Abstract
A member of the polyomavirus enhancer binding protein 2/core binding factor (PEBP2/CBF) is composed of PEBP2αB1/AML1 (as the α subunit) and a β subunit. It plays an essential role in definitive hematopoiesis and is frequently involved in the chromosomal abnormalities associated with leukemia. In the present study, we report functionally separable modular structures in PEBP2αB1 for DNA binding and for transcriptional activation. DNA binding through the Runt domain of PEBP2αB1 was hindered by the adjacent carboxy-terminal region, and this inhibition was relieved by interaction with the β subunit. Utilizing a reporter assay system in which both the α and β subunits are required to achieve strong transactivation, we uncovered the presence of transcriptional activation and inhibitory domains in PEBP2αB1 that were only apparent in the presence of the β subunit. The inhibitory domain keeps the full transactivation potential of full-length PEBP2αB1 below its maximum potential. Fusion of the transactivation domain of PEBP2αB1 to the yeast GAL4 DNA-binding domain conferred transactivation potential, but further addition of the inhibitory domain diminished the activity. These results suggest that the activity of the α subunit as a transcriptional activator is regulated intramolecularly as well as by the β subunit. PEBP2αB1 and the β subunit were targeted to the nuclear matrix via signals distinct from the nuclear localization signal. Moreover, the transactivation domain by itself was capable of associating with the nuclear matrix, which implies the existence of a relationship between transactivation and nuclear matrix attachment.
The polyomavirus enhancer binding protein 2 (PEBP2), also called core binding factor (CBF), is a transcription factor complex composed of α and β subunits (reviewed in references 21 and 51). The α subunit binds to DNA and harbors the transactivating activity, while the β subunit enhances the DNA binding activity of the α subunit. In mammals, members of the α subunit family are encoded by three genes, PEBP2αA/CBFA1/AML3, PEBP2αB/CBFA2/AML1, and PEBP2αC/ CBFA3/AML2, and all belong to the Runt domain gene family, which includes the Drosophila genes runt and lozenge. The β subunit is encoded by a single gene, PEBP2β/CBFB, whereas two genes, brother and big brother have been identified in Drosophila.
Among the three mammalian α subunit genes, PEBP2αB/CBFA2/AML1 (2, 36, 51) is disrupted in chromosomal translocations associated with several types of leukemia, including the M2 subtype of the French-American-British classification of leukemia, which is characterized by the 8-to-21 chromosome translocation [t(8;21)], and childhood acute lymphoblastic leukemia with the associated t(12;21) translocation. The t(8;21) and t(12;21) translocations produce the chimeric proteins, AML1/ETO(MTG8) and TEL-AML1, respectively (13, 19, 37). These proteins retain the entire Runt domain in their PEBP2αB/AML1 portions, which is essential and sufficient for dimerization with the β subunit and for DNA binding. In addition, it is likely that expression of AML1/ETO is subjected to the same regulatory controls as those of PEBP2αB. Therefore, the oncogenic mechanisms of these fusion proteins may involve deregulation of target gene expression that would otherwise be normally regulated by PEBP2. Along these lines, several investigators have reported that these proteins act as inhibitors of PEBP2-dependent transactivation (15, 20, 34), while others have reported that AML1/ETO acts as an activator of selected promoters (25, 48). Similarly, PEBP2β is disrupted in inv(16) of the FAB-M4Eo subtype, producing a chimeric protein, CBFβ/PEBP2β-SMMHC (30). Targeted disruptions in mice of either the PEBP2αB or the PEBP2β gene resulted in almost identical phenotypes: embryonic lethality with accompanying hemorrhage of the central nervous system and defects in definitive hematopoiesis (42, 46, 49, 54, 55). Therefore, cooperative functioning of the two subunits, PEBP2αB/AML1 and PEBP2β, seems essential for the development of definitive hematopoiesis. Mice with a targeted insertion (knock in) in one allele of either AML1/ETO or CBFB-MYH11 did not develop leukemia but had phenotypes similar to those of the targeted disruptions (7, 57). The results suggested that these chimeric proteins act as lethal dominant inhibitors during the early stages of normal hematopoietic development.
Another α subunit, PEBP2αA/CBFα1, has recently been discovered as a master regulator of bone formation (11) by targeted disruption studies (27, 47). In addition, the corresponding heterozygous mice displayed a phenotype that resembled that of human cleidocranial dysplasia syndrome, which has been linked to defect(s) in one allele of the PEBP2αA/CBFA1 gene (40, 62). These analyses in mice and humans present strong evidence in favor of PEBP2 involvement in multiple aspects of mammalian embryogenesis and suggest that PEBP2 acts in a specific way at each gene.
Molecular mechanisms of transactivation by PEBP2 have mostly been addressed through the analysis of cis-regulatory elements containing the PEBP2 consensus sequences PuACCPuCA (reviewed in reference 21). The cis elements have been identified in the regulatory regions of many genes, including the T-cell-receptor (TCR) alpha, beta, gamma, and delta chains, CD3ɛ, myeloperoxidase, neutrophil elastase, granzyme B, granulocyte-macrophage colony-stimulating factor, interleukin 3, macrophage colony-stimulating factor (M-CSF) receptor, osteocalcin, osteopontin (11), and Bcl-2 (25). Of these, the best characterized is the TCRα enhancer, in which binding sequences for CREB/ATF, LEF-1, PEBP2, and Ets-1 are arranged in such a way as to support context-dependent transactivation (18). LEF-1, an architectural factor, bends DNA, which enables a physical interaction between CREB/ATF and Ets-1. PEBP2 and Ets-1 physically interact, and PEBP2 facilitates DNA binding by Ets-1. Either phosphorylated CREB/ATF or a mixture of the other lymphoid factors (LEF-1, PEBP2, and Ets-1) is sufficient to induce transcription in vitro when present in excess, but strong synergistic activation can only be achieved when all these factors are added together (32). Recently, a non-DNA-binding coactivator termed ALY has been cloned and was found to interact with LEF-1 and PEBP2 independently (5). As such, PEBP2 is recruited in context-dependent transactivation by physically interacting with Ets-1 and ALY, and this contributes to the activation of the TCRα enhancer. Another well-characterized cis element is the myeloid-specific M-CSF receptor promoter, in which binding sequences for PEBP2, PU.1, and the CCAAT enhancer binding protein (C/EBP) were identified. Synergistic activation of the promoter by C/EBP and PEBP2 and a physical interaction between the two factors have been reported (60).
Little is known about the functional domains for transcription activation in PEBP2 subunits. The most characteristic and conserved structure of the α subunits is the Runt domain. The transactivation function has been vaguely assigned to the entire region that lies carboxy terminal to the Runt domain, which, however, does not contain any known transactivation motif (3).
In the present study, we established a luciferase reporter assay system using the M-CSF receptor promoter in Jurkat T cells, in which both the α and β subunits are required for transactivation. The system led to a detailed domain analysis of each of the two subunits and revealed the modular, functional structure of PEBP2αB1. We also found a new role for the β subunit in the unmasking of the DNA binding activity of the Runt domain, which explains why structural analysis of transactivation domains in the α subunit have not met with success up until now. Finally, we show that transactivation by PEBP2 takes place in a discrete subnuclear structure, the nuclear matrix.
MATERIALS AND METHODS
Plasmid construction.
