Abstract
The fields of precision and personalised medicine have led to promising advances in tailoring treatment to individual patients. Examples include genome/molecular alteration-guided drug selection, single-patient gene therapy design and synergy-based drug combination development, and these approaches can yield substantially diverse recommendations. Therefore, it is important to define each domain and delineate their commonalities and differences in an effort to develop novel clinical trial designs, streamline workflow development, rethink regulatory considerations, create value in healthcare and economics assessments, and other factors. These and other segments are essential to recognise the diversity within these domains to accelerate their respective workflows towards practice-changing healthcare. To emphasise these points, this article elaborates on the concept of digital health and digital medicine-enabled N-of-1 medicine, which individualises combination regimen and dosing using a patient’s own data. We will conclude with recommendations for consideration when developing novel workflows based on emerging digital-based platforms.
Keywords: Digital health, digital medicine, N-of-1, personalised medicine
INTRODUCTION
A foundational tenet of medicine has been the pursuit of care that is tailored to each patient. In assessing the repertoire that underpins each patient’s treatment, this includes the drugs (small molecule, biologics, targeted therapy, etc.) administered as monotherapies and combination therapies, immunotherapies (potentially as part of a combination regimen), radiotherapy and other approaches.[1,2,3,4] To realise this objective, the concept of N-of-1 medicine, which utilises a patient-specific avatar generated by the patient’s own data to guide treatment, has been gaining substantial traction at the interface of innovation, personalised medicine, clinical trial design, policy and other domains.[5]
Tailored therapy has been extensively studied, with promising outcomes. For example, transcriptomics, epigenetic profiling and other approaches have been used to individualise patient drug selection.[6] Broader molecular profiling for the personalisation of regimens designed for patient treatment approaches and next-generation sequencing have also been used to select actionable regimens.[7] Major national initiatives, such as SG100K, will play a vital role in correlating genetic and phenotypic characteristics towards systematically enabling treatment recommendations at scale.[8] Pharmacogenomics represents another strategy that has made important strides in using genetic profiles to assess the potential patient response to treatment from a safety and efficacy standpoint. This information can serve as the basis for dose and drug selection.[9,10,11]
Through emerging methodologies of harnessing data to drive healthcare decision-making, the fields of precision and personalised medicine are being increasingly impacted by the fields of digital health and digital medicine.[12,13,14,15,16,17,18,19,20,21,22,23,24,25] Wearable devices are increasing our access to longitudinal data to drive digital biomarker development, and the rapid rise in actionable data is empowering new ways of driving clinical decision support.[26,27,28,29,30] Realising clinically actionable strategies to correlate this data, big or small, into optimal and personalised treatment guidance may profoundly impact the ability to sustainably deliver truly personalised N-of-1 medicine at the required scale for all stakeholders involved.
N-of-1 medicine can also be impacted by recent advances in artificial intelligence (AI) and digital medicine. As these fields undergo continued clinical validation, there has been a major increase in how data at population scale are used to develop models for image analysis in pathology, dermatology and hospital operations (e.g., readmission prediction, triage, etc).[31,32,33,34,35] Deep learning is also being increasingly used for drug discovery to design novel drug candidates or identify repurposing opportunities for a wide array of disorders.[36] Additional advances include the use of small data-based optimisation platforms to design patient-specific combination regimens.[37,38]
This article seeks to illuminate the concept of N-of-1 medicine as a potential strategy to modulate patient treatment in a sustained manner, so that it evolves alongside each patient. Of note, the article does not evaluate or define N-of-1 medicine as a means of deriving population-scale conclusions from a single patient or as an approach to solely limit the trialling of approaches on only one subject. Instead, it explores the use of clinically actionable approaches to harness only a patient’s own data to manage only his/her own intervention dynamically.
From a technical standpoint, this article addresses drug regimen design in clinical decision support and the importance of dynamic dose optimisation, emphasising the point that patients evolve over time. True optimisation of treatment should evolve alongside the patient. Specifically, this article outlines key considerations (e.g. implementation of N-of-1 medicine) that are often encountered, but are highly diverse in definition and application. This includes providing some specificity for the terms, ‘personalised medicine’ and ‘optimisation’ in the context of N-of-1 medicine as described in this article and addressing the role of synergy in combination design and administration. At the same time, the article also discusses practical challenges of implementing these approaches in clinical workflows, the need for engagement of key stakeholders in these workflows and providing a set of recommendations that may help with catalysing phenotypic- and genotypic-driven N-of-1 digital health, and medicine innovation with a comprehensive roadmap that spans patients, caregivers, doctors, nurses, pharmacists, allied health professionals, data scientists, regulators, policymakers, payors and beyond, as well as other considerations.
N-OF-1 REGIMEN DESIGN
Through the integration of genotypic and phenotypic N-of-1 medicine, many platforms exist for the personalisation of regimens designed for patient treatment. These have spanned the use of N-of-1 gene therapy design, which includes whole genome sequencing and Sanger sequencing, among others, with antisense oligonucleotides, organoid platforms for ovarian cancer combination therapy development,[39,40,41] guiding clinical decision support using in vitro and in vivo cancer models,[42] and using patient-specific ex vivo approaches to pinpoint drug candidates among others. In another study, exceptional responders were identified using single-cell functional precision medicine, which resulted in outcomes that were three-fold longer than conventionally driven approaches.[43]
Recently, phenotypic-driven approaches based on correlations discovered via neural networks have also been developed. Using these platforms, a second-order relationship between drug/dose inputs and efficacy/toxicity outputs markedly reduced the amount of data needed to design clinically actionable drug combinations from ex vivo patient samples. Importantly, this approach demonstrated that while small data could successfully achieve global optimisation of efficacy and safety from a massive parameter space of combinatorial permutations, a prospectively acquired and carefully designed set of experiments was critical to pinpoint a list of actionable regimens.[12,37,38,44,45,46,47,48,49,50,51,52]
Given the diversity of phenotypic and genotypic approaches towards N-of-1 treatment regimen development being explored, the types of clinical workflows needed to support the implementation of promising platforms will be diverse. They will require a substantial degree of stakeholder engagement, given the broad spectrum of classes of data (small data vs. big data), infrastructure and other factors that are required for healthcare integration. In addition to workflow considerations, continuously emerging evidence is suggesting that patient responses to regimens are highly individualised and additional considerations, such as dosing and modulating drug interactions, will be essential to pinpointing more responders to intervention. In addition to impacting platform deployment at the point of care, this may involve a rethink of clinical trial designs and regulatory considerations. These points will be addressed in subsequent discussions and recommendations.
