Abstract
Large DNA assembly methodologies underlie milestone achievements in synthetic prokaryotic and budding yeast chromosomes. While budding yeast control chromosome inheritance through ~125 bp DNA sequence-defined centromeres, mammals and many other eukaryotes use large, epigenetic centromeres. Harnessing centromere epigenetics permits human artificial chromosome (HAC) formation but is not sufficient to avoid rampant multimerization of the initial DNA molecule upon introduction to cells. Here, we describe an approach that efficiently forms single-copy HACs. It employs a ~750 kb construct that is sufficiently large to house the distinct chromatin types present at the inner and outer centromere, obviating the need to multimerize. Delivery to mammalian cells is streamlined by employing yeast spheroplast fusion. These developments permit faithful chromosome engineering in the context of metazoan cells.
One-Sentence Summary:
A quarter century after the first human artificial chromosomes, a solution to their uncontrolled multimerization is achieved.
Yeast artificial chromosomes (YACs) (1–3) are typically 0.1-1 Mb and permitted triumphs of molecular biology including the cloning of large disease genes (4) and the generation of entire synthetic prokaryotic genomes (5, 6). They also provided the foundation for the generation of entirely synthetic budding yeast chromosomes (7). ‘Writing’ new chromosomes, or even entire genomes, is an aspiration for synthetic biologists working in diverse eukaryotes, including in mammalian and plant systems (8, 9), because it would enable applications of genome engineering across research, biotechnology, and health-related landscapes (8).
Human artificial chromosomes (HACs) were developed ~25 years ago (10–12) and are on average 1-10 Mb in their functional form after their establishment in cells. They pave the way for advances and insights in eukaryotic systems where a specific key chromosomal locus, the centromere, is typically more than a thousand times larger than a budding yeast point centromere and is functionally defined not by a particular sequence but by an array of nucleosomes containing a histone H3 variant, CENP-A (13). Unlike YACs in yeast, an unintended byproduct of de novo HAC formation is multimerization of the initial input DNA construct—typically 100-200 kb bacterial artificial chromosomes (BACs)—creating functional HACs with a variable number of multimers (typically >40-fold) (14–16). The multimerization and the uncontrolled rearrangement of the input DNA that accompanies it during the early steps of HAC formation (often with equally unintended incorporation of portions of natural chromosomes) has severely hindered their development towards their broader promise for synthetic biology and therapeutic applications (17). We have now overhauled the design and delivery of HACs: instead of trying to control the multimerization process, we sought to bypass it completely by increasing the size of the input DNA. Here we report success in forming single-copy HACs at an overall efficiency of de novo establishment that surpasses all earlier versions.
Results
An overhauled platform for efficient HAC formation
We predicted that to remain single-copy and avoid multimerization, the initial construct would need to be larger than the BAC-based HAC constructs of earlier versions (14–16). This is based on the understanding that centromeres require multiple domains with distinct functions that are spatially separated at mitosis when cohered sister chromatids align on the microtubule-based spindle (18). While the centromeric region harboring CENP-A nucleosomes (the region for assembling the mitotic kinetochore) typically discontinuously spans ~75-300 kb (19–22), the inner centromere is another largely heterochromatic region that regulates sister chromatid cohesion and a quality control pathway (termed “error correction”) that monitors bipolar spindle attachment. We reasoned that BAC-based HAC constructs, which typically start in the 100-300 kb size-range (14–16), can likely only form when multimerization occurs. The larger size is presumably required to accommodate formation of both distinct chromatin domains that define a functional centromere. Conversely, we reasoned that starting with a larger initial construct will bypass this requirement, allowing HACs to form more frequently and without multimerization.
To test our prediction, we devised a scheme that employs three recent technical advances to build and test a single-copy HAC construct (fig. S1A). First, YAC constructs are readily generated in the 0.5-2 Mb size range (5, 23) through transformation-associated recombination (TAR) cloning (24). Second, bypassing the requirement for long (>40 kb) stretches of highly repetitive centromere DNA (α-satellite) for HAC formation (16) permits the use of non-repetitive DNA. This is critical since TAR cloning is not compatible with long repetitive sequences (25). Third, large YAC constructs can be efficiently delivered to mammalian cells via optimized fusion with yeast spheroplasts (23), potentially leading to a marked increase in independent HAC formation events relative to what has been achieved with low-efficiency transfection-based delivery of BAC-based HAC vectors in prior versions (14–16).
The HAC template was constructed through TAR assembly starting with a YAC harboring 550 kb of M. mycoides genomic DNA (6), 4q21 BACLacO (16), and linkers for recombination that also include a yeast auxotrophic marker and a mammalian expression cassette for mCherry (fig. S1A). M. mycoides genomic DNA was chosen because it represents a heterologous DNA sequence that is known to be readily propagated in budding yeast (6). It serves as a non-coding sequence in the context of a eukaryotic cell and is not expected to elicit unintended or detrimental impact on HAC formation or cell function. It was also attractive because the AT richness of the M. mycoides DNA (~75%) is anticipated to support the heterochromatic nature of the inner centromere. Further, M. Mycoides DNA has already been efficiently delivered to cultured human cells (23), and it allows for unambiguous detection of HACs because it is comprised of unique, non-human sequence. 4q21 BACLacO was chosen because it is the only HAC construct comprised of non-repetitive DNA (182 kb of DNA from human chromosome 4 at the 4q21 locus) that has been demonstrated to form functional HACs, instead of the 40-200 kb of highly repetitive α-satellite-based BAC constructs that prior HAC studies have used (14–16, 26). The LacO array on 4q21 BACLacO permits subsequent targeting of mCherry-LacI-HJURP for initial seeding of CENP-A nucleosomes (16). We termed the new construct, YAC-Mm-4q21LacO.
