ABSTRACT
Macroautophagy/autophagy is an evolutionarily conserved degradation process in eukaryotic cells that destroys obsolete proteins and damaged organelles via lysosome action. Autophagy is regulated by multiple ATG (autophagy related) proteins, which are activated in distinct steps. Although the mechanisms by which ATGs regulate autophagy have been comprehensively studied, an increasing number of studies have demonstrated that, in addition to their canonical roles in regulating autophagy, ATGs still show several nonautophagic functions that are required for the maintenance of cellular homeostasis in eukaryotes. Therefore, intensive exploration into the nonautophagic roles of ATGs is essential for a complete understanding of the biological functions of ATGs. In addition, the results of these explorations may contribute to the development of therapeutic strategies for disorders related to the dysfunctions of ATGs. Here, in this review, we specifically focus on nonautophagic functions of ATGs and comprehensively summarize the research progresses related to ATGs, especially their regulatory effects on cell proliferation, differentiation, senescence and death; cargo transport and phagocytosis; immune responses; metabolic homeostasis; gene transcription; genome stability; and signal transduction. We believe that this review meaningfully contributes to understanding the roles and mechanisms by which ATGs regulate physiological and pathological processes.
Abbreviations
ATG: autophagy related; BECN1: beclin 1; cAMP: cyclic adenosine monophosphate; dsDNA: double-stranded DNA; EMT: epithelial-mesenchymal transition; IFN: interferon; ISCs: intestinal stem cells; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MAPK/JNK: mitogen-activated protein kinase/c-Jun N-terminal kinases; MTOR: mechanistic target of rapamycin kinase; STING1: stimulator of interferon response cGAMP interactor 1; UVRAG: UV radiation resistance associated; VPS: vacuolar protein sorting.
KEYWORDS: autophagy-related proteins, nonautophagic functions, cargo transport, cell growth and senescence, metabolic homeostasis, signal transduction
Introduction
Autophagy is an evolutionarily conserved pathway that leads to the degradation of intracellular substances. Mechanically, cytoplasmic compounds are transported to lysosomes and degraded into basic components that can be recycled for use in cell construction. Depending on the form of intracellular substance transport, autophagy is categorized into three categories: macroautophagy (also referred to as autophagy), microautophagy, and chaperone-mediated autophagy [1]. The autophagy process is classified into four major stages: autophagy initiation, autophagosome formation, autophagosome-lysosome fusion, and autolysosome maturation [2,3].
ATG (autophagy related) proteins are highly conserved proteins that are extensively involved in autophagy regulation in distinct stages. Collectively, more than 30 autophagy-related proteins have been identified. For instance, by cooperating with ATG13, ATG101, RB1CC1/FIP200 (RB1 inducible coiled-coil 1), etc., ULK1 (unc-51 like autophagy activating kinase 1) plays essential roles in autophagy initiation by forming a protein kinase complex [4]. ATG7, ATG3, ATG10, ATG5 and ATG12 form one of the two ubiquitin-like conjugation systems that have been implicated in the formation and expansion of autophagosomes. ATG7 functions as an E1-like enzyme (ubiquitin-activating enzyme), while ATG3 and ATG10 function as an E2-like enzymes (ubiquitin-conjugating enzyme) to synergistically catalyze the conjugation of ATG12–ATG5. Another ubiquitin-like conjugation system involves Atg8-family proteins. Autophagosome formation and maturation depend on the ATG12–ATG5 and the MAP1LC3/LC3 (microtubule associated protein 1 light chain 3)-phosphatidylethanolamine (PE) complexes established via the ubiquitin-like conjugation system. Additionally, mammals carry a homolog of the yeast VPS30/ATG6 gene called BECN1 (beclin 1), the product of which interacts with a class III phosphatidylinositol 3-kinase to form the PIK3C3/VPS34 complex and participates in autophagic vesicle membrane elongation.
The roles and mechanisms of autophagy-related proteins in the regulation of autophagy have been extensively explored. However, an increasing number of studies have suggested that ATGs function without affecting autophagy to regulate distinct biological processes. For instance, Takacs-Vellai et al., for the first time, found that, in addition to regulating autophagy, BEC-1 (homolog to human BECN1) is involved in CED-3/caspase-dependent apoptotic cell death in Caenorhabditis elegans [5]. Similarly, Ogura and Goshima found that unc-51, encoding a serine/threonine kinase and homologous to yeast ATG1, is involved in neuronal development without affecting autophagy in Caenorhabditis elegans [6].
This review specifically focuses on the nonautophagic functions of autophagy-related proteins and summarizes the distinct autophagy-independent regulatory mechanisms of autophagy-related proteins in multiple physiological processes, such as metabolic homeostasis, cell development, immune regulation, cell growth, proliferation, and senescence. The categorization and summarization of these nonautophagic functions of autophagy-related proteins may meaningfully contribute to an increased understanding of the biological roles of these autophagy regulators and inform the translational potential of autophagy-related proteins for use in clinical treatments.
Overview of autophagy
Through autophagy in eukaryotes, lysosomes degrade intracellular cytoplasmic proteins and damaged organelles via the regulatory actions on ATGs. Autophagy is beneficial to the growth and development of cells and protects cells from metabolic stress and oxidative damage. However, excessive autophagy may lead to metabolic stress and even cell death. Autophagy involves several membrane rearrangement events in cells [1], in a process classified into the initiation state of autophagy, the formation of sequestration membranes and autophagosomes, the fusion of autophagosomes with lysosomes, and the degradation of cargo transported in autophagosomes (Figure 1).
Figure 1.

Overview of autophagy. The PIK3C3/VPS34 and ULK1 complexes are involved in the initiation of autophagy. ATG4 cleaves LC3 and participates in LC3 lipidation. ATG7 contributes to two ubiquitin-like processes, which are formed by the ATG12–ATG5 system and LC3 system, respectively. Both ubiquitin-like systems are involved in the elongation and maturation of autophagosomes, which are fused with lysosomes and delivering their encapsulated contents for degradation ultimately.
Autophagy initiation and phagosome formation involve several protein complexes including the PIK3C3/VPS34 complex and the ULK1 complex [7]. The PIK3C3/VPS34 complex comprises PIK3C3/VPS34 (phosphatidylinositol 3-kinase catalytic subunit type 3), BECN1, ATG14, PIK3R4/VPS15/p150 and NRBF2, and the ULK1 complex is an important positive regulator of autophagosome formation. Expansion of a phagophore leads to the formation of an autophagosome, which is characterized by a unique double-membrane structure. This step requires the participation of two ubiquitin-like conjugating pathways, both of which are catalyzed by ATG7. When autophagosome formation is complete, LC3-II on the outer membrane is cleaved from its PE cognate via ATG4 and released back into the cytoplasm [8]. After fusion, a series of acid hydrolases degrade the materials sequestered within the cytoplasm. Small molecules produced via this degradation process, particularly amino acids, are transported back to the cytoplasm and reused for protein synthesis and to maintain normal cellular functions when nutrient levels are low or absent.
The nonautophagic functions of ATGs
As mentioned above, an increasing number of studies have indicated that ATGs regulate distinct biological processes without affecting autophagy. Next, we specifically focus on the nonautophagic functions of ATGs in the following sections, and multiple aspects of ATGs’ nonautophagic functions have been summarized (Figure 2).
Figure 2.
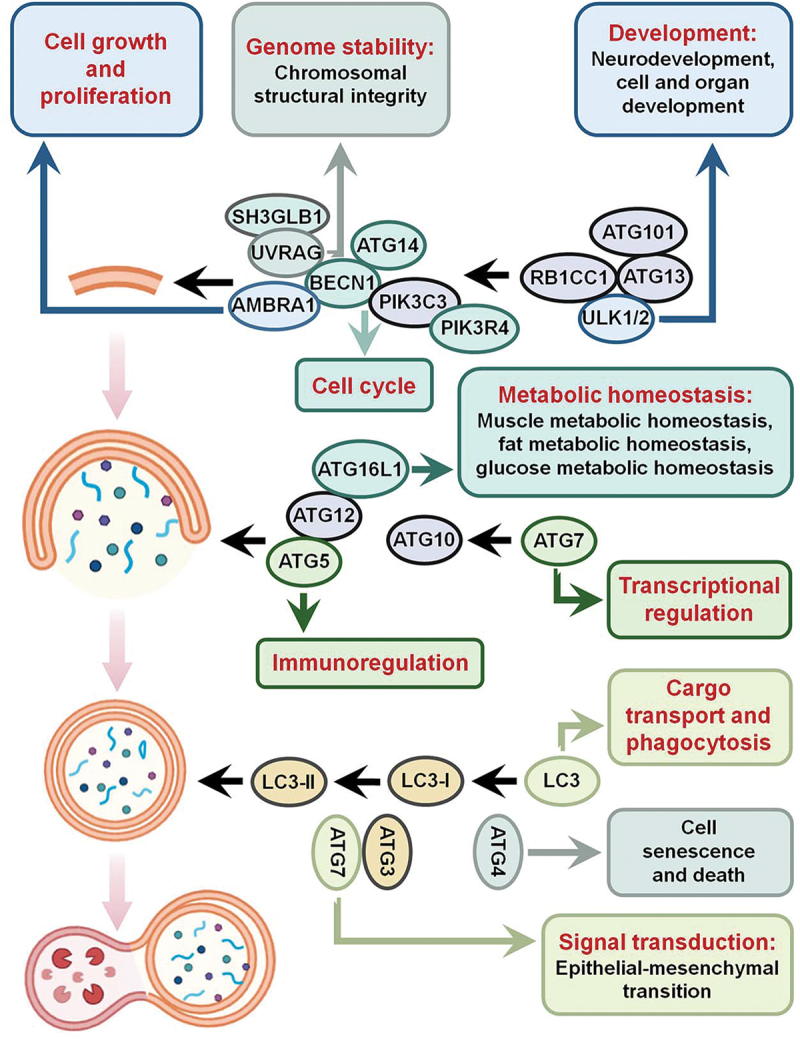
Overview of the nonautophagic functions of ATGs. The ULK1-ATG13-RB1CC1/FIP200-ATG101 complex activates autophagy, the ATG12–ATG5-ATG16L1 complex contributes to the extension of autophagic vesicles, and LC3 is involved the formation of autophagosomes. However, ATGs function in many nonautophagic physiological roles. For instance, ULK1 is involved in neurodevelopment as well as cell and organ development; UVRAG regulates genomic stability; AMBRA1 is involved in cell growth and proliferation; ATG4 regulates cell senescence and cell death; ATG5 regulates the immune response; BECN1 affects the cell cycle; ATG7 regulates gene transcription; LC3 is involved in cargo transport and phagocytosis; and ATG16L1 is involved in the regulation of cell metabolic homeostasis.
