Summary:
Spatial transcriptomics maps RNA molecules to the location in a tissue where they are expressed. Here we document the use of Slide-SeqV2 to visualize gene expression in the mouse olfactory bulb (OB). This approach relies on spatially identified beads to locate and quantify individual transcripts. The expression profiles associated with the beads are used to identify and localize individual cell types in an unbiased manner. We demonstrate the various cell types and subtypes with distinct spatial locations in the olfactory bulb that are identified using Slide-SeqV2.
Keywords: Slide-SeqV2, olfactory bulb, neural development, GABAergic interneurons, mitral and tufted cells, spatial transcriptome, mouse
1. Introduction:
Advances in next-generation sequencing (NGS) technologies have revolutionized how cells are identified and characterized. Bulk and single cell transcriptome profiling has enabled studies of gene expression in the olfactory tissues of vertebrate species to elucidate developmental changes and identify various cell types (1–9). In single cell or single nucleus transcriptomics, RNA molecules from dissociated cells or nuclei are tagged, amplified, and sequenced to determine expression profiles of individual cells (10,11). Cells sharing similar gene expression profiles can be clustered statistically and identified as specific types based on known markers (12). The dissociation process, however, has led to the loss of spatial information of cells. Moreover, rare cells may not be efficiently captured from the dissociated cell population, which can result in the underrepresentation, or the complete absence, of these cells in the final analysis.
To overcome these limitations, multiple technology platforms have been developed to preserve information of cells in the tissue context. For example, several single cell, high-plex imaging platforms have been used to identify cell clusters and reveal spatial relationships between cells (13–21). However, these platforms are limited in the number of genes to be quantified, which must be manually curated. Spatial transcriptomics, on the other hand, relies on localized RNA capturing followed by sequencing to provides a comprehensive survey of all transcripts in the cells (22–24). In Slide-SeqV2, for example, beads coated with oligonucleotides are used to capture and process mRNA by tethering mRNA molecules onto the beads for reverse transcription, followed by amplification and sequencing. Once sequenced, the spatial barcodes on the oligo allow post-hoc assignment of the sequenced molecules to individual beads, such that the mRNA species can be quantified and mapped to specific locations. This method not only preserves the spatial information, but also provides an unbiased analysis of transcriptomes.
In the developing olfactory system, olfactory sensory axons project into the olfactory bulb (OB) to innervate their target glomeruli (25–27). Sensory axons undergo a change in the level of plasticity during the first postnatal week, defined as the critical period (28,3,29,30). Coincidentally, the OB also undergoes a dramatic change during the first postnatal week. For example, the mitral tufted cells prune their primary dendrites (28,31–33). Different types of GABAergic interneurons mature during this period (34,35). Whereas multiple types of interneuron types have been described, the molecular composition that defines these cells remain unknow. Therefore, we use Slide-SeqV2 to perform unbiased cell clustering and map the cell clusters onto the olfactory bulb (36,37).We have used this approach to identify a new class of neurons in the olfactory epithelium (38). Here we describe the procedure to perform Slide-SeqV2 on a P7 OB section and the analysis that follows.
2. Materials:
-
Anesthetic reagent: dissolve 2g urethane in 10mL 1x PBS (pH = 7.4). Store at room temperature.
(Reagents 2–7 must be RNase free.)
Hank’s Balanced Salt Solution (HBSS, 1x), without Calcium, Magnesium, or Phenol red. (VWR life science)
O.C.T. (Sakura), embedding medium for frozen tissue specimens.
Pre-sequenced Slide-seq pucks. Pucks used in this study were generated and sequenced at the Broad Institute (Cambridge, MA) as described in the Methods and Supplementary Information section of the Slide-seqV2 publication (37). The pucks were received on small glass coverslips in 1.7mL LoBind tubes. Pucks need to be stored at 4°C after receiving.
SSC buffer (20X concentrate): dissolve 175.3 g NaCl and 88.2 g sodium citrate•2H2O in 800 mL DEPC-H2O. Use citric acid to adjust the pH to 5. Adjust the volume to 1 L with DEPC -H2O. Autoclave and store at room temperature.
Hybridization buffer: add 2unit/μL RNase inhibitor (Lucigen NxGen F83923–1) into 6x SSC buffer. 200 μL per puck. Prepare before use,
-
Reverse Transcription (RT) solution: 115 μL water, 40 μL 5X Maxima RT buffer (ThermoFisher, EP0753), 20 μL 10 mM dNTPs (NEB N0477L), 5 μL RNase Inhibitor (Lucigen NxGen F83923–1), 10 μL 50 μM Template Switch Oligo (Custom from IDT [see sequence below on 19]), 10 μL Maxima H-RTase (ThermoFisher, EP0753). 200 μL per puck. Prepare before use.
