Abstract
Wastewater sequencing has become a powerful supplement to clinical testing in monitoring SARS-CoV-2 infections in the post-COVID-19 pandemic era. While its applications in measuring the viral burden and main circulating lineages in the community have proved their efficacy, the variations in sequencing quality and coverage across the different regions of the SARS-CoV-2 genome are not well understood. Furthermore, it is unclear how different sample origins, viral extraction and concentration methods and environmental factors impact the reads sequenced from wastewater. Using high-coverage, amplicon-based, paired-end read sequencing of viral RNA extracted from wastewater collected directly from aircraft, pooled from different aircraft and airport buildings or from regular wastewater plants, we assessed the genome coverage across the sample groups with a focus on the 5′-end region covering the leader sequence and investigated whether it was possible to detect subgenomic RNA from viral material recovered from wastewater. We identified distinct patterns in the persistence of the different genomic regions across the different types of wastewaters and the existence of chimeric reads mapping to non-amplified regions. Our findings suggest that preservation of the 5′-end of the genome and the ability to detect subgenomic RNA reads, though highly susceptible to environment and sample processing conditions, may be indicative of the quality and amount of the viral RNA present in wastewater.
Keywords: SARS-CoV-2, Subgenomic RNA, Wastewater, Transcription/replication
1. Introduction
As SARS-CoV-2 infections diminish worldwide, wastewater sequencing has become an important option for the continuous monitoring of the dissemination of COVID-19. Coronaviruses are enveloped single-stranded positive-strand RNA viruses, that may have a primary transmission route via aerial droplets or by transfer from a contaminated surface to the mouth, nose or eyes. Some coronaviruses primarily infect the intestine and may have a preferred route of transmission via the fecal-oral route. While no solid evidence of waterborne transmission has been presented for SARS-CoV-2 [1], fragments of coronavirus RNA have been reported in feces and in wastewater [[2], [3], [4], [5], [6]] [[2], [3], [4], [5], [6]] [[2], [3], [4], [5], [6]], possibly following excretion via infected individuals' fecal material, or excreted from the respiratory tract into the water sewage systems. Wastewater from aircraft and airport buildings may constitute a relatively harsh environment, for example with added detergents and other chemicals used in the cleaning of aircraft toilets [7], often including highly alkaline chemicals that together degrade viruses and bacterial pathogens. The exposure to disinfectants and cleaning agents can lyse the virions and disrupt the double membrane vesicles, exposing the RNA freely to the liquid environment. It has been shown that membrane integrity is essential for viral RNA replication in coronaviruses and that the RNA present in the vesicles would be exposed to nucleases in the presence of detergent [8,9]. Exposed viral RNA may suffer environment-related damage, including alkaline hydrolysis or nuclease processing of RNA not protected by either an intact virion envelope or the so-called double-membrane vesicles produced during coronavirus replication/transcription. Wastewater could be efficiently treated with a wide range of detergents and disinfectants such as household bleach, ethanol, chlorhexidine, or sodium dioxide to eliminate the presence of SARS-CoV-2 [[10], [11], [12], [13]].
Furthermore, many studies [[12], [13], [14], [15], [16]] [[12], [13], [14], [15], [16]] [[12], [13], [14], [15], [16]] have evaluated the resistance of coronaviruses in wastewater environments. Wang et al. [12] showed that SARS-CoV could persist in hospital wastewater, domestic sewage, and non-chlorinated tap water for two days at 20 °C and up to 14 days at 4 °C, illustrating that coronavirus can persist in wastewater at both room and low temperatures and that survival time is reduced as temperature increases. Other factors such as microorganisms, pH, and encapsulation in solid debris present in the water have also been suggested as factors influencing the kinetics of virus degradation in untreated water [10,14,16] [[10], [14], [16], [17], [18], [19], [20], [21]] [[10], [14], [16], [17], [18], [19], [20], [21]]. It has been described that the intact enveloped SARS-CoV-2 virus is stable at room temperature in a wide pH range (pH from 3 to 10) [10], and that the SARS-CoV and SARS-CoV-2 S protein receptor binding domains (RBD) are stable within pH values ranging from 7.5 to 9 [22]. Interestingly, a study on stool samples [18] showed that SARS-CoV could survive up to 4 days in an alkaline pH of 9 while viruses in acidic stool samples with a pH of 6 are undetectable within 3 h. The conventional SARS-CoV-2 wastewater surveillance strategy relies predominantly on rapid surveys of the viral burden in the community [4,5] using a reverse transcription (RT)-qPCR-based approach. Wastewater sequencing [[23], [24], [25], [26]] [[23], [24], [25], [26]] [[23], [24], [25], [26]] can complement this approach and allow the deciphering and detection of small-scale community spread. In addition, wastewater sequencing approaches allow in-depth studies of all kinds of genomic features of the SARS-CoV-2 RNA that persist in wastewater, e.g. UTRs, genes, subgenomic RNAs, not only mutations or probe-based targets. Multiple parameters are crucial to ensure the quality of the sequencing output from a wastewater sample. Sample handling prior to the sequencing step involves multiple steps such as sample collection, transport, viral concentration, PCR amplification, which could individually contribute as a source of variation and affect the interpretation of the result. Seasonal effects such as the weather and viral circulation can impact the amount of virus that can be recovered from the wastewater, and therefore, studies consisting of samples collected within a large time spread should ensure that measurements are comparable. Viral particles can be concentrated in different ways for example by membrane filtration [27], using polyethylene glycol (PEG) [28], by centrifugation [29], or by using Nanotrap A microbiome particles (CeresNano) [[30], [31], [32]]. A number of comparative studies have been performed to compare concentration Nanotrap with other concentration methods [33,34], showing that all methods are applicable, and the choice of the method relies on many factors such as cost, resource availability and the actual set-up of the wastewater surveillance. Furthermore, higher amount of fecal material in a sample can make it more prone to PCR inhibition [35].
While wastewater monitoring serves as a powerful complement to community-based sequencing of nasopharyngeal swabs, showing similar trends of SARS-CoV-2 lineage circulation, there are very few applications of the methodology in the literature describing traces of virus active or prior replication in the wastewater environment. In a recent study [36] assessing respiratory and rectal shedding of SARS-CoV-2, no culturable virus or insignificant levels of subgenomic RNA reads could be found in rectal swab samples, suggesting that, if detectable, traces of active virus in wastewater could be challenging to find. In addition, while sequencing approaches have robustly proved to sufficiently capture the main circulating variants and mutations in wastewater, few comprehensive studies have described the degradation patterns of the different genomic features of SARS-CoV-2 in wastewater [37], available studies providing, for example, breadth of coverage as a metric [38].
Subgenomic RNA detection in SARS-CoV-2-positive samples is of particular interest as it is thought to provide an indication of active or previous virus replication and transcription. Presently, sequencing-based subgenomic RNA detection has been performed on routine naso- and oropharyngeal swabs [[39], [40], [41], [42]] [[39], [40], [41], [42]] [[39], [40], [41], [42]], while no available studies have provided evidence of the presence of these in wastewater. Identification of subgenomic RNA using paired-end sequence reads is a challenging task as it requires both a good coverage of the 5′-end of the genome, which contains the leader sequence and the transcription-regulating sequence (TRS) located at the 3′-end of the leader, TRS-L, and a good coverage of the regions around the TRS-B in the region preceding each of the ORFs in the 3’ -region of the virus genome. Many approaches to detect subgenomic RNA using paired-end sequence reads rely on finding read pairs spanning across these two regions [9,39,41,43,44]. It is therefore essential to understand the patterns of variation and degradation of the RNA obtained from wastewater, in particular in the vicinity the 5′-end region of the SARS-CoV-2 genome and to understand the possible factors, e.g. environmental, methodological that could contribute to this variation.
In our study, we collected (I) wastewater samples directly from six incoming aircraft, (II) wastewater pooled from samples multiple aircraft and airport buildings at the Copenhagen Airport, Denmark, in the period January–February 2023 and (III and |IV) wastewater from 29 treatment plants across Denmark [45] in June 2023 and November 2023. We mapped sequence reads obtained from the samples with a focus on the SARS-CoV-2 genomic 5′-end and detection of subgenomic RNAs. Using ORF-level sequence coverage and mapping summary statistics, we described the patterns of variation of sequence reads recovered from viral RNA in wastewater. Our results showed that subgenomic RNA can persist in wastewater but are infrequently detectable. Positive detection of the genomic 5′-end of the SARS-CoV-2 genome could be driven by multiple factors such as the viral concentration, RNA degradation in the wastewater environment and methodological aspects.
2. Methods
2.1. Sample collection and processing
Wastewater samples from aircraft were collected either (I) directly from six incoming long-haul flights arriving at Copenhagen Airport during the period 2023-01-12 to 2023-02-10 [46], or (II) from 10 samples from water tanks collecting water from different airport sources during the period 2023-01-02 to 2023-02-01. Wastewater collected from the aircraft was collected upon arrival as a grab sample from a service truck after emptying the tank of the aircraft. These samples were stored at <5 °C and directly transported to Statens Serum Institut, Copenhagen, Denmark, for analysis. Two types of pooled water samples were collected: first, wastewater pooled from several aircraft labelled as 'Pooled A′, and pooled water from aircraft and airport buildings such as hangars and terminals, labelled as 'Pooled B'. Sampling for the pools was conducted as 24-h composite time-proportional sampling. Pooled A samples were collected downstream the triturator, and Pooled B samples consisted of waters from downstream the triturator and other different sources. The samples were stored at ≤5 °C, collected from each site and transported on ice to a commercial laboratory for analysis. In addition, samples from 29 wastewater treatment plants [45] across Denmark were collected in 2023-06-13 week 24 (III), and 2023-11-23 week 47 (IV) as primarily flow proportional 24hr composites for comparison. The water was taken from 10L containers at the beginning of the inlet of the treatment plant and transported to Statens Serum Institut for analysis. The collection time periods were chosen so that wastewater collected during week 47 would have a similar viral concentration to wastewater collected in January 2023 [47]. A summary of the different types of samples is shown in Table 1.
