Abstract
Application of an immunomagnetic enrichment method selective for Vibrio parahaemolyticus serovar K6 allowed isolation of a strain belonging to the pandemic O3:K6 clone of V. parahaemolyticus from fresh shellfish not implicated in a clinical case in southern Thailand. Arbitrarily primed PCR profiles of this strain, clinical O3:K6 strains isolated from sporadic diarrhea cases in the same area, and a standard pandemic O3:K6 strain were indistinguishable.
Vibrio parahaemolyticus is a marine bacterium, and it can cause gastroenteritis in humans through consumption of seafood. However, not all strains are considered pathogenic. Almost all clinical strains and rare environmental strains were shown to exhibit the Kanagawa phenomenon (KP) in early epidemiological studies (6, 15). KP is a beta-type hemolysis induced by thermostable direct hemolysin in a special blood agar medium, Wagatsuma agar. Thermostable direct hemolysin is encoded by the tdh gene. KP-positive strains carry two tdh genes, and KP is due to high-level expression of one of the tdh genes (10, 14). Enterotoxicity of thermostable direct hemolysin was demonstrated using a mutant strain derived from a KP-positive strain in which both tdh genes were specifically inactivated (8). Some clinical strains are KP negative and carry the trh gene (11). The trh gene is 68 to 69% homologous to the tdh gene and encodes a thermostable direct hemolysin-related hemolysin (11). The tdh and trh genes in KP-negative strains are expressed at low levels, but molecular epidemiological studies revealed a strong association of clinical strains with possession of the tdh gene, the trh gene, or both genes (4, 16). In contrast, the tdh and trh genes were rarely detected in the environmental strains of V. parahaemolyticus (4, 9, 16).
Serotyping based on the O and K antigens of V. parahaemolyticus is often applied to epidemiological investigations. However, the dominance of a single serovar in outbreaks in a wide area had not been noted until recently; infections due to O4:K12 strains were reported on the western coasts of the United States and Mexico (1, 12). Emergence of a new O3:K6 clone in 1995 and its pandemic spread have been shown to be responsible for recent infections in seven Asian countries and in the United States (5, 12). In addition, serovariants of this clone emerged and followed a spreading pattern similar to that of the new O3:K6 clone (5). This O3:K6 clone and its variants could be distinguished from other strains by possession of the tdh gene but not the trh gene, by unique profiles in an arbitrarily primed PCR (AP-PCR) analysis, and by a new PCR method (5). The new PCR method was targeted to the toxRS operon, encoding a transcriptional regulator. The target sequences that are specific to the pandemic O3:K6 clone and its variants were selected in this PCR, and thus this PCR method was named GS-PCR for group-specific PCR (5).
It is not known which geographical region of the marine environment is the origin of the pandemic O3:K6 clone and how the clone spread. Isolation of the environmental strains belonging to this clone and comparative analysis of the environmental and clinical strains might allow us to elucidate these points. Some screening method is needed to isolate the pandemic clone from the environment. Isolation of tdh-bearing strains from the environmental samples not implicated in clinical cases is very rare. In this study, we demonstrated by an artificial contamination experiment that an immunomagnetic enrichment method can be helpful to selectively isolate the pandemic O3:K6 strain from seafood. We then examined by this method the fresh seafoods marketed in a city in southern Thailand where the incidence of V. parahaemolyticus infection is frequently recorded year round and where the pandemic O3:K6 clone has been isolated from the patients (5). As a result, we isolated a strain belonging to the pandemic O3:K6 clone from fresh, local seafood not implicated in a clinical case. We compared this environmental strain with the clinical strains isolated in the same geographical region.
Evaluation of K serovar-specific immunomagnetic enrichment method.
