Abstract
A new subgroup of avian leukosis virus (ALV) that includes a unique env gene, designated J, was identified recently in England. Sequence analysis of prototype English isolate HPRS-103 revealed several other unique genetic characteristics of this strain and provided information that it arose by recombination between exogenous and endogenous virus sequences. In the past several years, ALV J type viruses (ALV-J) have been isolated from broiler breeder flocks in the United States. We were interested in determining the relationship between the U.S. and English isolates of ALV-J. Based on sequence data from two independently derived U.S. field isolates, we conclude that the U.S. and English isolates of ALV-J derive from a common ancestor and are not the result of independent recombination events.
Members of the leukosis/sarcoma group of avian retroviruses are divided into subgroups based on the identity of their envelope genes. The mature env gene products are the gp85 surface glycoprotein (SU), which directs receptor binding, and the gp37 transmembrane protein (TM), which is linked to SU by disulfide bonds and which anchors the complex to the viral membrane (reviewed in reference 19). Five major subgroups of avian retroviruses (A to E), which differ in host range, viral interference, and cross-neutralization, properties that are determined by the portion of env encoding gp85 have been identified (7, 8, 13). The envelope genes for subgroups A to D are found in exogenous viruses, while the E subgroup is encoded by the env gene of the ev family of endogenous proviruses (3, 6, 12, 17, 18). The gp85 proteins of subgroup A to E viruses are approximately 85% identical to each other; subgroup-determining regions map to discrete variable and hypervariable regions of SU (7, 8, 14, 33).
Several years ago, a number of nonacute avian leukosis viruses were identified in England; these viruses exhibited a novel subgroup specificity, designated J, that differed from those of previously characterized avian virus subgroups A to E based on patterns of viral interference, cross-neutralization, and host range (2, 4, 5, 22–26, 35). These viruses were originally identified based on their ability to induce myelocytic myeloid leukosis (2, 23, 26, 27). Sequence analyses of several type J avian leukosis virus (ALV-J) isolates have demonstrated that the subgroup J gp85 genes show only 40% overall identity to the gp85 genes of subgroup A to E viruses (2, 23, 26, 27). In particular, the amino-terminal 43 amino acids and a region between residues 251 and 289 (Fig. 1) of the subgroup J gp85 protein each show a high degree of identity (84%) to subgroup A to E gp85 proteins; the remainder of the subgroup J protein shows no significant homology to those of the other subgroups. In addition, while the ALV-J SU proteins are over 90% identical to each other, they exhibit localized regions of sequence alterations and also show antigenic variation (35). Although only weakly related to the SU proteins of the subgroup A to E viruses, the ALV-J gp85 protein does include several regions between 6 and 21 amino acids in length that are over 90% identical to the gp85 protein of the ancient endogenous avian proviruses (EAVs) (4, 5; unpublished observations). Members of the EAV family of avian endogenous viruses are distinct from the well-characterized subgroup E endogenous viruses encoded by the ev loci (10, 11, 13); the subgroup specificity of the EAVs is unknown since all proviruses of this family identified to date include a defective env gene (10, 11, 13). The finding that the HPRS-103 env gene contains sequences related to those of EAVs, together with the genome structure of the prototype English ALV-J strain, HPRS-103 (see below), has led to the suggestion that HPRS-103 arose by the recombination of one or more exogenous viruses with other viruses (or a single virus), at least one of which was related to the EAVs (4, 5).
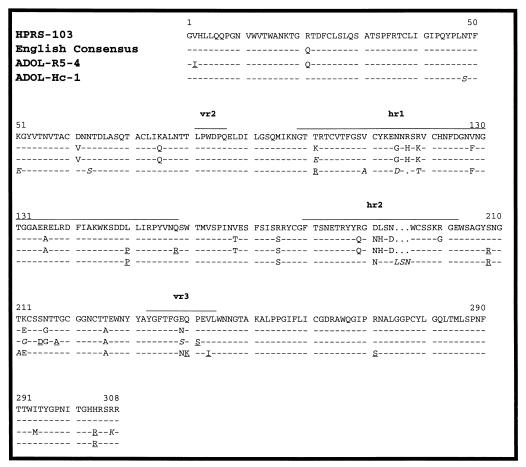
FIG. 1.