Table 1.
Type of samples collected for the study.
| Sample type | Number of samples | Collection time window | Description | Collection method |
|---|---|---|---|---|
| Aircraft | 6 | Week 3–5 (2023-01-16 to 2023-02-10) | Wastewater collected directly from the aircraft toilets upon arrival at Copenhagen airport | Grab sample |
| Pooled A | 4 | Week 1–5 (2023-01-04 to 2023-02-01) | Wastewater collected at Copenhagen airport, pooled from multiple aircraft | 24-h composite time-proportional sampling |
| Pooled B | 6 | Week 1–5 (2023-01-02 to 2023-02-01) | Wastewater collected at Copenhagen airport, pooled from multiple aircraft, hangars and terminals | 24-h composite time-proportional sampling |
| WWTP_wk24 and WWTP_wk47 |
58 | Week 24 (2023-06-13) and week 47 (2023-11-23) | Wastewater collected at the inlet of 29 wastewater plants across Denmark | Flow proportional 24-h composite |
’Pooled A′ and ‘Pooled B’ samples were processed by mixing the homogenized and centrifuged wastewater liquid phase with a 25 % polyethylene glycol (PEG) and 5.6 % saline solution (NaCl). Extraction of nucleic acids was performed using magnetic beads using the VIRSeek Extractor kit (Genescan Technologies) followed by lysis on a thermoshaker at 700 rpm at 50 °C for 10 min. The final extraction was performed on a PurePrep nucleic acid extraction platform (Molgen). Individual aircraft samples were centrifuged to pellet toilet paper and pH adjusted to 7–9 with HCl prior to processing. Aircraft samples and samples from treatments plants were concentrated using Nanotrap Microbiome A Particles (CeresNano). Total nucleic acid was extracted using Maxwell HT kit (Promega) on a Hamilton liquid handler. Information on the collection dates and extraction procedures are shown in Supplementary Table 1. In all cases, RT-qPCR quantification was performed using a TaqMan assay with CDC N2 primer/probes.
2.2. Genome sequencing
Sequencing of the nucleic acids extracted from the wastewater samples was performed using slightly modified ARTIC [48] protocol. Extracted RNA was reverse transcribed using random hexamers and the SuperScript IV Reverse Transcriptase kit (Invitrogen) at 42 °C for 50 min, 70 °C for 10 min and brought to 4 °C. Subsequently, SARS-CoV-2 genomes were PCR amplified using primer set v4.0 from the ARTIC Network for multiplexed PCR for samples (I), (II) and (III) and v.5.3.2 for samples (IV), which was set up in two separate pools. Each PCR reaction contained 5 μL of 10X Q5 Reaction Buffer, 0.5 μL of 10 mM dNTPs, 3.6 μL of each primer pool, 0.25 μL of Q5 High-Fidelity DNA Polymerase (NEB), 13.15 μl of nuclease-free water, and 2.5 μL of the cDNA. Samples were amplified with the following conditions, initial activation at 98 °C for 30 s, followed by 30 cycles of denaturation at 98 °C for 15 s, annealing at 65 °C for 5 min, and a final extension at 65 °C for 5 min. For library preparation using the Nextera XT DNA Library Prep Kit (Illumina), amplicons were first tagmented at 55 °C for 10 min, followed by a 15-cycle PCR with the following conditions, 72 °C for 3 min, 95 °C for 30 s and 15 cycles of 95 °C for 10 s, 50 °C for 30 s and 72 °C for 60 s before a single step at 72 °C for 5 min. Library size distribution and quality were verified on a 4200 Tapestation (Agilent), and concentration was quantified using the Qubit dsDNA HS Assay Kit (Invitrogen). Libraries were sequenced on a MiSeq Illumina platform with a paired-end read length of 150 nucleotides for samples (I), (II) and (III) and using a paired-end read length of 74 nucleotides for samples (IV).
Raw reads from different extractions of the same wastewater sample were pooled together into a merged FASTQ file and then processed as follows: Quality trimming was performed using trim_galore (v. 0.6.7) using a stringency setting of 1. Trimmed reads were paired using the reformat.sh accessory script from BBMap (v. 39.01). Human reads were removed by aligning trimmed reads to the human genome (build version GRCh38) using BWA-MEM (v. 0.7.17-r1188) [49] with default settings; unmapped reads from the resulting SAM file were extracted using samtools [50] (v. 1.15). The trimmed and host-removed reads were subsequently aligned with the Wuhan-Hu-1 reference SARS-CoV-2 genome (GenBank Accession: NC_045512.2). Primers were trimmed using iVar [51] (v. 1.3.1) with appropriate settings and using an ARTIC v. 4 BED file for samples (I), (II) and (III) and v. 5.3.2 for samples (IV) (https://github.com/artic-network/primer-schemes).
2.3. Identification of subgenomic RNAs
Sequence reads derived from subgenomic RNAs were identified using the methodology described in our previous study [39]. The first step consists of extracting all read pairs having their forward reads mapped to the expected positions of the leader motif, nucleotides 52–67 in the Wuhan-Hu-1 NC_045512/MN908947.3 reference. Subgenomic RNA reads were subsequently as reads with an insert size larger than 21,000 nucleotides and then further classified into the different Orfs based on the start position of the mate read anchored to the 3′-end of the subgenomic RNA fragment. We used the assumed 3′-end position of the potential subgenomic mRNAs as previously described [43]; S: 21,552, Orf3a: 25,385, E: 26,237, M: 26,469, Orf6: 27,041, Orf7a: 27,388, Orf8: 27,884, and Orf9: 28,256.
For S, E, M and ORFs 6, 7a, and 8, we allowed a tolerance window up to the position of the AUG codon of the following open reading frame. For Orf9 (Nucleocapsid/3′ open reading frames), Orf9a and 9b were pooled together, and all positions between position 28,250 and the 3′-end position were considered as representative of Orf9 since no evidence of Orf10 subgenomic reads were detected in the data. No filtering on the start position on the 5′-end was applied so that all reads starting down from position 1 were considered in the analyses. A pseudocount of 1 subgenomic RNA was added to each identified read to account for missingness due to the lower limit of detection.
Analysis of 5′-end extension of the sequenced viral RNA Reads with start positions upstream to the ARTIC v4 forward primer 1 (position 25–50, sequence AACAAACCAACCAACTTTCGATCTC) were selected for further analysis, during which soft and hard-clipped bases on the 5′ side to the read were extracted. These sub-sequences were then matched to the Wuhan-Hu1 reference to determine from which part of the reference genome they originated.
3. Principal component analysis
The structure of the sample collection was assessed using principal component analysis (PCA) on a dataset consisting of sample summary information (Supplementary Table 1) and mean coverage values within the genomic features of the SARS-CoV-2 genome (Supplementary Table 2). The variables considered in the analyses were the following: mean coverage in the different genomic features, Leader positions 52–67, Orf1a, Orf1ab, S, Orf3a, E, M, Orf6, Orf7a, Orf7b, Orf8, Orf9, Orf10, the presence/absence of the 5′-end first 300 nucleotides, the total numbers of sequenced reads and mapped reads, the fraction of the genome covered by 10X and above and N2 Ct value. The PCA was performed using data scaling and visualized as a biplot using the R packages ggfortify [52] v. 0.4.14 and cluster [53] v. 2.1.4.
The following variable transformations and curation steps were applied: the presence/absence of the first 300 nucleotides in the 5′-end of the genome was encoded as a binary variable (Yes/No). for a mean coverage cut-off over 10X. A pseudo count of 1 was added to all the mean coverage values to address the absence of coverage.
3.1. Statistical tests
The statistical tests performed in this study, i.e. non-parametric Wilcoxon rank-sum tests, Kruskal-Wallis test and Fisher's exact test were performed using R v. 3.6.3.
4. Results
4.1. Coverage profile of the wastewater samples
Wastewater was collected at Copenhagen Airport during the period January–February 2023 and from wastewater treatment plants in June 2023 and November 2023. Samples originated from (I) six incoming flights [46], (II) from water tanks containing water pooled from different aircraft (‘Pooled A’), and from aircraft and airport hangars and terminals (‘Pooled B’). In addition, samples from 29 wastewater treatment plants across Denmark were collected during the summer (III) and in the winter (IV). The samples from November 2023 were collected during a period with comparable viral concentration in the wastewater as in January 2023 to allow better comparison with (I) and (II), both collected in January 2023. Detailed information on the collection date and extraction procedure is shown in Supplementary Table 1. Illumina paired-end read sequencing was performed before reads were mapped to the Wuhan-Hu1 genome reference. Read depth information was collected for each sample and visualized as a coverage profile plot shown in Fig. 1 and the mean coverage of genomic features of interest - i.e., each open reading frame and the 5′-UTR region - are summarized in Supplementary Table 2.
