preenrichment in King's medium B did not improve the detection of P. savastanoi by PCR in knot tissues from naturally infected plants because P. savastanoi was detected in only 13 samples. However, a preenrichment in PVF-1 medium improved the detection of P. savastanoi by PCR to up to 29 of the 37 samples (Table 2). P. savastanoi was isolated by plating in some samples without enrichment (eight samples); however, the detection efficiency of bacterial isolation was not improved by any preenrichment (Table 2).
TABLE 2.
Detection of P. savastanoi in knots from inoculated and naturally infected plants
| Plant group | Host | No. of knot samples analyzed |
P. savastanoi detection (no. of positive samples)a:
|
|||||
|---|---|---|---|---|---|---|---|---|
| Without preenrichment
|
With preenrichment
|
|||||||
| King's B
|
PVF-1
|
|||||||
| Bacterial isolation | PCR and restriction analysis | Bacterial isolation | PCR and restriction analysis | Bacterial isolation | PCR and restriction analysis | |||
| Inoculated plants (109 CFU/ml) | Olive | 15 | 15 | 15 | ND | ND | ND | ND |
| Naturally infected plants | Olive | 26 | 5 | 16b | 3 | 10 | 3 | 19 |
| Oleander | 3 | 0 | 1 | 0 | 0 | 0 | 2 | |
| Broom | 8 | 3 | 4 | 0 | 3 | 0 | 8 | |
| Total no. of analyzed samplesc | 37 | 8 | 21 | 3 | 13 | 3 | 29 | |
That is, the number of samples in which P. savastanoi was detected. ND, not determined. Bacterial isolations were performed by plating serial dilutions on King's B and PVF-1 media, and after bacterial purifications, isolates were identified as described in Materials and Methods. PCR amplification was done by using the primer pair IAALF-IAALR, and subsequently, the amplified product was digested with the restriction enzyme HaeIII as described in Materials and Methods.
One of these amplified fragments was double strand sequenced, thus verifying iaaL detection.
Analyzed samples from naturally infected plants.
To evaluate the detection of P. savastanoi in asymptomatic olive tissues, stem samples were collected from branches without knots taken from naturally infected plants. A total of 1 to 10 g of stem tissues was processed, and 0.5 ml of the obtained samples was analyzed as described above. Uninfected greenhouse-grown plants were used as negative controls. When analyzing 38 asymptomatic samples, P. savastanoi was detected by the developed PCR and subsequent restriction analysis in only 4 of the 38 samples (Table 3). One of these amplified fragments was double strand sequenced showing that it corresponded to the iaaL gene. A preenrichment using either King's B (9) or PVF-1 (17) medium greatly improved the detection of P. savastanoi by PCR in symptomless stem samples (Table 3). The highest detection level was found with the enrichment-PCR method using PVF-1, which allowed the detection of P. savastanoi in 23 of the 38 samples. P. savastanoi was isolated by plating in 10 samples without preenrichment; however, the detection level obtained by plating was not improved by any preenrichment in these asymptomatic samples (Table 3).
TABLE 3.
Detection of P. savastanoi pv. savastanoi in asymptomatic stem tissues from infected olive plants
| Variety | No. of samples analyzed |
P. savastanoi pv. savastanoi detection (no. of positive samples)a
|
|||||
|---|---|---|---|---|---|---|---|
| Without preenrichment
|
With preenrichment
|
||||||
| King's B
|
PVF-1
|
||||||
| Bacterial isolation | PCR and restriction analysis | Bacterial isolation | PCR and restriction analysis | Bacterial isolation | PCR and restriction analysis | ||
| Blanqueta | 6 | 1 | 0 | 0 | 1 | 0 | 3 |
| Gordal | 3 | 2 | 0 | 0 | 3 | 1 | 2 |
| Manzanilla | 6 | 2 | 0 | 3 | 4 | 0 | 5 |
| Picual | 20 | 5 | 4b | 4 | 6 | 4 | 10 |
| Picudo | 3 | 0 | 0 | 1 | 2 | 1 | 3 |
| Total no. of samples | 38 | 10 | 4 | 8 | 16 | 6 | 23 |
That is, the number of samples in which P. savastanoi pv. savastanoi was detected. Bacterial isolations were performed by plating serial dilutions on King's B and PVF-1 media, and after bacterial purifications, isolates were identified as described in Materials and Methods. PCR amplification was done by using the primer pair IAALF-IAALR, and subsequently, the amplified product was digested with the restriction enzyme HaeIII as described in Materials and Methods.
One of these amplified fragments was double strand sequenced, thus verifying iaaL detection.
