A new single-cell whole-genome amplification method allows the genome-wide characterization of DNA damage and gene vulnerability.
Abstract
We report a novel single-cell whole-genome amplification method (LCS-WGA) that can efficiently capture spontaneous DNA damage existing in single cells. We refer to these damage-associated single-nucleotide variants as “damSNVs,” and the whole-genome distribution of damSNVs as the damagenome. We observed that in single human neurons, the damagenome distribution was significantly correlated with three-dimensional genome structures. This nonuniform distribution indicates different degrees of DNA damage effects on different genes. Next, we identified the functionals that were significantly enriched in the high-damage genes. Similar functionals were also enriched in the differentially expressed genes (DEGs) detected by single-cell transcriptome of both Alzheimer’s disease (AD) and autism spectrum disorder (ASD). This result can be explained by the significant enrichment of high-damage genes in the DEGs of neurons for both AD and ASD. The discovery of high-damage genes sheds new lights on the important roles of DNA damage in human diseases and disorders.
INTRODUCTION
As one of the major sources of DNA lesions, spontaneous DNA damage is known to be associated with mutagenesis (1–4). Recent studies have also suggested that spontaneous DNA damage could alter the epigenetic landscape and gene expression (5, 6). The levels of DNA damage on the genome are likely not uniform (7–9). As a result, we expect that different genes bear varying burdens of DNA damage. Hence, different genes come to have different degrees of vulnerability toward mutations and epigenetic instability, which could lead to the large-scale perturbations to gene functions and trigger the development of diseases. It is greatly desired that we can accurately profile the distribution of DNA damage in the human genome and identify different vulnerable genes in different types of cells.
To date, the main approaches for measuring DNA damage are high-performance liquid chromatography–based and chromatin immunoprecipitation–based methods (1, 10–13). High-performance liquid chromatography provides a global assessment of nucleotide modifications; however, its accuracy is largely limited by the artificial damage introduced during the step of mononucleotide preparation. On the other hand, chromatin immunoprecipitation–based methods could provide sequence-based measurements of the damage distribution; however, DNA shearing and decross-linking can introduce artificial damage to DNA templates, which directly limits the accuracy of measurement. We continue to lack a method that allows accurate damage profiling.
Here, instead of taking the bulk sample approach, we focus on developing a single-cell whole-genome amplification (WGA) assay to detect the DNA damage in single cells. So far, various types of single-cell WGA methods have been developed with the main focus on the detection of permanent genome changes including mutations, copy number variations, and large structure variations (14–20). We believe that DNA damage could also be detected by a single-cell WGA method as damaged base can cause base misincorporations during the amplification process, which can then be detected as de novo variants in the single-cell sequencing data. It is worth noting that detection of misincorporated bases caused by DNA damage has been successfully demonstrated in studies using bulk DNA samples (21–23).
The major advantage of using a single-cell WGA–based approach to profile DNA damage is that we can avoid the need for invasive treatments of DNA templates such as hydrolysis or decross-linking, leading to a substantial reduction in technical artifacts. However, it is worth pointing out that the major technical hurdle of the single-cell WGA approach is the amplification errors, and these technical artifacts have kept the accuracy in the detection of de novo mutations in single cells under intensive debate (18, 24–26). Therefore, for both accurate profiling of DNA damage and de novo mutations, a single-cell WGA method that allows for the efficient filtering of amplification errors remains greatly desired.
Here, we report a novel single-cell WGA method, termed “linear copy and split–based WGA” (LCS-WGA), that allows for nearly complete filtering of amplification errors. As a result, we achieved genome-wide detection of DNA damage at a single-cell resolution. For convenience, we denote damage-associated “de novo” single-nucleotide variants as damSNVs, and the entire genome characterization of damSNVs as the damagenome. It is worth noting that, with the single-cell WGA approach, the efficiency of detecting damaged bases depends on the misincorporation rate. With the requirement of misincorporation for damage detection, our method mainly focuses on the types of DNA damage that are prone to base misincorporation. For DNA modifications that do not cause base misincorporations, it is not feasible to detect them using our method. It is worth emphasizing that, for the major types of DNA damage that are commonly detected in bulk assays, such as oxidized cytosine and 8-oxoguanine, their misincorporation rate is high, which permits the reliable detection of these DNA damage.
With the ability to characterize DNA damagenome in single cells, we investigated age-dependent changes in DNA damage levels in single human neurons. Next, using the damagenome data of single human neurons, we successfully identified the genes with high levels of DNA damage in the human genome. We showed that these high-damage genes are significantly enriched in the differentially expressed genes (DEGs) of both individuals with Alzheimer’s disease (AD) and autism spectrum disorder (ASD), indicating that DNA damage is closely associated with the abnormal gene expression in these neurological conditions.
RESULTS
Reaction scheme of single-cell damagenome profiling
During LCS-WGA, preamplification was first performed to generate linear DNA copies from the original genomic DNA of the single cell (Fig. 1A). Specifically, we used a DNA polymerase without proofreading activity (i.e., large fragment of Bst polymerase) in the preamplification step to boost the rate of base misincorporation while using the damaged base as the template. During preamplification, three cycles of annealing and extension reactions were performed. The semiamplicons were linearly copied from the genomic DNA, and full amplicons were nonlinearly produced from semiamplicons (fig. S1A). In this context, the amplification errors in semiamplicons were all independently produced as the semiamplicons are directly copied from the original genome template of the single cell. In contrast, amplification errors in the full amplicons are not independent because multiple copies of full amplicons can be produced from the same semiamplicons; therefore, different full amplicons may carry the same errors (fig. S1A).
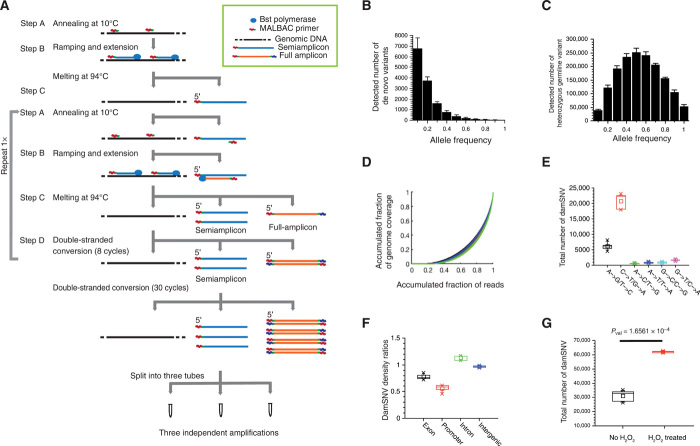
Fig. 1. Detection of damSNVs by LCS-WGA.
(A) Reaction scheme of LCS-WGA. (B) Histogram of AF distribution of de novo variants in single MCF10A cells. The distribution center at the low AF value supports the detection of damage-associated single-nucleotide variants (damSNVs). (C) Histogram of AF distribution of heterozygous germline variants in single MCF10A cells. The Gaussian distribution centered at AF = 0.5 indicates the evenness in our amplification. (D) Gini plots for the three split samples of MCF10A SC1–5, respectively. The average value of the Gini coefficients is 0.624. The similar curves of the three split samples of each cell support that similar amplification performance can be achieved for MDA using pipetting mixing–based approach (Materials and Methods). (E) Total number of different types of damSNVs of single MCF10A cells. (F) DamSNV density ratios of the exon, promoter, intron, and intergenic regions over the total genome in the MCF10A cells. (G) Comparison of the total number of damSNVs between MCF10A cells with and without hydrogen peroxide treatment.
A critical step of LCS-WGA is to split preamplification products into multiple tubes and conduct independent multiple displacement amplification (MDA) using only the linearly copied semiamplicons produced in the preamplification described above and the original genomic DNA as templates for amplification (Fig. 1A). However, before splitting the preamplification product for independent MDA reactions, we needed to perform 30 cycles of polymerase chain reaction (PCR) without the melting steps (which is denoted as the double-stranded conversion step in Fig. 1A). The reason for performing the double-stranded conversion step is to convert all the nonlinearly copied products (i.e., full amplicons produced in multiple annealing and extension cycles) into double-stranded DNA. Because double-stranded DNA cannot be primed by hexamers from the MDA reaction, the amplification of these nonlinear products is completely quenched during the MDA reaction. We validated that the double-stranded full amplicons were effectively prohibited from amplification in MDA (please refer to Materials and Methods for detailed information).
