Abstract
Purpose:
Head and neck cancer (HNC) improvements are stagnant, even with advances in immunotherapy. Our previous clinical trial data show that altered fatty acid (FA) metabolism correlates with outcome. We hypothesized that pharmacologic and dietary modulation of FA catabolism will affect therapeutic efficacy.
Experimental Design:
We performed in vivo and in vitro experiments using PPARα agonism with fenofibrate (FF) or high oleic acid diets (OAD) with radiotherapy, generating metabolomic, proteomic, stable isotope tracing, extracellular flux analysis, and flow-cytometric data to investigate these alterations.
Results:
FF improved antitumor efficacy of high dose per fraction radiotherapy in HNC murine models, whereas the OAD reversed this effect. FF-treated mice on the control diet had evidence of increased FA catabolism. Stable isotope tracing showed less glycolytic utilization by ex vivo CD8+ T cells. Improved efficacy correlated with intratumoral alterations in eicosanoid metabolism and downregulated mTOR and CD36.
Conclusions:
Metabolic intervention with increased FA catabolism improves the efficacy of HNC therapy and enhances antitumoral immune response.
Translational Relevance.
While the employment of immunotherapies has had success in improving outcomes in the metastatic and recurrent setting for head and neck cancers, progress in definitive management has been rather stagnant leaving a potential for improvement in radiotherapy efficacy. A current major area of interest is the intersection of the immune system and metabolism. This work expands on metabolomic findings from a recently published radioimmunotherapy phase I/Ib trial that shows alterations in fatty acid catabolism correlated with response. We employ an already FDA-approved pharmacologic metabolic intervention (fenofibrate) that upregulates fatty acid catabolism and improves radiotherapy efficacy. We also investigate how dietary supplementation of oleic acid factors into this, as dietary interventions are often brought up by patients in the clinic and there are reports of improved radiotherapy outcomes in other cancer subsites with oleic acid supplementation. Efforts to translate this into a clinical trial are ongoing.
Introduction
Head and neck cancer (HNC) remains a disease site with only modest benefit from recent immunotherapeutic advances (1–5). Prognosis in HNC has been linked to the ratio of tumoricidal effector T cells (Teff) to suppressive regulatory T cells (Treg; ref. 6) and the composition of the immunologic tumor microenvironment (TME) is highly dependent on both etiology of the carcinogenesis and anatomic location (7). Patients with tumors enriched for proinflammatory phenotype have improved prognosis (8). However, the majority of HPV-unrelated HNC is considered immunologically cold with relatively lower levels of tumor-infiltrating lymphocytes, specifically Teff cells (9).
Radiotherapy (RT) is utilized routinely for HNC in localized, recurrent, and metastatic settings (10). Through its intrinsic cytotoxic properties, RT can augment the immunologic response by recruiting effector cells through exposure of neoantigens and increased proinflammatory cytokine signaling (11, 12). Resistance to RT remains a major obstacle to the curative treatment for HPV-unrelated HNC (13), and the intersection of metabolism with the immune system represents an important sector that warrants further investigation. Furthermore, there is an increased interest in investigating high dose per fraction RT (HRT) in the context of HNC, known as stereotactic body radiotherapy (SBRT; ref. 14), which has differing radiobiological attributes in comparison with conventional RT, including potentially less immunosuppression and increased endothelial damage. Cancer cells commonly adapt metabolic pathways to take on an enriched glycolytic phenotype, which is often exacerbated by a dysfunctional tumor vasculature and associated hypoxic TMEs (11, 15, 16). This can enhance therapeutic resistance due to the scarcity of glucose, a carbon source that immune cells prefer for effector function. Concomitantly, the relative increase in the abundance of fatty acids (FA) favors Treg and other immunosuppressive cells that catabolize these substrates for long-term function (17, 18).
Despite a growing body of literature in support of tumor immunometabolic reprogramming, most conclusions are drawn from in vitro work, and can overlook the in vivo complexities (19). Our previous clinical trial incorporating neoadjuvant anti–PD-L1 therapy with SBRT in HNC had shown a positive correlation between metabolites involving FA catabolism and response to therapy, and further metabolomic interrogation revealed that higher serum oleic acid (OA) correlated with pathologic response (20). Following up on these observations in humans, here we investigated the interactions between dietary and pharmacologic modulation of FA catabolism in the setting of orthotopic, syngeneic murine models of HNC and the downstream effect on anticancer efficacy of HRT.
Materials and Methods
Cell lines
For in vivo and in vitro experiments, MOC2 (RRID:CVCL_ZD33), MEER (RRID:CVCL_B6J2), and LY2 (RRID:CVCL_Z594) murine cancer cells were used (MOC2 and LY2 cell lines are wild-type TP53 and EGFR). MOC2 was derived from C57Bl/6 mice (RRID:MGI:2159769, Jax Labs) who developed squamous cell carcinoma after exposure to 7,12-Dimethylbenz(a)anthracene (DMBA) and were obtained from Ravindra Uppaluri (Dana-Farber Cancer Institute). The MEER cell line was obtained from Dr. John Lee (Sanford Health). The LY2 cell line was derived from lymph node metastases in BALB/c (RRID:IMSR_APB:4790, Charles River) mice originating from PAM 212 squamous cell carcinoma and obtained from Nadarajah Vigneswaran (University of Texas Health Science Center). In culture, low (<20) passage cells were maintained at 37°C and at 5% CO2 and grown in media containing IMDM (MOC2) or DMEM-F12 (LY2) supplemented with 10% FBS and 1% primocin/fungin. The MOC2 media were also supplemented with 1.75 μg EGF, 20 μg hydrocortisone, and 0.1% insulin solution (human). Cell lines were periodically tested for Mycoplasma.
Animal models of HNC
Murine models of HPV-unrelated HNC cell lines MOC2 (105 cells) and LY2 (106 cells) or HPV-driven MEER (106 cells) were implanted into the buccal mucosa of anesthetized C57BL/6 (MOC2 or MEER) and BALB/c (LY2) mice as previously described (21, 22). The appropriate cell counts were suspended in 50:50 volumes of Matrigel (10 mg/mL, BD Biosciences) and culture media and then injected (100 μL) submucosally into the right buccal tissue. HRT was initiated (with n = 8–10 mice per group) when the average tumor volume or groups reached 100 mm3 (approximately 7 days for MOC2, 9 days for MEER, and 14 days for LY2). Tumor measurements were recorded at least twice weekly and calculated using the formula (V = short diameter × long diameter2 / 2 mm3). Mice that were experiencing treatment toxicity, significant weight loss, ulceration, or malocclusion were euthanized according to guidelines from the Institutional Animal Care and Use Committee (IACUC), and all protocols for experiments were approved by the IACUC of the University of Colorado. Specimens including tumors, draining lymph nodes, and serum were obtained for analysis from the mice after euthanization.
Radiotherapy
The X-RAD SmART image-guided irradiator (Precision X-ray) was used to deliver HRT treatments. Mice were anesthetized with isoflurane, positioned in the prone orientation, and monitored closely during treatments. The buccal tumors were identified via fluoroscopy and positioned to minimize the dose to surrounding structures. Mice were irradiated using an AP beam to 8 Gy per fraction (at 5.6 Gy/minute) for a total of 24 Gy (3 fractions) with 225 keV X-rays through a 0.3 mm Cu filter and a square collimator. Mice implanted with MEER cells were irradiated with 10 Gy in a single fraction. Dosimetry with the Monte-Carlo method and treatment plans were verified using the SmART-ATP software (SmART Scientific Solutions). Cancer cells irradiated in vitro received 8 Gy via open collimation while plated in a 6-well plate.
Antibodies and drugs
For in vivo experiments, fenofibrate (FF; Sigma-Aldrich F6020) was given at a dose of 100 mg/kg/day either by oral gavage or intraperitoneal injection (IP). FF was started on day 5 after the first fraction of HRT. Anti–PD-L1 antibodies were obtained from Bio X Cell and injected IP at 10 mg/kg given during the first and third fractions of HRT. For in vitro experiments, OA (Sigma-Aldrich O1383) was diluted in dimethylsulfoxide (DMSO). The activated form of FF, fenofibric acid (FFA), was obtained from Thermo Fisher (466170010) and diluted in DMSO for in vitro experiments. Drugs used for extracellular flux assays are described below in the Seahorse section.
Antibodies used for in vitro CD4 conversion flow cytometry: BV711-CD44 (BioLegend, cat # 103057, RRID:AB_2564214), BV785-CD62 L (BioLegend, cat # 104440, RRID:AB_2629685), BV750 TNF-alpha (BioLegend, cat # 506358, RRID:AB_2801090), AF532-FOXP3 [Thermo Fisher Scientific, cat # 58-5773-82 (also 58-5773), RRID:AB_11218870], PE/Dazzle594 CD25 [BioLegend, cat # 101920 (also 101919), RRID:AB_2721702], APC-CD36 (Thermo Fisher Scientific, cat # 17-0362-82, RRID:AB_2734967), PerCP-eF710-PD-1 [Thermo Fisher Scientific, cat # 46-9981-82 (also 46-9981), RRID:AB_11151142], APC-eF 780-Ki-67 (Thermo Fisher Scientific, cat # 47-5698-82, RRID:AB_2688065), Pe-Cy7-IFN gamma (Thermo Fisher Scientific, cat # 25-7311-82, RRID:AB_469680), BV605-IL-4 [BioLegend, cat # 504126 (also 504125), RRID:AB_2686971], PB-Granzyme B [BioLegend, cat # 515407 (also 515408), RRID:AB_2562195], PerCP/Cy5.5-CTLA4 [BioLegend, cat # 106315 (also 106316), RRID:AB_2564473].
For in vivo flow cytometry, antibodies used were (if not listed above): BV786-CD25 (Thermo Fisher Scientific, cat # 417-0251-80, RRID:AB_2925723), PerCP-Cy5.5-CD3 (BD Biosciences, cat # 340949, RRID:AB_400190), BUV805-CD3 (Thermo Fisher Scientific, cat # 368-0037-42, RRID:AB_2896068), BUV496-CD4 (Thermo Fisher Scientific, cat # 364-0042-80, RRID:AB_2920953), eF450-CD4 [Thermo Fisher Scientific, cat # 48-0041-82 (also 48-0041), RRID:AB_10718983], BV570-CD44 (BioLegend, cat # 103037, RRID:AB_10900641), BV421-CD44 (Thermo Fisher Scientific, cat # 404-0441-82, RRID:AB_2925505), eF506-CD45 (Thermo Fisher Scientific, cat # 69-0459-42, RRID:AB_2637382), AF700-CD45 [BioLegend, cat # 157210 (also 157209), RRID:AB_2860730], SB436-CD62 L (Thermo Fisher Scientific, cat # 62-0621-82, RRID:AB_2762739), BV570-CD62 L (BioLegend, cat # 104433, RRID:AB_10900262), BUV805-CD8 (BD Biosciences, cat # 564920, RRID:AB_2716856), BUV737-IFN gamma (BD Biosciences, cat # 564693, RRID:AB_2722494), BV605-IL10 (BioLegend, cat # 505031, RRID:AB_2563146), PE-Cy7-NKp46 [Thermo Fisher Scientific, cat # 25-3359-41 (also 25-3359), RRID:AB_2573443], PE-phospho-4E-BP1 (Cell Signaling Technology, cat # 7547, RRID:AB_10949897), AF488-phospho-S6 ribosomal protein (Cell Signaling Technology, cat # 5018, RRID:AB_10695861), PerCP-PPAR alpha (StressMarq Biosciences, cat # SPC-1309D-PCP, RRID:AB_2713364), AF488-Pan CK (Thermo Fisher Scientific, cat # 53-9003-80, RRID:AB_1834351), SB645-PD-L1 (Thermo Fisher Scientific, cat # 64-5982-80, RRID:AB_2688083), and BV650-MHCII (Thermo Fisher Scientific, cat # 416-5321-82, RRID:AB_2937214).
High OA diet
To supplement dietary OA, a customized diet (high OA diet, OAD) with 20% kcal from protein, 35% kcal from carbohydrates, and 45% kcal from fat was designed with the source of fat from high OA sunflower oil (37.6% of total kcal from OA; Research Diets D22083002 Rodent Diet with 45% kcal fat from High Oleic Sunflower Oil and Wheat Starch with Green dye; Supplementary Fig. S2). Mice were started on the high OA for at least 3 weeks prior to tumor implantation. The control diet (CD) used was the Teklad Diets Global Soy Protein-Free Extruded Rodent Diet 2020X (Supplementary Fig. S2).