A mammalian expression vector, pEF-BOS (39), was used to make a series of expression plasmids. pEF-αB1, pEF-αB2, pEF-αB1(1–243), and pEF-αB1(1–183) have already been described (3). pEF-αB1(1–446) and pEF-β2 have also been described (31), and pEF-AML1(453) was described previously as pEF-AML1b (61). For other αB1-deletion constructs [αB1(1–411), αB1(1–371), αB1(1–331), αB1(1–291), αB1(1–177), αB1(1–173), αB1(27–451), and αB1(50–451)], site-directed mutagenesis (Transformer site-directed mutagenesis kit; Clontech, Palo Alto, Calif.) was carried out on pEF-αB1 to make each deletion. pEF-AML1/ETO was constructed by inserting the coding region for AML1/ETO (clone 35 in reference 12) into pEF-BOS. pEF-GAL4-DBD was constructed by inserting PCR-amplified GAL4-DBD (amino acids 1 to 147) into an XbaI site. An XbaI site and a BamHI site were synthetically introduced before and after the stop codon, respectively, to serve as cloning sites for pEF-GAL4-αB1 fusion constructs. For pEF-GAL4-αB1 fusion constructs [pEF-GAL4-αB1(291–331), pEF-GAL4-αB1(291–371), pEF-GAL4-αB1(291–411), pEF-GAL4-αB1(262–371), pEF-GAL4-αB1(262–411), and pEF-GAL4-αB1(371–411)], the corresponding regions of αB1 were PCR amplified and cloned into XbaI-BamHI sites of pEF-GAL4-DBD so as to keep coding regions in frame with GAL4-DBD. pSG-GAL4-DBD was constructed by removing VP16 portion from pSG-GAL4-VP16 (16) by HindIII-BamHI digestion. Similarly, pSG-GAL4-αB1 fusion constructs [pSG-GAL4-αB1(178–451), pSG-GAL4-αB1(178–291), pSG-GAL4-αB1(292–371), and pSG-GAL4-αB1(372–451)] were prepared by replacing the VP16 portion of pSG-GAL4-VP16 with corresponding regions of αB1 aligned in frame with GAL4-DBD. Throughout the process, the authenticity of PCR-amplified sequences was always confirmed by sequencing. Expression vectors for C/EBPα (MSV-C/EBPα in reference 6) and PU.1 (PUpECE in reference 26) were as described. For luciferase reporters, pM-CSF-R-luc (59) and tk-GALpx3-LUC were used. tk-GALpx3-LUC was constructed by cloning three copies of the GAL4 binding site (17MX in reference 56) into HindIII site of tk-LUC as described by Forman et al. (14). Tβ3W4W-tkCAT was as described previously (44).
Transfection and reporter assays.
Jurkat human T cells and U937 human monocytes were maintained in RPMI 1640 supplemented with 10% fetal calf serum (FCS) and antibiotics. These cells (5 × 106 cells in 150 μl of the culture medium in a 0.4-cm cuvette) were transfected with a luciferase-reporter plus expression plasmids via electroporation by using Gene Pulser (Bio-Rad Laboratories, Hercules, Calif.) at a setting of 500 μF/250 V and at room temperature. The compositions of transfected plasmids are described in the figure legends. The total amount of transfected DNA was always kept at 10 μg by using a backbone plasmid, pEF-BOS. After 24 h, cells were harvested, and luciferase activity was assayed with a luciferase assay system from Promega (Madison, Wis.) according to the manufacturer’s instructions. Relative luciferase activity was measured with Lumat LB 9507 (EG&G Berthold, Bad Wildbad, Germany) and normalized by using a protein concentration assayed with the Protein Assay Kit from Bio-Rad (Hercules, Calif.).
Indirect immunofluorescence staining.
REF52 rat fibroblasts were cultured in Dulbecco modified Eagle medium supplemented with 10% FCS. Cells were trypsinized and electroporated as described above with 15 μg of the expression plasmids. The transfected cells were seeded onto chamber slides (Nalge Nunc, Naperville, Ill.) and after 48 h were fixed and stained with an antibody raised against the Escherichia coli-expressed αB1 prepared by E. Ogawa and J. Lu as described previously (31).
Electrophoretic mobility shift assays (EMSA).
Full-length αB1 and its deletion derivatives were translated in vitro and labeled with [35S]methionine using the TNT reticulocyte-lysate system (Promega, Madison, Wis.). The products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the radioactivity was quantified with a phosphorimager (BAS 2000; Fuji). An equal amount of translated product, as judged by the level of radioactivity, was used in a reaction with a 32P-labeled probe containing the PEBP2 binding site from the Tβ3 enhancer sequence: GATCTAACAGGATGTGGTTTGACATTTA (29). The total volume of reticulocyte-lysate was adjusted by use of a mock-translated mixture. Escherichia coli-produced β2 (43) was added when indicated, and the DNA-binding reaction and electrophoresis were carried out as described previously (3). Autoradiograms were obtained with two sheets of X-ray films to eliminate signals from 35S, and the images on the distal films are shown.
Whole-cell extracts and nuclear matrix fraction preparation.
P19 mouse embryonal carcinoma cells were cultured in Dulbecco modified Eagle medium and F12 medium supplemented with 10% FCS and antibiotics. P19 cells in 10-cm-diameter culture dishes were transfected by a modified calcium phosphate precipitation method (8). For experiments with full-length αB1 and its deletion constructs, 0.5 μg of the corresponding pEF-BOS-based expression plasmid was used together with 14.5 μg of a backbone plasmid, pEF-BOS, per transfection. For GAL4 fusion constructs, 5 μg of pSG5 (Stratagene)-based expression plasmids were used together with 10 μg of pEF-BOS. All the transfections were performed in duplicate in each experiment, and after 40 h the cells were harvested by scraping. One set of samples was freeze-thawed in a buffer (20 mM HEPES [pH 7.9], 0.4 M NaCl, 25% glycerol, 1 mM EDTA, 2.5 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride, and the supernatant was used as whole-cell extract. One 24th of each sample was loaded onto an SDS-polyacrylamide gel. The other set was sequentially extracted with the CSK, RSB-Majik, and digestion buffers and 0.25 M ammonium sulfate as described previously (33). The remaining insoluble material was defined as a nuclear matrix fraction and was solubilized in standard SDS-gel loading buffer, and one-sixth of each sample was separated by SDS-PAGE. The antibodies used for Western blotting were either a rabbit anti-peptide antibody raised against 16 amino acids following the initiating methionine of αB1 (produced by Research Genetics, Huntsville, Ala.) for the detection of αB1 carboxy-terminal deletion constructs or an anti-GAL4 DNA-binding domain antibody (Upstate Biotechnology, Lake Placid, N.Y.) for detecting the GAL4 fusion constructs. The ECL system from Amersham (Buckinghamshire, England) was used for detection.
Jurkat cells (107 cells) were transfected with 35 μg of pEF-β2 plus 35 μg of pEF-BOS, pEF-αB1, pEF-αB1(1–371), or pEF-αB1(1–291) by electroporation (960 μF/250 V). After 48 h, cells were harvested and divided into two parts. One part was freeze-thawed in a buffer containing 20 mM HEPES (pH 7.9), 0.4 M KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1 mM EGTA, 25% glycerol, and protease inhibitors (Boehringer Mannheim, Mannheim, Germany). Soluble fractions and insoluble pellets were separated by centrifugation, and the pellets were further extracted with the same buffer and then centrifuged. Only the results obtained with the first supernatants were taken to be the soluble fractions, since the second supernatant essentially did not exhibit any significant protein bands. Pellets were solubilized in a standard SDS-gel loading buffer. The other half of the samples was directly lysed in a standard SDS-gel loading buffer and used as a total fraction. Antibodies used for Western blotting were a rabbit antibody against the whole αB1 protein and a hamster antibody raised against the β subunit (31).
RESULTS
Establishment of an experimental system to analyze transcription activation by PEBP2.