Genome stability
The integrity of the genome is intimately correlated with multiple autophagy-related proteins, such as AMBRA1 (autophagy and beclin 1 regulator 1), BECN1, and UVRAG (UV radiation resistance associated). Xu et al. demonstrated that BECN1 is gradually localized to the nucleus during postnatal development in mice. When ATG7 is absent, BECN1 is still recruited to sites of DNA damage via its interactions with TOP2B (DNA topoisomerase II beta), thereby enhancing DNA double-strand break repair and maintaining genomic integrity [9,10]. BECN1, but not ATG5, regulates centrosome number, suggesting that BECN1 May maintain genome stability independent of its autophagic function [9]. By knocking out Atg5 and Atg7 in mouse bone marrow ITGAM/CD11b+ LY6G− myeloid cells, Fang et al. found that the loss of ATG7 resulted in the abnormal accumulation and aberrant cytoplasmic localization of histone H3C3/H3.1 in a nonautophagic manner, possibly because of suppressed heat-shock protein DNAJ expression. Moreover, ATG7-regulated histone H3C3/H3.1 further controls chaotic nucleosome/chromatin assembly [11]. Thus, BECN1 and ATG7 maintain genome stability via two distinct pathways, DNA replication and damage repair, and it is worth investigating whether BECN1 and ATG7 act synergistically.
UVRAG is an autophagy-related protein and tumor suppressor that is involved in maintaining the structural integrity and proper separation of chromosomes [12]. In addition to interacting with CEP63 (centrosomal protein 63) [12], an important component in the centrosome that maintains centrosome stability and euploidy, UVRAG also interacts with the DNA double-strand break-binding protein XRCC5/KU80 (X-ray repair cross complementing 5) to promote DNA double-strand break repair [13]. Combined with the supposition that BECN1 is involved in DNA damage repair [9,10] and reports showing that UVRAG interacts with BECN1 to activate autophagy, we suspect that UVRAG and BECN1 could function in correlation during DNA double-strand break repair. In addition, a mutation of UVRAG, leading to a truncated protein product, has been closely associates with microsatellite unstable colon cancer without affecting autophagy [14]. Thus, UVRAG is an attractive target gene for cancer therapy since its depletion may render cells more susceptible to chromosomal damage [15]. In summary, autophagy proteins are extensively involved in the regulation of genomic stability.
Transcriptional regulation
ULK1, ATG5, ATG7, and ATG16L1 affect mRNA levels and gene expression by modulating transcription stages independent of autophagy (Table 1). ULK1 is the mammalian homolog of the Saccharomyces cerevisiae autophagy protein Atg1, which is the sole protein with serine/threonine kinase activity in the autophagy signaling pathway [89]. Kalie et al. showed that ULK1 inhibits TYR (tyrosinase) transcription and melanogenesis independent of its autophagic function. Interestingly, WIPI1 (WD repeat domain, phosphoinositide interacting 1), a mammalian analog of yeast Atg18, enhances MITF (melanocyte inducing transcription factor) activation as well as the expression of downstream target gene TYR by blocking MTOR (mechanistic target of rapamycin kinase) complex 1 (MTORC1) signaling activity to regulate melanosome formation [16,82]. Therefore, whether ULK1 and WIPI1 coregulate MITF gene transcription to affect melanogenesis is worthy of attention.
Table 1.
Summary of the nonautophagic roles of ATGs.
| ATG proteins | Nonautophagic roles | References |
|---|---|---|
| ULK1/UNC-51/ATG1 | affects transcription, endocytosis and synaptic vesicle trafficking; regulates lipid metabolism, glucose metabolism and other metabolism; maintains ER homeostasis; regulates; suppresses cell growth; regulates development, cell senescence and cell death | Kalie et al. [16], Toda et al. [17], Wang et al. [18], Wang et al. [19], Park et al. [20], Sinha et al. [21], Li et al. [22], Joo et al. [23], Kuhn et al. [24], Lee et al. [25], Tyra et al. [26], Tekinay et al. [27], Tomoda et al. [28], Wairkar et al. [29], Xing et al. [30], Mizushima [31], Joshi et al. [32] |
| ATG3 | regulates mitochondrial activity and lipid content; affects cell death and chorion biogenesis | Da Silva Lima et al. [33], Santos and Ramos. [34], Radoshevich et al. [35] |
| ATG4 | regulates epidermal homeostasis and cell death | Ramkumar et al. [36] |
| ATG5 | affects protein post-translational modification and DNA methylation; regulates EMT, immune signaling pathways and phagocytosis; restraints tumor cell proliferation; promotes caspase-dependent cell death | Sun et al. [37], Huang et al. [38], Yang et al. [39], Takeshita et al. [40], Oh and Lee. [41], Li et al. [42], Li et al. [42], Bell et al. [43], Oh et al. [44], Kimmey et al. [45], Zhao et al. [46] |
| BECN1/ATG6 | promotes DNA double-strand break repair and enhances genome stability; modulates the transcriptome shift, intracellular vesicle transport, pollen germination and endocytosis; impacts the growth factor signaling transduction and type I IFN signaling; regulates chorion biogenesis; affects cell cycle and cell death; | Takacs-Vellai et al. [5], Park et al. [9], Xu et al. [10], Wang et al. [47], Rohatgi et al. [48], Qin et al. [49], Fujiki et al. [50], Shravage et al. [51], Lőrincz et al. [52], Bork et al, 2022 45]; Jin et al. [53] |
| ATG7 | maintain proper nucleosome assembly; enhances the stability of CTNNB1/β-catenin and regulates gene transcription; modulates EMT activity, the cAMP pathway, the c-JUN/PTEN pathway and the NF-κB pathway; regulates cell cycle arrest and development | Fang et al. [11], Yamaguchi et al. [54], Huang et al. [55], Feng et al. [56], Liu et al. [57], Levine and Kroemer. [58], Lin et al. [59], Zhuang et al. [60] |
| ATG8/LC3 | affects neural signal transduction and immune signal transduction; participates in non-autophagic phagocytosis and COPII-dependent endoplasmic reticulum output; affects development | Wesch et al. [61], Hawkins and Klionsky. [62], Martens and Fracchiolla. [63], Galluzzi and Green. [64], Jipa et al. [65], Al-Younes et al. [66] |
| ATG9 | regulates the MAPK/JNK signaling pathway and neurite outgrowth; regulates necrotizing programmed death and the formation of actin cytoskeleton; inhibits dsDNA-induced innate immune response | Tang et al. [67], Xiong et al. [68], Wen et al. [69], Kiss et al. [70], Imagawa et al. [71], Saitoh et al. [72], Saitoh et al. [73] |
| ATG10 | may act as a transcription factor | Zhao et al. [74], Zhang et al. [75] |
| ATG12 | regulates mitochondrial biogenesis and cellular respiration; regulates energy metabolism and cell death | Malhotra et al. [76], Liu et al. [77], Radoshevich et al. [35] |
| ATG16L1 | regulates gene transcription; promotes plasma membrane repair and maintains cholesterol homeostasis; involved in spermatogenesis and flagella formation | Li et al. [78], Tan et al. [79], Tan et al. [80], Boukhalfa et al. [81] |
| ATG18/WIPI1 | promotes MITF-M and TYR transcription | Ho et al. [82] |
| AMBRA1 | affects cell proliferation | Cianfanelli et al. [83], Cianfanelli et al. [84] |
| RB1CC1/FIP200 | involved in embryogenesis | Chen et al. [85] |
| UVRAG | preserves centrosome stability and euploidy; regulates EMT, cell proliferation and melanosome biogenesis; | Zhao et al. [12], Son et al. [13], Knaevelsrud et al. [14], He et al. [86], Afzal et al. [87], Li et al. [88] |
Additionally, ATG5 decreases the transcription of the mismatch repair gene MLH1 (mutL homolog 1) by enhancing the methylation of the MLH1 promoter, and it also interacts with EIF4A, a key component of the translation initiation complex, to regulate RNF20 translation and thus affect histone H2Bub1 modification [37,38]. In conclusion, ATG5 affects protein posttranslational modification and DNA methylation independent of its autophagic function.
Moreover, BECN1 suppresses the efficiency of induced cardiomyocytes in an autophagy-independent manner. BECN1 modulates the transcriptome shift during the early stage of induced cardiomyocyte reprogramming without altering chromatin accessibility. In addition, depletion of BECN1 enhances CTNNB1/β-catenin (catenin beta 1) nuclear translocation and activates the canonical WNT-CTNNB1/β-catenin signaling pathway during the induction of induced cardiomyocytes [47]. Through the genetic ablation of Atg7, but not Atg5, Yamaguchi et al. demonstrated that ATG7 interacts with TRP53/p53 (tumor protein p53) to increase Bbc3/Puma (BCL2 binding component 3) and Ripk3 (receptor-interacting serine-threonine kinase 3) transcription, which are essential regulators of cell necrosis, while the CCR4-NOT deadenylase complex diminishes the stability of Atg7 mRNA [54]. Moreover, ATG7 stimulates POU5F1/OCT4 (POU class 5 homeobox 1) transcription and maintains cancer stem cell identities by affecting the stability of CTNNB1/β-catenin [55]. In Epinephelus coioides, the Atg16 homolog EcATG16L1 blocks the functions of IFN (interferon), IFN-stimulated response elements, the NFKB (nuclear factor kappa B) promoter and proinflammatory cytokines, and negatively regulates gene transcription [78]. In addition, Atg16 affects the transcription or mRNA stability of the neuropeptide hormone Crz (Corazonin), whose expression is regulated by the transcription factor apt (apontic) during the alcohol-induced sedation response in Drosophila melanogaster. Thus, determining whether Atg16 affects the transcription factor apt, thereby influencing Crz transcription, may be a valuable line of inquiry [90].
Mutations to the autophagy protein ATG10 has been implicated in transcriptional regulatory effects. Two isoforms of the ATG10 protein have been identified, wild-type ATG10 and a short isoform, ATG10S, which lacks a 36-amino acid fragment. In addition to regulating autolysosome formation, ATG10S is translocated to the nucleus where it activates the expression of IFNL2/IL28A (interferon lambda 2) and IFNL3/IL28B (interferon lambda 3) and a series of immune-related genes, including RIGI/DDX58 (RNA sensor RIG-I), TLR3 (toll like receptor 3), TLR7 (toll like receptor 7), IRF3 (interferon regulatory factor 3) and IRF7 (interferon regulatory factor 7), in response to hepatitis C virus (HCV) subgenomic replication [74]. Moreover, two amino acids, cysteine 44 and 135 (Cys44 and Cys135, respectively), in ATG10 are profoundly involved in the regulatory roles played by ATG10 during transcription in response to HCV replication. ATG10 in which Cys44 and Cys135 have been replaced translocated to the nucleus, bound to the promoter of the IFNL2/IL28A gene, and eventually activated the expression of IFNL2/IL28A, suggesting that ATG10 with Cys44 and/or Cys135 mutations might function as a transcription factor to trigger the expression of specific immune-related genes during the anti-HCV response [75]. Additionally, ATG7 is expressed as a short isoform named ATG7(2), which has lost the canonical roles of ATG7 in regulating autophagy. ATG7(2) fails to bind LC3B to mediate its lipidation, but the roles of ATG7(2) remain to be investigated [91]. In summary, autophagy-related proteins can influence gene transcription, epigenetic modifications, and transcriptomic changes in an autophagy-independent manner.