For recipient cells, we used the HT1080Dox-inducible mCherry-LacI-HJURP line in which 4q21 BACLacO-based HACs were seeded with CENP-A nucleosomes (16). The HT1080 background, in general, was chosen because it is the one in which HAC formation has historically been performed (11, 12, 14–16, 26) due to its chromatin state that is permissive to occasional centromere formation (27). We also generated a second recipient line, U2OSDox-inducible mCherry-LacI-HJURP, since the U2OS background are established as an efficient recipient of YACs via spheroplast fusion (23). Both of our chosen recipient lines were first optimized for spheroplast fusion conditions (fig. S2) and then subjected to HAC formation assays with YAC-Mm-4q21LacO (Fig. 1; fig. S2; fig. S3). Following spheroplast fusion, we noted that, unlike prior HAC assays where only ~40 surviving colonies emerge in 2-3 weeks, a nearly confluent monolayer of G418-S-resistant cells was present after 8 days of selection. For both recipient cell types, a substantial proportion (42 +/− 9% and 46 +/− 5%) of the neomycin-resistant cells harbor HACs. Most or all of these are substantially smaller in size (<1 μm) (Fig. 1, B–D) than the multimerized HACs of prior generations (~2 μm) (14–16). Without induction of mCherry-LacI-HJURP, there was only a very small proportion of HACs, with the majority of cells with detectable FISH signal coming from an integration into a natural chromosome of the recipient cell (Fig. 1C). Our initial findings, therefore, strongly indicate exceedingly high efficiency of YAC delivery, robust HAC formation rates upon seeding CENP-A nucleosome assembly, essentially uniform avoidance of the stochastic and highly unlikely multimerization that has accompanied prior systems for de novo HAC formation, and no restriction to the specific cell line (HT1080) to which prior generations of HACs were confined.
Fig. 1. 760 kb HAC constructs efficiently acquire centromeres and exist as autonomous chromosomes.
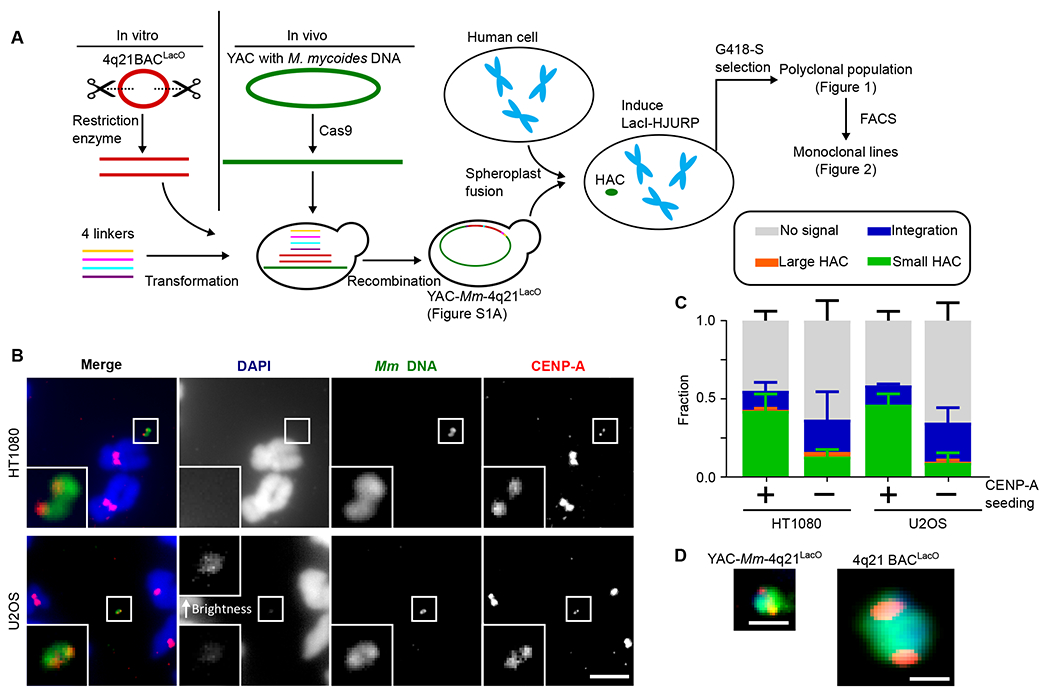
(A) Schematic of approach to generate a HAC. (B) Representative images of a single-copy HAC generated in HT1080 and U2OS cells. Insets: 5x magnification. Bar, 5 μm. See also Table S1. (C) Quantification of proportion of “small HACs” (FISH signal spans less than 1 μm), “large HACs” (FISH signal spans greater than 1 μm), “integrations” and “no signal” spreads generated from HAC formation assays. The mean (+/− SD) is shown. (D) Comparison of size of a HAC made from YAC-Mm-4q21LacO and a multimerized HAC made from 4q21 BACLacO. Both HACs are shown at the same scale. Coloring is identical to what is shown in the merged images in panel B. Bar, 1 μm.
YAC-Mm-4q21LacO HACs harbor multi-domain centromeres for faithful inheritance
We next sought to test the degree to which the centromeres on the HACs could support mitotic function. Starting from the polyclonal population of cells that survived G418-S selection (Fig. 1), we screened 24 monoclonal isolates and identified 8 lines harboring HACs (the other lines included 2 in which YAC-Mm-4q21LacO had integrated into a natural chromosome and 14 with no detectable HAC) and measured the proportion of cells in each harboring a detectable HAC (Fig. 2, A and B). Sequencing analysis of four of the monoclonal isolates revealed the presence of the HAC, but no evidence of any integration of the endogenous yeast chromosomes that were also introduced during the initial spheroplast fusion step (fig. S4). Our findings are consistent with prior studies where yeast chromosome segregation after spheroplast fusion was only observed after strong selection for rare genome integration events (28). Overall, the majority of cells upon isolation of a clonal line harbor HACs, matching this property of our prior generations of HACs (11, 12, 14, 16). Four of the lines (colored data points in graph in Fig. 2B) were subjected to three independent HAC stability assays over a month of growth in the absence of antibiotic selection (Fig. 2C). The average daily HAC loss rate of 0.011 +/− 0.006 (Fig. 2C) was similarly low as prior generations of HACs (12, 16, 29).
Fig. 2. YAC-Mm-4q21LacO-based HACs are inherited as autonomous chromosomes with functional kinetochores and robust CPC recruitment.