We have been studying the transactivating properties of PEBP2 by using the TCRβ-chain distal core enhancer Tβ3-Tβ4 linked to the thymidine kinase promoter in P19 cells in which expression of endogenous PEBP2α is very low, if it exists at all (3, 44). In this system, expression of only the α-subunit gene was sufficient to transactivate the reporter gene. Coexpression of the β subunit showed almost no effect, or inhibited the activity slightly, depending on the conditions (45). Under these conditions, a progressive carboxy-terminal truncation of the α subunit resulted in a gradual decrease of the activity and no discrete region responsible for transcription activation could be identified (see below). In sharp contrast, we found in the present study that transactivation of a reporter gene can be observed in Jurkat T cells only when the two subunits were coexpressed. The presence of intrinsic transactivation domain(s) in the α subunit was revealed for the first time with this cell system.
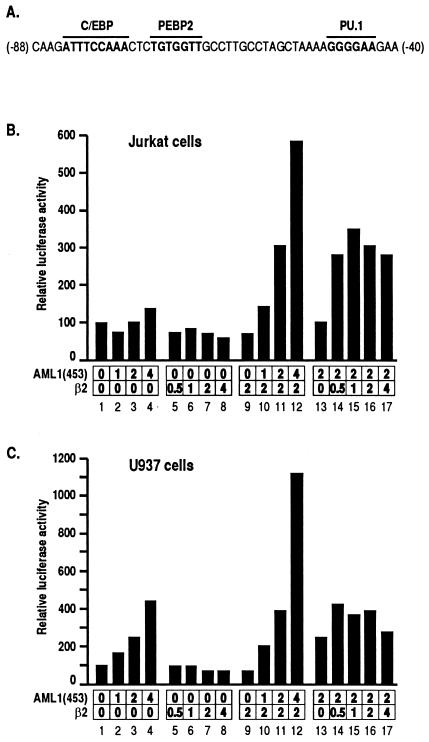
We used the M-CSF receptor promoter linked to the luciferase gene as a reporter, essential elements of which are illustrated in Fig. 1A (according to reference 60). In Jurkat T cells, the exogenous expression of AML1(453) transactivated the reporter activity, but only marginally (Fig. 1B, lanes 1 to 4). [Murine PEBP2αB1 is the 451 amino acid (aa)-long major product of PEBP2αB (3). Human PEBP2αB1 is referred to here as AML1(453).] As for the β subunit, we used the β2 isoform in the present study because β2 is the most abundantly expressed isoform of the β subunit in many cells (43). β2 did not activate the promoter at all when expressed alone (Fig. 1B, lanes 5 to 8). However, when increasing amounts of AML1(453) were cotransfected with a fixed amount of β2, strong transactivation of the promoter was observed in a dose-dependent manner (Fig. 1B, lanes 9 to 12). A similar level of functional cooperation between the two subunits was observed over a wide range of β2 concentrations, suggesting that the effect of β2 was saturating above a certain concentration (Fig. 1B, lanes 13 to 17). Thus, transactivation studies with the M-CSF receptor promoter in Jurkat cells provide a unique experimental system for the analysis of the contribution of each subunit of PEBP2.
FIG. 1.
Cooperative activation of the M-CSF receptor promoter by PEBP2 α and β subunits. (A) An essential element in the M-CSF receptor promoter, which contains binding sites for C/EBP, PEBP2, and PU.1. (B and C) Jurkat T cells or U937 monocytes were transfected with 4 μg of pM-CSF-R-luc and the indicated amounts (in micrograms) of pEF-AML1(453) and pEF-β2. Luciferase activities relative to those obtained with pM-CSF-R-luc and backbone plasmid pEF-BOS (set as 100) are shown. For presentation, lanes 9, 13, and 16 are duplicated from lanes 7, 3, and 11, respectively.
Similar experiments performed with U937 monocytic cells showed that AML1(453) by itself transactivated the M-CSF receptor promoter in this cell line (Fig. 1C, lanes 1 to 4). In accordance with the results obtained with Jurkat cells, however, this transactivation was significantly increased by the coexpression of β2 (Fig. 1C, lanes 9 to 12). This difference between Jurkat and U937 cells may partly reflect the availability of the endogenous β subunit in these cells. Another possibility may be that C/EBP and PU.1 are expressed in monocytes but not in T cells (26, 50, 59). However, coexpression of AML1(453) together with either C/EBPα or PU.1 in Jurkat cells did not result in a significant transactivation in the absence of the β subunit (data not shown).
Deletion analysis of PEBP2αB1 revealed intrinsic domains for transactivation and inhibition.
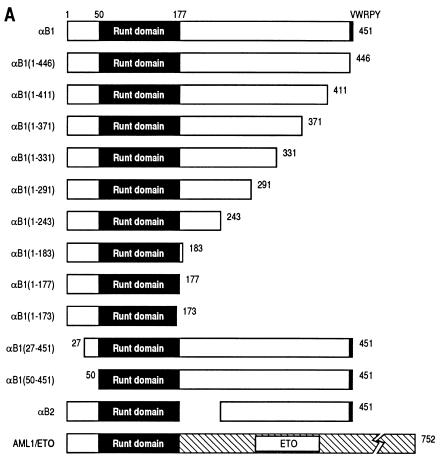
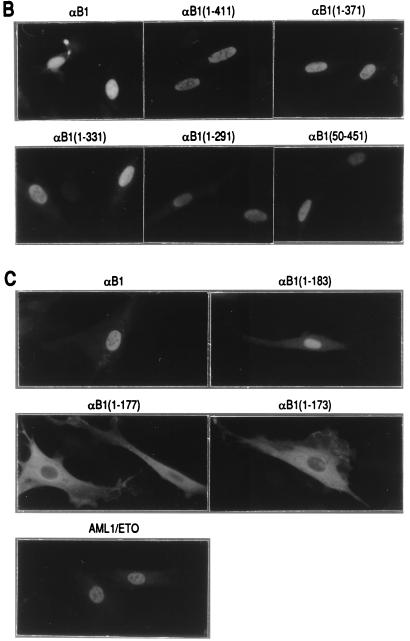
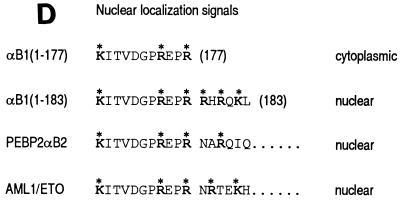
To analyze the regions required for transactivation, we prepared a series of deletion constructs of αB1 (for the sake of simplicity, we will refer to the PEBP2αB1 isoform as αB1 in this study) in expression plasmids as shown in Fig. 2A and tested their transactivation potential in the presence of β2 on the M-CSF receptor promoter. All the constructs except αB1(1–173) retained the whole Runt domain. We first checked the subcellular localization of the products of these constructs by transfecting them into REF52 fibroblasts and by immunostaining the expressed proteins. Full-length αB1, as well as all of the deletion derivatives except αB1(1–177) and αB1(1–173), was entirely localized to the nucleus (Fig. 2, panels B and C). AML1/ETO was also localized to the nucleus, although the corresponding αB1 portion αB1(1–177) did not enter the nucleus. Considering that αB1(1–183) was localized to the nucleus, the region of aa 178 to 183 must be critically important for nuclear translocation, and the ETO portion of AML1/ETO seems to compensate for the loss of these critical amino acids (Fig. 2D, discussed below).
FIG. 2.
PEBP2αB1 deletion derivatives and their nuclear localization. (A) Schematic illustration of the structures of full-length αB1 and its deletion derivatives. Numbers denote the positions of amino acids. The Runt domain is from aa 50 to 177. The structures of αB2 and AML1/ETO are also shown. (B and C) Indirect immunofluorescence of REF52 cells transfected with pEF-BOS-based expression plasmids coding for proteins which are indicated above the panels. Results in panels B and C were from separate experiments. (D) Amino acid sequences around the carboxy-terminal border of the Runt domain in αB1, αB2, and AML1/ETO. Boldface letters with asterisks constitute putative nuclear localization signals.