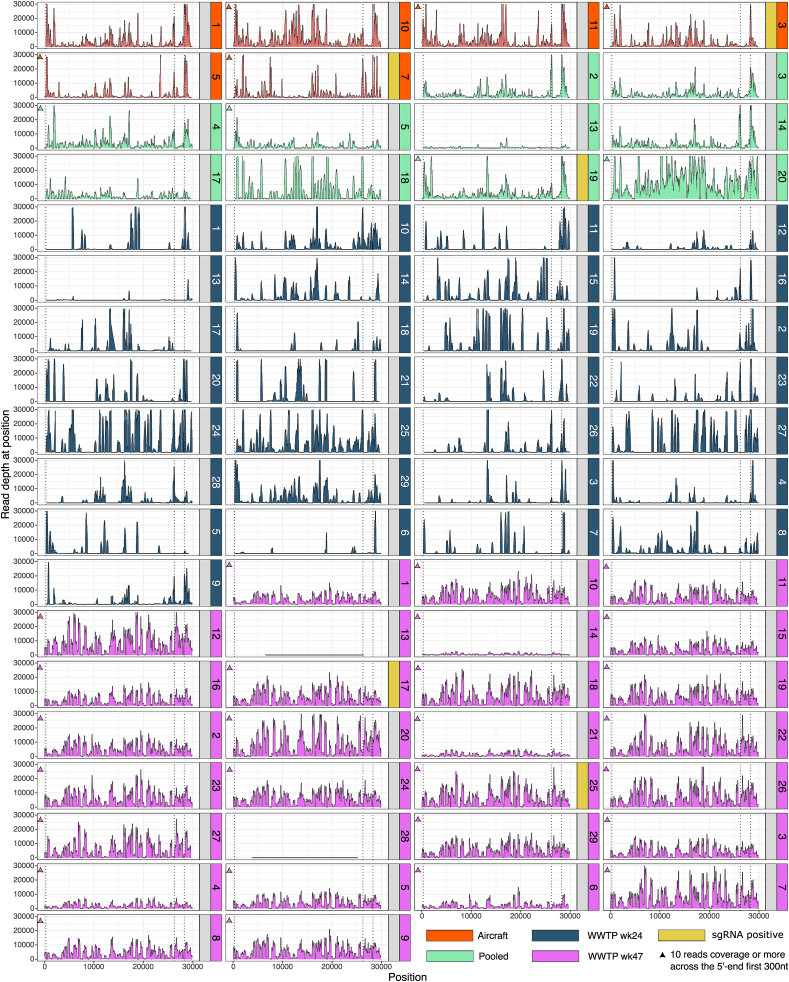
Fig. 1.
Genome coverage profile of the analyzed wastewater samples. The coverage profiles of reads sequenced from 74 wastewater samples collected in 2023 as shown. The samples consisted of wastewater collected directly from six long-distance flights arriving at Copenhagen airport (I, orange), 10 pooled samples from aircraft and airport buildings (II, green), and samples collected from 29 wastewater treatment plants in Denmark in week 24 (III, dark blue) and week 47 (IV, purple). The first vertical dotted line marks position 300, the second vertical dotted line the start position of subgenomic E (position 26,237) and the third vertical dotted line the start position of subgenomic Orf9 (position 28,255). The y-axis was limited to 30,000 reads per position. Samples covered by more than 10 reads in the 5′-end first 300-nucleotide region are marked with a triangle above the 300-nucletide region. Samples in which subgenomic RNAs were detected are marked in yellow. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
While looking at the coverage profiles, it was clearly noticeable that the sequenced wastewater samples were not uniformly covered, exhibiting a sawtooth pattern with distinct presence and absence of reads within regularly spaced large intervals, which could suggest signs of RNA degradation in the samples or failure of one of the two amplicon pools (e.g. sample WGSL18, AC7 or WWTP27_wk24, Fig. 1).
The 5′-end of the SARS-CoV-2 genome seemed to show important variation in coverage across the different types of samples. We found that the first 300 nucleotides were not covered in six out of 10 (60 %) wastewater samples pooled from aircraft and aircraft buildings while only one out of six (17 %) wastewater samples from individual aircraft toilet tanks did not have any coverage of the first 300 nucleotides. Interestingly, none of the samples from the wastewater plants collected in week 24 had coverage in this region (100 % with no coverage in this region) while 27 out of 29 (93 %) samples collected in week 47 had read coverage. We found generally that WWTP_wk47 samples covered the entire SARS-CoV-2 genome more extensively than WWTP_wk24 samples.
When coverage was available in the first 300 positions of the 5′-end, read depth was marginally lower than that of Orf1ab (depth range [3–8,535] vs. [10–13,354], Wilcoxon rank-sum test p = 0.03872), and significantly lower between the region covering the leader sequence (positions 52–67) and Orf1ab (depth range [1–2,729] vs. [10–13,354], Wilcoxon rank-sum test p = 5.483e-15).
Looking at the two sample groups with the most samples covering the first 300 nucleotide regions, i.e. the aircraft and WWTP_wk47 samples, we noted that WWTP_wk47 samples had little or no coverage in the leader region, 3 out 29 (10 %) samples having more than 10X coverage in this region compared to 4 out of 6 (66 %) of the aircraft samples (Fisher's exact test p = 0.008517). While the depth profiles and summary statistics showed that WWTP_wk47 had comparable genome coverage to aircraft samples, this result showed a depletion towards the very 5′-end of the genome.
The first 300 nucleotide region is covered by the first amplicon of the ARTIC v4 primer set (amplicon 1 left start 25 - amplicon 1 right end 431).
The 5′-end region of Orf9 was covered over 3,000X in 62 out of 74 (84 %) samples, mean depth range [6–23,422], median 7,085 while the 5′-end region of M was depleted in many samples, mean read depth range [1–14,998], median 2,150. Remarkably, while the first 300 nucleotides are covered in 9 out of 16 samples (56 %) from aircraft and pooled from aircraft and airport buildings, we found reads in one aircraft wastewater sample covering the region 1–25 nucleotides upstream of the first amplicon with a similar mean coverage of that of the first 300 bases (mean read depth 57 vs 92). In the wastewater samples, 3 samples out of 58 had reads covering the first 25 nucleotides, all of them in the WWTP_wk47 sample group.
Such reads upstream of the first amplicon, that is 5′-end extended reads, are covered in more detail in a subsequent section.
4.2. Subgenomic RNA identified in wastewater
We searched whether it was possible to find subgenomic RNA reads in the wastewater samples processed by Illumina sequencing. We used an analytic approach18 that relies on the paired-end reads spanning across the leader and the start position of the subgenomic ORFs. Out of 74 samples, we were able to identify subgenomic RNA reads in five (7 %) of them, two directly from (I) the six (33 %) aircraft wastewater samples and one from (II) the 10 (10 %) pooled wastewater from airport buildings and aircraft, two (7 %) from the wastewater collected in treatment plants in November 2023 (IV), with a majority of these reads mapping to subgenomic Orf9. No subgenomic RNA could be found in (III) wastewater from treatment plants during the summer sampling (0 out of 29 samples, or 0 %) since the 5′-end of the SARS-CoV-2 genome was not covered by any reads. A summary of the read counts and coverage is shown in Table 2.
Table 2.
Overview of the detected subgenomic RNA reads.
| Sample | Type of sample | Site | Count (incl. 1 pseudo-count) | Mapped reads | Counts per 2 million mapped reads | Mean coverage (Positions 52–67) |
|---|---|---|---|---|---|---|
| AC3 | Aircraft | E | 8 | 728,855 | 19 | 122 |
| AC3 | Aircraft | Orf9 | 52 | 728,855 | 140 | 122 |
| AC7 | Aircraft | Orf9 | 5 | 965,808 | 8 | 6 |
| WGSL19 | PooledB | Orf9 | 5 | 834,104 | 10 | 7 |
| WWTP17_wk47 | WWTP_wk47 | ORF3a | 31 | 1,869,738 | 32 | 70 |
| WWTP17_wk47 | WWTP_wk47 | M | 8 | 1,869,738 | 8 | 70 |
| WWTP25_wk47 | WWTP_wk47 | ORF3a | 2 | 2,132,648 | 1 | 4 |
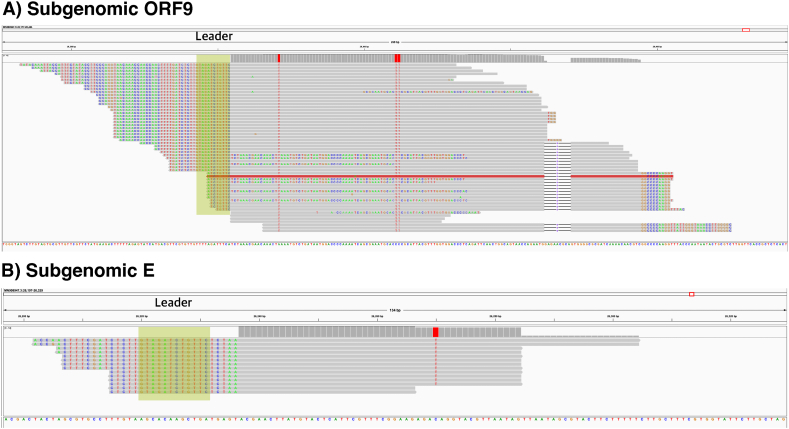
Given the coverage in the positions overlapping with the leader sequence, the subgenomic reads were relatively abundant when detected representing between 50 and 80 % of the local read depth. In terms of normalized read counts, the abundance was within the upper quarter of that found in our previous study [39] from 7 to 153 counts per 2 million total reads, suggesting that when found, subgenomic RNA was abundant in the wastewater samples. Furthermore, of the samples only one, AC3, had a deeply covered 5′-end, which allowed us to detect two different subgenomic RNAs (E and Orf9), both in relatively high abundance (Fig. 2A). Further investigation showed that the C28311T and C28312T substitutions were observed in all reads covering the Orf9 subgenomic RNA, which is consistent with the lineage call obtained using the program Freyja [23] in our previous study on wastewater collected from aircraft [46] for this sample which reported BQ.1 as the dominant sublineage (Fig. 2A). The observation of the C26270T substitution was similarly consistent for subgenomic RNA E (Fig. 2B).