Signal transduction
The autophagy-related proteins ATG5, ATG7, AMBRA1, and UVRAG affect the epithelial-mesenchymal transition (EMT) in different ways [39,56,86,92]. Li et al. found that the endogenous MIR30D-sponging circular RNA circHECW2 binds to miR30D to inhibit its activity, resulting in increased ATG5 expression without influencing autophagic flux. The elevated ATG5 further accelerates the endothelial-mesenchymal transition (EndoMT) via the action of the NOTCH1 signaling pathway [39]. In contrast, Feng et al. found that ATG7, by interacting with PKM2 (pyruvate kinase M1/2), a protein critical for the Warburg effect during tumorigenesis, reduces the phosphorylation of PKM2 at Tyr105 by preventing PKM2 binding to its upstream kinase, thereby dampening the Warburg effect and impairing the EMT activity [56]. The pluripotent stem cell marker POU5F1/OCT4 inhibits the EMT, and POU5F1/OCT4 is leveraged by ATG7 to regulate the EMT. ATG7 stabilizes CTNNB1/β-catenin, a mesenchymal marker that binds to the POU5F1/OCT4 promoter region and thus stimulates transcription of POU5F1/OCT4, further affecting the EMT progression [55]. The precise mechanisms by which ATG7 influences the EMT still need to be studied in depth. ATG7 triggers the expression of endothelial FN1 (fibronectin 1) by regulating the phosphorylation of CREB1/CREB (cAMP responsive element binding protein 1) mediated via protein kinase A [57]. Moreover, UVRAG has also been reported to participate in the regulation of the EMT by inhibiting the activation of RAC1 (Rac family small GTPase 1) [86]. A variety of autophagy-related proteins exert different effects on the EMT through distinct mechanisms, but whether there is crosstalk between these mechanisms is still unclear.
Autophagy proteins have been implicated in immune signaling pathways, growth factor signaling pathways, and neural signal transduction [40,48]. Specifically, previous studies on the roles of ATGs in regulating CTNNB1/β-catenin nuclear translocation have yielded conflicting results. BECN1 inhibits the nuclear translocation of CTNNB1/β-catenin, thus regulating the WNT signaling pathway, but Huang et al. found that ATG7 promotes CTNNB1/β-catenin stability and nuclear translocation [47,55]. Whether these contradictory effects on CTNNB1/β-catenin nuclear translocation by distinct ATGs lead to opposite effects on the WNT signaling pathway is a worthy line of inquiry. Tang et al. reported that ATG9/Atg9/dAtg9 regulates the MAPK/JNK (mitogen-activated protein kinase) signaling pathway in both mammals and Drosophila by interacting with TRAF6/Traf6/dTRAF2 (TNF receptor associated factor 6). Moreover, reactive oxygen species (ROS)-induced autophagy establishes as a negative feedback loop that regulates MAPK/JNK activity by disrupting the association between ATG9/Atg9/dAtg9 and TRAF6/Traf6/dTRAF2 [67]. Although ATG9 and autophagy play different roles in oxidative stress-induced MAPK/JNK activation, their synergistic regulation maintains the MAPK/JNK signaling pathway in a stable state.
AMBRA1 prevents PTK2/FAK1 (protein tyrosine kinase 2) overactivation in several signaling pathways and affects melanoma growth and invasion [92]. In addition to autophagy, Atg8-family proteins participate in multiple signaling pathways, such as pathways involved in neural signal transduction [61]. In summary, the nonautophagic activities of autophagy-related proteins are intimately tied to the regulation of intracellular signaling pathways.
Cargo transport and phagocytosis
Recently, cargo transport and phagocytosis have been reported to be regulated by multiple ATG proteins without affecting autophagy. Atg1 binds and phosphorylates the Khc (Kinesin heavy chain) adapter protein Unc-76 (Uncoordinated 76) at serine 143 and increases its affinity to Syt1 (Synaptotagmin 1), which is a major transmembrane protein in synaptic vesicles, thereby promoting synaptic vesicle transport in a phosphorylation-dependent manner in Drosophila melanogaster [17]. Studies have indicated that Drosophila ULK1/Atg1 plays a conserved role in regulating kinesin-mediated vesicle transport, thereby promoting the transport of axonal guidance molecules [93]. Furthermore, ULK1 and ULK2 participate in the endocytosis and transport of NTRK1 (neurotrophic receptor tyrosine kinase 1) receptor complexes, thus regulating RAB5 activity [18]. ATG6 (BECN1 in mammals), an Arabidopsis homolog of yeast Vps30/Atg6, is the core component of the PIK3C3/VPS34 complex, which is required for autophagy and important to the sporophyte and gametophyte stages in plant development. In Arabidopsis, ATG6 is involved in vesicle transport and pollen germination [49,50]. Atg6 has been reported to be essential for endocytosis and protein secretion in Drosophila melanogaster, and Atg6 mutants show hematopoietic abnormalities and enhanced formation of melanotic blood cell mass due to defects in several vesicle transport pathways [51]. Similarly, Lőrincz et al. found that endocytosis is severely impaired in Atg6-depleted pupal wing cells, late endosomes form abnormally, and multilayer bodies accumulate in dense vesicular bodies [52]. Additionally, Bork et al. recently reported that loss of BECN1 in podocytes results in aberrant vesicle formation in the trans-Golgi network (TGN), leading to marked vesicle accumulation and disrupted patterns of intracellular vesicle trafficking and membrane dynamics [94].
Immune cells, such as macrophages and microglia, consume and degrade the extracellular matrix during phagocytosis, a process that is essential for pathogen elimination and inflammation control by clearing extracellular debris. Once internalized, phagosomes enter the endosome transport system and eventually fuse with the lysosomes, a process similar to that of phagocytosis and autophagy. Key distinctions are that during phagocytosis, cells engulf extracellular materials in single-membrane vesicles, while in autophagy, cells engulf intracellular materials in double-membrane vesicles. O’Brien et al. found that the phagocytosis of apoptotic cells mediated by BECN1 May be involved in the development of Alzheimer disease (AD) [95]. ATG9 is a transmembrane protein that is important for autophagosome membrane extension because it affects the transportation of membrane lipids to growing autophagosomes. Xiong et al. compared the phenotypes of Dictyostelium discoideum after atg16(-) mutation and atg9(-) atg16(-) double mutations, and compared their phenotypes to that of atg9(-) mutant reported previously. Their observations indicated that ATG9 and ATG16 May be involved in pinocytosis and phagocytosis in an autophagy-independent manner [68]. In addition to the typical functions of ATG16L1 in regulating LC3 lipidation and autophagosome formation, ATG16L1 promotes lysosome-mediated plasma membrane repair and inhibits bacterial transmission independent of the roles it plays in autophagy [79,80].
Atg8/LC3-family proteins are usually recognized as autophagosome markers and reporters for autophagy. In addition to regulating autophagy, Atg8-family proteins participate in LC3-related phagocytosis, LC3-associated endocytosis, and antiviral immune signal transduction [62]. Thus, the noncanonical functions of Atg8-family proteins mainly include LC3-related phagocytosis, endocytosis, the interactions of Atg8-family proteins with viruses, and the unconventional secretion pathway mediated by Atg8-family proteins through autophagosomes or extracellular vesicles [96]. It has been well-known that Atg8/LC3 lipidation is a hallmark of autophagy. However, Atg8/LC3 lipidation can occur independently autophagy. Moreover, by interacting with TECPR2 (tectonin beta-propeller repeat containing 2), MAP1LC3C/LC3C promotes COPII-dependent endoplasmic reticulum output independent of its functions in autophagy [63]. Similarly, Oh and Lee found that loss of ATG5 in dendritic cells enhances their phagocytosis, whereas loss of ATG7 or ATG16L1 does not exert a similar effect [41]. Moreover, ATG3, ATG4B, and ATG7 have also been reported to be associated with nonautophagic phagocytosis [64]. Taken together, studies suggest that autophagy-related proteins can affect phagocytosis in a nonautophagic manner through different mechanisms.
Metabolic homeostasis
Metabolism involves a series of chemical processes that sustain life in living creatures. Maintenance of metabolic homeostasis is required for the normal functions of cells, tissues, and organisms. ULK1 is a direct target of energy- and nutrient-sensing kinases and interacts with MTOR to regulate energy metabolism. ULK1 plays atypical roles that have been linked to metabolic signaling networks and are vital for cellular metabolic homeostasis [97]. In addition, ULK1 is involved in muscle tissue metabolic balance and lipid metabolic balance independent of its roles in autophagy [19–21]. Interestingly, although ULK1 and ULK2 are mammalian homologs of Drosophila Atg1, they exhibit redundant functions [98]. For instance, studies have indicated that ULK1 positively regulates β-oxidation/fatty acid oxidation and negatively regulates fatty acid uptake and synthesis, whereas ULK2 positively regulates fatty acid uptake [99]. How these two proteins regulate fatty acid absorption in concert is unclear. Moreover, in addition to regulating autophagy, ULK1/2 function in glucose metabolism as a bifurcate node, thus preserving the glucose metabolism pathway [22].
An increasing number of studies have shown that multiple autophagy-related proteins contribute to the regulation of cellular and metabolic homeostasis. Autophagy plays a protective role against ER stress, but Woo et al. found that knocking down ULK1 and ATG13 expression increases cell viability, suggesting that ULK1 and ATG13 play different roles in ER stress [23]. Furthermore, Joo et al. previously reported that SEC16A (SEC16 homolog A, endoplasmic reticulum export factor), a key regulator controlling cargo transport from the ER to the Golgi apparatus, binds to and is phosphorylated by ULK1 [100]. Thus, ULK1 and ATG13 May be involved in regulating ER stress independently of their roles in autophagy. In addition, several other autophagy-related proteins, such as ATG3, ATG4B, ATG5, BECN2 (beclin 2), and ATG7, have also been reported to be involved in the regulation of cell homeostasis. For example, ATG3 relies on SIRT1 (sirtuin 1) and MAPK8/JNK1 to control mitochondrial activity and lipid content, whereas MAPK8/JNK1 degrades SIRT1 [33]. ATG4B and LC3B are involved in the transfer of melanosomes to keratinocytes, which is essential for epidermal homeostasis [36]. Similarly, RAB7, an essential regulator of osteoclast function, localizes to the ruffled border of osteoclasts in an ATG5-dependent manner and participates in the polarized secretion of lysosomal contents into the extracellular space by mediating the fusion of lysosomes with the plasma membrane [101]. Several autophagy proteins, including ATG4B, ATG5, ATG7, and LC3, are required for the production of the osteoclast ruffled border, secretory function of osteoclasts, and bone resorption in vitro and in vivo. Notably, the N-terminal region of BECN2 interacts with the C-terminal region of GPRASP1 (G protein-coupled receptor associated sorting protein 1), thereby facilitating the ligand-induced endolysosomal transport and degradation of several G protein-coupled receptors/GPCRs to maintain cell homeostasis. However, in contrast to BECN2, BECN1 does not interact with GPRASP1 [102].
