Abstract
Plant inflorescence architecture is determined by inflorescence meristem (IM) activity and controlled by genetic mechanisms associated with environmental factors. In Arabidopsis (Arabidopsis thaliana), TERMINAL FLOWER1 (TFL1) is expressed in the IM and is required to maintain indeterminate growth, whereas LEAFY (LFY) is expressed in the floral meristems (FMs) formed at the periphery of the IM and is required to activate determinate floral development. Here, we address how Arabidopsis indeterminate inflorescence growth is determined. We show that the 26S proteasome subunit REGULATORY PARTICLE AAA-ATPASE 2a (RPT2a) is required to maintain the indeterminate inflorescence architecture in Arabidopsis. rpt2a mutants display reduced TFL1 expression levels and ectopic LFY expression in the IM and develop a determinate zigzag-shaped inflorescence. We further found that RPT2a promotes DNA METHYLTRANSFERASE1 degradation, leading to DNA hypomethylation upstream of TFL1 and high TFL1 expression levels in the wild-type IM. Overall, our work reveals that proteolytic input into the epigenetic regulation of TFL1 expression directs inflorescence architecture in Arabidopsis, adding an additional layer to stem cell regulation.
The proteasome subunit RPT2a promotes DNA METHYLTRANSFERASE1 degradation, causing DNA hypomethylation upstream of TERMINAL FLOWER1 (TFL1) and high TFL1 levels in the Arabidopsis inflorescence meristem.
IN A NUTSHELL.
Background: While there is a rich diversity of shapes in the plant kingdom, inflorescence architecture can generally be classified into a limited number of types. For example, Brassicaceae members, such as Arabidopsis (Arabidopsis thaliana) or cabbage (Brassica oleracea), exhibit an indeterminate inflorescence architecture, whereas nightshade species, such as tomato (Solanum lycopersicum) or potato (Solanum tuberosum), display determinate inflorescences. These distinct architectures are associated with the indeterminacy of the inflorescence meristem (IM). The precise regulation of TERMINAL FLOWER1 (TFL1) expression is pivotal for governing both the architecture of the inflorescence and the indeterminacy of the IM. However, the mechanisms responsible for controlling TFL1 expression in this context have remained elusive.
Question: What are the mechanisms controlling the activity of the IM identity gene TFL1, thereby specifying the indeterminate nature of the Arabidopsis inflorescence? What is the role of protein degradation in directing inflorescence architecture in Arabidopsis?
Findings: We present insights into the mechanisms that fine-tune the activity of TFL1, a key regulator of indeterminate inflorescence growth in Arabidopsis. Our findings demonstrate that TFL1 levels in the IM are regulated by DNA methylation mediated by the DNA methyltransferase MET1. Low MET1 levels are maintained through proteolysis via the 26S proteasome subunit RPT2a, which keeps methylation at the TFL1 promoter low, thus promoting IM indeterminacy and indeterminate inflorescence architecture. Conversely, when RPT2a-mediated proteolysis of MET1 is hindered, TFL1 expression levels decrease, leading to a determinate zigzag-shaped inflorescence architecture. Our findings propose an integrative model that explains how proteolytic activity balances the epigenetic regulation of TFL1 expression, ultimately leading to indeterminate inflorescence architecture in Arabidopsis.
Next Steps: We plan to explore these diverse models in other plant species, aiming to expand our knowledge regarding the genetic mechanisms governing the wide range of inflorescence architectures observed in nature.
Introduction
Flowering plants display an enormous morphological diversity of inflorescence architectures, depending on the identity of distinct meristems in the inflorescence apex (Zhu and Wagner 2020; Vernoux et al. 2021). A primary inflorescence may produce many secondary inflorescences and flowers that are initiated from the main shoot axis. They are arranged on a compound architecture or as solitary flowers (Rickett 1954). In the indeterminate inflorescence of Arabidopsis (Arabidopsis thaliana), the apical inflorescence meristems (IMs) maintain indeterminacy and continuously produce flowers from lateral floral meristems (FMs) along a single main shoot axis. By contrast, determinate inflorescences of nightshade species, such as tomato (Solanum lycopersicum) and petunia (Petunia hybrida), arise from repeated cycles of determinate IMs that differentiate into a terminal flower and continuous shoot development by lateral “sympodial” IMs (Claßen-Bockhoff and Bull-Hereñu 2013).
Several regulators, including TERMINAL FLOWER1 (TFL1), LEAFY (LFY), and APETALA1 (AP1), determine the identity of IM and floral meristem (FM) and consequently the inflorescence morphology in Arabidopsis (Shannon and Meeks-Wagner 1993; Liljegren et al. 1999; Benlloch et al. 2015). The TFL1 gene encodes a phosphatidyl-ethanolamine binding protein and is specifically expressed in the apical IM of Arabidopsis to maintain its indeterminate meristematic fate (Bradley et al. 1997; Ratcliffe et al. 1999). The IMs of tfl1 loss-of-function mutants terminate after initiations of 1 or 2 lateral FMs in a central flower occupying the position of the stem cell niche (Bradley et al. 1997). The LFY and AP1 genes are both expressed in the FMs that arise at the flanks of the IM and are essential for the specification of FM identity in the Arabidopsis wild type (Weigel et al. 1992; Maizel et al. 2005). Loss-of-function mutants in these genes exhibit a partial conversion of determinate flowers into indeterminate IMs (Weigel et al. 1992; Shannon and Meeks-Wagner 1993). Previous studies reported that TFL1 maintains IM indeterminacy by repressing the expression of LFY and AP1 genes in the IM (Hanano and Goto 2011). Conversely, LFY and AP1 repress TFL1 expression in newly formed FM (Parcy et al. 1998; Liljegren et al. 1999; Wagner et al. 1999). The spatially separated antagonistic actions of TFL1 to LFY and AP1 regulate the balance between the indeterminate IM and determinate FM fates and thus shape the inflorescence development of Arabidopsis.
Ubiquitin–proteasome-mediated protein degradation is indispensable for regulating the levels and functions of many intracellular proteins. Polyubiquitinated proteins are degraded through the 26S proteasome (Collins and Goldberg 2017). Mutation of the 26S proteasome subunit REGULATORY PARTICLE AAA-ATPASE2a (RPT2a) leads to reduced meristematic activity in shoot and root apical meristems, increased cell size and decreased cell numbers in organs, and lethality in male and female gametophytes (Ueda et al. 2004; Kurepa et al. 2009; Sonoda et al. 2009; Lee et al. 2011; Ueda et al. 2011; Sako et al. 2012; Kim et al. 2019). However, how RPT2a affects these developmental processes has remained unclear.
Here we show that RPT2a is required to maintain the indeterminate growth mode of the IM in Arabidopsis. Knockout of RPT2a leads to the transformation of the indeterminate into a determinate inflorescence. Loss of RPT2a function results in excessive DNA METHYLTRANSFERASE1 (MET1) accumulation in IM associated with DNA hypermethylation and reduced TFL1 expression and in turn, ectopic LFY expression. Our results demonstrate that RPT2a-mediated proteolysis adjusts epigenetic regulation of TFL1 expression and thus IM indeterminacy and inflorescence architecture in Arabidopsis.
Results
Rpt2a mutants display a determinate inflorescence
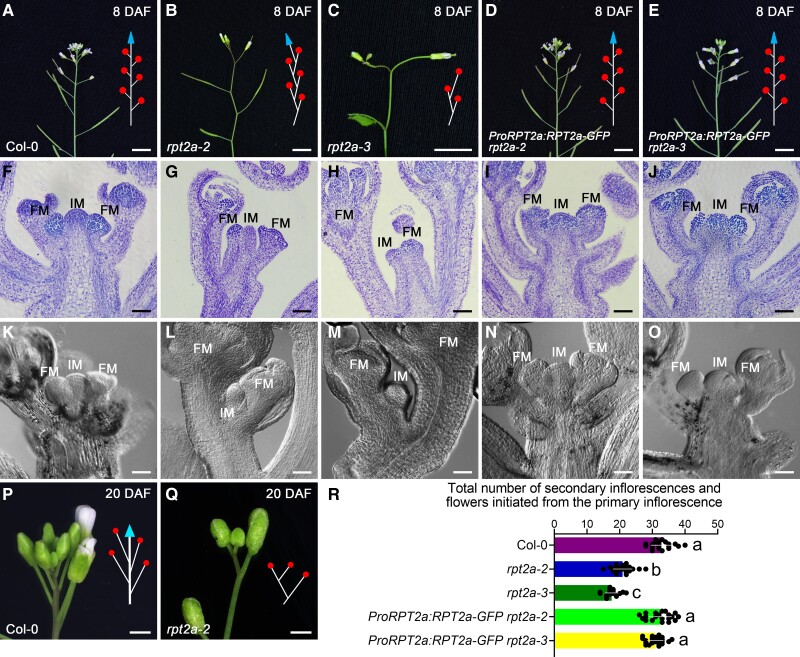
When examining the inflorescence architecture of Arabidopsis rpt2a mutants (Supplemental Fig. S1A to C), we noticed a determinate zigzag-shaped inflorescence phenotype at 8 days after flowering (DAF), unlike the indeterminate inflorescences of wild type (Fig. 1A, B, and R; Supplemental Fig. S2 and Data Set S1). Later in development, at 20 DAF, rpt2a-2 mutants displayed a terminal flower phenotype, whereas wild-type inflorescences were indeterminate (Fig. 1, P and Q). In wild type, the IM located at the apex of the inflorescence grew indeterminately (until senescence) and continuously produced lateral FMs along the main plant axis (Fig. 1A, F, K, and R; Supplemental Fig. S2A and Data Set S1).
Figure 1.
Knockout of RPT2a displays a determinate zigzag-shaped phenotype of inflorescence. A to E) Indeterminate inflorescences of the Col-0 (A; 93.8%, n = 97), ProRPT2a:RPT2a-GFP rpt2a-2 (D; 87.1%, n = 70), and ProRPT2a:RPT2a-GFP rpt2a-3 plants (E; 83.9%, n = 62); determinate inflorescences of the rpt2a-2 (B; 92.9%, n = 99) and rpt2a-3 mutants (C; 88%, n = 108) at 8 DAF. Schematic representations of the indeterminate inflorescences and zigzag-shaped inflorescence are on the right side, depicting flowers as circles and inflorescence meristems (IMs) as triangles. Bar = 5 mm. F to J) Longitudinal sections of inflorescences of the Col-0 (F; 90.9%, n = 44), rpt2a-2 (G; 87.2%, n = 39), rpt2a-3 (H; 87.5%, n = 40), ProRPT2a:RPT2a-GFP rpt2a-2 (I; 81.8%, n = 33), and ProRPT2a:RPT2a-GFP rpt2a-3 plants (J; 80%, n = 30) at 8 DAF. FM, floral meristem. Bar = 200 μm. K to O) Differential interference contrast microscope images of the inflorescences in Col-0 (K; 91.7%, n = 36), rpt2a-2 (L; 83.8%, n = 37), rpt2a-3 (M; 82.2%, n = 45), ProRPT2a:RPT2a-GFP rpt2a-2 (N; 85.3%, n = 34), and ProRPT2a:RPT2a-GFP rpt2a-3 plants (O; 86.2%, n = 29) at 8 DAF. Bar = 200 μm. P) The closeup of the indeterminate phenotype of Col-0 inflorescence at 20 DAF. Bar = 1 mm. Q) The closeup of the terminal flower phenotype of rpt2a-2 inflorescence at 20 DAF. Bar = 1 mm. R) Total number of secondary inflorescences and flowers initiated from the primary inflorescence in Col-0, rpt2a-2, rpt2a-3, ProRPT2a:RPT2a-GFP rpt2a-2, and ProRPT2a:RPT2a-GFP rpt2a-3 plants at 30 DAF. The different letters indicate significant differences (The data represent means ± SD; 1-way ANOVA, Tukey's multiple comparison tests; P < 0.05).