Fig. 2.
View of the subgenomic reads in ORF9 and E for the wastewater sample taken from an aircraft (AC3). A snapshot taken with IGV-2.16.2 depicting all subgenomic reads mapping to ORF9. Soft-clipped bases on the leftmost positions map to the region of the leader (position 52–67) containing the GTAGATCTGTTC sequence (green). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
While we observed a good coverage of the first 300 nucleotides, the leader region and Orf9 for AC3, it was surprising that we succeeded in detecting subgenomic E in this sample given the poor coverage of E (Supplementary Table 2). However, the detected reads were almost identical, suggesting that they have most likely been amplified from the same molecule (Fig. 2B), and it is probable that we were able to catch the very few subgenomic E molecules that survived in that sample. In addition, we found subgenomic Orf3a in two treatment plant samples, in high abundance in sample WWTP17_wk47, 30 reads. The mean coverage of the first 300 nucleotides, the leader region and Orf3a were all high (3,517, 70 and 4,218X respectively). We therefore observed in both AC3 and WWTP17_wk47 cases, that a high abundance of subgenomic reads coincided with strong read support of both the 5'-end region covering the leader sequence and the 3′ part of the subgenomic RNA.
4.3. Coverage profiles of reads recovered from different wastewater sources form distinct clusters
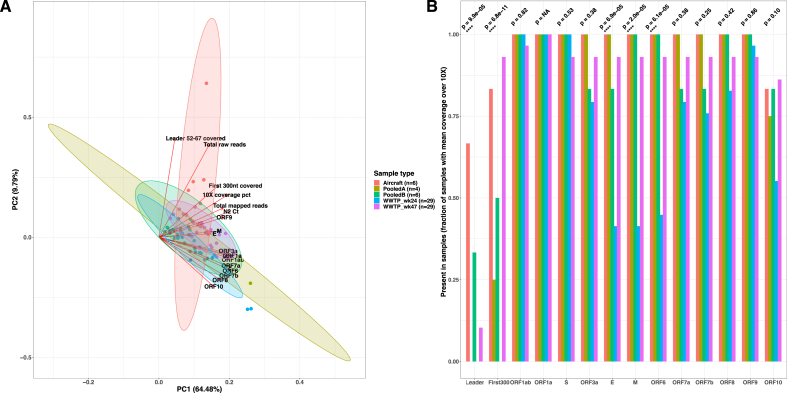
To investigate potential patterns in the variability of sequence coverage between samples and types of samples, we performed a principal component analysis (PCA) on the mean coverage values per genomic features such as the region covering the leader sequence, Orf1a, Orf1ab, S, Orf3a, E, M, Orf6, Orf7a, Orf7b, Orf8, Orf9, Orf10, a binary indicator of the presence/absence of the first 300 nucleotides region, the total number of raw and mapped reads, the fraction of the genome covered by more than 10X and the N2 Ct value. The structure of the dataset is shown as a biplot in Fig. 3A. The ellipses show the 95 % CI for belonging to each group and the arrows show the direction of increasing value of each variable. The first two principal components recapitulated 74.3 % of the explained variance.
Fig. 3.
Sequence read coverage variation across wastewater sample groups.
Panel A: Principal Component Analysis biplot on mean sequence coverage per region and the total number of raw and mapped reads, coverage at 10X coverage, presence/absence of the first 300 nucleotides of the 5′-UTR, and N2 Ct value. Panel B: Barplot showing the proportion of samples with each genomic region with at least 10X mean coverage. The samples are colored based on whether they originated directly from an aircraft (red), from pooled wastewater from aircraft and airport hangars and terminals (green) or from a wastewater treatment plant collected in week 24 (blue) and week 47 (purple). Statistical differences between sample groups in panel B were assessed using a Kruskal-Wallis test. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Five distinct clusters were observed corresponding to the different collections of wastewater. The red cluster is represented by the six wastewater samples collected directly from aircraft (I). The most representative variables of this cluster were the presence of the 5′ first 300 nucleotides, fraction of the genome over 10X coverage, and mean coverage in Orf9 and the leader region, N2 Ct and the number of raw and mapped reads.
The other sample types formed clusters with similar properties: pooled A (II light green) and pooled B (II dark green) had little separation, with the pooled A showing more variation, most probably since only 4 samples of this type were included in the study.
And finally, the blue and purple clusters consisted of all the samples collected from wastewater plants in week 24 (III) or week 47 (IV), were slightly separated, principally due to better coverage in E and M in WWTP_wk47 samples compared to WWTP_wk24.
All the wastewater plant and pooled samples were generally characterized by lower coverage of shorter ORFs such as 3a, E, M, 6, 7a, 7b, and 8. It is interesting to see that Pooled A samples, consisting of wastewater from multiple aircraft collected in a 24 composite manner, grouped with pooled B and the wastewater plant samples, instead of being grouped with aircraft samples.
We did not include variables of Supplementary Table 1 describing the extraction conditions, such as the use of centrifugation, use of Nanotrap particles or PEG-based extraction since they would trivially separate the different sample groups, PEG being for example applied only in Pooled A and B samples. In addition, since the total number of raw reads is larger for the aircraft samples, we repeated the PCA using the mean coverage values per genomic feature only to avoid any confounding bias (Supplementary Fig. 1). We again found three distinct clusters corresponding to each type of wastewater and WWTP_wk47 samples were more similar in terms of coverage to aircraft samples, with strong loadings in E and M.
To further characterize the differences in breadth of coverage between the different types of samples, we compared the fraction of samples covering each genomic region with at least 10 reads on average. The findings are summarized in Fig. 3B. The main genomic features such as Orf1a, Orf1ab, S and Orf9 are extensively covered in all types of wastewater samples, fulfilling the standard minimum 10X coverage usually required by variant calling or lineage designation tools in the routine analyses commonly performed for wastewater surveillance of SARS-CoV-2. In addition, while aircraft and pooled wastewater from aircraft and airport buildings showed similar breadth of coverage across ORFs, we observe a strong coverage depletion of the 5′-end first 300 nucleotide region and of smaller ORFs such as E, M, Orf6 in wastewater from treatment plants collected in week 24 (Kruskal-Wallis test p < 0.01) also reflected in the PCA. This analysis also supports that the leader and first 300 nucleotides region were more frequently found in aircraft samples and that despite having comparable proportion of samples covering the first 300, WWTP_wk47 samples had fewer samples covering the leader than aircraft samples (Kruskal-Wallis test p < 0.0001).
4.4. 5′-end extension of the subgenomic RNA reads
We investigated in detail the 5′-end extension of the subgenomic forward reads covering subgenomic Orf9 of the aircraft wastewater sample AC3, since this was the only sample with good coverage of the first 300 nucleotides in which we detected subgenomic RNAs.
Most of the reads (40 out of 51) covered a region outside the start position of the first primer (ARTIC v4 amplicon 1 left primer positions 25–50), extending as far upstream as position 11. These 150-nucleotide long reads all had either a hard or soft clipped sequence on their 5′-ends, matching a fragment of Orf1ab (Supplementary Table 3). The most frequent chimeric read occurrence consisted of this clipped sequence ending with the 4-mer TTTC, attached to the mapped part of the read starting at position 11 with the 4-mer TATA (35 out of 40 reads). These reads were nearly identical most of the time, suggesting that they have been amplified from the same molecule, which is intriguing since the first ARTIC primer starts at position 25 and these reads cover a region outside that region.
To better understand the mechanisms leading to the formation of these chimeric reads and estimate the extent of their occurrence, we performed an additional analysis investigating sequence reads from the 12,324 SARS-CoV-2 genomes collected by the Danish national mass test system [54] used in our previous study [39]. Reads were obtained from a similar sequencing approach, i.e. Illumina paired-end read sequencing using ARTIC primers, with the exception that RNA was extracted from routine surveillance oropharyngeal swabs and that the read length was shorter (74 bp instead of 150 bp for the wastewater study). Out of 23,192,194 reads covering the leader positions 52–67, we found a relatively large number of reads, (271,919 in total 1.2 %) with a start position before position 50 (expected first mapped position of amplicon 1) and among these 110,213 reads (0,5 %) with a 5′-end extension preceding the start position of the first ARTIC amplicon primer and a soft clipped sequence attached to their 5′-ends. Among these 5′-end extensions, 78,082 (0,3 %) were of length greater than 9 and about half of these were subgenomic (43,164 in total, 55.2 %). Looking at the location of the soft-clipped sequence attached upstream of the mapped bases, we performed a BLAST search against the Wuhan-Hu1 reference and found that most of these fragments belong to Orf1ab and Orf9, consistent with our findings from reads extracted from wastewater (Supplementary Fig. 2).