CD4 and CD8 T-cell isolations and culture
Spleens from experimental mice (ex vivo CD8 data Seahorse and heavy glucose assays) or donor C57BL/6 mice (CD4 conversion flow assay and Treg Seahorse assays) were harvested in order to isolate T cells of interest with EasySep Mouse CD8+ T-cell Isolation Kit (Stemcell), EasySep CD4+ T-cell Isolation Kit (Stemcell), or CD4+CD25+ Regulatory T-cell Isolation Kit (Miltenyi Biotec). Briefly, spleens were converted into a single-cell suspension with a 70-mm strainer and then incubated with the corresponding isolation cocktail followed by magnetic coating. To increase specificity for CD8, CD4 T cells, and Tregs, unwanted cells were then separated from the media with a magnet three times. In culture, Tregs were plated in a 24-well plate coated with 1 μg/mL anti-CD3 (clone 2C11, Invitrogen) and 5 μg/mL anti-CD28 (clonePV-1, Bio X Cell); on the first day of activation 500 U/mL recombinant IL2 (Biological Resources Branch, National Cancer Institute, USA) was added and then split as needed to a concentration of 1×106 cells/mL and replated on a coated plate with an additional 100 U/mL recombinant IL2. CD4+ T cells were plated on a 24-well plate coated with 2 μg/mL anti-CD3 and 2 μg/mL anti-CD28 and split as needed; 30 U/mL recombinant IL2 was given initially and on days of activation. For heavy glucose tracing experiments and upon isolation, the ex vivo CD8 T cells were cultured for 6 hours in glucose-free RPMI supplemented with 13C6-glucose in order to trace glycolytic metabolism with mass spectrometry. Cell pellets and media samples were taken at 5 minutes, 1 hour, 2 hours, and 6 hours for metabolomic analysis.
Flow cytometry
For flow-cytometric analysis of tumor tissue, tumors were digested into single-cell suspension as previously reported (23). Briefly, tumors were finely cut and incubated in HBSS solution with Collagenase III (Worthington) at 37°C. After incubation, tumors were passed through a 70-mm nylon mesh. The resulting cell suspension was centrifuged and resuspended in red blood cell (RBC) lysis buffer for 5 minutes. RBC lysis buffer was deactivated, cell suspensions were centrifuged, resuspended, and counted using an automated cell counter. Tumor-draining inguinal lymph nodes and spleens were processed into single-cell suspensions as above. For flow-cytometric analysis, cells were plated in 24-well plates and cultured for 4 hours in the presence of monensin, PMA, and ionomycin to stimulate cytokine production and block Golgi transport. Cells were then blocked with anti-CD16/32 antibody. Where necessary, cells were fixed and permeabilized prior to staining using the FoxP3 Fixation/Permeabilization protocol (eBioscience). Samples were run on the Cytek Aurora Spectral Analyzer (RRID:SCR_019826) at the Barbara Davis Center at the University of Colorado Diabetes Research Center (NIDDK grant # P30-DK116073). Data were analyzed using FlowJo Analysis software (RRID:SCR_008520, v10.8.1).
Serum metabolomics and heavy glucose tracing in CD8+ T cells
High-throughput metabolomics analysis was performed at the University of Colorado Cancer Center Mass Spectrometry Shared Resource on murine serum samples and CD8+ T cells upon ex vivo incubation with 11.1 mmol/L 13C6-glucose (Sigma-Aldrich) for 6 hours in glucose-free RPMI. Samples were thawed on ice and metabolites extracted from serum and CD8+ T cells by adding ice-cold 5:3:2 methanol:acetonitrile:water (v/v/v) to each tube at 1:24 ratio or 1 million cells/mL of solution equivalent, followed by 30 minutes of vortexing and 10 minutes of centrifugation, both at 4°C. All extracts were analyzed twice (10 mL injections each) by ultra-high performance liquid chromatography using a Thermo Vanquish UHPLC coupled to a Thermo Q Exactive mass spectrometer in negative and positive polarity modes, as described (24, 25). For each method, the UHPLC utilized a Phenomenex C18 column at a flow rate of 0.45 mL/minute with a 5-minute gradient (24, 25), whereas a separate UHPLC-MS method was used for oxylipins (25). Data analysis and peak picking (including 13C-labeled isotopologues) were performed via Maven (RRID:SCR_022491, 1.4.20-dev-772). MetaboAnalyst (RRID:SCR_015539, 5.0) was used to perform multivariate analyses including partial least square–discriminant analysis (PLS-DA), and hierarchical clustering analyses with heat map representation of the top 50 metabolites by ANOVA (26).
Bulk tumor proteomics
Approximately 500 μg of lyophilized tissue were resuspended in 8 mol/L urea, 0.1 mol/L Tris (pH 8.5), 5 mmol/L TCEP (tris(2-carboxyethyl)phosphine) and incubated with constant agitation (1,400 rpm) for 2 hours at 37°C (27, 28). Samples were then alkylated with 50 mmol/L 2-chloroacetamide for 30 minutes in the dark at room temperature. The solutions were diluted with four volumes of 100 mmol/L Tris-HCl (pH 8.5) before digestion with Lys-C (1:100; PierceTM) for 2 hours at 37°C with constant shaking (1,400 rpm). Samples were then digested overnight with trypsin (1:100) followed by a second treatment of trypsin for 2 hours. Both steps were carried out with continuous agitation (1,400 rpm) at 37°C. Following trypsin digestion, samples were acidified with formic acid to a final concentration of 1%, centrifuged at 16,000 × g for 5 minutes at room temperature, and the supernatant was collected. Aliquots containing 10 μg of digested peptides were purified using PierceTM C18 Spin Tips (Thermo Scientific) according to the manufacturer's protocol, dried in a vacuum centrifuge, and resuspended in 0.1% FA in mass spectrometry–grade water.
Liquid chromatography–tandem mass spectrometry (LC/MS-MS) was performed using an Easy nLC 1200 instrument coupled to an Orbitrap Fusion Lumos Tribrid mass spectrometer (all from Thermo Fisher Scientific). Proteolytic peptides were separated on a C18 column (100 μmol/L inner diameter × 20 cm) packed in-house with 2.7 μmol/L Cortecs C18 resin. The flow rate was set to 0.4 μL/minute and the column was developed with a linear gradient of 0.1% formic acid in ddH2O (solution A) and 0.1% formic acid in 80% ACN (solution B) at 6% B for 3 minutes, followed by 6%−42% B for 102 minutes, 42%−60% B for 5 minutes, 60%−95% B for 1 minute, isocratic at 95% B for 9 minutes. The Orbitrap Fusion Lumos was set to 120 K resolution, and the top N precursor ions in a 3-second cycle time (within a scan range of 300–1,800 m/z) were subjected to high-energy collision dissociation with 30% collision energy for peptide sequencing using a 15 K resolution setting. MS/MS was performed on the most abundant precursors exhibiting a charge state from 2 to 7 and an intensity threshold of 2 × 104. Dynamic exclusion was set to 45 seconds with a 10 ppm mass tolerance.
Fragmentation spectra were interpreted against the UniProt mouse proteome database (RRID:SCR_002380) using the MSFragger-based FragPipe computational platform (ref. 29; RRID:SCR_022864). Contaminants and reverse decoys were added to the database automatically. The precursor-ion mass tolerance and fragment-ion mass tolerance were set to 10 ppm and 0.2 Da, respectively. Fixed modifications were set as carbamidomethyl (C), and variable modifications were set as oxidation (M), oxidation (P; hydroxyproline), Gln→pyro- Glu (N-term), deamidated (NQ), and acetyl (Protein N-term). Two missed tryptic cleavages were allowed, and the protein-level false discovery rate was ≤ 1%.
Seahorse extracellular flux assays
Extracellular flux assays were run on the Agilent Seahorse XFe96 Analyzer (RRID:SCR_019545). Kits purchased from Agilent included the Seahorse XFe96/XF Pro FluxPak Mini and the Real-Time ATP rate Assay (containing oligomycin, rotenone and antimycin A). Additional reagents used included oligomycin (Cayman Chemical 11341), 2-deoxy-D-glucose (2-DG, Sigma-Aldrich D6134), and FCCP (Cayman Chemical 15218). Depending on the assay, instructions were followed based on the manufacturer's recommendations. Briefly, cancer cells were plated the night before (2 × 104 MOC2 cells/well or 1 × 104 LY2 cells/well) in appropriate tissue media and then washed on the day of the assay and replaced with Agilent Seahorse media. For T-cell experiments, 1.5 × 105ex vivo CD8 T-cell/well or 1 × 105 cultured Tregs/well were suspended in Agilent Seahorse media after isolation on the day of the assay. The Seahorse XF DMEM assay medium pack with appropriate dilution of 1.0 mol/L glucose, 100 mmol/L pyruvate, and 200 mmol/L glutamine was used for the assays. Final dilution was 2 mmol/L glutamine alone for the glycolytic stress test and 10 mmol/L glucose, 1 mmol/L pyruvate, and 2 mmol/L glutamine for all other assays. ATP rate assay final injection concentrations included 1.5 μmol/L oligomycin followed by 0.5 μmol/L rotenone with antimycin A; glycolytic rate assay included a final injection concentration of 10 mmol/L glucose, followed by 1.5 μmol/L oligomycin, and then 50 mmol/L 2-DG; the mitochondrial stress test included 1.5 μmol/L oligomycin followed by 1.5 μmol/L FCCP, and then 0.5 μmol/L rotenone with antimycin A. For the extracellular flux assays investigating the in vitro effect of experimental drugs on Tregs, drugs were injected prior to the glycolytic/mitochondrial assay as injection “A” (final concentration FFA 100 μmol/L, OA 100 μmol/L, and 100 μmol/L of combined FFA/OA). Hoescht stain (Thermo 622449) was used as a final injection and cells were counted with the Agilent BioTek Cytation-1 (RRID:SCR_019730) for normalization purposes.
Trypan blue viability assay
MOC2 cancer cells were seeded onto a 24-well plate (0.5×105 cells/mL media) and incubated for 24 hours at 37°C and 5% CO2 with the addition of 0.1% DMSO as control, or 100 μmol/L of OA, FF, or both diluted in 0.1% DMSO (6 replicates per group). After staining with Trypan Blue Solution, 0.4% (Thermo Fisher 15250061), cells were then counted twice with a Bio-Rad TC20 Automated Cell Counter. This experiment was repeated for confirmation, and the data shown are a compilation of both experiments.
Trial data
Human metabolomic and RNA sequencing (RNA-seq) data were used from the recently published phase I/Ib trial using combination hypofractionated SBRT with single dose durvalumab (anti–PD-L1) in the neoadjuvant setting (NCT03635164; ref. 20). Briefly, enrolled patients (>18 years old, n = 21) with high intermediate- or high-risk (stage III or IV and M0) HPV-unrelated HNC (squamous cell carcinoma of the oral cavity, oropharynx, larynx, or hypopharynx) who were deemed either resectable or borderline resectable by an otolaryngologist, underwent therapy at one of three locations (University of Colorado Hospital, Aurora, CO; Memorial Hospital Central, Colorado Springs, CO; Poudre Valley Hospital, Fort Collins, CO). Patients received one dose of neoadjuvant durvalumab (1,500 mg) with concurrent SBRT (12–18 Gy in 2–3 fractions) followed by standard-of-care therapy. Pathologic response was based on a review by two independent board-certified pathologists of the surgical specimen and determined to be either a complete response, microscopic residual tumor, or macroscopic residual tumor. For RNA-seq, please refer to the methods on publication of the trial (20). A heat map was generated from RNA-seq data described in Darragh and colleagues (20). Log2-transformed CPM values were used for time of surgery time points in responders and nonresponders to therapy. Expression values were z-scaled and visualized with the ComplexHeatmap package (v.2.14.0; ref. 30) in R (RRID:SCR_001905, v.4.2.3, https://www.R-project.org/).
The Cancer Genome Atlas program analysis
The HNSCC (head and neck squamous cell carcinoma) data set was generated from The Cancer Genome Atlas (TCGA; RRID:SCR_003193, https://www.cancer.gov/ccg/research/genome-sequencing/tcga); in order to make a subgroup of HPV-negative HNC that underwent RT, patients with oropharyngeal primaries, HPV-positive samples by ISH, and those that did not receive radiotherapy were removed from the cohort (final n = 268). Gene set enrichment was determined from RNA-seq data regarding the genetic expression of PPARs, CPTs, ACOXs, or CD36. Survival data were generated based on dichotomizing the cohorts into groups based on high or low expression with Xena (RRID:SCR_018938, University of California, Santa Cruz; ref. 31). These data were extracted and then analyzed using Prism GraphPad v9 (RRID:SCR_002798), with differences in survival analyzed using the log-rank test.
