Abstract
Reduced parasitic infection rates in the developed world are suspected to underlie the rising prevalence of autoimmune disorders. However, the long-term evolutionary consequences of decreased parasite exposure on an immune system are not well understood. We used the Mexican tetra Astyanax mexicanus to understand how loss of parasite diversity influences the evolutionary trajectory of the vertebrate immune system, by comparing river with cave morphotypes. Here, we present field data affirming a strong reduction in parasite diversity in the cave ecosystem, and show that cavefish immune cells display a more sensitive pro-inflammatory response towards bacterial endotoxins. Surprisingly, other innate cellular immune responses, such as phagocytosis, are drastically decreased in cavefish. Using two independent single-cell approaches, we identified a shift in the overall immune cell composition in cavefish as the underlying cellular mechanism, indicating strong differences in the immune investment strategy. While surface fish invest evenly into the innate and adaptive immune systems, cavefish shifted immune investment to the adaptive immune system, and here, mainly towards specific T-cell populations that promote homeostasis. Additionally, inflammatory responses and immunopathological phenotypes in visceral adipose tissue are drastically reduced in cavefish. Our data indicate that long-term adaptation to low parasite diversity coincides with a more sensitive immune system in cavefish, which is accompanied by a reduction in the immune cells that play a role in mediating the pro-inflammatory response.
Important efforts in hygiene and medical treatment in most industrialized countries have reduced microbial and parasitic infections considerably1. While this indisputably improves health and increases life expectancy, the diverse effects on the immune system are not well understood. Host–parasite interactions are the major driving force in the evolution of the immune system2. From an evolutionary perspective, the maintenance and control of the immune system is associated with costs2,3 and, given the harm of a parasitic infection on the host, a reduced parasite diversity that results in a reduction of parasitic infections is generally associated with an increased fitness of the host4.
In recent years, however, the reduction of parasite diversity has also been associated with negative consequences for host fitness. Based on a number of studies, it has been hypothesized that a decreased parasite diversity or the loss of biodiversity in general has contributed to the rising numbers of autoimmune diseases in the developed world5–8. This phenomenon has been described as the ‘Old Friends hypothesis’9, which argues that the reactivity of the vertebrate immune system depends on exposure to macro-parasites (for example, helminths) and microparasites (for example, bacteria, fungi and viruses). The hypothesis argues further that host–parasite interactions are important for the host to develop a proper functional immune response since co-evolved parasites help the host to maintain and establish an optimal baseline immunity, which minimizes the risk of developing autoimmune reactions that potentially result in immunopathology (for example, type 1 diabetes or atherosclerosis)9.
Despite important insights into the physiological underpinnings of autoimmune diseases10, we still lack fundamental knowledge of how autoimmune diseases initially develop. Human populations have been confronted with this decreased parasite diversity for only a couple of generations—very recently in evolutionary terms. This raises the question of how the immune system adapts to such environmental changes in the long term. Given the major impact on fitness of autoimmune disorders10, evolutionary adaptations of the immune system to environments with low biodiversity—and thereby low parasite diversity11,12—are likely to have been deployed.
The vertebrate immune system is composed of two main systems, the innate and the adaptive immune systems. The former is essential for the initial response against pathogens. Given the short lifetime of innate immune cells (such as granulocytes) and their high complexity, the innate immune system is thought to be very costly for the host13. The adaptive immune system of vertebrates is defined by its long-term protection against pathogens (for example, through the production of pathogen-specific antibodies) and is thought to be less costly for the host since its cells, such as B and T cells, have low complexity and are known for their longevity13.
Given the differences in costs, it has been suggested that the vertebrate immune system is capable of adjusting its immune investment strategy14–16. The host can invest to different degrees in either innate or adaptive immune cells depending on parasite abundance in the host environment14–16. Accordingly, these different immune investment strategies result in specific differences in immune responses17. Based on these phenotypic responses, adaption to environments that differ in parasite abundance result in distinct fixed immune investment strategies that are optimized for host fitness in the given environment. To explore this idea, we used an eco-immunological approach in the Mexican tetra Astyanax mexicanus to study how local adaptation of one host species to environments with a stark difference in parasite diversity affects the immune system of the host.
There exist both cave- and surface-adapted populations of this species that have adapted to their respective environments for approximately 50,000–200,000 years18,19. One important hallmark of cave environments is an overall decrease in biodiversity, including parasite diversity20,21. Here we present field data from a cavefish population (Pachón) and one surface fish population (Río Choy) confirming a stark difference in macro-parasite abundance between these two habitats and indicating a higher immune activity of surface fish compared to cavefish under natural conditions. Both cavefish and surface fish populations can be bred and raised for generations in the laboratory under identical environmental conditions, which readily facilitates the identification of heritable changes. Therefore we used laboratory populations derived from wild-type Pachón and Río Choy populations, and an additional cavefish population (Tinaja), to investigate the immunological consequences deriving from adaptational processes to environments with low parasite diversity. We demonstrate that cavefish immune cells display a more sensitive and prolonged immune response of pro-inflammatory cytokines towards bacterial endotoxins in vitro, similar to other vertebrate host species in environments with low biodiversity22,23. Using an image-based immune cell-clustering approach (Image3C) and single-cell RNA sequencing (scRNA-seq), we show that the observed differences in cellular immune responses are accompanied by differences in immune investment strategy, where cavefish produce more lymphoid cells (adaptive immunity) than myeloid cells (innate immunity). We demonstrate that this altered immune investment strategy does not generally affect all lymphocytes, but mainly leads to an over-representation of T cells in cavefish.
Further scRNA-seq analysis of the acute inflammatory response in fish treated with lipopolysaccharides (LPS) revealed transcriptional changes in innate and adaptive immune cells, as well as in haematopoietic stem cells (HSCs), that may drive the observed changes in immune investment strategy. In addition, we observed differences in the adaptive response of T and B cells, where cavefish display a higher activation response than surface fish. Finally, we show that reduction of granulocytic and monocytic cells in cavefish leads to reduced immunopathological consequences for visceral fat storage, which has been described as an adaptational response towards low food supply in the cave environment24,25.
Results and discussion
Examining parasite abundance in wild cavefish populations.
We started our investigation by collecting A. mexicanus in their natural habitat to study the differences in parasite abundance between river and cave environments, which have not been studied in detail and are based mainly on assumptions that derive from theoretical models11. We collected 15 surface fish (Río Choy) and 15 cavefish (Pachón), respectively (Fig. 1a), and examined them for parasitic infections as described before16. We found varying numbers of endo- and ectoparasites in surface fish (Fig. 1b; see also Supplementary Fig. 1 for mean numbers of parasite per fish, Supplementary Fig. 2 for a gallery of parasites found in surface fish and Supplementary Table 1 for prevalence and mean intensity of parasite classes found in surface fish). Interestingly, we did not detect infection with macro-parasites in the sampled cavefish (Fig. 1b). While we cannot exclude the possibility of viral or bacterial infections in the cavefish population, we did not detect any obvious signs of systemic or tissue-specific infections during the parasitological examination. However, although unlikely since we did not test the water bodies in the river and cave environment for potential fish parasites, there is the possibility that cavefish might have cleared or resisted potential parasitic infections. Here, future studies are needed to obtain more precise description of the fauna in these habitats. The lower parasitic infection rate in wild cavefish, however, is also reflected in a significantly lower spleen somatic index (an elevated immune activity in fish coincides with a swelling of the spleen and increases spleen somatic index26) in wild cavefish samples compared to surface fish (Fig. 1c; mean spleen somatic index of 0.663 and 0.304 in surface fish and cavefish, respectively (P = 0.0047)). In addition, we used a published transcriptome dataset from liver tissue isolated from three wild Pachón cavefish and Río Choy surface fish27, respectively, to compare differences in immune status. Given the high abundance of immune cells, the liver can be considered as an immunological organ in teleost fish and the expression of immune-related genes can be used to quantify infection status28. We used the available datasets from this study27 to compare the immune status in the wild fish. Gene Ontology (GO) enrichment analysis of downregulated genes when comparing wild Pachón with wild Rio Choy revealed a significant enrichment of genes in GO terms, including ‘regulation of cytokine production involved in immune response’, ‘positive regulation of production of molecular mediator of immune response’ and ‘positive regulation of immune response’ (see Supplementary Data 1). Here it is noteworthy that this was not observed when comparing laboratory strains from the same populations, indicating that the wild surface fish population is confronted with higher parasitic infections than is the wild cavefish population (see Supplementary Data 1).
Fig. 1 |. Adaptation to river and cave habitats with marked differences in parasite diversity results in changes of cellular immune response.
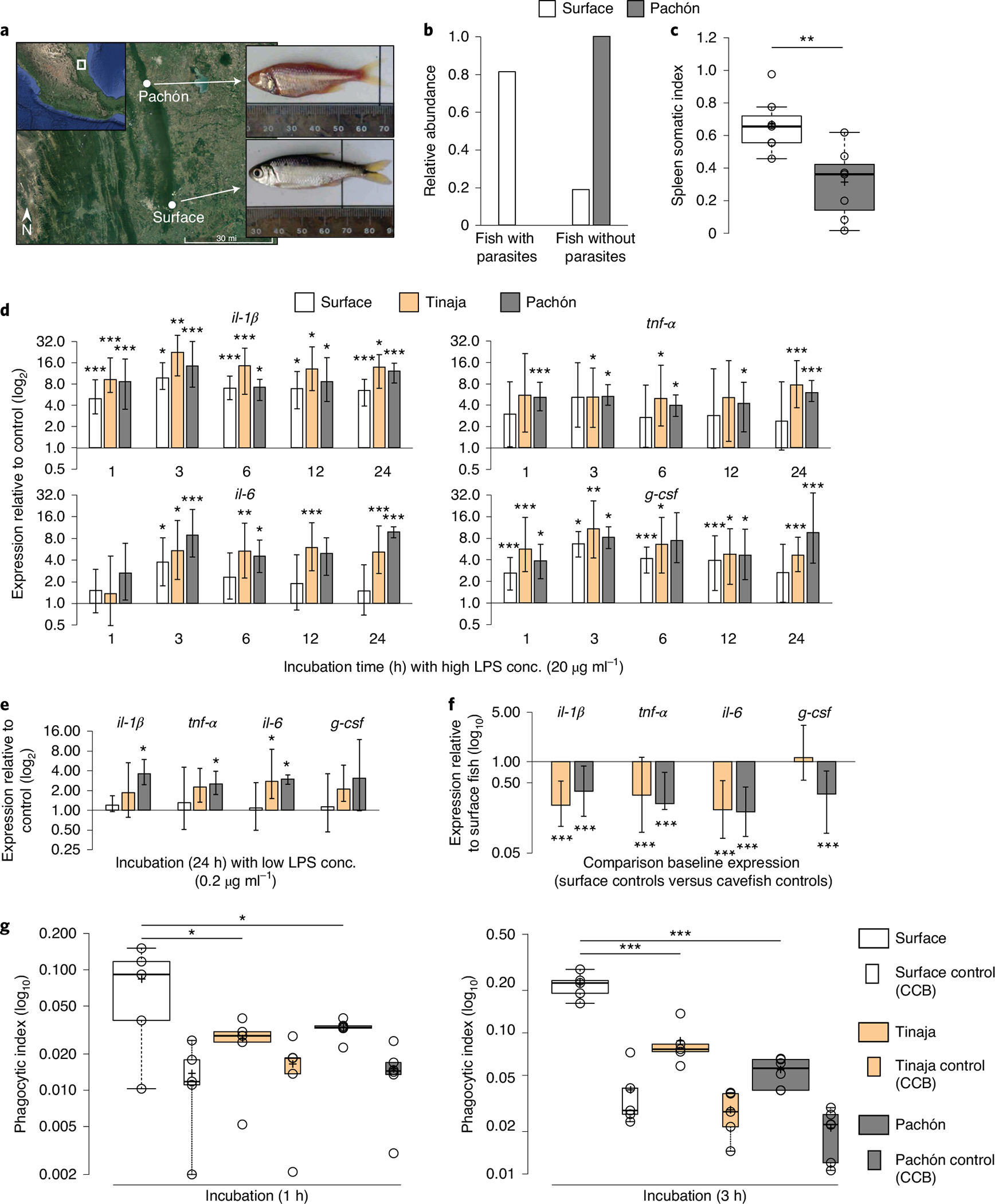
a, Collection sites of A. mexicanus surface fish (Río Choy) and cavefish (Pachón). Credit: Google Earth 2019 Landsat/Copernicus (map source); Google 2019, INEGI (map data). b, Number of fish with and without visible ecto- and endoparasites. c, Immune activity in wild surface and cavefish (each n = 7) using spleen somatic index: (weight (mg) (spleen)/weight (mg) (fish)) × 1,000. Significances were determined by one-way ANOVA. d,e, RT–qPCR analysis of pro-inflammatory cytokines, il-1β, tnf-α, il-6 and g-csf of HK cells from surface fish and cavefish after incubation with 20 μg ml−1 LPS at various time points (d) or 0.2 μg ml−1 LPS after 24 h (e) relative to HK cells incubated with PBS for the given time point. Plotted is the mean of three independent experiments with s.e.m. PBS control samples from each time point and sample were used as the reference to calculate relative expression of target genes for each time point and fish, respectively. f, RT–qPCR-based expression analysis of pro-inflammatory cytokines il-1β, tnf-α, il-6 and g-csf of cavefish relative to surface fish of naïve HK samples across all time points as shown in d (n = 18, error bar indicates s.e.m.). Significance values were determined by pairwise fixed reallocation randomization test using REST2009 software71. g, Box plot presentation of relative phagocytic rate of HK cells from surface fish and cavefish incubated with Alexa-488-coupled S. aureus. Control samples (HK cells incubated with Alexa-488-coupled S. aureus + 80 μg CCB) are presented in boxes. Significant differences between surface fish (n = 5), Tinaja (n = 5) and Pachón (n = 6) for each time point were determined by two-way ANOVA (see Supplementary Data 2 for statistical details). For all box plots, centre lines show the medians, crosses show means, box limits indicate the 25th and 75th percentiles as determined by R software80, whiskers extend 1.5× the interquartile range from the 25th and 75th percentiles and data points are represented by circles.
