Zeb2 controls neocortex structure by regulating migration onset and neuronal orientation via Nrp1 and Cdh6 adhesive pathways.
Abstract
The neocortex is stereotypically organized into layers of excitatory neurons arranged in a precise parallel orientation. Here we show that dynamic adhesion both preceding and following radial migration is essential for this organization. Neuronal adhesion is regulated by the Mowat-Wilson syndrome-associated transcription factor Zeb2 (Sip1/Zfhx1b) through direct repression of independent adhesion pathways controlled by Neuropilin-1 (Nrp1) and Cadherin-6 (Cdh6). We reveal that to initiate radial migration, neurons must first suppress adhesion to the extracellular matrix. Zeb2 regulates the multipolar stage by transcriptional repression of Nrp1 and thereby downstream inhibition of integrin signaling. Upon completion of migration, neurons undergo an orientation process that is independent of migration. The parallel organization of neurons within the neocortex is controlled by Cdh6 through atypical regulation of integrin signaling via its RGD motif. Our data shed light on the mechanisms that regulate initiation of radial migration and the postmigratory orientation of neurons during neocortical development.
INTRODUCTION
The neocortex regulates higher brain functions including cognition, language, and voluntary movement. The most abundant neuronal cell type in the neocortex is the pyramidal excitatory neuron, organized in stereotypic six layers. These neurons are precisely positioned within these layers, so that apical dendrites extend parallel to each other and perpendicular to the pia. This strict arrangement allows spatial compartmentalization of connectivity to both different layers and to distinct portions of the dendritic arbor. For example, long-range cortico-cortical input to layer II/III [upper layer (UL)] neurons occurs primarily onto distal portions of the apical dendrites, while short-range input from layer IV and the neighboring layer is primarily made onto basal dendrites (1). Alterations in neocortical cytoarchitecture or morphological deficits in dendritic arborizations of excitatory neurons are tightly associated with cognitive impairment (2–4). Despite the importance of acquiring precise anatomical and functional organization within the neocortex, the molecular mechanisms involved are not fully understood.
During early neurogenesis [embryonic day 11 (E11)], postmitotic neurons migrate mainly through somal translocation to form the preplate (5). This transient structure is split into subplate and marginal zone (MZ) by the subsequent migration of neurons, the first of which form the deepest layer (layer VI) of the cortical plate (CP). Subsequently born neurons must migrate past the earlier generated neurons to form more superficial layers, thereby building up the CP in an inside-out fashion (6). During this process, newborn neurons first adopt a multipolar morphology in the subventricular zone (SVZ) characterized by multiple processes (7). This multipolar stage can last up to 24 hours, after which neurons acquire a bipolar morphology, attach to radial glial fibers, and move along these fibers toward the MZ (8–10). After detachment from the radial glia fiber at the MZ, neurons undergo terminal somal translocation to reach their final position and subsequently initiate dendritic arborization (3, 5, 8, 11).
Many questions concerning this complex behavior of developing cortical neurons remain to be answered. What is the reason for the initial multipolar state and what determines when neurons start radial migration? Does neuronal polarity or other spatial information acquired before or during radial migration direct their final parallel orientation after detachment from radial fibers? Or do neurons undergo an independent reorientation process after completion of migration?
Heterozygous mutations in the gene encoding the transcriptional repressor ZEB2 (Zinc finger E-box–binding homeobox 2, also known as ZFHX1B or SIP1) are reported to cause Mowat-Wilson syndrome, a human condition characterized by severe intellectual disability, distinct facial features, and seizures (12). Zeb2 expression in the neocortex coincides with the appearance of postmitotic neurons from E12.5 onward and remains high in the CP throughout adulthood (13–15). Studies using Zeb2-deficient mice have addressed the cellular mechanisms underlying some of these congenital defects. These include analysis of the deficiencies in neural crest cell development that result in altered craniofacial structures (16, 17) and in defective development of the enteric nervous system (18), as well as of the disturbances in axonal outgrowth and myelination that may underlie agenesis of the corpus callosum observed in some Mowat-Wilson patients (19). Zeb2 has also been shown to regulate the correct proportion of pyramidal neurons and glia by controlling the onset of generation of UL neurons (14) and early hippocampal development (13). However, the role of Zeb2 in the establishment of neocortical cytoarchitecture or dendritic arborization has not yet been addressed.
Here, we show that organization of UL neurons in the neocortex, including their position, final orientation, and dendritic arborization, is regulated by dynamic changes in adhesive properties, under the master control of Zeb2. Specifically, we show that Zeb2-mediated repression of Neuropilin-1 (Nrp1) and the consequent suppression of downstream integrin-mediated adhesion are necessary for UL neurons to timely initiate radial migration. This pathway also controls the complexity of the dendritic arbor. In addition, we demonstrate that a second Zeb2-regulated pathway involving the cell adhesion receptor, Cadherin-6 (Cdh6), is required for the correct orientation and final positioning of UL neurons within the neocortex. Last, we show that Cdh6 regulates neuronal orientation through the Cdh6 integrin-binding arginine–glycine–aspartic acid (RGD) motif.
RESULTS
The onset of radial migration is controlled by Zeb2
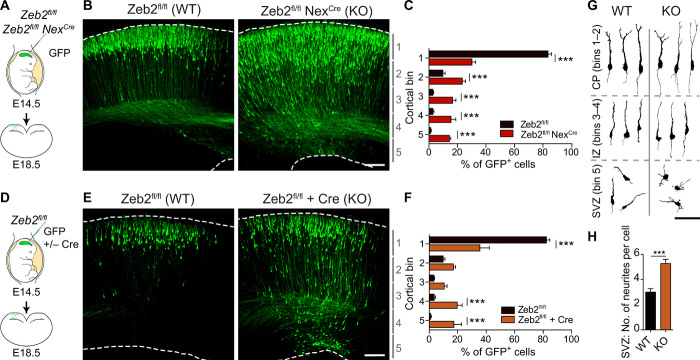
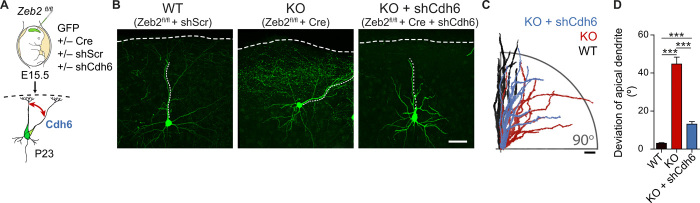
Radial migration of UL neurons to their correct laminar position is an essential step for the organization of the neocortex. Multiple neocortical defects associated with aberrant neuronal migration, including microcephaly, cortical dysplasia, heterotopias, pachygyria, and polymicrogyria, have been observed in Mowat-Wilson syndrome patients (20, 21). We therefore addressed whether Zeb2 regulates the positioning of neurons in the neocortex, by conditionally mutating Zeb2 in postmitotic neurons of the dorsal telencephalon. This was achieved through crosses between a floxed Zeb2 line [Zeb2fl/fl; (22)] and the Nex1Cre/+ mouse line [also called Neurod6Cre; (23)]. UL neurons were labeled by in utero electroporation (IUE) of green fluorescent protein (GFP) at E14.5 and analyzed 4 days later at E18.5 (Fig. 1, A to C). At this time point, most neurons in the wild-type (WT) brains have completed migration and were found in the upper cortical layers (bin 1; Fig. 1, B and C). In Zeb2-deficient mice, however, a substantial number of GFP+ cells were found accumulated in deeper layers of the developing cortex (Fig. 1, B and C). Zeb2-deficient displaced neurons retained expression of the UL-specific marker Satb2 and lacked expression of the deep layer marker Ctip2, suggesting that loss of Zeb2 function does not alter the cell identity of UL neurons (fig. S1, A and B).
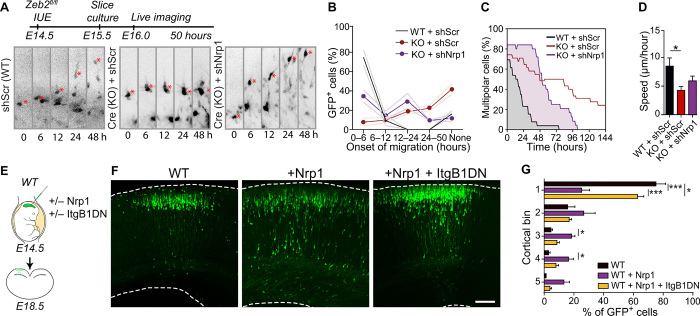
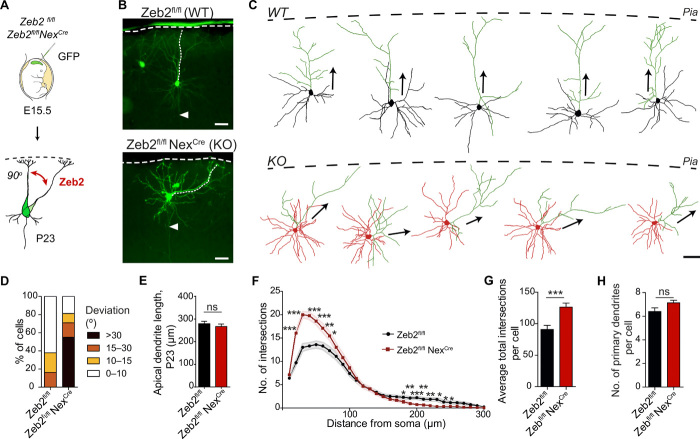
Fig. 1. Zeb2 promotes the onset of radial migration.
(A to C) Loss of Zeb2 causes laminar displacement of UL neurons. (A) Control (Zeb2fl/fl) and Zeb2-deficient (Zeb2fl/fl NexCre) littermate animals were in utero electroporated at E14.5 with a GFP expression construct and analyzed at E18.5. (B) Representative images of GFP+ neurons in neocortical slices. Scale bar, 100 μm. (C) Laminar distribution of GFP+ cells in each cortical bin. The division of the neocortex into five equally sized bins is shown on the right. N = 12 Zeb2fl/fl and 5 Zeb2fl/fl NexCre animals. Two-way analysis of variance (ANOVA) with Bonferroni post hoc test. (D to F) Zeb2 regulates laminar position in a cell-intrinsic fashion. (D) Zeb2fl/fl animals were in utero electroporated at E14.5 with GFP in the presence or absence of Cre recombinase expressed under the postmitotic promoter Neurod1 (Cre) and analyzed at E18.5. (E) Representative images of GFP+ neurons in neocortical slices. Scale bar, 100 μm. (F) Laminar distribution of GFP+ neurons in vivo. N = 11 control and 6 Cre-expressing brains. Two-way ANOVA with Bonferroni post hoc test. (G) Morphology of GFP+ neurons in the CP, intermediate zone (IZ), or SVZ as indicated, at E18.5. Scale bar, 100 μm. (H) Number of neurites per cell for GFP+ neurons in the SVZ at E18.5. N = 15 per condition. Unpaired t test. KO, knockout; WT, wild type.
In the cerebral cortex, Zeb2 can act cell intrinsically and extrinsically (24). To test whether the observed lamination deficits are due to the cell-intrinsic loss of Zeb2 function in UL neurons, we carried out mosaic deletion of Zeb2 by IUE of GFP and a plasmid encoding Cre recombinase under the postmitotic Neurod1 promoter (Cre), into the developing cortex of E14.5 Zeb2fl/fl mice. This approach induces Zeb2 deletion in a subset of UL neurons embedded in the WT tissue environment. In keeping with our previous observations, Neurod1-Cre–mediated mutation of Zeb2 in UL neurons led to the aberrant localization of these cells to deeper neocortical layers at E18.5 (Fig. 1, D to F, compared to Fig. 1, A to C). Mislocalization of late-born neurons upon loss of Zeb2 is thus a cell-intrinsic effect, independent of Zeb2 functions in other cell types or in earlier born neurons.
A closer inspection of the SVZ of Zeb2-deficient (Zeb2fl/fl Neurod1Cre) brains at E18.5 showed that Zeb2-deficient neurons accumulated here displayed multipolar morphologies that lacked clear polarity as compared to the few control counterparts still present in this region at this time point (Fig. 1, G and H, and fig. S1, C and D). On the other hand, Zeb2-deficient neurons that managed to progress into the intermediate zone, a transit region of migrating neurons en route to the CP, had a relatively normal bipolar morphology and orientation (Fig. 1, G and H, and fig. S1, C and D).