In this study an enrichment-PCR assay was developed for the detection of P. savastanoi in olive plants, and its efficiency was compared by isolation plating. We evaluated the specificity of the PCR assay by testing for amplification of the 454-bp DNA in a collection of P. savastanoi strains and other bacteria. Genomic DNA from all of the P. savastanoi strains tested was amplified with the iaaL primers, and subsequent restriction analysis of the amplified product yielded only the expected fragmentation pattern. It shows that this gene is present among the isolates of P. savastanoi from olive, oleander, ash, and jasmine hosts. Genomic DNA from all non-P. savastanoi strains tested, including outgroup species and different bacteria associated with olive plants, failed to produce the 454-bp PCR product upon amplification under the conditions described above.
A bacterial enrichment prior to serological and molecular techniques has been reported to improve the sensitivity of detection of other plant pathogenic bacteria (11, 15). For P. savastanoi detection in amended plant samples, knots, and symptomless stem tissues from naturally infected plants, a bacterial enrichment prior to PCR analysis in PVF-1 liquid medium (but not in King's medium B) greatly improved the detection level. This shows the higher efficiency of selective enrichment versus bacterial enrichment in common medium. With the developed enrichment-PCR assay it is possible to detect living cells of P. savastanoi at low population levels in plant material, thus avoiding the need to obtain bacterial cultures as required for the conventional detection techniques.
In knot samples from inoculated plants the developed PCR assay was as efficient as plating isolation because we recovered the inoculated P. savastanoi strain in all of the samples that were also positive by PCR. These samples came from fairly young knots because the analyses were done 8 months after the inoculations, when the knot tissues retain a high population of viable cells as shown by the isolation studies. In contrast, in knot samples from naturally infected plants the developed PCR assay was more efficient than bacterial isolation in detecting P. savastanoi. This finding could be due to lower populations of viable bacteria in old cracked knots. For bacterial isolations in these samples, an enrichment step, even using the semiselective PVF-1 medium, decreased the plating efficiency due to the growth of bacteria other than P. savastanoi in the plates. This finding suggests that the enrichment allows the multiplication of the P. savastanoi living cells; however, the rapid development of the colonies of the microbiota from the olive tissues on the isolation plates (mainly Pantoea agglomerans and fluorescent pseudomonads) interfered with the growth of P. savastanoi colonies but not with their detection in the enriched culture by PCR. Other authors (1, 17) suggested that fairly young olive knots should be used for the isolation of P. savastanoi because the presence of a number of other bacteria and low viable populations of the pathogen in the diseased tissues from old, cracked knots often made isolations very difficult. Our work agrees with these findings and extends it to oleander and broom knots.
The semiselective medium PVF-1 was more effective than King's medium B for P. savastanoi enrichment and subsequent PCR detection, probably due to the relative low growth rate of P. savastanoi compared to that of some other saprophytic bacteria present in plant samples. This finding also suggests that the enrichment effect was not primarily due to the dilution effect of the potential PCR inhibitors present in plant material, as previously suggested (15).
With the developed enrichment-PCR method we detected P. savastanoi in many samples in which we were not able to isolate the bacterium. We were then concerned about the possibility of false-positives with the PCR technique. To help detect cross-contamination among samples and PCR reagents, multiple controls were used in all steps of the protocol as described in Materials and Methods. Negative controls from every experiment were consistently negative. On the other hand, the designed primers were highly specific, and confirmation that the PCR product is from amplification of the target DNA was always made by restriction analysis and by DNA sequencing in two samples. Besides the conventional isolation methods, no other serological or molecular techniques are available for further confirmation of P. savastanoi detection directly in plant samples.
In conclusion, this study indicates that PCR amplification of P. savastanoi with the iaaL-derived primers and the developed preenrichment assay is a sensitive and specific method for the detection of this phytopathogenic bacterium in plant material. This enrichment-PCR method has advantages over all presently used detection methods for P. savastanoi in time, sensitivity, and cost. This assay may be useful for the early detection of low levels of P. savastanoi in olive plants during plant material propagation for certification purposes or epidemiological studies.
Acknowledgments
We thank P. G. Psallidas and N. Iacobellis for kindly providing bacterial strains and J. Piquer for technical support with plant management. We are grateful to A. Olmos for technical advice.
R. Penyalver is a recipient of a contract from the Spanish MEC (Programa de Incorporación de Doctores a Grupos de Investigación en España). This work was supported by grant OLI96-2179 from the CICYT of Spain.