Pathway analysis
GSEA was performed similarly as described (20). For pathway analyses of FF-based experiments, differential genetic expression from proteomic data were generated using the statistical analysis module from MetaboAnalyst 5.0 (26) after removing proteins with less than half of nonzero values (reduction of total proteins from 3,227 to 2,137). A volcano plot was used from this analysis to visualize up or downregulated genes. Differential expression was then determined by generating up- or downregulated genes based on the experiment for those values with a P < 0.15. KEGG pathways and GO terms for biological processes were evaluated with Metascape (RRID:SCR_016620; ref. 32) by entering the differentially expressed genes with background correction (and visualizing P value of pathways <0.10). For the MOC2 and MEER comparisons, differentially expressed proteins were detected using the R package DEP (Differential Enrichment analysis of Proteomics data, RRID:SCR_023090; ref. 33) after dividing by the sum of intensities over all proteins detected in each sample. GSEA was then performed using clusterProfiler (RRID:SCR_016884; ref. 34) and Hallmark, KEGG, or GO gene sets.
Figure graphics and statistical analyses
BioRender.com was used to generate Fig. 1B, schematics for experiments (Figs. 2A, 2E, 2G, and 3A), and for images that show metabolic reactions (Figs. 3C and 3D). GraphPad Prism v9 was used to generate other graphs in the figures, including survival plots, extracellular flux graphs, genetic/metabolic expression comparisons, tumor volume curves, and flow-cytometric data. Quantitative analyses were performed using the Mann–Whitney test, one-way ANOVA with multiple comparisons, or log-rank test for survival using GraphPad Prism. P values less than 0.05 were considered statistically significant. Generally, P values greater than 0.25 were not shown in the figures.
Figure 1.
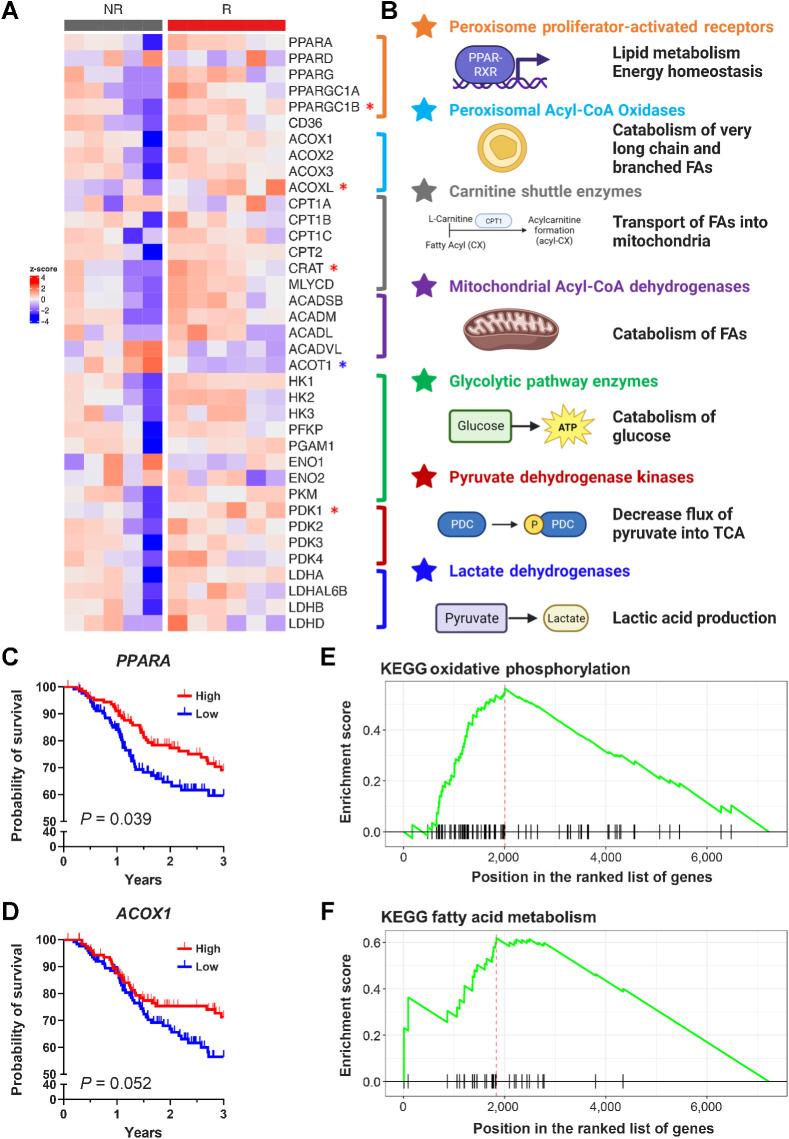
FA metabolism is altered in HPV-unrelated HNC and upregulated FA catabolism correlates with response to neoadjuvant SBRT with checkpoint blockade. A, Z-score of genetic expression of selected genes derived from RNA-seq from human samples from Darragh and colleagues (20). Significantly upregulated in responders by those shown with the red asterisk, whereas significantly downregulated shown with the blue asterisk (Mann–Whitley test, P < 0.05). B, Selected genes coded by color: PPARs (orange), peroxisomal acyl-CoA oxidases (light blue), carnitine shuttle enzymes (gray), mitochondrial acyl-CoA dehydrogenases (purple), glycolytic enzymes (green), pyruvate dehydrogenase kinases (red), and lactate dehydrogenases (dark blue). C and D, Overall survival of patients from TCGA H&N cohort split into high or low genetic expression PPARA (C; log2 normalized count cut point of 9.214) and ACOX1 (D; log2 normalized count cut point of 10.38). P values calculated with the log-rank test. E and F, GSEA of KEGG pathways showing enrichment of oxidative phosphorylation (E) and FA metabolism (F) in the MEER responder cell line in comparison with the MOC2 nonresponder cell line. (B, Created with BioRender.com.)
Figure 2.
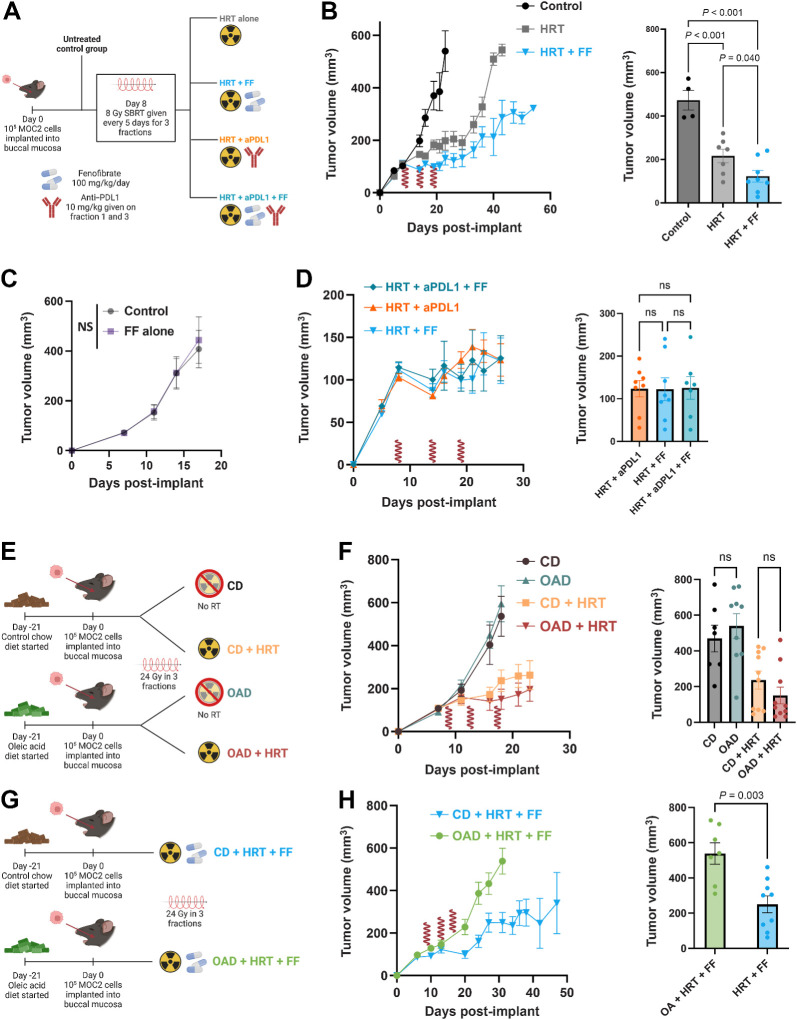
FF enhances antitumor efficacy of HRT, whereas OAD results in effect reversal. A, Schematic for experimental design with the following MOC2 groups (n = 8 per group): no treatment control (black), HRT alone (gray), HRT with FF (blue), HRT with aPDL1 (orange), HRT with FF and aPDL1 (teal). FF was started on day 5 after the first HRT fraction. HRT delivered as 24 Gy in 3 fractions (shown as red waves). All mice were on the CD. B, Tumor volume curve with day 26 mean volume. HRT fractions are denoted with a red wave. P values generated with an ordinary one-way ANOVA with multiple comparisons correction. HRT fractions are denoted with a red wave symbol. C, Tumor volume curve comparing control to daily FF (started on day 12, 100 mg/kg/day) alone. There was no difference between groups on day 17 (P = 0.721). D, Tumor growth curve and day 26 mean tumor volume showing HRT with FF vs. HRT with aPDL1 vs. HRT with both. There were no significant differences between groups (all P > 0.05 one-way ANOVA; P = 0.9961). FF was started on day 5 after the first HRT fraction. HRT delivered as 24 Gy in 3 fractions (shown as red waves). E, Schematic for experiment comparing CD vs. OAD with or without HRT (n = 10 mice in each group). F, Mean tumor volume curve with SEM for mice with buccal tumors irradiated to 24 Gy in 3 fractions who were fed the CD versus the OAD. Mann–Whitney test of mean tumor volume at day 18 was nonsignificant (P = 0.255). Mean tumor volume with SEM curve for mice fed the CD versus the OAD. Mann–Whitney test of mean tumor volume at day 18 was nonsignificant (P = 0.573). G, Schematic for experimental design with the following MOC2 groups (n = 10 mice in each group): OAD with HRT and FF (green) and control diet with HRT and FF (blue). H, Tumor volume curve for the OAD experiment with day 31 average tumor volumes and average weight over time. FF was started on day 5 after the first HRT fraction. HRT delivered as 24 Gy in 3 fractions (shown as red waves). Mean tumor volume at day 31 in the OAD (green) vs. the CD group (blue). P value generated from the Mann–Whitney test. (A, E, G, Created with BioRender.com.)
Figure 3.
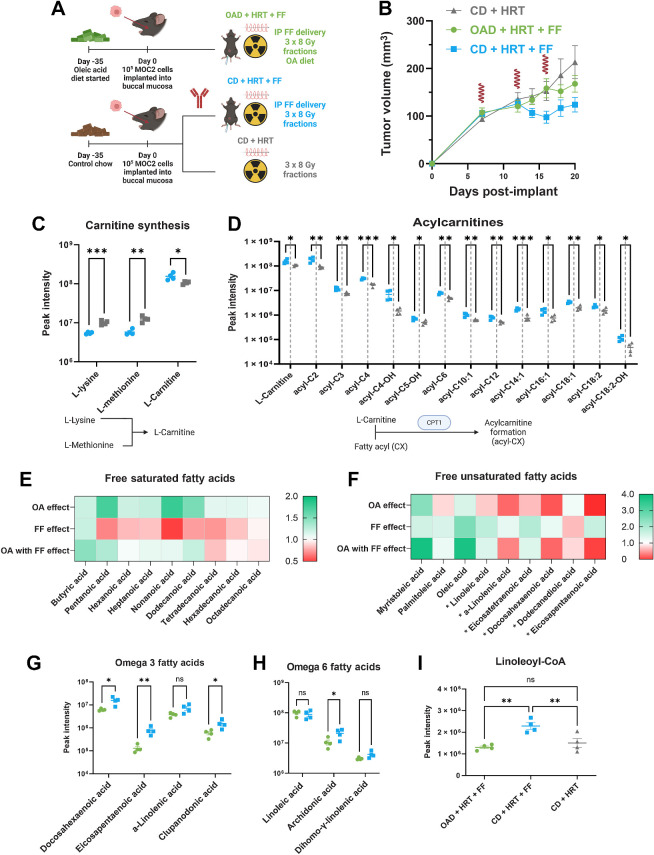
FF upregulated acyl-carnitine formation and fatty acid catabolism whereas the OAD results in significantly lower serum omega 3 and 6 fatty acids. A, Schematic for in vivo experiment for mechanistic evaluation (n = 10 mice per group). The mice were euthanized on day 20 for metabolomics, proteomics, and flow cytometry. The following graphics show groups: CD + HRT (gray), CD + HRT + FF (blue), OAD + HRT + FF (green). B, MOC2 tumor volume curve with 24 Gy HRT in 3 fractions given on days 7, 12, and 16. FF (100 mg/kg/day) was started on day 12. HRT fractions are shown as red waves. C and D, L-Carnitine is increased in FF compared with mice not on FF (C); acyl-carnitines are generally upregulated in FF-treated mice (D). Below each graph shows a schematic outlining the metabolic pathway involved. P values generated using multiple unpaired t tests (*, P <0.05; **, < 0.01; ***, <0.001). E and F, Fold change heat map of saturated and unsaturated FAs comparing mice on the OAD with HRT/FF vs. control diet with HRT/FF (OA effect); mice on the control diet treated with HRT/FF vs. HRT alone (FF effect); and mice on the OAD treated with HRT/FF vs. mice on the control diet treated with HRT alone (OA with FF effect). The FAs denoted with an asterisk (*) are PUFAs. G–I, Omega 3 FAs (G) and omega 6 FAs (H) are decreased in the OAD + HRT + FF group (green) in comparison with the CD + HRT + FF group (blue); linoleoyl-CoA (I) was higher in the HRT + FF group (blue) in comparison with the OA + HRT + FF group (green) and the CD + HRT group (gray). P values generated using multiple unpaired t tests or ordinary one-way ANOVA for 3 groups without multiple comparison correction (*, P < 0.05; **, P < 0.01). PUFA, polyunsaturated fatty acid. (A, Created with BioRender.com.)