(The following reagents need to be DNase free.)
2X Tissue Digestion Buffer: 100 mM Tris pH 8.0, 200 mM NaCl, 2% SDS, 5 mM EDTA, 32 unit / μL Proteinase K (NEB P8107S). Prepare before use.
Wash Buffer: 10 mM Tris pH 8.0, 1mM EDTA, 0.01% Tween-20.
10mM Tris-HCl (pH 8.0) (ThermoFisher, 15568025) For 10mM Tris-HCl, dilute 1M solution 1:100 with DEPC water. Prepare before use.
Exonuclease I solution: Add 20 μL ExoI buffer and 10 μL ExoI (NEB, M0568L) to 170 μL water. Prepare before use.
0.1N NaOH (Fisher Scientific, S25549). Store at room temperature.
Tris-EDTA buffer (TE buffer; ThermoFisher, AM9849).
Second Strand Mix: 133 μL water, 40 μL 5X Maxima RT buffer, 20 μL 10 mM dNTPs, 2 μL 1mM dN-SMRT oligo, 5 μL Klenow Enzyme (NEB, M0210). Prepare before use.
cDNA PCR mix: 88 μL water, 100 μL Terra PCR Direct Buffer (Takara Biosciences, 639270), 4 μL Terra Polymerase (Takara Biosciences, 639270), 4 μL 100 μM Truseq PCR handle primer (IDT), 4 μL 100 μM SMART PCR primer (IDT). Prepare before use.
AMPure XP beads (Beckman Coulter, A63881).
Library prep reaction mix: 10 μL Nextera TD Buffer (Illumina, FC-131–1096), 5uL Amplicon Tagmentation Enzyme (Illumina, FC-131–1096), add a total of 600pg cDNA from the cDNA synthesis step, adjust the total volume to 20 μL with water,
Nextera PCR Master Mix: 15 μL Nextera PCR mix (Illumina, FC-131–1096), 8uL water, 1 μL 10 μM P5-Truseq PCR hybrid oligo (IDT), 1 μL 10 μM Nextera N70X oligo (Illumina, FC-131–2001).
Sequencing kit: NextSeq2000 P3 (100 cycle) flow cells (Illumina, 20040559).
-
Template Switch Oligo: AAGCAGTGGTATCAACGCAGAGTGAATrG+GrG
Truseq_PCR_Handle: CTACACGACGCTCTTCCGATCT
SMART_PCR_Primer: AAGCAGTGGTATCAACGCAGAGT
dN-SMRT oligo: AAGCAGTGGTATCAACGCAGAGTGANNNGGNNNB
Truseq_P5: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT
3. Methods:
3.1. Tissue dissection and embedding:
Before experiment, inspect the pucks under stereomicroscope. Only pucks without obvious space between beads will be used for experiments.
Pre-chilled HBSS and O.C.T. on ice (over 10min).
Inject the mice intraperitoneally with urethane (10 μL /g body weight). Under full anesthesia, decapitate the mice, remove the brains from the skull, and rinse brains with ice-cold HBSS to remove blood (Note 1).
Separate the olfactory bulbs from the brain by a vertical cut along the dorsal-ventral axis slightly behind the OBs, using a scalpel. Transfer the tissues into pre-cooled O.C.T. in a small petri dish (351007, Falcon@ 60mm × 15mm, Corning) to remove liquid and then transfer the tissue to embedding molds (4566, Tissue-Tek Cryonold, Sakura).
Embed the OBs with pre-chilled O.C.T. and adjust their orientation under a stereomicroscope such that the rostral part of the OBs points downward. Flash freeze at −70°C (HistoChill, Novec™ 7000) (Note 2). Tissues can be kept at −70°C for up to 1 month. Stored tissues need to be wrapped with aluminum foil and kept in sealed plastic bags to prevent dehydrate.
3.2. Tissue sectioning:
On the day of tissue sectioning, remove the tissue blocks from −70°C storage and warm it inside a Cryostat microtome (CryoStar, NX70) to approximately −20°C.
Mount the block on the cutting stage and section it at 10μm thickness. Temperature setting: Chamber Temperature: −12°C, Object Temperature: −10°C.
Ensure slices are flat, no tear or damage on it. Place slices on the pucks (Note 3). Collect slices around 1.6mm from the anterior OB boundary. Adjacent slices should be also collected on normal slides for H&E and DAPI staining to check for tissue integrity (Figure 1A – 1C).
Immediately after the tissues are collected on the pucks, immerse the puck in 200 μL hybridization buffer for approximately 30min at room temperature to facilitate the binding of tissue mRNA to the spatially barcoded beads of the puck (Figure 1D; Note 4).