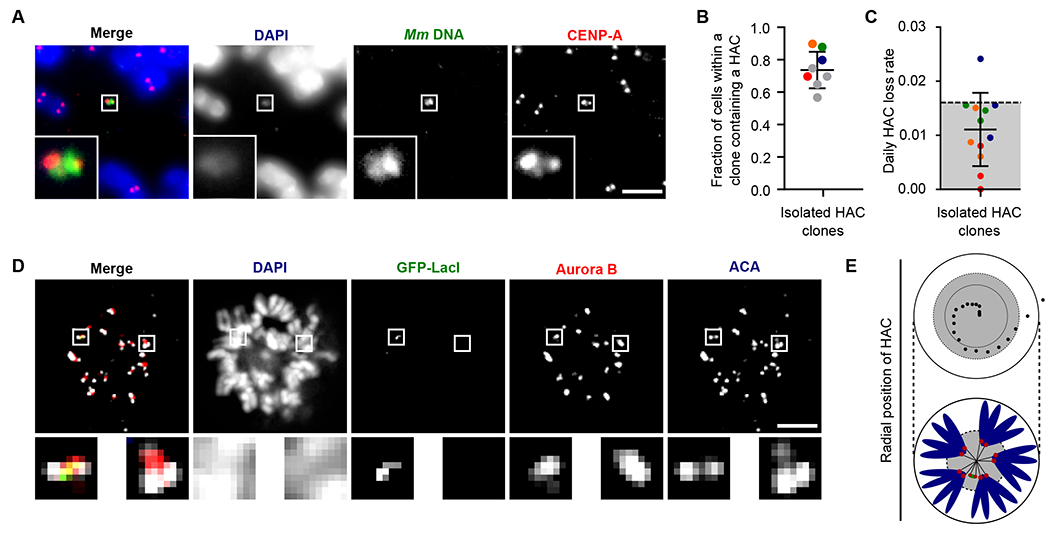
(A) Representative image of a single copy HAC that has been isolated in a monoclonal cell line. Inset: 5x magnification. Bar, 5 μm. (B) Quantification of fraction of spreads with a HAC in monoclonal cell lines. The mean (+/− SD) is shown. Colors indicate individual HAC lines that were assessed in HAC retention assays. Red, orange, green and blue correspond to HAC clones 1, 2, 3, and 5. (C) Quantification of HAC loss rate after culturing without selection for 30 days. The mean (+/−SD) is shown. Experiments are color coded to correspond to the clones shown in panel B. Grey shading indicates the range of loss rates for prior generations of HACs (12, 16, 29). (D) Representative image of HACs synchronized in mitosis showing Aurora B and ACA. The image shows 8 0.2 μm z-projected stacks (see also Fig. S4 for centromere delineation in the z-dimension). Inset: 5x magnification. Bar, 5 μm. (E) The radial position of HACs was measured relative to endogenous centromeres. The position of 20 HACs, each endogenous centromere and the center of DNA mass was measured. The distance between HAC or endogenous centromere and the center of DNA mass was calculated. The distance of each HAC from the center was normalized based on the total length across (i.e. the diameter) of mitotic chromosomes. The inner black circle represents the mean radial position of endogenous centromeres, while the dotted line represents one standard deviation from the mean. An illustration is shown below the graph.
To further interrogate our initial hypothesis that a larger initial HAC construct would confer full centromere function, we assessed the mitotic recruitment of a key component of the inner centromeric error correction mechanism, the Aurora B kinase (18). The inner centromere includes chromatin marked with H3T3phos and H2AT121phos modifications for recruiting the Chromosome Passenger Complex (CPC), containing Aurora B (30–32). Inner centromeric chromatin spans several Mb on natural chromosomes (33). The inner centromere in metazoans is not a single thread of chromatin but rather thought to be a densely packed region that spans a linear distance of 500-1000 nm between sister centromeres (34, 35). Given the high-fidelity transmission of our HACs (Fig. 2C), we predicted that they are sufficiently large to generate a robust inner centromere that recruits Aurora B. In order to have the necessary dispersion of chromosomes in the mitotic cell for robust detection of the HAC via expression of GFP-LacI, we induced the formation of monopolar spindles and assessed both the kinetochore forming part of the paired sister HACs (with anti-centromere antibodies; ACA) and the inner centromere (with antibodies to Aurora B) (Fig. 2, D and E; fig. S5). Aurora B was found in 33/35 HACs. Thus, our findings suggest that the increased size of YAC-Mm-4q21LacO relative to prior HAC constructs permits a robust inner centromere without the need to undergo large-scale multimerization. This is critical since it endows YAC-Mm-4q21LacO HACs with the ability to segregate in mitosis at high fidelity alongside natural counterparts.
Single-copy HACs
The small physical size of HACs formed from YAC-Mm-4q21LacO (Fig. 1) raised the possibility that they can form without any multimerization at all. To test this notion, we sought a cytological approach that reports on copy number without the deformations that happen naturally when chromosomes are attached to and stretched by the spindle or otherwise confounded by mitotic chromosome condensation. Fortuitously, we found that nuclear envelope lysis during isolation of nuclei releases the small HACs formed with YAC-Mm-4q21LacO that are subsequently efficiently separated from the rest of the genome via centrifugation (Fig. 3A; fig. S6). We harvested the top gradient fractions in the 10% sucrose layer (i.e. above the visible cell debris), and determined the location of CENP-A and the LacO sequences (Fig. 3B). We anticipated a single CENP-A focus on interphase HACs, even after replication, since sister centromeres are not separated into distinct foci on natural chromosomes until just prior to nuclear envelope breakdown near mitotic onset (36). LacO arrays, on the other hand, when present on repeated HAC constructs do not coalesce into a single focus (16). HACs were readily identified in these fractions, permitting us to visualize an individualized and functional metazoan chromosome in its decondensed, interphase form (Fig. 3B). In the vast majority of HACs, CENP-A and LacO staining each produced a single focus (Fig. 3B). We did not observe any HACs with a single CENP-A focus and more than one LacO focus. A small number of HACs harbored two CENP-A foci, consistent with them coming from cells that were in late G2 or early mitosis (i.e. prior to sister chromatid separation) at the time of isolation. Each of these also had precisely two LacO foci (Fig. 3B). Unlike the single-copy YAC-Mm-4q21LacO HACs, prior generations of HACs are large multimers that do not separate from endogenous chromosomes during nuclei isolation (16). In these HACs, CENP-A and LacO arrays are readily visualized on mitotic HACs in chromosome spreads (Fig. 3C). For the prior generation of HACs, the paired, replicated centromeres are visible as ‘double dots’ of CENP-A, whereas the LacO arrays are visible as numerous foci (Fig. 3C). Taken together with the earlier detection from uniformly small-sized HACs from populations of cells with nascent YAC-Mm-4q21LacO HACs (Fig. 1, B–D), our interphase HAC experiments (Fig. 3B) indicate that the new HACs are formed and maintained without multimerization.
