α-Actinin-3 deficiency alters muscle mass regulation and protects against dexamethasone-induced muscle wasting.
Abstract
Homozygosity for the common ACTN3 null polymorphism (ACTN3 577X) results in α-actinin-3 deficiency in ~20% of humans worldwide and is linked to reduced sprint and power performance in both elite athletes and the general population. α-Actinin-3 deficiency is also associated with reduced muscle mass, increased risk of sarcopenia, and altered muscle wasting response induced by denervation and immobilization. Here, we show that α-actinin-3 plays a key role in the regulation of protein synthesis and breakdown signaling in skeletal muscle and influences muscle mass from early postnatal development. We also show that α-actinin-3 deficiency reduces the atrophic and anti-inflammatory response to the glucocorticoid dexamethasone in muscle and protects against dexamethasone-induced muscle wasting in female but not male mice. The effects of α-actinin-3 deficiency on muscle mass regulation and response to muscle wasting provide an additional mechanistic explanation for the positive selection of the ACTN3 577X allele in recent human history.
INTRODUCTION
Homozygosity for a common null polymorphism in ACTN3 (R577X) results in complete absence of α-actinin-3 in more than 1.5 billion people worldwide (1). α-Actinin-3 deficiency (ACTN3 577XX) does not cause muscle disease due to expression of the highly homologous α-actinin-2 in skeletal muscle but is associated with significantly reduced sprint and muscle power performance in elite athletes and in the general population (2–4). The absence of α-actinin-3 is also associated with reduced skeletal muscle mass that persists through to old age, with an increased risk of sarcopenia, frailty, and loss of function in the elderly (5–8). In a case-control analysis of two large independent cohorts of Caucasian postmenopausal women (each n > 1200), the carriage of the ACTN3 577X allele was associated with 33% increased risk of falling (9). A recent study in an elderly Chinese population (ages 70 to 79, n = 1031) also showed significantly reduced strength and increased frailty score associated with ACTN3 577XX genotype (10). ACTN3 genotype has been shown to modify the clinical severity of various chronic disorders; it contributes to interpatient variability in disease onset and progression in muscle diseases such as McArdle’s disease (11), myositis (12), Pompe disease (13), and Duchenne muscular dystrophy (DMD) (14). Moreover, ACTN3 genotype is correlated with survival in patients with congestive heart failure; patients carrying the X allele have 1.72 times higher mortality than patients with ACTN3 577RR genotype (P = 0.01) (15).
Phenotypic analysis of an Actn3 knockout (KO) mouse model (16, 17) provides some mechanistic explanations for the effect of α-actinin-3 deficiency on skeletal muscle traits and performance. Compared to wild-type (WT) littermates, Actn3 KO muscles show significant reductions in type 2, fast-twitch glycolytic muscle fiber size, decreased anaerobic activity, and increased oxidative phosphorylation (16, 18). This “slowing” of the metabolic properties of fast glycolytic muscle fibers is driven by reduced glycogen phosphorylase activity (19) and is reversed by “rescue” or replacement of α-actinin-3 postnatally in skeletal muscle (20). α-Actinin-3 deficient fast-twitch fibers also show an increase in calcineurin activity (18) and alterations in contractile characteristics, with increased rate of decay of twitch transients and changes in calcium signaling caused by increased calcium leak from the sarcoplasmic reticulum and reuptake via the up-regulated expression of sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (SERCA1), calsequestrin, and sarcalumenin (21).
It is not yet fully understood how α-actinin-3 deficiency influences muscle mass at baseline and in response to atrophic stimuli. We have previously shown that α-actinin-3 deficiency is associated with reduced muscle wasting and alterations in fiber type switching (compared to WT) in response to hindlimb immobilization and denervation; this was mediated by elevated baseline calcineurin activity (22). Although calcineurin-dependent pathways are implicated in muscle growth and adaptation to functional overload (23, 24), the α-actinins also directly interact with soluble signaling factors, phosphatidylinositol 3-kinase p85 (PI3Kp85) (25), phosphatidylinositol 4,5-bisphosphate (26), and phosphatidylinositol 3,4,5-trisphosphate (27). All of these factors drive the downstream pathways that regulate a number of cellular functions, including the PI3K/Akt/mTOR (mechanistic target of rapamycin) signaling cascade that regulates protein synthesis and the insulin-like growth factor 1 (IGF1)–mediated hypertrophic response (28). In cardiomyocyte Z-discs, α-actinin-2 has also been shown to interact with atrogin-1 (Fbxo32), an E3 ubiquitin ligase that is up-regulated during muscle atrophy (29). In combination, these data suggest that α-actinin-3 deficiency alters muscle mass regulation by modifying key signaling molecules associated with protein synthesis and degradation.
Glucocorticoids are commonly prescribed to treat a variety of conditions associated with chronic inflammation such as DMD but are also known to cause adverse effects such as muscle wasting and osteoporosis (30). In skeletal muscle, glucocorticoids specifically cause atrophy of fast-twitch muscle fibers and elicit muscle wasting by increasing the rate of protein catabolism by the ubiquitin-proteasome and autophagy lysosome system and by suppressing the protein synthesis at the level of translation (31, 32). There is increasing biochemical evidence that members of the α-actinin family play a role in regulating glucocorticoid receptor (GR) activity, raising the possibility that α-actinin-3 deficiency may alter the skeletal muscle response to glucocorticoid treatment (32, 33). α-Actinin-4 (a nonmuscle α-actinin) interacts with GR in the presence of the glucocorticoid dexamethasone via the nuclear receptor–interacting motif LXXLL, which is conserved across all α-actinin family proteins (34–36) and potentiates GR activity in a dose-dependent manner (34–36). Similarly, both α-actinin-4 and α-actinin-2 [which is 80% identical to α-actinin-3 (37)] directly interact with and promote the activity of other nuclear receptors such as the estrogen receptor, androgen receptor, and thyroid receptor via the LXXLL motif (34, 36). α-Actinin-2 also binds the GR interacting protein (GRIP1) and increases its transactivation activities, as well as synergistically enhancing its nuclear receptor coactivator functions (34). These results suggest that deficiency of α-actinin-3, which increases α-actinin-2 expression (38), may likewise alter GR activity and response to glucocorticoids in skeletal muscle.
In this study, we aim to examine the effect of α-actinin-3 deficiency on pathways associated with protein synthesis and degradation in mature skeletal muscle and during development and then determine whether this influences the muscle wasting response to treatment with dexamethasone. We demonstrate that the effect of α-actinin-3 deficiency on muscle mass and downstream mTOR signaling occurs early during postnatal development and before full muscle maturation. We further show that α-actinin-3 deficiency reduces the atrophic response induced by dexamethasone and that α-actinin-3 expression directly influences the expression of key genes and pathways associated with muscle adaptation and inflammatory response following dexamethasone treatment. Last, we demonstrate that α-actinin-3 deficiency protects against muscle wasting induced by dexamethasone in female but not male mice and that this is mediated by increased protein synthesis and reduced expression of muscle atrophy–related genes.
RESULTS
α-Actinin-3 deficiency alters muscle mass and signaling pathways involved in protein synthesis and breakdown from early postnatal development
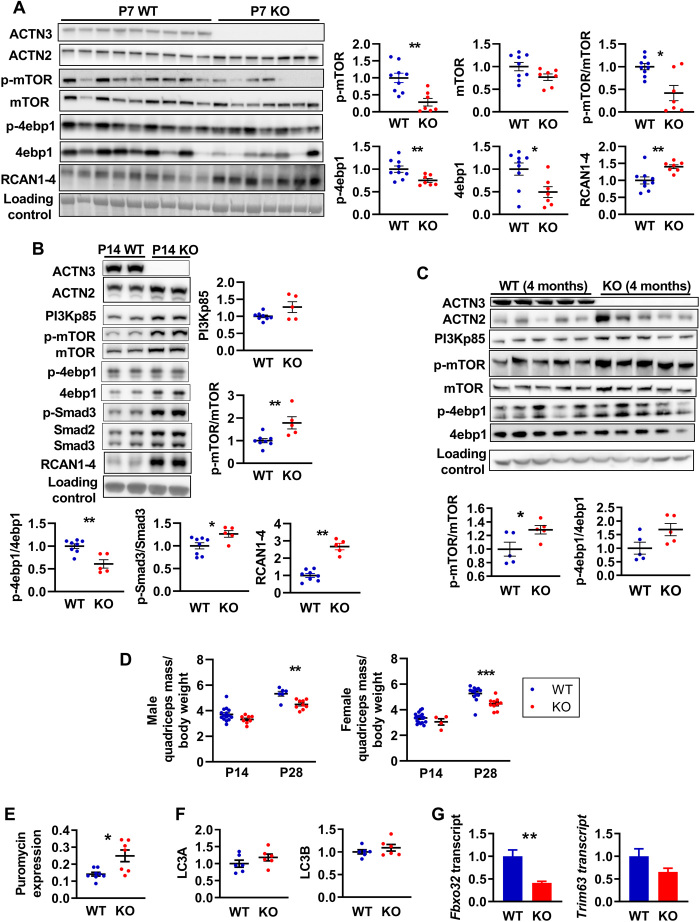
Signaling pathways responsible for muscle growth during development, regeneration, and overload-induced hypertrophy converge on mTOR and its downstream effectors that control protein synthesis (39). Since the first 3 weeks of postnatal mouse development is known to be a period of intense growth characterized by an increase in myofiber number (hyperplasia) and myofiber size (hypertrophy) (40), we examined the activation of mTOR via PI3K/Akt as well as the Smad3, which mediates myostatin/activin A signaling and cross-talks with mTOR signaling via Akt (39) at postnatal day 0 (P0), P7, P14, and P28. We also examined the expression of RCAN1-4 as a marker of calcineurin activity at these developmental time points, since we have previously observed an increased calcineurin activity in mature α-actinin-3–deficient skeletal muscle (18). Western blot analysis showed no differences in the activation of these pathways between genotypes at P0, although Actn3 KO muscles showed significant reductions in phosphorylated mTOR (p-mTOR) and mTOR compared to WT (fig. S1). At P7, Actn3 KO muscles again showed reductions in p-mTOR and mTOR as well as reduced 4ebp1 and p-4ebp1 and PI3Kp85; expression of Akt, p-Akt, Smad2/3, p-Smad3, S6RP, and p-S6RP was not different between genotypes. RCAN1-4 expression was increased in Actn3 KO muscles compared to WT (Fig. 1A and fig. S2A). In contrast, at P14, mTOR activity and total 4ebp1, but not p-4ebp1, were increased in Actn3 KO muscles compared to WT; similarly, PI3Kp85 and RCAN1-4 expression were also increased in Actn3 KO muscles (Fig. 1B and fig. S2B). Increased activation of Smad3, which is associated with suppression of protein synthesis, was also observed in Actn3 KO muscles relative to WT at P14. Similar trends were observed at P28 (fig. S3) and also in adult, mature skeletal muscles in both male and female mice (Fig. 1C and fig. S4).
