Abstract
The drill monkey has been shown by serology and PCR to harbor a unique simian immunodeficiency virus (SIVdrl). A pol sequence, amplified from uncultured peripheral blood cells, is most closely related to the equivalent SIV sequences from the red-capped mangabey (SIVrcm), the sabaeus African green monkey (SIVagmSAB), and the chimpanzee (SIVcpz) and to the human immunodeficiency virus type 1 (HIV-1) sequence of humans. It is as yet unclear whether SIVdrl has a mosaic genome like SIVrcm and SIVagmSAB, is a member of the SIVcpz/HIV-1 lineage, or represents a novel primate lentivirus lineage.
Several species of African nonhuman primates are known to be infected with simian immunodeficiency viruses (SIVs) (18, 31, 36). Most recently, the red-capped mangabey (Cercocebus torquatus torquatus) has been shown to harbor a novel lentivirus, SIVrcm (16). It is thought that these viruses only rarely cause disease in their natural hosts, suggesting that they have been associated with them for a long time (1, 13, 19, 21). However, if cross-species transmission occurs, the virus may be pathogenic for the new host. For example, in captivity, different species of macaques have apparently been infected with SIVsm from sooty mangabeys (Cercocebus atys) and have gone on to develop a simian AIDS-like syndrome, as have rhesus macaques when experimentally inoculated (21, 29, 34, 36). Furthermore, the human immunodeficiency viruses types 1 and 2 (HIV-1 and HIV-2, respectively) are thought by some to have been introduced into humans by at least two independent transmissions of SIVs from primates in Africa (9, 35, 42). The genome sequence of HIV-2 is closely related to the sequence of SIVsm (22), and natural infection with SIVsm of humans has been found (15), indicating that HIV-2 could have arisen from cross-species transmission of SIVsm to humans in west Africa.
Phylogenetic analysis of HIV-1 sequences shows that they can be considered as a major (M) cluster with outlying (O) variants (28). The genotypic and phenotypic differences between group O and M group isolates are consistent with separate introductions into the human population (4, 17, 30, 40, 43, 48). This introduction may have been with two distinct SIVs from chimpanzees (Pan troglodytes), one related to SIVcpz-gab (23, 24) and the other to SIVcpz-ant (46). It has also been suggested that cross-species transmission from humans to chimpanzees gave rise to SIVcpz (10, 33). Alternatively, SIVs from other African primates may have infected humans, chimpanzees, or both.
The drill, Mandrillus leucophaeus, is a large short-tailed monkey that exists only in several blocks of forest between the Sanaga River in southwestern Cameroon and the Cross River in southeastern Nigeria and on the offshore island of Bioko (Equatorial Guinea). Due to continuing habitat loss, forest fragmentation, and indiscriminate hunting for bushmeat, the drill has become endangered. It is recognized by the International Union for Conservation of Nature as the highest-ranked primate species for conservation action in Africa.
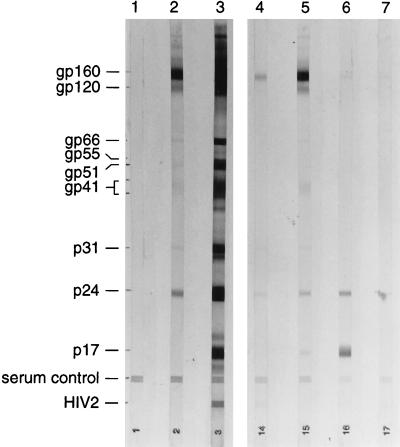
Blood samples were collected during their quarantine from drills officially donated to the Drill Rehabilitation and Breeding Center (14). These animals were wild born and had been illegally captured for the bushmeat trade. Cross-reactions observed during enzyme-linked immunosorbent assay (ELISA) screening of serum samples from 50 drills suggested that 16 of them were infected with a lentivirus, possibly an SIV related to HIV-1. The five samples most strongly reactive by ELISA were confirmed to be positive by Western blotting; the results from four of these tested at the same time are shown in Fig. 1.
FIG. 1.
