Abstract
Lack of permissive and productive cell cultures for the human papillomaviruses (HPVs) has hindered the study of virus-neutralizing antibodies and infection. We developed a cell-free system generating infectious HPV16 pseudovirions. HPV16 L1/L2 capsids, which had been self-assembled in insect cells (Sf9) expressing virion proteins L1 and L2, were disassembled with 2-mercaptoethanol (2-ME), a reducing agent, and reassembled by removal of 2-ME in the presence of a β-galactosidase expression plasmid. Plasmid DNA purified together with the reassembled capsids was resistant to DNase I digestion. The reassembled pseudovirions mediated DNA transfer to COS-1 cells, as monitored by induced β-galactosidase activity. Transfer was inhibited by anti-HPV16 L1 antiserum but not by antisera against L1s of HPV6 and HPV18. Construction in vitro of HPV pseudovirions containing marker plasmids would be potentially useful in developing methods to assay virus-neutralizing antibodies and to transfer exogenous genes to HPV-susceptible cells.
Human papillomaviruses (HPVs), which cause a variety of proliferating epithelial lesions including skin and cervical cancers, are classified into more than 70 types based on nucleotide sequence homology of viral DNA. Despite overall similarity in genomic organization, HPVs have rather diverse tissue specificities and malignant potentials (5, 13). Therefore, analyses of antigenic relationships among HPVs are needed to study viral pathogenicity and to develop a prophylactic vaccine strategy. Studies on neutralizing antibodies against HPVs have been hampered by the lack of cell culture systems for producing HPV particles and for monitoring infection with HPVs.
In order to overcome this problem, methods have been devised (9–11) to encapsidate DNA into L1/L2 capsids, which are self-assembled particles in eukaryotic cells infected with recombinant viruses, for expression of major (L1) and minor (L2) structural proteins of HPVs (1, 3). Pseudovirions of HPV16 were generated in hamster BPHE-1 cells by using a recombinant Semliki Forest virus (9), and those of HPV33 were produced in monkey COS-1 cells by using a recombinant vaccinia virus (10). These studies indicated that L2 is required for efficient encapsidation of DNA and that specific DNA sequences are not required for packaging (12). Infection of cells with pseudovirions is neutralized by anti-L1 antiserum in a type-specific manner (9, 10). In this report, we describe the encapsidation of plasmid DNA into HPV16 L1/L2 capsids in a cell-free system, producing infectious pseudovirions in vitro.
HPV16 L1/L2 capsids that had been disassembled with the reducing agent 2-mercaptoethanol (2-ME) were allowed to reassemble by the removal of 2-ME (4, 7) in the presence of closed circular (form I) plasmid DNA of pSV/β-gal (Promega Corp., Madison, Wis.), an expression plasmid (6.8 kb) for β-galactosidase, purified by the ethidium bromide-CsCl buoyant density method (8). The L1/L2 capsids produced in Sf9 cells infected with recombinant baculovirus capable of expressing HPV16 L1 and L2 were purified by CsCl equilibrium density gradient centrifugation, as described previously (2), and dialyzed extensively against phosphate-buffered saline (PBS). Purified L1/L2 capsids (1 mg) were incubated in 1 ml of PBS containing 2-ME (5%) for 16 h at 4°C to disassemble, under the conditions optimized by McCarthy et al. for disassembly of HPV11 L1-capsids (7). The disassembled capsids were mixed with ethanol-precipitated pSV/β-gal (2 mg), and the mixture was dialyzed against 4 liters of phosphate buffer (pH 7.4) containing NaCl (0.5 M) and CaCl2 (2 mM) for 24 h at 4°C. The solution was then centrifuged at 45,000 rpm for 14 h in an NVT65.2 rotor (Beckman) after the density of the solution was adjusted to 1.31 g/ml with PBS and CsCl. The resultant gradient was fractionated (120 μl/fraction) for determination of viral proteins and plasmid DNA.