Fig. 1. α-Actinin-3 deficiency alters protein synthesis and breakdown signaling in skeletal muscle from early postnatal development.
(A) At P7, Actn3 KO quadriceps muscles showed significantly reduced activation of mTOR and 4ebp1 but increased expression of RCAN1-4 compared to WT. (B) At P14, Actn3 KO muscles showed increased PI3Kp85, RCAN1-4, and activation of mTOR and Smad3 compared to WT but reduced p-4ebp1/4ebp1 ratio due to increased total 4ebp1 in Actn3 KO muscles. (C) Increased activation of mTOR and 4ebp1 is maintained in adult Actn3 KO muscles relative to WT. (D) At P28, Actn3 KO muscles showed significant reductions in quadriceps mass relative to body weight compared to WT. (E) Puromycin incorporation (indicator of rate of protein synthesis) is increased significantly higher in male Actn3 KO muscles compared to WT. (F) Protein expression of autophagy markers LC3A and LC3B is similar between WT and Actn3 KO muscles; however, (G) transcript expression of E3 ubiquitin ligase genes Fbxo32 and Trim63 is reduced in Actn3 KO. *P < 0.05, **P < 0.01, and ***P < 0.001, Mann Whitney U test. N = 4 to 9 for all experiments.
We have previously shown that α-actinin-3 deficiency is associated with reduced muscle mass in adult mice (16). To determine how changes in protein synthesis with α-actinin-3 deficiency during development correlate temporally with Actn3 genotype effects on muscle mass, we examined the quadriceps mass of WT and Actn3 KO mice during development. Muscle mass comparisons between genotypes were performed with muscle mass normalized to body weight to account for differences in litter sizes. Genotype difference in muscle mass relative to body weight was not detected at P14 but was significant by P28 in both male and female cohorts (Fig. 1D), suggesting that temporal changes in protein synthesis precede the effects of α-actinin-3 deficiency on skeletal muscle mass.
To further examine the impact of these signaling changes associated with α-actinin-3 deficiency on muscle mass regulation in adult skeletal muscle, we compared the rates of protein synthesis between WT and Actn3 KO muscles in vivo using the nonisotopic surface sensing of translation (SUnSET) technique (41). Analyses of puromycin incorporation over 30 min demonstrated increased protein synthesis in Actn3 KO muscles at baseline in males (Fig. 1E), although this did not reach significance in females (fig. S4). We also examined other markers of autophagy and ubiquitin-proteasome system and found no genotype difference in expression of autophagy markers LC3A and LC3B (Fig. 1F); however, Actn3 KO muscles show a marked down-regulation of Fbxo32 (P = 0.0022) and Trim63 (P = 0.0649) compared to WT, which encode for the E3 ubiquitin ligases atrogin-1 and muscle-specific RING finger protein 1 (MuRF1), respectively (Fig. 1G). Overall, these results suggest that the α-actinin-3 deficiency influences muscle mass and protein synthesis and degradation early during postnatal muscle development, and perturbations of these pathways are maintained in mature adult skeletal muscles in mice.
Actn3 KO muscles show reduced dexamethasone-induced muscle atrophy signaling
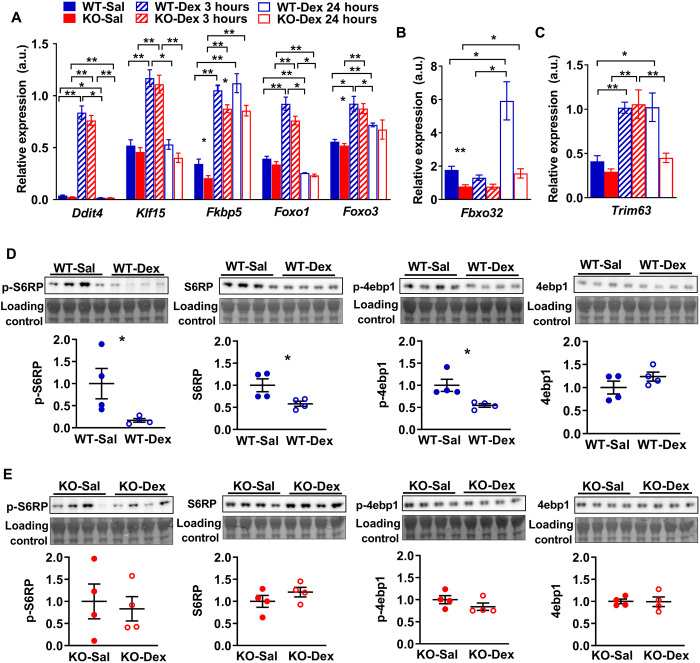
To determine whether the baseline alterations in protein synthesis and breakdown signaling could alter response to muscle wasting in α-actinin-3–deficient muscles, we administered male WT and Actn3 KO animals with a single pulse of dexamethasone (20 mg/kg) by intraperitoneal injection to activate muscle atrophy signaling and examined it at 3 and 24 hours after injection. Since α-actinins reportedly influence the activity of GR, we first assessed the transcriptional response of upstream GR target genes by reverse transcription quantitative polymerase chain reaction (RT-qPCR) in WT and Actn3 KO muscles after GR activation. In both WT and Actn3 KO muscles, treatment with dexamethasone (WT-Dex and KO-Dex) markedly increased the expression of upstream GR target genes Ddit4, Klf15, and Fkbp5, as well as Foxo1 and Foxo3 after 3 hours (Fig. 2A). There was no genotype difference in gene expression for the majority of these genes except for Fkbp5, where Actn3 KO muscles showed lower expression relative to WT muscles regardless of treatment. Despite similar upstream response to GR activation, dexamethasone failed to trigger increases in expression for the E3 ubiquitin ligase gene Fbxo32 in Actn3 KO-Dex muscles 24 hours after injection, while WT-Dex muscles showed ~4-fold increase in Fbxo32 transcript expression relative to WT mice that were given saline (WT-Sal) (Fig. 2B). Similarly, WT-Dex muscles show ~2.5-fold increase in Trim63 transcripts 3 and 24 hours after dexamethasone injection relative to WT-Sal; while KO-Dex muscles also show 3.6-fold increase in Trim63 relative to KO mice given saline (KO-Sal) after 3 hours; the increase in Trim63 transcript expression is not sustained after 24 hours (Fig. 2C). Two-way analysis of variance (ANOVA) showed significant genotype effect on dexamethasone-induced response of Fbxo32 (P = 0.029) and Trim63 (P = 0.007). These data suggest that α-actinin-3–deficient muscles are less responsive to dexamethasone-induced signaling for protein degradation. We further examined the expression of major downstream effectors of mTOR that promote protein synthesis. At 24 hours after dexamethasone injection, WT-Dex muscles showed significant down-regulation of p-S6RP, S6RP, and p-4ebp1 compared to WT-Sal (Fig. 2D), consistent with suppression of protein synthesis signaling. In contrast, KO-Dex muscles showed no significant changes in the expression of these markers relative to KO-Sal (Fig. 2E). In combination, these data indicate that α-actinin-3 deficiency results in reduced signaling for muscle atrophy in response to dexamethasone.
Fig. 2. Acute response to dexamethasone in WT and Actn3 KO mice.
(A) Actn3 KO mice show normal transcriptional response to GR activation at 3 and 24 hours after a single bolus injection of dexamethasone but fail to activate downstream E3 ubiquitin ligase genes (B) Fbxo32 or (C) maintain Trim63 up-regulation at 24 hours after dexamethasone treatment. a.u., arbitrary units. (D) Relative to WT-Sal controls, WT-Dex mice show significant down-regulation of p-S6RP, total S6RP, and p-4ebp1 and no changes to the total 4ebp1 at 24 hours after dexamethasone treatment, consistent with the dexamethasone-induced suppression of protein synthesis signaling. (E) In contrast, Actn3 KO-Dex mice are resistant to these changes and show similar levels relative to KO-Sal controls over the same time period. All transcriptional and protein analyses were performed in the quadriceps muscle. *P < 0.05 and **P < 0.01, Mann Whitney U test. N = 4 to 7 for all experiments.
