Abstract
In situ tissue engineering strategies are a promising approach to activate the endogenous regenerative potential of the cardiac tissue helping the heart to heal itself after an injury. However, the current use of complex reprogramming vectors for the activation of reparative pathways challenges the easy translation of these therapies into the clinic. Here, we evaluated the response of mouse neonatal and human induced pluripotent stem cell-derived cardiomyocytes to the presence of exogenous lactate, thus mimicking the metabolic environment of the fetal heart. An increase in cardiomyocyte cell cycle activity was observed in the presence of lactate, as determined through Ki67 and Aurora-B kinase. Gene expression and RNA-sequencing data revealed that cardiomyocytes incubated with lactate showed upregulation of BMP10, LIN28 or TCIM in tandem with downregulation of GRIK1 or DGKK among others. Lactate also demonstrated a capability to modulate the production of inflammatory cytokines on cardiac fibroblasts, reducing the production of Fas, Fraktalkine or IL-12p40, while stimulating IL-13 and SDF1a. In addition, the generation of a lactate-rich environment improved ex vivo neonatal heart culture, by affecting the contractile activity and sarcomeric structures and inhibiting epicardial cell spreading. Our results also suggested a common link between the effect of lactate and the activation of hypoxia signaling pathways. These findings support a novel use of lactate in cardiac tissue engineering, modulating the metabolic environment of the heart and thus paving the way to the development of lactate-releasing platforms for in situ cardiac regeneration.
Keywords: Cardiomyocytes, Lactate, Induced pluripotent stem cells, Cell cycle, Cardiac tissue engineering, Metabolic environment
1. Introduction
Cardiovascular disease (CVD) is the leading cause of death in the developed world. Myocardial infarction and cardiac failure, two prevalent forms of CVDs, are among the most important causes of mortality and morbidity worldwide [1]. During myocardial infarction, a significant portion of the myocardium dies and is replaced by fibrotic tissue, gradually leading to heart failure and eventually death. Therapeutic strategies with varying degrees of success have been proposed to stop this progression [2–4]. However, despite these efforts, cardiac regeneration and full functional recovery have not been achieved yet [5,6]. The design and development of cardiac constructs in vitro may fail when implanted in vivo due to the specific ischemic environment of the heart, with adverse conditions for cells that have been cultured under highly controlled in vitro environments. In this context, the use of an in situ tissue engineering approach, activating regenerative programs of the failing heart, offers the opportunity to bypass some of the major regulatory issues of tissue engineering strategies, as well as to overcome their expensive production costs and low scalability [7]. Current in situ tissue engineering approaches aim to stimulate cardiac regeneration using recombinant virus, interfering RNAs or other complex reprogramming methods. However, despite significant progress over the past years, these strategies are largely ineffective and further research is still needed to improve clinical efficacy and safety [8,9]. In contrast, recent advances in cell metabolism have attracted increasing attention in regenerative medicine due to its ability to influence cell behavior [10–12]. The exploitation of cell metabolism modulation through the use of biomaterials has thus the potential to inspire the development of innovative regenerative therapies that may encourage the next generation of in situ tissue engineering [13].
Interestingly, the heart is capable of using all classes of energy substrates when abundantly available, including glucose, lactate, ketones or amino acids [14]. Cardiac metabolism also changes during development and maturation. In early development, the fetus resides in a low oxygen and lactate-rich environment [15], where the heart is highly dependent on glycolysis and uses lactate as a primary metabolic substrate [16]. After birth, glycolysis and lactate levels are greatly reduced and fatty acid β-oxidation increases to become the predominant energy source.
Coincidentally, fetal cardiomyocytes are very proliferative and present a poorly differentiated phenotype, while changes in the metabolic environment after birth facilitates cardiomyocyte switch from hyperplasia to hypertrophy, ceasing proliferation and increasing cell size in an oxygen-rich environment. Thus, it is increasingly recognized that the metabolic environment of cardiac cells can become a critical regulator of cell function, including contraction, growth or survival [17]. Importantly, alterations in cardiac metabolism contribute to a large number of diseases and cardiomyopathies [14,18,19].
The role of metabolism in modulating cardiomyocyte behavior is thus gaining increasing attention in the last years, as demonstrated by the use of fatty acids to enhance hPSC (human pluripotent stem cells)-derived cardiomyocyte maturation [20]. Therefore, in this work, we hypothesized that exogenous lactate might also be an important signaling molecule on cardiomyocyte maturation and cardiac regeneration. Previous studies have shown that during mammalian heart regeneration, dedifferentiated cardiomyocytes that appear de novo from adult cardiomyocytes shift to a glycolytic metabolism and are thus more reliant on lactate than mature cardiomyocytes [3]. In this study, we investigated the use of exogenous lactate on in vitro mouse neonatal and human pluripotent stem cell-derived cardiomyocytes, evaluating their cell cycle activity and gene expression, as well as the effect of lactate on cardiac fibroblasts, one of the main cell types involved in cardiac tissue repair and remodeling. In addition, we also studied the use of lactate on three-dimensional cardiac tissue using ex vivo culture of neonatal hearts. Our data provide new roles for lactate in cardiovascular biology and development, and paves the way for the development of novel in situ cardiac tissue engineering approaches using lactate-releasing platforms.
Materials and methods
2.1. Isolation and in vitro culture of mouse cardiac cells
Cardiac primary cells were obtained from CD1 neonatal mice (Charles River Laboratories) following a protocol based on Ehler, et al [21]. The procedure was approved by the Animal Experimentation Committee (CEA #10725) from the Government of Catalonia. Hearts from 1 to 3-day-old mice were extracted and transferred into ice-cold phosphate-buffered saline (PBS) containing 20 mM of 2,3-Butanedione monoxime (BDM, Sigma), where they were cleaned and minced into small pieces using curved scissors (approximately 0.5–1 mm3 or smaller). Then, tissue fragments underwent predigestion through the use of a solution containing trypsin-EDTA (Ethylenediaminetetraacetic acid) 0.25 % (Sigma), 4 μg/mL of DNase I and 20 mM of BDM. The samples were then subjected to 20–25 cycles of enzymatic digestion in L-15 medium (Sigma) containing collagenase II (Gibco), Dispase II (Sigma) and 20 mM of BDM. Pooled supernatants were collected through a 70 μm nylon cell strainer (Corning) and centrifuged at 200G for 10 min. The pellet was resuspended in DMEM (Dulbecco’s Modified Eagle Medium) containing 1 g/L glucose (Gibco) supplemented with 19% M-199 medium (Sigma), 10 % horse serum (Sigma), 5 % fetal bovine serum (FBS, Sigma) and 1 % penicillin and streptomycin (Gibco). Cell suspension was plated into a cell culture dish in order to separate the non-myocytic cell fraction of the heart, and after 1 h of incubation the supernatant containing a purified population of cardiomyocytes was collected and centrifuged again. Cardiomyocytes were seeded on 0.5 % gelatin-coated multiwell plates at 80-150 103 cells/cm2 and cultured in DMEM (Gibco) supplemented with 17 % M-199 medium (Sigma), 4 % horse serum and 1 % penicillin and streptomycin (Gibco). All experiments were performed with DMEM containing 4.5 g/L glucose (25 mM) unless otherwise specified. DMEM without glucose (Gibco) was used to perform experiments in glucose-depleted conditions. L-( )-lactic acid solution (Sigma) was supplemented to the culture media at a concentration range from 0 to 20 mM according to the experiment. Cell media was replaced every 2–3 days.
Cardiac fibroblasts were obtained from the non-myocytic cell fraction of the digested hearts and cultured in DMEM supplemented with 10% FBS, 1 % L-glutamine and 1 % penicillin and streptomycin. All experiments were performed with DMEM containing 4.5 g/L glucose (25 mM) unless otherwise specified. DMEM without glucose (Gibco) was used to perform experiments in glucose-depleted conditions. L-( )-lactic acid solution (Sigma) was supplemented to the culture media at a concentration range from 0 to 20 mM according to the experiment.
2.2. hiPSCs culture and cardiomyocyte differentiation
Human induced pluripotent stem cell (hiPSCs) lines iPSC-L1 and iPSC-L2 were obtained from the Aguirre Lab (Michigan State University, USA). These two cell lines had been validated for pluripotency and lack of karyotypic abnormalities. hiPSCs were cultured in chemically defined growth media, Essential 8 Flex (Life Technologies), on growth factor-reduced Matrigel® (Corning)-coated plates and routinely passed with ReLeSR (Stem Cell Technologies). For cardiomyocyte differentiation, 60–80 % subconfluent hiPSCs were treated with Accutase (Sigma) and re-plated on a new Matrigel®-coated plate. Cells were differentiated to cardiomyocytes using CHIR99021 (Selleck Chemicals) and Wnt-C59 (Selleck Chemicals) in RPMI 1640 media (Life Technologies) supplemented with B27 (Life Technologies) with or without insulin following previously established protocols [22]. Cardiomyocytes were replated at a cell density of 50–200 103 cells/cm2 and L-lactic acid solution (Sigma) was added to the (glucose-containing) cell culture media at differentiation day 15. Cells were further cultured for 7 additional days.
For MCT1 inhibition, α-Cyano-4-hydroxycinnamic acid (Sigma) was added at a concentration of 1 mM. All cell lines were maintained in an incubator (37 °C and 5 % CO2) with media changes as necessary.
2.3. Ex vivo heart culture
Neonatal mouse hearts were obtained and cultured following an adapted protocol [23]. Briefly, hearts from neonatal mouse were extracted and transferred to PBS containing BDM. The excised hearts were then placed into Matrigel® solution combined 1:1 with cardiomyocyte culture medium. After gelation at 37 °C, pre-warmed culture medium (1 mL) was added to each well containing a heart. Glucose-containing culture media was supplemented with L-lactate at a concentration of 20 mM and half volume of media was changed every 2–3 days for an overall culture time of 9 days. Beating movements of muscle tissue were evaluated daily under an inverted microscope. A cohort of 50 neonatal hearts was used.
2.4. Immunostainings
Cell samples were fixed in 4 % paraformaldehyde (PFA, EMS) and washed in cold PBS containing glycine. They were permeabilized in 0.05 % Triton™ X-100 (Sigma) and blocked in 5 % BSA (Bovine Serum Albumin) or serum for 1 h. Samples were incubated with primary antibodies against Ki67 (Abcam ab16667 or ab15580), Aurora-B kinase (Abcam ab2254), vimentin (Abcam ab24525), alpha smooth muscle actin (Invitrogen PA5-18292) or cardiac Troponin T (cTnT, Abcam ab8295) in 1 % BSA overnight at 4 °C. Secondary antibodies conjugated with AlexaFluor® 488, 594 or 647 were used. Acti-stain™ Fluorescent Phalloidin (Cytoskeleton, Inc.) and DAPI (4′,6-diamidino-2-phenyl-indole, Sigma) were used as actin and nuclear counterstaining respectively. Images were taken using epifluorescence (Leica AF7000 and Olympus CKX53) and confocal microscopy (Zeiss LSM 800). Quantification and analysis were performed using Fiji software [24] and as detailed in Supplementary Material.
Immunohistochemistry of neonatal mice hearts was performed after 9 days of ex vivo culture in the presence of lactate treatment. Hearts were included in paraffin, cut into sections of around 5 μm using a microtome Leica RM 2155 and deparaffinized in xylene and ethanol, followed by antigen retrieval using citrate buffer at 95 °C for 20 min. A 4 % goat serum (Abcam) in Tris-buffered saline solution containing 0.1 % Tween® 20 (TBST) was used as blocking agent. Endogenous peroxidase was quenched with 3 % hydrogen peroxide. Incubation with primary antibody against Ki67 (Abcam) was performed overnight at 4 °C, followed by incubation with biotinylated secondary antibody (Abcam ab64256), streptavidin peroxidase (Abcam ab64269) and DAB Substrate kit (Abcam ab64238). Images were acquired in a Nikon E600 and processed using Fiji software.