Examining cellular immune responses of laboratory cavefish population.
Given the strong impact of host–parasite interaction on the evolution of the immune system2, we speculated that these extreme differences in parasite abundance between cavefish and surface fish environment would result in functional and/or physiological changes to the cavefish immune system. To investigate these potential differences under controlled environmental conditions, we used laboratory strains that originated from wild populations (Pachón and Río Choy) for all subsequent experiments. To test whether functional differences of the immune system also appear in other cavefish populations we included, where feasible, a second, independently derived, laboratory cavefish population (Tinaja) in the experimental set-up. It is noteworthy that while we obtained parasite data from only one cavefish population (Pachón), we reasoned that different cave habitats share similar environmental features specifically in terms of biodiversity (and therefore parasite biodiversity11) given the absence of sunlight as a source of primary energy production20.
To trigger a cellular pro-inflammatory immune response we used bacterial endotoxins, LPS, in cultures with extracted leukocytes. We focused on the pronephros (head kidney, HK) (see Extended Data Fig. 1), the main haematopoietic and lymphoid organ and a site of antigen representation in teleost fish29. We incubated HK cells from surface, Pachón and Tinaja fish with LPS (20 μg ml−1) for 1, 3, 6, 12 and 24 h and measured gene expression of pro-inflammatory cytokines interleukin-1-beta (il-1β), tumour necrosis factor alpha (tnf-α), interleukin-6 (il-6) and granulocyte colony-stimulating factor (g-csf) in relation to control samples (saline (PBS)) using quantitative PCR with reverse transcription (RT–qPCR) (Fig. 1d; see Methods and Supplementary Table 2 for details). HK cells from cavefish populations showed an overall greater inducible response following LPS treatment than HK cells from surface fish in vitro over time (Fig. 1d). Specifically, the gene expression of only il-1β remained significantly elevated in surface fish after 24 h (Fig. 1d). In contrast, cavefish expression of all pro-inflammatory cytokines tested remained significantly upregulated after 24 h (Fig. 1d). Since the cavefish response was saturated at this LPS concentration, we repeated the analysis with 100-fold lower LPS exposure (Fig. 1e). Here, LPS-treated HK cells from surface fish no longer displayed a significant response of any pro-inflammatory cytokine while Pachón cavefish cells still showed significant expression for il-1β, tnf-α and il-6 compared to untreated cells (Fig. 1e). This increased sensitivity was not present to the same degree in the Tinaja cave population, since we found an increase in the expression of il-6 only (Fig. 1e). This increased sensitivity of cavefish HK cells towards LPS in vitro is supported by previous findings of an increased immune and scarring response after wounding of Pachón cavefish compared to surface fish30.
Given that the HK is composed of many different cell types, no direct comparison was made of the response following LPS stimulation between different A. mexicanus populations, since it is known that cell-averaged transcriptional responses of heterogenous cell populations can be highly misleading when compared with one another31. To account for the possibility that there might be differences in the number of cells expressing specific cytokines, we directly compared baseline expression of il-1β, tnf-α, il-6 and g-csf in naïve HK cells of Pachón and Tinaja to surface fish (Fig. 1f). Surprisingly, the expression of all pro-inflammatory cytokines tested was significantly reduced in Pachón cavefish samples relative to surface fish cells (Fig. 1f). In the case of the pro-inflammatory cytokine il-1β, for example, naïve Pachón cavefish HK cells produced 61% less transcript than surface fish cells (relative expression, Pachón versus surface fish il-1β = 0.383, P < 0.001; Fig. 1f). Like the Pachón cavefish population, the Tinaja cave population differed in the expression of il-1β, tnf-α and il-6 compared to surface fish but not in the expression of g-csf (Fig. 1f).
In addition, we conducted a phagocytosis experiment in which we quantified the ability of HK cells from cavefish (Pachón and Tinaja) and surface fish to phagocytize Alexa-488-tagged Staphylococcus aureus cells in vitro at different time points (Fig. 1g; see Supplementary Fig. 3 for gating strategy). Using pairwise comparison, we found a significant decrease in phagocytic rate of both cavefish populations at both time points compared to surface fish (Fig. 1g; see Supplementary Data 2 for detailed statistics report).
Immune cell composition analysis reveals differences in immune investment strategy between cavefish and surface fish.
The decreased baseline expression of pro-inflammatory cytokines and decreased phagocytosis rate in cavefish could have been the result of changes in immune cell composition, since both of these cellular immune functions are mainly fulfilled by cells with a myelomonocytic origin (such as granulocytes and monocytes) in teleost fish32–34. To assess immune cell composition, we analysed scatter information from HK-derived single-cell suspensions from surface, Tinaja and Pachón fish. Using similar analytical approaches previously described for zebrafish33,35, we identified four distinct cell clusters: an erythroid, a myelomonocyte, a progenitor and a lymphoid/progenitor (Fig. 2a). These cells were stained with May–Grünwald Giemsa stain and, based on comparative morphological analysis29,33,35–38, we identified (1) erythrocytes, (2) promyelocytes, (3) eosinophils, (4) neutrophils, (5) monocytes, (6) macrophages, (7) erythroblasts, (8) myeloblasts, (9) erythroid progenitors, (10) lymphocytes and (11) undifferentiated progenitors (that is, haematopoietic stem cells, common lymphoid progenitors and common myeloid progenitors) within the four cell clusters (Fig. 2a). When we compared relative abundances of the three immune cell clusters, we identified fewer myelomonocytic cells in both cavefish populations (mean relative abundance of cells in myelomonocyte cluster in surface fish, 0.37 versus 0.32 in Tinaja (P < 0.05), versus 0.24 in Pachón (P < 0.01; Fig. 2b) and an increased number of cells in the lymphocyte/progenitor cluster (mean relative abundance of cells in lymphocyte/progenitor cluster in surface fish, 0.36 versus 0.44 in Tinaja (P < 0.05) and versus 0.53 in Pachón (P < 0.05; Fig. 2b). We used these relative abundances to calculate the myelomonocyte/lymphocyte (M/L) ratio. This ratio is an indicator of an individual’s relative investment in either innate (myelomonocyte) or adaptive (lymphocyte) immune cell populations13,39. While surface fish have a relatively balanced investment in myelomonocyte and lymphoid immune cells, cavefish invest less into myelomonocytic cells than into lymphoid immune cell populations (mean M/L ratio in surface fish, 1.06 versus 0.72 in Tinaja (P < 0.05) and versus 0.50 in Pachón (P < 0.01; Fig. 2c). Since all fish were raised under identical laboratory conditions, the observed differences in immune investment strategy point towards a genetic basis for this trait. To further study this, we analysed surface × Pachón hybrids (F1 hybrid embryos were pooled from a minimum of five families of surface females and cavefish males) and found a M/L ratio similar to that in the parental Pachón population, indicating that the change in immune investment strategy of the Pachón population is a dominant trait (mean M/L ratio in surface × Pachón F1, 0.52; Fig. 2c).
Fig. 2 |. Cellular composition analysis of HK reveals differences in immune investment strategy between cavefish and surface fish.
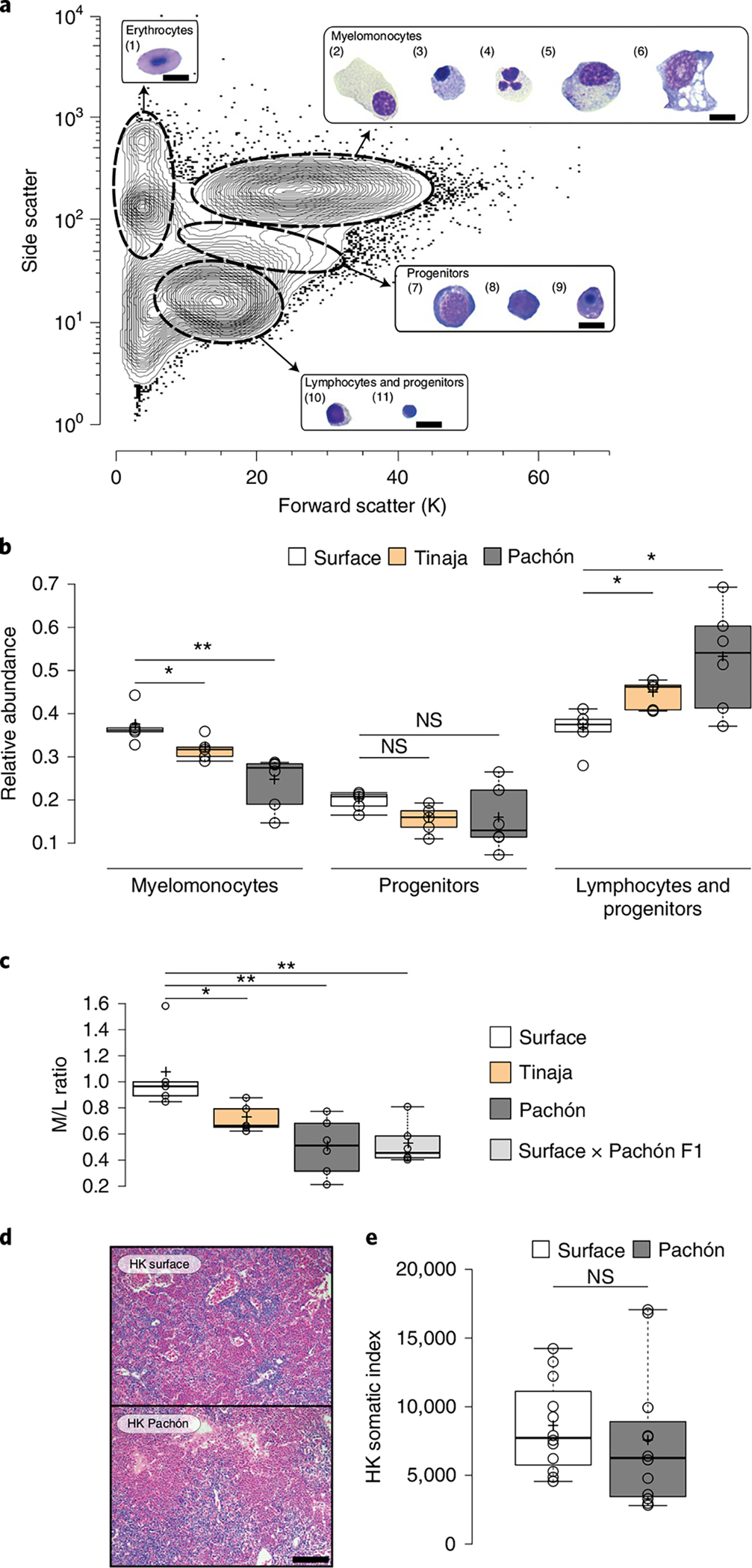
a, Representative contour plot of HK cells from three surface A. mexicanus after FACS analysis using scatter characteristics showing 99% of all events. Four different cell populations (erythrocytes, myelomonocytes, progenitors and lymphocytes/progenitors) were identified and sorted for May–Grünwald Giemsa staining. Images of representative cells found in each population are shown and identified based on similar approaches in zebrafish33,35 as (1) mature erythrocytes, (2) promyelocytes, (3) eosinophils, (4) neutrophils, (5) monocytes, (6) macrophages, (7) erythroblasts, (8) myeloblasts, (9) erythroid progenitors, (10) lymphocytes and (11) undifferentiated progenitors. Scale bars, 10 μm. b, Box plots of relative abundances of HK cells from surface and cavefish within the immune populations as defined by scatter characteristics in a. Significances between surface (n = 5), Tinaja (n = 5) and Pachón (n = 6) were determined by one-way ANOVA and subsequent FDR. c, Box plot representation of M/L ratio from surface (n = 5), Tinaja (n = 5), Pachón (n = 6) and surface × Pachón F1 hybrids (n = 6). Significances were determined by one-way ANOVA and subsequent FDR. d, Haematoxylin and eosin (H & E)-stained section of HK from surface fish and Pachón cavefish (scale bar, 100 μm). e, HK somatic index (mean number of HK cells per body weight (mg) fish) is shown for n = 12 surface fish and Pachón cavefish, respectively. Testing for significant differences was done using Mann–Whitney U-test. For all box plots, centre lines show medians, crosses show means, box limits indicate the 25th and 75th percentiles as determined by R software80, whiskers extend 1.5× interquartile range from the 25th and 75th percentiles and data points are represented by circles. *P < 0.05, **P < 0.01 for all experiments. NS, not significant.
Given the strong difference in parasite abundance and resource availibillity24 between cave and surface environments, cavefish potentially benefited from a change in the immune investment that reduces resource allocation to the immune system (see ref. 13 for a review on costs of innate and adaptive immune defences). To rule out the possibility that changes in HK morphology and/or total numbers of cells in the HK are responsible (and could potentially compensate) for observed differences in the M/L ratio, we compared HK morphology and total cell numbers from surface fish and Pachón cavefish. We observed no general differences in tissue morphology (see Fig. 2d) and no significant changes in absolute cell number from the entire HK between fish populations (median HK somatic index for surface fish and cavefish, 7,728 and 6,262, respectively (P = 0.265; Fig. 2e).