Comparison of the predicted amino acid sequences of ALV-J gp85 proteins. Shown are the predicted amino acid sequence of gp85 from HPRS-103 (4), the consensus sequence of the 13 English ALV-J strains (35), and the sequences of the two U.S. field isolates ADOL-R5-4 and ADOL-Hc-1. The consensus sequence is defined as the sequence found in seven or more of the sequenced English isolates. Dashes indicate identical residues, while letters indicate amino acid substitutions. The locations of the variable (vr) and hypervariable (hr) regions are based on alignments with gp85 proteins of subgroups A to E and do not necessarily reflect variation among the subgroup J proteins. Underlined residues in the U.S. field isolate sequences identify amino acids in the U.S. field isolates that, although not identical to those of the consensus sequence, are also found in one or more of the English isolates and therefore do not represent substitutions unique to U.S. strains. Amino acid substitutions in italics are unique to ADOL-R5-4 (residues 101, 212, 239, and 307) or ADOL-Hc-1 (residues 48, 51, 63, 110, 115, 117, 119, 211, and a three-amino-acid substitution after residue 194).
Over the last several years, several commercial breeders in the United States have reported the appearance of myeloid tumors similar to those induced by ALV-J viruses identified in England (15, 16). Biological testing (including viral interference assays and serological screening) of viruses obtained from these tumors indicated that the U.S. field isolates were subgroup J viruses (15, 16). Since these viruses had not been detected previously in the United States, we were interested in investigating the genetic content of the ALV-J U.S. field isolates to determine whether the U.S. isolates and the English prototype strain, HPRS-103, appeared to derive from a common ancestor or if they more likely arose from independent events. To approach this question, two field isolates of ALV-J, termed ADOL-R5-4 and ADOL-Hc-1 (15, 16), were subjected to partial sequence analysis. For the ADOL-R5-4 isolate, the sequence was obtained from an infectious molecular clone generated from infected-cell DNA, while the ADOL-Hc-1 sequence was obtained from cloned DNA.
env gene.
Figure 1 shows the predicted amino acid sequences of the SU portions of the ADOL-R5-4 and ADOL-Hc-1 env gene products aligned with that of HPRS-103 (4). Also included is the consensus sequence derived from the 13 sequenced English isolates of ALV-J (35). These data revealed that the ADOL-R5-4 SU protein showed 90.9% identity with the SU protein of HPRS-103, differing at only 28 residues. A comparison with the consensus sequence obtained from all sequenced English isolates revealed an even higher level of identity (95.1%). Similarly, the ADOL-Hc-1 strain SU proteins showed a 92.2 and 90.3% identity with the SU proteins of HPRS-103 and the consensus sequence, respectively. The level of variation between the two U.S. isolates was 11%. These results demonstrate that the HPRS-103, ADOL-R5-4, and ADOL-Hc-1 SU proteins are highly related and that the degrees of variation between the two U.S. isolates and between these isolates and the English isolates are similar to those seen among the group of English isolates (35). Interestingly, most of the amino acid substitutions seen in the U.S. field isolates were also seen in at least one of the English ALV-J isolates sequenced (35). There were in fact only 4 substitutions in the ADOL-R5-4 sequence and 11 substitutions in the ADOL-Hc-1 sequence that were not seen in any of the English isolates; none of these were shared between the two U.S. isolates. Of note was the finding that the ADOL-Hc-1 sequence showed a one-amino-acid deletion within hypervariable region 1 (hr1), a finding also reported for one of the English isolates (X12) (35). ADOL-Hc-1 was unique among all isolates sequenced in having a three-amino-acid insertion within the hr2 region; the significance of these findings are currently being investigated. Together, these data demonstrate that, while each SU gene is unique, they are all very highly related and are likely to have arisen from a common source.
pol gene.
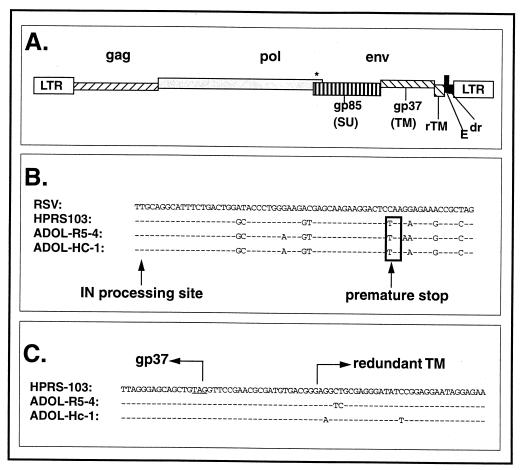
As mentioned above, the HPRS-103 genome exhibits several features that have not been reported previously for any other avian retrovirus (Fig. 2A). One such feature is the presence of a premature stop codon near the end of the pol gene (4). This mutation is unlikely to affect either the mature Pol proteins or virus growth. This conclusion is based on the fact that the codon resides in a region of pol that in other avian leukosis/sarcoma viruses (ALSVs) encodes the carboxy-terminal portion of the unprocessed Pol protein; this portion of Pol is removed during maturation to generate the mature integrase protein and is therefore not required for Pol or IN function. It has in fact been demonstrated that this carboxy-terminal tail is not essential for virus growth, at least in vitro (20). In our search of the GenBank database (which includes 16 different avian retrovirus sequences that span this region), we have not found another example of this mutation, suggesting that it might serve as a diagnostic for ALV-J and/or the virus(es) that might have given rise to ALV-J. We therefore sequenced this region from the two U.S. isolates; the data obtained are shown in Fig. 2B. As shown, the mutation that gives rise to the premature stop codon is found in both U.S. field isolates of ALV-J. These data indicate that the presence of the premature stop codon is not unique to the HPRS-103 strain but instead is a common feature of all three of the ALV-J strains. This finding, therefore, supports the hypothesis that either each of the ALV-J isolates analyzed traces to a common ALV-J ancestor or that one of the virus parents that gave rise to all of these isolates contained the pol gene mutation.