2.5. Transmission electron microscopy ultrastructural analysis
After 4 days of culture, mouse neonatal hearts were fixed in 2.5 % glutaraldehyde and 2 % paraformaldehyde in 0.1 M phosphate buffer overnight at 4 °C. They were rinsed in phosphate buffer and post-fixed with 1 % osmium tetroxide (EMS) for 2 h. Samples were then dehydrated in acetone series from 50 to 100 % and infiltrated and embedded in Epon resin (EMS). Ultrathin sections of 60 nm were obtained using a UC6 ultramicrotome (Leica Microsystems) and stained with 2 % uranyl acetate and lead citrate. Sections were observed in a Tecnai™ Spirit (FEI) transmission electron microscope equipped with a LaB6 cathode and images were acquired at 120 kV with a CCD Megaviwe 1kx1k. Images were processed using Fiji software and sarcomere width was manually measured using the “Measure” tool from over 800 sarcomeres from 4 different hearts per group.
2.6. Real-time quantitative PCR
All reagents were obtained from Qiagen. RNA from mouse cells was extracted at days 2, 6, 7, 9, and 10 with lactate treatment using the RNeasy® Plus Mini Kit, and cDNA synthesis was carried out using the RT2 First Strand Kit. The real-time PCR was performed with RT2 SYBR Green Mastermix using commercial RT2 qPCR Primer Assays in a StepOnePlus™ Real-Time PCR system (Applied Biosystems). RNA from human cells was extracted after 3 days of lactate treatment using the RNeasy® Mini Kit and reversed transcribed with the QuantiTect Reverse
Transcription Kit. The QuantiTect SYBR Green PCR Kit was used for the RT-PCR in a QuantStudio™ 5 Real-Time PCR System (Applied Bio-systems), using commercial primers from IDT. The results were analyzed by the ΔΔCt method using 18S ribosomal RNA and Hprt1 as house-keeping genes for mouse and human samples respectively.
2.7. RNA-seq and transcriptomic analysis
RNA was extracted following RNeasy kit manufacturer’s instructions (Qiagen) from hiPSC-derived cardiomyocytes cultured with 0 or 6 mM of L-lactate during 7 days as detailed in Section 2.2. Samples were processed using TruSeq RNA-seq sample prep kit from Illumina. Three independent samples per condition were loaded on an Illumina flowcell and clusters were generated by an Illumina cBot. The clusters were sequenced in an Illumina HiSeq 4000 (Michigan State University’s Genomics Core Facility, Michigan, USA). Read quality was assessed with FASTQC and reads were aligned using HISAT2 and the hg38 genome reference. EdgeR was used for differential gene expression analysis; values were reported as counts per million reads (cpm) or log2 FC. Gene Ontology functional enrichment and clustering calculations were obtained from iPathwayGuide (Advaita Bioinformatics) and DAVID with a fold-enrichment of 1.5.
2.8. Inflammation antibody array
Cardiac fibroblasts were isolated as previously described in Section 2.1. Approximately 500.000 cells were seeded in T75 Nunc™ EasY-Flasks™ and cultured for 6 days in culture media with a progressive reduction of serum content (FBS), from 10 % to 4 %. Cells were then trypsinized and seeded in 12-well plates at a cell density of 30,000 cells/well. After 2 days, cells were incubated in cell culture media containing 1 % FBS and 0 or 20 mM of L-( )-lactic acid. After 24 h, culture media was collected and cells were lysed in M-PER® mammalian protein extraction reagent (Thermo Scientific). Cell media from 4 replicates was pooled as a single sample for analysis using a mouse inflammation antibody array (RayBio®, AAM-INF-1-8). Briefly, antibody array membranes were blocked and incubated with culture media samples overnight at 4 °C. Then, membranes were incubated with biotinylated antibody for 2 h at room temperature, followed by incubation with Streptavidin conjugated to horseradish-peroxidase (HRP) for 2 h. Chemiluminescent detection was performed using an ImageQuant LAS4000 mini Biomolecular Imager (GE Healthcare Life Sciences). Intensity quantification was performed using Fiji software and the obtained values were corrected by culture media background (without having been in contact with cells) and total protein content measured using the Pierce™ BCA Protein Assay Kit (Thermo Scientific). Values were further normalized by internal negative and positive controls from the antibody assay, as 0 and 100 expression values respectively. The experiment was repeated twice with independent non-myocytic populations coming from different mouse hearts isolations and results are shown as mean ± SD of these two independent experiments.
2.9. Collagen quantification
Quantification of total collagen was performed on cardiac fibroblasts after 3, 6 and 10 days of culture with 0 or 20 mM of lactate supplemented in culture media. Briefly, cells were collected in M-PER Mammalian Protein Extraction Reagent (Thermo Scientific) and hydrolyzed in 6 M Hydrochloric acid (HCl) for 24 h at 110 °C. After hydrolysis, samples were dried and reconstituted in distilled water. They were then diluted in isopropanol and mixed with a Chloramine-T hydrate (Sigma) solution as oxidant agent and a 4-(Dimethylamino)benzaldehyde (Sigma) solution with perchloric acid as a colour reagent. The colorimetric detection (absorbance) of hydroxyproline content was measured using an Infinite M200 PRO Multimode Microplate (Tecan).
2.10. Wound scratch assay
Cardiac fibroblasts were cultured at an initial density of 250.000 cells per well in 12-well plates for 7 days until confluence. A scratch of around 900 μm was then made using a sterile p200 micropipette tip.
Cells were washed in PBS and incubated in DMEM media containing 5 % FBS and supplemented with 0 or 20 mM of L-( )-lactic acid solution. Four images per well were taken along the wound using a Nikon TE200 microscope after the scratching and after 24 h of incubation. Recovered area was calculated by measuring the scratched area of the same region before and after the incubation period using Fiji software.
2.11. Live/dead assay
Lactate toxicity was evaluated on primary mouse cardiomyocytes using calcein-AM (Sigma) and Propidium iodide (Fluka) for a live/dead assay. After culture, cells were washed in DPBS (Gibco) and incubated in a solution of 2 μM calcein and 4 μM propidium iodide in DPBS (Dulbecco’s phosphate-buffered saline) for 20 min at 37 °C. Cells were then washed in DPBS and immediately imaged using a Leica DMIRBE epifluorescence microscope.
2.12. LDH detection
Lactate dehydrogenase (LDH) detection was performed following Cytotoxicity Detection KitPLUS manufacturer’s instructions (Roche).
Cardiomyocytes were seeded in a 48-well plate (Nunc) at a cell density of 60,000 cells/well. After 24 h of incubation for cell attachment, media was supplemented with 20 mM of L-( )-lactic acid solution (Sigma). After 1, 3, 5, 7, 9, 12 and 15 days of culture, cell media was collected and stored at 20 °C until analysis, and new cell culture media supplemented with lactate was added to cells. At the end of the experiment, cells were lysed for 15 min and collected for maximum LDH release determination (100 %). Thawed cell culture media was pipetted into a 96-well plate in duplicates and mixed with kit’s reaction mixture. After 30 mins of incubation, the reaction was stopped and the absorbance was measured using a TECAN Infinite M200 Pro microplate reader set at 492 nm with wavelength correction at 680 nm. Culture media incubated without cells was used as a background control.
2.13. Cell viability
Cell viability was evaluated on hiPSC-derived cardiomyocytes (hiPSC-CM) using a colorimetric MTS Cell Proliferation Assay Kit (Abcam). Briefly, hiPSC were differentiated to cardiomyocytes in a 48-well plate and cells were incubated in a 10 % MTS solution in culture media for 2 h (day 0). Media was then transferred in duplicates to a 96-well plate for analysis of the absorbance at 490 nm in a microplate reader. Cells were washed in cell media and replaced by fresh RPMI/B27 (with insulin) media supplemented with 0, 5, 10, 15 or 20 mM of L-lactate solution. MTS incubation was repeated after 1, 3 and 6 days of culture. The MTS solution in cell culture media with different concentrations of lactate was used as a control for background signal. Measured values from each timepoint were corrected by values from day 0. Three biological replicates (wells) were used for each measurement and condition.
2.14. BMP10 ELISA
Neonatal mouse cardiomyocytes were seeded in 24-well plates at a cell density of 200.000 cells/well. After 24 h of incubation, cell media was supplemented with 0 or 20 mM of L-( )-lactic acid solution (Sigma). After 2, 6 and 10 days of culture, cells were washed in PBS and collected in M-PER® mammalian protein extraction reagent (Thermo Scientific) in triplicates. The presence of BMP10 (Bone Morphogenetic Protein-10) on cell lysate was analyzed using a DuoSet® ELISA Development System (R&D Systems, bio-techne). Briefly, a 96-well plate was incubated overnight with a mouse anti-mouse BMP10 antibody and blocked with a 1 % BSA solution in PBS for 1 h at room temperature. Samples were then loaded into the plate and incubated for 2 h, following detection with a biotinylated mouse anti-mouse BMP10 antibody for 2 more hours. HRP-Streptavidin was added and incubated for 20 min avoiding direct light contact. The reaction was stopped with H2SO4 and the optical density was determined using a microplate reader (Infinite M200 Pro, TECAN) set to 450 nm with wavelength correction at 540 nm. An 8 point standard curve was used for analysis. BMP10 data were corrected to total cell protein quantified following Pierce™ BCA Protein Assay Kit (Thermo Scientific) instructions.
2.15. Statistical analysis
Data are presented as mean and error bars represent standard deviation (SD). At least three independent cell cultures (n) were used for data analysis. GraphPad Prism 8.3 was used as analysis and graphical software. Student’s t-test (two-tailed distribution) was generally used to compare two samples, and one-way ANOVA followed by post-hoc Tukey’s test was used for multiple samples, unless otherwise specified. A p-value <0.05 was considered statistically significant.
Results
3.1. Effect of lactate on neonatal mouse cardiomyocyte cell cycle progression
To evaluate the effect of lactate on neonatal mouse cardiomyocytes, we added different concentrations of L-lactate, from 0 to 20 mM, to the cell culture media (which contains approx. 20 mM of D-glucose) and cells were evaluated periodically. These concentrations of lactate are actually within the range of concentrations found in blood circulation after intense exercise [25]. Higher concentrations were not considered because of excessive media acidification (data not shown). After 3 days of culture, the evaluation of cardiac cell viability did not reveal any detrimental effect of lactate, yet even suggested a positive effect of this metabolite on mouse cardiomyocytes (Sup. Fig. 1A). A live/death assay performed after 7 days of culture also showed no effect on cardiomyocyte viability when incubated with up to 20 mM of lactate (Figs. 1A and Sup. Fig. 1B). This lack of toxicity was further confirmed by the detection of LDH (Lactate dehydrogenase) in cell media, which revealed that the incubation of cardiac cells with 20 mM of lactate for up to 15 days did not affect cardiac cell death, as compared to the control (Fig. 1B). With these results, we decided to use a 20 mM concentration of L-lactate for the following experiments on neonatal mouse cardiac cells. The number of cardiomyocytes (cardiac Troponin T or cTnT+ cells) expressing the proliferation marker Ki67 was then evaluated (Fig. 1C).
Fig. 1. Effect of lactate on mouse cardiomyocyte cell cycle activity.