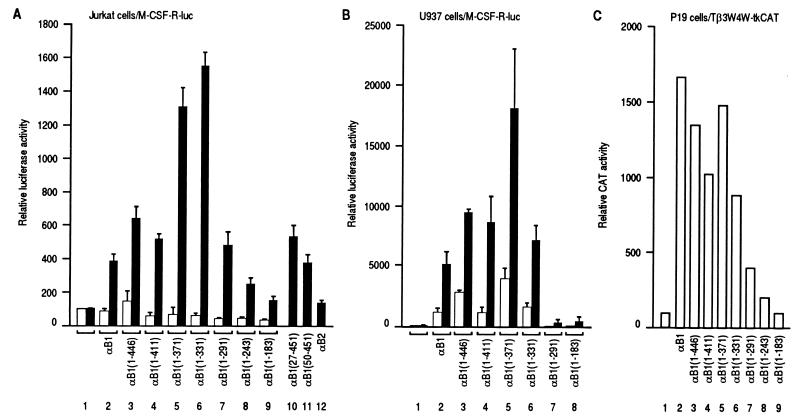
When assayed in the presence of β2, a carboxy-terminal truncation of αB1 increased its transactivating ability, but further truncation diminished it. As shown by filled bars in Fig. 3A, transactivation in Jurkat cells peaked with the constructs αB1(1–371) and αB1(1–331), suggesting the presence of a transactivation domain between aa 291 and 331 and the presence of an inhibitory domain between aa 371 and 411. It is worth noting that αB1(1–291) retained an activity that was significantly higher than the background level. αB1(1–243) possesses reduced, but still significant transactivating ability. On the other hand, αB1(1–183) did not activate the promoter to any significant degree. Deletion of the regions on the amino-terminal side of the Runt domain (the aa 1-to-26 region, which is conserved among the mammalian PEBP2α subunits, and the region of aa 1 to 49) did not significantly change the transactivation levels (lanes 10 and 11). PEBP2αB2 (referred to as αB2 here), a naturally occurring variant of αB1, which lacks the region of aa 178 to 242 (3), exhibited almost no activity (lane 12), implying that the missing region may be critically important. However, the region obviously does not harbor considerable transactivating activity by itself, as judged by the weak activity of αB1(1–243). Another set of analysis was done using Tβ3-Tβ4 linked to the thymidine kinase promoter-luciferase (TCRβ-tk-Luc) as a reporter. This gave similar results to those obtained with the M-CSF receptor promoter (data not shown). We checked the levels of protein expression in the transfected cells and confirmed that the apparent difference in transactivating ability between the truncated constructs was not due to differences in the stabilities of the protein products (data not shown).
FIG. 3.
Transactivation activity of full-length PEBP2αB1 and its deletion derivatives in the absence (open bars) or presence (filled bars) of β2. (A) For filled bars, Jurkat cells were transfected with 4 μg of pM-CSF-R-luc, 1.5 μg of pEF-β2, and 4.5 μg of a backbone vector pEF-BOS (lane 1) or pEF-BOS-based expression plasmids for αB1 and its deletion derivatives as indicated (lanes 2 to 12). For open bars, pEF-β2 was not included and was replaced by pEF-BOS. Luciferase activities relative to those obtained with cells transfected with 4 μg of pM-CSF-R-luc and 6 μg of EF-BOS (set as 100) are shown. Means and standard deviations of nine independent experiments with β2 and of three independent experiments without β2 are shown. (B) U937 cells were transfected and analyzed as in panel A. Results represent three independent experiments. (C) P19 cells were transfected with Tβ3W4W-tkCAT and pEF-BOS-based expression plasmids for αB1 and its deletion derivatives as indicated (lanes 2 to 9). CAT activities relative to those obtained with cells transfected with Tβ3W4W-tkCAT (lane 1, set as 100) are shown.
In the absence of β2, the transactivation levels seen with these αB1-derivatives were less than twofold the level obtained with the luciferase reporter alone (Fig. 3A). Although very low, we can see that the activity was highest with αB1(1–446) and decreased with the shorter constructs. Therefore, we emphasize here that coexpression of the β subunit not only increased the overall activity but also changed the profile of activities among the deletion constructs. Curiously, removal of VWRPY at the very extreme carboxyl terminus consistently gave a slight increase in the activity. A similar observation was reported recently by others (1).
When U937 monocytes were transfected without the β subunit, moderate transactivation of the M-CSF receptor promoter was observed with full-length αB1 and its deletion derivatives (Fig. 3B). These levels of transactivation were considerably increased on addition of β2 (Fig. 3B), a finding consistent with the observation in Fig. 1C. In the presence of transfected β2, the most prominent difference in the levels of transactivation among the deletion derivatives was observed between αB1(1–291) and αB1(1–331), and the carboxy-terminal extension up to aa 371 [αB1(1–371)] led to greater transactivation. This suggests the presence of a transactivation domain between aa 291 and 331 in agreement with the results obtained with Jurkat cells and the presence of an additional transactivation domain between aa 331 and 371. Further addition of more carboxy-terminal regions resulted in a decrease in transactivation ability, also suggesting the presence of inhibitory domain(s) in the carboxy-terminal region beyond aa 371.
In contrast, the P19 cell system with the Tβ3W4W-tkCAT reporter exhibited transactivation by the α subunit alone but failed to reveal apparent transactivation domains (Fig. 3C). A possible reason for this will be described below.
Regions responsible for transactivation and inhibition conferred the activities on the GAL4-DBD.
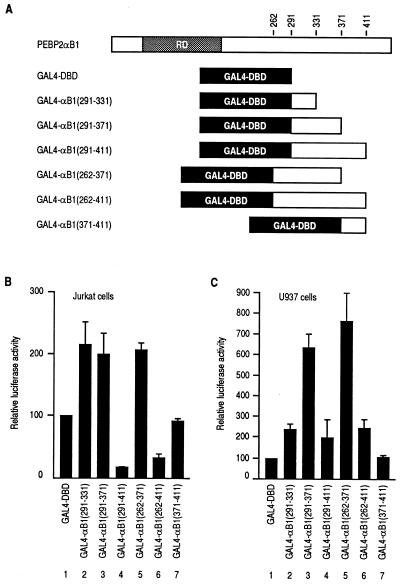
To address more directly the functional domains described above, we prepared constructs in which the yeast GAL4 DNA-binding domain (GAL4-DBD) was fused to the putative functional regions from αB1, as shown in Fig. 4A. The intrinsic stimulatory and inhibitory functions of the fused fragments were assayed as GAL4-binding-site-dependent transactivation. Regions corresponding to aa 291 to 331, 291 to 371, and 262 to 371 of αB1 conferred transactivation potential on the GAL4-DBD, a heterologous DNA-binding domain, in both Jurkat and U937 cells (Fig. 4B and C, lanes 2, 3, and 5). In U937 cells the presence of additional transactivation potential in the region of aa 331 to 371 was evident. Extension of these constructs to aa 411 always reduced the transactivating ability (Fig. 4B and C, lanes 4 and 6), indicating the presence of an inhibitory activity between aa 371 and 411. However, the region of aa 371 to 411 by itself did not confer a transcriptional inhibitory effect on GAL4-DBD (Fig. 4B and C, lane 7). In summary, these findings obtained with the GAL4-fusion constructs are entirely consistent with those obtained with the αB1 deletion constructs on the natural M-CSF receptor promoter and indicate that the activities observed on these two different promoters reflect those intrinsic to αB1.
FIG. 4.
Analysis of intrinsic transactivating activities of various regions from PEBP2αB1 fused to the yeast GAL4 DNA-binding domain. (A) Schematic illustration of the GAL4 fusion constructs. Full-length αB1 is shown for reference. (B and C) Jurkat cells or U937 cells were transfected with 5 μg of tk-GALpx3-LUC and 5 μg of the pEF-BOS-based expression plasmids as indicated. Luciferase activities relative to those obtained with GAL4-DBD (set as 100) are shown. The results represent three independent experiments.