Tomoyasu (19) reported an enrichment method to selectively isolate V. parahaemolyticus strains belonging to a specific K serovar from seafood by using immunomagnetic beads and antiserum specific to a K antigen. We determined by use of artificial contamination experiments whether Tomoyasu's method can be useful for isolation of the pandemic O3:K6 strains from seafood. First, seafood samples that did not contain V. parahaemolyticus were sought among the frozen seafood imported from Southeast Asian countries into Japan. Seafood samples of 10 to 30 g were added into 100-ml alkaline peptone water (APW; 1% peptone, 1% NaCl) at pH 9.2. The mixture was vigorously shaken, and 50 ml of the broth culture was transferred into a sterile container and incubated without shaking for 7 h at 37°C. One loopful of the enrichment broth was then streaked onto each of five thiosulfate-citrate-bile salts-sucrose (TCBS) agar (Eiken Chemical, Co., Ltd., Tokyo, Japan) plates, and five green colonies, if any, per plate were examined by the toxR-targeted PCR method for identification of V. parahaemolyticus as described previously (3). In addition, the alkaline peptone enrichment broth was examined directly by this PCR method with minor modifications. Briefly, 1.5 μl of the 1:10 diluted supernatant of the boiled broth culture was examined by using 35 cycles of the PCR amplification. The detection limit of this PCR method was 1.25 CFU per μl (A. Chowdhury and M. Nishibuchi, unpublished data). Seafood samples that gave V. parahaemolyticus-negative results in both the isolation and the direct PCR tests were used in the artificial contamination experiments. A 20-g portion of the seafood sample was added to each of seven tubes containing 40 ml of APW (pH 9.2). Strain VP47, a pandemic O3:K6 strain isolated from a patient with diarrhea in India in 1996 (13), was grown in Luria-Bertani broth containing 1% NaCl with shaking (160 rpm) overnight at 37°C. The culture was diluted appropriately in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KHPO4; pH 7.2) and inoculated into the APW containing the V. parahaemolyticus-negative seafood at various concentrations (shown in Table 1) and mixed well. The seafood in the culture was allowed to sediment for a few minutes. Then, 1 ml of the supernatant was transferred into a microtube, and 20 μl of anti-K6 antiserum (V. parahaemolyticus K antiserum K6; Denka Seiken, Co., Ltd., Tokyo, Japan) was added and incubated for 20 min at room temperature with gentle mixing. Bacterial cells were collected by centrifugation (2,040 × g, 10 min), washed with 1 ml of PBS twice, and then suspended in 1 ml of PBS. A 10-μl portion of anti-rabbit immunoglobulin G (IgG)-coated magnetic particle suspension (Dynabeads M-280 Sheep Anti-Rabbit IgG; Dynal A. S., Oslo, Norway) was added to the cell suspension, and the mixture was incubated for 20 min at room temperature with intermittent mixing. The immunomagnetic particles (IMP) were collected with a magnetic particle concentrator (Dynal MPC-M, Dynal A. S.), washed four times with 1 ml of PBS, and then suspended in 1 ml of PBS.
TABLE 1.
Recovery of V. parahaemolyticus VP47 inoculated into APW containing seafood by the K6-specific immunomagnetic separation method with or without enrichment culture of the immunomagnetic particles (IMP)
| Seafood sample and enrichment culture | Recoverya (no. of colonies recovered) of inoculated VP47 cells at:
|
||||||
|---|---|---|---|---|---|---|---|
| 2.3 × 104 CFU/ml | 2.3 × 103 CFU/ml | 2.3 × 102 CFU/ml | 23 CFU/ml | 2.3 CFU/ml | 0.23 CFU/ml | 0 CFU/ml | |
| Squid | |||||||
| Direct platingb | + (223) | + (46.6) | + (0.669) | − | − | − | − |
| Enrichment culture in APWc | + (37.0) | + (16.0) | + (21.7) | + (16.0) | + (23.7) | − | − |
| Enrichment culture in SPBd | NT | NT | NT | + (128) | + (78.5) | − | − |
| Fish (Nemipterus virgatus) | |||||||
| Direct plating | + (TNTC) | + (92.0) | + (17.0) | + (0.666) | − | − | − |
| Enrichment culture in APW | + (24.3) | + (21.3) | + (24.7) | + (36.0) | + (15.0) | − | − |
| Enrichment culture in SPB | NT | NT | NT | + (73.9) | + (117) | − | − |
| Shrimp (Penaeus monodon) | |||||||
| Direct plating | + (TNTC) | + (125) | + (6.00) | + (1.30) | + (0.33) | − | − |
| Enrichment culture in APW | + (36.0) | + (25.3) | + (31.0) | + (23.0) | + (39.7) | − | − |
| Enrichment culture in SPB | NT | NT | NT | + (16.0) | + (79.2) | − | − |
+, VP47 was recovered on TCBS agar; −, VP47 was not recovered on TCBS agar; NT, not tested. The numbers of VP47 colonies recovered per TCBS agar plate (average of three plates) are shown in the parentheses. TNTC, too numerous to count.