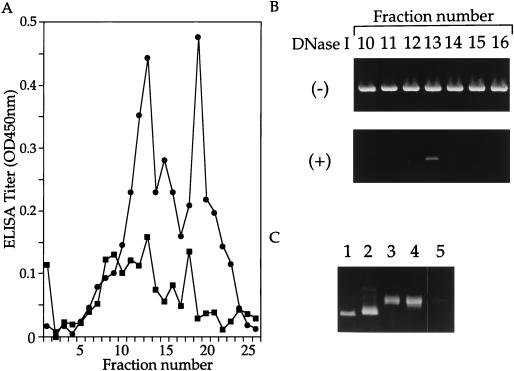
Virion proteins L1 and L2 in the fractions were assayed by enzyme-linked immunosorbent assay (ELISA). Aliquots (1 μl) from the fractions were incubated in a carbonate buffer (pH 9.6) for 16 h at 4°C to disrupt capsids and fixed in wells of ELISA plates. L1 and L2 were detected by using mouse antibodies against HPV-16 L1 or L2, as described previously (2). Distributions of L1 and L2 in the gradient are shown in Fig. 1A. The L1 distribution showed two peaks around fractions 13 and 19, whereas the L2 distribution had less-distinctive peaks. Fractions 11 to 14, containing both L1 and L2, corresponded to a density of 1.31 g/ml in the gradient.
FIG. 1.
(A) Distributions of L1 and L2 in CsCl equilibrium density gradient centrifugation of reassembled HPV-16 L1/L2 capsids were examined by ELISA using anti-L1 and anti-L2 antibodies. The ELISA titer (optical density [OD] at 450 nm) for L1 (•) and for L2 (■) of each fraction was plotted. (B) Plasmid DNA in fractions was examined by PCR with primers specific for the β-galactosidase gene and template DNA extracted from the fractions with or without DNase I digestion. An amplified DNA fragment (838 bp) was electrophoresed on a 1% agarose gel and stained with ethidium bromide. (C) The linear form of pSV/β-gal obtained by digestion with HindIII (lane 1), the closed circular form (lane 2), the open circular form obtained by treatment with 10 units of topoisomerase I for 20 min at 37°C (lane 3), pSV/β-gal incubated in PBS containing 2-ME (5%) for 16 h at 4°C (lane 4), and DNA extracted from fraction 13 (lane 5) were electrophoresed on a 0.7% agarose gel and stained with ethidium bromide.
The presence of plasmid DNA in the selected fractions was examined by PCR with primers specific for the β-galactosidase gene. Ten-microliter aliquots of fractions 9 to 16 were diluted to 360 μl with PBS containing 2.5 mM MgCl2 and digested with DNase I (250 U/ml) for 45 min at room temperature. For comparison, 10-μl aliquots of fractions 9 to 16 were similarly processed without DNase I. DNA extracted with phenol from the samples incubated with or without DNase I was used as the template for PCR. With the DNase I-digested samples, a DNA fragment with an expected size of 838 bp was amplified only from fraction 13 (Fig. 1B), whereas DNA fragments were amplified from all of the undigested fractions. Thus, it was concluded that fraction 13 contained plasmid DNA resistant to digestion with DNase I. The form of DNA in fraction 13 was examined by agarose gel electrophoresis (Fig. 1C). The mobility of phenol-extracted DNA from DNase I-digested fraction 13 was compared with those of closed circular (form I), open circular (form II), and linear (form III) DNA of pSV/β-gal. Both the DNA incubated in PBS containing 2-ME (5%) for 16 h at 4°C and the DNA extracted from fraction 13 showed mobilities similar to that of open circular DNA. The DNA data (Fig. 1B), along with the protein data (Fig. 1A), strongly suggest that fraction 13 (and probably fractions around 13) contained reassembled HPV16 pseudovirions encapsidating plasmid DNA of the open circular form.
