Abstract
Autosomal dominant sensorineural hearing loss (ADSNHL) is a genetically heterogeneous disorder caused by pathogenic variants in various genes, including MYH14. However, the interpretation of pathogenicity for MYH14 variants remains a challenge due to incomplete penetrance and the lack of functional studies and large families. In this study, we performed exome sequencing in six unrelated families with ADSNHL and identified five MYH14 variants, including three novel variants. Two of the novel variants, c.571G>C (p.Asp191His) and c.571G>A (p.Asp191Asn), were classified as likely pathogenic using ACMG and Hearing Loss Expert panel guidelines. In silico modeling demonstrated that these variants, along with p.Gly1794Arg, can alter protein stability and interactions among neighboring molecules. Our findings suggest that MYH14 causative variants may be more contributory and emphasize the importance of considering this gene in patients with non-syndromic mainly post-lingual severe form of hearing loss. However, further functional studies are needed to confirm the pathogenicity of these variants.
Keywords: Hearing Loss, Pathogenic variant, MYH14, Gene
1. INTRODUCTION
Hearing loss (HL) is a prevalent sensory disorder that affects over 5% of the global population, according to the World Health Organization. In 75% of cases, HL is the only clinical finding, which is referred to as non-syndromic hearing loss (NSHL). NSHL is typically congenital or prelingual in onset when inherited with autosomal recessive inheritance. In contrast, autosomal dominant non-syndromic hearing loss (ADNSHL) typically has post-lingual onset and is often progressive. While ADNSHL accounts for a smaller proportion of NSHL cases, it can still have a significant impact on affected individuals and their families (Alde et al. 2023; Smith et al. 2005).
MYH14 (MIM 608568) is among the 50 genes that have been implicated in causing ADNSHL (Donaudy et al. 2004). This gene is highly expressed in the Organ of Corti, where it plays a critical role in protecting against overstimulation by acoustic signals. Loss of this protective function can result in progressive hearing loss (Fu et al. 2016). Therefore, MYH14 is included in gene panels for clinical testing (Abou Tayoun et al. 2016; Peart et al. 2023).
To date, 86 variants reported in MYH14, at Human Gene Mutation Database (HGMD) (https://www.hgmd.cf.ac.uk/ac/index.php; last accessed:1/3/2024). Out of 86 variants, 59 of them associated with hearing loss. Among these variants, only 13 pathogenic or likely pathogenic missense and nonsense variants have been reported in families from various regions of the world (Choi et al. 2011; Donaudy et al. 2004; Hiramatsu et al. 2021; Kim et al. 2017; Qing et al. 2014; Shearer et al. 2010; Smith et al. 2005; Yang et al. 2005). The remaining variants have been classified as conflicting or benign due to incomplete penetrance and lack of functional or segregation data. There are 1036 entries for MYH14 in ClinVar ( https://www.ncbi.nlm.nih.gov/clinvar/; last accessed: 1/3/2024) and 13 of them are interpreted as pathogenic or likely pathogenic without conflict.
In this study, we identified five MYH14 variants in six families. We carefully evaluated the variants and classified them according to American College of Medical Genetics (ACMG) and Hearing Loss Expert panel (HLEP) guidelines (Oza et al. 2018; Richards et al. 2015). Our findings support the role of MYH14 in ADNSHL and broaden the spectrum of this gene as a contributor to post-lingual severe to profound deafness.
2. METHODS
2.1. Ethics and Consent
This study followed the principles of the Declaration of Helsinki and was approved by the University of Miami Institutional Review Board (USA) and the Ankara University Medical School Ethics Committee (Turkiye). Local ethics committee of Istanbul Medeniyet University Goztepe Training and Research Hospital, Istanbul, Turkiye (Decision date: 27.04.2022, Number: 2022/0281). A signed informed consent form was obtained from each participant or, in the case of a minor, from the parents.
2.2. Subjects
Our larger study cohort consists of 626 GJB2 pathogenic variant-negative multiplex and simplex hearing loss families of diverse ethnicity. The diagnosis of SNHL was established via standard audiometry in a soundproofed room according to the current clinical standards. Repeated audiometry was performed to check the progression of HL in the available families. Clinical evaluation included a thorough physical examination and otoscopy in all cases.
Electrocardiograms, urinalysis, and high-resolution computed tomography scan of the temporal bone to identify inner ear anomalies were obtained where possible. These clinical investigations performed to reveal syndromic features that can be seen in patients with hearing loss.