To identify more specific differences in the immune cell composition of laboratory cavefish and surface fish, we clustered HK cells based on cell morphological features using Image3C40. This tool uses image-based flow cytometry and advanced clustering algorithms to cluster cells based on their morphology and cellular features such as granularity of the cytoplasm or nucleus, independently of an observer bias that has been reported for such analysis41. This makes it an effective method for use in organisms lacking established transgenic lines or antibodies to identify specific immune cell populations.
Using Image3C we identified 21 distinct cell clusters (Fig. 3a,b). The identity of each cluster was determined based on cell image galleries from each cluster (see Supplementary Data 3 for complete cell gallery) in comparison to the histological staining of sorted cells, as presented in Fig. 2a and comparative morphological analysis29,33,35–38 (for details see Methods). In addition, to verify certain cellular features (for example, complexity of nuclei and cell shape, see Supplementary Table 3 for feature details) within a certain cluster, we used a feature intensity/cluster correlation analysis (Supplementary Fig. 4). In line with the scatter analysis, we found a significant reduction of cells within the myelomonocyte category in cavefish compared to surface fish (mean relative abundance, 0.468 and 0.320 cells, respectively (P < 0.001; Fig. 3c).
Fig. 3 |. Cell composition analysis of A. mexicanus HK surface and cave morphotypes using cell morphological and genetic features.
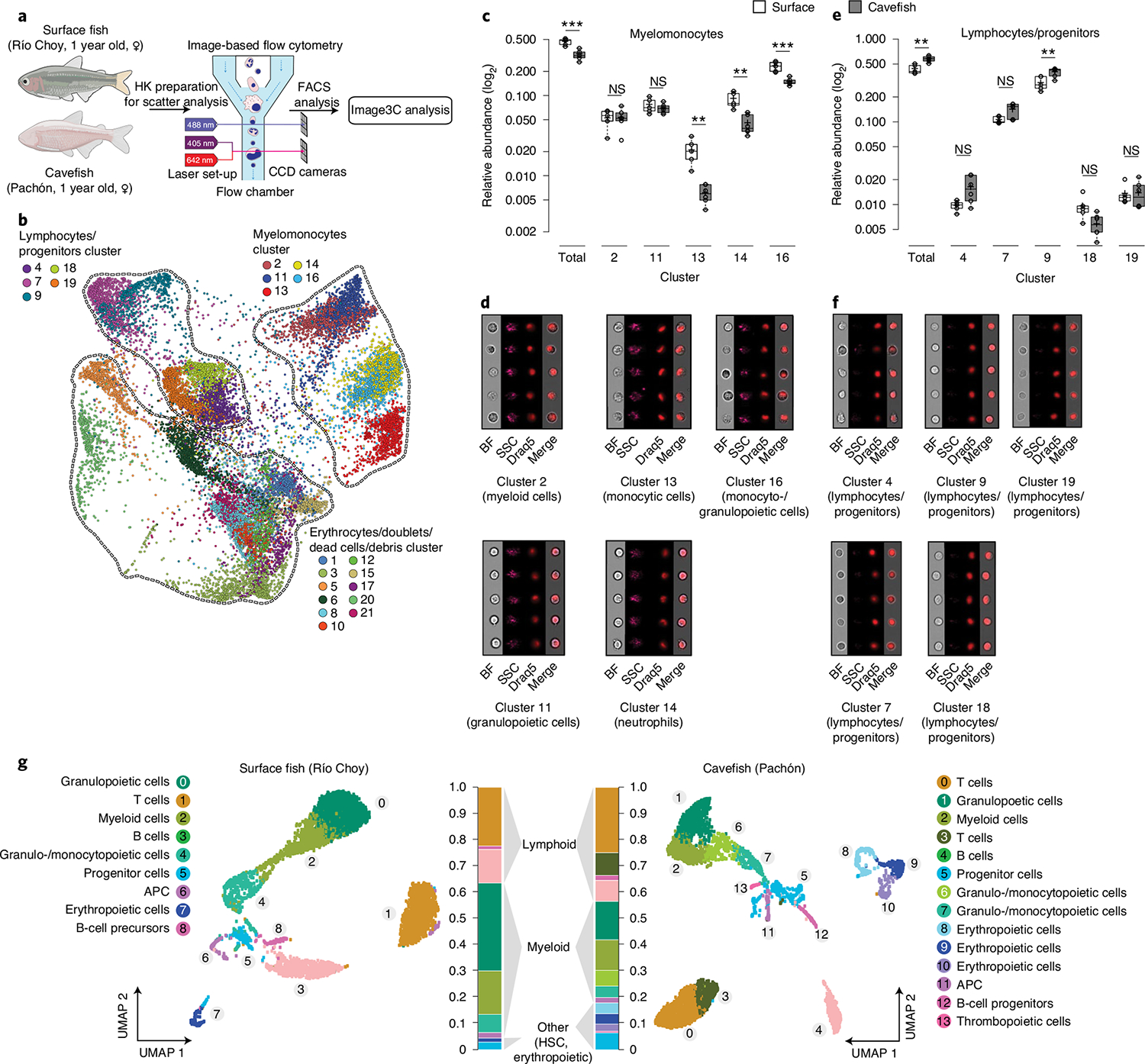
a, Experimental set-up for semi-unsupervised cell morphological clustering using the Image3C pipeline. CCD, charge-coupled device. b, Force-directed layout graph of cell clusters based on morphological feature intensities from HK cells of surface fish (n = 5) and Pachón cavefish (n = 6). Each dot represents one cell and each colour represents a unique cluster. Clusters were combined into three categories (myelomonocytes, lymphocytes/progenitors and mature erythrocytes/doublets/debris) based on their morphology (see Supplementary Data 3 for cell galleries for each cluster). c, Relative abundances of cells within each cluster of the myelomonocyte category. d, Image galleries of each cluster. e, Relative abundances of cells within each cluster of lymphocyte/progenitor category. f, Image galleries of each cluster. Significant differences in the relative abundance of cells within each category (total) or cluster were determined by one-way ANOVA and subsequent FDR. **P < 0.01, ***P < 0.001. In box plots, centre lines show the medians, crosses show means, box limits indicate the 25th and 75th percentiles as determined by R software, whiskers extend 1.5× interquartile range from the 25th and 75th percentiles and data points are represented by circles. Cell galleries show images of brightfield (BF), side scatter (SSC), nuclei (visualized by nuclear dye DRAQ5) and merged images of BF and DRAQ5 (Merge). g, UMAP plot of scRNA sequencing analysis of HK from one 1-year-old female surface fish (Río Choy) and an age-, sex- and weight-matched cavefish (Pachón), where each dot represents one cell and each colour represents a unique cell cluster as shown in the legend. Overall relative abundances are given for each cluster shown in the respective colours. See Extended Data Fig. 2 for expression of marker genes for specific cell clusters and their relative abundances. APC, antigen-presenting cells.
More specifically, we identified differences in the relative abundance of monocytic cells between surface fish and cavefish (Fig. 3c,d; cluster 13: mean relative abundance of 0.021 cells in surface fish versus 0.006 cells in Pachón cavefish; P < 0.01); of neutrophils (Fig. 3c,d; cluster 14: mean relative abundance of 0.088 cells in surface fish versus 0.044 cells in Pachón cavefish; P < 0.01); and monocytic, granulocytic and promyelocytic cells (Fig. 3c,d; cluster 16: mean relative abundance of 0.231 cells in surface fish versus 0.148 cells in Pachón cavefish; P < 0.01).
A reduction in almost all myeloid cell populations suggests that there is an overall reduced investment in the innate immune system in Pachón cavefish. The resulting reduction of granulocytes and monocytes in cavefish HK is in line with the observed decreased baseline expression of pro-inflammatory cytokines and phagocytic rate. Furthermore, we found that cells within the lymphocyte/progenitor category (Fig. 3e,f) are generally over-represented in cavefish when compared to surface fish (mean relative abundance of cells within lymphocytes/progenitor category: surface fish 0.433 versus cavefish 0.580; P < 0.01; Fig. 3e). The majority of clustering in the lymphocyte/progenitor category did not differ significantly between surface fish and cavefish, with the exception of cluster 9 (surface fish 0.295 versus cavefish 0.399; P < 0.01; Fig. 3e,f), which is the most abundant cluster in this category.
Here it is noteworthy that the M/L ratio we obtained for surface and cavefish with the Image3C approach is similar to that we obtained previously (Fig. 2c) using standard scatter information (M/L ratio surface fish 1.10 versus 0.56 in cavefish, P < 0.001; Supplementary Fig. 5). However, given the morphological similarities (see Supplementary Data 3) of early progenitor cells from haematopoietic lineages and specific lymphocyte cell types (B cells, T cells), we were unable to further resolve the identity of these clusters. Therefore, we took a genetic approach.
We performed scRNA sequencing (scRNA-seq) of HK cells. We used one female adult (1 year old) surface fish (Río Choy) and one age-, size- and sex-matched cavefish (Pachón) from our laboratory populations (Fig. 3g; see Methods for details). Cluster identification was done using a comparative approach with gene expression data from other teleost fish species (for details see Methods and Supplementary Data 4 for overall gene enrichment in each cluster). Consistent with morphological analyses, we found an overall reduction in all cells of myeloid linage in cavefish compared to surface fish. In more detail, cluster analysis revealed a reduction in myeloid cells (relative abundance of spi1b (pu.1) + mpx cells in surface fish, 0.221 versus 0.132 in cavefish; Extended Data Fig. 2a) and granulo- and monocytopoietic cells (relative abundance of ptprc + cebp1 + lyz + mpx cells in surface fish, 0.167 versus 0.100 in cavefish; Extended Data Fig. 2a) in cavefish. Furthermore, we verified a reduction in mature neutrophils (relative abundance of ptprc (cd45) + cebp1 + mmp9 cells in surface fish, 0.0586 versus 0.0191 in cavefish; Extended Data Fig. 2a) and monocytes (relative abundance of ptprc (cd45) + csf3r + cd74a cells in surface fish, 0.0165 versus 0.010 in cavefish; Extended Data Fig. 2a) in cavefish. Importantly, analysis of lymphoid cell linage revealed that there are distinct differences in specific lymphoid cell populations between cavefish and surface fish, and no overall increase in lymphoid cells in cavefish as morphological analysis might suggest (Extended Data Fig. 2b).
While we found an over-representation in the relative abundance of lymphocytes in cavefish compared to surface fish (Fig. 3g), we found almost identical relative abundances of B lymphocytes (relative abundance of igkc + cd74a cells in surface fish 0.119 versus 0.098 in cavefish; Extended Data Fig. 2b). In contrast, we found clear differences in the numbers of HK resident T cells (relative abundance of cd3e cells in surface fish, 0.136 versus 0.265 in cavefish; Extended Data Fig. 2b). We identified increased numbers of CD4+ T cells in cavefish (relative abundances of cd3e + tcrα + tcrβ + cd4–1 cells in surface fish, 0.002 versus 0.031 in cavefish; Extended Data Fig. 2b). Interestingly, we observed that γ+δ+CD4− CD8− (γδ) T cells reside in higher proportions in the HK of cavefish than in surface fish (relative abundances of cd3e + tcrγ cells in surface fish, 0.007 versus 0.028 in cavefish; Extended Data Fig. 2b). γδ T cells are a lymphoid cell population that potentially functions as a bridge between the innate and adaptive immune system, due to its ability to recognize antigens in a major histocompatibility complex-independent manner42, and has been discovered only recently in other teleost species43. Furthermore, γδ T cells are reported to play a significant role in the development of autoimmune diseases44, and in homeostasis and inflammation of mammalian adipose tissue45. Through their ability to directly recruit myeloid cells that drive inflammation and activation of other T-cell populations, γδ T cells have been described as being critical in the early stages of inflammation and autoimmune diseases46. In contrast to this, γδ T cells have been described as protecting tissue (such as intestinal epithelial cells) by regulation of tissue homeostasis, tissue repair and release of anti-inflammatory cytokines that suppress inflammation47. In zebrafish, γδ T cells are described as potential phagocytic cells that participate in full activation of the systematic adaptive humoral immunity43.