By contrast, in rpt2a-2 mutants, the IM was located at the lateral of the inflorescence and produced only a few apical FMs along the main axis (Fig. 1B, G, L, and R; Supplemental Fig. S2B and Data Set S1). This growth pattern repeated for several rounds, ultimately resulting in the zigzag-shaped inflorescence with the uppermost IM terminating in a central flower (Fig. 1, B and Q; Supplemental Fig. S2B). The majority of plants of the rpt2a-3 mutant allele displayed a similar determinate inflorescence growth as rpt2a-2 at 8 DAF (Fig. 1C, H, M, and R; Supplemental Data Set S1). However, in 5.56% (6/108) of the rpt2a-3 mutants, the primary IM terminated into an apical flower immediately at 3 DAF (Supplemental Fig. S1, D and E). The phenotypes of both mutant alleles were completely suppressed by ProRPT2a:RPT2a-GFP (Fig. 1D, E, I, J, N, O, and R; Supplemental Data Set S1), indicating that the RPT2a mutations caused the phenotypic changes. Thus, RPT2a is an essential regulator of IM indeterminacy and inflorescence architecture in Arabidopsis.
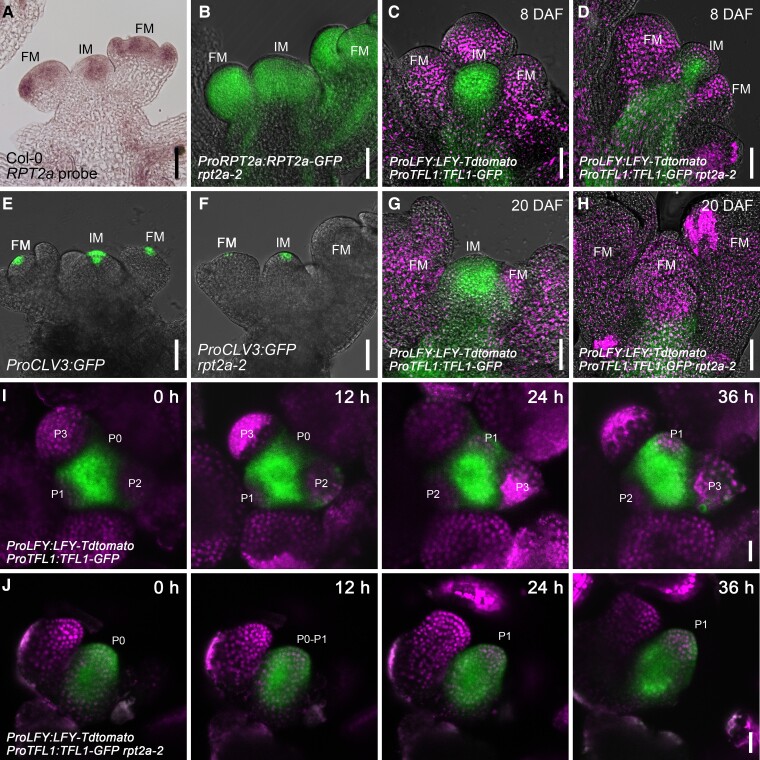
We then examined RPT2a expression patterns. At 8 DAF, we detected expression of RPT2a mRNA by in situ hybridization (Fig. 2A; Supplemental Fig. S4D) and a ProRPT2a:RPT2a-GFP reporter gene (Fig. 2B) in the IMs and the FMs. RPT2a is a subunit of the 19S regulatory particle of the 26S proteasome. To determine whether other subunits of the 19S regulatory particle are required to maintain IM indeterminacy in Arabidopsis, we analyzed the inflorescences in mutants of other 19S regulatory particle genes, including RPN12a, RPN1a, and RPT4a. However, rpn12a-2, rpn1a-2, and rpt4a-1 single mutants all showed indeterminate inflorescences similar to wild type at 8 DAF (Supplemental Fig. S3A, C, E, G, I and Dataset S2). To examine potential functional redundancy, we crossed these mutants with rpt2a-2 to generate double mutants. The IMs of all the double mutants produced fewer flowers and terminated earlier than the rpt2a-2 single mutants (Supplemental Fig. S3, B, D, F, H, I and Data Set S2). These enhanced phenotypes suggest that several subunits of the 19S complex participate in the maintenance of IM indeterminacy, with RPT2a being the main contributor.
Figure 2.
RPT2a regulates the expression of TFL1 and LFY. A)In situ hybridization analysis shows RPT2a mRNA expression in IM and FM at 8 DAF (88.4%, n = 43). Bar = 200 μm. B) Expression pattern of RPT2a-GFP in IM and FM of the ProRPT2a:RPT2a-GFP rpt2a-2 plants at 8 DAF (85.4%, n = 48). Bar = 200 μm. C and D) The expression of TFL1-GFP is detected in the IM, and the expression of LFY-Tdtomato is detected in the FM of the ProLFY:LFY-Tdtomato/ProTFL1:TFL1-GFP plants (C; 87.1%, n = 31) at 8 DAF. But TFL1-GFP is much weaker, and LFY-Tdtomato is ectopically expressed in the IM of the ProLFY:LFY-Tdtomato/ProTFL1:TFL1-GFP rpt2a-2 plants (D; 84.8%, n = 33). Bar = 200 μm. E and F) The expression of CLV3 is detected in the IM of ProCLV3:GFP plants (E; 85.7%, n = 21) but narrowed down and much weaker in the IM of ProCLV3:GFP rpt2a-2 plants (F; 84%, n = 25) at 8 DAF. Bar = 200 μm. G and H) The expression of TFL1-GFP and LFY-Tdtomato at 20 DAF is consistent with these at 8 DAF in the ProLFY:LFY-Tdtomato/ProTFL1:TFL1-GFP plants (G; 81.8%, n = 22). But at 20 DAF, TFL1-GFP is rarely detected in inflorescence, and LFY-Tdtomato is ectopically expressed in the terminal flower of ProLFY:LFY-Tdtomato/ProTFL1:TFL1-GFP rpt2a-2 plants (H; 79.2%, n = 24). Bar = 200 μm. I and J) The time-course analysis of the FM initiation in the same IM of the ProLFY:LFY-Tdtomato/ProTFL1:TFL1-GFP plants (I; 100%, n = 4) and ProLFY:LFY-Tdtomato/ProTFL1:TFL1-GFP rpt2a-2 plants (J; 83.3%, n = 6) from 2 DAF (0 h) during the next 36 h. Flower primordia at different stages are marked as P0, P1, P2, and P3. Bar = 100 μm.
RPT2a regulates inflorescence meristem indeterminacy through TFL1
Because IMs terminated into a flower in rpt2a-2 (Fig. 1Q), we investigated the expression patterns of the IM identity gene TFL1 (Bradley et al. 1997) and the FM identity gene LFY (Weigel et al. 1992). We introduced functional ProTFL1:TFL1-GFP and ProLFY:LFY-Tdtomato constructs (Supplemental Fig. S4A and Data Set S3) into wild type and the rpt2a-2 mutant to assess the expression of TFL1 and LFY proteins and studied their expression in apices at 8 DAF and 20 DAF. At 8 DAF, the TFL1 protein signals were much weaker in lateral IMs of rpt2a-2 compared to the apical IM of the wild type (Fig. 2, C and D). Furthermore, we detected an ectopic LFY-Tdtomato signal in the center of the rpt2a-2 apex but not the wild-type (Figs. 2C, D, and 7G; Supplemental Fig. S4B and Data Set S4).
Figure 7.
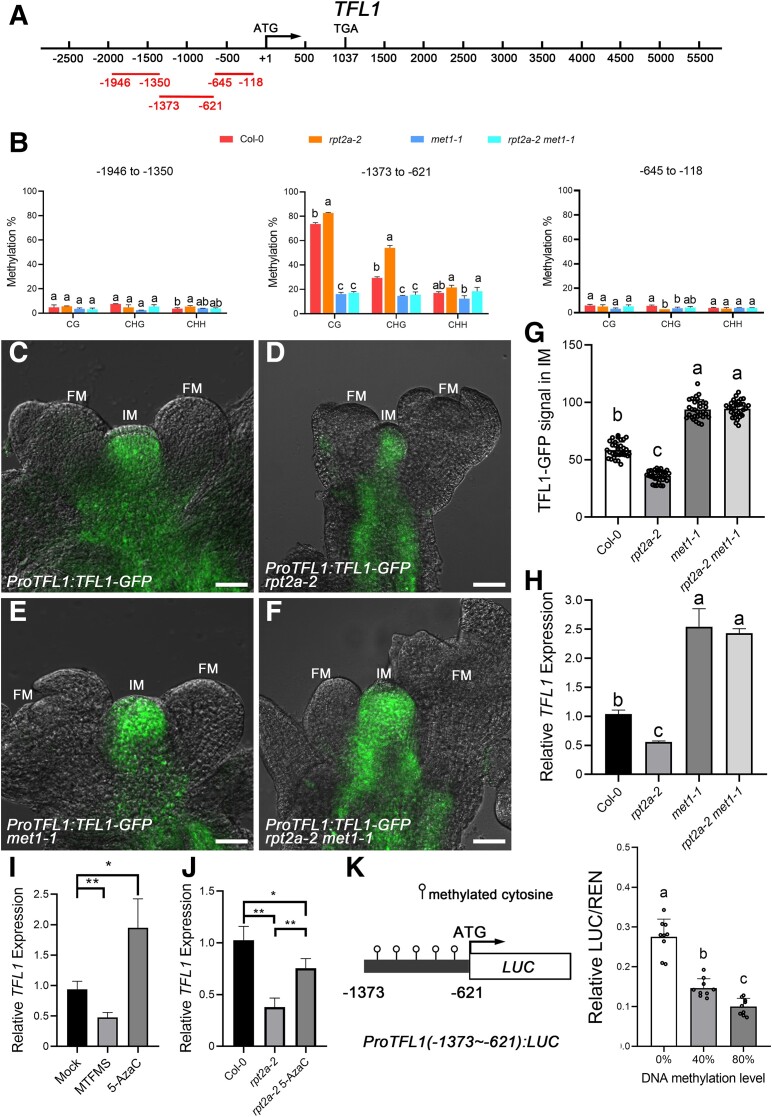
Accumulation of MET1 in rpt2a-2 inflorescences leads to hypermethylation of TFL1 promoter and decreased TFL1 expression. A) Diagram of the TFL1 gene structure, with ATG as the translation start site, TGA as the translation stop site, and the lines representing the detection regions. The regions −1,946 to −1350, −1,373 to −621, and −645 to −118 indicate the distance of the upstream regions from the TFL1 translation start site. B) The cytosine methylation levels of the TFL1 loci are assessed using bisulfite sequencing. The DNA methylation statuses in different regions (−1,946 to −1350, −1,373 to −621, and −645 to −118) of the TFL1 loci are shown in inflorescences of the Col-0, rpt2a-2, met1-1, and rpt2a-2 met1-1 mutants at 8 d after flowering (DAF). The fragment −1,373 to −621 of the TFL1 promoter is hypermethylation in rpt2a-2 mutants and hypomethylation in met1-1 and rpt2a-2 met1-1 mutants. The data are calculated from 3 biological replicates. The different letters indicate significant differences (The data represent means ± SD; 1-way ANOVA, Tukey's multiple comparison tests; P < 0.05). C to F) The expression of TFL1-GFP in IM of Col-0 (C; 81.8%, n = 33) is higher than that in rpt2a-2 mutants (D; 87.5%, n = 32), and weaker than those in met1-1 (E; 83.3%, n = 30) and rpt2a-2 met1-1 mutants (F; 80.6%, n = 31) at 8 DAF. IM, inflorescence meristem. FM, floral meristem. Bar = 200 μm. G) Fluorescence intensity profiles of TFL1-GFP in the IMs of Col-0, rpt2a-2, met1-1, and rpt2a-2 met1-1 plants at 8 DAF. The different letters indicate significant differences (The data represent means ± SD; 1-way ANOVA, Tukey's multiple comparison tests; P < 0.05). H) Relative expression of TFL1 in inflorescences of the Col-0, rpt2a-2, met1-1, and rpt2a-2 met1-1 mutants at 8 DAF. The data are calculated from 3 biological replicates. The different letters indicate significant differences (The data represent means ± SD; 1-way ANOVA, Tukey's multiple comparison tests; P < 0.05). I) Relative expression of TFL1 in inflorescences of the Col-0, MTFMS treatment, and 5-AzaC treatment. The data are calculated from 3 biological replicates. The significance tests for TFL1 expression are performed using 2-tailed Student's t-tests (*P < 0.05, **P < 0.01). The data represent means ± SD. J) Relative expression of TFL1 in inflorescences of the Col-0, rpt2a-2 mutants, and rpt2a-2 mutants with 5-AzaC treatment. The data are calculated from 3 biological replicates. The significance tests for TFL1 expression are performed using 2-tailed Student's t-tests (*P < 0.05, **P < 0.01). The data represent means ± SD. K) TFL1 expression is repressed in the hypermethylation ProTFL1(−1373∼−621):LUC plasmid. Left, Diagram of ProTFL1(−1373∼−621):LUC construct, and the promoter region (−1,373 to −621) of TFL1 is methylated in vitro. Right, Relative ratio of firefly LUC to Renilla LUC (REN) activity in Arabidopsis protoplasts transformed with ProTFL1(−1373∼−621):LUC plasmid with different methylation levels (0%, 40%, or 80%). The data are calculated from 3 biological replicates, and each biological replicate contains 3 technical replicates. The different letters indicate significant differences (The data represent means ± SD; 1-way ANOVA, Tukey's multiple comparison tests; P < 0.05).