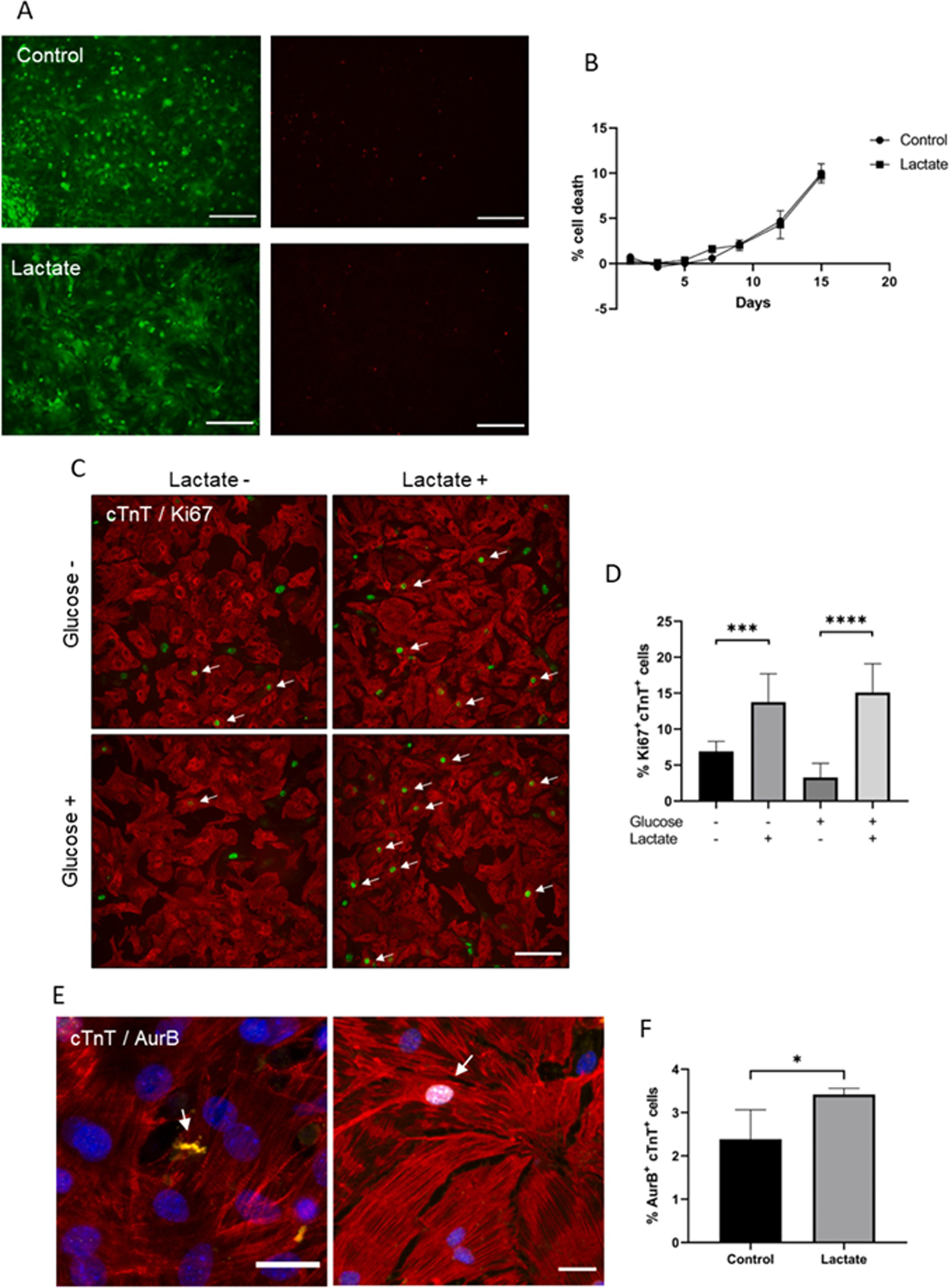
(A) Live/dead assay of cardiomyocytes after 7 days of incubation with 0 or 20 mM of lactate. Green (left) and red (right) channels showing alive and death cells respectively; scale bar: 200 μm. (B) LDH detection on culture media of cardiomyocytes incubated with 0 or 20 mM of lactate. Data are expressed as % of cell death compared to maximum LDH released by lysed cells (100 %). n = 3. (C) Ki67 immunostaining on cardiac cells after 3 days of culture with (+) or without (−) lactate. In this case, glucose-containing (Glucose +) and glucose-depleted (Glucose -) cell culture media was used. Red: cardiac Troponin T (cTnT), green: Ki67. Only Ki67+cTnT+ cells (white arrows) were quantified; scale bar: 100 μm. (D) Quantification of Ki67+cTnT+ cells after 3 days of culture with (+) or without (−) lactate in glucose-containing (Glucose +) or glucose-depleted (Glucose -) cell culture media. Data are represented as % of Ki67+cTnT+ cells to the total number of cTnT+ cells. Results are from 3 independent cell cultures and around 1200 cardiomyocytes were analyzed for each group. One-way ANOVA, ***p < 0.001, ****p < 0.0001. (E) Aurora-B kinase immunostaining on mouse cardiomyocytes. White arrows indicate AurB+cTnT+ cells. Yellow: AurB, red: cTnT, blue: nuclei; scale bar: 25 μm. (F) Quantification of AurB-positive cardiomyocytes after 10 days of culture with 20 mM of lactate supplemented into glucose-containing cell media. Data are represented as % of AurB+cTnT+ to the total number of cTnT+ cells. Results are from 4 independent cell cultures and around 4000 cardiomyocytes were analyzed for each group. Student’s t-test, *p < 0.05.
As glycolysis is the major energy source for proliferating cardiomyocytes [14], we tested whether glucose availability could influence the effect of lactate on cardiomyocyte cell cycle activity by adding 20 mM of L-lactate to glucose-containing cell culture media (approx. 20 mM, Glucose ) or glucose-depleted media (<1 mM, Glucose-). As observed in Fig. 1D, lactate significantly increased the number of Ki67+cTnT+ cells after 3 days of culture, an effect that was not affected by the presence or absence of glucose. For this reason, the following experiments of the present work were always performed using glucose-containing cell culture media as detailed in Materials and Methods section.
Interestingly, after 7 days of culture, the increase in the number of Ki67+ cells observed with lactate could not be detected if this metabolite was removed from culture media after 3 days (Sup. Fig. 1C), suggesting that lactate could be continuously required for stimulating cell cycle activity. The expression of Aurora-B kinase (ARK-2 or AurB), a key regulator for cytokinesis during mitosis, was also evaluated through immunostaining (Fig. 1E). Consistently, the number of AurB+cTnT+ cells was significantly increased in the presence of lactate (Fig. 1F). Interestingly, many cardiomyocytes showing disorganized sarcomeres (Sup. Fig. 1D and E) were also observed in the presence of lactate (approximately 5.5 ‰ cardiomyocytes, in contrast to only 1 ‰ found on control cells), a phenotype typically associated to dedifferentiated cardiomyocytes.
3.2. Gene expression of cardiomyocytes exposed to lactate
We investigated changes in gene expression on mouse cardiomyocytes exposed to 20 mM of L-lactate (Fig. 2A). Some of the genes analyzed were actually selected according to RNA-sequencing data from hiPSC-cardiomyocytes (Fig. 5C). Interestingly, we found that the continued exposure of mouse cardiomyocytes to lactate promoted an increasing expression of bone morphogenetic protein-10 (Bmp10) and transcription factor P63, reaching over a 4 times-fold expression after 10 days of culture compared to control without lactate. Additionally, the expression of Tcim, a positive regulator of the Wnt/β-catenin pathway associated to a poor differentiation state [26], was also increased in the presence of lactate (Fig. 2A). In a lower extent, lactate also stimulated the expression of Gata4 and c-kit, typically associated to cardiac immature cells. Furthermore, we also observed a reduction in the expression of Grik1, which belongs to the family of glutamate receptors [27] and has been recently suggested to inhibit cell proliferation [28]; and Dgkk, involved in diacylglycerol signaling, important for cardiac hypertrophy responses [29]. The effect of lactate was not only observed at the transcriptional level, but we also detected higher protein levels of BMP10 on neonatal mouse cardiomyocytes exposed to L-lactate (Fig. 2B).
Fig. 2. Gene expression of mouse cardiomyocytes exposed to lactate.
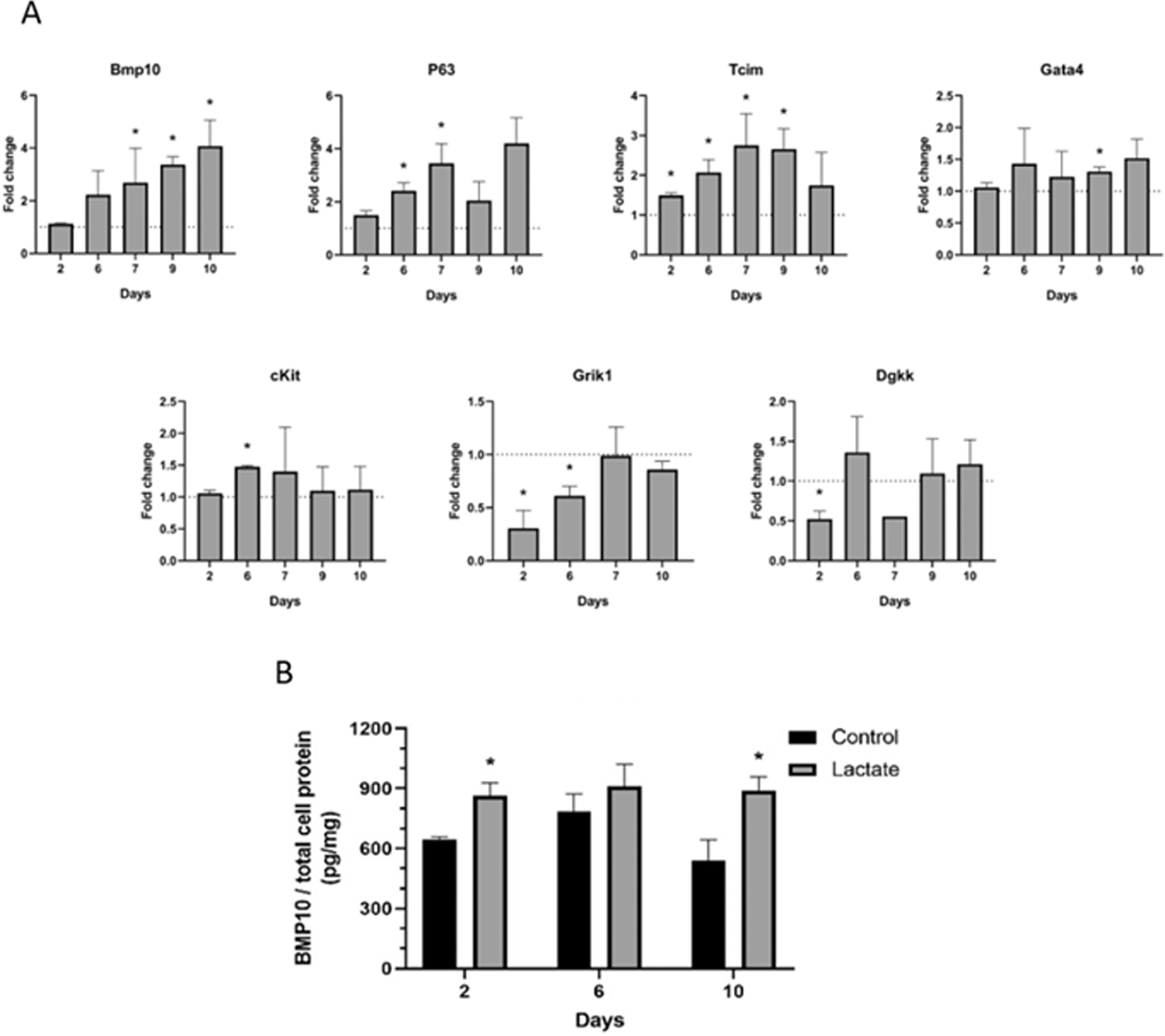
(A) Relative gene expression on cardiomyocytes cultured with 20 mM lactate as compared to the gene expression levels in the absence of lactate (dashed line). Results are from 1 to 4 independent experiments performed on different days. Data were analyzed using Student’s t-test compared to control without lactate for each timepoint, *p < 0.05. (B) BMP10 protein quantification from cell lysate at different timepoints. Student’s t-test compared to control without lactate of the same timepoint, *p < 0.05; n = 3. All experiments were performed in glucose-containing cell culture media.
