Abstract
Little is known about the ability of simian virus 40 (SV40) T antigen to bind single-stranded DNA. We demonstrate here that a mutant (259-708) missing the first 258 amino acids of T antigen and its origin-binding domain bound single-stranded DNA at close to normal levels, whereas a mutant containing only the first 259 amino acids failed to bind any single-stranded DNA. The 259-708 mutant also assembled into high-molecular-weight oligomers in the presence of single-stranded DNA. Its ATPase activity was stimulated by single-stranded DNA similarly to the wild type (WT). Furthermore, WT T antigen’s ability to bind to single-stranded DNA was inhibited by the binding of two monoclonal antibodies that recognize a region after residue 362. These results show that the domain responsible for binding to single-stranded DNA is completely separate from the origin-binding domain.
Simian virus 40 (SV40) large T antigen is a multifunctional protein required for the initiation of virus DNA replication in infected cells. Many of its functional domains and binding sites for cellular proteins have been mapped by genetic and biochemical approaches (see reviews by Fanning and Knippers [15] and Bullock [6]). One activity that has been poorly characterized is its ability to bind nonspecifically to single-stranded DNA (2, 16, 34). The function of this activity is not known, but since T antigen is a 3′-to-5′ helicase (49), one assumption is that it is required for interacting with single-stranded DNA during unwinding.
T antigen is the only viral protein required for SV40 DNA replication (46); all other factors are supplied by the cell. In the presence of ATP, the protein forms a double hexamer on the replication origin (23), melts the EP region, and untwists the A/T track within the origin (3, 4, 11, 29). The enzyme then unwinds the DNA bidirectionally (10, 12, 17, 51) by using its helicase activity (43, 44). The cellular proteins RPA (8, 14, 18, 26, 47, 50), DNA polymerase α/primase (13, 14, 24, 26), and topoisomerase I (38, 39) may be recruited to the origin to form a replication complex.
The origin-binding domain of T antigen has been well characterized. It maps approximately to residues 147 to 247 (1, 25, 36, 37, 41, 45). It can, by itself, bind specifically to the origin (1, 19, 25, 45) and is required but is not sufficient for nonspecific binding to double-stranded DNA (21). It is also essential for T antigen’s helicase activity (52). However, it remains unclear whether it also possesses the ability to bind single-stranded DNA. McVey et al. (25) reported that a fragment consisting of residues 132 to 246 has a measurable single-stranded DNA-binding activity, and Wun-Kim and Simmons (52) found that the smallest proteolytic fragment with which they could demonstrate single-stranded DNA binding contained the origin-binding region. However, both of those studies actually measured binding to a helicase substrate, a partially double-stranded molecule. Mohr et al. (27) found that a mutation of residue 522 affects single-stranded DNA binding without changing the ability to bind the origin, suggesting that the C-terminal region of T antigen may be involved in this activity. More recently, Joo et al. (19) reported that the origin-binding domain (131-260) bound single-stranded DNA only weakly.
To resolve this controversy and to begin to characterize the single-stranded DNA-binding activity of T antigen, we generated two deletion mutants separating the N-terminal region with its origin-binding domain from the C-terminal region of the molecule. We constructed deletion mutants 1-259 and 259-708 by PCR amplification of those regions of the large T antigen cDNA gene using appropriate primers followed by cloning into baculovirus transfer vector p1393 (Pharmingen). Recombinant baculoviruses were made according to the manufacturer’s directions, and the recombinant proteins were purified by immunoaffinity chromatography with PAb419 for 1-259 and PAb101 for 259-708 as previously described (35, 40). Wild-type (WT) T antigen was purified in the same way with either antibody. The purified protein was the major species detected by silver staining of acrylamide gels. A common contaminant was antibody that had eluted from the immunoaffinity column. It does not appear to interfere with any of T antigen’s biochemical activities (38–41).