Unmasking of the DNA binding activity of the Runt domain by the β subunit.
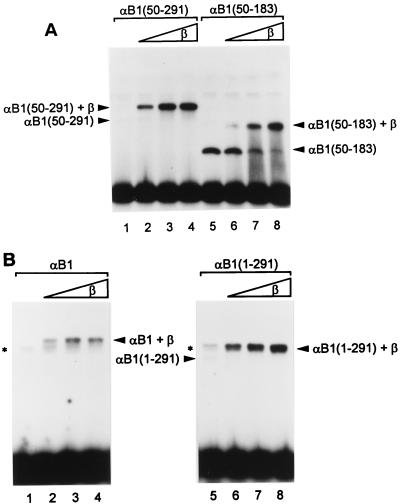
The reason why the intrinsic transactivation domain in the α subunit manifests itself only when the β subunit is present was hinted at by earlier observations on DNA binding (22, 43). The full-length α subunit exhibited weak DNA binding, whereas the Runt domain alone, devoid of its amino- and carboxy-terminal regions, showed strong DNA binding. In the presence of the β subunit, the DNA-binding activities of the Runt domain and especially of the entire α subunit were enhanced. To evaluate the effect of the β subunit on DNA binding in more detail, we performed experiments using partially truncated α subunits (Fig. 5). Like full-length αB1, truncated αB1(50–291) alone did not bind to DNA very well (Fig. 5A, lane 1). Upon removal of a further 108 aa, αB1(50–183) bound to DNA very well by itself (lane 5). Addition of the β subunit readily supershifted the bands, showing that heterodimers had been formed (lanes 2 to 4 and 6 to 8). It is important to note that the intensity of the αB1(50–291) band was strongly increased after addition of the β subunit, whereas that of αB1(50–183) was increased only mildly. This suggests that the region between aa 183 and 291 masks the surface of the Runt domain responsible for interaction with DNA and that binding of the β subunit changes the conformation in such a way as to unmask this surface. The region amino-terminal to the Runt domain, aa 1 to 49, also had an inhibitory effect on DNA binding by the Runt domain, a feature that will be described elsewhere (24). Thus, the inhibitory effect of the region between aa 183 and 291 was most dramatically exhibited by using the constructs starting at aa 50. Similarly, we analyzed the DNA-binding properties of a series of carboxy-terminal deletion constructs that are shown in Fig. 2A. The constructs were analyzed either alone or as heterodimers with the β subunit. The constructs having carboxyl termini beyond aa 291 were barely able to bind to DNA by themselves. Among these, the constructs that did not extend beyond aa 411 exhibited heterodimer binding as strong as that of αB1(50–291) (data not shown). However, longer constructs, including full-length αB1, showed less-effective heterodimer binding than αB1(50–291) and αB1(1–291) even in the presence of the same amount of β subunit (Fig. 5B), suggesting that dimerization with the β subunit may be blocked by the extreme carboxy-terminal region of αB1. In spite of repeated attempts, we could not precisely delineate the region that blocks dimerization in EMSA because the binding activities of these constructs turned out to be quite variable, suggesting that artificially truncated proteins are functionally unstable in vitro. Thus, we tentatively assign the region aa 411 to 451 to be responsible for preventing β-subunit interaction with the Runt domain.
FIG. 5.
Strong and comparable DNA binding by PEBP2αB1 and its deletion derivatives requires the β subunit. EMSAs were performed with in vitro-translated αB1 and its deletion derivatives with or without E. coli-produced β2 subunit. (A) Comparison between αB1(50–291) (lanes 1 to 4) and αB1(50–183) (lanes 5 to 8). The amounts of β2 added were 1.25 ng (lanes 2 and 6), 2.5 ng (lanes 3 and 7), and 5 ng (lanes 4 and 8). The positions of each complex are indicated. (B) Comparison between full-length αB1 (lanes 1 to 4) and αB1(1–291) (lanes 5 to 8). The amounts of β2 added were 0.5 ng (lanes 2 and 6), 1 ng (lanes 3 and 7), and 2 ng (lanes 4 and 8). The positions of each complex are indicated. Asterisks indicate the positions of nonspecific bands, which can be seen in lanes 1 to 4 and lane 5. These bands overlap with the bands corresponding to αB1(1–291)+β2 in lanes 6 to 8. The band corresponding to αB1 alone cannot be seen at this exposure.
Transactivation by PEBP2αB1 and β2 takes place in the nuclear matrix.
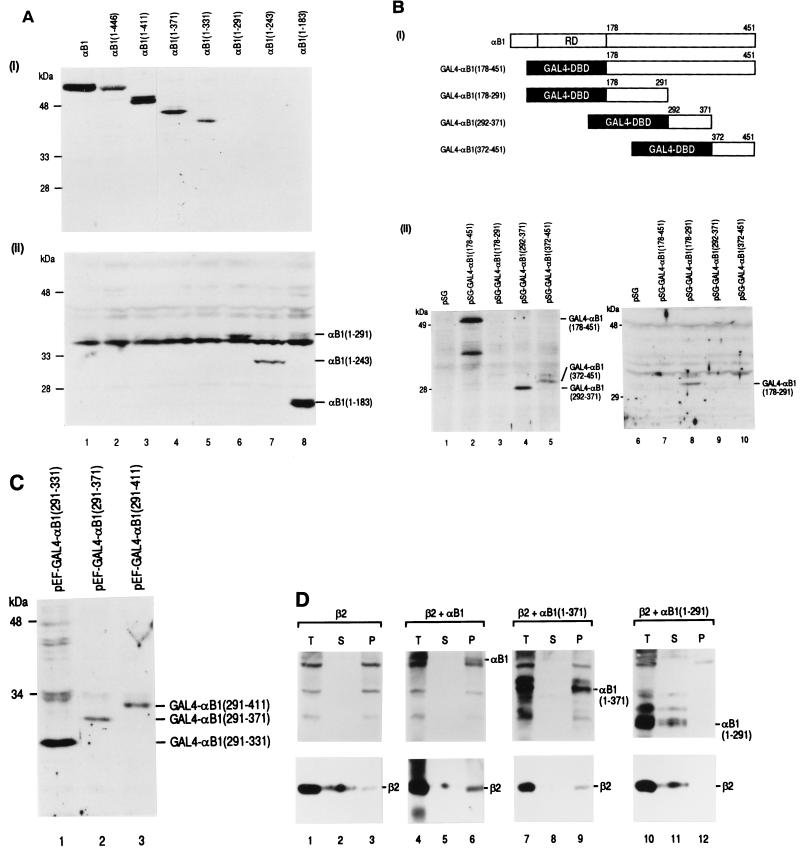
All the αB1 constructs used in the present study except αB1(1–177) and αB1(1–173) were localized to the nucleus. It has recently been reported that the PEBP2α subunit-related protein NMP-2, which is bone specific, is firmly attached to the nuclear matrix, a filamentous ribonucleoprotein network of the nucleus (33). We had also noticed in a variety of transfection experiments that αB1 was not easily extractable in salt solutions. We therefore examined the subnuclear distribution of full-length αB1 and its carboxy-terminal deletion derivatives after they were transfected into P19 cells devoid of the endogenous PEBP2α subunits. Whole-cell lysates were extracted with a high-concentration salt solution (soluble fractions), and the remaining compartments were treated with nucleases and further extracted with ammonium sulfate. This treatment resulted in pellets containing the insoluble nuclear matrix compartments. These fractions were analyzed by SDS-PAGE and Western blotting. The constructs with the carboxyl termini beyond aa 331 were recovered from the nuclear matrix compartments (Fig. 6A, panel i, lanes 1 to 5), whereas those that do not extend beyond aa 291 were not (lanes 6 to 8). Analysis of the soluble fractions exhibited the reciprocal presence of these constructs (Fig. 6A, panel ii), clearly demonstrating subnuclear compartmentalization. The results implied that the region aa 291 to 331 was involved in nuclear matrix attachment and suggested that there exists a close correlation between transactivating activity and nuclear matrix binding. While the GAL4-DBD did not exhibit nuclear matrix binding, its fusion with the region containing the αB1 transactivation domain (aa 292 to 371) conferred the ability to associate with the nuclear matrix (Fig. 6B). However, nuclear matrix targeting activity was not confined to the region of aa 291 to 331. The region of aa 372 to 451, but not the region of aa 178 to 291, also conferred nuclear matrix binding activity (Fig. 6B). Furthermore, the GAL4 fusion constructs containing the minimal transactivating element (aa 291 to 331 and aa 291 to 371), the transactivation and inhibition domains (aa 291 to 411), or the inhibition domain alone (aa 371 to 411) all exhibited nuclear matrix binding (Fig. 6C and data not shown). These results together suggest that nuclear matrix binding of αB1 might be necessary for transactivation but also that levels of transactivating activity may not exactly correlate with nuclear matrix binding per se. Nevertheless, possible roles of nuclear matrix attachment might include the concentration and compartmentalization of nuclear factors in a discrete structure in the nucleus. If effective transactivation by PEBP2 takes place in a manner associated with the nuclear matrix, the β subunit, which does not associate with the nuclear matrix by itself, should also colocalize to that compartment. To test this prediction, we transfected Jurkat cells with an expression plasmid for β2 and an expression plasmid for full-length αB1, αB1(1–371), or αB1(1–291) and prepared salt-extractable soluble fractions (S) and insoluble nuclear matrix fractions (P). As shown in Fig. 6D, β2 was recovered from the insoluble nuclear matrix compartment when coexpressed with αB1(1–371) (compare lanes 8 and 9) or with full-length αB1 (compare lanes 5 and 6), both of which were also present in the nuclear matrix fraction. This was not the case with αB1(1–291) (compare lanes 11 and 12), which did not attach to the nuclear matrix. When β2 alone was transfected, only a trace amount of β2 was detected in the matrix fraction (compare lanes 2 and 3). This occurred most likely through binding to endogenous PEBP2α subunits.