Global gene expression changes in WT and Actn3 KO muscles after single bolus dexamethasone injection
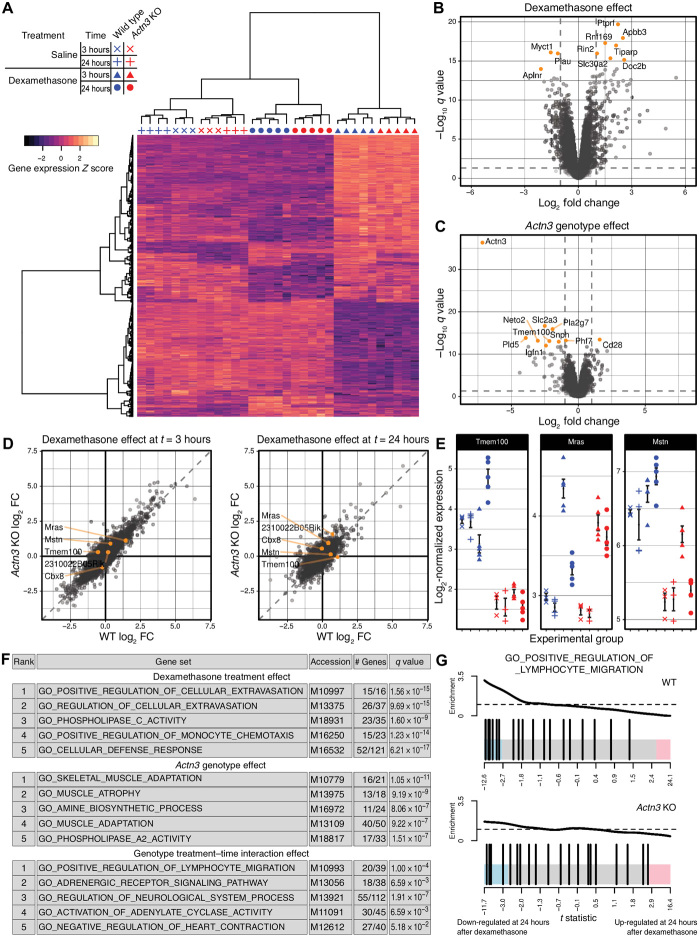
We further performed unbiased transcriptomic profiling to determine the acute global gene expression changes in response to dexamethasone injection at 3 and 24 hours that are influenced by Actn3 genotype in vivo. Multidimensional scaling analysis of the filtered and processed count data confirmed the experimental grouping of gene expression by genotype, treatment, and time after dexamethasone treatment (fig. S5). Differential expression testing for the effects of treatment, time after injection, genotype, and genotype interactions with either of the former factors detected a total of 8653 differentially expressed genes with q < 0.05 across at least one test. Of these, 1416 genes were considered robustly regulated, with both q < 0.05 and a fold change greater than 2 for at least one test. Testing confirmed effective transcript-level knockout (KO) of Actn3 expression in Actn3 KO relative to WT (q = 4.44 × 10−37; fig. S6).
The differences in expression of the 1416 robustly regulated genes are presented on a heatmap using genewise centring and scaling and showed that dexamethasone exhibits its largest influence upon transcription at 3 hours after injection in both WT and Actn3 KO (Fig. 3A). Volcano plots showed that both dexamethasone treatment (Fig. 3B) and Actn3 genotype (Fig. 3C) individually influence the transcriptional response. Although correlation plots of dexamethasone response at 3 and 24 hours suggest that WT and Actn3 KO mice have a broadly similar response to dexamethasone treatment (Fig. 3D), specific differential expression testing for interaction effects of Actn3 genotype on dexamethasone response detected five genes of interest, all of which are related to signal transduction pathways associated with muscle growth or wasting or immune phenotypes: Tmem100 (q = 0.00623), Cbx8 (q = 0.049), Mras (q = 0.049), 2310022B05Rik (q = 0.0550), and Mstn (q = 0.1238). Plotting of the normalized expression of Tmem100, Mras, and Mstn (which encodes for myostatin) highlight the Actn3-mediated difference in expression of these genes in response to dexamethasone (Fig. 3E).
Fig. 3. Transcriptomic analysis of quadriceps muscles from male WT and Actn3 KO mice injected with saline or dexamethasone after 3 or 24 hours.
(A) Heatmap of robustly regulated genes highlights the magnitude of response to dexamethasone treatment after 3 hours. Volcano plots of the effects of dexamethasone treatment (B) and Actn3 KO (C) demonstrate the efficacy of both interventions and the scale of transcriptional responses. Vertical dashed lines represent a log2 fold change (FC) of −1 or 1, and horizontal dashed lines represent a value of q = 0.05. (D) Correlation plots show that both Actn3 KO and WT mice exhibit broadly similar transcriptional responses to dexamethasone at 3 and 24 hours after injection. Genes with observed q < 0.15 for an interaction test between the treatment, time after injection, and genotype factors are highlighted in orange. (E) The log2-normalized expression of a selection of these genes highlights the difference in dexamethasone response between WT and Actn3 KO (means ± SEM). Symbols represent individual mice. (F) Gene set enrichment analysis describes a broadly immunosuppressive characterization of the dexamethasone response and known skeletal muscle phenotypes of Actn3 KO; gene sets showed the down-regulation in the dexamethasone-treated and Actn3 KO animals, respectively. (G) Barcode plots suggest greater down-regulation within GO_POSITIVE_REGULATION_OF_LYMPHOCYTE_ MIGRATION, the top ranked gene set for the interaction effect, in WT but not Actn3 KO. Enrichment is a relative measure compared to a uniform distribution of t statistics within the gene set, represented by the dashed horizontal line. (For each genotype, n = 3 for saline and n = 5 for dexamethasone-treated groups)
Gene set enrichment analysis following differential expression testing further characterized the overall response to dexamethasone with respect to Actn3 genotype (Fig. 3F). Analysis showed that enrichment of gene ontology terms linked to immune response (“regulation of cellular extravasation,” “regulation of monocyte chemotaxis,” and “cellular defence response”) were associated with the dexamethasone effect, while gene ontology terms such as “muscle adaptation” and “muscle atrophy” were associated with the Actn3 genotype effect. There was also evidence for a differential response to dexamethasone with Actn3 genotype over time, with gene ontology terms “positive regulation of lymphocyte migration” and “adrenergic receptor signaling pathway” highlighted as ranked by strength of evidence. Barcode plots for “positive regulation of lymphocyte migration” for WT and Actn3 KO (Fig. 3G) indicate suppression of genes related to this pathway in the WT but not in Actn3 KO, suggesting that, in the absence of α-actinin-3, skeletal muscles exhibit a diminished immunosuppressive response to dexamethasone.
Acute dexamethasone treatment increases expression of muscle atrophy–related genes in Actn3 KO muscles following ACTN3 replacement
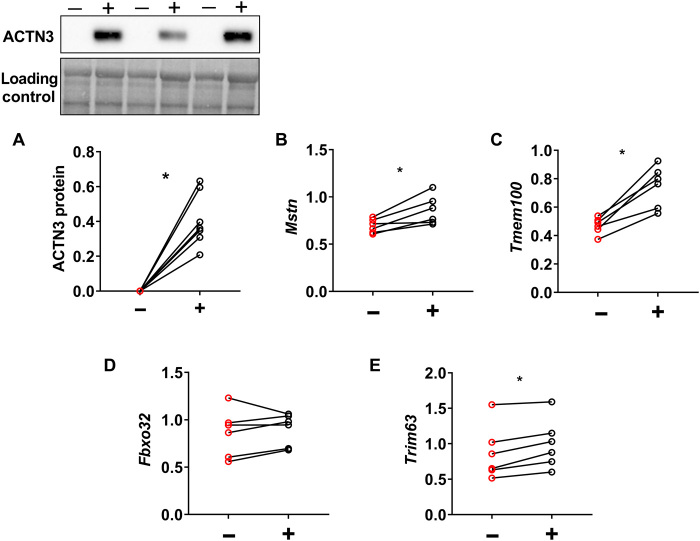
To confirm the role of α-actinin-3 in the altered response to dexamethasone in skeletal muscle, we assessed the dexamethasone-induced changes in Mstn, Tmem100, Fbxo32, and Trim63 expression in Actn3 KO muscles following replacement of the ACTN3 gene (rescue). ACTN3 replacement was achieved via recombinant adeno-associated viral vector mediated delivery, driven by the cytomegalovirus promoter (rAAV-CMV-ACTN3); this was injected into the tibialis anterior (TA) muscle of Actn3 KO mice of one leg and an empty control vector into the TA of the contralateral leg. When stable ACTN3 expression has been achieved after 4 weeks, mice were further treated with dexamethasone by intraperitoneal injection (20 mg/kg), and TA muscles were harvested after 24 hours (Fig. 4A). Comparison of control (−) and corresponding ACTN3 expressing muscles (+) within each Actn3 KO mouse by RT-qPCR showed that α-actinin-3–expressing muscles significantly increased the transcriptional expression of Mstn and Tmem100 24 hours after dexamethasone injection (Fig. 4, B and C), confirming that these genes mediate the altered dexamethasone response associated with α-actinin-3 deficiency. Similarly, expression of Fbxo32 and Trim63 were also up-regulated following α-actinin-3 replacement in Actn3 KO mouse muscles (Fig. 4, D and E), while protein synthesis marker p-4ebp1 and p-Akt were down-regulated (fig. S7).