During the double-stranded conversion step, the semiamplicons remained in single-stranded form. As a result, the amplification products in the MDA reaction were only amplified from the linearly produced semiamplicons and the original genomic DNA. Because the amplification errors in semiamplicons are independently produced, we can cross-compare the variants detected in different splits. By requiring the detection of the same variants in at least two split samples, we efficiently filter out the amplification errors existing in semiamplicons and, therefore, achieve an accurate detection of de novo variants in single cells (fig. S1B).
To evaluate the performance of LCS-WGA, we sequenced single cells isolated from a normal diploid breast epithelial cell line (MCF10A). Five single MCF10A cells were isolated from the culture in a serum-free medium for LCS-WGA. We first combined the sequencing data of three split samples and used the standard Genome Analysis Toolkit (GATK) pipeline (27) for variant calling. Next, we applied a two-split detection criterion (the variants need to be detected in at least two splits) to filter out amplification errors and identify the true variants. On the basis of the detected de novo variants using the combined splits, we determined that the average amplification error rate is ~5 × 10−5 per base, consistent with the error rate of Bst polymerase used in our preamplification reaction (4 × 10−5 per base) (18). When we conducted a cross-comparison of different splits and only kept the variants that were detected in at least two splits as the criterion for variant calling, we estimated that the error rate in our variant calling was approximately (5 × 10−5)2 × 0.333 ≈ 8.3 × 10−10 per base, where 5 × 10−5 is the amplification error rate described above and 0.333 is the probability for the two split samples to have the same type of error, respectively. Notably, the error rate of 8.3 × 10−10 per base corresponds to only five false positives per genome. Therefore, we concluded that LCS-WGA can achieve nearly complete filtering of amplification errors.
For the detection of de novo variants, we defined the following stringent criterion: Only when the variants detected in single-cell data were not detected by any reads in the bulk sequencing data, were they defined as de novo variants in the single cell (i.e., the zero-read criterion) (Supplementary Materials). Next, we evaluated the levels of de novo variants existing in single MCF10A cells and detected 13,467 ± 1160 de novo variants per MCF10A cell (table S1). We expected that the vast majority of these de novo variants originated from the damaged DNA bases in the genomic DNA of the single cell because a single normal MCF10A cell likely cannot have such a high load of de novo mutations. To show that the vast majority of the large number of de novo variants were not true de novo mutations, we plotted the numbers for de novo variants on the axis of the allele frequency (AF) and, indeed, observed that the histogram peaks at a low AF value (Fig. 1B). As a control for the AF distribution of true mutations, when we plotted the distribution of the heterozygous germline variants, we observed that the histogram was centered at AF = 0.5 (Fig. 1C and table S2). With the drastic differences in AF distributions, we concluded that the vast majority of the detected de novo variants are not true mutations, instead representing the misincorporated bases caused by DNA damage.
In Fig. 1B, we noticed that the AF distribution of de novo variants peaked between AF = 0.1 and AF = 0.2. With the damaged bases occurring on one strand, the theoretical AF value should be 0.25. However, considering the boundary effect (i.e., half of the damSNVs have AF values less than 0.25) and the misincorporation rate, we expected instead that the AF distribution of damSNVs would peak at a lower value, as observed in Fig. 1B. It is also worth noting that the requirement of the detection of SNVs in at least two split samples could also introduce a minor skewness at the left-side boundary of AF distribution (i.e., AF = 0.1). To minimize this effect, we adopted AF = 0.03 as the cutoff for calling SNVs (Supplementary Materials).
We would like to point out that the consistent performance of MDA reactions for each split is also important for achieving efficient detection of damSNV, especially considering that MDA is notorious for abrupt large bias. Here, we achieved the robust performance of MDA by limiting the amplification time together with frequent pipetting during the reaction. In Fig. 1D, the consistent Gini curves between different splits and cells show the robust performance of MDA. For 15 splits from five cells, we achieved an average genome coverage of ~84% per split. On the basis of the genome coverage in individual split samples, we could readily determine the strand dropout rate and the detection rate of damSNVs (table S1). The total level of damSNVs was then calculated for individual cells (please refer to the Supplementary Materials for detailed normalization procedure).
Next, we plotted the complete array of different types of damSNVs for MCF10A (Fig. 1E). Among all six types of damSNVs, the major contributions to damSNVs stemmed from the transition variants as follows: C➔T/G➔A and A➔G/T➔C. On the basis of prior knowledge of the major forms of DNA damage that occur to the genome, we reasoned that the damSNVs of the C➔T/G➔A type are associated with cytosine oxidation (28, 29); the damSNVs of the A➔G/T➔C type are associated with adenine oxidation, followed by deamination (1); and the damSNVs of the G➔T/C➔A type are associated with 8-oxoguanine (30, 31).
In Fig. 1F, we demonstrate that different functional regions (e.g., promoter, exon, intron, and intergenic regions) have different degrees of damSNV abundance, which further supports that the damSNVs detected in LCS-WGA are biologically authentic. The potential mechanisms for these differences include different levels of genotoxic exposure and variable DNA repair efficiency in different functional regions of the genome.
Here, we choose alkaline lysis to lyse single cells to avoid the various types of oxidative DNA damage caused by heating at high temperatures. While alkaline lysis will not induce various oxidation-related damages, the main drawback of this lysis is that it could induce cytosine deamination in the genomic template, which then leads to the detection of C➔T damSNVs. To remove this technical artifact, we performed uracil DNA glycosylase (UDG) treatment before preamplification to convert the deaminated C into AP sites and therefore prevent them from downstream amplification. The efficiency of this treatment has been validated (fig. S2 and Materials and Methods). To quantify the level of deaminated cytosine removed by the UDG treatment, we performed LCS-WGA for a single MCF10A cell without UDG treatment and observed a significant increase in C➔T damSNVs (fig. S3).
It is worth pointing out that, because of the UDG treatment, detection of biologically authentic deaminated cytosines becomes infeasible. However, on the basis of a recent study (32), the frequency of cytosine deamination events is low at only 400 to 600 lesions per cell. In contrast, most C➔T damSNVs are derived from oxidized cytosines. Therefore, the exclusion of the deaminated cytosines will not substantially alter our quantification of the total level of C➔T damSNVs.
For further validation of damage detection, we also applied LCS-WGA to the MCF10A cells treated with hydrogen peroxide (H2O2), and we observed that DNA damage levels drastically increased in the H2O2-treated cells (Fig. 1G). Here, we only performed a 5-min treatment of H2O2 to avoid introducing notable mutagenesis. The increase in damSNVs in all six subtypes of variants is also consistent with the known DNA lesion pattern caused by H2O2 treatment (fig. S4) (33).
Damagenome profiling of single human cortical and hippocampus neurons
To investigate the age-dependent changes of DNA damage levels, next we profiled the damagenome of postmortem human neurons. Here, we performed LCS-WGA on a total of 52 prefrontal cortical neurons isolated from 10 brain samples in three age groups (3 in the young- and middle-age groups and 4 in the old-age group; table S3) and 17 hippocampus neurons from six samples in the three age groups (two brain samples per age group; table S3). Each cell was sequenced at 30× depth with ~10× for each split sample (table S4). On average, we achieved genome coverage of ~83% per split, similar to MCF10A result. For single neurons, the distribution of germline variants and damSNVs was similar to that of MCF10A cells, in that the AF distribution of germline variants was centered at AF = 0.5 (Fig. 2A and fig. S5), while the AF distribution of de novo variants was centered between AF = 0.1 and AF = 0.2 (Fig. 2B and fig. S6). We also observed that the distributions of different types of damSNVs were similar to those of MCF10A cells (Fig. 2C and fig. S7).
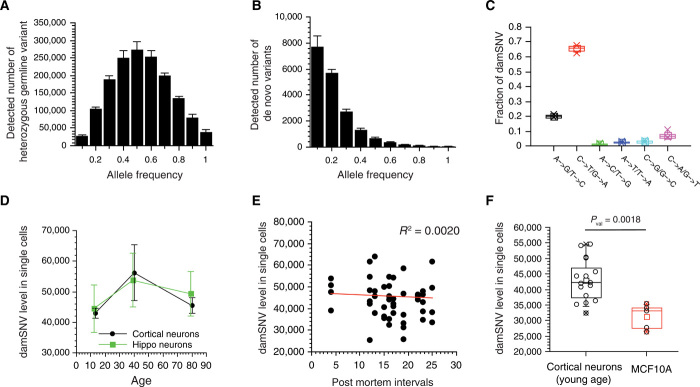
Fig. 2. Levels of damSNVs in individual neurons at different ages.