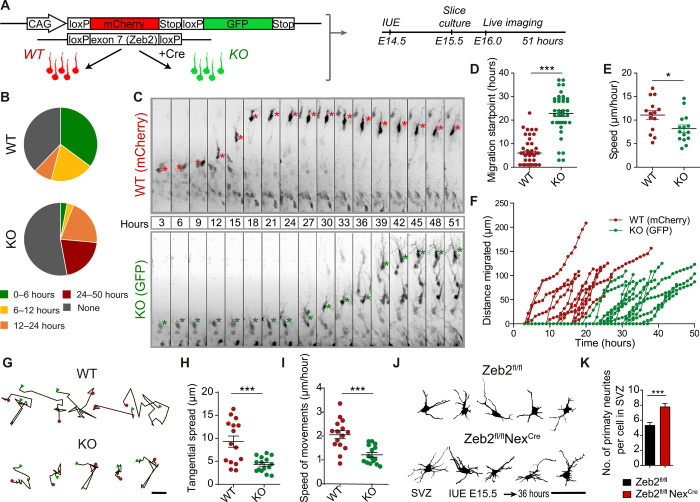
We investigated in more detail how Zeb2 regulates radial migration by live imaging of in utero labeled neurons in live brain slices. To analyze the behavior of WT and Zeb2-deficient UL neurons “side by side” in the same brain section, we coelectroporated a fl-mCherry-Stop-fl-GFP reporter construct with limiting amounts of a Cre recombinase into E14.5 Zeb2fl/fl mice and prepared live brain slices 1 day later. This approach generates a mixed population of WT control (mCherry positive) and Zeb2-deficient (GFP+) neurons in the same brain slice. Electroporated cells that incorporate only the reporter construct express mCherry (WT, Zeb2fl/fl control cells) and those that incorporate both constructs undergo Cre-mediated recombination of both the Zeb2fl/fl alleles and the loxP sites flanking the mCherry-Stop cassette of the reporter construct and thereby now express GFP and are Zeb2 deficient (Fig. 2A). We confirmed that coexpression of the fl-mCherry-Stop-fl-GFP reporter and limiting amounts of Cre recombinase in WT animals does not by itself affect the radial migration of WT UL neurons (fig. S2).
Fig. 2. Zeb2 regulates the multipolar stage.
(A to I) Zeb2 regulates onset of radial migration and restricts tangential movement of multipolar cells. (A) E14.5 Zeb2fl/fl animals were in utero electroporated with a flox-mCherry-Stop-flox-GFP reporter and limiting amounts of Cre. Cre recombination excises Zeb2 and enables GFP expression (KO). Unrecombined cells express mCherry (WT). Brain slices were imaged for 51 hours, 1 day after IUE. (B) Distribution of radial migration onset. N = 37 mCherry+ and 34 GFP+ cells. (C) Clips from a time-lapse movie showing migration of WT and KO cells. Asterisks mark the soma. Migration onset (D) and speed (E) of neurons undergoing migration. N = 46 mCherry+ and 41 GFP+ cells. Mann-Whitney and unpaired t test. (F) Traces of individual neurons undergoing migration. (G) Movement of multipolar cells in the SVZ (0 to 20 hours). Scale bar, 5 μm. A red dot marks the start and a green triangle marks the end position. Tangential spread (H) and speed (I) of neurons in the SVZ. N = 15 WT and KO cells. Unpaired t test. (J and K) Zeb2 regulates multipolar morphology. (J) Zeb2fl/fl and Zeb2fl/fl NexCre animals were in utero electroporated at E15.5 with GFP and their morphology in the SVZ was analyzed 36 hours later. Scale bar, 50 μm. (K) Number of primary neurites per cell. N = 20 Zeb2fl/fl and Zeb2fl/fl NexCre cells. Unpaired t test.
Analysis of coelectroporated Zeb2fl/fl brain slices showed that loss of Zeb2 function led to a large decrease in the proportion of UL neurons that initiated their radial migration during the imaged 51-hour period (Fig. 2B and movie S1). Furthermore, Zeb2-deficient UL neurons that began migration during imaging do so with a substantial delay of around 24 hours when compared to control cells in the same slice (Fig. 2, C and D). Once migration was initiated, however, Zeb2-deficient neurons showed normal cell polarity, direction, and progression of movement with only marginally slower speed in comparison to WT neurons (Fig. 2, E and F). This suggests that Zeb2 controls neuronal laminar position by regulating the onset of radial migration.
We thus next analyzed the multipolar stage of neuronal differentiation more closely. During the multipolar stage, neurons spread sideways in the SVZ away from their position of birth (tangential spread) and characteristically show dynamic extension and retraction of multiple neurites (8, 25). Analysis of the movement of individual cells within the VZ/SVZ before initiation of radial migration showed that loss of Zeb2 reduces the tangential spread and speed of movement of neurons during the multipolar stage (Fig. 2, G to I). To further investigate the morphology of neurons during this stage, we electroporated GFP into Zeb2-deficient (Zeb2fl/fl NexCre) and control animals at E15.5 and analyzed the cells 36 hours later. This revealed that Zeb2-deficient multipolar neurons in the SVZ produce more neurites than WT neurons (Fig. 2, J and K).
Together, these data indicate that Zeb2 alters the multipolar stage of development itself to promote initiation of radial migration.
Suppression of adhesion to the extracellular matrix is necessary for onset of radial migration
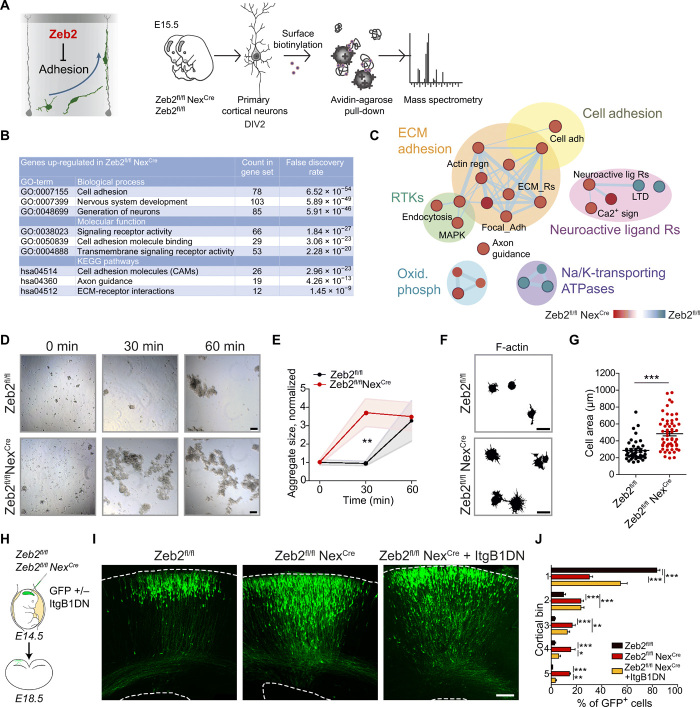
We hypothesized that Zeb2 might regulate the onset of radial migration by altering the interactions of young cortical neurons with their extracellular environment (Fig. 3A). We thus searched for receptors whose expression at the plasma membrane is altered by loss of Zeb2 using surface biotinylation of primary cortical neurons isolated from E15.5 control (Zeb2fl/f) and Zeb2-deficient (Zeb2fl/fl NexCre) embryos. Biotinylated proteins were isolated and subsequently processed for identification by mass spectrometry (MS) (Fig. 3, A to C). Using this approach, we identified over 200 proteins whose surface expression was elevated upon loss of Zeb2 (Fig. 3, A to C, and table S1). These included a number of known targets of Zeb2, such as the fibroblast growth factor receptor (Fgfr1) and the NT-3 growth factor receptor (Ntrk3) (14, 26, 27). Gene set enrichment analysis (GSEA) indicated strong enrichment of adhesion-related proteins in Zeb2-deficient neurons. These included integrins, which are involved in interactions with the extracellular matrix, as well as cadherins that are primarily involved in adhesion to other cells (Fig. 3, A to C, and table S1).
Fig. 3. Zeb2 represses cell adhesion.
(A to C) Zeb2 represses neuronal adhesion. (A) Surface proteins on Zeb2fl/fl NexCre and Zeb2fl/fl cortical neurons prepared from E15.5 embryos were identified by biotin-linked mass spectrometry. DIV, days in vitro. (B) Top Gene Ontology (GO) terms for surface proteins up-regulated ≥1.5× upon loss of Zeb2. ECM, extracellular matrix; KEGG, Kyoto Encyclopedia of Genes and Genomes. (C) GO of surface-expressed proteins. Red nodes are up-regulated in Zeb2fl/fl NexCre and blue nodes are up-regulated in Zeb2fl/fl animals. ATPase, adenosine triphosphatase; MAPK, mitogen-activated protein kinase; RTK, receptor tyrosine kinase. (D and E) Zeb2 suppresses neuronal aggregation. (D) Aggregation of single-cell suspensions over time. Scale bars, 100 μm. (E) Average cell aggregate size. N = 15, 10, and 7 Zeb2fl/fl and 12, 12, and 15 Zeb2fl/fl NexCre aggregates at 0, 30, and 60 min, respectively. One-way ANOVA with Kruskal-Wallis test. (F and G) Zeb2 inhibits adhesion to the extracellular matrix. (F) Attachment of neuronal suspensions to laminin-coated surfaces after 2 hours. Scale bars, 15 μm. (G) Lamellipodial spreading of attached cells. N = 42 Zeb2fl/fl and 56 Zeb2fl/fl NexCre cells. Mann-Whitney test. (H to J) Zeb2 regulates radial migration through suppression of integrin signaling. (H) IUE of GFP and ItgB1DN into Zeb2fl/fl and Zeb2fl/fl NexCre E14.5 littermate animals. (I) GFP+ neurons at E18.5. Scale bar, 100 μm. (J) Laminar distribution of GFP+ neurons. N = 12 Zeb2fl/fl, 5 Zeb2fl/fl NexCre, and 5 Zeb2fl/fl NexCre + ItgB1DN animals. Two-way ANOVA with Bonferroni post hoc test.
The increase in surface expression of these molecules suggests that Zeb2 regulates the adhesive behavior of neurons. To test this hypothesis, we carried out in vitro cell aggregation (Fig. 3, D and E) and adherence assays (Fig. 3, F and G) using single-cell suspensions prepared from control (Zeb2fl/fl) and Zeb2-deficient (Zeb2fl/fl NexCre) E15.5 littermate embryos. Zeb2-deficient neurons showed much faster cell aggregation in suspension as compared to control cells (Fig. 3, D and E). Similarly, Zeb2-deficient neurons specified more extensive lamellipodia on laminin, an integrin ligand (Fig. 3, F and G). Together, these data indicate that, in neurons, Zeb2 suppresses both cell-cell adhesion and adhesion to the extracellular matrix.
Our MS data showed increased surface expression of several integrin receptors, including the integrin β1 (Itgb1), in Zeb2-deficient neurons (table S1). Integrins function as heterodimeric αβ complexes and are the main mediators of adhesion to the extracellular matrix (28). We hypothesized that increased levels of integrin receptors at the surface of Zeb2-deficient UL neurons delay the onset of their migration. To test this, we generated a construct encoding a dominant negative mutant of Itgb1 (called here ItgB1DN). ItgB1DN lacks the cytoplasmic domain and thereby interferes with integrin signaling by forming unproductive complexes with multiple integrin α subunits (29). Because integrin signaling is required for progenitor cell attachment, proliferation, and survival (30), we expressed ItgB1DN specifically in postmitotic cells (under the control of the Neurod1 promoter). Coelectroporation of this Neurod1-driven ItgB1DN construct and GFP in E14.5 WT embryos did not interfere with the radial migration of WT UL neurons (fig. S3). However, inhibition of integrin signaling significantly improved the laminar position of Zeb2-deficient neurons (Fig. 3, H to J). Suppression of integrin-mediated adhesion by Zeb2 is thus necessary for UL neurons to initiate their migration away from the SVZ on time.
