Abstract
Myeloid neoplasms arise from preexisting clonal hematopoiesis (CH); however, the role of CH in the pathogenesis of acute lymphoblastic leukemia (ALL) is unknown. We found that 18% of adult ALL cases harbored TP53, and 16% had myeloid CH-associated gene mutations. ALL with myeloid mutations (MyM) had distinct genetic and clinical characteristics, associated with inferior survival. By using single-cell proteogenomic analysis, we demonstrated that myeloid mutations were present years before the diagnosis of ALL, and a subset of these clones expanded over time to manifest as dominant clones in ALL. Single-cell RNA sequencing revealed upregulation of genes associated with cell survival and resistance to apoptosis in B-ALL with MyM, which responds better to newer immunotherapeutic approaches. These findings define ALL with MyM as a high-risk disease that can arise from antecedent CH and offer new mechanistic insights to develop better therapeutic and preventative strategies.
Significance:
CH is a precursor lesion for lymphoblastic leukemogenesis. ALL with MyM has distinct genetic and clinical characteristics, associated with adverse survival outcomes after chemotherapy. CH can precede ALL years before diagnosis, and ALL with MyM is enriched with activated T cells that respond to immunotherapies such as blinatumomab.
INTRODUCTION
Acute lymphoblastic leukemia (ALL) represents 20% of adult leukemias (1). Although the cure rate of pediatric ALL is >90%, older adults with ALL have inferior long-term survival due to the adverse molecular characteristics of their disease and the overall frailty that precludes the use of intensive pediatric therapy (2, 3). It is imperative to understand the unique leukemogenic pathways driving ALL in adults in order to lessen the outcome disparity noted with increasing age. Yet, existing data on the genetics of ALL are biased toward ALL in children or young adults (<40 years) due to the underrepresentation of older adults in prospective therapeutic trials (4–12).
Clonal hematopoiesis (CH) is characterized by the presence of somatic genomic alterations in the blood cells of individuals without a hematologic malignancy (13, 14). The most frequent somatic mutations involve genes that are drivers of myeloid neoplasms (e.g., DNMT3A, TET2, ASXL1, TP53; ref. 15). CH is a well-characterized precursor lesion for the development of myeloid neoplasms (15–19). Recently, a different set of somatic gene mutations, defined as lymphoid CH, were shown to predict risk for lymphoid malignancies such as chronic lymphocytic leukemia or lymphomas (20). The prevalence of both myeloid and lymphoid CH increases with aging, but their role in the pathogenesis of ALL is unknown. We have previously shown that therapy-related ALL arising after exposure to genotoxic therapies (i.e., chemotherapy or radiation used for the treatment of solid tumors or autoimmunity) is enriched for mutations associated with myeloid CH, similar to what has been observed in therapy-related myeloid neoplasms (21). Based on these preliminary data, we hypothesized that CH constitutes a fertile genomic background for acute lymphoblastic leukemogenesis. Here, we performed comprehensive genetic profiling of 400 adult ALL cases and found that 18% of patients harbored TP53, and 16% of patients harbored at least one myeloid gene mutation. Through a more detailed characterization of cellular subtypes and clonal evolution with single-cell DNA and protein multiomics analysis, we showed that myeloid CH mutations were present in both myeloid cells and malignant lymphoblasts. Single-cell transcriptomic profiling of pretreatment samples revealed upregulation of genes associated with cell survival and resistance to apoptosis in B-ALL with myeloid mutations (MyM) when compared with B-ALL without MyM. Clinically, ALL with MyM had lower remission rates after cytotoxic chemotherapy, accounting for dismal survival rates. Newer immunotherapy approaches (e.g., blinatumomab) were more effective in B-ALL with MyM, which correlated with the activated T-cell signatures that we identified in single-cell studies.
RESULTS
ALL with MyM Is a Distinct Clinicopathologic Entity with Adverse Outcomes
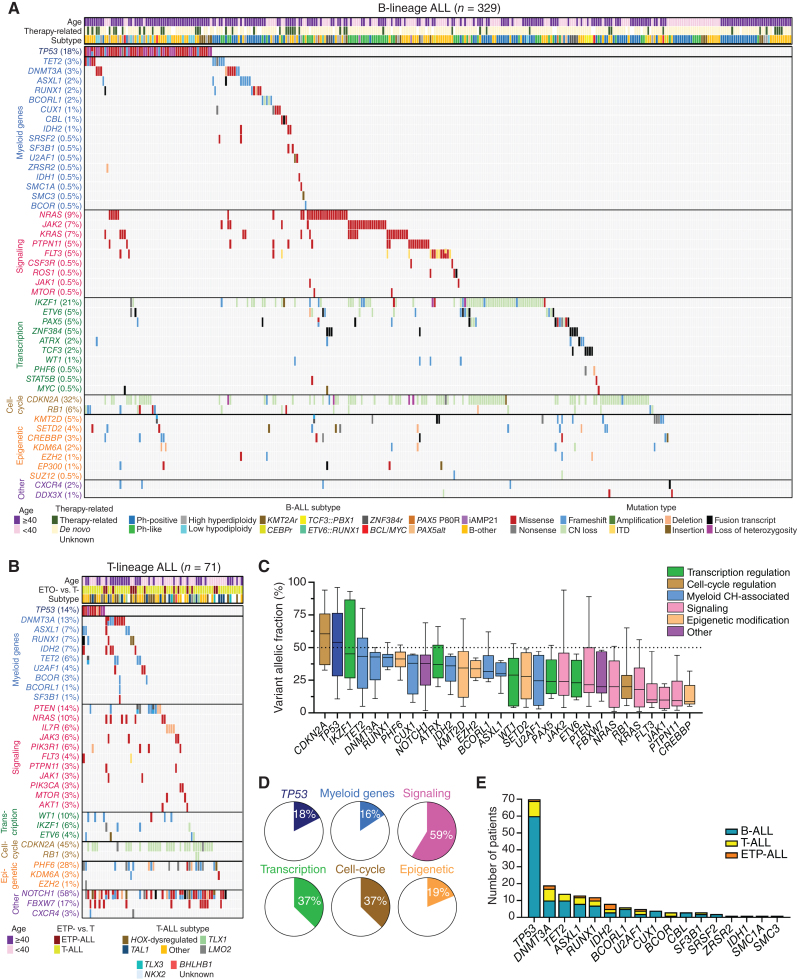
We studied the genetics and clinical outcomes of 400 adult ALL cases (329 with B-lineage ALL and 71 with T-lineage ALL). Using cytogenetic, fluorescence in-situ hybridization (FISH), DNA-seq, RNA sequencing (RNA-seq), and chromosome microarray analysis (CMA) data, we classified B- and T-lineage ALL into different subtypes, as recommended by the 2022 World Health Organization (WHO) and International Consensus Classification (ICC) criteria, excluding provisional entities that were not captured with our clinical assays (Supplementary Fig. S1; Supplementary Tables S1 and S2; refs. 22, 23). Our cohort was enriched with older adults since 60% of patients were ≥40 years of age (Supplementary Tables S3 and S4). Most common B-ALL subtypes included BCR::ABL1 (29%), BCR::ABL1-like (16%), low hypodiploid/near haploid (LH/NH, <40 chromosomes; 10%), KMT2A-rearranged (4%), and high hyperdiploid (>50 chromosomes; 3%), whereas 30% of cases were classified as B-other (Supplementary Table S5). Most common T-lineage ALL subtypes included HOX-dysregulated (24%), TAL1-rearranged (16%), and TLX1-rearranged (6%) T-ALL, whereas 34% of cases were classified as T-other (Supplementary Table S6). Bulk sequencing of pretreatment blood or bone marrow samples identified somatic TP53 mutations in 18% of B-ALL and 14% of T-lineage ALL cases. In addition, 13% of B-ALL and 27% of T-lineage ALL cases had at least one mutation in 16 myeloid CH-associated genes (Fig. 1A and B). Mutations involving lymphoid CH genes were detected in 7% of ALL cases (Supplementary Fig. S2). The median variant allelic frequency (VAF) of myeloid mutations across all ALL cases was 36% (Fig. 1C). Given the very high blast percentage of ALL diagnostic bone marrow samples (≥45% blasts in all cases, and ≥70% blasts in 80% of the cases), this high VAF suggested that myeloid mutations were present in most lymphoblasts (Supplementary Fig. S3). In contrast, mutations involving signaling pathway genes showed considerably lower VAFs, indicating that they were often present only in a subpopulation of the sequenced cells. The most frequently mutated myeloid genes were DNMT3A (5%), TET2 (4%), ASXL1 (3%), RUNX1 (3%), and IDH2 (2%; Fig. 1D and E).
Figure 1.
Myeloid mutations are frequent in adult patients with ALL. A and B, Oncoprints showing the spectrum of mutations and cytogenetic abnormalities in adult patients with B-lineage and T-lineage ALL. C, The box plot shows the median, 25th and 75th percentiles, and minimum and maximum variant allelic frequency (VAF) observed across the entire cohort of 400 patients. Boxes are colored according to the functional category assigned to each gene. The black dashed line marks an allele frequency of 50%, the expected VAF for a heterozygous variant present in all cells in the specimen. D, The frequencies of mutations for the six main categories of gene groups for the entire cohort. E, The distribution of mutations in 17 myeloid genes across the ALL cases of different lineages.
We also performed a skin biopsy and fibroblast culture for germline testing in 58 patients who had a clinical indication (Supplementary Fig. S4A–S4F). The most frequently detected pathogenic/likely pathogenic (P/LP) inherited variants involved CHEK2 (n = 8), MUTYH (n = 3), and BRCA2 (n = 2) genes (Supplementary Table S7). Among 70 patients with TP53-mutated ALL, only one was confirmed to have it in germline.