FIG. 6.
Nuclear matrix attachment of PEBP2αB1. (A) Western blotting showing nuclear matrix association of full-length and deletion derivatives of αB1. P19 cells were transfected with the pEF-BOS-based expression plasmids as indicated. Nuclear matrix fractions (section i) and soluble whole-cell extracts (section ii) were prepared, separated on 12.5% SDS-gel, and stained with a peptide-antibody against αB1. Positions of αB1(1–291), αB1(1–243), and αB1(1–183) are indicated on the right. The bands immediately beneath the position of αB1(1–291) are nonspecific and recognizable in all lanes. The amounts loaded represented the same proportion of samples obtained. (B) P19 cells were transfected with pSG5-based expression plasmids for the GAL4 fusion constructs as shown in section i. (ii) Nuclear matrix (lanes 1 to 5) and soluble fractions (lanes 6 to 10) were prepared, separated by SDS-PAGE, and stained with an antibody against GAL4-DBD. Positions of the detected proteins are indicated on the right. (C) P19 cells were transfected with pEF-BOS-based expression plasmids as shown in Fig. 4A. Nuclear matrix fractions were analyzed as in panel B, and the positions of the detected proteins are indicated on the right. (D) Jurkat cells were transfected with pEF-BOS-based expression plasmids for PEBP2 proteins in the indicated combinations. Total (T), soluble (S), and insoluble fractions (P) were prepared, separated by SDS-PAGE, and stained with an antibody against the whole αB1 protein or an anti-β antibody. Positions of αB1, αB1(1–371), αB1(1–291), and β2 are indicated on the right. Lanes 4 to 6 for β2 staining were exposed longer than others.
DISCUSSION
The M-CSF receptor promoter required both the α and β subunits of PEBP2 to achieve strong transcription in Jurkat T cells. This system allowed us to investigate functional regions in each of the two subunits in vivo. In the present study, we have described the α-subunit functional domains, which only became apparent in the presence of the β subunit.
The β subunit is required for the strong and equal DNA binding by the α-subunit derivatives.
The results presented in the present study, as well as those to be described later (24), suggest that the Runt domain of the α subunit is intramolecularly “masked” in two ways. Interaction with the β subunit is blocked by the carboxy-terminal region of aa 411 to 451, and the DNA-binding activity is inhibited by the region of aa 183 to 291. Although our analysis was not extensive enough to precisely determine the boundaries, we have termed the latter region the negative regulatory region of DNA binding (NRDB). When the β subunit associates with the Runt domain, the conformation of the α subunit is changed in such a way that the Runt domain can now bind to DNA strongly. We assume that this process reflects two separate events. After the β subunit binds to the Runt domain, the effect of NRBD is eliminated and the DNA-binding domain is exposed on the surface where it can interact directly with DNA. The second event is to increase the affinity of the Runt domain to DNA as described earlier (22, 43, 53). Although this assumption has to be proved by structural studies, we believe that the concept of two separate steps is a reasonable working hypothesis. From this premise, we propose that one of the main roles of the β subunit is to unmask the DNA binding surface of the Runt domain by eliminating the negative regulatory effect of NRDB.
Transactivation assay of the M-CSF receptor promoter in Jurkat cells is suitable for the analysis of PEBP2 subunits.
The requirement for both the α and β subunits in vivo has been shown most dramatically by gene disruption studies, in which the phenotypes of the PEBP2αB/AML1 knockout and β-gene knockout mice were found to be nearly identical (42, 46, 49, 54, 55). Therefore, it was to be expected that the transcription in the in vivo transcription assays would also require the presence of the β subunit, as was clearly shown in the present study. In contrast, we and others have reported transactivations that occur only with the α subunit (3, 11, 15, 20, 34, 44, 52). We assume that the endogenous β subunit was utilized in such cases, but there may be other possibilities. We have recently discovered that some transcription factors enhance DNA binding of the α subunit in the absence of the β subunit (24). Thus, at least for the analysis of intrinsic functional regions in either subunit, the system in which both the α and the β subunits are required for transactivation must be used.
The NRDB of the α subunit is located more proximally to the Runt domain than the transcription activation domain (AD, see below). Without the β subunit, progressive carboxy-terminal truncations would remove AD before eliminating NRDB, resulting in exposure of the DNA binding domain. This explains well why expression of the αB1 deletion derivatives alone in Jurkat cells and P19 cells did not allow clear resolution of the intrinsic functional regions.
PEBP2αB1 has modular structures.
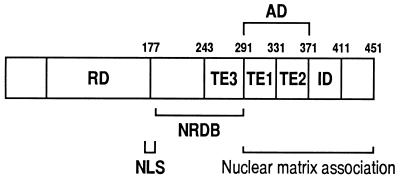
Using the serial deletion constructs as well as the GAL4-fusion constructs, we identified a major AD of αB1 in the region containing aa 291 to 371 (Fig. 7). Interestingly, the transactivation activity due to the AD was inhibited by the addition of the adjacent domain, i.e., aa 371 to 411, which we refer to as the inhibitory domain (ID). Accordingly, the presence of the ID makes the AD “cryptic” in the full-length αB1 protein. In T cells, the region of aa 291 to 331 (TE1 in Fig. 7) within the AD represented most of the transactivation function, and the additional region of aa 332 to 371 (TE2 in Fig. 7) considerably contributed to the transactivation activity in monocytes. Thus, the AD seems to consist of at least two transactivation elements (TE1 and TE2), and the activity of TE2 seems to be affected by cellular factors that are cell-type specific. Also, the region of aa 243 to 291 may constitute the third transactivation element (TE3) because this region is responsible for the relatively small but significant transactivation ability of αB1(1–291) compared with αB1(1–243). The possibility of a complexity in domain organization is raised by the finding that αB2, which lacks the region between aa 178 to 242 but retains all three transactivation elements and IDs, cannot transactivate. The positive transactivating ability of the AD is likely to be canceled by the ID in αB2 as well. Since αB1(1–243) showed only marginal transactivating ability, the missing region, i.e., aa 178 to 242 in αB2, may not contain a strong transactivation element by itself but may play an important role together with TE3 within the context of αB1. This may account for the difference between αB1 and αB2.