Western blot analysis of serum samples from four wild-born drill monkeys (lanes 4 to 7). Lanes: 1, negative control; 2, weak positive control; 3, strong positive control. Cross-reactions were observed with p24 only (lane 7); with p24 and gp160 (lane 4); with p17, p24, and gp160 (lane 6); and with p17, p24, gp41, gp120, and gp160 (lane 5). The blot was HIV blot 2.2 from Genelabs.
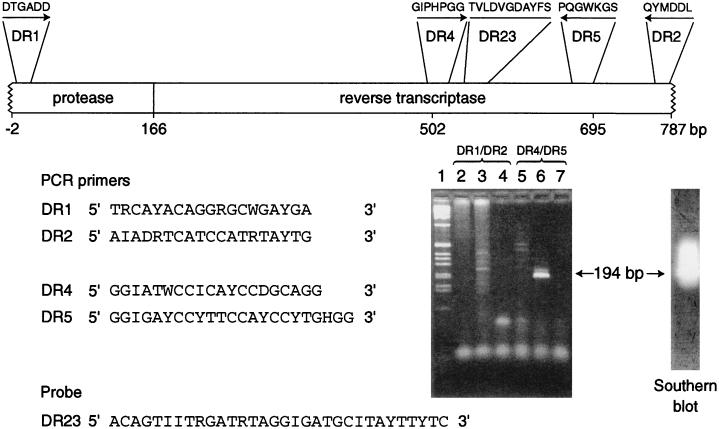
Degenerate, generic primers in the pol gene were designed by using a multiple alignment of HIV-1, HIV-2, and SIV sequences and the Primegen program (37). Regions identified as suitable included conserved motifs previously used for amplification of diverse retroviral sequences (8, 45, 47). The sequences of the primers are shown in Fig. 2. They are DR1, located in the LLDTGA motif of the protease gene, equivalent to bases 2601 to 2619 of SIVMM251 (GenBank/EMBL accession no. M19499) (12, 27); DR2, located in the YMDDLY motif of the reverse transcriptase gene, equivalent to bases 3371 to 3389 of SIVMM251; DR4, equivalent to bases 3104 to 3123 of SIVMM251; and DR5, from the PQGWK motif in the reverse transcriptase gene, equivalent to bases 3275 to 3297 of SIVMM251. An inosine residue was substituted at positions of maximum degeneracy in the primers and probe. A digoxigenin-labelled oligonucleotide probe (DR23 [Fig. 2]) was used to confirm the specificity of the amplicon produced by primers DR4 and DR5. This was equivalent to bases 3146 to 3177 of SIVMM251.
FIG. 2.
PCR amplification of SIV sequences from PBMC lysates from two drill monkeys. The nested primers used are shown relative to their location on the pol gene of the primate lentiviruses. The conserved amino acid motifs are indicated above the primer locations. The numbering relates to the sequence determined in this work. The ethidium bromide-stained agarose gel (3:1 NuSieve agarose; FMC Bioproducts, Flowgen, Lichfield, United Kingdom) was used for the analysis of the products of amplification; the specific product of 194 bp is arrowed. Lanes: 1, 1-kb marker (Life Technologies); 2 to 4, products of the primary amplification with outer primers DR1 and DR2 from two drills (lanes 2 and 3) and water (lane 4); 5 to 7, products of the secondary amplifications of the reactions shown in lanes 2 to 4 with the inner primers DR4 and DR5. A Southern blot probed with a digoxigenin (Boehringer Mannheim, Lewes, United Kingdom) probe, DR23, is also shown.
Amplifications were performed in 100 μl of mixture with Taq polymerase and reaction buffer, including detergent W1 (Life Technologies, Paisley, United Kingdom), with a model 9600 thermal cycler (Perkin-Elmer, Warrington, United Kingdom). Primer DR1 was used at 0.8 pmol per μl, and DR2, DR4, and DR5 were used at 2.5 pmol per μl. Touchdown PCR was used with primers DR1, DR2, DR4, and DR5 (7). After an initial denaturation at 94°C for 2 min, there were 30 cycles of 94°C for 15 s, 50°C decreasing by 0.5°C per cycle to 35°C, and 72°C for 1 min; this was followed by 15 cycles of 94°C for 15 s, 50°C for 30 s, and 72°C for 1 min, with a final elongation at 72°C for 5 min before cooling to 4°C. DR1 and DR2 were used in primary amplification reactions, and DR4 and DR5 were used in secondary, nested amplifications.