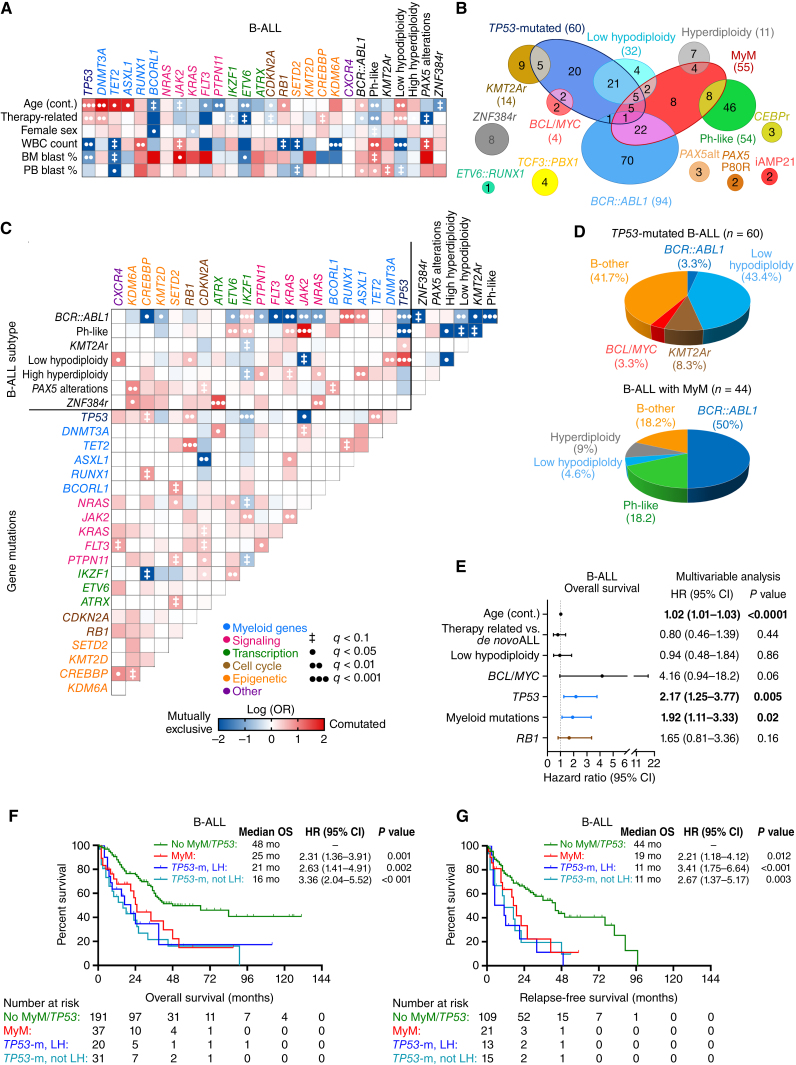
We investigated the associations of gene mutations occurring in five or more patients with baseline clinical characteristics, WHO/ICC subtype, and other gene mutations in B-ALL. Myeloid mutations were more common in older adults and patients with therapy-related B-ALL (Fig. 2A). In contrast, mutations involving signaling pathways or transcription factors implicated in pediatric ALL were less common in older adults and those with therapy-related B-ALL. Mutations in RUNX1 and ASXL1 co-occurred with BCR::ABL1 positivity, which was associated with the p210 transcript in 30% of these cases (Fig. 2B and C). Among 32 patients with LH/NH, 26 (81%) had concomitant TP53 mutation, but 57% of TP53-mutated ALL cases did not have LH/NH (Fig. 2B and D; Supplementary Fig. S5A and S5B). TP53 mutations frequently cooccurred with mutations in TET2 and RB1 genes. B-ALL subtypes that were mutually exclusive with myeloid mutations included ZNF384r, TCF3::PBX1, PAX5-alt and PAX5 P80R, CEBPr, iAMP21, and ETV6::RUNX1.
Figure 2.
Clinical and molecular characterization of B-lineage ALL with MyM. A, Associations between gene mutations and clinical characteristics were studied for gene mutations found in ≥5 patients, and statistical significance was assessed using the Fisher exact test for categorical variables and the Wilcoxon rank-sum test for continuous variables, with adjustment for multiple testing. Only those pairings that were significant at an adjusted q < 0.1 are shown. The OR of the association is color-coded, and the significance level is indicated by the symbol in each field. Shades of red indicate a positive association (i.e., two characteristics that frequently occurred together, or for the association between a mutation and a continuous variable, a higher value in those carrying the mutation). Shades of blue indicate a negative association (i.e., two characteristics that rarely occurred together, or for the association between a mutation and a continuous variable, a lower value in those carrying the mutation). B, Venn diagram showing the relationship between WHO/ICC-established B-ALL subtypes and myeloid mutations. C, Pairwise associations between gene mutations and cytogenetic abnormalities. D, Pie charts showing the distribution of WHO/ICC-established B-ALL subtypes in TP53-mutated B-ALL and B-ALL with MyM. E, Multivariable Cox regression analysis of OS in B-ALL, adjusting for the variables that were significant in univariable analysis (age, therapy-related ALL, TP53 mutation, MyM, and RB1 mutation). F and G, Kaplan–Meier OS and relapse-free survival (RFS) analysis of B-ALL patients stratified into three groups based on their MyM and low hypodiploidy (LH) status. Cox proportional hazards ratios were calculated. BM, bone marrow; PB, peripheral blood; WBC, white blood cell.
Given the high frequency of TP53 mutations in adults with ALL, and its role as both a CH mutation and a common cancer tumor suppressor gene, we stratified patients with ALL into three categories: TP53-mutated ALL, ALL with MyM, and ALL without MyM/TP53. In univariable analysis of overall survival (OS) in B-ALL, age, therapy-related B-ALL, low hypodiploidy, BCL/MYC subtype, TP53 mutation status, presence of MyM, and RB1 mutation were associated with lower OS (Supplementary Fig. S6A). In multivariable Cox regression analysis, age (HR for each year: 1.02 with age as a continuous variable; 95% CI, 1.01–1.03), TP53 mutation (HR: 1.95; 95% CI, 1.2–3.1), and MyM (HR: 1.95; 95% CI, 1.1–3.3) were independent predictors of poor OS in B-ALL (Fig. 2E). When stratified based on age ≥40 or ≥60 years, older adults with B-ALL and MyM had worse OS than patients without MyM/TP53 (Supplementary Fig. S6B and S6C). Given the association between TP53 mutation and LH/NH, we further divided TP53-mutated B-ALL into LH/NH and no LH/NH subgroups. Both TP53-mutated B-ALL groups (with or without LH/NH), and B-ALL with MyM had inferior survival when compared with patients without MyM/TP53 (Fig. 2F and G). The adverse prognostic impact of TP53 and MyM was observed in Ph-negative B-ALL patients treated with different chemotherapy regimens (Supplementary Fig. S7A–S7D). The independent prognostic significance of age, TP53, and MyM was evident in multivariable analysis after adjusting for the type of treatment received (Supplementary Table S8).
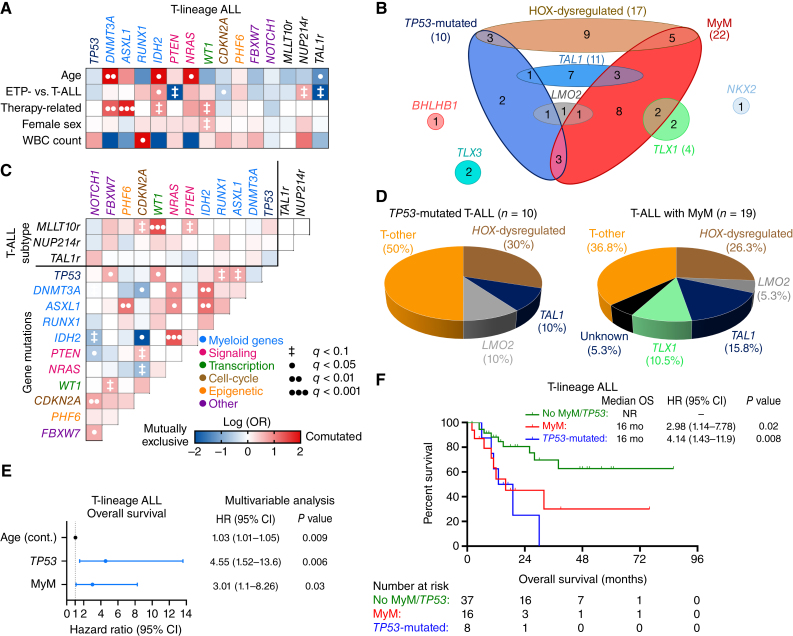
In patients with T-lineage ALL, MyM were observed in 27% of cases, and most common mutations involved DNMT3A (13%), ASXL1 (7%), RUNX1 (7%), and IDH2 (7%) genes (Fig. 1B). These mutations often coexisted, and were more prevalent in older adults and patients with therapy-related T-lineage ALL (Fig. 3A–C). TP53 and MyM were seen in 30% of HOX-dysregulated T-lineage ALL cases, but the majority of these cases were classified as T-other (Fig. 3D). T-lineage ALL with MyM had lower rates of complete remission (CR) without measurable residual disease (MRD) and OS (Supplementary Fig. S8A–S8D). Mutations in TP53 and myeloid genes were independent predictors of poor OS, whereas T-lineage ALL patients without MyM/TP53 had better OS (Fig. 3E and F).
Figure 3.
Clinical and molecular characterization of T-lineage ALL with MyM. A, Associations between gene mutations and clinical characteristics were studied for gene mutations found in ≥5 patients. B, Venn diagram showing the relationship between WHO/ICC-established T-lineage ALL subtypes and MyM. C, Pairwise associations between gene mutations. D, Pie charts showing the distribution of WHO/ICC-established T-lineage ALL subtypes in TP53-mutated T-ALL and T-ALL with MyM. E, Multivariable Cox regression analysis of OS in T-lineage ALL, adjusting for the variables that were significant in univariable analysis (age, TP53, and DNMT3A mutations). F, Kaplan–Meier OS analysis of T-lineage ALL patients stratified into three groups based on their MyM status. Cox proportional hazards ratios were calculated. BM, bone marrow; PB, peripheral blood; WBC, white blood cell.
Clonal Architecture of ALL with MyM
Based on the data showing high allelic frequency of MyM in ALL, we next investigated whether these mutations were present in the dominant (or founder) clone. Using simultaneous single-cell DNA and protein sequencing, we investigated the distribution of mutations in different cellular subsets in five different ALL samples (Supplementary Table S9). For these analyses, cells were stained with the oligo-conjugated antibody panel, and sequenced with the Tapestri Myeloid Panel (Methods). Cells were clustered by immunophenotype, and pathogenic mutations were overlayed onto individual cells of these clusters (Fig. 4A). For example, data from 4,180 cells of B-ALL1 were clustered based on surface marker expression, and two pathogenic mutations involving IDH2 and PTPN11 genes were overlayed onto the cells of 8 clusters (Fig. 4B and C; Supplementary Fig. S9A). Most of the mature lymphoid cells residing in T-, B-, and NK-cell clusters were wild-type, whereas B-lymphoblasts and myeloid cells shared the IDH2 p.R140Q mutation (Fig. 4D). PTPN11 mutation was more prevalent in B-lymphoblasts. Similarly, in additional B-ALL, T-ALL, and early T-precursor (ETP)-ALL cases, myeloid or TP53 mutations were shared between lymphoblasts and myeloid cells (Fig. 4E–M; Supplementary Figs. S9B–S9D and S10A–S10C). Mature lymphoid cells of these patients were often wild-type for the respective genes, but mutations could be detected at an allelic frequency <5%, as confirmed by DNA sequencing of sorted cell populations (Supplementary Fig. S11A and S11B). These data indicate that myeloid mutations are ancestral events for ALL with MyM, and these lesions arise in hematopoietic stem or progenitor cells that give rise to both lymphoblasts and myeloid cells.