Fig. 4. Acute dexamethasone treatment increases expression of muscle atrophy-related genes in Actn3 KO muscles following ACTN3 replacement.
Male Actn3 KO mice were injected with rAAV-CMV-ACTN3 into the TA muscle on one leg (+) and empty vector (−) into contralateral leg. After 4 weeks of vector expression, mice were given a single dose of dexamethasone at 20 mg/kg. TA muscles were harvested for RT-qPCR analyses after 24 hours. (A) Replacement of ACTN3 in Actn3 KO muscles increased expression of (B) Mstn, (C) Tmem100, (D) Fbxo32 (in five of the six mice), and (E) Trim63 24 hours after dexamethasone administration. *P < 0.05, Wilcoxon test. n = 6.
α-Actinin-3 deficiency protects against dexamethasone-induced muscle wasting in female mice
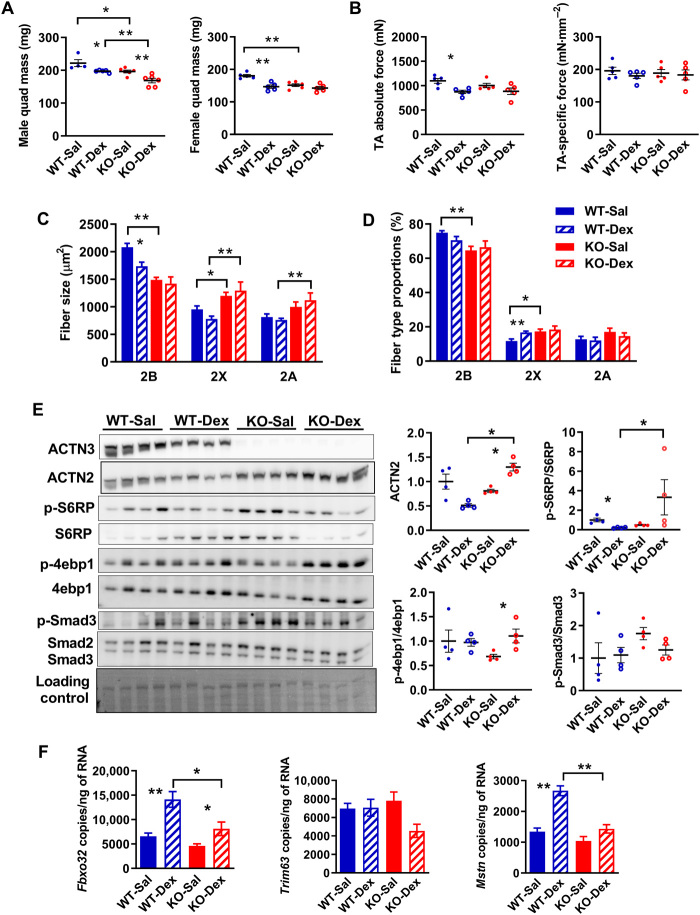
To determine whether the differential acute response to dexamethasone alters the induction of muscle wasting in α-actinin-3–deficient muscles, we administered WT and Actn3 KO animals with dexamethasone (20 mg/kg per day) by intraperitoneal injection daily for 2 weeks to induce muscle atrophy. Since glucocorticoids can have sexually dimorphic actions (42), both male and female mice were examined. In males, WT and Actn3 KO mice showed a similar atrophic response to dexamethasone in the quadriceps muscles; WT-Dex mice showed 11.1% atrophy relative to saline-treated mice, while KO-Dex showed 14.1% atrophy (Fig. 5A). In contrast, a differential genotype response in muscle atrophy was observed in female mice, with female WT-Dex showing significant muscle atrophy relative to WT-Sal in (−18.6%, P = 0.0079), while female KO-Dex showed only minimal loss of quadriceps mass relative to KO-Sal (−5.8%, P = 0.1775). These results were replicated in separate male and female cohorts (fig. S8). Two-way ANOVA confirmed the presence of a significant genotype effect on response to dexamethasone for quadriceps mass in females (P = 0.0045).
Fig. 5. Female Actn3 KO mice show resistance to muscle wasting induced by prolonged treatment with dexamethasone.
(A) Male WT and Actn3 KO mice show similar levels of muscle atrophy in the quadriceps following 2 weeks of daily dexamethasone administration. In contrast, female Actn3 KO mice showed minimal muscle wasting compared to WT following dexamethasone treatment. (B) Female WT-Dex TA muscles showed significant decrease in maximal force relative to WT-Sal; specific force is similar regardless of genotype and treatment. (C) Female WT-Dex muscles showed reduced fast 2B fiber size compared to WT-Sal and a small increase in 2× fiber proportion (D); there was no change in fiber size or proportions in KO-Dex relative to KO-Sal. (E) KO-Dex muscles show increased activation of protein synthesis markers S6RP and 4ebp1 and a trend for decreased Smad3 activity relative to KO-Sal. (F) Transcriptional changes in Fbxo32, Trim63, and Mstn were assessed by ddPCR in WT and Actn3 KO mice following 7 days of daily dexamethasone injection. WT-Dex muscles showed increased activation of Fbxo32 and Mstn relative to WT-Sal; KO-Dex muscles showed reduced expression of these genes compared to WT-Dex. Scale bar, 50 μm. *P < 0.05 and **P < 0.01 (Mann-Whitney U test). Western blot quantitation values are normalized to the total protein and expressed relative to WT-Sal.
Assessment of force generation in the TA muscles of female mice also showed significant reductions in WT-Dex relative to WT-Sal (−20.5%, P = 0.0159), while KO-Dex showed only minimal decrease in maximal force (−7.1%, P = 0.4286) (Fig. 5B). Fiber size analyses of the quadriceps from female mice showed that muscle atrophy in response to dexamethasone was restricted to fast 2B fibers in both WT and Actn3 KO muscles, with the decrease in fast 2B fiber size proportional to the loss in quadriceps mass (WT: −18.9%, P = 0.0317; KO: −6.6%, P = 0.5368) (Fig. 5C). There was no change in the total fiber number (fig. S9), but WT-Dex muscles showed a small increase in 2X fiber proportion (1.65%, P = 0.0079) relative to WT-Sal (Fig. 5D).
Markers of protein synthesis signaling (4ebp1 and S6RP) and myostatin activation (Smad3) were further analyzed (Fig. 5E). Compared to WT-Sal, WT-Dex muscles show a small but significant reduction in S6RP phosphorylation, suggesting a reduced protein synthesis signaling. In contrast, KO-Dex muscles show marked and significant increases in 4ebp1 phosphorylation, as well as a trend for decreased Smad3 phosphorylation.
Since myostatin expression returns to basal levels after 10 days of dexamethasone administration (43), a separate cohort of female mice were injected with dexamethasone daily for 7 days and assessed for transcriptional changes in atrophic signaling (Fig. 5F). Compared to WT-Sal, WT-Dex muscles showed significant increases in Fbxo32 and Mstn expression but not Trim63, while KO-Dex muscles showed a reduced activation of these genes compared to WT-Dex and a trend for reduced Trim63 compared to KO-Sal. Significant genotype effect on dexamethasone-induced response was demonstrated for Mstn (P = 0.004), consistent with decreased dexamethasone-induced muscle wasting associated with α-actinin-3 deficiency.
DISCUSSION
α-Actinin-3 regulates protein synthesis and breakdown in skeletal muscle from early development
Maintenance of muscle mass requires a balance of protein synthesis and degradation (44). In this study, we demonstrate that the loss of α-actinin-3 alters some of the major pathways involved from early postnatal development through to maturity and thereby regulates skeletal muscle mass during growth and adaptation (Fig. 6). The first 3 weeks of postnatal mouse development is a pivotal period of intense muscle growth (40). Our results show initial decreases in p-mTOR and total mTOR at P0 and P7 and the downstream protein synthesis effector 4ebp1 at P7 in α-actinin-3–deficient muscles. Increased calcineurin activity due to α-actinin-3 deficiency (and α-actinin-2 up-regulation) (18) is also detectable from P7. However, from P14, mTOR activation is increased in Actn3 KO muscles, as is the phosphorylation of the transcription factor Smad3, which mediates myostatin-induced atrophic signaling (39). The emergence of this switch in mTOR and Smad3 signaling coincides with the rapid increase in myosin heavy chain 2B expression in mouse skeletal muscle between P7 and P14 (45) and increased myofibrillar protein expression with α-actinin-3 deficiency at P14 (38), suggesting that structural changes associated with α-actinin-3 deficiency may be driven in part by changes in these pathways at this critical juncture in muscle development. The changes seen at P14 persist through to P28 and into maturity, leading to the significant reduction in muscle mass in both male and female α-actinin-3–deficient mice that is detectable from P28.
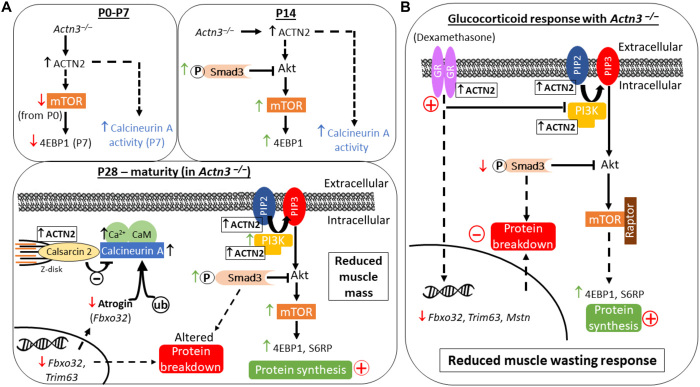
Fig. 6. Summary schematic of the effect of α-actinin-3 deficiency on muscle mass regulation and the adaptive response to dexamethasone-induced muscle wasting.
