Abstract
Background & Aims
Chronic liver disease (CLD) remains a global health issue associated with a significant disease burden. Liver fibrosis, a hallmark of CLD, is characterised by the activation of hepatic stellate cells (HSCs) that gain profibrotic characteristics including increased production of extracellular matrix protein. Currently, no antifibrotic therapies are available clinically, in part because of the lack of HSC-specific drug targets. Here, we aimed to identify HSC-specific membrane proteins that can serve as targets for antifibrotic drug development.
Methods
Small interfering RNA-mediated knockdown of GPR176 was used to assess the in vitro function of GPR176 in HSCs and in precision cut liver slices (PCLS). The in vivo role of GPR176 was assessed using the carbon tetrachloride (CCl4) and common bile duct ligation (BDL) models in wild-type and GPR176 knockout mice. GPR176 in human CLD was assessed by immunohistochemistry of diseased human livers and RNA expression analysis in human primary HSCs and transcriptomic data sets.
Results
We identified Gpr176, an orphan G-protein coupled receptor, as an HSC-enriched activation associated gene. In vitro, Gpr176 is strongly induced upon culture-induced and hepatocyte-damage-induced activation of primary HSCs. Knockdown of GPR176 in primary mouse HSCs or PCLS cultures resulted in reduced fibrogenic characteristics. Absence of GPR176 did not influence liver homeostasis, but Gpr176-/- mice developed less severe fibrosis in CCl4 and BDL fibrosis models. In humans, GPR176 expression was correlated with in vitro HSC activation and with fibrosis stage in patients with CLD.
Conclusions
GPR176 is a functional protein during liver fibrosis and reducing its activity attenuates fibrogenesis. These results highlight the potential of GPR176 as an HSC-specific antifibrotic candidate to treat CLD.
Impact and implications
The lack of effective antifibrotic drugs is partly attributed to the insufficient knowledge about the mechanisms involved in the development of liver fibrosis. We demonstrate that the G-protein coupled receptor GPR176 contributes to fibrosis development. Since GPR176 is specifically expressed on the membrane of activated hepatic stellate cells and is linked with fibrosis progression in humans, it opens new avenues for the development of targeted interventions.
Keywords: Fibrosis, G-protein-coupled receptors, GPR176, Liver cirrhosis, Myofibroblasts
Graphical abstract
Highlights
-
•
GPR176 is highly enriched in activated mouse HSCs in different models of chronic liver injury.
-
•
GPR176 shows early and sustained induction during HSC activation, preceding the expression of canonical myofibroblast markers.
-
•
Targeted Gpr176 mRNA in vitro knockdown and in vivo knockout results in downregulation of fibrotic markers.
-
•
GPR176 expression in human livers correlates with fibrosis stage, indicating its relevance to human liver disease.
Introduction
Chronic liver disease (CLD) can be defined as a spectrum of disease states of the liver characterised by disturbed hepatocyte regeneration, inflammation, and fibrosis.1 The main causes of CLD globally are HBV and HCV infections, alcohol abuse and metabolic dysfunction-associated steatotic liver disease (MASLD). CLD is marked by a high global burden of disease as it is estimated to account for 1.8% of global disability-adjusted life years and mainly affects working aged people.2 Its burden of disease can be explained by the occurrence of cirrhosis and hepatocellular carcinoma (HCC) in CLD patients. Cirrhosis is considered the end stage of CLD and is characterised by different clinical entities including the incidence of oesophageal varices and decompensation events such as ascites, encephalopathy, or jaundice.3 HCC is estimated to occur in 2–8% of cirrhotic patients each year, and arises in a background of CLD in 90% of diagnosed cases, and is the fourth most common cause of cancer-related death and the second most lethal tumour in terms of 5-year survival.4 Cirrhosis and HCC are estimated to account for 2 million deaths per year worldwide.5,6
Currently, targeting the underlying aetiology is the only therapeutic option for CLD which is not always feasible, certainly when looking at CLD as a multifactorial process instead of a single disease entity (e.g. the co-occurrence of MASLD and excessive alcohol intake).2 Since the progression of fibrosis to cirrhosis in CLD is associated with increased morbidity and mortality, developing antifibrotic therapies is an important strategy for tackling CLD and several drugs are already in clinical trials.7 Liver fibrosis is marked by a disbalance of extracellular matrix (ECM) turnover resulting in the pathological accumulation of ECM proteins which are produced by myofibroblasts.1 Multiple studies have shown that hepatic stellate cells (HSCs) are the main source of myofibroblasts during liver fibrosis, irrespective of the underlying disease aetiology.8,9 During fibrosis, retinol-storing quiescent HSCs (qHSCs) activate to an activated myofibroblast-like phenotype (aHSC) with proliferative, contractile, inflammatory and chemotactic characteristics as well as enhanced ECM production.10 Given their pivotal role in progression of CLD, HSCs and the mechanisms involved in their activation can be regarded as putative therapeutic targets for treatment of fibrosis and CLD. An example of one such putative HSC-based therapy is the inhibition of TGF-β signalling, which is one of the most important mechanisms stimulating the activation of HSC. However, TGF-β inhibitors must also address the fact that TGF-β signalling is crucial for homeostasis and diseases across multiple organs and cells. Thus, targeting the TGF-β pathway can lead to undesired side effects such as neoplasms.11 Because of this, not only should drugs target crucial mechanisms of HSC activation, but they should also do this in an HSC-specific manner.