Several autophagic proteins have been reported to regulate yolk catabolism [24] and energy metabolism [76] in an autophagy-independent manner. The Atg1 complex maintains normal organelles during the embryonic stage by promoting yolk degradation via the Atg1-mTor pathway in Drosophila melanogaster [24]. Although ATG12 and ATG5 form a covalent complex which is essential for autophagy, ATG12 exerts distinct functions from ATG5 in regulating diet-induced obesity. Deletion of ATG12, but not ATG5, specifically in POMC (proopiomelanocortin)-expressing neurons, accelerates the high-fat diet-induced adiposity and reduced energy expenditure, suggesting nonautophagic functions for ATG12 in POMC neurons in regulating lipid and energy metabolism [76,77]. In addition, ATG12 has been reported to be partially localized in mitochondria and regulate mitochondrial biogenesis in cancer cells. Moreover, depletion of ATG12, but not ATG5, in cancer cells leads to decreased cell proliferation and increases oncotic cell death, due to reduced ATP availability [77].
The autophagy-related protein ATG16L1 significantly impacts the integrity of the cell membrane, promotes lysosomal-mediated plasma membrane repair and preserves cholesterol homeostasis [62,80]. Boukhalfa et al. demonstrated that the interaction between ATG16L1 and IFT20 (intraflagellar transport 20), a ciliary protein, affects primary cilium biogenesis, and further studies are needed to determine whether ATG16L1-IFT20 is involved in the repair of damaged plasma membranes [103]. Altogether, autophagic proteins are involved in cellular metabolism and preserve intracellular homeostasis and metabolic equilibrium.
Cell growth, proliferation and cell cycle regulation
An increasing number of studies have demonstrated that Atg1/ULK1 inhibits cell growth through its kinase activity. Lee et al. found that Atg1/ULK1 inhibits S6k/RPS6KB (Ribosomal protein S6 kinase) activity by reducing the S6k/RPS6KB-specific kinase function and subsequent phosphorylation of S6k at threonine 389 in both Drosophila and mammals [25]. Similarly, Tyra et al. found that Atg1 promotes yki (yorkie) phosphorylation at serine 74 and serine 97 in Drosophila melanogaster. The phosphorylation of yki by Atg1 inhibits the growth-promoting activity of yki [26]. Furthermore, in Drosophila, Atg1 functions as a downstream target of bsk/MAPK/JNK to block apoptosis-induced compensatory proliferation without triggering autophagy [104]. In conclusion, the kinase activity of Atg1 can exert biological functions without affecting autophagy.
The autophagy-related protein ATG9 has also been reported to be involved in the regulation of cell growth. For instance, Atg9 regulates midgut growth by blocking the mTor signaling pathway independently of its autophagic function and its interaction with gig/TSC2 in Drosophila melanogaster [69]. Although multiple autophagy-related regulators interact with the Tor signaling pathway factors, whether other autophagy-related proteins in addition to Atg9 influence cell growth by modulating the Tor signaling pathway remains unclear. Therefore, more studies are needed. In addition, the ATG9A subtype of the ATG9 protein is linked to neurite growth regulation in mice. Neurite outgrowth is significantly impaired in atg9a-knockout (KO) brains, and this tendency is maintained in cells with an atg7-KO background, indicating that the role of ATG9A in the regulation of neurite outgrowth is likely mediated independent of its autophagic functions [105].
The nonautophagic mechanism by which ATGs regulate cell proliferation remains a mystery. Recently, we discovered that the autophagy-related protein ATG5 regulates cell proliferation and embryonic stem cell differentiation by modulating the protein degradation of the oncoprotein MYC/c-Myc (MYC proto-oncogene, bHLH transcription factor). In addition to participating in cargo degradation via autophagy pathways, ATG5 promotes the degradation of MYC/c-Myc via the ubiquitin‒proteasome pathway. ATG5 functions as a scaffold protein to recruit the E3 ubiquitin ligase FBXW7 (F-box and WD repeat domain containing 7) and further promotes the ubiquitination and degradation of MYC/c-Myc, thereby preventing tumor cell proliferation under standard culture conditions [42]. Moreover, Pua et al. showed that ATG5 May regulate T-cell proliferation independent of its autophagic function [106]. Correspondingly, when Atg5 is overexpressed in osteoblasts, the number of cells in the G1 phase is decreased, while the number of those in the S phase is increased; however, no discernible difference in the total cell number is detected, possibly due to increased apoptosis occurring when Atg5 is overexpressed [107]. In addition, depletion of ATG4 inhibits proliferation, enhances apoptosis, and increases the protein levels of CASP3 (caspase 3) in hepatocellular carcinoma cells, possibly mediated via the AKT-CASP3 pathway [108].
The roles of UVRAG, a gene related to UV radiation resistance, in the regulation of autophagosome maturation and endocytosis has been established. Moreover, UVRAG regulates cell proliferation in a cell-type-specific manner independent of its autophagic functions. For example, UVRAG has been reported to be a regulator of innate peripheral T-cell homeostasis, likely by regulating peripheral innate T-cell proliferation [87]. Simultaneously, AMBRA1 is a pro-autophagy protein that influences carcinogenesis and cell proliferation mainly by facilitating the dephosphorylation and degradation of the proto-oncoprotein MYC/c-Myc [83]. In brief, AMBRA1 preferentially promotes the interaction between MYC/c-Myc and its phosphatase PPP2/PP2A, increasing PPP2/PP2A activity to enhance MYC/c-Myc degradation, thereby reducing the cell division rate and further reducing both cell proliferation and tumorigenesis [83]. In addition, BECN1 regulates MYC/c-Myc activity but through a different pathway from that involving AMBRA1. Although BECN1, AMBRA1 and PP2A-C are components of the same macromolecular complex, BECN1 indirectly modulates MYC/c-Myc phosphorylation via the EGFR pathway, whereas AMBRA1 is more directly engaged in the dephosphorylation of MYC/c-Myc, leading to its eventual degradation via the endocytic pathway [84]. In conclusion, autophagy-related regulators control cell growth and proliferation through various strategies independent of their roles in autophagy.
ATG7 functions independent of its E1-like enzyme activity to regulate the cell cycle. For instance, ATG7 inhibits the DNA damage response and regulates TRP53/p53-dependent cell cycle arrest and apoptosis [109]. Notably, deletion of the protein kinase CHEK2/CHK2 (checkpoint kinase 2) partially rescues neonatal lethality phenotype in atg7-knockout mice, supporting the aforementioned notion that ATG7 regulates the cell cycle independently of its roles in autophagy [58]. Simultaneously, ATG7-deficient cells showed an impairment in TRP53/p53-mediated cell cycle arrest under low-nutrient conditions, but an increase in the TRP53/p53-mediated cell death rate was observed after ATG7 depletion under sustained metabolic stress conditions [109]. In addition, BECN1 has been reported to interact directly with ZWINT/ZWINT-1, a component of the KMN (KNL1-MIS12-NDC80) complex, and is required for the centromere-microtubule interaction [110]. Subsequently, functioning downstream of the KMN complex, BECN1 further affects the recruitment of exocentromere proteins and facilitates the precise anchoring of centromeres to the spindle during mitosis, and this function of BECN1 is likely achieved through its regulatory effect on the RZZ (ROD-ZW10-ZWILCH) complex. Therefore, BECN1 May be involved in cell cycle regulation [110].
Cell development and neurodevelopment
Autophagy is essential for energy homeostasis during multiple stages of development. However, research suggests that autophagy-related proteins may impact germ cell development independent of their roles in regulating autophagy. In Rhodnius prolixus, the biogenesis of the chorion in oocytes depends on ULK1, ATG3, and ATG6 [34,111,112]. In Drosophila, autophagy-related proteins are involved in reproductive system development independent of their roles in autophagy. For instance, Atg8b, an isoform of Atg8, is involved in male development, and Atg9 is required for female fertility [65,70]. Furthermore, Boukhalfa et al. found that ATG16L1 is necessary for spermatogenesis and male development in mice. ATG16L1 interacts with IFT20 via its WD40 domain, thereby altering flagellum formation [81]. Simultaneously, RB1CC1/FIP200’s nonautophagic function has been shown to be adequate to support complete embryogenesis by retaining cell resistance to TNF-induced apoptosis [85]. In addition, UVRAG has been linked to melanosome development. In general, UVRAG is a direct target of MITF, a melanocyte-specific transcription factor, and the alpha-melanocyte-stimulating hormone cAMP regulates the expression of UVRAG by regulating MITF activity. Furthermore, an increasing amounts of UVRAG interact with BLOC-1 (biogenesis of lysosome-related organelle complex-1) to regulate melanosome biogenesis without affecting autophagy [88].
Several studies have suggested that autophagy-related proteins contribute significantly to tissue and organ development, which is another role for these proteins worth highlighting. For instance, Atg1 has been reported to be involved in mouse forebrain and corpus callosum development [18], while Atg7 regulates eye morphology in Drosophila [113]. Additionally, another Atg8 subtype, Atg8a, has been reported to be involved in midgut development [65]. More intriguingly, Plasmodium falciparum Atg18 and Atg8 homologs (PfATG18 and PfATG8) are required for the inheritance of the nonphotosynthetic plastid-like organelle apicoplast, and this function is independent of their roles in autophagy, and is likely mediated by their interactions with 3’-triphosphorylated phosphoinositides, which regulate various critical processes in Plasmodium falciparum parasites [114,115].
Recently, autophagy-related proteins have been shown to be essential for the development of certain microorganisms. For instance, Atg1, Atg7, and Atg8/LC3 specifically affect the development of Dictyostelium discoideum, Beauveria bassiana, and Chlamydia trachomatis [27,59,66]. First, atg1 deletion caused abnormal Dictyostelium discoideum development, which was reversible [27]. Second, Atg7 May participate in the sporulation of Beauveria bassiana [59]. The cytoplasmic nonlipidated form of LC3 interacts with Chlamydia trachomatis clathrates, allowing LC3 to function as a microtubule-associated protein rather than an autophagosome-associated component, and this interaction is essential for completion of the developmental cycle of Chlamydia trachomatis [66]. In summary, autophagy-related proteins regulate cell and organ development in multiple species in an autophagy-independent manner.