Figure 4.
Single-cell DNA and protein sequencing to study the clonal architecture of ALL with MyM. A, Surface protein heat map of the canonical cell type–specific markers as measured by the Wilcoxon rank-sum test. B, Uniform manifold approximation and projection (UMAP) plot of B-ALL1 sample with cells clustered by immunophenotype. C, Gene mutations overlayed into the individual cells of immunophenotypically defined clusters. Genotype of a given cell is color-coded to indicate wild-type (WT) vs. homozygous (HOM) or heterozygous (HET) mutation status. Black dots indicate cells with unknown genotype. D, Graph showing the fractions of mutated cells in each cluster of B-ALL1. E–G, UMAP plots and mutation distributions in different immunophenotypic compartments of B-ALL3. H–J, UMAP plots and mutation distributions in different immunophenotypic compartments of ETP-ALL1. K–M, UMAP plots and mutation distributions in different immunophenotypic compartments of ETP-ALL2.
ALL Evolves from Antecedent CH
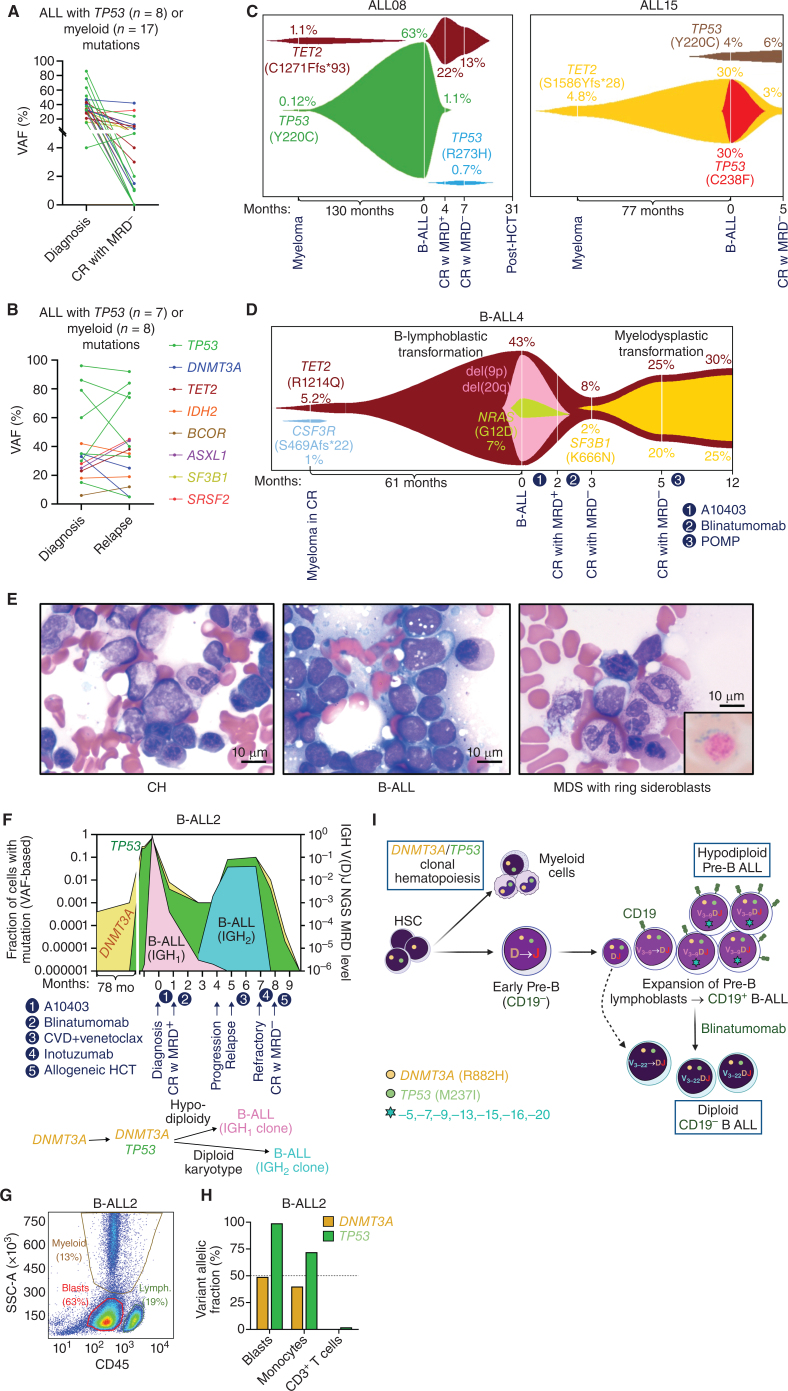
We next sought to determine whether myeloid CH mutations can be detected after eradication of ALL or as a precursor lesion years before the diagnosis of ALL. First, we showed that both MyM and TP53 mutations persist as CH mutations in patients who achieve MRD-negative CR after treatment of their ALL (Fig. 5A). In addition, these mutations persist or expand when patients experience relapse of their ALL, indicating their stability over the course of disease evolution (Fig. 5B; Supplementary Fig. S12A–S12F).
Figure 5.
Evolution of ALL from preexisting CH. Analysis of serial diagnosis and MRD-negative CR (A), as well as diagnosis and relapse (B) bone marrow samples revealed the persistence of myeloid CH mutations. C, FISH plots demonstrate that myeloid CH mutations were detectable years before the diagnosis of ALL and presented as dominant clones at ALL diagnosis. D, FISH plot showing the clonal evolution of therapy-related B-ALL and therapy-related myelodysplastic syndrome (MDS) from the same TET2 CH clone at different time points. E, Bone marrow aspirate smears for CH, B-ALL, and MDS with ring sideroblasts. Scale bar, 10 μm. F, Clonal evolution of B-ALL2 from preexisting DNMT3A CH clone, which was detectable 6 years before the diagnosis of DNMT3A/TP53-mutant ALL. G and H, Flow-cytometric analysis of different cellular compartments in B-ALL2, which were genotyped after cell sorting. CH mutations (DNMT3A and TP53) were shared between lymphoblasts and myeloid cells. I,DNMT3A/TP53-mutated, D-J rearranged early pro-B cell gave rise to hypodiploid CD19+ B-ALL2. After treatment with blinatumomab, a CD19-negative clone with a different V(D)J sequence emerged from the earlier diploid pro-B cell with CH mutations. CR, complete remission; HSC, hematopoietic stem cell; MRD, measurable residual disease. (I, Created with BioRender.com.)
We performed DNA sequencing on serial bone marrow samples from patients with therapy-related B-ALL, which were obtained more than five years before diagnosis of ALL (as part of myeloma surveillance after autologous stem cell transplantation), at the time of ALL diagnosis, and at subsequent follow-up (Fig. 5C; Supplementary Fig. S13). In ALL08, TP53 p.Y220C and TET2 p.C1271Ffs*93 CH clones were detectable in the marrow sample obtained 10 years before the diagnosis of ALL. The TP53 clone was the dominant clone in subsequent therapy-related B-ALL, whereas the TET2 clone was not detectable at 0.01% sensitivity. Following treatment, the B-ALL entered MRD-negative remission, and the TET2 clone reemerged in myeloid cells in the form of CH. In ALL15, TET2 p.S1586Yfs*28 CH clone was detectable 6 years before the diagnosis of ALL. This clone acquired a TP53 C238F mutation as it evolved into therapy-related B-ALL. After CR without MRD, a different TP53 clone was detectable in the form of myeloid CH.
We analyzed serial pre- and post-leukemia samples to decipher the evolution of sequential therapy-related ALL and myelodysplastic syndrome (MDS) diagnosed in one patient (B-ALL4; Fig. 5D and E). We were able to detect two different CH clones with TET2 and CSF3R mutations 5 years before the diagnosis of B-ALL. TET2 CH clone progressed to acquire additional genetic events including del(9p), del(20q), and NRAS mutation, resulting in Ph-negative B-ALL. Interestingly, following ALL-directed therapy resulting in CR without MRD, a TET2/SF3B1-mutant myeloid clone expanded, giving rise to MDS with ring sideroblasts.
In another patient (B-ALL2) with DNMT3A/TP53-mutant therapy-related B-ALL, the myeloid mutations were founder events and ALL was characterized by low hypodiploidy, including loss of heterozygosity for TP53 mutation (Fig. 5F). A DNMT3A p.R882H CH clone was detectable 6 years prior to the diagnosis. B-lymphoblast, CD14+ monocyte, and CD3+ T-cell populations were isolated by flow cytometry and cell sorting of the diagnostic peripheral blood sample (Fig. 5G; Supplementary Fig. S14A). As expected, lymphoblast and myeloid compartments shared the DNMT3A and TP53 mutations, whereas mature lymphoid cells were wild-type (Fig. 5H). When this patient experienced a recurrence of their ALL, DNMT3A and TP53 mutations were persistent, but the disease was diploid without del(17p), and blasts harbored a different IGH V-DJ sequence from original diagnosis (Supplementary Fig. S14B–S14D). High-sensitivity NGS analysis showed that diagnosis and relapse clones shared the same DJ sequence, rearranged with V3–9 at the diagnostic sample (IGH1) and V3–22 at the relapse sample (IGH2). The CD19-negative IGH2 clone was not detectable at diagnosis at 10−6 sensitivity, but became the dominant clone after blinatumomab therapy. These data suggest the emergence of two separate B-ALL clones from an early progenitor B cell with DJ rearrangement, which originated from a DNMT3A/TP53 mutated hematopoietic stem cell (Fig. 5I).