In this study, we demonstrate that the orphan G-protein coupled receptor (GPCR) GPR176 has enriched expression in human and mouse HSCs and its transcription is increased upon HSC activation. Furthermore, reduced expression of Gpr176 results in the reduction of fibrotic markers in primary HSC cultures and Gpr176-/- knockout mice exhibited less fibrosis in two mouse models of liver fibrosis. Finally, GPR176 expression correlated with the fibrosis stage in livers of patients suffering with MASLD and chronic HBV and HCV infection.
Materials and methods
Detailed materials and methods can be found in the supporting documents.
Animal models
All in vitro experiments were performed using healthy male BALB/c mice. Gpr176-/- and Gpr176+/+ mice12 were used with C57BL/6N background for the induction of liver fibrosis by either carbon tetrachloride (CCl4) or by common bile duct ligation. Cell isolation and treatment protocols were approved by the Animal Care and Use Committee of Vrije Universiteit Brussel in permits 15-212-5, 18-212-1, 19-212-1, 20-212-3, and 20-212-5.
Human liver samples
Patient liver tissue was obtained from surgically resected specimens from the Department of Hepatobiliary Surgery of the University Hospital of Brussels (UZ Brussel), Belgium. The study protocol was approved by the local ethical committee of the UZ Brussel and Vrije Universiteit Brussel (reference number 2015/278; B.U.N. 143201525406) and the study was performed in accordance with the Declaration of Helsinki.
In vitro models of liver fibrosis
Mouse precision cut liver slices (PCLS) were generated as previously described.13 In brief, the median lobe of healthy BALB/c mice was processed into 250 μm-thick slices with a 3 mm diameter. The PCLS were cultured in Wiiliam’s E (WME) medium (Gibco) for up to 5 days, with daily refreshments of medium. Spheroids were generated using primary male BALB/c hepatocytes and HSCs.14 In brief, 2000 purified cells (667 hepatocytes and 1333 HSCs) were seeded per well of a cell-repellent, U-bottom, 96-well plate. On day 6 of culture, cells were exposed to 5 ng/ml TGF-β1 or 2 mM acetaminophen (APAP) for 48 h. For each culture, six spheroids were pooled per condition for RNA analysis.
Results
Screening for putative HSC targets revealed Gpr176 as a surface marker of activated HSCs
In our search for potential HSC-specific therapeutic candidates, we defined features indicative of interesting targets. First, the candidate target should be involved in HSC activation and, second, it should have enriched expression in HSCs when compared with other liver cell types. Third, for pharmaceutical purposes, expression of the protein at the cellular membrane is preferable as this can facilitate easier targeting of activated HSCs. We have recently defined a fibrogenic aHSC transcriptional program that strongly correlates with ECM producing aHSCs in both human and mouse models (Table S1).15 We screened this gene set for genes encoding proteins expressed at the cellular membrane and identified 42 genes (Fig. 1A). Next, to select HSC-specific genes among these 42 genes, we screened transcriptome data sets, including HSCs isolated from healthy livers and those from livers subjected to common bile duct ligation (BDL) or carbon tetrachloride (CCl4) as well as data sets of liver tissue (foetal) and other cell types including erythroblasts, platelets, hepatocytes, and cells with an immunological (liver), endothelial (liver), and epithelial background (Fig. 1B; Table S2). From this screening, we identified Ptchd4 and Gpr176 to cluster together as markers that were highly enriched in HSCs. Because Gpr176 also had a more pronounced upregulation in BDL and CCl4 models when compared with qHSC, and since the function of Gpr176 under physiological and pathological states was elusive, we focused on Gpr176. As confirmation, we analysed Gpr176 mRNA expression in HSCs in three independent data sets consisting of HSCs isolated from BDL, CCl4, and MASLD models.[16], [17], [18] Gpr176 was significantly upregulated in all three murine models of CLD (Fig. 1C). Additionally, we also found Gpr176 to be downregulated in HSCs isolated from livers that had recovered for 4 weeks after an 8-week fibrosis induction by CCl4 (Fig. 1C; Fig. S1). In conclusion, we determined that Gpr176 could serve as a marker of activated mouse HSCs as it is highly enriched in activated HSCs in several mouse models of chronic liver injury.
Fig. 1.
Gpr176 is a selective marker of HSC activation.
(A) Cellular location of a 173 fibrogenic gene signature described previously (GSE176042):15 Forty-two genes encode for proteins expressed at the plasma membrane. (B) Gene expression of plasma membrane genes on microarray data of indicated cells and tissues showing Ptchd4 and Gpr176 is aHSC-specific. The values are depicted as the normalised expression value for each gene, subtracted by the mean expression value of that gene across all samples. Table S2 gives an overview of the 96 samples included. (C) Gpr176 mRNA levels in mouse models of CLD: BDL (GSE34640),16 MASLD (GSE134512)17 and CCl4 (GSE153703).18 qPCR analysis of Gpr176 mrRNA levels in HSCs isolated from healthy, 8 weeks CCl4-treated and 4 weeks recovery mice. Gapdh served as reference gene (n = 2–3), two sample t-test and one-way ANOVA with post-hoc Tukey multiple comparisons test. BDL, common bile duct ligation; CCl4, carbon tetrachloride; CLD, chronic liver disease; HSC, hepatic stellate cell; MASLD, metabolic dysfunction-associated steatotic liver disease.