Fig. 5. Gene expression and transcriptome analysis of hiPSC-CM.
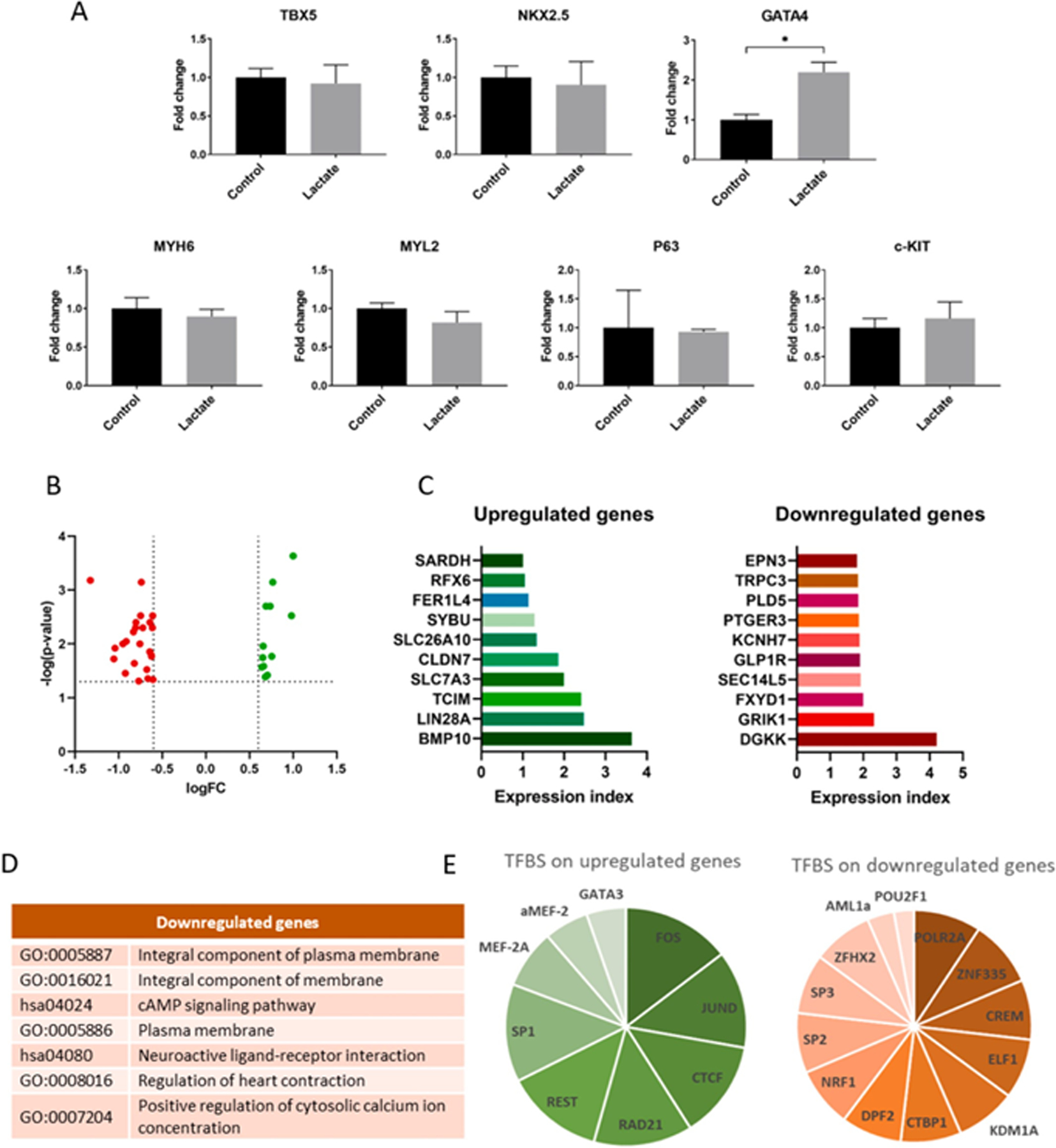
(A) Relative gene expression of cardiomyocytes cultured with 0 or 6 mM of lactate for 3 days. Results are from 3 independent cell cultures and data were analyzed using a student’s t-test, *p < 0.05. (B) Volcano plot showing differentially expressed (DE) genes from transcriptome sequencing from cardiomyocytes cultured with 0 or 6 mM of lactate during 7 days. Dashed lines represent DE thresholds used (0.6 log fold change cutoff and 0.05 p-value). Green: upregulated genes; red: downregulated genes; FC: fold change. (C) Top 10 up- and downregulated DE genes according to the expression index, the product of log(p-value) and logFC. (D) Gene ontologies (GO) and KEGG pathways associated to top downregulated genes (DAVID database, p < 0.05). (E) Summary of common transcription factors binding sites (TFBS) found on upregulated and downregulated genes, according to the number of genes in which the TFBS is found. All upregulated DE genes were used for the analysis, while only top downregulated genes were used. Data obtained from GeneCards® database. See also Sup.Tables 3 and 4.
Changes in cardiomyocyte gene expression observed in the presence of lactate were further confirmed by higher levels of cell division cycle genes (Cdc7, Cdc25c or Cdc20) and genes associated to cancer and dedifferentiation (P63, Ddit3, Mdm2, Sfn, Brca2 or Ppm1d), together with a downregulation of some tumor suppressor genes and genes involved in cell cycle arrest (Chek2, Cdkn2a or Atr) (Sup. Fig. 2).
3.3. Effect of lactate on cardiac fibroblast phenotype
Next, we wondered whether lactate could also affect other cardiac populations. Cardiac fibroblasts are the key drivers of cardiac fibrosis and tissue repair after cardiac injury, and their excessive stimulation is known to be detrimental [30]. Therefore, we studied the effect of lactate on mouse cardiac fibroblasts isolated as detailed in Materials and Methods section. These cells showed to be vimentin+, being about half of them also positive for alpha smooth muscle actin (α-sma), which is consistent with a myofibroblast phenotype (Sup. Fig. 3A and B). The number of Ki67+ cells was evaluated with lactate and/or glucose as described in Materials and Methods section (Fig. 3A). EdU (5-ethynyl-2′-deoxyuridine) incorporation was also assessed after incubation with different concentrations of lactate (Sup. Fig. 3C and D). Contrary to cardiomyocytes, lactate showed no effect on the number of proliferating fibroblasts. No significant differences were neither observed on collagen production after 3, 6 or 10 days of incubation with lactate (Fig. 3B) nor in their migration activity as compared to control (Fig. 3C). The fraction of myofibroblasts present in the culture was assessed by flow cytometry (Sup. Fig. 3E), indicating no significant difference in the number of vimentin+ and α-sma+ cells in the presence of lactate.
Fig. 3. Effect of lactate on cardiac fibroblasts.
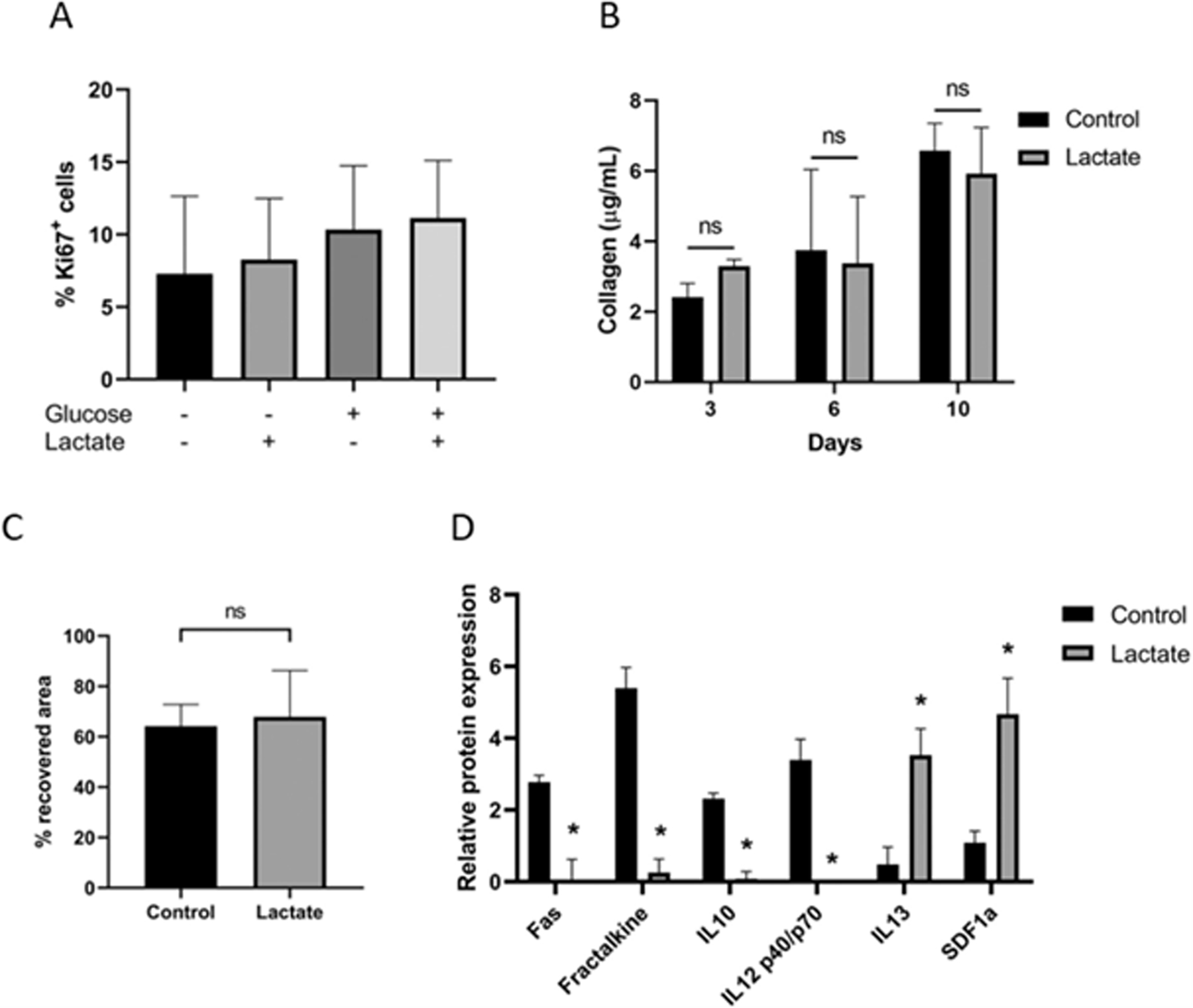
(A) Quantification of Ki67-positive cells after 3 days of culture with (+) or without (−) lactate in glucose-containing (+) and glucose-depleted (−) cell culture media. Data were analyzed using a one-way ANOVA and represented as % of Ki67+ cells to the total number of cells. Results are from 3 independent cell cultures and at least 214 Ki67+ cells were counted per group. (B) Quantification of total collagen content on cardiac fibroblasts after 3, 6 and 10 days of culture with 0 or 20 mM of lactate. Data were analyzed with a two-way ANOVA, ns: not significant; n = 2. (C) Wound closure of cells cultured with 0 or 20 mM of lactate. Data were measured as % of recovered area after 24 h of scratching and analyzed with a Student’s t-test. Results are from 8 different regions from 2 independent cell cultures. (D) Quantification of inflammatory cytokines (only significant differences are shown, see also Sup. Table 1). Signal expression of each sample was calibrated according to total protein. Internal positive (100) and negative controls (0) were used to generate numerical values of each sample. Results are from 2 independent experiments performed on different days. Student’s t-test of lactate compared to control condition, *p < 0.05.
In addition, the expression of different inflammatory cytokines was evaluated upon exposure to lactate (Sup. Table 1). Interestingly, lactate significantly reduced the expression of Fas, Fraktalkine, IL-10 and IL-12p40/p70, known to be prejudicial for cardiac recovery after injury [31,32], while increased the expression of IL-13 and SDF1a, known to improve healing and remodeling after a myocardial infarction [33,34] (Fig. 3D).