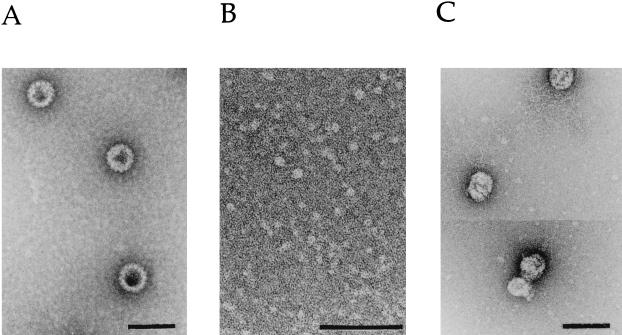
Electron microscopy confirmed the presence of pseudovirions in fraction 13 (Fig. 2). Particles purified from Sf9 cells expressing L1 and L2 (Fig. 2A) were broken down to the level of capsomeres (Fig. 2B), from which particles were reassembled (Fig. 2C). The reassembled capsids were morphologically similar to the particles assembled in Sf9 cells. The concentration of the reassembled capsids was estimated from the amount of L1 protein in fraction 13. Approximately 10 μg of pseudovirions were obtained from 1 mg of disassembled capsids.
FIG. 2.
Electron microscopy of purified HPV-16 L1/L2 capsids (A), disassembled capsids (B), and reassembled capsids (C). Bars, 100 nm.
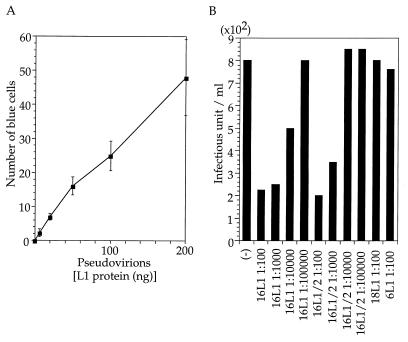
The pseudovirions were examined for the capacity to mediate the transfer of plasmid DNA to COS-1 cells. Fraction 13 was dialyzed against PBS to remove CsCl and incubated with COS-1 cells (4 × 105), which were dispersed with PBS containing EDTA (2.5 mM) and washed with Dulbecco’s modified Eagle’s medium in PBS (pH 6.8) containing bovine serum albumin (0.01%) for 2 h at 4°C with constant agitation. The cells were seeded into six-well plates, grown for 36 h, and fixed with glutaraldehyde. Cells expressing β-galactosidase were stained by using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as the substrate (In Situ β-Galactosidase staining kit; Promega Corp.). The number of blue cells was linearly dependent on the dilution of the pseudovirions (Fig. 3A). Blue cells were not detected in cells incubated with a mixture of L1/L2 capsid (50 ng) and pSV/β-gal (10 μg). DNase I treatment of the pseudovirions (50 ng), under conditions in which 20 μg of the plasmid was hydrolyzed completely in the presence of L1/L2 capsids (50 ng), did not reduce the number of blue cells; 17 blue cells were detected. The efficiency of DNA transfer by pseudovirions, approximately 50 blue cells per 200 ng of L1 of fraction 13, was comparable to the HPV33 pseudovirions generated with a recombinant vaccinia virus (10). The numbers of blue cells in CV-1 cell cultures infected with the pseudovirions were less than 1/10 of those in COS-1 cultures, indicating that autonomous replication of pSV/β-gal mediated by simian virus 40 T antigen was required for efficient detection of infected cells.
FIG. 3.
(A) Amount of L1 protein present in inoculum plotted against the number of blue cells. (B) Neutralization of pseudoinfection by antisera against HPV16 L1 capsids (16L1), HPV16 L1/L2 capsids (16L1/2), HPV18 L1 capsids (18L1), and HPV6 L1 capsids (6L1). Pseudovirions (80 ng) were mixed with PBS (−) or mouse antiserum diluted with PBS (indicated by, e.g., 1:100) and used to inoculate COS-1 cells. Infectivity was indicated by the number of blue cells (infectious unit) per ml.
Pseudovirion-mediated DNA transfer was inhibited by anti-HPV 16 L1/L2 or L1 antiserum (Fig. 3B). Pseudovirions were incubated at 4°C for 1 h with serial dilutions of a mouse antiserum obtained by immunizing BALB/c mice with L1/L2 (2) or L1 capsids (6) and then mixed with COS-1 cells, which were processed as described above. The numbers of blue cells were reduced by the antisera at dilutions of up to 1:1,000. Similarly prepared mouse antisera against HPV6 and HPV18 L1 capsids showed much weaker inhibitory effects on the transfer of DNA than did the anti-HPV16 antiserum (Fig. 3B), indicating that inhibition was specific to the HPV type.
