Abstract
Mythology is replete with good and evil shapeshifters, who, by definition, display great adaptability and assume many different forms—with several even turning themselves into trees. Cell walls certainly fit this definition as they can undergo subtle or dramatic changes in structure, assume many shapes, and perform many functions. In this review, we cover the evolution of knowledge of the structures, biosynthesis, and functions of the 5 major cell wall polymer types that range from deceptively simple to fiendishly complex. Along the way, we recognize some of the colorful historical figures who shaped cell wall research over the past 100 years. The shapeshifter analogy emerges more clearly as we examine the evolving proposals for how cell walls are constructed to allow growth while remaining strong, the complex signaling involved in maintaining cell wall integrity and defense against disease, and the ways cell walls adapt as they progress from birth, through growth to maturation, and in the end, often function long after cell death. We predict the next century of progress will include deciphering cell type–specific wall polymers; regulation at all levels of polymer production, crosslinks, and architecture; and how walls respond to developmental and environmental signals to drive plant success in diverse environments.
This review provides a historical context for the processes by which plant cell wall structures are created, assemble, sense, and respond to signals and change during progression from birth to death.
Introduction
In 1665, Robert Hooke peered through his primitive microscope at a slice of cork and described little boxes he called “cellula”—rooms that monks inhabited. These “cellula” were dead cells, and all that remained visible to him were their cell walls. For centuries after, much of what we knew about cell walls was derived from what was visible with our own eyes, either viewed directly or through the ever-improving technique of light microscopy. Form helps predict function, and we now know that cell walls play a key role in determining cell, tissue, and organ shapes. Botanists, amazed by the diverse types of cells found in plants, watched the ways in which cells with thin “primary” cell walls (PCWs) first begin to form a new wall at the time of cell division and how cell and overall plant growth is tied to the expansion of such thin walls. Cell diversity is also intimately linked to the way cells mature, building “secondary” cell walls (SCWs) that thicken to form structures that provide strength to stems to withstand loss of turgor, protect against water loss and diseases, and function in transport of water and minerals from roots to leaves. The overall shape of a plant and its organs, in particular flowers, was found to be one of best predicters of ancestry, and taxonomy became one of the most popular (and controversial) fields for botanists extending well into the early 20th century. Smith (1962) provides an absolutely delightful and colorful history of botany including the hilarious struggles to develop the field of botanical taxonomy.
In this centennial celebration year recognizing the founding of ASPB, we have aimed to provide a historical context to the process of discovery and to emphasize milestones that made a difference in moving the field forward. The first part of the review presents the progression of learning about the structure, biosynthesis, and functions of the 5 major types of wall polymers: cellulose, hemicelluloses, pectins, wall-associated proteins, and lignin. The second part aims to integrate this knowledge of individual polymers and delve deeper into some of the fascinating challenges faced by plant cell walls. We examine models that have attempted to explain the still-challenging problem of how the polymers found in PCWs can be assembled into a structure strong enough to resist high turgor pressure while allowing expansion to permit growth. Recognizing that walls are not passive structures but constantly communicating and adapting to serve the needs of the cells they protect, we then discuss the rapidly expanding field of cell wall signaling. We end with an examination of the life of the cell wall from birth to survival even beyond the death of their parent cells.
Because each of us has focused most of their career on one of the major wall polymers, we consider this a rare opportunity to integrate our collective knowledge gathered over many years. Aware that new ideas do not always pan out, we also take this opportunity to offer a few new ideas of our own and end with our assessment of major challenges for the future of cell wall research.
Not all cells that build walls do so the same way, so we must emphasize the importance of using a variety of model systems (Fig. 1), Arabidopsis thaliana, certainly, but our discussion just might include some of the botanists’ favorite very odd-shaped cells.
Figure 1.
Some model systems that are especially useful for the study of plant cell walls. A) Arabidopsis thaliana; its small size, short generation time, small genome, and ease of mutant selection make this a favorite for genetic studies (Liepman et al. 2010). B) Fiber cells that have SCWs that are cellulose-rich and nearly free—or with much reduced levels—of hemicelluloses and lignin include cotton fibers (actually epidermal hairs), bast fibers (especially from flax and ramie), and fibers of tension wood and are favorites for studying cellulose. Ramie (Boehemeria nivea) cellulose is favored by x-ray crystallographers for its clear diffraction patterns; cotton fibers are single cells that show synchronous development within a single boll, and their mature SCWs have >90% cellulose (Haigler et al. 2012). C) Suspension-cultured cells from the C4 plant sugarcane (Saccharum hybrid spp; example shown taken from Simões et al., 2020) or C3 plants such as Zinnia elegans (reviewed by Demura, 2013) can be induced to differentiate into tracheary elements, making them good models for study of xylem biogenesis. D) Hybrids of poplar (Populus) have become models of choice for trees due to ease of propagation, molecular breeding, transformation, and large genome resources (Tuskan et al. 2004). E) Onion epidermal cells (shown here are surface cellulose microfibrils [MFs]) can be easily isolated as a living, hormone-responsive monolayer, especially useful for studying PCW expansion (Suslov et al. 2009; Zhang et al. 2021c). F) A popular model for grass walls of very different Type II PCW structure is the maize coleoptile (Carpita 1984). G) Pollen tubes of Lily have been used to elucidate the oscillating nature of tip growth (McKenna et al. 2009) and discovery of key signaling cascades (Li et al. 2016a). H) Charophycean algae have played a key role in studying the role of pectins in diffusive cell wall elongation (Proseus and Boyer 2012b). I) Some mucilage-secreting cells (MSCs) such as those from Arabidopsis, are rich in pectins, whereas others are rich in only one kind of hemicellulose such as xyloglucan, xylan, or glucomannan, making MSCs valuable for studies on biosynthesis and wall polymer interactions (Arsovski et al. 2010). All figures are licensed through the Creative Commons and used with permision.
The 5 major polymers of the plant cell wall
Cellulose
Common claims that cellulose “is the most abundant organic compound on earth” and that taking density differences into consideration, it is “stronger than steel” are actually true (see McNamara et al. 2015). Yet cellulose, the cell wall polymer with the simplest basic structure—one type of sugar joined by 1 type of linkage—is anything but simple given the variety of structures that can be assembled from individual β-1,4-glucan chains. We will see that this feature allows it to interact, not only with itself but with other noncellulosic polysaccharides, and its presence is so important for wall structure that a decrease in cellulose level in the wall sends out distress signals that alert the cell to quickly do whatever possible to correct this situation. Beyond its role in the plant, all these properties make cellulose a key component of timber, textiles, paper, and a major feedstock for chemicals; and the fact that it is difficult to digest influences agriculture and global nutrition.
Structure of cellulose
The early studies
Early botanists often equated the wall with the term cellulose because of its abundance in many cell types. Anselme Payen (1838), “the father of cellulose chemistry,” isolated the major sugar of cell walls and was surprised to find that its elemental composition was the same as that of starch. Payen recognized that cellulose could be broken down to glucose but had no idea of its structure nor how the monomers were aggregated to create a polymer so very different from starch. Decades later, the sequential efforts of some towering figures in the emerging fields of sugar and polymer chemistry led to the conclusion that cellulose is a polymer of thousands of glucose residues linked in β-1–4 configuration (Fischer 1902; Haworth et al. 1927, 1931; Staudinger 1932). The concept of a polymer was novel to chemistry at the time, as the length of 1 type of polymer could vary considerably even as the chemical formula stayed the same; but by 1936, the idea became generally accepted (Kanaya and Kaji 2016). Estimates of chain lengths ranged from 100s to many thousands of residues depending on the source and how the cellulose was isolated (Delmer 1983).
The Swiss scientist Albert Frey-Wyssling defined a new field he termed “submicroscopic morphology” that led to several insightful predictions concerning the highly oriented and crystalline properties of cellulose (Frey-Wyssling and Ambronn 1927; Frey-Wyssling 1939). Using the newly introduced electron microscope, Frey-Wyssling's predictions were confirmed, and he and Fritz Muhlethaler provided our best early images of cellulose fibrils in native cell walls. One major conclusion was that fibrils of varying widths consisted of smaller fibrils of about 35 Å width, corresponding roughly to a 6×6 array of chains (Mühlethaler 1960; Frey-Wyssling and Mühlethaler 1963). This so-called “elementary fibril” (or microfibril [MF]) is now considered a fundamental unit of cellulose, although the predicted number of chains has been reduced over time. The Federal Institute of Technology Zurich under Frey-Wyssling also pioneered the technique of freeze-fracture (Moor et al. 1961) that led much later to the first images of cellulose synthase complexes (CSCs).
It took more than 60 years to answer the key question of whether the chains of native cellulose (cellulose I) were parallel (reducing ends all in the same direction) or anti-parallel (Kroon-Batenburg and Kroon 1997; French 2000; Zugenmaier 2021). Better computer modeling improved analyses of diffraction patterns, and, working independently with the same sample of the alga Valonia native cellulose I, Gardner and Blackwell (1974) and Sarko and Muggli (1974)—and later Woodcock and Sarko (1980) with fibers of ramie (Boehmeria nivea)—all concluded that the chains were indeed parallel. This was confirmed by other methods (Hieta et al. 1984; Chanzy and Henrissat 1983), and the parallel chain arrangement for cellulose I is now the accepted model. Cellulose II, produced by alkali treatment of native cellulose, is a more stable form, and the early findings of Meyer and Misch (1937) that the chains are in anti-parallel arrangement were confirmed later by Kolpak and Blackwell (1976) and Buleon and Chanzy (1978).
New technologies provide new insights
It took crystallographers so long to agree on the crystal structure of native cellulose largely because native cellulose can exist in many crystalline forms that show only subtle differences in x-ray diffraction. There has since been a dramatic rise in the number of new technologies that can be applied to study cellulose structure (Jarvis 2013, 2018). According to Jarvis (2018), “Broadly, NMR spectroscopy is most informative about chain conformation; crystallography, about chain packing; Fourier transform infrared spectroscopy, about hydrogen bonding; atomic force microscopy (AFM), about microfibril dimensions; small-angle scattering, about the aggregation of MFs.” Using cross-polarization magic angle NMR, Atalla and Vanderhart (1984) demonstrated that native cellulose can exist in at least 2 crystalline forms: Iα, found in native bacterial and algal cellulose, and Iβ, found in tunicates and higher plants. Different conformations at C-6 in the crystalline core (Fig. 2A) versus surface MFs (Fig. 2B) can create altered patterns for hydrogen bonding that alter the potential for surface chains to interact with other MFs or other polymers.
Figure 2.
Hydrogen bonding schemes for cellulose. A) A 2-chain segment of “crystalline” cellulose with all C-6 in the tg conformation, permitting a line of intramolecular hydrogen bonds (shaded arrows) that run along each side of each cellulose chain (lightly shaded horizontal bands). Intermolecular hydrogen bonds are shown as open arrows. B) Two-chain segment of “surface” cellulose with the edge of the upper chain having C-6 in the gt conformation, so that the line of intramolecular hydrogen bonding is interrupted and there is an increased number of transversely oriented, intermolecular hydrogen bond sites (top row of open arrows) that allows for interaction with water, other MFs or other polymers. Modified from Jarvis (2018) by permission of the author.
The widely cited work of Nishiyama et al. (2002) showed 2 parallel chains in pure Iβ having slightly different conformations, and neutron scattering supported 2 different types of intrachain hydrogen bonding between 1 glucose residue with each of its neighbors that stabilizes a co-planar orientation of glucose. Strikingly, no hint of inter-sheet O—H…O bonds was found, indicating that sheets of cellulose must be held together primarily by van der Waals forces along with some weak C—H…O bonds (see also Notley et al. 2004; McNamara et al. 2015). Thus, the glucan chains have a hydrophilic and a hydrophobic face that allow for very strong interchain interaction, although each individual interaction is weak.
Surprisingly, the cellulose of maize (Zea mays) PCWs consists of up to 7 different allomorphs, only 1 of which is almost identical to either Iα or Iβ (Wang et al. 2016b). Some allomorphs show differences on the surface of the MF and were suggested to be possible targets for interaction with other polymers such as pectins. Others show interior chains associated with surface chains, whereas another is embedded in the core of the MF. Yet another is predicted to be on the surface but poorly hydrated and was suggested as a possible target for expansins (see below). How the surfaces of MFs interact with water, with themselves to create larger bundles or with other wall polymers, is critical to overall wall structure and mode of expansion (see below) and is still an ongoing field of investigation (Cosgrove 2022; Jarvis 2023).
Biosynthesis of cellulose
It is hard to document all the progress that is being made in this field, so it is fortunate that many details have been well-reviewed by others (Wallace and Somerville 2014; McNamara et al. 2015; Turner and Kumar 2018; Wilson et al. 2021). In particular, we leave detailed discussion of CESA (cellulose synthase) trafficking, some aspects of regulation, and the evolution of the genes/proteins involved to others (Haigler and Roberts 2019; Lampugnani et al. 2019; Polko and Kieber 2019). Hopefully our historical perspective provides examples that might inform future work.
The early years
Luis Leloir, an Argentine biochemist, physician, and pioneer of carbohydrate biosynthesis, made several truly milestone discoveries. Key was the discovery in the 1950s of nucleoside diphosphate sugars and recognition that the free energy of hydrolysis releasing the sugar was sufficient for them to serve as donors of sugars for polysaccharide synthesis. Discovery of UDP-Glc in 1950 was only part of many that led to the Nobel Prize (see Leloir 1983). UDP-Glc is a key substrate in polysaccharide biosynthesis, either directly for cellulose, callose (β-1,3-glucan), and hemicellulosic glucans or indirectly as precursor to other NDP-sugars that are themselves substrates for synthesis of noncellulosic polysaccharides (Bar-Peled and O’Neill 2011).
The obligate aerobic bacterium Acetobacter xylinum, lacking flagellae, secretes cellulose in order to float and gain oxygen in liquid environments (Hestrin 1962). Glaser (1958) showed that it uses UDP-Glc as substrate for cellulose synthesis in vitro, albeit at quite low rates. Moshe Benziman's group later achieved high rates in vitro through the important discovery of cyclic-di-GMP as a potent activator (Aloni et al. 1982; Ross et al. 1987). One gene (BcsA) in a 4-gene operon encodes the catalytic subunit (Saxena et al. 1990); the other genes (Saxena et al. 1994; Römling and Galperin 2015) encode proteins that assist in binding of c-di-GMP to BcsA (BcsB), create a pore for MF extrusion through the outer membrane (BcsC), or have endoglucanase activity (BcsD) that, along with other proteins (Abidi et al. 2022), are important for determining crystallinity of the product.
Another prominent biochemist, Zev Hassid, believed in 1957 that it would only be a short time before his laboratory showed how cellulose was synthesized in plants. He set 2 of his students to work using mung beans expecting to find that UDP-Glc was substrate. The students returned with news that the product was not cellulose but callose (β-1-3-glucan). Hassid could not believe it, and the poor students had to repeat their results several more times before it was published (Feingold et al. 1958; the students, Liz Neufeld and David Feingold, became very well-known polysaccharide biochemists in spite of abandoning cellulose synthesis as a target!). This frustration was repeated by many other laboratories, finding that the bulk of the product in vitro was all or mostly callose (Robinson and Quader 1981; Delmer 1983). Callose synthesis in vitro generally requires 2 activators; micromolar levels of Ca2+ and any 1 of a variety of β-glucosides (Hayashi et al. 1987), and assays for cellulose synthase often inadvertently contained both because cellobiose was often supplied and micromolar Ca2+ is present in plant extracts if not specifically chelated. Vincent Bulone stands out as one who often reported successful in vitro synthesis of cellulose and deserves much credit for providing protocols for assays of glycosyltransferases (GTs) in plants (Brown et al. 2012). Callose synthases are very stable enzymes in contrast to the quite labile plant CSCs. More recently, Oehme et al. (2019) and Purushotham et al. (2022) showed that only minor changes in amino acid sequences and conformation of glycosyltransferase-2 (GT2) enzymes as classified by the Carbohydrate-Active Enzyme Database (CAZy) (Cantarel et al. 2009; Drula et al. 2022) can lead to β-1,3, as opposed to β-1,4-linkages. One wonders why plants, having evolved CSLF and CSLH enzymes (also GT2 proteins, see Hemicellulose section) that can make glucan backbones with both linkages, have never evolved a callose synthase analogous to the GT2 bacterial β-1,3-glucan (curdlan) synthase (Oehme et al. 2019). Instead, plant callose synthases evolved from yeast/fungi and are quite different in structure (Schneider et al. 2016).
Early contributions from microscopy
Preston (1964)—another force in the world of cell wall research—speculated that a plant CSC is a complex of catalytic subunits in which the number of subunits dictates the number of chains produced in a single MF. Malcolm Brown—a genius at capturing images of the process of cellulose synthesis—showed pictures of A. xylinum extruding cellulose from a linear array of pores (Brown et al. 1976). Other freeze-fracture images supported Preston's vision, showing huge multi-subunit “terminal complexes” (TCs) at the ends of very large growing MFs in algae like Oocystis and Valonia (Montezinos and Brown 1976; Itoh and Brown 1984; Brown 1996). Surprisingly, some of these TCs were aligned next to each other and were clearly moving in opposite directions, creating an arrangement that could lead to more stable antiparallel interactions between the MFs; this bi-directional mobility was also later seen for plant CSCs (Paredez et al. 2006). Arabidopsis mutants defective in phosphorylation of the complex lose this property, which affects MF bundling and stem phenotype (Chen et al. 2016). This raises the question of whether anti-parallel regions of MF bundling might be the “hot-spots” for expansin function proposed by Cosgrove (2022) (see later). Jarvis (2023) comments that it is not known to what extent MFs bundle in either the parallel or more stable antiparallel orientations, but the issue would certainly seem to deserve further study (Makarem et al. 2020). For plants, a more modest complex was observed in the plasma membrane of maize (Mueller and Brown 1980) and tracheary elements of cress (Herth 1985) showing 6-fold symmetry and was termed a rosette (probably by Andrew Staehelin according to Nick Carpita). These generally appear as solitary structures in plants, and Giddings et al. (1980) demonstrated remarkable arrays of up to 175 rosettes in the plasma membrane of the alga Micrasterias.
CesA genes in plants
The first plant CESA gene was not identified until 1996 (Pear et al. 1996). In cotton (Gossypium hirsutum) fibers, the rate of cellulose synthesis increases more than 100-fold during the transition from PCW to SCW (Meinert and Delmer 1977), and random cDNA sequencing of a transition stage cDNA library found 2 very similar cDNAs containing scattered sequences characteristic of processive GT2 enzymes and other regions showing strong sequence similarity to the transmembrane helices (TMHs) of the BcsA protein (Pear et al. 1996). But the cotton genes also possessed an N-terminal domain (NTD) encoding a ring-finger domain, a highly plant-conserved region (PCR) and another region of variable sequence (HVR, later called CSR for class-specific region). The latter 2 insertions split the conserved sequences required for catalytic activity, thus explaining why the genes had not been discovered earlier. The cotton genes were strongly induced at the onset of SCW synthesis (Pear et al. 1996), but no genetic evidence was presented to confirm that they encoded cellulose synthases.
Chris Somerville and Herman Hofte had the vision to identify Arabidopsis (Fig. 1) as a model plant in which specific mutations in cellulose synthesis could be easily selected. Williamson's laboratory in Australia isolated a temperature-sensitive mutant of Arabidopsis termed rsw1 impaired in the synthesis of PCW cellulose (Arioli et al. 1998). It quickly became evident that the RSW1 locus had a very close relationship to that of the cotton fiber genes and encoded what we now refer to as AtCESA1 that functions in synthesis of PCW cellulose. And so…off to the races! Richmond and Somerville (2000) showed that Arabidopsis has 10 CESA genes. Also identified were a number of related cellulose-synthase-like (CSL) families, some of which function in hemicellulose synthesis (see below). The CSLD family members are closest to the CESAs (Doblin et al. 2002), and reports increasingly suggest that at least some CSLD proteins serve an alternate function to the CESAs for synthesis of cellulose (see below). CSLD3 was recently verified to possess β-1,4-glucan synthase activity and is most similar in structure to CESA6 (Yang et al. 2020b).
A complete CSC in vivo involves more than just CESA proteins (Lampugnani et al. 2019). An endoglucanase, KORRIGAN, analogous to BcsD, may play a role in cleaving disordered chains during synthesis (Nicol et al. 1998; Vain et al. 2014), although its role remains enigmatic because a mutation in the predicted catalytic site still complemented the kor mutant phenotype (Lei et al. 2014). Also unclear is the exact function of COBRA, a glycosylphosphatidylinositol (GPI)-anchored protein that is apparently associated with the CSC and affects crystallinity (Schindelman et al. 2001; Roudier et al. 2005). A better understanding of the functions of KOR and COBRA is needed. Furthermore, many scientists seem to have forgotten that freeze-fracture studies of rosettes showed protein globules on the opposite face of the PM that matched in position the central core of the rosettes on the opposite membrane face (Giddings et al. 1980; Mueller and Brown 1980), suggesting the central globule as 1 possible location for accessory proteins. CESAs may also be regulated through phosphorylation and S-acylation that may affect activity, rosette structure, and/or recycling (Kumar et al. 2016; Polko and Kieber 2019).
Comparisons of bacterial and plant CESA structures
Study of CESA mutants of Arabidopsis has led to 2 surprising but important conclusions: the rosette responsible for synthesis of the fundamental MF is comprised of 2 classes defined largely, but not exclusively, by the homology of their CSRs; and each class contains 3 necessary but non-identical subunits, usually (but not always) in 1:1:1 ratio. Class I proteins (CESA1, CESA3, and any one of CESA 2, 5, 6, or 9) are used primarily for PCW synthesis, whereas Class 2 (CESA 4, 7, and 8) are necessary for SCW synthesis (Turner and Kumar 2018). For clarity, the names of all plant homologs of the Arabidopsis CESAs in other plants have been changed in this review to match those used for Arabidopsis.
A significant milestone came when Jochen Zimmer's group crystallized a catalytically active bacterial BcsA-BcsB protein complex and determined its structure to 3.25-A resolution (Morgan et al. 2013; McNamara et al. 2015); a cello-oligosaccharide remained in the structure during crystallization. In this proposed structure, BcsA contains 4 TMHs in the amino-terminal region and 4 in the carboxy-terminal region, separated by a cytoplasmic intracellular loop that forms the catalytic domain that is protected by a cyclic-di-GMP–regulated gating loop that controls access to the active site (Morgan et al. 2014). The channel formed by TMHs 3–8 accommodates the translocating glucan. The overall structure resembles other membrane-integrated, processive GT2s such as hyaluronic acid synthase and chitin synthase (Bi et al. 2015).
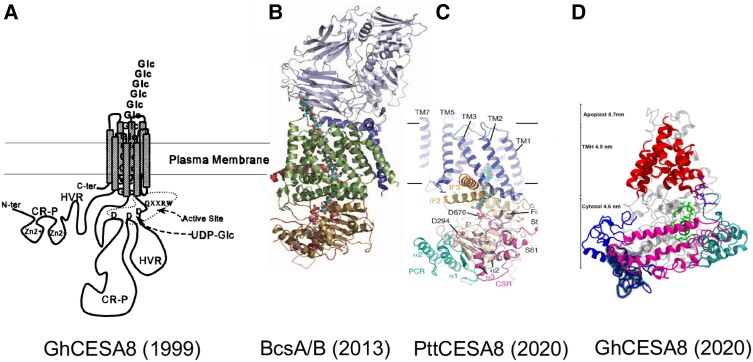
Significant advances have been made in producing catalytically active recombinant CESAs; PpCESA5 from moss (Physcomitrella patens) and PttCESA8 from hybrid poplar (Populus tremula × tremuloides) synthesized very thin β-1,4-glucan fibrils in vitro in the absence of added primer (Purushotham et al. 2016; Cho et al. 2017). When isolated from insect cells, PttCESA8 reconstituted into structures that largely ranged from monomer to trimer, and a homotrimer fraction was size-selected and analyzed by cryo-electron microscopy (Purushotham et al. 2020). In Fig. 3, a model of the proposed monomer of PttCESA8 is compared with that of other proposed catalytic subunit structures, including BcsA, and, for historical purposes, the original crude model (Delmer 1999) that was based on simple domain structure analyses.
Figure 3.
Comparison of models of cellulose synthase catalytic subunits. A) Cartoon model taken from Delmer (1999) shows how the active site in plants is recreated to resemble that of BcsA by looping out of CSR (HVR) and PCR (CR-P) domains. B) BcsA/B structure determined by X-ray diffraction from its crystal structure (Morgan et al. 2013). C and D) PttCESA8 modeled from cryo-electron microscopy of reconstituted recombinant protein (Purushotham et al. 2020) and cotton fiber GhCESA8 (original name GhCESA-1) computationally modeled from the sequence (Singh et al. 2020). All models possess an elongated shape with a central catalytic domain (brown in BcsA; wheat in PttCESA8; grey and green in GhCESA) and a channel comprised of TMHs (green, blue, red, moving L to R) through which the growing glucan chain is extruded through the inner/plasma membrane. The PCR (aqua in PttCESA8; magenta in GhCESA8), CSR domains (magenta in poplar; aqua in cotton), and NTD (shown in blue only in GhCESA8) are plant specific and not found in BcsA. The BcsB domain associated with BcsA (blue gray) is not found in plants.
Although differing in many details, all models show that the TMHs form transmembrane channels, and the catalytic domain of plant CESAs is reconstructed by looping out into the cytoplasm of the PCR and CSR domains that are proposed to function as sites for CESA-CESA interactions and/or necessary accessory proteins. Possible modes of regulation of the plant gating loop are under study (Olek et al. 2023; Verma et al. 2023). Binding of c-di-GMP regulates catalytic activity through the gating loop in bacteria but not plants (Morgan et al. 2014), and this loop is predicted to collapse into the region of the active site when the CSC is inactive in order to maintain PM membrane potential. Older work by Bacic and Delmer (1981) and Delmer et al. (1982) indicated that glucan synthesis in membrane vesicles of cotton or intact A. xylinum is inhibited by collapsing the delta psi (electric component), but not delta, pH component of the membrane potential and perhaps offers some clues in this regard.
Initiation of synthesis without added primer (see above) may put to rest the suggestion that sitosterol glucoside can serve as the primer for cellulose synthesis in cotton fibers (Peng et al. 2002). Cotton fibers clearly make sterol-di-tri- and tetra-cello-oligosaccharides, but attempts to demonstrate their initiator activity in Arabidopsis have failed. However, it does appear that CSCs function in sterol-rich portions of the plasma membrane (Schrick et al. 2012; Turner and Kumar 2018), another promising area for further study.
MF structure predicts rosette structure (or vice versa)?
Current thinking considers 2 different models for the way in which the 3 different CESAs required for synthesis of PCW or SCW assemble into rosettes. One model proposes that each of 2 of the 6 lobes are comprised of CESA homotrimers that alternate positions within the rosette; alternatively, each of the 6 lobes is assembled into identical heterotrimers, wherein each CESA in the lobe has a unique role. As of this writing, there is no clear answer, but homotrimers may be more likely because recombinant SCW CESA8 (and more recently CESA7; Zhang et al. 2021b) can so easily form active homotrimers, although no complete rosette has yet been assembled. Confusingly, recombinant rice (Oryza sativa) CESA8 CatD domain can show clear redox-dependent dimerization (Olek et al. 2014), whereas Vandavasi et al. (2016) obtained trimers from a similar recombinant AtCESA1. Analysis of the PCR crystal structure (Rushton et al. 2017; Zhang et al. 2021b) fitted well with the trimeric structure of PttCESA8s proposed by Purushotham et al. (2020). However, studies of the CSR domain by Scavuzzo-Duggan et al. (2018) and Olek et al. (2023) suggest dimerization through key cysteine residues. Kurek et al. (2002) showed redox-dependent formation of either homo- or heterodimers of the ring fingers of cotton fiber CESAs, and the NTD domain is clearly essential for activity, although it has not proven possible to model trimers with dimerized ring fingers.
The answers may lie in the way trimers are assembled into rosettes. Wilson et al. (2021) further examined the idea that redox-dependent CSR dimerization is one stabilizing link between trimers. Examining their models for a dimer of trimers, one can see several possibilities where ring-finger dimers could also form and act as stabilizing trimer-trimer associations, a possibility that deserves further consideration. Dimers clearly predominate in decomposing CSCs from some cells such as cotton fibers (Kurek et al. 2002; Wen et al. 2022), and it was reported that when GhCESA7 was knocked out, the CSC degraded to GhCESA4–8 dimers, while knocking out GhCESA8 led to GhCESA4–7 dimers (Wen et al. 2022). CESAs 4, 7, and 8 can dimerize with each other in a redox-dependent manner, and this interaction was proposed to occur before assembly into a complete rosette (Altanassov et al. 2009). Whether one gets dimers or trimers in decomposing rosettes may depend on the relative stability of the redox-dependent linkages found in trimers vs possible dimerizations that link trimers into rosettes.
The current conclusion is that the basic PCW MF is usually comprised of 18 chains (Jarvis 2013; Newman et al. 2013; Nixon et al. 2016; Ye et al. 2018; Haigler and Roberts 2019; Song et al. 2020). Models suggest assembly into MFs might be based on 5 layers of chains in a 34,443 arrangement (Kubicki et al. 2018) or 6 layers of 234,432 chains (Song et al. 2020). However, a 24-chain model has been convincingly proposed for spruce (Fernandes et al. 2011), and recent work strongly supports this model for other wood SCW MFs as well (Tai et al. 2023). Haigler and Roberts (2019) have written an excellent analysis of the evolution of rosettes and TCs and the relationship between structure of the MFs and the rosettes.
The role of cortical microtubules in orientation of MFs
Perhaps the most exciting contribution from microscopy that opened a whole new approach to studying cellulose synthesis is the early work of Paredez et al. (2006). A DNA construct encoding AtCESA6 was fused to a fluorescent protein and was active when transformed into a mutant cesA6 background, and mobile CSC complexes were visualized in hypocotyl epidermal cells. They moved at a constant speed corresponding to an addition of 300 to 1,000 Glc residues per minute and displayed a short residence time in the PM of 7 to 10 min—roughly the amount of time to synthesize 1 PCW glucan chain. Movement was often bidirectional and paralleled the orientation of aligned cortical microtubules beneath the PM. One can only imagine how the early pioneers of cytoskeletal biology would have reacted to actually watching the process of MT-aligned MF synthesis in real time. Although the residence time of PCW CesAs is short, other work indicates that the CesA proteins are quite stable, suggesting they may be recycled (Hill et al. 2018). In contrast, SCW CesAs of cotton fibers showed half-lives of less than 30 minutes (Jacob-Wilk et al. 2006).
The residence time of a CSC was calculated to exceed that of the underlying dynamic MTs, and it was observed often that the MT would disappear, but the CSC would continue its forward motion in the same direction, confirming that CSC movement is not assisted by some MT-motor protein mechanism but rather by the force of polymerization of the MFs. Later, Chan and Coen (2020) studied CSC movement after MT disappearance and confirmed that CSCs continued in the same direction for the residence time of the CSC, after which new CSCs appeared that followed the same trails as previous ones; this pattern continued apparently guided by the orientation of the nascent MFs in the wall. This kind of behavior has been seen in other situations in vivo; for example, the pattern of cellulose deposition during xylem vessel development is initially determined by MT orientation but persists long after MT disruption (Roberts et al. 2004; Schneider et al. 2017; Watanabe et al. 2018). Such observations have occasionally been used as an argument that MTs may not be so important for directing CSC motion. However, the MT-directed path takes preference because, when a CSC encounters an MT, the CSC preferentially begins following the new MT orientation. These studies clearly support the notion that the primary mechanism for orientation of MFs, a major event in the anisotropic elongation of plant cells, is controlled by the orientation of cortical MTs. But what controls reorientation of the MTs? One intriguing finding is that the localized de-methylesterification of pectins in the wall predicts and precedes the pattern adopted by the MTs (Peaucelle et al. 2015). Other evidence indicates that actin microfilaments interact with MTs and may play a special role in targeting of CSC and MF orientation (especially in SCW synthesis) (Lei et al. 2012). MT orientation may occur by sensing the direction of tensile stress (Hamant et al. 2019), and recent work indicates that the tethering of CSCs to MTs disturbs this stress-induced MT reorganization (Schneider et al. 2022).