Data availability
The RNA-seq data (20) have been previously deposited in a repository and is publicly available through GEO accession number GSE210287 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE210287). Other nonsequencing data generated in this study are available upon request from the authors.
Results
FA metabolism is altered in HPV-unrelated HNC and upregulated FA catabolism correlates with response to neoadjuvant HRT with checkpoint blockade.
Our phase I/Ib clinical trial using combination checkpoint blockade with neoadjuvant SBRT for HPV-unrelated advanced HNC (20) showed that responders to treatment had increased tumor-infiltrating effector T cells, decreased immunosuppressive cells, and increased antigen presentation with corresponding alterations in FA metabolites. Specifically, plasma metabolomic analyses revealed increased circulating free FAs in nonresponders as well as decreased acyl-carnitines, a potential signal for increased FA synthesis and/or decreased FA catabolism (Extended Data from figure 7 of Darragh and colleagues; ref. 20). Further investigation with transcriptomic analysis revealed that responders had enrichment in FA metabolism and oxidative phosphorylation along with a generalized pattern of upregulated FA catabolism (Fig. 1A and B). Specifically, we observed increased expression relating to peroxisome proliferator-activated receptors (PPAR), the nuclear receptors that regulate FA homeostasis and metabolic phenotype of cells (Supplementary Fig. S1A).
Because PPARs serve as a barometer of metabolite availability through FAs acting as their natural ligands, we examined the expression of downstream enzymes involved in FA catabolism and beta-oxidation as a readout of PPAR activity. These included enzymes that catalyze acyl-carnitine formation and shuttling of FAs into the mitochondria (35). Responders had higher levels of rate-limiting carnitine shuttle enzymes (ref. 36; Supplementary Fig. S1B), CD36 (the FA membrane translocase; Supplementary Fig. S1C), and rate-limiting FA oxidation enzymes, including both peroxisomal Acyl-CoA oxidases (ref. 37; Supplementary Fig. S1D) and acyl-CoA dehydrogenases (ref. 38; Supplementary Fig. S1E). Concordantly, expression of ACOT1, an enzyme that hydrolyzes coenzyme A from fatty acyl-esters resulting in decreased FA catabolism, was significantly lower in responders (Supplementary Fig. S1F). PDK1, an enzyme involved in inactivating the pyruvate dehydrogenase complex and polarizing the citric acid cycle away from glycolysis and toward utilizing FAs as the source of acetyl-CoA, was also upregulated in responders (Fig. 1A). To validate these findings, we utilized the TCGA HNC cohort data set and generated a subgroup of HPV-negative HNC patients who underwent RT. Overall survival (OS) was significantly higher in those with higher expression of PPARA (P = 0.039; Fig. 1C) and trended toward improved OS in those with higher ACOX1 expression (P = 0.052; Fig. 1D). Other related or downstream genes were concordant with similar survival analyses findings, including PPARGC1A (PPARγ cofactor; Supplementary Fig. S1G), CPT1C (the carnitine shuttle; Supplementary Fig. S1H), and PDK1 (shunts the citric acid cycle toward FA use; Supplementary Fig. S1I). To further explore this finding, we compared proteomic enrichment pathways between mice implanted with the MEER cell line that responds to HRT (responders) to those implanted with the MOC2 cell line (nonresponders; Supplementary Fig. S2A). KEGG and Hallmark pathways for oxidative phosphorylation and fatty acid metabolism were upregulated in the responder mice (Fig. 1E and F; Supplementary Fig. S2B–S2D), as well as the GO Biological Process for lipid oxidation (Supplementary Fig. S2E). Overall, these data are consistent with the upregulated expression of fatty acid catabolism and correlated with improved outcomes in our clinical trial data.
To investigate the potential link between metabolism and immune response, we interrogated circulating metabolites in trial patients’ plasma. Serum OA, a main constituent FA in olive oil (39–41), was significantly upregulated in pretreatment sera of responders (Supplementary Fig. S1J) and correlated with response (ROC curve with AUC 0.885; Supplementary Fig. S1K). Cell viability assays showed increased intrinsic cancer cell toxicity in vitro upon treatment with FF or OA (Supplementary Fig. S1L). Extracellular acidification rate (ECAR), a surrogate to glycolysis, also decreased in cancer cells in response to acute injections of FF, OA, or both (Supplementary Fig. S1M and S1N). Furthermore, measurement of extracellular flux 24 hours after cancer cell irradiation with 8 Gy showed that RT results in an augmented glycolytic phenotype (Supplementary Fig. S1O). These data suggest that pharmacologic treatment with either FF or OA could increase cancer cell cytotoxicity and synergize with RT in concordance with previous literature (42, 43). Given these collective findings, we aimed to investigate the relationship between RT, immune response, and metabolic changes in murine models of HNC fed a high concentration OA diet (37.6% kcal from OA; Supplementary Fig. S3) with and without pharmacologic upregulation of FA catabolism with PPARα agonist fenofibrate (FF).
FF enhances antitumor efficacy of HRT whereas OAD results in effect reversal
Given the in vitro findings of enhanced cancer cell kill and glycolytic activity, we hypothesized that enhancing FA catabolism either through a high OA diet (Supplementary Fig. S3) or pharmacologic PPARα agonism with FF would improve the efficacy of RT in combination with checkpoint blockade in murine orthotopic models of HNC (Fig. 2A; Supplementary Fig. S4A). At day 26 after tumor implantation, FF combined with HRT significantly improved antitumor effect in comparison with HRT alone or control (Fig. 2B), an effect that was only present synergistically with HRT (Fig. 2C). Notably, mice treated with combination HRT and FF had equivalent outcomes in mean tumor volume in comparison with HRT with anti–PD-L1 therapy and HRT with both FF and anti–PD-L1 therapy (Fig. 2D). Upon measuring tumor volume response in mice fed either the OAD or CD for 3 weeks prior to cancer cell implantation, there were no differences between groups based on either diet or the addition of radiotherapy (Fig. 2E and F). However, in stark contrast to the HRT + FF combination treatment, the addition of OAD to this combination resulted in negation of the antitumoral growth effect observed with HRT and FF (Fig. 2G and H; Supplementary Fig. S4B). Validation of increased OA with OAD was confirmed with bulk serum metabolomics showing upregulated serum-free OA and the acyl-carnitine form of OA in mice fed the OAD (Supplementary Fig. S4C and S4D). This reversal of effect with the OAD was replicated with the LY2 murine HNC model (Supplementary Fig. S4E and S4F). To explore the potential reason why there was no synergy of FF and anti–PD-L1, flow cytometry performed on tumors removed from mice on either the OAD or CD and treated with HRT and FF or HRT alone showed that PD-L1 expression was overall lower for all cells and CD45-negative cells in mice treated with FF (Supplementary Fig. S4G). Pertinently, the reversal of the antitumoral benefit of FF + HRT observed with OAD was unique to this combination as it was not observed in the context of HRT and anti–PD-L1 (Supplementary Fig. S4H–S4K). These results collectively suggest that when combined with HRT, FF is equally efficacious to immune checkpoint inhibition in reducing tumor growth, an effect that is negated with the addition of OAD.
FF upregulates acyl-carnitine formation and FA catabolism whereas the OAD results in significantly lower serum omega 3 and 6 FAs
For metabolic exploration of the interaction of the OAD, FF, and HRT, mice were started on either the OAD or CD prior to implantation and treated them with CD + HRT, CD + HRT + FF, or OAD + HRT + FF (Fig. 3A). Consistent with our previous findings, treatment with FF improved HRT efficacy, whereas the OAD counteracted this effect (Fig. 3B). Serum metabolomic analysis revealed that mice from the CD + HRT + FF group compared with HRT alone were consistent with upregulated carnitine production, where the amino acids used to synthesize L-carnitine were significantly lower, whereas L-carnitine itself was higher in the combination HRT + FF–treated group (Fig. 3C). Several fatty acyls (FAs attached to L-carnitine to shuttle into the mitochondria for beta oxidation), including short, medium, and long-chain fatty acyls were also higher in the combination CD + HRT + FF group in comparison with the CD + HRT group (Fig. 3D). These data are consistent with upregulated enzymes involved in the carnitine shuttle pathway, including CPT1, and imply that FA catabolism increased with FF treatment.
To assess the effect of FF on FA catabolism, serum FA comparison between the CD + HRT + FF group and the CD + HRT group showed lower free saturated FA with relatively unaffected free unsaturated FAs (Fig. 3E and F, shown as “FF effect”). The addition of OAD to the HRT + FF group increased free saturated FAs and lowered unsaturated FAs (“OA effect,” Fig. 3E and F) in comparison with the HRT + FF group on the CD. When the OAD was combined with FF, the free saturated FAs remained increased or near baseline. Lastly, upon a comparison of the OAD + HRT + FF group with the CD + HRT group, the decrease in saturated FAs noted from the FF effect was negated (“OA with FF effect,” Fig. 3E and F). This shows that when OA and FF were combined, the FA profile appeared to be less affected by the PPARα agonism with FF and more so by the supplementation with OA. Subclassification by monounsaturated fatty acids (MUFA) and polyunsaturated fatty acids (PUFA) revealed that the FF effect resulted in predominantly increased PUFAs, whereas the OAD with FF resulted in increased MUFAs (PUFAs shown with asterisks in Fig. 3F). These findings were corroborated with significant reduction in omega-3 FAs and omega-6 FAs (key PUFAs) when OA was added to HRT+ FF (Fig. 3G–I). Specifically, the omega-6 FA arachidonic acid and linoleoyl-CoA (activated form of linoleic acid) were higher in the CD + HRT + FF group. In summary, these data show that FF lowers serum free saturated FAs and increases PUFA, whereas the addition of OAD increased free saturated FAs and lowered free unsaturated FAs. The combination of OAD and FF resulted in an FA profile similar to OAD alone.
Proteomic analyses reveal that in addition to HRT upregulation of antigen presentation, FF alters arachidonic acid metabolism
As PUFAs, particularly omega-3 and omega-6 FAs, are relevant in the synthesis of eicosanoids (prostaglandins, leukotrienes, hydroxyeicosatetraenoic acids, hydroxyoctadecadienoic acids, etc.), which play a role in the regulation of inflammation and the immune response (44), we next examined the differential expression of these proteins using proteomic analyses with mass spectrometry. Pathway analyses of tumors taken from the in vivo experiments (Figs. 2 and 3) showed that HRT increased proteins related to pathways involved in antigen processing and presentation, regulation of response to cytokines, and extracellular matrix organization. These features are consistent with previous literature (refs. 45–47; Supplementary Fig. S5A). Downregulated features in the HRT group included regulation of DNA metabolism, protein assembly, and positive cell-cycle regulation (Supplementary Fig. S5B).