FIG. 2.
Sequence comparison of selected regions of the HPRS-103 and U.S. field isolates. (A) Line drawing of the genome of HPRS-103 (see text for a description of each region). An asterisk indicates a premature stop codon near the end of the pol gene. (B) Sequence comparison of the 3′ ends of the pol genes from the indicated viruses. The IN processing site is the site in the Pol polyprotein that is subject to proteolytic processing to generate the carboxy-terminal end of the mature IN protein. The boxed TAA sequence is the location of the premature stop codon found in HPRS-103 and the two U.S. ALV-J field isolates. This sequence has not been found in other avian retrovirus sequences in the GenBank database (unpublished observations). The coding sequence for the normal Pol processing site in Rous sarcoma virus (RSV) is 66 nucleotides downstream from the premature stop codon shown in this figure. (C) Shown is the junction between the portion of the ALV-J env gene that encodes the J-specific gp37 protein and that which encodes the partially duplicated copy of TM (rTM) that is related to the gp37 from subgroup A to E viruses.
rTM.
A third unusual feature of the HPRS-103 isolate is the presence of a partially duplicated copy of the transmembrane (TM)-encoding portion of the env gene (the redundant TM or rTM). As originally noted by Bai et al. (5), the rTM is over 90% identical to that of the TM-encoding portions of the env genes of ALSVs of the A to E subgroups but shows only approximately 60% identity to that of the HPRS-103 TM-encoding portion of the env gene. It is therefore presumed to have arisen from the exogenous virus parent of HPRS-103 after an illegitimate recombination event with one or more other exogenous or endogenous viruses. As with the pol mutation, the partial duplication of the TM-encoding portion of the env gene found in HPRS-103 has not been described previously in other avian viruses of any subgroup. The retention of the TM-rTM junction, therefore, would be highly suggestive of a common recombinant having given rise to the U.S. and English isolates of ALV-J. Figure 2C shows a sequence alignment of this region from ADOL-R5-4, ADOL-Hc-1, and HPRS-103. As shown, each of the U.S. field isolates shows the same junction sequence as HPRS-103 between the end of the subgroup J gp37/TM-encoding portion of the env gene and the rTM.
In addition to the sequence data shown here, we also have sequenced the 3′-untranslated regions (3′-UTRs) and the long terminal repeats (LTRs) of the two U.S. field isolates and find that they are virtually identical to those of the HPRS-103 strain (data not shown). In particular, both the ADOL-R5-4 and the ADOL-Hc-1 viruses contain 150-bp sequences within their 3′-UTRs, called E elements, that are also found in several strains of Rous sarcoma virus (21, 28, 34) but that have not been reported previously in other strains of ALV. All isolates also contain a direct repeat (dr) sequence in the 3′-UTR. In addition, the two U.S. field isolates, like HPRS-103, contain only one copy of an enhancer motif, called EFII, that is found in two copies in other avian retroviral LTRs (9, 29–32). Together, these data support the hypothesis that all ALV-J viruses analyzed to date arose from a common ancestor that was generated by one or more rare recombination events between exogenous and endogenous virus sequences. We are currently extending these analyses to identify the origin of the ALV-J env gene and have identified novel endogenous virus sequences as the likely source.
Nucleotide sequence accession numbers.
The env sequences used to generate the predicted amino acid sequences presented here have been submitted to GenBank. The ADOL-R5-4 sequence has been assigned accession no. AF076887, and the ADOL-Hc-1 sequence has been assigned accession no. AF097731.