The approach conceived in 2006 to follow rosette movement in vivo has also revealed important information on CSC trafficking (Lampugnani et al. 2019). CSCs assemble in the Golgi and are transported to the PM, where they become tethered to cortical MTs through binding of the CESA NTD to POM-POM2/Cellulose Synthase Interacting proteins (Gu et al. 2010; Bringmann et al. 2012). Once activated, the CSCs stall after 7 to 10 min and then are endocytosed. CESAs then transiently accumulate in small CESA compartments or are degraded in the lytic vacuole. While active, the forward movement of the CSC might be expected to displace the MTs from their PM location, but this has been shown to be prevented by cortical MT uncoupling proteins that regulate this interaction (Liu et al. 2016). Other older data may still be relevant: the cellulose synthesis inhibitor 2,6-dichlorobenzonitrile (DCB) somehow may act through disruption of MT organization, and an 18- to 20-kD MT-associated protein (MAP20) binds an active photo-affinity analog of DCB (Delmer et al 1987; Rajangam et al. 2008), consistent with a complex interaction between synthesis of cellulose and orientation of MTs (Schneider et al. 2017). However, genetic evidence that MAP20 is the target of DCB action in vivo is still lacking. Also fascinating is the finding that although kinesins do not control CSC movement, they are involved in movement on MTs of noncellulosic polysaccharide cargos from the Golgi to the PM, and 1 specific kinesin, referred to as Fragile Fiber1, has recently been shown to interact with cortical MT uncoupling proteins. This interaction is proposed to facilitate the localized deposition of matrix polymers in proximity to the sites where cellulose is deposited, representing a potentially important way to coordinate the deposition of cellulose with that of noncellulosic polymers (Ganguly et al. 2020).
Hemicelluloses
Structures of hemicelluloses
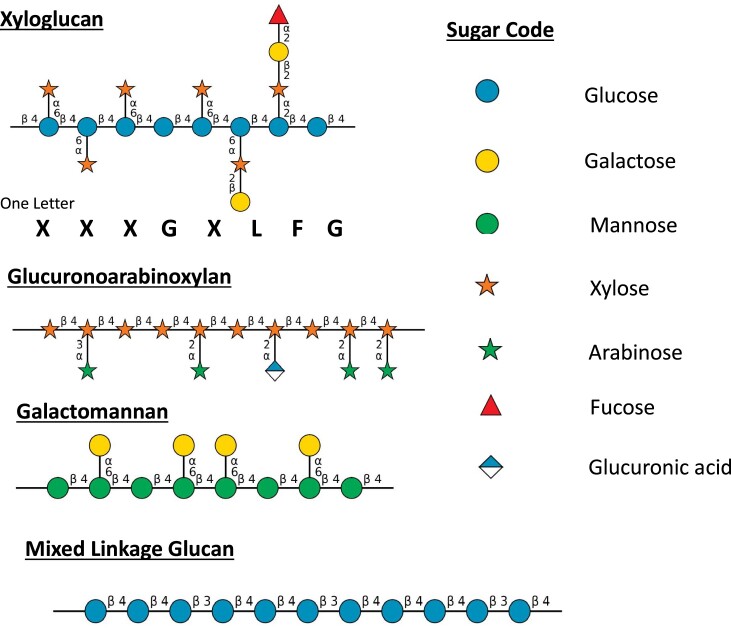
Hemicellulosic polysaccharides consist of a diverse collection of different polymers including xyloglucans (XyG), xylans, mannans, and mixed-linkage glucans (MLGs) (Fig. 4). Providing a precise definition of a hemicellulose is difficult. The common feature is that they bind to cellulose. Most have a backbone with β-1,4-linked sugars, but as the name implies, MLG also has β-1,3-linkages in the backbone. The backbones of most hemicelluloses have additional sugars and/or acetyl groups attached, but again MLG is the exception (Fig. 4).
Figure 4.
Representative structures of hemicelluloses. The xyloglucan fragment is typical of dicot PCWs. The galactomannan is typical of those found in legume seeds. The glucuronoxyloarabinan is typical of those found in grass PCWs. The structures were drawn using software described by Cheng et al. (2017).
When considering matrix polysaccharides, both hemicelluloses and pectins, it is important to remember that the structures depicted in figures are a representation of many different molecules. Polysaccharides are different from nucleic acids and proteins, where the sequence of monomers is determined by the genetic code. In contrast, the structure of polysaccharides is determined by the specificity of the enzymes that synthesize them, and not all individual molecules in a collection of polysaccharides have the same structure. Another difference is that sugars have multiple hydroxyl residues where a neighboring sugar, or an acetyl residue, can be attached. Thus, determining the structure of a polysaccharide requires complex chemical and analytical methods.
Xyloglucans
Among hemicelluloses, XyGs have the most complex structures (Fig. 4). Like cellulose, they have a backbone of β-1,4–linked glucosyl residues but with α-linked xylosyl residues attached in a regular pattern to the 6-position of most glucosyl residues (Fig. 4). Many xylose residues are further substituted, most frequently with galactosyl or galactosyl plus fucose residues. A shorthand notation for the substitution pattern of each location along the chain uses a single letter designation (Fig. 4) (Fry et al. 1993). Other substitution patterns are found in XyGs from various plant species, with the patterns sometimes varying among different tissues within a species (Hayashi 1989; Scheller and Ulvskov 2010; Schultink et al. 2014; Pauly and Keegstra 2016). XyG is the most abundant hemicellulose in the PCWs of gymnosperms, dicots, and noncommelinid monocots. Plants with XyG as the most abundant hemicellulose are said to have Type I primary walls (Carpita and Gibeaut 1993).
Xylans
Xylans have a pentose-based backbone of β-1,4–linked xylosyl residues linked in a way that allows binding to cellulose. The backbone is substituted with α-linked arabinose residues, α-linked glucuronic acid resides, and variable numbers of acetyl esters (Fig. 4). Xylans, as opposed to XyGs, are the most abundant hemicellulose in the PCWs of grasses; these walls are categorized as Type II (Carpita and Gibeaut 1993). The distribution of the side-chain residues varies depending on the plant species, the plant tissue, and the developmental stage of the tissue (Scheller and Ulvskov 2010). In addition, xylans from gymnosperms and selected angiosperms have a unique tetrasaccharide located at the reducing end of the polysaccharide, although the function of this sequence of 4 sugars is unknown (Smith et al. 2017). The distribution and patterns of substitution along the xylan backbone has functional significance. For example, the backbone substitution patterns of SCW xylans impact their interactions with cellulose (Busse-Wicher et al. 2016; Grantham et al. 2017). More recently, Tryfona et al. (2023) demonstrated that grass cell walls have xylans with at least 3 different substitution patterns and suggest that these patterns influence the way in which xylans interact with other wall components. For example, an arabinoxylan with evenly distributed arabinofuranosyl residues and lacking glucuronic acid facilitates binding to cellulose while other substitution patterns may facilitate interactions with other wall components such as lignin (Tryfona et al. 2023). Xylans are the most abundant hemicellulose in the SCWs of all angiosperms. Because of their abundance in SCWs of woody plants, there are massive quantities of xylans in the biosphere; it has been estimated that 10 billion tons of carbon are incorporated into xylans annually (Smith et al. 2017).
Mannans
Mannans are taxonomically the most widely distributed and evolutionarily the most ancient of the hemicelluloses (Voiniciuc 2022). They contain either a β-1,4–linked mannan backbone or a backbone consisting of both β-1,4–linked mannose and glucose. Some have been found to have alternating mannose and glucose in the backbone (Yu et al. 2022). The backbone sugars are substituted with α-linked galactose attached to the 6-position of backbone mannose (Fig. 4). Some mannans have very few or no side chains, whereas other galactomannans are heavily substituted with galactose, thereby producing polysaccharides with very different physical properties. For example, the unsubstituted mannans in ivory nut (from Phytelephas species) are insoluble and can be used as carving material for artwork. In some algae, unsubstituted mannans can substitute for cellulose (Scheller and Ulvskov 2010). On the other hand, highly substituted galactomannans are water soluble and can produce viscous solutions with food product and industrial applications (Sharma et al. 2022). Mannans are generally very minor components of PCWs and SCWs of most angiosperms but are frequently the most abundant hemicellulose in the SCWs of gymnosperms.
Mixed-linked glucans (MLGs)
MLG has a simple structure with no side chain residues (Fig. 4). It is less widely distributed in the plant kingdom than other hemicelluloses, being found mainly in the primary cell walls of the grasses plus in the cell walls of a few lower plants and some fungi (Burton and Fincher 2009). The β-1,3 linkages create kinks in an otherwise straight polymer so that the frequency and spatial distribution of β-1,3 linkages determines the physical properties of the polymers (Burton and Fincher 2009).
Hemicelluloses as storage compounds in plant seeds
Grant Reid and his colleagues significantly contributed to our understanding of plants that use hemicelluloses as reserve polysaccharides in their seeds. In his 1985 review, he pointed out that studies began late in the 19th century, when “…it was clearly recognized by botanists that the massively thickened cell walls present in many seeds contained reserve substances” (Reid 1985). Different plant species utilize different polysaccharides as storage reserves; for example, tamarind (Tamarindus indica), nasturtium (Tropaeolum majus), and many other species store XyG as a reserve polysaccharide. On the other hand, fenugreek (Trigonella foenum-graecum), coffee (Coffea arabica), and many other species store polymers in the mannan family, especially galactomannans that have received significant attention because of their industrial and medical applications (Sharma et al. 2022). Their unique rheological properties cause them to have wide use in the food, pharmaceutical, and cosmetic industries, as well as in hydraulic fracturing fluid (Sharma et al. 2022). Dendrobium catenatum, containing large quantities of glucomannan, has been extensively studied for centuries and utilized as both food and a Chinese herbal medicine (Qi et al. 2022).
MLG is also found as a reserve polysaccharide in the seeds of cereal grains (Morrall and Briggs 1978). Use of MLG as a reserve polysaccharide has been studied in detail in the model grass Brachypodium distachyon, where it is present in thick endosperm cell walls and is mobilized during seedling germination (Francin-Allami et al. 2023). Although xylans are also found in seeds of some plants, the arabinoxylan from the seed husks of Plantago ovata has received the most attention (Belorio and Gomez 2022). This psyllium mucilage consists of a highly branched β-1,4–linked xylan backbone (Fischer et al. 2004) and assists P. ovata seeds with hydration and gemination. It has been widely used as fiber in the human diet.
As methods for investigating polysaccharide structure were developed in the middle of the 20th century, it was recognized that storage polysaccharides, primarily from seeds (Kooiman 1957) but also from other organs, were similar in structure to hemicelluloses from plant cell walls. For example, XyG from seeds (Kooiman 1967), extracellular polysaccharides of suspension-cultured sycamore (Acer pseudoplatanus) cells, and PCWs (Becker et al. 1964; Aspinall et al. 1969; Bauer et al. 1973) all had similar structures.
Because seed reserve and mucilage polysaccharides can be isolated more easily and using milder conditions, they have provided valuable insight into the structure of the various hemicelluloses. As described below, these seed systems have been exploited to identify the genes and proteins involved in the biosynthesis of cell wall polysaccharides. Because the presence of wall polysaccharides as reserve polymers has arisen independently several times during evolution, analysis of the seed systems provides an opportunity to understand how polysaccharide synthesis is regulated. For example, the entire pathway from sucrose to galactomannan in fenugreek is upregulated during seed formation (Wang et al. 2012). Such systems should provide opportunities to identify transcription factors controlling this process.
Biosynthesis of hemicelluloses
The biosynthesis of matrix polysaccharides, both hemicelluloses and pectins, takes place in the Golgi. It involves a series of complex processes that generally require a different biosynthetic enzyme for every different linkage found in each polymer (Lerouxel et al. 2006). Results over the last 2 decades have revealed that plant cells employ 3 different strategies for the synthesis of matrix polysaccharide backbones. The first is exemplified by MLG that is synthesized by CSLFs, proteins that are closely related to CESA and CSLD proteins (Yin et al. 2014; Little et al. 2018). CSLF is a glycan synthase (Purushotham et al. 2022) that operates similarly to the CESA proteins described earlier. The second strategy is illustrated by mannans and XyG, which are synthesized by CSLA and CSLC, respectively (Dhugga et al. 2004; Cocuron et al. 2007). These proteins are also glycan synthases but are distant relatives of CESA (Yin et al. 2014; Little et al. 2018). They also differ from the CESA, CSLD, and CSLF proteins in that they operate in concert with GTs that add side chains to the growing backbone (Pauly and Keegstra 2016; Zabotina et al. 2021; Voiniciuc 2022). The third strategy is fundamentally different in that the backbone of xylans and pectic polysaccharides are synthesized by complexes of glycosyltransferase that operate in the lumen of the Golgi (Ye and Zhong 2022; Anders et al. 2023). In these cases, complexes of GTs assemble the backbone (Engle et al. 2022; Ye and Zhong 2022; Anders et al. 2023) while different GTs add the side chain residues when present (Smith et al. 2017; Ye and Zhong 2022).
Over the past 2 decades, investigators have exploited the systems that produce large quantities of individual polysaccharides described above to identify the genes and proteins responsible for their synthesis in a manner analogous to the studies with cotton that led to the identification of the CESA proteins (Pear et al. 1996). Dhugga and colleagues (2004) used developing guar (Cyamopsis tetragonoloba) seeds that store large quantities of galactomannan to identify the CSLA genes responsible for the synthesis of the mannan backbone. Cocuron et al (2007) used developing nasturtium (Tropaeolum spp.) seeds that produce large quantities of XyG to identify the CSLC genes responsible for XyG backbone synthesis. For the synthesis of the backbone of rhamnogalacturonan I (RG-I) (see section on pectins below), Takenaka et al (2018) investigated mutant Arabidopsis plants that had defects in mucilage production to identify a rhamnosyltransferase required for RG-I synthesis. This strategy has also been used to identify the GTs required for the addition of side chain sugar residues (Edwards et al. 1999; Jensen et al. 2012, 2013).
Because GTs are mostly type II membrane proteins where the active site is in the Golgi lumen, they utilize nucleotide sugars present inside the lumen. Thus, Golgi-localized sugar nucleotide transporters are required during polysaccharide synthesis. As noted earlier, the various NDP sugars are made from UDP-Glc via a complex set of pathways present in the cytoplasm or the Golgi membranes (Bar-Peled and O’Neill 2011). On the other hand, glycan synthases, including most CSL proteins, have multiple membrane spans; these enzymes accept sugars from nucleotide sugars in the cytosol and, during synthesis, transport the nascent polymer to the lumen of the Golgi, where GTs add side chain sugars as needed. The newly synthesized polysaccharides move from the Golgi lumen to the cell surface via membrane vesicles, which fuse with the plasma membrane, thereby releasing the polymers into the wall matrix. However, very little is known about how the Golgi-synthesized polysaccharides are assembled into a functional cell wall. A summary of the biosynthesis of each type of hemicellulose is presented below.
Mannans
Soon after the discovery of mannan synthase (Dhugga et al. 2004), Liepman et al. (2005) demonstrated that several members of the Arabidopsis CSLA gene family had mannan or glucomannan synthase activity when expressed in cultured insect cells. Subsequently, CSLA gene family members from other species were shown to be responsible for mannan biosynthesis, and it has been postulated that all CSLA genes encode mannan synthase (Liepman and Cavalier 2012; Voiniciuc 2022). Of note is that GDP-Man and GDP-Glc, as opposed to UDP-Glc, are used for backbone synthesis.
Xyloglucan
Expression of cDNA clones from both nasturtium and Arabidopsis CSLC genes in Pichia pastoris resulted in the cells producing β-1,4–linked glucan (Cocuron et al. 2007), leading to the conclusion that both likely encode the glucan synthase that makes the XyG backbone. Because Pichia does not produce UDP-xyl, it was not possible to confirm these glucan synthases were responsible for XyG biosynthesis, although several lines of evidence supported this conclusion. Later, Kim et al. (2020) demonstrated that a quintuple mutant with disruptions in all 5 Arabidopsis CSLC genes has undetectable XyG levels, thereby providing compelling evidence that CSLC genes are responsible for XyG biosynthesis. The phenotypes of these and other mutants lacking XyG are described below.
The biosynthesis of mannans and XyGs is similar in many ways (Liepman and Cavalier 2012). With respect to backbone synthesis, the CSLA and CSLC gene families have sequence similarities and a common evolutionary origin while being evolutionarily distinct from other subgroups in the CESA superfamily (Yin et al. 2014; Little et al. 2018). Little et al. (2018) suggest this lack of connection reflects “…the likely dual endosymbiotic origin of the superfamily” with CSLA/CSLC coming from 1 event, whereas CESA, CSLD, and CSLF came from a different event. Side chains attached to the 6-position of each polymer are added by enzymes present in CAZy family GT34 that have both sequence and biochemical similarities (Edwards et al. 1999; Faik et al. 2002; Cavalier and Keegstra 2006). Yu et al. (2022) have elaborated on the similarities in the structure and biosynthesis of the 2 polymers, with evidence that they may have similar functions in that mannans may be able to substitute for XyG in mutants lacking XyG.
As noted above, XyG has a complex array of side chain substitutions. Work in the last 2 decades has produced considerable knowledge about the genes and proteins responsible for the addition of these side chains. Because this work is well documented in several recent reviews (Scheller and Ulvskov 2010; Schultink et al. 2014; Pauly and Keegstra 2016; Julian and Zabotina 2022), here we briefly highlight the important role played by genetics, both forward and reverse, in identifying not only the genes and proteins responsible for side chain addition but also the functions of the polysaccharides and their side chains.
The story begins in Chris Somerville's laboratory, where several Arabidopsis mutants with alterations in cell wall composition were identified (Reiter et al. 1997). One of them, mur2, a mutant with reduced levels of fucose, led to identification of the gene responsible for the addition of the fucose side chains on XyG (Perrin et al. 1999; Vanzin et al. 2002). A second, the mur3 mutant, was later shown to encode a galactosyltransferase that adds galactose to XyG at the location where fucose is attached (Fig. 4) (Madson et al. 2003; Kong et al. 2015). Reverse genetics was also used to isolate mutant plants with disruption of the genes encoding XyG xylosyltransfereases (XXT) (Cavalier et al. 2008; Zabotina et al. 2012). As described in more detail in a later section, these mutant lines also aided in evaluating the function of XyG.
Mixed-linked glucans (MLGs)
Early studies on the biosynthesis of MLG focused on confirming that it was made by a unique enzyme. As noted above, early studies on cellulose biosynthesis identified enzyme preparations that made both β-1,3 and β-1,4 linkages. The question was whether MLG was made by the combined action of 2 different enzymes or a unique enzyme that made both linkages. Bruce Stone was a memorable contributor to plant cell wall biochemistry. Both funny and feisty, he possessed encyclopedic knowledge of β-glucans in plants and managed to exhaust 2 sets of authors who worked with him writing 2 volumes on the subject. Bruce and his colleagues resolved the question of 1 or 2 enzymes by identifying and partially characterizing an enzyme activity that produced MLG (Smith and Stone 1973; Henry and Stone 1982a, 1982b). Many years later, Stone was coauthor of a manuscript reporting the identification of the CSLF genes and proteins responsible for MLG biosynthesis (Burton et al. 2006). Doblin et al. (2009) provided evidence that the grass-specific CSLH genes also encode proteins capable of MLG synthesis. Both the CSLF and CSLH genes are more closely related to CESA genes than to CLSA and CSLC genes (Yin et al. 2014; Little et al. 2018). Although many questions remain about the details of MLG biosynthesis, recent in vitro studies using the barley (Hordeum vulgare) CSLF protein have demonstrated that 1 enzyme can synthesize both linkages found in MLG. Molecular modeling and mutagenesis identified a region of the protein near the cytoplasmic side of the glucan channel that controls the frequency of β-1,3 links in the polymer product (Purushotham et al. 2022).
Xylan
As noted above, xylan biosynthesis is very different from the biosynthesis of the other hemicelluloses. Most of the genes encoding xylan synthase proteins were identified through forward genetic screens as mutations that altered xylem function, called irregular xylem or irx mutants. The xylan backbone is synthesized by a complex of GTs and related proteins that either have a single transmembrane domain or that lack a transmembrane domain. The proteins encoded by these genes belong to families GT47 and GT43 in the CAZy classification scheme. Xylan backbone synthesis requires the cooperation of 3 different nonredundant proteins that make up the xylan synthase complex. They include IRX10, or related proteins such as IRX10L, belonging to CAZy family GT47 plus IRX9 and IRX14, or related proteins belonging to CAZy family GT43 (Smith et al. 2017; Ye and Zhong 2022; Anders et al. 2023). The exact role of each of these proteins in the xylan synthase complex is still not completely resolved. The emerging consensus is that IRX10 and related proteins have xylosyltransferase activity (Urbanowicz et al. 2014; Jensen et al. 2018) and provide the active site that links together the xylosyl resides in the backbone. Most, or maybe all, IRX10 and related proteins lack a transmembrane domain. They are thought to associate with the IRX9 and/or IRX14 proteins, which are type II integral Golgi membrane proteins with a single transmembrane domain. It is not yet clear whether the family GT43 proteins have xylosyltransferase activity or whether they have other functions in the xylan synthase complex. Various members of these protein families are utilized depending on the plant species, the plant tissue, and whether primary or secondary wall xylan is being synthesized (Smith et al. 2017; Ye and Zhong 2022; Anders et al. 2023). Anders et al. (2023) postulate that different members of the gene families are involved in xylan synthesis in PCWs than those in SCWs, similar to the situation with the CESA family members.
In addition to the various proteins involved in backbone synthesis, Golgi membranes contain several different GTs involved in adding the side chains found on xylans. These include glucuronosyltransferases in CAZy family GT8 (Mortimer et al. 2010; Oikawa et al. 2010) and arabinosyltransferases in GT61 (Ye and Zhong 2022). Because xylans, especially those in SCWs, are heavily acetylated, Golgi membranes also contain several different enzymes, including acetyltransferases, involved in xylan acetyl esterification as well as acetyl esterification of other matrix polysaccharides (Smith et al. 2017; Pauly and Ramirez 2018; Julian and Zabotina 2022), including pectins as described in the next section.
Pectins
Structures of pectins
History of pectin and its complexity
Pectin (pectique), from the Greek πηκτικοί (to coagulate), was first used by Henri Braconnot to describe the acidic viscous substances with gelling properties isolated from multiple tissues of more than 15 plant species (Braconnot 1825a, 1825b; Wisniak 2007). Braconnot noted that the ability of a very small amount of pectin to gelatinize a large mass of sugar water provided it industrial uses such as in the confectionary business (Braconnot 1825b; Wisniak 2007). These properties led to the establishment of the pectin industry in the early 1900s (Kertesz 1951), and numerous studies on the chemical basis of the gelling properties of pectin from commercial sources such as citrus and apple fruit followed. Braconnot also recognized that pectin's ability to bind and remove toxic metals imparted medicinal properties (Braconnot 1825a), and increasing numbers of industrial, food, and medical applications for pectin have since been explored, including its use in edible coatings on foods, antimicrobial bio-based films, nanoparticles, and as healing agents and cancer treatments (Willats et al. 2006; Freitas et al. 2021). The diverse functional characteristics of pectins led Braconnot to propose that a substance so “universally distributed in plants” “must play a role of great importance in the plant and should be studied by plant physiologists” (Braconnot 1825b, p178). Since then, plant biologists and chemists have confirmed multiple functions for this most structurally complex of the plant cell wall glycans, including roles in plant growth and cell expansion (Proseus and Boyer 2012a, 2012b; Daher et al. 2018; Haas et al. 2020), cell shape (Amsbury et al. 2016; Haas et al. 2020; Lin et al. 2022b), organ size and texture (Zhang et al. 2021a), cell-to-cell adhesion (Marry et al. 2006; Yang et al. 2020a), cell wall porosity (Baron-Epel et al. 1988; Fleischer et al. 1999), cell wall structure (Pérez García 2011), signaling (Feng et al. 2018; Lin et al. 2022b; Du et al. 2022), fruit ripening (De Vries et al. 1981; Paniagua et al. 2014), organ abscission (Daher and Braybrook 2015), pollen development (Mollet et al. 2013), and plant defense (Bacete et al. 2018; Molina et al. 2021; Liu et al. 2023). How does a single class of cell wall glycan provide such diverse functions? This question continues to beset plant biologists.
Understanding pectins’ multiple functions requires knowledge of (1) their primary, secondary, tertiary, and quaternary structure (Diener et al. 2019); (2) covalent and noncovalent interactions between individual pectic polymers and between pectins and other wall polymers; (3) pectins’ roles in cell type-specific wall architecture; and (4) the specific pectic polymeric structure associated with each functional response. In no case is this information currently known. While we know a great deal about the different general types of pectic glycans present in cell wall extracts from different tissues (reviewed by Mohnen et al. 2024), we lack information about the number and structure of pectic polymers in different specific cell types and how they are integrated into cell wall architecture. Furthermore, pectins that are synthesized and inserted into the wall are subsequently modified during development and in response to biotic and abiotic stress by large families of pectin-modifying enzymes, including pectin hydrolases (Yang et al. 2018), lyases (Molina-Hidalgo et al. 2013; Leng et al. 2017), methylesterases (Willats et al. 2006; Wolf et al. 2009), acetylesterases (Philippe et al. 2017), and proteins that modify or inhibit these enzymes (Federici et al. 2006; Hocq et al. 2017b). Importantly, the different classes of enzymes include proteins that uniquely target the different pectic backbones homogalacturonan (HG) and rhamnogalacturonan (see below).
The complexity of pectin structure
All pectic polymers have 1 or 2 backbones: HG and/or rhamnogalacturonan. HG is a homopolymer of α-D-1,4–linked galacturonic acid that may have low, partial, or almost full methyl-esterification of the C-6 carboxyl (Bosch and Hepler 2005) (Fig. 5A). It has been proposed that in vivo HG is synthesized in a highly methyl-esterified form; however, the extent and mechanism of esterification of HG synthesized by the different HG biosynthetic enzymes in vivo remain to be determined. HG biosynthetic GALACTURONOSYLTRANSFERASES (GAUTs) can synthesize polymeric HG with no methyl-esterification in vitro (Amos et al. 2018; Engle et al. 2022), indicating that HG methyl-esterification is not a requirement for enzymatic synthesis of HG in vitro (reviewed by Mohnen et al. 2024). Yet, many biological effects are associated with the methyl-esterification status of pectins, and immunolabeling studies indicate that many cell walls have high or changing levels of HG methylester content during development (Wolf et al. 2009).
Figure 5.
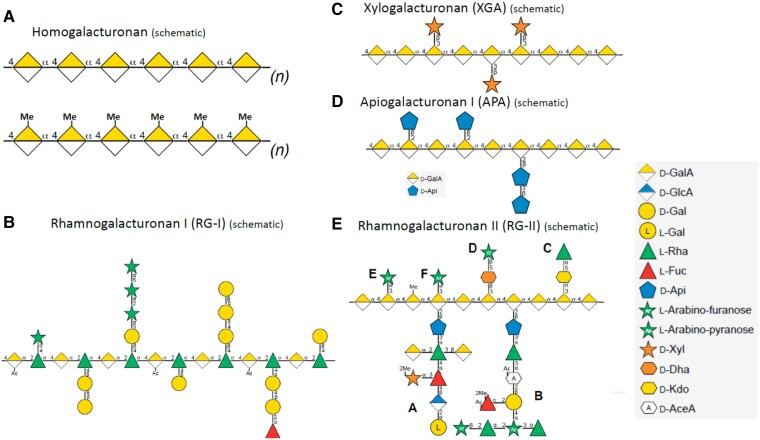
Glycan symbol structure representations of the main pectic glycan domains. A) HG. B) RG-I. C) XGA. D) APA. E) RG-II. The structures were drawn using Symbol Nomenclature for Glycans from (SNFG) https://www.ncbi.nlm.nih.gov/glycans/snfg.html#nomn.
Each GalA residue in HG has a carboxylic acid group at the C6 position that will be highly ionized and negatively charged at pH above 3.5. Thus, polymeric HG is a polyanion and can interact with positive ions such as calcium or positive patches on proteins. Methylesterification of the carboxyl group by pectin methyltransferases (PMTs) reduces the charge and increases the hydrophobicity of each GalA residue and of methylesterified regions of HG. De-esterification of HG has been associated with changes in wall structure as regions of non-esterified HG can interact to yield HG-Ca++-HG crosslinks and such cross-linking can affect the structure and conformation of the HG polymer and of the pectin network in the wall. An understanding of HG structure during development will require knowledge of the extent and pattern of methyl (and acetyl) esterification in different HG-containing polymers as they are inserted into the wall and subsequent modification of HG once it is deposited in the wall. The degree and pattern of HG esterification and HG size in the wall is dependent on cell type, developmental stage, and status of biotic or abiotic stress because these affect the expression of HG degradative and modifying enzymes. These HG-modifying enzymes include pectin methylesterases (PMEs) (Pelloux et al. 2007), PME inhibitors (PMEIs) (Coculo and Lionetti 2022), acetyl esterases (Sénéchal et al. 2014), and polygalacturonases (Kim et al. 2006), which hydrolyze the methyl/acetyl esters and/or the HG backbone, respectively (Amsbury et al. 2016; Liu et al. 2023). Changes in HG methyl esterification and/or HG size are associated with numerous HG functions, including cell:cell adhesion (Kohorn et al. 2021a), cell elongation/shape/growth (Daher et al. 2018; Haas et al. 2020), and fruit ripening (Paniagua et al. 2014).
There is no definitive evidence for pure HG as a separate polymer in plant cell walls, although pure HG is synthesized in vitro (Fig. 6A) (Sterling et al. 2006; Atmodjo et al. 2011; Amos et al. 2018; Engle et al. 2022). Rather, HG is generally isolated covalently attached to RG-I (Fig. 6B) or to the substituted HGs rhamnogalacturonan II (RG-II) (Fig. 6E), xylogalacturonan (XGA) (Fig. 6C), and apiogalacturonan (Fig. 5D). All substituted HGs have an HG backbone and the requirement for endopolygalacturonase (EPGase) digestion of cell walls to release RG-II and RG-I from the walls has often been interpreted as evidence that RG-II and RG-I exist within an HG polymer and possibly as 1 covalently interconnected heteropolymer in the wall. However, this has not been structurally confirmed, and the architecture of pectins and the different pectic heteroglycans and glycoconjugates in the wall remains a critical question.
Figure 6.
Examples of structurally confirmed homoglycans, heteroglycans, and glycoconjugates that contain the different pectic glycan regions. A) HG. B) The heteroglycan HG-RG-I-HG. C) The heteroglycan HG-RG-II-HG. D) The proteoglycan pectic AGP APAP1. E) The proteoglycan pectic AGP AGP-RG-I.
RG-II (Fig. 5E) is the most structurally complex of the pectic polymers, with 13 different sugars (Darvill et al. 1978; Kobayashi et al. 1996; Bar-Peled et al. 2012; Ndeh et al. 2017) organized into 6 highly conserved side branches on an HG backbone. Why do plants spend so much energy to make this complex glycan? At least part of the answer is likely the structural role it played during the transition of plants from an aqueous environment onto land (Matsunaga et al. 2004), including wall strengthening via RG-II dimers formed by borate diester complex formation between the side chain A apiosyl residues (Kobayashi et al. 1996; O’Neill et al. 1996; Begum and Fry 2022). Indeed, the dwarf (Reuhs et al. 2004) and cell wall structural (Shi et al. 2017) phenotypes of RG-II mutants support a necessary role for RG-II in cell wall architecture and cell/plant growth (O’Neill et al. 2001).
RG-I is the only pectic polymer not built on an HG backbone (Fig. 6B). RG-I has a GalA-Rha disaccharide repeat backbone [−α−1,4-D-GalA-α−1,2-L-Rha-](n) with about one-half of the rhamnose backbone residues substituted by single or branched galactose, arabinose, or other sugars (Lau et al. 1985) (Fig. 5B). The degree and type of branching is tissue and developmental stage specific (Kaczmarska et al. 2022, 2023). For example, Arabidopsis seed mucilage RG-I is unbranched (Fabrissin et al. 2019; McGee et al. 2021), whereas RG-I in pea cotyledons is initially arabinosylated and acquires significant β-1,4-galactan side branching only in late development (McCartney et al. 2000). The precise order and location of the side branches is not known for any of the pectic polymers, a point that should be kept in mind when viewing schematic representations of pectic heteroglycan structures.