FIG. 7.
Schematic illustration of functional regions in PEBP2αB1. Abbreviations: AD, activation domain; TE, transactivation element; ID, inhibitory domain; NLS, nuclear localization signal; NRDB, negative regulatory region of DNA binding; RD, Runt domain.
These putative transactivation elements do not contain the well-known motifs of classic transactivation domains such as acidic amino acids or such as proline- or glutamine-rich stretches of amino acid sequence. Transactivating and inhibitory modular structures have also been reported in C/EBP (28, 41), c-Fos (4), c-Myb (10), and Brca2 (35). The regulatory states of these proteins could be modulated by phosphorylation of either or both structures. For instance, the suppressive activity of inhibitory elements in C/EBPβ may be deactivated by signal-induced phosphorylation (28). It is noteworthy that the TE3 element of αB1 can be phosphorylated by ERK, a member of the MAP kinase family (52).
As for mechanisms of inhibition by the ID, these may include masking of the activation surface on the transactivation domain as in c-Myb (10) and binding of active inhibitors as in c-Fos (4). The result showing that the GAL4 fusion construct containing only the ID did not exhibit inhibitory effect by itself supports the intramolecular masking model. However, the inhibitor-binding model cannot be entirely excluded especially in T cells, since GAL4 fusion constructs containing both the AD and the ID significantly reduced the transactivation activity below the basal activity.
Nuclear localization signal of PEBP2αB1.
We previously reported that one of the regions responsible for the nuclear localization of PEBP2αA1 and -αB1 resides in the Runt domain (31). In the present study, we found that αB1(1–177), which ends at the exact border of the Runt domain, was localized in the cytoplasm. Because αB1(1–183) can enter the nucleus, the last six amino acids (RHRQKL) must be critically important for nuclear import (Fig. 2D). As a cluster of basic amino acids is important for nuclear localization (9), we speculate that the motif KXXXXXXRXXRRXRXKX (where X is any amino acid) in aa 167 to 183 may constitute a nuclear localization signal (NLS). AML1/ETO was also localized to the nucleus despite the fact that its αB1/AML1 portion ends at aa 177. The amino acid sequence at the beginning of the ETO portion contains the sequence NRTEKH, which may reconstitute the NLS by supplying arginine and lysine residues. In a previous study, we concluded that the region between aa 178 and 242 does not contain an NLS because of the observed nuclear localization of αB2 which lacks this region (31). However, a compensatory effect similar to that of AML1/ETO may also work for αB2 nuclear localization, in which the arginine residue of the sequence NARQIQ beginning at aa 243 becomes juxtaposed by alternative splicing. We also noticed poor nuclear localization ability of the region carboxy terminal to the Runt domain when it was expressed as a separate protein (23).
Nuclear matrix association and transactivation.
We found that αB1 associates with the nuclear matrix in a region that is distinct from that containing the NLS. What might be the influence of nuclear matrix association on transactivation? If the association is to be a prerequisite for αB1 to transactivate, it would be reasonable to speculate that an essential coactivating factor(s) for αB1 is also tightly associated with the nuclear matrix. Consistent with this hypothesis, we found that the AD containing region of αB1 has the ability to associate with the nuclear matrix. During the preparation of this manuscript, Zeng et al. (58) reported the identification of a nuclear matrix targeting signal (NMTS) in the region between aa 351 and 381 of AML1B [also defined as AML1c (38), which has a different amino terminus from αB1/AML1(453) due to differential promoter usage (17)]. This region corresponds to aa 324 to 353 of αB1, an area which resides well within the AD identified in the present study. Also, we found separable nuclear matrix targeting activities spread over a broader region between aa 292 and 451 in αB1 (Fig. 7), suggesting the presence of multiple NMTSs in αB1.
Interestingly, a certain amount of the β subunit was confined to the nuclear matrix through dimerization with αB1, strongly suggesting that efficient PEBP2-dependent transactivation occurs in this specific compartment. Likewise, it is possible that PEBP2 may participate in context-dependent transactivation by recruiting other transcription factors, which lack their own intrinsic NMTS, to this specific, subnuclear compartment. We are currently investigating the mechanism of synergistic transactivation of the M-CSF receptor promoter by PEBP2, C/EBPα, and PU.1 (59, 60). The structure and function of the nuclear matrix in transcription will be important subjects for future study.
ACKNOWLEDGMENTS
We thank K. Umesono for tk-GALpx3-LUC, and D.-E. Zhang for pM-CSF-R-luc.
This work was partly supported by the New Energy and Industrial Technology Development Organization (FY1995, B-333), and by a grant-in-aid for Priority Area on Cancer Research from the Minister of Education, Science and Culture, Japan (no. 0925322).
REFERENCES
- 1.Aronson B D, Fisher A L, Blechman K, Caudy M, Gergen J P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae S C, Yamaguchi-Iwai Y, Ogawa E, Maruyama M, Inuzuka M, Kagoshima H, Shigesada K, Satake M, Ito Y. Isolation of PEBP2αB cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993;8:809–814. [PubMed] [Google Scholar]
- 3.Bae S C, Ogawa E, Maruyama M, Oka H, Satake M, Shigesada K, Jenkins N A, Gilbert D J, Copeland N G, Ito Y. PEBP2αB/mouse AML1 consists of multiple isoforms that possess differential transactivation potentials. Mol Cell Biol. 1994;14:3242–3252. doi: 10.1128/mcb.14.5.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown H J, Sutherland J A, Cook A, Bannister A J, Kouzarides T. An inhibitor domain in c-Fos regulates activation domains containing the HOB1 motif. EMBO J. 1995;14:124–131. doi: 10.1002/j.1460-2075.1995.tb06982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruhn L, Munnerlyn A, Grosschedl R. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 6.Cao Z, Umek R M, MacKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 7.Castilla L H, Wijmenga C, Wang Q, Stacy T, Speck N A, Eckhaus M, Marin-Padilla M, Collins F S, Wynshaw-Boris A, Liu P P. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 10.Dubendorff J W, Whitaker L J, Eltman J T, Lipsick J S. Carboxy-terminal elements of c-Myb negatively regulate transactivational activation in cis and in trans. Genes Dev. 1992;6:2524–2535. doi: 10.1101/gad.6.12b.2524. [DOI] [PubMed] [Google Scholar]
- 11.Ducy P, Zhang R, Geoffroy V, Ridall A L, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 12.Era T, Asou N, Kunisada T, Yamasaki H, Asou H, Kamada N, Nishikawa S, Yamaguchi K, Takatsuki K. Identification of two transcripts of AML1/ETO-fused gene in t(8;21) leukemic cells and expression of wild-type ETO gene in hematopoietic cells. Genes Chromosomes Cancer. 1995;13:25–33. doi: 10.1002/gcc.2870130105. [DOI] [PubMed] [Google Scholar]
- 13.Erickson P, Gao J, Chang K S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 14.Forman B M, Umesono K, Chen J, Evans R M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 15.Frank R, Zhang J, Uchida H, Meyers S, Hiebert S W, Nimer S D. The AML1/ETO fusion protein blocks transactivation of the GM-CSF promoter by AML1B. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 16.Fujii M, Tsuchiya H, Seiki M. HTLV-1 Tax has distinct but overlapping domains for transcriptional activation and for enhancer specificity. Oncogene. 1991;6:2349–2352. [PubMed] [Google Scholar]
- 17.Ghozi M C, Bernstein Y, Negreanu V, Levanon D, Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc Natl Acad Sci USA. 1996;93:1935–1940. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 19.Golub T R, Barker G F, Bohlander S K, Hiebert S W, Ward D C, Bray-Ward P, Morgan E, Raimondi S C, Rowley J D, Gilliland D G. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiebert S W, Sun W, Davis J N, Golub T, Shurtleff S, Buijs A, Downing J R, Grosveld G, Roussel M F, Gilliland D G, Lenny N, Meyers S. The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol Cell Biol. 1996;16:1349–1355. doi: 10.1128/mcb.16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito Y, Bae S-C. The Runt domain transcription factor, PEBP2/CBF, and its involvement in human leukemia. In: Yaniv M, Ghysdael J, editors. Oncogenes as transcriptional regulators. Vol. 2. Basel, Switzerland: Birkhauser Verlag; 1997. pp. 107–132. [Google Scholar]
- 22.Kagoshima H, Akamatsu Y, Ito Y, Shigesada K. Functional dissection of the alpha and beta subunits of transcription factor PEBP2 and the redox susceptibility of its DNA binding activity. J Biol Chem. 1996;271:33074–33082. doi: 10.1074/jbc.271.51.33074. [DOI] [PubMed] [Google Scholar]
- 23.Kanno, Y., T. Kanno, and Y. Ito. Unpublished data.