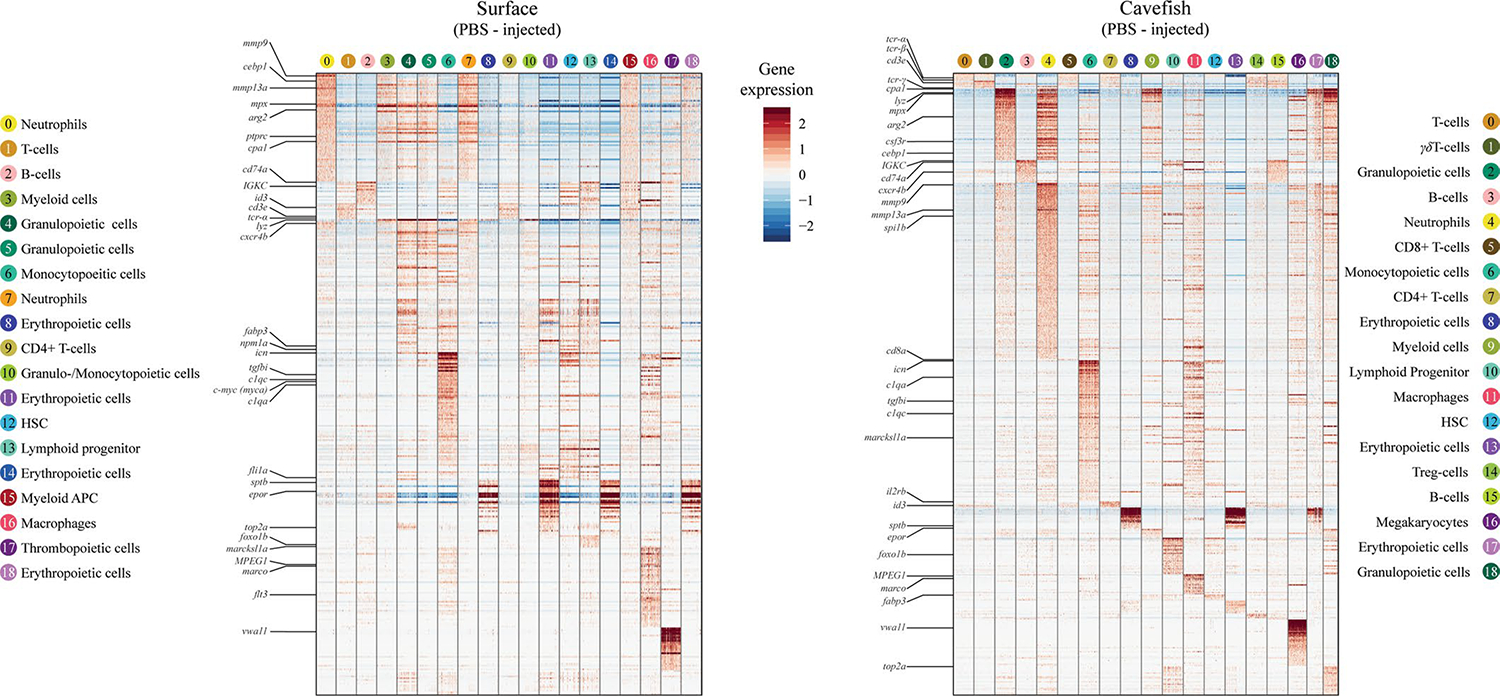
Changes in the immune investment strategy of cavefish suggest that the inflammatory response of Pachón cavefish could be affected, since numbers of cells that drive pro-inflammatory responses (monocytes and neutrophils) are decreased while cells that can potentially affect tissue homeostasis (γδ T cells) are increased in Pachón cavefish. In addition, the dominance of the Pachón cavefish immune investment phenotype suggests that there are genetic differences driving these changes. To address these questions, we designed another scRNA-seq experiment where we injected Pachón cavefish and Río Choy surface fish with either LPS or PBS (Fig. 4a). We found higher numbers of myeloid cells in both treatment groups of surface fish, which also resulted in higher numbers of granulopoietic (granulocytes and their precursor cells) and monocytopoietic (monocytes and their precursor cells) cell clusters (Fig. 4b). We were not able to identify a specific cluster of eosinophils, but this is mainly due to the lack of a suitable genetic marker for this cell type. In contrast to the increased numbers of myeloid cells in surface fish, we found an increased number of T cells in both treatment groups of cavefish, similar to the previous experiment (see Figs. 4b and 3g). Based on gene expression profiles, we found naïve T cells, CD8+ T cells, Treg cells, CD4+ T cells and γδ T cells in cavefish while we mainly found naïve and CD4+ T cells in surface fish (Fig. 4b and Extended Data Fig. 3). Even though we found T cells with the same identity in surface fish, the low abundance of these T cell populations in HK probably prevented their clustering into a unique T-cell cluster in surface fish. Differences in the abundance of T cells underline differences in the immune investment strategy between surface fish and Pachón cavefish, where the former invest more into innate immune cells and the latter more into adaptive immune cells, specifically T cells.
Fig. 4 |. Cellular analysis of acute inflammatory response following LPS injection of A. mexicanus HK cells.
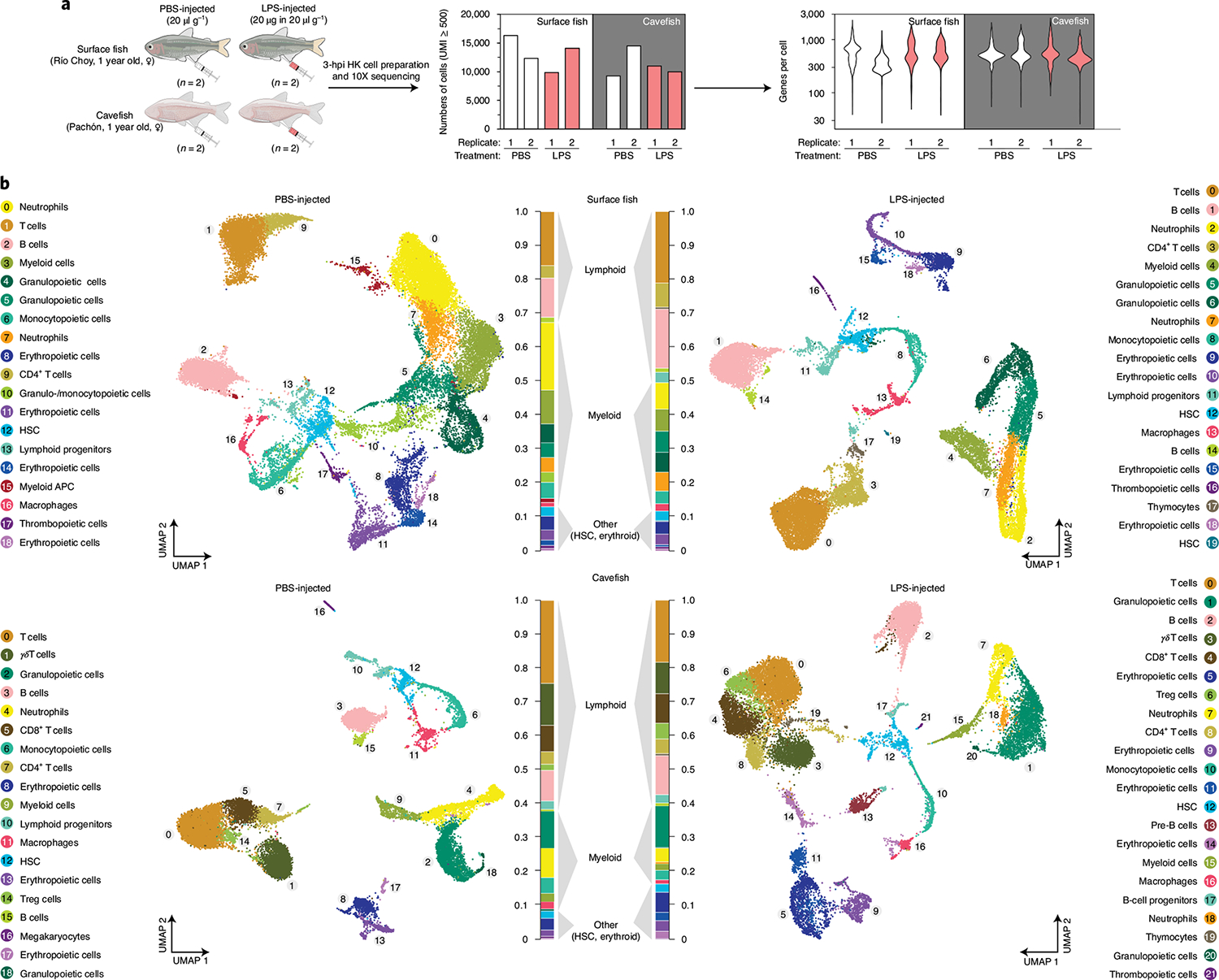
a, Experimental set-up of the scRNA-seq experiment using HK from surface fish (Río Choy) and cavefish (Pachón) 3 h postinjection (3 hpi) of either PBS or an LPS mix in the concentrations shown. After processing (for example, removal of mature erythrocytes; see Methods for details), cells were sequenced using 10X Genomics technology (see Methods for details), which resulted in given cell numbers and average genes per cell. b, UMAP projection of PBS- and LPS-injected surface and cavefish. Cell clusters are based on gene enrichment analysis for each cluster (Supplementary Data 5).
Transcriptional changes in haematopoietic stem cells accompany differences in immune investment strategies.
We speculated that changes in the immune investment strategy might be driven by genetic changes that could affect the development, maturation and/or maintenance of HSCs. While we found no changes in the relative abundance of HSCs across all samples (0.02–0.03; see Fig. 4b), we found distinct changes in the transcriptional profiles of HSCs between surface fish and cavefish control samples that could affect lineage fate decision, the level of quiescence and self-renewal capacity (for a complete list of differential expressed genes in HSCs, see Supplementary Data 10). For example, we found that cavefish cells show increased expression of bcl3 (log2fold change (FC) = 3.87), a marker for lymphoid progenitor cells48, and decreased expression of both npm1a (log2FC = −1.22) and mpx (log2FC = −2.22), which are both key markers for early myeloid progenitor cells49,50. While further genetical analysis is needed to verify the genetic shift towards lymphoid progenitor cells in cavefish HSCs, the transcriptional changes suggest that the different immune investment strategies in A. mexicanus could indeed be affected by genetic changes in cavefish HSCs.
In addition, we found transcriptional changes that indicate differences in self-renewal capacity and quiescence of HSCs between surface fish and cavefish. We detected increased expression of c-myc (myca, log2FC = 1.66) in cavefish HSC cells, a positive regulator of HSC quiescence and self-renewal51. Furthermore, we found genes such as fscna (log2FC = 3.056) and pim1 (log2FC = 3.29)—genetic markers for LT (long-term)-HSC—significantly increased and genetic markers—such as top2a (log2FC = −4.72), kif2c (log2FC = −4.04) and kif4 (log2FC = −3.05)—for multipotent progenitor cells significantly decreased in cavefish control samples compared to surface fish controls52. Interestingly, we found no expression of cd34, a common marker for mature HSCs and progenitor cells53,54, in any of the Pachón HSC cells while we found expression of this marker in HSCs of surface fish samples. Cd34− stem cells have been described as immature and quiescent, with lower self-renewal capacity54. These findings point towards an increased proportion of immature LT-HSCs in a more quiescent state in Pachón cavefish compared to surface fish.
Differences in immune investment strategy lead to changes in Th1 response.
The expression of interferon-γ (ifng) of activated T cells and natural killer (NK) cells is a well-established response following bacterial or viral infection. Although Pachón cavefish possess a high abundance of T cells, we found an increased abundance of ifng-expressing cells in surface fish following LPS injection (Fig. 5a,b; relative abundance of ifn-γ-expressing cells in surface fish, 0.041 versus 0.016 in cavefish).
Fig. 5 |. Adaptive immune response in HK and spleen of surface and cavefish A. mexicanus following LPS injection.
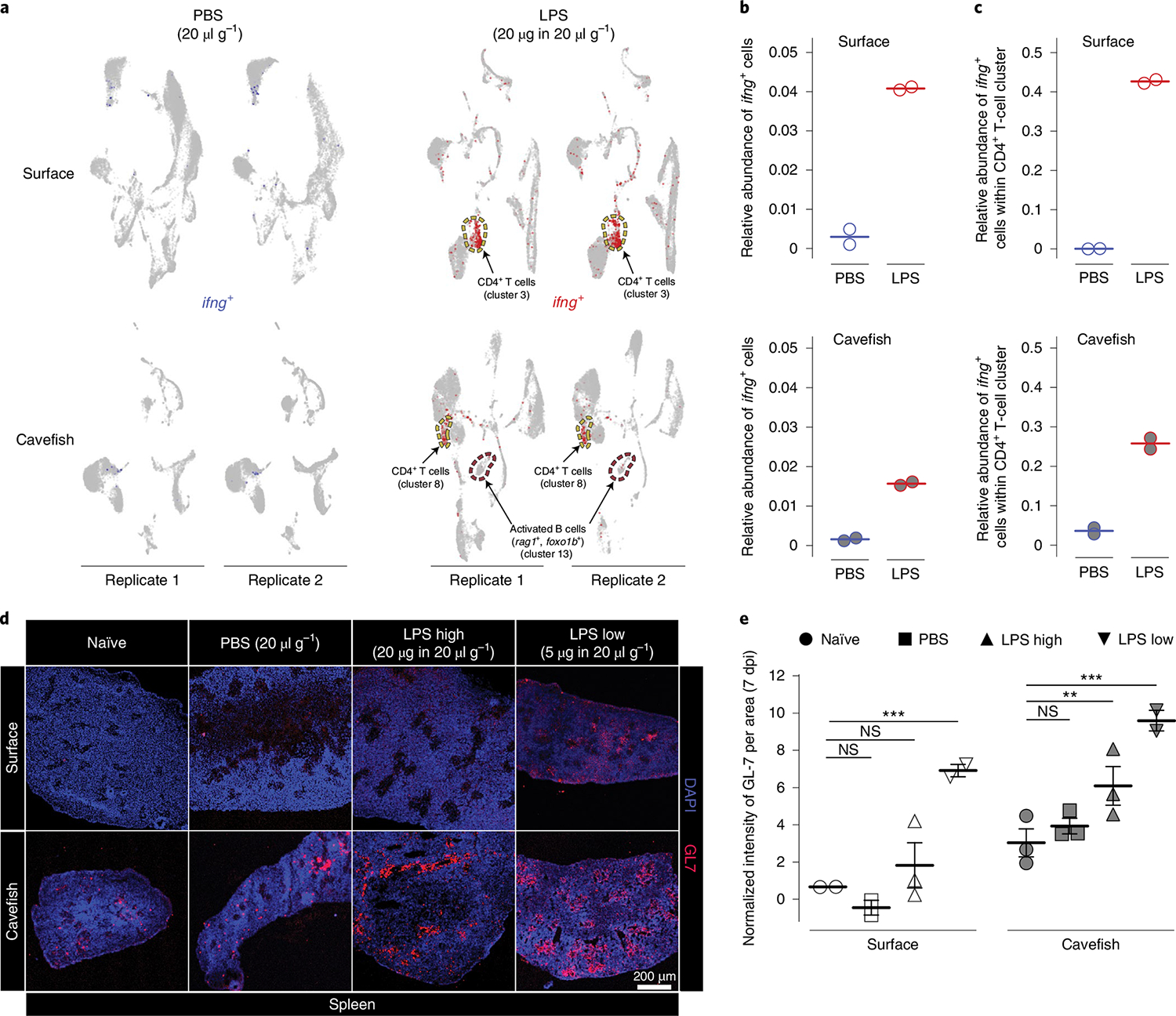
a, ifng expression 3 h following PBS (blue) or LPS (red) injection in HK cells. Following LPS injection, an activated B-cell cluster emerged in cavefish expressing marker genes foxo1b and rag1. This cluster (cluster 13 in LPS-injected cavefish) is absent in all other treatment groups. b, Overall relative abundances of ifng-expressing cells from scRNA-seq experiments for each treatment group. c, Relative abundances of ifng-expressing cells from scRNA-seq experiments with CD4+ T cells for each treatment group. d, Antibody staining (rat anti-GL-7 Alexa Fluor 647 of activated B and T cells in spleen 7 dpi of either PBS (20 μl g−1 fish bodyweight), a high dose of LPS (LPS high; 20 μg in 20 μl g−1 fish bodyweight) or a low dose of LPS (LPS low; 5 μg in 20 μl g−1 fish bodyweight) or left naïve (not injected) as a control group. e, Results of intensity analysis from d for 7 dpi. Images were analysed from the four treatment groups at two time points (7 and 14 dpi; for 14-dpi analysis results, see Supplementary Fig. 6), with n = 2–3 per treatment × population × time point sample. GL-7 signal was quantified per area as defined by the DAPI signal using Fiji (see Methods for details), and intensities were normalized using the respective isotype control (see Methods for details). Empty symbols represent surface fish, and filled symbols represent cavefish. Two-way ANOVA and subsequent multiple testing with FDR correction (Benjamini–Hochberg81) was used to detect differences between injected and control groups (for a complete statistical report see Supplementary Data 12). **P < 0.01, ***P < 0.001. NS, not significant.