Conformation of pectins
Structure dictates function. Beyond pectins’ primary structures (which are already very complex), their secondary (e.g. degree and type of helix formation), tertiary (single molecule 3D structure), and quaternary (polymer-polymer association) structures lead to their vast diversity of biological and industrial functions. Yet, our understanding of these higher levels of pectin structure is unclear (Zdunek et al. 2021). In the mid-1930s, Bonner concluded—based on X-ray diffraction and viscometry studies—that pectins existed as long chains (Bonner 1936). It was known that pectins isolated from plants ranged from high to low methyl esterification content and that the amount of free carboxyl groups affected pectins’ colloidal properties (Bonner 1936). Subsequent x-ray diffraction analysis of isolated methyl esterified and de-esterified pectate in the absence and presence of calcium revealed a fibrillar 31 (3 residues per turn) helical quaternary conformation for various forms of HG, including sodium pectate, pectic acid, methylesterified HG (pectinic acid), and calcium salt–bridged HG (Walkinshaw and Arnott 1981a, 1981b). Shortly thereafter, Rees and colleagues presented evidence for a 21 conformation (2 residues per turn) of calcium pectate dimers, naming this conformation the egg box structure (Morris et al. 1982). More recent modeling suggests further conformational complexity (Braccini and Pérez 2001; Pérez et al. 2003; Braccini et al 2005), and the conformation of HG in vivo remains a matter of debate. Regarding the substituted HG RG-II, Pérez and colleagues provided evidence for a compact, flat, disc-like RG-II monomer structure with compaction of each monomer during dimerization yielding RG-II dimer conformations akin to 2 flattened, parallel disks stacked on top of each other (Pérez et al. 2003). Recent studies of RG-I backbone oligosaccharides indicated a 31 right-handed helical structure with 2 backbone disaccharide repeats per turn (Scanlan et al. 2022). Clearly, future studies to clarify the in vivo and in vitro conformation(s) of pectins are critical to delineate pectin structure/function relationships.
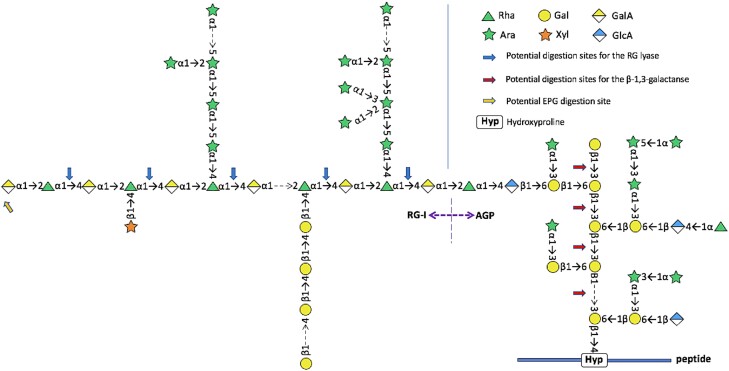
Numerous reports over many years have indicated that pectins may be attached to proteins or hemicelluloses (reviewed in Mohnen et al. 2024). Selvendran (1985) presented evidence for pectin-hydroxyproline and serine-rich protein complexes in cell walls of parenchymatous tissues from multiple species. Later, Mort and colleagues reported pectin-protein cross-links in cotton that appeared to be between RG-I and extensin (Qi et al. 1995). The identification of a covalent connection between the arabinogalactan domain of AGP57C and the backbone of RG-I in the pectic AGP APAP1 (arabinoxylan pectin arabinogalactan protein 1) isolated from the medium of Arabidopsis suspension cultures (Tan et al. 2013) provided a biochemical confirmation of a covalent linkage between pectins and protein. APAP1 had short xylan chains linked to the arabinogalactan domain and to Rha in the RG-I backbone. The RG-I backbone was elongated on the nonreducing end with HG (Fig. 6, D and E). Importantly, in a recent study of RG-I isolated from the cell walls of Arabidopsis suspension cells, a pectic AGP named RG-I-AGP was identified as the most abundant form of RG-I present in those walls (Tan et al. 2023b). Because the pectic AGP was isolated by the classical EPGase digestion of the cell walls followed by size exclusion chromatography, the results indicated that the bulk of classically defined RG-I in the walls was covalently linked to protein (Tan et al. 2023b) (Fig. 7). The RG-I-AGP was similar to APAP1; however, it did not have covalently linked xylan chains but rather single xylose residues attached to the RG-I backbone. Efforts to delineate whether all RG-I is synthesized as pectic AGP and, if so, how much is connected to the plasma membrane via GPI-anchoring of the AGP protein core versus how much is synthesized as a free heteroglycan not attached to protein [e.g. existing between HG domains in an HG-RG-HG glycan (Nakamura et al. 2002)] is critical to understanding pectin structure and cell wall architecture. It is noteworthy that the negatively charged family of pectic heteroglycans and glycoconjugates have structural and functional analogies to animal glycosaminoglycans and proteoglycans (Varki et al. 2022).
Figure 7.
Glycan symbol structure representation of pectic AGP isolated from cell walls of Arabidopsis suspension culture cells. Pectic AGP RG-I-AGP from suspension culture cell walls (from Tan et al. 2023b). The structures were drawn using Symbol Nomenclature for Glycans from (SNFG) as described in Fig. 5.
Biosynthesis of pectins
HG is synthesized by the CAZy GT8 GAUT family in Arabidopsis with GAUTs 1, 4, 10, 11, 13, 14, and possibly more functioning as catalytic HG biosynthetic GAUTs and 3 (GAUTs 5, 6, 7) functioning as Golgi-anchoring subunits of GAUT1 in GAUT1 protein complexes (Lund et al. 2020; Engle et al. 2022). All catalytic GAUTs synthesize polymeric HG in vitro by acceptor-dependent mechanisms, but some such as GAUTs 13, 14, and 1 can also de novo synthesize HG using only UDP-GalA as both the donor and acceptor substrate (Sterling et al. 2006; Atmodjo et al. 2011; Amos et al. 2018; Engle et al. 2022). The role of GAUT4 in wood formation in trees and secondary wall formation in grasses was probed due to its high expression levels in switchgrass (Panicun virgatum) stems and during transition from primary wall to wood formation in poplar (Populus trichocarpa). Downregulation of GAUT4 in switchgrass, poplar, and rice resulted in greatly reduced amounts of HG and RG-II (Biswal et al. 2018), indicating that in herbaceous dicots, grasses, and trees, GAUT4 may synthesize the bulk of HG and the HG backbone for RG-II. These results provide support for the existence of heteroglycan HG-RG-II-HG glycans (Fig. 6C). Interestingly, there was no reduction in RG-I in these plants, indicating that the synthesis of RG-I is not dependent on the synthesis of the bulk of HG, nor on the synthesis of RG-II. Interrogation of GAUT4 activity in a species with a single GAUT4 homolog, such as Arabidopsis, is challenging due to apparent lethality upon loss of gene function (Caffall et al. 2009). However, expansion of the GAUT family in poplar and switchgrass to 2 and 6 GAUT4 homologs, respectively, allowed knockdown expression of the most highly expressed GAUT4 homolog and analysis of phenotypes. The increase in cell size in the GAUT4 knockdown plants (Biswal et al. 2018) supports proposed roles for the pectin cross-linked matrix in plant cell growth and expansion (Proseus and Boyer 2012a, 2012b; Haas et al. 2020, 2021) (see Models of Cell Wall Organization section below).
Progress has been made in identifying some enzymes involved in synthesis of the substituted HGs. Xylogalacturonan Deficient 1 was identified as a GT47 xylosyltransferase that adds β-Xyl onto the 3-position of GalA in the HG backbone (Jensen et al. 2008), providing tools to explore the biological significance of XGA substitution within the HG backbone. Although originally believed to be most prevalent in fruits, XGA has been identified in many cell types (Zandleven et al. 2007). Immunological evidence suggests it may be associated with plant cell separation and detachment (Willats et al. 2004); however, a complete characterization of the epitope recognized by the antibody will be required to substantiate the functional interpretation of the immunolabeling results, a caution associated with many immunolabeling results. Little is known about the synthesis of the most universal substituted HG, RG-II. Three xylosyltransferases that add α-xylose to the 3-position of L-Fuc (RGXT) (GT77) in RG-II chain A have been identified (Egelund et al. 2006, 2008), but genes encoding the other RG-II biosynthetic enzymes remain to be identified.
The methylesterification state of HG dramatically affects its in vivo and industrial properties, with the pattern and degree of esterification affecting pectins’ gelling properties, ability to form pectate calcium ionic crosslinks, and sensitivity to enzymatic cleavage by polygalacturonases (Willats et al. 2006; Wolf et al. 2009). Correspondingly, considerable effort has gone into searches for the methyltransferases that esterify the HG backbone. However, although many mutants affected in methyl ester content have been identified, only with the recent biochemical confirmation of the HG methyltransferase activity of Quasimodo2 has the door been opened to understanding this crucial process (Du et al. 2020a). Critical experiments for the future include determining whether methyltransferases function in complexes with HG biosynthetic enzymes and the mechanisms for HG methyl esterification levels and patterning during synthesis.
The identification of the biosynthetic RG-I backbone rhamnosyltransferase (RRT) (GT106) (Takenaka et al. 2018) and galacturonosyltransferase (RGGAT) (GT116) (Amos et al. 2022) families that function together to synthesize the RG-I rhamnogalacturonan backbone (Fig. 6B) opens the door to in vitro synthesis of complex pectic heteroglycans and pectic AGP glycoconjugates such as APAP1 (Tan et al. 2013) and RG-I-AGP (Tan et al. 2023b) (Fig. 7D), as well as to efforts to reconstitute the galactose- and arabinose-rich side chains.
Complex RG-I synthesis will require identification of the enzymes that initiate side branch formation on the backbone, enzymes that thus far have remained elusive. The identification of the β-1,4 galactan synthases (GALS) (GT92) that extend the galactose side branches (Liwanag et al. 2012; Ebert et al. 2018; Laursen et al. 2018) was an important step forward and provided catalysts for study of type II galactan synthesis and structure. Interestingly, GALS1 was demonstrated to be a bifunctional enzyme that also can add arabinose to β-1,4-galactan chains, an activity hypothesized to function in chain termination (Laursen et al. 2018; Prabhakar et al. 2023). The 3D crystal structure of GalS1 from P. trichocarpa revealed a modular protein structure with an N-terminal CBM95 RG-I backbone binding domain, C-terminal GT92 GT-A fold catalytic domain, and stem region involved in homodimer formation (Prabhakar et al. 2023). Mutagenesis informed by comparison of GT-A fold sequences to the GT92 family identified a novel histidine as the catalytic base and other amino acids involved in β-1,4-galactan synthesis and acceptor binding. The approaches used in this study, including docking with molecular dynamics simulations, exemplify structure-function and acceptor specificity tools that may accelerate future progress on identifying and characterizing the enzymes involved in plant cell wall synthesis.
The combination of genetic loss-of-function phenotype with enzyme activity determined by in vitro heterologous expression of encoded protein sequence is the most definitive evidence for biochemical function of cell wall GTs (Amos and Mohnen 2019). Two putative arabinosyltransferases (ARAD1 and ARAD2) (GT47) that may add α−1,5-linked Ara onto RG-I side branches have been identified via studies of the arabinan deficient 1 mutant and molecular complementation (Harholt et al. 2006, 2012). Although biochemical confirmation of enzymatic activity has not yet been provided, these enzymes provide tools to attempt in vitro reconstitution of complex RG-I via coexpression with RG-I backbone synthesizing enzymes.
The identification of pectin biosynthetic genes has enabled the generation of cell wall glycan–modified mutant/variants and their probing using nanoscale imaging to test the hypothesis that pectins interact with cellulose and that hemicelluloses and pectins share load-bearing functions in the wall (Pérez García 2011). Coexpression in Arabidopsis stems of cytosolic UDP-Glc epimerase, Golgi localized UDP-Rha/UDP-Gal transporter, and RG-I GALS1 increased RG-I galactan content by 50% (Aznar et al. 2018). Multidimensional solid-state NMR analysis of C13-labeled high galactan-expressing Arabidopsis stems showed an increased proximity between galactan and cellulose in the secondary walls, leading the authors to hypothesize that increased galactan chain lengths led to increased mobile contacts between galactan and cellulose and longer spatial contacts between xylan and pectins in Arabidopsis SCWs (Gao et al. 2023). More research is required to understand the degree and importance of molecular interactions between pectins and cellulose in the cell wall, critical information for generation of meaningful cell wall models (see below).
Functions of pectins in vivo
Hypothetical model for pectin structure in the wall
A biologically meaningful interpretation of pectin mutant phenotypes requires, at minimum, knowing how many different types of pectic polymers exist in the wall and how they are covalently and noncovalently connected to each other and to other polymers. At present, pectic polymer models do not provide this information. Thus, a conundrum confronts us. On the one hand, making a model that is structurally inaccurate can lead to erroneous molecular genetic conclusions. On the other hand, without a model, functional hypotheses cannot be made.
Testing hypotheses of pectin function requires knowing which pectic glycan (Fig. 6A), heteroglycan (Fig. 6, B and C), or glycoconjugate (Fig. 6, D and E) is affected in the mutant/variant of interest. For example, the homogalacturon structure depicted in Fig. 6A has been shown to be synthesized in vitro as a nonmethylesterified homopolymer by GAUTs 10, 11, 13, 14, and the GAUT1:7 complex (Amos et al. 2018; Engle et al. 2022; Mohnen et al. 2024), and yet, there is scant evidence that pure homogalacturon exists as an isolated polysaccharide in the wall.
To develop a more pectin polymer-based vocabulary for discussing pectin structures and pectin mutant phenotypes, a working hypothetical model for pectin polymeric structures in plant cell walls is proposed. The pectic heteroglycan and glycoconjugate model (Fig. 8) depicts pectins as consisting of 5 different possible glycans, heteroglycans, and glycoconjguates. These include the covalently linked pectic heteroglycan HG:RG-I-HG-RG-II-HG, the separate heteroglycans HG-RG-I-HG and HG-RG-II-HG, the glycan HG by itself, and the pectic glycoconjugate pectic-AGP. The pectic polymers interact via ionic HG-calcium salt bridges and covalent RG-II borate diester cross-links. The pectic AGP is bound to the plasma membrane via a GPI anchor on the AGP protein core (although cleavage by phosphodiesterases may solubilize this into the wall). Representative methyl and acetyl esterification of the HG backbone is shown, but no specific pattern is implied. It is likely that the degree and pattern of methyl and acetyl esterification is development-, tissue-, cell-type-, and stress-level dependent. This model is supported by the observation that RG-I and RG-II are released from cell walls by treatment with EPGase and the supposition that although it is possible that they may all be covalently connected in a single network, it is also possible that they exist as separate heteropolymers in the wall (reviewed in Mohnen et al. 2024). Some of the RG-I and HG are also depicted as pectic AGPs due to the observation that RG-I in Arabidopsis exists in this form (Tan et al. 2013, Tan et al. 2023b). The backbones of the polymers are depicted, but it is understood that RG-I will have diverse side chains depending on the tissue and developmental stage. Future studies of pectin synthesis and mutant phenotypes are required to test the hypothesized pectic polymeric structures.
Figure 8.
Pectic heteroglycan and glycoconjugate model. In this testable hypothetical model, pectin in the primary wall is proposed to consist of 5 different pectic polymeric structures: a small amount of HG, the pectic heteroglycan HG-RG-II-HG; the pectic heteroglycan HG-RG-I-HG, the pectic glycoconjugate pectic-AGP, and the possible complex pectic heteroglycan HG-RG-I-HG-RG-II-HG representing the covalent connection of the separate glycan and heteroglycan polymers. These 5 pectic polymers may interact via ionic interactions and covalent boron diester cross-linking. Representative methyl and acetyl esterification of the HG backbone is shown but no specific pattern is implied. No side glycan or single sugar side branching is shown but is implied as in Fig. 5. RG-II dimers could occur between any 2 adjacent RG-II molecules. The model is not meant to be quantitatively accurate but rather to denote possible heteroglycan and glycoconjugate structures.
Pectin location
Plant biologists soon recognized that pectins were the main components in the middle lamella between cells and were also present in the PCW, along with cellulose, hemicelluloses, and to a lesser extent lignin (Mangin 1893a, 1893b; Kerr and Bailey 1934). Some, including Mangin (1893b) and Bonner (1936), suggested that pectins in the middle lamella existed as calcium pectate, whereas others concluded that they existed as “protopectin” and/or a pectin-protein complex (Sloep 1928; Kertesz 1951). More recent results with antibodies recognizing RG-I (Moore and Staehelin 1988) and transgenic plants expressing reduced levels of pectin metabolic enzymes clearly show that the pectin in the middle lamella contains not only HG but also RG-I (Molina-Hidalgo et al. 2013).
Although less appreciated, considerable data indicate a function for pectins in walls that span the primary-secondary wall definition. There are multiple examples of alternating layers of pectins and cellulose in so-called collenchymatous secondary walls (Anderson 1927; Bonner 1936; Chen et al. 2019), a location consistent with GAUT1 expression in both primary and secondary (xylem) walls in Arabidopsis (Atmodjo et al. 2011). A microscopic study of freshly cut tomato (Solanum lycopersicum) (Anderson 1927) provided early evidence for a lamellar wall structure and cellulose-pectin interactions, including alternating layers of protopectin and cellulose in collenchymatic secondary walls (Fig. 9A), a finding confirmed in celery collenchyma walls, walls defined by the authors as thickened PCWs because the cells were still elongating (Chen et al. 2019). Solid-state NMR revealed reduced mobility for pectins and cellulose as the walls matured and expansion ceased (Chen et al. 2019) (Fig. 9B).
Figure 9.
Microscopic evidence for alternating pectin-cellulose layers in collenchyma cells and fibrillary nature of pectin. A) A single tomato collenchyma cell wall after treatment with potassium iodide and sulfuric acid to remove pectin and expose cellulose lamellae, ×300 magnification (from Fig. 10 from Anderson 1927). B) TEM images of longitudinal sections of celery petiole collenchyma cells showing lamellae of cellulose MFs stained with uranyl acetate and lead citrate. Pm: plasma membrane (from Fig. 1i from Chen et al. 2019). C) Fast-freeze, deep-etch rotary shadowed (FDR) replica image of tomato suspension cells adapted to growth on 12 µM DCB showing pectin fibers in wall (from Fig. 3E from Wells et al. 1994).
It has been proposed that the specialized secondary walls in fibers in G layers of tension wood, extraxylary flax (Linum usitatissimum), and hemp (Cannabis sativa) fibers, and fibers of climbing plants are different from other secondary walls in that they have a very high cellulose content (85% to 90%), a very low amount of lignin and xylan, and the presence of a unique RG-I highly substituted with galactan (Gorshkova et al. 2022). In such fibers, RG-I is first synthesized with long galactan side chains and inserted between cellulose MFs, preventing lateral association of neighboring MFs. Over time, β-1,4-galactosidase cleaves the galactan chains and RG-I lyase cleaves the backbone, enabling lateral association of the cellulose fibrils and leading to tension that can result in contractile properties of the fibers. These results indicate that pectins have roles in unique secondary walls and in promoting stems to bend and climb.
Pectins in cell:cell adhesion and fruit ripening
Fremy (1847, 1848) proposed the existence of pectose/protopectin in vegetables, fruits, and roots as an insoluble pectin that could be converted into the soluble pectins (reviewed in Bonner 1936) and concluded that what most people studied as pectin was a portion of the original protopectin in plant tissues. Fremy (1848) and Carre (1922) also attributed the thinning of fruit cell walls during ripening to the “decomposition of the insoluble protopectin and the formation of soluble pectin” (Bonner 1936; Kertesz 1951), and enzymes were identified that catalyzed such degradation. Thus, already in the 1800s and early 1900s, functions of pectins were recognized in cell:cell adhesion and fruit texture, and it was understood that easily extractable pectins represented only a portion of pectins in the wall. Both HG (Shedletzsky et al. 1992; Liners et al. 1994; Sobry et al. 2005) and RG-I (Yang et al. 2020a) have since been confirmed to function in cell:cell adhesion, leading to the question of whether they function together in a given cell type or have unique roles in different cell types and tissues. The differential abundance of HG and RG-I in growing vs nongrowing cells, respectively, would support the latter (Mohnen et al. 2024).
Pectin fibers
For more than 50 years, extensive evidence has indicated the existence of pectin fibrils or filaments, yet most modern models of pectins do not represent such structures. In 1951, Roelofsen and Kreger showed, using polarization microscopy, diffraction, and electron microscopy, that pectins in collenchyma cell walls of butterbur (Petasites hybridus) existed as oriented fibrils and not, as had been previously held, as amorphous and optically isotropic (Roelofsen and Kreger 1951). Staehelin and Pickett-Heaps (1975) reported pectinase-sensitive fibrils with a 3- to 13-nm diameter in the Chlorophyceaen green alga Scenedesmus. Shortly thereafter, Leppard and Colvin (1972) identified electron opaque ruthenium red-staining pectin fibrils in root epidermal cells. More recently, pectic nanofibers have been demonstrated in Arabidopsis epidermal pavement cells, and roles for the fibers in cell expansion have been proposed (Haas et al. 2020). The evidence for pectic fibers highlights the question of the tertiary and quaternary structure of pectins, and more specifically HG. As there is increasing evidence for pectins associating with and between cellulose MFs and fibers (Zykwinska et al. 2005; Lin et al. 2016), the importance of understanding pectin structure and architecture in the wall and in association with cellulose is increasingly important. Recently, resonant X-ray scattering of onion epidermal cell walls revealed a 20-nm spacing of cellulose MFs with a pectin matrix between the fibrils (Ye et al. 2018).
Pectins in cell growth and expansion
Tomato cells adapted to growth on DCB are able to divide and expand with a “virtual absence of a cellulose-xyloglucan network” (Shedletzky et al. 1990). Subsequent studies of the DCB-adapted cells revealed enhanced wall-membrane connections and an accompanying reduced ability to plasmolyze the cells, an ability that could be recovered by treatment of the cells with 50 mm CDTA and a pure endopolygalacturonase. These results implicated calcium-homogalacturonan salt bridges as load-bearing components that could support growth of the DCB-treated cells (Shedletzky et al. 1992). Imaging of the DCB-adapted cells provided a visualization of a fibrillar pectin network in the absence of the cellulose-xyloglucan network (Wells et al. 1994) (Fig. 9C). The pectin load-bearing hypothesis was supported by lysis of tobacco and tomato DCB-adapted cells by overnight treatment with CDTA, a lysis that did not occur in non-adapted cells. Although the tensile strength of the DCB-adapted cells was less than one-half that of normal cells, as expected due to the drastic reduction in cellulose, the ability of the calcium cross-linked pectate (i.e. HG) to provide sufficient load-bearing to keep the cell intact and enable growth provided strong evidence for a role of a pectin cross-linked matrix in cell expansion and growth in dicot cells (Shedletzky et al 1992). Further evidence of a role for pectins in cell expansion is presented below (see Models of Cell Wall Organization section).
Wall-associated proteins
Types of cell wall proteins
Plant cell walls contain a diverse collection of proteins, glycoproteins, and proteoglycans that perform an array of different functions (Albersheim et al. 2011). These can be organized into 2 general classes, insoluble and soluble wall proteins, with each class further subdivided into multiple categories (Albersheim et al. 2011). The insoluble proteins are tightly bound to the cell wall and are generally considered to play structural roles; these include large families of hydroxyproline-rich proteins (HRGPs), extensins (EXTs), proline-rich proteins (PRPs), and glycine-rich proteins (Dougall and Shimbayashi 1960; Lamport and Northcote 1960) and have been extensively reviewed (Varner and Lin 1989; Showalter 1993; Kieliszewski and Lamport 1994; Ringli et al. 2001; Showalter et al. 2010; Lamport et al. 2011; Moussu and Ingram 2023). The soluble proteins include enzymes, lectins, transport- and defense-related proteins, as well as hydroxyproline-rich arabinogalactan proteins (AGPs) (Silva et al. 2020). The AGPs may be insoluble or soluble, depending on whether they are covalently attached to wall polysaccharides (reviewed in Tan et al. 2023a, 2023b). Cell wall enzymes include glycan exo- and endo-hydrolases and lyases, transglycosidases, methyl and acetyl esterases, peroxidases and laccases involved in lignin polymerization, enzyme inhibitors, lipid transport proteins functioning in cutin and wax synthesis, and proteases.
Another way to classify the large superfamily of plant cell wall HRGP proteins is via their level of glycosylation, with AGPs being hyperglycosylated, extensins moderately glycosylated, and PRPs lightly glycosylated. The AGP proteins contain noncontiguous Hyp residues and are about 90% carbohydrate by mass, while the extensins have regions of contiguous Hyp residues such as Ser-Pro-Pro-Pro-Pro motifs that become hydroxylated and arabinosylated with 1 to 4 Araf residues (Kieliszewski et al 1992a, 1992b; Petersen et al. 2021). The “Hyp-contiguity hypothesis” developed by Kieliszewski and Lamport (1994) proposes that contiguous Hyp residues [Ser-Hyp4] (as found in extensins) are sites for short arabinose oligosaccharide addition, whereas repetitive noncontiguous Hyp residues (as found in AGPs) are sites for Type II arabinogalactan attachment.
Because it is difficult to identify the large classes of insoluble proteins by BLAST analyses of amino acid sequence due to overlapping protein sequence motifs and arrangements, Showalter et al. (2010) developed a multifaceted bioinformatics approach and identified 166 Arabidopsis HRGPs that grouped into 85 AGPs, 59 EXTs, 18 PRPs, and 4 AGP/EXT hybrids. In poplar, with its expanded genome and large amount of woody secondary walls, a similar approach identified 271 HRGPs, including 162 AGPs, 60 EXTs, and 49 PRPs. The bioinformatics analyses identified and distinguished EXTs with variations of the X-Hyp4 sequences including SP3, SP4, and SP5. There are also EXT hybrid proteins with additional domains such as the PERKS (proline-rich extension-like receptor kinases), formins with an EXT motif and actin binding domain, hybrid-EXTs such as the LRR-EXTs (leucine rich repeat EXTs), and hybrid AGP-EXT (Borassi et al. 2016). Many of the prolines in the repetitive domains of HRGPs are hydroxylated, and these may, depending on the repetitive amino acid motif, be heavily or partially glycosylated. Pro-Hyp-Val-Tyr-Lys regions in HRGPs appear to be involved in intermolecular crosslinking, adhesion, and cohesion, and Tyr-X-Tyr-X in isodityrosine and pulcherosine covalent cross-linking and self-assembly catalyzed by class III peroxidases (Kieliszewski and Lamport 1994; Showalter et al. 2010; Mishler-Elmore et al. 2021). Analysis of coexpressed GTs, prolyl 4-hydroxylases, and peroxidases have provided candidate enzymes involved in glycosylation, proline hydroxylation, and crosslinking, respectively, and progress is being made in delineating the biosynthetic pathways for these post-translational modifications that are critical for HRGP function (Silva et al. 2020).
The AGPs are a diverse, highly glycosylated, multi-functional subgroup of HRGPs. Tony Bacic and his colleagues in Australia were major contributors to our understanding of this interesting group of complex glycoproteins (Ellis et al. 2010). They are found across all plant species, including algae, bryophytes, and ferns (Seifert and Roberts 2006; Silva et al. 2020; Ma and Johnson 2023; Tan et al. 2023a). AGPS are classified as classical AGPs, lysine-rich AGPs, arabinogalactan peptides, fasciclin-like AGPs, plastocyanin AGPs, and chimeric AGPs (Showalter et al. 2010; Shafee et al. 2020). Of the 85 AGPs in Arabidopsis, 56 were predicted to have a C-terminal GPI anchor sequence (Showalter et al. 2010) directing them to the outer leaflet of the plasma membrane, where it is hypothesized that they remain tethered unless cleaved from the wall by phospholipases. The term GPI anchor is a misnomer because the plant glycolipid contains an inositol ceramide rather than phosphatidylinositol as found in animals (Silva et al. 2020).
The AGPs are glycosylated on selected Hyp residues by large β-1,3-galactan and β-1,6 branched side chains (Type II arabinogalactans) that can be further decorated with sugars such as Araf, Rha, Gal, Fuc, and GlcA. Some AGPs, such as Arabidopsis AGP57C, also have covalently attached pectic glycan RG-I, which is further elongated with HG producing so-called pectic AGPs, and some of these also contain covalently linked xylan (Tan et al. 2013, Tan et al. 2023b) (see Fig. 6). Classic AGPs can be precipitated with β-Yariv reagent, which has been used to detect AGPs in the wall, but pectic AGPs are not reactive with Yariv reagent due to their complex glycosylation. AGP glycosylation is species and tissue dependent with potential multiple AG chains per protein core and with 65 to 142 sugar residues per AG chain reported (Tan et al. 2023a). Although progress continues in defining specific AGP glycosyl structures, we know very little about the variation of specific AG side chain structures within a given cell type or in different tissues or differences in glycosylation of different AGP protein cores. AGP glycan structural diversity is due to the specificity of different GTs and to any glycan processing by hydrolases that may occur within the Golgi complex, as recently suggested for Golgi localized CAZy GH43 β-1,3-galactosyl hydrolase (Nibbering et al. 2020).
Functions of cell wall proteins in vivo
The AGPs are the largest family of cell wall proteins and perhaps the most multi-functional, with roles including plant growth and development, cell expansion, division, signaling, embryogenesis, vascular and gametophyte development, and biotic and abiotic stress response (Seifert and Roberts 2006; Silva et al. 2020; Lin et al. 2022a; Leszczuk et al. 2023; Ma and Johnson 2023; Tan et al. 2023a). Mechanisms for how they may carry out these functions include mediation of information between the cell wall, plasma membrane, and cytoplasm in part due to their anchoring to the plasma membrane and covalent and/or noncovalent interactions with wall carbohydrates and/or proteins. The amount of glucuronidation of the AGP has been associated with calcium binding, signaling, and salt tolerance (Lopez-Hernandez et al. 2020); covalent connection of AGPs to pectin has been implicated in the establishment and maintenance of wall structure and cell expansion (Tan et al. 2013; Biswal et al. 2018; Tan et al. 2023b). AGP signaling mechanisms include indirect signaling through effects on wall properties, action as co-receptors, and activity of the arabinogalactan carbohydrates as signaling molecules (reviewed in Seifert and Roberts 2006; Ma and Johnson 2023). For example, an arabinogalactan 4-methyl-GlcA-β-1,6-Gal terminal AGP epitope from Torenia fournieri mature ovules, known as AMOR, has been shown to induce competency of pollen tubes to respond to attractant peptide signals from the synergid cell, thereby identifying specific AGP glycans as signals required for guidance of pollen tubes and pollination (Mizukami et al. 2016), although the specific AGP(s) on which the AG signaling glycan is present remain to be determined. A mechanistic understanding of the roles of AGPs in cell wall structural integrity and cell wall signaling, however, remains fragmentary for most AGPs. A key area of research for the future is to understand how different AGPs and other HRGPs interact in the wall with each other and with carbohydrates, lignin, and proteins, and in those cases where the AGPs are direct signaling molecules to identify their ligand and receptor/co-receptor partners and signaling pathways. Physiological roles for some of the AGPs are just now being identified through molecular genetic analyses of biosynthetic mutants (reviewed in Moreira et al. 2023).
As a group, the structural wall proteins are thought to be part of a cell wall network that functions within or is associated with the cellulose-hemicellulose-pectin carbohydrate network, thereby providing or contributing to mechanical strength and possibly to assembly or architecture of the wall (Fig. 10). It has been proposed that EXTs with their positive charge may interact with pectins to form a protein-carbohydrate polymer gel-like matrix and possible scaffold with the potential to respond to pH and ionic conditions in the wall (Tierney and Varner 1987; Lamport et al. 2011). The repetitive motifs and structures of the EXTs (and other HRGPs) certainly support structural roles (Mishler-Elmore et al. 2021), but the difficulty of extracting them from walls due to cross-linking with the protein, carbohydrate, and lignin components has made this difficult to prove (Qi et al. 1995; Nuñez et al. 2009). In no case has a specific structural role of EXTs in the wall been unambiguously confirmed (Moussu and Ingram 2023). However, recently LEUCINE-RICH REPEAT EXTENSIN (LRX8) in pollen tubes was shown to form a complex with the peptide RAPID ALKANIZATION FACTOR 4 (RALF4), thereby exposing a positive patch on RALF4 that bound HG and stabilized a reticulate cell wall architecture (Moussu et al. 2023) that strengthened the pollen tube wall during growth. A similar case for root hairs involving LRX1 and RALF22 is discussed in more detail in the signaling section (Schoenaers et al. 2023).