2.3. DNA Sequencing
Exome (ES) and Sanger sequencing were performed using our previously reported protocol (Bademci et al. 2016a; Bademci et al. 2016b) . PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/), Sorting Intolerant from Tolerant (SIFT) (http://sift.jcvi.org/), Combined Annotation Dependent Depletion (CADD GRCh37-v1.6) (https://cadd.gs.washington.edu/), Rare Exome Variant Ensemble Learner (REVEL) (https://sites.google.com/site/revelgenomics/), MutationAssesor (http://mutationassessor.org/r3/) and MutationTaster (http://www.mutationtaster.org/) scores were considered for pathogenicity analysis, Genomic Evolutionary Rate Profiling (GERP) (http://mendel.stanford.edu/SidowLab/downloads/gerp/index.html) score was used to evaluate the conservation of the variants. The population frequency of each variation was evaluated using data from the gnomAD database (https://gnomad.broadinstitute.org/) and 1000 Genome Project database (https://www.internationalgenome.org/data). ACMG guidelines were followed for variant interpretation (Richards et al. 2015).
Probands who are heterozygous for MYH14 variants underwent Sanger sequencing for confirmation. Other available family members were added for segregation analysis.
2.4. Structural Modeling
The effect of novel missense variants on protein structure and stability was analyzed. Partial protein sequences were retrieved from Uniprot. The 3D structures of proteins containing wild type and mutated residues at respective positions were obtained using I-TASSER (Zhou et al. 2022). Protein models were verified at SAVES (http://servicesn.mbi.ucla.edu/SAVES/) The best model with each variant was selected for further analysis.
3. RESULTS
We found a total of five variants in MYH14 in probands of six families after ES (Table 1) out of which three were novel. The genetic findings in all affected individuals are shown in Supplementary Table 1. Pedigree analysis of families with novel MYH14 variants suggested ADNSHL (Figure 1 and Supplementary Figure 1). No other clinical findings were detected in affected individuals. Audiometric re-evaluation showed stable HL in all affected individuals from family 1. All the affected individuals from this family had post-lingual, severe, non-progressive HL (Supplementary Figure 2). Family 2 had eight affected individuals, we only had mother’s (II:3) and proband’s (III:2) DNA. Mother had severe while the proband presented with moderate HL. We were not able to access the audiograms for the rest of the families. Family 1 and 2 have different substitutions at the same nucleotide position (c.571G>C and c.571G>A, respectively), which co-segregated with the phenotype in family 1 and 2. These two variants were classified as likely pathogenic. Moreover, the protein residue at position 191 is highly conserved among different vertebrates except two species, Scarlet macaw (Ara macao) and Spiny softshell turtle (Apalone spinifera) (Figure 1B). A novel missense variant c.5380G>A, lying in the tail domain of MYH14 was identified in a simplex family 3 and classified as VUS. We were not able to confirm this variant as a de novo since parents were not available (Supplementary Figure 1). For family 4 the variant c.526G>A was confirmed by Sanger sequencing in the proband. She has moderate hearing loss. However, there were no additional DNA samples from the family available to test segregation. In ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/) this variant is classified as variant of uncertain significance (VUS) and together with the in-silico predictions and our analyses, it still remains as VUS (Chen et al. 2016). In family 5 there were two affected individuals, and the variant c.547C>T in MYH14 co-segregated with HL (Supplementary Figure 3). The similar variant was identified in the proband of family 6 along with a common mitochondrial pathogenic variant m.1555A>G.
Table 1.