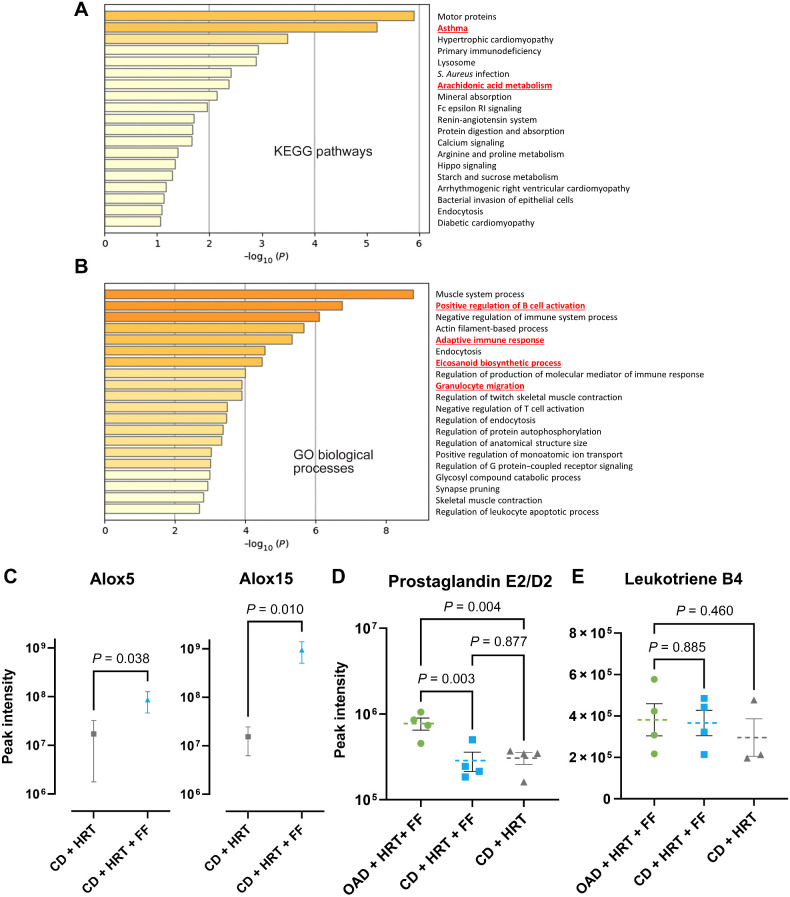
To examine the impact of pharmacologic treatment of FF on proteomic profiles, we compared enrichment patterns of HRT + FF to HRT alone and observed upregulation of KEGG pathways for asthma and arachidonic acid metabolism (Fig. 4A). GO biological processes for B-cell activation, adaptive immune response, and granulocyte migration and eicosanoid biosynthetic processes were also increased (Fig. 4B). Upon comparison of the proteomic profiles of OAD + HRT + FF and CD + HRT + FF, unsupervised clustering shows well-defined groups (Supplementary Fig. S5C) with upregulated GO pathways including interleukin-4 production, cellular response to oxygen levels, and positive regulation of apoptotic signaling pathways in the OAD group (Supplementary Fig. S5D). In connection with the metabolomic findings, lipoxygenase enzymes commonly expressed in leukocytes and involved in catalyzing leukotriene production (ALOX5 and ALOX15) were increased when FF was added to HRT (Fig. 5C). Given that arachidonate 5-lipoxygenase (ALOX5) converts arachidonic acid into leukotrienes, whereas arachidonate 15-lipoxygenase (ALOX15) converts polyunsaturated omega FAs into various metabolites (15 S-HETE and 13S-HODE; ref. 48), these data collectively suggest the pharmacologic treatment with FF increases FA catabolism and leukotriene production.
Figure 4.
Proteomic analyses reveal that in addition to HRT upregulation of antigen presentation, FF alters arachidonic acid metabolism. A and B, Bulk proteomic pathway analyses showing KEGG pathways (A) or GO processes (B) that were upregulated in FF + HRT compared with HRT alone. DGE terms with P < 0.15 and pathway analyses with P ≤ 0.1. C, Spectral intensity of Alox5 and Alox15 proteins. Mann–Whitney test was used to compare means. D and E, Oxylipin analysis with expression of prostaglandin E2/D2 (D) and leukotriene B4 (E) within the OAD + HRT + FF (green), CD + HRT + FF (blue), and CD + HRT (gray) groups.
Figure 5.
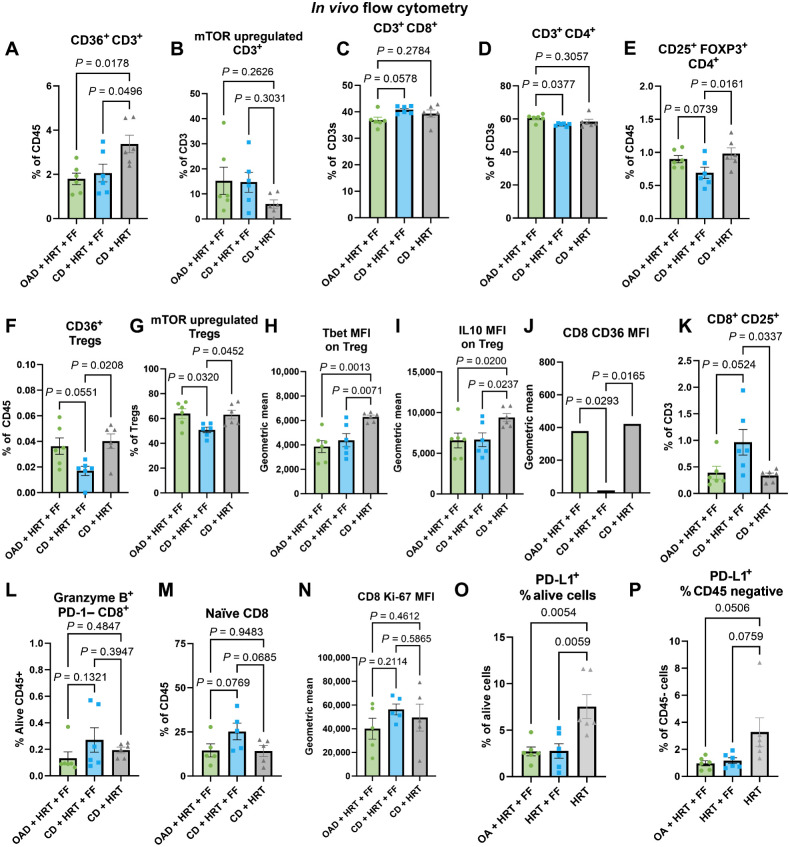
In vivo flow-cytometric characterization of the immunologic effects of FF and OA. A and B, Flow cytometry results from the excised tumors from the in vivo experiment comparing OAD + HRT + FF (green) with CD + HRT + FF (blue) and CD + HRT (gray), including CD36+ CD3+ T cells (A) and mTOR upregulated CD3+ T cells (B). C–I, Flow cytometry results from the draining lymph node from the in vivo experiment comparing OAD + HRT + FF (green) with CD + HRT + FF (blue) and CD + HRT (gray). Included gating: CD8+ T cells (C), CD4+ T cells (D), CD25+ FOXP3+ regulatory T cells (E), CD36+ regulatory T cells (F), and mTOR upregulated (p4EBP1 and pS6 ribosomal protein) regulatory T cells (G). Geometric MFI is shown for Tbet and IL10 expression on regulatory T cells (H–I). J–L, Flow cytometry results from the draining lymph node from the in vivo experiment comparing OAD + HRT + FF (green) with CD + HRT + FF (blue) and CD + HRT (gray). Included MFI of CD36 on CD8s (J), CD25+ CD8+ T cells (K), and granzyme B+ PD-1– CD8+ T cells (L). M–P, Flow cytometry results from the excised tumors from the in vivo experiment comparing OAD + HRT + FF (green) with CD + HRT + FF (blue) and CD + HRT (gray), including naïve CD44− CD62 L high CD8 T cells (J), MFI of Ki-67 on CD8 T cells (K), PD-L1+ alive cells (O), and PD-L1+ CD45-negative cells (P).
To further substantiate the association of FF treatment with enhanced PUFA catabolism, we compared the levels of oxylipins, lipophilic signaling molecules that are derived from the oxidation of PUFAs (ref. 49; Supplementary Fig. S6A and S6B). Oxylipin analysis revealed a significantly higher expression of Prostaglandin E2/D2 within OAD + HRT + FF mice in comparison with both CD groups (Fig. 4D). However, there was no significant difference in leukotriene B4 between groups (Fig. 4E). Prostaglandins such as E2 have been identified to be involved in the progression, invasion, and modulation of immune signaling in epithelial cancers (50, 51). Finally, to examine the impact of FF treatment on protein acetylation patterns, which have been shown to correlate with tumor progression and metastasis (52–54), acetylome analysis was conducted. Of relevance, N-terminal acetylation showed a decreased trend in FF-treated groups relative to other groups (Supplementary Fig. S6C). These data suggest that N-acetylation could serve as a potential future biomarker to evaluate the efficacy of FF-based treatment.
In vivo flow-cytometric characterization of the immunologic effects of FF and OA
Given our data demonstrating that the addition of FF acts to catalyze the increased FA catabolism (Fig. 3), the observed increase in the leukotrienes (Fig. 4D), and previous literature associating FF with CD8 T-cell stimulation via increased FA catabolism (55), we sought to examine the immune effects of FF within the TME and regional lymph nodes.
Multispectral flow cytometry was utilized to investigate the in vivo immunologic profile of the tumor and tumor-draining lymph node (LN) of mice from the experiment comparing CD + HRT, CD + HRT + FF, and OAD + HRT + FF. Within the TME, there was a lower proportion of CD36+ T cells in the FF groups (Fig. 5A). Accordingly, the HRT alone group had less overall mTOR upregulated T cells compared with the FF groups, but this was not significant (Fig. 5B). Within the LN compartment, there was a trend toward an increase in CD8 T cells observed in the CD + HRT + FF group relative to other groups (Fig. 5C), whereas a corresponding significant decrease was seen of CD4 T cells when comparing the CD + HRT + FF group to the OAD + HRT + FF group (Fig. 5D). This decrease corresponded with a significant reduction in Tregs in mice treated with CD + HRT + FF relative to other groups, most prominently in the LN (Fig. 5E). In the CD + HRT + FF group, Tregs also had less CD36 expression, and less mTOR upregulation (defined as phosphorylated 4EBP1 and s6 ribosomal protein; Fig. 5F and G). Tregs from the CD + HRT group had a significantly higher expression of T-box transcription factor TBX21 (Tbet) and IL10 on MFI in contrast to the other two groups (Fig. 5H and I). In contrast, CD8 activation markers were highest in the CD + HRT + FF group in both the tumor and the LN (Fig. 5J–N). Additionally, PD-L1 expression was significantly lower in both alive cells and CD45-negative cells in groups treated with FF (Fig. 5O–P). These data collectively suggest that within the LN compartment, CD + HRT + FF mice had more activation of CD8+ T cells and less CD36+ expressing Tregs with less mTOR activation, indicative of a dampened activation profile.
FF favors a th1 phenotype while the addition of OA polarizes CD4 T cells to a Treg phenotype
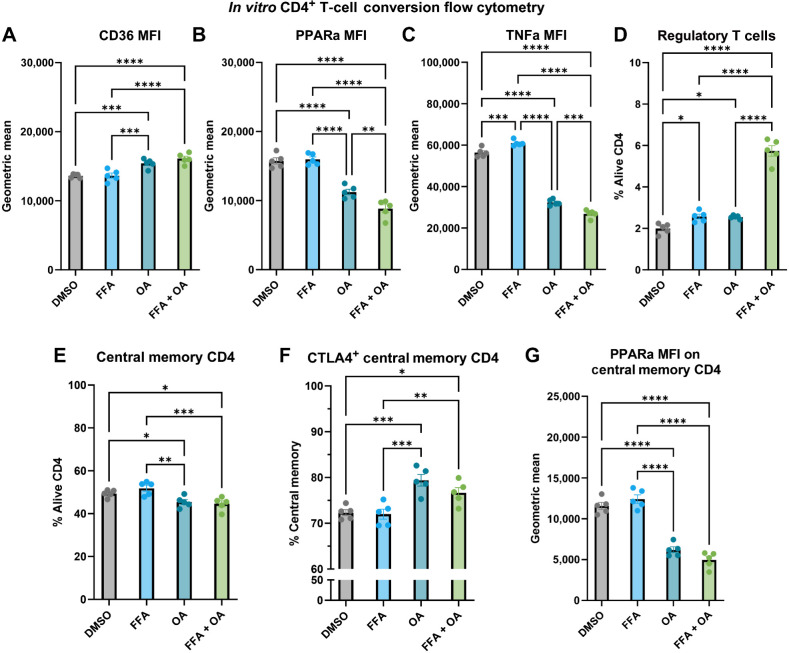
With the increase in Tregs that was observed when OA was added to HRT + FF, we hypothesized that cytotoxic effector T-cell function is possibly mitigated by this uprise in Tregs. To test this hypothesis, we performed a CD4+ T-cell conversion assay where isolated CD4+ T cells from tumor-free C57BL/6 mice were cultured for 24 hours with DMSO control, FFA, OA, or a combination of FFA + OA. On flow-cytometric analysis, treating the CD4+ T cells with either OA or FFA + OA combination resulted in significantly upregulated CD36, the FA translocase (Fig. 6A). Contrarily, PPARα was downregulated in CD4 T cells cultured with OA or a combination FFA + OA (Fig. 6B). Average expression of TNFα was higher in CD4+ T cells cultured in FFA alone in comparison with the other three groups (Fig. 6C) and a marked increase in polarization to Tregs was observed with the addition of OA to HRT and FF (Fig. 6D).