Figure 1. Illustration of Slide-Seq procedure.
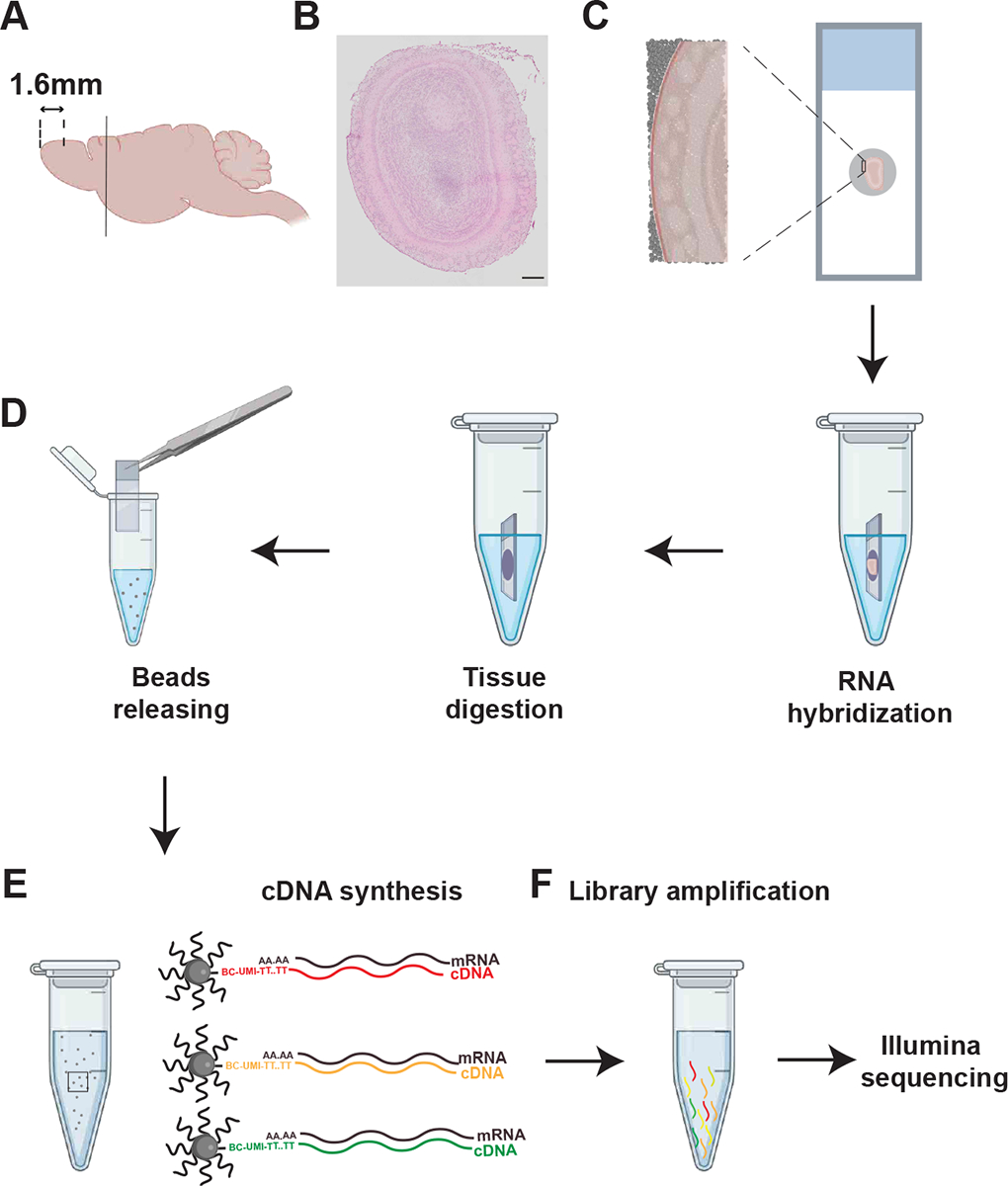
(A) The mouse brain is cut behind OB (black line) to remove excess tissue. Coronal sections are collected at the 1.6mm from the anterior edge of olfactory bulb.
(B) H&E staining showing the morphology of the section. Only slices from tissues without damage or malformation are used for downstream experiments.
(C) Illustration of placement of the tissue section on the puck. Magnified view illustrates how the tissue is laid on top of the beads.
(D) Illustration of the pucks with section immersed in hybridization buffer for mRNA capturing, tissue digestion, and beads releasing.
(E) Process for cDNA synthesis.
(F) Process for library amplification and sequencing.