NEED FOR DYNAMIC AND OPTIMAL TREATMENT AT THE N-OF-1 LEVEL
There are a number of terms and factors spanning drug synergy to dose optimisation at the foundation of N-of-1 regimen design and dosing that represent a diverse set of strategies to personalise patient treatment. Traditionally, the standard of care (SOC) treatment often relies on dose escalation to achieve maximum tolerated dose (MTD), which dictates the appropriate dosing range from population-based medicine. However, treatment outcomes are often patient specific and a one-size-fits-all dosing regimen obtained from subpopulation studies may sometimes result in unsatisfactory clinical outcomes. Similar to MTD-based dosing, drug synergy has also been a key criterion for combination therapy design.[53,54,55] However, recent work has shown that independent drug activity without synergy can also result in positive treatment outcomes.[56,57] Therefore, these and other drug combination design studies based on the concept of drug resistance that is non-overlapping versus synergy have added important insight that spans beyond conventional MTD- and synergy-based regimen development.[58]
With regards to drug synergy, the aforementioned and other works have shown that drug interactions can be patient specific, time dependent and/or dose dependent.[59,60] Further adding to these points, the term ‘optimisation’ with regards to patient treatment also merits attention as it can sometimes be defined through dose escalation to find MTD. In other studies, Bayesian optimisation has been used to predict MTD for Phase I studies.[61] Other work has sought to assess the context of MTD in terms of dose optimisation and to explore if other methodologies may be appropriate.[62] Recently, there have been emerging studies suggesting that MTD may not be optimal for certain therapies or circumstances.[63,64] In sum, it is becoming clearer that drug interactions, optimisation and other determinants of patient outcomes in pharmacological intervention may be patient specific, dictated by a number of controllable parameters (e.g. dose), and dynamic. These findings may serve as a critical impetus towards the need for treatment to evolve dynamically alongside patients.
It is vital to note that patients differ from themselves over time. The right dose for a patient on a Monday might be very different 1 hour, 1 day, 1 week or 1 month later for the same patient. Therefore, personalised medicine, where possible and practical, should evolve alongside the patient. Examples of dynamically adjusted dosing have previously been demonstrated in a clinical setting. These approaches were developed to confront the challenges of traditional high-dose treatment and may end up eliminating a vast majority of the drug-sensitive population, leaving the drug-resistant population behind, which can lead to complete treatment resistance and failure, and eventually, metastatic disease. To overcome this challenge, evolutionary dosing and game theory-based approaches operate by the principle of tumour management based on therapy that manages the population of drug-sensitive tumour cells versus drug-resistant cells.[65,66,67,68,69,70,71,72,73,74,75,76] Other strategies have included adaptive therapeutic approaches. For instance, optimal adaptive therapeutic schedules were identified through the combination of mathematical modelling and dynamic optimisation, and this approach can precisely pinpoint the optimal personal drug doses and respective schedules for prostate cancer.[77] In a pilot clinical trial with 17 subjects, evolutionary-based models were incorporated into the dosing of abiraterone for metastatic castrate-resistant prostate cancer.[78] This approach led to improved median time to progression and overall survival rate. The integration of evolutionary- and adaptive-based therapeutic approaches may lead to significantly improved clinical outcomes.
For the purposes of this article, we will define personalised medicine as a dynamic process based upon the platforms and trials described. We also acknowledge that there are other definitions in the field based upon important findings and trials with other platforms. It is well established that patients vary substantially from one another. Furthermore, the aforementioned and other approaches towards modulated drug administration will play an increasing role in ensuring that treatment modalities evolve alongside patients based on longitudinal assessment of treatment response. The dynamic and patient-specific nature of this treatment response, coupled with emerging approaches that can harness this data at the population- or patient-specific level to adjust care, could open up doors to hyper-personalised intervention.
CURATE.AI AND N-OF-1 MEDICINE: USE CASES
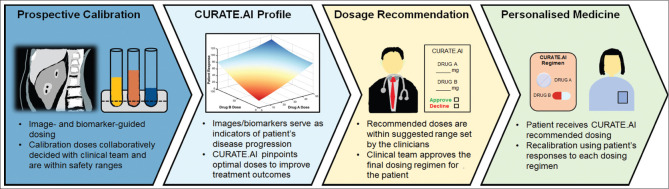
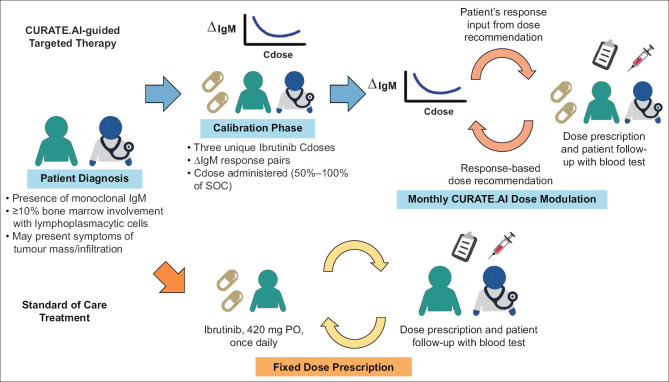
CURATE.AI is based on a neural network-discovered correlation that relates the inputs and outputs of a biological system using a second-order algebraic series. Inputs can include the drugs in a regimen as well as corresponding drug doses. Other inputs can include digital, electrical or mechanical stimulation, among other approaches, as well as corresponding analogues to dosing such as magnitude. Outputs can include clinically actionable biomarkers that reflect therapeutic efficacy and toxicity, imaging and other quantifiable measures of treatment outcomes [Figures 1 and 2]. The biological system can include preclinical (in vitro, in vivo, ex vivo) and clinical (patient) contexts. The small-scale experiments mentioned in earlier sections have paved the way for CURATE.AI-based retrospective studies over the years.[79,80,81] The retrospective studies serve to assess the feasibility of prospective trials for CURATE.AI, help formulate hypotheses, identify potential risk factors and assist in trial designs. Some of these studies have progressed to clinical trials for further validation [Table 1].