REFERENCES
- 1.Azad H R, Cooksey D A. A semiselective medium for detecting epiphytic and systemic populations of Pseudomonas savastanoi from oleander. Phytopathology. 1995;85:740–745. [Google Scholar]
- 2.Casano F J, Hung S Y, Wells J M. Differentiation of some pathovars of Pseudomonas syringae with monoclonal antibodies. Eur Mediterr Plant Prot Organ Bull. 1987;17:173–176. [Google Scholar]
- 3.Comai L, Kosuge T. Involvement of plasmid deoxyribonucleic acid in indoleacetic acid synthesis in Pseudomonas savastanoi. J Bacteriol. 1980;143:950–957. doi: 10.1128/jb.143.2.950-957.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gardan L, Bollet C, Abu Ghorrah M, Grimont F, Grimont P A D, Grimont P A D. DNA relatedness among the pathovar strains of Pseudomonas syringae subsp. savastanoi Janse (1982) and proposal of Pseudomonas savastanoi sp. nov. Int J Syst Bacteriol. 1992;42:606–612. [Google Scholar]
- 5.Glass N L, Kosuge T. Cloning of the gene for indoleacetic acid-lysine synthetase from Pseudomonas syringae subsp. savastanoi. J Bacteriol. 1986;166:598–603. doi: 10.1128/jb.166.2.598-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutzinger O, Kosuge T. 3-Indoleacetic-l-lysine, a new conjugate of 3-indoleacetic acid produced by Pseudomonas savastanoi. In: Wightman F, Setterfield G, editors. Biochemistry and physiology of plant growth substances. Ottawa, Ontario, Canada: The Runge Press, Ltd.; 1968. pp. 183–194. [Google Scholar]
- 7.Iacobellis N S, Sisto A, Surico G, Evidente A, Di Maio E. Pathogenicity of Pseudomonas syringae pv. savastanoi mutants defective in phytohormone production. J Phytopathol. 1994;140:238–248. [Google Scholar]
- 8.Janse J D. Pathovars discrimination within Pseudomonas syringae subsp. savastanoi using whole-cell fatty acids and pathogenicity as criteria. Syst Appl Microbiol. 1991;13:79–84. [Google Scholar]
- 9.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 10.Llop P, Caruso P, Cubero J, Morente C, López M M. A simple extraction procedure for efficient routine detection of pathogenic bacteria in plant material by polymerase chain reaction. J Microbiol Methods. 1999;37:23–31. doi: 10.1016/s0167-7012(99)00033-0. [DOI] [PubMed] [Google Scholar]
- 11.López M M, Gorris M T, Llop P, Cubero J, Vicedo B, Cambra M. Selective enrichment improves isolation, serological and molecular detection of plant pathogenic bacteria. In: Dehne H W, et al., editors. Diagnosis and identification of plant pathogens. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 117–121. [Google Scholar]
- 12.Maggie A R, Wilson E E, Kosuge T. Indoleacetamide as an intermediate in the synthesis of indoleacetic acid in Pseudomonas savastanoi. Science. 1963;141:1281–1282. doi: 10.1126/science.141.3587.1281. [DOI] [PubMed] [Google Scholar]
- 13.Mazzola M, White F F. A mutation in the indole-3-acetic acid biosynthesis pathway of Pseudomonas syringae pv. syringae affects growth in Phaseolus vulgaris and syringomycin production. J Bacteriol. 1994;176:1374–1382. doi: 10.1128/jb.176.5.1374-1382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberto F F, Klee H, White F, Nordeen R, Kosuge T. Expression and fine structure of the gene encoding Nɛ-(indole-3-acetyl)-l-lysine synthetase from Pseudomonas savastanoi. Proc Natl Acad Sci USA. 1990;87:5797–5801. doi: 10.1073/pnas.87.15.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaad N W, Cheong S S, Tamaki S, Hatziloukas E, Panopoulos N J. A combined biological and enzymatic amplification (BIO-PCR) technique to detect Pseudomonas syringae pv. phaseolicola in bean seed extracts. Phytopathology. 1995;85:243–248. [Google Scholar]
- 16.Smidt M, Kosuge T. The role of indole-3-acetic acid accumulation by alpha-methyl tryptophan-resistant mutants of Pseudomonas savastanoi in gall formation in oleander. Physiol Plant Pathol. 1978;13:203–214. [Google Scholar]
- 17.Surico G, Lavermicocca P. A semiselective medium for the isolation of Pseudomonas syringae pv. savastanoi. Phytopathology. 1989;79:185–190. [Google Scholar]
- 18.Surico G, Iacobellis N S, Sisto S. Studies on the role of indole-3-acetic acid and cytokinins in the formation of knots on olive and oleander plants by Pseudomonas syringae pv. savastanoi. Physiol Plant Pathol. 1985;26:309–320. [Google Scholar]
- 19.Wilson E E. Pathological histogenesis in oleander tumors induced by Pseudomonas savastanoi. Phytopathology. 1965;55:1244–1249. [Google Scholar]
- 20.Yamada T, Palm C J, Brooks B, Kosuge T. Nucleotide sequences in the Pseudomonas savastanoi indolacetic acid genes show homology with Agrobacterium tumefaciens T-DNA. Proc Natl Acad Sci USA. 1985;82:6522–6526. doi: 10.1073/pnas.82.19.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young J M, Triggs C M. Evaluation of determinative tests for pathovars of Pseudomonas syringae van Hall 1902. J Appl Bacteriol. 1994;77:195–207. doi: 10.1111/j.1365-2672.1994.tb03064.x. [DOI] [PubMed] [Google Scholar]