Arrows from C to F indicate experimental flow. Scale bar: 200 μm in panel (B)
3.3. Library preparation:
After hybridization, transfer the pucks into 200 μL reverse transcription solution, incubate for 30 minutes at room temperature followed by 1.5 hours at 52°C. This reverse transcription step allows the first strand cDNA to be synthesized from RNA molecules captured on the beads.
After reverse transcription, digest the tissue on the pucks by adding 200 μL 2x Tissue Digestion Buffer directly into RT buffer and incubating at 37°C for 30min (Figure 1D).
Add 200 μL of Wash Buffer following the digestion. Pipetted up and down several times to release the beads from the coverslip. Once the beads have been released, remove the coverslip with forceps (Figure 1D; Note 5).
Centrifuge the tubes under 3000 RCF for 3min, discard the supernatant, and resuspend the precipitated beads with 200 μL Wash Buffer.
Repeat step 4 twice to remove residual enzymes and dNTPs.
After the third centrifugation step, add 200 μL 10mM Tris-HCl (pH 8.0) and centrifuge the tube at 3000 RCF for 3 minutes. Remove the supernatant.
Resuspend the samples with exonuclease I solution and incubate for 50 minutes at 37°C to degrade free-floating single-strand DNA and reduce background.
Wash the beads using 200 μL Wash Buffer, centrifuge for 3 minutes at 3000 RCF, removing the supernatant, and resuspending the beads. Repeat once.
After the last wash, resuspend the beads with 0.1N NaOH and incubated for 5 minutes at room temperature. NaOH incubation would denature the RNA-DNA complex and release the RNA molecules.
Wash twice with 200 μL Wash Buffer as in Step 8 (Note 6).
Resuspend the beads in 200 μL TE, pellet down, remove the supernatant, resuspend them in 200 μL Second Strand Mix, and incubated for 1 hour at 37°C. This step generates second strand cDNA from the first strand on the beads.
Wash the beads 3 times with 200 μL Wash Buffer as in Step 8. Perform a final wash in 200 μL of water, pellet down, remove the supernatant, and resuspend them in cDNA PCR mix. Transfer the mix into PCR tubes for PCR (Note 7).
- cDNA was amplified by PCR using the following program (Figure 1E):
- 98°C 3 minutes
- 4 cycles of
- 98°C, 20 seconds
- 65°C, 45 seconds
- 72°C, 3 minutes
- 9 cycles of
- 98°C, 20 seconds
- 67°C, 20 seconds
- 72°C, 3 minutes
- 72°C, 5 minutes
- 4°C, forever
Purify the PCR products with 0.6X AMPure XP beads twice, resuspend in 20 μL water, and check for quality and quantity using the Bioanalyzer (Agilent) and Qubit Fluorometer (ThermoFisher). Expected cDNA concentration should be above 0.15ng/ μL. The size of cDNA is organism dependent, typically between 1000bp-2000bp.
Perform library preparation using the Nextera XT kit (Illumina, FC-131–1096) reagents. Incubate 600pg of cDNA with the library prep reaction mixes at 55°C for 5minutes. After incubation, add 5 μL of neutralization buffer (Illumina, FC-131–1096), mix with pipetting, then incubate at room temperature for 5 minutes. Add Nextera PCR MasterMix to the solution and mix by pipetting (Note 8)
- Perform library amplification by PCR using the following program (Figure 1F):
- 72°C 3 minutes
- 95°C, 30 seconds
- 12 cycles of
- 95°C, 10 seconds
- 55°C, 30 seconds
- 72°C, 30 seconds
- 72°C, 5 minutes
- 10°C, forever
Purify the PCR product with 0.6X AMPure XP beads once, resuspend in 10 μL water, and check for quality and quantity using the Bioanalyzer (Agilent) and Qubit Fluorometer (ThermoFisher). Libraries are normally above 3ng/ μL in concentration and between 400–700bp in length.
3.4. Sequencing
Perform sequencing. We used a P3 flow cell on an Illumina NextSeq 2000 instrument controlled by the NextSeq 2000 1.4.0.39521 software with the following paired read length: 42 bp Read1, 8 bp i7 Index, and 60 bp Read2. Illumina Primary Analysis version NextSeq2K RTA 3.9.2 and Secondary Analysis version bcl2fastq2 v2.20 were run to demultiplex reads for all libraries and generate FASTQ files (Figure 1F).
3.5. Sequencing data analysis
Single cell RNA-Seq data can be leveraged to enhance cell clustering. Before we divide the analysis into two parts.
scRNA-Seq Data Analysis
We aligned scRNA-Seq reads to mm10 mouse genome reference from UCSC using CellRanger (v4.0.0; 10X Genomics). Annotation was from Ensembl 102.