Figure 10.
Evidence for roles of structural cell wall proteins in wall structure, assembly, and architecture. A to C: Confocal microscopy 3D reconstruction of protoxylem (PX) in apical region of soybean hypocotyl immunolocalized with anti-GRP1.8 polyclonal rabbit antibody (green fluorescence) and lignin (red fluorescence). A) PX with ring-shaped lignified SCW and GRP structure. B) PX with helical lignified SCW and GRP structure present during passive PX elongation. C) Two PXs with younger one (left) with closely spaced SCW and no GRP structure and older one (right) with more widely spaced SCW and GRP structure (taken from Ryser et al. 2003, Fig. 1, A–C). D) In vitro self-assembled Arabidopsis EXT3 network imaged by AFM (taken from Cannon et al. 2008, Fig. 5).
Transcript expression analyses suggest that most EXTs, about one-half the AGPs, and a few of the PRPs have organ-specific expression, suggesting that some HRGPs may have roles within most cells whereas others have cell-, tissue-, or organ-specific function. Immunolabeling of wall proteins has revealed cell type and sub-cell wall–specific labeling for some HRGPs. For example, the Arabidopsis proline-rich cell wall protein AtPRP3 is preferentially expressed in root hair–bearing epidermal cells at the root/shoot junction and in the root differentiation zone of light-grown seedlings (Bernhardt and Tierney 2000). Molecular genetic analyses support a role for AtPRP3 in cell wall structure in differentiating root hairs (Bernhardt and Tierney 2000; Albersheim et al. 2011).
Other HRGPs are localized to SCW-rich tissues. In etiolated soybean hypocotyls, bean GRP 1.8 localizes to protoxylem, the water-conducting tissue of young and elongating tissues (Keller et al. 1989; Ryser 2003). Deposition of GRP 1.8 begins in cell corners between protoxylem elements and develops further to interconnect secondary walls between adjacent protoxylem and form a GRP structural wall between 2 dead protoxylem cells. It also connects protoxylem elements and xylem parenchyma cells in a 3-dimensional GRP network that stabilizes the whole protoxylem by interconnecting ring and spiral-shaped secondary wall thickenings of the protoxylem and the middle lamella of living xylem parenchyma cells, preventing premature collapse of the protoxylem. Confocal laser scanning microscopy of pectinase-softened hypocotyls using anti-GRP1.8 antibodies identified the GRP structure as a distinct structural element arranged in the longitudinal axis of the protoxylem elements (Fig. 10, A–C) (Ryser et al. 2003) that is proposed to function in stabilizing cell corners and anchoring and stabilizing protoxylem elements (Ryser 2003). The GRP structural element is closely associated with an RG-I and HG pectin network (Ryser 2003; Ryser et al. 2003), providing further evidence for a function of pectins in secondary walls and a role for passive elongation of protoxylem cells (Fig. 10B).
In addition to providing mechanical strength to walls, EXTs are proposed to self-assemble into a network structure that may serve as a positively charged scaffold for pectin in the wall (Fig. 10D) (Cannon et al. 2008) (see “Life of a Cell Wall” below), be involved in wall repair and response to microbial interactions, and be induced when plants are wounded and/or attacked by pathogens (Petersen et al. 2021). Although the name extensin was coined by Lamport (1963) with the proposition that such proteins may be involved in cell expansion, there are many reports in which secretion of extensin into the walls is correlated with the cessation of cell growth (Sadava et al. 1973; Mishler-Elmore et al. 2021). EXTs have also been associated with stresses, including ethylene, wounding, and cold and hot temperatures (Swords and Staehelin 1989).
Lignin
Structure and function of lignin
Lignin is a major structural component of plant SCWs. As mentioned above, the cell walls Robert Hooke first described in the 1660s were from cork, and their shape was immortalized by the recalcitrant layers of what was later shown to be suberin, a complex polymer of polyunsaturated fatty acids esterified to phenolic units that is likely anchored to lignin. Suberin makes up 40% of the cork cell wall and lignin comprises another 20%. The term lignin was coined by the Swiss taxonomist Augustin Pyramus de Candolle in 1813. He had isolated a fibrous plant material that was insoluble in water but soluble in alkaline solutions and named it lignin based on the Latin name for wood (lignum). The underlying chemical structure of lignin as a biopolymer of coniferyl alcohol units joined through ether linkages was proposed by Peter Klason in 1897, and the residue after total acid hydrolysis of the carbohydrate components of wood is still termed Klason lignin. In a series of seminal papers between 1940 and 1970, Freudenberg and colleagues established, through chemical analysis and precursor labeling experiments, that lignin was formed primarily from hydroxyphenyl, coniferyl, and sinapyl alcohols (Fig. 11) derived from the amino acid L-phenylalanine linked through free radical coupling (dehydrogenation) (Freudenberg 1959; Freudenberg and Neish 1968). Studies in spruce and magnolia showed that gymnosperm lignins are composed almost exclusively of guaiacyl (G) units, whereas angiosperm lignins are primarily derived from G and syringyl (S) units, with a much lower proportion of hydroxyphenyl (H) units. As a result of their assembly by non-enzymatic polymerization, there is no exact structure for lignin chains. Earlier models suggested very large 3-dimensional structures with a high degree of cross-linking (Freudenberg and Neish 1968), whereas later models favor shorter, more linear, less branched chains (Ralph et al. 2019). The extent of cross-linking will depend on the S/G ratio based on chemical coupling preferences that can be illustrated with in vitro monolignol polymerization systems (van Parijs et al. 2010).
Figure 11.
Structure, monomer composition, and linkages of lignin. A) A generic lignin molecule showing the major monomer types and the varieties of linkage type. β-O-4 linkages are the most common and the easiest to break. Carbon-carbon linkages such as 5-5´ are hard to break and contribute to the recalcitrance of lignin toward degradation. Reproduced from Zakzeski et al. (2010). B) The 4 naturally occurring monolignols.
The linkages between the H, G, and S monolignols can be determined by analysis of partial degradation products or, more conveniently, by NMR (Marita et al. 1999). The most common linkage is the β-O-4 linkage between the free 4-OH group of the H, G, or S units and the side chain C2 position of another unit (Fig. 11A), and cleavage at this linkage is the basis for the determination of lignin content and composition by the commonly used technique of thioacidolysis (Lapierre et al. 1985). The several other linkages in lignin are not cleaved by this method. This highlights the problems in the quantitative measurement of lignin; the methods with the most chemical precision are less able to sample the whole molecule, whereas those that rely on simple gravimetric or UV/VIS spectroscopic measurement are prone to interference from contaminants. Conveniently, lignin shows blue autofluorescence under UV light, although similar fluorescence is shown by wall-bound hydroxycinnamic acids. Lignin localization in plant tissues can also be determined by Weisner (phloroglucinol) or Maule (permanganate) staining, the latter showing some specificity for S-lignin, although neither is fully specific or quantitative (Lewis and Yamamoto 1990). Raman spectroscopy provides a means to both visualize and quantify lignin under the microscope (Agarwal and Ralph 1997; Zeng et al. 2010). These various approaches to lignin visualization, along with examples of lignification patterns in stems of both monocots and dicots, are shown in Fig. 12.
Figure 12.
Lignin detection in situ. A) Cross section of an Arabidopsis stem viewed under UV light. Lignin autofluorescence is blue, chlorophyll autofluorescence red. B) Higher magnification of a cross-section of an Arabidopsis stem stained with phloroglucinol-HCl. C) As in B) but stained with Maule-reagent. The S-lignin–rich interfascicular fibers stain red, whereas the G-lignin–rich vessel elements stain yellow-brown. A to C are from Wang et al. (2010). D) Phloroglucinol staining of a cross-section of a transgenic maize stem with ectopic lignification, showing the different organization of fibers compared with the dicot Arabidopsis. E) Cross section of a switchgrass stem viewed under UV light. F) Higher magnification of the vascular tissue of a switchgrass stem stained with phloroglucinol-HCl. Bs, bundle sheath cell; Ph, phloem; Ve, vascular element; xy, xylem. D to F are from Gallego-Giraldo et al. (2015). G) Forward coherent anti-Stokes Raman scattering (F-CARS) microscopy of a cross section through the epidermal tissue, cortex, and inter-fascicular fiber region of a wild-type alfalfa (Medicago sativa) stem. The highest CARS signal is in vascular tissue and can be quantified. From Zeng et al. (2010). H) Cross-section through the stem of an alfalfa line undergoing ectopic lignification as a result of downregulation of a WRKY transcription factor. Arrows show lignification in the cell corners of lignified and lignifying cells (L. Gallego-Giraldo and R. A. Dixon, unpublished results).
It is perhaps surprising that more is not known of the interactions between lignin and the other cell wall components. The current picture is informed by both direct measurements and inferences from indirect approaches such as genetic modification of lignin or other cell wall polysaccharides. It has been generally assumed that lignin assembles on hemicelluloses, and sequential subcritical water extraction of hemicelluloses from birchwood revealed the presence of specific xylan domains that appeared to interact with lignin (Martínez-Abad et al. 2018). This has been confirmed by solid-state NMR measurements of maize stems, where the lignin was shown to be present in hydrophobic nanodomains with electrostatic interactions with the polar motifs of xylans that are not strongly associated with cellulose (Kang et al. 2019).
Sequential glycome profiling of cell wall residues of stems of transgenic plants with altered lignin content and/or composition has shown that both rhamnogalacturonan and xylan epitopes become more water-soluble upon reducing lignin content (Gallego-Giraldo et al. 2018, 2020). Furthermore, biomass from stems of Arabidopsis with reduced lignin content becomes an effective substrate for growth of strains of the thermophilic bacterium Caldicellulosiruptor bescii lacking the pectinase gene cluster that is normally required for growth on plant biomass (Gallego-Giraldo et al. 2020). These studies suggest that either lignin is directly associated with pectins and xylan, thereby protecting their accessibility to mild solvent, or that lignin modification brings about active cell wall remodeling leading to greater solubility of other wall polymers, as further discussed below.
It is generally accepted that lignin is found in all higher plants but not in the mosses. Lignin has been identified in at least 1 red alga, where it has been ascribed a role in strengthening cell walls in the genicular tissue linking articulated fronds (Martone et al. 2009). Through studies on delignification of fibers, it has been shown that lignin contributes to tensile strength but has limited impact on bending strength (Wang et al. 2017). Other than during the early stages of lignin deposition in the middle lamella, only plant SCWs are lignified, and such walls are found in xylem, interfascicular fibers, Casparian strip in roots, and some seed coats. Lignin is also found in various other cell types in smaller amounts, including in the walls of anthers and in cotton fibers (Gao et al. 2019). The cell biology of lignification has been well reviewed elsewhere (Barros et al. 2015). Based on the location of lignified walls and more recent studies demonstrating the impairments resulting from genetically engineered reductions in lignin content, the primary functions of lignin are presumed to be the imparting of physical strength, allowing plants to maintain an upright habit or to permit dehiscence of fruits and seeds; preserving a hydrophobic environment to allow passage of water in vessels; and sealing of damaged tissues to prevent pathogen ingress.
The chemical recalcitrance of lignin associated with these functions has been an impediment to the utilization of lignocellulosic materials for conversion of the cellulosic and hemicellulosic cell wall components to liquid biofuels (Li et al. 2016a, 2016b, 2016c; De Meester et al. 2022) and for the digestibility of forage crops (Barros-Rios et al. 2018). At the same time, the realization that lignin could replace petroleum products as a feedstock for the synthesis of biomaterials, high value chemicals, and fuels has led to the concept of lignin valorization (Ragauskas et al. 2014; Martins et al. 2022; Ullah et al. 2022). Together, these concepts have inspired great effort to better understand lignin biosynthesis and its regulation over the past 20 years, which has been the subject of a number of review articles (Dixon and Barros 2019; Ohtani and Demura 2019; Ralph et al. 2019; Vanholme et al. 2019; De Meester et al. 2022).
Biosynthesis of lignin
Early models of lignin biosynthesis envisaged a metabolic grid encompassing multiple routes from the first committed precursor, trans-cinnamic acid, to the H, G, and S monolignols. Subsequent models, based primarily on the substrate specificities of recombinant pathway enzymes, suggested a more defined set of routes through the grid (Humphreys et al. 1999), but the complexity and flexibility of the pathway has become more apparent recently through new genetic evidence and the application of kinetic and metabolic flux analysis (Vanholme et al. 2013; Wang et al. 2018; Barros et al. 2022). It is now clear that, in grasses, a significant proportion of the flux into lignin originates from tyrosine rather than phenylalanine (Barros et al. 2016). Figure 13 illustrates the currently accepted routes to the monolignols. The genes encoding all the enzymatic steps have been isolated and most of their functions validated through genetic approaches (Dixon and Barros 2019; Vanholme et al. 2019).
Figure 13.
Biosynthetic origins of lignin building blocks, showing enzymatic reactions involved in lignin biosynthesis from primary metabolism (phenylalanine and tyrosine). Arrows are color-coded to show different classes of reactions: blue, deamination; green, hydroxylation; red, O-methylation; brown, CoA activation; purple, reduction. Enzymes are PAL, L-phenylalanine ammonia-lyase; TAL, L-tyrosine ammonia-lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, (hydroxy)cinnamate CoA ligase; HCT, hydroxycinnamoyl CoA: shikimate hydroxycinnamoyl transferase (HCT); C3´H, coumaroyl shikimate 3´-hydroxylase; CSE, caffeoyl shikimate esterase, C3H, coumarate 3-hydroxylase; COMT, caffeic acid/5-hydroxyconiferaldehyde 3/5-O-methyltransferase; CCoAOMT, caffeoyl CoA 3-O-methyltransferase; CAD, cinnamoyl CoA reductase (CCR); CAD, cinnamyl alcohol dehydrogenase; F5H, ferulic acid/coniferaldehyde 5-hydroxylase.
Unlike the other cell wall polymers, lignin shows a degree of flexibility as to its monomeric constituents. This first became apparent from the discovery of noncanonical units such the flavanol tricin, stilbenes, or feruloyl putresceine in the lignin from some species (Ralph et al. 2019), stimulating attempts to introduce additional units with more labile bonds to allow for easier cell wall processing. This was achieved via the engineering of pathways for the formation of artificial monolignol ferulate and the diferuloyl methane curcumin, which were transported to the apoplast and incorporated into lignin, resulting in enhanced saccharification of biomass (Wilkerson et al. 2014; Oyarce et al. 2019). Incorporation of a pathway for formation of p-hydroxybenzoate (pHBA) resulted in a large increase in esterified pHBA in poplar lignin, with potential as a “clip-off” molecule for processing to downstream high value chemicals (Mottiar et al. 2023).
Lignin is polymerized in the apoplast after the pectic, hemicellulosic, and cellulosic components have been laid down following their transport through the Golgi system (pectin and hemicellulose) or synthesis by a trans-membrane molecular “machine” (cellulose). This raises a number of questions concerning the transport of lignin monomers and the initiation of their polymerization. Based on the presence of high levels of coniferin (coniferyl alcohol β-D-glucoside) in xylem sap and its temporal association with lignification in the developing xylem of coniferous trees, along with results of radiolabeling experiments (Terashima and Fukushima 1988), it was proposed that monolignols were transported to the apoplast as their glycosides utilizing membrane transporters and then underwent conversion back to the aglycones through the activity of an apoplastic glycosidase (Tsuyama and Takabe 2015). Biochemical studies suggested the presence of ATP-dependent transporters for coniferin and other monolignol glycosides in membrane preparations from a number of woody species as well as Arabidopsis (Miao and Liu 2010; Tsuyama et al. 2013), but, to date, the only transporter characterized at the molecular level is a low-affinity H-monomer transporter (Alejandro et al. 2012). There is still no genetic evidence for the existence of G- or S-unit transporters, and an alternative hypothesis of passive diffusion down a concentration gradient has been proposed, supported by theoretical thermodynamic models (Vermaas et al. 2019) and by the observation that polymerization to lignin appears to be required for movement of monolignols out of the cytosol to the apoplast (Perkins et al. 2019, 2022; Zhuo et al. 2022). Glycosylation of small monolignol oligomers appears to be required for their intracellular transport from cytosol to vacuole (Dima et al. 2015). A passive diffusion mechanism, or highly promiscuous transporters, are consistent with the ability of “unnatural” monolignols to pass to the apoplast and be incorporated into lignin (Wilkerson et al. 2014; Ralph et al. 2019). The “good neighbor” hypothesis suggests that monolignols may be generated in parenchyma cells adjacent to lignifying xylem elements, then diffusing into and through the apoplast, where lignification is initiated at the cell corners (Fig. 12H) and/or in the middle lamella. In contrast, lignification in interfascicular fibers appears to be cell autonomous (Smith et al. 2013).
The polymerization of monolignols was at first thought to be initiated by the activity of cell wall peroxidases (Freudenberg and Neish 1968), consistent with the cellular localization of these enzymes and the well-studied formation of lignin “dehydrogenation polymers” generated in vitro from monolignols, peroxidase and hydrogen peroxide (e.g. van Parijs et al. 2010). Because the peroxidases are encoded by large gene families (73 members in Arabidopsis) with potential redundancy, it has been difficult to ascribe functions by genetic experiments. It was later proposed that another group of copper oxidases, the laccases, were also likely involved in monolignol polymerization (Sterjiades et al. 1992; Bao et al. 1993). This suggestion has received considerable support from genetic loss of function experiments. For example, lignification is completely absent in stems of the Arabidopsis lac4 lac11 lac17 triple mutant (Zhao et al. 2013). Genetic analysis has, however, now confirmed roles in lignification for some peroxidases (e.g. Zhang et al. 2022a), and the current model proposes involvement of both enzymes in most lignin biosynthesis, with an exception in the Casparian strip of the root, where peroxidase alone is believed to be necessary (Lee et al. 2013).
Assuming passive transport of monolignols to the apoplast, the localization of the enzymes that initiate free radical formation for polymerization could be critical for determining the site of initiation of lignification. Lignification often starts in the cell corners (Turlapati et al. 2011), and Arabidopsis PEROXIDASE 64 localizes to this region, whereas LACCASE 4 is immobile and localized to the thick secondary wall (Chou et al. 2018). In view of the lack of lignin in the lac triple mutant, this suggests that simple initiation of polymerization in the cell corners is insufficient to prime growing lignin chains throughout the wall and that other initiation sites are likely present. Understanding how lignification is initiated in different cell types is a critical question for the future. The pioneering work of Niko Geldner's laboratory has defined the Casparian strip as model for understanding the cell biology and biochemistry of lignin initiation (Barbosa et al. 2019).
Unlike most other plant polymers, lignin is assembled non-enzymatically. The potential lack of any enzymatic control over such an important process led some experts in the field to suggest that the free-radical-based chain elongation might require some kind of template to allow for correct assembly (Davin and Lewis 2000). This view was inspired by the seminal finding of Norman Lewis’ group that oxidative coupling of 2 coniferyl alcohol units to form dimeric lignans was under strict stereochemical control in most plants and that this control was engendered by the co-action of a so-called dirigent protein with the laccase that generated the monolignol radicals (Davin et al. 1997). Dirigent proteins are trimeric proteins with no identified catalytic reaction, but which direct the stereochemistry of radical-based coupling (Davin et al. 1997). They have been found in nearly all plant species, localize to the cell wall, and are often associated with disease resistance (Paniagua et al. 2017). For several years, a heated debate took place as to whether monolignol assembly for lignin (as opposed to lignans) was under chemical control only, as promoted by John Ralph, or required some kind of template that was composed of an array of dirigent sites (supported by Norman Lewis, Simo Sarkannen, and others). The arguments on both sides can be found in Ralph et al. (2008). Although the pervading view now is that lignin chain assembly is essentially purely chemical (and lignin clearly does not possess any recognizable stereochemistry), the idea of a function for dirigent proteins in some aspects of lignification (initiation or localization) is gaining support. For example, maize mutants lacking a specific dirigent protein displayed impaired deposition of lignin in the Casparian strip, leading to reduced salt tolerance (Wang et al. 2022a, 2022b).
The template hypothesis also posited that the H, G, and S monolignols were the only components that could be assembled into “true” lignin, consistent with steric limitations for a template-driven process. The recent discovery that a wide variety of chemically enabled components (tricin, stilbenes, coniferyl, ferulate, curcumin, etc.) can be incorporated into lignin appears to undermine this tenet of the template hypothesis but still leaves open the possibility of specific proteinaceous initiation sites for lignin chain growth. The flavonol tricin has been proposed to serve as an initiation site for lignification in the grasses, although the effects of manipulating its levels through genetic means differ depending on species (Lam et al. 2021). Nevertheless, the flexibility of lignin structure provides an unprecedented opportunity to fine-tune the composition of a major cell wall structural polymer to either assist biomass degradation or promote its valorization.
Multiple structures involved in many functions
Models of cell wall organization
Studies of cell wall architecture by Roberts and McCann in the early 1990s produced a series of fast-freeze, deep etch, rotary-shadowing images of onion parenchymatous cell walls (McCann et al 1990) that greatly influenced thinking about how the cell wall is arranged. Such images revealed multi-lamellate 100-nm-thick primary walls with 10-nm pores and cellulose MFs (possibly with associated hemicellulose) of 5- to 12-nm diameter. Importantly, the measured lengths of the isolated polysaccharides were in the 700 nm range, enough to span the wall and enable cell to cell wall cross-links. Already it was recognized that the “spaghetti type” models of the primary cell wall were problematic. “It is clear that the classical description of cellulose microfibrils embedded in an amorphous matrix (Varner and Lin 1989) is conceptually problematic; microfibrils are cross-linked together; all wall components are long rod-like fibres; only removal of cellulose with cellulase gives an image that could be described as an amorphous matrix. It is the nature of the cross-links that remains to be determined” (McCann et al. 1990, p330).
Any comprehensive model of wall structure must answer the crucial unsolved problem of how the various components found in plant cell walls are organized into a matrix that carries out the diverse functions that walls perform. Despite many efforts, with most models focusing on how growth is accomplished, the current “models” are inadequate. Most are descriptive diagrams that aim to depict important structure-function relationships. To be useful, such models should be sufficiently specific that they allow interpretation of experimental data and allow the formulation of predictions that can be experimentally tested. Only recently have models become sufficiently sophisticated that they allow quantitative explanations and predictions (Zhang et al. 2021c).
Over the years, one of the major uses for cell wall models was to explain the reorganization of wall components during elongation growth, especially auxin stimulated growth (see Du et al. 2020b for a recent review). More than half a century ago, it was demonstrated that elongation could be stimulated in many systems by acidic conditions (Rayle and Cleland 1970). It is now understood that auxin stimulates a plasma membrane–localized proton pump that acidifies the cell wall (Du et al. 2020b). What is still not completely resolved is how low pH reorganizes wall components to allow growth.
One of the first efforts to describe such structure/function relationships was the “multi-net” hypothesis put forward in the early 1950s by Roelofsen and Houwink (1953). Its main emphasis was to explain the changes in orientation of cellulose MFs as plant cells undergo anisotropic elongation. They observed that elementary fibrils near the plasma membrane of cotton hairs, and other cells, were oriented transversely, but as cells elongated, the MFs gradually changed their orientation and were mainly axial in outer layers of the primary walls.
A significant step forward came in the early 1970s with the models emerging from the laboratory of Peter Albersheim, one of the great pioneers of cell wall biochemistry; they incorporated growing knowledge of the structures of wall matrix polysaccharides. One model emphasized the cellulose/XyG/RG-I network as a major source of wall strength that resists the forces of turgor pressure but can be reorganized to allow expansion as cells grow (Albersheim 1975). An early version of this model also included experimentally supported connections of pectic molecules to cell wall proteins (Keegstra et al. 1973). However, the proposed XyG to RG-I connection could not be identified, so the emphasis in subsequent models switched to a cellulose/hemicellulose network (Hayashi 1989). Further refinements were made by others, including describing the differences between Type I and Type II walls (Carpita and Gibeaut 1993), by making the dimensions of the primary wall thickness (50–100 nm) proportional to that of the plasma membrane (7–8 nm) (McCann and Roberts 1991), or by showing the various components in the proper proportions to cellulose content (Somerville et al. 2004). All these models postulate that the strength of the wall is determined by a cellulose/hemicellulose network. Many authors have called this model the tethered network hypothesis (Fig. 14A) (reviewed in Cosgrove 2022). But, as described below, several lines of evidence now argue against its validity (Cosgrove 2014, 2022), so new ideas are needed.
Figure 14.
Variations of the tethered network hypothesis using different matrix polysaccharides. A) The tethered network hypothesis shows CMFs (thick horizontal rods) to be well spaced and connected by XyG (thin lines). Taken from Cosgrove (2022). B) CMFs (thick horizontal rods) linked by both XyG (thin undecorated lines) and pectin (thin line decorated with side chains). C) CMFs (thick horizontal rods) linked via a network of pectic polysaccharides (thin lines decorated with side chains). Panels B and C are taken from Zykwinska et al. (2007).
The tethered network hypothesis popularly featured XyG, the most abundant hemicellulose in the primary walls of many plant species (Keegstra et al. 1973; Hayashi 1989; Albersheim et al. 2011; Hayashi and Kaida 2011). In the context of XyG tethers connecting MFs (Fig. 14A), several ideas were considered as to how the network was reorganized during growth. One popular idea was the action of XyG transglucosylase hydrolase (XTH) proteins that cleave or reorganize the XyG tethers (Rose et al. 2002). More recent evidence argues against a role for XTHs in cell expansion, but they may have roles in plant responses to biotic and abiotic stresses (Ishida and Yokoyama 2022). Another option for reorganizing the network came with the breakthrough discovery of expansins, proteins that stimulate cell expansion in a pH-dependent manner without breaking covalent bonds (McQueen-Mason et al. 1992). Expansin proteins are encoded by a large family of genes in plants (Sampedro and Cosgrove 2005). Despite a great deal of molecular information about expansin proteins, the molecular details of how they achieve elongation growth remains unclear (Cosgrove 2015, 2016).
An Arabidopsis double mutant (xxt1/xxt2; Cavalier et al. 2008) and a triple mutant (xxt1/xxt2/xxt5; Zabotina et al. 2012) with disruptions of genes responsible for adding xylose to the XyG backbone have XyG levels below the limit of detection. But these mutant plants grow and develop with only minor defects, casting doubt on the validity of the hypothesis that XyG plays an important role in defining the mechanical properties of primary cell walls. More recently, an Arabidopsis quintuple mutant with disruptions of all 5 CSLC genes, encoding proteins that synthesize the glucan backbone of XyG, has undetectable levels of XyG but can grow and develop with minor altered phenotypes, including altered root hairs and pollen tube growth (Kim et al. 2020).
It is noteworthy that the presence of aberrant forms of XyG is more detrimental to plant growth and development than the lack of XyG. The Arabidopsis mur3-3 mutant that lacks fucose and has very low levels of galactose on XyG exhibits severe dwarfism and other growth phenotypes (Kong et al. 2015; Xu et al. 2017). But crossing the mur3-3 mutant to the xxt1/xxt2 double mutant lacking XyG restores the normal size of the triple mutant plants (Kong et al. 2015). These findings emphasize the importance of the physical properties of wall polysaccharides in determining polymer interactions.
Several groups have utilized the Arabidopsis mutants lacking XyG to investigate cell wall functions related to expansion growth. The walls in xxt1/xxt2 mutant plants were examined in detail and found to be more extensible than wild-type walls using stress/strain assays but less extensible in creep mediated by α-expansin (Park and Cosgrove 2012b). The same group used multiple different endoglucanases to treat tissues from wild type and xxt1/xxt2 mutant plants followed by examination of wall biomechanical properties. Based on their observations, they concluded: “Our results are incompatible with the common depiction of xyloglucan as a load-bearing tether spanning the 20- to 40-nm spacing between cellulose microfibrils….” To explain their observations, they postulated that “…XyG-mediated control of wall extension may be restricted to relatively inaccessible contact surfaces between microfibrils of the primary cell wall” (Park and Cosgrove 2012a, p1933. Later publications referred to these contact surfaces as “biomechanical hotspots” (Cosgrove 2022). One version of the hypothesis assumes that residual XyG is present in the walls of the mutant plants, which has not been demonstrated. Another possibility for the “biomechanical hotspots” is that they consist of direct interactions between cellulose MFs as described by Cosgrove (2022). In this case, the role of matrix polysaccharides, whether XyG, other hemicelluloses, or pectins, is to prevent MF from coalescing into bundles that form a rigid inflexible network. A different strategy was used by Kuki et al. (2020), who prepared protoplasts from the leaves of Arabidopsis xxt1/xxt2 mutant plants. As protoplasts regenerated cell walls, they observed “…only a slight difference in the structure of cellulose microfibril network between xxt1/xxt2 and wild-type (WT) protoplasts.” From these and other observations, they concluded that “…xyloglucan is not essential for the initial assembly of the cellulose network, and the cellulose network formed in the absence of xyloglucan provides sufficient tensile strength to the primary cell wall regenerated from protoplasts.” (Kuki et al. 2020, p. 1)
One possible explanation of the ability of plants to survive the lack of XyG is that other wall molecules can substitute for it, that is, functional redundancy. Sowinski et al. (2022) explored this possibility by performing a detailed analysis of the composition of the xxt1/xxt2 double and xxt1/xxt2/xxt5 triple mutants. They observed a small increase in the levels of glucomannan, but significant increases in the levels of pectic polysaccharides. Interestingly, these increases were not caused by any major increases in transcripts responsible for biosynthesis of these polymers. Yu et al (2022) have suggested that a specific galactoglucomannan may function like XyG. Mutants lacking the ability to make this mannan are nearly normal, but when crossed with the xxt1/xxt2 mutant, the triple mutant has more severe phenotypes than mutants lacking either polymer (Yu et al. 2022) but is still able to grow and complete its life cycle.
Despite the evidence against the tethered network involving cellulose and XyG, there are several reasons why it has proven difficult to abandon this model. First, the structure of XyG is highly conserved in all land plants, arguing that it has an important conserved function. Second, XyG is abundant in plants with Type I PCWs, although its abundance varies greatly from one species to another and sometimes within the tissues of a single species, and its abundance is low in plants with Type II PCWs. Yet despite the abundance of XyG and its conserved structure, no one (including the authors) have yet proposed a plausible hypothesis for its role in PCWs.
Another version of the tethered network hypothesis proposes that pectic polysaccharides either share duties with XyG (Fig. 14B) or replace it (Fig. 14C) in forming tethers between cellulose MFs (Zykwinska et al. 2007). Although this hypothesis has not received the same level of attention, we argue that this hypothesis, or some variation of it, deserves attention considering the phenotypes of the XyG-deficient mutants.
During his thesis work in the laboratory of James Bonner at Caltech, Peter Albersheim explored the possibility that auxin enhances cell elongation by changes in the metabolism of pectins (Albersheim and Bonner 1959; Jansen et al. 1960). They focused on changes in the extent that galacturonic acid is methyl esterified during auxin treatment of oat coleoptiles. Although primitive by today's standards, this early work examined some of the same ideas for how pectins might regulate cell expansion that are being pursued today.
Boyer and colleagues studied the growing internodes of Chara corallina, a green alga with a higher plant-like cell wall (Domozych et al. 2010). In a series of experiments spanning 13 years, they provided compelling evidence that in Chara calcium pectate wall polymers provide the critical interactions that regulate cell expansion (Proseus et al. 1999, 2000; Proseus and Boyer 2005, 2007, 2012a, 2012b). They proposed a “calcium pectate cycle” model describing calcium-pectate (HG) as the load-bearing component in the Chara wall.
In their review of cell wall mechanics and plant cell growth, Peaucelle et al. (2012) made the compelling argument that the mechanism of cell wall extensibility in Charophycean algae, that is, the coupling of cell wall synthesis to cell wall extensibility through calcium-exchange between existing and newly synthesized calcium-pectate, also exists in terrestrial plants and called that mechanism the “ancient process.” They further proposed that land plants have at least 2 load-bearing constituents, a calcium-pectate component, and a cellulose-xyloglucan network and that the latter is associated with an expansin-dependent process. They proposed that the more recent expansin-based process “amplifies” the calcium-pectate mechanism and “allows faster growth.”