Furthermore, we looked at the 4-mer occurrences on both sides of the junction, i.e. the last 4 soft-clipped RNA sequences with first 4 mapped bases in the SARS-CoV-2 genome in all the chimeric reads extracted from the samples used in our previous study (Supplementary Table 4), and in the subgenomic reads only (Supplementary Table 5). Overall, we found that the most frequent junctions were TCCT_AGGT (4,927 times), GATC_TAAA (4,275 times), TCTA_AACA (1,718 times), TCAC_TAAA (1,000 times), GTAC_CAAA (792 times), while the combination TTTC_TATA seen in the wastewater sample AC3 occurred 37 times (expectation would be 19 if occurring at random). In contrast, when looking at the subgenomic reads only, the most frequent junctions were the same as the ones observed in all reads, with the exception of GATC_TAAA, which was less frequently observed in subgenomic reads (612 occurrences in contrast with the 4,275 occurrences in all reads, Fisher's two-sided test p < 2.2e-16). The 5′-end mapped sequence corresponding to the 4-mer TAAA starts at position 3 which corresponds to the very beginning of the genomic RNA sequence, which is possibly more difficult to anneal. Finally, to elucidate the sequence context allowing template switching at the junction, we looked at the 2 nucleotides following the 3′-end of the clipped sequences (Supplementary Tables 4 and 5). We found that in the 10 most frequent junctions, the di-nucleotide consisted of a A/T combination (i.e. AA/AT/TA/TT) or contained at least a T or an A, e.g. GT, CT. The template switching sequence context seen in the reads obtained from swabs was similar to that observed in the wastewater AC3 sample.
5. Discussion
In this study, we analyzed SARS-CoV-2 RNA sequence reads obtained from wastewater collected directly from aircraft and from tanks of wastewater pooled from aircraft and airport buildings. We compared these samples with regular wastewater collected from 29 treatment plants across Denmark at two time points. Our objective was to assess the difference in coverage and depth of the sequenced aggregate SARS-CoV-2 genomes obtained from such samples, focusing in particular on the 5′-end, and to determine whether it was possible to identify SARS-CoV-2 subgenomic RNAs from reads covering this region (the leader sequence) in particular. The secondary objective was to understand factors contributing to the sequencing variation and describe the favourable conditions for detecting subgenomic RNA reads and the implication of the ability to detect subgenomic RNA in wastewater.
Only five out of 74 samples (7 %) contained subgenomic RNA reads, mostly mapping to subgenomic Orf9. This is consistent with previous work reporting that N1 and N2 were the sequence features of SARS-CoV-2 most likely to be recovered from wastewater [37]. We also found subgenomic Orf3a in abundance in one wastewater sample, a subgenomic RNA for which RT-qPCR assays [55] are also available, demonstrating the ability of wastewater sequencing to detect such RNA molecules. While looking at the mean coverage of the first 300 nucleotides on the 5′-end of the genome, we found that most samples directly collected from aircraft had this region covered (5 out of 6), while only 4 out of 10 samples had reads covering these regions in samples obtained from water tanks pooled from various sources. None of the samples from wastewater plants collected in the summer had coverage in the 5′-end, while 27 out of 29 wastewaters collected in the sample plants in the winter had reads covering this region. This result illustrates the variability of the sequencing output and, in the case of the two WWTP sample groups, collected at the same sites and processed in the same way, the difference in viral circulation between summer and winter as well as weather conditions, e.g. heat and precipitation, could be possible contributors to the observed difference.
The PCA (Fig. 3A) showed that the sequence coverage metrics of the samples clustered into the five groups based on the different types of water collections and that the 5′-end persistence was strongly associated with the aircraft wastewater.
While aircraft wastewater strongly separated from the other types of samples, we observed thatthan both WWTP and pooled A and B samples were quite similar in terms of reads covering the SARS-CoV-2 regions, although viral RNA concentration was done differently: Nanotrap was used for WWTP samples and a PEG-based method for pooled A and B.
We also found little difference between aircraft and WWTP_wk47 samples which were deeply covered across the genome where both sample types had been processed using a PEG-based method. The only difference between these two were the read support in the leader. Aircraft samples are saturated with fecal material compared to WWTP samples, which made them more susceptible to PCR inhibition. However, these samples are more concentrated in terms of viral material and have a shorter retention time compared to WWTP samples. Longer retention time of WWTP samples compared to aircraft waste could also contribute to the observed difference in preservation of the 5′-end. In vitro experiments would be needed to confirm this. Finally, pooled samples were processed using PEG unlike aircraft and WWTP samples. To better decipher the sources of variation of coverage between pooled and aircraft samples, in particular pooled A, future experiments could be added to the study by processing the pooled wastewater with Nanotrap particles.
Many factors influence SARS-CoV-2 RNA degradation in wastewater, including temperature, pH, elapsed time, and the presence of disinfecting chemicals or detergent that would break the membrane structures and expose the viral RNA to the hostile environment. Unlike the samples directly taken from aircraft, pooled samples were obtained from water tanks used to collect wastewater from multiple sources, and the material in these tanks has been stored for longer and at room temperature before samples were collected, allowing environmental factors to cause more damage to the viral RNA. Wastewater from treatment plants was collected at ambient temperature (between 13.5 and 20 °C) and exposed to climate conditions such as rain.
Due to incomplete data collection, we were not able to fully assess the impact of pH on the quality of the sequenced viral RNA in the samples. Although the literature seems to support that the enveloped SARS-CoV-2 virus is stable in a wide range of pH [10,22], the available data we have on this, reveal a high pH value (9–10) in samples directly from aircraft due to addition of Idu-flight [7] disinfectant and alkaline cleaning agents, which have been described to be very efficient in eliminating the virus within a few hours [10,11], including the negative-strand RNAs, sheltered in double-membrane vesicles which are also susceptible to detergents [8].
When we investigated the completeness of the genomes across samples (Fig. 3B and Supplementary Table 2) we first noted that all sample types had the main regions of interest well covered, allowing reliable genomic surveillance tasks such as variant calling or lineage designation. However, our data shows that coverage completeness had deteriorated more in pooled wastewater from aircraft and airport buildings and in wastewater from treatment plants. Not only the overall fraction of the genome supported by more than 10 reads decreases (90 % covered over 10X on average for aircraft, 81 % for pooled A, 75 % for pooled B and 36 % for wastewater plants week 24, 91 % for week 47 samples) but deterioration and lack of coverage was focused in a few regions, in particular the 5′-end first 300 nucleotides and small ORFs such as E, M, Orf6, Orf7a, Orf7b, Orf8 and Orf10, with significant differences between groups for the 5′-end, E, M and Orf6. This could have implications on the type of wastewater to use to perform analyses requiring deep and extensive coverage across all regions of the SARS-CoV-2 genome.
Altogether, since our higher quality samples clustered based on being directly collected from an aircraft and that despite good coverage across the genome, winter wastewaters were lacking reads covering the very 5′-end, we can hypothesize that time may be a strong factor in degrading the 5′-end of the genome and decrease the ability to detect e.g. subgenomic RNA, as WWTP samples undergo longer retention (several days vs. hours), longer collection time (24hr vs. grab sampling) and longer transportation time (15 min from the airport to the processing lab vs. hours of transportation time from the treatment plants).