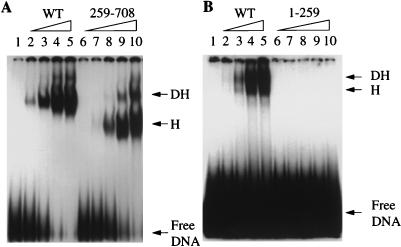
To test the single-stranded DNA-binding activities of these two truncated proteins, we carried out a gel shift assay using a 5′ end-labeled, 55-nucleotide, single-stranded oligonucleotide corresponding to the bottom strand of the fork substrate described by SenGupta and Borowiec (34). Increasing amounts of WT T antigen and 259-708 were first reacted with the labeled DNA for 30 min at 37°C under replication buffer conditions (40). The DNA-protein complexes were cross-linked with glutaraldehyde and subjected to electrophoresis on a nondenaturing 4% acrylamide gel (Fig. 1A). It is apparent that the deletion mutant missing the origin-binding domain (259-708) could bind to single-stranded DNA, although binding activity was slightly lower than that of WT. It should be noted that we used proportionate molar amounts of WT and 259-708 in lanes 2 through 5 and 6 through 9, respectively (Fig. 1A); lane 10 contained a higher amount of the mutant protein to demonstrate that binding activity was not at saturation. We should also note that ATP was included in the replication buffer, although its addition had no measurable effect on single-stranded DNA binding under our conditions (data not shown).
FIG. 1.
Single-stranded DNA binding activity of WT and mutant T antigens. Purified T antigens were incubated with end-labeled, single-stranded DNA, cross-linked to the DNA with glutaraldehyde, and applied to a 4% acrylamide gel in Tris-borate-EDTA buffer. (A) Binding activity of WT and 259-708 T antigens. T antigens were purified by immunoaffinity chromatography with monoclonal antibody PAb101 as described elsewhere (39). Lanes: 1, no T antigen; 2 through 5, 3, 7.5, 15, and 22.5 pmol of WT T antigen, respectively. Lanes 6 through 10 contained 3, 7.5, 15, 22.5, and 37.5 pmol of 259-708, respectively. (B) Binding activities of WT and 1-259 T antigens. Proteins were purified by using PAb419 antibody. Lanes: 1, no T antigen; 2 through 5, 0.84, 2.1, 4.2, and 6.3 pmol of WT T antigen, respectively; 6 through 10, 0.84, 2.1, 4.2, 6.3, and 21 pmol of 1-259, respectively. The positions of the free DNA and DNA bound in presumed hexamers (H) and double hexamers (DH) are indicated.
In contrast to these results with 259-708, deletion mutant 1-259 did not bind any single-stranded DNA (Fig. 1B). As expected, this truncated protein could still bind to origin DNA (data not shown), since it contains the origin-binding domain (residues 147 to 246). These results demonstrate that the single-stranded DNA-binding region is located after residue 259 and is not part of the origin-binding domain. The results also show that both WT and 259-708 T antigens form two complexes with single-stranded DNA (Fig. 1A), presumably consisting of hexamers and double-hexamers. This indicates that 259-708 can form oligomers in the presence of single-stranded DNA, just like WT.
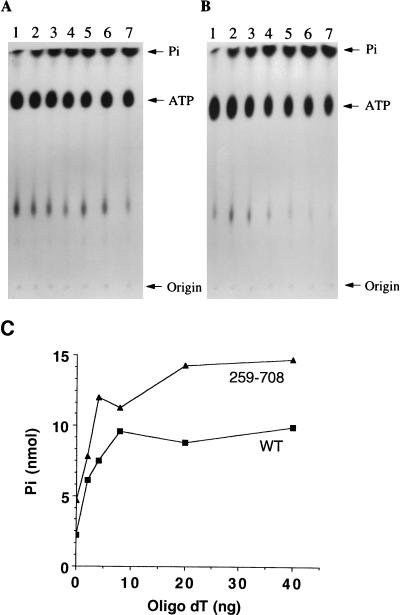
It has been shown that single-stranded DNA can stimulate the ATPase activity of T antigen (16, 31). Here, we investigated whether single-stranded DNA also stimulates the ATPase activity of mutant 259-708. Increasing amounts of a 32-mer dT oligonucleotide were added to an ATPase reaction containing 20 pmol of WT (Fig. 2A) or 259-708 (Fig. 2B) T antigens. The reactions were performed as described by Clark et al. (7). Free inorganic phosphate was separated from the [γ-32P]ATP substrate by ascending chromatography on polyethyleneimine-cellulose plates as described elsewhere (5). Reaction products were quantitated by scintillation counting. It is apparent that the ATPase activity of 259-708 could be stimulated by oligo(dT) (Fig. 2B) in a way similar to that of WT T antigen (Fig. 2A and C). The mutant T antigen had just as much if not more ATPase activity compared to that of WT, and its activity was stimulated by about 3-fold by oligo(dT) (Fig. 2C). WT T antigen was stimulated maximally by 4.5-fold (Fig. 2C). These results are consistent with the observation that the single-stranded DNA binding region is located between residues 259 and 708.