A recent study on neurodevelopment revealed that Atg1/Unc-51 is expressed in many neurons and is necessary for neuronal growth in Drosophila melanogaster [93]. Additionally, in Caenorhabditis elegans, UNC-51/ATG-1 is involved in axon development [28], in mouse, ULK1 and ULK2 are required for proper axons guidance [18], and in Drosophila, Atg1 is essential for proper synaptic density, structure and function in neurons [29]. By interacting with LET-92, a catalytic subunit of Caenorhabditis elegans PP2A (protein phosphatase 2A), UNC-51 affects the axonal guidance phenotype of neurons and negatively controls dorsally directed dorsal D (DD) and ventral D (VD) axonal guidance. Moreover, the PP2A catalytic subunit positively regulates UNC-51 action [116]. Furthermore, Atg1/UNC-51 phosphorylates the kinesin microtubule-associated complex Unc-76 in Drosophila melanogaster. Phosphorylated Unc-76 then controls synaptic vesicle transport by interacting with the synaptic vesicle protein Syt1 (Synaptotagmin 1) [17]. Moreover, ULK1/UNC51.1 regulates axonal growth and suppresses SYNGAP1 (synaptic Ras GTPase activating protein 1 homolog (rat)) activity through the RAS and RAB5 signaling pathways in house mouse [28]. Atg1/Unc-51 is required for synapses to reach the appropriate density in motoneurons to function properly in the apposition of active zones and glutamate receptors in Drosophila melanogaster [29]. UNC-51/Atg1 regulates axon guidance in DD/VD neurons by controlling the subcellular localization of the netrin receptor UNC-5 in Caenorhabditis elegans [6]. In addition, ATG7 has been reported to impact the development of the central nervous system and the polar decoupling of the Golgi apparatus and dendrites during dendritic development [60]. Together, these studies suggest that autophagy-related proteins, particularly UNC-51/Atg1/ULK1 in distinct species, play critical roles in neurodevelopment, likely in an autophagy-independent manner.
Cell senescence and death
ULK1 is the mammalian homolog of Atg1 and has been reported to participate in the regulation of cellular senescence in an autophagy-independent manner. Xing et al. found that ULK1 negatively regulates 26S proteasome activity through an O-linked N-acetylglucosamine (GlcNAc) transferase (OGT)-dependent pathway, thereby affecting the protein degradation of SIRT1 [30,31]. SIRT1 is a key regulator of cellular senescence that functions mainly through regulating the deacetylation of histones or nonhistones, including transcription factors [117]. Therefore, ULK1 likely regulates cell senescence by controlling SIRT1 protein levels. Additionally, loss of Atg6 in Drosophila intestinal stem cells (ISCs) results in the acquisition of an aging phenotype in ISCs; this phenotype manifests as excessive ISC proliferation, centrosome amplification, and increased DNA damage. However, depletion of intestinal progenitor cell-specific Atg6 does not affect the fate of the ISCs. This outcome suggests that Atg6 May be necessary to maintain the integrity of ISCs in the midgut of Drosophila [118].
Several proteins required for autophagy also play critical roles in apoptosis regulation. For example, ULK1 accelerates cell death by promoting the excessive accumulation of ROS. Moreover, hyperoxidants or DNA damage reagents activate the nuclear translocation of ULK1, promote its interaction with PARP1 (poly(ADP-ribose) polymerase 1), and thus increase the activity of PARP1 to accelerate ATP consumption and cell death [32,97]. Studies have shown that disruption of the ATG12–ATG3 complex leads to an increase in mitochondrial mass, destroy of the mitochondrial reticular system, and inhibition of the mitochondrial pathway-mediated cell death. Moreover, the regulation of mitochondrial homeostasis and cell death mediated by the ATG12–ATG3 complex is irrelevant to their roles in regulating autophagy [35].
ATG4D is a caspase substrate whose cleavage produces truncated products that enhance GABARAPL1 priming activity in vitro, and ATG4D overexpression usually induces cell death in human cells [119]. ATG4D cleaved by caspase is highly cytotoxic, and its toxicity is not related to enhanced autophagy, suggesting an autophagy-independent role for ATG4D in controlling cell death. However, the cytotoxicity of cleaved ATG4D is likely achieved through its C-terminal BH3 domain, which is essential for the transient binding of ATG4D to mitochondria [120]. Similarly, ATG5 induces high levels of cytotoxicity after being cleaved by calpain under stress conditions but does not promote autophagy [121]. Moreover, ATG5 directly interacts with the FADD (Fas associated via death domain) protein to promote caspase-dependent cell death [43]. Therefore, the regulation of cell death by ATG4D and ATG5 is protease sensitive and autophagy independent, and distinct proteases (calpain for ATG5 and CASP3 for ATG4D) are involved [120].
Interestingly, a previous study showed that several autophagosomal membrane-associated proteins, including ATG5, ATG8, and ATG16, recruit CASP8 (caspase 8) and are essential for CASP8 activation and apoptosis, allowing the autophagosomal membrane to serve as a platform for the intracellular death-inducing signaling complex/iDISC [122]. ATG5 and ATG7 regulate apoptosis and promote cell proliferation through the EIF2AK3/PERK (eukaryotic translation initiation factor 2 alpha kinase 3)-related pathway. The protective effect of ATG5 and ATG7 overexpression on chondrocyte survival depends on the presence and activity of EIF2AK3/PERK [123]. ATG6 is cleaved by multiple caspases (including CASP3, CASP6, CASP9, and CASP10) during TNFSF10/TRAIL (TNF superfamily member 10)-induced apoptosis and shows a protective effect on TNFSF10/TRAIL-induced cell death [124]. Furthermore, ectopic cell death resulting from BEC-1 (Caenorhabditis elegans homolog of mammalian BECN1 and yeast Vps30/Atg6) inactivation is closely related to the typical CED-3/caspase-mediated cell death pathway. Adult worms with BEC-1 knocked down presented with various developmental defects, including large intestinal vacuoles, impaired vulval processes or vulval ectoprocesses, and extension of the outer nuclear membrane of germ cells [5]. In addition, ATG6/BECN1 exerts distinct effects on the endogenous (e.g., induced by glucocorticoid or BCL2 inhibition) and exogenous (e.g., induced by anti-FAS treatment or BCL2 resistance) apoptotic death pathways. Overexpression of BECN1-GFP in thymocytes at high levels accelerates glucocorticoid-induced apoptosis. However, no detectable difference in cell sensitivity to anti-FAS or anti-TCR antibody was observed in cells with and without BECN1-GFP high overexpression. Thus, ATG6/BECN1 likely augments cell death induced by dexamethasone, but exerts no significant impact on apoptosis induced by anti-FAS or anti-TCR antibody [125]. Independent of its E1-like enzyme activity and autophagy function, ATG7 regulates TRP53/p53-dependent cell cycle arrest and apoptosis and inhibits DNA damage responses without affecting protein kinase CHEK2/CHK2 activity [58]. Necrotic programmed cell death during bone morphogenesis requires ATG9A action but not its autophagic activity. ATG9A-dependent necrosis is important for bone mineralization/calcification, and atg9a-knockout mice show bone surface malformations. However, whether ATG9A directly regulates cell death and whether this cell death induces bone calcification is unclear, and further investigation is needed [71].
Immunoregulation
Autophagy-related proteins are also involved in immune response regulation. For instance, Atg8/LC3-family members regulate antiviral immune responses by mediating the action of pattern recognition receptors (PRRs) and IFN production during viral infection [96]. Moreover, ATG5 is involved in innate and adaptive immunity. ATG5 regulates B-cell differentiation and plays key roles in cytokine secretion, pathogen clearance, and antigen presentation. Notably, atg5-knockout mice and human eosinophils with low levels of ATG5 expression show greater degranulation [126]. During respiratory syncytial virus (RSV) infection, ATG5 activates the anti-RSV immune response by regulating glycolytic reprogramming in dendritic cells, which highlights the metabolic roles of ATG5 in dendritic cells [44]. In addition, ATG5 does not rely on autophagy to defend against Mycobacterium tuberculosis infection but plays a unique and autophagy-independent protective role by preventing polymorphonuclear neutrophil-mediated immunopathology [45]. Furthermore, ATG5 has been demonstrated to be necessary for defense against the pathogens Listeria monocytogenes and Toxoplasma gondii in granulosa cells and macrophages. In primary macrophages, ATG5 promotes IFNG (interferon gamma) and lipopolysaccharide-induced injury to the Toxoplasma gondii parasitophorous vacuole membrane and parasite clearance [46]. STING1 (stimulator of interferon response cGAMP interactor 1), a key regulator in the dsDNA-induced innate immune response, is transported from the endoplasmic reticulum to the Golgi apparatus to ultimately localize to the cell particles where it binds TBK1 (an IRF3 kinase) during the response to viral dsDNA. Interestingly, STING1 and ATG9 colocalize to the Golgi apparatus and form typical puncta. However, when ATG9A is depleted, the number of punctate structures formed by STING1 in the Golgi apparatus are increased, and the interaction between STING1 and TBK1 is enhanced. Therefore, ATG9A inhibits the dsDNA-induced innate immune response, likely by regulating the STING1 and TBK1 interaction [72,73]. Moreover, a cyclic dinucleotide triggers ULK1/Atg1 to phosphorylate STING1, resulting in the suppression of STING1 activity. The impaired STING1 activity leads to suppressed expression of a series of innate immunity-related genes (such as IRF3) [127]. ULK1/Atg1 regulates the transcriptional activation of type I IFN-stimulating genes by recognizing the IFN stimulation response element and IFNG activation site. Therefore, ULK1/Atg1 activity is required for type I IFN-dependent antiviral and antitumor effects. Furthermore, ULK1/Atg1 is an upstream regulator of MAPK14/p38α and is necessary for the antiproliferative response and type I IFN-induced antitumor effect on bone marrow and external bone marrow proliferative tumor cells [128].
The levels of ATG1, ATG7, ATG8, and ATG9 are closely related to different degrees of Cryptococcus neoformans virulence, irrespective of their roles in regulating autophagy [129]. After dsDNA stimulation, CGAS (cyclic GMP-AMP synthase) interacts with BECN1 via the NTase (nucleotidyltransferase) domain in CGAS and the CCD (coiled-coil domain) in BECN1. Then, BECN1 inhibits the NTase activity of CGAS and reduces the synthesis of cGAMP (cyclic GMP-AMP), thereby decreasing IFNB (interferon beta) production and negatively regulating the IFN response. Thus, at least two independent negative feedback regulatory mechanisms mediate CGAS-STING1 pathway activation, namely, the BECN1-mediated inhibition of CGAS activity and the cGAMP-mediated inhibition of STING1 activity, through which the host immune system is ultimately maintained at an appropriate level [130]. Additionally, USP19 has been reported to negatively regulate the type I IFN signaling pathway by blocking the RIGI-MAVS interaction through a BECN1-mediated nonautophagic pathway. Lack of USP19 or BECN1 results in enhanced type I IFN signaling and antiviral responses following RNA virus infection [53]. Moreover, BECN1 and RB1CC1/FIP200 play key roles in the regulation of immune homeostasis. For instance, BECN1 and RB1CC1/FIP200 inhibit systemic IFNG effects, thereby regulating the quiescence of tissue-resident macrophages. Mice lacking BECN1 or RB1CC1/FIP200 in myeloid cells showed spontaneous immune activation and resistance to Listeria monocytogenes infection [131].