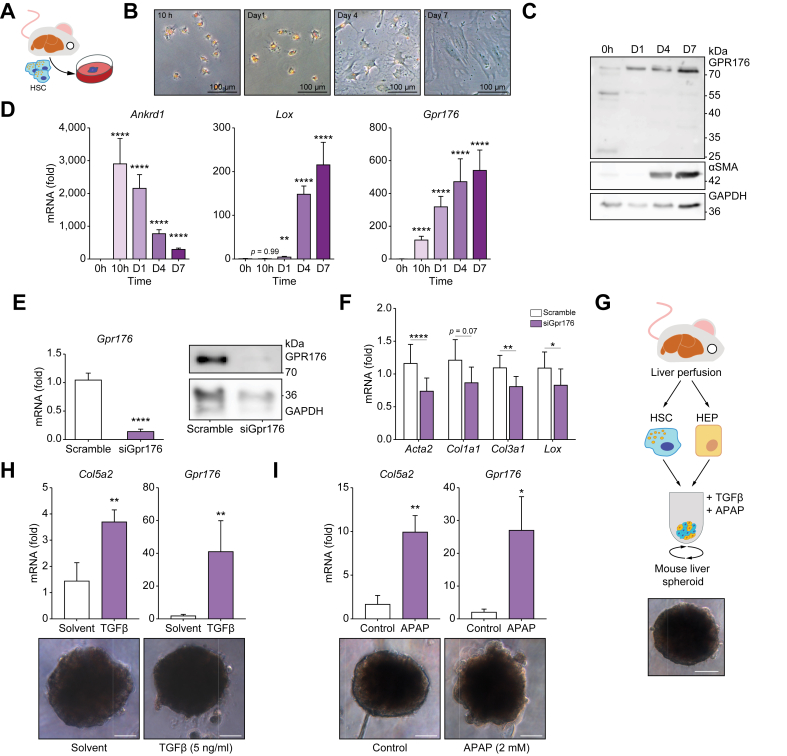
Gpr176 is induced upon HSC activation and knockdown of Gpr176 prevents full activation
Next, we investigated the time-dependent expression of Gpr176 during the process of HSC activation. To achieve this, we used two-dimensional (2D) in vitro cultures of primary mouse HSCs to allow for controlled monitoring of HSC activation (Fig. 2A). In this model, HSCs phenotypically start differentiating into myofibroblasts by day 4 and reach a fully differentiated state by day 7 (Fig. 2B). At both the mRNA and protein level we noted an early and sustained induction of GPR176 expression. Elevated levels of GPR176 protein can be identified prior to the appearance of phenotypic changes or the expression of α-smooth muscle actin (α-SMA) protein (Fig. 2C). Additionally, we noted that in activating HSCs, GPR176 was expressed at ±75 kDa, whereas additional bands were detected at 55 kDa and 25 kDa in freshly isolated qHSCs. As a positive control we used protein extracts from mouse heart tissue which revealed a band at ±55 kDa, which was in line with the predicted 56 kDa (Fig. S2A). We believe that the 75 kDa band is the N-glycosylated form of GPR176, which was shown in previous studies to be a requirement for proper expression and functionality.19 At the mRNA level, Gpr176 mRNA is increased at a similar timepoint as Ankrd1, a known YAP-downstream gene and HSC initiation marker,18 and preceded the expression of Lox, a canonical myofibroblast differentiation marker (Fig. 2D). We validated the expression of Gpr176 during in vivo and in vitro HSC activation by analysing HSC RNASeq data. In these data sets, HSC activation was induced either via acute liver injury by a single CCl4 injection or via 2D culture of HSCs. In vivo, we noted an early (24 h) and sustained (7 days) upregulation of Gpr176 after a single injection of CCl4, whereas in vitro, Gpr176 was induced after 12 h which is in line with our qPCR findings (Fig. S2B and C). Finally, single cell RNA analysis of liver cells isolated from mice subjected to a single injection of CCl4 showed that Gpr176 was exclusively expressed in activated HSCs/myofibroblasts, whereas Gpr176 exhibited negligible expression in healthy liver tissue (Fig. S3). Given its marked upregulation during HSC activation both in vivo and in vitro, we investigated whether Gpr176 mRNA could be an antifibrotic target. To achieve this, we used siRNAs targeting mouse Gpr176 during in vitro activation of primary mouse HSCs. Following mRNA interference, a clear downregulation of both Gpr176 mRNA and 75 kDa GPR176 protein was accompanied by the downregulation of Acta2, Col3a1, and Lox mRNA levels, with only a tendency for a decrease in Col1a1 mRNA (Fig. 2E and F). However, we were unable to show clear phenotypical alterations or reduction in α-SMA protein expression in GPR176 reduced cultures (data not shown). Lastly, because of the hard substrate to which HSCs are exposed during 2D monoculture, the 2D in vitro HSC activation does not properly mimic in vivo activation.14,20 This might confound the value of Gpr176 gene expression as a marker of HSC activation in vitro. To bypass this, we used a 3D spheroid co-culture model of hepatocytes and HSCs that maintains hepatocytes and HSCs in a non-injured state (Fig. 2G).14 A fibrogenic response was induced by activating HSCs either in a direct manner (TGF-β) or indirectly by inducing hepatocyte damage via APAP. While APAP-treated cultures were marked by blebbing of cells, indicative of hepatocyte damage, this was less the case in TGF-β treated cultures (Fig. 2H and I). Both treatments were marked by an upregulation of Col5a2 (indicating HSC activation) as well as Gpr176 mRNA (Fig. 2H and I). In conclusion, Gpr176 expression was induced during the initiation of HSC activation and maintained this upregulation throughout the HSC activation process. Furthermore, this upregulation was not an artefact of the 2D monolayer culture but rather represented both direct and indirect activation of HSCs. Finally, GPR176 was, at least partially, responsible for the fibrogenic reprogramming of HSCs.