Fig. 3. YAC-Mm-4q21LacO HACs are functional as single copy DNA.
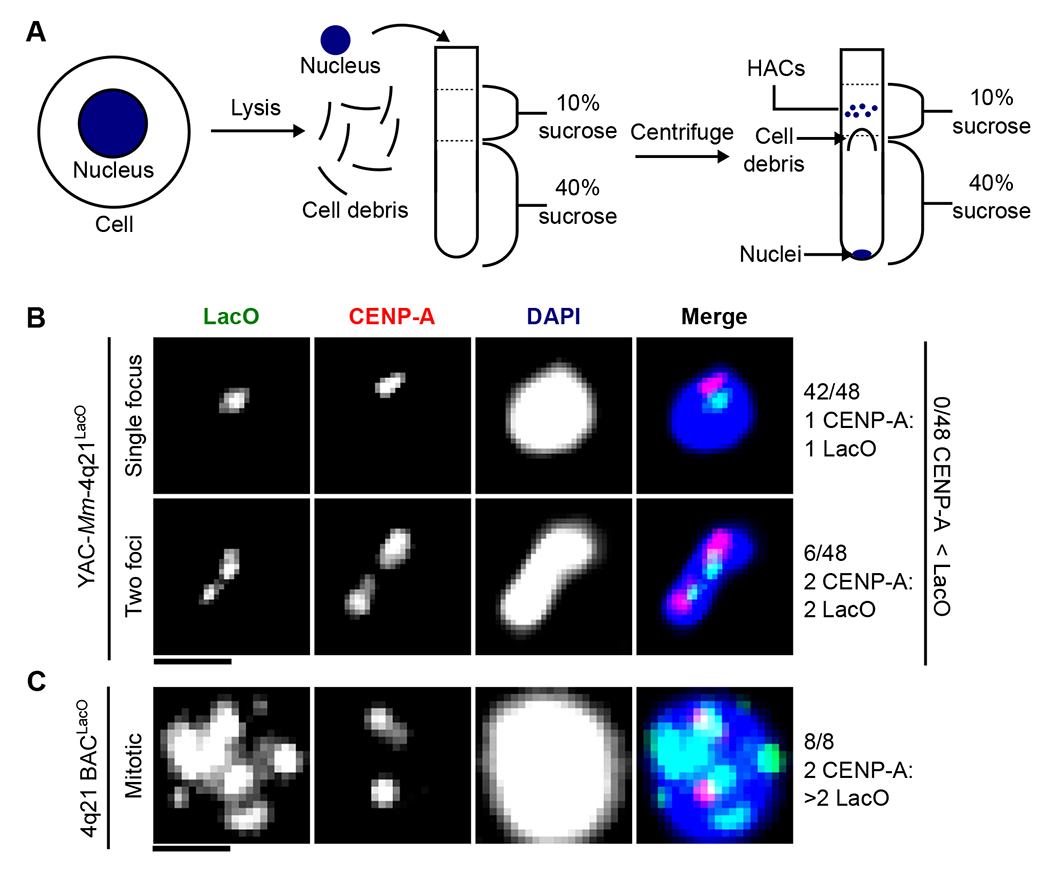
(A) Schematic of approach used to enrich single copy HACs. (B) Representative image of HACs isolated by sucrose gradient with either a single or two foci of LacO. The proportion of HACs with a single or two foci is noted. HACs with two LacO foci also had two CENP-A foci suggesting that they are mitotic. Bar, 1 μm. (C) Representative image of a multimerized HAC (Clone 27 from (16)) from mitotic chromosome spreads. Bar, 1 μm.
We next assessed the size and topology of functional YAC-Mm-4q21LacO-based HACs (Fig. 4). This is important because earlier generations of HACs typically formed in a manner accompanied by large-scale DNA sequence multimerization and even acquisition of portions (>100 kb) of host cell chromosomal DNA (16, 37, 38). YAC-Mm-4q21LacO contains a single FseI site (fig. S1), and we found that two isolated HAC-containing cell lines required FseI digestion to enter a pulse-field gel (Figs. 4A; fig. S7). This is consistent with well-established topological trapping of circular chromosomes prior to digestion (39). The mobility of the linearized HAC suggests that it has not lost or gained large fragments of DNA (Fig. 4A; fig. S7). We compared this to a circular, multimerized BAC-based HAC (16) that has one FseI site per repeating ~200 kb monomer (Figs. 4A; fig. S7). Along with our cytological data indicating the HACs are single copy (Fig. 3), their behavior on pulse-field gels (Figs. 4A; fig. S5) support the notion that they function and are inherited through cell divisions with the same single-copy circular nature in which they were initially constructed in yeast.
Fig. 4. YAC-Mm-4q21LacO-based HACs are intact 760 kb circles with similar chromatin stretching properties as natural chromosomes.