(A) Histogram of AF distribution of heterozygous germline variants in single cortical neurons of sample p5554. (B) Histogram of the AF distribution of damSNVs in individual cortical neurons of sample p5554. The distribution center at a low AF value supports the detection of damSNVs. (C) Fraction of different types of damSNVs for single neurons of p5554. (D) Plot of the average level of damSNVs of individual neurons on the axis of age. The damage level is high in middle age. (E) Plot of the damSNV levels of all single cells on the axis of the corresponding postmortem intervals. (F) The levels of damSNV levels of single neurons are significantly higher than that of MCF10A cells.
In Fig. 2D, we plotted the average levels of damSNVs for different age groups. We observed that the average level of damSNVs increases from young age to middle age. The average level of damSNVs in the middle-age samples was slightly higher than that in the old-age samples. We also observed a similar trend in the mean levels of DNA damage changes along with advancing age in hippocampus neurons (Fig. 2D). Although a widely held concept is that DNA damage accumulates with age, reports in the scientific literature remain inconsistent (34), and here, our results suggest that the level of DNA damage in the brain may reach to the highest levels in middle age. It is worth noting that, despite that consistent trends were observed in both cortical and hippocampus neurons, because of the large variations in damSNVs among single cells, the differences between different age groups did not reach statistical significance.
It is known that mosaic copy number variations frequently occur in frontal cortical neurons (35), which could limit the accuracy in our estimation of the total level of damSNVs in single cells. However, we noticed that the regions with mosaic copy number variations are relatively limited (i.e., <10% of the genome size for the vast majority of neurons with mosaic copy number variations). Therefore, their effect on our quantification of damSNV levels is likely minor. On the other hand, we suspect that these genomic mosaicisms could affect the physiology of the neurons, which directly contributes to the large cell-to-cell variations in terms of the damage levels observed in single cells.
Furthermore, we did not observe any correlation between the number of damSNVs and the postmortem intervals (Fig. 2E). Therefore, we concluded that the damSNVs we detected were biologically authentic and were not generated during the sample collection process. We also noticed that the damage level in neurons was significantly higher than that of the cultured MCF10A cells (Fig. 2F).
Nonuniform distribution of the damagenome is associated with three-dimensional genome topology
Beyond comparing the total DNA damage levels in different brain samples, sequencing-based measurement also allows us to investigate local DNA damage distribution in the genome. We first sought to examine whether significant changes in the local damSNV densities exist because of aging. We calculated genome-wide damSNV densities in 200-kb windows for the young-age, middle-age, and old-age neuron groups, respectively. The local coverage in each window was used for the normalization of damSNVs. Next, we plotted scatterplots between different age groups and observed significant correlations (Fig. 3, A to C). No local regions with statistically significant changes in DNA damage levels between different age groups were identified. Because there were no significant changes in damagenome distribution with age, we combined the cortical neurons at different ages to generate the damagenome profile of cortical neurons. We also generated the damagenome profile of hippocampus neurons. When we compared these two damagenome profiles, we also did not observe any significant difference between them. Supported by this result, we combined all the neurons and plotted the distribution of damSNV densities in the genome (Fig. 3D and table S5). The histogram of damSNV densities is shown in fig. S8.
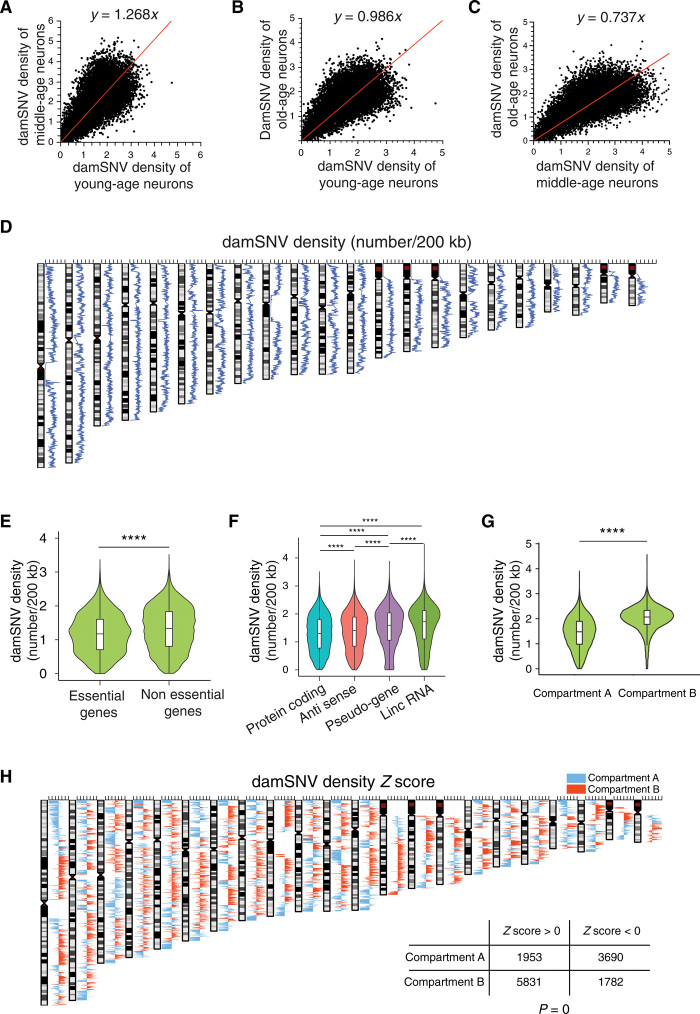
Fig. 3. Distribution of damSNV on the genome.
(A) The scatter plot between damSNV densities of the young-age neurons and the middle-age neurons. (B) The scatter plot between damSNV densities of the young-age neurons and the old-age neurons. (C) The scatter plot between damSNV densities of the middle-age neurons and the old-age neurons. (D) Distribution of damSNV density in 200-kb binning windows on the genome. (E) Comparison of damSNV densities of essential genes versus nonessential genes. ****P < 0.0001. (F) Comparison of damSNV densities of different types of genes, including protein-coding genes, antisense genes, pseudogenes, and lincRNA. (G) Comparison of damSNV densities in A compartment versus B compartment of three-dimensional genome topology. (H) The Z score of the damSNV density in 200-kb windows along the genome. The A compartments are colored blue, and the B compartments are colored red. The significant correlation between the damagenome distribution and genome topological compartments is shown in the inset table.
In Fig. 3D, we observed that damSNV density is nonuniformly distributed in the genome, which suggests that different damage burdens could exist for different genes. To investigate gene-specific effects, we adopted the local damSNV density to represent the damage burden of the genes from the same window. In Fig. 3E, we found that the damSNV densities associated with the essential genes (36–39) were significantly lower than those associated with nonessential genes. Furthermore, we showed that protein-coding genes have the lowest damSNV densities, followed by antisense genes and pseudogenes, while long intergenic noncoding RNAs (lincRNAs) have the highest damSNV densities (Fig. 3F).
Recently, Ochs et al. (40) have shown that genome topology plays a vital role in safeguarding genome integrity in response to double-stranded breaks. To investigate the possible association between the genome topology and single-nucleotide DNA damage, we compared the damSNV densities in A compartments versus B compartments identified previously by Hi-C experiments (41). Note that the A compartments are associated with open chromatin, and the B compartments are associated with more closed chromatin. Here, we observed that the damSNV densities in B compartments were significantly higher than those in A compartments (Fig. 3G). To further demonstrate the local association between the topological compartments and damSNV abundance, next we calculated the Z score of the damSNV density for each 200-kb binning window of the genome (Fig. 3H). We colored each window based on the compartment (A compartments are colored blue, and B compartments are colored red). As a result, we observed that 75% of the binning windows with positive Z scores (high damSNV density) belong to B compartments, and 69% of the binning windows with negative Z scores (low damSNV density) belong to A compartments. The odds ratio for the association between the damSNV Z scores and topological compartments was 6.63, and the P value was less than 2.2 × 10−16 according to Fisher’s exact test. This result confirms that there is a significant association between the density of spontaneous DNA damage and the topological compartments of the genome.