Figure 1.
CURATE.AI workflow for personalised dosing.
Figure 2.
Workflow of CURATE.AI-guided targeted therapy for Waldenström macroglobulinaemia in comparison with the workflow of standard of care (SOC) targeted therapy. AI: artificial intelligence, IgM: immunoglobulin M, PO: per os
Table 1.
Registered clinical trials on ClinicalTrials.gov.
| Registration no. | Clinical trial description |
|---|---|
| NCT03832101 | Digital therapeutics for cognitive training |
|
| |
| NCT03759093 | Haematologic cancer personalised therapy |
|
| |
| NCT04357691 | CURATE.AI cardiorespiratory performance optimisation |
|
| |
| NCT04522284 | PRECISE CURATE.AI: solid cancer therapy |
|
| |
| NCT05376683 | Hypertension personalised therapy |
|
| |
| NCT04848935 | COR-Tx trial for post brain radiotherapy patients |
|
| |
| NCT05532397 | Neuro-oncology combination therapy |
|
| |
| NCT05381038 | Hybrid combo and dose optimisation (gastric cancer) |
|
| |
| NCT05175235 | CURATE.AI immunotherapy optimisation |
|
| |
| NCT06240897 | CURATE.Dtx as a diagnostic and management tool for delirium |
CURATE.AI is a hyper-personalised approach and treats each patient as a discrete case. Therefore, it does not use population-based big data to train an AI model that is then applied to each subsequent patient. Instead, each patient’s treatment (e.g. dose) is modulated using only their own data. The initial dataset for each patient is based on small data and is prospectively calibrated from scratch by the clinician through an established dose modulation and biomarker-monitoring protocol from which a patient-specific digital avatar is created to guide subsequent dose recommendations [Figures 1 and 2]. In the initial stage, the CURATE.AI team communicates the foundation of the platform with the clinical team to formulate the design of experiment and trial, and the feedback from the clinical team and other stakeholders (e.g. regulator, behavioural/implementation scientist) is considered when refining such a multidisciplinary study.[82] The CURATE.AI clinical trials are divided into two phases: calibration and efficacy driven. In the calibration phase, predetermined drug doses are paired with their associated clinical responses within the same treatment cycle. The dose–response pairs will be used to generate the initial patient-specific avatar to guide doses in subsequent cycles during the efficacy-driven phase. In the efficacy-driven phase, recommended modulated doses leading to responses identified by the treatment intent are obtained from the generated avatar. As the treatment progresses longitudinally, the patient-specific avatar evolves dynamically when it is fed with new dose–response-paired data collected from preceding cycles. The CURATE.AI dosing cycles are tailored to match the dosing frequency in SOC. For example, in Figure 2, the ongoing Personalised, Rational, Efficacy-Driven Cancer Drug Dosing via an Artificial Intelligence SystEm (PRECISE) clinical trial (NCT04522284) illustrates how a CURATE.AI treatment is conducted for Waldenström macroglobulinaemia with ibrutinib.[22] The avatar provides a total dose recommendation for a 28-day cycle. The clinician divides that total dose across the 28 days and prescribes the daily dose before the commencement of the next cycle. The patient will take the prescribed dose once daily according to the SOC dosing frequency. CURATE.AI has been trialled for various indications that span oncology to digital therapy, hypertension and transplant.[12,18,19,21,22,23,60,81,83,84,85,86] It is important to note that the CURATE.AI workflow varies substantially from what one might perceive an AI-driven treatment protocol to entail [Figure 3]. For example, CURATE.AI implementation is not characterised as an AI-driven process that operates independently from the clinical treatment team followed by re-engagement of the clinical team to approve or reject a treatment recommendation. Instead, the clinical team builds its own small dataset for each patient during the prospective calibration workflow. The calculations that drive the treatment recommendation are available at all times to the clinical team, and the clinical team makes the final decision on whether or not to accept the recommendation. This highly intertwined process ensures that the decision-making process is predominantly driven by the human in the workflow.
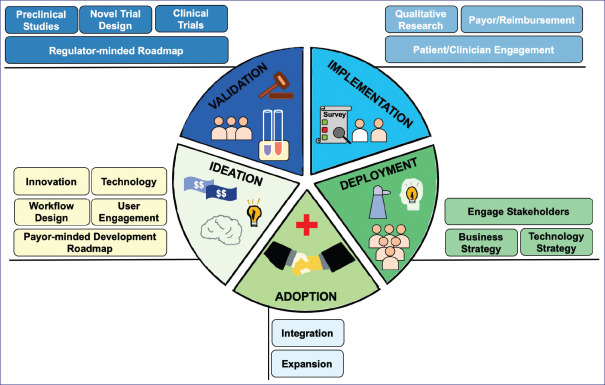
Figure 3.
Disciplinary workflow for successful translation of ideas into real-world practice.
In standard care, dose modification under clinical oncology guidelines is patient toxicity guided. Harnessing CURATE.AI, our previous work has shown that patient efficacy-guided dosing can potentially be possible through initial clinical pilot studies. For example, in a previous study on a metastatic prostate cancer patient, a small dataset of modulated dosing and corresponding prostate-specific antigen (PSA) was used to construct a patient-specific digital avatar.[84] This avatar subsequently recommended a substantial dose reduction to increase treatment efficacy. A corresponding reduction in PSA, downstream dynamically modulated dosing and imaging enabled and confirmed a durable response. Importantly, this work represented a shift towards efficacy-guided dose adjustment versus the clinical standard of toxicity-guided dose adjustment. This also represented a paradigm shift in the clinical oncology treatment workflow. Subsequent validation has since expanded towards cohort-level studies for solid cancer and blood cancer, immunotherapy, digital therapeutics, hypertension management and other indications [Table 1]. The CURATE.AI workflow has since advanced into clinical education resources.[21]
IMPORTANCE OF USER ENGAGEMENT
Understanding the patients or users that a technology intends to serve is a vital component of the innovation roadmap.[87,88,89,90] Too often, user engagement is conducted very late in the development lifecycle or not conducted at all. Technology-centred development can realise and validate new innovations that can potentially outperform standard care. However, actively addressing user experience and interface through broader disciplines such as industrial design and, more specifically, interaction design, among other disciplines, can play a major role in achieving robust and sustainable healthcare system-wide adoption of an innovation [Figure 3].