Nrp1 is a direct target of Zeb2 involved in the regulation of integrin-mediated adhesion
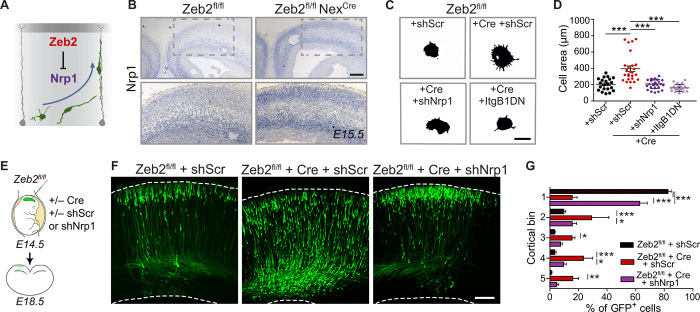
Although we observed the elevated plasma membrane localization of several integrin subunits upon loss of Zeb2 (table S1), these were not up-regulated at the mRNA level in Zeb2-deficient mice [fig. S4; (14)]. We searched for a direct Zeb2 target that could regulate neuronal adhesion and migration, by cross-referencing our MS list of up-regulated surface proteins against our previously published microarray data generated from Zeb2-deficient brains (14). This led to the identification of Nrp1 as a potential direct Zeb2 target. In situ hybridization for Nrp1 confirmed its increased expression in Zeb2-deficient (Zeb2fl/fl NexCre) brains (Fig. 4, A and B). The expression of Sema3A or 3C, which can function as ligands for Nrp1, or their plexin co-receptor was not altered upon loss of Zeb2 (fig. S4, D to F).
Fig. 4. Nrp1, a novel Zeb2-target, regulates the laminar position of UL neurons.
(A) Zeb2 represses Nrp1 expression. (B) ISH for Nrp1 in Zeb2fl/fl and Zeb2fl/fl NexCre animals at E15.5. Scale bar, 50 μm. (C and D) Nrp1 and integrins mediate enhanced adhesion of Zeb2-deficient neurons. (C) Adherence of Zeb2fl/fl primary cortical neurons transfected with scrambled short hairpin–mediated RNA (shScr), an Nrp1 shRNA (shNrp1) or ItgB1DN, and Cre as indicated to laminin-coated surfaces for 2 hours. Scale bar, 15 μm. (D) Lamellipodial spreading. N = 21 Zeb2fl/fl + shScr, 26 Zeb2fl/fl + Cre + shScr, 26 Zeb2fl/fl Cre + shNrp1, and 22 Zeb2fl/fl + Cre + ItgB1DN. One-way ANOVA (Kruskal-Wallis test) with Dunn’s multiple comparison test. (E to G) Zeb2 regulates laminar position through repression of Nrp1. (E) Zeb2fl/fl animals were in utero electroporated at E14.5 with shScr, shNrp1, and Cre as indicated and analyzed at E18.5. (F) GFP+ neurons in the neocortex. Scale bar, 100 μm. (G) Laminar distribution of GFP+ neurons at E18.5. N = 11 Zeb2fl/fl + shScr, 5 Zeb2fl/fl + shScr and Cre, and 7 Zeb2fl/fl + Cre + shNrp1 animals. Two-way ANOVA with Bonferroni post hoc test.
We next asked whether Nrp1 might be a direct target of Zeb2. We used the FANTOM5 database (https://fantom.gsc.riken.jp/5/) to locate the potential promoter region in the locus of the Nrp1 gene. Promoter regions often show enrichment in histone H3K27 acetylation levels, CpG islands, and deoxyribonuclease (DNase) I hypersensitivity clusters. We identified three such regions around 6 kb upstream from the Nrp1 transcriptional start site (RR1 to RR3) and found 11 Zeb2 consensus-binding sequences, CACCT(G) (31), within them (fig. S5). We assessed whether these sites represent genuine Zeb2 binding by chromatin immunoprecipitation from chromatin prepared from E15.5 WT neocortices using Zeb2-specific antibodies followed by quantitative polymerase chain reaction (ChIP-qPCR). This showed strong enrichment of Zeb2 at the analyzed sites, suggesting that Nrp1 is a direct target of Zeb2 (fig. S5C).
Although an adhesion-related function for Nrp1 has not been described in neurons, Nrp1 is reported to enhance integrin signaling in other cell types (32). We addressed whether Nrp1 also regulates integrin-mediated adhesion in neurons using a specific short hairpin–mediated RNA (shRNA) to down-regulate Nrp1 expression in Zeb2-deficient neurons (shNrp1; fig. S4G). Down-regulation of Nrp1 normalized the adherence of Zeb2-deficient neurons to laminin-coated plates to a similar extent as the dominant negative Itgb1 mutant, ItgB1DN (Fig. 4, C and D). Together, these data establish Nrp1 as a direct target of Zeb2 and as a previously unidentified regulator of integrin-mediated adhesion to the extracellular matrix in neurons.
Nrp1 delays initiation of radial migration through regulation of integrin signaling
Next, we asked whether repression of Nrp1 by Zeb2 is necessary for radial migration. To test this, we down-regulated Nrp1 expression in Zeb2-deficient (Zeb2fl/fl NexCre) UL neurons by IUE of shNrp1 at E14.5 (Fig. 4, E to G). Down-regulation of Nrp1 expression in Zeb2-deficient UL neurons restored their correct laminar position at E18.5. To investigate whether, like Zeb2, Nrp1 specifically influences the onset of radial migration, we conducted the live imaging of Zeb2-deficient neurons in brain slices freshly prepared from E14.5 Zeb2fl/fl animals electroporated in utero with Cre and either a control scrambled shRNA (shScr) or an shRNA against Nrp1 (shNrp1). A systematic analysis of the time-lapse recordings showed that upon down-regulation of Nrp1, Zeb2-deficient neurons initiate their migration earlier and have a faster transition to bipolar morphology (Fig. 5, A to C, and movie S2). Down-regulation of Nrp1 did not affect the speed of movement of cells once migration was initiated (Fig. 5D).
Fig. 5. Nrp1 controls the onset of migration in an integrin-dependent manner.
(A to D) Nrp1 regulates migration onset downstream of Zeb2. (A) Littermate Zeb2fl/fl animals were in utero electroporated at E14.5 with GFP, shScr, shNrp1, or Cre as indicated. Selected clips from the 50-hour time-lapse movie are shown. (B) Percentage of GFP+ cells starting migration during the indicated time windows. None = cells that do not initiate migration during imaging. N = 3 animals per condition. Two-way ANOVA with Bonferroni post hoc tests. (C) Proportion of multipolar GFP+ cells at different time points. N = 29 cells, 3 brains for WT; 33 cells, 3 brains for KO; and 25 cells, 3 brains KO + shNrp1. (D) Speed of radial migration. N = 9 WT + shScr, 9 KO + shScr, and 19 KO + shNrp1. One-way ANOVA (Kruskal-Wallis) with Dunn’s multiple comparison test. (E to G) Nrp1 induces laminar displacement through integrins. (E) WT animals were in utero electroporated at E14.5 with an internal ribosomal entry site–driven GFP construct, the same construct containing Nrp1 or with ItgB1DN as indicated. (F) GFP+ neurons in brain sections at E18.5. Scale bar, 100 μm. (G) Laminar distribution of GFP+ neurons. N = 7 control WT, 7 WT + Nrp1, and 9 WT + Nrp1 + ItgB1DN animals.
We then asked whether enhanced Nrp1 signaling is sufficient to induce laminar displacement of UL neurons. Overexpression of Nrp1 in the neocortex at E14.5 caused the laminar displacement of UL neurons to deeper positions in a fashion reminiscent of the phenotype displayed by Zeb2-deficient UL neurons (Fig. 5, E and F; see also Fig. 1, B to F). Last, we asked whether Nrp1 controls neuronal position in vivo through regulation of integrin signaling, by coexpressing ItgB1DN with Nrp1 in UL neurons. Inhibition of integrin signaling in Nrp1-overexpressing UL neurons restored their normal laminar position at E18.5 (Fig. 5, E to G). In other systems, Nrp1 has been shown to form a protein complex with various integrin subunits (32). To address whether this protein interaction is conserved in neurons, we carried out a proximity ligation assay for endogenous Nrp1 and Itgb1 in primary cortical neurons from E15.5 littermate Zeb2fl/fl and Zeb2fl/fl NexCre embryos. This showed that the interaction of endogenous Nrp1 and Itgb1 proteins in cortical neurons is enhanced upon loss of Zeb2 (fig. S6).
Together, these data demonstrate that Zeb2 controls the onset of radial migration by repression of Nrp1 and downstream adhesion. Nrp1 forms protein complexes with integrins to promote the integrin-driven adhesion of young neurons to the extracellular matrix.
Zeb2 regulates the postnatal orientation of neurons and dendritic arborization
We then assessed the consequences of the delayed onset of migration for the postnatal cytoarchitecture of the neocortex. Neurons in Zeb2-deficient (Zeb2fl/fl NexCre) and control (Zeb2fl/fl) brains were stained using the Golgi-Cox method at postnatal day 23 (P23), a time point at which neuronal maturity has been reached. Notably, we observed a strong disorganization and loss of the correct parallel orientation of pyramidal neurons located in upper neocortical layers, whereas neurons found in deeper layers appeared to be correctly organized (fig. S7, A and B).
To explore in greater detail the altered neuronal organization observed in the neocortex of Zeb2-deficient animals, we labeled a few UL neurons by IUE of GFP at E15.5 and analyzed their dendritic morphology postnatally in mature animals (P23; Fig. 6A). In agreement with the Golgi staining, the majority of Zeb2-deficient UL neurons were incorrectly oriented with respect to the pial surface (Fig. 6, B to D, and fig. S7C). This approach also confirmed the developmental origin of the aberrantly oriented neurons as excitatory late-born neurons. Following radial migration, UL neurons normally extend their apical dendrites perpendicularly to the pia and parallel to other neurons in the layer. Zeb2-deficient UL neurons, in contrast, are tilted with respect to the pia and display a deviation in the angle of the apical dendrite with respect to the pia of over 30° from norm (Fig. 6D). In addition, Zeb2-deficient UL neurons were positioned closer to the pia than control neurons (Fig. 6B and fig. S7, C and D). The length of the apical dendrite, however, was not altered upon loss of Zeb2 (Fig. 6E), indicating that the altered orientation did not result from defective elongation of the apical dendrite. The Golgi apparatus and centrosome were still observed to locate to the base of the apical dendrite of neurons showing defective orientation in vivo and in cultured neurons, respectively, indicating that the apical dendrite was correctly specified (fig. S7, E and F). Furthermore, axons were observed to project at a correct perpendicular orientation with respect to the pia in Zeb2-deficient UL neurons (Fig. 6B, arrows).
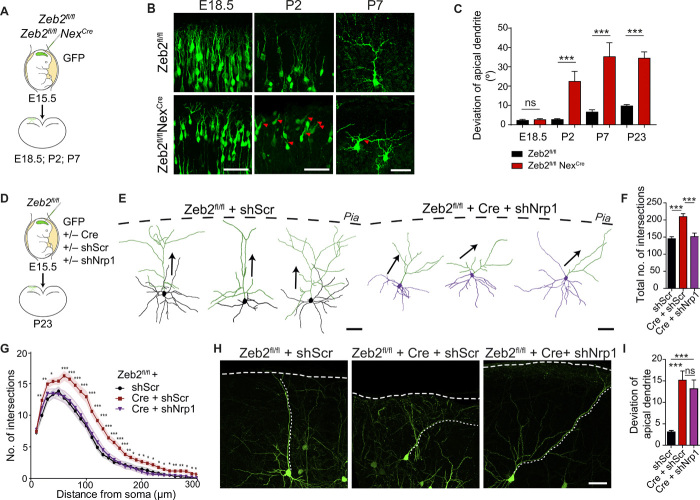
Fig. 6. Zeb2 regulates the orientation and dendritic arborization of UL neurons.
(A) Zeb2fl/fl and littermate Zeb2fl/fl NexCre animals were in utero electroporated at E15.5 with GFP. (B) Representative images of GFP+ UL neurons showing the orientation of the dendritic tree with respect to the pia (marked by a dashed line) at P23. Arrowheads mark the axon. Scale bars, 50 μm. (C) Tracings of P23 GFP+ WT and KO neurons. The dashed line indicates the position of the pia. Apical dendrites are shaded green. Arrows indicate orientation of the apical dendrite. Scale bar, 50 μm. (D) Proportion of cells with aberrant orientation at P23. N = 65 cells per condition. (E) Average length of the apical dendrite at P23. N = 65 cells per condition. Welch’s t test. (F to H) Sholl analysis of the complexity of the dendritic arbor of WT (Zeb2fl/fl) and KO (Zeb2fl/fl NexCre) neurons at P23. (F) Sholl curve showing the number of dendritic intersections at increasing distance from the soma. N = 30 neurons, 10 neurons from three different animals per condition. Unpaired t test with Welch’s correction. (G) Average number of dendritic intersections per cell. Mann-Whitney test. (H) Average number of primary dendrites per cell. Unpaired t test with Welch’s correction.