FIG. 2.
Stimulation of the ATPase activities of WT and mutant T antigens by (oligo)dT. WT (A) and 259-708 (B) T antigens were purified by immunoaffinity with PAb101 as described elsewhere (35) and assayed for ATPase activity in the presence of different amounts of oligo(dT). The free inorganic phosphate (Pi) was separated from the ATP substrate by ascending thin-layer chromatography. The origin is shown on the right. Lanes 1, no T antigen; 2 through 7, 0, 2, 4, 8, 20, and 40 ng of oligo(dT), respectively. (C) The amounts of released inorganic phosphate were quantitated by scintillation counting and plotted against the amount of oligo(dT) in each reaction.
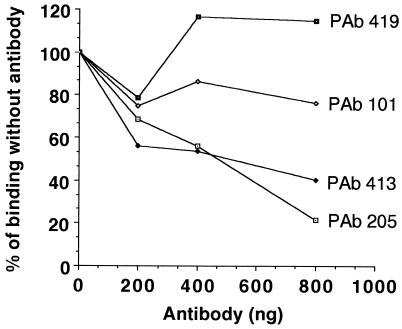
To obtain additional data that the single-stranded DNA-binding region is present after residue 259, we performed antibody inhibition assays with monoclonal antibodies known to bind to specific sites on T antigen. A 400-ng amount of WT T antigen (4.4 pmol) was preincubated with various amounts of monoclonal antibodies (PAb419, PAb205, PAb413, or PAb101) which bind to the N-terminal end (PAb419), central region (PAb205 and PAb413), and C-terminal end (PAb101) of T antigen. After a 20-min preincubation at room temperature, labeled single-stranded DNA was added and incubation continued at 37°C for 30 min. Complexes were separated on acrylamide gels and quantitated with a PhosphorImager, and the results were plotted as a percentage of binding without antibody (Fig. 3). Both PAb205 and PAb413 inhibited binding, while PAb419 and PAb101 did not. Addition of the latter two antibodies resulted in a supershifted band as expected (data not shown). The epitopes on T antigen for PAb205 and PAb413 are maximally located between residues 362 and 708 (28), although a smaller region mapping between residues 362 and 447 may be the major binding site for these antibodies (20). Therefore, the association of this region with antibodies interferes with single-stranded DNA binding. It should be noted that neither of these two antibodies inhibits binding to the origin (21).
FIG. 3.
Inhibition of single-stranded DNA-binding activity by monoclonal antibodies. Increasing amounts of purified monoclonal antibodies (as shown) were preincubated with WT T antigen purified as described elsewhere (35) with PAb101 antibody. Labeled single-stranded DNA was added, and incubation was continued at 37°C. The labeled bound DNA was analyzed as described in the legend to Fig. 1, and the values were plotted as percentages of binding without antibody.
We attempted to map the single-stranded DNA-binding domain more closely by constructing a deletion mutant containing the 362-to-447 region close to the N-terminal end (e.g., 325-708) and a mutant with most of this region at the C-terminal end (e.g., 1-432), but neither of these mutants was stable. However, we did find that deletion mutant 246-627 bound single-stranded DNA at levels close to that of the WT and formed oligomerized complexes similar to (but smaller than) that of the WT (data not shown), demonstrating that the single-stranded DNA-binding domain does not extend beyond residue 627. Taken together, the data show that this domain is located between residues 259 and 627. The ability of proteins 259-708 and 246-627 to assemble into large-molecular-weight complexes with DNA indicates that the oligomerization domain must also be localized to residues 259 to 627.