It is important to recognise that the user of an innovation is not always solely the patient or subject for whom an innovation was designed to diagnose or treat. The user can also be their caregiver, doctor, nurse, pharmacist, allied health professional or a combination of these stakeholders.[82,87,89,91] Properly identifying these stakeholders, developing workflows that are aligned with theirs and behavioural change consideration are essential. These factors should be considered early and not solely at the implementation phase, as it could substantially delay or preclude translation and/or adoption [Figure 3].
TECHNOLOGY ALONE CANNOT CHANGE HEALTHCARE
Empowering N-of-1 medicine at scale will not be catalysed solely through technology. Importantly, innovation and adoption are substantially different objectives, which require an expansive range of disciplines, policy advances and other factors to achieve the level of behavioural change needed to drive changes in practice at the clinical and user stages, not to mention the validated economics and value outcomes needed at the reimbursement and subsidy stages.[92,93]
Nonetheless, there are a number of key considerations to be made when developing digital health and medicine innovation portfolios. A recurring theme in these considerations is addressing or, at a minimum, engagement with subject matter experts relatively early on during the development lifecycle [Figure 3]. The following list is not exhaustive, but summarises the key points experientially derived at the Institute for Digital Medicine:
Effective communication of the foundation (e.g. mathematical, technological, etc.) that drives the innovation should be clearly made to workflow stakeholders at the outset of development, validation and onwards.
Workflow development should consider feedback and viewpoints (when appropriate) from the clinical team (doctors, nurses, pharmacists, allied health professionals), user (patient, subject), caregiver, regulator and payor, among other relevant stakeholders.
Formal user engagement studies are foundational towards obtaining actionable guidance that can drive sustained user adherence and adoption.
Design is and will continue to be a critical catalyst for behavioural change and adoption. User experience and user interface may be essential elements to address in digital health/medicine innovation development. More specifically, interaction design and relevant domains should be considered early during technology development.
Exploring applicable implementation sciences models to facilitate real-world integration, the approaches and frameworks that influence endpoints and strategies for assessment may be essential during the technology development lifecycle.[94]
Digital health and digital medicine platforms will be essential to driving healthy behaviour adherence, mental health solutions, access to healthcare, data sharing and other facets of next-generation well-being. Bridging these platforms with home use will be critical. These aforementioned points have already catalysed and will continue to catalyse a reimagination of our definition of interdisciplinarity. The importance of engaging disciplines, such as behavioural economics, implementation sciences, architecture, built environment, urban planning, communications and new media, public policy, sociology, nursing, social work, regulatory sciences, health economics and others, will be vital.
When possible, regulator engagement to understand regulatory pathways and potential innovation in clinical trial design should be considered. A lot of guidance is available, and depending on the agency being engaged, procedures for requesting regulator meetings can be found via respective resources.
In addition to trial registration in established online databases, trial protocols should be published for transparency and, where applicable, to contribute to ongoing discussions to advance novel trial designs.
Familiarisation with relevant clinical trial reporting standards (e.g. Consolidated Standards of Reporting Trials [CONSORT], CONSORT-AI) will be important for downstream sharing of study findings, as well as for transparency and compliance.
With technology development, deprioritising programmes can be at least as important as prioritising programmes to advance. Adherence, utility to stakeholders, economics, alignment with value-based healthcare priorities and other factors may serve as pivotal criteria from which to base development objectives.
Innovation in trial design should continue to be explored. Trial and adherence incentives should also be explored using emerging platforms.
Financial support and sponsorship
Ho D receives funding from the following: Institute for Digital Medicine Translational Research Programme (grant number A-0001319-00-00), Yong Loo Lin School of Medicine, NUS; AI Singapore Programme (award number: AISG-GC-2019-002), Singapore National Research Foundation; Open Fund-Large Collaborative Grant (grant number MOH-OFLCG18May-0028), National Medical Research Council, Ministry of Health; Tier 1 FRC Grant (grant number R-397-000-333-114), Ministry of Education; Next-Generation Brain-Computer-Brain Platform – A Holistic Solution for the Restoration & Enhancement of Brain Functions (NOURISH) project from the RIE2020 Advanced Manufacturing And Engineering (Ame) Programmatic Fund [grant number A20G8b0102/A-0002199-02-00]; Micron Foundation and Sun Life Singapore.
Conflicts of interest
Ho D, Blasiak A, Kumar SK and Tan L are coinventors of a pending patent pertaining to personalised regimen dosing. Ho D is a scientific cofounder and shareholder of KYAN Therapeutics, which is developing personalised oncology platforms.
Acknowledgement
Ho D gratefully acknowledges support from the Office of the President, Office of the Senior Deputy President and Provost, and Office of the Deputy President for Research and Technology at National University of Singapore (NUS).
The authors acknowledge the impactful work being conducted in the fields of precision and personalised medicine by a growing community of researchers. Our team gratefully acknowledges collaborations with Prof Chng Wee Joo, A/Prof Christopher L Asplund, A/Prof Jason Kai Wei Lee, A/Prof BT Thomas Yeo, A/Prof Edward Kai-Hua Chow, A/Prof Mark Chan, Dr Laureen Wang, Dr Raghav Sundar, Dr Sanjay de Mel and Dr Balamurugan A Vellayappan, as well as many other colleagues from: Department of Biomedical Engineering, College of Design and Engineering, NUS; Department of Gastroenterology, Department of Obstetrics & Gynaecology and Department of Paediatrics, Yong Loo Lin School of Medicine, NUS; National University Hospital; National University Cancer Institute; DSO National Laboratories; National Centre for Infectious Diseases; Alexandra Hospital; Nursing Transformation, Research & Informatics, Singapore General Hospital; Nursing Quality, Research & Transformation, SingHealth Group Nursing; and many other institutions. We also thank Ms Poonam Rai (The Institute for Digital Medicine [WisDM] and the N.1 Institute for Health, NUS), Ms Si Ying Khew (The N.1 Institute for Health, NUS), Ms Noor Julihaty Binte Jufri (The N.1 Institute for Health, NUS) and Ms Phoebe Koh (The Institute for Digital Medicine [WisDM], NUS) for their dedication and inspiring contributions to the team.