While we did not detect a specific NK cluster in any of the treatment groups, we observed that mainly CD4+ cells express ifng in the acute pro-inflammatory response following LPS injection (for a complete list of differential expressed genes in the CD4+ T-cell cluster, see Supplementary Data 11). We also compared the relative abundance of ifng-expressing cells within the CD4+ T-cell cluster and, again, found an increased abundance of ifng-expressing cells in surface fish (Fig. 5c; relative abundance of ifng-expressing cells in the CD4+ T-cell cluster in surface fish, 0.43 versus 0.26 in cavefish). This decreased inflammatory response of CD4+ T cells (Th1 response) in cavefish suggests that cavefish lymphoid cells possess a different mode of response to that of surface fish following bacterial recognition. We noticed a B-cell population in LPS-injected cavefish that is absent in cavefish PBS-treated groups and in both surface fish treatment groups (Figs. 4b and 5a). The main characteristic of this B-cell population is the expression of foxo1b, rag1 (Fig. 5a) and, to a lesser extent, rag-2, which is characteristic of activated B cells55,56. Given that HK is the primary lymphoid organ in teleost fish29 and that expression of foxo1b is also a marker for developing B cells55, this B-cell population could also present a new emerging B cell following LPS exposure in cavefish. The presence of this B-cell population in Pachón LPS-treated groups and its absence in all other treatment groups, however, points towards differences in the activation of the adaptive immune response between cavefish and surface fish.
Based on this, we hypothesized that Pachón cavefish display increased activation of B and T cells in lymphoid organs, such as the spleen, after injection with LPS compared to surface fish. To visualize activated B and T cells we used an antibody against the GL-7 antigen that specifically stains activated B and T cells in the spleen germinal centre (GC) of mammals57,58. Although GCs have not been described in the spleen of teleost fish, related structures, the so-called melanomacrophage centre, have been reported as having similar functions (for example, antigen processing) to mammalian GCs59,60. In surface fish, we found a significant increase in only the GL-7 signal in the ‘LPS low’ group compared to the naïve group at 7 d postinjection (dpi) (P < 0.0001; Fig. 5d,e; see Methods for details and Supplementary Data 12 for detailed statistical analysis). For cavefish, however, we found a significant increase in the GL-7 signal in the ‘LPS high’ and LPS low groups compared to the naïve group at 7 dpi (P < 0.01 and P < 0.0001, respectively; Fig. 5d,e). Here it is noteworthy that we observed a higher baseline expression of the GL-7 antigen in Pachón cavefish in the naïve control group, which may have contributed to the increased signal intensity in Pachón when compared to surface fish. Since GL-7 is also expressed to a lesser degree in a variety of different B-cell lineages in an inactivated state61, we hypothesize that the observed baseline difference is possibly due to differences in the abundance of different B-cell lineages between surface fish and cavefish, which needs further exploration. We did not find a significant response following LPS injection after 14 dpi in either surface or cavefish (see Supplementary Fig. 6 and Supplementary Data 12 for detailed statistical analysis). These findings indicate that Pachón cavefish mount a more lymphoid (adaptive)-driven immune response following bacterial recognition than surface fish.
Reduction in innate immune cells reduces pro-inflammatory response and formation of crown-like structures (CLS) in cavefish.
Finally, we asked whether reduced investment in innate immune cells, such as granulocytes and monocytes, in cavefish is accompanied by genetic changes in these cells during homeostasis and/or during the pro-inflammatory response (for a complete list of differential expressed genes in neutrophil and macrophage clusters, see Supplementary Data 13 and 14, respectively). Here we found that neutrophils and macrophages from cavefish showed increased expression of csf3r (log2FC=2.85 and 1.14 compared to the surface fish PBS group, respectively). This increased expression of csf3r in neutrophils and macrophages indicates a higher sensitivity for its ligand csf3 (g-csf), which is produced by a variety of different immune cells to stimulate the release of Csf3r+ cells into the bloodstream during inflammation.
Based on the overall reduced investment of cavefish in innate immune cells, we asked whether there is also a reduction in cells that mediate pro-inflammatory responses or whether reduced investment is compensated by an increased proportion of cells responding to a pro-inflammatory stimulus. To test this, we used expression of the cytokine Il-1β as a readout, because this cytokine is described as a major regulator of pro-inflammatory responses in teleost fish62. We detected induced expression in granulopoietic cells (mainly mature neutrophils), monocytopoietic cells (mainly mature monocytes) and macrophages in surface fish and cavefish following LPS injection (Fig. 6a). We found a 2.3-fold increase in il-1β-expressing cells in surface fish 3 h following LPS injection compared to cavefish (mean overall relative abundance of cells expressing il-1β in surface fish, 0.065 versus 0.028 in cavefish; see Fig. 6b). Interestingly, cavefish seemed highly variable following PBS injection (Fig. 6b) but, when we compared expression of il-1β within each cell cluster, only cavefish neutrophils showed elevated il-1β expression in both replicates of the PBS-injected group comparable to induced expression after LPS injection (Fig. 6c). This, however, is a cavefish neutrophil-specific phenomenon, since monocytopoietic cells and macrophages did not express il-1β in the PBS-injected Pachón samples to the same degree as the LPS-injected Pachón samples (Fig. 6c). It is noteworthy that macrophages from surface fish and cavefish showed the highest increase in il-1β-expressing cells and, presumably, represent the main producer of Il-1β following LPS injection in A. mexicanus (Fig. 6c).
Fig. 6 |. Reduced immune investment in myeloid cells alters inflammatory and immunopathological responses of A. mexicanus.
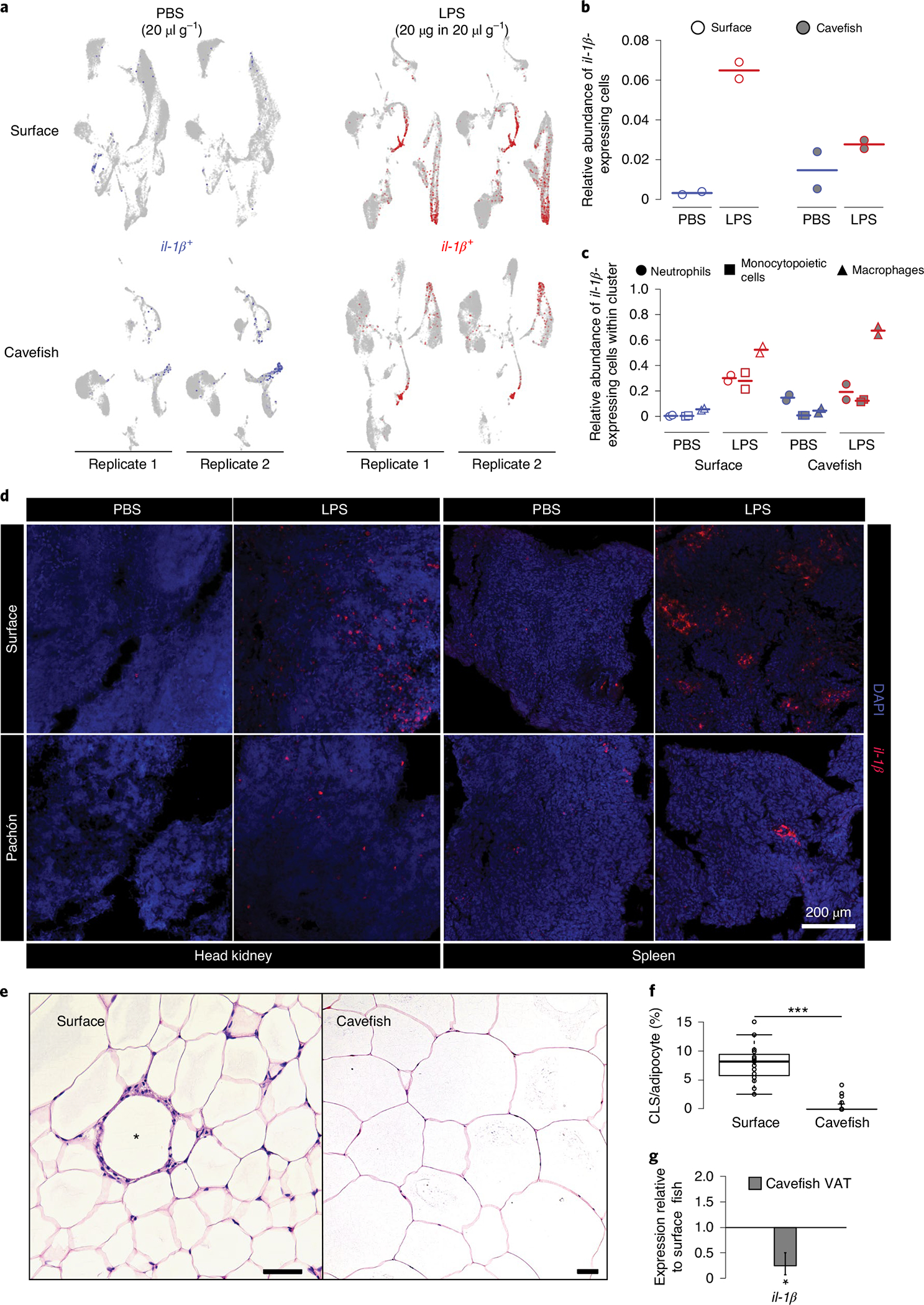
a, il-1β expression 3 h following PBS or LPS injection in HK cells. b, Overall relative abundances of il-1β-expressing cells from scRNA-seq experiments for each treatment group. c, Relative abundances of il-1β-expressing cells from scRNA-seq experiments within the main il-1β-expressing cell cluster for each treatment group. d, In vivo inflammatory response displayed by in situ hybridization of il-1β using RNA Scope in HK and spleen of surface fish and cavefish 3 h following intraperitoneal injection of 20 μg in 20 μl g−1 (bodyweight) LPS. Images are representative of two independent experiments. e, H&E staining of VAT of surface fish and cavefish. CLS is indicated by an asterisk in surface VAT. Scale bars, 50 μm. f, CLS count per 100 adipocytes in VAT of surface fish and cavefish in at least three fields of view for each fish (n = 3). Significance values were determined by Mann–Whitney U-test. For all box plots, centre lines show the medians, crosses show means, box limits indicate the 25th and 75th percentiles as determined by R software80, whiskers extend 1.5× interquartile range from the 25th and 75th percentiles and data points are represented by circles. ***P < 0.001. g, Gene expression of il-1β in cavefish VAT relative to surface fish (same fish that were used in f). Significance values were determined by a pairwise fixed reallocation randomization test using REST2009 software71; *P < 0.05. Error bar represents s.e.m.
To verify the reduction in neutrophils and monocytes/macrophages that can initiate a pro-inflammatory response, we designed an il-1β in situ RNA Scope probe to visualize il-1β expression in HK and spleen following LPS stimulation (see Methods for details). In line with the scRNA-seq analysis, LPS-injected surface fish showed an increased number of il-1β-positive cells in HK compared to cavefish (Fig. 6d). We also detected considerably fewer cells expressing il-1β after injection with LPS in the spleen from cavefish compared to surface fish (Fig. 6d). In teleost fish, the spleen contains high numbers of mononuclear phagocytes—for example, macrophages33,60—but is not generally a haematopoietic tissue for such cell types59. In addition, we also used the il-1β RNA Scope probe on dissociated HK cells from surface fish 3 h after injection with LPS, and were able to validate that mainly cells with monocytic and neutrophilic characteristics (multi-lobed nuclei) express il-1β (Supplementary Fig. 7).