Next, we analyzed the complexity of the dendritic arbor of UL neurons using a modified Sholl analysis (3). Loss of Zeb2 function led to a marked increase in the complexity of UL dendritic arborization as indicated by a large increase in the number of dendritic intersections and upward shift in the Sholl curve, in particular within 100 μm from the soma (Fig. 6, F to H). This was primarily due to increased branching of the basal dendrites with no change in the number of primary dendrites (Fig. 6H and fig. S7G).
Last, we investigated the temporal onset of neuronal orientation by measuring the angle of the apical dendrite with respect to the pia at different time points following IUE of GFP into control (Zeb2fl/fl) and Zeb2-deficient (Zeb2fl/fl NexCre) mice at E15.5 (Fig. 7, A to C). Zeb2-deficient UL neurons showed normal orientation of their apical process at E18.5. Tilting of the soma and apical dendrite upon loss of Zeb2 was observed only from P2 onward, suggesting that Zeb2 controls post-migratory specification of neuronal orientation.
Fig. 7. Zeb2 regulates neuronal orientation independently of Nrp1.
(A and B) Zeb2 regulates neuronal orientation from P2. (A) IUE of Zeb2fl/fl and Zeb2fl/fl NexCre littermate animals with GFP at E15.5. (B) GFP+ neurons were analyzed at different time points. Arrowheads indicate aberrantly oriented neurons. Scale bars, 50 μm. (C) Deviation of the apical dendrite. N = 40, 36, 33, and 65 Zeb2fl/fl and 33, 56, 22, and 65 Zeb2fl/fl NexCre neurons at E18.5, P2, P7, and P23, respectively. Mann-Whitney test. (D to I) Nrp1 regulates dendritic complexity but not neuronal orientation. (D) IUE of Zeb2fl/fl animals at E15.5 with shScr, Cre, or shNrp1 as indicated. (E) Traces of GFP+ neurons at P23. Apical dendrites are shaded green. Arrows indicate orientation of the apical dendrite. Scale bars, 50 μm. (F and G) Sholl analysis at P23. N = 30 Zeb2fl/fl + shScr cells (from five animals), 39 Zeb2fl/fl + Cre + shScr (six animals), and 30 Zeb2fl/fl + Cre + shNrp1 (seven animals). (F) Average number of dendritic intersections per cell. One-way ANOVA with Bonferroni’s multiple comparison test. (G) Dendritic complexity at P23. Unpaired Student’s t test. (H) Representative GFP+ neurons at P23. (I) Deviation of the apical dendrite. N = 143 Zeb2fl/fl + shScr (five animals), 49 Zeb2fl/fl + Cre + shScr (six animals), and 64 Zeb2fl/fl + Cre + shNrp1 neurons (seven animals). One-way ANOVA (Kruskal-Wallis) with Dunn’s multiple comparison tests.
Nrp1 signaling regulates dendritic arbor complexity but not neuronal orientation
Next, we assessed whether the final orientation of neurons in the postnatal neocortex was dependent on Nrp1 signaling and on the correct onset of radial migration. We generated mosaic deletion of Zeb2 by IUE of Cre into Zeb2fl/fl animals at E15.5 and combined this with down-regulation of Nrp1 expression by IUE of shNrp1 (Fig. 7, D to I). Similar to our observations in Zeb2fl/fl NexCre mice, mosaic deletion of Zeb2 in UL neurons disrupted the orientation of neurons in the neocortex and increased dendritic complexity, indicating that these functions are cell intrinsic (Fig. 7, F to I). Down-regulation of Nrp1 in Zeb2-deficient UL neurons reduced dendritic arbor complexity to levels comparable to those displayed by control neurons (Fig. 7, E to G). However, Nrp1 down-regulation failed to restore correct apical orientation in Zeb2-deficient neurons (Fig. 7, H and I).
We also asked whether Nrp1 overexpression disturbs the orientation of UL neurons in the mature neocortex. We overexpressed Nrp1 by IUE in WT UL neurons at E15.5 and analyzed at P23 (fig. S8). Sholl analysis revealed that overexpression of Nrp1 in WT UL neurons increased dendritic branching and the formation of an overly complex dendritic tree, a phenotype reminiscent of Zeb2-deficient UL neurons (Fig. 6F and fig. S8, A to D). Overexpression of Nrp1 had no effect on the orientation of UL neurons with respect to the pia (fig. S8E).
Together, these data demonstrate that Nrp1 regulates the complexity of the dendritic arbor downstream of Zeb2 but not the orientation of neurons in the neocortex. These data also indicate that the initiation of radial migration and the post-migratory orientation of neurons in the neocortex are controlled by independent signaling events.
Cdh6 regulates cell aggregation and adhesion to the extracellular matrix downstream of Zeb2
Our biotinylation-linked MS screen identified another class of adhesion molecules whose surface expression was increased in Zeb2-deficient neurons, the cadherin family (Fig. 3C). Cadherins mediate cell adhesion through homophilic and heterophilic interactions with other cadherins (33). We observed the highest increase in the surface expression of Cdh6 and Cdh10 upon loss of Zeb2 (both 3.1-fold up-regulated in Zeb2fl/fl NexCre neurons; table S1). We focused on Cdh6 as cross-referencing against our published microarray data from Zeb2-deficient brains (14) showed that only Cdh6 exhibited increased mRNA levels in the neocortex of Zeb2 mice. In situ hybridization for Cdh6 confirmed the increased mRNA expression of this cadherin in the CP of Zeb2-deficient (Zeb2fl/fl NexCre) animals at E15.5 (Fig. 8A).
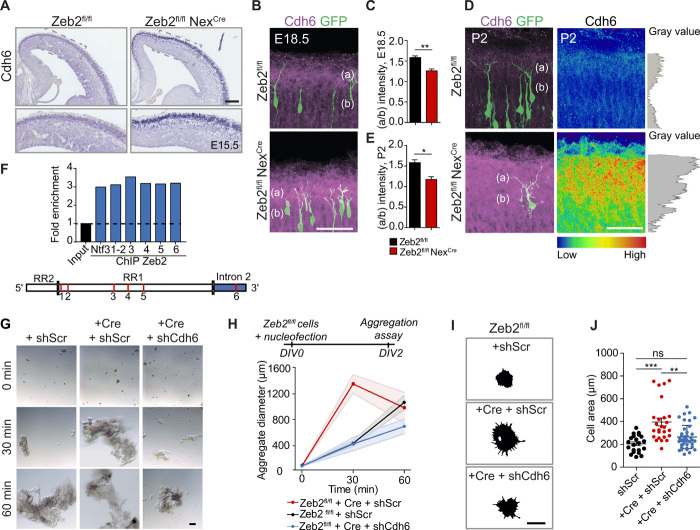
Fig. 8. Cdh6 regulates adhesion downstream of Zeb2.
(A) ISH for Cdh6 in E15.5 Zeb2fl/fl and Zeb2fl/fl NexCre brains. Scale bar, 50 μm. (B to E) Up-regulation and redistribution of Cdh6 protein upon loss of Zeb2. E14.5 Zeb2fl/fl and Zeb2fl/fl NexCre animals were in utero electroporated with GFP and stained for Cdh6 (magenta) at E18.5 (B) and P2 (D). Cdh6 intensity at P2 is shown as a heatmap and gray plot. Scale bar, 50 μm. (C and E) Ratio of Cdh6 in layer I (a) versus CP (b) at E18.5 (C) and P2 (E). N = 3 brains per condition. Unpaired t test. (F) ChIP from E15.5 neocortex shows Zeb2 occupancy at Cdh6 regulatory regions. Red lines mark analyzed sites. Positive control = Ntf3 (fig. S9). (G and H) Cdh6 promotes cell adhesion downstream of Zeb2. (G) Aggregation of E15.5 Zeb2fl/fl and Zeb2fl/fl NexCre neurons transfected with shScr, shCdh6, and Cre as indicated. Scale bar, 100 μm. (H) Average aggregate size. N = 15, 10, and 7 Zeb2fl/fl + shScr; 12, 12, and 15 Zeb2fl/fl NexCre + shScr; and 9, 7, and 4 Zeb2fl/fl NexCre + shCdh6 aggregates at 0, 30, and 60 min. (I and J) Cdh6 regulates adhesion to the extracellular matrix downstream of Zeb2. (I) Attachment of Zeb2fl/fl neurons, transfected with shScr, shCdh6, or Cre as indicated, to laminin-coated surfaces. Adhering cells were visualized after 2 hours by F-actin staining. Scale bar, 15 μm. (J) Lamellipodial spreading. N = 21 Zeb2fl/fl + shScr, 36 Zeb2fl/fl NexCre + shScr, and 41 Zeb2fl/fl NexCre + shCdh6 cells. One-way ANOVA (Kruskal-Wallis test) with Dunn’s multiple comparison test (H and J).
We analyzed the levels and localization of Cdh6 protein in the brains of control (Zeb2fl/fl) and littermate Zeb2-deficient (Zeb2fl/fl NexCre) animals at E18.5 and P2. IUE of GFP at E14.5 was used to visualize the position of neurons in the neocortex (Fig. 8, B to E). Endogenous Cdh6 protein was up-regulated in Zeb2-deficient brains at both ages (Fig. 8, B and D). In addition, we observed a change in the distribution of Cdh6 protein in the neocortex upon loss of Zeb2 (Fig. 8, C and E). In control animals at both ages, Cdh6 shows the highest expression levels in layer 1, an area occupied by the apical dendrite tufts, and the lowest in an area occupied by the shaft region of the apical dendrite in the CP {ratio of intensity in layer 1 [region (a)] versus CP [region (b)] > 1; Fig. 8, C and E}. In contrast, upon loss of Zeb2, elevated Cdh6 protein levels were observed in both of these regions (Fig. 8, C and E).
To address whether Cdh6 is a direct target of Zeb2, we performed ChIP-qPCR assays using a Zeb2-specific antibody and chromatin prepared from E15.5 WT neocortices as before. In silico analysis for regions showing enriched histone H3K27 acetylation, CpG islands, and DNase I hypersensitivity clusters was used to define an approximately 4kb region upstream (RR1) and a 2-kb region downstream (Intron) from the transcriptional start site of Cdh6 as likely regulatory regions. We identified six Zeb2 CACCT(G) consensus motifs within these regions that displayed strong enrichment for Zeb2 in our ChIP-qPCR analyses, indicating that Cdh6 is a novel direct target of Zeb2 (Fig. 8F and fig. S9).
Next, we asked whether up-regulation of Cdh6 was responsible for increased neuronal adhesion observed upon loss of Zeb2 function. Cell aggregation assays were carried out using single-cell suspensions of Zeb2-deficient cortical neurons after down-regulation of Cdh6 with shRNA (shCdh6). Down-regulation of Cdh6 restored cell-to-cell adhesion in Zeb2-deficient neurons to control levels, indicating that Cdh6 mediates neuronal cell adhesion downstream of Zeb2 (Fig. 8, G and H).
Cdh6 is unusual among cadherins in containing an RGD motif in the first extracellular domain that may be involved in interactions with integrins (34). We therefore asked whether Cdh6 also regulates adhesion to the extracellular matrix. This was done by measuring the lamellipodia spread of Zeb2-deficient neurons on laminin following down-regulation of Cdh6. Down-regulation of Cdh6 partially suppressed the enhanced adhesion of Zeb2-deficient neurons to the extracellular matrix, suggesting that Cdh6 may also regulate integrin function downstream of Zeb2 in neurons (Fig. 8, I and J).
We also tested whether Cdh6, like Nrp1, regulates the onset of radial migration. For this, we overexpressed Cdh6 in UL neurons by IUE at E14.5 and assessed the laminar position of modified neurons at E18.5 (fig. S10A). We observed no effect of Cdh6 overexpression on the laminar distribution of UL neurons, indicating that Cdh6 does not regulate radial migration (fig. S10, B and C).