A specific band of the predicted size of approximately 194 bp was produced when these primers were used with human lymphocytes previously shown to have been infected with HIV-1 subtype A, HIV-1 group O, and HIV-2. A band of similar size was observed (Fig. 2) when this primer set was used with a lymphocyte lysate from a drill monkey. This band, which was shown to hybridize with the probe by Southern blotting (Fig. 2), was purified and sequenced. Specific primers were designed with the program OLIGO (41) from the SIVdrl sequence obtained. Uneven PCR was used in reactions with one specific primer and one degenerate primer (5). Specific primers were used at 0.1 pmol per μl for primary amplifications and at 0.5 pmol per μl for secondary (nested) amplifications. After an initial denaturation at 94°C for 1 min, there were 3 cycles of 94°C for 30 s, 55°C for 1 min, 72°C for 1 min, 94°C for 15 s, 42°C for 1 min, and 72°C for 1 min, and then there were 20 cycles of 94°C for 15 s, 57°C for 30 s, 72°C for 30 s, 94°C for 15 s, 45°C for 30 s, and 72°C for 30 s, with a final elongation at 72°C for 5 min before cooling to 4°C. PCR products were purified with Centricon columns (Amicon, Stonehouse, United Kingdom), or by extraction from gels, and sequenced with the primers used for their amplification with Perkin-Elmer–Applied Biosystems reagents and apparatus (3). A sequence of 787 bp between DR1 and DR2 was obtained by the methods described above. In a comparison with database sequences (Table 1 and Fig. 3), the 787-bp sequence was shown to be related to HIV and SIV pol sequences. As shown in Table 1 and Fig. 4, the virus identified as having the most similar pol sequence to SIVdrl was SIVrcm (14a).
TABLE 1.
Genetic distances between SIVdrl and primate lentiviruses for a partial pol sequence
| Virus | % Genetic distance from virus:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIVdrl | SIVrcm | HIVlant70 | SIVagmSAB | SIVcpza | SIVcpz | HIV-1 | SIVmnd | SIVsm | SIVagmTAN | HIV-2 | SIVagmGRI | SIVagmVER | |
| SIVrcm | 72.5 | ||||||||||||
| HIV1ant70 | 69.3 | 66.3 | |||||||||||
| SIVagmSAB | 68.2 | 66.8 | 65.0 | ||||||||||
| SIVcpza | 66.3 | 66.3 | 71.5 | 64.2 | |||||||||
| SIVcpz | 66.0 | 69.0 | 77.1 | 63.4 | 71.6 | ||||||||
| HIV-1 | 64.8 | 66.8 | 73.2 | 62.6 | 70.8 | 79.0 | |||||||
| SIVmnd | 62.2 | 61.7 | 61.7 | 59.1 | 56.6 | 60.8 | 59.1 | ||||||
| SIVsm | 61.8 | 58.9 | 59.9 | 60.5 | 59.4 | 61.2 | 61.2 | 57.8 | |||||
| SIVagmTAN | 60.9 | 61.1 | 60.8 | 59.5 | 58.3 | 61.6 | 59.6 | 62.2 | 55.3 | ||||
| HIV-2 | 59.6 | 60.0 | 59.4 | 59.4 | 58.6 | 60.0 | 60.7 | 58.6 | 78.4 | 56.0 | |||
| SIVagmGRI | 59.4 | 61.3 | 60.5 | 62.0 | 56.9 | 60.0 | 61.2 | 56.4 | 57.9 | 65.5 | 58.9 | ||
| SIVagmVER | 57.7 | 59.1 | 60.7 | 62.4 | 58.7 | 59.6 | 60.3 | 63.9 | 54.9 | 65.0 | 53.9 | 66.9 | |
| SIVsyk | 56.6 | 57.4 | 56.4 | 57.4 | 52.3 | 60.4 | 55.3 | 57.3 | 58.9 | 57.2 | 58.1 | 54.0 | 53.9 |
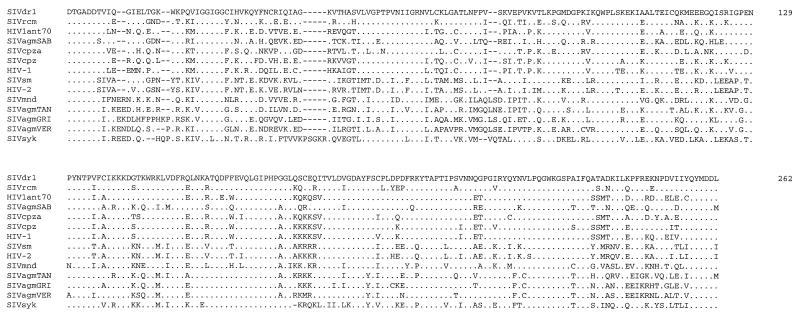
FIG. 3.