Based on these results, we hypothesized that the lack of cells initiating a systemic pro-inflammatory response in cavefish following exposure to an immune stimulant (for example, LPS) could potentially lead to a reduced presence of immunopathological phenotypes resulting from such pro-inflammatory responses. Cavefish produce substantially more visceral adipose tissue (VAT) than surface fish25. In mammals, the amount of VAT is positively correlated with the number of monocytes infiltrating adipose tissue and mediating pro-inflammatory processes, resulting in the formation of crown-like structures (CLS)63. Therefore, we tested whether A. mexicanus VAT shows signs of CLS and whether laboratory surface fish and cavefish differ in their occurrence. We detected CLS in the VAT of surface fish (Fig. 6e) but not in cavefish, despite the prevalence of large, hypertrophic adipocytes (median numbers of CLS in 100 adipocytes, 8.197 for surface versus 0 for cavefish, P < 0.001; Fig. 6f). In addition, we detected reduced expression of il-1β in cavefish VAT relative to surface fish (mean relative expression of cavefish compared with surface fish, 0.249, P < 0.05; Fig. 6g). In combination with the reduced number of CLS, our data indicate a reduction in pro-inflammatory granulocytes and macrophages in cavefish VAT, potentially enabling increased VAT storage in cavefish with no immunopathological consequences.
Conclusion
Our study elucidates how adaptation to low biodiversity and parasite diversity in caves affects the immune investment strategy of a vertebrate host. We cannot, however, rule out the possibility that other environmental components (for example, microbiome, diet, circadian rhythm) contribute to differences in immune investment strategy between cavefish and surface fish, since these factors are known to influence haematopoiesis in mammals and thus the development of immunopathological phenotypes64–66. A more intriguing possibility is that the lower investment in cells that mediate pro-inflammatory processes is an adaption to the increased level of VAT in cavefish to lowering the risk of inflammation that increases with increased VAT. Future experiments are needed to distinguish between these possibilities63. Furthermore, it is possible that founder effects—that is, the specific immune phenotype of the founder population in the two caves (Pachón and Tinaja)—could potentially contribute to the differences that we observed between cavefish and surface fish populations. Given the marked differences in parasitic infections, however, we interpret the changes in the immune investment strategy and the reduction of innate immune cells in cavefish, which mediate pro-inflammatory processes and act against parasites, mainly as an adaptation to decreasing auto-aggressive immunopathology from a hypersensitive immune system in an environment with very low parasite diversity. With A. mexicanus we present a vertebrate system that lost parasite diversity for thousands of generations and presents immunological adaptations to such an environment that prevent immunopathology.
Methods
Field sample collection.
Collection for this study was conducted under permit no. SGPA/DGVS/03634/19 granted by the Secretaría de Medio Ambiente y Recursos Naturales to Ernesto Maldonado. Study sites are located in the Sierra de El Abra region of northeastern Mexico in the states of San Luis Potosí and Tamaulipas. The El Abra region experienced repeated uplift and erosional events that carved the underground limestone caverns67. We collected samples from Pachón cave, one of 30 in the region with known cave-dwelling Astyanax populations67,68. We also collected samples of the surface morphotype from Nacimiento Río Choy, approximately 95 km south of Pachón cave.
We collected fish daily from Pachón cave and Nacimiento Río Choy on 12–14 July 2019 during the rainy season. Pachón fish were collected in the morning of 12 July using handheld nets. Río Choy fish were collected during the daytime on 13/14 July using a combination of handheld nets, net traps and a modified plastic bottle trap. Captured fish were placed in their environmental water and euthanized on the day of capture. Fish were weighed, measured and immediately screened for parasitic infections as described previously16. Briefly, fish were screened for ectoparasites before dissection. After screening of the body cavity for the presence of parasites, the gills, liver and gut were removed and screened for the presence of parasites using a compressorium (Hauptner–Herberholz).
Fish husbandry.
The aquatic animal programme at the Stowers Institute meets all federal regulations and has been fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care since 2005. Surface morphs of A. mexicanus were reared from Mexican surface fish (Río Choy), and cavefish originating from the Pachón and Tinaja caves were kept in the laboratory for around 10–15 generations in total and were established in the facility using at least six families imported as adults in 2015 from the Tabin laboratory, Harvard, and housed on an independent isolation rack. All experiments with laboratory fish used adult female fish aged 12–16 months that originated from bleached embryos pooled from at least six families (which were housed on a special breeding rack) and were raised and maintained on a separate rack. Bleaching (embryo surface sanitation) was done as described previously69. Each rack uses an independent recirculating aquaculture system with mechanical, chemical and biological filtration and ultraviolet disinfection. Astyanax were housed in glass fish tanks on racks (Pentair) with a 14/10 h light/dark photoperiod. All laboratory populations used for experiments were housed on the same rack at a density of approximately two fish per litre. For each experiment, 24 h before the procedure one fish per population and treatment was individualized and fasted to avoid potential confounding tank effects, and to reduce stress for the fish. For all experiments, fish were euthanized by placing them in 500 mg l−1 MS-222 for 5 min and processed immediately. For more information on fish husbandry please see Supplementary methods.
In vitro gene expression analysis.
Single-cell suspensions from freshly dissected HK tissue were produced by forcing through a 40-μM cell strainer into L-15 medium (Sigma) containing 10% water, 5 mM HEPES buffer (pH 7.2) and 20 U ml−1 heparin (L-90). The strainer was washed once with L-90 and cells were washed once by spinning at 500g and 4 °C for 5 min. Supernatant was discarded and cells were resuspended in 1 ml of L-90 medium (L-15 containing 10% water, 5 mM HEPES pH 7.2), 5% fetal calf serum, 4 mM l-glutamine and penicillin/streptomycin mix (both 10,000 U ml−1). Cells were counted using a EC800 analyser (Sony Biotechnology), and 1 × 106 cells were plated on 48-well plates in volumes of 500 μl and incubated overnight at 21 °C. At time point 0, either 20 or 0.2 μg ml−1 LPS mix in PBS (Escherichia coli O55:B5 and E. coli O111:B4 (both Sigma), 1 mg ml−1 each) or PBS per se as a control was added to the cells, respectively. After 1, 3, 6, 12 and 24 h, cells were harvested, immediately snap-frozen in liquid nitrogen and RNA was isolated as described previously70. RNA (100 ng; the concentration was measured using the Qubit system (Thermo Fisher)) from each sample was used for complementary DNA synthesis using the SuperScript III First-Strand Synthesis System kit (Invitrogen), following the manufacturer’s instructions. The resulting cDNA was used for RT–qPCR with the PerfeCTa SYBR Green FastMix (Low ROX; Quantabio), following the manufacturer’s instructions. Gene-specific primers (see Supplementary Table 2) were used for amplification of the target and the two housekeeping genes (rpl32 and rpl13a; see Supplementary Table 2 for details). Where possible, gene-specific primers were designed to span one exon–exon junction. Samples were pipetted in a 384-well plate using a Tecan EVO PCR Workstation, and samples were run in technical triplicate on a QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher). Quality control for each sample was performed using QuantStudio Real-Time PCR software (Thermo Fisher), and data were exported for analysis in REST2009 (ref. 71) as described previously70. PBS control samples from each time point and sample were used as the reference to calculate the relative expression of target genes for each time point and fish, respectively.
Phagocytosis assay.
Phagocytosis was measured as previously described72. Briefly, a single-cell solution from freshly dissected HK tissue was prepared as described above; 4 × 105 cells were pipetted into a 96-well flat-bottom plate and Alexa-488-tagged Staphylococcus aureus (Thermo Fisher) was added at a ratio of 1:50 cells:bacteria. To control for cell viability a sample without bacteria was included, and to control for active phagocytosis a sample with cells containing bacteria and cytochalasin B (CCB, 0.08 mg ml−1, Sigma), an actin polymerization inhibitor, for each individual sample was included. Cells were incubated in 200 μl of L-90 medium at 21 °C for 1 and 3 h, respectively. To exclude dead cells and signal from non-phagocytosed particles, cells were stained with Hoechst and all samples were quenched using 50 μl of Trypan blue (0.4% solution, Sigma) before measurement. Samples were measured on a EC800 analyser (Sony Biotechnology). Cells were gated for live and Alexa-488 positive, and phagocytosis rate was calculated as the ratio of live (Hoechst-positive, excitation 352 nm, emission 461 nm) phagocytes (Alexa-488-positive, excitation 495 nm, emission 519 nm) versus live cells.
Scatter analysis of HK.
Single HK cells from adult surface fish were extracted as described above. Cells were stained with 4,6-diamidino-2-phenylindole (DAPI) to exclude dead cells, and live cells were sorted based on populations as described in Fig. 2a using the forward scatter (FSC) and side scatter (SSC) characteristics of cells with an Influx System (BD). One thousand cells per population were sorted on Thermo Scientific Shandon Polysine Slides and incubated for 30 min at 21 °C, so that they could settle and adhere to the slides. Cells were then fixed with 4% paraformaldehyde (PFA) and washed three times in PBS. Cells were stained using the May–Grünwald Giemsa protocol. Briefly, slides were stained for 10 min with a 1:2 solution of May–Grünwald (in phosphate buffer pH 6.5, filtered), the excess stain was drained off and slides were stained for 40 min with a 1:10 solution of Giemsa (in phosphate buffer pH 6.5, filtered). The slides were then rinsed in double-distilled water (ddH20) by passage ten times under running ddH20. For differentiation, a drop of 0.05% acid water (5 ml glacial acetic acid/95 ml ddH2O) was applied to the slides for ~4 s and immediately rinsed off. Slides were rinsed well, air dried and coverslipped using Micromount Mounting Medium (Leica). Only cells that could clearly be identified based on studies in closely related organisms using a similar approach29,33 were used as representative images for Fig. 2a.
Image-based cluster analysis of HK.
For this analysis, haematopoietic HK cells were presorted to remove mature erythrocyte clusters using the S3 Cell Sorter (BioRad) with scatter features (see Fig. 2a). This was necessary, since mature erythrocytes account for about 8 and 7% in surface fish and Pachón fish, respectively, of the entire HK cell count. Based on the scatter sorting approach described before, we were able to identify mature erythrocytes using scatter per se (see Fig. 2a). However, due to their biconcave morphology, we found erythrocytes in all populations that were separated through scattering to a certain degree since different orientations in erythrocyte flow resulted in different properties being revealed by FSC/SSC analysis40. In Image3C analysis we also found that a high abundance of mature erythrocytes increased the number of clusters containing erythrocytes in different orientations. Reduction in mature erythrocytes through sorting of only myeloid, progenitor and lymphoid cell clusters based on scatter was used successfully to reduce the amount of over-clustering using the Image3C pipeline40. Sorted cells were stained with 5 μM DRAQ5 for 10 min, and 10,000 nucleated, single events were acquired from samples on ImageStream X Mark II at 60×, slow flow speed, with 633-nm laser excitation. Bright field was acquired on channels 1 and 9, and DRAQ5 on channel 11. SSC was acquired on channel 6. Intensities from 25 unique morphological features were extracted (see Supplementary Table 3 for details). Further analysis was done as described in ref. 40, which also includes the open source code and complete R documentation. For a detailed protocol of the Image3C pipeline, please see Supplementary methods. Cell clusters were assigned an identity based on the following morphological characteristics: myelomonocytes (relatively large cells with medium to high granularity, irregularly shaped nuclei and high cytoplasm/nuclei ratio; clusters 2, 11, 13, 14, 16); lymphocytes/progenitors (relatively small cells with low granularity and low cytoplasm/nuclei ratio; clusters 4, 7, 9, 18, 19); and mature erythrocytes/doublets/debris (clusters 1, 3, 5, 6, 8, 10, 12, 15, 17, 20, 21) (see Fig. 3b). More specifically: monocytic cells (cluster 13) were defined as relatively large cells with cytoplasm of complex structure, high granularity and kidney-shaped nuclei; neutrophils (cluster 14) were defined as medium-sized cells with evenly distributed cytoplasm, high granularity and multi-lobed nuclei; monocytic, granulocytic and promyelocytic cells (cluster 16) were defined as medium to large cells with high granularity.
Intraperitoneal injection of LPS.
Fish were individualized and fasted the day before treatment (naïve, PBS- or LPS-injected). After 24 h they were anaesthetized using ice-cold tap water and either PBS (20 μ g−1 bodyweight) or a LPS mix (E. coli O55:B5 and E. coli O111:B4; both Sigma, 20 or 5 μg at 20 μl g−1 bodyweight) was injected intraperitoneally using an insulin syringe (0.33 ml, length 8 mm, 31G, BD Pharmingen). At given time points, fish were euthanized using buffered Tricaine solution (500 mg l−1) and respective organs were dissected and either dissociated (scRNA sequencing) or immediately fixed in 4% PFA/diethyl pyrocarbonate (DEPC) water (RNA Scope analysis) for subsequent analysis.
Single-cell RNA-seq of naïve fish.