Identified MYH14 variants in our cohort and in silico prediction scores (all variants are indicated in NM_001145809.2)
| Family ID | Chromosomal Position (hg19) | Protein change | c.DNA change | Variant Type | MAF (1000 Genome) | gnomAD v2–v3(TAF) | CADD GRCh37-v1.6 | GERP | REVEL | PolyPhen2 | Mutation Taster | Mutation Assesor | ClinVar | Variant Classification | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Chr19: 50726348 | p.Asp191His | c.571G>C | Missense | - | - | 28.40 | 4.5 | 0.833 | PsD | DC | Medium | - | PM2,PP3,PM5,PS4 Likely Pathogenic | This study |
| 2 | Chr19: 50726348 | p.Asp191Asn | c.571G>A | Missense | - | - | 24.20 | 4.51 | 0.357 | B | DC | Neutral | - | PM2,PM5,PS4 Likely Pathogenic | This study |
| 3 | Chr19: 50796855 | p.Gly1794Arg | c.5380G>A | Missense | - | - | 27.30 | 3.61 | 0.48 | PsD | DC | Low | - | PM2 VUS | This study |
| 4 | Chr19: 50720992 | p.Ala176Thr | c.526G>A | Missense | 0.0004 | 0.00018–0.00013 | 24.6 | 4.63 | 0.71 | PD | DC | Medium | VUS | PP3,BS1 VUS | Chen et al 2016 |
| 5 & 6 | Chr19: 50721013 | p.Arg183Trp | c.547C>T | Missense | - | 0.0000082–0.000014 | 27.5 | 2.40 | 0.71 | PD | DC | High | VUS | PM2,PP3 VUS | Retterer et. al 2016 |
VUS:Variant Unknown Significance, B: Benign, D: Damaging, T: Tolerated, PD: Probably Damaging, PsD:Possibly Damaging, DC:Disease Causing
Figure 1.

Pedigrees, audiograms and chromatograms of identified novel likely pathogenic variants in MYH14. (A) Pedigrees of families. (B) The protein alignment shows conservation across 24 vertebrates including five diverse classes for the 191Asp residue by using Multiz Alignments at https://genome.ucsc.edu/. (C) Chromatograms showing the identified variants.
For three novel p.Asp191His (Family 1), p.Asp191Asn (Family 2) and p.Gly1794Arg (Family 3) variants we performed the structural modeling (Figure 2). In silico analysis revealed that these changes are unfavorable for protein stability. The substitution of Histidine at position 191 produces a long protruding chain affecting the proper folding of the protein. Similarly, Glycine is a small neutral amino acid substituted for an acidic residue at position 191. It disrupts hydrogen bonding and ionic interactions with the neighboring molecules. Glycine at position 1794 is changed to arginine which is a basic amino acid thus altering the interaction with the adjacent molecules in the chain.
Figure 2.
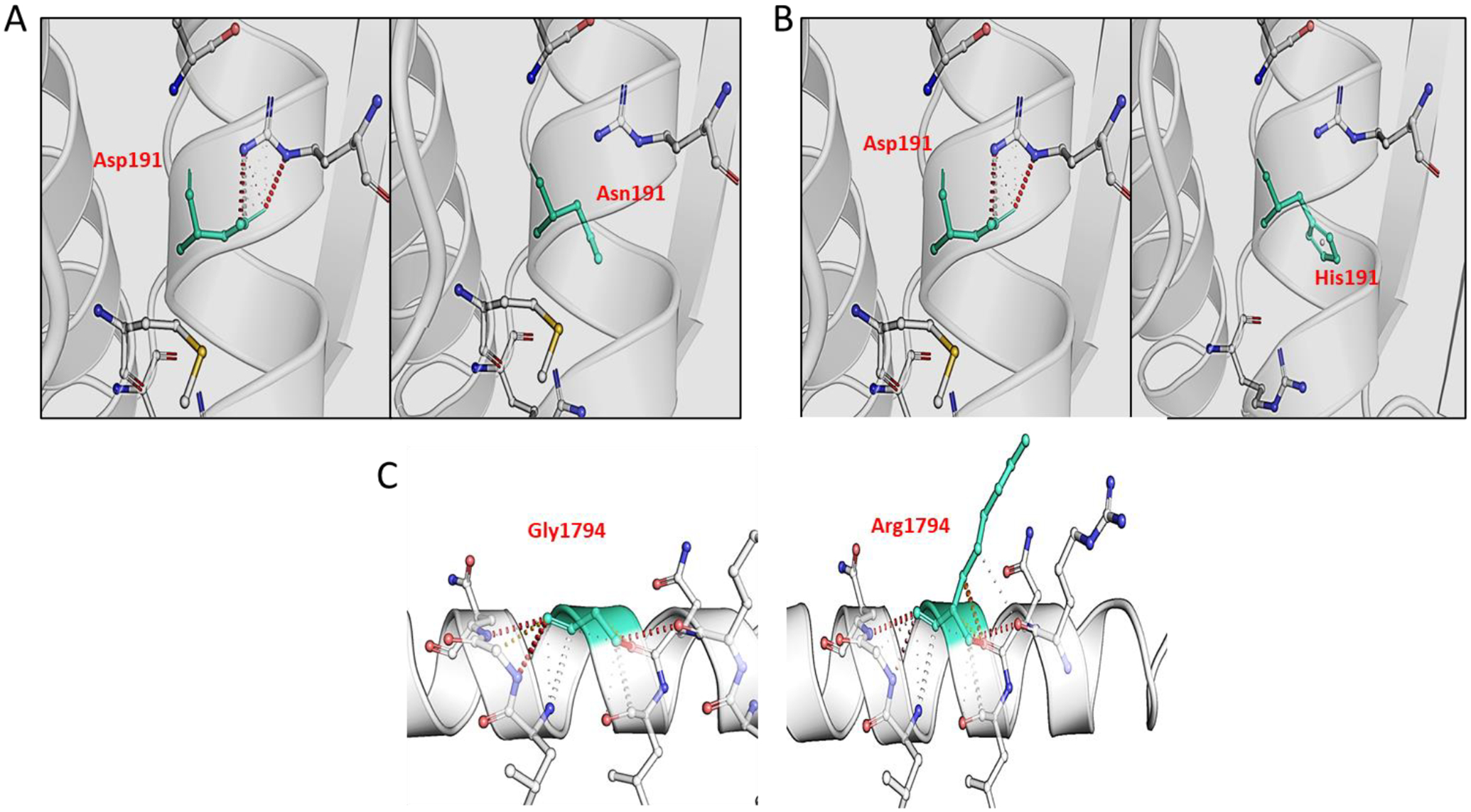
Structural modelling of novel pathogenic variants in MYH14. A) The substitution of Asp to Asn at position 191 disrupts the interaction with neighboring amino acid. B) Asp at position 191 is replaced with Histidine; a basic polar amino acid which has interrupted the interactions with adjacent amino acid in the chain. C) Glycine is a small amino acid which is replaced by Arginine at position 1794. Arginine being a large hydrophilic amino acid has developed additional bonding with the amino acids in the vicinity which may cause additional constraint on the structure. Mutated amino acid residues are indicated in green. Red dotted lines show hydrogen bonding, grey dotted lines represent aromatic contacts and yellow dotted lines represent ionic interactions.