Figure 6.
FF favors a Th1 phenotype whereas the addition of OA polarizes CD4 T cells to a Treg phenotype. A–C, Flow cytometry results of the CD4 conversion assay showing the MFI of CD36 (A), PPARα (B), and TNFα (C) as a geometric mean of the four groups: DMSO control (gray), 100 μmol/L FFA (blue), 100 μmol/L OA (teal), combination 100 μmol/L FFA + OA (light green). D–G, Flow cytometry results of the CD4 conversion assay showing the polarization of cells to FOXP3+ CD25+ regulatory T cells (D), CD44+ CD62 L high central memory CD4 T cells (E), CTLA4+ expressing central memory CD4 T cells (F), PPARα MFI on central memory CD4 T cells (G).
Consistent with increased Tregs with OA addition, both OA alone and combination FFA + OA also had proportionally lower central memory CD4+ T cells (Fig. 6E; gated as CD62Lhigh and CD44+) with higher CTLA4 positivity (Fig. 6F) and with lower PPARα expression (Supplementary Fig. S7A). Accordingly, PPARα was significantly lower in the OA and combination FFA + OA groups (Fig. 6G). Overall, these data suggest that FF increased the proportion of central memory CD4+ T-cell polarization in comparison with OA groups, whereas OA downregulated PPARα signaling within all CD4+ T cells and memory CD4+ T cells; the combination FFA + OA group increased the proportion of Tregs in comparison with the other three groups.
Ex vivo isolated CD8 T cells from mice treated with FF have decreased glycolytic activity
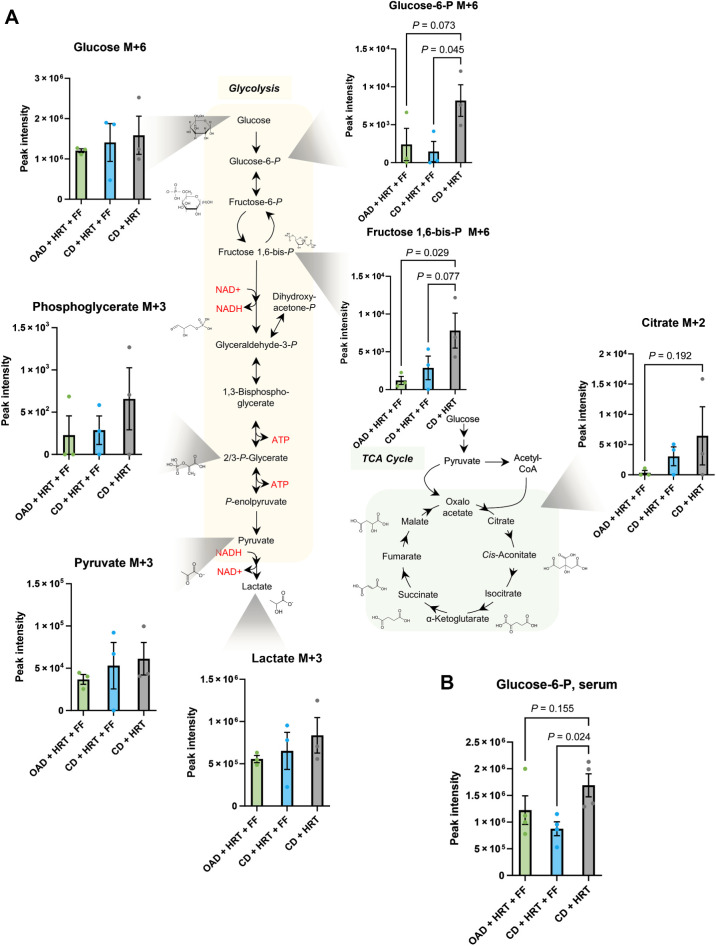
Supported by our flow cytometry data, we predicted that FF-induced upregulation of FA catabolism within the irradiated TME would allow higher glucose availability specifically for CD8 T cells to perform their effector functions (56). To test this hypothesis, CD8 T cells were isolated from the groups in Fig. 4A for ex vivo13C6-glucose labeling studies and tracing of their glycolytic metabolism. CD8 T cells in CD + FF + HRT mice had decreased glycolytic activity, and mice on the OAD had even lower glycolytic activity (Fig. 7A). Specifically, the initial glycolytic intermediate after the rate-limiting step of hexokinase (glucose-6-phosphate) was lower in the two FF groups compared with CD + HRT and this trend continued throughout the glycolytic pathway. This corresponded with metabolomic data on the serum of tumor-bearing mice sacrificed at the same time point that revealed lower glucose 6-phosphate levels in mice treated with HRT and FF on the CD in comparison with HRT alone (Fig. 7B).
Figure 7.
Ex vivo isolated CD8 T cells from mice treated with fenofibrate had decreased glycolytic activity. A, Heavy glucose labeling of ex vivo CD8 T cells from mice on the control diet with HRT alone (gray) or HRT with FF (blue), and mice on the high OAD treated with HRT and FF (green). Mice treated with fenofibrate results in decreased glycolytic flux, which was further decreased by the addition of OA. B, Correlative serum glucose 6-phosphate from HRT alone mice (gray), HRT with FF mice (blue), and mice on the OAD treated with HRT and FF (green). One-way ANOVA with multiple comparisons testing (uncorrected Fisher LSD) was used to determine P values (P values greater than 0.25 not shown).
To further examine the metabolic effects on CD8 T cells, we performed extracellular metabolic flux analysis on ex vivo CD8 T cells from murine models of HNC who were fed the CD or OAD and treated with HRT with or without FF. A glycolytic stress test (Supplementary Fig. S7A) revealed that ex vivo CD8 T cells from mice treated with FF and regardless of diet had decreased glycolytic capacity in comparison with control mice on either the CD or OAD. A mitochondrial stress test (Supplementary Fig. S7B) did not show a significant difference in mitochondrial respiration. Alternatively, to characterize how Tregs respond in vitro to PPARα agonism and OA, we performed similar extracellular flux assays including a mitochondrial stress test and a glycolytic stress test with drug injections of FFA (activated FF), OA, or both (Supplementary Fig. S7C and S7D). These assays revealed that Tregs increase respiratory capacity in response to FFA, OA, and both; however, FF alone did not increase glycolytic capacity whereas OA or FFA/OA combination did result in an increase in glycolytic capacity.
Discussion
This study expanded on potential avenues of immunometabolic interactions uncovered from our phase I/Ib clinical trial using anti–PD-L1 immunotherapy and HRT for HPV-unrelated HNC (20) that showed increased FA catabolism in responders. We evaluated the interaction of PPARα agonism (FF), supplemental dietary OA (with the OAD), and HRT on murine models of HNC. The combination of HRT with FF improved local control of tumors in comparison with HRT alone, and mice on the OAD showed a reversal of this effect. The combination FF and anti–PD-L1 therapy group did not result in significant tumor volume reduction in comparison with single-agent FF, which we postulate may be related to suppression of PD-L1 expression on tumor cells. Metabolomic exploration showed that FF-treated CD mice had lower free saturated fats and higher acyl-carnitine metabolite production with higher PUFAs whereas FF-treated OAD mice had lower free unsaturated fats and importantly, higher MUFAs. Stable isotope tracing showed less glycolytic utilization by ex vivo CD8+ T cells from mice treated with FF or OA, and extracellular flux analyses were consistent with these findings. Bulk tumor proteomic analysis suggested that the beneficial effect of PPARα signaling through FF is potentially related to arachidonic metabolism, eicosanoid biosynthetic processes, and the adaptive immune response. Characterization of immunologic changes from these metabolic modulators showed that increased FA catabolism or supplementation of OA resulted in enhanced memory phenotypes of memory lymphocytes and that the combination of the two may enhance immunosuppressive Tregs and result in decreased antitumor efficacy. The FF group without OA supplementation resulted in a Treg phenotype within the draining LN with downregulated mTOR and CD36, which could explain differences in tumor growth.
Modulating the metabolic conditions of the TME is of increasing interest as the link between the immune response and the metabolic environment has been further elucidated (57–59). The TME of HNC is typically hypoxic, which correlates with treatment resistance and oxidative stress (43, 60–62). The result is a cascade of metabolic changes that shift the tumor cell to a glycolytic phenotype characterized by higher expression of glucose transporters and glycolytic enzymes (7, 42). These metabolic changes result in a hypoglycemic TME that favors immunosuppressive and memory cells, but one that is poorly conducive to Teff cells maintaining effector functions. Previous work on murine sarcoma models showed that tumor competition for glucose consumption restricts Teffs function by dampening mTOR activity, glycolytic capacity, and interferon gamma (IFNγ) production. In these models, PD-L1 inhibition on the cancer cell inhibited mTOR activity and decreased glycolytic enzyme expression (63) whereas the effector function of T cells, specifically IFNγ and granzyme production, was restored with increased glucose availability. A similar effect was observed in a murine melanoma model where Teff cells showed increased PPARα signaling in hypoglycemic and hypoxic conditions but were rescued with FF (PPARα agonism), reprogramming them to instead utilize FAs subsequently resulting in improved response to immunotherapy (55).
Our results are consistent with previous literature in that FF-treated CD8+ T cells showed decreased glucose utilization ex vivo and increased FA catabolism with lower free saturated FAs and increased FA shuttling with acyl-carnitines were observed with FF treatment in vivo. Flow-cytometric results substantiated these concepts through enhanced mTOR expression in lymphocytes in the TME with decreased CD36 expression. Recent immunometabolism work has shown that CD36 expression in CD8+ T cells is associated with tumor progression and poor survival, as CD36 is integral for lipid peroxidation, ferroptosis, and dampening of tumor-infiltrating effector cells (64). Our data also showed that CD36 was downregulated in Tregs in the CD + HRT + FF group in comparison with the OAD + HRT + FF group and CD + HRT group, and in conversion assay, the addition of OA to FF markedly enhances Treg polarization phenotype. These data are indicative of enhanced Treg-suppressive function with the addition of OA as it has been recently shown that CD36 expression through PPARβ signaling is a characteristic feature of activated and highly suppressive Tregs and that lipid uptake via CD36 is vital for maintaining suppressive function (65). Likewise, the CD4 conversion assay (Fig. 6) showed polarization of CD4s to Treg in both the FFA and OA groups, but with the most significant polarization within the combination FFA + OA groups, which may be related to suppressed mTOR activity (66) from PPAR agonism. Likewise, extracellular flux analyses of cultured Tregs showed enhanced glycolytic capacity only in OA groups but did show enhanced mitochondrial respiration in FF groups. Although suppression of the mTOR pathway can result in polarization of naïve CD4 T cells to favor Treg phenotypes, there is evidence that mTORC1 (p4EBP1 and pS6 ribosomal protein pathway) is necessary to maintain suppressor function (67–69), and that downregulation of mTOR in already activated Tregs may dampen their suppressor functions. This could explain how there was more Treg polarization in the in vitro assay in FFA groups, whereas the in vivo flow data showed potentially a benefit of mTOR downregulated Tregs in the CD + FF + HRT group with less CD36 expression.
Additionally, various PUFAs including docosahexaenoic acid and arachidonic acid were upregulated in the CD + FF + HRT group in comparison with the OAD + FF + HRT group, which is notable due to previous work showing that these metabolites are associated with antiproliferative effects (70). Furthermore, arachidonic acid is critical for eicosanoid biosynthetic pathways, which was shown to be upregulated in FF-treated mice in the proteomics data. Specifically, alox5 and alox15 were upregulated in FF-treated mice which are involved in the conversion of arachidonic acid into leukotrienes. There are data that alox15 expression can lead to cancer cell apoptosis and is downregulated in many cancer subtypes, and increasing flux through these pathways with cyclooxygenase inhibitors could have antitumor efficacy (71, 72); likewise, alox15 may promote tumor ferroptosis (73), which could also explain improved HRT efficacy. Additionally, mice sera of the poorer performing OAD + HRT + FF group had increased levels of prostaglandin E2/D2, which has been linked to stimulated proliferation, stabilization of B-catenin, activation of EGFR, and stimulation of tumor angiogenesis (74). This difference in prostaglandin levels between dietary groups treated with FF may be explained by increased linoleoyl-CoA in the CD + FF + HRT group in comparison with the OAD group, as linoleoyl-CoA can inhibit prostaglandin synthesis (75).