To address whether these different protein levels were due to differential transcriptional regulation, we analyzed mRNA expression by in situ hybridization. Similar to the TFL1 protein signals, we detected strong TFL1 mRNA signals in the wild-type IM (Supplemental Fig. S4, H and K). In contrast, much weaker TFL1 mRNA was detected in the IM of rpt2a-2, and the signals were present only in a central subset of cells (Supplemental Figs. S4I and S5A). Likewise, we detected ectopic LFY mRNA at the position of the IM in rpt2a-2 mutants, where the TFL1 expression was reduced compared to the wild type (Supplemental Fig. S4L, M, and O). Furthermore, in the rpt2a-3 allele, LFY was ectopically expressed in the apex of those lines, where the IM differentiated into a flower (Supplemental Fig. S4N). These results suggest that TFL1 and LFY were differentially regulated in rpt2a-2 at the transcript level. To assess whether stem cell maintenance was altered in rpt2a-2, we analyzed the expression of the stem cell marker CLAVATA3 (CLV3) in the IM at 8 DAF. We found that the expression of CLV3 was noticeably weaker in the IM of rpt2a-2 mutants compared to the wild type (Fig. 2, E and F; Supplemental Fig. S4, C, E, F, G and Data Set S5), suggesting a decrease in stem cell activity, which corresponds to the diminished formation of secondary inflorescences and flowers originating from the primary inflorescence in rpt2a-2 mutants.
At 20 DAF, the presence of TFL1 signals in the rpt2a-2 inflorescence was scarce (Fig. 2H), displaying a substantial difference compared to the wild type (Fig. 2G), and the expression of LFY signals was ectopically extended throughout the terminal FM (Fig. 2, G and H). This observation aligns with the terminal flower phenotype observed in rpt2a-2 inflorescences (Fig. 1, P and Q). Thus, TFL1 is more weakly expressed, and LFY is ectopically expressed in the IM of rpt2a-2 compared to the wild type, which is consistent with the rpt2a-2 inflorescence phenotypes showing differentiation into terminal flowers.
In order to obtain a more detailed understanding of rpt2a-2 inflorescence development, we performed a detailed time-course analysis using ProTFL1:TFL1-GFP and ProLFY:LFY-Tdtomato reporters in the inflorescences of wild-type and rpt2a-2 mutants from 2 DAF (0 h) during the next 36 h (Fig. 2, I and J). TFL1 expression in rpt2a-2 inflorescences is substantially weaker than the wild type during the initiation of new FMs. Notably, ectopic expression of LFY was observed in the IM, while TFL1 expression persisted in the same IM (Fig. 2J) until the occurrence of a terminal flower phenotype in rpt2a-2 inflorescences (Fig. 2H). These results indicate that in rpt2a-2 mutants, the apical FMs initiated from a single lateral IM rather than a lateral meristem became the new IM instead of an FM.
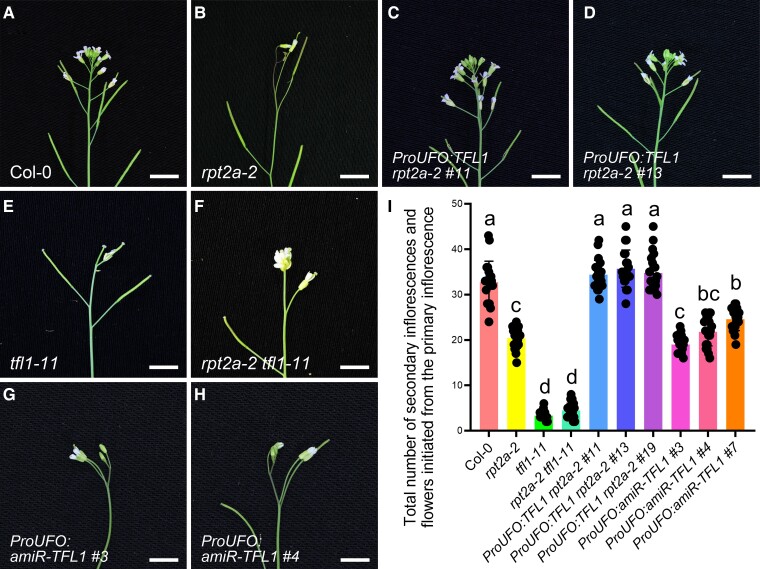
Given our finding that TFL1 expression was reduced in the IMs of the rpt2a-2 mutants, we attempted to suppress the phenotype by ectopically expressing TFL1. To this end, we expressed TFL1 driven by either the shoot apical meristem specific UNUSUAL FLORAL ORGANS (UFO) promoter (Samach et al. 1999) or the ubiquitously expressed 35S promoter in the rpt2a-2 mutants (Supplemental Figs. S4J and S5A). Both constructs completely suppressed the rpt2a-2 inflorescence phenotypes at 8 DAF (Fig. 3A to D, and I; Supplemental Fig. S6 and Data Set S6 and S7). The TFL1 null mutant tfl1-11 (Hou and Yang 2009), showed conversion of the IM into an FM, and thus termination of the main inflorescence stem in a terminal flower (Fig. 3E). We generated rpt2a-2 tfl1-11 double mutants and found that they were phenotypically indistinguishable from single homozygous tfl1-11 inflorescences (Fig. 3A, B, E, F, and I; Supplemental Data Set S6). Additionally, TFL1 knock-down lines with the amiR-TFL1 construct driven by the UFO promoter displayed a determinate inflorescence phenotype as shown in the rpt2a-2 mutants (Fig. 3G to I; Supplemental Fig. S5B and Data Set S6). Together, these data strongly suggest that IM termination in rpt2a-2 is caused by the reduced expression of the TFL1 gene in the IM and upregulation of LFY.
Figure 3.
Reduced expression of TFL1 in rpt2a-2 mutants results in determinate inflorescences. A to H) Indeterminate inflorescences of Col-0 (A; 90.2%, n = 61), ProUFO:TFL1 rpt2a-2 #11 (C; 85.4%, n = 41), and ProUFO:TFL1 rpt2a-2 #13 lines (D; 83.7%, n = 43); determinate inflorescences of rpt2a-2 (B; 91.4%, n = 58), ProUFO:amiR-TFL1 #3 (G; 81.1%, n = 37), and ProUFO:amiR-TFL1 #4 plants (H; 78.8%, n = 33); terminal flowers of tfl1-11 (E; 87.2%, n = 47) and rpt2a-2 tfl1-11 mutants (F; 90%, n = 50) at 8 d after flowering (DAF). Bar = 5 mm. I) Total number of secondary inflorescences and flowers initiated from the primary inflorescence in Col-0, rpt2a-2, tfl1-11, rpt2a-2 tfl1-11 mutants, ProUFO:TFL1 rpt2a-2 #11, ProUFO:TFL1 rpt2a-2 #13, ProUFO:TFL1 rpt2a-2 #19, ProUFO:amiR-TFL1 #3, ProUFO:amiR-TFL1 #4, and ProUFO:amiR-TFL1 #7 lines at 30 DAF. The different letters indicate significant differences (The data represent means ± SD; 1-way ANOVA, Tukey's multiple comparison tests; P < 0.05).
RPT2a-mediated degradation of MET1 controls inflorescence architecture
Given that RPT2a encodes a subunit of the 26S proteasome and that the rate of ubiquitination-dependent proteolysis is reduced upon loss of RPT2a function (Kurepa et al. 2008), we considered that the inflorescence phenotypes of the rpt2a mutants might result from aberrant protein accumulation. As a plausible candidate, we considered the LFY protein that was ectopically expressed in rpt2a-2 IM based on our reporter gene results (Fig. 2, C and D). However, recombinant His-tagged LFY was degraded at nearly the same rate in total protein extracts from wild-type and rpt2a-2 inflorescences at 8 DAF by in vitro degradation assays (Supplemental Fig. S7 and Data Set S8), suggesting that RPT2a is not required for LFY degradation in this system.
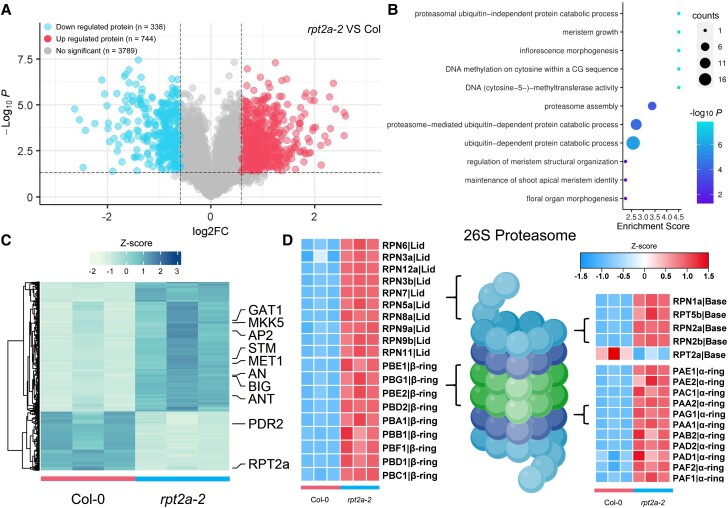
We, therefore, used nontargeted LC-MS/MS to examine protein accumulation in wild-type and rpt2a-2 inflorescences at 8 DAF. Using a 1.5-fold cutoff, we detected 1082 differentially abundant proteins, among which 744 showed increased abundance, and 338 decreased abundance in rpt2a-2 compared to wild-type inflorescences (Fig. 4A; Supplemental Data Set S9 and S10). Gene Ontology (GO) analysis indicated enrichment for proteins related to inflorescence morphogenesis, meristem growth, as well as proteasome-related processes and DNA methylation (Fig. 4, B and C; Supplemental Data Set S11). In addition, a large number of subunits except RPT2a in the 26S proteasome (Fig. 4D) and some E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases (Supplemental Fig. S8 and Data Set S10) were significantly increased in rpt2a-2 mutant compared to wild type, suggesting that there might be a compensation mechanism to reduce the effects of loss of RPT2a function in the 26S proteasome.
Figure 4.
Proteome analysis shows that proteins involved in inflorescence development and DNA methylation accumulate in rpt2a-2 mutants. A) Volcano plot shows the changes in the levels of proteins in rpt2a-2 inflorescence compared to wild-type inflorescence. The significant threshold here is log 2 (fold change) > 0.58 or < −0.58; P-value < 0.05. B) GO analysis shows differentially expressed proteins involved in inflorescence morphogenesis, proteasome, and DNA methylation in rpt2a-2 compared to Col-0 inflorescences. C) Expression heatmap illustrating the differentially-expressed proteins in Col-0 and rpt2a-2 inflorescences. Proteins associated with inflorescence morphogenesis, meristem development, and DNA methylation are shown. The differentially expressed proteins are clustered by expression levels. D) Expression heatmap of differentially expressed proteins involved in a large number of 26S proteasome subunits in Col-0 and rpt2a-2 inflorescences.