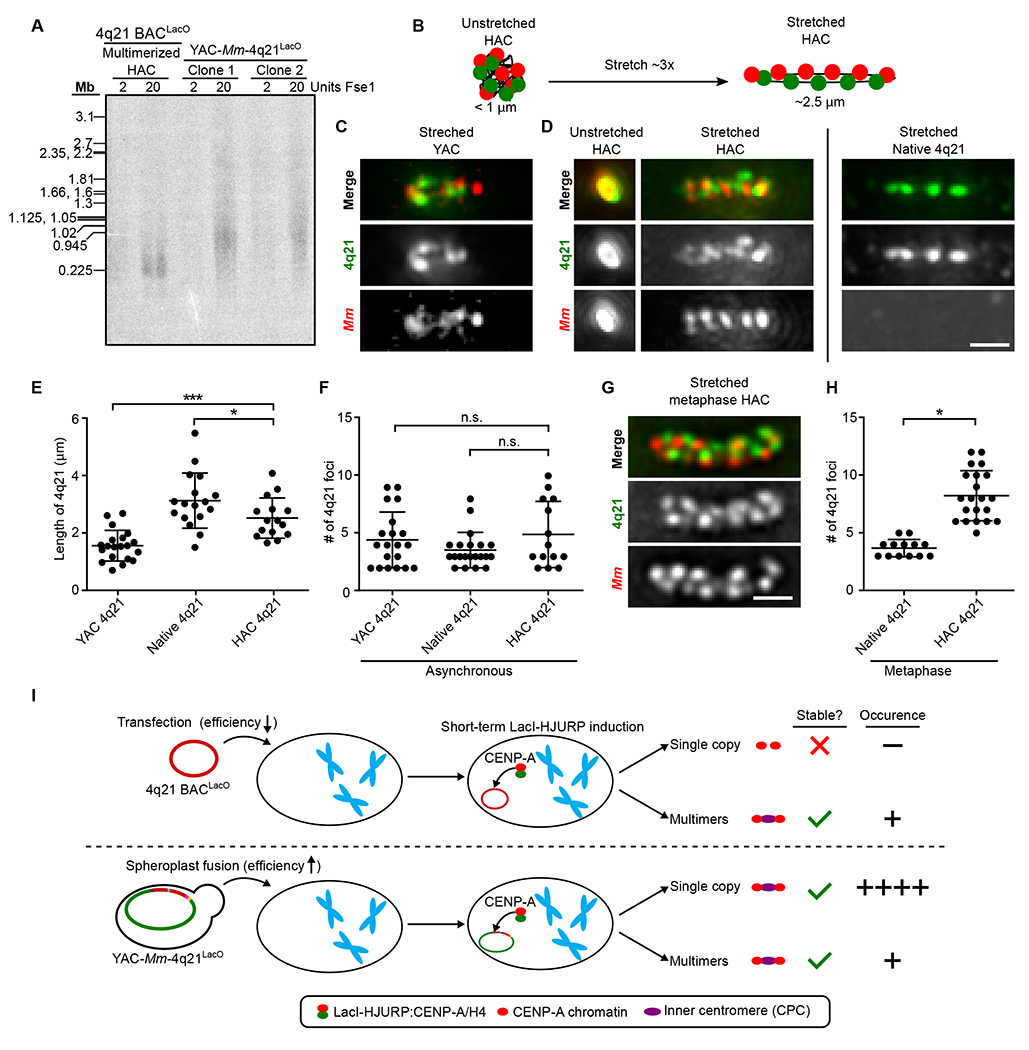
(A) Southern bot analysis of the indicated HAC lines using a LacO probe. The multimerized HAC line was diluted 20-fold in HT1080 cells to reduce signal relative to the YAC-Mm-4q21LacO-based HAC so that they could be in a similar range of detection. (B) Schematic showing extent of stretching HACs in our experiments (panels D-H), with indicated regions detected by FISH. The number of foci shown is in the range predicted by prior stretching experiments with natural chromosomes, with actual outcomes measured in panels C-G and fig. S6. (C) Representative images of a stretched YAC with both the 4q21 and M. mycoides sequence labeled via FISH compared to endogenous 4q21 in asynchronous cells. Bar, 1 μm. (D) Representative images of an unstretched and stretched HAC with both the 4q21 and M. mycoides sequence labeled via FISH compared to endogenous 4q21 in asynchronous cells. Bar, 1 μm. (E) Quantification of the length of 4q21 FISH in the YAC, HAC and the endogenous chromosome after stretching chromatin in asynchronous cells. The mean (+/− SD) is shown. p< 0.001 and p < 0.05 based on an unpaired, two-tailed t-test. (F) Quantification of the number of foci from 4q21 FISH in the YAC, HAC and in the endogenous chromosome after stretching chromatin in asynchronous cells. The mean (+/− SD) is shown. p values > 0.05 based on an unpaired, two-tailed t-test and is marked as not significant (n.s.). (G) Representative images of a stretched HAC with both the 4q21 and M. mycoides sequence labeled via FISH after enriching for cells in metaphase. Bar, 1 μm. (H) Quantification of the number of foci from 4q21 FISH in the HAC and the endogenous chromosome after stretching chromatin and enriching for cells in metaphase. The mean (+/− SD) is shown. p < 0.0001 based on an unpaired, two-tailed t-test. (I) Model illustrating how construct size influences HAC formation outcomes. Single-copy BAC-based HACs have never been isolated and are shown as too small to harbor a functional inner centromere.
To cytologically examine YAC-Mm-4q21LacO as it exists in yeast and then after it forms a functional HAC in human cells, we employed a well-established chromatin stretching approach (34, 40) (Fig. 4, B–H; fig. S8). This permits us to monitor the 182 kb region of 4q21 present on the HACs in comparison to the initial YAC construct or the endogenous locus on chromosome 4 (Fig. 4, B–H). The degree of stretching we achieved is about 3-fold, making the circle fold back upon itself (Figs. 4, B–D; fig. S8). Stretching maintains large blocks (roughly 40 kb) of chromatin linked by highly stretched regions with little or no detectable FISH signal (Fig. 4, C and D and G; fig. S8). The overall length of the HAC in human cells was greater than the YAC in yeast, but smaller than native 4q21 (Fig. 4E). The number of 4q21 foci produced by stretching of the YAC in yeast or at the native locus and HAC in mammalian cells is similar (Fig. 4F). We conclude that the similarity of YACs and HACs reflects the absence of rampant multimerization or other substantial molecular changes (Fig. 4, C–F; fig. S1; fig. S4), with relatively subtle changes in length and foci number likely reflecting differences in chromatin properties between budding yeast and human cells (41, 42).