Defining the Interplay Between Lymphoblasts and the Immune Microenvironment to Study Chemotherapy Resistance in ALL with MyM
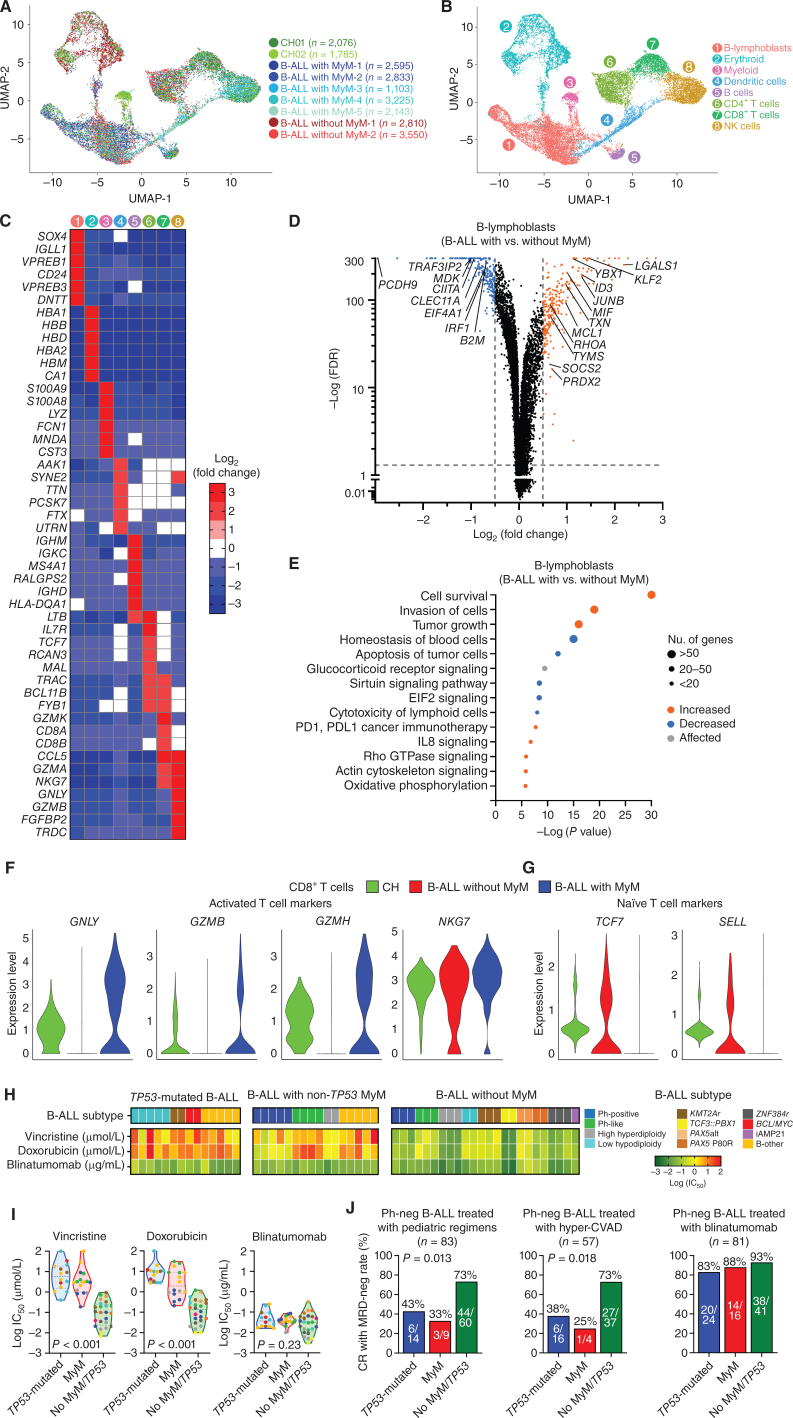
To understand the mechanisms driving therapeutic resistance in Ph-negative B-ALL with MyM, we performed single-cell RNA-seq (scRNA-seq) of mononuclear bone marrow specimens taken at disease diagnosis from age/sex-matched five patients with B-ALL with MyM, two patients with B-ALL without MyM, and two individuals with myeloid CH (Fig. 6A; Supplementary Fig. S15A, Supplementary Table S10). We analyzed 22,120 cells that were processed using the Seurat toolkit utilizing its anchoring-based integration method in order to account for the biological and technical variation between samples, followed by uniform manifold approximation and projection (UMAP) dimensionality reduction for visualization (Methods; refs. 24, 25). We performed extensive unbiased clustering of all bone marrow cells to identify 8 broad hematopoietic cell clusters: B-lymphoblasts, erythroid cells, myeloid cells, dendritic cells, B cells, CD4+ T cells, CD8+ T cells, and NK cells (Fig. 6B). These clusters were classified based on the relative expression levels of gene signatures encompassing lineage-specific transcription factors, surface markers, and effector genes (Fig. 6C; Supplementary Fig. S15B).
Figure 6.
B-lymphoblasts are intrinsically resistant to cytotoxic chemotherapy, but susceptible to immunotherapies in B-ALL with MyM. A, Uniform manifold approximation and projection (UMAP) visualization of 22,120 individual cells from 9 individual primary thawed mononuclear bone marrow samples taken from patients with CH (n = 2), B-ALL with MyM (n = 5), and B-ALL without MyM (n = 2). B, Marker-based cell-type identification analysis allowed the prediction of eight broad hematopoietic cell types across all profiled single cells. C, Gene-expression heat map of the top six cell type–specific marker genes as measured by Wilcoxon rank-sum test. D, Volcano plot showing differential expression of genes in B-lymphoblasts from B-ALL with MyM vs. B-ALL without MyM. E, Major pathways predicted to be differentially regulated in B-lymphoblasts from B-ALL with MyM vs. B-ALL without MyM, assessed by Ingenuity Pathway Analysis. F and G, Relative expression of activated vs. naïve T-cell markers between patients with CH, B-ALL without MyM, and B-ALL with MyM. H and I, Heat maps and violin plots showing log(IC50) values for primary human B-ALL samples with TP53 mutation (n = 14), non-TP53 MyM (n = 16), and no MyM (n = 24), treated with vincristine, doxorubicin, and blinatumomab at escalating doses. J, Rates of measurable residual disease (MRD)-negative complete remission (CR) in B-ALL patients with vs. without MyM treated with cytotoxic chemotherapy (vincristine and doxorubicin) and blinatumomab. HyperCVAD, combination chemotherapy consisting of cyclophosphamide, vincristine, adriamycin, and dexamethasone.
We interrogated transcriptomic differences of B-lymphoblasts between B-ALL with MyM and B-ALL without MyM (Fig. 6D). Genes with a log2 fold change >0.5 or < −0.5, and an adjusted P value <0.05 were considered as differentially expressed and were investigated by Ingenuity Pathway Analysis (Ingenuity Systems Inc.). In B-lymphoblasts with MyM, genes regulating cell survival, invasion, and tumor growth were upregulated, whereas genes regulating apoptosis, blood cell homeostasis, sirtuin signaling, EIF2 signaling, and lymphoid cell cytotoxicity were downregulated (Fig. 6E; Supplementary Fig. S15C and S15D, Supplementary Table S11).
Within the T-cell populations of three groups (CH, B-ALL with MyM, B-ALL without MyM), the gene signature of activated T cells was most pronounced in B-ALL with MyM, highlighted by the upregulation of GNLY, GZMB, GZMH, and NKG7 in their cytotoxic T lymphocytes (Fig. 6F). In contrast, naïve T-cell markers (e.g., TCF7 and SELL) were more prominent in B-ALL patients without MyM and individuals with CH (Fig. 6G). By in vitro cell viability assay of 54 adult B-ALL samples, we demonstrated higher IC50 values for cytotoxic chemotherapy agents (vincristine and doxorubicin) in B-ALL with MyM when compared with B-ALL without MyM/TP53 (Fig. 6H and I; Supplementary Fig. S16). Blinatumomab was effective in a MyM/TP53-agnostic manner (Supplementary Figs. S16 and S17). Similarly, CR with MRD-negativity rates after standard cytotoxic chemotherapy (e.g., pediatric-inspired or hyper-CVAD) were higher in Ph-negative B-ALL patients without MyM/TP53, and lower in B-ALL patients with MyM or TP53-mutated B-ALL (Fig. 6J). The inferior chemotherapy responses in B-ALL with MyM were independent of age, gender, or study cohort site (Supplementary Fig. S18A and S18B). However, B-ALL with MyM cases had high CR with MRD-negativity rates when treated with blinatumomab or inotuzumab (Fig. 6J; Supplementary Fig. S18C).
To investigate the mechanisms of similar blinatumomab sensitivity between B-ALL with MyM, TP53-mutated B-ALL, and B-ALL without MyM/TP53, we compared the transcriptomes of blasts by using validated blinatumomab response signatures (26). Among MHC class I antigens, HLA-A and HLA-C levels were similar between B-ALL with and without MyM/TP53 (Supplementary Fig. S19A). HLA-B levels and certain class II antigens (HLA-DP, HLA-DQB1, HLA-DRA, and HLA-DRB1) were downregulated in B-ALL with MyM. Using a previously validated gene signature of blinatumomab resistance in adult B-ALL, we compared B-ALL blasts with and without MyM/TP53 (Supplementary Fig. S19B). We did not observe any significant differences in these genes, which supports the observed in vitro and clinical efficacy of blinatumomab in all B-ALL groups.
These data suggest that in B-ALL with MyM, lymphoblasts are intrinsically more resistant to cytotoxic chemotherapy due to the upregulated cell survival and apoptotic resistance pathways, whereas T cells have an activated phenotype which translates into increased efficacy of antibody-based therapies such as blinatumomab and inotuzumab.
DISCUSSION
Our findings establish ALL arising from myeloid CH as a unique entity with distinct genetic and clinical characteristics. ALL with MyM is more frequent in older adults and patients with therapy-related ALL and is associated with poor survival outcomes. Using a comprehensive single-cell multiomics approach (DNA, RNA, and protein), we showed that TP53 and non-TP53 CH can precede ALL years before diagnosis and that the founder clone for ALL with MyM is a multipotent stem cell that can also differentiate into myeloid cells with CH mutations. By providing insights into the potential of CH-associated lymphoblastic leukemogenesis, we bring new perspectives for future studies investigating leukemia prevention in persons with CH, as well as novel treatment approaches for this high-risk ALL subset.