The crosstalk between the nonautophagic and autophagic functions of ATGs
Studies have shown that the autophagic and nonautophagic functions of ATGs are interrelated. On the one hand, these proteins lead to opposite outcomes or exhibit different regulatory patterns in specific processes. For instance, in addition to regulating protein degradation through autophagy, ATGs also modulate protein degradation through nonautophagic pathways. Specifically, ULK1 inhibits SIRT1 degradation through the proteasome pathway, thereby promoting cell senescence [30]. As mentioned above, both in mammals and Drosophila, ATG9/Atg9/dAtg9 activates MAPK/JNK signaling pathway and triggers ROS responses in an autophagy-independent manner, but this activation is hindered by the induction autophagy, forming negative feedback [67]. Similarly, ATG12 has been reported to enhance apoptosis by activating CASP3 in an autophagy-independent manner, while the activated CASP3 promotes the cleavage of BECN1, resulting in the inhibition of autophagy [132]. Through its nonautophagic activity, ATG7 inhibits the apoptosis-related function of CASP9 by directly binding to CASP9, while the ATG7-CASP9 complex promotes the formation of LC3-II, resulting in the enhancement of autophagy [133]. In summary, several studies have shown crosstalk between autophagy and nonautophagic processes mediated by ATGs. However, the circumstances under which ATGs play autophagic or nonautophagic roles needs to be further studied.
On the other hand, the autophagic and nonautophagic regulatory mechanisms of ATGs lead to similar biological consequences. For instance, autophagy mainly provides raw materials for cells and maintains cellular homeostasis by degrading senescent or damaged organelles and misfolded proteins. Multiple nonautophagic functions of ATGs are also involved in maintaining cellular homeostasis. For example, autophagy-related proteins, such as BECN1 and UVRAG, improve DNA damage repair without affecting autophagy, and ATG1 promotes yolk catabolism to provide energy for cells independently of its autophagic function [24]. In summary, the autophagic and nonautophagic functions of ATGs are closely related and cross-regulated, but the exact mechanisms still need to be examined in depth.
Conclusion
Autophagy-related studies have been a hotspot in the field of biomedical research. The physiological importance of autophagy is clearly recognized. Autophagy is regulated by multiple autophagy-related proteins, and the mechanisms by which ATGs regulate autophagy are clearly understood. However, ATGs also play numerous underappreciated and novel roles that are irrelevant to their autophagic functions. The identification of these autophagy-independent functions of ATGs are essential for a complete understanding of the biological roles of ATGs. This review specifically summarizes the progresses of the nonautophagic functions of autophagy-related proteins and contributes meaningfully to understanding the molecular basis underlying life processes especially in which ATGs are involved. However, a large number of unknown autophagy-independent functions of ATGs still need to be further explored.
Acknowledgements
The authors would like to thank all members of the Chen lab for discussions and supports.
The authors apologize to colleagues whose work could not be cited due to space constraints.
Funding Statement
This review was supported by the National Natural Science Foundation of China [Grant No.: 81972650 and 82273067], the Henan Provincial Natural Science Foundation [Grant No.: 222300420028 and 222300420120].
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
No data was used for the research described in the article.
References
- [1].Parzych KR, Klionsky DJ.. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signaling. 2014;20(3):460–473. doi: 10.1089/ars.2013.5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mehrpour M, Esclatine A, Beau I, et al. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20(7):748–762. doi: 10.1038/cr.2010.82 [DOI] [PubMed] [Google Scholar]
- [3].Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124–131. doi: 10.1016/j.ceb.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19(1):12. doi: 10.1186/s12943-020-1138-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Takacs-Vellai K, Vellai T, Puoti A, et al. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol. 2005;15(16):1513–1517. doi: 10.1016/j.cub.2005.07.035 [DOI] [PubMed] [Google Scholar]
- [6].Ogura K, Goshima Y. The autophagy-related kinase UNC-51 and its binding partner UNC-14 regulate the subcellular localization of the Netrin receptor UNC-5 in Caenorhabditis elegans. Development. 2006;133(17):3441–3450. doi: 10.1242/dev.02503 [DOI] [PubMed] [Google Scholar]
- [7].Lin MG, Hurley JH. Structure and function of the ULK1 complex in autophagy. Curr Opin Cell Biol. 2016;39:61–68. doi: 10.1016/j.ceb.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kaizuka T, Morishita H, Hama Y, et al. An autophagic flux probe that releases an internal control. Mol Cell. 2016;64(4):835–849. doi: 10.1016/j.molcel.2016.09.037 [DOI] [PubMed] [Google Scholar]
- [9].Park JM, Tougeron D, Huang S, et al. Beclin 1 and UVRAG confer protection from radiation-induced DNA damage and maintain centrosome stability in colorectal cancer cells. PLoS One. 2014;9(6):e100819. doi: 10.1371/journal.pone.0100819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Xu F, Fang Y, Yan L, et al. Nuclear localization of Beclin 1 promotes radiation-induced DNA damage repair independent of autophagy. Sci Rep. 2017;7(1):45385. doi: 10.1038/srep45385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fang Y, Gu Y, Li L, et al. Loss of Atg7 causes chaotic nucleosome assembly of mouse bone marrow CD11b + Ly6G - myeloid cells. Aging. 2020;12(24):25673–25683. doi: 10.18632/aging.104176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhao Z, Oh S, Li D, et al. A dual role for UVRAG in maintaining chromosomal stability independent of autophagy. Dev Cell. 2012;22(5):1001–1016. doi: 10.1016/j.devcel.2011.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Son JH, Hwang EC, Kim J. Systematic analyses of the ultraviolet radiation resistance-associated gene product (UVRAG) protein interactome by tandem affinity purification. Arch Pharm Res. 2016;39(3):370–379. doi: 10.1007/s12272-015-0689-1 [DOI] [PubMed] [Google Scholar]
- [14].Knaevelsrud H, Ahlquist T, Merok MA, et al. UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy. 2010;6(7):863–870. doi: 10.4161/auto.6.7.13033 [DOI] [PubMed] [Google Scholar]
- [15].Zhao Z, Ni D, Ghozalli I, et al. UVRAG: at the crossroad of autophagy and genomic stability. Autophagy. 2012;8(9):1392–1393. doi: 10.4161/auto.21035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kalie E, Razi M, Tooze SA, et al. ULK1 regulates melanin levels in MNT-1 cells independently of mTORC1. PLoS One. 2013;8(9):e75313. doi: 10.1371/journal.pone.0075313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Toda H, Mochizuki H, Flores R 3rd, et al. UNC-51/ATG1 kinase regulates axonal transport by mediating motor-cargo assembly. Genes Dev. 2008;22(23):3292–3307. doi: 10.1101/gad.1734608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang B, Iyengar R, Li-Harms X, et al. The autophagy-inducing kinases, ULK1 and ULK2, regulate axon guidance in the developing mouse forebrain via a noncanonical pathway. Autophagy. 2018;14(5):796–811. doi: 10.1080/15548627.2017.1386820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang B, Maxwell BA, Joo JH, et al. ULK1 and ULK2 regulate stress granule disassembly through phosphorylation and activation of VCP/p97. Mol Cell. 2019;74(4):742–57 e8. doi: 10.1016/j.molcel.2019.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Park JS, Lee DH, Lee YS, et al. Dual roles of ULK1 (unc-51 like autophagy activating kinase 1) in cytoprotection against lipotoxicity. Autophagy. 2020;16(1):86–105. doi: 10.1080/15548627.2019.1598751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sinha RA, Singh BK, Zhou J, et al. Loss of ULK1 increases RPS6KB1-NCOR1 repression of NR1H/LXR-mediated Scd1 transcription and augments lipotoxicity in hepatic cells. Autophagy. 2017;13(1):169–186. doi: 10.1080/15548627.2016.1235123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li TY, Sun Y, Liang Y, et al. ULK1/2 Constitute a Bifurcate Node controlling glucose metabolic Fluxes in addition to autophagy. Mol Cell. 2016;62(3):359–370. doi: 10.1016/j.molcel.2016.04.009 [DOI] [PubMed] [Google Scholar]
- [23].Woo M, Choi HI, Park SH, et al. The unc-51 like autophagy activating kinase 1-autophagy related 13 complex has distinct functions in tunicamycin-treated cells. Biochem Biophys Res Commun. 2020;524(3):744–749. doi: 10.1016/j.bbrc.2020.01.160 [DOI] [PubMed] [Google Scholar]
- [24].Kuhn H, Sopko R, Coughlin M, et al. The Atg1-Tor pathway regulates yolk catabolism in Drosophila embryos. Development. 2015;142(22):3869–3878. doi: 10.1242/dev.125419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee SB, Kim S, Lee J, et al. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 2007;8(4):360–365. doi: 10.1038/sj.embor.7400917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tyra LK, Nandi N, Tracy C, et al. Yorkie growth-promoting activity is limited by Atg1-mediated phosphorylation. Dev Cell. 2020;52(5):605–16 e7. doi: 10.1016/j.devcel.2020.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tekinay T, Wu MY, Otto GP, et al. Function of the Dictyostelium discoideum Atg1 kinase during autophagy and development. Eukaryot Cell. 2006;5(10):1797–1806. doi: 10.1128/EC.00342-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tomoda T, Kim JH, Zhan C, et al. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 2004;18(5):541–558. doi: 10.1101/gad.1151204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wairkar YP, Toda H, Mochizuki H, et al. Unc-51 controls active zone density and protein composition by downregulating ERK signaling. J Neurosci. 2009;29(2):517–528. doi: 10.1523/JNEUROSCI.3848-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xing J, Liu H, Yang H, et al. Upregulation of Unc-51-like kinase 1 by nitric oxide stabilizes SIRT1, independent of autophagy. PLoS One. 2014;9(12):e116165. doi: 10.1371/journal.pone.0116165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22(2):132–139. doi: 10.1016/j.ceb.2009.12.004 [DOI] [PubMed] [Google Scholar]
- [32].Joshi A, Iyengar R, Joo JH, et al. Nuclear ULK1 promotes cell death in response to oxidative stress through PARP1. Cell Death Differ. 2016;23(2):216–230. doi: 10.1038/cdd.2015.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].da Silva Lima N, Fondevila MF, Novoa E, et al. Inhibition of ATG3 ameliorates liver steatosis by increasing mitochondrial function. J Hepatol. 2022;76(1):11–24. doi: 10.1016/j.jhep.2021.09.008 [DOI] [PubMed] [Google Scholar]