3.4. Cell cycle activity of human iPSC-derived cardiomyocytes exposed to lactate
Since our results showed that lactate stimulated mouse cardiomyocyte cell cycle progression, we further explored the effect of this metabolite on human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CM). First, we tested the viability of cells at different concentrations of lactate, from 0 to 20 mM, using an MTS assay (Fig. 4A). Lactate concentrations above 10 mM demonstrated to reduce hiPSC-CM viability, showing up tolerance differences between mouse and human cells. For this reason, a lower range of concentrations up to 6 mM was used to evaluate human cardiomyocyte proliferation. Our results showed a concentration-dependent effect of lactate on the number of Ki67+cTnT+ cells after only 24 h of incubation (Fig. 4B and C), with slightly higher numbers (p-value 0.077) of cardiomyocytes in karyokinesis (Sup. Fig. 4A and B). After 4 days of culture, the positive effect of lactate on Ki67 expression was still observed (Fig. 4D).
Fig. 4. Effect of lactate on hiPSC-cardiomyocyte proliferation.
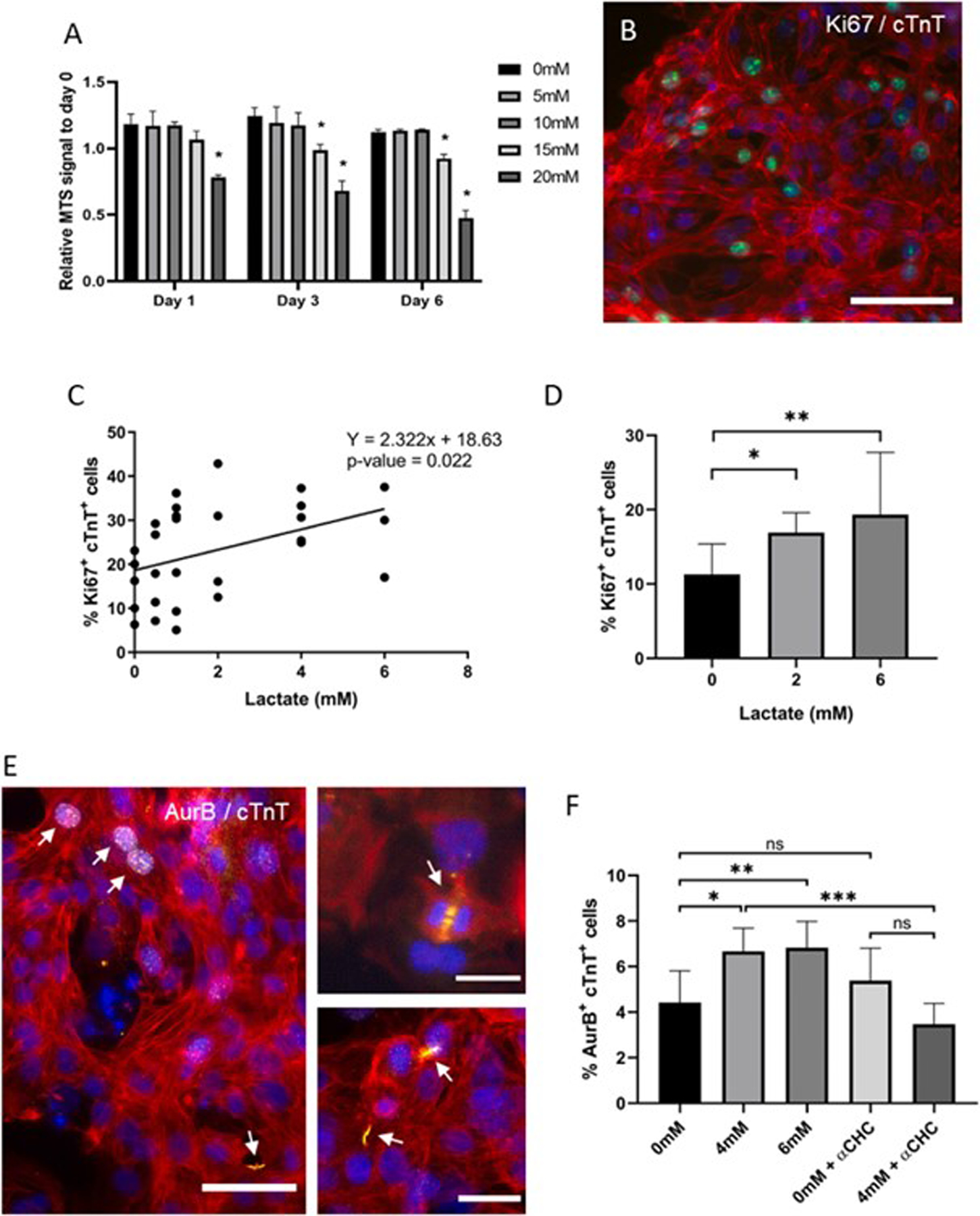
(A) Cell viability in the presence of different concentrations of lactate. MTS signal intensity was corrected to values from day 0. Data were analyzed using a two-way ANOVA, *p < 0.01, n = 3. (B) Immunofluorescent staining of Ki67 on hiPSC-CM. Green: Ki67, blue: nuclei, red: cTnT; scale bar: 150 μm. (C) Quantification of Ki67+cTnT+ cells following 24 h of culture with different concentrations of lactate. Data are represented as % of Ki67+cTnT+ to the total number of cTnT+ cells and was analyzed using simple linear regression. Results are from at least 3 independent cell cultures and an average of 1500 cardiomyocytes were analyzed for each condition. (D) Quantification of Ki67+cTnT+ cells following 4 days of culture with different concentrations of lactate. Data are represented as % of Ki67+cTnT+ to the total number of cTnT+ cells and was analyzed using a one-way ANOVA, *p < 0.05, **p < 0.01. Results are from at least 3 independent cell cultures and at least 582 cardiomyocytes were counted per group. (E) Aurora-B immunostaining on hiPSC-cardiomyocytes. White arrows indicate AurB+cTnT+ cells. Red: cardiac Troponin T (cTnT), yellow: Aurora-B, blue: nuclei; scale bars: 50 μm (left) and 25 μm (top and bottom right). (F) Quantification of AurB+cTnT+ cells after 7 days of incubation with lactate at 0, 4 or 6 mM concentration. 1 mM of α-cyano-4-hydroxycinnamic acid (αCHC) was used as inhibitor of MCT1. Data are represented as % of AurB+cTnT+ to the total number of cTnT+ cells and it was analyzed using a one-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001. Results are from at least 6 independent cell cultures and an average of 4400 cardiomyocytes were counted per group.
The number of AurB+cTnT+ cells (Fig. 4E), even though not significantly affected after only 2 days of culture (Sup. Fig. 4C), was increased after 7 days of incubation with 4 and 6 mM of lactate (Fig. 4F). In addition, we blocked lactate uptake into the cell using 1 mM of α-Cyano-4-hydroxycinnamic acid (αCHC), a reversible inhibitor of the monocarboxylate transporter MCT1, the primary lactate transporter [35]. As shown in Fig. 4F, the addition of αCHC inhibited the increase in the number of AurB+ cardiomyocytes resulting from lactate supplementation.
3.5. Gene expression and transcriptomic analysis on human iPSC-derived cardiomyocytes
Gene expression analysis of hiPSC-CM exposed to 6 mM of L-lactate revealed that GATA4 was the main cardiomyogenic transcription factor affected by lactate supplementation (Fig. 5A). Consistently, GATA4 is required for cardiomyocyte proliferation and the expression of cell cycle regulators [36], and it is a primary contributor to zebrafish heart regeneration [37]. Other cardiac markers, such as myosin heavy chain α (MYH6) and myosin light chain 2 (MYL2), predominantly expressed in cardiac atria and ventricle respectively, were not affected by lactate. Surprisingly, P63 or c-KIT were also not affected on hiPSC-CM exposed to lactate.
To provide further insights into gene expression changes, hiPSC-CM were incubated with L-lactate and analyzed via RNA sequencing. Transcriptomic analysis revealed a small number of differentially expressed (DE) genes (36 genes from 12,388 genes with measured expression, using a log fold change of expression with absolute value of at least 0.6 and a threshold of 0.05 for statistical significance): 12 upregulated and 24 downregulated genes (Sup. Table 2 and Fig. 5B). This suggested a remarkably specific impact of lactate and the activation of a targeted molecular pathway. To simplify the analysis, an expression index was calculated for each gene as a result of the product of log [Fold change] and log [p-value]. A list of the top 10 upregulated and downregulated genes by expression index is shown in Fig. 5C. Genes associated to stemness and proliferation could be identified among the upregulated genes, specially BMP10, LIN28A, TCIM and CLDN7. Regarding the downregulated genes, we confirmed the repression of GRIK1 and DGKK as the principal downregulated genes, but we could also identify genes encoding for potassium (KCNH7) and calcium (TRPC3) ion channels, typically associated to mature and contractile cardiomyocytes. Gene ontologies (GO) and KEGG pathways associated to the most relevant downregulated genes (Fig. 5D) included “cAMP signaling pathway”, “neuroactive ligand-receptor interaction”, “regulation of heart contraction” or “positive regulation of cytosolic calcium ion concentration” (DAVID database, p < 0.05). No common GO were found for the upregulated genes.
With the aim to shed some light into the regulatory mechanism by which lactate was exerting its effect on cardiomyocytes, we used the human gene database GeneCards® to identify the transcription factor binding sites (TFBS) associated to the DE genes. Sup. Tables 3 and 4 show the top TFBS that are recurrently found on the upregulated (Sup. Table 3) or downregulated (Sup. Table 4) DE genes. Although upregulated and downregulated DE genes were evaluated separately, they were both included in each table, as some TFBS were found on both gene clusters. A summary of the common TFBS is shown in Fig. 5E according to the number of up- or downregulated genes in which the TFBS was found. Interestingly, most of these transcription factors were related to hypoxia pathways [38]. Some of them have already demonstrated to play important roles during cardiac hypoxia, such as SP1 [39], CREM [40], CREB1 [41] or MEF2A [42], involved in cardioprotection or cardiac stem cell proliferation under low-oxygen conditions.
3.6. Ex vivo heart culture
Despite in vitro culture of cardiomyocytes is a versatile technique to study cardiac cells, it is an artificial environment far from the native conditions of the heart. Different ex vivo heart culture systems can be found in the literature to maintain the three-dimensional morphology of the heart while allowing contraction and cross-talking between different regions and cardiac cell types. These culture systems, however, present some limitations. Cardiac tissue can only be maintained without significant necrosis for a few days and epicardial cells rapidly spread and attach to the surrounding matrix [23]. Given the effect of lactate on two-dimensional culture of cardiac cells, we decided to test whether the presence of a lactate-rich environment may also have any effect on ex vivo culture of three-dimensional native cardiac tissue (Fig. 6A).
Fig. 6. Ex vivo heart culture.