Next, we examined whether the variations of damSNV densities depend on the GC (Guanine and Cytosine) percentage variations on the genome. We observed a negative correlation between the GC percentage and damSNV density in 200-kb windows on the genome (fig. S9). Because the A compartments had a significantly higher GC percentage than the B compartments (fig. S10), when we normalized the damSNV densities by the corresponding GC percentages, the difference in damSNV density between the A and B compartments became become further enlarged (fig. S11). In addition, we showed that, for both GC- and AT-related damSNVs, the average damage density in the A compartment group was significantly lower than that in the B compartment group (figs. S12 and S13), indicating that the differences in damSNV densities between compartment A and B did not depend on damage types.
García-Nieto et al. (9) recently profiled the genome-wide susceptibility of the human genome to ultraviolet damage and observed that the susceptibility to ultraviolet damage in gene-enriched and euchromatic regions is significantly lower than that in heterochromatin with repressed transcription activity. Consistent with this observation, we also detected a negative correlation between gene density and DNA damage abundance (fig. S14). This result further supports that the damagenome we characterized is biologically genuine.
Similar functional enrichments exist in high-damage genes and DEGs in AD and ASD
Given the nonuniformly distributed damagenome, we aimed to examine the potential enrichment of any functionals among the high-damage genes. Here, we ranked genes based on their local damSNV density and then used the top 2000 genes to perform functional enrichment analysis using the UniProtKB Knowledgebase in DAVID algorithm (42). For convenience, we refer to these high–damage density genes as the high-damage genes hereafter. The total group of protein-coding genes was used as the gene background in the functional enrichment analysis. We observed significant enrichment of different functionals (Fig. 4A). Next, we conducted STRING analysis (43) and observed a large protein interaction network (fig. S15). We observed that, among the high-damage genes, protein-protein interactions were significantly more common than the average level of protein-protein interactions in the human genome. This result suggests that the effects of DNA damage on cells are not exerted in small isolated networks but instead through a large and well-connected protein-protein interaction network.
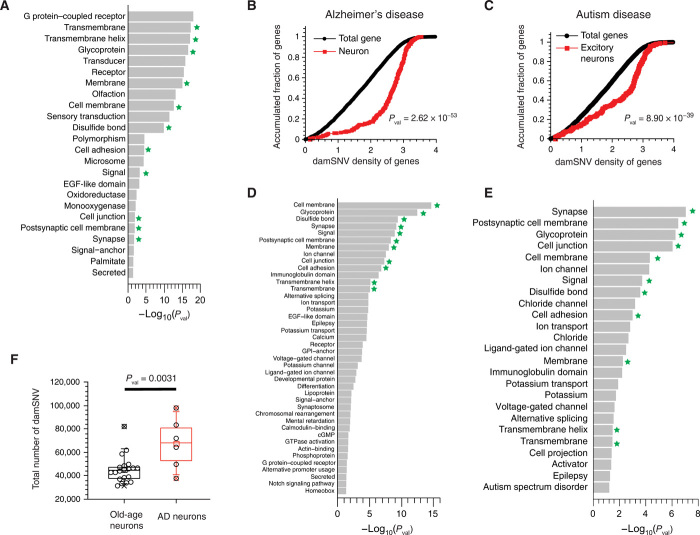
Fig. 4. Functional enrichment analyses of the high-damage genes.
(A) Significant functional enrichment for the top 2000 high-damage genes ranked by damage density. (B) The KS tests of the damSNV distributions of the DEGs (DEGs between AD and normal brain samples) and the total genes. (C) The KS tests of the damSNV distributions of the DEGs (DEGs between ASD and normal brain samples) and the total genes. (D) Significant enrichment of protein function annotation for the DEGs between AD and normal brain samples using UniProt Knowledgebase. (E) Significant enrichment of protein function annotation for the DEGs between ASD and normal brain samples using UniProt Knowledgebase. (F) The damSNV levels in cortical neurons isolated from AD brains are significantly higher than the levels in normal neurons.
Given the recent endeavors of high-throughput, single-cell transcriptome profiling of AD (44, 45) and ASD (46), we investigated the potential connection between high-damage genes and gene expression alternations that occur in the neurons of patients with neurological conditions. To do so, we performed the Kolmogorov-Smirnov (KS) test to examine whether the high-damage genes are significantly enriched among the DEGs in the neurons between the diseased samples and the normal samples. The lists of DEGs of both AD and ASD were downloaded from the data of Grubman et al. (44) and Velmeshev et al. (46) (table S6). In the KS plots for AD (Fig. 4B) and ASD (Fig. 4C), we observed large gaps between two cumulative curves, indicating that significant enrichment of the high-damage genes existed among the DEGs. This result directly shows that the alternation of gene expression in patients with neurological conditions is significantly associated with DNA damage density.
For the DEGs of AD and ASD, we conducted functional enrichment analysis using the UniProt Knowledgebase in DAVID algorithm, and we used the total detected genes in normal samples as the gene set background in the analysis. We observed that many enriched functionals (see Fig. 4D for AD and Fig. 4E for ASD, indicated by the star) overlap with the enriched functionals from the high-damage gene–based analysis (Fig. 4A). Specifically, the top seven hits for AD and the top five hits for ASD are among the enriched functionals from the high-damage gene–based analysis. The significant overlap of enriched functionals is consistent with the result of the KS test, indicating that high-damage genes contribute to differential gene expressions in both diseases.
The damage density–based functional enrichment analysis described above corresponds to the scenario of that the DNA damage effect is mainly exerted on promoter regions, assuming that the sizes of promoter regions are similar between different genes. It is worth noting that Lu et al. (47) showed that damage that occurs in promoter regions affects the gene expression. On the other hand, we reason that the effect of DNA damage may also be exerted through gene bodies. Next, we ranked the genes based on the DNA damage load (which corresponds to the product of damage density and gene length). We also used the top 2000 genes to perform a functional enrichment analysis using the DAVID algorithm (48), and the total group of protein-coding genes was used as the background. We observed significant enrichment of splicing and phosphoproteins in terms of protein functions of the UniProt Knowledgebase (42) (fig. S16). This result indicates that DNA damage could play important roles by perturbing the functions of alternative splicing and phosphoproteins through the high–damage load genes. Consistent with this analysis, Raj et al. (49) have successfully identified hundreds of aberrant pre–messenger RNA splicing events in AD that are reproducibly associated with the disease. Hsieh et al. (50) also observed that cryptic splicing errors are associated with neurofibrillary tangle burden. In the protein-protein interaction analysis using the STRING algorithm, we also observed a well-connected network, indicating a large-scale, systems-level effect on the cells (fig. S17). It is worth noting that the functionals of alternative splicing and phosphoproteins were not enriched in the high–damage density genes, indicating that the large-size genes are enriched in these functionals.
While the damage density– and damage load–based analyses suggest different mechanisms for DNA damage’s effects on gene expression, these two mechanisms do not need to be mutually exclusive. As described above, we observed evidence supporting both models of damage effect. We reason that, for genes of small or medium length, the effects of DNA damage on the regulatory regions are important, while for genes of substantially large sizes, the total damage load will weigh more in terms of the effects on changes in gene expression.
In AD, it is well known that amyloid formation can cause neurotoxicity and lead to an increase in oxidative radicals and other free radicals (51). Other potential mechanisms include mitochondria dysfunction and metal homeostasis caused by amyloid formation (52–54). We performed LCS-WGA for single neurons isolated from diseased samples to examine whether the DNA damage levels in these cells are indeed elevated. As a result, we observed that the DNA damage levels in the AD neurons were significantly higher than those of normal old-age neurons (Fig. 4F).