We made a cutoff to maintain cells with 2000–5000 genes and less than 5% mitochondrial counts for downstream analysis
We used DoubletFinder (v2.0.03) (39) to remove doublets from each individual dataset assuming a 7.5% doublet formation rate.
We used Seurat SCTransform V2 to process each individual sample and merge them into a single Seurat object. We then performed PCA dimensional reduction and ran FindClusters using 50 PCs with 0.3 resolution.
We annotated the clusters with marker genes expression patterns: mitral/tufted cell: Cdhr1, Uchl1, Emoes; GABAergic interneurons: Gad1, Gad2, Meis2; Oligodendrocytes: Olig2 & Cspg4; Astrocytes: Slc1a3 & Gfap; Microglia: Ctss & Hexb; Olfactory ensheathing cells (OECs): Plp1 & Npy; Meninges: Slc6a13 & Cxcl12; Pericyte: Myl9; Endothelial cells: Pecam1 & Cd34.
Slide-Seq Data Analysis
We aligned sequencing reads to mm10 mouse genome reference from UCSC using CellRanger (v4.0.0; 10X Genomics) Annotation was from Ensembl 102. Only beads with barcode mismatch <= 1 and UMI counts >= 10 were kept for future analysis.
After generating the expression matrix, we assigned each bead to a location on the puck according to the original spatial barcode information. Files containing the expression matrix and coordinate location were exported for further analysis.
Using prior knowledge regarding the anatomical structure, we used the R package gatepoints to manually select the cells in the olfactory bulb. Other spurious cells around the periphery were discarded from further analysis. (Figure 2A). Most beads can detect 100–580 UMIs and 75–450 genes with around 1.5 UMIs/Genes ratio (Figure 2B).
We used Seurat functions FindTransferAnchors and TransferData to transfer cluster labels in the scRNA-seq data to the Slide-Seq data (40). For each individual Slide-Seq pixel (bead), the probability that it belongs to the same cell-type as the single-cell clusters is calculated, with probabilities summing up to 1 across all cell types. The single cell cluster with the highest probability was used as the assigned label for each pixel. If no individual single-cell cluster receives above a 0.25 probability, the pixel is labeled as “Unassigned”. (Figure 2E)
We annotate the cluster using a combination of marker gene expression patterns and spatial patterns of single-cell clusters across the tissue. Using known marker genes of specific cell types, single cell cluster annotations were assigned and transferred over to the slide-seq data. For clusters with no strong known marker genes, previous anatomical knowledge along with the spatial pattern of the Slide-Seq pixels in the olfactory bulb of a given cluster was used to inform cluster annotations. (Figure 2C, 2D, & 2E).
Figure 2. P7 OB slide-seq result revealed distinguished cell types.
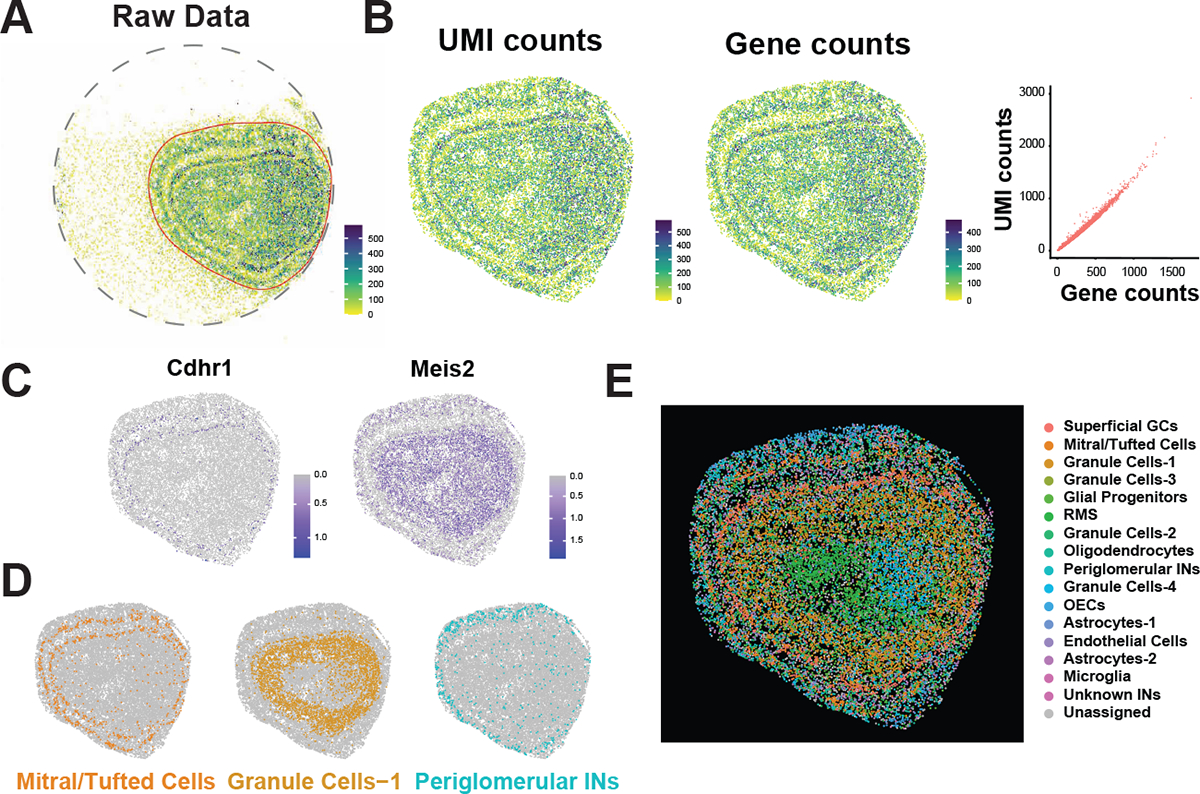
(A) Total unique molecule index (UMI) count mapped onto the puck. Tissue area was circle (red lined area) to remove signals in the background.
(B) Heatmaps showing the total UMI (left) and gene (right) counts mapped to the beads. Scatter plot between UMI counts and gene counts indicates relative low depth of Slide-Seq.
(C) Neuronal markers detected by Slide-Seq data are found in the expected locations. Cdhr1 is located to the mitral/tufted cell layer. Meis2 is located to the inner plexiform layer where GABAergic interneurons reside.
(D & E) Multiple cell types are identified based on single cell cluster features and located in the section based on the spatial information. Cell types are marked based on marker gene expression and are color coded. Abbreviations: GC, granule cell; RMS, rostral migratory stream; IN, interneuron; OEC, olfactory ensheathing cell.
4. Notes:
Note 1: Dissection for each animal should be finished in 2–3min. All dissection process should be performed on ice.