Enrichment culture of IMP was not performed. A 1-ml portion of the supernatant of the APW-seafood mixture containing VP47 was treated with IMS, and one-tenth of the IMS suspension was directly streaked onto TCBS agar.
The IMP suspension was prepared as for the direct plating method. Four-tenths of the IMS suspension was incubated in 5 ml of APW at pH 8.6 overnight at 37°C. A 50-μl portion of the 105-fold dilution of the enrichment culture was inoculated onto TCBS agar so that VP47 could be recovered as isolated colonies.
The IMP suspension was prepared as for the direct plating method. Four-tenths of the IMS suspension was incubated in 5 ml of salt polymyxin broth overnight at 37°C. A 50-μl portion of the 105-fold dilution of the enrichment culture was inoculated onto TCBS agar so that VP47 could be recovered as isolated colonies.
Tomoyasu's procedure included overnight incubation of the washed IMP in an enrichment broth before inoculation onto TCBS agar. We examined whether this enrichment culture of the washed IMP is necessary or not. Recovery of the inoculated O3:K6 cells from the IMP with or without the enrichment step was compared. Tomoyasu (19) employed salt polymyxin broth (SPB) for enrichment of the IMP suspension. APW was superior to SPB for selective isolation of V. parahaemolyticus from seafood in our past surveys (Chowdhury and Nishibuchi, unpublished). We therefore compared APW and SPB in the initial experiment (Table 1) and mainly used APW in the subsequent experiment (Table 2). We employed APW at pH 9.2 for incubation of seafood samples because the pH usually changes to ca. 8.6 after the addition of the seafood samples. APW at pH 8.6 was used for incubation of the IMP suspension. A 100-μl portion of the washed IMP was directly streaked onto TCBS agar in triplicate without enrichment culture. This modification is referred to below as the direct plating method. A 400-μl portion of the IMP suspension was inoculated into 5 ml of APW at pH 8.6, SPB (Nissui Seiyaku Co., Tokyo, Japan), or both and incubated overnight at 37°C. Three 50-μl aliquots of the 105-fold dilution of the broth culture were then streaked onto TCBS agar so that VP47 absorbed onto the IMP and subsequently grown to high densities (>107 CFU/ml) would be recovered as isolated colonies. This method was named the enrichment culture method. Up to 20 representative green colonies, if any, were selected from those that appeared on the TCBS agar plate; these were examined by the slide agglutination test with anti-K6 antiserum according to the manufacturer's specification (Denka Seiken) and by the PCR methods to detect the toxR gene (3), the tdh gene (18) (primers D3 and D5), and the trh gene (18) (primers R2 and R6). All examined colonies gave K6 antigen-positive, toxR gene-positive, tdh gene-positive, and trh gene-negative results, and therefore all green colonies were assumed to be VP47. The results of the recovery of VP47 from various inoculated seafoods are summarized in Table 1. The lowest limit of VP47 detection by the direct plating method was 2.3 to 2.3 × 102 CFU/ml (1.15 to 1.15 × 102 CFU/g of seafood) and that found by the enrichment culture method was 2.3 CFU/ml (1.15 CFU/g of seafood); the latter value was not influenced by the kind of enrichment broth used (APW or SPB).
TABLE 2.