REFERENCES
- 1.McCarty TR, Bazarbashi AN, Hathorn KE, Thompson CC, Ryou M. Combination therapy versus monotherapy for EUS-guided management of gastric varices: A systematic review and meta-analysis. Endosc Ultrasound. 2020;9:6. doi: 10.4103/eus.eus_37_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Melero I, Castanon E, Alvarez M, Champiat S, Marabelle A. Intratumoural administration and tumour tissue targeting of cancer immunotherapies. Nat Rev Clin Oncol. 2021;18:558–76. doi: 10.1038/s41571-021-00507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmid A, Wolfensberger A, Nemeth J, Schreiber PW, Sax H, Kuster SP. Monotherapy versus combination therapy for multidrug-resistant Gram-negative infections: Systematic Review and Meta-Analysis. Sci Rep. 2019;9:15290. doi: 10.1038/s41598-019-51711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K, Tepper JE. Radiation therapy-associated toxicity: Etiology, management, and prevention. CA Cancer J Clin. 2021;71:437–54. doi: 10.3322/caac.21689. [DOI] [PubMed] [Google Scholar]
- 5.Schork NJ. Personalized medicine: Time for one-person trials. Nature. 2015;520:609–11. doi: 10.1038/520609a. [DOI] [PubMed] [Google Scholar]
- 6.Malone ER, Oliva M, Sabatini PJ, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med. 2020;12:1–19. doi: 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartzberg L, Kim ES, Liu D, Schrag D. Precision oncology: Who, how, what, when, and when not? Am Soc Clin Oncol Educ Book. 2017;37:160–9. doi: 10.1200/EDBK_174176. [DOI] [PubMed] [Google Scholar]
- 8.Wong E, Bertin N, Hebrard M, Tirado-Magallanes R, Bellis C, Lim WK, et al. The Singapore national precision medicine strategy. Nat Genet. 2023;55:178–86. doi: 10.1038/s41588-022-01274-x. [DOI] [PubMed] [Google Scholar]
- 9.Jeibouei S, Akbari ME, Kalbasi A, Aref AR, Ajoudanian M, Rezvani A, et al. Personalized medicine in breast cancer: Pharmacogenomics approaches. Pharmacogenomics Pers Med. 2019;12:59–73. doi: 10.2147/PGPM.S167886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morganti S, Tarantino P, Ferraro E, D’Amico P, Duso BA, Curigliano G. Next generation sequencing (NGS): A revolutionary technology in pharmacogenomics and personalized medicine in cancer. Adv Exp Med Biol. 2019;1168:9–30. doi: 10.1007/978-3-030-24100-1_2. [DOI] [PubMed] [Google Scholar]
- 11.Radhakrishnan A, Kuppusamy G, Ponnusankar S, Shanmukhan NK. Pharmacogenomic phase transition from personalized medicine to patient-centric customized delivery. Pharmacogenomics J. 2020;20:1–18. doi: 10.1038/s41397-019-0135-8. [DOI] [PubMed] [Google Scholar]
- 12.Blasiak A, Truong A, Jeit W, Tan L, Kumar KS, Tan SB, et al. PRECISE CURATE. AI: A prospective feasibility trial to dynamically modulate personalized chemotherapy dose with artificial intelligence. Am Soc Clin Oncol. 2022;40 doi: 10.1200/JCO.2022.40.16_suppl. 1574. [Google Scholar]
- 13.de Hond AA, Leeuwenberg AM, Hooft L, Kant IM, Nijman SW, van Os HJ, et al. Guidelines and quality criteria for artificial intelligence-based prediction models in healthcare: A scoping review. NPJ Digit Med. 2022;5:2. doi: 10.1038/s41746-021-00549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho D. Artificial intelligence in cancer therapy. Science. 2020;367:982–3. doi: 10.1126/science.aaz3023. [DOI] [PubMed] [Google Scholar]
- 15.Ho D, Sapanel Y, Blasiak A. Baltimore: Johns Hopkins University Press; 2023. Medicine without Meds: Transforming Patient Care with Digital Therapies. [Google Scholar]
- 16.Huckvale K, Venkatesh S, Christensen H. Toward clinical digital phenotyping: A timely opportunity to consider purpose, quality, and safety. NPJ Digit Med. 2019;2:1–11. doi: 10.1038/s41746-019-0166-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathews SC, McShea MJ, Hanley CL, Ravitz A, Labrique AB, Cohen AB. Digital health: A path to validation. NPJ Digit Med. 2019;2:38. doi: 10.1038/s41746-019-0111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukhopadhyay A, Sumner J, Ling LH, Quek RHC, Tan ATH, Teng GG, et al. Personalised dosing using the CURATE. AI algorithm: Protocol for a feasibility study in patients with hypertension and type II diabetes mellitus. Int J Environ Res Public Health. 2022;19:8979. doi: 10.3390/ijerph19158979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Remus A, Tadeo X, Ng GSK, Blasiak A, Kee T, Vijayakumar S, et al. CURATE. AI COR-Tx platform as a digital therapy and digital diagnostic for cognitive function in brain tumour patients post-radiotherapy treatment: Protocol for a prospective mixed-methods feasibility clinical trial. BMJ Open. 2023;13:e077219. doi: 10.1136/bmjopen-2023-077219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rieke N, Hancox J, Li W, Milletari F, Roth HR, Albarqouni S, et al. The future of digital health with federated learning. NPJ Digit Med. 2020;3:119. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senthil Kumar K, Miskovic V, Blasiak A, Sundar R, Pedrocchi ALG, Pearson AT, et al. Artificial intelligence in clinical oncology: From data to digital pathology and treatment. Am Soc Clin Oncol Educ Book. 2023;43:e390084. doi: 10.1200/EDBK_390084. [DOI] [PubMed] [Google Scholar]
- 22.Tan BKJ, Teo CB, Tadeo X, Peng S, Soh HPL, Du SDX, et al. Personalised, rational, efficacy-driven cancer drug dosing via an artificial intelligence SystEm (PRECISE): A protocol for the PRECISE CURATE. AI pilot clinical trial. Front Digit Health. 2021;3:635524. doi: 10.3389/fdgth.2021.635524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan S-B, Tan J, Raczkowska MN, Lee JCW, Rai B, Remus A, et al. Digital game-based interventions for cognitive training in healthy adults and adults with cognitive impairment: Protocol for a two-part systematic review and meta-analysis. BMJ Open. 2023;13:e071059. doi: 10.1136/bmjopen-2022-071059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Ho D. Deep learning and drug discovery for healthy aging. ACS Cent Sci. 2023;9:1860–3. doi: 10.1021/acscentsci.3c01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.You K, Wang P, Ho D. N-of-1 healthcare: Challenges and prospects for the future of personalized medicine. Front Digit Health. 2022;4:830656. doi: 10.3389/fdgth.2022.830656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeVore AD, Wosik J, Hernandez AF. The future of wearables in heart failure patients. JACC Heart Fail. 2019;7:922–32. doi: 10.1016/j.jchf.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Hong W, Lee WG. Wearable sensors for continuous oral cavity and dietary monitoring toward personalized healthcare and digital medicine. Analyst. 2020;145:7796–808. doi: 10.1039/d0an01484b. [DOI] [PubMed] [Google Scholar]