- [34].Santos A, Ramos I. ATG3 is important for the chorion ultrastructure during oogenesis in the insect vector rhodnius prolixus. Front Physiol. 2021;12:638026. doi: 10.3389/fphys.2021.638026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Radoshevich L, Murrow L, Chen N, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142(4):590–600. doi: 10.1016/j.cell.2010.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ramkumar A, Murthy D, Raja DA, et al. Classical autophagy proteins LC3B and ATG4B facilitate melanosome movement on cytoskeletal tracks. Autophagy. 2017;13(8):1331–1347. doi: 10.1080/15548627.2017.1327509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sun SY, Hu XT, Yu XF, et al. Nuclear translocation of ATG5 induces DNA mismatch repair deficiency (MMR-D)/microsatellite instability (MSI) via interacting with Mis18alpha in colorectal cancer. Br J Pharmacol. 2021;178(11):2351–2369. doi: 10.1111/bph.15422 [DOI] [PubMed] [Google Scholar]
- [38].Huang X, Yang L, Cai FF, et al. Autophagy-related protein ATG5 regulates histone H2B mono-ubiquitylation by translational control of RNF20. J Genet Genomics. 2017;44(10):503–506. doi: 10.1016/j.jgg.2017.08.004 [DOI] [PubMed] [Google Scholar]
- [39].Yang L, Han B, Zhang Y, et al. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy. 2018;14(3):404–418. doi: 10.1080/15548627.2017.1414755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Takeshita F, Kobiyama K, Miyawaki A, et al. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy. 2008;4(1):67–69. doi: 10.4161/auto.5055 [DOI] [PubMed] [Google Scholar]
- [41].Oh DS, Lee HK. Autophagy protein ATG5 regulates CD36 expression and anti-tumor MHC class II antigen presentation in dendritic cells. Autophagy. 2019;15(12):2091–2106. doi: 10.1080/15548627.2019.1596493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li S, Zhang L, Zhang G, et al. A nonautophagic role of ATG5 in regulating cell growth by targeting c-Myc for proteasome-mediated degradation. iScience. 2021;24(11):103296. doi: 10.1016/j.isci.2021.103296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bell BD, Leverrier S, Weist BM, et al. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci U S A. 2008;105(43):16677–16682. doi: 10.1073/pnas.0808597105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Oh DS, Park JH, Jung HE, et al. Autophagic protein ATG5 controls antiviral immunity via glycolytic reprogramming of dendritic cells against respiratory syncytial virus infection. Autophagy. 2020;17(9):2111–2127. doi: 10.1080/15548627.2020.1812218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kimmey JM, Huynh JP, Weiss LA, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015;528(7583):565–569. doi: 10.1038/nature16451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhao Z, Fux B, Goodwin M, et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4(5):458–469. doi: 10.1016/j.chom.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang L, Ma H, Huang P, et al. Down-regulation of Beclin1 promotes direct cardiac reprogramming. Sci Transl Med. 2020;12(566). doi: 10.1126/scitranslmed.aay7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rohatgi RA, Janusis J, Leonard D, et al. Beclin 1 regulates growth factor receptor signaling in breast cancer. Oncogene. 2015;34(42):5352–5362. doi: 10.1038/onc.2014.454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Qin G, Ma Z, Zhang L, et al. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 2007;17(3):249–263. doi: 10.1038/cr.2007.7 [DOI] [PubMed] [Google Scholar]
- [50].Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007;143(3):1132–1139. doi: 10.1104/pp.106.093864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Shravage BV, Hill JH, Powers CM, et al. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013;140(6):1321–1329. doi: 10.1242/dev.089490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lorincz P, Lakatos Z, Maruzs T, et al. Atg6/UVRAG/Vps34-containing lipid kinase complex is required for receptor downregulation through endolysosomal degradation and epithelial polarity during Drosophila wing development. Biomed Res Int. 2014;2014:851349. doi: 10.1155/2014/851349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jin S, Tian S, Chen Y, et al. USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1. EMBO J. 2016;35(8):866–880. doi: 10.15252/embj.201593596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yamaguchi T, Suzuki T, Sato T, et al. The CCR4-NOT deadenylase complex controls Atg7-dependent cell death and heart function. Sci Signaling. 2018;11(516). doi: 10.1126/scisignal.aan3638 [DOI] [PubMed] [Google Scholar]
- [55].Huang H, Wang C, Liu F, et al. Reciprocal network between cancer stem-like cells and macrophages facilitates the progression and androgen deprivation therapy resistance of prostate cancer. Clin Cancer Res. 2018;24(18):4612–4626. doi: 10.1158/1078-0432.CCR-18-0461 [DOI] [PubMed] [Google Scholar]
- [56].Feng Y, Liu J, Guo W, et al. Atg7 inhibits Warburg effect by suppressing PKM2 phosphorylation resulting reduced epithelial-mesenchymal transition. Int J Biol Sci. 2018;14(7):775–783. doi: 10.7150/ijbs.26077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liu H, Wei JY, Li Y, et al. Endothelial depletion of Atg7 triggers astrocyte-microvascular disassociation at blood-brain barrier. J Cell Bio. 2023;222(5). doi: 10.1083/jcb.202103098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi: 10.1016/j.cell.2018.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lin HY, Wang JJ, Feng MG, et al. Autophagy-related gene ATG7 participates in the asexual development, stress response and virulence of filamentous insect pathogenic fungus Beauveria bassiana. Curr Genet. 2019;65(4):1015–1024. doi: 10.1007/s00294-019-00955-1 [DOI] [PubMed] [Google Scholar]
- [60].Zhuang SF, Liu DX, Wang HJ, et al. Atg7 regulates brain angiogenesis via NF-κB-Dependent IL-6 production. Int J Mol Sci. 2017;18(5):968. doi: 10.3390/ijms18050968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wesch N, Kirkin V, Rogov VV. Atg8-family proteins—Structural features and molecular interactions in autophagy and beyond. Cells. 2020;9(9):2008. doi: 10.3390/cells9092008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hawkins WD, Klionsky DJ. The expanding role of Atg8. Autophagy. 2021;17(11):3273–3274. doi: 10.1080/15548627.2021.1967566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Martens S, Fracchiolla D. Activation and targeting of ATG8 protein lipidation. Cell Discov. 2020;6(1):23. doi: 10.1038/s41421-020-0155-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Galluzzi L, Green DR. Autophagy-independent functions of the autophagy machinery. Cell. 2019;177(7):1682–1699. doi: 10.1016/j.cell.2019.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jipa A, Vedelek V, Merenyi Z, et al. Analysis of Drosophila Atg8 proteins reveals multiple lipidation-independent roles. Autophagy. 2021;17(9):2565–2575. doi: 10.1080/15548627.2020.1856494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Al-Younes HM, Al-Zeer MA, Khalil H, et al. Autophagy-independent function of MAP-LC3 during intracellular propagation of Chlamydia trachomatis. Autophagy. 2011;7(8):814–828. doi: 10.4161/auto.7.8.15597 [DOI] [PubMed] [Google Scholar]
- [67].Tang H-W, Liao H-M, Peng W-H, et al. Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev Cell. 2013;27(5):489–503. doi: 10.1016/j.devcel.2013.10.017 [DOI] [PubMed] [Google Scholar]
- [68].Xiong Q, Unal C, Matthias J, et al. The phenotypes of ATG9, ATG16 and ATG9/16 knock-out mutants imply autophagy-dependent and -independent functions. Open Biol. 2015;5(4):150008. doi: 10.1098/rsob.150008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wen JK, Wang YT, Chan CC, et al. Atg9 antagonizes TOR signaling to regulate intestinal cell growth and epithelial homeostasis in Drosophila. Elife. 2017;6:6. doi: 10.7554/eLife.29338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kiss V, Jipa A, Varga K, et al. Drosophila Atg9 regulates the actin cytoskeleton via interactions with profilin and ena. Cell Death Differ. 2020;27(5):1677–1692. doi: 10.1038/s41418-019-0452-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Imagawa Y, Saitoh T, Tsujimoto Y. Vital staining for cell death identifies Atg9a-dependent necrosis in developmental bone formation in mouse. Nat Commun. 2016;7(1):13391. doi: 10.1038/ncomms13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Saitoh T, Fujita N, Yoshimori T, et al. Regulation of dsDNA-induced innate immune responses by membrane trafficking. Autophagy. 2010;6(3):430–432. doi: 10.4161/auto.6.3.11611 [DOI] [PubMed] [Google Scholar]
- [73].Saitoh T, Fujita N, Hayashi T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106(49):20842–20846. doi: 10.1073/pnas.0911267106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhao Q, Hu ZY, Zhang JP, et al. Dual roles of two isoforms of autophagy-related gene ATG10 in HCV-Subgenomic replicon mediated autophagy flux and innate immunity. Sci Rep. 2017;7(1):11250. doi: 10.1038/s41598-017-11105-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zhang MQ, Li JR, Peng ZG, et al. Differential effects of autophagy-related 10 protein on HCV replication and autophagy flux are mediated by its Cysteine(44) and Cysteine(135). Front Immunol. 2018;9:2176. doi: 10.3389/fimmu.2018.02176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Malhotra R, Warne JP, Salas E, et al. Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy. 2015;11(1):145–154. doi: 10.1080/15548627.2014.998917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Liu H, He Z, Germic N, et al. ATG12 deficiency leads to tumor cell oncosis owing to diminished mitochondrial biogenesis and reduced cellular bioenergetics. Cell Death Differ. 2020;27(6):1965–1980. doi: 10.1038/s41418-019-0476-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Li C, Wang L, Zhang X, et al. Molecular cloning, expression and functional analysis of Atg16L1 from orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 2019;94:113–121. doi: 10.1016/j.fsi.2019.09.004 [DOI] [PubMed] [Google Scholar]
- [79].Tan JMJ, Mellouk N, Osborne SE, et al. An ATG16L1-dependent pathway promotes plasma membrane repair and limits Listeria monocytogenes cell-to-cell spread. Nat Microbiol. 2018;3(12):1472–1485. doi: 10.1038/s41564-018-0293-5 [DOI] [PubMed] [Google Scholar]
- [80].Tan JMJ, Mellouk N, Brumell JH. An autophagy-independent role for ATG16L1: promoting lysosome-mediated plasma membrane repair. Autophagy. 2019;15(5):932–933. doi: 10.1080/15548627.2019.1586261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Boukhalfa A, Roccio F, Dupont N, et al. The autophagy protein ATG16L1 cooperates with IFT20 and INPP5E to regulate the turnover of phosphoinositides at the primary cilium. Cell Rep. 2021;35(4):109045. doi: 10.1016/j.celrep.2021.109045 [DOI] [PubMed] [Google Scholar]