The pinnacle of modern oncology has been the cure of pediatric ALL with multiagent cytotoxic chemotherapy. Unfortunately, this approach has not made an impact on a similar scale for older adults with ALL. It is well recognized that older adults and patients with therapy-related ALL have lower response and inferior OS when treated with cytotoxic chemotherapy (2, 27–29). However, the introduction of new immunotherapies such as antibody–drug conjugates (inotuzumab), bispecific T-cell engagers (blinatumomab), and chimeric antigen receptor T (CAR-T) cell therapies improved outcomes in adults with B-ALL (30–32). Our results indicate that when compared with B-ALL patients without MyM/TP53, tumor cells from B-ALL patients with MyM are intrinsically resistant to chemotherapy, which is characterized by a prosurvival gene-expression signature that translates into high rates of first-line treatment failure. These patients may benefit from newer immunotherapy or chemoimmunotherapy combination approaches, as well as therapies targeted toward CH mutations (e.g., IDH inhibitors).
There is an emerging field of leukemia prevention that resulted from our improved understanding of CH and its long-term consequences (33). To that end, findings from our study can make a timely impact by establishing CH as a risk factor for ALL (in addition to myeloid neoplasms). Data on precursor lesions for ALL are limited, but recent reports showed that TP53 mutations can be detected as preleukemic events in adults with ALL (34–36). TP53 mutations frequently co-occur with low hypodiploidy in B-ALL, a subset with poor OS. We identified a subgroup of TP53-mutated ALL cases without low hypodiploidy, which had equally poor long-term OS when compared with TP53-mutated low hypodiploid cases. Similarly, preexisting aneuploid cells could be found in cord blood samples from pediatric cases of ALL (37). We also showed evidence for CH as a precursor lesion to two acute leukemias (AML and ALL, or two different ALL clones) in the same individual. This was also described in a case report of IDH1-mutated AML and ALL arising consecutively in the same patient (38). These data suggest that CH can be a reservoir for lymphoblastic leukemogenesis in adults. Therefore, future studies should explore therapeutic interventions for individuals with CH who are at high risk for ALL (e.g., myeloma patients receiving long-term lenalidomide therapy; ref. 39) that may potentially delay or even prevent the development of ALL.
METHODS
Study Cohort
Adult patients with ALL treated at the University of Chicago (n = 183) and Moffitt Cancer Center (n = 217) between 2014 and 2021 were studied. We also studied serial pre- and post-ALL samples of additional patients from the City of Hope and Dana-Farber Cancer Institute. Diagnosis, relapse, and disease status were confirmed and assigned according to WHO criteria (40). Patients with mixed phenotype acute leukemia were excluded. Therapy-related ALL was defined as ALL occurring after exposure to genotoxic therapy (i.e., chemotherapy or radiotherapy given for management of solid tumor, lymphoma, multiple myeloma, or autoimmune disease). The early T-precursor (ETP)-ALL immunophenotype was defined as follows: (i) absent (<5% positive cells) CD1a and CD8 expression, (ii) absent or dim (<75% positive cells) CD5 expression, and (iii) expression (>25% positive cells) of 1 or more stem cell (CD34 and HLA-DR) or myeloid (CD11b, CD13, CD33, and CD117) antigens. CR was defined as <5% bone marrow blasts, no circulating blasts or extramedullary disease, peripheral blood neutrophil counts > 1 × 109/L, and platelet counts > 100 × 109/L. MRD was assessed with three different methods as applicable: (i) multiparameter flow cytometry (sensitivity of 0.01%; ref. 41); (ii) reverse transcriptase polymerase chain reaction to detect BCR::ABL1 transcripts in patients with Ph+ B-ALL (sensitivity of 0.001%); (iii) V(D)J-based NGS MRD to detect IGH and TR sequences (sensitivity of 0.0001%) was performed with the ClonoSEQ platform (Adaptive Technologies) as previously reported (42). CR with MRD negativity was defined as the absence of any detectable disease with any of these three methods. MRD was assessed postinduction and postconsolidation, followed by every 3 months monitoring for 2 years for patients in remission. The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Boards of participating sites. Written informed consent was obtained from patients.
Genomic Profiling and Molecular Subtyping
Subtype classification for 400 ALL cases was based on the use of multiple assays, including cytogenetics, FISH, PCR for known fusions, RNA-seq, and CMA for copy-number abnormalities. These assays were performed as part of routine clinical care in CLIA-certified laboratories. Patient samples from the University of Chicago underwent high-throughput genetic sequencing with a targeted deep sequencing assay of 178 genes (OncoPlus; ref. 43). Concomitantly, a hybrid capture–based RNA-seq assay was used to detect known and novel fusions. DNA and RNA of samples from Moffitt Cancer Center were sequenced by using the Foundation One Heme assay (Supplementary Tables S1 and S2). Ph-like gene signature was assessed by FISH for common fusions (CRLF2, ABL1, ABL2, JAK2, CSF1R, and PDGFRB) and RNA-seq. Karyotyping of diagnostic bone marrow aspirate specimens was performed by counting 20 metaphase cells. The overall schema for molecular classification of B-ALL is shown in Supplementary Fig. S1.
DNA was prepared from blood or bone marrow aspirate specimens using the QIAmp DNA Blood Mini Kit (Qiagen) per the manufacturer's instructions. For the University of Chicago OncoPlus assay, DNA was quantified using the Qubit fluorometric assay (Thermo Fisher Scientific). DNA was subjected to ultrasonic fragmentation and subsequent library preparation using xGen9 cfDNA and FFPE DNA Library Preparation Kit (IDT), followed by library amplification with patient-specific indexing primers (IDT). After library amplification, quantification, and pooling, fragments originating from targeted genomic regions were enriched using a panel of biotinylated oligonucleotides (xGen Lockdown probes, IDT) supplemented with additional probes to assist with genome-wide copy-number assessment (xGen CNV Backbone Panel, IDT). After subsequent amplification and pooled library quantification, libraries were sequenced in rapid run mode on a NovaSeq 6000 system (Illumina) to produce 2 × 101 bp paired-end sequencing reads. Sequencing data were analyzed via custom-designed bioinformatics pipelines on a University of Chicago HIPAA-compliant high-performance computing system, using the hg19 (GRCh37) human genome reference sequence for alignment. Variants were filtered for quality (Phred quality score >30) and depth (>100×) with variant signal >5% retained for analysis. Variant calls were annotated and classified for pathogenicity using the American College of Medical Genetics and Genomics/Association for Molecular Pathology (44).
We also performed copy-number analysis (CNA) by evaluating the average exon interval depths recorded via the Genome Analysis Tool Kit DepthofCoverage module (45). Using 24 nonmalignant clinical samples, a historical normalized baseline was generated for each interval. Test sample data were subjected to a normalization algorithm to control for individual gene profile run-specific variability. To detect the potential copy-number regions, fold change and Z-scores were calculated for each interval. Thresholds were set at >200% (gain) or <66% (loss) with Z-score >3 or < −2, respectively. Genes with more than half the intervals showing copy-number changes in the same direction were identified.
RNA-seq of diagnostic blood and bone marrow samples was performed with the University of Chicago's RNA gene fusion analysis, which is a hybrid capture–based RNA-sequencing assay for detecting known and novel fusions involving any of the 1,005 targeted cancer-associated genes. RNA was extracted from peripheral blood or bone marrow collected in EDTA tubes using the simplyRNA Blood Kit on Maxwell RSC instrument (Promega) and quantified using a Qubit fluorometric assay (Thermo Fisher Scientific), adjusted for the percentage of fragments greater than 100 bp using a TapeStation system (Agilent). 300 ng RNA was subjected to library prep using the KAPA stranded RNA-seq Kit with RiboErase (Kapa Biosystems), followed by quantitation using the KAPA library quantification kit (Kapa Biosystems). Pooled libraries were captured using a panel of biotinylated oligonucleotides (xGen Lockdown probes, IDT). Amplified pooled captured libraries were sequenced in rapid run mode on a NovaSeq 6000 system (Illumina) to produce 2 × 101 bp paired-end sequencing reads. Sequence data were aligned to the hg19 human reference transcriptome using STAR aligner (46), and fusions were detected using a combination of in-house developed Python software and STAR fusion software.
Moffitt Cancer Center patients underwent genomic profiling with FoundationOne Heme DNA + RNA assay as described before (47). Variants were filtered for depth (>150×). For single-nucleotide variations, the VAF cutoff was 1% for known somatic variants and 5% for novel somatic variants. For indels, the VAF cutoff was 3% for known somatic variants and 10% for novel variants. For fusion detection from RNA-seq, raw sequence reads were aligned to whole transcriptome (refSeq), and reads with suboptimal mapping were aligned to whole-genome references. Rearrangements were detected by identifying clusters of chimeric read pairs from both DNA and RNA.
Supplementary Tables S1 and S2 summarize all the genes that were analyzed for all 400 patients (University of Chicago and Moffitt Cancer Center combined) for DNA mutations and RNA fusions, respectively. RNA-seq could not be performed on 8 T-lineage ALL patients, for whom the T-ALL subtype was designated as “unknown.”
In addition to analyzing DNA sequencing data for CNA, we also performed CMA for patients with aneuploidy, including patients with suspected masked hypodiploidy and IKZF1 deletion. This was performed using Affymetrix CytoScan HD microarray. This microarray consists of 2,696,550 oligonucleotide probes across the genome, including 1,953,246 unique nonpolymorphic probes and 743,304 SNP (single-nucleotide polymorphism) probes. Patient hybridization parameters were compared with data derived from phenotypically normal individuals. Detected CNVs were reported when found to have clear or suspected clinical relevance; CNVs devoid of relevant gene content or reported as common findings in the general population were not reported. Long stretches of homozygosity were reported only if they most likely represented somatic events in tumor cells, and when their size exceeded 3,000 kb. Genomic linear positions were given relative to NCBI build 37 (hg19). This microarray and associated software (Chromosome Analysis Suite, v4.2) were manufactured by Affymetrix and used for the purpose of identifying DNA copy-number gains and losses associated with large chromosomal imbalances.
For high-sensitivity NGS of preleukemic samples, we performed targeted sequencing of 11 CH genes by using a single molecule molecular inversion probe panel to detect low-frequency variants in hematopoietic samples obtained years before ALL diagnosis, as described before (48). The panel covered full exons of 11 genes plus four mutational hotspots (accounting for 90% of CH mutations). These studies were performed at Broad Institute.