These ideas were explored in a series of studies examining the elongation of dark-grown Arabidopsis hypocotyls (Pelletier et al. 2010; Peaucelle et al. 2015; Daher et al. 2018). In one study, AFM was used along with antibodies that distinguish ME-HG from de-esterified HG (DE-HG) in plants where expression of PME and PMEIs had been manipulated. Although space constraints preclude a detailed explanation of the study, the authors concluded that “…growth symmetry breaking is controlled at a cellular scale by bipolar pectin de-methylesterification…” (Peaucelle et al. 2015, p. 1746. In a follow-up study using the same experimental system, Daher et al. (2018) also found asymmetry in wall elasticity in cells that was coincident with growth anisotropy and correlated with changes in pectin HG de-methylesterification. They concluded that pectin asymmetry in the hypocotyl epidermis contributes to anisotropic growth.
Jönsson et al. (2021) studied the bending of the apical hook after emergence of the hypocotyl. This process is known to be regulated by auxin and involves differential growth on the 2 sides of the hypocotyl. Again, space constraints preclude a complete description of the experiments, but the authors found a “…spatial correlation between asymmetric auxin distribution, methyl esterified HG pectin, and mechanical properties of the epidermal layer of the hypocotyl in Arabidopsis.” (Jönsson et al. 2021, p. 1154)
In a ground-breaking study of expansion in Arabidopsis epidermal pavement cells, Haas et al. (2020) used super-resolution 3-dimensional direct stochastic optical reconstruction microscopy, cryo-scanning electron microscopy, and HG-specific antibodies that distinguish ME-HG versus DE-HG to show that in cotyledon anticlinal walls HG is organized as HG nanofilaments proposed to be similar to the pectate multichain helical crystalline fibers described by Walkinshaw and Arnott (1981a, 1981b). They further showed that the dimensions of DE-HG nanofilaments are 1.4 times greater than the ME-HG nanofilaments and proposed that de-methylesterification of HG nanofilament quaternary structure leads to pavement cell expansion. They call this the “expanding beam” model of pavement cell expansion. They further tested this model through development of a 3D nonlinear finite element method model, which predicted the pavement cell growth observed. Further tests of the model via in silico variation in level of methyl esters on HG by overexpression of PME and PMEI showed that, as expected, overexpression of PMEI led to growth inhibition.
More recently, Haas et al. (2021) hypothesized that enzymatic de-methylesterification of HG to induce calcium-crosslinked HG in the cell wall, in addition to turgor pressure, drives phase separation in cell wall architecture and associated plant cell expansion. Depending on the methylesterification state, HG is either highly partially or relatively poorly charged. As HG appears to be highly methylesterified in vivo when it is synthesized and inserted into the wall, its de-esterification by wall-located PMEs can lead to increases in charge and, when calcium is present, salt bridges between calcium and adjacent HG chains. This HG de-methylesterification leads to associated changes in HG quaternary structure and volume transition (Haas et al. 2020) akin to phase separation.
Based on the evidence summarized above and explained in more detail in various reviews (Peaucelle et al. 2012; Daher and Braybrook 2015; Braybrook and Jönsson 2016; Chebli and Geitmann 2017; Hocq et al. 2017a; Haas et al. 2021; Höfte 2023), pectins play an important role in defining cell shape and regulating growth. In addition, increasing amounts of NMR data provide evidence for abundant interactions between cellulose and pectic polysaccharides (Pérez García 2011; Wang et al. 2015; Wang and Hong 2016; Phyo et al. 2019; Temple et al. 2022). Furthermore, genetic evidence supports an important role for pectins. In contrast to the various mutants lacking XyG that have only minor phenotypes, several pectin-deficient mutants have severe phenotypes (Bouton et al. 2002; Mouille et al. 2007; Du et al. 2020a) and others may be lethal (Caffall et al. 2009). The important conclusion is that pectic polysaccharides must be included in models that attempt to explain how wall structure is reorganized during growth. More detailed knowledge about pectin structure as it exists in PCWs is critical to understanding cell growth.
Current efforts to generate a model of the PCW have similarities to the parable of a group of blind people trying to describe an elephant with each person describing their part: a leg, a trunk, a tusk, etc. The problem is made worse by the recognition that PCWs are dynamic structures with many different functions and stages in their life, as described in the following sections.
Cell wall signaling
Perhaps the biggest surprise over the past few decades has been the realization that the cell wall is not the passive structure we assumed but is actively engaged in signaling processes. Plants are constantly under changing stresses, be they abiotic (e.g. salinity, drought, tensile) or biotic. Because degradation of plant cell walls is often a necessity for establishing infection, it seems intuitive that the cell wall would possess sensing mechanisms for cell wall integrity (CWI) that could recognize damage associated with either pathogen ingress or wounding. Even the act of cell elongation involves processes of wall loosening that may create tensile stress (Wolf 2022). Historically, the field of cell wall signaling developed from 3 distinct approaches: the identification of mobile signals within the cell wall such as cell wall–derived oligosaccharide elicitors of plant defense responses (e.g. Ayers et al. 1976); the manipulation of the level of specific wall polymer types that trigger compensatory changes in wall composition (e.g. Hematy et al. 2007); and genetic screens for genes underlying developmental phenotypes, which revealed several key plasma membrane signaling receptors (e.g. Boisson-Dernier et al. 2009; Guo et al. 2009, Duan et al. 2010).
Communication between the extracellular matrix and the cytosol is largely brought about through the binding of ligands that activate PM-localized receptor-like kinases (RLKs), with their ligand-binding domains located within the wall matrix (Wolf 2022). Plants contain large numbers of such transmembrane receptors, many of which, such as the LRR-receptor kinase resistance (R) genes or wall-associated kinases (WAKs), are associated with the recognition of pathogens (McHale et al. 2006; Cho et al. 2022). Other mechano-sensitive receptors sense physical features of the PM-wall continuum such as turgor or tensile stress (Hamant et al. 2019; Bacete and Hamann 2020), whereas others, such as the class referred to as CrRLK1 receptors, physically interact with cell wall components such as pectin or peptides (Bacete et al. 2018; Zhu et al 2021). Remodeling of the cell wall is the process that closes out the cycle of cell wall signaling. The involvement of jasmonic acid (JA) and salicylic acid (SA), and their cross-talk with other hormones such as ABA and ethylene in control of cell wall remodeling, is well-reviewed elsewhere (Vlot et al. 2009; Tenhaken 2014; Shigenaga et al. 2017; Ali and Baek 2020; Anderson and Kieber 2020; Liu and Timko 2021).
With no intention of covering this complex field in depth, we focus primarily on initial events that occur within the cell wall and the cell wall-PM interface. We discuss causes of loss of CWI, examples of the roles of tensile stresses in growth/CWI signaling, elicitors and some key receptors and co-receptors, and end with examples of signaling in SCWs. A number of recent reviews cover the broader field in more detail (Bacete et al. 2018; Rui and Dinneny 2019; Vaahtera et al. 2019; Anderson and Kieber 2020; Bacete and Hamann 2020; Wolf 2022; Colin et al. 2023). Unless indicated otherwise, studies were carried out in Arabidopsis and focused on PCWs. Because the field is moving so fast, it is almost guaranteed that, by the time this review appears, there will have been exciting new breakthroughs.
Causes of loss of CWI
One of the most dramatic early examples of the amazing plasticity of cell walls in response to stress is that of Shedletzky et al. (1990, 1992) who showed that cultured cells of tomato, tobacco, or the grass Lolium can slowly adapt and compensate for the loss of cellulose caused by growth on the cellulose synthesis inhibitor DCB. The dicot cells compensated by creating walls comprised almost exclusively of pectin and proteins, while the grass, lacking much pectin, largely enhanced phenolic cross-linking of arabinoxylan. Similar studies with cultured Arabidopsis cells using ISX [that inhibits CSC activity but not that of the CSLD proteins that also synthesize cellulose (Yang et al. 2020b; Wu et al. 2023)], showed less dramatic decreases in cellulose, similar increases in pectin, but also upregulation of CSLD5 and a collagen-like, gly-rich protein (Manfield et al. 2004).
Because cellulose is a major load-bearing polymer in PCWs, it is not surprising that short-term inhibition of its synthesis also has notable consequences (Wang et al. 2016a; Kesten et al. 2017; Wolf 2022). These include swelling of expanding cells, inhibition of elongation, changes in levels and composition of pectins, and sometimes but not always, deposition of callose or ectopic lignin. Natural causes of cellulose synthesis inhibition might include tensile stresses that affect mechanosensitive calcium channels (MCAs), disruption of CSC-MT associations, salt/osmotic stress, or many poorly understood conditions relating to the extreme lability of rosette CSCs. Plants can also detect stress-induced alterations in pectin content or structures due to the presence of heavy metals (Muschitz et al. 2015) or high salinity, which can trigger de-methylesterification of loosely bound pectins, MT degradation, and loss of cellulose (Gigli-Bisceglia et al. 2022; Colin et al. 2023). The middle lamella is rich in DE-HG, and mutations in qua1, a GAUT, and qua2, a PMT, can affect cellulose synthesis and other phenotypes due to increased cell separation (Du et al. 2020a). Cell wall–degrading enzymes from pathogens play a key role in releasing small signaling molecules and altering CWI, especially through attack on cellulose and pectins. There is less evidence for sensing in response to altered levels of PCW XyG or structural proteins, although LRXs and Gly-rich proteins have been implicated in signaling (Park et al. 2001; Manfield et al., 2004; Herger et al. 2019). As described later, engineered changes to lignin in SCWs can surprisingly also lead to large effects.
Oligosaccharin elicitors as mobile signal molecules
Studies on defense signaling began with the pioneering work of Peter Albersheim and his students and post-docs on elicitor-active oligosaccharides from plants and pathogenic fungi (reviewed by Côté and Hahn 1994). Not particularly welcomed coming from a young intruder from the cell wall world into the phytopathology community, the Albersheim hypothesis has nevertheless proven largely valid. This work developed from the elicitor hypothesis for induction of plant defense responses by fungal cell wall components but was extended to include plant developmental responses (Augur et al. 1992) and evidence that pectin-derived fragments signal loss of CWI beyond that caused by pathogens (Wolf 2022). Elicitor-active cell wall–derived oligosaccharides include hepta-glucosides from the cell walls of Phytophthora sojae, a range of oligogalacturonides (OGAs) from various sources, xyloglucan oligosaccharides (Ayers et al. 1976; Davis et al. 1986; Fry 1986, 1994), and even a grass-specific MLG-derived elicitor (Yang et al. 2021). Depending on how they are generated, such elicitor molecules are now generally referred to as Damage Associated Molecular Patterns or Pathogen Associated Molecular Patterns (Boutrot and Zipfel 2017; Bacete et al. 2018). The early studies on oligosaccharins were essentially pharmacological, and, although providing reagents for studying defense gene induction, did not provide understanding of their biological significance or mechanism of action. Furthermore, methods for accurately determining the complete repertoire of wall-released molecules were in their infancy. Recent improvements in liquid chromatography-mass spectrometry are now making it easier to find in vivo players and link them to their in vivo bioactivities (Voxeur et al. 2019; Liu et al. 2023).
Recent work on oligosaccharins showed that a family of enzymes called lytic monosaccharide oxygenases (LPMOs) found in pathogens can cleave recalcitrant and crystalline polysaccharides such as cellulose through oxidative attack. One group of LPMOs (AA9) was shown to produce oxidized cellobiose and larger oxidized cellodextrins, and a recent study demonstrated that plants perceive the unoxidized and oxidized forms through different signaling pathways and that a combination of both cellobiose and oxidized cellobiose provided better resistance to necrotrophic infection than either alone (Zarattini et al. 2021).
Peptide elicitors
One major class of peptide elicitors that has been recently recognized as central to cell wall signaling is a family of peptides called RALFs. The first of these was discovered accidentally by Clarence “Bud” Ryan during isolation of the small systemin peptide that induces wound responses in Solanaceous plants; during isolation, a different, larger 5-KDa peptide was also discovered that caused even more rapid alkalinization of the apoplast than systemin (Pearce et al. 2001). RALF peptides are now known to have diverse signaling roles related to alteration of apoplastic pH and growth and/or defense (Haruta et al. 2014; Xiao et al. 2019; Blackburn et al. 2020). Some RALF peptides are generated and activated by proteolysis of propeptides, whereas others have no propeptide sequence or cleavage site, characteristics that can affect the nature of their signaling activity (Stegmann et al. 2017). Another small family of peptides in Arabidopsis referred to as Peps also plays a key role in immune and other types of signaling (Bartels and Boller 2015; Dressano et al. 2020) leading to classic responses such as elevation of intracellular Ca2+, ROS, and production of defense-related hormones such as JA and SA. In a positive feedback loop, they are generated from propeptides by Ca2+-dependent metacaspases (Shen et al. 2019).
Plasma membrane receptors
To understand the receptors for CWI signaling that have been discovered in recent years, we first need to meet a cast of classical Greek and Etruscan characters, starting with FERONIA (FER), named after the Etruscan goddess of fertility, and others to be discussed later such as THESEUS1 (THE1), ERULUS (ERU), and HERCULES (HERK). These are members of a clade of 16 PM-localized proteins referred to as CrRLK1Ls that are within the superfamily of RLKs (Cheung and Wu 2011; Nissen et al. 2016; Zhu et al. 2021). CrRLK1L PM-localized receptors are characterized by their extracellular motifs resembling sugar-binding domains called malectins coupled to an intracellular kinase domain (Nissen et al. 2016). Depending on what ligands are bound, the receptors often interact through phosphorylation of guanine nucleotide exchange factors (GEFs) that enhance the downstream activity of plant-specific small GTPases called ROPs (Berken 2006). Further ROP signaling can lead to many other possible downstream events such as activation or inhibition (depending on the CrRL1K/GEF/ROP combination) of NADPH oxidase to produce ROS and of Ca2+ channels that when activated elevate [Ca2+cyt] that can promote exocytosis of wall precursors and activate other downstream signals that lead to production of JA or SA (Chen et al. 2020a, 2020b; Tang et al. 2022). In turn, these hormones can lead to upregulated expression of proteins involved in defense responses or cell wall remodeling (Vlot et al. 2009; Ali and Baek 2020; Liu and Timko 2021).
FERONIA—a scaffold on which to create diverse signaling pathways
FER was first discovered in studies of pollen/stigma interactions (Huck et al. 2003). Because it has since been implicated in signaling for a multitude of pathways, FER has been described as a “scaffold” that can interact with different ligands (e.g. RALF peptides, OGAs) and/or co-receptor combinations to control different signaling pathways. The remarkable number of pleiotropic responses signaled by 1 FER protein and its partners are reviewed in depth elsewhere (Cheung and Wu 2011; Li et al. 2016a; Vaahtera et al. 2019; Chen et al. 2020a, 2020b; Xie et al. 2022). FER can play a role in inhibiting immune signaling in several ways. Figure 15A shows 1 example of how certain ligand and protein partners inhibit the outcome of PAMP-mediated ROS production that is signaled through a FLS2/BR11/BAK1 complex. Inhibition occurs when the pro-peptide of RALF23 is cleaved, and the resulting RALF23 interacts with the GPI-anchored co-receptor protein LLG1 (Li et al. 2015) that in turn interacts with FER, and this complex in turn inhibits PAMP-induced ROS production and subsequent immune signaling (Stegmann et al. 2017; Xiao et al. 2019) (Fig. 15A).
Figure 15.
Examples of how variation in ligands and receptor/co-receptor combinations control cell wall signaling outcomes. A) A FER-LLG1-RALF23 combination inhibits a PAMP-mediated ROS production and subsequent immunity signaling (Stegmann et al. 2017; Xiao et al. 2019). B) The work of Haruta et al. (2014) demonstrated that RALF1 binding to FER complex (that was later shown to include co-receptor LLG1) leads to phosphorylation and inhibition of AHA2 PM-ATPase. This leads to alkalinization of the cell wall and inhibition of root cell elongation. Later work indicates other RALF1-binding proteins may also be involved (not shown, see Dressano et al. 2017). C) The role of FER and an LRX1-DE-HG complex in regulating root hair elongation. Loss of function of ERULUS also regulates FER and AHA10 phosphorylation (not shown). Relevant references are Duan et al. 2010, Li et al. 2015, Schoenaers et al. 2018 and 2023. D) Examples of different roles for ligands and/or mechano-sensing by THE1, FER, and Ca2+ channels for responses to CWI and for the complex regulation involving RLP12 of hypo- (through JA) and hyper-osmotic stresses (through ABA). Relevant references are Hematy et al. (2007), Engelsdorf et al. (2018), Feng et al. (2018), Hamant et al. (2019), and Bacete et al. (2022). E) Induction of PR proteins as a result of engineered reduction of lignin content. FER senses altered wall integrity to induce the polygalacturonase ADG1, which releases GalA3 and other elicitors from pectin; these are recognized by WAKs resulting in induction of PR1 and PR10 (Gallego-Giraldo et al. 2020; Liu et al. 2023).
Two other examples compare FER's partner combinations for control of the auxin-regulated growth of elongating root cells with the complex and differing configurations used to regulate tip growth of root hairs. A study by Mike Sussman's group showed that when FER-LLG1 associates with RALF1, it leads to suppression of cell elongation through alkalinization of the cell wall through FER phosphorylation-induced inhibition of the PM H+-ATPase AHA2 (Haruta et al. 2014; Li et al. 2015) (Fig. 15B). Root hair tip growth is a somewhat different process that shows oscillations in which the growth phase is promoted by lowered apoplastic pH and elevated ROS gradients as well as the creation of a tip-focused [Ca2+cyt] gradient stimulating actin reorganization and polar exocytosis of wall polymers to the tip (Zhang et al. 2022a, 2022b). One of FER's roles involves its association with LLG1 in root hair tips, activation of a specific associated ROP that further activates NADPH oxidase leading to apoplastic ROS production (Duan et al. 2010; Li et al. 2015; Zhang et al. 2022a, 2022b) (Fig. 15C; right). Another CrRKL1 root hair receptor called ERULUS is auxin induced, positively controls the phosphorylation of FER, and affects pectin dynamics through activation of PME activities (Schoenaers et al. 2018). Ground-breaking work that followed (Schoenaers et al. 2023) showed that RALF22 is expressed specifically in root hairs and primarily located in rings in the cell wall attached to a LRX1 (Fig. 15C, right). These studies also indicate that RALF 22 is not only a signaling peptide but also functions as part of a structural component that regulates the state of HG condensation. From their results, the authors envision an oscillation in which ME-HG, RALF22, and LRX1 are secreted at the tip during the low-pH wall-loosening phase, then RALF22 interacts with FER-LLG1 to inhibit the H + ATPase, and wall pH rises leading to the growth inhibition phase with increased PME activity that leads to a rise in DE-HG. DE-HG then binds to a separate complex of RALF22 with LRX1, leading to wall stiffening and formation of the ring structure that fixes the direction of expansion. The authors hypothesize that the wall stiffening may be a mechanosensory signal to the FER-LLG1-RALF22 complex as part of the growth cycle. More recently, a process strikingly analogous to the novel work described by Schoenaers et al. (2023) for Arabidopsis root hairs was reported for pollen tubes by Moussu et al. (2023; see also Mohnen 2023). In pollen tubes, 2 molecules of RALF4 were shown to interact with 2 LRX8 molecules to form a positively charged heterotetramer at the pollen tube tip that can then interact with DE-HG to form organized structural complexes in the shank wall that play a very similar role to that in root hairs determining wall strength and shape.
Only in retrospect, through collectively re-examining the examples chosen for Fig. 15, did it occur to us that the pleiotropic character of FER must relate to its key role in balancing the strength of the outcomes of signaling pathways for growth (Fig. 15, B and C), hypo-osmotic (tensile) versus hyper-osmotic stress (Fig. 15D), and defense (Fig. 15, A and E). In a recent thought-provoking paper, Malivert and Hamant (2023) discuss the many different phenotypes generated by fer mutants to try to identify a unifying feature that could underlie FER functions. They discuss many interesting options but seem to favor the idea that FER is fundamentally a mechanosensor modulated by different co-receptors and ligands such as pectins/OGAs and RALF peptides leading to changes in pH, Ca2+ and/or ROS that function to modulate the mechanical integrity of the cell.
Other receptors involved in CWI signaling
In a study that has defined an important experimental system for interrogating cell wall signaling, Herman Hofte and colleagues recognized that a number of different mutants of Arabidopsis impaired in cellulose synthesis either through mutations in CesAs or associated proteins such as KORRIGAN or by use of the cellulose synthase inhibitor ISX all showed a set of characteristic responses that include production of ectopic lignin, alterations in levels and structure of pectins, reduction in cell elongation, as well as increases in JA production. In a search for suppressor mutations of Procuste (cesA6), they identified a new, widely expressed CrRLKL1 that they termed THESEUS1 (THE1), “a character from Greek mythology who slew the brigand Procustes” (Hematy et al. 2007) (Fig. 15D). Importantly, they found that loss of THE1 sensing could attenuate the growth effects and production of ectopic lignin caused by the cesA6 mutation and some other, but not all, cesA mutations (in 2007, it was not known that ISX does not inhibit the CSLD CSCs, and this needs to be kept in mind when reading papers that assume ISX is completely inhibiting all the cellulose synthesis). Gonneau et al. (2018) have shown that binding of RALF34 to THE1 promotes cytosolic [Ca2+cyt] transients and cell wall alkalinization similar to that observed for RALF1 and FER (Fig. 15D). The fer 1-4 or the the1/fer 1-4 double mutant lacked such responses to both RALF1 and RALF34 as well as the same effects of both peptides on upregulation of specific genes, suggesting THE1 may act upstream of FER or in parallel in a FER-dependent way.
Mechano-sensing is increasingly recognized as playing a key and perhaps essential role in CWI sensing (Hamant et al. 2019; Bacete and Hamann 2020). Loss of cellulose synthesis can create tensile stress that can affect mechano-sensitive Ca2+ channels, and THE1 acts upstream of the MCA1 ion channel that senses hypo-osmotic stress (Engelsdorf et al. 2018), while FER, through binding of OGAs to its malectin-like domains, promotes activity of a different Ca2+ channel that opens in response to hyper-osmotic conditions (Feng et al. 2018) (Fig. 15D). Such stress can also lead to microtubule disorganization that is controlled by FER-dependent and -independent pathways (Hamant et al. 2019; Malivert et al. 2021) (Fig. 15D). An exciting new study has tied together the functioning of THE1 with the interactions between sensing of CWI affected by swelling and PM stretching akin to hypo-osmotic stress, as opposed to drought or salinity that cause hyper-osmotic stress that causes membrane contraction and plasmolysis. Bacete et al. (2022) showed that in ISX-treated cells, THE1 modulates both cell wall stiffness as measured by Brillouin microscopy and synthesis of JA, associated with CWI sensing. In contrast, treatment with sorbitol (osmotic stress) stimulated not JA but ABA production, while, with both ISX and sorbitol, JA synthesis occurred but ABA synthesis was inhibited. Stimulation of JA synthesis was under control of a protein called RLP12 that was only expressed in the presence of both sorbitol and ISX (Fig. 15D). Also important was that sorbitol-induced ABA production was not observed in protoplasts, indicating that it involves PM wall connections—most likely the Hecht's threads observed in plasmolyzed cells (Yoneda et al. 2020). Overall, it appears that THE1 is a positive regulator of JA synthesis and negative regulator of ABA synthesis (Fig. 15D). THE1 can also work in concert with another receptor kinase, HERCULES, and FER to mitigate the stress of heavy metals on cell elongation (Richter et al. 2017) and with the LRR kinase FEI2 to induce JA synthesis associated with CWI. Because THE1 is not required for pattern-triggered immunity, it is likely specifically involved in CWI signaling (Engelsdorf et al. 2018). However, Pogorelko et al. (2013) and Bacete et al. (2018) have made clear in their reviews that there are many examples of significant cross-talk between CWI and defense signaling that are not reviewed here.
A direct role of oligosaccharins in defense signaling first became apparent from the important early discovery that they are sensed through their interaction with wall-associated receptor kinases called WAKs (Kohorn et al. 2006, 2021b), leading to induced defenses through activation of downstream MAPK1 signaling pathways (Fig. 15E). In addition to WAKs, many other receptors (including FER; see Fig. 15A) are involved in immune signaling (Narváez-Barragán et al. 2022). The pathogen-related expression pattern of a cluster of 5 other CrRLK1L genes suggests a specific role for other CrRLK1Ls in the interaction with pathogens (Lindner et al. 2012). Another interesting example involves the Arabidopsis immune peptides called Peps that are widely distributed in the plant kingdom (Lori et al. 2015); the PROPEP genes are induced by signals such as OGAs, Peps, and defense hormones. Peps bind to a receptor complex of PEPR and other co-receptors, setting off a series of classic immune responses and/or repressing CWI signaling (Shen et al. 2019; Dressano et al. 2020).
Signaling involving SCWs
Although mostly studied in the context of PCWs, signaling may also be triggered following modification of SCWs. For example, reduced growth associated with ectopic activation of defense responses, particularly synthesis of pathogenesis-related (PR) proteins, has sometimes confounded attempts to reduce recalcitrance of SCWs to digestion through reduction of lignin content or modification of lignin composition (Gallego-Giraldo et al. 2011, 2018). Defense trade-offs have been suggested as 1 possible explanation. This is not always the case, because some suppressor mutants of the growth reduction in plants with reduced lignin levels still exhibit defense gene expression (reviewed in Ha et al. 2021). The recent demonstration that some PR proteins are themselves cell wall localized, and function via generating small peptide defense signals (Chen et al. 2020a, 2020b), suggests that PR protein induction could be viewed as part of a broader program of cell wall remodeling that occurs following genetic modification of cell wall polymers.
Although pectin is considered a quantitatively minor component in SCWs and xylan the main wall component interacting with lignin (Kang et al. 2019), emerging evidence points to the possibility that pectin is a critical part of an underlying SCW structural scaffold that encompasses multiple wall components, including xylan and lignin (Hao and Mohnen 2014; Li et al. 2019). Modification of pectin through downregulation of GAUT4 increases both growth of and saccharification efficiency in switchgrass, the latter being a characteristic result of lignin reduction in SCWs (Biswal et al. 2018). Conversely, reduced lignification (e.g. in the cinnamoyl CoA reductase 1 mutant of Arabidopsis) causes solubilization of pectic elicitors that activate production of a suite of PR genes (Gallego-Giraldo et al. 2020). This process involves the FERONIA-dependent transcriptional activation of ADPG1 (Liu et al. 2023), a polygalacturonase normally associated with anther dehiscence, a process that is also dependent on correct lignin deposition (Dai et al. 2019). The ectopic expression of ADPG1 is not itself responsible for the initial release of pectic material from the wall but rather with its processing to elicitor-active moieties, which are then recognized by WAK receptors leading to induced defenses (Gallego-Giraldo et al. 2020; Liu et al. 2023) (Fig. 15E).
Coordinating signal pathways for growth, development, and stress
Clearly activities that occur within the cell wall sit at 1 nexus that controls the regulation of growth, development, and responses to stress. All the processes of cell wall biosynthesis and secretion and polymer modifications and interactions, when functioning normally, are essential to carry out the complex programs involved in plant growth and development. This section has addressed the ways in which cell walls sense when normal growth is disrupted and set in motion processes designed to mitigate such stresses and that these processes rely on similar or sometimes identical ligands, receptors, co-receptors, and downstream processes used for growth. The wall as a shapeshifter is made even more clear in the final section that will examine the changing nature of the cell wall as it plays a key role in coordinated patterns of cell division, expansion, maturation, and programmed cell death (PCD). Again, we shall see that hormones play key roles as input and output signals, and the levels of both intracellular and extracellular Ca2+, ROS, and pH are recurring key actors. Understanding how the signals involved in all these events are coordinated is certainly one of the future challenges facing cell wall signaling.
The life of a cell wall
Although not often discussed, cell walls have 4 distinct life stages: birth at the cell plate, tip, or diffusive growth, SCW maturation, and sometimes prolonged death long after that of their parent cells. Cell plate, tip growth, and maturation show some interesting and rather surprising similarities in terms of wall deposition. The outlier—the process of expansion growth—is one of most-studied processes in all of plant biology and has always seemed different and perhaps the most complex. This section examines these stages, placing emphasis on similarities and differences in wall structure and function between the life stages.
Birth at the cell plate
Cell walls are born during the process of cell division which involves building a new wall where none existed before. Obvious places of birth are stem and root apical meristems, but there are other important meristems such as those in leaf primordia and the cambium. Work by Peter Hepler on the dynamic behavior of the cytoskeleton recognized the importance of temporal and spatial changes in intracellular Ca2+ to cell plate formation and tip growth (Hepler 2005). Andrew Staehelin, who coupled a strong intellect to interpretation of his outstanding EM images, contributed greatly to our understanding of membrane dynamics and secretory processes. Hepler and Staehelin's work and several seminal reviews laid the foundation for our current understanding of cell plate formation (Samuels et al. 1995; Staehelin and Hepler 1996).
The initial establishment of a Ca2+ gradient focused on the developing cell plate is essential to the process, and conditions that dissipate this gradient disrupt cell plate formation (Staehelin and Hepler 1996). Ca2+ is important for cytoskeletal dynamics, vesicle fusion, callose synthesis, and pectin structure, all relevant to cell birth. During anaphase the spindle MT array gives way to a new MT array that defines the central position of the cell plate; actin microfilaments also form in this region and both are oriented with their plus ends to the center. This growing structure, called the phragmoplast, becomes a complex array of MTs, actin microfilaments, ER- and Golgi-derived vesicles (Samuels et al. 1995), and/or PM-derived endocytotic vesicles (Baluska et al. 2003) that delivers materials necessary for wall formation and is dismantled upon completion of the new wall. Vesicles fuse to form tubular structures that become the new PMs separated by the newly deposited wall.
Extensin3 (EXT3) was proposed to be deposited and cross-linked to form a scaffold on which to build the cell plate (Cannon et al. 2008). This has recently been questioned (Doll et al. 2022), although it is clear that, in the presence of extensin peroxidase, EXT3 undergoes self-recognition and polymerization into rope-like and cross-linked dendritic assemblies via end-on and lateral association, as revealed by AFM (Fig. 10D). ME-HG is also deposited early and is gradually demethylated, but whether DE-HG interacts with the positively charged EXT3 or some other similarly charged protein to form a “scaffold” requires further examination. Moore and Staehelin (1988) detected xyloglucan showing abundant labeling early in vesicles surrounding the developing plate and also within the plate. Callose is then detected in the well-developed tubular structures in a rare case where it serves as a wall component instead of defense polymer. Callose is by far the most abundant polymer of cell plates, and studies with an inhibitor of synthesis indicate that it is absolutely essential (Park et al. 2014; Drakakaki 2015). Figure 16 shows an impressive image of a developing plate from Arabidopsis that we suggest appears to show 2 different layers that might represent the early scaffold followed by a callose-rich layer.
Figure 16.
The developing cell plate in Arabidopsis. A) The direction of the Ca2+ gradient. B) An expanded version of A). Modified from Fujita and Wasteneys (2014) to indicate the calcium gradient and the positioning of a proposed location of a callose layer and a scaffold.
Cellulose deposition begins sometime after callose synthesis, and the process differs from that in elongating cells because it involves CSLD proteins. In Arabidopsis cell plates, Gu et al. (2016) reported early deposition of cellulose by AtCSLD5 while AtCESA3 was more active at a much later stage, and once the new wall merges with the parent wall, CesA proteins can be seen migrating to the new wall; other reports indicate some overlap in CSLD and CESA expression (Miart et al. 2014; Chen et al. 2018). Wu et al. (2023) have shown that, in moss protonema that display both tip growth and cell divisions, PpCSLD6 (and not CesA) is active in both tips and cell plates; it was also sensitive to DCB but not ISX providing an easy way to distinguish CSLD from CESA activity.