Our results demonstrate that the origin-binding and single-stranded DNA-binding regions are separate. This finding has implications for the mechanism of DNA unwinding. T antigen binds to the origin of replication as a double hexamer (23) and functions in this form as a helicase (42, 48). It is not known where the double-stranded origin DNA is located relative to the hexameric helicase, but there is evidence that the hexamer is a sixfold ring structure with a channel large enough for double-stranded or single-stranded DNA (30). T antigen can hexamerize without DNA (22, 23), but preformed hexamers have low affinity for DNA (9). When inactive hexamers are incubated without ATP, they dissociate into DNA-binding monomers, suggesting that T antigen first binds origin DNA as a monomer and subsequently assembles into hexamers (9). One interpretation of these data is that the origin DNA occupies the hexameric channel after binding. Furthermore, there is evidence that during DNA unwinding, at least one strand of DNA is present within the hexameric channel. Electron microscopy studies reveal that the DNA is threaded through the T-antigen hexameric complexes during unwinding (48). During this reaction, T antigen appears to bind to only one strand of the unwound DNA (32–34), in agreement with its function as a 3′-to-5′ helicase (49). It has been proposed (30) that one single strand fits into the six-sided channel during unwinding and that the other is excluded. If this is correct, it would imply that one strand must be displaced from the channel during or after ori opening. How this might happen is unknown, but our observation that the domains for single-stranded and origin DNA binding are distinct imply that the DNA is tethered to different regions of the protein before and after origin melting. Furthermore, our result that the 259-708 mutant has ATPase activity enhanced by single-stranded DNA suggests that this region of the protein interacts with at least one single-strand during DNA unwinding.
In an earlier study (21), we reported that nonspecific double-stranded DNA binding by T antigen requires the presence of the origin-binding region and of a second region mapping between residues 269 and 522. The relationship between this second region and single-stranded DNA binding is unknown. However, all evidence (see references 21 and 52 and the results presented here) points to the likelihood that nonspecific double-stranded and single-stranded DNA-binding activities are partially separable. This suggests that T antigen has three distinct DNA-binding activities, and it is reasonable to propose that all three activities are utilized during the binding and subsequent unwinding of SV40 DNA.
Acknowledgments
This work was supported by grant CA36118 from the National Cancer Institute.
REFERENCES
- 1.Arthur A K, Höss A, Fanning E. Expression of simian virus 40 T antigen in Escherichia coli: localization of T-antigen origin DNA-binding domain to within 129 amino acids. J Virol. 1988;62:1999–2006. doi: 10.1128/jvi.62.6.1999-2006.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auborn K J, Markowitz R B, Wang E, Yu Y T, Prives C. Simian virus 40 (SV40) T antigen binds specifically to double-stranded DNA but not to single-stranded DNA or DNA/RNA hybrids containing the SV40 regulatory sequences. J Virol. 1988;62:2204–2208. doi: 10.1128/jvi.62.6.2204-2208.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borowiec J A, Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988;7:3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borowiec J A, Dean F B, Hurwitz J. Differential induction of structural changes in the simian virus 40 origin of replication by T antigen. J Virol. 1991;65:1228–1235. doi: 10.1128/jvi.65.3.1228-1235.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley M K, Griffin J, Livingston D M. Relationship of oligomerization to enzymatic and DNA binding properties of the SV40 large T antigen. Cell. 1982;28:125–134. doi: 10.1016/0092-8674(82)90382-8. [DOI] [PubMed] [Google Scholar]
- 6.Bullock P A. The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 7.Clark R, Lane D P, Tjian R. Use of monoclonal antibodies as probes of simian virus 40 T antigen ATPase activity. J Biol Chem. 1981;256:11854–11858. [PubMed] [Google Scholar]