Germline Testing
Germline testing was performed by skin biopsy in individuals with a clinical indication for testing: a personal history of two cancers, family history of another hematopoietic malignancy within two generations of the proband, family history of a nonhematopoietic tumor diagnosed at age 50 or younger within two generations of the proband, and/or the identification of gene variants on somatic mutation testing (OncoPlus) associated with hereditary hematologic malignancies (Supplementary Fig. S4; refs. 49, 50). Skin biopsy specimens were cultured in RPMI medium to grow fibroblasts, from which DNA was extracted for exome sequencing. Exome sequencing was performed with the Agilent SureSelect Clinical Research Exome kit (Agilent Technologies) that targets the exome, augmented with spike-in probes covering noncoding regions of select genes (i.e., ANKRD26, DKC1, TERC, TERT, GATA2, and RTEL1). Sequencing was performed as described before, and variants were interpreted by a team of board-certified geneticists and genetic counselors (50).
Single-Cell DNA and Protein Sequencing
Single-cell DNA and protein library preparation and sequencing of ALL samples were performed by using Mission Bio, lnc Tapestri platform as described before (51). Briefly, bone marrow aspirate specimens from the time of ALL diagnosis were centrifuged on Ficoll, and mononuclear cells were viably frozen. For Tapestri studies, 1 × 106 viable cells were incubated with TruStain FcX and Tapestri staining buffer (#420201, BioLegend) at room temperature. The pool of 45 oligo-conjugated antibodies from BioLegend TotalSeq-D Heme Oncology Cocktail (#399906, BioLegend) was then added and incubated for 30 minutes on ice. Cells were then washed and resuspended in Tapestri cell buffer, requantified, and loaded into a Tapestri microfluidics cartridge. Single cells were encapsulated, lysed, and barcoded. DNA PCR products were isolated from individual droplets and purified with Ampure XP beads. Protein PCR products were incubated with Tapestri pullout oligo (5 μmol/L) at 96°C, followed by incubation on ice. Protein PCR products were purified with Invitrogen Streptavidin C1 beads and beads were used as a PCR template for the incorporation of i5/i7 Illumina indices. Both DNA and protein libraries were quantified using an Agilent Bioanalyzer and pooled for sequencing on the Illumina NovaSeq 6000 from the University of Chicago Genomics Core.
In order to have a comprehensive coverage of myeloid mutations implicated in CH, we used the Tapestri Single-Cell DNA Myeloid Panel, which was designed against 1,208 targets across 45 genes, covering 64.4 kb of the human genome. The panel has a total of 312 amplicons (range of amplicon length is 125–375 bp), 92.7% coverage, >90% uniformity, and requires ∼188 million read pairs per sample (https://missionbio.com/wp-content/uploads/2020/03/DataSheet_MissionBio_Myeloid_RevC.pdf).
FASTQ files for single-cell DNA and protein libraries were uploaded onto and processed by the TapestriV2 pipeline, which goes through a QC module, trims adaptor sequences, extracts barcodes, aligns reads to the human genome (hg19), assigns sequence reads to cell barcodes and performs genotype calling with GATKv3.7. Initial QC and filtering low-quality genotypes or cells were performed in Tapestri Insights with default parameters. After pipeline processing, data for each run were aggregated into h5 files, which were downloaded and read into the Jupyter Notebook where Python codes were executed for analysis (IPython, RRID:SCR_001658). This pipeline determines the genotype (homozygous, heterozygous, or wild-type) by analyzing the amplified DNA segment, and the phenotype by reading the oligo-tag conjugated to antibody in the same cell that is indexed by i5/i7 Illumina sequences. Cells were then projected in two-dimensional space by using PCA followed by UMAP. Cells were clustered by their surface immunophenotype, and the genotypes of DNA variants were projected onto them.
Single-Cell RNA Sequencing and Data Analysis
Bone marrow aspirate specimens from age- and sex-matched CH, ALL with MyM, and ALL without MyM patients were tagged with cell hashing oligo-tagged antibodies (BioLegend) according to the manufacturer's instructions. The libraries were prepared using the Chromium Single-Cell 3′ Reagent kits (v2 and v3). Libraries were run on an Illumina NovaSeq. Sequencing data were demultiplexed and converted to FASTQ format. The Cell Ranger Single-Cell Software Suite was used to perform demultiplexing and barcode processing. The cDNA insert was aligned to the hg38/GRCh38 reference genome. All further analyses including the dimensionality reduction, unsupervised clustering algorithms, and the assessment of differentially expressed genes were performed using the Seurat R package (52).
To exclude low-quality cells, we removed cells with more than 20% of the transcripts coming from mitochondrial genes. The data were normalized by the total expression, multiplied this by a scale factor of 100,000, and log-transformed. We used the Seurat anchor-based integration method for merging data sets in order to account for the biological and technical batch differences between the patients and libraries (24). The nine samples were processed, genes that were detected in fewer than 10 cells were filtered out. The filtered data set included 22,120 cells. To project the cells in two-dimensional space, the dimensionality of the scaled integrated data matrix was further reduced by using PCA followed by UMAP. A range of resolutions was utilized to establish a sufficient number of clusters to identify known populations based on the levels of canonical markers. The final unsupervised clusters were assigned known hematopoietic cell types based on the expression of lineage-specific markers. Next, cluster marker genes were determined by performing differential gene-expression analysis using the Wilcoxon rank-sum test. Bonferroni multiple-comparison correction was used to correct for cells within each cluster against all other cells within the cell type.
Ingenuity Pathway Analysis (Qiagen, Inc) was used to analyze gene networks for differentially expressed genes with a log2 fold change >0.5 or < −0.5, and an adjusted P < 0.05 (RRID:SCR_008653).
Flow Cytometry and Cell Sorting
Primary human B-ALL cells at a concentration of 1–2 million cells/mL were assessed by flow cytometry and sorted on BD FACSAria II to isolate B- or T-lymphoblasts (CD45dim Ssclow), CD14+ monocytes, CD3+ T cells and CD19+ CD34– CD45+ B cells. The antibodies used for FACS analysis were BV650-conjugated CD3 (1:100; BD Biosciences, BDB740562), PE-Cy7-conjugated CD14 (1:100, BD Biosciences, BDB560919), PE-conjugated CD19 (1:100, BD Biosciences, BDB561741), BB700-conjugated CD34 (1:100, BD Biosciences, BDB745835), and APC-conjugated CD45 (1:100, BD Biosciences, BDB555485). Appropriate isotype controls were used to set gates. Data analysis was performed using FlowJo software (Tree Star, RRID:SCR_008520). Sorted cells were subjected to DNA extraction and sequencing on OncoPlus DNA platform as described above.
Cell Viability Assays for Vincristine, Doxorubicin, and Blinatumomab
For in vitro cell viability assays, freshly cryopreserved primary human B-ALL cells were thawed and cultured in RPMI-1640 supplemented with 2 mmol/L L-glutamine, 10% FBS, and insulin–transferrin–selenium. Cells were incubated for 48 hours in the presence of increasing concentrations of vincristine and doxorubicin treatments. Cell viability was assessed by adding the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc) to the cell culture for the last 4 hours, and quantitated using Bio Tek Synergy H4 plate reader and Gen5 software. IC50 dose was defined as the half-maximal inhibitory concentration, calculated using nonlinear fitted dose–response curves in GraphPad Prism v.9.0. Experiments were done in triplicate.
To assess the cytotoxicity of blinatumomab, primary human B-ALL peripheral blood samples containing >20% blasts and >20% lymphocytes were treated with increasing doses of blinatumomab (1 to 10,000 ng/mL). At 48 hours posttreatment, cells were assessed by flow cytometry to quantify B-lymphoblast (CD45dim SSclow), CD3+ T-cell and CD14+ monocyte populations. Cell viability in each dosage was calculated by dividing the live B-lymphoblast cell population in each dosage by the live B-lymphoblast cell population in the control group (without blinatumomab). Percent survival of B-lymphoblasts was plotted by using nonlinear fitted dose–response curves in GraphPad Prism v.9.0 (RRID:SCR_002798).
Statistical Analysis
All statistical analyses were performed using R v.3.4.4. The figures were generated with the ggplot2 package (RRID:SCR_014601) in R and GraphPad Prism version 8 (GraphPad Software, RRID: SCR_002798). Other packages used in data processing include dplyr, RColorbrewer, and cowplot. Clinical data for patients were available from medical records. The associations between different classes of mutations and clinical variables were investigated by calculating the odds ratios with the Fisher exact test for categorical variables and the Wilcoxon rank-sum test for continuous variables. Adjustment for multiple testing was done by the Benjamini–Hochberg method. Univariable and multivariable Cox regression analyses were used to identify the impact of potential factors on survival outcomes. OS was defined as the time from diagnosis until death or last follow-up. RFS was defined as the time from first CR until disease relapse, death, or last follow-up. OS and RFS estimates were calculated using the Kaplan–Meier method, and differences between the curves were assessed using the log-rank test.
Data Sharing Statement
Clinical data, somatic mutation data detected by clinical sequencing, and raw data for scRNA-seq are provided in Supplementary Raw Data Tables. Single-cell sequencing raw data files can be found in the NCBI Sequence Read Archive at PRJNA945186 (RRID: SCR_001370).
Supplementary Material
Raw patient and scRNA-seq data
Supplemental Figure 1. WHO/ICC subtypes for the B-ALL cohort.
Supplemental Figure 2. Lymphoid clonal hematopoiesis (CH) mutations are less common in adults with ALL.
Supplemental Figure 3. Blast percentage in the pre-treatment sample and detected variant allelic frequencies (VAF) of mutations in ALL patients with myeloid mutations.
Supplemental Figure 4. Germline testing in the adult ALL cohort.
Supplemental Figure 5. Associations of gene mutations and WHO/ICC disease subtypes in B-ALL.
Supplemental Figure 6. Overall survival in adults with B-ALL.
Supplemental Figure 7. Kaplan-Meier overall survival curves for Ph-negative B-ALL patients stratified based on the presence of TP53 and non-TP53 MyM (A), and treatment with different chemotherapy protocols (B, C, D). LH, low hypodiploidy.
Supplemental Figure 8. T-lineage ALL with myeloid mutations (MyM).
Supplemental Figure 9. Expression levels of lineage-specific surface markers.
Supplemental Figure 10. Single cell DNA and protein sequencing to study the clonal architecture of Tlineage ALL with MyM.
Supplemental Figure 11. A, Flow plots showing ETP-lymphoblast, myeloid, and mature lymphocytic compartments of ETP-ALL1, which were sorted for DNA extraction and sequencing. B, Distribution of variant allelic frequencies (VAFs) for myeloid (IDH2, DNMT3A) and NRAS mutations across subpopulations of cells in ETP-ALL1.
Supplemental Figure 12. Clonal dynamics of ALL with MyM.
Supplemental Figure 13. Two CH mutations (TP53 and DNMT3A) were detectable seven years before the diagnosis of therapy-related ALL22.