Fig. 2.
Induction of GPR176 during HSC activation is partially responsible for induction of fibrogenic genes.
(A) Mouse HSCs were isolated by Nycodenz from healthy male BALB/c mice. (B) Light microscopy of 2D cultured mouse HSCs shows myofibroblast differentiation at day 4 of culture. (C) Western blot analysis for GPR176 and α-SMA protein reveals early induction of GPR176 (n = 1). (D) qPCR analysis of 2D cultured mouse HSCs for initiation marker Ankrd1, activation marker Lox, and Gpr176. Gtf2b and Ywhaz served as reference genes (n = 3), type III Analysis of Variance Table with Satterthwaite's method. (E, F) Primary mouse HSCs transfected with Gpr176 siRNAs or scramble sequences. qPCR (n = 7) and Western blotting (n = 1) confirmed suppression of GPR176 expression and HSC activation markers at day 8. Gapdh served as reference gene, paired t-test. (G) Hepatocyte-HSC spheroids at day 6 of culture generated in cell repellent 96-well plates. Scalebar: 100 μm. (H and I) 48 h exposure of spheroids to TGF-β (H) or APAP (I) results in induction of both Col5a2 and Gpr176 mRNA (n = 4-5), two sample t-test. Scalebar: 100 μm. Gtf2b and Des served as reference genes. Data are reported as mean ± SEM. Levels of significance: ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001, ∗∗∗∗p <0.0001. 2D, two-dimensional; APAP, acetaminophen; HSC, hepatic stellate cell.
Gpr176 upregulation is implicated in the fibrogenic response of precision-cut liver slice cultures
We evaluated the potential of Gpr176 mRNA interference in a second multicellular 3D in vitro model (i.e. mouse PCLS cultures), a model of hepatocyte injury and of subsequent HSC activation. PCLS are generated by slicing mouse liver tissue with a vibratome and punching the slices into 3 mm discs. Although maintaining liver tissue integrity, the slicing and punching induces tissue damage with a subsequent regenerative and fibrogenic response (Fig. 3A and B).13 As shown by qPCR, this induced tissue led to the upregulation of fibrogenic markers Acta2, Col1a1, and Lox after the second day of culture, which was also accompanied by upregulation of Gpr176 mRNA (Fig. 3C). Next, we tested the potential of Gpr176 mRNA interference in mPLCS cultures. Gpr176 RNA interference in PCLS cultures led to reduced expression of Gpr176 mRNA as well as downregulation of Acta2 and Col1a1 expression, whereas a downward trend was observed for Lox expression (Fig. 3D and E). Taken together these data suggest that Gpr176 mRNA levels can serve as a suitable fibrogenic marker in multicellular liver cultures and Gpr176 mRNA targeting has potential as an antifibrotic therapy.
Fig. 3.
Expression and profibrogenic role of Gpr176 in precision-cut liver slice cultures.
(A) PCLS were generated by slicing and punching livers from healthy mice. (B) Macroscopic image of a PCLS. (C) qPCR for HSC activation markers and Gpr176. Statistics were calculated comparing cultured PCLS at day 1 (n = 4), one-way ANOVA with post-hoc Tukey multiple comparisons test. (D, E) PCLS transfected at day 0 with Gpr176 siRNAs or scrambled sequences. qPCR confirmed suppression of Gpr176 mRNA and indicated HSC activation markers at day 2. Gtf2b and Ywhaz served as reference genes (n = 7), paired t-test. Data are mean ± SEM. Levels of significance: ∗p <0.05, ∗∗ p <0.01, ∗∗∗p <0.001, ∗∗∗∗p <0.0001. HSC, hepatic stellate cell; PCLS, precision-cut liver slice.
Reduced Gpr176 expression alleviates murine liver fibrosis
Next, we investigated the potential profibrotic role of GPR176 in vivo using Gpr176-/- knockout mice.12 Homozygous deletion of Gpr176 did not cause any gross abnormalities, lethality, or infertility,12 allowing us to study liver fibrosis induction in these murine model. We could not observe any difference in liver damage, HSC activation status, collagen crosslinking, or in the amount of α-SMA-positive cells when comparing healthy Gpr176+/+ to Gpr176-/- knockout livers (Fig. 4 A–C, healthy conditions). To investigate the role of GPR176 during CLD, we treated Gpr176+/+ and Gpr176-/- mice with CCl4 for a total duration of 4 weeks. This resulted in liver damage, marked by the increase in alanine aminotransferase (ALT) levels, and fibrosis as evidenced by an increased collagen deposition (Fig. 4A and C). No differences in ALT levels were noted between Gpr176+/+ or Gpr176-/- with a similar level of liver injury in both groups (Fig. 4A). HSCs isolated from fibrotic livers of Gpr176+/+ and Gpr176-/- mice showed significant induction of activation markers Acta2, Col1a1, and Lox when compared with HSCs isolated from healthy livers, marking their activation during CLD. Interestingly, we noticed a downward trend for Col1a1 and Lox in HSCs isolated from Gpr176-/- mice when compared with HSCs isolated from Gpr176+/+ mice. The opposite was noted for Acta2 expression. However, none of these trends were statistically significant (Fig. 4B). Additionally, we compared liver sections from Gpr176+/+ or Gpr176-/- mice for collagen crosslinking and the proportion of α-SMA positive cells. A significant reduction in collagen deposition in Gpr176-/- vs Gpr176+/+ mice treated with CCl4 was observed. The decrease in α-SMA positive cells in Gpr176-/- mice was not significant (Fig. 4C). Lastly, we assessed the impact of reduced GPR176 expression on cholestatic liver disease by performing BDL in both Gpr176+/+ and Gpr176-/- mice models. BDL led to increased levels of bilirubin, ALT, and aspartate aminotransferase (AST) levels (Fig. 5A) as well as an increase in the amount of intrahepatic bile ducts, collagen deposition, and the amount of α-SMA-positive cells (Fig. 5B). Although no differences were observed in circulating liver injury markers in the blood between Gpr176+/+ or Gpr176-/- mice (Fig. 5A), Gpr176-/- mice showed a clear reduction of collagen crosslinking, mainly at the perisinusoidal level (Fig. 5B). No differences were observed in α-SMA-positive cells in the Gpr176-/- BDL mouse model (Fig. 5B). These results indicate that GPR176 is not implicated in homeostasis of healthy livers and that it stimulates fibrogenesis via, at least partially, stimulation of HSC activation and their production of collagens.