Alignment of the SIVdrl partial pol amino acid sequence with the equivalent sequences of other representative HIVs and SIVs. Identities to the SIVdrl sequence are shown by a dot. The accession numbers of the sequences used are given in parentheses as follows: SIVsyk (L06042); HIV1ant70 (L20587); SIVcpz (X52154); SIVcpza (U42720); HIVz2, HIV-1 subtype D (M22639); SIVagmSAB (U04005); SIVmnd (M27470); SIVagmGRI from the grivet (M66437); SIVagmVER from the vervet (X07805); HIV2st, HIV-2 subtype A (M31113); SIVsm (X14307); and SIVrcm (14a).
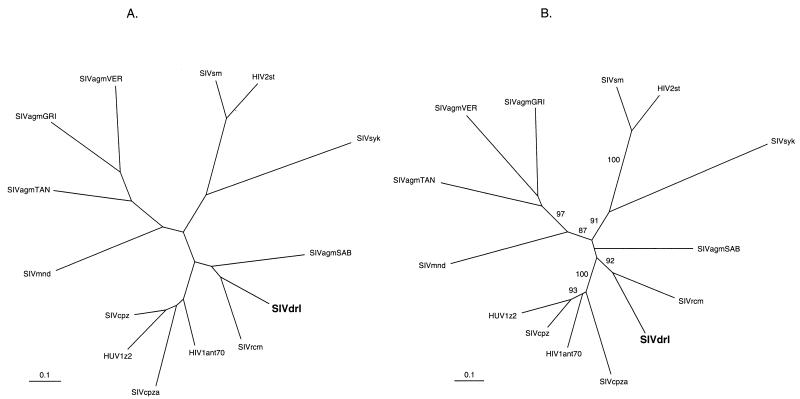
FIG. 4.
Phylogenetic trees showing the relationship of the SIVdrl pol nucleotide sequence to the equivalent sequence from other SIV and HIV genomes. (A) Maximum likelihood. (B) Distance (neighbor joining). An in-frame nucleotide alignment was made with the Clustal algorithm (44) in Megalign (Lasergene, DNAStar) with SIVdrl and the equivalent region from representatives of the available primate lentivirus sequences. Gaps were excluded from the alignment. For the maximum-likelihood analysis, the alignment was used in PAUP version 4.0 (D. L. Swofford) with the F84 model of evolution, and rates were assumed to follow a gamma distribution (alpha was estimated to be 0.48, and the transition/transversion ratio was estimated to be 1.16). For the neighbor-joining analysis, the alignment was used in DNAdist and Fitch (11) with the Jin-Nei model of evolution (25). The reliability of the neighbor-joining tree was estimated by 1,000 bootstrap replicates, and values greater than 70% are shown in panel B. Trees were displayed with Treeview (38). The accession numbers of the sequences used are given in the legend to Fig. 3. The scale bar indicates the number of nucleotide substitutions per site.