Dissociated haematopoietic cells were stained with DAPI to exclude dead cells, and live cells were sorted based on population as described in Fig. 2a, where only myelomonocyte, lymphocyte and progenitor populations were sorted in L-90 medium, to reduce the relative abundance of mature erythrocytes. Sorted cells were spun down (500 × relative centrifugal force (r.c.f.), 4 °C, 5 min), the supernatant was discarded and the cells were resuspended in L-90 medium and run again on an InfluxDB system to ensure removal of mature erythrocyte clusters and to measure cell viability (percentage of live cells after sorting: surface fish and cavefish, 82.2 and 88.9, respectively). Cells were loaded on a Chromium Single Cell Controller (10X Genomics), based on live cell concentration, with a target of 6,000 cells per sample. Libraries were prepared using the Chromium Single Cell 3’ Library & Gel Bead Kit v.2 (10X Genomics) according to the manufacturer’s directions. Resulting short-fragment libraries were checked for quality and quantity using an Agilent 2100 Bioanalyzer and Invitrogen Qubit Fluorometer. Libraries were sequenced individually to a depth of ~330 M reads each on an Illumina HiSeq 2500 instrument, using Rapid SBS v.2 chemistry with the following paired read lengths: 26 bp read1, 8 bp I7 index and 98 bp read2. Raw sequencing data were processed using the 10X Genomics Cell Ranger pipeline (v.2.1.1). Reads were de-multiplexed into Fastq file format using cellranger mkfastq. The genome index was built by cellranger mkref using the cavefish genome astMex1, ensembl 87 gene model. Data were aligned by STAR aligner and cell count tables were generated using the cellranger count function with default parameters. Cells with at least 500 unique molecular identifier (UMI) counts were loaded into the R package Seurat (v.2.3.4) for clustering and trajectory analysis. For Pachón cavefish, 5,874 and 4,717 cells were used for surface and downstream analysis, respectively. The UMI count matrix was log normalized to find variable genes. The first 12 principal components were selected for dimension reduction and t-distributed stochastic neighbour embedding plots. Marker genes were used to classify clusters into lymphocyte, myelomonocyte and progenitor types. The results generated by Cell Ranger can be retrieved from the GEO database with accession number GSE128306. The assignment of cell identities is based on their transcription profile determined by similar approaches in zebrafish33,73–76.
Single-cell RNA-seq of PBS- and LPS-injected fish.
We injected laboratory surface fish (Río Choy) and cavefish (Pachón) with either PBS (20 μl g−1 bodyweight) or LPS (20 μg at 20 μl g−1 bodyweight) and dissected HK 3 h later. We removed most mature erythrocytes through fluorescent activated cell sorter (FACS) sorting as described before. Sorted cells were spun down (500× r.c.f., 4 °C, 5 min), the supernatant was discarded and cells were resuspended in L-90 medium and run again on an Influx system to ensure removal of mature erythrocyte clusters and to measure cell viability (percentage live cells after sorting: surface fish PBS replicates 1 and 2, 89 and 83%, respectively; surface fish LPS replicates 1 and 2, 86 and 90%, respectively; cavefish PBS replicates 1 and 2, 88 and 82%, respectively; cavefish LPS replicates 1 and 2, 89 and 91%, respectively). Cells were loaded on a Chromium Single Cell Controller (10X Genomics) based on live cell concentration. Using a unique molecular identifier count of ≥500, we obtained a mean of 12,128 cells for each of the eight samples with a mean number of 667 genes per cell in each sample. Libraries were paired-end sequenced using an Illumina Novaseq S2 flowcell. Raw data were processed using Cell Ranger pipeline (v.3.0) and de-multiplexed into Fastq file format using cellranger mkfastq. Data were aligned to the astMex1, ensemble 87 gene model by STAR aligner, and cell count tables were generated using cellranger count function with default parameters. Cells with a count of at least 500 UMI were loaded into the R package Seurat (v.3.0) for clustering and trajectory analysis. The UMI count matrix was log normalized to find the top 2,000 variable genes using the vst selection method from Seurat. Replicates were then integrated using SCTtransform function FindIntegrationAnchors based on Seurat vignettes. Principal component cut-offs were selected based on Jackstraw and Elbowplot function for dimension reduction and uniform manifold approximation and projection (UMAP) plots. De novo marker genes were generated using FindAllMarkers function, and to plot the heatmaps in Extended Data Fig. 3. Trajectory analyses were computed using the R package slingshot. All scRNA-seq data can be retrieved from the GEO database with accession number GSE128306.
Il-1β RNA Scope assay.
Section preparation and RNA in situ hybridization were performed as previously reported25,77. Briefly, for tissue sections, respective tissues (HK, spleen) were dissected from surface fish and cavefish followed by immediate immersion in 4% PFA in DEPC H2O (diluted from 16% (w/v) aqueous solution, Electron Microscopy Sciences, no. 15710) for 24 h at 4 °C to fix the tissue, then rinsed well with 1× PBS, dehydrated through graded ethanol (30, 50, 70%) and processed with a PATHOS Delta hybrid tissue processor (Milestone Medical Technologies). Paraffin sections (8 μm) were cut using a Leica RM2255 microtome (Leica Biosystems) and mounted on Superfrost Plus microscope slides (no. 12-550-15, Thermo Fisher). For single-cell solutions, HK and spleen were dissected, and solutions were produced as described above; approximately 20 μl of the suspension was pipetted on Superfrost Plus microscope slides. Cells were allowed to settle for 30 min and fixed using 4% PFA (diluted from 16% (w/v) aqueous solution, Electron Microscopy Sciences, no. 15710) for 1 h at room temperature, then rinsed well with 1× PBS and dehydrated through graded ethanol (30, 50, 70%). RNA in situ hybridization was performed using the RNA Scope multiplex fluorescent detection v.2 kit according to the manufacturer’s instructions (Advanced Cell Diagnostics). The RNA Scope probe used for Il-1β was a 16ZZ probe (Ame-LOC103026214-C2 targeting 217–953 of XM_022680751.1).
Images of sections were acquired from a Nikon 3PO spinning disc on a Nikon Ti Eclipse base, outfitted with a W1 disc. A CFI Plan Apochromat Lambda ×20/0.75 numerical aperture air objective was used. DAPI and AF647 were excited with a 405- and 640-nm laser, respectively, with a 405/488/561/640-nm main dichroic. Emission was collected by an ORCA-Flash 4.0 V2 digital sCMOS camera, through 700/75- and 455/50-nm filters for the far-red and DAPI channel, respectively. Z-step spacing was 1.5 μ. All microscope parameters and acquisitions were controlled with Nikon Elements software. Identical camera exposure time and laser power were used across samples. All image processing was done with an open source version of FIJI78 with standard commands. A Gaussian blur with a radius of 1 was applied, and a rolling-ball background subtraction with a radius of 200 pixels was applied to every channel except for DAPI. Following that, a max. projection across the slice was applied. For direct comparison, images shown are contrasted identically in the far-red channel (Il-1β).
GL-7 analysis of spleen following LPS injection.
Fish were dissected 3 h after treatment and the spleen was immediately embedded with OCT compound (Tissue-Tek) and frozen at −70 °C. Cryo-sections of 12-μm thickness were cut using a Leica CM3050S cryostat (Leica Biosystems) and mounted on glass slides. Sections were maintained in the cryostat for 2 h before fixing with pre-chilled 75% acetone/25% ethanol at room temperature for 30 min. Immunofluorescence assay was performed manually using an Alexa Fluor 647-conjugated rat anti-mouse T- and B-cell activation antigen (BD Pharmingen, GL-7 clone, no. 561529) and a matched isotype control (BD Pharmingen, R4–22 clone, no. 560892). Immunofluorescence experiments were repeated three times with different populations and time points, which accumulated a total of 48 animals. In brief, sections were rehydrated with 1× PBS and the background was blocked by incubating sections in Background Buster solution (no. NB306, Innovex Biosciences) for 30 min. The antibody was diluted 1:500 in Antibody Diluting Reagent (no. 003118, Invitrogen) and incubated overnight at 4 °C. Sections were further stained with DAPI (1:1,000) for 10 min and then washed in Tris-buffered saline (25 mM Tris, 0.15 mM NaCl, pH 7.2) with 0.05% Tween-20 and coverslipped using ProLong Gold Antifade Mountant (Thermo Fisher) before imaging. Images of sections were acquired on a Zeiss LSM 700 upright microscope. A 5× air objective was used. DAPI and AF647 were excited with a 405- and 640-nm laser, respectively, with a 405/488/561/640-nm main dichroic. Emission was collected by an ORCA-Flash 4.0 v.2 digital sCMOS camera, through a 700/75- and 455/50-nm filter for the far-red and DAPI channel, respectively. All image processing was done with an open source version of FIJI78 with standard commands. For GL-7 intensity analysis we used a Fiji macro that is publicly available under https://github.com/jouyun/smc-macros/blob/master/ROP_IntensityMeasurement.ijm.
VAT analysis.
We dissected VAT from the abdominal cavity as described previously25. In short, we manually removed the intestinal sac and carefully isolated a piece of fat tissue located around the gut for RT–qPCR as described above. The remainder of the sample was immediately fixed in 4% PFA for 18 h at 4 °C and embedded in JB-4 embedding solution (Electron Microscopy Sciences, no. 14270-00) while following the kit instructions for dehydration, infiltration and embedding. After sectioning at 5 μm, we dried the slides for 1 h in an oven at 60 °C and stained them with haematoxylin for 40 min. After rinsing the slides in PBS, semi-dried slides were stained with eosin (3% in de-salted water) for 3 min. Slides were then washed with de-salted water and air dried. At least three images from VAT of each fish were taken at similar locations of the gut, and CLS were scored as described previously79. Images were obtained using the 10× objective on a Zeiss Axioplan2 upright microscope, and adipocytes and CLS were counted using Adobe Photoshop CC (v.19.1.0).
Statistical analysis.
Graphical data and statistics were produced using R80 except where otherwise stated. Data were tested, where appropriate, for normal distribution. For comparison between populations, normal distributed data were analysed using one-way analysis of variance (ANOVA) and corrected for multiple testing against the same control group (false discovery rate, FDR) with Benjamini–Hochberg testing81. For non-normally distributed data, the Mann–Whitney U-test was used. For analysis of RT–qPCR data we used REST2009 software, where significant differences between two groups were determined by a pairwise fixed reallocation randomization test71. Two-way ANOVA analysis was done using Graph Pad Prism Software (v.8.0.2). Multiple testing against the same control group was corrected with FDR and Benjamini–Hochberg testing81.
Animal experiments.
Research and animal care were approved by the Institutional Animal Care and Use Committee of the Stowers Institute for Medical Research.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. A. mexicanus anatomy.
Cartoon of adult A. mexicanus indicates anatomical position of the main hematopoietic and lymphoid organ, the head kidney (HK) that was used for subsequent in vitro experiments from surface fish and cavefish lab strains.
Extended Data Fig. 2. |. Gene expression of specific head kidney cell types in cavefish and surface fish.
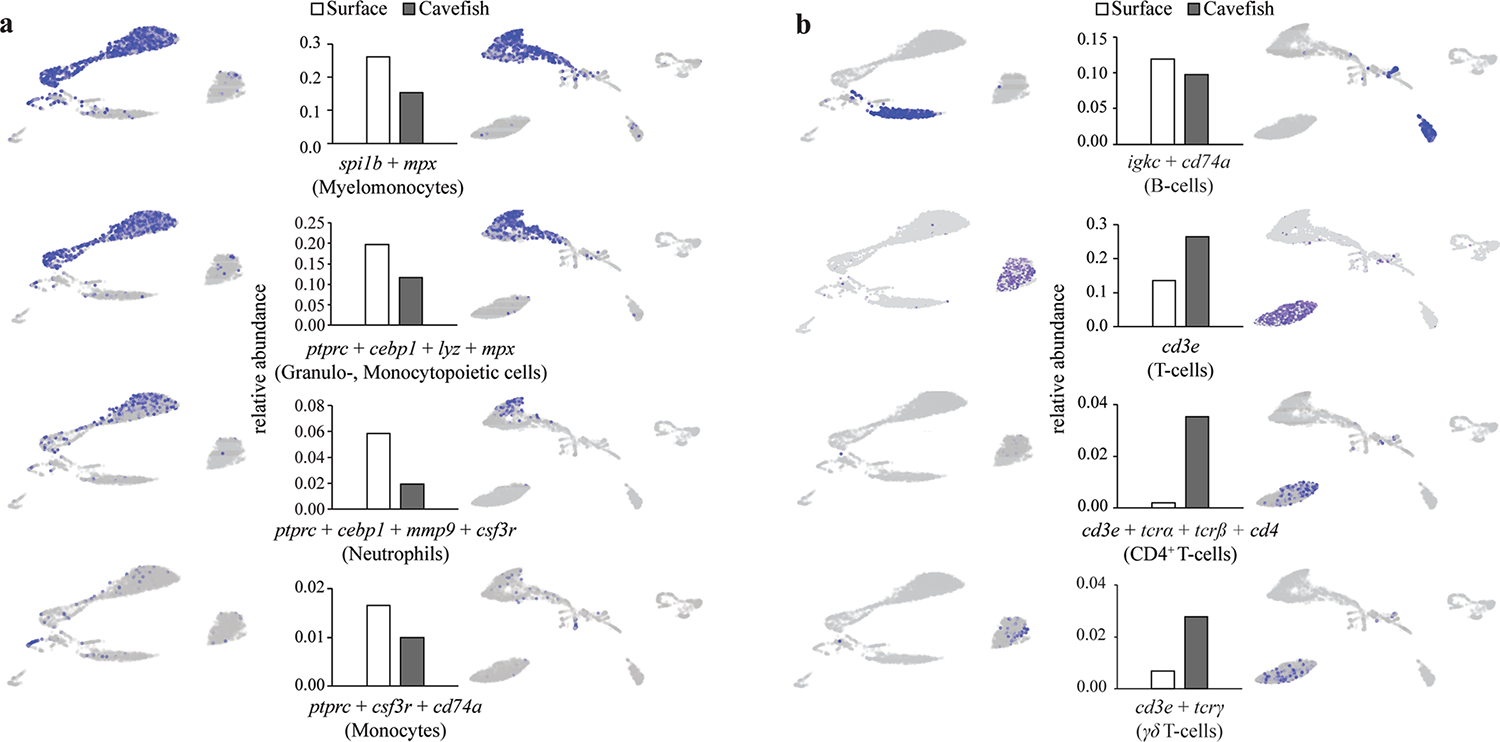
Relative abundance of specific cell populations of surface fish and cavefish and their location within UMAP representation of specific cell types from, a, myelomonocytes and, b, lymphocytes based on the expression of given gene(s). See Supplementary Data 4 for gene enrichment in each cluster.