Note 2: Fresh tissues with O.C.T. need to be frozen rapidly for preventing crystals formation.
Note 3: Slices need to be placed in the middle of the puck, where the beads are (Figure 1C). Double confirm slices are fully attached to the beads without bubbles.
Note 4: The timing for this incubation step is kept consistent between all samples, following the Broad Institutes recommended 15–30minute hybridization time.
Note 5: Beads can be visually observed dislocating from the puck.
Note 6: Insufficient wash may result in high background for sequencing data.
Note 7: Each 200 μL sample is split into two PCR tubes prior to cDNA amplification.
Note 8: The library preparation described is performed according to manufacturer’s directions for the Nextera XT kit (Illumina, FC-131–1096), modifying the starting input of material to 600pg of cDNA and using a specific P5-Truseq PCR hybrid oligo in place of the Nextera XT i5 adapter.
Acknowledgement:
We thank Dr. Fei Chen and Mr. Evan Murray at Broad Institute for providing the pre-sequenced pucks. We thank Lab Animal Service Facility, Histology, Sequencing, and Microscopy Center of Stowers Institute for their technical assistance. We also thank Yu lab members for their insightful discussions. This work was supported by funding from NIH (R01DC016696 and R01DC014701) and Stowers Institute for Medical Research to C.R.Y.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References:
- 1.Fletcher RB, Das D, Gadye L, Street KN, Baudhuin A, Wagner A, Cole MB, Flores Q, Choi YG, Yosef N, Purdom E, Dudoit S, Risso D, Ngai J (2017) Deconstructing Olfactory Stem Cell Trajectories at Single-Cell Resolution. Cell stem cell 20 (6):817–830 e818. doi: 10.1016/j.stem.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tepe B, Hill MC, Pekarek BT, Hunt PJ, Martin TJ, Martin JF, Arenkiel BR (2018) Single-Cell RNA-Seq of Mouse Olfactory Bulb Reveals Cellular Heterogeneity and Activity-Dependent Molecular Census of Adult-Born Neurons. Cell reports 25 (10):2689–2703 e2683. doi: 10.1016/j.celrep.2018.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Y, Ma L, Duyck K, Long CC, Moran A, Scheerer H, Blanck J, Peak A, Box A, Perera A, Yu CR (2018) A Population of Navigator Neurons Is Essential for Olfactory Map Formation during the Critical Period. Neuron 100 (5):1066–1082 e1066. doi: 10.1016/j.neuron.2018.09.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanchate NK, Kondoh K, Lu Z, Kuang D, Ye X, Qiu X, Pachter L, Trapnell C, Buck LB (2015) Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science 350 (6265):1251–1255. doi: 10.1126/science.aad2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukahara T, Brann DH, Pashkovski SL, Guitchounts G, Bozza T, Datta SR (2021) A transcriptional rheostat couples past activity to future sensory responses. Cell 184 (26):6326–6343 e6332. doi: 10.1016/j.cell.2021.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vihani A, Hu XS, Gundala S, Koyama S, Block E, Matsunami H (2020) Semiochemical responsive olfactory sensory neurons are sexually dimorphic and plastic. eLife 9. doi: 10.7554/eLife.54501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horgue LF, Assens A, Fodoulian L, Marconi L, Tuberosa J, Haider A, Boillat M, Carleton A, Rodriguez I (2022) Transcriptional adaptation of olfactory sensory neurons to GPCR identity and activity. Nature communications 13 (1):2929. doi: 10.1038/s41467-022-30511-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duyck K, DuTell V, Ma L, Paulson A, Yu CR (2017) Pronounced strain-specific chemosensory receptor gene expression in the mouse vomeronasal organ. BMC genomics 18 (1):965. doi: 10.1186/s12864-017-4364-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Jiang H, Yang L (2020) Transcriptome Analysis of Zebrafish Olfactory Epithelium Reveal Sexual Differences in Odorant Detection. Genes 11 (6). doi: 10.3390/genes11060592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nature methods 6 (5):377–382. doi: 10.1038/nmeth.1315 [DOI] [PubMed] [Google Scholar]