The phylogenetic relationship of the SIVdrl sequence to other primate lentivirus sequences was investigated by maximum-likelihood and neighbor-joining methods (Fig. 4A and B). Several conclusions were drawn from these analyses (Table 1 and Fig. 4). First, the previously recognized five primate lentivirus lineages (20, 42) can be seen, supported by bootstrap values of greater than 70% in the neighbor-joining tree (Fig. 4B). These lineages are (i) HIV-1 and SIVcpz; (ii) SIV from sooty mangabeys (SIVsm) and HIV-2; (iii) SIV from African green monkeys, Cercopithecus aethiops (SIVagm); (iv) SIV from mandrills, Mandrillus sphinx (SIVmnd); and (v) SIV from Sykes’ monkey, Cercopithecus mitis albogularis (SIVsyk). In addition, two recombinant genomes have been described, SIVagmSAB from Cercopithecus aethiops sabeus, the sabaeus African green monkey (2, 26), and SIVrcm (16), which do not fall consistently into these lineages when different regions of their genomes are analyzed. Second, whichever method was used, and whether the nucleotide or predicted amino acid sequences were compared, the phylogenetic trees obtained were congruent, with the exception of the SIVagmSAB sequence. In trees made by maximum-likelihood analysis, the SIVagmSAB pol sequence fell on the same branch as SIVdrl and SIVrcm (Fig. 4A), whereas by distance methods (neighbor joining), it fell on a separate branch (Fig. 4B), albeit with bootstrap values of less than 70%. This is consistent with the recombinant nature of the SIVagmSAB genome. Third, the SIVdrl pol sequence did not cluster with the SIVmnd pol sequence from M. sphinx, suggesting that these congeners are not infected with a similar SIV. Fourth, in all of the trees found, the SIVdrl sequence was most closely related to the SIVrcm sequence. Fifth, these two viruses appear to be related to the HIV-1–SIVcpz lineage of viruses. However, the SIVrcm genome is only similar to this lineage in pol; in gag it falls onto a separate branch (16), and, overall, it has a mosaic genome, presumably the result of recombination between distinct SIVs (14a, 16). Analysis of the exact relationship between these two genomes will only be possible when the SIVdrl genome has been completely sequenced. It is of interest that drills and red-capped mangabeys are sympatric, and exchange of viruses between them is therefore possible. More complete SIVdrl genome sequence should also reveal whether it has a mosaic genome or constitutes a sixth lineage of the primate lentiviruses.
The specific primers designed from the SIVdrl sequence were used to test peripheral blood mononuclear cells (PBMCs) from 11 of the 16 ELISA-reactive drills. By PCR, all 11 were found to be infected. One animal that was ELISA negative was found to be negative for SIVdrl by PCR testing. The drills that were PCR positive appeared to be healthy and were over 3 years old. An important question is whether drills are naturally infected with this virus. Since the drills are only ever kept with other drills after confiscation, they could not have been infected by other species except before arrival at the rehabilitation center. The animals were bled with single-use needles at various times after arrival. One drill which was sampled on the day of its arrival was ELISA reactive, as were two that were bled within 2 months, indicating that they were infected before admission. Three others were seronegative when first tested, between 1 day and 6 months after arrival. A fourth was seronegative 2 years after arrival. These four animals all subsequently seroconverted. Thus, SIVdrl appears to be a natural infection that has also spread between drills in captivity. This conclusion is supported by 207 bases (69 amino acids) of pol sequence data from three other drills, including one of the animals identified as being infected on arrival at the rehabilitation center. None of these three sequences, or the prototype sequence shown in this paper, are identical: one differs from the prototype by three amino acids (4.5% amino acid divergence and 14% nucleotide divergence), and two others each differ by four amino acids from the prototype (6% amino acid divergence and 13.2% nucleotide divergence and 6% amino acid divergence and 3% nucleotide divergence, respectively). Over the same region, SIVrcm differs from the SIVdrl by 12 amino acids (19.8% amino acid divergence and 33.4% nucleotide divergence). These data are consistent with SIVdrl being a natural infection, acquired with increasing age, although the route of transmission is unclear from the current studies. We note that it has been suggested that sexual transmission of SIV among mandrills is rare (6).
In the recent past, the drill was more numerous. Today, although fewer in number, drills inhabit areas of Cameroon and Nigeria with other nonhuman primates, including chimpanzees, forest guenons, colobines, mangabeys, and gorillas; exchange of SIVs between these species has, therefore, been possible. It is probable that there has been ample opportunity for any SIV from a species eaten as bushmeat to infect humans when these animals are butchered for food. The HIV-1 group O variant viruses have their highest prevalence in west Africa, including Cameroon and Nigeria (32, 39). Further characterization of the SIVdrl genome and of SIVs from the more than 22 other simian species that inhabit this particular region is required to unravel their relationships to the human lentiviruses.