4. DISCUSSION AND CONCLUSIONS
In this study, we identified five MYH14 variants two of which were novel and interpreted as likely pathogenic in patients with ADNSHL. Interpretation of the autosomal dominant variants is difficult due to incomplete penetrance, lack of functional studies, and large families. To date only 13 variants have been reported in HGMD in families with more than two affected individuals. Our findings expand the knowledge of MYH14 variants associated with HL.
The likely pathogenic p.Asp191His (c.571G>C) variant in Family 1 and p.Asp191Asn (c.571G>A) variant in Family 2, affects the aspartic acid 191 residue in the motor domain of MYH14. This residue is highly conserved between species indicating its importance for the protein’s function. Only two species, Scarlet macaw (Ara macao) and, Spiny softshell turtle (Apalone spinifera) has Asn instead Asp in the same residue. The in-silico structural analysis of the p.Asp191His and p.Asp191Asn variants revealed that both variants cause the disruption of the hydrogen and ionic bonding between the neighboring amino acids in the chain. As aspartic acid is polar and acidic, and histidine is polar and basic, while asparagine is polar and neutral, it is possible that the phenotype of the affected individuals with these variants may differ due to the varying physicochemical properties of the amino acids. A p. Asp191Gly variant affecting same codon has been reported in a proband and affected father as a cause of prelingual severe and progressive autosomal dominant NSHL (Kim et al. 2017).
We have identified a novel p.Gly1794Arg variant in proband of the Family 3. The variant is rare and amino acid residue is not conserved in many vertebrates (Supplemental Figure 1). In the Family 4 we identified a p.Ala176Thr variant in proband (Supplementary Figure 3) which was previously reported in a case with hereditary hearing loss (Chen et al. 2016). In ClinVar database this variant reported as VUS and our interpretation remained same.
We have identified the p.Arg183Trp variant in Family 5 and 6 which was previously reported by Retterer et al (Retterer et al. 2016). In family 5 it was segregated in affected father and son with hearing loss. In family 6 proband and unaffected father has the variant. Proband also carries m.1555A>G variant which is inherited from unaffected mother. The proband does not have aminoglycoside usage history. We have interpreted the variant p.Arg183Trp as VUS.