The translational implications of this work are significant and pave the way for integrating FF as a novel, widely available and inexpensive, immunotherapeutic agent in HNC. Our data showed that FF had an equivalent benefit to immune-checkpoint blockade when combined with HRT. Historically, PPARα agonists (fibrates) have been used in the treatment of dyslipidemia, whereas PPARγ agonists (glitazones) have been used to increase insulin sensitivity in the management of diabetes mellitus (35). FF is commonly used and is FDA-approved to treat various forms of dyslipidemia (generally as second-line therapy; ref. 76), as its main mechanism for this indication is lowering triglycerides and increasing high-density lipoprotein cholesterol (HDL). It has a favorable toxicity profile compared with most oncologic therapies which can be easily monitored (77). Oncologic studies involving PPARγ agonists have shown dichotomous effects, with some studies showing inhibition of cancer cell proliferation and others showing anti-inflammatory effects that may dampen the immune response (78). In HNC, and in agreement with the beneficial effects of FF, others have shown that FF can have a direct effect on decreasing cancer cell proliferation, survival, and metastasis through the NFKB pathway (79), reactivation of the p53 pathway (80), or enhancing cancer cell apoptosis by G2–M arrest especially when combined with RT (81).
There has been considerable debate about the optimal diet while undergoing anticancer therapies and pharmacologic metabolic interventions are in the early stage of employment in clinical trials (82). Early work evaluating nutritional status in the setting of chemotherapy and radiotherapy revolved around support to optimize the diet, considering the relevance of anorexia related to treatment side effects and cancer-related cachexia (83). Several retrospective reviews of HNC show improved OS and improved distant control with higher pretreatment BMI (84–86). Dietary modifications are so far inconclusive as a general means of improving oncologic outcomes (87).
There have been conflicting studies regarding the oncologic benefits or lack thereof for OA. Curiously, when comparing the OAD to CD when treating with HRT without FF, there was a trend for improvement in tumor volume control for mice on the OAD which aligned with our original hypothesis (Fig. 2F). The OAD only had a negative effect in comparison with CD when fenofibrate was added to HRT (Fig. 2H). A recent review (88) highlights OA as a dietary MUFA from vegetables with several health benefits that exerts its effect through upregulated FA oxidation via PPARγ coactivator 1-a (PGC-1a) whereas other common long-chain FAs do not (89). The benefit of oleic acid in a nononcologic context could be related to resistance to ferroptosis; however, exogenous MUFAs such as OA can promote an advantage for cancer cells to resist RT through downregulated ferroptosis by displacing PUFAs from plasma membranes and resulting in decreased reactive oxygen species accumulation (90). Therefore, the OAD may provide a protective advantage to tumor cells through resistance to ferroptosis (91). The CD + HRT + FF group showed a higher proportion of PUFAs in comparison with MUFAs, which may be related, in part, to the advantage of FF alone. Research supporting potential oncologic benefits for OA is related to the suppression of the HER2/EGFR/ERBB family or contrarily, induction of apoptosis from intracellular reactive oxygen species production (92, 93). Alternatively, the induction of immunosuppression results from the feedback loop from overinflammation of the TME to regain homeostasis by recruiting Tregs. Further in vivo investigation of these agents is warranted as a potential avenue to modulate immunometabolism with diet or drugs of long approval status and known tolerability and adverse effects.
There are several limitations of this study that should be considered before drawing concluding interpretations of this study. The RNA-seq data from human samples included a relatively low number of samples for hypothesis generation, and the overall interpretation of the data in Fig. 1 should be approached with caution due to low numbers. The TCGA survival analyses almost certainly contained a cohort of conventionally fractionated patients as opposed to SBRT. However, this specific treatment information is not included in the database. It is also important to note that the small animal linear accelerator used in this study is not capable of delivering SBRT but did deliver HRT as a surrogate under fluoroscopic guidance, which has a similar dose and fractionation as SBRT. The bulk mass spectrometry–based analyses also have inherent limitations including the measurement of relative protein/metabolite concentrations instead of absolute quantifications, the limited detection of low-abundance proteins/metabolites, and the lack of spatial or temporal resolution (94, 95). For these reasons, statistically significant findings in the reported data may not always imply biological significance. Another consideration is that OA was not administered as a single agent for the in vivo experiments and otherwise was supplemented with a diet that included other ingredients that could influence the results.
Supplementary Material
Supplemental figures
Acknowledgments
Dr. Karam is funded by NIH 1R01DE028282-01, 1R01DE028529-01, 1P50CA261605-01, and 1R01CA284651-01. The Mass Spectrometry Shared Resource receives support from the University of Colorado Cancer Center via NCI P30CA046934.
Dr. Ross is funded by the Radiological Society of North America, RSNA #RR2337 and the Cancer League of Colorado, CLC AWD-232571-RR.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
R.B. Ross reports grants from the Radiological Society of North America (RSNA) and the Cancer League of Colorado during the conduct of the study. S.D. Karam reports grants from R01DE028282, R01DE028529, R01CA284651, and P50CA261605 during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
R.B. Ross: Conceptualization, resources, data curation, software, formal analysis, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J. Gadwa: Conceptualization, formal analysis, investigation, methodology, writing–review and editing. J. Yu: Conceptualization, data curation, investigation, methodology, writing–review and editing. L.B. Darragh: Conceptualization, investigation, methodology, writing–review and editing. M.W. Knitz: Conceptualization, data curation, software, formal analysis, validation, visualization, methodology, writing–review and editing. D. Nguyen: Investigation, methodology, writing–review and editing. N.A. Olimpo: Conceptualization, methodology, writing–review and editing. K.N.M. Abdelazeem: Conceptualization, investigation, methodology, writing–review and editing. A. Nguyen: Investigation, writing–review and editing. S. Corbo: Investigation, methodology, writing–review and editing. B. Van Court: Software, investigation, methodology, writing–review and editing. J. Beynor: Investigation, writing–review and editing. B. Neupert: Software, investigation, writing–review and editing. A.J. Saviola: Data curation, formal analysis, investigation, methodology, writing–review and editing. A. D'Alessandro: Conceptualization, data curation, formal analysis, supervision, validation, investigation, methodology, writing–review and editing. S.D. Karam: Conceptualization, resources, data curation, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft.
References
- 1. Lee NY, Ferris RL, Psyrri A, Haddad RI, Tahara M, Bourhis J, et al. Avelumab plus standard-of-care chemoradiotherapy versus chemoradiotherapy alone in patients with locally advanced squamous cell carcinoma of the head and neck: a randomised, double-blind, placebo-controlled, multicentre, phase 3 trial. Lancet Oncol 2021;22:450–62. [DOI] [PubMed] [Google Scholar]
- 2. Haddad RI, Harrington K, Tahara M, Ferris RL, Gillison M, Fayette J, et al. Nivolumab plus ipilimumab versus EXTREME regimen as first-line treatment for recurrent/metastatic squamous cell carcinoma of the head and neck: the final results of CheckMate 651. J Clin Oncol 2023;41:2166–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 2019;394:1915–28. [DOI] [PubMed] [Google Scholar]
- 4. Ferris RL, Blumenschein G, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 2016;375:1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruffin AT, Li H, Vujanovic L, Zandberg DP, Ferris RL, Bruno TC. Improving head and neck cancer therapies by immunomodulation of the tumour microenvironment. Nat Rev Cancer 2023;23:173–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen N, Bellile E, Thomas D, McHugh J, Rozek L, Virani S, et al. Tumor infiltrating lymphocytes and survival in patients with head and neck squamous cell carcinoma. Head Neck 2016;38:1074–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers 2020;6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Y-P, Wang Y-Q, Lv J-W, Li Y-Q, Chua MLK, Le Q-T, et al. Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann Oncol 2019;30:68–75. [DOI] [PubMed] [Google Scholar]
- 9. Partlová S, Bouček J, Kloudová K, Lukešová E, Zábrodský M, Grega M, et al. Distinct patterns of intratumoral immune cell infiltrates in patients with HPV-associated compared to non-virally induced head and neck squamous cell carcinoma. Oncoimmunology 2015;4:e965570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mody MD, Rocco JW, Yom SS, Haddad RI, Saba NF. Head and neck cancer. Lancet 2021;398:2289–99. [DOI] [PubMed] [Google Scholar]
- 11. Barker HE, Paget JT, Khan AA, Harrington KJ. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer 2015;15:409–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lhuillier C, Rudqvist NP, Elemento O, Formenti SC, Demaria S. Radiation therapy and anti-tumor immunity: exposing immunogenic mutations to the immune system. Genome Med 2019;11:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alsahafi E, Begg K, Amelio I, Raulf N, Lucarelli P, Sauter T, et al. Clinical update on head and neck cancer: molecular biology and ongoing challenges. Cell Death Dis 2019;10:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mohamad I, Karam I, El-Sehemy A, Abu-Gheida I, Al-Ibraheem A, AL-Assaf H, et al. The evolving role of stereotactic body radiation therapy for head and neck cancer: where do we stand? Cancers (Basel) 2023;15:5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mittal A, Nenwani M, Sarangi I, Achreja A, Lawrence TS, Nagrath D. Radiotherapy-induced metabolic hallmarks in the tumor microenvironment. Trends Cancer 2022;8:855–69. [DOI] [PubMed] [Google Scholar]
- 16. Lim AR, Rathmell WK, Rathmell JC. The tumor microenvironment as a metabolic barrier to effector T cells and immunotherapy. eLife 2020;9:e55185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blagih J, Coulombe F, Vincent EE, Dupuy F, Galicia-Vázquez G, Yurchenko E, et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 2015;42:41–54. [DOI] [PubMed] [Google Scholar]
- 18. Prendeville H, Lynch L. Diet, lipids, and antitumor immunity. Cell Mol Immunol 2022;19:432–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaymak I, Williams KS, Cantor JR, Jones RG. Immunometabolic interplay in the tumor microenvironment. Cancer Cell 2021;39:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Darragh LB, Knitz MM, Hu J, Clambey ET, Backus J, Dumit A, et al. A phase I/Ib trial and biological correlate analysis of neoadjuvant SBRT with single-dose durvalumab in HPV-unrelated locally advanced HNSCC. Nat Cancer 2022;3:1300–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oweida AJ, Bhatia S, Van Court B, Darragh L, Serkova N, Karam SD. Intramucosal inoculation of squamous cell carcinoma cells in mice for tumor immune profiling and treatment response assessment. J Vis Exp 2019(146):10.3791/59195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhatia S, Nguyen D, Darragh LB, Van Court B, Sharma J, Knitz MW, et al. EphB4 and ephrinB2 act in opposition in the head and neck tumor microenvironment. Nat Commun 2022;13:3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petit V, Massonnet G, Maciorowski Z, Touhami J, Thuleau A, Némati F, et al. Optimization of tumor xenograft dissociation for the profiling of cell surface markers and nutrient transporters. Lab Invest 2013;93:611–21. [DOI] [PubMed] [Google Scholar]
- 24. Nemkov T, Reisz JA, Gehrke S, Hansen KC, D'Allesandro A. High-throughput metabolomics: isocratic and gradient mass spectrometry-based methods. Methods Mol Biol 2019;1978:13–26. [DOI] [PubMed] [Google Scholar]
- 25. Reisz JA, Zheng C, D'Alessandro A, Nemkov T. Untargeted and semi-targeted lipid analysis of biological samples using mass spectrometry-based metabolomics. Methods Mol Biol 2019;1978:121–35. [DOI] [PubMed] [Google Scholar]
- 26. Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 2021;49:W388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCabe MC, Schmitt LR, Hill RC, Dzieciatkowska M, Maslanka M, Daamen WF, et al. Evaluation and refinement of sample preparation methods for extracellular matrix proteome coverage. Mol Cell Proteomics 2021;20:100079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Naba A, Clauser KR, Hynes RO. Enrichment of extracellular matrix proteins from tissues and digestion into peptides for mass spectrometry analysis. J Vis Exp 2015:e53057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat Methods 2017;14:513–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 31. Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang X, Smits AH, van Tilburg GB, Ovaa H, Huber W, Vermeulen M. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat Protoc 2018;13:530–50. [DOI] [PubMed] [Google Scholar]