(A) In the absence of α-actinin-3, α-actinin-2 is up-regulated to maintain the total sarcomeric α-actinin pool in fast-twitch muscle fibers (38). Differences in α-actinin-2 expression between genotypes are significant from P7. Decreases in p-mTOR and the total mTOR occur from P0, which subsequently lead to decreases in p-4ebp1 and the total 4ebp1 at P7. These are reversed at P14, and increased p-Smad3 is also observed. Increased calcineurin activity [as shown by increased RCAN1-4 expression (18)] occurs from P7. Changes in downstream mTOR and Smad3 signaling in α-actinin-3–deficient muscles persist through to P28 into maturity, resulting in an overall increase in rates of protein synthesis as measured by puromycin incorporation. α-Actinin-3 deficiency also decreases the transcriptional expression of E3 ubiquitin ligase genes Fbxo32 and Trim63 in muscle. Decreased Fbxo32 levels with α-actinin-3 deficiency are consistent with increased calcineurin signaling (18), since atrogin-1 targets calcineurin for ubiquitin-mediated proteolysis. These alterations in various protein breakdown pathways along with increased protein synthesis culminate in reduced muscle mass with α-actinin-3 deficiency, which is detectable from P28. (B) Despite similar activation of GR by dexamethasone, α-actinin-3–deficient muscles show reduced muscle wasting response due to the inherent baseline effects on protein synthesis and breakdown pathways.
Transcript expression of the E3 ubiquitin ligase genes, atrogin-1 and MuRF1, which promote protein degradation, is also reduced with α-actinin-3 deficiency, consistent with the observed increase in calcineurin activity from P7. In striated muscle, atrogin-1 is known to repress calcineurin activity by targeting calcineurin for ubiquitin-mediated proteolysis, and suppression of atrogin-1 has been shown to increase calcineurin activity in cardiomyocytes (29). α-Actinin-2 also directly associates with atrogin-1 at the Z-disk; although α-actinin-2 and calcineurin bind to atrogin-1 on topologically adjacent interaction domains, only calcineurin is targeted for ubiquitination (29), suggesting that α-actinin-2 serves as a dock for recruiting atrogin-1 into the protein complex. In combination, our data suggest that sarcomeric α-actinins play a critical role in regulating muscle protein synthesis and degradation and the homeostatic maintenance of healthy muscle mass from early development through a combination of their effects on downstream mTOR and Smad3 signaling, reduced atrogin-1 and MuRF1, and regulation of calcineurin activity.
α-Actinin-3 deficiency attenuates the dexamethasone-induced atrophic response independently of GR activation and protects against muscle wasting
Evaluation of the GR target genes Ddit4, Klf15, and Fkbp5 following acute dexamethasone (32) and downstream transcription factors Foxo1 and Foxo3, which act cooperatively with Klf15 to up-regulate atrogin-1 and MuRF-1 (32, 46), indicates that initial GR activation is comparable between genotypes. However, the transactivation response for atrogin-1 and MuRF-1 was different between WT and Actn3 KO muscles at 24 hours, with α-actinin-3–deficient muscles showing markedly lower expression of both genes. These results suggest that, in the absence of α-actinin-3, muscles are unable to sustain the dexamethasone-induced transcriptional activation of these E3 ubiquitin ligase genes and that this occurs independently of GR activation. We verified this to be the case by the postnatal replacement of ACTN3 in Actn3 KO mice, which led to increased expression of atrogin-1 and MuRF1 24 hours after dexamethasone treatment. In addition, there was no suppression of downstream mTOR protein synthesis effectors in muscles from dexamethasone-treated Actn3 KO mice, consistent with an attenuated atrophic response. Muscles from female Actn3 KO mice, given prolonged dexamethasone treatment likewise, showed reduced atrophic signaling (Smad3, Fbxo32, Trim63, and Mstn) and enhanced protein synthesis and were protected from muscle wasting.
Protection against muscle wasting with α-actinin-3 deficiency is also mediated by the suppression of myostatin signaling
Myostatin is known to mediate dexamethasone-induced muscle atrophy (43), which involves synergistic activation of Akt-FoxO signaling, inhibition of IGF1-PI3K-Akt signaling, and increased expression of atrogin-1 mediated by enhanced Smad2/3 signaling (47). Deletion of myostatin prevents muscle atrophy in glucocorticoid-treated mice and blunts induction of proteolytic genes atrogin-1 and MuRF1 (48). In this study, we showed that α-actinin-3 expression is required for the maintenance of myostatin induction. Acute dexamethasone treatment induced increases in Mstn in WT muscles that were sustained at 24 hours, while Mstn levels returned to basal levels in Actn3 KO muscles after 24 hours. Similar results were observed following 7 days of daily treatment. This direct association between the α-actinin-3 expression and the maintenance of Mstn induction was confirmed by increased Mstn levels after 24 hours following rescue in of Actn3 KO muscle with ACTN3. Unbiased transcriptomic profiling of WT and Actn3 KO muscles following dexamethasone injection showed similar results for Tmem100, a gene encoding an intracellular transmembrane protein that is up-regulated during activin-A–induced muscle wasting (which acts via the myostatin signaling pathway) (49).
Sexual dimorphic effect of α-actinin-3 deficiency on the response to dexamethasone-induced muscle wasting
The protection against dexamethasone-induced muscle wasting with α-actinin-3 deficiency specifically in female, but not male, mice is intriguing, given that similar changes in baseline protein synthesis and breakdown signaling were observed in both male and female Actn3 KO mice relative to WT. Dexamethasone has been shown to differentially affect gene expression in the livers of male and female rats (42), and this may be due to cross-talk between GR and other members of the steroid nuclear receptor subfamily, since both estrogen and androgen signaling modulates GR activity in various cell types and tissues (50, 51). α-Actinin-2, which is up-regulated in α-actinin-3 deficiency, has been shown to influence androgen and estrogen receptor activities in vitro (34). The effects of ACTN3 R577X on muscle performance in humans also vary with gender; the impact of α-actinin-3 deficiency is more pronounced in elite female athletes compared to males (2). The effect of α-actinin-3 deficiency on sex hormone signaling in skeletal muscle, and how this intersects with atrophic response, requires further study. Gender differences in muscle wasting response have previously been reported in other models. A transgenic mouse model that specifically overexpressed myostatin in skeletal muscle resulted in moderate muscle atrophy (20%) only in males but not females (52).
ACTN3 genotype influences the anti-inflammatory and muscle wasting response to glucocorticoid
In addition to reduced atrophic signaling, gene set testing demonstrated a diminished immunosuppressive response to dexamethasone with α-actinin-3 deficiency. Two of the five genes of interest that showed significant differential response to dexamethasone between Actn3 genotypes—2310022B05Rik and Cbx8 (chromobox homolog 8)—are associated with immune phenotypes. In particular, Cbx8 is a known GR-responsive gene. In human lung adenocarcinoma cells, activation of GR with dexamethasone results in down-regulation (transrepression) of Cbx8 through direct binding of GR to its negative glucocorticoid response element (53). Given that GR transrepression activity is generally associated with anti-inflammatory activity and clinical efficacy of glucocorticoids (54, 55), these results suggest that ACTN3 R577X acts as a pharmacogenetic variant influencing both the anti-inflammatory and muscle wasting response to glucocorticoids in skeletal muscles.
Implications for ACTN3 577X–positive selection
The ACTN3 577X allele is thought to have arisen before the appearance of modern humans in Europe and Asia between 40,000 and 60,000 years ago and positively selected due to its enhancing effect on muscle metabolic efficiency (17). Phylogenetic analyses indicated that the ACTN3 577X allele rapidly increased in frequency in various populations in adaptation to increased species richness and temperature related to latitudinal changes (56). Recent studies in the Actn3 KO mouse further suggested that α-actinin-3 deficiency enhances muscle fatigue resistance and thermogenic mechanisms through greater calcium pumping and release as a result of increased SERCA expression, and this could be advantageous for populations that migrate to and reside in cold environments during recent evolution (21).
The present finding that α-actinin-3 deficiency results in reduced muscle expression of atrogin-1 and MuRF1 at baseline and decreased activation following dexamethasone treatment suggests that protection of muscle mass may also contribute to the positive selection of the ACTN3 577X allele. Since up-regulation of these atrogenes is common in various muscle disuse models such as denervation, immobilization, sepsis, and fasting/starvation (57), reduced activation of these genes in response to various environmental challenges could be advantageous during modern human migration. An altered muscle adaptive response to denervation and immobilization with α-actinin-3 deficiency, in part mediated by enhanced calcineurin activity, has previously been shown in Actn3 KO mice (22). Similarly, reduced activation of myostatin, which is functionally conserved through evolution (58), is also consistent with increased preservation of muscle mass with α-actinin-3 deficiency that could be beneficial for modern humans. Together, these data provide an additional mechanistic explanation for the drive toward the increased prevalence of the ACTN3 577X allele in recent human history.
In conclusion, this study shows that α-actinin-3 plays a role in the regulation of muscle mass through modulating muscle protein synthesis and breakdown signaling and that ACTN3 R577X may influence the glucocorticoid-induced anti-inflammatory and muscle atrophy signaling response. α-Actinin-3 deficiency, which occurs in ~ 1 in 5 people worldwide (1), protects against dexamethasone-induced muscle wasting, but only in females, suggesting that sex hormone signaling also influences the effect of ACTN3 genotype on the response to prolonged glucocorticoid use.