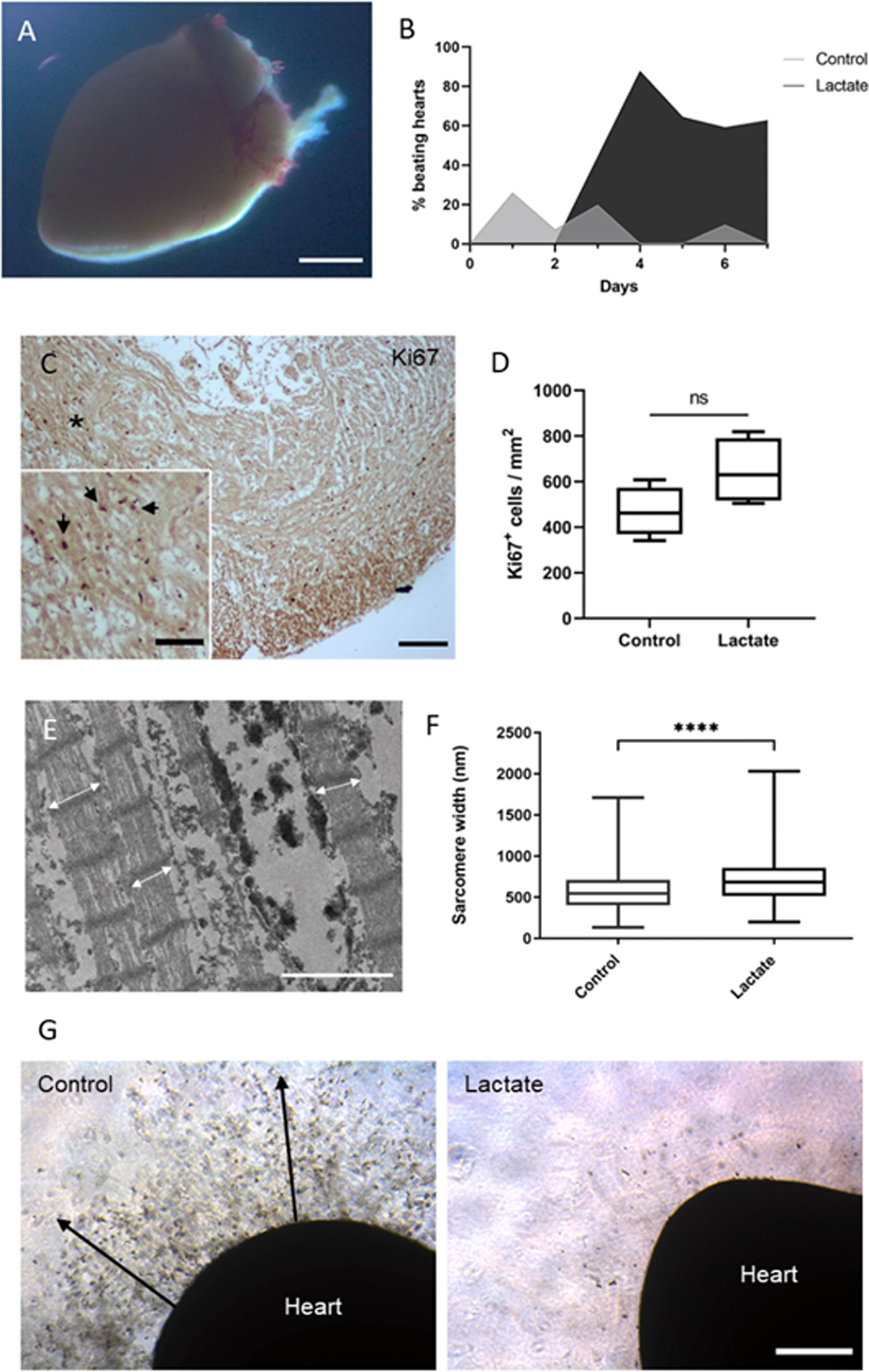
(A) Stereomicroscope image of a cultured neonatal mouse heart embedded in Matrigel® matrix. Scale bar: 1 mm. (B) Fraction of ex vivo hearts showing spontaneous contraction (beating) activity at any region of the organ. 50 neonatal mouse hearts were evaluated. (C) Immunohistochemistry staining of Ki67 on a cross-section of a neonatal mouse heart cultured ex vivo for 9 days. Black arrows indicate positive Ki67 nuclei on a higher magnification image of the (*) region. Scale bar: 100 μm and 50 μm (magnification). (D) Quantification of Ki67-positive cells on tissue sections from ex vivo hearts incubated with 0 or 20 mM of lactate for 9 days. Data were analyzed using a student’s t-test, ns: non-significant, p = 0.097, n = 4 hearts. (E) Transmission electron microscopy image of a cardiac tissue section from an ex vivo cultured heart after 4 days. White arrows indicate sarcomere width. Scale bar: 2 μm. (F) Quantification of sarcomere width from ex vivo hearts incubated with 0 or 20 mM of lactate for 4 days. Data were analyzed with a student’s t-test with Welch’s correction, ****p < 0.0001. n = 4 hearts, and over 800 sarcomeres were measured per group. (G) Optical microscopy images of ex vivo hearts cultured with 0 or 20 mM of lactate for 9 days. Heart cells spread to the surrounding matrix on control hearts (black arrows). Scale bar: 400 μm.
Interestingly, some regions of the ex vivo heart tissue still showed spontaneous beating activity during in vitro culture (Sup. Video 1). Around 20 % of hearts in culture media without lactate (control) exhibited some spontaneous contractions during the first 2 days, while none of the hearts cultured with lactate showed beating activity (Fig. 6B). The fraction of control beating hearts decreased over time, as expected from this type of ex vivo culture. However, mouse hearts supplemented with lactate showed increasing contractile abilities after 3 days of culture, with almost 90 % of these hearts exhibiting beating activity at day 4 and still 60 % after 7 days. As determined by histological analysis, cardiac cells from hearts incubated with lactate showed an apparent higher expression (p-value 0.09) of the proliferation marker Ki67 as compared to control hearts (Fig. 6C and D) and presented significantly wider sarcomeres (Fig. 6E and F), likely caused by a lower tissue necrosis and disorganization of the structures. As expected, epicardial cells from control hearts clearly colonized the surrounding matrix after 9 days of culture (Fig. 6G). Interestingly, this event was not observed on lactate-supplemented ex vivo hearts.
Discussion
The control of the metabolic environment is arising as an interesting approach for in situ tissue regeneration strategies and the development of new biomaterials [13], as the metabolic state of a cell is shown to be a key modulator of important biological processes, specially linked to pluripotency and stem cell differentiation [12,17]. In the last years, some studies showed the huge potential of metabolic control on the in vitro culture of cardiac cells [20,43,44], but little to no studies have proposed an effective strategy for in situ control of the cardiac metabolic microenvironment for the activation of endogenous regenerative programs. Lactate, a typical glycolytic metabolite, has also arisen as an important molecule for different cellular processes, such as angiogenesis, wound healing or neurogenesis [45,46]. In this context, our study demonstrates a novel use of lactate on mammalian cardiac tissue engineering, promoting cell cycle progression and modulating the expression of different genes involved in development and maturation of cardiac cells. Apparently, this effect is restricted to cardiomyocytes, as neither cardiac fibroblasts (as shown in this work) nor undifferentiated stem cells [47] respond to the presence of lactate in a similar way. Tohyama et al. already showed distinct metabolic differences between cardiomyocytes and other proliferating cells, indicating the preferential use of lactate in cardiomyocyte metabolism [43]. Their study became the basis for a widely used approach to obtain enriched cardiomyocyte populations from human pluripotent stem cells. However, their approach is based on the elimination of stem cells in glucose-free conditions. In this work we have demonstrated that indeed, lactate promotes changes in cardiomyocyte phenotype, independently of glucose availability. Our results also suggest that this process requires the uptake of lactate through MCT1, as also shown in the work of Tohyama et al. The ability of lactate to reprogram cardiomyocytes may thus have been masked until now by the more evident effect of glucose depletion on non-cardiomyocyte cells. This study has thus uncovered a crucial link between metabolism and maturation of mammalian cardiomyocytes.
We showed that lactate activates a specific genetic program associated to immature cells. This reprogramming is driven by the expression of LIN28, BMP10 and TCIM among others, along with the downregulation of DGKK, GRIK1 and different ion channels. Importantly, Lin28 is a primal regulator of growth and metabolism in stem cells [10], while it also protects cardiomyocytes against apoptosis and regulates cardiac progenitor activation [48,49]. We have also shown upregulation of BMP10, not only transcriptionally but also at a protein level. Bmp10 is a secreted protein that induces cardiomyocyte proliferation and prevents maturation of cardiac cells [50,51]. Thus, the production of Bmp10 may help to prevent maturation and stimulate proliferation on cardiomyocytes in the presence of lactate.
While most of the regenerative approaches for cardiac tissue are focused on cardiomyocytes, cardiac fibroblasts are, in fact, the predominant cell type present in a myocardial infarction and they must be highly considered for the design of new therapies. Our results demonstrated that cardiac fibroblasts exposed to lactate modulate the production of inflammatory cytokines, reducing the production of cytokines involved in hypertrophy [31] or heart failure [32] while stimulating the production of cytokines beneficial for cardiac healing and remodeling [33,34]. Therefore, the synergistic response of cardiomyocytes and cardiac fibroblasts to the presence of lactate could potentially improve tissue regeneration and help the development of novel metabolic therapies for cardiovascular diseases. In this context, we have preliminary shown that the presence of lactate can improve the ex vivo heart tissue culture by overcoming some of its major limitations, such as epicardial cell spreading or the short viability of the tissue [23]. The presence of lactate has shown to maintain the contractile activity and sarcomeric integrity of ex vivo cardiac tissue for over a week of culture in the presence of lactate. Nevertheless, this was a preliminary study on the effect of lactate on ex vivo tissue, to show the potential application of lactate in cardiac tissue culture and developmental studies, but more data would be needed to understand some of the events observed here. Nevertheless, the mechanisms of action of lactate are yet unclear.
Some studies support the idea that lactate is directly affecting epigenetic modulation by affecting histone acetylation [52] or even by the direct lactylation of histones [53]. In this work, we suggest a common relationship between the observed changes in gene expression and important regulators of the hypoxic response, a hypothesis that is supported by the work of others [54,55]. It has been demonstrated that hypoxia is a key regulator of cardiomyocyte dedifferentiation and proliferation [44,56–58]. Taken together, lactate may be a crucial intermediate of the cardiac response to hypoxia, as it is accumulated due to HIF1-mediated increase in glycolysis [55]. In the same way, lactate may explain the enhancement in adult cardiomyocyte proliferation caused by exercise [59] and observed after ischemic injury [60]. In these two events, an increase in cardiomyocyte lactate uptake has been reported [61,62].
Even though some differences have been identified on the effect of lactate between mouse and human cells (such as different concentration tolerances or the expression of P63), both cell types showed a similar response to lactate, supporting the activation of a conserved pathway. In addition, although neonatal and iPSC-CM differ from adult cardiomyocytes in their regenerative abilities [63,64], different studies have shown that endogenous regenerative programs of the heart can be reactivated [3,65]. The activation of Lin28 [11,49] or hypoxia-induced responses [57,58] have already shown to enhance tissue repair in adults. Our work supports that the sole addition of L-lactate into cell environment is capable of reactivating similar regenerative mechanisms on cardiomyocytes, avoiding the use of complex recombinant and transfection methods, the manipulation of genetically modified cells or the use of interfering RNAs, which can, in some cases, produce uncontrolled effects in an in vivo environment [66]. This offers a simple and easy approach for an effective control of cardiomyocyte phenotype based on the manipulation of the metabolic environment, which can be, for instance, achieved through the use of smart biomaterials [67]. The use of lactate-releasing scaffolds has already proven to be useful in many applications other than cardiac repair, such as angiogenesis, wound healing, cartilage repair or neurogenesis among others [46,68–70]. Most of these strategies are based on poly (lactic-co-glycolic acid) (PLGA) or poly-l-lactate acid (PLLA) materials, which have typically been recognized as being detrimental to cell function because of their acid degradation products. The recent studies on the biological effects of lactate, including this work, offer new implications on the potential use of these materials for the future design of scaffolds. We are, however, aware of the limitations of our study regarding the use of not-fully mature cardiomyocytes, and studies on adult cardiac tissue are ongoing in our laboratory. Despite these limitations, we consider that the findings in this work are very promising as a novel tool to change the heart microenvironment toward a pro-regenerative milieu. The challenge is now to develop smart platforms that can properly recreate a regenerative ecosystem in adult cardiac tissue in terms of mechanical properties, degradation rate and lactate-releasing abilities among others.