Cdh6 controls the post-migratory orientation of neurons
Last, we evaluated the potential role of Cdh6 in the postnatal orientation of neurons. First, we investigated whether enhanced Cdh6 levels in Zeb2-deficient UL neurons account for their aberrant orientation within the neocortex. We down-regulated Cdh6 expression in Zeb2-deficient neurons by IUE of a Cdh6-specific shRNA (shCdh6) at E15.5 (Fig. 9A). Down-regulation of Cdh6 almost completely restored the orientation of Zeb2-deficient neurons at P23 (Fig. 9, B to D).
Fig. 9. Cdh6 regulates apical dendrite orientation downstream of Zeb2.
(A) Zeb2fl/fl animals were in utero electroporated at E15.5 with GFP, Cre, shScr, or shCdh6 as indicated. (B) Representative images of GFP+ neurons in brain slices at P23. The apical dendrite is marked by a fine parallel tracing; the pia is marked by a thicker dashed line. Scale bar, 50 μm. (C) Tracings of 10 apical dendrites per condition showing their orientation with respect to the pia. Apical dendrites were superimposed so that all deviations face the right-hand side of the image. Scale bar, 50 μm. (D) Average deviation of the apical dendrite. N = 143 Zeb2fl/fl + shScr cells (seven animals), 57 Zeb2fl/fl NexCre + Cre cells (five animals), and 85 Zeb2fl/fl NexCre + shCdh6 cells (four animals). One-way ANOVA (Kruskal-Wallis test) with Dunn’s multiple comparison test.
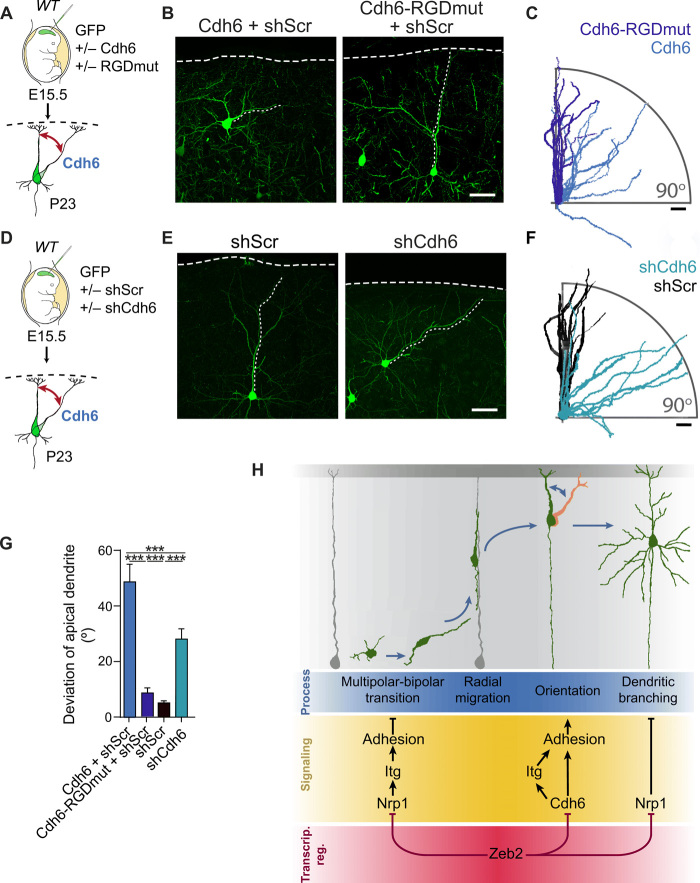
Conversely, up-regulation of Cdh6 signaling in WT neurons, by IUE of Cdh6 at E15.5, strongly disrupted the orientation of UL neurons at both P23 and P7 (Fig. 10, A to C, and fig. S10, D to F). In addition, reminiscent of our observations in Zeb2-deficient neurons, Cdh6 overexpressing UL neurons show normal orientation and morphology until E18.5 (fig. S10, D to F). Together, these data show that enhanced Cdh6 signaling is responsible and sufficient for the aberrant post-migratory orientation of neurons in vivo.
Fig. 10. A balance of Cdh6 signaling regulates neuronal orientation.
(A to C) Cdh6 regulates neuronal orientation through its RGD motif. (A) WT E15.5 animals were in utero electroporated with Cdh6 or an RGD mutant of Cdh6 (Cdh6-RGDmut). (B) GFP+ neurons at P23. Scale bar, 50 μm. (C) Tracings of 10 apical dendrites showing their orientation with respect to the pia. Apical dendrites were superimposed to face the right-hand side of the image. Scale bar, 50 μm. (D to F) Cdh6 is necessary for neuronal orientation. (D) E15.5 WT animals were in utero electroporated with shScr or shCdh6. (E) GFP+ neurons at P23. Scale bar, 50 μm. (F) Tracings of 10 apical dendrites per condition. Scale bar, 50 μm. (G) Average deviation of the apical dendrite for experiments (A to F). N = 25 Cdh6 + shScr cells (from three animals), 53 Cdh6-RGDmut + shScr cells (four animals), 31 shScr cells (seven animals), and 66 shCdh6 cells (five animals). One-way ANOVA (Kruskal-Wallis test) with Dunn’s multiple comparison test. (H) Zeb2 shapes neocortical cytoarchitecture by regulating adhesion at distinct time points of development through repression of Nrp1 and Cdh6. Repression of Nrp1 and downstream integrin signaling by Zeb2 promotes multipolar-bipolar transition and initiation of radial migration. Following migration, Zeb2 regulates Cdh6 expression for correct neuronal orientation. Subsequently, Zeb2 determines dendritic complexity through repression of Nrp1.
We then addressed whether the ability of Cdh6 to interact with integrins is required for the orientation of UL neurons in the neocortex. To address this, we engineered an expression vector encoding Cdh6 with a mutated RGD motif (called here Cdh6-RGDmut) by changing the sequence for residues p.84G>A and p.85D>E of Cdh6. Equivalent mutations in CDH6 and other RGD-containing cadherins, such as the human CDH17 and CDH5, have been shown to specifically disrupt association with integrins and thus adhesion to the extracellular matrix, but not homophilic interactions or cell adhesion (35). Overexpression of Cdh6-RGDmut in WT UL neurons by IUE did not alter their orientation (Fig. 10, A to C and G), demonstrating that Cdh6 controls the orientation of neurons via its RGD motif.
Last, we investigated whether normal levels of Cdh6 signaling are also involved in orienting UL neurons during neocortical development by down-regulating Cdh6 expression in WT brains by IUE of shCdh6 or a control shRNA (shScr) at E15.5. Down-regulation of Cdh6 in WT neurons disrupted the orientation of the apical dendrite and soma of UL neurons. This was corrected by re-expression of Cdh6 (Fig. 10, D to G, and fig. S10, G to J). Together, these data suggest that following migration, a tight balance of Cdh6 signaling is required for the correct orientation of UL neurons in the neocortex.
DISCUSSION
Precise organization of excitatory neurons within the neocortex enables compartmentalization and sorting of different connections to distinct layers and to distinct dendritic compartments (1, 36). Our data show that this organization is dependent on dynamic changes in the adhesive properties of UL neurons. The transcriptional repressor Zeb2 plays an important role in suppressing adhesion to influence two distinct stages in the organization of the neocortex: first, the onset of radial migration of UL neurons and, second, their orientation and final positioning within the UL upon termination of migration as well as dendritic branching (Fig. 10H).
Onset of migration requires suppression of Nrp1 and integrin-mediated adhesion
UL neurons have been shown to sojourn around a day in a multipolar state in the SVZ before initiating migration (8). The reason for this is still unclear but may reflect a temporal state necessary for changes in the proteome of newborn neurons. Several steps are likely involved in this multipolar-to-bipolar transition. One critical step, for example, appears to be the expression of N-cadherin (Cdh2), a cadherin required for the interaction of newborn neurons with radial glia and thus for their motility along the radial glial fibers (37). In addition, radial glia and neuronal cytoskeletal integrity are essential for radial migration and their disruption also results in accumulation of multipolar neurons in the VZ/SVZ (38, 39).
Our data reveal that the decision to initiate migration is controlled independently of migration itself, through regulation of adhesive pathways. The transcriptional repressor Zeb2, whose own expression is initiated shortly after neurogenesis (14), directly regulates the multipolar stage of neuronal development by restricting neurite formation and promoting increased tangential movement of multipolar neurons in the SVZ. This function is dependent on Zeb2-mediated suppression of adhesion to the extracellular matrix through inhibition of integrin signaling. Zeb2 suppresses adhesion by repression of Nrp1 expression, a novel direct target of Zeb2 (Figs. 3 to 5). An adhesive role for Nrp1 in the neocortex had not previously been described, but Nrp1 directly interacts with and promotes integrin signaling in a variety of cancer and endothelial cells (32). Furthermore, Nrp1 has been shown to regulate the internalization of integrin receptors in endothelial cells (40), providing a possible mechanism for the enhanced presence of integrins at the plasma membrane in the absence of Zeb2 (table S1). We observe enhanced association of Nrp1 and Itgb1 in Zeb2-deficient neurons (fig. S6). Moreover, inhibition of integrin signaling in vivo restores the laminar position of Zeb2-deficient or Nrp1-overexpressing neurons (Figs. 3, H to J, and 5, E to G). Thus, our data show the existence of a conserved signaling relationship between Nrp1 and integrins in neurons and demonstrate its function in regulating the onset of radial migration downstream of Zeb2 (Fig. 10H).
In the neocortex, Nrp1 is known to act as a receptor for semaphorin-3 (Sema3) ligands during, for example, axon repulsion (41). Sema3A has been shown to regulate laminar positioning in the neocortex, but it was unclear whether Nrp1 is involved (42, 43). Here, we show that enhanced Nrp1 signaling leads to laminar mispositioning of UL neurons at E18.5, specifically by delaying the start of migration and without altering cell movement itself (Figs. 4 and 5). This is unlikely due to enhanced Sema3A signaling as expression of Sema3A and 3C was not altered in Zeb2-deficient mice (fig. S4) and enhanced Sema3A signaling would be expected to promote migration (42, 44). We have also uncovered an additional role for Nrp1 later in development, in the promotion of dendritic arbor complexity (Fig. 7, E to G, and fig. S8).
Cdh6 regulates the post-migratory orientation of neurons
Although the parallel organization of neurons is an essential feature of neocortical architecture and function, we do not understand how this is achieved or whether this orientation is predetermined by events during radial migration or, for example, by inherent differences in the growth of apical and basal dendrites.
Our data show that parallel orientation of UL neurons occurs after completion of radial migration, around P2, and that it is independent of radial migration or dendritic growth. Furthermore, our data indicate that the orientation of apical dendrites and their soma is under the control of adhesive pathways that are distinct from those involved in radial migration and early polarization events. Nrp1 signaling regulates the onset of radial migration but is not involved in controlling neuronal orientation (Fig. 7, H and I). This is in line with observations made in Nrp1-deficient mice where neurons show normal orientation in the neocortex (43).
Aberrant orientation of the apical dendrite has been reported upon loss of the chemo-attractants Reelin or Sema3A, or their downstream signaling effectors (45, 46). For Sema3A, these effects are restricted to layer V neurons, raising the possibility that different orientation cues operate in different cortical layers (46, 47). We did not observe orientation defects in the proximal region of the apical dendrite of layer V Zeb2-deficient neurons (fig. S7, A and B), although we cannot rule out that distal portions of the apical dendrite of these neurons are incorrectly oriented.
Both Reelin and Sema3A, however, are potent and necessary promoters of dendritic growth and cytoskeletal remodulation (44, 45, 48, 49). Loss of either Reelin or Sema3A is associated with laminar displacement of neurons and profound morphological defects during and following radial migration, including severely underdeveloped dendritic arbors and loss of a clear apical dendrite (46, 50). Reelin and Sema3A thus likely regulate neuron orientation principally by directing and promoting the growth and stabilization of their leading process and apical dendrite (45, 51, 52).
Here, we show that signaling by the cadherin, Cdh6, directs UL neuron orientation in the neocortex, independently of radial migration or of dendritic growth. Enhanced Cdh6 signaling results in a marked aberrant orientation of UL apical dendrites without affecting apical dendrite length or radial migration (Figs. 9 and 10 and fig. S10, A to C). Loss of Zeb2 is associated with both defective orientation of UL neurons and with increased dendritic outgrowth. Arbor complexity, but not orientation, can be restored by down-regulation of Nrp1 (Fig. 6), again demonstrating that postnatal neuronal orientation is independent of dendritic outgrowth. To the best of our knowledge, this is the first report that events independent of radial migration or dendritic growth direct the orientation of neurons in the neocortex.