- 28.Knight SR, Ng N, Tsanas A, Mclean K, Pagliari C, Harrison EM. Mobile devices and wearable technology for measuring patient outcomes after surgery: A systematic review. NPJ Digit Med. 2021;4:157. doi: 10.1038/s41746-021-00525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwan JL, Lo L, Ferguson J, Goldberg H, Diaz-Martinez JP, Tomlinson G, et al. Computerised clinical decision support systems and absolute improvements in care: Meta-analysis of controlled clinical trials. BMJ. 2020;370:m3216. doi: 10.1136/bmj.m3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutton RT, Pincock D, Baumgart DC, Sadowski DC, Fedorak RN, Kroeker KI. An overview of clinical decision support systems: Benefits, risks, and strategies for success. NPJ Digit Med. 2020;3:17. doi: 10.1038/s41746-020-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baxi V, Edwards R, Montalto M, Saha S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol. 2022;35:23–32. doi: 10.1038/s41379-021-00919-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esteva A, Chou K, Yeung S, Naik N, Madani A, Mottaghi A, et al. Deep learning-enabled medical computer vision. NPJ Digit Med. 2021;4:5. doi: 10.1038/s41746-020-00376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hilton CB, Milinovich A, Felix C, Vakharia N, Crone T, Donovan C, et al. Personalized predictions of patient outcomes during and after hospitalization using artificial intelligence. NPJ Digit Med. 2020;3:51. doi: 10.1038/s41746-020-0249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabir T, Syn NL, Shaw V, Tan YHA, Chua HW, Ong LWL, et al. Defining the optimal time to appendectomy: A step toward precision surgery. Surgery. 2022;172:798–806. doi: 10.1016/j.surg.2022.04.021. [DOI] [PubMed] [Google Scholar]
- 35.Kim CK, Choi JW, Jiao Z, Wang D, Wu J, Yi TY, et al. An automated COVID-19 triage pipeline using artificial intelligence based on chest radiographs and clinical data. NPJ Digit Med. 2022;5:5. doi: 10.1038/s41746-021-00546-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pun FW, Leung GHD, Leung HW, Liu BHM, Long X, Ozerov IV, et al. Hallmarks of aging-based dual-purpose disease and age-associated targets predicted using PandaOmics AI-powered discovery engine. Aging (Albany NY) 2022;14:2475. doi: 10.18632/aging.203960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goh J, De Mel S, Hoppe MM, Mohd Abdul Rashid MB, Zhang XY, Jaynes P, et al. An ex vivo platform to guide drug combination treatment in relapsed/refractory lymphoma. Sci Transl Med. 2022;14:eabn7824. doi: 10.1126/scitranslmed.abn7824. [DOI] [PubMed] [Google Scholar]
- 38.Rashid MBMA, Toh TB, Hooi L, Silva A, Zhang Y, Tan PF, et al. Optimizing drug combinations against multiple myeloma using a quadratic phenotypic optimization platform (QPOP) Sci Transl Med. 2018;10:eaan0941. doi: 10.1126/scitranslmed.aan0941. [DOI] [PubMed] [Google Scholar]
- 39.Hill SF, Meisler MH. Antisense oligonucleotide therapy for neurodevelopmental disorders. Dev Neurosci. 2021;43:247–52. doi: 10.1159/000517686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Hu C, Moufawad El Achkar C, Black LE, Douville J, Larson A, et al. Patient-customized oligonucleotide therapy for a rare genetic disease. N Engl J Med. 2019;381:1644–52. doi: 10.1056/NEJMoa1813279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang S, Iyer S, Ran H, Dolgalev I, Gu S, Wei W, et al. Genetically defined, syngeneic organoid platform for developing combination therapies for ovarian cancer. Cancer Discov. 2021;11:362–83. doi: 10.1158/2159-8290.CD-20-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–77. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kornauth C, Pemovska T, Vladimer GI, Bayer G, Bergmann M, Eder S, et al. Functional precision medicine provides clinical benefit in advanced aggressive hematologic cancers and identifies exceptional responders. Cancer Discov. 2022;12:372–87. doi: 10.1158/2159-8290.CD-21-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abdulla A, Wang B, Qian F, Kee T, Blasiak A, Ong YH, et al. Project IDentif. AI: Harnessing artificial intelligence to rapidly optimize combination therapy development for infectious disease intervention. Adv Ther. 2020;3:2000034. doi: 10.1002/adtp.202000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blasiak A, Lim JJ, Seah SGK, Kee T, Remus A, Chye DH, et al. IDentif. AI: Rapidly optimizing combination therapy design against severe Acute Respiratory Syndrome Coronavirus 2 (SARS-Cov-2) with digital drug development. Bioeng Transl Med. 2021;6:e10196. doi: 10.1002/btm2.10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blasiak A, Truong AT, Remus A, Hooi L, Seah SGK, Wang P, et al. The IDentif. AI-x pandemic readiness platform: Rapid prioritization of optimized COVID-19 combination therapy regimens. NPJ Digit Med. 2022;5:83. doi: 10.1038/s41746-022-00627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blasiak A, Truong AT, Wang P, Hooi L, Chye DH, Tan S-B, et al. IDentif. AI-Omicron: Harnessing an AI-derived and disease-agnostic platform to pinpoint combinatorial therapies for clinically actionable anti-SARS-CoV-2 intervention. ACS Nano. 2022;16:15141–54. doi: 10.1021/acsnano.2c06366. [DOI] [PubMed] [Google Scholar]