Recovery of V. parahaemolyticus VP47, VP87, and VP41 inoculated into APW containing seafood by the K6-specific immunomagnetic separation method with or without enrichment culture of the IMP
| Seafood sample and enrichment culture | Recoverya (no. of colonies) of inoculated test strain(s):
|
||||||
|---|---|---|---|---|---|---|---|
| VP47 | VP87 | VP41 | VP47 and VP87 | VP47 and VP41 | VP47, VP87, and VP41 | None | |
| Squid | |||||||
| Direct platingb | + (7.33) | − | − | + (6.67) | + (1.33) | + (2.67) | − |
| Enrichment culture in APWc | + (29.3) | + (26.3) | + (54.7) | + (45.3) | + (65.6) | + (38.3) | − |
| Fish (Nemipterus virgatus) | |||||||
| Direct plating | + (26.0) | − | − | + (38.0) | + (20.0) | + (14.0) | − |
| Enrichment culture in APW | + (31.7) | + (7.33) | + (12.3) | + (11.0) | + (6.67) | + (38.3) | − |
| Shrimp (Penaeus monodon) | |||||||
| Direct plating | + (282) | + (0.333) | + (4.00) | + (383) | + (522) | + (285) | − |
| Enrichment culture in APW | + (10.0) | + (7.67) | + (22.3) | + (86.0) | + (56.0) | + (10.3) | − |
| Enrichment culture in SPBd | + (119) | + (28.2) | + (52.1) | + (16.9) | + (14.7) | + (92.9) | − |
+, Test strain (green colony) was recovered on TCBS agar; −, test strain (green colony) was not recovered on TCBS agar. The numbers of green colonies recovered per TCBS agar plate (average of three plates) are shown in the parentheses. When mixed test strains were inoculated, all green colonies were found to be VP47 (as explained in the text).
Enrichment culture of IMP was not performed. A 1-ml portion of the supernatant of the APW-seafood mixture containing VP47 was treated with IMS, and one-tenth of the IMS suspension was directly streaked onto TCBS agar.
The IMP suspension was prepared as for the direct plating method. Four-tenths of the IMS suspension was incubated in 5 ml of APW at pH 8.6 overnight at 37°C. A 50-μl portion of the 105-fold dilution of the enrichment culture was inoculated onto TCBS agar so that VP47 could be recovered as isolated colonies.
The IMP suspension was prepared as described for the direct plating method. Four-tenths of the IMS suspension was incubated in 5 ml of salt polymyxin broth overnight at 37°C. A 50-μl portion of the 105-fold dilution of the enrichment culture was inoculated onto TCBS agar so that VP47 could be recovered as isolated colonies.
The specificity of the direct plating and enrichment culture methods was examined next. The seafood sample was inoculated with the test strain(s), and recovery of the inoculated strains was determined as described above with the following modifications. The test strains were VP47, VP87 (an O1:K1 strain carrying the tdh and trh gene, isolated from a patient in 1996 in India [13]) and VP41 (an O4:K4 strain lacking both tdh and trh genes, isolated from the environment in 1998 in Thailand); for some inoculations, a mixture of two or three of the test strains was used. Each of the test strains was inoculated into the APW containing the seafood sample to achieve a final concentration of 103 CFU/ml (5 × 102 CFU/g of seafood). The numbers of the green colonies recovered on TCBS agar plates were counted as described above, and the results are summarized in Table 2. The representative green colonies (up to 20 colonies, if any, per TCBS agar plate) were examined for the K serovar by the agglutination method and for presence or absence of the toxR, tdh, and trh genes by the PCR methods described above. The green colonies resulting from inoculation of a single test strain were confirmed to be the spiked strain. All green colonies resulting from the simultaneous inoculation of VP47 with other test strain(s) were judged to be VP47 and not the other test strains. The results indicate that VP47 could be recovered specifically by the immunomagnetic enrichment method. The non-K6 strains inoculated singly could be recovered by the direct plating method only in the examination of shrimp, and the numbers of the colonies on TCBS agar were 0.1 and 1.4% of that found with VP47. However, non-K6 strains inoculated singly were recovered by the enrichment culture method in all examinations. The adsorbed non-K6 strains, without VP47, multiplied to levels similar to that of VP47 (on the order of 107 or 108 CFU/ml). The results indicate that the non-K6 strains were also adsorbed to the immunomagnetic beads at very low frequencies. This suggests a possibility that organisms other than the K6 strains contained in the environmental sample can be competitors of K6 strains for adsorption onto the immunomagnetic beads and for subsequent growth. This drawback can decrease the specificity of immunomagnetic separation unless the concentration of the O3:K6 strain in the natural environment is high, which does not appear to be the case. The simplicity of the direct plating method and the possible low specificity of the enrichment culture method led us to conclude that the direct plating method is the method of choice, although the sensitivity of the direct plating method may be lower (up to a 2-log difference) than that of the enrichment culture method. The reduction in the sensitivity of the direct plating method is probably due to adsorption of the bacterial cells in part to seafood materials, and the degree of the adsorption appears to vary among seafood materials.