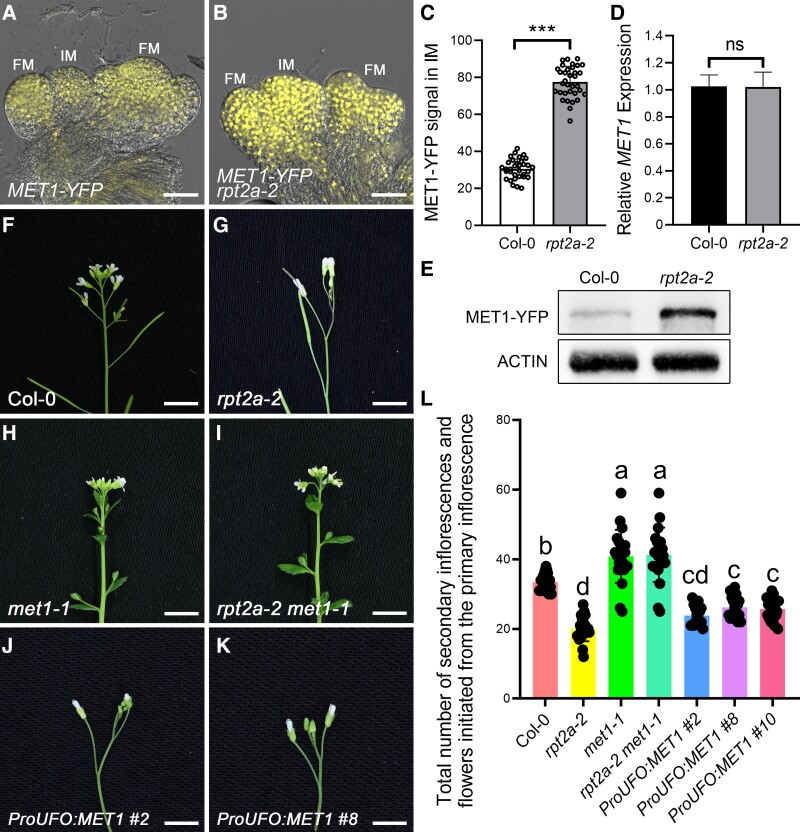
Among proteins with increased abundance in rpt2a-2 inflorescences (Fig. 4C), we chose the MET1 protein, which is essential for genome-wide CG methylation in Arabidopsis, for further analysis because a previous study reported that the primary inflorescences of the strong MET1-antisense plants produced much more secondary inflorescences before flower formation than wild type (Ronemus et al. 1996). We confirmed that also met1-1 mutants produced more secondary inflorescences than wild type grown in short days (Supplemental Fig. S9A and Data Set S12), suggesting that the increase in secondary inflorescences in met1-1 mutants cannot be solely attributed to delayed flowering (Kankel et al. 2003), emphasizing the essential role of MET1 in the development of inflorescences. We observed that expression of a MET1-YFP (yellow fluorescent protein) fusion protein driven from its native promoter was elevated in rpt2a-2 compared to wild-type inflorescences at 8 DAF (Fig. 5A to C; Supplemental Data Set S13), validating our MS data. Importantly, the MET1 mRNA levels were similar in rpt2a-2 and wild-type inflorescences (Fig. 5D), whereas MET1-YFP protein levels were elevated in rpt2a-2 inflorescences compared to the wild type (Fig. 5E; Supplemental Fig. S10A). This result indicates that RPT2a affects MET1 expression at the protein level.
Figure 5.
Excessive accumulation of MET1 leads to determinate inflorescences in rpt2a-2 mutants. A and B) The expression of MET1-YFP is much stronger in the inflorescences of rpt2a-2 mutants (B; 80%, n = 35) compared to Col-0 (A; 87.9%, n = 33) at 8 d after flowering (DAF). IM, inflorescence meristem. FM, floral meristem. Bar = 200 μm. C) Fluorescence intensity profiles of MET1-YFP in the inflorescences of Col-0 and rpt2a-2 mutants at 8 DAF. The significance tests for MET1-YFP signals are performed using 2-tailed Student's t-tests (***P < 0.001). The data represent means ± SD. D) Relative levels of MET1 mRNA in inflorescences of the Col-0 and rpt2a-2 mutants at 8 DAF are not significantly different from each other. The data are calculated from 3 biological replicates (The data represent means ± SD; 2-tailed Student's t-tests; ns, not significant). E) Immunoblot analysis shows that the level of MET1-YFP protein in rpt2a-2 inflorescences at 8 DAF is much higher than that in Col-0 inflorescences with anti-GFP antibody. ACTIN is used as a control. Three independent replicates show similar results. F to K) Indeterminate inflorescences of the Col-0 (F; 89.5%, n = 57), met1-1 (H; 84.8%, n = 46), and rpt2a-2 met1-1 mutants (I; 85.7%, n = 49), and determinate inflorescences of the rpt2a-2 (G; 93.6%, n = 47), ProUFO:MET1 #2 (J; 84%, n = 25), and ProUFO:MET1 #8 plants (K; 81.5%, n = 27) at 8 DAF. Bar = 5 mm. L) Total number of secondary inflorescences and flowers initiated from the primary inflorescence in Col-0, rpt2a-2, met1-1, rpt2a-2 met1-1 mutants, ProUFO:MET1 #2, ProUFO:MET1 #8, and ProUFO:MET1 #10 plants at 30 DAF. The different letters indicate significant differences (The data represent means ± SD; 1-way ANOVA, Tukey’s multiple comparison tests; P < 0.05).
To investigate the role of MET1 in the rpt2a-2 inflorescence phenotypes, we generated the double mutants of met1-1, a strong loss-of-function allele, and rpt2a-2. In met1-1, flowers were partially transformed into indeterminate shoot meristems (Fig. 5, F and H; Supplemental Fig. S9A and Data Set S12), consistent with the previous study (Ronemus et al. 1996). Strikingly, rpt2a-2 met1-1 double mutants had an indeterminate inflorescence that was indistinguishable from met1-1 mutants at 8 DAF (Fig. 5, F-I, and L; Supplemental Data Set S14), indicating that inflorescence termination in rpt2a-2 mutants requires MET1 function. We further observed that when MET1 is overexpressed under the control of the UFO promoter (ProUFO:MET1), it also exhibits a determinate inflorescence phenotype, resembling that of the rpt2a-2 mutants (Fig. 5, J to L; Supplemental Fig. S9B and Data Set S14). These findings indicate that elevating the levels of MET1 in the rpt2a-2 mutants results in IM termination.
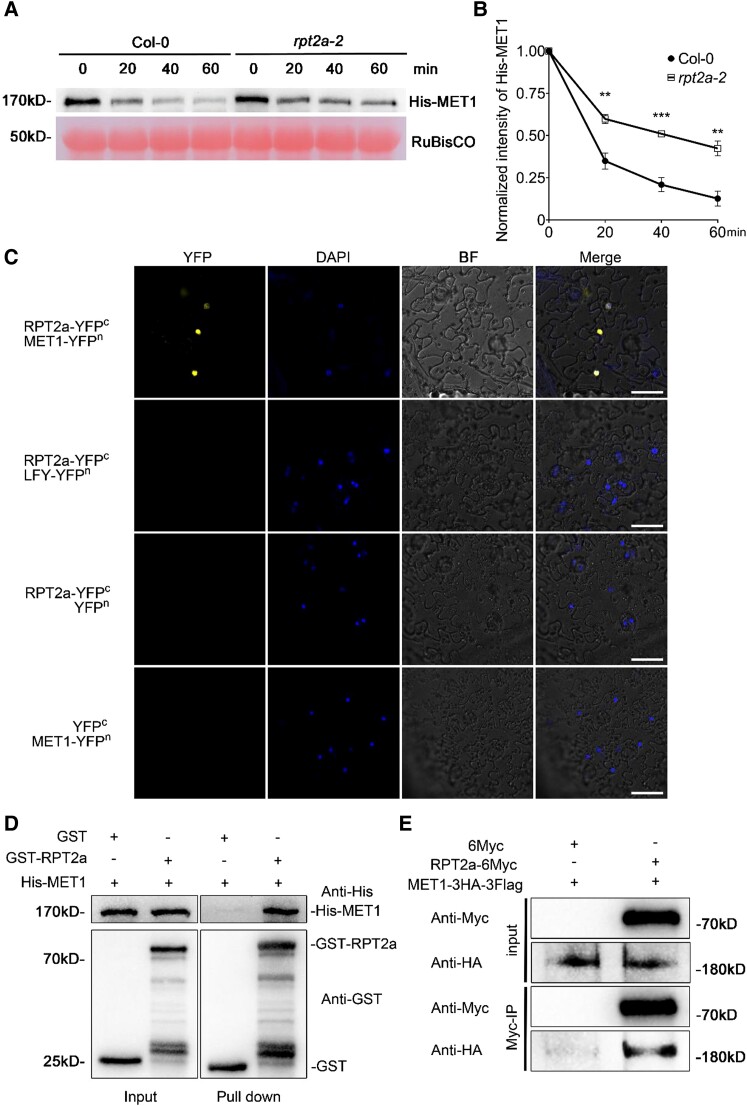
Given the reported function of RPT2a in protein degradation, we considered that the elevated levels of MET1 protein in rpt2a-2 inflorescences might be due to diminished RPT2a-mediated degradation of MET1. Consistent with this hypothesis, recombinant His-tagged MET1 was degraded more slowly in in vitro degradation assays with total protein extracts from rpt2a-2 inflorescences than wild type (Fig. 6, A and B; Supplemental Fig. S10B and Data Set S15). Next, we tested whether RPT2a interacted directly with MET1 and found an interaction between transiently expressed RPT2a-cYFP and MET1-nYFP by bimolecular fluorescence complementation (BiFC) in Nicotiana benthamiana (Fig. 6C). Furthermore, GST-RPT2a interacted with His-MET1 in in vitro pull-down assays (Fig. 6D; Supplemental Fig. S10C). Finally, MET1-3HA-3Flag co-immunoprecipitated with RPT2a-6Myc from extracts of Arabidopsis protoplasts co-expressing both plasmids (Fig. 6E; Supplemental Fig. S10D). These findings suggest that RPT2a directly interacts with MET1 leading to its degradation, which is required to maintain IM indeterminacy and the indeterminate inflorescence architecture of Arabidopsis.
Figure 6.
RPT2a-MET1 interaction promotes MET1 degradation. A) Cell-free degradation assays show that RPT2a promotes MET1 degradation. Equal amounts of total proteins extracted from Col-0 and rpt2a-2 inflorescences at 8 d after flowering (DAF) are incubated with recombinant His-MET1 protein in the presence of ATP. MET1 is detected with an anti-His antibody. RuBisCO is used as a loading control. Three independent replicates show similar results. B) Quantitative analysis of recombinant His-MET1 protein degradation rate in the Col-0 and rpt2a-2 inflorescences at 8 DAF in 3 independent experiments. The significance tests for His-MET1 protein degradation are performed. There are significant differences in 3 time points (20, 40, and 60 mins; The data represent means ± SD; 2-tailed Student's t-tests; **P < 0.01, ***P < 0.001). C) BiFC assays in Nicotiana benthamiana epidermal cells show that RPT2a interacts with MET1. LFY protein, which degradation is not promoted by RPT2a, is used as a negative control. BF represents bright field. Bar = 50 μm. D) Pull-down assays with GST-RPT2a immobilized on a Glutathione sepharose show that His-MET1 is pulled down. Three independent replicates show similar results. E) RPT2a is co-immunoprecipitated with MET1. RPT2a-6Myc, and MET1-3HA-3Flag are transiently co-expressed in Arabidopsis protoplasts; controls include transformation with 6Myc and MET1-3HA-3Flag. Three independent replicates show similar results.