The number of 4q21 foci observed on the HAC varied greatly, revealing the possible existence of two HAC populations (Fig. 4F). We reasoned that the HAC would be visualized as a single chromatid early in the cell cycle (G1 and early S phase) but would be visualized as paired chromatids later in the cell cycle (late S, G2, and M). On the other hand, the native 4q21 locus would be visualized in stretching experiments as a single chromatid, even late in the cell cycle, since each natural chromosome arm location would make its own chromatin fiber. To test this notion, we enriched cells in mitosis, prior to sister chromatid separation, and found that the average number of 4q21 foci in the HAC increased from 4.9 (+/− 2.9) to 8.2 (+/− 2.1) (Fig. 4H) relative to those from asynchronous cell populations (Fig. 4F), whereas, the endogenous 4q21 locus has a similar number of foci after stretching in both instances (Fig. 4, F and H). We note that the M. mycoides chromatin appears to be relatively resistant to stretching, since there is a similar number of foci and length (Fig. 4, C and D; fig. S8) despite its three-fold greater length relative to 4q21 on YAC-Mm-4q21LacO. A likely explanation is that the AT-rich M. mycoides sequence has denser chromatin relative to the human 4q21 sequence, which could be rendered by histone modifications and/or AT-hook protein-mediated compaction. Indeed, in addition to CENP-A-containing chromatin, a portion of the HAC DNA contains chromatin harboring H3K9me3 (Fig. S6), suggesting it harbors the heterochromatin typically found in pericentromeric regions and offering a means through which differences in chromatin stretching could be achieved (43, 44). HAC stretching supports the notion of a chromatinized circular topology supporting propagation and inheritance in dividing cells.
Discussion
De novo HACs, like the ones we advance in this paper, are the only viable platform to generate a new mammalian chromosome where the entire DNA sequence can be designed in the lab. This presents an extremely wide horizon of possibilities for downstream biological and applied uses, and we report a system to create HACs that faithfully exist in their functional form as a single copy. The advances are centered on the portion of the chromosome, the centromere, that controls segregation of the HAC at cell division. Single-copy HAC formation requires establishment of a high local density of CENP-A nucleosomes that can self-propagate alongside natural centromeres (16), but this is not sufficient for centromere function. Rather, an entirely different chromatin domain, the inner centromere, must function in mitotic quality control and control of sister chromatid cohesion. The HACs formed from the YAC-Mm-4q21LacO are large enough to harbor robust CENP-A arrays and inner centromeric chromatin (Fig. 2; Fig. 4I; fig. S9).
The overhauled HAC cell delivery approach via spheroplast fusion was necessary because the initial construct is too large to reliably purify. It further has the benefit of being far more efficient than HAC construct transfections. YACs from spheroplasts will be packaged into chromatin, which may also contribute to the high rate of HAC formation upon introduction into mammalian cells. The overall high efficiency of HAC delivery and formation upon moving to spheroplast fusion-based delivery has important practical implications for the development and testing of specific features of HACs. In prior generations of HACs, rigorous testing of a modest number of constructs or cell lines required the isolation and subsequent cytological assessment of hundreds of cell lines that are cloned weeks after initial HAC construct transfection (16, 27), since the initial selected cell populations are so sparse and HAC formation is so inefficient. With the YAC-Mm-4q21LacO platform, we can measure high HAC formation efficiency in rapidly generated cell populations, obviating the need to isolate clonal lines. To quantify the proportion of HACs with the replicates shown in Fig. 1D, we would have needed to generate a minimum of hundreds of cell lines with our prior approach (16). One can easily envision a multitude of HAC features –including their genetic cargo and recipient cell types—that are attractive to test in the future. There are many potential opportunities for further optimization: some common to prior generations of HACs and some that arise because of the single copy nature of YAC-Mm-4q21LacO. For instance, gene silencing on HACs is a known challenge (45) that confounds expression of selection markers, reporters, and eventually other useful genetic payloads. The instability of all generations of HACs, including YAC-Mm-4q21LacO HACs (Fig. 2C) relative to natural chromosome could be impacted by several modifiable factors. These include, but aren’t limited to, HAC topology (i.e., circular versus linear), size of pericentromeric heterochromatin (fig. S6), and the choice of stuffer DNA. For the latter, we note that while the extreme AT-richness in the stuffer M. mycoides DNA present in YAC-Mm-4q21LacO may positively contribute to forming a functional inner centromere (Fig. 2), it causes barriers to routine high-throughput sequence-based approaches (fig. S4) that could otherwise be employed to map distinct chromatin features. YAC-Mm-4q21LacO represents an attractive platform for HAC debugging—using synthetic budding yeast chromosomes as a model (46)—and for gaining new insights into mammalian chromosome biology while doing so. The central innovations with YAC-Mm-4q21LacO are that it avoids the rampant multimerization that has complicated past HAC efforts and that it permits rapid assessment of HAC function (Fig. 1). The latter is bound to accelerate the pace of future HAC debugging.
Artificial chromosomes, more broadly, could deliver useful cargos for biomedical and industrial applications in a variety of eukaryotes. Since both the CENP-A-based epigenetic centromere specification mechanism and the inner centromeric dimensions and molecular constituency (e.g. sister centromere cohesion components and CPC) are common to diverse eukaryotic species, including in many agriculturally important plants, we envision that YAC-Mm-4q21LacO-based artificial chromosomes will be readily modified and extended into many useful biological systems. For advancing the understanding of chromosomes, HACs can be used as testing grounds for designing what nature has evolved to ensure the stability of our genome, as well as designing the functional features that govern chromosome “outputs” in gene expression programs and epigenetic regulation. In summary, the advancements made in this study to HAC design, delivery, formation, and function will expedite both discovery-based and applied genome science.
Supplementary Material
Acknowledgments:
We thank our UPenn colleagues M. Lampson (for comments on the manuscript) and M. Gerace (for assistance with preparing reagents). We also thank E. Makeyev (King’s College) for reagents.
Funding:
National Institutes of Health grant GM130302 (B.E.B.)
National Institutes of Health grant HG012445 (B.E.B. and J.I.G.)
National Institutes of Health grant CA261198 (K.K.)
National Institutes of Health grant GM007229 (G.J.B.)
Footnotes
Competing interests:
C.W.G., D.M.B., J.I.G., and B.E.B. are inventors on a provisional patent application submitted by UPenn related to this work.
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the paper or the supplementary materials or have been uploaded to SRA (PRJNA985068). The materials used in this study are available from commercial sources or from the corresponding authors on reasonable request upon publication of the study.