- 8.Collins K, Kelly T J. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase α-primase. Mol Cell Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean F B, Borowiec J A, Eki T, Hurwitz J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 10.Dean F B, Bullock P, Murakami Y, Wobbe C R, Weissbach L, Hurwitz J. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc Natl Acad Sci USA. 1987;84:16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean F B, Hurwitz J. Simian virus 40 large T antigen untwists DNA at the origin of DNA replication. J Biol Chem. 1991;266:5062–5071. [PubMed] [Google Scholar]
- 12.Dodson M, Dean F B, Bullock P, Echols H, Hurwitz J. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science. 1987;238:964–967. doi: 10.1126/science.2823389. [DOI] [PubMed] [Google Scholar]
- 13.Dornreiter I, Copeland W C, Wang T S. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase alpha with large T antigen. Mol Cell Biol. 1993;13:809–820. doi: 10.1128/mcb.13.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dornreiter I, Erdile L F, Gilbert I U, von Winkler D, Kelly T J, Fanning E. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 16.Giacherio D, Hager L P. A poly(dT)-stimulated ATPase activity associated with simian virus 40 large T antigen. J Biol Chem. 1979;254:8113–8116. [PubMed] [Google Scholar]
- 17.Goetz G S, Dean F B, Hurwitz J, Matson S W. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J Biol Chem. 1988;263:383–392. [PubMed] [Google Scholar]
- 18.Iftode C, Borowiec J A. Denaturation of the simian virus 40 origin of replication mediated by human replication protein A. Mol Cell Biol. 1997;17:3876–3883. doi: 10.1128/mcb.17.7.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joo W S, Luo X, Denis D, Kim H Y, Rainey G J, Jones C, Sreekumar K R, Bullock P A. Purification of the simian virus 40 (SV40) T antigen DNA-binding domain and characterization of its interactions with the SV40 origin. J Virol. 1997;71:3972–3985. doi: 10.1128/jvi.71.5.3972-3985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane D P, Gannon J. Monoclonal antibody analysis of the SV40 large T antigen-p53 complex. Cancer Cells. 1986;4:387–393. [Google Scholar]
- 21.Lin H-J L, Upson R, Simmons D T. Nonspecific DNA binding activity of simian virus 40 large T antigen: evidence for the cooperation of two regions for full activity. J Virol. 1992;66:5443–5452. doi: 10.1128/jvi.66.9.5443-5452.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loeber G, Stenger J E, Ray S, Parsons R E, Anderson M E, Tegtmeyer P. The zinc finger region of simian virus 40 large T antigen is needed for hexamer assembly and origin melting. J Virol. 1991;65:3167–3174. doi: 10.1128/jvi.65.6.3167-3174.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature (London) 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto T, Eki T, Hurwitz J. Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. Proc Natl Acad Sci USA. 1990;87:9712–9716. doi: 10.1073/pnas.87.24.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McVey D, Strauss M, Gluzman Y. Properties of the DNA-binding domain of the simian virus 40 large T antigen. Mol Cell Biol. 1989;9:5525–5536. doi: 10.1128/mcb.9.12.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 27.Mohr I J, Fairman M P, Stillman B, Gluzman Y. Large T-antigen mutants define multiple steps in the initiation of simian virus 40 DNA replication. J Virol. 1989;63:4181–4188. doi: 10.1128/jvi.63.10.4181-4188.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mole S E, Gannon J V, Ford M J, Lane D P. Structure and function of large T antigen. Phil Trans R Soc Lond Ser B. 1987;317:455–469. doi: 10.1098/rstb.1987.0072. [DOI] [PubMed] [Google Scholar]
- 29.Roberts J. Simian virus 40 (SV40) large tumor antigen causes stepwise changes in SV40 origin structure during initiation of DNA replication. Proc Natl Acad Sci USA. 1989;86:3939–3943. doi: 10.1073/pnas.86.11.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.San Martin M C, Gruss C, Carazo J M. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J Mol Biol. 1997;268:15–20. doi: 10.1006/jmbi.1997.0952. [DOI] [PubMed] [Google Scholar]
- 31.Scheffner M, Knippers R, Stahl H. RNA unwinding activity of SV40 large T antigen. Cell. 1989;57:955–963. doi: 10.1016/0092-8674(89)90334-6. [DOI] [PubMed] [Google Scholar]