- 48.Ho D. Addressing COVID-19 drug development with artificial intelligence. Adv Intell Syst. 2020;2:2000070. doi: 10.1002/aisy.202000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho D. Digital nanomedicine: A new frontier for drug development. ACS Nano. 2022;16:3435–7. [Google Scholar]
- 50.Mukherjee D, Wang P, Hooi L, Sandhu V, You K, Blasiak A, et al. Addressing antimicrobial resistance with the IDentif. AI platform: Rapidly optimizing clinically actionable combination therapy regimens against nontuberculous mycobacteria. Theranostics. 2022;12:6848. doi: 10.7150/thno.73078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, Lee D-K, Chen K-Y, Chen J-Y, Zhang K, Silva A, et al. Mechanism-independent optimization of combinatorial nanodiamond and unmodified drug delivery using a phenotypically driven platform technology. ACS Nano. 2015;9:3332–44. doi: 10.1021/acsnano.5b00638. [DOI] [PubMed] [Google Scholar]
- 52.Li M, You K, Wang P, Hooi L, Chen Y, Siah A, et al. Discovery of broad-spectrum repurposed drug combinations against Carbapenem-Resistant Enterobacteriaceae (CRE) through Artificial Intelligence (AI)-driven platform. Adv Ther. 2024 doi: 10.1002/adtp. 202300332. [Google Scholar]
- 53.Duarte D, Vale N. Evaluation of synergism in drug combinations and reference models for future orientations in oncology. Curr Res Pharmacol Drug Discov. 2022;3:100110. doi: 10.1016/j.crphar.2022.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.García-Fuente A, Vázquez F, Viéitez JM, García Alonso FJ, Martín JI, Ferrer J. CISNE: An accurate description of dose-effect and synergism in combination therapies. Sci Rep. 2018;8:4964. doi: 10.1038/s41598-018-23321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin W, Stokes JM, Eastman RT, Itkin Z, Zakharov AV, Collins JJ, et al. Deep learning identifies synergistic drug combinations for treating COVID-19. Proc Natl Acad Sci U. S. A. 2021;118:e2105070118. doi: 10.1073/pnas.2105070118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palmer AC, Chidley C, Sorger PK. A curative combination cancer therapy achieves high fractional cell killing through low cross-resistance and drug additivity. Elife. 2019;8:e50036. doi: 10.7554/eLife.50036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palmer AC, Sorger PK. Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell. 2017;171:1678–91.e13. doi: 10.1016/j.cell.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer AC, Chidley C, Sorger PK. Drugs in a curative combination therapy for lymphoma exhibit low cross-resistance but not pharmacological synergy. Biorxiv. 2018 462184. doi: 10.1101/462184. [Google Scholar]
- 59.Ding X, Chang VH, Li Y, Li X, Xu H, Ho CM, et al. Harnessing an artificial intelligence platform to dynamically individualize combination therapy for treating colorectal carcinoma in a rat model. Adv Ther. 2020;3:1900127. [Google Scholar]
- 60.Zarrinpar A, Lee D-K, Silva A, Datta N, Kee T, Eriksen C, et al. Individualizing liver transplant immunosuppression using a phenotypic personalized medicine platform. Sci Transl Med. 2016;8:333ra49. doi: 10.1126/scitranslmed.aac5954. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi A, Suzuki T. Bayesian optimization for estimating the maximum tolerated dose in Phase I clinical trials. Contemp Clin Trials Commun. 2021;21:100753. doi: 10.1016/j.conctc.2021.100753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fourie Zirkelbach J, Shah M, Vallejo J, Cheng J, Ayyoub A, Liu J, et al. Improving dose-optimization processes used in oncology drug development to minimize toxicity and maximize benefit to patients. J Clin Oncol. 2022;40:3489–500. doi: 10.1200/JCO.22.00371. [DOI] [PubMed] [Google Scholar]
- 63.FDA. Optimizing the Dosage of Human Prescription Drugs and Biological Products for the Treatment of Oncologic Diseases. 2023. [[Last accessed on 2023 Oct 16]]. Available from: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/optimizing-dosage-human-prescription-drugs-and-biological-products-treatment-oncologic-diseases .
- 64.Moon H. FDA initiatives to support dose optimization in oncology drug development: The less may be the better. Transl Clin Pharmacol. 2022;30:71. doi: 10.12793/tcp.2022.30.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Basanta D, Gatenby RA, Anderson AR. Exploiting evolution to treat drug resistance: Combination therapy and the double bind. Mol Pharm. 2012;9:914–21. doi: 10.1021/mp200458e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diniz MA, Kim S, Tighiouart M. A Bayesian adaptive design in cancer phase I trials using dose combinations with ordinal toxicity grades. Stats. 2020;3:221–38. doi: 10.3390/stats3030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donagher J, Martin JH, Barras MA. Individualised medicine: Why we need Bayesian dosing. Intern Med J. 2017;47:593–600. doi: 10.1111/imj.13412. [DOI] [PubMed] [Google Scholar]
- 68.Gatenby R, Cunningham J, Brown J. Evolutionary triage governs fitness in driver and passenger mutations and suggests targeting never mutations. Nat Commun. 2014;5:5499. doi: 10.1038/ncomms6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gluzman M, Scott JG, Vladimirsky A. Optimizing adaptive cancer therapy: Dynamic programming and evolutionary game theory. Proc Royal Soc B. 2020;287:20192454. doi: 10.1098/rspb.2019.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mathur D, Barnett E, Scher HI, Xavier JB. Optimizing the future: How mathematical models inform treatment schedules for cancer. Trends Cancer. 2022;8:506–16. doi: 10.1016/j.trecan.2022.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Razaee ZS, Cook-Wiens G, Tighiouart M. A nonparametric Bayesian method for dose finding in drug combinations cancer trials. Stat Med. 2022;41:1059–80. doi: 10.1002/sim.9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun J, Xu S, Liu Y, Zhang H. Combination therapy-based adaptive control for organism using medicine dosage regulation mechanism. Adaptive Dynamic Programming: For Chemotherapy Drug Delivery. Springer. 2023:93–113. [Google Scholar]