Isolation of V. parahaemolyticus.
We employed the direct plating method to examine local, fresh seafood samples in Thailand. A total of 114 fresh seafood samples were examined during the period between December 1998 and January 1999 in Songkla Province, Thailand. The samples consisted of 54 shellfish samples (bloody clam [Anadara granosa], oyster [Crassostrea belehen], and green mussel [Perna viridis]), 30 shrimp samples (tiger shrimp [Penaeus monodon] and banana shrimp [Penaeus merguiensis]), and 30 fish (red snapper [Lutianus sanguineus], seabass [Lates calcarifer], chub mackerel [Rastrelliger negletus], and fusillier [Caesio tile]). These fresh seafood samples were purchased in a fresh market in Hat Yai City, Songkla Province. The majority of these seafoods were harvested in Songkla Lake, a brackish-water lake that is connected to the Gulf of Thailand. The seafood samples were transferred to the laboratory at room temperature and examined within 2 h of sample collection. Shellfish was shucked aseptically and subjected to examination. The seafood samples were examined essentially as described above for the direct plating method. Portions of 10 to 30 g of each seafood sample were inoculated into 100 ml of APW at pH 9.2 and mixed using a stomacher for 30 s, and the contents of the mixture were allowed to settle down for a few minutes. Only the supernatant was transferred into a sterile flask and incubated without shaking at 37°C for 7 h. After incubation of the seafood in APW, 2 ml of the broth culture was mixed with 20 μl of the anti-K6 antiserum, and then the mixture was incubated for 20 min at room temperature with gentle mixing. Bacterial cells were harvested by centrifugation and washed with 2 ml of PBS, and the cells were then suspended in 1 ml of PBS. Next, 10 μl of the anti-rabbit IgG-coated Dynabead suspension was added to the cell suspension followed by incubation for 20 min with intermittent mixing. The IMP were collected, washed twice with 2 ml of PBS, and suspended in 10 μl of PBS, and then the entire IMP suspension was streaked onto a TCBS agar (Difco Laboratories, Detroit, Mich.) plate. After an 18- to 24-h incubation at 37°C, five green colonies were selected and examined for biochemical characteristics to screen for V. parahaemolyticus. The strains showing motility in a nutrient agar-based semisolid medium and positive results in oxidase, lysine decarboxylase, and indole tests were selected. The strains thus screened were examined by the PCR method for detection of the V. parahaemolyticus toxR gene as described previously (3). The strains that gave a positive result in this species-specific PCR assay were identified as V. parahaemolyticus. V. parahaemolyticus strains were isolated from 51 (94%) of 54 shellfish samples, 25 (83%) of 30 shrimp samples, and 22 (73%) of 30 fish samples and totaled 296.
Fecal samples collected from the patients of sporadic diarrhea cases in Hat Yai Hospital, Songkla Province, were examined in September and October 1998. Each fecal sample was streaked directly onto a TCBS agar (Eiken Chemical) plate. Green colonies were examined by the standard biochemical tests for identification of V. parahaemolyticus (2). The identification was confirmed by the toxR-specific PCR method (3). Twenty-three clinical strains thus identified were obtained.
Characterization of isolated strains.