- [82].Ho H, Kapadia R, Al-Tahan S, et al. WIPI1 coordinates melanogenic gene transcription and melanosome formation via TORC1 inhibition. J Biol Chem. 2011;286(14):12509–12523. doi: 10.1074/jbc.M110.200543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Cianfanelli V, Fuoco C, Lorente M, et al. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol. 2015;17(1):20–30. doi: 10.1038/ncb3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Cianfanelli V, D’Orazio M, Cecconi F. AMBRA1 and BECLIN 1 interplay in the crosstalk between autophagy and cell proliferation. Cell Cycle. 2015;14(7):959–963. doi: 10.1080/15384101.2015.1021526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chen S, Wang C, Yeo S, et al. Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model. Genes Dev. 2016;30(7):856–869. doi: 10.1101/gad.276428.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].He S, Zhao Z, Yang Y, et al. Truncating mutation in the autophagy gene UVRAG confers oncogenic properties and chemosensitivity in colorectal cancers. Nat Commun. 2015;6(1):7839. doi: 10.1038/ncomms8839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Afzal S, Hao Z, Itsumi M, et al. Autophagy-independent functions of UVRAG are essential for peripheral naive T-cell homeostasis. Proc Natl Acad Sci U S A. 2015;112(4):1119–1124. doi: 10.1073/pnas.1423588112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Li S, Jang GB, Quach C, et al. Darkening with UVRAG. Autophagy. 2019;15(2):366–367. doi: 10.1080/15548627.2018.1522911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Deng R, Zhang HL, Huang JH, et al. MAPK1/3 kinase-dependent ULK1 degradation attenuates mitophagy and promotes breast cancer bone metastasis. Autophagy. 2021;17(10):3011–3029. doi: 10.1080/15548627.2020.1850609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Varga K, Nagy P, Arsikin Csordas K, et al. Loss of Atg16 delays the alcohol-induced sedation response via regulation of Corazonin neuropeptide production in Drosophila. Sci Rep. 2016;6(1):34641. doi: 10.1038/srep34641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ogmundsdottir MH, Fock V, Sooman L, et al. A short isoform of ATG7 fails to lipidate LC3/GABARAP. Sci Rep. 2018;8(1). doi: 10.1038/s41598-018-32694-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Di Leo L, Bodemeyer V, Bosisio FM, et al. Loss of Ambra1 promotes melanoma growth and invasion. Nat Commun. 2021;12(1). doi: 10.1038/s41467-021-22772-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Mochizuki H, Toda H, Ando M, et al. Unc-51/ATG1 controls axonal and dendritic development via kinesin-mediated vesicle transport in the Drosophila brain. PLoS One. 2011;6(5):e19632. doi: 10.1371/journal.pone.0019632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Bork T, Liang W, Kretz O, et al. BECLIN1 is essential for podocyte secretory pathways mediating vegf secretion and podocyte-endothelial crosstalk. Int J Mol Sci. 2022;23(7):3825. doi: 10.3390/ijms23073825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].O’Brien CE, Wyss-Coray T. Sorting through the roles of beclin 1 in microglia and neurodegeneration. J Neuroimmune Pharmacol. 2014;9(3):285–292. doi: 10.1007/s11481-013-9519-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Nieto-Torres JL, Leidal AM, Debnath J, et al. Beyond autophagy: the expanding roles of ATG8 proteins. Trends Biochem Sci. 2021;46(8):673–686. doi: 10.1016/j.tibs.2021.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wang B, Kundu M. Canonical and noncanonical functions of ULK/Atg1. Curr Opin Cell Biol. 2017;45:47–54. doi: 10.1016/j.ceb.2017.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Demeter A, Romero-Mulero MC, Csabai L, et al. ULK1 and ULK2 are less redundant than previously thought: computational analysis uncovers distinct regulation and functions of these autophagy induction proteins. Sci Rep. 2020;10(1):10940. doi: 10.1038/s41598-020-67780-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ro SH, Jung CH, Hahn WS, et al. Distinct functions of Ulk1 and Ulk2 in the regulation of lipid metabolism in adipocytes. Autophagy. 2013;9(12):2103–2114. doi: 10.4161/auto.26563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Joo JH, Wang B, Frankel E, et al. The noncanonical role of ULK/ATG1 in ER-to-Golgi trafficking is essential for cellular homeostasis. Mol Cell. 2016;62(4):491–506. doi: 10.1016/j.molcel.2016.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].DeSelm CJ, Miller BC, Zou W, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011;21(5):966–974. doi: 10.1016/j.devcel.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].He C, Wei Y, Sun K, et al. Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism. Cell. 2013;154(5):1085–1099. doi: 10.1016/j.cell.2013.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Boukhalfa A, Roccio F, Dupont N, et al. When the autophagy protein ATG16L1 met the ciliary protein IFT20. Autophagy. 2021;17(7):1791–1793. doi: 10.1080/15548627.2021.1935004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Li M, Lindblad JL, Perez E, et al. Autophagy-independent function of Atg1 for apoptosis-induced compensatory proliferation. BMC Biol. 2016;14(1). doi: 10.1186/s12915-016-0293-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Yamaguchi J, Suzuki C, Nanao T, et al. Atg9a deficiency causes axon-specific lesions including neuronal circuit dysgenesis. Autophagy. 2018;14(5):764–777. doi: 10.1080/15548627.2017.1314897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Pua HH, Dzhagalov I, Chuck M, et al. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204(1):25–31. doi: 10.1084/jem.20061303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Weng YM, Ke CR, Kong JZ, et al. The significant role of ATG5 in the maintenance of normal functions of Mc3T3-E1 osteoblast. Eur Rev Med Pharmacol Sci. 2018;22(5):1224–1232. doi: 10.26355/eurrev_201803_14462 [DOI] [PubMed] [Google Scholar]
- [108].Zhao JY, Li XY, Liu TD, et al. Silencing of ATG4D suppressed proliferation and enhanced cisplatin-induced apoptosis in hepatocellular carcinoma through Akt/Caspase-3 pathway. Mol Cell Biochem. 2021;476(11):4153–4159. doi: 10.1007/s11010-021-04224-z [DOI] [PubMed] [Google Scholar]
- [109].Lee IH, Kawai Y, Fergusson MM, et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336(6078):225–228. doi: 10.1126/science.1218395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Fremont S, Gerard A, Galloux M, et al. Beclin-1 is required for chromosome congression and proper outer kinetochore assembly. EMBO Rep. 2013;14(4):364–372. doi: 10.1038/embor.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Bomfim L, Ramos I. Deficiency of ULK1/ATG1 in the follicle cells disturbs ER homeostasis and causes defective chorion deposition in the vector Rhodnius prolixus. FASEB J. 2020;34(10):13561–13572. doi: 10.1096/fj.202001396R [DOI] [PubMed] [Google Scholar]
- [112].Vieira PH, Bomfim L, Atella GC, et al. Silencing of RpATG6 impaired the yolk accumulation and the biogenesis of the yolk organelles in the insect vector R. prolixus. PLoS Negl Trop Dis. 2018;12(5):e0006507. doi: 10.1371/journal.pntd.0006507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Chen SF, Kang ML, Chen YC, et al. Autophagy-related gene 7 is downstream of heat shock protein 27 in the regulation of eye morphology, polyglutamine toxicity, and lifespan in Drosophila. J Biomed Sci. 2012;19(1):52. doi: 10.1186/1423-0127-19-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Bansal P, Tripathi A, Thakur V, et al. Autophagy-related protein ATG18 regulates apicoplast biogenesis in apicomplexan parasites. MBio. 2017;8(5). doi: 10.1128/mBio.01468-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Walczak M, Ganesan SM, Niles JC, et al. ATG8 is essential specifically for an autophagy-independent function in apicoplast biogenesis in blood-stage malaria parasites. MBio. 2018;9(1). doi: 10.1128/mBio.02021-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Ogura K, Okada T, Mitani S, et al. Protein phosphatase 2A cooperates with the autophagy-related kinase UNC-51 to regulate axon guidance in Caenorhabditis elegans. Development. 2010;137(10):1657–1667. doi: 10.1242/dev.050708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Chung S, Yao H, Caito S, et al. Regulation of SIRT1 in cellular functions: role of polyphenols. Arch Biochem Biophys. 2010;501(1):79–90. doi: 10.1016/j.abb.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Na HJ, Pyo JH, Jeon HJ, et al. Deficiency of Atg6 impairs beneficial effect of metformin on intestinal stem cell aging in Drosophila. Biochem Biophys Res Commun. 2018;498(1):18–24. doi: 10.1016/j.bbrc.2018.02.191 [DOI] [PubMed] [Google Scholar]
- [119].Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci. 2009;122(Pt 14):2554–2566. doi: 10.1242/jcs.046250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Betin VM, Lane JD. Atg4D at the interface between autophagy and apoptosis. Autophagy. 2009;5(7):1057–1059. doi: 10.4161/auto.5.7.9684 [DOI] [PubMed] [Google Scholar]
- [121].Yousefi S, Perozzo R, Schmid I, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8(10):1124–1132. doi: 10.1038/ncb1482 [DOI] [PubMed] [Google Scholar]
- [122].Young MM, Takahashi Y, Khan O, et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012;287(15):12455–12468. doi: 10.1074/jbc.M111.309104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zheng W, Xie W, Yin D, et al. ATG5 and ATG7 induced autophagy interplays with UPR via PERK signaling. Cell Commun Signal. 2019;17(1):42. doi: 10.1186/s12964-019-0353-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Cho DH, Jo YK, Hwang JJ, et al. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett. 2009;274(1):95–100. doi: 10.1016/j.canlet.2008.09.004 [DOI] [PubMed] [Google Scholar]
- [125].Arsov I, Li X, Matthews G, et al. BAC-mediated transgenic expression of fluorescent autophagic protein beclin 1 reveals a role for beclin 1 in lymphocyte development. Cell Death Differ. 2008;15(9):1385–1395. doi: 10.1038/cdd.2008.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Germic N, Hosseini A, Stojkov D, et al. ATG5 promotes eosinopoiesis but inhibits eosinophil effector functions. Blood. 2021;137(21):2958–2969. doi: 10.1182/blood.2020010208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155(3):688–698. doi: 10.1016/j.cell.2013.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Saleiro D, Mehrotra S, Kroczynska B, et al. Central role of ULK1 in type I interferon signaling. Cell Rep. 2015;11(4):605–617. doi: 10.1016/j.celrep.2015.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Ding H, Caza M, Dong Y, et al. ATG genes influence the virulence of Cryptococcus neoformans through contributions beyond core autophagy functions. Infect Immun. 2018;86(9). doi: 10.1128/IAI.00069-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Liang Q, Seo GJ, Choi YJ, et al. Crosstalk between the cGAS DNA sensor and beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 2014;15(2):228–238. doi: 10.1016/j.chom.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Wang YT, Zaitsev K, Lu Q, et al. Select autophagy genes maintain quiescence of tissue-resident macrophages and increase susceptibility to Listeria monocytogenes. Nat Microbiol. 2020;5(2):272–281. doi: 10.1038/s41564-019-0633-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Ni Chonghaile T, Letai A. Who put the “A” in Atg12: autophagy or apoptosis? Mol Cell. 2011;44(6):844–845. doi: 10.1016/j.molcel.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Han J, Hou W, Goldstein LA, et al. A complex between Atg7 and caspase-9: a novel mechanism of cross-regulation between autophagy and apoptosis. J Biol Chem. 2014;289(10):6485–6497. doi: 10.1074/jbc.M113.536854 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.