Fig. 4.
Gpr176 knockout is marked by reduced collagen deposition during chronic hepatotoxic liver injury.
(A) Plasma ALT levels in healthy and diseased (CCl4) Gpr176+/+ or Gpr176-/- mice (n = 7-10), Kruskal–Wallis rank sum test with Dunn multiple comparisons. (B) FACS-sorted HSCs from healthy or diseased (CCl4) Gpr176+/+ or Gpr176-/- mice were subjected to mRNA analysis for indicated HSC activation markers and Gpr176. Gtf2b and Ywhaz serve as reference genes (n = 3-4), one-way ANOVA with post-hoc Tukey multiple comparisons test. (C) Sirius Red and α-SMA staining show fibrosis development in diseased (CCl4) Gpr176+/+ and Gpr176-/- mice. Sirius Red staining was quantified as percentage of positively stained area, α-SMA staining was quantified as percentage of α-SMA positive cells. Quantifications are represented as bar plots in the right panels (n = 7-10), one-way ANOVA with post-hoc Tukey multiple comparisons test. Scale bar: 200 μm. Data are mean ± SEM. Levels of significance: ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001, ∗∗∗∗p <0.0001.ALT, alanine transaminase; CCl4, carbon tetrachloride; FACS, fluorescence-activated cell sorting; HSC, hepatic stellate cell; α-SMA, α-smooth muscle actin.
Fig. 5.
Gpr176 knockout is marked by reduced collagen deposition in obstructive cholestasis.
(A) Plasma total bilirubin, ALT, and AST levels in healthy and diseased (BDL) Gpr176+/+ and Gpr176-/- mice (n = 5-7), Kruskal-Wallis rank sum test with Dunn multiple comparisons. (B) Sirius Red and α-SMA staining show fibrosis development in diseased Gpr176+/+ and Gpr176-/- mice. Sirius Red staining was quantified as percentage of positively stained area, α-SMA staining was quantified as percentage of α-SMA positive cells. Quantifications are represented as bar plots in the right panels (n = 7–9), one-way ANOVA with post-hoc Tukey multiple comparisons test. Scalebar: 200 μm. Data are mean ± SEM. Levels of significance: ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001, ∗∗∗∗p <0.0001. ALT, alanine transaminase; AST, aspartate transaminase; BDL, common bile duct ligation; α-SMA, α-smooth muscle actin.
Gpr176 expression in human livers correlates with CLD
Next, we investigated the relevance of our findings on GPR176 for human liver diseases. To determine whether GPR176 is specifically expressed in HSCs in human livers, liver cell types were isolated from liver resection pieces by means of fluorescence-activated cell sorting (FACS) and subjected to qPCR (Fig. 6A, Fig. S4A–C and Table S3). We confirmed enriched expression of GPR176 in HSCs when compared with hepatocytes, Kupffer cells (KC) and liver sinusoidal endothelial cells (LSEC). Although HSCs also have increased expression of GPR176 when compared with cholangiocytes (CHOL), this was not statistically significant, implying that CHOL might also express GPR176 at intermediate levels (Fig. 6B). To determine whether human HSCs (hHSC) upregulate GPR176 upon activation, we isolated hHSCs by using a Nycodenz-based isolation protocol and cultured them to allow culture-induced 2D in vitro activation (Fig. 6A). Human HSC lines at passage 3 (P3), were compared with their status immediately after Nycodenz-based isolation (fresh cells). The myofibroblast phenotype was evident at P3 as shown by qPCR for ACTA2, COL5A2, and LOXL2 as well as by α-SMA staining (Fig. 6C and D). In these activated hHSC, we clearly detected GPR176 protein at 75 kDa and observed an increase in GPR176 mRNA when compared with freshly isolated hHSC (Fig. 6C and D). Lastly, we investigated whether GPR176 expression was correlated with human CLD. Currently, MASLD, alcoholic liver disease (ALD) and chronic HBV and HCV infection are the main causes of cirrhosis and CLD complications, which led us to analyse transcriptome data of liver tissue derived from these diseases. Here, we clearly correlated liver GPR176 mRNA expression with human liver fibrosis caused by MASLD and chronic HBV and HCV infection but not with ALD (Fig. 6E). Additionally, we showed that, although GPR176 expression correlated with fibrosis stage, it did not correlate with the NAFLD Activity Score in livers of patients with MASLD (Fig. S5A and B). Finally, we stained human liver tissue, obtained from liver resection pieces with either mild fibrosis (F0–F1) or cirrhosis (F4), for VIM (hHSC marker) and GPR176. Although GPR176-positive cells were scarce, GPR176 protein expression was only present in F4 liver tissue and all GPR176 expressing cells also co-expressed VIM (Fig. 6F). Altogether, activated human HSCs have enriched expression of GPR176 and expression of GPR176 in human livers correlates with fibrosis stage at least in MASLD and chronic HBV and HCV infection.