Some MYH14 variants may be mediated by environmental influences, while others may be tolerated due to compensatory mechanisms. In a knockout mice study by Fu et al., 2016, Myh14 knockout mice did not exhibit significant HL until five months of age, but outer hair cell loss was observed after acoustic trauma. The authors suggested that MYH10 can compensate for most of the functions of MYH14 in the cochlea and could explain the milder phenotypes in mice. Additionally, Myh14 was shown to play a protective role in noise-induced damage of outer hair cells, suggesting that some MYH14 variants may be more susceptible to environmental factors (Fu et al. 2016).
In conclusion, this study broadens the spectrum of MYH14 gene variants associated with ADNSHL and supports its role as a contributor to post-lingual severe to profound deafness. The identified variants can aid in genetic counseling and facilitate early diagnosis and intervention for affected individuals and their families. Further functional studies are warranted to elucidate the pathogenic mechanisms underlying MYH14-related HL.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all patients for their participation.
FUNDING INFORMATION
This work was supported by National Institutes of Health grants R01DC009645 and R01DC012836 to M.T and by TUBITAK (1059B191801475) to D.D . Authors declare that there is no conflict of interest to report.
Footnotes
CONFLICT OF INTEREST STATAMENT
The authors declare no conflict of interest.
DATA AVAILABILITY STATEMENT
The analyzed variants have been submitted to the ClinVar. The data can be accessed at https://www.ncbi.nlm.nih.gov/clinvar/ via following ClinVar IDs: Variant c.571G>C (SCV003928150), c.571G>A (SCV003928156), c.5380G>A (SCV003928162), c.526G>A (SCV003928163), and c.547C>T (SCV003928164)
REFERENCES
- Abou Tayoun AN, Al Turki SH, Oza AM, Bowser MJ, Hernandez AL, Funke BH, Rehm HL, Amr SS (2016) Improving hearing loss gene testing: a systematic review of gene evidence toward more efficient next-generation sequencing-based diagnostic testing and interpretation. Genet Med 18: 545–53. doi: 10.1038/gim.2015.141 [DOI] [PubMed] [Google Scholar]
- Alde M, Cantarella G, Zanetti D, Pignataro L, La Mantia I, Maiolino L, Ferlito S, Di Mauro P, Cocuzza S, Lechien JR, Iannella G, Simon F, Maniaci A (2023) Autosomal Dominant Non-Syndromic Hearing Loss (DFNA): A Comprehensive Narrative Review. Biomedicines 11. doi: 10.3390/biomedicines11061616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bademci G, Cengiz FB, Foster Ii J, Duman D, Sennaroglu L, Diaz-Horta O, Atik T, Kirazli T, Olgun L, Alper H, Menendez I, Loclar I, Sennaroglu G, Tokgoz-Yilmaz S, Guo S, Olgun Y, Mahdieh N, Bonyadi M, Bozan N, Ayral A, Ozkinay F, Yildirim-Baylan M, Blanton SH, Tekin M (2016a) Variations in Multiple Syndromic Deafness Genes Mimic Non-syndromic Hearing Loss. Sci Rep 6: 31622. doi: 10.1038/srep31622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bademci G, Foster J 2nd, Mahdieh N, Bonyadi M, Duman D, Cengiz FB, Menendez I, Diaz-Horta O, Shirkavand A, Zeinali S, Subasioglu A, Tokgoz-Yilmaz S, Huesca-Hernandez F, de la Luz Arenas-Sordo M, Dominguez-Aburto J, Hernandez-Zamora E, Montenegro P, Paredes R, Moreta G, Vinueza R, Villegas F, Mendoza-Benitez S, Guo S, Bozan N, Tos T, Incesulu A, Sennaroglu G, Blanton SH, Ozturkmen-Akay H, Yildirim-Baylan M, Tekin M (2016b) Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med 18: 364–71. doi: 10.1038/gim.2015.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Dong C, Wang Q, Zhong Z, Qi Y, Ke X, Liu Y (2016) Targeted Next-Generation Sequencing Successfully Detects Causative Genes in Chinese Patients with Hereditary Hearing Loss. Genet Test Mol Biomarkers 20: 660–665. doi: 10.1089/gtmb.2016.0051 [DOI] [PubMed] [Google Scholar]