Conclusions
The metabolic environment arises as an important modulator of the phenotype of cardiac cells, able to stimulate cell cycle progression and modulate the expression of key developmental genes. The use of lactate can be consolidated as a powerful tool to control cardiomyocyte fate, opening new possibilities for the potential use of this molecule on cardiac tissue engineering with important implications for advanced biomaterial design and the treatment of cardiovascular diseases.
Supplementary Material
Acknowledgements
We would like to thank Lourdes Sánchez-Cid and Sara Calatayud for animal care. Epifluorescence microscopy and image analysis was possible thanks to the Molecular Imaging Platform (MIP IBMB-PCB) and Dr. Elena Rebollo. We thank CCiTUB for the help with sample preparation and TEM imaging, as well as for flow cytometry analysis; the Histopathology Facility (IRB) for heart tissue sample preparation; and MSU Genomics Core for sequencing services. We also wish to thank all members of the Engel and Aguirre labs for valuable comments and advice. The content of this manuscript has been submitted to the preprint server bioRxiv [71].
Funding
This work has been funded with the support of the European Regional Development Fund (FEDER) and Spanish Ministry of Science and Innovation (MICINN) with the Projects MICINN BES-2015-071997, MAT2015-62725-ERC, RTI2018-096320-B-C21, the European Commission-Euronanomed nAngioderm Project (JTC2018-103) and the Spanish network of cell therapy (TERCEL). Researchers also thank Programme/Generalitat de Catalunya (2017-SGR-359) and the Severo Ochoa Programme of the Spanish Ministry of Science and Innovation (MICINN—Grant SEV-2014-0425, 2015-2019 and CEX2018-000789-S, 2019-2023). Research in the Aguirre Laboratory was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under award number HL135464 and the American Heart Association under award number 19IPLOI34660342. J.O. was additionally supported by Daniel Bravo Andreu Private Foundation and CIBER.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioadv.2022.213035.
CRediT authorship contribution statement
Jesús Ordono: Conceptualization, Investigation, Methodology, Formal analysis, Writing - Original Draft. Soledad Perez-Amodio: Conceptualization, Supervision. Kristen Ball: Investigation, Resources. Aitor Aguirre: Conceptualization. Elisabeth Engel: Conceptualization, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
RNA sequencing data are available at the National Center for Biotechnology Information Gene Expression Omnibus repository under accession number GSE153938.
Data will be made available on request.
References
- [1].Mendis S, Puska P, Norrving B, Global atlas on cardiovascular disease prevention and control, World Heal. Organ (2011) 2–14, https://doi.org/NLM classification: WG 120. [Google Scholar]
- [2].Bobi J, Solanes N, Fernández-Jiménez R, Galán-Arriola C, Dantas AP, Fernández-Friera L, Gálvez-Montó C, Rigol-Monzó E, Agüero J, Ramírez J, Roque M, Bayes-Genís A, Sanchez-Gonzalez J, García-Alvarez A, Sabate M, Roura S, Ibanez B, Rigol M, Intracoronary administration of allogeneic adipose tissue-derived mesenchymal stem cells improves myocardial perfusion but not left ventricle function, in a translational model of acute myocardial infarction, J. Am. Heart Assoc 6 (2017), 10.1161/JAHA.117.005771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aguirre A, Montserrat N, Zacchigna S, Nivet E, Hishida T, Krause MN, Kurian L, Ocampo A, Vazquez-Ferrer E, Rodriguez-Esteban C, Kumar S, Moresco JJ, Yates JR, Campistol JM, Sancho-Martinez I, Giacca M, Izpisua Belmonte JC, In vivo activation of a conserved microRNA program induces mammalian heart regeneration, Cell Stem Cell 15 (2014) 589–604, 10.1016/j.stem.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Liu B, Lee BW, Nakanishi K, Villasante A, Williamson R, Metz J, Kim J, Kanai M, Bi L, Brown K, Di Paolo G, Homma S, Sims PA, Topkara VK, Vunjak-Novakovic G, Cardiac recovery via extended cell-free delivery of extracellular vesicles secreted by cardiomyocytes derived from induced pluripotent stem cells, Nat. Biomed. Eng 2 (2018) 293–303, 10.1038/s41551-018-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tzahor E, Poss KD, Cardiac regeneration strategies: staying young at heart, Science (80-.) 356 (2017) 1035–1039, 10.1126/science.aam5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Aguirre A, Sancho-Martinez I, Izpisua Belmonte JC, Reprogramming toward heart regeneration: stem cells and beyond, Cell Stem Cell 12 (2013) 275–284, 10.1016/j.stem.2013.02.008. [DOI] [PubMed] [Google Scholar]
- [7].Sengupta D, Waldman SD, Li S, From in vitro to in situ tissue engineering, Ann. Biomed. Eng 42 (2014) 1537–1545, 10.1007/s10439-014-1022-8. [DOI] [PubMed] [Google Scholar]
- [8].Sadahiro T, Cardiac regeneration with pluripotent stem cell-derived cardiomyocytes and direct cardiac reprogramming, Regen. Ther 11 (2019) 95–100, 10.1016/j.reth.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nam Y-J, Munshi NV, The promise of cardiac regeneration by in situ lineage conversion, Circulation 135 (2017) 914–916, 10.1161/CIRCULATIONAHA.116.025830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C, Ross CA, Cacchiarelli D, Xia Q, Seligson M, Shinoda G, Xie W, Cahan P, Wang L, Ng S-C, Tintara S, Trapnell C, Onder T, Loh Y-H, Mikkelsen T, Sliz P, Teitell MA, Asara JM, Marto JA, Li H, Collins JJ, Daley GQ, LIN28 regulates stem cell metabolism and conversion to primed pluripotency, Cell Stem Cell 19 (2016) 66–80, 10.1016/j.stem.2016.05.009. [DOI] [PubMed] [Google Scholar]
- [11].Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ, Lin28 enhances tissue repair by reprogramming cellular metabolism, Cell 155 (2013) 778–792, 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, Bomze D, Elena-Herrmann B, Scherf T, Nissim-Rafinia M, Kempa S, Itskovitz-Eldor J, Meshorer E,Aberdam D, Nahmias Y, Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells, Cell Metab 21 (2015) 392–402, 10.1016/j.cmet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- [13].Ma C, Kuzma ML, Bai X, Yang J, Biomaterial-based metabolic regulation in regenerative engineering, Adv. Sci (2019), 10.1002/advs.201900819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ritterhoff J, Tian R, Metabolism in cardiomyopathy: every substrate matters, Cardiovasc. Res 113 (2017) 411–421, 10.1093/cvr/cvx017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Burd LI, Jones MD, Simmons MA, Makowski EL, Meschia G, Battaglia FC, Placental production and foetal utilisation of lactate and pyruvate, Nature 254 (1975) 710–711, 10.1038/254710a0. [DOI] [PubMed] [Google Scholar]
- [16].Werner JC, Sicard RE, Lactate metabolism of isolated, perfused fetal, and newborn pig hearts, Pediatr. Res 22 (1987) 552–556, 10.1203/00006450-198711000-00016. [DOI] [PubMed] [Google Scholar]
- [17].Kolwicz SC, Purohit S, Tian R, Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes, Circ. Res 113 (2013) 603–616, 10.1161/CIRCRESAHA.113.302095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ritterhoff J, Young S, Villet O, Shao D, Neto FC, Bettcher LF, Hsu Y-WA, Kolwicz SC, Raftery D, Tian R, Metabolic remodeling promotes cardiac hypertrophy by directing glucose to aspartate biosynthesis, Circ. Res 126 (2020) 182–196, 10.1161/CIRCRESAHA.119.315483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lopaschuk GD, Karwi QG, Tian R, Wende AR, Abel ED, Cardiac energy metabolism in heart failure, Circ. Res 128 (2021) 1487–1513, 10.1161/CIRCRESAHA.121.318241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang X, Rodriguez ML, Leonard A, Sun L, Fischer KA, Wang Y, Ritterhoff J, Zhao L, Kolwicz SC, Pabon L, Reinecke H, Sniadecki NJ, Tian R, Ruohola-Baker H, Xu H, Murry CE, Fatty acids enhance the maturation of cardiomyocytes derived from human pluripotent stem cells, Stem Cell Rep 13 (2019) 657–668, 10.1016/j.stemcr.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ehler E, Moore-Morris T, Lange S, Isolation and culture of neonatal mouse cardiomyocytes, J. Vis. Exp (2013), 10.3791/50154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP, Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions, Nat. Protoc 8 (2013) 162–175, 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dyer LA, Patterson C, A novel ex vivo culture method for the embryonic mouse heart, J. Vis. Exp (2013), 10.3791/50359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: an open-source platform for biological-image analysis, Nat. Methods 9 (2012) 676–682, 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lacour JR, Bouvat E, Barthelemy JC, Post-competition blood lactate concentrations as indicators of anaerobic energy expenditure during 400-m and 800-m races, Eur. J. Appl. Physiol. Occup. Physiol 61 (1990) 172–176, 10.1007/BF00357594. [DOI] [PubMed] [Google Scholar]
- [26].Zheng Y-W, Zhang L, Wang Y, Chen S-Y, Lei L, Tang N, Yang D-L, Bai L-L, Zhang X-P, Jiang G-Y, Yang L-H, Xu H-T, Li Q-C, Qiu X-S, Wang E-H, Thyroid cancer 1 (C8orf4) shows high expression, no mutation and reduced methylation level in lung cancers, and its expression correlates with b-catenin and DNMT1 expression and poor prognosis, Oncotarget 8 (2017) 405–496, 10.18632/oncotarget.16877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gill S, Veinot J, Kavanagh M, Pulido O, Human heart glutamate receptors—implications for toxicology, food safety, and drug discovery, Toxicol. Pathol 35 (2007) 411–417, 10.1080/01926230701230361. [DOI] [PubMed] [Google Scholar]
- [28].Ren Z, Liu J, Yao L, Li J, Qi Z, Li B, Glutamate receptor ionotropic, kainate 1 serves as a novel tumor suppressor of colorectal carcinoma and predicts clinical prognosis, Exp. Ther. Med 20 (2020), 10.3892/etm.2020.9296, 1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Deisl C, Fine M, Moe OW, Hilgemann DW, Hypertrophy of human embryonic stem cell–derived cardiomyocytes supported by positive feedback between Ca2 and diacylglycerol signals, Pflugers Arch. - Eur. J. Physiol 471 (2019) 1143–1157, 10.1007/s00424-019-02293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kong P, Christia P, Frangogiannis NG, The pathogenesis of cardiac fibrosis, Cell. Mol. Life Sci 71 (2014) 549–574, 10.1007/s00018-013-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Feng Q-Z, Zhao Y-S, Abdelwahid E, The role of fas in the progression of ischemic heart failure: prohypertrophy or proapoptosis, Coron. Artery Dis 19 (2008) 527–534, 10.1097/MCA.0b013e3283093707. [DOI] [PubMed] [Google Scholar]
- [32].Xuan W, Liao Y, Chen B, Huang Q, Xu D, Liu Y, Bin J, Kitakaze M, Detrimental effect of fractalkine on myocardial ischaemia and heart failure, Cardiovasc. Res 92 (2011) 385–393, 10.1093/cvr/cvr221. [DOI] [PubMed] [Google Scholar]