- 11.Nawy T (2014) Single-cell sequencing. Nature methods 11 (1):18. doi: 10.1038/nmeth.2771 [DOI] [PubMed] [Google Scholar]
- 12.Shapiro E, Biezuner T, Linnarsson S (2013) Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature reviews Genetics 14 (9):618–630. doi: 10.1038/nrg3542 [DOI] [PubMed] [Google Scholar]
- 13.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X (2015) RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348 (6233):aaa6090. doi: 10.1126/science.aaa6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X (2016) High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proceedings of the National Academy of Sciences of the United States of America 113 (39):11046–11051. doi: 10.1073/pnas.1612826113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C, Zhuang X (2016) High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proceedings of the National Academy of Sciences of the United States of America 113 (50):14456–14461. doi: 10.1073/pnas.1617699113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilliams M, Bonnardel J, Haest B, Vanderborght B, Wagner C, Remmerie A, Bujko A, Martens L, Thone T, Browaeys R, De Ponti FF, Vanneste B, Zwicker C, Svedberg FR, Vanhalewyn T, Goncalves A, Lippens S, Devriendt B, Cox E, Ferrero G, Wittamer V, Willaert A, Kaptein SJF, Neyts J, Dallmeier K, Geldhof P, Casaert S, Deplancke B, Ten Dijke P, Hoorens A, Vanlander A, Berrevoet F, Van Nieuwenhove Y, Saeys Y, Saelens W, Van Vlierberghe H, Devisscher L, Scott CL (2022) Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185 (2):379–396 e338. doi: 10.1016/j.cell.2021.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, Peters DT, Turczyk BM, Marblestone AH, Inverso SA, Bernard A, Mali P, Rios X, Aach J, Church GM (2014) Highly multiplexed subcellular RNA sequencing in situ. Science 343 (6177):1360–1363. doi: 10.1126/science.1250212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, Zhang K, Church GM (2015) Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nature protocols 10 (3):442–458. doi: 10.1038/nprot.2014.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, Cai L (2019) Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568 (7751):235–239. doi: 10.1038/s41586-019-1049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, Nolan GP, Bava FA, Deisseroth K (2018) Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361 (6400). doi: 10.1126/science.aat5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S, Nolan GP (2018) Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 174 (4):968–981 e915. doi: 10.1016/j.cell.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lein E, Borm LE, Linnarsson S (2017) The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358 (6359):64–69. doi: 10.1126/science.aan6827 [DOI] [PubMed] [Google Scholar]
- 23.Stahl PL, Salmen F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg A, Ponten F, Costea PI, Sahlen P, Mulder J, Bergmann O, Lundeberg J, Frisen J (2016) Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353 (6294):78–82. doi: 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- 24.Chen A, Liao S, Cheng M, Ma K, Wu L, Lai Y, Qiu X, Yang J, Xu J, Hao S, Wang X, Lu H, Chen X, Liu X, Huang X, Li Z, Hong Y, Jiang Y, Peng J, Liu S, Shen M, Liu C, Li Q, Yuan Y, Wei X, Zheng H, Feng W, Wang Z, Liu Y, Wang Z, Yang Y, Xiang H, Han L, Qin B, Guo P, Lai G, Munoz-Canoves P, Maxwell PH, Thiery JP, Wu QF, Zhao F, Chen B, Li M, Dai X, Wang S, Kuang H, Hui J, Wang L, Fei JF, Wang O, Wei X, Lu H, Wang B, Liu S, Gu Y, Ni M, Zhang W, Mu F, Yin Y, Yang H, Lisby M, Cornall RJ, Mulder J, Uhlen M, Esteban MA, Li Y, Liu L, Xu X, Wang J (2022) Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185 (10):1777–1792 e1721. doi: 10.1016/j.cell.2022.04.003 [DOI] [PubMed] [Google Scholar]