Nucleotide sequence accession number.
The EMBL accession number of the sequence reported in this study is AJ011017.
Acknowledgments
We thank J. V. Parry, C. Arnold, and L. Metherell for help, advice, and encouragement and C. King of Vetdiagnostics for initial ELISA testing. We are grateful to B. H. Hahn for making the sequence of SIVrcm available before publication.
REFERENCES
- 1.Allan J S, Kanda P, Kennedy R C, Cobb E K, Anthony M, Eichberg J W. Isolation and characterization of simian immunodeficiency viruses from two subspecies of African green monkeys. AIDS Res Hum Retroviruses. 1990;6:275–285. doi: 10.1089/aid.1990.6.275. [DOI] [PubMed] [Google Scholar]
- 2.Allan J S, Short M, Taylor M E, Su S, Hirsch V M, Johnson P R, Shaw G M, Hahn B H. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65:2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlow K L, Tosswill J H C, Parry J V, Clewley J P. Performance of the Amplicor human immunodeficiency virus type 1 PCR and analysis of specimens with false-negative results. J Clin Microbiol. 1997;35:2846–2853. doi: 10.1128/jcm.35.11.2846-2853.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braaten D, Franke E K, Luban J. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIVCPZGAB but not group O HIV-1 or other primate immunodeficiency viruses. J Virol. 1996;70:4220–4227. doi: 10.1128/jvi.70.7.4220-4227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Wu R. Direct amplification of unknown genes and fragments by uneven polymerase chain reaction. Gene. 1997;185:195–199. doi: 10.1016/s0378-1119(96)00637-3. [DOI] [PubMed] [Google Scholar]
- 6.Cooper R, Feistner A, Evans S, Tsujimoto H, Hayami M. A lack of evidence of sexual transmission of a simian immunodeficiency agent in a semifree-ranging group of mandrills. AIDS. 1989;3:764. [PubMed] [Google Scholar]
- 7.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donehower L A, Bohannon R C, Ford R J, Gibbs R A. The use of primers from highly conserved pol regions to identify uncharacterized retroviruses by the polymerase chain reaction. J Virol Methods. 1990;28:33–46. doi: 10.1016/0166-0934(90)90085-t. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle R F, Feng D-F, Johnson M S, McClure M A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 10.Ewald P W. Evolution of infectious disease. Oxford, United Kingdom: Oxford University Press; 1994. [Google Scholar]
- 11.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 12.Franchini G, Gurgo C, Guo H G, Gallo R C, Collalti E, Fargnoli K A, Hall L F, Wong-Staal F, Reitz M S. Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature. 1987;328:539–543. doi: 10.1038/328539a0. [DOI] [PubMed] [Google Scholar]
- 13.Fultz P N, McClure H M, Anderson D C, Swenson R B, Ananad R, Srinvasan A. Isolation of a T-lymphotropic retrovirus from naturally-infected sooty mangabey monkeys (Cerocebus atys) Proc Natl Acad Sci USA. 1986;83:5286–5290. doi: 10.1073/pnas.83.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gadsby E L, Jenkins P D, Feistner A T C. Coordinating conservation for the drill (Mandrillus leucophaeus): endangered in forest and zoo. In: Olney P J S, Mace G M, Feistner A T C, editors. Creative conservation: interactive management of wild and captive animals. London, United Kingdom: Chapman and Hall; 1994. pp. 439–454. [Google Scholar]
- 14a.Gao, F., and B. H. Hahn. Personal communication.