Fig. 6.
Correlation of GPR176 expression with human HSC activation status and fibrosis.
(A) Human liver resection specimens were subjected to one of two cell isolation protocols. Either FACS-based isolation of liver cell types was performed or a Nycodenz-based protocol was used for isolation of hHSC and subsequent in vitro culture. (B) GPR176 mRNA expression in the indicated human liver cell types (FACS-based isolation). GAPDH and YWHAZ serve as reference genes (n = 3-5), one-way ANOVA with post-hoc Tukey multiple comparisons test. (C) qPCR analysis comparing HSC activation markers and GPR176 in either freshly Nycodenz-based isolated hHSC or passage 3 activated hHSC. HPRT1 and YWHAZ serve as reference genes (n = 4), two sample t-test or Wilcoxon rank-sum. (D) Protein expression and phenotype of culture activated hHSC as shown by immunocytochemistry and Western blotting for α-SMA and light microscopy. These aHSCs also express GPR176 protein (n = 2). (E) Microarray expression data of GPR176 in livers from patients with chronic liver diseases, two sample t-test. (F) Sirius Red staining and VIM-GPR176 co-staining of human liver tissue obtained from liver resection pieces. Upper panel: F0–F1 liver tissue obtained from a patient with colorectal carcinoma liver metastasis. Mid-panel: liver tissue obtained from a patient with MASLD related cirrhosis (F4) and HCC. Lower panel: liver tissue obtained from a patient with primary cholangiocarcinoma and concomitant cirrhosis (F4) of undetermined aetiology. Levels of significance: ∗p <0.05, ∗∗ p <0.01, ∗∗∗p <0.001, ∗∗∗∗p <0.0001. ALD, alcoholic liver disease; FACS, fluorescence-activated cell sorting; HCC, hepatocellular carcinoma; HSC, hepatic stellate cell; MASLD, metabolic dysfunction-associated steatotic liver disease.
Discussion
CLD remains a global health issue. Additionally, the incidence of cirrhosis and HCC is estimated to increase in the following years which, at least in part, may be explained by an increase in the incidence of MASLD.2 Because liver fibrosis is a hallmark of CLD and it is strongly correlated with CLD prognosis, antifibrotics represent an attractive strategy for treating CLD. In particular, antifibrotic candidates that target HSC activation are of great interest because of the pivotal contribution of HSCs to the myofibroblast pool.11,21 To identify new putative targets for antifibrotic drugs, we have recently adopted a transcriptional dysregulation standpoint15 and defined a fibrogenic aHSC transcriptional profile that is strongly associated with fibrogenesis and ECM deposition. We hypothesised that a putative antifibrotic candidate should be present within this transcriptional profile, have enriched expression in HSCs and, for pharmaceutical targeting purposes, encode for a protein expressed at the cellular membrane. In this study, we demonstrated that Gpr176 fulfils these criteria as it (i) strongly correlated with fibrosis, (ii) had enriched expression in HSCs, (iii) encoded for a protein expressed at the cellular membrane, (iv) had no phenotype change when deleted in mice, but its absence resulted in a reduction of liver fibrosis. Altogether, this demonstrated that Gpr176 inhibition has potential as a treatment against CLD.
The identification of Gpr176 as HSC activation marker is not completely novel, as two studies have previously indicated that Gpr176 forms a part of large gene sets enriched in HSC activation: one as part of an HSC-specific gene signature (122 genes) that correlates with outcomes of HCC and hepatitis C and the other as part of a transcriptional program (236 genes) of HSC activation conserved between CCl4 and the western diet.22,23 However, these studies did not look further into Gpr176. In this study, we identified Gpr176 as an HSC initiation marker as its upregulation was detected in HSCs as early as 10 h after in vitro culture and 24 h after a single injection of CCl4 in mice. We also showed that culture-activated human HSCs expressed GPR176 protein and that GPR176/VIM double positive cells were only present in livers of patients with fibrosis. Thus, to the best of our knowledge, this study is the first to thoroughly describe the correlation between Gpr176 expression and its potential involvement in both liver fibrosis and HSC activation.
By adopting a similar transcriptome screening approach as that described by Zhang et al.,22 we identify the enriched expression of Gpr176 in mouse HSCs compared with other liver cell types and tissues. Using FACS-based isolation of human liver cell types, we also demonstrated the enriched expression of GPR176 in human HSCs when compared to LSECs, macrophages, cholangiocytes, and hepatocytes. Nonetheless, human cholangiocytes had intermediate levels of GPR176 when compared with non-HSC liver cell types. Of note, Doi et al. described a low expression of Gpr176 in many non-diseased mouse organs, including the liver.12 We acknowledge these results as, compared with aHSCs, qHSC and healthy liver tissue, exhibit very low expression of Gpr176.