References and Notes
- 1.Burke DT, Carle GF, Olson MV, Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science 236, 806–812 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Murray AW, Szostak JW, Construction of artificial chromosomes in yeast. Nature 305, 189–193 (1983). [DOI] [PubMed] [Google Scholar]
- 3.Traver CN, Klapholz S, Hyman RW, Davis RW, Rapid screening of a human genomic library in yeast artificial chromosomes for single-copy sequences. Proc. Natl. Acad. Sci 86, 5898–5902 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green ED, Olson MV, Chromosomal region of the cystic fibrosis gene in yeast artificial chromosomes: a model for human genome mapping. Science 250, 94–99 (1990). [DOI] [PubMed] [Google Scholar]
- 5.Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA, Smith HO, Complete chemical synthesis, assembly, and cloning of a mycoplasma genitalium genome. Science. 319, 1215–1220 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang R-Y, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi Z-Q, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison I. I. I. Clyde A., Smith HO, Venter JC, Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 329, 52–56 (2010) [DOI] [PubMed] [Google Scholar]
- 7.Annaluru N, Muller H, Mitchell LA, Ramalingam S, Stracquadanio G, Richardson SM, Dymond JS, Kuang Z, Scheifele LZ, Cooper EM, Cai Y, Zeller K, Agmon N, Han JS, Hadjithomas M, Tullman J, Caravelli K, Cirelli K, Guo Z, London V, Yeluru A, Murugan S, Kandavelou K, Agier N, Fischer G, Yang K, Martin JA, Bilgel M, Bohutski P, Boulier KM, Capaldo BJ, Chang J, Charoen K, Choi WJ, Deng P, DiCarlo JE, Doong J, Dunn J, Feinberg JI, Fernandez C, Floria CE, Gladowski D, Hadidi P, Ishizuka I, Jabbari J, Lau CYL, Lee PA, Li S, Lin D, Linder ME, Ling J, Liu J, Liu J, London M, Ma H, Mao J, McDade JE, McMillan A, Moore AM, Oh WC, Ouyang Y, Patel R, Paul M, Paulsen LC, Qiu J, Rhee A, Rubashkin MG, Soh IY, Sotuyo NE, Srinivas V, Suarez A, Wong A, Wong R, Xie WR, Xu Y, Yu AT, Koszul R, Bader JS, Boeke JD, Chandrasegaran S, Total synthesis of a functional designer eukaryotic chromosome. Science. 344, 55–58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke JD, Church G, Hessel A, Kelley NJ, Arkin A, Cai Y, Carlson R, Chakravarti A, Cornish VW, Holt L, Isaacs FJ, Kuiken T, Lajoie M, Lessor T, Lunshof J, Maurano MT, Mitchell LA, Rine J, Rosser S, Sanjana NE, Silver PA, Valle D, Wang H, Way JC, Yang L, The Genome Project-Write. Science 353, 126–127 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Dawe RK, Charting the path to fully synthetic plant chromosomes. Exp. Cell Res 390, 111951 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Barrey EJ, Heun P, Artificial chromosomes and strategies to initiate epigenetic centromere establishment. Prog. Mol. Subcell. Biol 56, 193–212 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Harrington JJ, Bokkelen GV, Mays RW, Gustashaw K, Willard HF, Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet 15, 345–355 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H, Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol 16, 431–439 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Kixmoeller K, Allu PK, Black BE, The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle. Open Biol. 10, 200051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayden KE, Strome ED, Merrett SL, Lee H-R, Rudd MK, Willard HF, Sequences associated with centromere competency in the human genome. Mol. Cell. Biol 33, 763–772 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouprina N, Samoshkin A, Erliandri I, Nakano M, Lee H-S, Fu H, Iida Y, Aladjem M, Oshimura M, Masumoto H, Earnshaw WC, Larionov V, Organization of synthetic alphoid DNA array in human artificial chromosome (HAC) with a conditional centromere. ACS Synth. Biol 1, 590–601 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logsdon GA, Gambogi CW, Liskovykh MA, Barrey EJ, Larionov V, Miga KH, Heun P, Black BE, Human artificial chromosomes that bypass centromeric DNA. Cell. 178, 624–639.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basu J, Willard HF, Artificial and engineered chromosomes: non-integrating vectors for gene therapy. Trends Mol. Med 11, 251–258 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Carmena M, Wheelock M, Funabiki H, Earnshaw WC, The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol 13, 789–803 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altemose N, Logsdon GA, Bzikadze AV, Sidhwani P, Langley SA, Caldas GV, Hoyt SJ, Uralsky L, Ryabov FD, Shew CJ, Sauria MEG, Borchers M, Gershman A, Mikheenko A, Shepelev VA, Dvorkina T, Kunyavskaya O, Vollger MR, Rhie A, McCartney AM, Asri M, Lorig-Roach R, Shafin K, Lucas JK, Aganezov S, Olson D, de Lima LG, Potapova T, Hartley GA, Haukness M, Kerpedjiev P, Gusev F, Tigyi K, Brooks S, Young A, Nurk S, Koren S, Salama SR, Paten B, Rogaev EI, Streets A, Karpen GH, Dernburg AF, Sullivan BA, Straight AF, Wheeler TJ, Gerton JL, Eichler EE, Phillippy AM, Timp W, Dennis MY, O’Neill RJ, Zook JM, Schatz MC, Pevzner PA, Diekhans M, Langley CH, Alexandrov IA, Miga KH, Complete genomic and epigenetic maps of human centromeres. Science 376, eabl4178 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altemose N, Maslan A, Smith OK, Sundararajan K, Brown RR, Mishra R, Detweiler AM, Neff N, Miga KH, Straight AF, Streets A, DiMeLo-seq: a long-read, single-molecule method for mapping protein-DNA interactions genome wide. Nat. Methods 19, 711–723 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE, The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol 20, 687–695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Logsdon GA, Vollger MR, Hsieh P, Mao Y, Liskovykh MA, Koren S, Nurk S, Mercuri L, Dishuck PC, Rhie A, de Lima LG, Dvorkina T, Porubsky D, Harvey WT, Mikheenko A, Bzikadze AV, Kremitzki M, Graves-Lindsay TA, Jain C, Hoekzema K, Murali SC, Munson KM, Baker C, Sorensen M, Lewis AM, Surti U, Gerton JL, Larionov V, Ventura M, Miga KH, Phillippy AM, Eichler EE, The structure, function and evolution of a complete human chromosome 8. Nature 593, 101–107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown DM, Chan YA, Desai PJ, Grzesik P, Oldfield LM, Vashee S, Way JC, Silver PA, Glass JI, Efficient size-independent chromosome delivery from yeast to cultured cell lines. Nucleic Acids Res. 45, e50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouprina N, Larionov V, Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma 125, 621–632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song J, Dong F, Lilly JW, Stupar RM, Jiang J, Instability of bacterial artificial chromosome (BAC) clones containing tandemly repeated DNA sequences. Genome 44, 463–469 (2001). [PubMed] [Google Scholar]