Cdh6 has been poorly studied in the nervous system, and a role in neocortical development remained uncharted. Like other cadherins, Cdh6 can signal through homophilic and heterophilic interactions with other cadherins (33). Heterophilic interactions of Cdh6 with multiple cadherins are thought to be involved in axon-target matching in a subset of retinal cells (53, 54) and the regulation of high-magnitude LTP in the hippocampus (55) and, like other cadherins, to cause the disruption of the overall morphology of the murine barrel cortex upon exogenous overexpression (56). In addition, Cdh6 is one of five cadherins known to have a highly conserved RGD motif (34, 35), a classic integrin-binding motif, in the extracellular domain. We show that Cdh6 regulates the adhesion of neurons not only to each other but also to integrin substrates (Fig. 8, G to J). Moreover, we show that the Cdh6 RGD motif is necessary for determining the orientation of neurons in vivo (Fig. 10, A to C). Our data thus indicate that interactions of Cdh6 with integrin receptors and downstream modulation of the adhesion to the extracellular matrix provide positional information during neuronal orientation.
Both enhancement and down-regulation of Cdh6 signaling interfere with the post-migratory orientation of UL neurons (Fig. 10 and fig. S10). This suggests that it is essential to achieve the right balance of Cdh6-dependent adhesion for neurons to correctly orient within the neocortex. The role of Zeb2, therefore, is principally to modulate this balance by restricting Cdh6 expression level. Moreover, our finding that, under normal circumstances, Cdh6 protein expression is elevated in layer 1 may indicate that a gradient of the subcellular localization of this receptor provides positional information. In keeping with this notion, layer 1 is rich in various integrin substrates and activation of Itgb1 during perinatal stages has been specifically observed in this region (57, 58).
The stereotypical organization of the neocortex is a critical requirement for its function. In summary, we show here that dynamic changes in the adhesion of UL neurons enable both multipolar-bipolar transition and initiation of radial migration and, subsequently, the orientation of UL neurons within the neocortex. Furthermore, we have identified two novel adhesion pathways downstream of the transcriptional repressor Zeb2 involved in these processes, namely, an Nrp1/integrin pathway involved in prolonging the multipolar stage and delaying migration and a Cdh6/integrin pathway required for the correct post-migratory parallel orientation of neurons within the neocortical layer.
MATERIALS AND METHODS
Experimental design
This study aims to understand how higher cellular organization is achieved in the mammalian neocortex. Multiple in vivo and in vitro manipulations of signaling pathways in the mouse brain, including in utero electroporation into WT and genetically modified mouse lines and live imaging of neurons in live brain slices as well as novel candidate identification by MS-linked screens, are used for this purpose.
Animals
All animal experiments were performed at the Charité—Universitätsmedizin Berlin in accordance with German Law using protocols approved by the Landesamt für Gesundheit und Soziales, Berlin. To inactivate Zeb2 in developing cortex, we crossed Zeb2fl/fl mice with NexCre line in which Cre recombinase expression is driven by Nex (Neurod6) gene allele (22, 23). Please see table S4 for genotyping primers. WT mice (NMRI strain) for overexpression studies were obtained from Charles River Laboratories. Experimental ages used are indicated in the figure legends.
In/ex utero electroporation
In utero and ex utero electroporations were carried out as previously described (3, 59). Briefly, the DNA mixture was prepared in endotoxin-free water and 0.1% Fast Green FCF (Sigma-Aldrich) and injected into the lateral ventricle of the embryo using micropipettes prepared from 1.5 to 1.8 × 100 mm borosilicate glass capillaries (Kimble and Chase). DNA uptake by neuronal progenitor cells was induced by the application of six pulses of 35 to 40 V across the uterine wall using platinum electrodes. For the analysis of the laminar distribution of the modified cells, electroporation was performed at E14.5 embryos and analyzed at E18.5. For live imaging, embryos were electroporated in utero at E14.5 and cortical slice cultures were prepared after 24 hours. For the morphological analysis of postnatal neurons, embryos were electroporated in utero at E15.5 and animals were collected at P2, P7, or P23. Primary cortical cell culture cells were prepared from embryos electroporated at E15.5.
Live imaging of slice culture
Slice cultures were prepared as previously described with small changes (60). Brains of electroporated embryos were isolated and embedded in 4% low-melting point agarose (Promega) in complete Hanks’ balanced salt solution (HBSS) on ice. Coronal vibratome sections (250 μm) (Leica, VT1200S) were collected on laminin/poly-l-lysine–coated (1 μm/μl) (Sigma-Aldrich) cell culture inserts (Millicell, Merck Millipore) placed inside FluoroDishes (WPI) containing slice culture medium (60). Sections were allowed to recover at 37°C, 5% CO2 for 6 hours. Imaging was performed using a spinning disk confocal microscope (Zeiss ZEN 2012) equipped with a humidified incubation chamber at 37°C, 5% CO2. Brain slices of different conditions under comparison were imaged in parallel. Imaging was performed using a spinning disk confocal microscope (Zeiss ZEN 2012) equipped with a humidified incubation chamber at 37°C, 5% CO2. z-stack images were taken once every 20 min for a maximum of 80 hours. Movies were analyzed using ImageJ software and the TrackMate plugin.
Primary cortical cell cultures, cell aggregation, and adhesion assays
Primary cortical neurons were prepared from E15.5 embryos. Brains were dissected in ice-cold HBSS under a stereomicroscope. Where necessary, cells were transfected before plating using NucleofectorTM technology (Lonza) according to the manufacturer’s instructions. Primary neurons were grown on laminin/poly-l-lysine–coated plates or coverslips in complete BrainPhys medium (STEMCELL Technologies) at 37°C in the presence of 5% carbon dioxide. Transfected neurons were grown for 2 days to enable expression of the nucleofected DNA constructs before preparation of single-cell suspensions. Where transfection was not required, single-cell suspensions were prepared directly from dissected E15.5 cortices. All cell suspensions were prepared using 1 mM EDTA/HBSS.
For the aggregation assay, primary cortical neuron suspensions with equalized cell numbers were placed in uncoated wells filled with serum-free medium under gentle agitation in a humidified incubator at 37°C, 5% CO2. Wells were imaged and the size of cell aggregates was determined at 0, 30, and 60 min.
For the adhesion assay, primary cortical neuron suspensions with equalized cell numbers were plated on laminin/poly-l-lysine–coated coverslips in serum-free medium and allowed to settle for 2 hours. Aggregation and adhesion assays were both imaged using an Axiovert 40 CFL microscope.
Histology
Perfusion
Mice were injected with a lethal dose of pentobarbital (Narcoren, Boehringer Ingelheim). Mouse unresponsiveness was checked by toe-pinch response. Intracardiac perfusion with 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) was performed manually on animals older than P2. Brains were collected in ice-cold PBS and fixed in 4% PFA/PBS overnight at 4°C. Brains were stored for later use in 0.01% sodium azide/PBS (Sigma-Aldrich) at 4°C.
Sectioning
Fixed brains were prepared for cryosectioning by incubation through a sucrose/PBS gradient and subsequently snap-frozen in isopentane (Roth) cooled to around 70° to 80°C using dry ice. Fifty-micrometer sections of E16.5-P2 brains were prepared on a Leica CM3050S cryostat and collected in either wells filled with 0.01% sodium azide/PBS or directly on Superfrost Plus glass slides.
For in situ hybridization, E15.5 brains were isolated in PBS/diethyl pyrocarbonate (DEPC) and fixed overnight in 4% PFA/15% sucrose/PBS/DEPC at 4°C. Fixed brains were processed through a sucrose/PBS/DEPC gradient, embedded in Tissue-Tek O.C.T. TM Compound (Sakura) at around −70°C, and sectioned at a thickness of 16 μm on a Leica CM3050S cryostat. P7 to P23 brains were sectioned at 100 μm on a vibratome (Microm, HM650V) and collected in 0.01% sodium azide/PBS.
Immunohistochemistry
All stainings were carried out in parallel on samples derived from littermate animals. Brain sections of P23 and P7 perfused animals were incubated for 5 min with sodium borohydride (1 mg/ml) (Sigma-Aldrich) and then washed three times in PBS. Sections were incubated for 30 to 60 min in blocking solution (5% horse serum and 0.05% Triton X-100 in PBS) and then overnight with primary antibodies and nuclear stain (DRAQ5, Invitrogen or DAPI, Sigma-Aldrich) diluted in blocking solution at +4°C. Next, sections were washed twice with PBS, incubated for 2 to 4 hours with secondary antibodies diluted in blocking solution, washed twice, and mounted on Superfrost Plus glass slides (Thermo Fisher Scientific) with a cover glass (Menzel-Gläser) and Immu-Mount mounting medium (Shandon, Thermo Fisher Scientific). Please see table S5 for a list of antibodies used. Brain slices were imaged as 2-μm-spaced z stacks using Leica SL and Sp8 confocal microscopes.
Biotinylation-linked MS
Biotinylation and isolation of surface proteins
Primary cortical neuron cultures were prepared from littermate Zeb2fl/fl and Zeb2fl/fl NexCre E15.5 embryos and cultured for 2 days on laminin/poly-l-lysine–coated six-well plates. Cells were gently rinsed with ice-cold rinsing solution [PBS (pH 7.5), containing 0.1 mM CaCl2 and 1 mM MgCl2] and then incubated on ice with EZ-Link Sulfo-NHS-SS-biotin (1 mg/ml) (Thermo Fisher Scientific) in rinsing solution for 30 min. Biotinylation was ended by washes in quenching solution (rinsing solution supplemented with 100 mM glycine) on ice followed by a short wash in rinsing solution. Labeled cells were lysed in lysis buffer [50 mM tris (pH 7.4), 100 mM NaCl, 1 mM EDTA, and 1% v/v Triton X-100] containing protease and phosphatase inhibitors [40 mM Na3VO4 (Sigma-Aldrich), pepstatin (10 μg/ml) (Sigma-Aldrich), leupeptin (10 μg/ml) (Sigma-Aldrich), aprotinin (10 μg/ml) (Sigma-Aldrich), 1× PhosStop (Roche), and 1× protease inhibitor cocktail (Sigma-Aldrich)] and clarified by centrifugation at 13,500 rpm at 4°C, and the protein concentrations were determined using Bradford assay.
Lysates from five embryos were pooled per genotype and biotinylated proteins from equal amounts of total protein were isolated by incubation with Pierce NeutrAvidin agarose beads (Thermo Fisher Scientific) for 2 hours at 4°C under rotation. Beads were washed thoroughly in lysis buffer and bound proteins were eluted by boiling in sample buffer.
SDS-PAGE and Coomassie staining of gels for MS
Samples were subjected to SDS–polyacrylamide gel electrophoresis (PAGE), and the resolved gels were stained overnight with colloid Coomassie Brilliant Blue G-250 (CBB G-250) dye [0.02% w/v CBB G-250, 5% aluminum sulfate-(14-18)-hydrate, and 10% v/v ethanol 2% orthophosphoric acid] as previously described (61). Gels were destained in 10% v/v ethanol 2% orthophosphoric acid and subsequently washed in deionized H2O. Gel regions containing the separated proteins were cut and subjected to tryptic digestion, as previously described (62).
Mass spectrometry
Tryptic peptides were analyzed by online nanoflow liquid chromatography tandem MS (LC–MS/MS) using a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Peptide mixtures were fractionated by an Ultimate 3000 RSLCnano (Thermo Fisher Scientific) with a two-linear-column system. Digests were concentrated onto a trapping guard column (PepMap C18, 5 mm × 300 μm × 5 μm, 100 Ǻ; Thermo Fisher Scientific). Samples were eluted from the analytical column, a 75-μm inside diameter × 250 mm nano LC column (Acclaim PepMap C18, 2 μm; 100 Å; Thermo Fisher Scientific). Separation was achieved using a mobile phase from 0.1% formic acid (FA, buffer A) and 80% acetonitrile with 0.1% FA (buffer B) and by applying a linear gradient from 8 to 28% of buffer B for 60 min at a flow rate of 300 nl/min. The Q Exactive instrument was operated in the data-dependent mode to automatically switch between full scan MS and MS/MS acquisition. Survey full scan MS spectra [350 to 1650 mass/charge ratio (m/z)] were acquired in the Orbitrap with 70,000 resolution (200 m/z) after 50-ms accumulation of ions to a 3 × 106 target value. Dynamic exclusion was set to 10 s. The 10 most intense multiply charged ions (z ≥ 2) were sequentially isolated and fragmented by higher-energy collisional dissociation (HCD) with a maximal injection time of 100 ms, automatic gain control 5 × 104, and a resolution of 17,500. Typical mass spectrometric conditions were as follows: spray voltage, 2.1 kV; no sheath and auxiliary gas flow; heated capillary temperature, 275°C; and normalized HCD collision energy 27%. In addition, the background ion m/z 445.1200 acted as lock mass.