The strains identified as V. parahaemolyticus were examined for the presence or absence of the tdh and trh genes by the PCR methods described above. Of the environmental strains, one tdh gene-positive strain and one trh gene-positive strain were detected. Both strains were isolated from bloody clam. Of the 23 clinical strains, 21 carried the tdh gene and none had the trh gene. The 2 environmental strains carrying the tdh or trh gene, the 27 strains randomly selected from toxin gene-negative environmental strains (isolated from 7 shellfish samples, 6 shrimp samples, and 13 fish samples), and all of the clinical strains were examined for the O:K serovar as described previously (17) and for the toxRS sequence unique to the pandemic clone by the GS-PCR as described previously (5). The results are summarized in Table 3. Although the K6-specific immunomagnetic separation method was employed for the isolation of environmental strains, the isolated strains belonging to various K serovars and 2 of the 29 environmental strains belonged to the K6 serovar. The results suggest that K6 strains, including the pandemic O3:K6 clone, may be scarcely distributed in this environment. However, we did isolate one pandemic O3:K6 strain, as evidenced by the O3:K6 serovar, the tdh-positive and trh-negative gene profile, and the GS-PCR-positive result. This strain was designated E1. On the other hand, 20 of 23 clinical strains were pandemic O3:K6 strains. Apparently, a low frequency of isolation of the pandemic O3:K6 strain from the environment may possibly be due to the fact that the IMP-based isolation method was designed for and evaluated with rapidly growing cells in culture media. V. parahaemolyticus can exist as viable but nonculturable cells under a stressed condition, and it can be resuscitated to culturable cells by using stress-reducing reagents (7). If a considerable proportion of the environmental O3:K6 pandemic clone persists as nonculturable cells not producing enough K6 antigens, the IMP-based isolation method without the resuscitation step is not useful. We are planning a future environmental study to examine this possibility.
TABLE 3.
Characteristics of V. parahaemolyticus strains isolated in Songkla Province, Thailand
| Source of isolate | No. of strains | O:K serovara | Presence of geneb:
|
Results of GS-PCRc | |
|---|---|---|---|---|---|
| tdh | trh | ||||
| Environmentald | |||||
| Shellfish | 1 | 1:56 | − | − | − |
| 1 | 3:6 | + | − | + | |
| 2 | 4:12 | − | − | − | |
| 1 | 4:13 | − | − | − | |
| 1 | 4:68 | − | − | − | |
| 1 | 4:UT | − | − | − | |
| 1 | 5:UT | − | − | − | |
| 1 | 11:36 | − | + | − | |
| Prawn | 1 | 1:32 | − | − | − |
| 1 | 2:22 | − | − | − | |
| 2 | 3:12 | − | − | − | |
| 1 | 10:24 | − | − | − | |
| 1 | 11:30 | − | − | − | |
| Fish | 1 | 2:UT | − | − | − |
| 1 | 3:5 | − | − | − | |
| 1 | 3:6 | − | − | − | |
| 1 | 3:7 | − | − | − | |
| 1 | 3:12 | − | − | − | |
| 1 | 3:45 | − | − | − | |
| 1 | 4:34 | − | − | − | |
| 1 | 4:UT | − | − | − | |
| 1 | 5:UT | − | − | − | |
| 1 | 6:UT | − | − | − | |
| 1 | 8:UT | − | − | − | |
| 3 | 11:UT | − | − | − | |
| Clinicale | 20 | 3:6 | + | − | + |
| 1 | 3:72 | + | − | − | |
| 1 | 3:UT | − | − | − | |
| 1 | 4:4 | − | − | − | |
UT, untypeable.
+, Present; −, absent.
+, Positive; −, negative.
Isolated in September and October 1998.
Isolated in December 1998 and January 1999.