- 32.SenGupta D J, Blackwell L J, Gillette T, Borowiec J A. Recognition of model DNA replication forks by the SV40 large tumor antigen. Chromosoma. 1992;102:S46–S51. doi: 10.1007/BF02451785. [DOI] [PubMed] [Google Scholar]
- 33.SenGupta D J, Borowiec J A. Strand and face: the topography of interactions between the SV40 origin of replication and T-antigen during the initiation of replication. EMBO J. 1994;13:982–992. doi: 10.1002/j.1460-2075.1994.tb06343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.SenGupta D J, Borowiec J A. Strand-specific recognition of a synthetic DNA replication fork by the SV40 large tumor antigen. Science. 1992;256:1656–1661. doi: 10.1126/science.256.5064.1656. [DOI] [PubMed] [Google Scholar]
- 35.Simanis V, Lane D P. An immunoaffinity purification procedure for SV40 large T antigen. Virology. 1985;144:88–97. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 36.Simmons D T. Geometry of the simian virus 40 large tumor antigen-DNA complex as probed by protease digestion. Proc Natl Acad Sci USA. 1988;85:2086–2090. doi: 10.1073/pnas.85.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmons D T, Loeber G, Tegtmeyer P. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J Virol. 1990;64:1973–1983. doi: 10.1128/jvi.64.5.1973-1983.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmons D T, Roy R, Chen L, Gai D, Trowbridge P W. The activity of topoisomerase I is modulated by large T antigen during unwinding of the SV40 origin. J Biol Chem. 1998;273:20390–20396. doi: 10.1074/jbc.273.32.20390. [DOI] [PubMed] [Google Scholar]
- 39.Simmons D T, Trowbridge P W, Roy R. Topoisomerase I stimulates SV40 T antigen-mediated DNA replication and inhibits T antigen’s ability to unwind DNA at nonorigin sites. Virology. 1998;242:435–443. doi: 10.1006/viro.1997.9024. [DOI] [PubMed] [Google Scholar]
- 40.Simmons D T, Upson R, Wun-Kim K, Young W. Biochemical analysis of mutants with changes in the DNA-binding domain of simian virus 40 tumor antigen. J Virol. 1993;67:4227–4236. doi: 10.1128/jvi.67.7.4227-4236.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmons D T, Wun-Kim K, Young W. Identification of simian virus 40 T antigen residues important for specific and nonspecific binding to DNA and for helicase activity. J Virol. 1990;64:4858–4865. doi: 10.1128/jvi.64.10.4858-4865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smelkova N V, Borowiec J A. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J Virol. 1997;71:8766–8773. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stahl H, Droge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986;5:1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahl H, Scheffner M, Wiekowski M, Knippers R. DNA unwinding function of the SV40 large tumor antigen. Cancer Cells. 1988;6:105–112. [Google Scholar]
- 45.Strauss M, Argani P, Mohr I J, Gluzman Y. Studies on the origin-specific DNA-binding domain of simian virus 40 large T antigen. J Virol. 1987;61:3326–3330. doi: 10.1128/jvi.61.10.3326-3330.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tegtmeyer P. Simian virus 40 deoxyribonucleic acid synthesis: The viral replicon. J Virol. 1972;10:591–598. doi: 10.1128/jvi.10.4.591-598.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Treuner K, Ramsperger U, Knippers R. Replication protein A induces the unwinding of long double-stranded DNA regions. J Mol Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 48.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiekowski M, Schwarz M W, Stahl H. Simian virus 40 large T antigen DNA helicase. Characterization of the ATPase-dependent DNA unwinding activity and its substrate requirements. J Biol Chem. 1988;263:436–442. [PubMed] [Google Scholar]
- 50.Wold M S, Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wold M S, Li J J, Kelly T J. Initiation of simian virus 40 DNA replication in vitro: large-tumor-antigen- and origin-dependent unwinding of the template. Proc Natl Acad Sci USA. 1987;84:3643–3647. doi: 10.1073/pnas.84.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wun-Kim K, Simmons D T. Mapping of helicase and helicase substrate binding domains on simian virus 40 large T antigen. J Virol. 1990;64:2014–2020. doi: 10.1128/jvi.64.5.2014-2020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]