Supplemental Figure 14. Clonal evolution of B-ALL2.
Supplemental Figure 15. Lymphoblasts from B-ALL with MyM are characterized by their resistance to cytotoxic chemotherapy.
Supplemental Figure 16. Curves showing viability of primary human B-ALL samples with TP53 mutation (n= 14), MyM (n= 16) and no MyM/TP53 (n= 24), treated with vincristine, doxorubicin, and blinatumomab at escalating doses.
Supplemental Figure 17. Flow cytometry plots showing ALL22 (TP53-mutated B-ALL) sample treated with escalating doses of blinatumomab.
Supplemental Figure 18. A, CR with MRD negativity rates in Ph-negative B-ALL patients treated with pediatric regimens, stratified based on age, gender, and treatment site. B, CR with MRD negativity rates in Ph-negative B-ALL patients treated with hyper-CVAD, stratified based on age, gender, and treatment site. C, CR with MRD negativity rates in Ph-negative B-ALL patients treated with inotuzumab.
Supplemental Figure 19. A, MHC class I and II antigen expression in blasts from B-ALL with MyM vs B-ALL without MyM/TP53. B, Comparison of genes implicated in blinatumomab resistance in B-ALL with vs without MyM/TP53.
Supplemental Table 1. List of DNA gene mutations tested in diagnostic tumor samples of our ALL cohort.
Supplemental Table 2. List of 80 genes with RNA sequencing coverage.
Supplemental Table 3. Demographics of 400 adult ALL cases.
Supplemental Table 4. List of first-line (initial) therapies in Ph-positive B-ALL, Ph-negative B-ALL, and Tlineage ALL patients.
Supplemental Table 5. Baseline gene rearrangements and structural variants of 329 molecularly characterized adult B-ALL patients.
Supplemental Table 6. Baseline gene rearrangements and structural variants of 71 molecularly characterized adult T-lineage ALL patients.
Supplemental Table 7. List of pathogenic/likely pathogenic germline variants in our ALL cohort.
Supplementary Table 8. Multivariable analysis of overall survival (OS) in B-ALL patients.
Supplemental Table 9. Clinical characteristics of patients studied in scDNA + protein sequencing experiments.
Supplemental Table 10. Clinical characteristics of patients studied in scRNA-seq experiment.
Supplemental Table 11. Ingenuity pathway analysis comparing B-lymphoblasts from B-ALL with MyM vs B-ALL without MyM.
Acknowledgments
C. Saygin is supported by the Leukemia Lymphoma Society Special Fellow Award, Prevent Cancer Foundation Fellowship, and Cancer Research Foundation. This work was supported by NIH (R01HL082945, P01CA066996, and P50CA206963), the Howard Hughes Medical Institute, the Edward P. Evans Foundation, and the Leukemia and Lymphoma Society (to B.L. Ebert). A.S. Sperling was supported by grants from the National Cancer Institute (K08CA252174) and the U.S. Department of Defense (CA210827). The authors thank Dr. Ross Levine for the kind gift of Tet2fl/fl Vav-Cre mice, Dr. Pieter Faber and Ms. Sandy Arun from Genomics Core for their assistance with single-cell studies, and Janice Pacheco and Clinton Osei of the University of Chicago for their assistance with patient samples.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
C. Saygin reports grants from the Leukemia Lymphoma Society, Prevent Cancer Foundation, and Cancer Research Foundation during the conduct of the study. J. Stauber reports grants from National Heart, Lung, and Blood Institute during the conduct of the study. A.S. Sperling reports personal fees from Novartis and Roche outside the submitted work. L.D. Weeks reports grants from the American Society of Hematology and Edward P. Evans Foundation for MDS during the conduct of the study and personal fees from Vertex, Sobi, and AbbVie outside the submitted work. M.R. Luskin reports other support from Pfizer, Novartis, KITE, Jazz, and Servier outside the submitted work. T.C. Knepper reports personal fees from AstraZeneca outside the submitted work. S. Gurbuxani reports other support from Jazz Pharmaceutical outside the submitted work. A.A. Patel reports other support from Kronos Bio, grants from Pfizer, and personal fees from AbbVie, and Bristol Myers Squibb outside the submitted work. S. Umesh Nagalakshmi reports other support from MedGenome Inc. during the conduct of the study and other support from MedGenome Inc. outside the submitted work. R.A. Larson reports he has acted as a consultant or advisor to AbbVie, Amgen, Ariad/Takeda, Astellas, Celgene/BMS, Curis, CVS/Caremark, Epizyme, Immunogen, Jazz Pharmaceuticals, Kling Biotherapeutics, MedPace, MorphoSys, Novartis, and Servier, has received clinical research support to his institution from Astellas, Celgene, Cellectis, Daiichi Sankyo, Forty Seven/Gilead, Novartis, and Rafael Pharmaceuticals, and has received royalties from UpToDate. N.C. Munshi reports personal fees from BMS, J&J, Pfizer, Sebia, Oncopep, and Sanofi outside the submitted work. B.L. Ebert reports grants from Novartis and Calico, and personal fees from Neomorph, Inc, TenSixteen Bio, Skyhawk Therapeutics, and Exo Therapeutics outside the submitted work. U. Steidl reports grants from NIH/NCI during the conduct of the study; and grants from Aileron Therapeutics, Pfizer, Roche, Novartis, Smurf Therapeutics, and Trillium Therapeutics and personal fees and other support from Stelexis Therapeutics outside the submitted work. B.D. Shah reports grants, personal fees, and nonfinancial support from KITE/Gilead, personal fees from BMS, Amgen, Novartis, Pfizer, Precision Biosciences, Adaptive, Beigene, Century Therapeutics, Autolus, Deciphera, Lilly/Loxo, Takeda, and AstraZeneca, grants and personal fees from Jazz Pharmaceuticals and Servier, and other support from Pepromene BIo outside the submitted work. W. Stock reports personal fees from Newave, Amgen, Servier, and grants from Kura outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
C. Saygin: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, writing–review and editing. P. Zhang: Conceptualization, resources. J. Stauber: Data curation, methodology, writing–review and editing. I. Aldoss: Conceptualization, resources. A.S. Sperling: Conceptualization, formal analysis, writing–review and editing. L.D. Weeks: Conceptualization, resources, writing–review and editing. M.R. Luskin: Conceptualization, resources, writing–review and editing. T.C. Knepper: Conceptualization, resources, data curation, formal analysis, writing–review and editing. P. Wanjari: Conceptualization, resources, data curation, writing–review and editing. P. Wang: Conceptualization, resources, writing–review and editing. A.M. Lager: Conceptualization, resources, data curation, writing–review and editing. C. Fitzpatrick: Conceptualization, resources, data curation, writing–review and editing. J.P. Segal: Conceptualization, resources, writing–review and editing. M. Gharghabi: Conceptualization, resources, writing–review and editing. S. Gurbuxani: Conceptualization, resources, writing–review and editing. G. Venkataraman: Conceptualization, resources, data curation. J.X. Cheng: Conceptualization, resources, data curation. B.J. Eisfelder: Resources, data curation. O. Bohorquez: Resources, data curation. A.A. Patel: Conceptualization, resources, data curation, writing–review and editing. S. Umesh Nagalakshmi: Resources, data curation, formal analysis. S. Jayaram: Conceptualization, resources, data curation, software. O.M. Odenike: Conceptualization, resources, data curation. R.A. Larson: Resources, data curation, writing–review and editing. L.A. Godley: Resources, data curation. D.A. Arber: Resources, data curation, writing–review and editing. C.J. Gibson: Conceptualization, resources, data curation, writing–review and editing. N.C. Munshi: Conceptualization, resources, data curation. G. Marcucci: Conceptualization, resources, data curation. B.L. Ebert: Conceptualization, resources, data curation, writing–review and editing. J.M. Greally: Conceptualization, resources, data curation. U. Steidl: Conceptualization, resources, data curation, writing–review and editing. R. Lapalombella: Conceptualization, resources, data curation, writing–review and editing. B.D. Shah: Conceptualization, resources, data curation, software, writing–review and editing. W. Stock: Conceptualization, resources, data curation, software, formal analysis, supervision, writing–review and editing.