MATERIALS AND METHODS
Animals and ethics
Analyses were performed in muscles from male and female C57BL/6J mice aged P0, P7, P14, P28, 12 weeks, and 4 to 5 months. Experiments involving 1 or 2 weeks dexamethasone treatment used male and female mice on the C57BL/10ScSn background (aged 4 and 10 to 12 months). Mice were fed standard chow and water ad libitum and were maintained in a 12-hour light/12-hour dark cycle at ambient room temperature (∼22°C). All experiments were approved by the Animal Ethics Committee of the Murdoch Children’s Research Institute.
Dexamethasone administration
Mice were administered with dexamethasone at 20 mg/kg per day or saline by intraperitoneal injection either daily for 1 or 2 weeks (chronic response) or with a single bolus injection (acute response; mice were then euthanized at 3 or 24 hours after injection). Hindlimb muscles [TA, extensor digitorum longus (EDL), gastrocnemius, soleus, and quadriceps] were harvested and snap-frozen in liquid nitrogen or cryopreserved in optimal cutting temperature reagent.
rAAV
Eight adult male C57BL6 Actn3 KO mice were anesthetized (3.5% isoflurane in oxygen) and given temgesic (0.05 mg/kg) for pain management. Hamilton syringes (Hamilton) were used to deliver 30 μl of rAAV6-CMV-ACTN3 (20) or rAAV6-MCS (control empty vector with multiple cloning site) diluted in Hanks’ balanced salt solution at a dose of 1e10 vector genomes. Vectors were injected into the anterior compartment of the lower hindlimb targeting the TA and the EDL.
Muscle physiology
Force analyses were performed on the TA muscles from mice treated with dexamethasone or saline for 2 weeks using the Aurora Scientific Dual-Mode Lever System with supplied software (DMC5 4.5, DMA) and carried out at 37°C as previously described (20).
Immunoblotting
Snap-frozen quadriceps muscles were homogenized in 2% SDS lysis buffer and assessed for the total protein concentration using Direct Detect (Thermo Fisher Scientific). Proteins were separated by SDS–polyacrylamide gel electrophoresis using precast midi-criterion gels (Bio-Rad) and then transferred to polyvinylidene fluoride membranes (Millipore). These were blocked with 5% bovine serum albumin in 1× TBST (tris-buffered saline containing 0.1% Tween 20) and probed overnight at 4°C with primary antibodies against α-actinin-3, α-actinin-2 (ab68204 and ab68167, Abcam), PI3Kp85 [no. 4257, Cell Signaling Technology (CST)], Akt1 (no. C73H10, CST), p-Akt (Ser463) (no. 4060, CST), mTOR (no. 2983, CST), p-mTOR (no. 5536, CST), p-4ebp1 (no. 2855, CST), 4ebp1 (no. 9452, CST), S6RP (no. 2217, CST), p-S6RP (no. 4856, CST), Smad2/3 (no. 8685, CST), and p-Smad3 (ab52903; Abcam). Blots were washed, then probed with secondary antibodies at room temperature for 1 to 2 hours, and then developed with enhanced chemiluminescent reagents (Amersham Biosciences). Membranes were washed, developed, and imaged using ImageQuant (GE Healthcare). Densitometry was performed using the ImageJ image processing software (National Institutes of Health) and quantified using the area under the curve. Some analyses were performed or verified using an automated Western blotting technique (Wes, ProteinSimple). Results were normalized to loading control derived from GelCode Blue (Thermo Fisher Scientific) staining of membranes, or α-skeletal actin (5C5; Sigma-Aldrich), and presented relative to WT control.
Surface sensing of translation
SUnSET was performed as previously described previously (41). Male and female mice aged 12 weeks were given either a single bolus of puromycin dihydrochloride/phosphate-buffered saline (PBS) solution (0.04 μmol/g; Calbiochem) or PBS alone by intraperitoneal injection. Animals were euthanized at 30 min after injection. The TA muscles were dissected and immediately snap-frozen (right leg) and stored at −80°C. To determine the rate of protein synthesis, puromycin incorporation was determined by western blot using a puromycin specific antibody (12D10; Merck).
Fiber morphometry analysis
Fiber typing was performed as previously described (59). Sections were imaged on the VSlide Scanner (MetaSystems) and analyzed using MetaMorph software (Molecular Devices).
RNA extractions and cDNA synthesis
Total RNA was extracted from ~50 mg of mouse quadriceps by phenol chloroform extraction (1 ml of TRIsure solution; Bioline). RNA was purified using the RNeasy Mini Kit (QIAGEN) as per the company’s protocol and eluted in 30 μl of Milli-Q water. RNA integrities [RNA integrity number (RIN) value] and total RNA concentrations were then measured using TapeStation (Agilent Technologies 2200), and samples with RINs of 8.5 to 9.2 were used for further RT-qPCR, droplet digital polymerase chain reaction (ddPCR), and RNA sequencing analyses. Quantification of diluted RNA concentrations was performed using Qubit 3.0 fluorometer (Thermo Fisher Scientific). All RNA samples were further diluted in RNA (25 and 1 ng/μl) and 4 μl of RNA (25 ng/μl) or 2 μl of RNA (1 ng/μl) samples and reverse transcribed to synthesise cDNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) as per manufacturer guidelines.
ddPCR and analyses
ddPCR assay was conducted using 2× QX200 ddPCR EvaGreen Supermix (Bio-Rad) in a twin.tec 96-well plate (Bio-Rad) to a final volume of 24 μl for lipid droplet generation. The plate was heat-sealed at 180°C for 5 s with foil (PX1 Plate Sealer, Bio-Rad) and centrifuged at 4000 revolutions per minute for 1 min before loading into an Automated Droplet Generator (Bio-Rad), where each sample was partitioned into 20,000 uniform sized lipid droplets with the transcripts of interest and cDNA distributed randomly into the droplets. The sample plate of droplets was placed in a thermal cycler (T100, Bio-Rad) for subsequent PCR amplification within each droplet. The thermal cycling conditions were as follows: one activation cycle of 5 min at 95°C, 40 denaturation cycles of 30 s at 96°C and annealing cycles of 1 min at 55° to 60°C depending on the target gene of interest, and a post-cycling step of signal stabilization of 5 min at 4°C followed by 5 min at 90°C. All cycling steps were performed using a 2°C per second ramp rate. Following PCR amplification, the sample plate was loaded on the QX200 Droplet Reader (Bio-Rad), and the assay information was entered into the software QuantaSoft (Bio-Rad). Droplets were analyzed by the droplet reader in the EvaGreen fluorescent channel. The fraction of positive droplets in a sample were counted and fitted to a statistical Poisson algorithm to provide plot readouts of absolute quantification and concentration of the transcript of interest. Samples with lipid droplet counts less than 10,000 were rejected. The one-dimensional amplitude plot of fluorescence amplitude was used to determine the data quality based on positive and negative droplets separation, droplet scattering, and channel amplitude signals (minimum of ~5400). A baseline threshold was applied equally across all samples depending on the fluorescence amplitude to distinguish positive from negative droplets. Readings of the target were adjusted for RNA loading and presented as absolute values of RNA transcript copies/ng of RNA.
RT-qPCR
cDNA was generated using 500 ng of total RNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) as per manufacturer guidelines. RT-qPCR was performed in triplicate with the LightCycler 480 instrument (Roche Diagnostics) and LightCycler 384-well plates with sealing foil (Roche Diagnostics). Reaction volume of 10 μl contains 2× SensiFAST SYBR No-ROX mix (Bioline), 0.4 μl of each 10 μM forward and reverse primers, and 1 μl of 1:10 diluted cDNA. Amplification of the single PCR product was confirmed using the melting point dissociation curve, and the Cp values were calculated using the LightCycler 480 software. For each gene, a standard curve was generated using a WT-Sal sample (sample cDNA neat: 1 in 5, 1 in 10, and 1 in 100) and used to convert the Cp values to relative gene expression for all unknown samples. Gene expression is further normalized to the geomean of three housekeepers Aldoa, Rer1, and Rpl7L1 (60). Primers for qPCR and ddPCR reactions are as follows: Tmem100 (forward: CTTTCCCAGAAGTTGAACG and reverse: TGCAAGCTCACAGAAAGG), Klf15 (forward: CCCTTTGCCTGCACCTGG and reverse: TGGTACGGCTTCACACCC), Ddit4 (forward: CTGGAGAGCTCGGACTGC and reverse: CCCATCCAGGTATGAGGAG), Fkbp5 (forward: GCCATCGTGAAAGAGAAGGG and reverse: CAGCCAGGACACTATCTTCC), Foxo1 (forward: ACGAGTGGATGGTGAAGAGC and reverse: GGACAGATTGTGGCGAATTG), Foxo3 (forward: ATAAGGGCGACAGCAACAGC and reverse: CGTGCCTTCATTCTGAACG), Fbx032 (forward: ATTCTACACTGGCAGCAGCA and reverse: TCAGCCTCTGCATGATGTTC), Trim63 (forward: ACCTGCTGGTGGAAAACATC and reverse: AGGAGCAAGTAGGCACCTCA), Mstn (forward: GTTCATGCTGATTGCTGCTG and reverse: CACGCACATGCATTACACAG), Actn3 (forward: TTCAACCACTTTGACCGGAA and reverse: CACCATGGTCATGATTCGAG), Aldoa (forward: ACATTGCTGAAGCCCAACAT and reverse: ACAGGAAAGTGACCCCAGTG), Rer1 (forward: GCCTTGGGAATTTACCACCT and reverse: CTTCGAATGAAGGGACGAAA), and Rpl7L1 (forward: ACGGTGGAGCCTTATGTGAC and reverse: TCCGTCAGAGGGACTGTCTT).