At present, it is not clear why CSLDs are the preferred cellulose synthases for both cell plate and tip growth. One possibility is that CSLDs, but not CESAs, are tolerant of the high Ca2+ gradient at the center of cell plates and also at the tip of root hairs and pollen tubes. It may also be important that the products of CSLD synthesis are less oriented and/or less crystalline (Yang et al. 2020b). There can be interactions between β-1,3-glucans and cellulose that result in changed properties in hydrogels or aqueous solutions (Abou-Saleh et al. 2018). Such a hybrid network may be more easily produced when the cellulose MFs are more disordered and less crystalline and such a new composite may be more suitable for the new wall—an area rich for further investigation. As the cell plate matures and joins the parent wall, it is reported that callose is digested (Drakakaki 2015), although it is not entirely clear whether some or all may be masked by the beginning of “normal” wall synthesis.
Growth
Tip-growing cells
The major targets for study, the pollen tube of Lily and both the pollen tube and root hair of Arabidopsis, have been excellently reviewed (Cheung and Wu 2011; Rounds and Bezanilla 2013; Johnson et al. 2019; Çetinbaş-Genç et al. 2022). This discussion focuses mostly on root hairs and some of the interesting similarities with cell plate formation. The deposition of wall polymers at the tip also occurs where no wall exists and relies upon the establishment of a [Ca2+cyt] tip-focused gradient but with rapid (∼30 s) extracellular oscillations that parallel those of protons and ROS but lag slightly behind the auxin-induced oscillations in growth (Monshausen et al. 2007; Zhang et al. 2022b). Wall construction begins with deposition of ME-HG at the tip that gradually becomes DE-HG through the action of PMEs. Beat Keller's group (Baumberger et al. 2001) discovered that the leucine-rich extensin LRX1 becomes insolubilized in root hair cell walls, providing a very early suggestion that it may function as a scaffold along with DE-HG. Xyloglucan is also deposited early, and it is interesting that an unusual acidic xyloglucan (that potentially could also interact with LRX1) is deposited in root hairs but not pollen tubes (Pena et al. 2012). As for cell plates, callose deposition begins later, but with some differences: root hairs are relatively richer in cellulose, although they do deposit some callose, while pollen tubes and cell plate walls are very rich in callose [although some is found in callose plugs in pollen tubes (Kapoor and Geitmann 2023)]. Pollen tubes were one of the first places where CSLDs were shown to function (Doblin et al. 2001) but, in the same year, Favery et al. (2001) showed that the KOJAK gene that encodes AtCSLD3 is necessary for root hair growth. Subsequently it was shown that AtCSLD2 and AtCSLD3 but not CESAs function in cellulose synthesis in root hairs of Arabidopsis (Bernal et al. 2008: Park et al. 2011).
Signaling networks for tip growth are complex and involve many of the players described for CWI signaling as well as those used for diffusive growth (Kroeger et al. 2008; Zhang et al. 2022b). As previously discussed (see Fig. 15C), Schoenaers et al. (2023) provide perhaps the best description yet of oscillations in signals and wall structure that promote either wall-loosening or stiffening and correlate with oscillations in growth in root hairs.
Expansion (diffusive) growth
Studies of expansion growth began with the discovery of auxin by Fritz Went and have continued on for decades by other pioneers such as James Lockhart, Bob Cleland, and Dan Cosgrove (see Du et al. 2020b for recent review). To reiterate briefly, the driving force for growth is turgor, and, if cellulose MFs are aligned perpendicular to the axis of growth, the cell elongates. Auxin promotes wall acidification through activation of PM H+-ATPase (AHA2) that leads to “acid growth” through wall-loosening that creates new “space” that is filled in by deposition of new wall polymers. Wall remodeling (stiffening) that follows is assumed to involve mechanisms that work in opposition to wall-loosening.
To take a new look at expansion growth, we ask whether it is fundamentally not so very different from tip growth, as has also been implied by others (Wolf and Hofte 2014; Wolf et al. 2014; Höfte 2015; Jobert et al. 2023). It seems not too difficult to imagine such growth as an oscillation between a loose wall and a stiff wall. In a surely oversimplified scenario for diffusive growth, the loose wall has a lower pH, is relatively enriched in newly synthesized polymers (and therefore should initially have a high ratio of ME-HG to DE-HG), has a low level of cross-linking of pectic and/or protein polymers, and newly deposited cellulose is kept from interacting with itself and other polymers by expansin. The stiff wall follows in rapid succession as pH rises, ME-DG is converted to DE-HG promoted by PME, much more stiffening occurs through Ca2+-bridges and/or HG condensation through interaction with positively-charged proteins, peroxidase-mediated activity, and borate cross-links for RGII. The concept of this “loose wall” and the “stiff wall” could easily be consistent with oscillations in Ca2, ROS (for crosslinking and redox control), new wall polymer deposition, and wall pH (activity of PME vs PMEI, expansin, and THE1 and FER are also all pH regulated). This concept is consistent with FER and THE1 being positive regulators of cell wall stiffness (Höfte 2015; Bacete et al. 2022). But we do not see these 2 different types of walls when we isolate growing tissues because we isolate a mixture of loose and stiff walls.
If this oversimplified scenario has any truth to it, then the larger questions are what feedback loops involving auxin and/or other signals regulate the oscillations, what is the period, where are they spatially located, and why do we not see the process as a growth oscillation? Perhaps because it is not as localized as is tip growth? Perhaps it might be regulated by oscillations of only seconds to minutes and localized to a multitude of growth sites? There is 1 apparent oscillation occurring in multiple places that we can actually see at a microscopic level in elongating hypocotyls: CESAs have a residence time and period of activity in the PM of 7 to 10 minutes after which they become inactive and are internalized where they are targeted for degradation or recycling. Later new and recycled CESAs are inserted along the same MT-aligned path. Assuming deposition of other wall polymers is also locally coordinated with cellulose deposition (Ganguly et al. 2020), this is a highly localized oscillation in wall deposition that would occur in a multitude of sites that would be difficult to detect. Hepler and colleagues argued that a measured oscillation in exocytosis of wall materials in tip-growing pollen was a driving force for oscillations in wall thickness and growth (McKenna et al. 2009). A cycle driven by an oscillation in auxin action leading to oscillations in wall pH, internal Ca2+ levels and ROS, might fit with an assumption that CESAs are inhibited by Ca2+ (something that needs direct investigation) and would require the substitution of CSLDs for CESAs when the Ca2+ gradient is tip focused or high at the cell plate and also promote callose synthesis. For such diffuse growth there may be no need for an LRX1-DE-HG scaffold complex or callose. LRX1 is specific for tip growth (Carol and Dolan 2002), and perhaps there is some mechanism to keep callose synthesis inhibited, 1 possibility being its association with the abundant Ca2+ -binding annexin (Andrawis et al. 1993).
Exciting recent work indicates feedback loops ranging from seconds to minutes between auxin, wall pH, Ca2+, and pectin remodeling, reviewed by Jobert et al. (2023). They suggest a possibility of special regulation within microdomains wherein there are feedback loops between the state of auxin transporters and cell wall structures. It takes a few minutes for auxin's effects on transcription to show up [e.g. as upregulation of key regulators such as ERU, H+ ATPases, and small auxin up-regulated RNAs (Du et al. 2020b; Jobert et al. 2023)]. But auxin can also cause very rapid (seconds) fluxes in wall pH in roots that are in opposition to the canonical regulation of AHA2 by auxin carried out through transcriptional regulation (Li et al. 2021).
If we could stop the process at one point in the cycle, would we then see a wall structure that is largely loose or stiff? Perhaps DCB-adapted cells with their pectin-based walls (Shedletzky et al. 1990, 1992) or the alternating pectin/cellulose wall layers in celery collenchyma cells (see Fig. 9) offer clues. Or perhaps walls with disturbances on a larger spatial scale such as during hypocotyl hook formation or gravitropism (Jobert et al. 2023). Stay tuned; acid growth has never seemed so exciting!
Maturation
Cell wall maturation is defined here as the production of a SCW that is traditionally said to consist of S-1, S-2, and S-3 layers. The process is assumed to begin when elongation of a cell ceases, but there is sometimes an interesting overlap between these 2 processes during deposition of the S-1 layer. This transition cell wall (TCW) is the most interesting, and the one to which we give most attention. In view of space limitations, we leave discussion of many other aspects of SCW development, including regulation of lignification, to others (Turner et al. 2007; Hobson et al. 2010; Li et al. 2016c; Meents et al. 2018; Rao and Dixon 2018; Liu et al. 2021). In some ways, the making of a TCW is akin to deposition of a new wall because it is different from the PCW and is laid down without integration of wall material into the PCW. Thus, maturation might use the TCW as a scaffold upon which to deposit new material.
The cotton fiber is a unique cell type and has many advantages for study of the development of cell walls (Haigler et al. 2012). It originates from epidermal cells of the ovule. However, evidence indicates that it displays both tip growth and diffusive growth along the length of the fiber (Qin and Zhu 2011). Fiber cells within a boll develop synchronously from the day of anthesis; fibers elongate (up to about 2 cm) and deposit a typical dicot PCW comprised mostly of cellulose, xyloglucan and pectins (Meinert and Delmer 1977). At about 16 days post-anthesis, TCW synthesis begins. The fiber continues to elongate while beginning the deposition of this new TCW called the “winding layer” that is analogous to the S-1 layer of wood cells. From about 24 days onward, the fiber deposits almost exclusively cellulose in S-2 and S-3 layers resulting in a wall that is >90% cellulose by weight at maturity.
The idea that the TCW serves as a scaffold occurred to us because we realized that there are some interesting similarities to wall deposition in cell plates and tip-growing cells. The beginning of the TCW is characterized by a remodeling of pectin resulting in a net loss of uronic acids from walls that occurs at the same time the middle lamella between fibers degrades (Meinert and Delmer 1977; Singh et al. 2009). The expression of 2 Rac/ROP GTPases occurs precisely during this TCW phase of fiber growth (Delmer et al. 1995). Potikha et al. (1999) demonstrated H202 production associated with the TCW stage, and early addition of H202 caused premature initiation of TCW formation while an inhibitor of NADPH oxidase inhibited initiation. Strikingly, these observations are very similar to those observed for initiation of tracheary element differentiation in Zinnia cell cultures (Karlsson et al. 2005). Roberts and Haigler (1989) also presented evidence for a rise in Ca2+ levels in tracheary elements during the transition, and significant callose deposition is initiated early in the TCW stage in cotton fibers concomitant with first signs of SCW cellulose, a key component of the winding layer and showing no evidence of turnover (Maltby et al. 1979). As far as we can tell, there is no report of callose in S1-layers of wood, although it is found in tracheids of compression wood where it is called Larcinan (Boyd 1978) and in the G-layer of flax phloem fibers (Ibragimova et al. 2017). Primary wall CesAs apparently continue to be active with a gradual transition to secondary wall CesAs during the TCW transition (Tuttle et al. 2015; Watanabe et al. 2018). CslDs are expressed at all stages of fiber development and their role, if any, needs clarification (Li et al. 2017). In sum, it appears that the cotton fiber winding layer (as a scaffold?) is initiated by elevation of levels of intracellular Ca2+/ROS, production of DE-HG followed by a callose-rich layer mixed with cellulose perhaps made by both PCW and SCW CesAs. There is other very interesting evidence that there may be a pectin-rich scaffold in cells of wood since transgenic modification of poplar to make less HG leads to acceleration of growth and improvement in digestibility of the walls (Biswal et al. 2018). This may also relate to the finding that overexpression of an enzyme that degrades RGI enhances degradation of the middle lamella and also enhances growth in poplar (Yang et al. 2020a).
As for the later synthesis of the S-2 and S-3 layers, synthesis of xylans, glucomannans and lignin have already been discussed. CSCs increase significantly in numbers and velocity of movement and become more aggregated (Watanabe et al. 2015). Cotton fibers, bast fibers, and tracheary elements all share the same primary transcriptional regulation and the same set of genes is involved in SCW cellulose synthesis. An early transcriptome analysis to identify a common gene network for SCW synthesis in cells such as cotton fibers, xylem, and cellulose-rich tension wood identified 52 genes that met a very wide range of criteria (Ko et al. 2006). Among the genes on the list are NAC and myb TFs, genes encoding enzymes for all the major SCW polymers and, interestingly, also pectin synthesis. Not surprisingly, the cellulose-rich cotton and bast fibers all downregulate the transcription factors that induce deposition of lignin and hemicelluloses (Tuttle et al. 2015).
SCW synthesis represents a major carbon sink in maturing cells (Verbančič et al. 2018; Pottier et al. 2023). As discussed in those reviews, the source of the UDP-Glc that is the substrate for both cellulose and callose synthesis still remains controversial for some. For cotton fibers that make only callose and cellulose and little or no starch during SCW synthesis, the carbon comes from sucrose that enters the fibers through plasmodesmata. Amor et al. (1995) showed that 80% of the highly expressed sucrose synthase (SuSy) becomes associated with the plasma membrane fraction during SCW callose and cellulose synthesis, and Salnikov et al. (2001) also showed similar PM localization in developing zinnia tracheary elements. Amor et al. (1995) and Haigler et al. (2001) proposed this SuSy may channel carbon directly to cellulose or callose, a pathway that saves 1 ATP worth of energy for each Glc residue polymerized compared with the alternate pathway involving invertase and UDP-Glc pyrophosphorylase, In addition, recycling of the UDP (that inhibits glucan synthases) back to PM-SuSy would prevent its localized accumulation. This work also showed that supplied UDP-Glc can support callose but not cellulose synthesis in permeabilized fibers while sucrose can supply carbon for synthesis of both, another argument for possible in vivo channeling of UDP-Glc directly to the active site of CSCs. This PM-SuSy is most likely the same isoform found to be up-regulated by Arpat et al. (2004), and Brill et al. (2011) also showed strong up-regulation of a special SuSy in PM and wall fractions of cotton fibers starting during the TCW stage. However, as discussed by Verbančič et al. (2018) and Pottier et al. (2023), this role for SuSuy has been questioned, primarily due to mutant analyses showing that SuSy clearly is not absolutely necessary in Arabidopsis (Wang et al. 2022a, 2022b), and mutant analysis of cytoplasmic invertases, that may be important for more reasons than supplying carbon for cellulose synthesis, are essential for normal seedling growth (Barratt et al. 2009). However, both SuSy and UDP-Glc pyrophosphorylase are found in high levels during both PCW and SCW phases and both catalyze reversible reactions, suggesting important roles for each and that one should also be available to substitute when the other is absent. Verbančič et al. (2018) and Pottier et al. (2023) do conclude that there is certainly strong evidence to support an important role for PM-localized SuSy in SCWs. Winter and Huber (2010) have shown that SuSy becomes associated with actin upon Ca-dependent phosphorylation and, its association with actin directs CSCs to appropriate sites on the PM during SCW (Lei et al. 2012)]. Whether such associations might relate to possible channeling of carbon directly through the gating loop of CSCs in vivo deserves further study.
One key step has been missing from the life of the secondary wall—what are the signals that turn on the transcription factors? Clearly hormones are involved (Haigler et al. 2012; Demura 2013; Meents et al. 2018). Just as we were finishing writing this review, a landmark paper appeared that opens up new avenues for this area of research. Cai et al. (2023) have shown that different variants of WAK10, in combination with a brassinosteroid receptor and modulated by binding of OGAs of different sizes and affinities, control the expression of CesA 4,7, and 8 genes, but not lignin, in rice. This process leads to either taller or shorter rice with less or more secondary wall cellulose. Furthermore, breeding of rice has gradually selected a whole set of WAK10 variants leading to shorter or longer rice. This may tie in with the appearance of OGAs at the TCW transition that can bind to and activate WAKs. All of this invokes shades of the Green Revolution and suggests new approaches for regulation of synthesis of cell walls in other crops and particularly those used for biofuels.
Death
Just as the cell signaling community seems to revere Greek gods, the PCD community has fun asking “Was it murder or suicide? What were the murder weapons? Where did they put the corpse?” The emerging answers seem to be: “It was suicide, and, as we have seen for life, the chief weapons are Ca2+ and ROS (and probably FER as well) and, as for the corpse, it gets eaten.” Intracellular Ca2+ and ROS levels certainly rise as an early event of cell death (Roberts and Haigler 1989), and it may be that the regulation of SCW synthesis and PCD are coordinated. A 40-kD serine protease appears at initiation of SCW formation and builds up over time until it appears to serve as the same signal to induce PCD (Groover and Jones 1999). As for the corpse, in some cases the entire cell including the cell wall is digested, but the walls of cells like cotton fibers, bast fibers, vessels, and fiber cells of xylem all survive—sometimes for millennia—in the cotton robes and wooden caskets found in the tombs of the pharaohs.
There are 2 kinds of PCD: one that is developmentally programmed such as that seen for tracheary elements, and the other that is involved in defense against pathogens (pPCD). pPCD can be of the type that is involved in the hypersensitive response (HR-pPCD) to biotrophic and hemi-biotrophic pathogens, whereas another form might be called nec-pPCD in which necrotrophic pathogens digest everything, including walls. Being able to understand the differences between these is very important for devising proper defense strategies, but that gets beyond the scope of this review. The details of plant PCD are all still emerging; the reviews of Huysmans et al. (2017) and Ustun et al. (2017) summarize the overall state of our knowledge.
In sum, it is quite remarkable how the same signaling pathways can be modified in one way or another during the life of the plant and how important the cell wall is for that life. But the life story of most cell walls ends well. Most walls survive for a very long time, carrying out their many functions serving not only their parent plants but the entire global ecosystem.
Challenges for the next 100 years
What structures do the different wall polymer types have at the time they are synthesized and deposited in walls of specific cell types?
What are the interactions between and among polymer, and how do these interactions change, both temporally and spatially, for walls with different structures and functions?
How are all these processes regulated and coordinated?
Acknowledgments
The authors thank the cell wall community that has inspired the work described in this review and, of course, our patient spouses.
Contributor Information
Deborah Delmer, Section of Plant Biology, University of California Davis, Davis, CA 95616, USA.
Richard A Dixon, BioDiscovery Institute and Department of Biological Sciences, University of North Texas, Denton, TX 76203, USA.
Kenneth Keegstra, MSU-DOE Plant Research Laboratory, Michigan State University, East Lansing, MI 48823, USA.
Debra Mohnen, Complex Carbohydrate Research Center and Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA 30602, USA.
Author contributions
All authors contributed significanlty to the preparation of this review. D.D. was the primary author of the “cellulose” and “life of a cell wall” sections, R.A.D. was the primary author of the “lignin” section, K.K. was the primary author of the “hemicellulose” and “model” sections, and D.M. was the primary author of the “pectin” and “wall-associated protein“ sections. D.D. and R.A.D. were the primary authors of the “signaling” section.
Funding
Work in the laboratories of R.D. and D.M. was partially funded from The Center for Bioenergy Innovation, a US Department of Energy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science and, for D.M., partially from the US Department of Energy, Office of Science, Basic Energy Sciences, Chemical Sciences, Geosciences and Biosciences Division under award no. DE-SC0015662.
Data availability
This historical review contains no new data.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Abidi W, Decossas M, Torres-Sánchez L, Puygrenier L, Létoffé S, Ghigo J-M, Krasteva PV. Bacterial crystalline cellulose secretion via a supramolecular BcsHD scaffold. Sci Adv. 2022:8(50):eadd1170. 10.1126/sciadv.add1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou-Saleh RH, Hernandez-Gomez MC, Amsbury S, Paniagua C, Bourdon M, Miyashima S, Helariutta Y, Fuller M, Budtova T, Connell SD, et al. Interactions between callose and cellulose revealed through the analysis of biopolymer mixtures. Nat Commun. 2018:9(1):4538. 10.1038/s41467-018-06820-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal UP, Ralph SA. FT-Raman spectroscopy of wood: identifying contributions of lignin and carbohydrate polymers in the spectrum of black spruce (Picea mariana). Appl Spectroscopy. 1997:51(11):1648–1655. 10.1366/0003702971939316 [DOI] [Google Scholar]
- Albersheim P. The walls of growing plant cells. Sci Am. 1975:232(4):80–95. 10.1038/scientificamerican0475-80 [DOI] [PubMed] [Google Scholar]
- Albersheim P, Bonner J. Metabolism and hormonal control of pectic substances. J Biol Chem. 1959:234(12):3105–3108. 10.1016/S0021-9258(18)69630-9 [DOI] [PubMed] [Google Scholar]
- Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. Plant cell walls. London: Garland Science, Taylor & Francis; 2011, 430 pp. [Google Scholar]
- Alejandro S, Lee Y, Tohge T, Sudre D, Osorio S, Park J, Bovet L, Lee Y, Geldner N, Fernie AR, et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol. 2012:22(13):1207–1212. 10.1016/j.cub.2012.04.064 [DOI] [PubMed] [Google Scholar]
- Ali MS, Baek K-H. Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int J Mol Sci. 2020:21(2):621. 10.3390/ijms21020621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni Y, Delmer DP, Benziman M. Achievement of high rates of in vitro synthesis of 1, 4-beta-D-glucan: activation by cooperative interaction of the Acetobacter xylinum enzyme system with GTP, polyethylene glycol, and a protein factor. Proc Natl Acad Sci U S A. 1982:79(21):6448–6452. 10.1073/pnas.79.21.6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altanassov II, Pittman JK, Turner SR. Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J Biol Chem. 2009:284(6):3833–3841. 10.1074/jbc.M807456200 [DOI] [PubMed] [Google Scholar]
- Amor Y, Haigler CH, Johnson S, Wainscott M, Delmer DP. A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc Natl Acad Sci U S A. 1995:92(20):9353–9357. 10.1073/pnas.92.20.9353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos RA, Atmodjo MA, Huang C, Gao Z, Venkat A, Taujale R, Kannan N, Moremen KW, Mohnen D. Polymerization of the backbone of the pectic polysaccharide rhamnogalacturonan I. Nat Plants. 2022:8(11):1289–1303. 10.1038/s41477-022-01270-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos RA, Mohnen D. Critical review of plant cell wall matrix polysaccharide glycosyltransferase activities verified by heterologous protein expression. Front Plant Sci. 2019:10:915. 10.3389/fpls.2019.00915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos RA, Pattathil S, Yang JY, Atmodjo MA, Urbanowicz BR, Moremen KW, Mohnen D. A two-phase model for the non-processive biosynthesis of homogalacturonan polysaccharides by the GAUT1:GAUT7 complex. J Biol Chem. 2018:293(49):19047–19063. 10.1074/jbc.RA118.004463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsbury S, Hunt L, Elhaddad N, Baillie A, Lundgren M, Verhertbruggen Y, Scheller HV, Knox JP, Fleming AJ, Gray JE. Stomatal function requires pectin de-methyl-esterification of the guard cell wall. Curr Biol. 2016:26(21):2899–2906. 10.1016/j.cub.2016.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders N, Frederick L, Wilson L, Sorieul M, Nikolovski N, Dupree P. β-1,4-xylan backbone synthesis in higher plants: how complex can it be? Front Plant Sci. 2023:13:1076298. 10.3389/fpls.2022.1076298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DB. Uber die struktur der kollenchymzellwand auf grund mikrochemischer untersuchungen. Sitzungsberichte der Akademie der Wissenschaften Mathematisch-Naturwissenschaftliche Klasse. 1927:136:429–329. Available at: https://www.zobodat.at/pdf/SBAWW_136_0429-0439.pdf [Google Scholar]
- Anderson CT, Kieber JJ. Dynamic construction, perception, and remodeling of plant cell walls. Annu Rev Plant Biol. 2020:71(1):39–69. 10.1146/annurev-arplant-081519-035846 [DOI] [PubMed] [Google Scholar]
- Andrawis A, Solomon M, Delmer DP. Cotton fiber annexins: a potential role in the regulation of callose synthase. Plant J. 1993:3(6):763–772. 10.1111/j.1365-313X.1993.00763.x [DOI] [PubMed] [Google Scholar]
- Arioli A, Peng L, Betzner A, Burn J, Wittke W, Herth W, Camilleri C, Hofte H, Plazinski J, Birch R, et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 1998:279(5351):717–720. 10.1126/science.279.5351.717 [DOI] [PubMed] [Google Scholar]
- Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA. Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol. 2004:54(6):911–929. 10.1007/s11103-004-0392-y [DOI] [PubMed] [Google Scholar]
- Arsovski AA, Haughn GW, Western TL. Seed coat mucilage cells of Arabidopsis thaliana as a model for plant cell wall research. Plant Signal Behav. 2010:5(7):796–801. 10.4161/psb.5.7.11773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspinall GO, Molloy JA, Craig JWT. Extracellular polysaccharides from suspension-cultured sycamore cells. Can J Biochem. 1969:47(11):1063–1070. 10.1139/o69-170 [DOI] [PubMed] [Google Scholar]
- Atalla RH, Vanderhart DL. Native cellulose: a composite of two distinct crystalline forms. Science. 1984:223(4633):283–285. 10.1126/science.223.4633.283 [DOI] [PubMed] [Google Scholar]
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell JA, Mohanty SS, Atwood JA III, Orlando R, Scheller HV, Mohnen D. GAUT1:GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci U S A. 2011:108(50):20225–20230. 10.1073/pnas.1112816108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augur C, Yu L, Sakai K, Ogawa T, Sinaÿ P, Darvill AG, Albersheim P. Further studies of the ability of xyloglucan oligosaccharides to inhibit auxin-stimulated growth. Plant Physiol. 1992:99(1):180–185. 10.1104/pp.99.1.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers AR, Valent B, Ebel J, Albersheim P. Host-pathogen interactions. XI. Composition and structure of wall-released elicitor fractions. Plant Physiol. 1976:57(5):766–774. 10.1104/pp.57.5.766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar A, Chalvin C, Shih PM, Maimann M, Ebert B, Birdseye DS, Loqué D, Scheller HV. Gene stacking of multiple traits for high yield of fermentable sugars in plant biomass. Biotechnol Biofuels. 2018:11:2. 10.1186/s13068-017-1007-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacete L, Hamann T. The role of mechanoperception in plant cell wall integrity maintenance. Plants. 2020:9(5):574–592. 10.3390/plants9050574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacete L, Mélida H, Miedes E, Molina A. Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. 2018:93(4):614–636. 10.1111/tpj.13807 [DOI] [PubMed] [Google Scholar]
- Bacete L, Schulz J, Engelsdorf T, Bartosova Z, Vaahtera L, Yan G, Gerhold JM, Tichá T, Øvstebø C, Gigli-Bisceglia N, et al. THESEUS1 modulates cell wall stiffness and abscisic acid production in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2022:119(1):e2119258119. 10.1073/pnas.2119258119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacic A, Delmer DP. Stimulation of membrane-associated polysaccharide synthetases by a membrane potential in developing cotton fibers. Planta. 1981:152(4):346–351. 10.1007/BF00388260 [DOI] [PubMed] [Google Scholar]
- Baluska F, Samaj J, Wojtaszek P, Volkmann D, Menzel D. Cytoskeleton-plasma membrane-cell wall continuum in plants. Emerging links revisited. Plant Physiol. 2003:133(2):482–491. 10.1104/pp.103.027250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, O’Malley DM, Whetten R, Sederoff RR. A laccase associated with lignification in loblolly pine xylem. Science. 1993:260(5108):672–674. 10.1126/science.260.5108.672 [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, O'Neill MA. Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu Rev Plant Biol. 2011:62(1):127–155. 10.1146/annurev-arplant-042110-103918 [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, Urbanowicz BR, O'Neill MA. The synthesis and origin of the pectic polysaccharide rhamnogalacturonan II—insights from nucleotide sugar formation and diversity. Front Plant Sci. 2012:3:92. 10.3389/fpls.2012.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa ICR, Rojas-Murcia N, Geldner N. The Casparian strip—one ring to bring cell biology to lignification? Curr Opin Biotechnol. 2019:56:121–129. 10.1016/j.copbio.2018.10.004 [DOI] [PubMed] [Google Scholar]
- Baron-Epel O, Gharyal PK, Schindler M. Pectins as mediators of wall porosity in soybean cells. Planta. 1988:175(3):389–395. 10.1007/BF00396345 [DOI] [PubMed] [Google Scholar]
- Barratt DHP, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule AJ, Smith AM. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc Natl Acad Sci U S A. 2009:106(31):13124–13129. 10.1073/pnas.0900689106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros-Rios J, Temple S, Dixon RA. Development and commercialization of reduced lignin alfalfa. Curr Opin Biotechnol. 2018:56:48–54. 10.1016/j.copbio.2018.09.003 [DOI] [PubMed] [Google Scholar]
- Barros J, Serk H, Granlund I, Pesquet E. The cell biology of lignification in higher plants. Ann Bot. 2015:115(7):1053–1074. 10.1093/aob/mcv046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros J, Serrani-Yarce JC, Chen F, Baxter D, Venables BJ, Dixon RA. Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Nat Plants. 2016:2(6):16050. 10.1038/nplants.2016.50 [DOI] [PubMed] [Google Scholar]
- Barros J, Shrestha HK, Serrani-Yarce JC, Engle N, Abraham PE, Tschaplinski TJ, Hettich RL, Dixon RA. Proteomic and metabolic disturbances in lignin modified Brachypodium distachyon. Plant Cell. 2022:34(9):3339–3363. 10.1093/plcell/koac171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels S, Boller T. Quo vadis, pep? Plant elicitor peptides at the crossroads of immunity, stress, and development. J Exp Bot. 2015:66(17):5183–5193. 10.1093/jxb/erv180 [DOI] [PubMed] [Google Scholar]