REFERENCES
- 1. Brown PA, Shah B, Advani A, Aoun P, Boyer MW, Burke PW, et al. Acute lymphoblastic leukemia, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2021;19:1079–109. [DOI] [PubMed] [Google Scholar]
- 2. Advani AS. Novel strategies in the treatment of acute lymphoblastic leukaemia. Lancet Haematol 2022;9:e240–e1. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7–33. [DOI] [PubMed] [Google Scholar]
- 4. Paietta E, Roberts KG, Wang V, Gu Z, Buck GAN, Pei D, et al. Molecular classification improves risk assessment in adult BCR-ABL1-negative B-ALL. Blood 2021;138:948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kimura S, Mullighan CG. Molecular markers in ALL: clinical implications. Best Pract Res Clin Haematol 2020;33:101193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brady SW, Roberts KG, Gu Z, Shi L, Pounds S, Pei D, et al. The genomic landscape of pediatric acute lymphoblastic leukemia. Nat Genet 2022;54:1376–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 2014;371:1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lilljebjorn H, Fioretos T. New oncogenic subtypes in pediatric B-cell precursor acute lymphoblastic leukemia. Blood 2017;130:1395–401. [DOI] [PubMed] [Google Scholar]
- 9. Li JF, Dai YT, Lilljebjorn H, Shen SH, Cui BW, Bai L, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci U S A 2018;115:E11711–E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007;446:758–64. [DOI] [PubMed] [Google Scholar]
- 11. Stanulla M, Dagdan E, Zaliova M, Moricke A, Palmi C, Cazzaniga G, et al. IKZF1(plus) defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric B-cell precursor acute lymphoblastic leukemia. J Clin Oncol 2018;36:1240–9. [DOI] [PubMed] [Google Scholar]
- 12. Creasey T, Barretta E, Ryan SL, Butler E, Kirkwood AA, Leongamornlert D, et al. Genetic and genomic analysis of acute lymphoblastic leukemia in older adults reveals a distinct profile of abnormalities: analysis of 210 patients from the UKALL14 and UKALL60+ clinical trials. Haematologica 2022;107:2051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science 2019;366:eaan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 2018;559:400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desai P, Mencia-Trinchant N, Savenkov O, Simon MS, Cheang G, Lee S, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med 2018;24:1015–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bolton KL, Ptashkin RN, Gao T, Braunstein L, Devlin SM, Kelly D, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet 2020;52:1219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Voso MT, Falconi G, Fabiani E. What's new in the pathogenesis and treatment of therapy-related myeloid neoplasms. Blood 2021;138:749–57. [DOI] [PubMed] [Google Scholar]
- 19. Sperling AS, Guerra VA, Kennedy JA, Yan Y, Hsu JI, Wang F, et al. Lenalidomide promotes the development of TP53-mutated therapy-related myeloid neoplasms. Blood 2022;140:1753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Niroula A, Sekar A, Murakami MA, Trinder M, Agrawal M, Wong WJ, et al. Distinction of lymphoid and myeloid clonal hematopoiesis. Nat Med 2021;27:1921–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saygin C, Kishtagari A, Cassaday RD, Reizine N, Yurkiewicz I, Liedtke M, et al. Therapy-related acute lymphoblastic leukemia is a distinct entity with adverse genetic features and clinical outcomes. Blood Adv 2019;3:4228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: lymphoid neoplasms. Leukemia 2022;36:1720–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, et al. International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood 2022;140:1200–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2019;37:38–44. [DOI] [PubMed] [Google Scholar]
- 26. Zhao Y, Aldoss I, Qu C, Crawford JC, Gu Z, Allen EK, et al. Tumor-intrinsic and -extrinsic determinants of response to blinatumomab in adults with B-ALL. Blood 2021;137:471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pourhassan H, Yang D, Afkhami M, Pillai R, Ball B, Al Malki M, et al. High prevalence and inferior long-term outcomes for TP53 mutations in therapy-related acute lymphoblastic leukemia. Am J Hematol 2022;97:E171–E3. [DOI] [PubMed] [Google Scholar]
- 28. Barnea Slonim L, Gao J, Burkart M, Odetola OE, Kocherginsky M, Dinner SN, et al. Therapy-related B-cell acute lymphoblastic leukemia in adults has unique genetic profile with frequent loss of TP53 and inferior outcome. Leukemia 2021;35:2097–101. [DOI] [PubMed] [Google Scholar]
- 29. Geyer MB, Shaffer BC, Bhatnagar B, Mims AS, Klein V, Dilip D, et al. Lenalidomide-associated B-cell ALL: clinical and pathologic correlates and sensitivity to lenalidomide withdrawal. Blood Adv 2023;7:3087–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kantarjian H, Stein A, Gokbuget N, Fielding AK, Schuh AC, Ribera JM, et al. Blinatumomab versus chemotherapy for advanced acute lymphoblastic leukemia. N Engl J Med 2017;376:836–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kantarjian HM, DeAngelo DJ, Stelljes M, Martinelli G, Liedtke M, Stock W, et al. Inotuzumab Ozogamicin versus standard therapy for acute lymphoblastic leukemia. N Engl J Med 2016;375:740–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park JH, Riviere I, Gonen M, Wang X, Senechal B, Curran KJ, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N Engl J Med 2018;378:449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Steensma DP, Bolton KL. What to tell your patient with clonal hematopoiesis and why: insights from 2 specialized clinics. Blood 2020;136:1623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chitadze G, Stengel A, John-Klaua C, Bruckmuller J, Trautmann H, Kotrova M, et al. Somatic TP53 mutations are pre-leukemic events in acute lymphoblastic leukemia. Blood 2023;141:1640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim R, Bergugnat H, Larcher L, Duchmann M, Passet M, Gachet S, et al. Adult low-hypodiploid acute lymphoblastic leukemia emerges from preleukemic TP53-mutant clonal hematopoiesis. Blood Cancer Discov 2023;4:134–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barnell EK, Skidmore ZL, Newcomer KF, Chavez M, Campbell KM, Cotto KC, et al. Distinct clonal identities of B-ALLs arising after lenolidomide therapy for multiple myeloma. Blood Adv 2022;7:236–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maia AT, Tussiwand R, Cazzaniga G, Rebulla P, Colman S, Biondi A, et al. Identification of preleukemic precursors of hyperdiploid acute lymphoblastic leukemia in cord blood. Genes Chromosomes Cancer 2004;40:38–43. [DOI] [PubMed] [Google Scholar]
- 38. Raman HS, Shanmugam V, Li J, Steensma DP, Kim AS, Luskin MR. NPM1-mutant acute myeloid leukemia relapsing as acute lymphoblastic leukemia with clonal persistence of IDH1 mutation. Leuk Lymphoma 2021;62:1790–2. [DOI] [PubMed] [Google Scholar]
- 39. Richardson PG, Jacobus SJ, Weller EA, Hassoun H, Lonial S, Raje NS, et al. Triplet therapy, transplantation, and maintenance until progression in myeloma. N Engl J Med 2022;387:132–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 41. Bartram J, Patel B, Fielding AK. Monitoring MRD in ALL: methodologies, technical aspects and optimal time points for measurement. Semin Hematol 2020;57:142–8. [DOI] [PubMed] [Google Scholar]
- 42. Saygin C, Cannova J, Stock W, Muffly L. Measurable residual disease in acute lymphoblastic leukemia: methods and clinical context in adult patients. Haematologica 2022;107:2783–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kadri S, Long BC, Mujacic I, Zhen CJ, Wurst MN, Sharma S, et al. Clinical validation of a next-generation sequencing genomic oncology panel via cross-platform benchmarking against established amplicon sequencing assays. J Mol Diagn 2017;19:43–56. [DOI] [PubMed] [Google Scholar]
- 44. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. He J, Abdel-Wahab O, Nahas MK, Wang K, Rampal RK, Intlekofer AM, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 2016;127:3004–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miller PG, Fell GG, Foy BH, Scherer AK, Gibson CJ, Sperling AS, et al. Clonal hematopoiesis of indeterminate potential and risk of death from COVID-19. Blood 2022;140:1993–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Singhal D, Hahn CN, Feurstein S, Wee LYA, Moma L, Kutyna MM, et al. Targeted gene panels identify a high frequency of pathogenic germline variants in patients diagnosed with a hematological malignancy and at least one other independent cancer. Leukemia 2021;35:3245–56. [DOI] [PubMed] [Google Scholar]
- 50. Saygin C, Roloff GW, Hahn CN, Chhetri R, Gill SI, Elmariah H, et al. Allogeneic hematopoietic stem cell transplant outcomes in adults with inherited myeloid malignancies. Blood Adv 2023;7:549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Miles LA, Bowman RL, Merlinsky TR, Csete IS, Ooi AT, Durruthy-Durruthy R, et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 2020;587:477–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw patient and scRNA-seq data
Supplemental Figure 1. WHO/ICC subtypes for the B-ALL cohort.
Supplemental Figure 2. Lymphoid clonal hematopoiesis (CH) mutations are less common in adults with ALL.
Supplemental Figure 3. Blast percentage in the pre-treatment sample and detected variant allelic frequencies (VAF) of mutations in ALL patients with myeloid mutations.
Supplemental Figure 4. Germline testing in the adult ALL cohort.
Supplemental Figure 5. Associations of gene mutations and WHO/ICC disease subtypes in B-ALL.
Supplemental Figure 6. Overall survival in adults with B-ALL.
Supplemental Figure 7. Kaplan-Meier overall survival curves for Ph-negative B-ALL patients stratified based on the presence of TP53 and non-TP53 MyM (A), and treatment with different chemotherapy protocols (B, C, D). LH, low hypodiploidy.
Supplemental Figure 8. T-lineage ALL with myeloid mutations (MyM).
Supplemental Figure 9. Expression levels of lineage-specific surface markers.
Supplemental Figure 10. Single cell DNA and protein sequencing to study the clonal architecture of Tlineage ALL with MyM.
Supplemental Figure 11. A, Flow plots showing ETP-lymphoblast, myeloid, and mature lymphocytic compartments of ETP-ALL1, which were sorted for DNA extraction and sequencing. B, Distribution of variant allelic frequencies (VAFs) for myeloid (IDH2, DNMT3A) and NRAS mutations across subpopulations of cells in ETP-ALL1.
Supplemental Figure 12. Clonal dynamics of ALL with MyM.
Supplemental Figure 13. Two CH mutations (TP53 and DNMT3A) were detectable seven years before the diagnosis of therapy-related ALL22.
Supplemental Figure 14. Clonal evolution of B-ALL2.
Supplemental Figure 15. Lymphoblasts from B-ALL with MyM are characterized by their resistance to cytotoxic chemotherapy.
Supplemental Figure 16. Curves showing viability of primary human B-ALL samples with TP53 mutation (n= 14), MyM (n= 16) and no MyM/TP53 (n= 24), treated with vincristine, doxorubicin, and blinatumomab at escalating doses.
Supplemental Figure 17. Flow cytometry plots showing ALL22 (TP53-mutated B-ALL) sample treated with escalating doses of blinatumomab.
Supplemental Figure 18. A, CR with MRD negativity rates in Ph-negative B-ALL patients treated with pediatric regimens, stratified based on age, gender, and treatment site. B, CR with MRD negativity rates in Ph-negative B-ALL patients treated with hyper-CVAD, stratified based on age, gender, and treatment site. C, CR with MRD negativity rates in Ph-negative B-ALL patients treated with inotuzumab.
Supplemental Figure 19. A, MHC class I and II antigen expression in blasts from B-ALL with MyM vs B-ALL without MyM/TP53. B, Comparison of genes implicated in blinatumomab resistance in B-ALL with vs without MyM/TP53.
Supplemental Table 1. List of DNA gene mutations tested in diagnostic tumor samples of our ALL cohort.
Supplemental Table 2. List of 80 genes with RNA sequencing coverage.
Supplemental Table 3. Demographics of 400 adult ALL cases.
Supplemental Table 4. List of first-line (initial) therapies in Ph-positive B-ALL, Ph-negative B-ALL, and Tlineage ALL patients.
Supplemental Table 5. Baseline gene rearrangements and structural variants of 329 molecularly characterized adult B-ALL patients.
Supplemental Table 6. Baseline gene rearrangements and structural variants of 71 molecularly characterized adult T-lineage ALL patients.
Supplemental Table 7. List of pathogenic/likely pathogenic germline variants in our ALL cohort.
Supplementary Table 8. Multivariable analysis of overall survival (OS) in B-ALL patients.
Supplemental Table 9. Clinical characteristics of patients studied in scDNA + protein sequencing experiments.
Supplemental Table 10. Clinical characteristics of patients studied in scRNA-seq experiment.
Supplemental Table 11. Ingenuity pathway analysis comparing B-lymphoblasts from B-ALL with MyM vs B-ALL without MyM.