- 26.Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H, CENP-B controls centromere formation depending on the chromatin context. Cell 131, 1287–1300 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Ohzeki J, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H, Breaking the HAC barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 31, 2391–2402 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jülicher K, Vieten L, Bröcker F, Bardenheuer W, Schütte J, Opalka B, Yeast artificial chromosome transfer into human renal carcinoma cells by spheroplast fusion. Genomics 43, 95–98 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Ebersole TA, Ross A, Clark E, McGill N, Schindelhauer D, Cooke H, Grimes B, Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum. Mol. Genet. 9, 1623–1631 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H, Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330, 235–239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JMG, Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330, 231–235 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamagishi Y, Honda T, Tanno Y, Watanabe Y, Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330, 239–243 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Bassett EA, Wood S, Salimian KJ, Ajith S, Foltz DR, Black BE, Epigenetic centromere specification directs aurora B accumulation but is insufficient to efficiently correct mitotic errors. J. Cell Biol 190, 177–185 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blower MD, Sullivan BA, Karpen GH, Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2, 319–330 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC, A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci 107, 10484–10489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allu PK, Dawicki-McKenna JM, Van Eeuwen T, Slavin M, Braitbard M, Xu C, Kalisman N, Murakami K, Black BE, Structure of the human core centromeric nucleosome complex. Curr. Biol 29, 2625–2639.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erliandri I, Fu H, Nakano M, Kim J-H, Miga KH, Liskovykh M, Earnshaw WC, Masumoto H, Kouprina N, Aladjem MI, Larionov V, Replication of alpha-satellite DNA arrays in endogenous human centromeric regions and in human artificial chromosome. Nucleic Acids Res. 42, 11502–11516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pesenti E, Liskovykh M, Okazaki K, Mallozzi A, Reid C, Abad MA, Jeyaprakash AA, Kouprina N, Larionov V, Masumoto H, Earnshaw WC, Analysis of complex DNA rearrangements during early stages of HAC formation. ACS Synth. Biol 9, 3267–3287 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noskov VN, Chuang R-Y, Gibson DG, Leem S-H, Larionov V, Kouprina N, Isolation of circular yeast artificial chromosomes for synthetic biology and functional genomics studies. Nat. Protoc 6, 89–96 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyriacou E, Heun P, High-resolution mapping of centromeric protein association using APEX-chromatin fibers. Epigenetics Chromatin 11, 68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan ZY, Cai S, Noble AJ, Chen JK, Shi J, Gan L, Heterogeneous non-canonical nucleosomes predominate in yeast cells in situ. eLife 12, RP87672 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai S, Böck D, Pilhofer M, Gan L, The in situ structures of mono-, di-, and trinucleosomes in human heterochromatin. Mol. Biol. Cell 29, 2450–2457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters AHFM, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AAHA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JHA, Jenuwein T, Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD, Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12, 1591–1598 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Lee NCO, Kononenko AV, Lee H-S, Tolkunova EN, Liskovykh MA, Masumoto H, Earnshaw WC, Tomilin AN, Larionov V, Kouprina N, Protecting a transgene expression from the HAC-based vector by different chromatin insulators. Cell. Mol. Life Sci. CMLS 70, 3723–3737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y, Coelho C, Hughes AL, Lazar-Stefanita L, Yang S, Brooks AN, Walker RSK, Zhang W, Lauer S, Hernandez C, Cai J, Mitchell LA, Agmon N, Shen Y, Sall J, Fanfani V, Jalan A, Rivera J, Liang F-X, Bader JS, Stracquadanio G, Steinmetz LM, Cai Y, Boeke JD, Debugging and consolidating multiple synthetic chromosomes reveals combinatorial genetic interactions. Cell 186, 5220–5236.e16 (2023). [DOI] [PubMed] [Google Scholar]
- 47.Khandelia P, Yap K, Makeyev EV, Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc. Natl. Acad. Sci. U. S. A 108, 12799–12804 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS, In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol 135, 1685–1700 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gopalakrishnan R, Winston F, Whole genome sequencing of yeast cells. Curr. Protoc. Mol. Biol 128, e103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bickmore W, Chromosome structural analysis: a practical approach (Oxford University Press, 1999). [Google Scholar]
- 51.Morgenstern JP, Land H, Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18, 3587–3596 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sekulic N, Bassett EA, Rogers DJ, Black BE, The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature 467, 347–351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logsdon GA, Barrey EJ, Bassett EA, DeNizio JE, Guo LY, Panchenko T, Dawicki-McKenna JM, Heun P, Black BE, Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J Cell Biol 208, 521–531 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gotta M, Laroche T, Gasser SM, “Analysis of nuclear organization in Saccharomyces cerevisiae” in Methods in Enzymology (Academic Press, 1999; https://www.sciencedirect.com/science/article/pii/S0076687999040409)vol. 304 of Chromatin, pp. 663–672. [DOI] [PubMed] [Google Scholar]
- 55.Lorente-Leal V, Farrell D, Romero B, Álvarez J, de Juan L, Gordon SV, Performance and Agreement Between WGS Variant Calling Pipelines Used for Bovine Tuberculosis Control: Toward International Standardization. Front. Vet. Sci 8, 780018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the supplementary materials or have been uploaded to SRA (PRJNA985068). The materials used in this study are available from commercial sources or from the corresponding authors on reasonable request upon publication of the study.