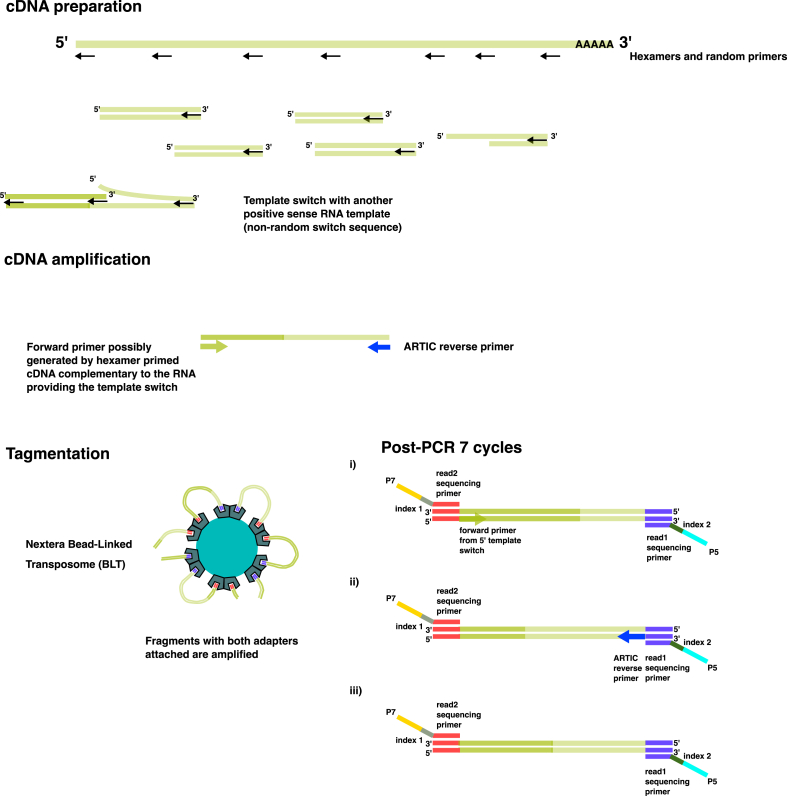
An important part of our study focused on investigating the sequenced reads mapping to the 5′-end of the SARS-CoV-2 genome. Sequencing was performed using amplicon-based Illumina paired-end sequencing using the ARTIC v4 primer set, which covers the SARS-CoV-2 genome between positions 25 to 29,854. Looking at the sequence reads mapping to the 5′-end, we found approximately 40 reads covering the region upstream of the first amplicon, extending as far as SARS-CoV-2 position 11. Interestingly, all the reads with a start position preceding the position of the first primer had either a soft or hard clipped sequence adjacent to the 5′ most first mapped bases of the reads, all mapping to RNA sequences located in Orf1ab. While comparing this result with the one we gathered from analyzing genomes from routine oropharyngeal swab samples collected by the Danish national mass test system [54] and used in our previous study [39] we found that most of these adjacent fragments belong to Orf1ab and Orf9. In addition, many of these reads were identical, suggesting that they have been amplified from the same molecule. While we demonstrated the existence of chimeric reads consisting of sequences mapping to different regions in the SARS-CoV-2 genome, their origin is still unclear. It has been previously described that next-generation sequencing-associated chimeric artefacts formed by annealing via short sequences are abundant in reads from FFPE (formalin-fixed paraffin-embedded) genome libraries [56]. While this example case affected single-stranded DNA fragments formed by degradation during FFPE storage, this phenomenon occurred this time on viral RNA and possibly during the cDNA synthesis step. In Fig. 4, we describe a possible scenario for the formation of these chimeric reads. First, random hexamers bind to the positive-sense viral RNA during cDNA formation, and double-stranded cDNA are synthesized. For some reads, in particular reads mapping to the 5′-end of the SARS-CoV-2 genome, template-switching occurs with another positive-strand RNA template sequence via non-random short sequence patterns and chimeric cDNA consisting of the sequences from the two RNA templates are formed. cDNA amplification occurs using the ARTIC first reverse primer and a forward primer possibly formed during cDNA synthesis. Some evidence of this amplification is shown by the high similarity of certain reads to one another, see Supplementary Table 3. Subsequently, some of these amplified fragments have sequencing adapters and primers attached and Illumina paired-end sequencing occurs. The analyses of the most frequent genomic locations of the 5′-end fragment attached to the mapped portion of the chimeric reads, together with the analysis of the most frequent 4-mers at the junction demonstrate that the template annealing mechanism is relatively frequent and importantly non-random. Based on the data collected from reads sequenced from wastewater and from swabs collected in a previous study, we found that the positive-strand RNA fragments could anneal via sequences as short as two nucleotides, consisting mostly of A/T combinations in the most frequently observed cases. The observed annealing mechanism resembles the mechanism described for Tn5/I50 recognition sites [57] which involve one A or T nucleotide spacer between both half-sites, with the difference that we did not see palindromic or symmetrical half-sites. While our study only looked at chimeric reads mapping to the 5′-end, it is highly plausible that it happens to sequence reads that map to other parts of the genome, especially since the template-switching mechanism occurs during the synthesis of subgenomic RNAs.
Fig. 4.
Schematic representation of the plausible template switching mechanism occurring to some 5′end reads.
Hexamers and random primers attach to the positive strand viral RNA, and the negative strand cDNA is generated by the reverse transcriptase included in the reaction. In some cases, non-random template switch occurs on the 5′-end with another positive-strand RNA while generating cDNA for positive-strand RNA template. The resulting cDNA is then amplified by the reverse ARTIC primer and a hexamer that reverse complements the positive strand sequence generated by template switching. In the tagmentation step, fragments containing i) the forward hexamer primer on the 5′-end, ii) the ARTIC reverse primer sequence on the 3′-end or iii) none of the primer sequences are amplified by post-PCR.
Chimeric reads consisting of sequences belonging to different parts of the SARS-CoV-2 genome have substantial implications in the analysis and mapping of the region upstream of the first ARTIC amplicon, as these could introduce of mapping errors or be misinterpreted as mutations. It has, for example, been suggested to mask the positions 1–55 and 29,804–29,903 to avoid potential miscalled nucleotides in the analyses of SARS-CoV-2 assemblies [58]. The issue should be of lesser importance or negligible in the regions well-amplified and covered by the ARTIC-based amplicons.
While the most plausible explanation of these chimeric sequences captured by reads mapping the 5′-end of the genome is from a technical origin, we cannot exclude that the formation of such sequences could occur during coronavirus replication under a process that needs to be clarified. It has been described in bovine coronavirus that subgenomic RNA undergo replication throughout infection, plus and minus strand synthesis happening simultaneously [59]. We could hypothesize that annealing occurs between two positive-strand fragments while negative-strand RNA is being replicated. Clearly, strand-switching is not new for coronavirus transcription as it plays a major role during generation of subgenomic RNAs. Consequently, envisioning that additional modes of strand-switching could be part of the coronavirus transcription and replication strategy is most certainly possible and may provide insight into replication of the very ends of the virus genome.
In summary, we have performed a next-generation sequencing analysis of SARS-CoV-2 RNA extracted from aircraft, airport and treatment plant wastewater. Our findings confirmed a high variability in genome coverage and depth, particularly in a region covering the first 300 nucleotides of the 5′-end of the genome and the leader, which appeared to be more degraded in wastewater tanks that collect waste from multiple sources including aircraft and buildings and in wastewater plants which are exposed to the weather. We did, however, succeed in identifying subgenomic reads in five of the samples at abundance levels similar to those we reported in a previous study using RNA samples collected from oropharyngeal swabs. The analysis of these subgenomic reads also uncovered the existence of chimeric reads consisting of a fragment of viral RNA attached to the 5′-extension of the SARS-CoV-2 genome that covers the leader.
While it is clear that coverage of sequence reads recovered from wastewater is subject to high variability due to multiple environmental and technical factors, the ability to detect subgenomic RNA in the wastewater seems to require certain conditions of preservation of the 5'-end, in particular the persistence of the first 25 nucleotides and the leader. We suggest that the ability to detect subgenomic RNA, could be used as an indicator of the abundance of viral particles, good sample handling and to a lower extent the degradation of the 5′-extension of the SARS-CoV-2 RNA. Further investigation and experiments will be needed to better clarify the origin of the chimeric 5′-end reads.
Ethical statement
Samples included in this study have been collected as part of Danish COVID-19 surveillance and according to the Danish law, ethical approval is not required for this type of study. The research is approved by the legal advisory board at SSI, a public research institute under the auspices of the Danish Ministry of Health. The study contains aggregated results without identifiable personal data and therefore complies with the European General Data Protection Regulations (GDPR).
Funding statement
The author(s) received no specific funding for this work.
Data availability statement
Raw sequence reads of the wastewaters (I), (II) and (III) have been deposited to the ENA database under the BioProject identifiers PRJEB66221 (I), PRJEB66497 (II), PRJEB66496 (III), and 29 samples deposited in project PRJEB65603, with accession numbers ranging from ERS17211[113–141] (IV).