- 15.Gao F, Yue L, White A T, Pappas P G, Barchue J, Hanson A P, Greene B M, Sharp P M, Shaw G M, Hahn B H. Human infection by genetically diverse SIVSM-related HIV-2 in West Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 16.Georges-Courbot M C, Lu C Y, Makuwa M, Telfer P, Onanga R, Dubreuil G, Chen Z, Smith S M, Georges A, Gao F, Hahn B H, Marx P A. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J Virol. 1998;72:600–608. doi: 10.1128/jvi.72.1.600-608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gürtler L G, Zekeng L, Tsague J M, van Brunn A, Afane Z E, Eberle J, Kaptue L. HIV-1 subtype O: epidemiology, pathogenesis, diagnosis, and perspectives of the evolution of HIV. Arch Virol Suppl. 1996;11:195–202. doi: 10.1007/978-3-7091-7482-1_17. [DOI] [PubMed] [Google Scholar]
- 18.Hatami M, Ido E, Miura T. Survey of simian immunodeficiency virus among nonhuman primate populations. Curr Top Microbiol Immunol. 1994;188:1–20. doi: 10.1007/978-3-642-78536-8_1. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch V M, Dapolito G, Johnson P R, Elkins W R, London W T, Montali R J, Goldstein S, Brown C. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J Virol. 1995;69:955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch V M, Dapolito G A, Goldstein S, McClure H, Emau P, Fultz P N, Isahakia M, Lenroot R, Myers G, Johnson P R. A distinct African lentivirus from Sykes’ monkeys. J Virol. 1993;67:1517–1528. doi: 10.1128/jvi.67.3.1517-1528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch V M, Johnson P R. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 1994;32:183–203. doi: 10.1016/0168-1702(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch V M, Olmstead R A, Murphey-Corb M, Purcell R H, Johnson P R. An African primate lentivirus (SIVSM) closely related to HIV-2. Nature. 1989;339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 23.Huet T, Cheynier R, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organisation of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–359. doi: 10.1038/345356a0. [DOI] [PubMed] [Google Scholar]
- 24.Janssens W, Fransen K, Peeters M, Heyndrickx L, Motte J, Bedjabaga L, Delaporte E, Piot P, van der Groen G. Phylogenetic analysis of a new chimpanzee lentivirus SIV cpz-gab2 from a wild captured chimpanzee from Gabon. AIDS Res Hum Retroviruses. 1994;10:1191–1192. doi: 10.1089/aid.1994.10.1191. [DOI] [PubMed] [Google Scholar]
- 25.Jin L, Nei M. Limitations of the evolutionary parsimony method of phylogenetic analysis. Mol Biol Evol. 1990;7:82–102. doi: 10.1093/oxfordjournals.molbev.a040588. [DOI] [PubMed] [Google Scholar]
- 26.Jin M J, Hui H, Robertson D L, Muller M C, Barre-Sinoussi F, Hirsch V M, Allan J S, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kestler H W I, Li Y, Naidu Y M, Butler C V, Ochs M F, Jaenel G, King N W, Daniel M D, Desrosiers R C. Comparison of simian immunodeficiency virus isolates. Nature. 1988;331:619–622. doi: 10.1038/331619a0. [DOI] [PubMed] [Google Scholar]
- 28.Leitner T. Genetic subtypes of HIV-1. In: Myers G, Korber B, Foley B, Jeang K-T, Mellors J W, Wain-Hobson S, editors. Human retroviruses and AIDS. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. pp. III-28–III-40. [Google Scholar]
- 29.Letvin N L, Daniel M D, Sehgal P K, Desrosiers R C, Hunt R D, Waldron L M, MacKey J J, Schmidt D K, Chalifoux L V, King N W. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science. 1985;230:71–73. doi: 10.1126/science.2412295. [DOI] [PubMed] [Google Scholar]
- 30.Loussert-Ajaka I, Chaix M-L, Korber B, Letourneur F, Gomas E, Allen E, Ly T-D, Brun-Vézinet F, Simon F, Saragosti S. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J Virol. 1995;69:5640–5649. doi: 10.1128/jvi.69.9.5640-5649.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowenstine L J, Pederson N C, Higgins J, Pallis K C, Uyeda A, Marx P, Lerche N W, Munn R J, Gardner M B. Seroepidemiologic survey of captive old-world primates for antibodies to human and simian retroviruses, and isolation of a lentivirus from a sooty mangabey (Cerocebus atys) Int J Cancer. 1986;38:563–574. doi: 10.1002/ijc.2910380417. [DOI] [PubMed] [Google Scholar]