Like HPV33 L1 capsids containing DNA (10), the HPV16 particles reassembled by the present procedure from L1 capsids and pSV/β-gal were much less infectious (data not shown) than the reassembled L1/L2 pseudovirions. Thus, L2 appears to be required for encapsidation of plasmids and/or expression of infectivity of virions.
In this study, we were able to reconstruct in vitro infectious HPV16 pseudovirions from purified and disassembled L1/L2 capsids and marker plasmid DNA. The reconstructed pseudovirions may be used for studies of the early events in HPV infection of cultured cells, because they are free of contaminating helper viruses. The fact that antiserum inhibition of DNA transfer is HPV-type specific provides a basis, with appropriate modifications of marker DNA and target cells in the present system, for development of a system to quantitate neutralizing antibodies of HPVs. It is also possible that packaging of plasmid DNA in vitro be used as a method to transfer exogenous genetic materials, which are protected from nucleases, to HPV-susceptible cells.
Acknowledgments
This work was supported by a grant-in-aid from the Ministry of Health and Welfare for the Second-Term Comprehensive 10-Year Strategy for Cancer Control and by a cancer research grant from the Ministry of Education, Science, and Culture.
We thank K. Suzuki for assistance in electron microscopy.
REFERENCES
- 1.Hagensee M E, Olson N H, Baker T S, Galloway D A. Three-dimensional structure of vaccinia virus-produced human papillomavirus type 1 capsids. J Virol. 1994;68:4503–4505. doi: 10.1128/jvi.68.7.4503-4505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawana K, Matsumoto K, Yoshikawa H, Taketani Y, Kawana T, Yoshiike K, Kanda T. A surface immunodeterminant of human papillomavirus type 16 minor capsid protein L2. Virology. 1998;245:353–359. doi: 10.1006/viro.1998.9168. [DOI] [PubMed] [Google Scholar]
- 3.Kirnbauer R, Taub J, Greenstone H, Roden R, Dürst M, Gissmann L, Lowy D R, Schiller J T. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li M, Beard P, Estes P A, Lyon M K, Garcea R L. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J Virol. 1998;72:2160–2167. doi: 10.1128/jvi.72.3.2160-2167.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorincz A T, Reid R, Jensen A B, Greenberg M D, Lancaster A W, Kurman R J. Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet Gynecol. 1991;79:328–337. doi: 10.1097/00006250-199203000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Matsumoto K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. Antibodies to human papillomavirus 16, 18, 58, and 6b major capsid proteins among Japanese females. Jpn J Cancer Res. 1997;88:369–375. doi: 10.1111/j.1349-7006.1997.tb00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy M P, White W I, Palmer-Hill F, Koenig S, Suzich J A. Quantitative disassembly and reassembly of human papillomavirus type 11 viruslike particles in vitro. J Virol. 1998;72:32–41. doi: 10.1128/jvi.72.1.32-41.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radloff R, Bauer W, Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci USA. 1967;57:1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roden R B S, Greenstone H L, Kirnbauer R, Booy F P, Jessie J, Lowy D R, Schiller J T. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J Virol. 1996;70:5878–5883. doi: 10.1128/jvi.70.9.5875-5883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unckell F, Streeck R E, Sapp M. Generation and neutralization of pseudovirions of human papillomavirus type 33. J Virol. 1997;71:2934–2939. doi: 10.1128/jvi.71.4.2934-2939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Stenzel D J, Sun X-Y, Frazer I H. Synthesis and assembly of infectious bovine papillomavirus particles in vitro. J Gen Virol. 1993;74:763–768. doi: 10.1099/0022-1317-74-4-763. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, Sun X-Y, Louis K, Frazer I H. Interaction of human papillomavirus (HPV) type 16 capsid proteins with HPV DNA requires an intact L2 N-terminal sequence. J Virol. 1994;68:619–625. doi: 10.1128/jvi.68.2.619-625.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288(Suppl. 2):F55–F78. [DOI] [PubMed]