- 34. Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Montaigne D, Butruille L, Staels B. PPAR control of metabolism and cardiovascular functions. Nat Rev Cardiol 2021;18:809–23. [DOI] [PubMed] [Google Scholar]
- 36. Talley JT, Mohiuddin SS. Biochemistry, fatty acid oxidation. Treasure Island (FL): StatPearls; 2023. [PubMed] [Google Scholar]
- 37. Zhang X, Li K, Jones RA, Bruner SD, Butcher RA. Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans. Proc Natl Acad Sci USA 2016;113:10055–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goetzman ES. The regulation of acyl-CoA dehydrogenases in adipose tissue by rosiglitazone. Obesity (Silver Spring) 2009;17:196–8. [DOI] [PubMed] [Google Scholar]
- 39. Sales-Campos H, Souza PR, Peghini BC, da Silva JS, Cardoso CR. An overview of the modulatory effects of oleic acid in health and disease. Mini Rev Med Chem 2013;13:201–10. [PubMed] [Google Scholar]
- 40. Pauwels EK. The protective effect of the Mediterranean diet: focus on cancer and cardiovascular risk. Med Princ Pract 2011;20:103–11. [DOI] [PubMed] [Google Scholar]
- 41. Menendez JA, Vellon L, Colomer R, Lupu R. Oleic acid, the main monounsaturated fatty acid of olive oil, suppresses Her-2/neu (erbB-2) expression and synergistically enhances the growth inhibitory effects of trastuzumab (Herceptin) in breast cancer cells with Her-2/neu oncogene amplification. Ann Oncol 2005;16:359–71. [DOI] [PubMed] [Google Scholar]
- 42. Zhong J, Rajaram N, Brizel DM, Frees AE, Ramanujam N, Batinic-Haberle I, et al. Radiation induces aerobic glycolysis through reactive oxygen species. Radiother Oncol 2013;106:390–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meijer TW, Kaanders JH, Span PN, Bussink J. Targeting hypoxia, HIF-1, and tumor glucose metabolism to improve radiotherapy efficacy. Clin Cancer Res 2012;18:5585–94. [DOI] [PubMed] [Google Scholar]
- 44. Johnson AM, Kleczko EK, Nemenoff RA. Eicosanoids in cancer: new roles in immunoregulation. Front Pharmacol 2020;11:595498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gupta A, Probst HC, Vuong V, Landshammer A, Muth S, Yagita H, et al. Radiotherapy promotes tumor-specific effector CD8+ T cells via dendritic cell activation. J Immunol 2012;189:558–66. [DOI] [PubMed] [Google Scholar]
- 46. Lugade AA, Moran JP, Gerber SA, Rose RC, Frelinger JG, Lord EM. Local radiation therapy of B16 melanoma tumors increases the generation of tumor antigen-specific effector cells that traffic to the tumor. J Immunol 2005;174:7516–23. [DOI] [PubMed] [Google Scholar]
- 47. Henke E, Nandigama R, Ergun S. Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front Mol Biosci 2019;6:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mashima R, Okuyama T. The role of lipoxygenases in pathophysiology; new insights and future perspectives. Redox Biol 2015;6:297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gabbs M, Leng S, Devassy JG, Monirujjaman M, Aukema HM. Advances in our understanding of oxylipins derived from dietary PUFAs. Adv Nutr 2015;6:513–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wilson DJ, DuBois RN. Role of prostaglandin E2 in the progression of gastrointestinal cancer. Cancer Prev Res (Phila) 2022;15:355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Echizen K, Hirose O, Maeda Y, Oshima M. Inflammation in gastric cancer: Interplay of the COX-2/prostaglandin E2 and Toll-like receptor/MyD88 pathways. Cancer Sci 2016;107:391–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Van Damme P, Lasa M, Polevoda B, Gazquez C, Elosegui-Artola A, Kim DS, et al. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc Natl Acad Sci USA 2012;109:12449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pavlou D, Kirmizis A. Depletion of histone N-terminal-acetyltransferase Naa40 induces p53-independent apoptosis in colorectal cancer cells via the mitochondrial pathway. Apoptosis 2016;21:298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. El Kawak M, Dhaini HR, Jabbour ME, Moussa MA, El Asmar K, Aoun M. Slow N-acetylation as a possible contributor to bladder carcinogenesis. Mol Carcinog 2020;59:1017–27. [DOI] [PubMed] [Google Scholar]
- 55. Zhang Y, Kurupati R, Liu L, Zhou XY, Zhang G, Hudaihed A, et al. Enhancing CD8(+) T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell 2017;32:377–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Palmer CS, Ostrowski M, Balderson B, Christian N, Crowe SM. Glucose metabolism regulates T cell activation, differentiation, and functions. Front Immunol 2015;6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang H, Franco F, Ho PC. Metabolic regulation of Tregs in cancer: opportunities for immunotherapy. Trends Cancer 2017;3:583–92. [DOI] [PubMed] [Google Scholar]
- 58. Riera-Domingo C, Audigé A, Granja S, Cheng W-C, Ho P-C, Baltazar F, et al. Immunity, hypoxia, and metabolism-the menage a trois of cancer: implications for immunotherapy. Physiol Rev 2020;100:1–102. [DOI] [PubMed] [Google Scholar]
- 59. Goliwas KF, Deshane JS, Elmets CA, Athar M. Moving immune therapy forward targeting TME. Physiol Rev 2021;101:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Swartz JE, Pothen AJ, van Kempen PMW, Stegeman I, Formsma FK, Cann EMV, et al. Poor prognosis in human papillomavirus-positive oropharyngeal squamous cell carcinomas that overexpress hypoxia inducible factor-1alpha. Head Neck 2016;38:1338–46. [DOI] [PubMed] [Google Scholar]
- 61. Brizel DM, Sibley GS, Prosnitz LR, Scher RL, Dewhirst MW. Tumor hypoxia adversely affects the prognosis of carcinoma of the head and neck. Int J Radiat Oncol Biol Phys 1997;38:285–9. [DOI] [PubMed] [Google Scholar]
- 62. Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell 2004;5:429–41. [DOI] [PubMed] [Google Scholar]
- 63. Chang C-H, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 2015;162:1229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ma X, Xiao L, Liu L, Ye L, Su P, Bi E, et al. CD36-mediated ferroptosis dampens intratumoral CD8(+) T cell effector function and impairs their antitumor ability. Cell Metab 2021;33:1001–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang H, Franco F, Tsui Y-C, Xie X, Trefny MP, Zappasodi R, et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat Immunol 2020;21:298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kurniawan H, Soriano-Baguet L, Brenner D. Regulatory T cell metabolism at the intersection between autoimmune diseases and cancer. Eur J Immunol 2020;50:1626–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu X, Mo W, Ye J, Li L, Zhang Y, Hsueh EC, et al. Regulatory T cells trigger effector T cell DNA damage and senescence caused by metabolic competition. Nat Commun 2018;9:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 2013;499:485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chapman NM, Zeng H, Nguyen T-LM, Wang Y, Vogel P, Dhungana Y, et al. mTOR coordinates transcriptional programs and mitochondrial metabolism of activated T(reg) subsets to protect tissue homeostasis. Nat Commun 2018;9:2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Trombetta A, Maggiora M, Martinasso G, Cotogni P, Canuto RA, Muzio G. Arachidonic and docosahexaenoic acids reduce the growth of A549 human lung-tumor cells increasing lipid peroxidation and PPARs. Chem Biol Interact 2007;165:239–50. [DOI] [PubMed] [Google Scholar]
- 71. Shureiqi I, Wu Y, Chen D, Yang XL, Guan B, Morris JS, et al. The critical role of 15-lipoxygenase-1 in colorectal epithelial cell terminal differentiation and tumorigenesis. Cancer Res 2005;65:11486–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Umar A. Is 15-LOX-1 a tumor suppressor? J Natl Cancer Inst 2012;104:645–7. [DOI] [PubMed] [Google Scholar]
- 73. Shintoku R, Takigawa Y, Yamada K, Kubota C, Yoshimoto Y, Takeuchi T, et al. Lipoxygenase-mediated generation of lipid peroxides enhances ferroptosis induced by erastin and RSL3. Cancer Sci 2017;108:2187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer 2020;122:4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fujimoto Y, Nakajima T, Murakami Y, Takami K, Nishida H, Sakuma S, et al. Effects of fatty acyl-coenzyme A esters on prostaglandin synthesis in rabbit kidney medulla microsomes. Prostaglandins Leukot Essent Fatty Acids 1992;47:265–8. [DOI] [PubMed] [Google Scholar]
- 76. Goldfine AB, Kaul S, Hiatt WR. Fibrates in the treatment of dyslipidemias–time for a reassessment. N Engl J Med 2011;365:481–4. [DOI] [PubMed] [Google Scholar]
- 77. Blane GF. Comparative toxicity and safety profile of fenofibrate and other fibric acid derivatives. Am J Med 1987;83:26–36. [DOI] [PubMed] [Google Scholar]
- 78. Mirza AZ, Althagafi II, Shamshad H. Role of PPAR receptor in different diseases and their ligands: physiological importance and clinical implications. Eur J Med Chem 2019;166:502–13. [DOI] [PubMed] [Google Scholar]
- 79. Su T-R, Yu C-C, Chao S-C, Huang C-C, Liao Y-W, Hsieh P-L, et al. Fenofibrate diminishes the self-renewal and metastasis potentials of oral carcinoma stem cells through NF-kappaB signaling. J Formos Med Assoc 2022;121:1900–7. [DOI] [PubMed] [Google Scholar]
- 80. O'Neill WQ, Xie X, Gui S, Yu H, Davenport J, Cartwright T, et al. Repositioning fenofibrate to reactivate p53 and reprogram the tumor-immune microenvironment in HPV+ head and neck squamous cell carcinoma. Cancers (Basel) 2022;14:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu J, Ge Y-Y, Zhu H-C, Yang X, Cai J, Zhang C, et al. Fenofibrate increases radiosensitivity in head and neck squamous cell carcinoma via inducing G2–M arrest and apoptosis. Asian Pac J Cancer Prev 2014;15:6649–55. [DOI] [PubMed] [Google Scholar]
- 82. Stine ZE, Schug ZT, Salvino JM, Dang CV. Targeting cancer metabolism in the era of precision oncology. Nat Rev Drug Discov 2022;21:141–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Donaldson SS, Lenon RA. Alterations of nutritional status: impact of chemotherapy and radiation therapy. Cancer 1979;43(5 Suppl):2036–52. [DOI] [PubMed] [Google Scholar]
- 84. Hicks DF, Bakst R, Doucette J, Kann BH, Miles B, Genden E, et al. Impact of obesity on outcomes for patients with head and neck cancer. Oral Oncol 2018;83:11–17. [DOI] [PubMed] [Google Scholar]
- 85. Fattouh M, Chang GY, Ow TJ, Shifteh K, Rosenblatt G, Patel VM, et al. Association between pretreatment obesity, sarcopenia, and survival in patients with head and neck cancer. Head Neck 2019;41:707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hollander D, Kampman E, van Herpen CM. Pretreatment body mass index and head and neck cancer outcome: a review of the literature. Crit Rev Oncol Hematol 2015;96:328–38. [DOI] [PubMed] [Google Scholar]
- 87. Kanarek N, Petrova B, Sabatini DM. Dietary modifications for enhanced cancer therapy. Nature 2020;579:507–17. [DOI] [PubMed] [Google Scholar]
- 88. Tutunchi H, Ostadrahimi A, Saghafi-Asl M. The effects of diets enriched in monounsaturated oleic acid on the management and prevention of obesity: a systematic review of human intervention studies. Adv Nutr 2020;11:864–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lim J-H, Gerhart-Hines Z, Dominy JE, Lee Y, Kim S, Tabata M, et al. Oleic acid stimulates complete oxidation of fatty acids through protein kinase A-dependent activation of SIRT1-PGC1alpha complex. J Biol Chem 2013;288:7117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Stockwell BR. Ferroptosis turns 10: emerging mechanisms, physiological functions, and therapeutic applications. Cell 2022;185:2401–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ubellacker JM, Tasdogan A, Ramesh V, Shen B, Mitchell EC, Martin-Sandoval MS, et al. Lymph protects metastasizing melanoma cells from ferroptosis. Nature 2020;585:113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Carrillo C, Cavia Mdel M, Alonso-Torre SR. Antitumor effect of oleic acid; mechanisms of action: a review. Nutr Hosp 2012;27:1860–5. [DOI] [PubMed] [Google Scholar]
- 93. Menendez JA, Papadimitropoulou A, Vellon L, Lupu R. A genomic explanation connecting "Mediterranean diet," olive oil and cancer: oleic acid, the main monounsaturated fatty acid of olive oil, induces formation of inhibitory "PEA3 transcription factor-PEA3 DNA binding site" complexes at the Her-2/neu (erbB-2) oncogene promoter in breast, ovarian and stomach cancer cells. Eur J Cancer 2006;42:2425–32. [DOI] [PubMed] [Google Scholar]
- 94. Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods 2012;9:555–66. [DOI] [PubMed] [Google Scholar]
- 95. Domon B, Aebersold R. Mass spectrometry and protein analysis. Science 2006;312:212–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures
Data Availability Statement
The RNA-seq data (20) have been previously deposited in a repository and is publicly available through GEO accession number GSE210287 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE210287). Other nonsequencing data generated in this study are available upon request from the authors.