- Choi BO, Kang SH, Hyun YS, Kanwal S, Park SW, Koo H, Kim SB, Choi YC, Yoo JH, Kim JW, Park KD, Choi KG, Kim SJ, Zuchner S, Chung KW (2011) A complex phenotype of peripheral neuropathy, myopathy, hoarseness, and hearing loss is linked to an autosomal dominant mutation in MYH14. Hum Mutat 32: 669–77. doi: 10.1002/humu.21488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Di Stazio M, Ferrara A, Lanzara C, Ficarella R, Declau F, Pusch CM, Nurnberg P, Melchionda S, Zelante L, Ballana E, Estivill X, Van Camp G, Gasparini P, Savoia A (2004) Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4). Am J Hum Genet 74: 770–6. doi: 10.1086/383285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Zhang L, Jin Y, Sun X, Zhang A, Wen Z, Zhou Y, Xia M, Gao J (2016) Loss of Myh14 Increases Susceptibility to Noise-Induced Hearing Loss in CBA/CaJ Mice. Neural Plast 2016: 6720420. doi: 10.1155/2016/6720420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu K, Nishio SY, Kitajiri SI, Kitano T, Moteki H, Usami SI, On Behalf Of The Deafness Gene Study C (2021) Prevalence and Clinical Characteristics of Hearing Loss Caused by MYH14 Variants. Genes (Basel) 12. doi: 10.3390/genes12101623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Kim AR, Han JH, Lee C, Oh DY, Choi BY (2017) Discovery of MYH14 as an important and unique deafness gene causing prelingually severe autosomal dominant nonsyndromic hearing loss. J Gene Med 19. doi: 10.1002/jgm.2950 [DOI] [PubMed] [Google Scholar]
- Oza AM, DiStefano MT, Hemphill SE, Cushman BJ, Grant AR, Siegert RK, Shen J, Chapin A, Boczek NJ, Schimmenti LA, Murry JB, Hasadsri L, Nara K, Kenna M, Booth KT, Azaiez H, Griffith A, Avraham KB, Kremer H, Rehm HL, Amr SS, Abou Tayoun AN, ClinGen Hearing Loss Clinical Domain Working G (2018) Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat 39: 1593–1613. doi: 10.1002/humu.23630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart L, Gonzalez J, Morel Swols D, Duman D, Saridogan T, Ramzan M, Zafeer MF, Liu XZ, Eshraghi AA, Hoffer ME, Angeli SI, Bademci G, Blanton S, Smith C, Telischi FF, Tekin M (2023) Dispersed DNA variants underlie hearing loss in South Florida’s minority population. Hum Genomics 17: 103. doi: 10.1186/s40246-023-00556-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing J, Yan D, Zhou Y, Liu Q, Wu W, Xiao Z, Liu Y, Liu J, Du L, Xie D, Liu XZ (2014) Whole-exome sequencing to decipher the genetic heterogeneity of hearing loss in a Chinese family with deaf by deaf mating. PLoS One 9: e109178. doi: 10.1371/journal.pone.0109178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, Vertino-Bell A, Smaoui N, Neidich J, Monaghan KG, McKnight D, Bai R, Suchy S, Friedman B, Tahiliani J, Pineda-Alvarez D, Richard G, Brandt T, Haverfield E, Chung WK, Bale S (2016) Clinical application of whole-exome sequencing across clinical indications. Genet Med 18: 696–704. doi: 10.1038/gim.2015.148 [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–24. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J 2nd, Scherer S, Scheetz TE, Smith RJ (2010) Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A 107: 21104–9. doi: 10.1073/pnas.1012989107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Bale JF Jr., White KR (2005) Sensorineural hearing loss in children. Lancet 365: 879–90. doi: 10.1016/S0140-6736(05)71047-3 [DOI] [PubMed] [Google Scholar]
- Yang T, Pfister M, Blin N, Zenner HP, Pusch CM, Smith RJ (2005) Genetic heterogeneity of deafness phenotypes linked to DFNA4. Am J Med Genet A 139: 9–12. doi: 10.1002/ajmg.a.30989 [DOI] [PubMed] [Google Scholar]
- Zhou X, Zheng W, Li Y, Pearce R, Zhang C, Bell EW, Zhang G, Zhang Y (2022) I-TASSER-MTD: a deep-learning-based platform for multi-domain protein structure and function prediction. Nat Protoc 17: 2326–2353. doi: 10.1038/s41596-022-00728-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analyzed variants have been submitted to the ClinVar. The data can be accessed at https://www.ncbi.nlm.nih.gov/clinvar/ via following ClinVar IDs: Variant c.571G>C (SCV003928150), c.571G>A (SCV003928156), c.5380G>A (SCV003928162), c.526G>A (SCV003928163), and c.547C>T (SCV003928164)