- Bauer WD, Talmadge KW, Keegstra K, Albersheim P. The structure of plant cell walls II. The hemicellulose of the walls of suspension-cultured sycamore cells. Plant Physiol. 1973:51(1):174–187. 10.1104/pp.51.1.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Ringli C, Keller B. The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 2001:5(9):1128–1139. 10.1101/gad.200201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker GE, Hui PA, Albersheim P. Synthesis of extracellular polysaccharides by suspensions of Acer pseudoplatanus cells. Plant Physiol. 1964:39(6):913–920. 10.1104/pp.39.6.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum RA, Fry SC. Boron bridging of rhamnogalacturonan-II in Rosa and Arabidopsis cell cultures occurs mainly in the endo-membrane system and continues at a reduced rate after secretion. Ann Bot. 2022:130(5):703–715. 10.1093/aob/mcac119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belorio M, Gomez M. Psyllium: a useful functional ingredient in food systems. Crit Rev Food Sci Nutr. 2022:62(2):527–538. 10.1080/10408398.2020.1822276 [DOI] [PubMed] [Google Scholar]
- Berken A. ROPs in the spotlight of plant signal transduction. Cell Mol Life Sci. 2006:63(21):2446–2459. 10.1007/s00018-006-6197-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal AJ, Yoo C-M, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sorensen I, Blancaflor EB, Scheller HV, Willats WGT. Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 2008:148(3):1238–1253. 10.1104/pp.108.121939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C, Tierney ML. Expression of AtPRP3, a proline-rich structural cell wall protein from Arabidopsis, is regulated by cell-type-specific developmental pathways involved in root hair formation. Plant Physiol. 2000:122(3):705–714. 10.1104/pp.122.3.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y, Hubbard C, Purushotham P, Zimmer J. Insights into the structure and function of membrane-integrated processive glycosyltransferases. Curr Opin Struct Biol. 2015:34:78–86. 10.1016/j.sbi.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal AK, Atmodjo MA, Li M, Baxter HL, Yoo CG, Pu Y, Lee Y-C, Mazarei M, Black IM, Zhang J-Y, et al. Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nat Biotechnol. 2018:36(3):249–257. 10.1038/nbt.4067 [DOI] [PubMed] [Google Scholar]
- Blackburn MR, Haruta M, Moura DS. Twenty years of progress in physiological and biochemical investigation of RALF peptides. Plant Physiol. 2020:182(4):1657–1666. 10.1104/pp.19.01310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Roy S, Kritsas K, Grobei MA, Jaciubek M, Schroeder JI, Grossniklaus U. Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development. 2009:136(19):3279–3288. 10.1242/dev.040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J. The chemistry and physiology of the pectins. Bot Rev. 1936:2(10):475–497. 10.1007/BF02869919 [DOI] [Google Scholar]
- Borassi C, Sede AR, Mecchia MA, Salter JDS, Marzol E, Muschietti JP, Estevez JM. An update on cell surface proteins containing extensin-motifs. J Exp Bot. 2016:67(2):477–487. 10.1093/jxb/erv455 [DOI] [PubMed] [Google Scholar]
- Bosch M, Hepler PK. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell. 2005:17(12):3219–3226. 10.1105/tpc.105.037473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton S, Leboeuf E, Mouille G, Leydecker M-T, Talbotec J, Granier F, Lahaye M, Höfte H, Truong H-N. QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell. 2002:14(10):2577–2590. 10.1105/tpc.004259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Zipfel C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol. 2017:55(1):257–286. 10.1146/annurev-phyto-080614-120106 [DOI] [PubMed] [Google Scholar]
- Boyd JD. Significance of larcinan in compression wood tracheids. Wood Sci Technol. 1978:12(1):23–35. 10.1007/BF00390008 [DOI] [Google Scholar]
- Braccini I, Pérez S. Molecular basis of Ca2+-induced gelation in alginates and pectins: the egg-box model revisited. Biomacromolecules. 2001:2(4):1089–1096. 10.1021/bm010008g [DOI] [PubMed] [Google Scholar]
- Braccini I, Rodriguez-Carvajal MA, Pérez S. Chain-chain interactions for methyl polygalacturonate: models for high methyl-esterified pectin junction zones. Biomacromolecules. 2005:6(3):1322–1328. 10.1021/bm049457h [DOI] [PubMed] [Google Scholar]
- Braconnot H. Nouvelles observations sur l’acide pectique. Ann Chim. 1825a:30:96–102. [Google Scholar]
- Braconnot H. Recherches sur un nouvel acide universellement répandu dans tous les végétaux. Ann Chim. 1825b:28(2):173–178. [Google Scholar]
- Braybrook SA, Jnsson H. Shifting foundations: the mechanical cell wall and development. Curr Opin Plant Biol. 2016:29:115–120. 10.1016/j.pbi.2015.12.009 [DOI] [PubMed] [Google Scholar]
- Brill E, van Thournout M, White RG, Llewellyn D, Campbell PM, Engelen S, Ruan YL, Arioli T, Furbank RT. A novel isoform of sucrose synthase is targeted to the cell wall during secondary cell wall synthesis in cotton fiber. Plant Physiol. 2011:157(1):40–54. 10.1104/pp.111.178574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann M, Li E, Sampathkumar A, Kocabek T, Hauser M-T, Persson S. POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell. 2012:24(1):163–177. 10.1105/tpc.111.093575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RM. The biosynthesis of cellulose. J Macromol Sci Part A. 1996:33(10):1345–1373. 10.1080/10601329608014912 [DOI] [Google Scholar]
- Brown C, Leijon F, Bulone V. Radiometric and spectrophotometric in vitro assays of glycosyltransferases involved in plant cell wall carbohydrate biosynthesis. Nat Protocols. 2012:7(9):1634–1650. 10.1038/nprot.2012.089 [DOI] [PubMed] [Google Scholar]
- Brown RM Jr, Wilison JHM, Richardson CL. Cellulose biosynthesis in Acetobacter xylinum: visualization of the site of synthesis and direct measurement of the in vivo process. Proc Natl Acad Sci U S A. 1976:73(12):4565–4569. 10.1073/pnas.73.12.4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buleon A, Chanzy H. Single crystals of cellulose II. J Polym Sci: Polym Phys Ed. 1978:16:833–839. 10.1002/pol.1978.180160508 [DOI] [Google Scholar]
- Burton RA, Fincher GB. (1,3;1,4)-β-D-glucan in cell walls of the Poaceae, lower plants, and fungi: a tale of two linkages. Mol Plant. 2009:2(5):873–882. 10.1093/mp/ssp063 [DOI] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin N, Bacic A, Fincher GB. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science. 2006:311(5769):1940–1942. 10.1126/science.1122975 [DOI] [PubMed] [Google Scholar]
- Busse-Wicher M, Li A, Silveira RL, Pereira CS, Tryfona T, Gomes TCF, Skaf MS, Dupree P. Evolution of xylan substitution patterns in gymnosperms and angiosperms: implications for xylan interaction with cellulose. Plant Physiol. 2016:171(4):2418–2431. 10.1104/pp.16.00539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall KH, Pattahil S, Phillips SE, Hahn MG, Mohnen D. Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol Plant. 2009:2(5):1000–1014. 10.1093/mp/ssp062 [DOI] [PubMed] [Google Scholar]
- Cai W, Hong J, Liu Z, Wang W, Zhang J, An G, Liang W, Persson S, Zhang D. A receptor-like kinase controls the amplitude of secondary cell wall synthesis in rice. Curr Biol. 2023:33(3):498–506.e6. 10.1016/j.cub.2022.12.035 [DOI] [PubMed] [Google Scholar]
- Cannon MC, Terneus K, Hall Q, Tan L, Wang Y, Wegenhart BL, Chen L, Lamport DT, Chen Y, Kieliszewski MJ. Self-assembly of the plant cell wall requires an extensin scaffold. Proc Natl Acad Sci U S A. 2008:105(6):2226–2231. 10.1073/pnas.0711980105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009:37(Database):D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol RJ, Dolan L. Building a hair: tip growth in Arabidopsis thaliana root hairs. Philos Trans R Soc Lond B Biol Sci. 2002:357(1422):815–821. 10.1098/rstb.2002.1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC. Cell wall development in maize coleoptiles. Plant Physiol. 1984:76(1):205–212. 10.1104/pp.76.1.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in the flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993:3(1):1–30. 10.1111/j.1365-313X.1993.tb00007.x [DOI] [PubMed] [Google Scholar]
- Carre M. An investigation of the pectic constituents of stored fruits. Biochem J. 1922:16(5):704. 10.1042/bj0160704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Keegstra K. Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J Biol Chem. 2006:281(45):34197–34207. 10.1074/jbc.M606379200 [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, Yamauchi K, Reinecke A, Freshour G, Zabotina OA, Hahn MG, Burgert I, Pauly M, et al. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell. 2008:20(6):1519–1537. 10.1105/tpc.108.059873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çetinbaş-Genç A, Conti V, Cai G. Let's shape again: the concerted molecular action that builds the pollen tube. Plant Reprod. 2022:35(2):77–103. 10.1007/s00497-022-00437-4 [DOI] [PubMed] [Google Scholar]
- Chan J, Coen E. Interaction between autonomous and microtubule guidance systems controls cellulose synthase trajectories. Curr Biol. 2020:30(5):941–947. 10.1016/j.cub.2019.12.066 [DOI] [PubMed] [Google Scholar]
- Chanzy H, Henrissat B. Electron microscopy study of the enzymic hydrolysis of Valonia cellulose. Carbohydr Polym. 1983:3(3):161–173. 10.1016/0144-8617(83)90016-4 [DOI] [Google Scholar]
- Chebli Y, Geitmann A. Cellular growth in plants requires regulation of cell wall biochemistry. Curr Opin Cell Biol. 2017:44:28–35. 10.1016/j.ceb.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Chen Y-L, Fan K-T, Hung S-C, Chen Y-R. The role of peptides cleaved from protein precursors in eliciting plant stress reactions. New Phytol. 2020b:225(6):2267–2282. 10.1111/nph.16241 [DOI] [PubMed] [Google Scholar]
- Chen S, Jia H, Zhao H, Liu D, Liu Y, Liu B, Bauer S, Somerville CR. Anisotropic cell expansion is affected through the bidirectional mobility of cellulose synthase complexes and phosphorylation at two critical residues on CESA3. Plant Physiol. 2016:171(1):242–250. 10.1104/pp.15.01874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Melton LD, McGillivray DJ, Ryan TM, Harris PJ. Changes in the orientations of cellulose microfibrils during the development of collenchyma cell walls of celery (Apium graveolens L). Planta. 2019:250(6):1819–1832. 10.1007/s00425-019-03262-8 [DOI] [PubMed] [Google Scholar]
- Chen HW, Persson S, Grebe M, McFarlane HE. Cellulose synthesis during cell plate assembly. Physiol Plant. 2018:164(1):17–26. 10.1111/ppl.12703 [DOI] [PubMed] [Google Scholar]
- Chen J, Zhu S, Ming Z, Liu X, Yu F. FERONIA cytoplasmic domain: node of varied signal outputs. aBIOTECH. 2020a:1(2):135–146. 10.1007/s42994-020-00017-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Zhou Y, Neelamegham S. DrawGlycan-SNFG: a robust tool to render glycans and glycopeptides with fragmentation information. Glycobiology. 2017:27(3):200–205. 10.1093/glycob/cww115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wu HM. THESEUS 1, FERONIA and relatives: a family of cell wall-sensing receptor kinases? Curr Opin Plant Biol. 2011:14(6):632–641. 10.1016/j.pbi.2011.09.001 [DOI] [PubMed] [Google Scholar]
- Cho H, Lee J, Oh E. Leucine-rich repeat receptor-like proteins in plants: structure, function, and signaling. J Plant Biol. 2022:66(2):99–107. 10.1007/s12374-022-09374-1 [DOI] [Google Scholar]
- Cho SH, Purushotham P, Fang C, Maranas C, Diaz-Moreno SM, Bulone V, Zimmer J, Kumar M, Nixon BT. Synthesis and self-assembly of cellulose microfibrils from reconstituted cellulose synthase. Plant Physiol. 2017:175(1):146–156. 10.1104/pp.17.00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou EY, Schuetz M, Hoffmann N, Watanabe Y, Sibout R, Samuels AL. Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J Exp Bot. 2018:69(8):1849–1859. 10.1093/jxb/ery067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coculo D, Lionetti V. The plant invertase/pectin methylesterase inhibitor superfamily. Front Plant Sci. 2022:13:863892. 10.3389/fpls.2022.863892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron J-C, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc Natl Acad Sci U S A. 2007:104(20):8550–8555. 10.1073/pnas.0703133104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin L, Ruhnow F, Zhu J-K, Zhao C, Zhao Y, Persson S. The cell biology of primary cell walls during salt stress. Plant Cell. 2023:35(1):201–217. 10.1093/plcell/koac292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Re-constructing our models of cellulose and primary cell wall assembly. Curr Opin Plant Biol. 2014:22:122–131. 10.1016/j.pbi.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol. 2015:25:162–172. 10.1016/j.pbi.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall modifying enzymes. J Exp Bot. 2016:67(2):463–476. 10.1093/jxb/erv511 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Building an extensible cell wall. Plant Physiol. 2022:189(3):1246–1277. 10.1093/plphys/kiac184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté F, Hahn MG. Oligosaccharins: structures and signal transduction. Plant Mol Biol. 1994:26(5):1379–1411. 10.1007/BF00016481 [DOI] [PubMed] [Google Scholar]
- Daher FB, Braybrook SA. How to let go: pectin and plant cell adhesion. Front Plant Sci. 2015:6:523. 10.3389/fpls.2015.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher FB, Chen Y, Bozorg B, Clough J, Jönsson H, Braybrook SA. Anisotropic growth is achieved through the additive mechanical effect of material anisotropy and elastic asymmetry. eLife. 2018:7:e38161. 10.7554/eLife.38161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S-Y, Hsu W-H, Yang C-H. The gene ANTHER DEHISCENCE REPRESSOR (ADR) controls male fertility by suppressing the ROS accumulation and anther cell wall thickening in Arabidopsis. Sci Rep. 2019:9(1):5112. 10.1038/s41598-019-41382-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvill AG, McNeil M, Albersheim P. Structure of plant cell walls: VIII. A new pectic polysaccharide. Plant Physiol. 1978:62(3):418–422. 10.1104/pp.62.3.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin LB, Lewis NG. Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 2000:123(2):453–462. 10.1104/pp.123.2.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin LB, Wang HB, Crowell AL, Bedgar DL, Martin DM, Sarkanen S, Lewis NG. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science. 1997:275(5298):362–366. 10.1126/science.275.5298.362 [DOI] [PubMed] [Google Scholar]
- Davis KR, Darvill AG, Albersheim P, Dell A. Host-pathogen interactions. XXIX. Oligogalacturonides released from sodium polypectate by endopolygalacturonic acid lyase are elicitors of phytoalexins in soybean. Plant Physiol. 1986:80(2):568–577. 10.1104/pp.80.2.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP. Biosynthesis of cellulose. Adv Carbohydrate Chem Biochem. 1983:41:105–153. 10.1016/S0065-2318(08)60057-8 [DOI] [PubMed] [Google Scholar]
- Delmer DP. Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Biol. 1999:50(1):245–276. 10.1146/annurev.arplant.50.1.245 [DOI] [PubMed] [Google Scholar]
- Delmer DP, Benziman M, Padan E. Requirement for a membrane potential for cellulose synthesis in intact cells of Acetobacter xylinum. Proc Natl Acad Sci U S A. 1982:79(17):5282–5286. 10.1073/pnas.79.17.5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer DP, Pear JR, Andrawis A, Stalker DM. Genes encoding small GTP-binding proteins analogous to mammalian rac are preferentially expressed in developing cotton fibers. Mol Gen Genet. 1995:248(1):43–51. 10.1007/BF02456612 [DOI] [PubMed] [Google Scholar]
- Delmer DP, Read SM, Cooper G. Identification of a receptor protein in cotton fibers for the herbicide 2, 6-dichlorobenzonitrile. Plant Physio. 1987:84:415–420. 10.1104/pp.84.2.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester B, Vanholme R, Mota T, Boerjan W. Lignin engineering in forest trees: from gene discovery to field trials. Plant Commun. 2022:3(6):100465. 10.1016/j.xplc.2022.100465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura T. Tracheary element differentiation. Plant Biotechnol Rep. 2013:8(1):17–21. 10.1007/s11816-013-0293-0 [DOI] [Google Scholar]
- De Vries JA, Voragen AGJ, Rombouts FM, Pilnik W. Extraction and purification of pectins from Alcohol Insoluble Solids from ripe and unripe apples. Carbohydr Polym. 1981:1(2):117–127. 10.1016/0144-8617(81)90004-7 [DOI] [Google Scholar]
- Dhugga KS, Barreirro R, Whistten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, et al. Guar seed (-mannan synthase is a member of the cellulose synthase super gene family. Science. 2004:303(5656):363–366. 10.1126/science.1090908 [DOI] [PubMed] [Google Scholar]
- Diener M, Adamcik J, Sánchez-Ferrer A, Jaedig F, Schefer L, Mezzenga R. Primary, secondary, tertiary and quaternary structure levels in linear polysaccharides: from random coil, to single helix to supramolecular assembly. Biomacromolecules. 2019:20(4):1731–1739. 10.1021/acs.biomac.9b00087 [DOI] [PubMed] [Google Scholar]
- Dima O, Morreel K, Vanholme B, Kim H, Ralph J, Boerjan W. Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell. 2015:27(3):695–710. 10.1105/tpc.114.134643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Barros J. Lignin biosynthesis- old roads revisited and new roads explored. Open Biol. 2019:9(12):190215. 10.1098/rsob.190215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM. Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol. 2001:125(4):2040–2052. 10.1104/pp.125.4.2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP. Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol. 2002:43(12):1407–1420. 10.1093/pcp/pcf164 [DOI] [PubMed] [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A. A barley cellulose synthase-like CSLH gene mediates (1,3:1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci U S A. 2009:106(14):5996–6001. 10.1073/pnas.0902019106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll NM, Berenguer E, Truskina J, Ingram G. AtEXT3 is not essential for early embryogenesis or plant viability in Arabidopsis. New Phytol. 2022:236(5):1629–1633. 10.1111/nph.18452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domozych DS, Sørensen I, Pettolino FA, Bacic A, Willats WGT. The cell wall polymers of the Charophycean green alga Chara corallina: immunobinding and biochemical screening. Int. J Plant Sci. 2010:171(4):345–361. 10.1086/651227 [DOI] [Google Scholar]
- Dougall DK, Shimbayashi K. Factors affecting growth of tobacco callus tissue and its incorporation of tyrosine. Plant Physiol. 1960:35(3):396–404. 10.1104/pp.35.3.396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G. Polysaccharide deposition during cytokinesis: challenges and future perspectives. Plant Sci. 2015:236:177–184. 10.1016/j.plantsci.2015.03.018 [DOI] [PubMed] [Google Scholar]
- Dressano K, Ceciliato PHO, Silva AL, Guerrero-Abad JC, Bergonci T, Ortiz-Morea FA, Burger M, Silva-Filho MC, Moura DS. BAK1 is involved in AtRALF1-induced inhibition of root cell expansion. PLoS Genet. 2017:13(10):e1007053. 10.1371/journal.pgen.1007053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressano K, Weckwerth PR, Poretsky E, Takahashi Y, Villarreal C, Shen Z, Schroeder JI, Briggs SP, Huffaker A. Dynamic regulation of Pep-induced immunity through post-translational control of defence transcript splicing. Nat Plants. 2020:6(8):1008–1019. 10.1038/s41477-020-0724-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drula E, Garron M-L, Dogan S, Lombard V, Henrissat B, Terrapon N. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022:50(D1):D571–D577. 10.1093/nar/gkab1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Anderson CT, Xiao C. Dynamics of pectic homogalacturonan in cellular morphogenesis and adhesion, wall integrity sensing and plant development. Nat Plants. 2022:8(4):332–340. 10.1038/s41477-022-01120-2 [DOI] [PubMed] [Google Scholar]
- Du J, Kirui A, Huang S, Wang L, Barnes WJ, Kiemle SN, Zheng Y, Rui Y, Ruan M, Qi S, et al. Mutations in the pectin methyltransferase QUASIMODO2 influence cellulose biosynthesis and wall integrity in Arabidopsis. Plant Cell. 2020a:32(11):3576–3597. 10.1105/tpc.20.00252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Spalding EP, Gray WM. Rapid auxin-mediated cell expansion. Ann Rev Plant Biol. 2020b:71(1):379–402. 10.1146/annurev-arplant-073019-025907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu H-M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci U S A. 2010:107(41):17821–17826. 10.1073/pnas.1005366107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert B, Birdseye D, Liwanag AJM, Laursen T, Rennie EA, Guo X, Catena M, Rautengarten C, Stonebloom SH, Gluza P, et al. The three members of the Arabidopsis glycosyltransferase family 92 are functional β-1,4-galactan synthases. Plant Cell Physiol. 2018:59(12):2624–2636. 10.1093/pcp/pcy180 [DOI] [PubMed] [Google Scholar]
- Edwards ME, Dickson CA, Chengappa S, Sidebottom C, Gidley MJ, Reid JSG. Molecular characterization of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 1999:19(6):691–697. 10.1046/j.1365-313x.1999.00566.x [DOI] [PubMed] [Google Scholar]
- Egelund J, Damager I, Faber K, Olsen CE, Ulvskov P, Petersen BL. Functional characterisation of a putative rhamnogalacturonan II specific xylosyltransferase. FEBS Lett. 2008:582(21-22):3217–3222. 10.1016/j.febslet.2008.08.015 [DOI] [PubMed] [Google Scholar]
- Egelund J, Petersen BL, Motawia MS, Damager I, Faik A, Olsen CE, Ishii T, Clausen H, Ulvskov P, Geshi N. Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-α-D-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell. 2006:18(10):2593–2607. 10.1105/tpc.105.036566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A. Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 2010:153(2):403–419. 10.1104/pp.110.156000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsdorf T, Gigli-Bisceglia N, Veerabagu M, McKenna JF, Vaahtera L, Augstein F, Van der Does D, Zipfel C, Hamann T. The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana. Sci Signal. 2018:11(536):eaao3070. 10.1126/scisignal.aao3070 [DOI] [PubMed] [Google Scholar]
- Engle KA, Amos RA, Yang JY, Glushka J, Atmodjo MA, Tan L, Huang C, Moremen KW, Mohnen D. Multiple Arabidopsis galacturonosyltransferases (GAUTs) synthesize polymeric homogalacturonan by oligosaccharide acceptor-dependent or de novo synthesis. Plant J. 2022:109(6):1441–1456. 10.1111/tpj.15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrissin I, Cueff G, Berger A, Granier F, Sallé C, Poulain D, Ralet MC, North HM. Natural variation reveals a key role for rhamnogalacturonan I in seed outer mucilage and underlying genes. Plant Physiol. 2019:181(4):1498–1518. 10.1104/pp.19.00763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K. An Arabidopsis gene encoding an (-xylosyltransferase involved in xyloglucan biosynthesis. Proc Natl Acad Sci U S A. 2002:99(11):7797–7802. 10.1073/pnas.102644799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Ryan E, Foreman J, Linstead P, Boudonck K, Steer M, Shaw P, Dolan L. KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 2001:15(1):79–89. 10.1101/gad.188801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici L, Matteo AD, Fernandez-Recio J, Tsernoglou D, Cervone F. Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci. 2006:11(2):65–70. 10.1016/j.tplants.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Feingold DS, Neufeld EF, Hassid WZ. Synthesis of a β-1, 3-linked glucan by extracts of Phaseolus aureus seedlings. J Biol Chem. 1958:233(4):783–788. 10.1016/S0021-9258(18)64655-1 [DOI] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu M-C, Maman J, Steinhorst L, Schmitz-Thom I, et al. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol. 2018:28(5):666–675.e5. 10.1016/j.cub.2018.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC. Nanostructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci U S A. 2011:108(47):E1195–E1203. 10.1073/pnas.1108942108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E. Syntheses in the purine and sugar group. Nobel Prize Lect. 1902:12:22–35. [Google Scholar]
- Fischer MH, Yu N, Gray GR, Ralph J, Anderson L, Marlett JA. The gel-forming polysaccharide of psyllium husk (Plantago ovata Forsk). Carbohydr Res. 2004:339(11):2009–2017. 10.1016/j.carres.2004.05.023 [DOI] [PubMed] [Google Scholar]
- Fleischer A, O'Neill MA, Ehwald R. The pore size of non-graminaceous plant cell walls is rapidly decreased by borate ester cross-linking of the pectic polysaccharide rhamnogalacturonan II. Plant Physiol. 1999:121(3):829–838. 10.1104/pp.121.3.829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francin-Allami M, Bouder A, Geairon A, Alvardo C, Le-Bot L, Daniel S, Shao M, Laudencia-Chingcuanco D, Vogel JP, Guillon F, et al. Mixed-linkage glucan is the main carbohydrate source and starch is an alternative source during Brachypodium grain germination. Int J Mol Sci. 2023:24(7):6821. 10.3390/ijms24076821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas CMP, Coimbra JSR, Souza VGL, Sousa RCS. Structure and applications of pectin in food, biomedical, and pharmaceutical industry: a review. Coatings. 2021:11(8):922. 10.3390/coatings11080922 [DOI] [Google Scholar]
- Fremy E. Recherches sur les matieres gelatineuses des fruits. Compt Rend. 1847:24:1076. [Google Scholar]
- Fremy ME. On the ripening of fruits and the gelatious bodies of vegetables. London Edinburgh Dublin Philos Mag J Sci. 1848:33(223):394–396. [Google Scholar]
- French AD. The structure and mechanism of biosynthesis of cellulose. Part I Structure. In: Kung S-D Yang S-F, editors. Discoveries in plant biology. Vol. 3. Singapore: World Scientific; 2000. p. 1975–2001. 10.1142/9789812813503_0008. [DOI] [Google Scholar]
- Freudenberg K. Biosynthesis and constitution of lignin. Nature. 1959:183(4669):1152–1155. 10.1038/1831152a0 [DOI] [PubMed] [Google Scholar]
- Freudenberg K, Neish AC. Constitution and biosynthesis of lignin (molecular biology, biochemistry, and biophysics). New York: Springer-Verlag; 1968. 132 pp. [Google Scholar]
- Frey-Wyssling A. The submicroscopic structure of cell walls. Sci Prog. 1939:34:249–262. [Google Scholar]
- Frey-Wyssling A, Ambronn H. Das polarisationmikroskop. Ber dt. Bot Ges. 1927:45:60–71. 10.1111/j.1438-8677.1927.tb03752.x [DOI] [Google Scholar]
- Frey-Wyssling A, Mühlethaler K. Die elementarfibrillen der cellulose. Die Makromolekulare Chemie. 1963:62(1):25–30. 10.1002/macp.1963.020620103 [DOI] [Google Scholar]
- Fry SC. In-vivo formation of xyloglucan nonasaccharide: a possible biologically active cell-wall fragment. Planta. 1986:169(3):443–453. 10.1007/BF00392143 [DOI] [PubMed] [Google Scholar]
- Fry SC. Oligosaccharins as plant growth regulators. Biochem Soc Symp. 1994:60:5–14. [PubMed] [Google Scholar]
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau J-P, Kato Y, Lorences EP, Maclachlan GA, McNeil M, et al. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant. 1993:89(1):1–3. 10.1111/j.1399-3054.1993.tb01778.x [DOI] [Google Scholar]
- Fujita M, Wasteneys GO. A survey of cellulose microfibril patterns in dividing, expanding, and differentiating cells of Arabidopsis thaliana. Protoplasma. 2014:251(3):687–698. 10.1007/s00709-013-0571-2 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Jikumaru Y, Kamiya Y, Tang Y, Dixon RA. Selective lignin down-regulation leads to constitutive defense response expression in alfalfa (Medicago sativa L.). New Phytol. 2011:190(3):627–639. 10.1111/j.1469-8137.2010.03621.x [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Liu C, Pose-Albacete S, Pattathil S, Peralta AG, Young J, Westphaling J, Hahn M, Rao X, de Meester B, et al. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1) releases latent defense signals in stems with reduced lignin content. Proc Natl Acad Sci U S A. 2020:117(6):3281–3290. 10.1073/pnas.1914422117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Pose-Albacete S, Pattathil S, Peralta AG, Hahn M, Ayre BG, Sunuwar H, Hernandez J, Patel M, Shah J, et al. Elicitors and defense gene induction in plants with altered lignin compositions. New Phytol. 2018:219(4):1235–1251. 10.1111/nph.15258 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L, Shadle G, Shen H, Barros-Rios J, Corrales SF, Wang H, Dixon RA. Combining enhanced biomass density with reduced lignin level for improved forage quality. Plant Biotech J. 2015:14(3):895–904. 10.1111/pbi.12439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Zhu C, Chen W, Dixit R. FRA1 kinesin modulates the lateral stability of cortical microtubules through cellulose synthase-microtubule uncoupling proteins. Plant Cell. 2020:32(8):2508–2524. 10.1105/tpc.19.00700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Lipton AS, Munson CR, Ma Y, Johnson KL, Murray DT, Scheller HV, Mortimer JC. Elongated galactan side-chains mediate cellulose-pectin interactions in engineered Arabidopsis secondary cell walls. Plant J. 2023:115(2):529–545. 10.1111/tpj.16242 [DOI] [PubMed] [Google Scholar]
- Gao Z, Sun W, Wang J, Zhao C, Zuo K. GhbHLH18 negatively regulates fiber strength and length by enhancing lignin biosynthesis in cotton fibers. Plant Sci. 2019:286:7–16. 10.1016/j.plantsci.2019.05.020 [DOI] [PubMed] [Google Scholar]
- Gardner K, Blackwell J. The structure of native cellulose. Biopolymers. 1974:13(10):1975–2001. 10.1002/bip.1974.360131005 [DOI] [Google Scholar]
- Giddings TH, Brower TL, Staehelin LA. Visualization of the particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J Cell Biol. 1980:84(2):327–339. 10.1083/jcb.84.2.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigli-Bisceglia N, van Zelm E, Huo W, Lamers J, Testerink C. Arabidopsis root responses to salinity depend on pectin modification and cell wall sensing. Development. 2022:149(12):dev200363. 10.1242/dev.200363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser L. The synthesis of cellulose in cell-free extracts of Acetobacter xylinum. J Biol Chem. 1958:232(2):627–636. 10.1016/S0021-9258(19)77383-9 [DOI] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Martin M, Doblas VG, Bacete L, Miart F, Sormani R, Hematy K, Renou J, Landrein B, et al. Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis. Curr Biol. 2018:28(15):2452–2458. 10.1016/j.cub.2018.05.075 [DOI] [PubMed] [Google Scholar]
- Gorshkova T, Petrova A, Mikshina P. Review: tertiary cell wall of plant fibers as a source of inspiration in material design. Carbohydr Polym. 2022:295:119849. 10.1016/j.carbpol.2022.119849 [DOI] [PubMed] [Google Scholar]
- Grantham NJ, Wurman-Rodrich J, Terrett OM, Lyczakowski JJ, Stott K, Iuga D, Simmons TJ, Durand-Tardif M, Brown SP, Dupree R, et al. An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat Plants. 2017:3(11):859–865. 10.1038/s41477-017-0030-8 [DOI] [PubMed] [Google Scholar]
- Groover A, Jones AM. Tracheary element differentiation uses a novel mechanism coordinating programmed cell death and secondary cell wall synthesis. Plant Physiol. 1999:119(2):375–384. 10.1104/pp.119.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Bringmann M, Combs JR, Yang J, Bergmann DC, Nielsen E. Arabidopsis CSLD5 functions in cell plate formation in a cell cycle-dependent manner. Plant Cell. 2016:28(7):1722–1737. 10.1105/tpc.16.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin T I, Persson S, Somerville CR. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc Natl Acad Sci U S A. 2010:107(29):12866–12871. 10.1073/pnas.1007092107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2009:106(18):7648–7653. 10.1073/pnas.0812346106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Rao X, Saxena G, Dixon RA. Growth-defense trade-offs as a result of lignin pathway engineering. New Phytol. 2021:231(1):60–74. 10.1111/nph.17383 [DOI] [PubMed] [Google Scholar]
- Haas KT, Wightman R, Meyerowitz EM, Peaucelle A. Pectin homogalacturonan nanofilament expansion drives morphogenesis in plant epidermal cells. Science. 2020:367(6481):1003–1007. 10.1126/science.aaz5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas KT, Wightman R, Peaucelle A, Höfte H. The role of pectin phase separation in plant cell wall assembly and growth. Cell Surface. 2021:7:100054. 10.106/j.tcsw.2021.100054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler CH, Betancur L, Stiff MR, Tuttle JR. Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front Plant Sci. 2012:3:104. 10.3389/fpls.2012.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler CH, Ivanova-Datcheva M, Hogan PS, Salnikov, Vv, Hwang S, Martin K, Delmer DP. Carbon partitioning to cellulose synthesis. Plant Mol Biol. 2001:47:29–51. 10.1007/978-94-010-0668-2_3. [DOI] [PubMed] [Google Scholar]