An RPT2a-MET1 axis regulates TFL1 expression via its promoter methylation
MET1 functions to maintain DNA methylation (Ronemus et al. 1996), which can be associated with transcriptional silencing (Finnegan et al. 1996). We therefore asked whether the reduced expression of TFL1 in the rpt2a-2 IM (Fig. 2, C and D) might be due to increased MET1-mediated DNA methylation at the TFL1 locus. We analyzed the published data from a genome-wide analysis of DNA methylation in the wild type and met1 mutants (Lister et al. 2008) and found that a fragment (from −1,350 to −600) within the TFL1 promoter was highly methylated in the wild type but not in met1 mutants (Supplemental Fig. S11). These data suggest that MET1 is required for methylation of this TFL1 upstream region.
We, therefore, examined DNA methylation of TFL1 in wild-type, rpt2a-2, met1-1, and rpt2a-2 met1-1 inflorescences at 8 DAF by bisulfite sequencing the regions (−1,946 to −1,350, −1,373 to −621, and −645 to −118) upstream of the TFL1 coding region (Fig. 7A) and its 3′ untranslated region (2,002 to 2449, 2,596 to 3,288, 3,799 to 4,365, and 4,286 to 4,680) (Serrano-Mislata et al. 2016; Supplemental Fig. S12A). We observed 1 region (−1,373 to −621) that displayed elevated 5-methylcytosine (5mC) levels of CG and CHG contexts in rpt2a-2 inflorescences compared to the wild type (Fig. 7B; Supplemental Figs. S12B, S13 and Data Set S16 and S17) but showed reduced 5mC levels in met1-1 inflorescences (Fig. 7B; Supplemental Fig. S13 and Data Set S16). Importantly, met1-1 suppressed elevated methylation levels of rpt2a-2 (Fig. 7B; Supplemental Fig. S13 and Data Set S16), indicating that RPT2a controls methylation of the TFL1 upstream region (−1,373 to −621) through MET1.
To determine if met1-1 suppressed the determinate rpt2a-2 inflorescence phenotype by restoring TFL1 expression, we transformed Arabidopsis with the ProTFL1:TFL1-GFP construct. Indeed, the TFL1-GFP signals were elevated in the IM of met1-1 and rpt2a-2 met1-1 mutants compared with the transformed wild type and rpt2a-2 mutants (Fig. 7C to G; Supplemental Data Set S18). We further observed the reduced levels of TFL1 transcription in rpt2a-2 inflorescences compared to wild type but elevated levels in the inflorescences of both met1-1 and rpt2a-2 met1-1 double mutants (Fig. 7H).
To further validate the role of DNA methylation in TFL1 expression, inflorescences were treated with DNA methylation accelerator methyl trifluoromethanesulfonate (MTFMS) and DNA methylation inhibitor 5-AzaC to manipulate the DNA methylation level in the TFL1 promoter (Supplemental Fig. S14, A and B). We found that inflorescences treated with MTFMS exhibited increased DNA methylation and decreased expression of TFL1, while those treated with 5-AzaC showed decreased DNA methylation and increased TFL1 expression compared to the wild type (Fig. 7I; Supplemental Fig. S14A). Furthermore, the rpt2a-2 inflorescences treated with 5-AzaC exhibited decreased DNA methylation and increased TFL1 expression compared to the rpt2a-2 inflorescences (Fig. 7J; Supplemental Fig. S14B).
Additionally, a specific fragment (−1,373 to −621) of the TFL1 promoter was cloned into the pGreenII 0800-LUC vector. Subsequently, this plasmid was subjected to varying levels of DNA methylation through in vitro methylation using the CpG methylase M.SssI (Fig. 7K; Supplemental Fig. S14C). Upon transformation into Arabidopsis protoplasts, TFL1 expression was reduced in the hypermethylated plasmid (Fig. 7K; Supplemental Data Set S19). Additionally, we found that tfl1-11 met1-1 double mutants displayed early terminated inflorescence phenotypes similar to those of the tfl1-11 single mutants (Supplemental Fig. S15 and Data Set S20), confirming that TFL1 is required for the inflorescence phenotypes in met1-1. In summary, these results suggest that the hypermethylation of the TFL1 promoter, induced by MET1 accumulation in the rpt2a-2 mutants, leads to decreased TFL1 expression, ultimately disrupting the indeterminacy of the IM.
Discussion
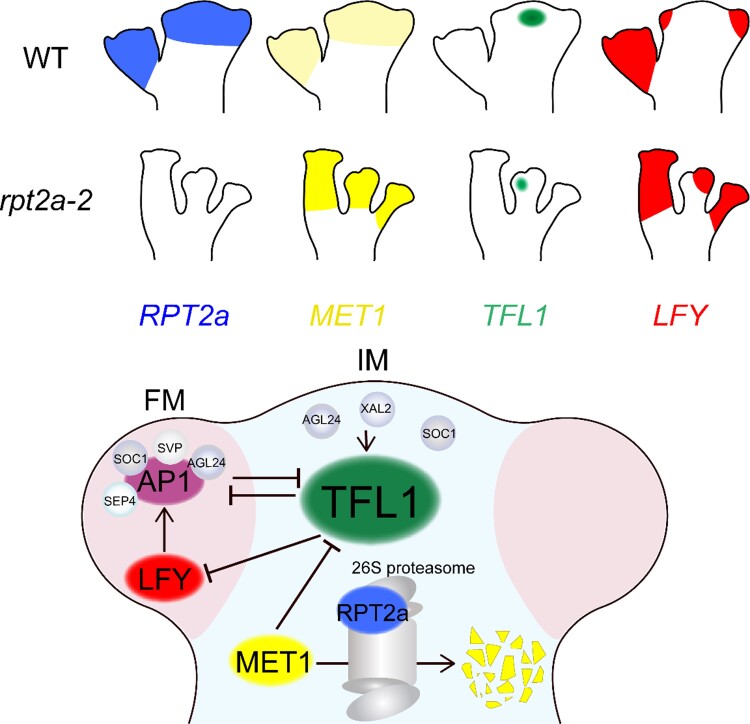
Despite a plethora of shapes in the plant kingdom, the architecture of inflorescences falls into a few main categories, such as racemes as in Brassicaceae, cymes as in nightshade species, or panicles as in grasses. The different inflorescence architectures are associated with different duration of stem cell activity in the IMs (McKim and Hay 2010; Wang et al. 2018). In this study, we show that in Arabidopsis, proteolytic activity balances the epigenetic input into the transcription of the key regulator of indeterminacy, TFL1, which in turn promotes indeterminate over determinate shoot architectures in Arabidopsis (Fig. 8).
Figure 8.
Model of RPT2a regulating the maintenance of the IM. Arabidopsis RPT2a promotes the degradation of MET1, leading to DNA hypomethylation upstream of the IM identity gene TFL1 and high TFL1 expression levels in the IM. TFL1 maintains IM indeterminacy by repressing the expression of LFY and AP1 genes. LFY can activate TFL1 expression in ap1 cal double mutants, but in wild-type, inflorescence can activate AP1 to repress TFL1 expression. In FM, MADS-box transcription factors, including SOC1, short vegetative phase, AGL24, and SEP4, directly suppress TFL1 in an AP1-dependent manner. In IM, SOC1, AGL24, and XAL2 also bind to and induce TFL1 expression (Hanano and Goto 2011; Winter et al. 2011; Liu et al. 2013; Pérez-Ruiz et al. 2015; Goslin et al. 2017; Azpeitia et al. 2021). The arrows represent activation and the T-ended arrows represent inhibition.
A key in this regulatory pathway maintaining IM indeterminacy is the regulation of TFL1 expression. Previously, using immature floral buds as source tissue, hypomethylation of the TFL1 locus was detected in the met1-3 loss-of-function mutants, suggesting that MET1 contributes to the silencing of TFL1 transcription outside the IM (Lister et al. 2008). However, MET1 is also expressed in the IM, where TFL1 expression is required for IM maintenance, raising the question of why TFL1 is not silenced there. Our results provide insight into this paradox by showing that in the wild-type IM, RPT2a-mediated MET1 protein degradation ensures high TFL1 expression levels and, thus, indeterminacy of the IM (Fig. 8). These findings are in line with previous reports that the RPT2a mutation leads to DNA hypermethylation and silencing of transgenes and selected endogenous genes in seedlings, but the mechanism was not revealed and the situation in meristems was not addressed (Sako et al. 2012).
The similarity between the determinate zigzag-shaped inflorescence in the Arabidopsis rpt2a mutants and natural nightshade plants with a cyme inflorescence raises the question of whether similar molecular mechanisms are in place. Detailed genetic and molecular studies provide convincing evidence that homologous factors are employed in the indeterminate IM of Arabidopsis and model nightshade species such as petunia and tomato (Pnueli et al. 1998; Souer et al. 1998; Molinero-Rosales et al. 1999; Lippman et al. 2008). Many of these Arabidopsis factors complement the corresponding mutants in petunia or tomato and vice versa, suggesting conserved protein functions (Lippman et al. 2008; Souer et al. 2008). However, the expression patterns and specific roles of these homologous genes in regulating meristem behavior vary considerably.
For example, in Arabidopsis, the reduction of TFL1 expression in the IM results in loss of IM indeterminacy. Considering that there is no indeterminate IM in tomato, it is not surprising that mutations in the TFL1 ortholog, SELF-PRUNING (SP), do not alter the overall sympodial growth mode of tomato. However, it reduces the number of sympodial units formed by transforming the sympodial meristem into a terminal inflorescence and this has been interpreted as loss of indeterminacy of the shoot system (Pnueli et al. 1998). While these SP functions might appear reminiscent of TFL1 function in maintaining indeterminacy and blocking floral fate, the expression patterns differ. Unlike its Arabidopsis counterpart, SP is expressed at similar levels in IM and FM (Pnueli et al. 1998). Therefore, the different inflorescence architectures between Arabidopsis and petunia or tomato appear to involve differential expression patterns and modified functions of homologous proteins.
Our results show that a graded input of TFL1 results in different IM phenotypes in Arabidopsis. In wild type, high TFL1 levels enable IM indeterminate growth. In the strong loss-of-function rpt2a-2 mutants, TFL1 expression is reduced but still present in the lateral IM until it transforms into a terminal flower. In the complete absence of TFL1 activity, the primary IM of the tfl1-11 mutant produces only 1 or 2 FMs before differentiating into a terminal flower. Thus, the duration of IM activity appears to depend on TFL1 levels, with decreasing TFL1 activities in wild type > rpt2a-2 > tfl1-11 causing decreasing IM indeterminacy.
In addition to the loss of indeterminacy of the IM in rpt2a-2 mutants, lateral meristems are specified as IM, rather than FM as in wild type. Although our proteome analysis shows that RPT2a affects the levels of many proteins, we consider it unlikely that deregulation of a yet undiscovered RPT2a client protein accounts for this difference because transgenic upregulation of TFL1 entirely restores shoot architecture in rpt2a. Therefore, we consider 2 possible scenarios of how the lateral meristems in rpt2a mutants might be specified as IMs.
First, a lateral suppression model implies that high levels of TFL1 in the primary IM are required to suppress the formation of lateral IMs in the wild-type shoot apex. Consequently, the reduction of TFL1 levels in the rpt2a primary IM might disturb this lateral suppression, thus enabling the formation of lateral IMs instead of FM. The presence of lateral inhibition mechanisms to separate functions of different meristem regions is supported by elegant experiments ablating the central cells of the shoot meristem, which caused the initiation of a new meristem center in neighboring cells (Reinhardt et al. 2003).
A second model holds that the rpt2a IM could have retained some vegetativeness and thus directly maintained a lateral “branch” IM rather than an FM. This view is in line with the “transient model” for the lateral shoot formation in nightshade species put forward previously (Prusinkiewicz et al. 2007; Castel et al. 2010). The model proposes that variations in the level of vegetativeness, which might encompass complex regulatory machinery, determine whether an apical meristem produces a side shoot or a flower. Along these lines, a fully generative IM in the wild type would impose floral fate in lateral buds, whereas a reduced vegetative IM in the rpt2a inflorescence would maintain a lateral IM.