Protein identification
Relative label-free quantification of the proteins was performed with MaxQuant software version 1.6.0.1 and default Andromeda LFQ parameter. Spectra were matched to a Mus musculus database (17,040 reviewed entries, downloaded from uniprot.org), a contaminant and decoy database. MS/MS spectra were searched with a precursor mass tolerance of 10 parts per million, a fragment tolerance of 0.05 Da, trypsin specificity with a maximum of two missed cleavages, cysteine carbamidomethylation set as fixed, and methionine oxidation as variable modification. Identifications were filtered at 1% false discovery rate (FDR) at the peptide level.
Analysis of MS data
String software version 11.0 was used to identify the top Gene Ontology terms of surface proteins up-regulated by at least 1.5-fold upon loss of Zeb2. GSEA (C2:KEGG database, max set size, 500; min set size, 5) and Cytoscape 3.7.2 software were used to build the network.
In situ hybridization
In situ hybridization was performed using E15.5 mouse brain cryosections. Tissue sections were dried using a vacuum chamber and incubated with the probe and hybridization solution [50% formamide (AppliChem), 5× SSC, 1% Boehringer block, DEPC-H2O, 5 mM EDTA, 0.1% Tween 20 (Sigma-Aldrich), 0.1% CHAPS (Sigma-Aldrich), Heparin (0.1 mg/ml), and yeast RNA (100 μg/ml) (Invitrogen)] overnight at 70°C. Slides were washed twice in wash solution (50% formamide/1× SSC pH 4.5/0.1% Tween 20) for 30 min at 65°C and twice in 1× maleic acid buffer containing Tween 20 (MABT) solution [100 mM maleic acid, 150 mM NaCl, and 0.1% Tween 20 (pH 7.5)] for 30 min at room temperature (RT). The slides were then incubated overnight in blocking solution (10% sheep serum in 1× MABT) with Digoxigenin-alkaline phosphatase antibody (Roche Diagnostics) at 4°C in a humidified chamber. Next, slides were washed in 1× MABT three times for 15 min at RT and in freshly prepared prestaining solution [5 M NaCl, 1 M MgCl2, 1 M tris (pH 9.5), 200 μl of Tween 20, and Milli-Q water] twice for 5 min at RT. Staining was carried out by incubation in staining solution (25 ml of prestaining solution containing nitro blue tetrazolium and bromochloroindolyl phosphate at a dilution of 1:1000; Roche) and 25 ml of 10% poly-vinylalcohol (Sigma-Aldrich) until color development. Last, slides were dried and mounted with Entellan medium (Merck) and imaged on an Olympus BX60 microscope.
ChIP-qPCR
The promoter regions of Nrp1 and Cdh6 gene were defined based on regions with increased H3K27ac modification, the presence of CpG islands, and DNase hypersensitivity clusters surrounding the start codon (using UCSC genome browser, FANTOM5). These regions were then searched for the presence of CACCT(G) sites. For ChIP analysis, we used E15.5 Zeb2fl/fl and Zeb2fl/fl NexCre embryonic cortices. ChIP was carried out using the ChIP-IT Express Kit (Active Motif) with slight changes. Briefly, cortices were lysed with 0.05% trypsin (Gibco), incubated 10 min at 37°C, spun down, and fixed with freshly prepared 1% PFA/PBS for 10 min at RT. Following centrifugation, pellets were then washed with ice-cold PBS and respun. Next, cells were lysed in lysis buffer containing protease and phenylmethylsulfonyl fluoride (both Active Motif), homogenized using a chilled dounce homogenizer, and spun down. Pellets were diluted in shearing buffer (Active Motif) and sonicated using a Bioruptor (Diagenode). The obtained chromatin was precleared with magnetic beads (Dynobeads Protein G, Invitrogen) and then incubated overnight with magnetic beads precoupled to anti-Zeb2 antibody (13). Bound DNA was eluted in elution buffer (Active Motif) and cleaned up according to the manufacturer’s instructions. The obtained DNA was used for a series of qPCRs using GoTaq qPCR Master Mix (Promega) and StepOnePlus Real-Time PCR System (Applied Biosystems). Please see table S4 for primer sequences.
Quantification and statistical analysis
Morphological analysis of postnatal neurons
Morphological analysis was carried out as previously described (3). Briefly, 1-μm-spaced z-stack images of brain slices were imaged on an Olympus BX60 microscope using AxioVision 4.8 (Zeiss) and CellSens Dimension software. Dendritic tree reconstructions were made using the Simple Neurite Tracer plugin in ImageJ. Sholl analysis was performed blindly on the z-stack images using a starting radius of 10 μm and a 10-μm step size increase in radius and the Sholl analysis plugin in ImageJ. The images of the neuronal reconstructions shown are maximal projections.
The angle of the apical dendrite was determined using the angle tool in ImageJ. The apical dendrite was defined as the dendrite extending from the Golgi-containing, triangular side of the soma that extends toward the MZ. Cells, where the identity of the apical dendrite was unclear, were not analyzed.
Quantification of distribution of cortical neurons
The laminar distribution of neurons and cell fate analysis was performed as described previously (59). Briefly, z stacks of 50-μm-thick in utero electroporated mouse cortices were acquired with a Leica Sp2 confocal microscope with 20× or 40× objective lens. Maximal projections were divided into five identically sized bins (as indicated in Fig. 1B). The number of electroporated neurons in each bin was manually counted with Cell Counter plug-in of ImageJ software and expressed relative to the total number of transfected cells.
Quantification of in vitro adhesion experiments
The average maximal diameter of cell aggregates was determined at the indicated time points and normalized to the average size of control aggregates at the zero time point. The area of adherent cells was determined after staining for F-actin using phalloidin–tetramethyl rhodamine isothiocyanate (Sigma-Aldrich) or GFP and using thresholded images and ImageJ software and the linewidth tool.
Quantification of live-imaging experiments
GFP+ or mCherry+ neurons were manually identified and followed throughout the imaged period using ImageJ software and the Simple Neurite Tracer plugin. Speed was determined as the distance traveled per hour during the period of cell migration. Multipolar cells were defined as cells containing more than two neurite projections and lacking a clear leading process.
Statistical analyses
All statistics were conducted using GraphPad Prism 5 software. Values and statistical details for all experiments are listed in the figure legends and in full detail in tables S2 and S3. Normality of distribution was analyzed using D’Agostino-Pearson and Shapiro-Wilk normality tests. For normally distributed data, we used unpaired two-tailed t test or one-/two-way analysis of variance (ANOVA) with Bonferroni post hoc test. For nonnormally distributed data, we applied Mann-Whitney test or Kruskal-Wallis test with Dunn’s multiple comparison. Probabilities were presented in graphs as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Acknowledgments
We thank M. Studer, L. R. Hernandez-Miranda, and S. Mayer for critical reading of the manuscript. We also thank A. Newman for help with GSEA and ChIP, J. Schueler for assistance with microscopy, D. Alatriste-Gonzalez for technical assistance, and D. Blottner for use of the Leica SP8 confocal microscope. Funding: This work was supported by the German Research Foundation DFG grant RO 3497/4-1 (to M.R.). Author contributions: E.E. performed, imaged, and quantified the majority of the experiments. V.S. carried out the in situ hybridizations and most dendritic reconstructions and supported E.E. during quantifications. V.S. and M.R. prepared samples for mass spectrometry. Initial characterization of the phenotype was carried out by T.N. and M.R. with the assistance of V.S., E.B., and O.B. K.T.-T. carried out the mass spectrometry. D.L. provided technical assistance throughout the project. O.B. and I.B. assisted in establishing live imaging of brain sections. S.H. carried out preliminary IUEs. T.S. cloned the Nrp1 construct. M.C.A. contributed technical and scientific feedback. M.R. and V.T. conceptualized and supervised the project. M.R. wrote the manuscript. Figures were prepared by E.E. and M.R. The manuscript was reviewed by all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabf1973/DC1
REFERENCES AND NOTES
- 1.Petreanu L., Mao T., Sternson S. M., Svoboda K., The subcellular organization of neocortical excitatory connections. Nature 457, 1142–1145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuster S., Rivalan M., Strauss U., Stoenica L., Trimbuch T., Rademacher N., Parthasarathy S., Lajkó D., Rosenmund C., Shoichet S. A., Winter Y., Tarabykin V., Rosário M., NOMA-GAP/ARHGAP33 regulates synapse development and autistic-like behavior in the mouse. Mol. Psychiatry 20, 1120–1131 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Rosario M., Schuster S., Juttner R., Parthasarathy S., Tarabykin V., Birchmeier W., Neocortical dendritic complexity is controlled during development by NOMA-GAP-dependent inhibition of Cdc42 and activation of cofilin. Genes Dev. 26, 1743–1757 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaufmann W. E., Moser H. W., Dendritic anomalies in disorders associated with mental retardation. Cereb. Cortex 10, 981–991 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Nadarajah B., Brunstrom J. E., Grutzendler J., Wong R. O., Pearlman A. L., Two modes of radial migration in early development of the cerebral cortex. Nat. Neurosci. 4, 143–150 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Angevine J. B. Jr., Sidman R. L., Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature 192, 766–768 (1961). [DOI] [PubMed] [Google Scholar]
- 7.Tabata H., Nakajima K., Multipolar migration: The third mode of radial neuronal migration in the developing cerebral cortex. J. Neurosci. 23, 9996–10001 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noctor S. C., Martinez-Cerdeno V., Ivic L., Kriegstein A. R., Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Tabata H., Kanatani S., Nakajima K., Differences of migratory behavior between direct progeny of apical progenitors and basal progenitors in the developing cerebral cortex. Cereb. Cortex 19, 2092–2105 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Hatanaka Y., Hisanaga S.-I., Heizmann C. W., Murakami F., Distinct migratory behavior of early- and late-born neurons derived from the cortical ventricular zone. J. Comp. Neurol. 479, 1–14 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Rakic P., Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 145, 61–83 (1972). [DOI] [PubMed] [Google Scholar]
- 12.Mowat D. R., Croaker G. D., Cass D. T., Kerr B. A., Chaitow J., Adès L. C., Chia N. L., Wilson M. J., Hirschsprung disease, microcephaly, mental retardation, and characteristic facial features: Delineation of a new syndrome and identification of a locus at chromosome 2q22-q23. J. Med. Genet. 35, 617–623 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miquelajauregui A., van de Putte T., Polyakov A., Nityanandam A., Boppana S., Seuntjens E., Karabinos A., Higashi Y., Huylebroeck D., Tarabykin V., Smad-interacting protein-1 (Zfhx1b) acts upstream of Wnt signaling in the mouse hippocampus and controls its formation. Proc. Natl. Acad. Sci. U.S.A. 104, 12919–12924 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seuntjens E., Nityanandam A., Miquelajauregui A., Debruyn J., Stryjewska A., Goebbels S., Nave K. A., Huylebroeck D., Tarabykin V., Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat. Neurosci. 12, 1373–1380 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., Chen L., Chen L., Chen T. M., Chi Chin M., Chong J., Crook B. E., Czaplinska A., Dang C. N., Datta S., Dee N. R., Desaki A. L., Desta T., Diep E., Dolbeare T. A., Donelan M. J., Dong H. W., Dougherty J. G., Duncan B. J., Ebbert A. J., Eichele G., Estin L. K., Faber C., Facer B. A., Fields R., Fischer S. R., Fliss T. P., Frensley C., Gates S. N., Glattfelder K. J., Halverson K. R., Hart M. R., Hohmann J. G., Howell M. P., Jeung D. P., Johnson R. A., Karr P. T., Kawal R., Kidney J. M., Knapik R. H., Kuan C. L., Lake J. H., Laramee A. R., Larsen K. D., Lau C., Lemon T. A., Liang A. J., Liu Y., Luong L. T., Michaels J., Morgan J. J., Morgan R. J., Mortrud M. T., Mosqueda N. F., Ng L. L., Ng R., Orta G. J., Overly C. C., Pak T. H., Parry S. E., Pathak S. D., Pearson O. C., Puchalski R. B., Riley Z. L., Rockett H. R., Rowland S. A., Royall J. J., Ruiz M. J., Sarno N. R., Schaffnit K., Shapovalova N. V., Sivisay T., Slaughterbeck C. R., Smith S. C., Smith K. A., Smith B. I., Sodt A. J., Stewart N. N., Stumpf K. R., Sunkin S. M., Sutram M., Tam A., Teemer C. D., Thaller C., Thompson C. L., Varnam L. R., Visel A., Whitlock R. M., Wohnoutka P. E., Wolkey C. K., Wong V. Y., Wood M., Yaylaoglu M. B., Young R. C., Youngstrom B. L., Feng Yuan X., Zhang B., Zwingman T. A., Jones A. R., Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Van de Putte T., Francis A., Nelles L., van Grunsven L. A., Huylebroeck D., Neural crest-specific removal of Zfhx1b in mouse leads to a wide range of neurocristopathies reminiscent of Mowat-Wilson syndrome. Hum. Mol. Genet. 