By performing in vitro RNA interference experiments, we show that in vitro HSCs, at least partially, depend on Gpr176 for proper induction of activation markers. This profibrotic role for Gpr176 was confirmed in PCLS cultures and in Gpr176 knockout mice. Surprisingly, the expression of Acta2 seems to be increased in HSCs isolated from CCl4-treated knockout mice and the amount of a-SMA positive cells was not altered in fibrotic Gpr176 knockout mice (Fig. 4C, Fig. 5B). Currently we do not have a good explanation for this inconsistency. However, analysis of spatial transcriptomic data from mouse livers subjected to a MASLD diet24 (Fig. S6A) indicated that, although Gpr176 expression was rather low (probably because of the limited sequencing depth), all Gpr176-positive (Gpr176+) cells expressed Dcn, an HSC specific marker (Fig. S6A). To our surprise, we noted that Gpr176+ cells generally expressed higher levels of Col1a1 and lower levels of Acta2 (Fig. S6A). These results suggested that Gpr176+ activated HSCs might characterise a subset of Col1a1-expressing activated HSCs, and that the knockout of Gpr176 mainly influenced Col1a1-expressing cells and not Acta2-expressing cells.
Although we propose a profibrogenic role for GPR176 in murine models of fibrogenesis, further studies should focus on the mechanisms driving its function in HSCs. GPR176 is a class A orphan GPCR with a constitutive inhibitory effect on cAMP-responsive element (CRE) transcriptional activity for which no known endogenous ligand exists.25 GPR176 has an agonist-independent basal activity that represses cAMP production by its interaction with the unique G-protein subclass Gz, but not the canonical Gi.12 The antifibrotic properties of cAMP, either via PKA or Epac, have been well established in the literature.26 Some known downstream effects of cAMP/PKA signalling include phosphorylation of CREB and inhibition of RhoA activity, whereas Epac-1 can alleviate the TGF-β response in cardiac fibroblasts.27,28 All of these pathways have been implicated in HSC activation. Furthermore, we show that although qHSCs expressed GPR176 at its predicted 56 kDa molecular mass, we noted that during initiation (after the first 24 h), HSCs induced GPR176 protein expression accompanied by a shift to a higher molecular mass at approximately 75 kDa. A more recent study showed that GPR176 undergoes N-glycosylation, which increases its mass from 56 kDa to approximately 75 kDa. This N-glycosylation was required for GPR176 protein stability and proper cell surface expression, but not per se for its agonist-independent basal activity of repressing cAMP levels.19 Our results thus suggest early induction of Gpr176 transcription, leading to translation of GPR176 protein with post translational N-glycosylation that stabilises GPR176 and leads to an increase in GPR176 protein and cell surface expression. The reduction of cAMP levels can thus be hypothesised to be the driver of the profibrogenic activity induced by GPR176 protein. Further mechanistic studies should focus on how Gpr176 expression is regulated, how early N-glycosylation is established, and whether the increase in GPR176 protein effectively reduces cAMP levels in aHSCs.
A clinical value for GPR176 can only be achieved by identifying pharmaceutical agents that can target GPR176 and alter its function. GPCRs constitute the most successful group of pharmaceutical targets; however, the lack of known modulators for orphan GPCRs remains an important challenge. Although the rate of “deorphanisation” is slowing, structure-based methods and virtual ligand screening can provide means to identify endogenous or surrogate ligands. Herein, we describe a profibrotic role for GPR176 in CLD. Combined with previous studies by researchers, a more fundamental view on the interaction of GPR176 with its suitable heterotrimeric G protein, its signalling pathways, and consequently, its role in health and disease is emerging.12,19 This knowledge warrants further investigation on GPR176 to find endogenous or surrogate ligands suitable for pharmaceutical targeting strategies.29,30
In conclusion, we identified enhanced expression of GPR176 in both mouse and human activated HSCs and demonstrated a profibrotic role for this orphan GPCR. Our findings provide new insight into an orphan GPCR for which, currently, only limited information exists. Nonetheless, within the context of CLD, we identify GPR176 as a potential novel target for treating liver fibrosis.
Financial support
Fonds Wetenschappelijk Onderzoek (FWO) PhD mandate 1192920N to VDS; FWO Post-doc mandate 117300/12N5415N LV to IM; Gilead Sciences BeLux Fellowship 2020 grant (ID: 08309) to HR; FWO Post-doc mandate 1243121N to SV; FWO G030616N, G042719N, G071922N to LAvG.
Authors’ contributions
Conceptualization, writing – original draft and review and editing, data curation, formal analysis, investigation, visualization: VDS. Data curation, investigation: EG. Methodology, investigation: NE, LD, AS. Investigation: MKD, PL. Resources: NM. Funding acquisition: HR. Formal analysis, investigation, supervision, writing – review and editing: SV. Methodology, investigation, supervision, writing – review and editing: IM. Conceptualization, writing – review and editing, funding acquisition, project administration, supervision, resources: LAvG.
Data availability
All data is available through 10.5281/zenodo. 10528667.
Conflict of interest
This work was partially funded by Gilead Sciences through a BeLux Fellowship 2020 grant (ID: 08309).