- 24.Kim, W.-Y., and Y. Ito. Unpublished data.
- 25.Klampfer L, Zhang J, Zelenetz A O, Uchida H, Nimer S D. The AML1/ETO fusion protein activates transcription of BCL-2. Proc Natl Acad Sci USA. 1996;93:14059–14064. doi: 10.1073/pnas.93.24.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 27.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R T, Gao Y H, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 28.Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Novel mechanism of C/EBPβ (NF-M) transactivational control: activation through derepression. Genes Dev. 1994;8:2781–2791. doi: 10.1101/gad.8.22.2781. [DOI] [PubMed] [Google Scholar]
- 29.Krimpenfort P, de Jong R, Uematsu Y, Dembic Z, Ryser S, von Boehmer H, Steinmetz M, Berns A. Transcription of T cell receptor β-chain genes is controlled by a downstream regulatory element. EMBO J. 1988;7:745–750. doi: 10.1002/j.1460-2075.1988.tb02871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P, Tarle S A, Hajra A, Claxton D F, Marlton P, Freedman M, Siciliano M J, Collins F S. Fusion between transcription factor CBFβ/PEBP2β and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 31.Lu J, Maruyama M, Satake M, Bae S C, Ogawa E, Kagoshima H, Shigesada K, Ito Y. Subcellular localization of the α and β subunits of the acute myeloid leukemia-linked transcription factor PEBP2/CBF. Mol Cell Biol. 1995;15:1651–1661. doi: 10.1128/mcb.15.3.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayall T P, Sheridan P L, Montminy M R, Jones K A. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 33.Merriman H L, van Wijnen A J, Hiebert S, Bidwell J P, Fey E, Lian J, Stein J, Stein G S. The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/Runt domain transcription factor family: interpretations with the osteocalcin gene promoter. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 34.Meyers S, Lenny N, Hiebert S W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milner J, Ponder B, Hughes-Davies L, Seltmann M, Kouzarides T. Transcriptional activation functions in BRCA2. Nature. 1997;386:772–773. doi: 10.1038/386772a0. [DOI] [PubMed] [Google Scholar]
- 36.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88:10432–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyoshi H, Ohira M, Shimizu K, Mitani K, Hirai H, Imai T, Yokoyama K, Soeda E, Ohki M. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mundlos S, Otto F, Mundlos C, Mulliken J B, Aylsworth A S, Albright S, Lindhout D, Cole W G, Henn W, Knoll J H, Owen M J, Mertelsmann R, Zabel B U, Olsen B R. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 41.Nerlov C, Ziff E B. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-α on the serum albumin promoter. Genes Dev. 1994;8:350–362. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- 42.Niki M, Okada H, Takano H, Kuno J, Tani K, Hibino H, Asano S, Ito Y, Satake M, Noda T. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcriptional factor, polyomavirus enhancer binding protein 2/core binding factor. Proc Natl Acad Sci USA. 1997;94:5697–5702. doi: 10.1073/pnas.94.11.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, Shigesada K. Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2α. Virology. 1993;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- 44.Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA. 1993;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogawa, E., and Y. Ito. Unpublished data.
- 46.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 47.Otto F, Thornell A P, Crompton T, Denzel A, Gilmour K C, Rosewell I R, Stamp G W H, Beddington R S P, Mundlos S, Olsen B R, Selbey P B, Owen M J. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 48.Rhoades K L, Hetherington C J, Rowley J D, Hiebert S W, Nucifora G, Tenen D G, Zhang D E. Synergistic up-regulation of the myeloid-specific promoter for the macrophage colony-stimulating factor receptor by AML1 and the t(8;21) fusion protein may contribute to leukemogenesis. Proc Natl Acad Sci USA. 1996;93:11895–11900. doi: 10.1073/pnas.93.21.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasaki K, Yagi H, Bronson R T, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott L M, Civin C I, Rorth P, Friedman A D. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 51.Speck N A, Stacy T. A new transcription factor family associated with human leukemias. Crit Rev Eukaryotic Gene Expr. 1995;5:337–364. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka T, Kurokawa M, Ueki K, Tanaka K, Imai Y, Mitani K, Okazaki K, Sagata N, Yazaki Y, Shibata Y, Kadowaki T, Hirai H. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol Cell Biol. 1996;16:3967–3979. doi: 10.1128/mcb.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S, Wang Q, Crute B E, Melnikova I N, Keller S R, Speck N A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol Cell Biol. 1993;13:3324–3339. doi: 10.1128/mcb.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Disruption of the cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T-L, Huang X, Bushweller J H, Bories J-C, Alt F W, Ryan G, Liu P P, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 56.Webster N, Jin J R, Green S, Hollis M, Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988;52:169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- 57.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- 58.Zeng C, van Wijnen A J, Stein J L, Meyers S, Sun W, Shopland L, Lawrence J B, Penman S, Lian J B, Stein G S, Hiebert S W. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-α transcription factors. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang D E, Hetherington C J, Chen H M, Tenen D G. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol. 1994;14:373–381. doi: 10.1128/mcb.14.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang D E, Hetherington C J, Meyers S, Rhoades K L, Larson C J, Chen H M, Hiebert S W, Tenen D G. CCAAT enhancer-binding protein (C/EBP) and AML1 (CBF alpha2) synergistically activate the macrophage colony-stimulating factor receptor promoter. Mol Cell Biol. 1996;16:1231–1240. doi: 10.1128/mcb.16.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y W, Bae S C, Huang G, Fu Y X, Lu J, Ahn M Y, Kanno Y, Kanno T, Ito Y. A novel transcript encoding an N-terminally truncated AML1/PEBP2αB protein interferes with transactivation and blocks granulocytic differentiation of 32Dcl3 myeloid cells. Mol Cell Biol. 1997;17:4133–4145. doi: 10.1128/mcb.17.7.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y W, Bae S-C, Takahashi E, Ito Y. The cDNA cloning of the transcripts of human PEBP2αA/CBFA1 mapped to 6p12.3–p12.1, the locus for cleidocranial dysplasia. Oncogene. 1997;15:367–371. doi: 10.1038/sj.onc.1201352. [DOI] [PubMed] [Google Scholar]