Extended Data Fig. 3 |. HK cell gene expression profiles of PBS injected fish.
Heatmap of enriched genes within each cell cluster of control groups from surface fish and cavefish. Genes that were used for cell cluster identification are shown. For a complete heatmaps for PBS and LPS injected groups see Supplementary Data 6–9.
Supplementary Material
Acknowledgements
We thank the cavefish facility staff at the Stowers Institute for support and husbandry of the fish. We thank the staff from the Histology core at the Stowers Institute for their technical support; J. Blanck from the Cytometry core for performing sorting of HK cells; M. Peterson, A. Peak and A. Perera for support with scRNA-seq; and M. Miller for support with the fish anatomy figure. The authors acknowledge the University of Kansas Medical Center Genomics Core for sequencing support. Furthermore, we thank S. A. McKinney for providing the ImageJ macro for GL-7 quantification. The authors also kindly acknowledge J. Kurtz for helpful discussions. N.R. is supported by institutional funding, funding from the JDRF, the Edward Mallinckrodt Foundation, NIH Grant R01 GM127872 and NSF IOS-1933428 and EDGE award 1923372. R.P. was supported by a grant (no. PE 2807/1-1) from Deutsche Forschungsgemeinschaft.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41559-020-1234-2.
Supplementary information is available for this paper at https://doi.org/10.1038/s41559-020-1234-2.
Data availability
Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb1391. The scRNA-seq data generated by Cell Ranger can be retrieved from the GEO database with accession number GSE128306.
References
- 1.The Global Burden of Disease: 2004 Update (WHO, 2004). [Google Scholar]
- 2.Sheldon BC & Verhulst S Ecological immunology: costly parasite defences and trade-offs in evolutionary ecology. Trends Ecol. Evol. 11, 317–321 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Schmid-Hempel P Variation in immune defence as a question of evolutionary ecology. Proc. R. Soc. B. 270, 357–366 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid-Hempel P Evolutionary Parasitology (Oxford Univ. Press, 2013). [Google Scholar]
- 5.Rook GA Regulation of the immune system by biodiversity from the natural environment: an ecosystem service essential to health. Proc. Natl Acad. Sci. USA 110, 18360–18367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Hertzen L, Hanski I & Haahtela T Natural immunity. Biodiversity loss and inflammatory diseases are two global megatrends that might be related. EMBO Rep. 12, 1089–1093 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belkaid Y & Hand TW Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambrecht BN & Hammad H The immunology of the allergy epidemic and the hygiene hypothesis. Nat. Immunol. 18, 1076–1083 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Rook GA, Martinelli R & Brunet LR Innate immune responses to mycobacteria and the downregulation of atopic responses. Curr. Opin. Allergy Clin. Immunol. 3, 337–342 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Rosenblum MD, Remedios KA & Abbas AK Mechanisms of human autoimmunity. J. Clin. Invest. 125, 2228–2233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lafferty KD Biodiversity loss decreases parasite diversity: theory and patterns. Philos. Trans. R. Soc. Lond. B 367, 2814–2827 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamiya T, O’Dwyer K, Nakagawa S & Poulin R Host diversity drives parasite diversity: meta-analytical insights into patterns and causal mechanisms. Ecography 37, 689–697 (2014). [Google Scholar]
- 13.McDade TW, Georgiev AV & Kuzawa CW Trade-offs between acquired and innate immune defenses in humans. Evol. Med. Public Health 2016, 1–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindstrom KM, Foufopoulos J, Parn H & Wikelski M Immunological investments reflect parasite abundance in island populations of Darwin’s finches. Proc. R. Soc. B 271, 1513–1519 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer A, Mora T, Rivoire O & Walczak AM Diversity of immune strategies explained by adaptation to pathogen statistics. Proc. Natl Acad. Sci. USA 113, 8630–8635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scharsack JP, Kalbe M, Harrod C & Rauch G Habitat-specific adaptation of immune responses of stickleback (Gasterosteus aculeatus) lake and river ecotypes. Proc. R. Soc. B 274, 1523–1532 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaczorowski KJ et al. Continuous immunotypes describe human immune variation and predict diverse responses. Proc. Natl Acad. Sci. USA 114, E6097–E6106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herman A et al. The role of gene flow in rapid and repeated evolution of cave-related traits in Mexican tetra, Astyanax mexicanus. Mol. Ecol. 27, 4397–4416 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fumey J et al. Evidence for late Pleistocene origin of Astyanax mexicanus cavefish. BMC Evol. Biol. 18, 43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibert J & Deharveng L Subterranean ecosystems: a truncated functional biodiversity. BioScience 52, 473–481 (2002). [Google Scholar]
- 21.Tabin JA et al. Temperature preference of cave and surface populations of Astyanax mexicanus. Dev. Biol. 441, 338–344 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abolins S et al. The comparative immunology of wild and laboratory mice, Mus musculus domesticus. Nat. Commun. 8, 14811 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trama AM et al. Lymphocyte phenotypes in wild-caught rats suggest potential mechanisms underlying increased immune sensitivity in post-industrial environments. Cell Mol. Immunol. 9, 163–174 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aspiras AC, Rohner N, Martineau B, Borowsky RL & Tabin CJ Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proc. Natl Acad. Sci. USA 112, 9668–9673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong S, Krishnan J, Peuss R & Rohner N Early adipogenesis contributes to excess fat accumulation in cave populations of Astyanax mexicanus. Dev. Biol. 441, 297–304 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Wiens GD & Vallejo RL Temporal and pathogen-load dependent changes in rainbow trout (Oncorhynchus mykiss) immune response traits following challenge with biotype 2 Yersinia ruckeri. Fish Shellfish Immunol. 29, 639–647 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Krishnan J et al. Comparative transcriptome analysis of wild and lab populations of Astyanax mexicanus uncovers differential effects of environment and morphotype on gene expression. J. Exp. Zool. B 10.1002/jez.b.22933 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Moller AM, Korytar T, Kollner B, Schmidt-Posthaus H & Segner H The teleostean liver as an immunological organ: intrahepatic immune cells (IHICs) in healthy and benzo[a]pyrene challenged rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 46, 518–529 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Traver D et al. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 4, 1238–1246 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Stockdale WT et al. Heart regeneration in the Mexican cavefish. Cell Rep. 25, 1997–2007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramsey S et al. Transcriptional noise and cellular heterogeneity in mammalian macrophages. Philos. Trans. R. Soc. Lond. B. 361, 495–506 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogryzko NV, Renshaw SA & Wilson HL The IL-1 family in fish: swimming through the muddy waters of inflammasome evolution. Dev. Comp. Immunol. 46, 53–62 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Wittamer V, Bertrand JY, Gutschow PW & Traver D Characterization of the mononuclear phagocyte system in zebrafish. Blood 117, 7126–7135 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Sunyer JO Evolutionary and functional relationships of B cells from fish and mammals: Insights into their novel roles in phagocytosis and presentation of particulate antigen. Infect. Disord. Drug Targets 12, 200–212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lugo-Villarino G et al. Identification of dendritic antigen-presenting cells in the zebrafish. Proc. Natl Acad. Sci. USA 107, 15850–15855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haugland GT et al. Phagocytosis and respiratory burst activity in lumpsucker (Cyclopterus lumpus L.) leucocytes analysed by flow cytometry. PLoS ONE 7, e47909 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieschke GJ & Trede NS Fish immunology. Curr. Biol. 19, R678–R682 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Balla KM et al. Eosinophils in the zebrafish: prospective isolation, characterization, and eosinophilia induction by helminth determinants. Blood 116, 3944–3954 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolnick DI, Shim KC, Schmerer M & Brock CD Population-specific covariation between immune function and color of nesting male threespine stickleback. PLoS ONE 10, e0126000 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peuß R et al. Label-independent flow cytometry and unsupervised neural network method for de novo clustering of cell populations. Preprint at bioRxiv 10.1101/603035 (2020). [DOI] [Google Scholar]
- 41.van der Meer W, Scott CS & de Keijzer MH Automated flagging influences the inconsistency and bias of band cell and atypical lymphocyte morphological differentials. Clin. Chem. Lab. Med. 42, 371–377 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Getz GS Thematic review series: the immune system and atherogenesis. Bridging the innate and adaptive immune systems. J. Lipid Res. 46, 619–622 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Wan F et al. Characterization of gammadelta T cells from zebrafish provides insights into their important role in adaptive humoral immunity. Front. Immunol. 7, 675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shilpi, Paul S & Lal G. Role of gamma-delta (gammadelta) T cells in autoimmunity. J. Leukoc. Biol. 97, 259–271 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Fan X & Rudensky AY Hallmarks of tissue-resident lymphocytes. Cell 164, 1198–1211 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papotto PH, Reinhardt A, Prinz I & Silva-Santos B Innately versatile: gammadelta17 T cells in inflammatory and autoimmune diseases. J. Autoimmun. 87, 26–37 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Fay NS, Larson EC & Jameson JM Chronic Inflammation and gammadelta T. Cells Front. Immunol. 7, 210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossi DJ et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc. Natl Acad. Sci. USA 102, 9194–9199 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolli N et al. Expression of the cytoplasmic NPM1 mutant (NPMc+) causes the expansion of hematopoietic cells in zebrafish. Blood 115, 3329–3340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stachura DL et al. Clonal analysis of hematopoietic progenitor cells in the zebrafish. Blood 118, 1274–1282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reavie L et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat. Immunol. 11, 207–215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cabezas-Wallscheid N et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15, 507–522 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Cheng J et al. Hematopoietic defects in mice lacking the sialomucin CD34. Blood 87, 479–490 (1996). [PubMed] [Google Scholar]
- 54.Anjos-Afonso F et al. CD34(−) cells at the apex of the human hematopoietic stem cell hierarchy have distinctive cellular and molecular signatures. Cell Stem Cell 13, 161–174 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Amin RH & Schlissel MS Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 9, 613–622 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han S, Zheng B, Schatz DG, Spanopoulou E & Kelsoe G Neoteny in lymphocytes: Rag1 and Rag2 expression in germinal center B cells. Science 274, 2094–2097 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Naito Y et al. Germinal center marker GL7 probes activation-dependent repression of N-glycolylneuraminic acid, a sialic acid species involved in the negative modulation of B-cell activation. Mol. Cell Biol. 27, 3008–3022 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laszlo G, Hathcock KS, Dickler HB & Hodes RJ Characterization of a novel cell-surface molecule expressed on subpopulations of activated T and B cells. J. Immunol. 150, 5252–5262 (1993). [PubMed] [Google Scholar]
- 59.Fänge R & Nilsson S The fish spleen: structure and function. Experientia 41, 152–158 (1985). [DOI] [PubMed] [Google Scholar]
- 60.Steinel NC & Bolnick DI Melanomacrophage centers as a histological indicator of immune function in fish and other poikilotherms. Front. Immunol. 8, 827 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cervenak L, Magyar A, Boja R & Laszlo G Differential expression of GL7 activation antigen on bone marrow B cell subpopulations and peripheral B cells. Immunol. Lett. 78, 89–96 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Secombes CJ, Wang T & Bird S The interleukins of fish. Dev. Comp. Immunol. 35, 1336–1345 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Weisberg SP et al. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 112, 1796–1808 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christ A et al. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell 172, 162–175 e114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McAlpine CS et al. Sleep modulates haematopoiesis and protects against atherosclerosis. Nature 566, 383–387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heidt T et al. Chronic variable stress activates hematopoietic stem cells. Nat. Med. 20, 754–758 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitchell RG, Russell WH & Elliott WR Mexican Eyeless Characin Fishes, Genus Astyanax: Environment, Distribution, and Evolution (Texas Tech Press, 1977). [Google Scholar]
- 68.Espinasa L et al. A new cave locality for Astyanax cavefish in Sierra de El Abra, Mexico. Subterr. Biol. 26, 39–53 (2018). [Google Scholar]
- 69.Embryo Surface Sanitation (Egg Bleaching) Protocol https://zebrafish.org/wiki/protocols/ess (ZIRC, 2019). [Google Scholar]
- 70.Peuß R, Eggert H, Armitage SA & Kurtz J Downregulation of the evolutionary capacitor Hsp90 is mediated by social cues. Proc. R. Soc. B 282, 20152041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pfaffl MW, Horgan GW & Dempfle L Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, 36e (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang YA et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 11, 827–835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rowe RG, Mandelbaum J, Zon LI & Daley GQ Engineering hematopoietic stem cells: lessons from development. Cell Stem Cell 18, 707–720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stachura DL et al. The zebrafish granulocyte colony-stimulating factors (Gcsfs): 2 paralogous cytokines and their roles in hematopoietic development and maintenance. Blood 122, 3918–3928 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Jong JL & Zon LI Use of the zebrafish system to study primitive and definitive hematopoiesis. Ann. Rev. Genet. 39, 481–501 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Athanasiadis EI et al. Single-cell RNA-sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat. Commun. 8, 2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeng A et al. Prospectively isolated tetraspanin(+) neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell 173, 1593–1608 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun K et al. Endotrophin triggers adipose tissue fibrosis and metabolic dysfunction. Nat. Commun. 5, 3485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.R Core Team R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2014). [Google Scholar]
- 81.Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data underlying this manuscript can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb1391. The scRNA-seq data generated by Cell Ranger can be retrieved from the GEO database with accession number GSE128306.