Significant enrichment of high-damage genes in DEGs of non-neuron cell types for both AD and ASD
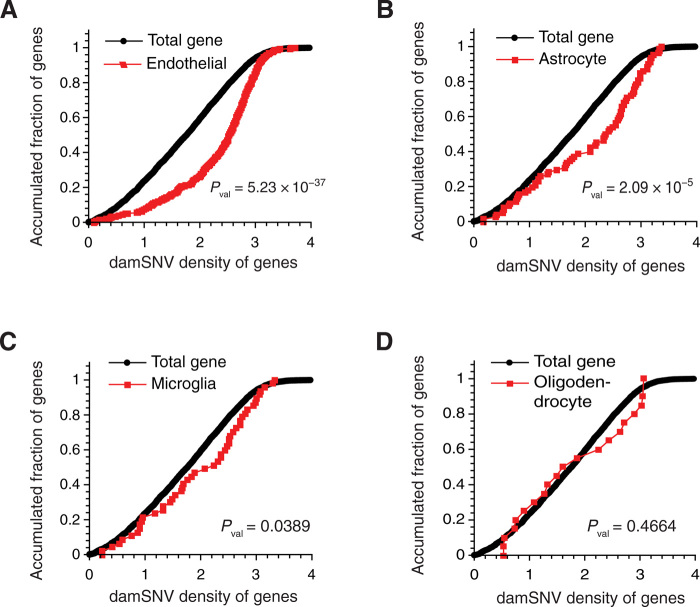
It has been shown that large-scale genome topological domains between different cell types share significant similarities (at the level of ~80% similarity) (41, 55–58). Considering the direct association between damSNV densities and genome topological structures, we reason that the damagenome distribution between different cell types could also be similar. Supported by this rationale, we performed the KS tests for the DEGs detected in oligodendrocytes, astrocytes, microglia, and endothelial cells. We observed statistically significant enrichment of high-damage genes in DEGs for endothelial cells (Fig. 5A), astrocytes (Fig. 5B), and microglia (Fig. 5C), indicating that the DNA damage could also exert effects on gene expression across different cell types. However, we did not observe enrichment of high-damage genes in the DEGs of oligodendrocytes (Fig. 5D). The different levels of statistical significance in different cell types could be due to variations in the damagenome distributions or the total damage levels, or both in different cell types. Therefore, direct characterization of the genome topology and the damagenome of different types of cells is highly desired in the future.
Fig. 5. Enrichment analysis of high-damage genes in the DEGs of different cell types in AD.
The KS tests of the damSNV distributions of the DEGs (DEGs between AD and normal brain samples) and the total genes for the endothelial cells (A), astrocytes (B), microglia (C), and oligodendrocyte (D).
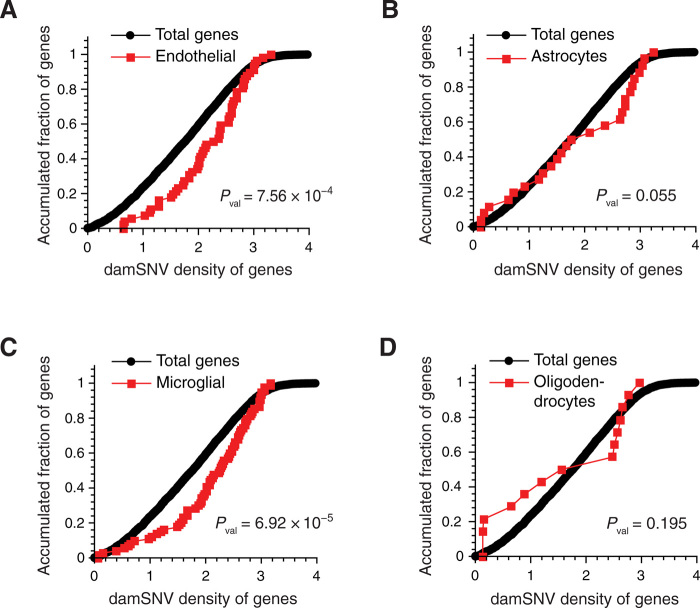
In ASD, we also observed significant enrichment in various cell types besides neurons (Fig. 6). Similar to that in AD, significant enrichment of high-damage genes was observed in endothelial cells, astrocytes, and microglia (Fig. 6, A to C). With the limited number of DEGs in oligodendrocytes, no significant enrichment of high-damage genes was observed (Fig. 6D). The consistent enrichment of high-damage genes in the DEGs of endothelial, astrocytes, and microglia for both AD and ASD indicates that the effects of DNA damage are not only genome-wide but also general across different cell types, which demands future investigation.
Fig. 6. Enrichment analysis of high-damage genes in the DEGs of different cell types in ASD.
KS tests of the damSNV distributions of the DEGs (DEGs between ASD and normal brain samples) and the total genes for the endothelial cells (A), astrocytes (B), microglia (C), and oligodendrocyte (D).
DISCUSSION
While it is well known that DNA damage could induce mutations, DNA damage can also influence gene expression. For example, Lu et al. (47) reported that an accumulation of oxidative lesions in the promoter regions can cause down-regulation of the corresponding gene expression. Changes in gene expression could be induced by an altered epigenetic landscape, which could be induced by frequent DNA damage and repair of genes with high damage frequency (5). Several recent studies have shown that DNA damage can cause changes in epigenetic markers and chromatin organization (59, 60). For example, frequent DNA damage could lead to the opening of compact chromatin and, as a result, changes in gene expression. Furthermore, the overall trend of reduction in epigenetic stability has also been proven in both aging and neural degeneration (6, 61–65). Recently, Klein et al. (66) observed genome-wide changes of histone acetylation driven by tau pathology.
To characterize the genome-wide effects of DNA damage in alternating gene expression and epigenetic landscape, we first needed to profile the distribution of DNA damage on the genome. Here, we report a new single-cell WGA method, which enables us to detect the DNA damage as de novo variants in the sequencing data with high accuracy. First, it is worth noting that this method allows us to directly quantify the levels of the major types of DNA damage existing in single human cells. Second, with this genome sequencing–based approach, we successfully characterized the damage distribution in the human genome. As a result, we observed that DNA damage is nonuniformly distributed within the genome, and this nonuniform distribution is significantly associated with the compartment distribution of three-dimensional genome structures.
With the uneven damagenome, we identified the genes with high damage density (i.e., high–damage density genes). Among the top 2000 most high-damage genes, we observed significant functional enrichments. Considering that these high-damage genes are the Achilles’ heel of the genome, we hypothesized that these genes and their associated functional vulnerability could play important roles in the development of neurological diseases. When we considered AD and ASD in concert with the recently published single-cell transcriptome data, we observed significant enrichment of high-damage genes in the DEGs of both diseases. Furthermore, the functionals enriched in the DEGs of neurons for both diseases are also similar to the functionals enriched in the high-damage genes. Considering the significant dependence of the damagenome on the large scale of genome topology, which exhibits around 80% similarity between different cell types, we also examined whether high-damage genes are also significantly enriched in the DEGs of other cell types. We confirmed significant enrichment in endothelial cells, microglia, and astrocytes, but not in oligodendrocytes. These findings strongly suggest that the effects of DNA damage are not confined to only neurons in both neurological conditions.
Overall, as the first single-cell, whole-genome sequencing method for accurate measurement of DNA damage existing in individual cells, we believe that LCS-WGA will be broadly applied in future biological research. The identification of high-damage genes as the potential Achilles’ heel of the human genome offers new insights into the potentially important roles of DNA damage in increasing individual’s susceptibility toward complex diseases and disorders such as AD and ASD. Furthermore, a dual-omics approach that combines LCS-WGA and single-cell transcriptome is under development, which can provide new mechanistic insights into the variations in damage levels among different single cells.
MATERIALS AND METHODS
Cell line and culture
MCF10A (CRL-10317; American Type Culture Collection, Manassas, VA, USA) cells were cultured in Dulbecco’s modified Eagle’s medium/F12 (no. 11330-032, Invitrogen, Carlsbad, CA, USA) supplemented with 5% horse serum (no. 16050-122, Invitrogen), epidermal growth factor (EGF) (20 ng/ml; PeproTech, Rocky Hill, NJ, USA), hydrocortisone (0.5 μg/ml; Sigma-Aldrich, no. H-0888), cholera toxin (100 ng/ml; no. C-8052; Sigma-Aldrich, St, Louis, MO, USA), insulin (10 μg/ml; no. I-1882, Sigma-Aldrich), penicillin (100 IU/ml), and streptomycin (100 μg/ml) at 37°C and 5% CO2. To induce cells into the G0 phase, we cultured MCF10A cells in serum-free medium (i.e., medium without horse serum, EGF, and insulin) for 48 hours.
Human tissues and DNA samples
All human tissues were obtained from the NeuroBioBank of the National Institutes of Health (Bethesda, MD, USA) under the supervision of the guidelines. The sample indices were 5554,1740, 4925, 5818, 5844, 4643, 4546, 5246, 5671, and 5182. Prefrontal cortex BA 8–10 regions were used.