Acknowledgments
This work was supported by Public Health Service grant R01-GM41571 from the National Institute of General Medical Sciences and by a grant from the Leukemia Task Force. S.J.B. was supported by training grant CA09138 from the National Institutes of Health.
REFERENCES
- 1.Alexander F, Leis J, Soltis D A, Crowl R M, Danho W, Poonian M S, Pan Y-C E, Skalka A M. Proteolytic processing of avian sarcoma and leukosis viruses pol-endo recombinant proteins reveals another pol gene domain. J Virol. 1987;61:534–542. doi: 10.1128/jvi.61.2.534-542.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arshad S S, Howes K, Barron G S, Smith L M, Russell P H, Payne L N. Tissue tropism of the HPRS-103 strain of J subgroup avian leukosis virus and of a derivative acutely transforming virus. Vet Pathol. 1997;34:127–137. doi: 10.1177/030098589703400205. [DOI] [PubMed] [Google Scholar]
- 3.Astrin S M, Robinson H L, Crittenden L B, Buss E G, Wyban J, Hayward W S. Ten genetic loci in the chicken that contain structural genes for endogenous avian leukosis viruses. Cold Spring Harbor Symp Quant Biol. 1980;44:1105–1109. doi: 10.1101/sqb.1980.044.01.119. [DOI] [PubMed] [Google Scholar]
- 4.Bai J, Howes K, Payne L N, Skinner M A. Sequence of host-range determinants in the env gene of a full-length, infectious proviral clone of exogenous avian leukosis virus HPRS-103 confirms that it represents a new subgroup (designated J) J Gen Virol. 1995;76:181–187. doi: 10.1099/0022-1317-76-1-181. [DOI] [PubMed] [Google Scholar]
- 5.Bai J, Payne L N, Skinner M A. HPRS-103 (exogenous avian leukosis virus, subgroup J) has an env gene related to those of endogenous elements EAV-0 and E51 and an E element found previously only in sarcoma viruses. J Virol. 1995;69:779–784. doi: 10.1128/jvi.69.2.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker B, Robison H, Varmus H E, Bishop J M. Analysis of endogenous avian retrovirus DNA and RNA: viral and cellular determinants of retrovirus gene expression. Virology. 1981;114:8–22. doi: 10.1016/0042-6822(81)90248-8. [DOI] [PubMed] [Google Scholar]
- 7.Bova C A, Manfredi J P, Swanstrom R. env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology. 1986;152:343–354. doi: 10.1016/0042-6822(86)90137-6. [DOI] [PubMed] [Google Scholar]
- 8.Bova C A, Olsen J C, Swanstrom R. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J Virol. 1988;62:75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowers W J, Ruddell A. a1/EBP: a leucine zipper protein that binds CCAAT/enhancer elements in the avian leukosis virus long terminal repeat enhancer. J Virol. 1992;66:6578–6586. doi: 10.1128/jvi.66.11.6578-6586.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyce-Jacino M T, O’Donoghue K, Faras A J. Multiple complex families of endogenous retroviruses are highly conserved in the genus Gallus. J Virol. 1992;66:4919–4929. doi: 10.1128/jvi.66.8.4919-4929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyce-Jacino M T, Resnick R, Faras A J. Structural and functional characterization of the unusually short long terminal repeats and their adjacent regions of a novel endogenous avian retrovirus. Virology. 1989;173:157–166. doi: 10.1016/0042-6822(89)90231-6. [DOI] [PubMed] [Google Scholar]
- 12.Coffin J M, Tsichlis P N, Conklin K F, Senior A, Robinson H L. Genomes of endogenous and exogenous avian retroviruses. Virology. 1983;126:51–72. doi: 10.1016/0042-6822(83)90461-0. [DOI] [PubMed] [Google Scholar]
- 13.Dorner A J, Coffin J M. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell. 1986;45:365–374. doi: 10.1016/0092-8674(86)90322-3. [DOI] [PubMed] [Google Scholar]
- 14.Dorner A J, Stoye J P, Coffin J M. Molecular basis of host range variation in avian retroviruses. J Virol. 1985;53:32–39. doi: 10.1128/jvi.53.1.32-39.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fadly A M, Smith E J. Proceedings of the Avian Tumor Viruses Symposium. Kennett Square, Pa: American Association of Avian Pathologists; 1997. An overview of subgroup J-like avian leukosis virus infection in broiler breeder flocks in the United States; pp. 54–57. [Google Scholar]
- 16.Fadly, A. M., and E. J. Smith. Isolation and some characteristics of a subgroup-J-like avian leukosis virus associated with myeloid leukosis in meat-type chickens in the United States. Avian Dis., in press. [PubMed]