RNA sequencing analysis
RNA extracted from quadriceps tissue from a total of 17 WT and 16 KO male mice across treatment groups (Sal/Dex; at t = 3 or 24 hours) underwent RNA sequencing using the Illumina HiSeq 2500 Sequencing System per manufacturer’s instructions. Raw read data were processed using the Illumina BaseSpace RNA Express application per standard operating instructions. Reads were aligned using STAR (spliced transcripts alignment to a reference) ultrafast RNA-seq aligner (61) and then counted using HTSeq (62). Analysis of the resultant genewise count data was performed using the R statistical programming language (version 3.5.0). The dataset was filtered for genes registering more than 0.5 counts per million in at least two samples. This resulted in retaining 13,097 genes for analysis. Count data were then normalized and processed for further analyses using the voom function in the Limma software package version 3.36.2 (63). Normalized gene expressions were fit to a linear model, and differential expression was tested using empirical Bayes methods. For the purposes of differential expression testing, samples taken from animals injected with saline at t = 3 and 24 hours after injection were treated as a single saline-injected grouping. Multidimensional scaling plots and hierarchical cluster analysis both failed to detect a robust difference between these time points, validating the decision to pool sample groups for increased statistical power. The false discovery rate of multiple comparisons was controlled using Benjamini and Hochberg’s correction (q value). Genes with a q <0.05 and a fold change of >2 for any contrast were considered robustly regulated and hierarchically clustered using a correlation distance metric. Expression data were characterized using the fast preranked implementation of the gene set enrichment analysis method (64). Gene set tests were conducted using the mouse ortholog conversions of Hallmark and C2-curated gene sets from the molecular signatures database (MSigDB 3.0) (65).
Statistics
Analyses were performed in Graphpad Prism (V7, Graphpad Software Inc.) and StataSE (StataCorp). As group sizes consisted of <12, two-sided, unpaired t tests using nonparametric statistics, Mann-Whitney U tests were applied using an α of 0.05 for all analyses involving independent samples and Wilcoxon signed-rank test for dependent samples. Data were presented as individual points with the calculated group mean (line) or as bar graphs with ± SEM error bars for each group.
Acknowledgments
We thank S. Donath for the assistance with the statistical analyses. Funding: This work is supported by the Australian National Health and Medical Research Council (NHMRC) project grant (APP1130215) awarded to J.T.S., P.G., and K.N.N. K.N.R. is supported by a NHMRC Dora Lush Postgraduate Scholarship (GNT1114935). M.S. is supported by an Australian Government Research Training Program (RTP) Scholarship. K.G.R.Q. is a Scientia Senior Lecturer at UNSW Sydney. Author contributions: J.T.S., P.G., and K.N.N. designed the study. J.T.S. performed all the animal injections. L.B. and P.J.H. assisted with the injections in replication cohorts. J.T.S., K.N.R., and L.R.M. optimized and performed the western blot experiments. L.R.M. and S.F.L. performed the developmental studies. J.T.S. and K.N.R. performed the puromycin experiments. H.D.W. performed the final transcriptomic analyses. M.S. and K.G.R.Q. performed the early transcriptomic analyses. J.T.S. performed the rAAV studies, muscle physiology, and fiber type analyses. J.T.S., P.J.H., and C.T. performed the RT-qPCR and ddPCR analyses. J.T.S. and K.N.N. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability statement: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials and have been deposited in NCBI’s Gene Expression Omnibus (66), which are accessible through the GEO Series accession number GSE169571 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169571). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabg0088/DC1
REFERENCES AND NOTES
- 1.North K. N., Yang N., Wattanasirichaigoon D., Mills M., Easteal S., Beggs A. H., A common nonsense mutation results in alpha-actinin-3 deficiency in the general population. Nat. Genet. 21, 353–354 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Yang N., MacArthur D. G., Gulbin J. P., Hahn A. G., Beggs A. H., Easteal S., North K., ACTN3 genotype is associated with human elite athletic performance. Am. J. Hum. Genet. 73, 627–631 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarkson P. M., Devaney J. M., Gordish-Dressman H., Thompson P. D., Hubal M. J., Urso M., Price T. B., Angelopoulos T. J., Gordon P. M., Moyna N. M., Pescatello L. S., Visich P. S., Zoeller R. F., Seip R. L., Hoffman E. P., ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. J. Appl. Physiol. 99, 154–163 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Lee F. X., Houweling P. J., North K. N., Quinlan K. G., How does α-actinin-3 deficiency alter muscle function? Mechanistic insights into ACTN3 the ‘gene for speed’. Biochim. Biophys. Acta 1863, 686–693 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Kikuchi N., Yoshida S., Min S.-k., Lee K., Sakamaki-Sunaga M., Okamoto T., Nakazato K., The ACTN3 R577X genotype is associated with muscle function in a Japanese population. Appl. Physiol. Nutr. Metab. 40, 316–322 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Walsh S., Liu D., Metter E. J., Ferrucci L., Roth S. M., ACTN3 genotype is associated with muscle phenotypes in women across the adult age span. J. Appl. Physiol. 105, 1486–1491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanabe K., Murakami H., Iemitsu M., Maeda S., Kuno S., ACTN3 polymorphism affects thigh muscle area. Int. J. Sports Med. 31, 138–142 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Cho J., Lee I., Kang H., ACTN3 gene and susceptibility to sarcopenia and osteoporotic status in older korean adults. Biomed. Res. Int. 2017, 4239648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Judson R. N., Wackerhage H., Hughes A., Mavroeidi A., Barr R. J., Macdonald H. M., Ratkevicius A., Reid D. M., Hocking L. J., The functional ACTN3 577X variant increases the risk of falling in older females: Results from two large independent cohort studies. J. Gerontol. A Biol. Sci. Med. Sci. 66, 130–135 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Ma T., Lu D., Zhu Y. S., Chu X. F., Wang Y., Shi G. P., Wang Z. D., Yu L., Jiang X. Y., Wang X. F., ACTN3 genotype and physical function and frailty in an elderly Chinese population: The Rugao longevity and ageing study. Age Ageing 47, 416–422 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Lucia A., Gomez-Gallego F., Santiago C., Perez M., Mate-Munoz J. L., Chamorro-Viña C., Nogales-Gadea G., Foster C., Rubio J. C., Andreu A. L., Martín M. A., Arenas J., The 577X allele of the ACTN3 gene is associated with improved exercise capacity in women with McArdle’s disease. Neuromuscul. Disord. 17, 603–610 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Sandoval-García F., Petri M. H., Saavedra M. A., Cruz-Reyes C. V., Jara-Quezada L. J., Dávalos-Rodriguez I. P., Salazar-Páramo M., Gámez-Nava J. I., González-López L., García-Iglesias T., Corona-Sánchez E. G., Zavaleta-Muñiz S., Vargas-Ramírez R., Mercado M. V.-D., Martín-Márquez B. T., The ACTN3 R577X polymorphism is associated with inflammatory myopathies in a Mexican population. Scand. J. Rheumatol. 41, 396–400 (2012). [DOI] [PubMed] [Google Scholar]
- 13.De Filippi P., Saeidi K., Ravaglia S., Dardis A., Angelini C., Mongini T., Morandi L., Moggio M., Muzio A. D., Filosto M., Bembi B., Giannini F., Marrosu G., Rigoldi M., Tonin P., Servidei S., Siciliano G., Carlucci A., Scotti C., Comelli M., Toscano A., Danesino C., Genotype-phenotype correlation in Pompe disease, a step forward. Orphanet J. Rare Dis. 9, 102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogarth M. W., Houweling P. J., Thomas K. C., Gordish-Dressman H., Bello L.; Cooperative International Neuromuscular Research Group (CINRG), Pegoraro E., Hoffman E. P., Head S. I., North K. N., Evidence for ACTN3 as a genetic modifier of Duchenne muscular dystrophy. Nat. Commun. 8, 14143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernardez-Pereira S., Santos P. C., Krieger J. E., Mansur A. J., Pereira A. C., ACTN3 R577X polymorphism and long-term survival in patients with chronic heart failure. BMC Cardiovasc. Disord. 14, 90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacArthur D. G., Seto J. T., Chan S., Quinlan K. G., Raftery J. M., Turner N., Nicholson M. D., Kee A. J., Hardeman E. C., Gunning P. W., Cooney G. J., Head S. I., Yang N., North K. N., An Actn3 knockout mouse provides mechanistic insights into the association between alpha-actinin-3 deficiency and human athletic performance. Hum. Mol. Genet. 17, 1076–1086 (2008). [DOI] [PubMed] [Google Scholar]