- 73.Tansey W, Tosh C, Blei DM. A Bayesian model of dose-response for cancer drug studies. Ann Appl Stat. 2022;16:680–705. [Google Scholar]
- 74.Wölfl B, Te Rietmole H, Salvioli M, Kaznatcheev A, Thuijsman F, Brown JS, et al. The contribution of evolutionary game theory to understanding and treating cancer. Dyn Games Appl. 2022;12:313–42. doi: 10.1007/s13235-021-00397-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye J, Reaman G, De Claro RA, Sridhara R. A Bayesian approach in design and analysis of pediatric cancer clinical trials. Pharm Stat. 2020;19:814–26. doi: 10.1002/pst.2039. [DOI] [PubMed] [Google Scholar]
- 76.Zhang J, Cunningham JJ, Brown JS, Gatenby RA. Integrating evolutionary dynamics into treatment of metastatic castrate-resistant prostate cancer. Nat Commun. 2017;8:1816. doi: 10.1038/s41467-017-01968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu R, Wang S, Tan X, Zou X. Identifying optimal adaptive therapeutic schedules for prostate cancer through combining mathematical modeling and dynamic optimization. Appl Math Model. 2022;107:688–700. [Google Scholar]
- 78.Zhang J, Cunningham J, Brown J, Gatenby R. Evolution-based mathematical models significantly prolong response to abiraterone in metastatic castrate-resistant prostate cancer and identify strategies to further improve outcomes. Elife. 2022;11:e76284. doi: 10.7554/eLife.76284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan SB, Kumar KS, Gan TRX, Tan LW, Truong AT, Blasiak A, et al. CURATE. AI–Artificial untelligence-derived personalized tacrolimus dosing for pediatric liver transplant: A retrospective study. Adv Ther. 2024 doi: 10.1002/adtp. 2300236. [Google Scholar]
- 80.Lee D-K, Chang VY, Kee T, Ho C-M, Ho D. Optimizing combination therapy for acute lymphoblastic leukemia using a phenotypic personalized medicine digital health platform: Retrospective optimization individualizes patient regimens to maximize efficacy and safety. SLAS Technol. 2017;22:276–88. doi: 10.1177/2211068216681979. [DOI] [PubMed] [Google Scholar]
- 81.Truong AT, Tan LW, Chew KA, Villaraza S, Siongco P, Blasiak A, et al. Harnessing CURATE. AI for N-of-1 optimization analysis of combination therapy in hypertension patients: A retrospective case series. Adv Ther. 2021;4:2100091. [Google Scholar]
- 82.Vijayakumar S, Lee VV, Leong QY, Hong SJ, Blasiak A, Ho D. Physicians’perspectives on AI in clinical decision support systems: Interview study of the CURATE. AI personalized dose optimization platform. JMIR Hum Factors. 2023;10:e48476. doi: 10.2196/48476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kee T, Weiyan C, Blasiak A, Wang P, Chong JK, Chen J, et al. Harnessing CURATE. AI as a digital therapeutics platform by identifying N-of-1 learning trajectory profiles. Adv Ther. 2019;2:1900023. [Google Scholar]
- 84.Pantuck AJ, Lee DK, Kee T, Wang P, Lakhotia S, Silverman MH, et al. Modulating BET bromodomain inhibitor ZEN-3694 and enzalutamide combination dosing in a metastatic prostate cancer patient using CURATE. AI, an artificial intelligence platform. Adv Ther. 2018;1:1800104. [Google Scholar]
- 85.Raczkowska MN, Remus A, Vijayakumar S, Kwek SP, Lee VV, Tadeo X, et al. Mixed-methods clinical trial to evaluate the feasibility of the CURATE. AI opimised digital cognitive rehabilitation therapeutic (COR-Tx) in patients post brain radiotherapy. J Clin Oncol. 2023;41 Number 16_suppl. doi: 10.1200/JCO.2023.41.16_suppl. TPS1615. [Google Scholar]
- 86.Truong AT, Tan S-B, Wang GZ, Yip AW, Egermark M, Yeung W, et al. CURATE. AI-assisted dose titration for anti-hypertensive personalised therapy: Study protoocl for a multi-arm, randomized, pilot feasibility trial using CURATE. AI (CURATE. AI ADAPT trial) Eur Heart J Digit Health. 2023:ztad063. doi: 10.1093/ehjdh/ztad063. doi: 10.1093/ehjdh/ztad063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee VV, Lau NY, Blasiak A, Siah KTH, Ho D. Involving patients in the process: Development of a constipation patient-reported outcome measure for symptoms and quality of life. Comput Struct Biotechnol J. 2023;22:41–9. doi: 10.1016/j.csbj.2023.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee VV, Lau NY, Xi DJ, Truong AT, Blasiak A, Siah KT, et al. A systematic review of the development and psychometric properties of constipation-related patient-reported outcome measures: Opportunities for digital health. J Neurogastroenterol Motil. 2022;28:376. doi: 10.5056/jnm22004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee VV, Vijayakumar S, Lau NY, Blasiak A, Siah KTH, Ho D. Understanding the user: Patients’perception, needs, and concerns of health apps for chronic constipation. Digit Health. 2022;8:20552076221104673. doi: 10.1177/20552076221104673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee VV, Vijayakumar S, Ng WY, Lau NY, Leong QY, Ooi DSQ, et al. Personalization and localization as key expectations of digital health intervention in women pre-to post-pregnancy. NPJ Digit Med. 2023;6:183. doi: 10.1038/s41746-023-00924-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blasiak A, Sapanel Y, Leitman D, Ng WY, De Nicola R, Lee VV, et al. Omnichannel communication to boost patient engagement and behavioral change with digital health interventions. J Med Internet Res. 2022;24:e41463. doi: 10.2196/41463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sapanel Y, Tadeo X, Brenna CT, Remus A, Koerber F, Cloutier LM, et al. Economic evaluation associated with clinical-grade mobile app–based digital therapeutic interventions: Systematic review. J Med Internet Res. 2023;25:e47094. doi: 10.2196/47094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.WHO. Overview of Regulatory Considerations on Artificial Intelligence for Health. 2021. Available from: https://www.itu.int/en/ITU-T/focusgroups/ai4h/Documents/all/FGAI4H-M-052.pdf .
- 94.Nilsen P. Making sense of implementation theories, models, and frameworks. Implement Sci. 2015;10:53–79. doi: 10.1186/s13012-015-0242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]