16, 1423–1436 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Van de Putte T., Maruhashi M., Francis A., Nelles L., Kondoh H., Huylebroeck D., Higashi Y., Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am. J. Hum. Genet. 72, 465–470 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanchina L., Van de Putte T., Goossens M., Huylebroeck D., Bondurand N., Genetic interaction between Sox10 and Zfhx1b during enteric nervous system development. Dev. Biol. 341, 416–428 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Srivatsa S., Parthasarathy S., Molnar Z., Tarabykin V., Sip1 downstream Effector ninein controls neocortical axonal growth, ipsilateral branching, and microtubule growth and stability. Neuron 85, 998–1012 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Conces M. R., Hughes A., Pierson C. R., Neuropathology of Mowat-Wilson syndrome. Pediatr. Dev. Pathol. 23, 322–325 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Garavelli L., Ivanovski I., Caraffi S. G., Santodirocco D., Pollazzon M., Cordelli D. M., Abdalla E., Accorsi P., Adam M. P., Baldo C., Bayat A., Belligni E., Bonvicini F., Breckpot J., Callewaert B., Cocchi G., Cuturilo G., Devriendt K., Dinulos M. B., Djuric O., Epifanio R., Faravelli F., Formisano D., Giordano L., Grasso M., Grønborg S., Iodice A., Iughetti L., Lacombe D., Maggi M., Malbora B., Mammi I., Moutton S., Møller R., Muschke P., Napoli M., Pantaleoni C., Pascarella R., Pellicciari A., Poch-Olive M. L., Raviglione F., Rivieri F., Russo C., Savasta S., Scarano G., Selicorni A., Silengo M., Sorge G., Tarani L., Tone L. G., Toutain A., Trimouille A., Valera E. T., Vergano S. S., Zanotta N., Zollino M., Dobyns W. B., Paciorkowski A. R., Neuroimaging findings in Mowat-Wilson syndrome: A study of 54 patients. Genet. Med. 19, 691–700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higashi Y., Maruhashi M., Nelles L., van de Putte T., Verschueren K., Miyoshi T., Yoshimoto A., Kondoh H., Huylebroeck D., Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis 32, 82–84 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Goebbels S., Bormuth I., Bode U., Hermanson O., Schwab M. H., Nave K. A., Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis 44, 611–621 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Epifanova E., Babaev A., Newman A. G., Tarabykin V., Role of Zeb2/Sip1 in neuronal development. Brain Res. 1705, 24–31 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Dimidschstein J., Passante L., Dufour A., van den Ameele J., Tiberi L., Hrechdakian T., Adams R., Klein R., Lie D. C., Jossin Y., Vanderhaeghen P., Ephrin-B1 controls the columnar distribution of cortical pyramidal neurons by restricting their tangential migration. Neuron 79, 1123–1135 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Parthasarathy S., Srivatsa S., Nityanandam A., Tarabykin V., Ntf3 acts downstream of Sip1 in cortical postmitotic neurons to control progenitor cell fate through feedback signaling. Development 141, 3324–3330 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Kok F. O., Shepherd I. T., Sirotkin H. I., Churchill and Sip1a repress fibroblast growth factor signaling during zebrafish somitogenesis. Dev. Dyn. 239, 548–558 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hynes R. O., Integrins: Bidirectional, allosteric signaling machines. Cell 110, 673–687 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Lee K. K., de Repentigny Y., Saulnier R., Rippstein P., Macklin W. B., Kothary R., Dominant-negative beta1 integrin mice have region-specific myelin defects accompanied by alterations in MAPK activity. Glia 53, 836–844 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Leone D. P., Relvas J.~. B., Campos L. S., Hemmi S., Brakebusch C., Fässler R., ffrench-Constant C., Suter U., Regulation of neural progenitor proliferation and survival by β1 integrins. J. Cell Sci. 118, 2589–2599 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Verschueren K., Remacle J. E., Collart C., Kraft H., Baker B. S., Tylzanowski P., Nelles L., Wuytens G., Su M. T., Bodmer R., Smith J. C., Huylebroeck D., SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J. Biol. Chem. 274, 20489–20498 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Niland S., Eble J. A., Neuropilin: Handyman and power broker in the tumor microenvironment. Adv. Exp. Med. Biol. 1223, 31–67 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Brasch J., Katsamba P. S., Harrison O. J., Ahlsén G., Troyanovsky R. B., Indra I., Kaczynska A., Kaeser B., Troyanovsky S., Honig B., Shapiro L., Homophilic and heterophilic interactions of type II cadherins identify specificity groups underlying cell-adhesive behavior. Cell Rep. 23, 1840–1852 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunne E., Spring C. M., Reheman A., Jin W., Berndt M. C., Newman D. K., Newman P. J., Ni H., Kenny D., Cadherin 6 has a functional role in platelet aggregation and thrombus formation. Arterioscler. Thromb. Vasc. Biol. 32, 1724–1731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casal J. I., Bartolome R. A., Beyond N-cadherin, relevance of cadherins 5, 6 and 17 in cancer progression and metastasis. Int. J. Mol. Sci. 20, 3373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeNardo L. A., Berns D. S., DeLoach K., Luo L., Connectivity of mouse somatosensory and prefrontal cortex examined with trans-synaptic tracing. Nat. Neurosci. 18, 1687–1697 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jossin Y., Cooper J. A., Reelin, Rap1 and N-cadherin orient the migration of multipolar neurons in the developing neocortex. Nat. Neurosci. 14, 697–703 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo G., Arnaud L., Kronstad-O’Brien P., Cooper J. A., Absence of Fyn and Src causes a reeler-like phenotype. J. Neurosci. 25, 8578–8586 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai J. W., Chen Y., Kriegstein A. R., Vallee R. B., LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 170, 935–945 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valdembri D., Caswell P. T., Anderson K. I., Schwarz J. P., König I., Astanina E., Caccavari F., Norman J. C., Humphries M. J., Bussolino F., Serini G., Neuropilin-1/GIPC1 signaling regulates α5β1 integrin traffic and function in endothelial cells. PLOS Biol. 7, e1000025 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polleux F., Giger R. J., Ginty D. D., Kolodkin A. L., Ghosh A., Patterning of cortical efferent projections by semaphorin-neuropilin interactions. Science 282, 1904–1906 (1998). [DOI] [PubMed] [Google Scholar]
- 42.Chen G., Sima J., Jin M., Wang K. Y., Xue X. J., Zheng W., Ding Y. Q., Yuan X. B., Semaphorin-3A guides radial migration of cortical neurons during development. Nat. Neurosci. 11, 36–44 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Hatanaka Y., Matsumoto T., Yanagawa Y., Fujisawa H., Murakami F., Masu M., Distinct roles of neuropilin 1 signaling for radial and tangential extension of callosal axons. J. Comp. Neurol. 514, 215–225 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Polleux F., Morrow T., Ghosh A., Semaphorin 3A is a chemoattractant for cortical apical dendrites. Nature 404, 567–573 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Forster E., Reelin, neuronal polarity and process orientation of cortical neurons. Neuroscience 269, 102–111 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Sasaki Y., Cheng C., Uchida Y., Nakajima O., Ohshima T., Yagi T., Taniguchi M., Nakayama T., Kishida R., Kudo Y., Ohno S., Nakamura F., Goshima Y., Fyn and Cdk5 mediate semaphorin-3A signaling, which is involved in regulation of dendrite orientation in cerebral cortex. Neuron 35, 907–920 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Demyanenko G. P., Schachner M., Anton E., Schmid R., Feng G., Sanes J., Maness P. F., Close homolog of L1 modulates area-specific neuronal positioning and dendrite orientation in the cerebral cortex. Neuron 44, 423–437 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Vastrik I., Eickholt B. J., Walsh F. S., Ridley A., Doherty P., Sema3A-induced growth-cone collapse is mediated by Rac1 amino acids 17–32. Curr. Biol. 9, 991–998 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Jossin Y., Goffinet A. M., Reelin signals through phosphatidylinositol 3-kinase and Akt to control cortical development and through mTor to regulate dendritic growth. Mol. Cell. Biol. 27, 7113–7124 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fenstermaker V., Chen Y., Ghosh A., Yuste R., Regulation of dendritic length and branching by semaphorin 3A. J. Neurobiol. 58, 403–412 (2004). [DOI] [PubMed] [Google Scholar]
- 51.O’Dell R. S., Cameron D. A., Zipfel W. R., Olson E. C., Reelin prevents apical neurite retraction during terminal translocation and dendrite initiation. J. Neurosci. 35, 10659–10674 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shelly M., Cancedda L., Lim B. K., Popescu A. T., Cheng P. L., Gao H., Poo M. M., Semaphorin3A regulates neuronal polarization by suppressing axon formation and promoting dendrite growth. Neuron 71, 433–446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duan X., Krishnaswamy A., Laboulaye M. A., Liu J., Peng Y.-R., Yamagata M., Toma K., Sanes J. R., Cadherin combinations recruit dendrites of distinct retinal neurons to a shared interneuronal scaffold. Neuron 99, 1145–1154.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osterhout J. A., Josten N., Yamada J., Pan F., Wu S. W., Nguyen P. L., Panagiotakos G., Inoue Y. U., Egusa S. F., Volgyi B., Inoue T., Bloomfield S. A., Barres B. A., Berson D. M., Feldheim D. A., Huberman A. D., Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron 71, 632–639 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basu R., Duan X., Taylor M. R., Martin E. A., Muralidhar S., Wang Y., Gangi-Wellman L., Das S. C., Yamagata M., West P. J., Sanes J. R., Williams M. E., Heterophilic type II cadherins are required for high-magnitude synaptic potentiation in the hippocampus. Neuron 96, 160–176.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terakawa Y. W., Inoue Y. U., Asami J., Hoshino M., Inoue T., A sharp cadherin-6 gene expression boundary in the developing mouse cortical plate demarcates the future functional areal border. Cereb. Cortex 23, 2293–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tachikawa K., Sasaki S., Maeda T., Nakajima K., Identification of molecules preferentially expressed beneath the marginal zone in the developing cerebral cortex. Neurosci. Res. 60, 135–146 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Sekine K., Kawauchi T., Kubo K. I., Honda T., Herz J., Hattori M., Kinashi T., Nakajima K., Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin α5β1. Neuron 76, 353–369 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ambrozkiewicz M. C., Schwark M., Kishimoto-Suga M., Borisova E., Hori K., Salazar-Lázaro A., Rusanova A., Altas B., Piepkorn L., Bessa P., Schaub T., Zhang X., Rabe T., Ripamonti S., Rosário M., Akiyama H., Jahn O., Kobayashi T., Hoshino M., Tarabykin V., Kawabe H., Polarity acquisition in cortical neurons is driven by synergistic action of Sox9-regulated Wwp1 and Wwp2 E3 ubiquitin ligases and intronic miR-140. Neuron 100, 1097–1115.e15 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Polleux F., Ghosh A., The slice overlay assay: A versatile tool to study the influence of extracellular signals on neuronal development. Sci. STKE 2002, pl9 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Dyballa N., Metzger S., Fast and sensitive colloidal coomassie G-250 staining for proteins in polyacrylamide gels. J. Vis. Exp., 1431 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lehmann A., Niewienda A., Jechow K., Janek K., Enenkel C., Ecm29 fulfils quality control functions in proteasome assembly. Mol. Cell 38, 879–888 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/27/eabf1973/DC1