Single-cell isolation for cell culture and alkaline lysis
Cultured cells were dissociated with 0.05% trypsin at 37°C for 15 min. Then, cell culture medium was added to inactivate trypsin. Following several washes with phosphate-buffered saline (PBS), single MCF10A cells were picked into a PCR tube containing 2 μl of alkaline lysis buffer [400 mM KOH, 100 mM dithiothreitol (DTT), 2 mM EDTA] by mouth pipetting. After briefly spinning down, the lysis was performed by incubating at 30°C for 1.5 hours. After cell lysis, 2 μl of stop solution [600 mM tris-HCl (pH 7.5) treated by ultraviolet light and 400 mM HCl] was added into each PCR tube to neutralize the lysis buffer. The lysed cells were ready for UDG treatment and LCS-WGA.
Preparation of neuronal nuclei from brain tissues
Nuclei were isolated using the protocol of Krishnaswami et al. (67). Briefly, frozen tissue was first sectioned on ice and was then transferred to a precooled Dounce homogenizer by 1500 μl of homogenization buffer [250 mM sucrose, 25 mM KCl, 5 mM MgCl2, 10 mM tris buffer (pH 8.0), 1 μM DTT, 1× Halt protease inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA, USA), and 0.1% Triton X-100]. The tissue section was then homogenized on ice with 5 strokes of the loose pestle and 12 strokes of the tight pestle. Next, the homogenate was passed through a 40-μm filter and transferred to a 1.7-ml tube. The isolated neuronal nuclei were then pelleted by centrifugation at 4°C (1000g for 8 min) and then resuspended in 500 μl of PBS with 0.5% bovine serum albumin (BSA) (166099A, Thermo Fisher Scientific).
Blocking of nonspecific binding was performed on ice for 15 min. Then, 100 μl of nuclei was transferred to a new tube as an isotype control, while the rest of the nuclei were incubated with anti-NeuN antibody (ab177487; Abcam, Cambridge, England) at room temperature for 30 min on a tube rotator. To wash the samples, we added 500 μl of PBS with 0.5% BSA into the samples and then performed centrifugation at 4°C (500g for 5 min). After removing the supernatant, the nuclei were resuspended in 500 μl of PBS with 0.5% BSA and then incubated with goat anti-rabbit Alexa Fluor 488–conjugated secondary antibody at room temperature for 30 min on a tube rotator. Following the incubation, the nuclei were washed with 500 μl of PBS with 0.5% BSA and pelleted by centrifugation at 4°C (500g for 5 min). We then resuspended the nuclei in cold PBS and used Hoechst 33342 for DNA staining. Neuronal nuclei were identifiable by green and blue fluorescence under a fluorescence microscope. Single neuronal nuclei were picked into PCR tubes with lysis buffer by mouth pipetting. After briefly spinning down, lysis of single cells was performed at 30°C for 1.5 hours. Then, 2 μl of stop solution was added to neutralize the lysis buffer. The lysed single cells were ready for UDG treatment and LCS-WGA.
UDG enzyme treatment
UDG enzyme treatment was performed before the preamplification as follows: 0.2 μl of UDG enzyme [New England Biolabs (NEB)] and 1× ThermoPol reaction buffer (NEB) were added into the single-cell lysate. The single-cell lysate was then incubated at 37°C for 30 min. To evaluate the efficiency of UDG treatment in converting U base to AP site, we have designed a U-containing oligo. DNA oligo was first treated with UDG at 37°C for 30 min—the same treatment used in LCS-WGA. Then, we designed a pair of primers at both ends of this DNA oligo for quantitative PCR (qPCR) quantification, as UDG will leave an AP site on the oligo after it removes U, and the Taq polymerase cannot read through the AP site. Therefore, we could measure the UDG efficiency based on the ΔCt value between UDG-treated and no-treatment groups. As a result, the oligos with UDG treatment was eight cycles behind relative to the control experiment (fig. S3), which corresponds to an efficiency of 99.5% (~200-fold change). The DNA oligo with a uracil base and the qPCR primers are given as follows: U-containing oligo, GATGTGAGTGATGGTTGAGGATGTGTGCTAGAGTUATGTGACCTGGATGTGAGTGAGATGAG; forward primer for qPCR, GATGTGAGTGATGGTTGAGGATGTGTGC; reverse primer for qPCR, CTCATCTCACTCACATCCAG.
Even if all the U are coming from deamination caused by cell lysis, there will be only 80,000 × 0.5% = 400 U that were not processed by UDG. Therefore, 400 U is the upper bound of artifact due to alkaline lysis. In comparison to the total level of C➔T based on the estimation from the MCF10A cells (20,845 ± 2520), this level of technical artifact due to the residual deaminated cytosine can be ignored. To further quantify the artifact levels of deaminated cytosine caused by alkaline lysis, we tested one MCF10A cell without UDG treatment (fig. S2). As a result, we observed the significant increase in the variants of C➔T/G➔A type (~80,000 C➔T de novo variants), indicating that a substantial number in cytosine deamination was introduced during the alkaline lysis. UDG treatment is therefore required to remove these technical artifacts.
Linear copy and split–based whole-genome amplification
The LCS-WGA starts with multiple annealing cycles as described below. Deoxynucleotide triphosphate (dNTP) (300 μM) and 380 nM GAT27NT/NG primers are added into each PCR tube containing the lysed cell: GAT27NG, GTG AGT GAT GGT TGA GGA TGT GTG GAG NNN NNG GG; GAT27NT, GTG AGT GAT GGT TGA GGA TGT GTG GAG NNN NNT TT.
In this first cycle, DNA was melted at 94°C for 50 s. Next, 2.8 U of Bst large fragment polymerase (NEB) was added at 65°C. We then transferred the reaction onto the ice for 30 s to allow the primer hybridization at low temperature. After putting back to the PCR machine, the reaction was performed with the following program: 10°C for 40 s, 20°C for 40 s, 25°C for 40 s, 30°C for 40 s, 40°C for 1 min, 45°C for 1 min, 55°C for 40 s, and 65°C for 4 min.
In the second cycle, DNA was denatured at 94°C for 20 s. Next, 2.8 U of Bst large fragment polymerase (NEB) and 0.25 μl of GAT27 primer (GTGAGTGATGGTTGAGGATGTGTGGAG; 10 μM) were added at 65°C. To convert the full amplicons into double-stranded DNA, eight cycles of double-stranded conversion (63°C for 15 s and 65°C for 20 s) were then performed. After incubating at 65°C for 1 min, the reaction was transferred onto the ice for 30 s for primer hybridization. The following program was then performed: 10°C for 40 s, 20°C for 40 s, 25°C for 40 s, 30°C for 40 s, 40°C for 1 min, 45°C for 1 min, 55°C for 40 s, 65°C for 4 min 30 s. In the third cycle, DNA was first denatured at 94°C for 20 s. Next, 2.8 U of Bst large fragment polymerase was then added at 65°C. The following procedures were the same as the second cycle.
After the three steps of multiple annealing and extension cycles, 0.2 μl of GAT27 primer (10 μM) and 3.8 μl of H2O were added at 78°C, and then DNA was denatured at 94°C for 20 s. After that, 3.6 U of Bst large fragment (NEB) polymerase was added into each tube at 65°C. Next, 30 cycles of double-stranded conversion (63°C for 15 s and 65°C for 20 s) were then performed, followed by one step of 2-min incubation at 65°C. This double stand conversion step warrants the efficient conversion of all full amplicons into the double-stranded DNA before the MDA reaction. Bst large fragment polymerase was inactivated at 72°C for 25 min.
After the preamplification step, qPCR [5 μl of iTaq Universal SYBR Green Supermix, 0.5 μl of GAT27 primers (10 μM), 4 μl of H2O, and 0.5 μl of preamplification products] was performed to quantify the preamplification yield of full amplicons. The Ct values and the melting curves of the qPCR result were used to validate the success of the preamplification. The qPCR program is as follows: 94°C for 2 min for denaturation, 28 cycles of 94°C for 20 s, 60°C for 25 s, and 72°C for 2 min 20 s.
After qPCR quantification, single-cell preamplified products were split into three tubes (about 4.7 μl per split) after 10-s vortex at 600 rpm. MDA master mix (24.6 μl), including 1X phi29 buffer, 1 mM dNTP, 100 μM random hexamer, 0.5% Tween 20, BSA (0.2 mg/ml), and 10 U of phi29 DNA polymerase, was added into each PCR tube. After brief pipetting mixing and centrifuging, the tubes were put on ice for 3 min to increase the efficiency of primer binding. MDA is performed at 30°C for 25 to 30 min with frequent mixing by pipetting. Phi29 DNA polymerase was then inactivated at 65°C for 10 min. After the MDA reaction, the products were purified by using 1X AMPure XP beads and eluted into 6.5 μl of H2O. The yield was quantified by Qubit High Sensitivity dsDNA kit (Invitrogen Life Science). The yield was expected to be around 1 ng per split reaction.