- 17.Hayward W S, Braverman S B, Astrin S M. Transcriptional products and DNA structure of endogenous avian proviruses. Cold Spring Harbor Symp Quant Biol. 1980;44:1111–1121. doi: 10.1101/sqb.1980.044.01.120. [DOI] [PubMed] [Google Scholar]
- 18.Hughes S H, Toyoshima K, Bishop J M, Varmus H E. Organization of the endogenous proviruses of chickens: implications for origin and expression. Virology. 1981;108:189–207. doi: 10.1016/0042-6822(81)90538-9. [DOI] [PubMed] [Google Scholar]
- 19.Hunter E. Viral entry and receptors. In: Coffin J M, Hughes S H, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 71–119. [PubMed] [Google Scholar]
- 20.Katz R A, Skalka A M. A C-terminal domain in the avian sarcoma-leukosis virus pol gene product is not essential for viral replication. J Virol. 1988;62:528–533. doi: 10.1128/jvi.62.2.528-533.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laimins L A, Tsichlis P, Khoury G. Multiple enhancer domains in the 3′ terminus of the Prague strain of Rous sarcoma virus. Nucleic Acids Res. 1984;12:6427–6442. doi: 10.1093/nar/12.16.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Payne L N. Developments in avian leukosis research. Leukemia. 1992;6:150S–152S. [PubMed] [Google Scholar]
- 23.Payne L N, Gillespie A M, Howes K. Myeloid leukaemogenicity and transmission of the HPRS-103 strain of avian leukosis virus. Leukemia. 1992;6:1167–1176. [PubMed] [Google Scholar]
- 24.Payne L N, Gillespie A M, Howes K. Recovery of acutely transforming viruses from myeloid leukosis induced by the HPRS-103 strain of avian leukosis virus. Avian Dis. 1993;37:438–450. [PubMed] [Google Scholar]
- 25.Payne L N, Howes K, Gillespie A M, Smith L M. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated J. J Gen Virol. 1992;73:2995–2997. doi: 10.1099/0022-1317-73-11-2995. [DOI] [PubMed] [Google Scholar]
- 26.Payne L N, Howes K, Smith L M, Venugopal K. Avian Tumor Viruses Symposium on the Diagnosis and Control of Neoplastic Diseases of Poultry, Reno, Nev. 1997. Current status of diagnosis, epidemiology and control of ALV-J; pp. 58–62. [Google Scholar]
- 27.Russell P H, Ahmad K, Howes K, Payne L N. Some chickens which are viraemic with subgroup J avian leukosis virus have antibody-forming cells but no circulating antibody. Res Vet Sci. 1997;63:81–83. doi: 10.1016/s0034-5288(97)90163-6. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz D E, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 29.Sealey L, Chalkley R. At least two nuclear proteins bind specifically to the Rous sarcoma virus long terminal repeat enhancer. Mol Cell Biol. 1987;7:787–798. doi: 10.1128/mcb.7.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sears R C, Sealy L. Characterization of nuclear proteins that bind the EFII enhancer sequence in the Rous sarcoma virus long terminal repeat. J Virol. 1992;66:6338–6352. doi: 10.1128/jvi.66.11.6338-6352.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sears R C, Sealy L. Multiple forms of C/EBPβ bind the EFII enhancer sequence in the Rous sarcoma virus long terminal repeat. Mol Cell Biol. 1994;14:4855–4871. doi: 10.1128/mcb.14.7.4855. . (Erratum, 14:5617.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith C D, Baglia L A, Curristin S M, Ruddell A. The VBP and a1/EBP leucine zipper factors bind overlapping subsets of avian retroviral long terminal repeat CCAAT/enhancer elements. J Virol. 1994;68:6232–6242. doi: 10.1128/jvi.68.10.6232-6242.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taplitz R A, Coffin J M. Selection of an avian retrovirus mutant with extended receptor usage. J Virol. 1997;71:7814–7819. doi: 10.1128/jvi.71.10.7814-7819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsichlis P N, Donehower L, Hager G, Zeller N, Malavarca R, Astrin S, Skalka A M. Sequence comparison in the crossover region of an oncogenic avian retrovirus recombinant and its nononcogenic parent: genetic regions that control growth rate and oncogenic potential. Mol Cell Biol. 1982;2:1331–1338. doi: 10.1128/mcb.2.11.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venugopal K, Smith L M, Howes K, Payne L N. Antigenic variants of J subgroup avian leukosis virus: sequence analysis reveals multiple changes in the env gene. J Gen Virol. 1998;79:757–766. doi: 10.1099/0022-1317-79-4-757. [DOI] [PubMed] [Google Scholar]