- 32.Mauclere P, Loussert-Ajaka I, Damond F, Fagot P, Souquieres S, Monny Lobe M, Mbopi Keou F X, Barre-Sinoussi F, Saragosti S, Brun-Vezinet F, Simon F. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS. 1997;11:445–453. doi: 10.1097/00002030-199704000-00007. [DOI] [PubMed] [Google Scholar]
- 33.Mindell D P. Positive selection and rates of evolution in immunodeficiency viruses from humans and chimpanzees. Proc Natl Acad Sci USA. 1996;93:3284–3288. doi: 10.1073/pnas.93.8.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphey-Corb M, Martin L N, Rangan S R, Baskin G, Gormus B J, Wolf R H, Andes W A, West M, Montelaro R C. Isolation of an HTLV-III-related retrovirus from macaques with simian AIDS and its possible origin in asymptomatic mangabeys. Nature. 1986;321:435–437. doi: 10.1038/321435a0. [DOI] [PubMed] [Google Scholar]
- 35.Myers G, MacInnes K, Korber B. The emergence of simian/human immunodeficiency viruses. AIDS Res Hum Retroviruses. 1992;3:373–386. doi: 10.1089/aid.1992.8.373. [DOI] [PubMed] [Google Scholar]
- 36.Novembre F J, Hirsch V M, McClure H M, Fultz P N, Johnson P R. SIV from stump-tailed macaques: molecular characterization of a highly transmissible primate lentivirus. Virology. 1992;186:783–787. doi: 10.1016/0042-6822(92)90047-s. [DOI] [PubMed] [Google Scholar]
- 37.O’Hara P J, Venezia D. PRIMEGEN, a tool for designing primers from multiple alignments. Comput Appl Biosci. 1991;7:533–534. doi: 10.1093/bioinformatics/7.4.533. [DOI] [PubMed] [Google Scholar]
- 38.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 39.Peeters M, Gueye A, Mboup S, Bibollet-Ruche F, Ekaza E, Mulanga C, Ouedrago R, Gandji R, Mpele P, Dibanga G, Koumare B, Saidou M, Esu-Williams E, Lombart J P, Badombena W, Luo N, Vanden Haesevelde M, Delaporte E. Geographical distribution of HIV-1 group O viruses in Africa. AIDS. 1997;11:493–498. doi: 10.1097/00002030-199704000-00013. [DOI] [PubMed] [Google Scholar]
- 40.Quiñones-Mateu M E, Soriano V, Domingo E, Menéndez-Arias L. Characterization of the reverse transcriptase of a human immunodeficiency virus type 1 group O isolate. Virology. 1997;236:364–373. doi: 10.1006/viro.1997.8748. [DOI] [PubMed] [Google Scholar]
- 41.Rychlik W, Rhoades R E. A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res. 1989;17:8543–8551. doi: 10.1093/nar/17.21.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharp, P. M., D. L. Robertson, F. Gao, and B. H. Hahn. 1994. Origins and diversity of human immunodeficiency viruses. AIDS 8(Suppl. 1):S27–S42.
- 43.Sharp P M, Robertson D L, Hahn B H. Cross-species transmission and recombination of ‘AIDS’ viruses. Phil Trans R Soc Lond B. 1995;349:41–47. doi: 10.1098/rstb.1995.0089. [DOI] [PubMed] [Google Scholar]
- 44.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tristem M, Marshall C, Karpas A, Hill F. Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 1992;11:3405–3412. doi: 10.1002/j.1460-2075.1992.tb05419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanden Haesevelde M M, Peeters M, Jannes G, Janssens W, van der Groen G, Sharp P M, Saman E. Sequence analysis of a highly divergent HIV-1-related lentivirus isolated from a wild captured chimpanzee. Virology. 1996;221:346–350. doi: 10.1006/viro.1996.0384. [DOI] [PubMed] [Google Scholar]
- 47.Wichman H A, Van Den Bussche R A. In search of retrotransposons: exploring the potential of the PCR. BioTechniques. 1992;13:258–264. [PubMed] [Google Scholar]
- 48.Zhu T, Korber B T, Nahmias A J, Hooper E, Sharp P M, Ho D D. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]