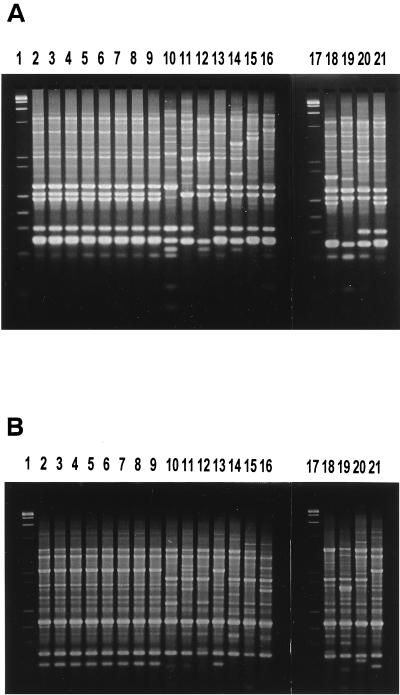
We also analyzed representative strains by the AP-PCR method. We reported previously that the pandemic O3:K6 strains isolated in other countries showed nearly identical AP-PCR profiles when we used six different primers designated primers 1 to 6 (5, 13). The representative strains and the control pandemic strain, VP47, were compared by this AP-PCR method. The results obtained with the six primers were essentially identical. Those obtained with primers 4 and 5 are shown in Fig. 1A and B, respectively. The AP-PCR profiles of the selected clinical O3:K6 strains (lanes 2 to 9) and the environmental strain, E1 (lane 13), were indistinguishable from that of the control strain (lane 20). The other strains exhibited various AP-PCR profiles that were distinct from these profiles. The results confirmed that the clinical O3:K6 strains and E1 belong to the pandemic O3:K6 clone.
FIG. 1.
Results of AP-PCR assay. The results obtained with primer 4 and primer 5 are presented in panels A and B, respectively. Lanes 1 and 17, molecular size markers (mixture of phage λ DNA digested with HindIII and phage φX174 DNA digested with HaeIII); lanes 2 to 9, clinical O3:K6 strains; lanes 10 to 12, clinical non-O3:K6 strains (serovars were O3:K untypeable, O3:K73, and O4:K4, respectively); lane 13, strain EV1 (an environmental O3:K6 strain carrying the tdh gene and lacking the trh gene); lane 14, an environmental O11:K36 strain lacking the tdh gene and carrying the trh gene; lane 15, an environmental O3:K6 strain lacking the tdh and trh genes; lanes 16, 18, 19, and 20, environmental non-O3:K6 strains (serovars are O3:K5, O3:K7, O4:K12, O3:K45, respectively); lane 21, strain VP47 (a control strain of the pandemic O3:K6 clone).
In conclusion, this work reports the isolation of the pandemic O3:K6 clone in local seafood not implicated in a clinical case. Distribution of this clone in the natural environment could be associated with clinical cases since the strains indistinguishable from the environmental strain were also isolated from many sporadic cases of diarrhea in the same geographical area.
Acknowledgments
This research was supported in part by the COE program on “Making Regions: Proto-Areas, Transformations and New Formations in Asia and Africa”; by a grant-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan; by the “Research for the Future” Program from The Japan Society for the Promotion of Science (JSPS-RFTF97L00706); by the U.S.-Japan Cooperative Medical Science Program (Japanese Panel); and by a fund from Faculty of Science and Medicine, Prince of Songkla University.
We are grateful to Yohko Takeda for technical assistance.