- [33].Wang K, Zhao X, Kuang C, Qian D, Wang H, Jiang H, Deng M, Huang L, Overexpression of SDF-1α enhanced migration and engraftment of cardiac stem cells and reduced infarcted size via CXCR4/PI3K pathway, PLoS One 7 (2012), e43922, 10.1371/journal.pone.0043922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hofmann U, Knorr S, Vogel B, Weirather J, Frey A, Ertl G, Frantz S, Interleukin-13 deficiency aggravates healing and remodeling in male mice after experimental myocardial infarction, Circ. Heart Fail 7 (2014) 822–830, 10.1161/CIRCHEARTFAILURE.113.001020. [DOI] [PubMed] [Google Scholar]
- [35].Sonveaux P, Copetti T, De Saedeleer CJ, Végran F, Verrax J, Kennedy KM, Moon EJ, Dhup S, Danhier P, Frérart F, Gallez B, Ribeiro A, Michiels C, Dewhirst MW, Feron O, Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis, PLoS One 7 (2012), e33418, 10.1371/journal.pone.0033418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL, GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium, Mol. Cell. Biol 28 (2008) 5420–5431, 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, MacRae CA, Stainier DYR, Poss KD, Primary contribution to zebrafish heart regeneration by gata4 cardiomyocytes, Nature 464 (2010) 601–605, 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cummins EP, Taylor CT, Hypoxia-responsive transcription factors, Pflugers Arch. - Eur. J. Physiol 450 (2005) 363–371, 10.1007/s00424-005-1413-7. [DOI] [PubMed] [Google Scholar]
- [39].Li R, Geng H, Xiao J, Qin X, Wang F, Xing J, Xia Y, Mao Y, Liang J, Ji X, miR-7a/b attenuates post-myocardial infarction remodeling and protects H9c2 cardiomyoblast against hypoxia-induced apoptosis involving Sp1 and PARP-1, Sci. Rep 6 (2016) 29082, 10.1038/srep29082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Oikawa M, Wu M, Lim S, Knight WE, Miller CL, Cai Y, Lu Y, Blaxall BC, Takeishi Y, Abe J, Yan C, Cyclic nucleotide phosphodiesterase 3A1 protects the heart against ischemia-reperfusion injury, J. Mol. Cell. Cardiol 64 (2013) 11–19, 10.1016/j.yjmcc.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lim B, Jung K, Gwon Y, Oh JG, Roh J, Hong S-H, Kho C, Park W-J, Lee H-W, Bae J-W, Jung Y-K, Cardioprotective role of APIP in myocardial infarction through ADORA2B, Cell Death Dis 10 (2019) 511, 10.1038/s41419-019-1746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang Q, Lu G, Chen Z, MALAT1 promoted cell proliferation and migration via MALAT1/miR-155/MEF2A pathway in hypoxia of cardiac stem cells, J. Cell. Biochem 120 (2019) 6384–6394, 10.1002/jcb.27925. [DOI] [PubMed] [Google Scholar]
- [43].Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, Egashira T, Seki T, Muraoka N, Yamakawa H, Ohgino Y, Tanaka T, Yoichi M, Yuasa S, Murata M, Suematsu M, Fukuda K, Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes, Cell Stem Cell 12 (2013) 127–137, 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- [44].Bon-Mathier A-C, Rignault-Clerc S, Bielmann C, Rosenblatt-Velin N, Oxygen as a key regulator of cardiomyocyte proliferation: new results about cell culture conditions!, Biochim. Biophys. Acta - Mol. Cell Res 1867 (2020), 118460, 10.1016/j.bbamcr.2019.03.007. [DOI] [PubMed] [Google Scholar]
- [45].Brooks GA, The science and translation of lactate shuttle theory, Cell Metab 27 (2018) 757–785, 10.1016/j.cmet.2018.03.008. [DOI] [PubMed] [Google Scholar]
- [46].Alvarez Z, Castano O, Castells AA, Mateos-Timoneda MA, Planell JA, Engel E, Alcantara S, Neurogenesis and vascularization of the damaged brain using a lactate-releasing biomimetic scaffold, Biomaterials 35 (2014) 4769–4781, 10.1016/j.biomaterials.2014.02.051. [DOI] [PubMed] [Google Scholar]
- [47].Gupta P, Hourigan K, Jadhav S, Bellare J, Verma P, Effect of lactate and pH on mouse pluripotent stem cells: importance of media analysis, Biochem. Eng. J 118 (2017) 25–33, 10.1016/j.bej.2016.11.005. [DOI] [Google Scholar]
- [48].Zhang M, Niu X, Hu J, Yuan Y, Sun S, Wang J, Yu W, Wang C, Sun D, Wang H, Lin28a protects against Hypoxia/Reoxygenation induced cardiomyocytes apoptosis by alleviating mitochondrial dysfunction under high glucose/high fat conditions, PLoS One 9 (2014), e110580, 10.1371/journal.pone.0110580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xiang Q, Yang B, Li L, Qiu B, Qiu C, Gao X, Zhou H.(Jenny), Min W, Critical role of Lin28-TNFR2 signalling in cardiac stem cell activation and differentiation, J. Cell. Mol. Med 23 (2019) 2943–2953, 10.1111/jcmm.14202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sun L, Yu J, Qi S, Hao Y, Liu Y, Li Z, Bone morphogenetic Protein-10 induces cardiomyocyte proliferation and improves cardiac function after myocardial infarction, J. Cell. Biochem (2014) 1868–1876, 10.1002/jcb.24856. [DOI] [PubMed] [Google Scholar]
- [51].Chen H, Yong W, Ren S, Shen W, He Y, Cox KA, Zhu W, Li W, Soonpaa M, Payne RM, Franco D, Field LJ, Rosen V, Wang Y, Shou W, Overexpression of bone morphogenetic protein 10 in myocardium disrupts cardiac postnatal hypertrophic growth, J. Biol. Chem 281 (2006) 27481–27491, 10.1074/jbc.M604818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ, Hayward L, Langridge-Smith P, Gilbert N, Ramsahoye BH, Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression, Nucleic Acids Res 40 (2012) 4794–4803, 10.1093/nar/gks066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, Ding J, Czyz D, Hu R, Ye Z, He M, Zheng YG, Shuman HA, Dai L, Ren B, Roeder RG, Becker L, Zhao Y, Metabolic regulation of gene expression by histone lactylation, Nature 574 (2019) 575–580, 10.1038/s41586-019-1678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].De Saedeleer CJ, Copetti T, Porporato PE, Verrax J, Feron O, Sonveaux P, Lactate activates HIF-1 in oxidative but not in Warburg-phenotype human tumor cells, PLoS One 7 (2012), e46571, 10.1371/journal.pone.0046571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lee DC, Sohn HA, Park Z-Y, Oh S, Kang YK, Lee K, Kang M, Jang YJ, Yang S-J, Hong YK, Noh H, Kim J-A, Kim DJ, Bae K-H, Kim DM, Chung SJ, Yoo HS, Yu D-Y, Park KC, Il Yeom Y, A lactate-induced response to hypoxia, Cell 161 (2015) 595–609, 10.1016/j.cell.2015.03.011. [DOI] [PubMed] [Google Scholar]
- [56].Jopling C, Suñé G, Faucherre A, Fabregat C, Izpisua Belmonte JC, Hypoxia induces myocardial regeneration in zebrafish, Circulation 126 (2012) 3017–3027, 10.1161/CIRCULATIONAHA.112.107888. [DOI] [PubMed] [Google Scholar]
- [57].Nakada Y, Canseco DC, Thet S, Abdisalaam S, Asaithamby A, Santos CX, Shah AM, Zhang H, Faber JE, Kinter MT, Szweda LI, Xing C, Hu Z, Deberardinis RJ, Schiattarella G, Hill JA, Oz O, Lu Z, Zhang CC, Kimura W, Sadek HA, Hypoxia induces heart regeneration in adult mice, Nature 541 (2017) 222–227, 10.1038/nature20173. [DOI] [PubMed] [Google Scholar]
- [58].Kimura W, Xiao F, Canseco DC, Muralidhar S, Thet S, Zhang HM, Abdulrahman Y, Chen R, Garcia JA, Shelton JM, Richardson JA, Ashour AM, Asaithamby A, Liang H, Xing C, Lu Z, Zhang CC, Sadek HA, Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart, Nature 523 (2015) 226–230, 10.1038/nature14582. [DOI] [PubMed] [Google Scholar]
- [59].Vujic A, Lerchenmüller C, Wu T-D, Guillermier C, Rabolli CP, Gonzalez E, Senyo SE, Liu X, Guerquin-Kern J-L, Steinhauser ML, Lee RT, Rosenzweig A, Exercise induces new cardiomyocyte generation in the adult mammalian heart, Nat. Commun 9 (2018) 1659, 10.1038/s41467-018-04083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wang WE, Li L, Xia X, Fu W, Liao Q, Lan C, Yang D, Chen H, Yue R, Zeng C, Zhou L, Zhou B, Duan DD, Chen X, Houser SR, Zeng C, Dedifferentiation, proliferation, and redifferentiation of adult mammalian cardiomyocytes after ischemic injury, Circulation 136 (2017) 834–848, 10.1161/CIRCULATIONAHA.116.024307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bergman BC, Tsvetkova T, Lowes B, Wolfel EE, Myocardial glucose and lactate metabolism during rest and atrial pacing in humans, J. Physiol 587 (2009) 2087–2099, 10.1113/jphysiol.2008.168286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Evans RK, Schwartz DD, Gladden LB, Effect of myocardial volume overload and heart failure on lactate transport into isolated cardiac myocytes, J. Appl. Physiol 94 (2003) 1169–1176, 10.1152/japplphysiol.00778.2002. [DOI] [PubMed] [Google Scholar]
- [63].Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA, Transient regenerative potential of the neonatal mouse heart, Science (80-.) 331 (2011) 1078–1080, 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Paradis AN, Gay MS, Zhang L, Binucleation of cardiomyocytes: the transition from a proliferative to a terminally differentiated state, Drug Discov. Today 19 (2014) 602–609, 10.1016/j.drudis.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu T-D, Guerquin-Kern J-L, Lechene CP, Lee RT, Mammalian heart renewal by pre-existing cardiomyocytes, Nature 493 (2013) 433–436, 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Gabisonia K, Prosdocimo G, Aquaro GD, Carlucci L, Zentilin L, Secco I, Ali H, Braga L, Gorgodze N, Bernini F, Burchielli S, Collesi C, Zandonà L, Sinagra G, Piacenti M, Zacchigna S, Bussani R, Recchia FA, Giacca M, MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs, Nature 569 (2019) 418–422, 10.1038/s41586-019-1191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Puiggalí-Jou A, Ordoño J, del Valle LJ, Pérez-Amodio S, Engel E, Aleman C, Tuning multilayered polymeric self-standing films for controlled release of L-lactate by electrical stimulation, J. Control. Release 330 (2021) 669–683, 10.1016/j.jconrel.2020.12.049. [DOI] [PubMed] [Google Scholar]
- [68].Porporato PE, Payen VL, De Saedeleer CJ, Préat V, Thissen J-P, Feron O, Sonveaux P, Lactate stimulates angiogenesis and accelerates the healing of superficial and ischemic wounds in mice, Angiogenesis 15 (2012) 581–592, 10.1007/s10456-012-9282-0. [DOI] [PubMed] [Google Scholar]
- [69].Chereddy KK, Lopes A, Koussoroplis S, Payen V, Moia C, Zhu H, Sonveaux P, Carmeliet P, des Rieux A, Vandermeulen G, Préat V, Combined effects of PLGA and vascular endothelial growth factor promote the healing of non-diabetic and diabetic wounds, Nanomed. Nanotechnol. Biol. Med 11 (2015) 1975–1984, 10.1016/j.nano.2015.07.006. [DOI] [PubMed] [Google Scholar]
- [70].Zhang X, Wu Y, Pan Z, Sun H, Wang J, Yu D, Zhu S, Dai J, Chen Y, Tian N, Heng BC, Coen ND, Xu H, Ouyang H, The effects of lactate and acid on articular chondrocytes function: implications for polymeric cartilage scaffold design, Acta Biomater 42 (2016) 329–340, 10.1016/j.actbio.2016.06.029. [DOI] [PubMed] [Google Scholar]
- [71].Ordono J, Perez-Amodio S, Ball K, Aguirre A, Engel E, Lactate Promotes Cardiomyocyte Dedifferentiation Through Metabolic Reprogramming, BioRxiv, 2020, 10.1101/2020.07.21.213736. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data are available at the National Center for Biotechnology Information Gene Expression Omnibus repository under accession number GSE153938.
Data will be made available on request.