Since the initial isolation of genes regulating development in Arabidopsis, transcription regulation has been studied most intensively. The developmental input at other regulatory levels, such as protein stability, has received less attention. Our results demonstrate that proteolytic adjustment of epigenetic regulation of TFL1 expression plays a major role in ensuring IM indeterminacy and indeterminate shoot architecture in Arabidopsis, adding an additional layer to stem cell regulation. Future studies will address these different models and increase our understanding of the genetic control of the variety of inflorescence architecture.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana accession Columbia-0 (Col-0) was used as the wild-type background in this study. For phenotypic and genetic analysis, T-DNA insertion lines in rpt2a-2 (SALK_005596), rpt2a-3 (SALK_130019), and rpn12a-2 (SALK_134934C) were obtained from the Arabidopsis Biological Resource Center (ABRC). The transgenic lines MET1-YFP (Li et al. 2017) and ProCLV3:GFP (Gordon et al. 2007), the mutants of rpn1a-2 (SALK_129604) (Yao et al. 2012), rpt4a-1 (SALK_052372) (Yao et al. 2012), tfl1-11 (Hou and Yang 2009), and met1-1 (Liu et al. 2018) were described previously. The double mutants rpt2a-2 met1-1, rpt2a-2 tfl1-11, tfl1-11 met1-1, rpt2a-2 rpn1a-2, rpt2a-2 rpt4a-1, and rpt2a-2 rpn12a-2 were generated by crossing single mutants. The transgenic lines MET1-YFP and ProCLV3:GFP were crossed with rpt2a-2 mutants. Mutants used in this study are also listed in Supplemental Data Set S21.
Arabidopsis seeds were surface sterilized (washed in 70% (v/v) ethanol for 5 min, 2.6% sodium hypochlorite for 10 min, and 5 times in sterile water) and grown on 1/2× Murashige and Skoog (MS) medium (Solarbio, China; M8521) containing 1% (w/v) agar and 1% (w/v) sucrose. The seedlings were stored at 4 °C for 2 days and grown at 22 °C, with a 16-h light/8-h dark photoperiod 10,000 l× light-emitting diode (LED)-light for 9 days before transplanting into the soil for further growth.
Plasmid construction and plant transformation
To generate the ProRPT2a:RPT2a-GFP rpt2a-2 lines, 4.1 kb RPT2a genomic fragment, and 0.5 kb 3′ UTR fragment were PCR-amplified from Arabidopsis inflorescence genomic DNA using Phanta Max Master Mix (Vazyme, China; P515-01), and cloned into the pROKII-GFP vector (Baulcombe et al. 1986) by ClonExpress II One Step Cloning Kit (Vazyme, China; C112-01). To generate the ProUFO:TFL1 and ProUFO:MET1 lines, 3.8 kb UFO promoter fragment, 0.5 kb TFL1 coding sequence, and 4.6 kb MET1 coding sequence were PCR-amplified from genomic DNA and inflorescence cDNA, respectively, and cloned into the pBI101 vector (Datia et al. 1992). To generate the Pro35S:TFL1 lines, the 0.5 kb TFL1 coding sequence was PCR-amplified from inflorescence cDNA and cloned into the pEarleyGate203 vector (Earley et al. 2006). To generate the ProUFO:amiR-TFL1 lines, a 3.8 kb UFO promoter fragment and 0.5 kb amiR-TFL1 fragment were PCR-amplified from genomic DNA and RS300 plasmid (Schwab et al. 2006), and cloned into the pBI101 vector (Datia et al. 1992).
To generate the ProTFL1:TFL1-GFP construct, a 3.8 kb TFL1 genomic fragment and a 4.6 kb 3′ UTR fragment were PCR-amplified from genomic DNA and cloned into the pBI101-GFP vector (Datia et al. 1992), respectively. To generate the ProLFY:LFY-Tdtomato construct, a 5.2 kb LFY genomic fragment was PCR-amplified from genomic DNA, and cloned into the pG2NBL-Tdtomato vector (Blilou et al. 2005). The constructs were introduced into the wild-type plants using the floral dip method, employing Agrobacterium tumefaciens infection as described by Clough and Bent (Clough and Bent 1998). This allowed us to obtain transgenic wild-type plants, which were subsequently crossed with rpt2a-2, met1-1, tfl1-11, and lfy mutants, respectively. Homozygous mutant plants carrying the transgene (F2 generation) were identified in this study. Primers used in this study are listed in Supplemental Data Set S22.
RNA in situ hybridization and RT-qPCR analysis
The coding sequences of RPT2a, TFL1, and LFY were PCR-amplified and cloned into the pEASY-Blunt3 vector (TransGen Biotech, China; CB301-01). The clones with forward or reverse sequences were screened by DNA sequencing; then the sequences were labeled as sense or antisense RNA probes in vitro using T7 RNA polymerases. Section preparation, in situ hybridization, and detection of hybridized signals were operated as previously described (Su et al. 2020).
Total RNA was extracted from the inflorescences using Ultrapure RNA Kit (CWbiotech, China; CW0581M). 5 μg RNA was used to synthesize first-strand cDNA using an EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, China; AE311-03). SYBR Green Real-time PCR Master Mix (Tiangen Biotech, China; FP205) was used for RT-qPCR, according to the manufacturer's protocol. The 60 inflorescences that were cultivated at the same time were collected as a biological replicate; 3 biological replicates were performed in each sample.
Live imaging and fluorescence analysis
Seeds were germinated on 1/2× MS medium and grown for 7 days. The plants were transferred into 3 cm deep glass dishes containing 1/2× MS medium and cultivated until bolting. When the shoot apex grew out from the rosette, older flower buds were removed carefully by a fine tweezer and dissecting needle to expose the shoot apex. After the operation above, the plants were continuously cultivated in a 22 °C, 24-h light 10,000 lx LED-light incubator when the plants were not imaging (Reddy et al. 2004).
The inflorescences of ProTFL1:TFL1-GFP, ProLFY:LFY-Tdtomato, ProCLV3:GFP, and MET1-YFP reporter lines were imaged by a Zeiss 810 laser scanning microscope using a 40× water dipping Achroplan lens. For GFP, YFP, and Tdtomato detection, we used 488, 514, and 561 nm laser excitation, 505 to 550 nm, 530 to 600 nm, and 580 to 650 nm emission spectra for each respective excitation. The glass dishes were filled with water to ensure the plants were submerged. After imaging, the water was removed, and the plants were returned to the incubator. Zeiss ZEN software was used to image, analyze, and export. To evaluate the fluorescence signal intensity, we employed Zeiss ZEN (blue edition) software. The GFP, Tdtomato, and YFP fluorescence area was delineated using a spline contour tool, and subsequently, the mean intensity value within the selected area was measured. To determine the fluorescence signal intensity, we evaluate about 20 to 30 inflorescences in each material for statistical analysis.
TMT-label proteomics analysis
For tandem mass tag (TMT)-label proteomics analysis, we utilized the inflorescence apex tissue collected at 8 DAF. Initially, we carefully removed most of the visible flower buds. Subsequently, we excised the inflorescences from the main plant axis and rapidly froze them in liquid nitrogen. Protein digestion, TMT labeling, HPLC fractionation, and liquid chromatograph (LC)-MS/MS analysis were performed by Shanghai OE Biotech Company (China). The raw MS/MS data were processed using Proteome Discover 2.3 (Thermo Fisher, USA), with settings described as follows: 2 missed cleavages, a mass tolerance of 10 ppm for peptide ions, and 0.02 Da for fragment ions, respectively. The P-value < 0.05 was assigned as a threshold to identify the significant differentially expressed proteins, together with a fold change ≥ 1.5. The 200 inflorescences were cultivated at the same time and were collected as a biological replicate; 3 biological replicates were performed for the TMT-label proteomics analysis. The detailed data of TMT-label proteomics is added in Supplemental Data Set S9 to S11.
Co-immunoprecipitation assays
For co-immunoprecipitation (Co-IP) assays, the coding sequences of RPT2a and MET1 were PCR-amplified and cloned into the transient expression vectors pSuper1300-Myc and pCAMBIA1307-C-HF (Ding et al. 2015), respectively. Co-IP assays were performed as previously described (Liu et al. 2021). In brief, the total proteins were extracted from Arabidopsis protoplasts expressing MET1-3HA-3Flag/RPT2a-6Myc, or MET1-3HA-3Flag/6Myc constructs with protein extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 20% glycerol (v/v), 0.1% Nonidet P 40 (NP-40) (v/v), 0.1% Triton X-100 (v/v), 5 mm DTT, and 1× protease inhibitor cocktail (Roche, Germany; 04693132001)) and then incubated with anti-Myc antibody-conjugated agarose (Sigma-Aldrich, USA; A7470) at 4 °C for 2 h. After washing by extraction buffer 5 times, the co-immunoprecipitated products were separated by SDS–PAGE and detected with anti-Myc (1:5,000 dilution, Sigma-Aldrich, USA; M4439) and anti-HA (1:5,000 dilution, Sigma-Aldrich, USA; H3663) antibodies.
In vitro pull-down assays
For in vitro pull-down assays, the coding sequence of RPT2a was cloned into the pGEX-4T-1 vector (GE Healthcare, USA; 27-4580-01). In vitro pull-down assays were performed as previously described (Su et al. 2020).10 μg purified GST-RPT2a or GST proteins were incubated with Glutathione Sepharose 4 Fast Flow (GE Healthcare, USA; 17-5132-01) in phosphate buffer saline buffer containing 0.3% NP-40, 1× protease inhibitor cocktail, and 5 mm DTT (named as pull-down binding buffer) at 4 °C for 2 h and then washed by pull-down binding buffer for 1 time. The immunoprecipitated GST-RPT2a or GST was incubated with 3 μg His-MET1 (Chen et al. 2020) at 4 °C for 2 h in a pull-down binding buffer. After 5 washes, the proteins were separated by SDS–PAGE. The GST and GST-RPT2a proteins were detected by anti-GST antibody (1:2,000 dilution, Beijing Protein Innovation, China; AbM59001-2H5-PU), and the His-MET1 protein was detected by anti-His antibody (1:2,000 dilution, Abbkine, China; ABT2050).
BiFC assays
The coding sequences of RPT2a and MET1 were PCR-amplified and cloned into the pFGC-YC155 and pFGC-YN173 vectors (Wang et al. 2017), which contain fragments encoding the C- or N-terminal fusions of YFP (YFPC or YFPN), respectively. The 2 resulting constructs, the control vectors pFGC-YC155 and pFGC-YN173, and the cotransformation vector Pro35S:P19 (Voinnet et al. 1999) were transformed into the Agrobacterium tumefaciens EHA105 strain through freezing-thawing method. The strains were incubated, harvested, and resuspended in infiltration buffer (10 mM MES-KOH pH 5.7, 150 μM acetosyringone, and 10 mM MgCl2) at a final concentration of OD600 = 0.6. Equal volumes of different combinations of strains were mixed and injected into 6-wk-old Nicotiana benthamiana leaves using a needleless syringe. The leaves were cultivated at 25 °C for 48 h before the detection of YFP fluorescence by Zeiss 810 laser scanning microscope with 514 nm laser excitation, 530 to 600 nm emission spectra.
Cell-free protein degradation assays
The coding sequence of LFY was PCR-amplified and cloned into the pET-28a vector (Novagen, USA; 69864-3). Cell-free protein degradation assays were performed as previously described (Ding et al. 2015). Purified His-LFY and His-MET1 (Chen et al. 2020) recombinant proteins were incubated with total proteins extracted from Col-0 and rpt2a-2 inflorescences in cell-free buffer (50 mm Tris–MES pH 8.0, 0.5 m Sucrose, 1 mM MgCl2, 10 mM EDTA pH 8.0, and 5 mM DTT) in the presence of ATP at 25 °C for different time courses. His-LFY and His-MET1 proteins were detected by immunoblotting with anti-His antibody (1:2,000 dilution, Abbkine, China; ABT2050).