To prove the efficiency of double-stranded conversion, we performed the MDA reaction using only double-stranded full amplicons. With a 100-pg pure full amplicon sample, no detectable DNA product was produced after MDA reaction.
Library construction procedures
We first carried out the tagmentation reaction as follows: 6.3 μl of 2X Illumina Tagment DNA buffer and 0.2 μl of 10-fold diluted TDE1 enzyme were added into the MDA product. The solution was incubated for 2 min at 55°C, followed by adding 1.5 μl of 0.2 M EDTA to release transposase at 50°C for 30 min. Mg(Ac)2 (0.2 M; 1.5 μl) was then added to quench EDTA. NEBNext Ultra II Q5 Master Mix (18.7 μl), 1.5 μl of Index 1 primers (N5), and 1.5 μl of Index 2 primers (N7) were then added to the reaction. The PCR was performed with the following steps: 5 min at 72°C and 30 s at 98°C and then 12 cycles of 10 s at 98°C, 30 s at 58°C, 1 min at 72°C, and 5 min at 72°C. The products were purified by 0.8X AMPure XP beads and eluted to 20 μl of tris-Cl buffer.
The library yield was quantified by using Qubit High Sensitivity dsDNA kit (Invitrogen Life Science), and the library size was examined by using the TapeStation (Agilent). In general, the library size ranges from 300 to 1500 base pairs (bp), and the yield is around 100 ng per split.
Loci test and whole-genome sequencing
Before performing whole-genome sequencing, we also perform qPCR of randomly selected loci to validate the amplification evenness. Here, we randomly select six loci from six chromosomes. For each split reaction, if two or more locus drop out, we will discard the cell. Paired-end sequencing (150 bp × 2) was performed on a HiSeq X10 instrument, with each split having around 10X sequencing depth.
Acknowledgments
We are grateful to the McNair family and C. Neblett for support. We would also like to thank C. Herman, C. Bradley, G. Ira, H. Dierick, W. Dang, H. Zheng, P. Hastings, S. Rosenberg, A. Beaudet, and B. Lee for the helpful discussions. We would also like to thank other Zong lab members for kind support. We would also like to thank the NIH NeuroBioBank for providing brain tissue samples. Funding: This work was supported by the NIH Director’s New Innovator Award (1DP2EB020399) and the McNair Scholarship. Author contributions: C.Z. designed the project and wrote the manuscript. Q.Z. and M.G. contributed to the development of linear preamplification. Q.Z. and Y.N. contributed to the development of splitting MDA strategy and its integration with the preamplification. Q.Z. and Y.N. performed the LCS-WGA experiments of MCF10A and human cortical neurons. C.Z. and Y.N. developed the bioinformatics pipeline and performed the data analysis. Competing interests: C.Z. and M.G are inventors on two patents related to this work filed by the Baylor College of Medicine (no. 16/407,032, filed 8 May 2019, published 29 August 2019; no. 15/308,592, filed 5 May 2015, published 28 May 2019). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabf3329/DC1
REFERENCES AND NOTES
- 1.De Bont R., van Larebeke N., Endogenous DNA damage in humans: A review of quantitative data. Mutagenesis 19, 169–185 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Dizdaroglu M., Coskun E., Jaruga P., Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques. Free Radic. Res. 49, 525–548 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Ames B. N., Shigenaga M. K., Hagen T. M., Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. U.S.A. 90, 7915–7922 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tudek B., Winczura A., Janik J., Siomek A., Foksinski M., Oliński R., Involvement of oxidatively damaged DNA and repair in cancer development and aging. Am. J. Transl. Res. 2, 254–284 (2010). [PMC free article] [PubMed] [Google Scholar]
- 5.Dabin J., Fortuny A., Polo S. E., Epigenome maintenance in response to DNA damage. Mol. Cell 62, 712–727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayano M., Yang J.-H., Bonkowski M. S., Amorim J. A., Ross J. M., Coppotelli G., Griffin P. T., Chew Y. C., Guo W., Yang X., Vera D. L., Salfati E. L., Das A., Thakur S., Kane A. E., Mitchell S. J., Mohri Y., Nishimura E. K., Schaevitz L., Garg N., Balta A.-M., Rego M. A., Gregory-Ksander M., Jakobs T. C., Zhong L., Wakimoto H., Mostoslavsky R., Wagers A. J., Tsubota K., Bonasera S. J., Palmeira C. M., Seidman J. G., Seidman C. E., Wolf N. S., Kreiling J. A., Sedivy J. M., Murphy G. F., Oberdoerffer P., Ksander B. R., Rajman L. A., Sinclair D. A., DNA break-induced epigenetic drift as a cause of mammalian aging. bioRxiv 2019, 808659 (2019). [Google Scholar]
- 7.Gonzalez-Perez A., Sabarinathan R., Lopez-Bigas N., Local determinants of the mutational landscape of the human genome. Cell 177, 101–114 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Tubbs A., Nussenzweig A., Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-Nieto P. E., Schwartz E. K., King D. A., Paulsen J., Collas P., Herrera R. E., Morrison A. J., Carcinogen susceptibility is regulated by genome architecture and predicts cancer mutagenesis. EMBO J. 36, 2829–2843 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshihara M., Jiang L., Akatsuka S., Suyama M., Toyokuni S., Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res. 21, 603–612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J., McKeague M., Sturla S. J., Nucleotide-resolution genome-wide mapping of oxidative DNA damage by click-code-Seq. J. Am. Chem. Soc. 140, 9783–9787 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Poetsch A. R., Boulton S. J., Luscombe N. M., Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis. Genome Biol. 19, 215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding Y., Fleming A. M., Burrows C. J., Sequencing the mouse genome for the oxidatively modified base 8-Oxo-7,8-dihydroguanine by OG-Seq. J. Am. Chem. Soc. 139, 2569–2572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lodato M. A., Rodin R. E., Bohrson C. L., Coulter M. E., Barton A. R., Kwon M., Sherman M. A., Vitzthum C. M., Luquette L. J., Yandava C. N., Yang P., Chittenden T. W., Hatem N. E., Ryu S. C., Woodworth M. B., Park P. J., Walsh C. A., Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodato M. A., Woodworth M. B., Lee S., Evrony G. D., Mehta B. K., Karger A., Lee S., Chittenden T. W., D’Gama A. M., Cai X., Luquette L. J., Lee E., Park P. J., Walsh C. A., Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Waters J., Leung M. L., Unruh A., Roh W., Shi X., Chen K., Scheet P., Vattathil S., Liang H., Multani A., Zhang H., Zhao R., Michor F., Meric-Bernstam F., Navin N. E., Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 512, 155–160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X., Hou Y., Yin X., Bao L., Tang A., Song L., Li F., Tsang S., Wu K., Wu H., He W., Zeng L., Xing M., Wu R., Jiang H., Liu X., Cao D., Guo G., Hu X., Gui Y., Li Z., Xie W., Sun X., Shi M., Cai Z., Wang B., Zhong M., Li J., Lu Z., Gu N., Zhang X., Goodman L., Bolund L., Wang J., Yang H., Kristiansen K., Dean M., Li Y., Wang J., Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell 148, 886–895 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zong C., Lu S., Chapman A. R., Xie X. S., Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338, 1622–1626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navin N., Kendall J., Troge J., Andrews P., Rodgers L., McIndoo J., Cook K., Stepansky A., Levy D., Esposito D., Muthuswamy L., Krasnitz A., McCombie W. R., Hicks J., Wigler M., Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean F. B., Nelson J. R., Giesler T. L., Lasken R. S., Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11, 1095–1099 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gehrke T. H., Lischke U., Gasteiger K. L., Schneider S., Arnold S., Müller H. C., Stephenson D. S., Zipse H., Carell T., Unexpected non-Hoogsteen-based mutagenicity mechanism of FaPy-DNA lesions. Nat. Chem. Biol. 9, 455–461 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Purmal A. A., Lampman G. W., Bond J. P., Hatahet Z., Wallace S. S., Enzymatic processing of uracil glycol, a major oxidative product of DNA cytosine. J. Biol. Chem. 273, 10026–10035 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Purmal A. A., Kow Y. W., Wallace S. S., Major oxidative products of cytosine, 5-hydroxycytosine and 5-hydroxyuracil, exhibit sequence context-dependent mispairing in vitro. Nucleic Acids Res. 22, 72–78 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong X., Zhang L., Milholland B., Lee M., Maslov A. Y., Wang T., Vijg J., Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat. Methods 14, 491–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luquette L. J., Bohrson C. L., Sherman M. A., Park P. J., Identification of somatic mutations in single cell DNA-seq using a spatial model of allelic imbalance. Nat. Commun. 10, 3908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C., Xing D., Tan L., Li H., Zhou G., Huang L., Xie X. S., Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI). Science 356, 189–194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreutzer D. A., Essigmann J. M., Oxidized, deaminated cytosines are a source of C → T transitions in vivo. Proc. Natl. Acad. Sci. U.S.A. 95, 3578–3582 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagner J. R., Hu C. C., Ames B. N., Endogenous oxidative damage of deoxycytidine in DNA. Proc. Natl. Acad. Sci. U.S.A. 89, 3380–3384 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neeley W. L., Essigmann J. M., Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 19, 491–505 (2006). [DOI] [PubMed] [Google Scholar]