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
We thank Jean Marc Lazou for flow cytometry cell sorting of the HSCs.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2024.101036.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Campana L., Esser H., Huch M., et al. Liver regeneration and inflammation: from fundamental science to clinical applications. Nat Rev Mol Cel Biol. 2021;22:608–624. doi: 10.1038/s41580-021-00373-7. [DOI] [PubMed] [Google Scholar]
- 2.Pimpin L., Cortez-Pinto H., Negro F., et al. Steering Committee. Burden of liver disease in Europe: epidemiology and analysis of risk factors to identify prevention policies. J Hepatol. 2018;69:718–735. doi: 10.1016/j.jhep.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 3.D’Amico G., Morabito A., D’Amico M., et al. Clinical states of cirrhosis and competing risks. J Hepatol. 2018;68:563–576. doi: 10.1016/j.jhep.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Llovet J.M., Kelley R.K., Villanueva A., et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;21(7):6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 5.GBD 2017 Cirrhosis Collaborators The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:245–266. doi: 10.1016/S2468-1253(19)30349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asrani S.K., Devarbhavi H., Eaton J., et al. Burden of liver diseases in the world. J Hepatol. 2019;70:151–171. doi: 10.1016/j.jhep.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Tacke F., Puengel T., Loomba R., et al. An integrated view of anti-inflammatory and antifibrotic targets for the treatment of NASH. J Hepatol. 2023;79:552–566. doi: 10.1016/j.jhep.2023.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwaisako K., Jiang C., Zhang M., et al. Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci U S A. 2014;111:E3297–E3305. doi: 10.1073/pnas.1400062111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mederacke I., Hsu C.C., Troeger J.S., et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trivedi P., Wang S., Friedman S.L. The power of plasticity-metabolic regulation of hepatic stellate cells. Cell Metab. 2021;33:242–257. doi: 10.1016/j.cmet.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kisseleva T., Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2021;18:151–166. doi: 10.1038/s41575-020-00372-7. [DOI] [PubMed] [Google Scholar]
- 12.Doi M., Murai I., Kunisue S., et al. Gpr176 is a Gz-linked orphan G-protein-coupled receptor that sets the pace of circadian behaviour. Nat Commun. 2016;7 doi: 10.1038/ncomms10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewyse L., De Smet V., Verhulst S., et al. Improved precision-cut liver slice cultures for testing drug-induced liver fibrosis. Front Med (Lausanne) 2022;9 doi: 10.3389/fmed.2022.862185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mannaerts I., Eysackers N., Anne van Os E., et al. The fibrotic response of primary liver spheroids recapitulates in vivo hepatic stellate cell activation. Biomaterials. 2020;261 doi: 10.1016/j.biomaterials.2020.120335. [DOI] [PubMed] [Google Scholar]
- 15.De Smet V., Eysackers N., Merens V., et al. Initiation of hepatic stellate cell activation extends into chronic liver disease. Cell Death Dis. 2021;12:1110. doi: 10.1038/s41419-021-04377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pradere J.P., Kluwe J., De Minicis S., et al. Hepatic macrophages but not dendritic cells contribute to liver fibrosis by promoting the survival of activated hepatic stellate cells in mice. Hepatology. 2013;58:1461–1473. doi: 10.1002/hep.26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asakawa M., Itoh M., Suganami T., et al. Upregulation of cancer-associated gene expression in activated fibroblasts in a mouse model of non-alcoholic steatohepatitis. Sci Rep. 2019;20(9) doi: 10.1038/s41598-019-56039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mannaerts I., Leite S.B., Verhulst S., et al. The Hippo pathway effector YAP controls mouse hepatic stellate cell activation. J Hepatol. 2015;63:679–688. doi: 10.1016/j.jhep.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Wang T., Nakagawa S., Miyake T., et al. Identification and functional characterisation of N-linked glycosylation of the orphan G protein-coupled receptor Gpr176. Sci Rep. 2020;10:4429. doi: 10.1038/s41598-020-61370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Grunsven L.A. 3D in vitro models of liver fibrosis. Adv Drug Deliv Rev. 2017;121:133–146. doi: 10.1016/j.addr.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Huisman T.M., Dieterich D.T., Friedman S.L. Experimental and investigational targeted therapies for the management of fibrosis in NASH: an update. J Exp Pharmacol. 2021;13:329–338. doi: 10.2147/JEP.S265286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D.Y., Goossens N., Guo J., et al. A hepatic stellate cell gene expression signature associated with outcomes in hepatitis C cirrhosis and hepatocellular carcinoma after curative resection. Gut. 2016;65:1754–1764. doi: 10.1136/gutjnl-2015-309655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcher A.B., Bendixen S.M., Terkelsen M.K., et al. Transcriptional regulation of hepatic stellate cell activation in NASH. Sci Rep. 2019;9:2324. doi: 10.1038/s41598-019-39112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guilliams M., Bonnardel J., Haest B., et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185:379–396.e38. doi: 10.1016/j.cell.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin A.L., Steurer M.A., Aronstam R.S. Constitutive activity among orphan class-a G protein coupled receptors. PLoS One. 2015;10 doi: 10.1371/journal.pone.0138463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Insel P.A., Murray F., Yokoyama U., et al. cAMP and Epac in the regulation of tissue fibrosis. Br J Pharmacol. 2012;166:447–456. doi: 10.1111/j.1476-5381.2012.01847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sohail M.A., Hashmi A.Z., Hakim W., et al. Adenosine induces loss of actin stress fibers and inhibits contraction in hepatic stellate cells via Rho inhibition. Hepatology. 2009;49:185–194. doi: 10.1002/hep.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yokoyama U., Patel H.H., Lai N.C. The cyclic AMP effector Epac integrates pro- and antifibrotic signals. Proc Natl Acad Sci U S A. 2008;105:6386–6391. doi: 10.1073/pnas.0801490105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngo T., Kufareva I., Coleman J., et al. Identifying ligands at orphan GPCRs: current status using structure-based approaches. Br J Pharmacol. 2016;173:2934–2951. doi: 10.1111/bph.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laschet C., Dupuis N., Hanson J. The G protein-coupled receptors deorphanization landscape. Biochem Pharmacol. 2018;153:62–74. doi: 10.1016/j.bcp.2018.02.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available through 10.5281/zenodo. 10528667.