- 25.Mombaerts P (2006) Axonal wiring in the mouse olfactory system. Annual review of cell and developmental biology 22:713–737. doi: 10.1146/annurev.cellbio.21.012804.093915 [DOI] [PubMed] [Google Scholar]
- 26.Sakano H (2020) Developmental regulation of olfactory circuit formation in mice. Development, growth & differentiation 62 (4):199–213. doi: 10.1111/dgd.12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakano H (2010) Neural map formation in the mouse olfactory system. Neuron 67 (4):530–542. doi: 10.1016/j.neuron.2010.07.003 [DOI] [PubMed] [Google Scholar]
- 28.Ma L, Wu Y, Qiu Q, Scheerer H, Moran A, Yu CR (2014) A developmental switch of axon targeting in the continuously regenerating mouse olfactory system. Science 344 (6180):194–197. doi: 10.1126/science.1248805 [DOI] [PubMed] [Google Scholar]
- 29.Tsai L, Barnea G (2014) A critical period defined by axon-targeting mechanisms in the murine olfactory bulb. Science 344 (6180):197–200. doi: 10.1126/science.1248806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheetham CEJ, Park U, Belluscio L (2016) Rapid and continuous activity-dependent plasticity of olfactory sensory input. Nature communications 7:10729. doi: 10.1038/ncomms10729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh DN, Nathaniel EJ (1977) Postnatal development of mitral cell perikaryon in the olfactory bulb of the rat. A light and ultrastructural study. The Anatomical record 189 (3):413–431. doi: 10.1002/ar.1091890303 [DOI] [PubMed] [Google Scholar]
- 32.Lin DM, Wang F, Lowe G, Gold GH, Axel R, Ngai J, Brunet L (2000) Formation of precise connections in the olfactory bulb occurs in the absence of odorant-evoked neuronal activity. Neuron 26 (1):69–80. doi: 10.1016/s0896-6273(00)81139-3 [DOI] [PubMed] [Google Scholar]
- 33.Cao L, Dhilla A, Mukai J, Blazeski R, Lodovichi C, Mason CA, Gogos JA (2007) Genetic modulation of BDNF signaling affects the outcome of axonal competition in vivo. Current biology : CB 17 (11):911–921. doi: 10.1016/j.cub.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batista-Brito R, Close J, Machold R, Fishell G (2008) The distinct temporal origins of olfactory bulb interneuron subtypes. The Journal of neuroscience : the official journal of the Society for Neuroscience 28 (15):3966–3975. doi: 10.1523/JNEUROSCI.5625-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angelova A, Tiveron MC, Cremer H, Beclin C (2018) Neuronal Subtype Generation During Postnatal Olfactory Bulb Neurogenesis. Journal of experimental neuroscience 12:1179069518755670. doi: 10.1177/1179069518755670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, Macosko EZ (2019) Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363 (6434):1463–1467. doi: 10.1126/science.aaw1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stickels RR, Murray E, Kumar P, Li J, Marshall JL, Di Bella DJ, Arlotta P, Macosko EZ, Chen F (2021) Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nature biotechnology 39 (3):313–319. doi: 10.1038/s41587-020-0739-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, Ma L, Qiu Q, Xu W, Misra A, Duyck K, Blanck J, Scott AR, Chen S, Hassan H, Corbin TJ, Moran A, Hall K, Li H, Perera A, Yu CR (2022) Molecular Control of Circuit Plasticity and the Permanence of Imprinted Odor Memory. bioRxiv:2022.2003.2029.486284. doi: 10.1101/2022.03.29.486284 [DOI] [Google Scholar]
- 39.McGinnis CS, Murrow LM, Gartner ZJ (2019) DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell systems 8 (4):329–337 e324. doi: 10.1016/j.cels.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd,Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R (2021) Integrated analysis of multimodal single-cell data. Cell 184 (13):3573–3587 e3529. doi: 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]