Bisulfite sequencing
Arabidopsis inflorescences of Col-0, rpt2a-2, met1-1, and rpt2a-2 met1-1 mutants were collected for bisulfite sequencing. Bisulfite treatment was carried out using the DNA Bisulfite Conversion Kit (Tiangen Biotech, China; DP215). PCR products were cloned into the pEASY-Blunt3 Vector, and at least 12 clones were sequenced to determine the DNA methylation level of TFL1 loci. Sequencing data were analyzed by the online CyMATE software (Hetzl et al. 2007). The 90 inflorescences that were cultivated at the same time were collected as biological replicates and 3 biological replicates were performed in each sample.
MTFMS and 5-AzaC treatment
DNA methylation accelerator MTFMS (TCI, Japan; T2029) or DNA methylation inhibitor 5-AzaC (Sigma-Aldrich, USA; A2385) were applied to promote or suppress DNA methylation, respectively. The inflorescences were dipped into the 500 μM MTFMS or 100 μM 5-AzaC solution containing 0.05% (v/v) Silwet L-77. Distilled water with Silwet L-77 was used as a control (Mock). In this treatment, inflorescences were dipped once every day for a total duration of 4 d. Each treatment included a minimum of 50 inflorescences.
DNA methylation in vitro and dual-luciferase
The fragment −1,373 to −621 of the TFL1 promoter was PCR-amplified and cloned into the pGreenII 0800-LUC vector (Hellens et al. 2005). The ProTFL1(−1373∼−621):LUC plasmid was methylated in vitro by CpG methylase M.SssI (NEB, USA; M0226S) at 37 °C. Different incubation times could obtain different levels of DNA methylation, which was verified by the DNA-methylation-sensitive enzyme HpaII (Thermo Fisher, USA; FD0514). The pGreenII 0800-LUC and ProTFL1(−1373∼−621):LUC plasmids of different methylation levels were transformed into Arabidopsis protoplasts, respectively. The protoplasts were lysed and detected by the Dual-Luciferase Reporter Gene Assay Kit (Yeasen Biotechnology, China; 11402ES60). The pGreenII 0800-LUC protoplasts were used as background (Devesa-Guerra et al. 2020).
Quantification and statistical analysis
To determine the total number of secondary inflorescences and flowers initiated from the primary inflorescence, we tested at least 20 plants in each material for statistical analysis. All t-test analyses and analysis of variance (ANOVA) analyses were conducted using GraphPad software. To determine statistical significance, we employed 2-tailed Student's t-tests between 2 groups and 1-way ANOVA with Tukey's multiple comparison tests among various genotypes. A value of P < 0.05 was considered statistically significant.
Accession numbers
Arabidopsis gene sequence data can be found in TAIR under the following accession numbers: RPT2a (AT4G29040), RPT4a (AT5G43010), RPN1a (AT2G20580), RPN12a (AT1G64520), TFL1 (AT5G03840), LFY (AT5G61850), CLV3 (AT2G27250), UFO (AT1G30950), and MET1 (AT5G49160). The nontargeted LC-MS/MS (TMT-label proteomics) data have been deposited in the Beijing Institute of Genomics Data Center (http://bigd.big.ac.cn) with the BioProject PRJCA011875.
Supplementary Material
Acknowledgments
We are grateful to Prof. Elliot M. Meyerowitz (California Institute of Technology) for providing the seeds of ProCLV3:GFP transgene lines, Dr. Huishan Guo (Institute of Microbiology, Chinese Academy of Sciences) for providing the vector of His-MET1, Prof. Dingzhong Tang (Fujian Agriculture and Forestry University) for providing the seeds of rpn1a-2 and rpt4a-1 mutants, and Prof. Binglian Zheng (Fudan University) for providing the seeds of MET1-YFP transgene lines.
Contributor Information
Wang Jinsong Yao, State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai'an, Shandong 271018, China.
Yi Peng Wang, State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai'an, Shandong 271018, China.
Jing Peng, State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai'an, Shandong 271018, China.
Pei Pei Yin, State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai'an, Shandong 271018, China.
Hengbin Gao, State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai'an, Shandong 271018, China.
Li Xu, State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai'an, Shandong 271018, China.
Thomas Laux, Sino-German Joint Research Center on Agricultural Biology, Shandong Agricultural University, Tai’an, Shandong 271018, China; Signalling Research Centres BIOSS and CIBSS, Faculty of Biology, University of Freiburg, Schänzlestrasse 1, 79104 Freiburg, Germany.
Xian Sheng Zhang, State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai'an, Shandong 271018, China.
Ying Hua Su, State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai'an, Shandong 271018, China; Sino-German Joint Research Center on Agricultural Biology, Shandong Agricultural University, Tai’an, Shandong 271018, China.
Author contributions
Y.H.S. designed the experiments. Y.H.S., X.S.Z., and T.L. wrote the manuscript. W.J.Y., Y.P.W., J.P., P.P.Y., H.B.G., and L.X. performed the experiments. W.J.Y. and Y.P.W. performed the data analysis.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Characterization of rpt2a mutants.
Supplemental Figure S2. The shoot apex is located at the lateral of determinate zigzag-shaped inflorescence in rpt2a-2 mutants.
Supplemental Figure S3. Functional redundancy among subunits of the 19S regulatory particle for the maintenance of IM indeterminacy.
Supplemental Figure S4. The expression of TFL1 and LFY is altered in rpt2a-2 mutants.
Supplemental Figure S5. TFL1 expression in ProUFO:TFL1 rpt2a-2, Pro35S:TFL1 rpt2a-2, and ProUFO:amiR-TFL1 transgenic lines.
Supplemental Figure S6. TFL1-overexpression can suppress the phenotype of the rpt2a-2 inflorescences.
Supplemental Figure S7. The degradation of LFY protein is not promoted by RPT2a.
Supplemental Figure S8. Many proteins in the ubiquitin system accumulate in rpt2a-2 inflorescence.
Supplemental Figure S9. MET1 affects inflorescence architecture.
Supplemental Figure S10. MET1 is degraded by RPT2a via interaction.
Supplemental Figure S11. Genome-wide analysis of DNA methylation in the TFL1 loci.
Supplemental Figure S12. Bisulfite sequencing in the TFL1 3′ untranslated regions.
Supplemental Figure S13. Methylation of cytosine residues in fragment −1,373 to −621 of TFL1 promoter.
Supplemental Figure S14. Manipulation of the DNA methylation level in fragment −1,373 to −621 of the TFL1 promoter.
Supplemental Figure S15. TFL1 is required in MET1 regulating IM indeterminacy.
Supplemental Data Set 1. Figure 1R data: Total number of secondary inflorescences and flowers initiated from the primary inflorescence in Col-0, rpt2a-2, rpt2a-3, ProRPT2a:RPT2a-GFP rpt2a-2, and ProRPT2a:RPT2a-GFP rpt2a-3 plants.
Supplemental Data Set 2. Supplemental Figure S3I data: Total number of secondary inflorescences and flowers initiated from the primary inflorescence in Col-0, rpt2a-2, rpn12a-2, rpt2a-2 rpn12a-2, rpn1a-2, rpt2a-2 rpn1a-2, rpt4a-1, and rpt2a-2 rpt4a-1 mutants.
Supplemental Data Set 3. Supplemental Figure S4A data: Total number of secondary inflorescences and flowers initiated from the primary inflorescence in Col-0, lfy, tfl1-11 mutants, ProLFY:LFY-Tdtomato lfy, and ProTFL1:TFL1-GFP tfl1-11 lines.
Supplemental Data Set 4. Supplemental Figure S4B data: Fluorescence intensity profiles of LFY-Tdtomato in inflorescences of Col-0 and rpt2a-2 mutants at 8 DAF.
Supplemental Data Set 5. Supplemental Figure S4C data: Fluorescence intensity profiles of ProCLV3:GFP in inflorescences of Col-0 and rpt2a-2 mutants.
Supplemental Data Set 6. Figure 3I data: Total number of secondary inflorescences and flowers initiated from the primary inflorescence in Col-0, rpt2a-2, tfl1-11, rpt2a-2 tfl1-11 mutants, ProUFO:TFL1 rpt2a-2 #11, ProUFO:TFL1 rpt2a-2 #13, ProUFO:TFL1 rpt2a-2 #19, ProUFO:amiR-TFL1 #3, ProUFO:amiR-TFL1 #4, and ProUFO:amiR-TFL1 #7 lines.
Supplemental Data Set 7. Supplemental Figure S6E data: Total number of secondary inflorescences and flowers initiated from the primary inflorescence in Col-0, rpt2a-2 mutants, Pro35S:TFL1 rpt2a-2 #10, Pro35S:TFL1 rpt2a-2 #17, and Pro35S:TFL1 rpt2a-2 #27 lines.
Supplemental Data Set 8. Supplemental Figure S7B data: Quantitative analysis of recombinant His-LFY protein degradation rate in the Col-0 and rpt2a-2 inflorescences.
Supplemental Data Set 9. All proteins detected by TMT-label proteomics.
Supplemental Data Set 10. All differentially abundant proteins detected by TMT-label proteomics.
Supplemental Data Set 11. All GO terms are shown in the TMT-label proteomics.
Supplemental Data Set 12. Supplemental Figure S9A data: Number of secondary inflorescences in the primary inflorescence in Col-0 under long-day (LD) condition, Col-0 under short-day (SD) condition, and met1-1 mutants under LD condition.
Supplemental Data Set 13. Figure 5C data: Fluorescence intensity profiles of MET1-YFP in inflorescences of Col-0 and rpt2a-2 mutants.
Supplemental Data Set 14. Figure 5L data: Total number of secondary inflorescences and flowers initiated from the primary inflorescence in Col-0, rpt2a-2, met1-1, rpt2a-2 met1-1 mutants, ProUFO:MET1 #2, ProUFO:MET1 #8, and ProUFO:MET1 #10 plants.
Supplemental Data Set 15. Figure 6B data: Quantitative analysis of recombinant His-MET1 protein degradation rate in the Col-0 and rpt2a-2 inflorescences.
Supplemental Data Set 16. Figure 7B data: The cytosine methylation levels of the TFL1 loci are assessed using bisulfite sequencing.
Supplemental Data Set 17. Supplemental Fig. S12B data: The cytosine methylation levels of the TFL1 loci are assessed using bisulfite sequencing.
Supplemental Data Set 18. Figure 7G data: Fluorescence intensity profiles of TFL1-GFP in the IMs of Col-0, rpt2a-2, met1-1, and rpt2a-2 met1-1 plants.
Supplemental Data Set 19. Figure 7K data: Relative ratio of firefly luciferase (LUC) to Renilla LUC (REN) activity in Arabidopsis protoplasts transformed with ProTFL1(−1373∼−621):LUC plasmid with different methylation levels (0%, 40%, or 80%).
Supplemental Data Set 20. Supplemental Fig. S15E data: Total number of secondary inflorescences and flowers initiated from the primary inflorescence in Col-0, tfl1-11, met1-1, and tfl1-11 met1-1 mutants.
Supplemental Data Set 21. Mutants were used in the study.
Supplemental Data Set 22. Primers used in the study.
Funding
This research was funded by the National Natural Science Foundation of China (32070199, 31872669, 31730008) and the Natural Science Foundation of Shandong Province (ZR2022JQ12).