CRediT authorship contribution statement
Man-Hung Eric Tang: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation. Marc Bennedbaek: Writing – review & editing, Validation, Investigation, Data curation. Vithiagaran Gunalan: Writing – review & editing, Validation, Methodology, Investigation, Formal analysis, Data curation. Amanda Gammelby Qvesel: Writing – review & editing, Validation, Investigation, Data curation. Theis Hass Thorsen: Writing – review & editing, Validation, Investigation, Formal analysis, Data curation, Conceptualization. Nicolai Balle Larsen: Writing – review & editing, Validation, Methodology, Investigation, Data curation. Lasse Dam Rasmussen: Writing – review & editing, Validation, Methodology, Formal analysis, Data curation. Lene Wulff Krogsgaard: Writing – review & editing, Methodology, Investigation. Morten Rasmussen: Writing – review & editing, Validation, Investigation, Formal analysis, Data curation. Marc Stegger: Writing – review & editing, Supervision, Project administration, Methodology, Investigation. Soren Alexandersen: Writing – review & editing, Supervision, Project administration, Investigation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank the Danish Covid-19 Genome Consortium for providing the sequencing data of SARS-CoV-2 positive swab and wastewater samples.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29703.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Termansen M.B., Frische S. Fecal-oral transmission of SARS-CoV-2: a systematic review of evidence from epidemiological and experimental studies. Am. J. Infect. Control. 2023 doi: 10.1016/j.ajic.2023.04.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toledo D.M., Robbins A.A., Gallagher T.L., Hershberger K.C., Barney R.E., Salmela S.M., Pilcher D., Cervinski M.A., Nerenz R.D., Szczepiorkowski Z.M., Tsongalis G.J., Lefferts J.A., Martin I.W., Hubbard J.A. Wastewater-based SARS-CoV-2 surveillance in northern new England. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.02207-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallejo J.A., Trigo-Tasende N., Rumbo-Feal S., Conde-Pérez K., López-Oriona Á., Barbeito I., Vaamonde M., Tarrío-Saavedra J., Reif R., Ladra S., Rodiño-Janeiro B.K., Nasser-Ali M., Cid Á., Veiga M., Acevedo A., Lamora C., Bou G., Cao R., Poza M. Modeling the number of people infected with SARS-COV-2 from wastewater viral load in Northwest Spain. Sci. Total Environ. 2022;811 doi: 10.1016/j.scitotenv.2021.152334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anand U., Adelodun B., Pivato A., Suresh S., Indari O., Jakhmola S., Jha H.C., Jha P.K., Tripathi V., Di Maria F. A review of the presence of SARS-CoV-2 RNA in wastewater and airborne particulates and its use for virus spreading surveillance. Environ. Res. 2021;196 doi: 10.1016/j.envres.2021.110929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordahl Petersen T., Rasmussen S., Hasman H., Carøe C., Bælum J., Charlotte Schultz A., Bergmark L., Svendsen C.A., Lund O., Sicheritz-Pontén T., Aarestrup F.M. Meta-genomic analysis of toilet waste from long distance flights; a step towards global surveillance of infectious diseases and antimicrobial resistance. Sci. Rep. 2015;5 doi: 10.1038/srep11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sethna P.B., Brian D.A. Coronavirus genomic and subgenomic minus-strand RNAs copartition in membrane-protected replication complexes. J. Virol. 1997;71:7744–7749. doi: 10.1128/jvi.71.10.7744-7749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamings A., Bhatta T.R., Alexandersen S. Subgenomic and negative sense RNAs are not markers of active replication of SARS-CoV-2 in nasopharyngeal swabs. medRxiv. 2021 doi: 10.1101/2021.06.29.21259511. 2021.06.29.21259511. [DOI] [Google Scholar]
- 10.Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.-L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1:e10. doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kampf G., Todt D., Pfaender S., Steinmann E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020;104:246–251. doi: 10.1016/j.jhin.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X.-W., Li J.-S., Jin M., Zhen B., Kong Q.-X., Song N., Xiao W.-J., Yin J., Wei W., Wang G.-J., Si B., Guo B.-Z., Liu C., Ou G.-R., Wang M.-N., Fang T.-Y., Chao F.-H., Li J.-W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J Virol Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatanaka N., Xu B., Yasugi M., Morino H., Tagishi H., Miura T., Shibata T., Yamasaki S. Chlorine dioxide is a more potent antiviral agent against SARS-CoV-2 than sodium hypochlorite. J. Hosp. Infect. 2021;118:20–26. doi: 10.1016/j.jhin.2021.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foladori P., Cutrupi F., Cadonna M., Manara S. Coronaviruses and SARS-CoV-2 in sewerage and their removal: step by step in wastewater treatment plants. Environ. Res. 2022;207 doi: 10.1016/j.envres.2021.112204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran H.N., Le G.T., Nguyen D.T., Juang R.-S., Rinklebe J., Bhatnagar A., Lima E.C., Iqbal H.M.N., Sarmah A.K., Chao H.-P. SARS-CoV-2 coronavirus in water and wastewater: a critical review about presence and concern. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai M.Y.Y., Cheng P.K.C., Lim W.W.L. Survival of severe acute respiratory syndrome coronavirus. Clin. Infect. Dis. 2005;41:e67–e71. doi: 10.1086/433186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amoah I.D., Abunama T., Awolusi O.O., Pillay L., Pillay K., Kumari S., Bux F. Effect of selected wastewater characteristics on estimation of SARS-CoV-2 viral load in wastewater. Environ. Res. 2022;203 doi: 10.1016/j.envres.2021.111877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varbanov M., Bertrand I., Philippot S., Retourney C., Gardette M., Hartard C., Jeulin H., Duval R.E., Loret J.-F., Schvoerer E., Gantzer C. Somatic coliphages are conservative indicators of SARS-CoV-2 inactivation during heat and alkaline pH treatments. Sci. Total Environ. 2021;797 doi: 10.1016/j.scitotenv.2021.149112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atoui A., Cordevant C., Chesnot T., Gassilloud B. SARS-CoV-2 in the environment: contamination routes, detection methods, persistence and removal in wastewater treatment plants. Sci. Total Environ. 2023;881 doi: 10.1016/j.scitotenv.2023.163453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Y., Guo W., Lopez-Hernadez A., Teng S., Li L. The pH effects on SARS-CoV and SARS-CoV-2 spike proteins in the process of binding to hACE2. Pathogens. 2022;11:238. doi: 10.3390/pathogens11020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karthikeyan S., Levy J.I., De Hoff P., Humphrey G., Birmingham A., Jepsen K., Farmer S., Tubb H.M., Valles T., Tribelhorn C.E., Tsai R., Aigner S., Sathe S., Moshiri N., Henson B., Mark A.M., Hakim A., Baer N.A., Barber T., Belda-Ferre P., Chacón M., Cheung W., Cresini E.S., Eisner E.R., Lastrella A.L., Lawrence E.S., Marotz C.A., Ngo T.T., Ostrander T., Plascencia A., Salido R.A., Seaver P., Smoot E.W., McDonald D., Neuhard R.M., Scioscia A.L., Satterlund A.M., Simmons E.H., Abelman D.B., Brenner D., Bruner J.C., Buckley A., Ellison M., Gattas J., Gonias S.L., Hale M., Hawkins F., Ikeda L., Jhaveri H., Johnson T., Kellen V., Kremer B., Matthews G., McLawhon R.W., Ouillet P., Park D., Pradenas A., Reed S., Riggs L., Sanders A., Sollenberger B., Song A., White B., Winbush T., Aceves C.M., Anderson C., Gangavarapu K., Hufbauer E., Kurzban E., Lee J., Matteson N.L., Parker E., Perkins S.A., Ramesh K.S., Robles-Sikisaka R., Schwab M.A., Spencer E., Wohl S., Nicholson L., McHardy I.H., Dimmock D.P., Hobbs C.A., Bakhtar O., Harding A., Mendoza A., Bolze A., Becker D., Cirulli E.T., Isaksson M., Schiabor Barrett K.M., Washington N.L., Malone J.D., Schafer A.M., Gurfield N., Stous S., Fielding-Miller R., Garfein R.S., Gaines T., Anderson C., Martin N.K., Schooley R., Austin B., MacCannell D.R., Kingsmore S.F., Lee W., Shah S., McDonald E., Yu A.T., Zeller M., Fisch K.M., Longhurst C., Maysent P., Pride D., Khosla P.K., Laurent L.C., Yeo G.W., Andersen K.G., Knight R. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature. 2022;609:101–108. doi: 10.1038/s41586-022-05049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agrawal S., Orschler L., Tavazzi S., Greither R., Gawlik B.M., Lackner S. Genome sequencing of wastewater confirms the arrival of the SARS-CoV-2 omicron variant at frankfurt airport but limited spread in the city of frankfurt, Germany, in november 2021. Microbiol Resour Announc. 2022;11 doi: 10.1128/MRA.01229-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smyth D.S., Trujillo M., Gregory D.A., Cheung K., Gao A., Graham M., Guan Y., Guldenpfennig C., Hoxie I., Kannoly S., Kubota N., Lyddon T.D., Markman M., Rushford C., San K.M., Sompanya G., Spagnolo F., Suarez R., Teixeiro E., Daniels M., Johnson M.C., Dennehy J.J. Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater. Nat. Commun. 2022;13:635. doi: 10.1038/s41467-022-28246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aßmann E., Agrawal S., Orschler L., Böttcher S., Lackner S., Hölzer M. Impact of reference design on estimating SARS-CoV-2 lineage abundances from wastewater sequencing data. bioRxiv. 2023 doi: 10.1101/2023.06.02.543047. 2023.06.02.543047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., McElroy K.A., Rhode S.F., Matus M., Wuertz S., Thompson J., Alm E.J. Wastewater surveillance of SARS-CoV-2 across 40 U.S. States from february to June 2020. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wurtzer S., Marechal V., Mouchel J., Maday Y., Teyssou R., Richard E., Almayrac J., Moulin L. Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.50.2000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karthikeyan S., Nguyen A., McDonald D., Zong Y., Ronquillo N., Ren J., Zou J., Farmer S., Humphrey G., Henderson D., Javidi T., Messer K., Anderson C., Schooley R., Martin N.K., Knight R. Rapid, large-scale wastewater surveillance and automated reporting system enable early detection of nearly 85% of COVID-19 cases on a university campus. mSystems. 2021;6 doi: 10.1128/mSystems.00793-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karthikeyan S., Ronquillo N., Belda-Ferre P., Alvarado D., Javidi T., Longhurst C.A., Knight R. High-throughput wastewater SARS-CoV-2 detection enables forecasting of community infection dynamics in San Diego county. mSystems. 2021;6 doi: 10.1128/mSystems.00045-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhan Q., Babler K.M., Sharkey M.E., Amirali A., Beaver C.C., Boone M.M., Comerford S., Cooper D., Cortizas E.M., Currall B.B., Foox J., Grills G.S., Kobetz E., Kumar N., Laine J., Lamar W.E., Mantero A.M.A., Mason C.E., Reding B.D., Robertson M., Roca M.A., Ryon K., Schürer S.C., Shukla B.S., Solle N.S., Stevenson M., Tallon J.J., Jr., Thomas C., Thomas T., Vidović D., Williams S.L., Yin X., Solo-Gabriele H.M. Relationships between SARS-CoV-2 in wastewater and COVID-19 clinical cases and hospitalizations, with and without normalization against indicators of human waste. ACS ES&T Water. 2022;2:1992–2003. doi: 10.1021/acsestwater.2c00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu P., Guo L., Cavallo M., Cantrell C., Hilton S.P., Nguyen A., Long A., Dunbar J., Barbero R., Barclay R., Sablon O., Wolfe M., Lepene B., Moe C. Comparison of Nanotrap® Microbiome A Particles, membrane filtration, and skim milk workflows for SARS-CoV-2 concentration in wastewater. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1215311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dehghan Banadaki M., Torabi S., Rockward A., Strike W.D., Noble A., Keck J.W., Berry S.M. Simple SARS-CoV-2 concentration methods for wastewater surveillance in low resource settings. Sci. Total Environ. 2024;912 doi: 10.1016/j.scitotenv.2023.168782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen R.M., Tornby D.S., Bang L.L., Madsen L.W., Skov M.N., Sydenham T.V., Steinke K., Jensen T.G., Johansen I.S., Andersen T.E. Rectally shed SARS-CoV-2 in COVID-19 inpatients is consistently lower than respiratory shedding and lacks infectivity. Clin. Microbiol. Infection. 2022;28:304.e1–304.e3. doi: 10.1016/j.cmi.2021.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hart J.J., Jamison M.N., McNair J.N., Szlag D.C. Frequency and degradation of SARS-CoV-2 markers N1, N2, and E in sewage. J. Water Health. 2023;21:514–524. doi: 10.2166/wh.2023.314. [DOI] [PubMed] [Google Scholar]
- 38.Lin X., Glier M., Kuchinski K., Ross-Van Mierlo T., McVea D., Tyson J.R., Prystajecky N., Ziels R.M. Assessing multiplex tiling PCR sequencing approaches for detecting genomic variants of SARS-CoV-2 in municipal wastewater. mSystems. 2021;6 doi: 10.1128/mSystems.01068-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang M.-H.E., Ng K.L., Edslev S.M., Ellegaard K., Stegger M., Alexandersen S. Comparative subgenomic mRNA profiles of SARS-CoV-2 Alpha, Delta and Omicron BA.1, BA.2 and BA.5 sub-lineages using Danish COVID-19 genomic surveillance data. EBioMedicine. 2023;93 doi: 10.1016/j.ebiom.2023.104669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dimcheff D.E., Blair C.N., Zhu Y., Chappell J.D., Gaglani M., McNeal T., Ghamande S., Steingrub J.S., Shapiro N.I., Duggal A., Busse L.W., Frosch A.E.P., Peltan I.D., Hager D.N., Gong M.N., Exline M.C., Khan A., Wilson J.G., Qadir N., Ginde A.A., Douin D.J., Mohr N.M., Mallow C., Martin E.T., Johnson N.J., Casey J.D., Stubblefield W.B., Gibbs K.W., Kwon J.H., Talbot H.K., Halasa N., Grijalva C.G., Baughman A., Womack K.N., Hart K.W., Swan S.A., Surie D., Thornburg N.J., McMorrow M.L., Self W.H., Lauring A.S. Total and subgenomic RNA viral load in patients infected with SARS-CoV-2 Alpha, Delta, and Omicron variants. J. Infect. Dis. 2023 doi: 10.1093/infdis/jiad061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agius J.E., Johnson-Mackinnon J.C., Fong W., Gall M., Lam C., Basile K., Kok J., Arnott A., Sintchenko V., Rockett R.J. SARS-CoV-2 within-host and in vitro genomic variability and sub-genomic RNA levels indicate differences in viral expression between clinical cohorts and in vitro culture. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.824217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.V Mears H., Young G.R., Sanderson T., Harvey R., Crawford M., Snell D.M., Fowler A.S., Hussain S., Nicod J., Peacock T.P., Emmott E., Finsterbusch K., Luptak J., Wall E., Williams B., Gandhi S., Swanton C., V Bauer D.L. Emergence of new subgenomic mRNAs in SARS-CoV-2. bioRxiv. 2022 doi: 10.1101/2022.04.20.488895. 2022.04.20.488895. [DOI] [Google Scholar]
- 43.Alexandersen S., Chamings A., Bhatta T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020;11:6059. doi: 10.1038/s41467-020-19883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong X., Penrice-Randal R., Goldswain H., Prince T., Randle N., Donovan-Banfield I., Salguero F.J., Tree J., Vamos E., Nelson C., Clark J., Ryan Y., Stewart J.P., Semple M.G., Baillie J.K., Openshaw P.J.M., Turtle L., Matthews D.A., Carroll M.W., Darby A.C., Hiscox J.A. Analysis of SARS-CoV-2 known and novel subgenomic mRNAs in cell culture, animal model, and clinical samples using LeTRS, a bioinformatic tool to identify unique sequence identifiers. GigaScience. 2022;11 doi: 10.1093/gigascience/giac045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krogsgaard L.W., Benedetti G., Gudde A., Richter S.R., Rasmussen L.D., Midgley S.E., Qvesel A.G., Nauta M., Bahrenscheer N.S., von Kappelgaard L., McManus O., Hansen N.C., Pedersen J.B., Haimes D., Gamst J., Nørgaard L.S., Jørgensen A.C.U., Ejegod D.M., Møller S.S., Clauson-Kaas J., Knudsen I.M., Franck K.T., Ethelberg S. Results from the SARS-CoV-2 wastewater-based surveillance system in Denmark, July 2021 to June 2022. Water Res. 2024 doi: 10.1016/j.watres.2024.121223. [DOI] [PubMed] [Google Scholar]
- 46.Qvesel A.G., Bennedbæk M., Larsen N.B., Gunalan V., Krogsgaard L.W., Rasmussen M., Rasmussen L.D. SARS-CoV-2 variants BQ.1 and XBB.1.5 in wastewater of aircraft flying from China to Denmark, 2023. Emerg. Infect. Dis. 2023;29 doi: 10.3201/eid2912.230717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.National surveillance of SARS-CoV-2 in wastewater. 2024. https://en.ssi.dk/covid-19/national-surveillance-of-sars-cov-2-in-wastewater
- 48.Quick J. nCoV-2019 sequencing protocol. 2020;3 https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bp2l6n26rgqe/v3?version_warning=no [Google Scholar]
- 49.Li H., Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J., Tan A.L., Paul L.M., Brackney D.E., Grewal S., Gurfield N., Van Rompay K.K.A., Isern S., Michael S.F., Coffey L.L., Loman N.J., Andersen K.G. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang Y., Horikoshi M., Li W. Ggfortify: unified interface to visualize statistical results of popular R packages. R J. 2016;8:474. doi: 10.32614/RJ-2016-060. [DOI] [Google Scholar]
- 53.Maechler M., Rousseeuw P., Struyf A., Hubert M., Hornik K. Cluster: cluster analysis basics and extensions. 2022. https://CRAN.R-project.org/package=cluster
- 54.Gram M.A., Steenhard N., Cohen A.S., Vangsted A.-M., Mølbak K., Jensen T.G., Hansen C.H., Ethelberg S. Patterns of testing in the extensive Danish national SARS-CoV-2 test set-up. PLoS One. 2023;18 doi: 10.1371/journal.pone.0281972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhosle S.M., Tran J.P., Yu S., Geiger J., Jackson J.D., Crozier I., Crane A., Wada J., Warren T.K., Kuhn J.H., Worwa G. Duplex one-step RT-qPCR assays for simultaneous detection of genomic and subgenomic RNAs of SARS-CoV-2 variants. Viruses. 2022;14:1066. doi: 10.3390/v14051066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haile S., Corbett R.D., Bilobram S., Bye M.H., Kirk H., Pandoh P., Trinh E., MacLeod T., McDonald H., Bala M., Miller D., Novik K., Coope R.J., Moore R.A., Zhao Y., Mungall A.J., Ma Y., Holt R.A., Jones S.J., Marra M.A. Sources of erroneous sequences and artifact chimeric reads in next generation sequencing of genomic DNA from formalin-fixed paraffin-embedded samples. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky1142. e12–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goryshin I.Y., Miller J.A., Kil Y.V., Lanzov V.A., Reznikoff W.S. Tn 5/IS 50 target recognition. Proc. Natl. Acad. Sci. USA. 1998;95:10716–10721. doi: 10.1073/pnas.95.18.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sapoval N., Mahmoud M., Jochum M.D., Liu Y., Elworth R.A.L., Wang Q., Albin D., Ogilvie H.A., Lee M.D., Villapol S., Hernandez K.M., Maljkovic Berry I., Foox J., Beheshti A., Ternus K., Aagaard K.M., Posada D., Mason C.E., Sedlazeck F.J., Treangen T.J. SARS-CoV-2 genomic diversity and the implications for qRT-PCR diagnostics and transmission. Genome Res. 2021;31:635–644. doi: 10.1101/gr.268961.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hofmann M.A., Sethna P.B., Brian D.A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 1990;64:4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence reads of the wastewaters (I), (II) and (III) have been deposited to the ENA database under the BioProject identifiers PRJEB66221 (I), PRJEB66497 (II), PRJEB66496 (III), and 29 samples deposited in project PRJEB65603, with accession numbers ranging from ERS17211[113–141] (IV).