- 31.David S. S., O’Shea V. L., Kundu S., Base-excision repair of oxidative DNA damage. Nature 447, 941–950 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galashevskaya A., Sarno A., Vågbø C. B., Aas P. A., Hagen L., Slupphaug G., Krokan H. E., A robust, sensitive assay for genomic uracil determination by LC/MS/MS reveals lower levels than previously reported. DNA Repair 12, 699–706 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Jaruga P., Dizdaroglu M., Repair of products of oxidative DNA base damage in human cells. Nucleic Acids Res. 24, 1389–1394 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soares J. P., Cortinhas A., Bento T., Leitão J. C., Collins A. R., Gaivã I., Mota M. P., Aging and DNA damage in humans: A meta-analysis study. Aging 6, 432–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McConnell M. J., Lindberg M. R., Brennand K. J., Piper J. C., Voet T., Cowing-Zitron C., Shumilina S., Lasken R. S., Vermeesch J. R., Hall I. M., Gage F. H., Mosaic copy number variation in human neurons. Science 342, 632–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T., Birsoy K., Hughes N. W., Krupczak K. M., Post Y., Wei J. J., Lander E. S., Sabatini D. M., Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blomen V. A., Majek P., Jae L. T., Bigenzahn J. W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F. R., Olk N., Stukalov A., Marceau C., Janssen H., Carette J. E., Bennett K. L., Colinge J., Superti-Furga G., Brummelkamp T. R., Gene essentiality and synthetic lethality in haploid human cells. Science 350, 1092–1096 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Bartha I., di Iulio J., Venter J. C., Telenti A., Human gene essentiality. Nat. Rev. Genet. 19, 51–62 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Hart T., Chandrashekhar M., Aregger M., Steinhart Z., Brown K. R., MacLeod G., Mis M., Zimmermann M., Fradet-Turcotte A., Sun S., Mero P., Dirks P., Sidhu S., Roth F. P., Rissland O. S., Durocher D., Angers S., Moffat J., High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Ochs F., Karemore G., Miron E., Brown J., Sedlackova H., Rask M. B., Lampe M., Buckle V., Schermelleh L., Lukas J., Lukas C., Stabilization of chromatin topology safeguards genome integrity. Nature 574, 571–574 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Schmitt A. D., Hu M., Jung I., Xu Z., Qiu Y., Tan C. L., Li Y., Lin S., Lin Y., Barr C. L., Ren B., A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 17, 2042–2059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N. T., Morris J. H., Bork P., Jensen L. J., Mering C., STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grubman A., Chew G., Ouyang J. F., Sun G., Choo X. Y., McLean C., Simmons R. K., Buckberry S., Vargas-Landin D. B., Poppe D., Pflueger J., Lister R., Rackham O. J. L., Petretto E., Polo J. M., A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22, 2087–2097 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J. Z., Menon M., He L., Abdurrob F., Jiang X., Martorell A. J., Ransohoff R. M., Hafler B. P., Bennett D. A., Kellis M., Tsai L. H., Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Velmeshev D., Schirmer L., Jung D., Haeussler M., Perez Y., Mayer S., Bhaduri A., Goyal N., Rowitch D. H., Kriegstein A. R., Single-cell genomics identifies cell type–specific molecular changes in autism. Science 364, 685–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu T., Pan Y., Kao S.-Y., Li C., Kohane I., Chan J., Yankner B. A., Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Huang D. W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raj T., Li Y. I., Wong G., Humphrey J., Wang M., Ramdhani S., Wang Y. C., Ng B., Gupta I., Haroutunian V., Schadt E. E., Young-Pearse T., Mostafavi S., Zhang B., Sklar P., Bennett D. A., de Jager P. L., Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet. 50, 1584–1592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsieh Y.-C., Guo C., Yalamanchili H. K., Abreha M., Al-Ouran R., Li Y., Dammer E. B., Lah J. J., Levey A. I., Bennett D. A., De Jager P. L., Seyfried N. T., Liu Z., Shulman J. M., Tau-mediated disruption of the spliceosome triggers cryptic RNA splicing and neurodegeneration in Alzheimer’s disease. Cell Rep. 29, 301–316.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mattson M. P., Cellular actions of beta-amyloid precursor protein and its soluble and fibrillogenic derivatives. Physiol. Rev. 77, 1081–1132 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Fang E. F., Hou Y., Palikaras K., Adriaanse B. A., Kerr J. S., Yang B., Lautrup S., Hasan-Olive M. M., Caponio D., Dan X., Rocktäschel P., Croteau D. L., Akbari M., Greig N. H., Fladby T., Nilsen H., Cader M. Z., Mattson M. P., Tavernarakis N., Bohr V. A., Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 22, 401–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manczak M., Reddy P. H., Abnormal interaction of VDAC1 with amyloid beta and phosphorylated tau causes mitochondrial dysfunction in Alzheimer’s disease. Hum. Mol. Genet. 21, 5131–5146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Busche M. A., Hyman B. T., Synergy between amyloid-β and tau in Alzheimer’s disease. Nat. Neurosci. 23, 1183–1193 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L. A., Lander E. S., Dekker J., Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao S. S., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., Robinson J. T., Sanborn A. L., Machol I., Omer A. D., Lander E. S., Aiden E. L., A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., Ren B., Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V. V., Ren B., A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oberdoerffer P., Sinclair D. A., The role of nuclear architecture in genomic instability and ageing. Nat. Rev. Mol. Cell Biol. 8, 692–702 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Tamburini B. A., Tyler J. K., Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25, 4903–4913 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peleg S., Sananbenesi F., Zovoilis A., Burkhardt S., Bahari-Javan S., Agis-Balboa R. C., Cota P., Wittnam J. L., Gogol-Doering A., Opitz L., Salinas-Riester G., Dettenhofer M., Kang H., Farinelli L., Chen W., Fischer A., Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Krishnan V., Chow M. Z. Y., Wang Z., Zhang L., Liu B., Liu X., Zhou Z., Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 108, 12325–12330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vijg J., Suh Y., Genome instability and aging. Annu. Rev. Physiol. 75, 645–668 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Madabhushi R., Pan L., Tsai L.-H., DNA damage and its links to neurodegeneration. Neuron 83, 266–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nativio R., Lan Y., Donahue G., Sidoli S., Berson A., Srinivasan A. R., Shcherbakova O., Amlie-Wolf A., Nie J., Cui X., He C., Wang L. S., Garcia B. A., Trojanowski J. Q., Bonini N. M., Berger S. L., An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet. 52, 1024–1035 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klein H. U., McCabe C., Gjoneska E., Sullivan S. E., Kaskow B. J., Tang A., Smith R. V., Xu J., Pfenning A. R., Bernstein B. E., Meissner A., Schneider J. A., Mostafavi S., Tsai L. H., Young-Pearse T. L., Bennett D. A., de Jager P. L., Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains. Nat. Neurosci. 22, 37–46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krishnaswami S. R., Grindberg R. V., Novotny M., Venepally P., Lacar B., Bhutani K., Linker S. B., Pham S., Erwin J. A., Miller J. A., Hodge R., McCarthy J. K., Kelder M., McCorrison J., Aevermann B. D., Fuertes F. D., Scheuermann R. H., Lee J., Lein E. S., Schork N., McConnell M. J., Gage F. H., Lasken R. S., Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc. 11, 499–524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabf3329/DC1