- 17.MacArthur D. G., Seto J. T., Raftery J. M., Quinlan K. G., Huttley G. A., Hook J. W., Lemckert F. A., Kee A. J., Edwards M. R., Berman Y., Hardeman E. C., Gunning P. W., Easteal S., Yang N., North K. N., Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat. Genet. 39, 1261–1265 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Seto J. T., Quinlan K. G. R., Lek M., Zheng X. F., Garton F., MacArthur D. G., Hogarth M. W., Houweling P. J., Gregorevic P., Turner N., Cooney G. J., Yang N., North K. N., ACTN3 genotype influences muscle performance through the regulation of calcineurin signaling. J. Clin. Invest. 123, 4255–4263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan K. G., Seto J. T., Turner N., Vandebrouck A., Floetenmeyer M., Macarthur D. G., Raftery J. M., Lek M., Yang N., Parton R. G., Cooney G. J., North K. N., Alpha-actinin-3 deficiency results in reduced glycogen phosphorylase activity and altered calcium handling in skeletal muscle. Hum. Mol. Genet. 19, 1335–1346 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Garton F. C., Houweling P. J., Vukcevic D., Meehan L. R., Lee F. X. Z., Lek M., Roeszler K. N., Hogarth M. W., Tiong C. F., Zannino D., Yang N., Leslie S., Gregorevic P., Head S. I., Seto J. T., North K. N., The effect of ACTN3 gene doping on skeletal muscle performance. Am. J. Hum. Genet. 102, 845–857 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Head S. I., Chan S., Houweling P. J., Quinlan K. G., Murphy R., Wagner S., Friedrich O., North K. N., Altered Ca2+ kinetics associated with α-actinin-3 deficiency may explain positive selection for ACTN3 null allele in human evolution. PLOS Genet. 11, e1004862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garton F. C., Seto J. T., Quinlan K. G., Yang N., Houweling P. J., North K. N., α-Actinin-3 deficiency alters muscle adaptation in response to denervation and immobilization. Hum. Mol. Genet. 23, 1879–1893 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Chin E. R., Olson E. N., Richardson J. A., Yang Q., Humphries C., Shelton J. M., Wu H., Zhu W., Bassel-Duby R., Williams R. S., A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 12, 2499–2509 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn S. E., Chin E. R., Michel R. N., Matching of calcineurin activity to upstream effectors is critical for skeletal muscle fiber growth. J. Cell Biol. 151, 663–672 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibasaki F., Fukami K., Fukui Y., Takenawa T., Phosphatidylinositol 3-kinase binds to alpha-actinin through the p85 subunit. Biochem. J. 302, 551–557 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukami K., Sawada N., Endo T., Takenawa T., Identification of a phosphatidylinositol 4,5-bisphosphate-binding site in chicken skeletal muscle α-actinin. J. Biol. Chem. 271, 2646–2650 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Sprague C. R., Fraley T. S., Jang H. S., Lal S., Greenwood J. A., Phosphoinositide binding to the substrate regulates susceptibility to proteolysis by calpain. J. Biol. Chem. 283, 9217–9223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rommel C., Bodine S. C., Clarke B. A., Rossman R., Nunez L., Stitt T. N., Yancopoulos G. D., Glass D. J., Mediation of IGF-1-induced skeletal myotube hypertrophy by PI, (3)K/Akt/mTOR and PI, (3)K/Akt/GSK3 pathways. Nat. Cell Biol. 3, 1009–1013 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Li H. H., Kedar V., Zhang C., McDonough H., Arya R., Wang D. Z., Patterson C., Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Invest. 114, 1058–1071 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein G. L., The effect of glucocorticoids on bone and muscle. Osteoporos. Sarcopenia. 1, 39–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braun T. P., Marks D. L., The regulation of muscle mass by endogenous glucocorticoids. Front. Physiol. 6, 12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu N., Yoshikawa N., Ito N., Maruyama T., Suzuki Y., Takeda S., Nakae J., Tagata Y., Nishitani S., Takehana K., Sano M., Fukuda K., Suematsu M., Morimoto C., Tanaka H., Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 13, 170–182 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Munck A., Guyre P. M., Holbrook N. J., Physiological functions of glucocorticoids in stress and their relation to pharmacological actions. Endocr. Rev. 5, 25–44 (1984). [DOI] [PubMed] [Google Scholar]
- 34.Huang S. M., Huang C. J., Wang W. M., Kang J. C., Hsu W. C., The enhancement of nuclear receptor transcriptional activation by a mouse actin-binding protein, alpha actinin 2. J. Mol. Endocrinol. 32, 481–496 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Zhao X., Khurana S., Charkraborty S., Tian Y., Sedor J. R., Bruggman L. A., Kao H. Y., α Actinin 4 (ACTN4) regulates glucocorticoid receptor-mediated transactivation and transrepression in podocytes. J. Biol. Chem. 292, 1637–1647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khurana S., Chakraborty S., Zhao X., Liu Y., Guan D., Lam M., Huang W., Yang S., Kao H. Y., Identification of a novel LXXLL motif in α-actinin 4-spliced isoform that is critical for its interaction with estrogen receptor α and co-activators. J. Biol. Chem. 287, 35418–35429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mills M., Yang N., Weinberger R., Vander Woude D. L., Beggs A. H., Easteal S., North K., Differential expression of the actin-binding proteins, alpha-actinin-2 and -3, in different species: Implications for the evolution of functional redundancy. Hum. Mol. Genet. 10, 1335–1346 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Seto J. T., Lek M., Quinlan K. G. R., Houweling P. J., Zheng X. F., Garton F., Arthur D. G. M., Raftery J. M., Garvey S. M., Hauser M. A., Yang N., Head S. I., North K. N., Deficiency of α-actinin-3 is associated with increased susceptibility to contraction-induced damage and skeletal muscle remodeling. Hum. Mol. Genet. 20, 2914–2927 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Schiaffino S., Dyar K. A., Ciciliot S., Blaauw B., Sandri M., Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 280, 4294–4314 (2013). [DOI] [PubMed] [Google Scholar]
- 40.White R. B., Bierinx A. S., Gnocchi V. F., Zammit P. S., Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev. Biol. 10, 21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodman C. A., Hornberger T. A., Measuring protein synthesis with SUnSET: A valid alternative to traditional techniques? Exerc. Sport Sci. Rev. 41, 107–115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duma D., Collins J. B., Chou J. W., Cidlowski J. A., Sexually dimorphic actions of glucocorticoids provide a link to inflammatory diseases with gender differences in prevalence. Sci. Signal. 3, ra74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma K., Mallidis C., Bhasin S., Mahabadi V., Artaza J., Gonzalez-Cadavid N., Arias J., Salehian B., Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. Am. J. Physiol. Endocrinol. Metab. 285, E363–E371 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Yoon M.-S., mTOR as a key regulator in maintaining skeletal muscle mass. Front. Physiol. 8, 788 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gokhin D. S., Ward S. R., Bremner S. N., Lieber R. L., Quantitative analysis of neonatal skeletal muscle functional improvement in the mouse. J. Exp. Biol. 211, 837–843 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L., Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonaldo P., Sandri M., Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 6, 25–39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilson H., Schakman O., Combaret L., Lause P., Grobet L., Attaix D., Ketelslegers J. M., Thissen J. P., Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology 148, 452–460 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Chen J. L., Walton K. L., Winbanks C. E., Murphy K. T., Thomson R. E., Makanji Y., Qian H., Lynch G. S., Harrison C. A., Gregorevic P., Elevated expression of activins promotes muscle wasting and cachexia. FASEB J. 28, 1711–1723 (2014). [DOI] [PubMed] [Google Scholar]
- 50.DCE S., Nixon M., Buurstede J. C., Sips H. C., Schilperoort M., Kuipers E. N., Backer E. A., Kooijman S., Rensen P. C. N., Homer N. Z. M., Walker B. R., Meijer O. C., Kroon J., Androgens modulate glucocorticoid receptor activity in adipose tissue and liver. J. Endocrinol. 1, JOE-18-0503 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y., Leung D. Y., Nordeen S. K., Goleva E., Estrogen inhibits glucocorticoid action via protein phosphatase 5 (PP5)-mediated glucocorticoid receptor dephosphorylation. J. Biol. Chem. 284, 24542–24552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reisz-Porszasz S., Bhasin S., Artaza J. N., Shen R., Sinha-Hikim I., Hogue A., Fielder T. J., Gonzalez-Cadavid N. F., Lower skeletal muscle mass in male transgenic mice with muscle-specific overexpression of myostatin. Am. J. Physiol. Endocrinol. Metab. 285, E876–E888 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Surjit M., Ganti K. P., Mukherji A., Ye T., Hua G., Metzger D., Li M., Chambon P., Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145, 224–241 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Newton R., Holden N. S., Separating transrepression and transactivation: A distressing divorce for the glucocorticoid receptor? Mol. Pharmacol. 72, 799–809 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Hoffman E. P., Reeves E., Damsker J., Nagaraju K., McCall J. M., Connor E. M., Bushby K., Novel approaches to corticosteroid treatment in Duchenne muscular dystrophy. Phys. Med. Rehabil. Clin. N. Am. 23, 821–828 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedlander S. M., Herrmann A. L., Lowry D. P., Mepham E. R., Lek M., North K. N., Organ C. L., ACTN3 allele frequency in humans covaries with global latitudinal gradient. PLOS ONE 8, e52282 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bodine S. C., Baehr L. M., Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol. Metab. 307, E469–E484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodgers B. D., Garikipati D. K., Clinical, agricultural, and evolutionary biology of myostatin: A comparative review. Endocr. Rev. 29, 513–534 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garton F., Seto J. T., North K. N., Yang N., Validation of an automated computational method for skeletal muscle fibre morphometry analysis. Neuromuscul. Disord. 20, 540–547 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Thomas K. C., Zheng X. F., Garces Suarez F., Raftery J. M., Quinlan K. G., Yang N., North K. N., Houweling P. J., Evidence based selection of commonly used RT-qPCR reference genes for the analysis of mouse skeletal muscle. PLOS ONE 9, e88653 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pyl P. T., Anders S., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J. P., Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabg0088/DC1