REFERENCES
- 1.Abbott S L, Powers C, Kaysner C A, Takeda Y, Ishibashi M, Joseph S W, Janda J M. Emergence of a restricted bioserovar of Vibrio parahaemolyticus as the predominant cause of Vibrio-associated gastroenteritis on the West Coast of the United States and Mexico. J Clin Microbiol. 1989;27:2891–2893. doi: 10.1128/jcm.27.12.2891-2893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly M T, Hickman-Brenner F W, Farmer J J., III . Vibrio. In: Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: American Society for Microbiology; 1991. pp. 384–395. [Google Scholar]
- 3.Kim Y B, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37:1173–1177. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kishishita M, Matsuoka N, Kumagai K, Yamasaki S, Takeda Y, Nishibuchi M. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl Environ Microbiol. 1992;58:2449–2457. doi: 10.1128/aem.58.8.2449-2457.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Rammamurthy T, Wong H-C, DePaola A, Kim Y B, Albert M J, Nishibuchi M. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol. 2000;38:578–585. doi: 10.1128/jcm.38.2.578-585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyamoto Y, Kato T, Obara Y, Akiyama S, Takizawa K, Yamai S. In vitro hemolytic characteristic of Vibrio parahaemolyticus: its close correlation with human pathogenicity. J Bacteriol. 1969;100:1147–1149. doi: 10.1128/jb.100.2.1147-1149.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizunoe, Y., S. N. Wai, T. Ishikawa, A. Takade, and S. Yoshida. Resuscitation of viable but nonculturable cells of Vibrio parahaemolyticus induced at low temperature under starvation. FEMS Microbiol. Lett., in press. [DOI] [PubMed]
- 8.Nishibuchi M, Fasano A, Russel R G, Kaper J B. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect Immun. 1992;60:3539–3545. doi: 10.1128/iai.60.9.3539-3545.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishibuchi M, Ishibashi M, Takeda Y, Kaper J B. Detection of the thermostable direct hemolysin gene and related DNA sequences in Vibrio parahaemolyticus and other Vibrio species by the DNA colony hybridization test. Infect Immun. 1985;49:481–486. doi: 10.1128/iai.49.3.481-486.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishibuchi M, Kaper J B. Duplication and variation of thermostable direct haemolysin (tdh) gene in V. parahaemolyticus. Mol Microbiol. 1990;4:87–99. doi: 10.1111/j.1365-2958.1990.tb02017.x. [DOI] [PubMed] [Google Scholar]
- 11.Nishibuchi M, Taniguchi T, Misawa T, Khaeomanee-iam V, Honda T, Miwatani T. Cloning and nucleotide sequence of the gene (trh) encoding the hemolysin related to the thermostable direct hemolysin of Vibrio parahaemolyticus. Infect Immun. 1989;57:2691–2697. doi: 10.1128/iai.57.9.2691-2697.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nolan C M, Ballard J, Kaysner C A, Lilja J L, Williams L P, Jr, Tenover F C. Vibrio parahaemolyticus gastroenteritis: an outbreak associated with raw oysters in the Pacific Northwest. Diagn Microbiol Infect Dis. 1984;2:119–128. doi: 10.1016/0732-8893(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 13.Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay A K, Garg S, Bhattacharya S K, Nair G B, Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol. 1997;35:3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okuda J, Nishibuchi M. Manifestation of the Kanagawa phenomenon, the virulence-associated phenotype, of Vibrio parahaemolyticus depends on a particular single base change in the promoter of the thermostable direct hemolysin. Mol Microbiol. 1998;30:499–511. doi: 10.1046/j.1365-2958.1998.01072.x. [DOI] [PubMed] [Google Scholar]
- 15.Sakazaki R, Tamura K, Kato T, Obara Y, Yamai S, Hobo K. Studies of the enteropathogenic, facultatively halophilic bacteria, Vibrio parahaemolyticus. III. Enteropathogenicity. Jpn J Med Sci Biol. 1968;21:325–331. doi: 10.7883/yoken1952.21.325. [DOI] [PubMed] [Google Scholar]
- 16.Shirai H, Ito H, Hirayama T, Nakabayashi Y, Kumagai K, Takeda Y, Nishibuchi M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun. 1990;58:3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suthienkul O, Ishibashi M, Iida T, Nettip N, Supavej S, Eampokalap B, Makino M, Honda T. Urease production correlates with possession of the trh gene in Vibrio parahaemolyticus strains isolated in Thailand. J Infect Dis. 1995;172:1405–1408. doi: 10.1093/infdis/172.5.1405. [DOI] [PubMed] [Google Scholar]
- 18.Tada J, Ohashi T, Nishimura N, Shirasaki Y, Ozaki H, Fukushima S, Takano J, Nishibuchi M, Takeda Y. Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes. 1992;6:477–487. doi: 10.1016/0890-8508(92)90044-x. [DOI] [PubMed] [Google Scholar]
- 19.Tomoyasu T. Development of the immunomagnetic enrichment method selective for Vibrio parahaemolyticus serotype K and its application to food poisoning study. Appl Environ Microbiol. 1992;58:2679–2682. doi: 10.1128/aem.58.8.2679-2682.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]