Data availability
All source data are provided with this article.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Azpeitia E, Tichtinsky G, Le Masson M, Serrano-Mislata A, Lucas J, Gregis V, Gimenez C, Prunet N, Farcot E, Kater MM, et al. Cauliflower fractal forms arise from perturbations of floral gene networks. Science. 2021:373(6551):192–197. 10.1126/science.abg5999 [DOI] [PubMed] [Google Scholar]
- Baulcombe DC, Saunders GR, Bevan MW, Mayo MA, Harrison BD. Expression of biologically active viral satellite RNA from the nuclear genome of transformed plants. Nature. 1986:321(6068):446–449. 10.1038/321446a0 [DOI] [Google Scholar]
- Benlloch R, Berbel A, Ali L, Gohari G, Millán T, Madueño F. Genetic control of inflorescence architecture in legumes. Front Plant Sci. 2015:6:543. 10.3389/fpls.2015.00543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005:433(7021):39–44. 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997:275(5296):80–83. 10.1126/science.275.5296.80 [DOI] [PubMed] [Google Scholar]
- Castel R, Kusters E, Koes R. Inflorescence development in petunia: through the maze of botanical terminology. J Exp Bot. 2010:61(9):2235–2246. 10.1093/jxb/erq061 [DOI] [PubMed] [Google Scholar]
- Chen ZQ, Zhao JH, Chen Q, Zhang ZH, Li J, Guo ZX, Xie Q, Ding SW, Guo HS. DNA Geminivirus infection induces an imprinted E3 ligase gene to epigenetically activate viral gene transcription. Plant Cell. 2020:32(10):3256–3272. 10.1105/tpc.20.00249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claßen-Bockhoff R, Bull-Hereñu K. Towards an ontogenetic understanding of inflorescence diversity. Ann Bot. 2013:112(8):1523–1542. 10.1093/aob/mct009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998:16(6):735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Collins GA, Goldberg AL. The logic of the 26S proteasome. Cell. 2017:169(5):792–806. 10.1016/j.cell.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datia RSS, Hammerlindl JK, Panchuk B, Pelcher LE, Keller W. Modified binary plant transformation vectors with the wild-type gene encoding NPTII. Gene. 1992:122(2):383–384. 10.1016/0378-1119(92)90232-E [DOI] [PubMed] [Google Scholar]
- Devesa-Guerra I, Morales-Ruiz T, Pérez-Roldán J, Parrilla-Doblas JT, Dorado-León M, García-Ortiz MV, Ariza RR, Roldán-Arjona T. DNA Methylation editing by CRISPR-guided excision of 5-methylcytosine. J Mol Biol. 2020:432(7):2204–2216. 10.1016/j.jmb.2020.02.007 [DOI] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S. OST1 Kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell. 2015:32(3):278–289. 10.1016/j.devcel.2014.12.023 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006:45(4):616–629. 10.1111/j.1365-313X.2005.02617.x [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci U S A. 1996:93(16):8449–8454. 10.1073/pnas.93.16.8449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Heisler MG, Reddy GV, Ohno C, Das P, Meyerowitz EM. Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development. 2007:134(19):3539–3548. 10.1242/dev.010298 [DOI] [PubMed] [Google Scholar]
- Goslin K, Zheng B, Serrano-Mislata A, Rae L, Ryan PT, Kwasniewska K, Thomson B, O’Maoileidigh DS, Madueno F, Wellmer F, et al. Transcription factor interplay between LEAFY and APETALA1/CAULIFLOWER during floral initiation. Plant Physiol. 2017:174(2):1097–1109. 10.1104/pp.17.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S, Goto K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell. 2011:23(9):3172–3184. 10.1105/tpc.111.088641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005:1(1):13. 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007:51(3):526–536. 10.1111/j.1365-313X.2007.03152.x [DOI] [PubMed] [Google Scholar]
- Hou CJ, Yang CH. Functional analysis of FT and TFL1 orthologs from orchid (oncidium gower ramsey) that regulate the vegetative to reproductive transition. Plant Cell Physiol. 2009:50(8):1544–1557. 10.1093/pcp/pcp099 [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003:163(3):1109–1122. 10.1093/genetics/163.3.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Jeon J, Lee S, Lee JH, Gao L, Lee BH, Park JM, Kim YJ, Kwak JM. Proteasome subunit RPT2a promotes PTGS through repressing RNA quality control in Arabidopsis. Nat Plants. 2019:5(12):1273–1282. 10.1038/s41477-019-0546-1 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Toh-e A, Smalle JA. 26S Proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 2008:53(1):102–114. 10.1111/j.1365-313X.2007.03322.x [DOI] [PubMed] [Google Scholar]
- Kurepa J, Wang S, Li Y, Zaitlin D, Pierce AJ, Smalle JA. Loss of 26S proteasome function leads to increased cell size and decreased cell number in Arabidopsis shoot organs. Plant Physiol. 2009:150(1):178–189. 10.1104/pp.109.135970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Minami A, Marshall RS, Book AJ, Farmer LM, Walker JM, Vierstra RD. The RPT2 subunit of the 26S proteasome directs complex assembly, histone dynamics, and gametophyte and sporophyte development in Arabidopsis. Plant Cell. 2011:23(12):4298–4317. 10.1105/tpc.111.089482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wu W, Zhao Y, Zheng B. A reciprocal inhibition between ARID1 and MET1 in male and female gametes in Arabidopsis. J Integr Plant Biol. 2017:59(9):657–668. 10.1111/jipb.12573 [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF. Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell. 1999:11(6):1007–1018. 10.1105/tpc.11.6.1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Cohen O, Alvarez JP, Abu-Abied M, Pekker I, Paran I, Eshed Y, Zamir D. The making of a compound inflorescence in tomato and related nightshades. PLoS Biol. 2008:6(11):e288. 10.1371/journal.pbio.0060288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008:133(3):523–536. 10.1016/j.cell.2008.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Teo ZWN, Bi Y, Song S, Xi W, Yang X, Yin Z, Yu H. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev Cell. 2013:24(6):612–622. 10.1016/j.devcel.2013.02.013 [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang H, Dong YX, Hao YJ, Zhang XS. DNA METHYLTRANSFERASE1-mediated shoot regeneration is regulated by cytokinin-induced cell cycle in Arabidopsis. New Phytol. 2018:217(1):219–232. 10.1111/nph.14814 [DOI] [PubMed] [Google Scholar]
- Liu Q, Ding Y, Shi Y, Ma L, Wang Y, Song C, Wilkins KA, Davies JM, Knight H, Knight MR, et al. The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. Embo J. 2021:40(2):e104559. 10.15252/embj.2020104559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D. The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science. 2005:308(5719):260–263. 10.1126/science.1108229 [DOI] [PubMed] [Google Scholar]
- McKim S, Hay A. Patterning and evolution of floral structures—marking time. Curr Opin Genet Dev. 2010:20(4):448–453. 10.1016/j.gde.2010.04.007 [DOI] [PubMed] [Google Scholar]
- Molinero-Rosales N, Jamilena M, Zurita S, Gómez P, Capel J, Lozano R. FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J. 1999:20(6):685–693. 10.1046/j.1365-313X.1999.00641.x [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. A genetic framework for floral patterning. Nature. 1998:395(6702):561–566. 10.1038/26903 [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz RV, García-Ponce B, Marsch-Martínez N, Ugartechea-Chirino Y, Villajuana-Bonequi M, de Folter S, Azpeitia E, Dávila-Velderrain J, Cruz-Sánchez D, Garay-Arroyo A, et al. XAANTAL2 (AGL14) is an important component of the complex gene regulatory network that underlies Arabidopsis shoot apical meristem transitions. Mol Plant. 2015:8(5):796–813. 10.1016/j.molp.2015.01.017 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E. The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development. 1998:125(11):1979–1989. 10.1242/dev.125.11.1979 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007:316(5830):1452–1456. 10.1126/science.1140429 [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Bradley DJ, Coen ES. Separation of shoot and floral identity in Arabidopsis. Development. 1999:126(6):1109–1120. 10.1242/dev.126.6.1109 [DOI] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004:131(17):4225–4237. 10.1242/dev.01261 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Frenz M, Mandel T, Kuhlemeier C. Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development. 2003:130(17):4073–4083. 10.1242/dev.00596 [DOI] [PubMed] [Google Scholar]
- Rickett HW. Materials for a dictionary of botanical terms-I. Bull Torrey Bot Club. 1954:81(1):1–15. 10.2307/2482165 [DOI] [Google Scholar]
- Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996:273(5275):654–657. 10.1126/science.273.5275.654 [DOI] [PubMed] [Google Scholar]
- Sako K, Maki Y, Kanai T, Kato E, Maekawa S, Yasuda S, Sato T, Watahiki MK, Yamaguchi J. Arabidopsis RPT2a, 19S proteasome subunit, regulates gene silencing via DNA methylation. PLoS One. 2012:7(5):e37086. 10.1371/journal.pone.0037086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Klenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Crosby WL. The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 1999:20(4):433–445. 10.1046/j.1365-313x.1999.00617.x [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006:18(5):1121–1133. 10.1105/tpc.105.039834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Mislata A, Fernández-Nohales P, Doménech MJ, Hanzawa Y, Bradley D, Madueño F. Separate elements of the TERMINAL FLOWER 1 cis-regulatory region integrate pathways to control flowering time and shoot meristem identity. Development. 2016:143(18):3315–3327. 10.1242/dev.135269 [DOI] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR. Genetic interactions that regulate inflorescence development in Arabidopsis. Plant Cell. 1993:5(6):639–655. 10.2307/3869807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda Y, Sako K, Maki Y, Yamazaki N, Yamamoto H, Ikeda A, Yamaguchi J. Regulation of leaf organ size by the Arabidopsis RPT2a 19S proteasome subunit. Plant J. 2009:60(1):68–78. 10.1111/j.1365-313X.2009.03932.x [DOI] [PubMed] [Google Scholar]
- Souer E, Krol A, Kloos D, Spelt C, Bliek M, Mol J, Koes R. Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development. 1998:125(4):733–742. 10.1242/dev.125.4.733 [DOI] [PubMed] [Google Scholar]
- Souer E, Rebocho AB, Bliek M, Kusters E, de Bruin RAM, Koes R. Patterning of inflorescences and flowers by the F-box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. Plant Cell. 2008:20(8):2033–2048. 10.1105/tpc.108.060871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YH, Zhou C, Li YJ, Yu Y, Tang LP, Zhang WJ, Yao WJ, Huang R, Laux T, Zhang XS. Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc Natl Acad Sci U S A. 2020:117(36):22561–22571. 10.1073/pnas.2015248117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Matsui K, Ishiguro S, Kato T, Tabata S, Kobayashi M, Seki M, Shinozaki K, Okada K. Arabidopsis RPT2a encoding the 26S proteasome subunit is required for various aspects of root meristem maintenance, and regulates gametogenesis redundantly with its homolog, RPT2b. Plant Cell Physiol. 2011:52(9):1628–1640. 10.1093/pcp/pcr093 [DOI] [PubMed] [Google Scholar]
- Ueda M, Matsui K, Ishiguro S, Sano R, Wada T, Paponov I, Palme K, Okada K. The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development. 2004:131(9):2101–2111. 10.1242/dev.01096 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Besnard F, Godin C. What shoots can teach about theories of plant form. Nat Plants. 2021:7(6):716–724. 10.1038/s41477-021-00930-0 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Pinto YM, Baulcombe DC. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci U S A. 1999:96(24):14147–14152. 10.1073/pnas.96.24.14147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Sablowski RW, Meyerowitz EM. Transcriptional activation of APETALA1 by LEAFY. Science. 1999:285(5427):582–584. 10.1126/science.285.5427.582 [DOI] [PubMed] [Google Scholar]
- Wang B, Smith SM, Li J. Genetic regulation of shoot architecture. Annu Rev Plant Biol. 2018:69(1):437–468. 10.1146/annurev-arplant-042817-040422 [DOI] [PubMed] [Google Scholar]
- Wang Z, Yang R, Devisetty UK, Maloof JN, Zuo Y, Li J, Shen Y, Zhao J, Bao M, Ning G. The divergence of flowering time modulated by FT/TFL1 is independent to their interaction and binding activities. Front Plant Sci. 2017:8:697. 10.3389/fpls.2017.00697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992:69(5):843–859. 10.1016/0092-8674(92)90295-N [DOI] [PubMed] [Google Scholar]
- Winter CM, Austin RS, Blanvillain-Baufumé S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker JE, et al. LEAFY Target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev Cell. 2011:20(4):430–443. 10.1016/j.devcel.2011.03.019 [DOI] [PubMed] [Google Scholar]
- Yao C, Wu Y, Nie H, Tang D. RPN1a, a 26S proteasome subunit, is required for innate immunity in Arabidopsis. Plant J. 2012:71(6):1015–1028. 10.1111/j.1365-313X.2012.05048.x [DOI] [PubMed] [Google Scholar]
- Zhu Y, Wagner D. Plant inflorescence architecture: the formation, activity, and fate of axillary meristems. Cold Spring Harb Perspect Biol. 2020:12(1):a034652. 10.1101/cshperspect.a034652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source data are provided with this article.