- Haigler CH, Roberts AW. Structure/function relationships in the rosette cellulose synthesis complex illuminated by an evolutionary perspective. Cellulose. 2019:26(1):227–247. 10.1007/s10570-018-2157-9 [DOI] [Google Scholar]
- Hamant O, Inoue D, Bouchez D, Dumais J, Mjolsness E. Are microtubules tension sensors? Nat Commun. 2019:10(1):2360–2373. 10.1038/s41467-019-10207-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Mohnen D. A review of xylan and lignin biosynthesis: foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes. Crit Rev Biochem Mol Biol. 2014:49(3):212–241. 10.3109/10409238.2014.889651 [DOI] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sorensen SO, Orfila C, Pauly M, Scheller HV. ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 2006:140(1):49–58. 10.1104/pp.105.072744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Verhertbruggen Y, Søgaard C, Bernard S, Nafisi M, Poulsen CP, Geshi N, Sakuragi Y, Driouich A, et al. ARAD proteins associated with pectic arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta. 2012:236(1):115–128. 10.1007/s00425-012-1592-3 [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman M. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science. 2014:343(6169):408–411. 10.1126/science.1244454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth WN, Hirst EL, Thomas HA. CXVI.—Polysaccharides. Part VII. Isolation of octamethyl cellobiose, hendecamethyl cellotriose, and a methylated cellodextrin (cellotetrose?) as crystalline products of the acetolysis of cellulose derivatives. J Chem Soc (Resumed). 1931:(0):824–829. 10.1039/JR9310000824 [DOI] [Google Scholar]
- Haworth WN, Long CW, Plant JHG. CCCLXXVI.—The constitution of the disaccharide. Part XVI. Cellobiose. J Chem Soc (Resumed). 1927:(0):2809–2814. 10.1039/JR9270002809 [DOI] [Google Scholar]
- Hayashi T. Xyloglucans in the primary cell wall. Ann Rev Plant Physiol Plant Mol Biol. 1989:40(1):139–168. 10.1146/annurev.pp.40.060189.001035 [DOI] [Google Scholar]
- Hayashi T, Kaida R. Functions of xyloglucan in plant cells. Mol Plant. 2011:4(1):17–24. 10.1093/mp/ssq063 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Read SM, Bussell J, Thelen M, Lin F-C, Brown RM Jr, Delmer DP. UDP-Glucose: (1-3)-β-glucan synthases from mung bean and cotton. Diifferential effects of Ca2+ and Mg2+ on enzyme properties and structure of the reaction product. Plant Physiol. 1987:83(4):1054–1062. 10.1104/pp.83.4.1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hematy K, Sado PE, Van Tuinen A, Rochange S, Desnos T, Balzergue S, Pelletier S, Renou JP, Hofte H. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol. 2007:17(11):922–931. 10.1016/j.cub.2007.05.018 [DOI] [PubMed] [Google Scholar]
- Henry RJ, Stone BA. Factors influencing β-glucan synthesis by particulate enzymes from suspension-cultured Lolium multiflorum endosperm cells. Plant Physiol. 1982a:69(3):632–636. 10.1104/pp.69.3.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RJ, Stone BA. Solubilization of β-glucan synthases from the membrane of cultured ryegrass endosperm cells. Biochem J. 1982b:203(3):629–636. 10.1042/bj2030629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler P. Calcium: a central regulator of plant growth and development. Plant Cell. 2005:17(8):2142–2155. 10.1105/tpc.105.032508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herger A, Dunser K, Kleine-Vehn J, Ringli C. Leucine-rich repeat extensin proteins and their role in cell wall sensing. Curr Biol. 2019:29(17):R851–R858. 10.1016/j.cub.2019.07.039 [DOI] [PubMed] [Google Scholar]
- Herth W. Plasma-membrane rosettes involved in localized wall thickening during xylem vessel formation of Lepidium sativum L. Planta. 1985:163(1):12–21. 10.1007/BF00391020 [DOI] [PubMed] [Google Scholar]
- Hestrin S. Synthesis of polymer homopolysaccharides. In: Gunsalus RY, Stanier IC, editors. The bacteria: A treatise on structure and function (Volume IIII Biosynthesis). iii. New York: Academic Press; 1962. p. 373–388. [Google Scholar]
- Hieta K, Kuga S, Usuda M. Electron staining of reducing ends evidences a parallel-chain structure in Valonia cellulose. Biopolymers. 1984:23(10):1807–1810. 10.1002/bip.360231002 [DOI] [Google Scholar]
- Hill JL Jr, Josephs C, Barnes WJ, Anderson CT, Tien M. Longevity in vivo of primary cell wall cellulose synthases. Plant Mol Biol. 2018:96(3):279–289. 10.1007/s11103-017-0695-4 [DOI] [PubMed] [Google Scholar]
- Hobson N, Roach MJ, Deyholos MK. Gene expression in tension wood and bast fibres. Russian J Plant Physiol. 2010:57(3):321–327. 10.1134/S1021443710030039 [DOI] [Google Scholar]
- Hocq L, Pelloux J, Lefebvre V. Connecting homogalacturonan-type pectin remodeling to acid growth. Trends Plant Sci. 2017a:22(1):20–29. 10.1016/j.tplants.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Hocq L, Sénéchal F, Lefebvre F, Lehner A, Domon JM, Mollet JC, Dehors J, Pageau K, Marcelo P, Guérineau F, et al. Combined experimental and computational approaches reveal distinct pH dependence of pectin methylesterase inhibitors. Plant Physiol. 2017b:173(2):1075–1093. 10.1104/pp.16.01790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfte H. The yin and yang of cell wall integrity control: brassinosteroid and FERONIA signaling. Plant Cell Physiol. 2015:56(2):224–231. 10.1093/pcp/pcu182 [DOI] [PubMed] [Google Scholar]
- Höfte H. A dive into the cell wall with Arabidopsis. Comp Rend Biol. 2023:345(4):1–20. 10.5802/crbiol.101 [DOI] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development. 2003:130(10):2149–2159. 10.1242/dev.00458 [DOI] [PubMed] [Google Scholar]
- Humphreys JM, Hemm MR, Chapple C. New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci U S A. 1999:96(18):10045–10050. 10.1073/pnas.96.18.10045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysmans M, Lema AS, Coll NS, Nowack MK. Dying two deaths—programmed cell death regulation in development and disease. Curr Opin Plant Biol. 2017:35:37–44. 10.1016/j.pbi.2016.11.005 [DOI] [PubMed] [Google Scholar]
- Ibragimova NN, Ageeva MV, Gorshkova TA. Development of gravitropic response: unusual behavior of flax phloem G-fibers. Protoplasma. 2017:254(2):749–762. 10.1007/s00709-016-0985-8 [DOI] [PubMed] [Google Scholar]
- Ishida K, Yokoyama R. Reconsidering the function of the xyloglucan endotransglucosyl/ hydrolase family. J Plant Res. 2022:135(2):145–156. 10.1007/s10265-021-01361-w [DOI] [PubMed] [Google Scholar]
- Itoh T, Brown RM Jr. The assembly of cellulose microfibrils in Valonia macrophysa Kutz. Planta. 1984:160(4):372–381. 10.1007/BF00393419 [DOI] [PubMed] [Google Scholar]
- Jacob-Wilk D, Kurek I, Hogan P, Delmer DP. The cotton fiber zinc-binding domain of cellulose synthase A1 from Gossypium hirsutum displays rapid turnover in vitro and in vivo. Proc Natl Acad Sci U S A. 2006:103(32):12191–12196. 10.1073/pnas.0605098103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen EF, Jang R, Albersheim P, Bonner J. Pectic metabolism of growing cell walls. Plant Physiol. 1960:35(1):87–97. 10.1104/pp.35.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis MC. Cellulose biosynthesis: counting the chains. Plant Physiol. 2013:163(4):1485–1486. 10.1104/pp.113.231092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis MC. Structure of native cellulose microfibrils, the starting point for nanocellulose manufacture. Philos Trans R Soc A. 2018:376(2112):2017045. 10.1098/rsta.2017.0045 [DOI] [PubMed] [Google Scholar]
- Jarvis MC. Hydrogen bonding and other non-covalent interactions at the surfaces of cellulose microfibrils. Cellulose. 2023:30(2):667–687. 10.1007/s10570-022-04954-3 [DOI] [Google Scholar]
- Jensen JK, Busse-Wicher M, Poulsen CP, Fangel JU, Smith PJ, Yang J-Y, Peña M-J, Dinesen MH, Martens HJ, Melkonian M, et al. Identification of an algal xylan synthase indicates that there is functional orthology between algal and plant cell wall biosynthesis. New Phytol. 2018:218(3):1049–1060. 10.1111/nph.15050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Johnson N, Wilkerson CG. Discovery of diversity in xylan biosynthesis genes by transcriptional profiling of a heteroxylan containing mucilaginous tissue. Front Plant Sci. 2013:4:183. 10.3389/fpls.2013.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Schultink A, Keegstra K, Wilkerson CG, Pauly M. RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol Plant. 2012:5(5):984–992. 10.1093/mp/sss032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Sorensen SO, Harholt J, Geshi N, Sakuragi Y, Moller I, Zandleven J, Bernal AJ, Jensen NB, Sørensen C, et al. Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell. 2008:20(5):1289–1302. 10.1105/tpc.107.050906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobert F, Yadav S, Robert S. Auxin as an architect of the pectin matrix. J Exp Bot. 2023:74(22):6933–:6949. 10.1093/jxb/erad174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Harper JF, Palanivelu R. A fruitful journey: pollen tube navigation from germination to fertilization. Annu Rev Plant Biol. 2019:70(1):809–837. 10.1146/annurev-arplant-050718-100133 [DOI] [PubMed] [Google Scholar]
- Jönsson K, Lathe RS, Kierzkowski D, Routier-Kierzkowska A-L, Hamant O, Bhalearo RP. Mechanochemical feedback mediates tissue bending required for seedling emergence. Curr Biol. 2021:31(6):1154–1164. 10.1016/j.cub.2020.12.016 [DOI] [PubMed] [Google Scholar]
- Julian JD, Zabotina OA. Xyloglucan biosynthesis: from genes to proteins and their functions. Front Pl Sci. 2022:13:920494. 10.3389/fpls.2022.920494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczmarska A, Pieczywek PM, Cybulska J, Zdunek A. Structure and functionality of rhamnogalacturonan I in the cell wall and in solution: a review. Carbohydr Polym. 2022:278:118909. 10.1016/j.carbpol.2021.118909 [DOI] [PubMed] [Google Scholar]
- Kaczmarska A, Pieczywek PM, Cybulska J, Zdunek A. A mini-review on the plant sources and methods for extraction of rhamnogalacturonan I. Food Chem. 2023:403:134378. 10.1016/j.foodchem.2022.134378 [DOI] [PubMed] [Google Scholar]
- Kanaya T, Kaji K. History of fiber structure. In: High-performance and specialty fibers. The Society of Fiber Science and Technology Japan , editors. Tokyo: Springer; 2016. p. 3–21. 10.1007/978-4-431-55203-1_1 [DOI] [Google Scholar]
- Kang X, Kirui A, Dickwella Widanage MC, Mentick-Vigier F, Cosgrove DJ, Wang T. Lignin-polysaccharide interactions in plant secondary cell walls revealed by solid-state NMR. Nat Commun. 2019:10(1):347. 10.1038/s41467-018-08252-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor K, Geitmann A. Pollen tube invasive growth is promoted by callose. Plant Reprod. 2023:36(2):157–171. 10.1007/s00497-023-00458-7 [DOI] [PubMed] [Google Scholar]
- Karlsson M, Melzer M, Prokhorenko I, Johansson T, Wingsle G. Hydrogen peroxide and expression of hipI-superoxide dismutase are associated with the development of secondary cell walls in Zinnia elegans. J Exp Bot. 2005:56(418):2085–2093. 10.1093/jxb/eri207 [DOI] [PubMed] [Google Scholar]
- Keegstra K, Talmadge KW, Bauer WD, Albersheim P. The structure of plant cell walls III. A model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiol. 1973:51(1):188–196. 10.1104/pp.51.1.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller B, Templeton MD, Lamb CJ. Specific localization of a plant cell wall glycine-rich protein in protoxylem cells of the vascular system. Proc Natl Acad Sci U S A. 1989:86(5):1529–1533. 10.1073/pnas.86.5.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr T, Bailey IW. The cambium and its derivative tissues: No. X. Structure, optical properties and chemical composition of the so-called middle lamella. J Arnold Arboretum (Harvard University). 1934:15(4):327–349. 10.5962/p.185316 [DOI] [Google Scholar]
- Kertesz ZI. The pectic substances. New York: Interscience Publishers; 1951, p. 1–628. [Google Scholar]
- Kesten C, Menna A, Sánchez-Rodríguez C. Regulation of cellulose synthesis in response to stress. Curr Opin Plant Biol. 2017:40:106–113. 10.1016/j.pbi.2017.08.010 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, de Zacks R, Leykam JF, Lamport DTA. A repetitive proline-rich protein from the gymnosperm Douglas fir is a hydroxyproline-rich glycoprotein. Plant Physiol. 1992a:98(3):919–926. 10.1104/pp.98.3.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski MJ, Kamyab A, Leykam JF, Lamport DTA. A histidine-rich extensin from Zea mays is an arabinogalactan protein. Plant Physiol. 1992b:99(2):538–547. 10.1104/pp.99.2.538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DTA. Extensive: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 1994:5(2):157–172. 10.1046/j.1365-313X.1994.05020157.x [DOI] [PubMed] [Google Scholar]
- Kim SJ, Chandrasekar B, Rea AC, Danhof L, Zemelis-Durfee S, Thrower N, Shepard ZS, Pauly M, Brandizzi F, Keegstra K. The synthesis of xyloglucan, an abundant plant cell wall polysaccharide, requires CSLC function. Proc Natl Acad Sci U S A. 2020:117(33):20316–20324. 10.1073/pnas.2007245117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Shiu SH, Thoma S, Li WH, Patterson SE. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2006:7(9):R87. 10.1186/gb-2006-7-9-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Beers EP, Han KH. Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Mol Genet Genomics. 2006:276(6):517–531. 10.1007/s00438-006-0157-1 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Matoh T, Azuma J. Two chains of rhamnogalacturonan II are cross-linked by borate-diol ester bonds in higher plant cell walls. Plant Physiol. 1996:110(3):1017–1020. 10.1104/pp.110.3.1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Dexter-Meldrum J, Zorensky FDH, Chabout S, Mouille G, Kohorn S. Pectin dependent cell adhesion restored by a mutant microtubule organizing membrane protein. Plants. 2021a:10(4):690. 10.3390/plants10040690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Greed BE, Mouille G, Verger S, Kohorn SL. Effects of Arabidopsis wall associated kinase mutations on ESMERALDA1 and elicitor induced ROS. PLoS One. 2021b:16(5):e0251922. 10.1371/journal.pone.0251922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang LF, Koch K, Fu S, Dotson A, Byers N. An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 2006:46(2):307–316. 10.1111/j.1365-313X.2006.02695.x [DOI] [PubMed] [Google Scholar]
- Kolpak F, Blackwell J. Determination of the structure of cellulose II. Macromolecules. 1976:9(2):273–278. 10.1021/ma60050a019 [DOI] [PubMed] [Google Scholar]
- Kong Y, Pena MJ, Renna L, Avci U, Pattathil S, Tuomivaara ST, Li X, Reiter W-D, Brandizzi F, Hahn M, et al. Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol. 2015:167(4):1296–1306. 10.1104/pp.114.255943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooiman P. Amyloids of seed plants. Nature. 1957:179(4550):107–109. 10.1038/179107a0 [DOI] [Google Scholar]
- Kooiman P. The constitution of the amyloid from seeds of Annona Muricata L. Phytochemistry. 1967:4(12):1665–1673. 10.1016/S0031-9422(00)82900-3 [DOI] [Google Scholar]
- Kroeger JH, Geitmann A, Grant M. Model for calcium dependent oscillatory growth in pollen tubes. J Theor Biol. 2008:253(2):363–374. 10.1016/j.jtbi.2008.02.042 [DOI] [PubMed] [Google Scholar]
- Kroon-Batenburg LMJ, Kroon J. The crystal and molecular structures of cellulose I and II. Glycoconjugate J. 1997:14(5):677–690. 10.1023/A:1018509231331 [DOI] [PubMed] [Google Scholar]
- Kubicki JD, Yang H, Sawada D, O'Neill H, Oehme D, Cosgrove D. The shape of native plant cellulose microfibrils. Sci Rep. 2018:8(1):13983. 10.1038/s41598-018-32211-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuki H, Yokoyama R, Kuroha T, Nishitani K. Xyloglucan is not essential for the formation and integrity of the cellulose network in the primary cell wall regenerated from Arabidopsis protoplasts. Plants. 2020:9(5):629. 10.3390/plants9050629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Wightman R, Atanassov I, Gupta A, Hurst CH, Hemsley PA, Turner S. S-Acylation of the celluose synthase complex is essential for its plasma membrane localization. Science. 2016:353(6295):166–169. 10.1126/science.aaf4009 [DOI] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Jacob-Wilk D, Doblin M, Delmer D. Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci U S A. 2022:99(17):11109–11114. 10.1073/pnas.162077099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Lui ACW, Wang L, Liu H, Umezawa T, Tobimatsu Y, Lo C. Tricin biosynthesis and bioengineering. Front Plant Sci. 2021:12:733198. 10.3389/fpls.2021.733198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DTA. Oxygen fixation into hydroxyproline of plant cell wall protein. J Biol Chem. 1963:238(4):1438–1440. 10.1016/S0021-9258(18)81202-9 [DOI] [PubMed] [Google Scholar]
- Lamport DTA, Kieliszewski MJ, Chen Y, Cannon MC. Role of the extension superfamily in primary cell wall architecture. Plant Physiol. 2011:156(1):11–19. 10.1104/pp.110.169011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport DTA, Northcote DH. Hydroxyproline in primary cell walls of higher plants. Nature. 1960:188(4751):665–666. 10.1038/188665b013757790 [DOI] [Google Scholar]
- Lampugnani ER, Flores-Sandoval E, Tan QW, Mutwil M, Bowman JL, Persson S. Cellulose synthesis—central components and their evolutionary relationships. Trends Plant Sci. 2019:24(5):402–412. 10.1016/j.tplants.2019.02.011 [DOI] [PubMed] [Google Scholar]
- Lapierre C, Monties B, Rolando C, de Chirale L. Thioacidolysis of lignin: comparison with acidolysis. J Wood Chem Technol. 1985:5(2):277–292. 10.1080/02773818508085193 [DOI] [Google Scholar]
- Lau JM, McNeil M, Darvill AG, Albersheim P. Structure of the backbone of rhamnogalacturonan I, a pectic polysaccharide in the primary cell walls of plants. Carbohydr Res. 1985:137:111–125. 10.1016/0008-6215(85)85153-3 [DOI] [Google Scholar]
- Laursen T, Stonebloom SH, Pidatala VR, Birdseye DS, Clausen MH, Mortimer JC, Scheller HV. Bifunctional glycosyltransferases catalyze both extension and termination of pectic galactan oligosaccharides. Plant J. 2018:94(2):340–351. 10.1111/tpj.13860 [DOI] [PubMed] [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013:153(2):402–412. 10.1016/j.cell.2013.02.045 [DOI] [PubMed] [Google Scholar]
- Lei L, Li S, Gu Y. Cellulose synthase complexes: composition and regulation. Front Plant Sci. 2012:3:75. 10.3389/fpls.2012.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Zhang T, Strasser R, Lee CM, Gonneau M, Mach L, Vernhettes S, Kim SH, Cosgrove DJ, Li S, et al. The jiaoyao1 mutant is an allele of korrigan1 that abolishes endoglucanase activity and affects the organization of both cellulose microfibrils and microtubules in Arabidopsis. Plant Cell. 2014:26(6):2601–2616. 10.1105/tpc.114.126193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leloir LF. Far away and long ago. Ann Rev Biochem. 1983:52(1):1–15. 10.1146/annurev.bi.52.070183.000245 [DOI] [PubMed] [Google Scholar]
- Leng Y, Yang Y, Ren D, Huang L, Dai L, Wang Y, Chen L, Tu Z, Gao Y, Li X, et al. A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol. 2017:172(2):1151–1166. 10.1104/pp.16.01625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppard GG, Colvin JR. Electron-opaque fibrils and granules in and between the cell walls of higher plants. J Cell Biol. 1972:53(3):695–703. 10.1083/jcb.53.3.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel O, Cavalier DM, Liepman AH, Keegstra K. Biosynthesis of plant cell wall polysaccharides-a complex process. Curr Opin Plant Biol. 2006:9(6):621–630. 10.1016/j.pbi.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Leszczuk A, Kalaitzis P, Kulik J, Zdunek A. Review: structure and modifications of arabinogalactan proteins (AGPs). BMC Plant Biol. 2023:23(1):45. 10.1186/s12870-023-04066-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NG, Yamamoto E. Lignin: occurrence, biogenesis and biodegradation. Annu Rev Plant Physiol Plant Mol Biol. 1990:41(1):455–496. 10.1146/annurev.pp.41.060190.002323 [DOI] [PubMed] [Google Scholar]
- Li Z, Fernie A R, Persson S. Transition of primary to secondary cell wall synthesis. Sci Bull. 2016c:61(11):838–846. 10.1007/s11434-016-1061-7 [DOI] [Google Scholar]
- Li M, Pu Y, Ragauskas AJ. Current understanding of the correlation of lignin structure with biomass recalcitrance. Front Chem. 2016b:4:45. 10.3389/fchem.2016.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Verstraeten I, Roosjen M, Takahashi K, Rodriguez L, Merrin J, Chen J, Shabala L, Smet W, Ren H, et al. Cell surface and intracellular auxin signalling for H(+) fluxes in root growth. Nature. 2021:599(7884):273–277. 10.1038/s41586-021-04037-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wu HM, Cheung AY. FERONIA and her pals: functions and mechanisms. Plant Physiol. 2016a:171(4):2379–2392. 10.1104/pp.16.00667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yang T, Dai D, Hu Y, Guo X, Guo H. Evolution, gene expression profiling and 3D modeling of CSLD proteins in cotton. BMC Plant Biol. 2017:17(1):119. 10.1186/s12870-017-1063-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yeh F-L, Cheung AY, Duan Q, Kita D, Liu M-C, Maman J, Luu EJ, Wu BW, Gates L, et al. Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis. Elife. 2015:4:e06587. 10.7554/eLife.06587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yoo CG, Pu Y, Biswal AK, Tolbert AK, Mohnen D, Ragauskas AJ. Downregulation of pectin biosynthesis gene GAUT4 leads to reduced ferulate and lignin-carbohydrate cross-linking in switchgrass. Commun Biol. 2019:2(1):22. 10.1038/s42003-018-0265-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Cavalier DM. The CELLULOSE SYNTHASE-LIKE A and CELLULOSE SYNTHASE-LIKE C families: recent advances and future perspectives. Front Plant Sci. 2012:3:109. 10.3389/fpls.2012.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Wightman R, Geshi N, Turner SR, Scheller HV. Arabidopsis—a powerful model system for plant cell wall research. Plant J. 2010:61(6):1107–1121. 10.1111/j.1365-313X.2010.04161.x [DOI] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci U S A. 2005:102(6):2221–2226. 10.1073/pnas.0409179102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Lopez-Sanchez P, Gidley MJ. Interactions of pectins with cellulose during its synthesis in the absence of calcium. Food Hydrocoll. 2016:52:57–68. 10.1016/j.foodhyd.2015.06.004 [DOI] [Google Scholar]
- Lin S, Miao Y, Huang H, Zhang Y, Huang L, Cao J. Arabinogalactan proteins: focus on the role in cellulose synthesis and deposition during plant cell wall biogenesis. Int J Mol Sci. 2022a:23(12):6578. 10.3390/ijms23126578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Tang W, Pan X, Huang A, Gao X, Anderson CT, Yang Z. Arabidopsis pavement cell morphogenesis requires FERONIA binding to pectin for activation of ROP GTPase signaling. Curr Biol. 2022b:32(3):497–507. 10.1016/j.cub.2021.11.030 [DOI] [PubMed] [Google Scholar]
- Lindner H, Müller LM, Boisson-Dernier A, Grossniklaus U. CrRLK1L receptor-like kinases: not just another brick in the wall. Curr Opin Plant Biol. 2012:15(6):659–669. 10.1016/j.pbi.2012.07.003 [DOI] [PubMed] [Google Scholar]
- Liners F, Gaspar T, Van Cutsem P. Acetyl- and methyl-esterification of pectins of friable and compact sugar-beet calli: consequences for intercellular adhesion. Planta. 1994:192(4):545–556. 10.1007/BF00203593 [DOI] [Google Scholar]
- Little A, Schwerdt JG, Shirley NJ, Khor SF, Neumann K, O’Donovan LA, Lahnstein J, Collins HM, Henderson M, Fincher GB, et al. Revised phylogeny of the Cellulose Synthase gene superfamily: insights into cell wall evolution. Plant Physiol. 2018:177(3):1124–1141. 10.1104/pp.17.01718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Cui H, Zhang B, Song M, Chen S, Xiao C, Tang Y, Liesche J. Reduced pectin content of cell walls prevents stress-induced root cell elongation in Arabidopsis. J Exp Bot. 2021:72(4):1073–1084. 10.1093/jxb/eraa533 [DOI] [PubMed] [Google Scholar]
- Liu Z, Schneider R, Kesten C, Zhang Y, Somssich M, Zhang Y, Fernie AR, Persson S. Cellulose-microtubule uncoupling proteins prevent lateral displacement of microtubules during cellulose synthesis in Arabidopsis. Dev Cell. 2016:38(3):305–315. 10.1016/j.devcel.2016.06.032 [DOI] [PubMed] [Google Scholar]
- Liu H, Timko MP. Jasmonic acid signaling and molecular crosstalk with other phytohormones. Int J Mol Sci. 2021:22(6):2914. 10.3390/ijms22062914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yu H, Voxeur A, Rao X, Dixon RA. FERONIA and wall-associated kinases coordinate the defense response induced by cell wall engineering in Arabidopsis. Sci Adv. 2023:9(10):eadf7714. 10.1126/sciadv.adf7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liwanag AJM, Ebert B, Verhertbruggen Y, Rennie EA, Rautengarten C, Oikawa A, Andersen MCF, Clausen MH, Scheller HV. Pectin biosynthesis: GALS1 in Arabidopsis thaliana is a β-1,4-galactan β-1,4-galactosyltransferase. Plant Cell. 2012:24(12):5024–5036. 10.1105/tpc.112.106625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Hernandez F, Tryfona T, Rizza A, Yu XL, Harris MOB, Webb AR, Kotake T, Dupree P. Calcium binding by arabinogalactan polysaccharides is important for normal plant development. Plant Cell. 2020:32(10):3346–3369. 10.1105/tpc.20.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori M, van Verk MC, Hander T, Schatowitz H, Klauser D, Flury P, Gehring CA, Boller T, Bartels S. Evolutionary divergence of the plant elicitor peptides (peps) and their receptors: interfamily incompatibility of perception but compatibility of downstream signalling. J Exp Bot. 2015:66(17):5315–5325. 10.1093/jxb/erv236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund CH, Stenbæk A, Atmodjo MA, Rasmussen RE, Moller IE, Erstad SM, Biswal AK, Mohnen D, Mravec J, Sakuragi Y. Pectin synthesis and pollen tube growth in Arabidopsis involves three GAUT1 Golgi-anchoring proteins: gAUT5, GAUT6 and GAUT7. Front Plant Sci. 2020:11:585774. 10.3389/fpls.2020.585774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Johnson K. Arabinogalactan proteins—multifunctional glycoproteins of the plant cell wall. Cell Surf. 2023:9:100102. 10.1016/j.tcsw.2023.100102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter W-D. The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell. 2003:15(7):1662–1670. 10.1105/tpc.009837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarem M, Nishiyama Y, Xin X, Durachko DM, Gu Y, Cosgrove DJ, Kim SH. Distinguishing mesoscale polar order (unidirectional vs bidirectional) of cellulose microfibrils in plant cell walls using sum frequency generation spectroscopy. J Phys Chem B. 2020:124(37):8071–8081. 10.1021/acs.jpcb.0c07076 [DOI] [PubMed] [Google Scholar]
- Malivert A, Erguvan O, Chevallie A, Dehem A, Friaud R, Liu M, Martin M, Peyraud T, Hamant O, Verger S. FERONIA and microtubules independently contribute to mechanical integrity in the Arabidopsis shoot. PloS Biol. 2021:19:e3001454. 10.1371/journal.pbio.3001454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malivert A, Hamant O. Why is FERONIA pleiotropic? Nat Plants. 2023:9(7):1018–1025. 10.1038/s41477-023-01434-9 [DOI] [PubMed] [Google Scholar]
- Maltby D, Carpita NC, Montezinos D, Kulow C, Delmer DP. β-1, 3-glucan in developing cotton fibers: structure, localization, and relationship of synthesis to that of secondary wall cellulose. Plant Physiol. 1979:63(6):1158–1164. 10.1104/pp.63.6.1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfield IW, Orfila C, Mccartney L, Harholt J, Bernal AJ, Scheller HV, Gilmartin PM, Mikkelsen JD, Paul Knox J, Willats WG. Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. Plant J. 2004:40:260–275. 10.1111/j.1365-313X.2004.02208.x [DOI] [PubMed] [Google Scholar]
- Mangin LA. Sur l'emploi du rouge de ruthénium en anatomie végétale. C R Acad Sci (Paris). 1893a:116:653–656. [Google Scholar]
- Mangin ML. Nouvelles observations sur la membrane. Bull Soc Bot de France. 1893b:40(7):273–280. 10.1080/00378941.1893.10828818 [DOI] [Google Scholar]
- Marita JM, Ralph J, Hatfield RD, Chapple C. NMR characterization of lignins in Arabidopsis altered in the activity of ferulate 5-hydroxylase. Proc Natl Acad Sci U S A. 1999:96(22):12328–12332. 10.1073/pnas.96.22.12328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marry M, Roberts K, Jopson SJ, Huxham IM, Jarvis MC, Corsar J, Robertson E, McCann MC. Cell–cell adhesion in fresh sugar-beet root parenchyma requires both pectin esters and calcium cross-links. Physiol Plant. 2006:126(2):243–256. 10.1111/j.1399-3054.2006.00591.x [DOI] [Google Scholar]
- Martins MM, Carvalheiro F, Gírio F. An overview of lignin pathways of valorization: from isolation to refining and conversion into value-added products. (Biomass Conv Bioref. 2022). Germany: Springer-Verlag GmbH; 2022. 10.1007/s13399-022-02701-z [DOI] [Google Scholar]
- Martínez-Abad A, Giummarella N, Lawoko M, Vilaplana F. Differences in extractability under subcritical water reveal interconnected hemicellulose and lignin recalcitrance in birch hardwoods. Green Chem. 2018:20(11):2534–2546. 10.1039/C8GC00385H [DOI] [Google Scholar]
- Martone PT, Estevez JM, Lu F, Ruel K, Denny MW, Somerville C, Ralph J. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr Biol. 2009:19(2):169–175. 10.1016/j.cub.2008.12.031 [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Ishii T, Matsumoto S, Higuchi M, Darvill A, Albersheim P, O'Neill MA. Occurrence of the primary cell wall polysaccharide rhamnogalacturonan II in pteridophytes, lycophytes, and bryophytes. Implications for the evolution of vascular plants. Plant Physiol. 2004:134(1):339–351. 10.1104/pp.103.030072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann M, Roberts K. Architecture of the primary cell wall. In: Lloyd CW, editor. Cytoskeletal basis of plant growth and form. London: Academic Press; 1991. p. 109–129. [Google Scholar]
- McCann M, Wells B, Roberts K. Direct visualization of cross-links in the primary cell wall. J Cell Sci. 1990:96(2):323–334. 10.1242/jcs.96.2.323 [DOI] [Google Scholar]
- McCartney L, Ormerod AP, Gidley MJ, Knox JP. Temporal and spatial regulation of pectic (1-->4)-(-d-galactan in cell walls of developing pea cotyledons: implications for mechanical properties. Plant J. 2000:22(2):105–113. 10.1046/j.1365-313x.2000.00719.x [DOI] [PubMed] [Google Scholar]
- McGee R, Dean GH, Wu D, Zhang Y, Mansfield SD, Haughn GW. Pectin modification in seed coat mucilage by in vivo expression of rhamnogalacturonan-I and homogalacturonan-degrading enzymes. Plant Cell Physiol. 2021:62(12):1912–1926. 10.1093/pcp/pcab077 [DOI] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006:7(4):212. 10.1186/gb-2006-7-4-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna ST, Kunkel JG, Bosch M, Rounds CM, Vidali L, Winship LJ, Hepler PK. Exocytosis precedes and predicts the increase in growth in oscillating pollen tubes. Plant Cell. 2009:21(10):3026–3040. 10.1105/tpc.109.069260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JT, Morgan JLW, Zimmer J. A molecular description of cellulose biosynthesis. Annu Rev Biochem. 2015:84(1):895–921. 10.1146/annurev-biochem-060614-033930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. Two endogenous proteins that induce cell wall expansion in plants. Plant Cell. 1992:4(11):1425–1433. 10.1105/tpc.4.11.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meents MJ, Watanabe Y, Samuels AL. The cell biology of secondary cell wall biosynthesis. Ann Bot. 2018:121(6):1107–1125. 10.1093/aob/mcy005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinert MC, Delmer DP. Changes in biochemical composition of the cell wall of the cotton fiber during development. Plant Physiol. 1977:59(6):1088–1097. 10.1104/pp.59.6.1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KH, Misch L. Positions des atomes dans le nouveau modele spatial de la cellulose. Helv Chim Acta. 1937:20(1):232–244. 10.1002/hlca.19370200134 [DOI] [Google Scholar]
- Miao YC, Liu CJ. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc Natl Acad Sci U S A. 2010:107(52):22728–22733. 10.1073/pnas.1007747108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miart F, Desprez T, Biot E, Morin H, Belcram K, Hofte H, Gonneau M, Vernhettes S. Spatio-temporal analysis of cellulose synthesis during cell plate formation in Arabidopsis. Plant J. 2014:77(1):71–84. 10.1111/tpj.12362 [DOI] [PubMed] [Google Scholar]
- Mishler-Elmore JW, Zhou Y, Sukul A, Oblak M, Tan L, Faik A, Held MA. Extensins: self-assembly crosslinking, and the role of peroxidases. Front Plant Sci. 2021:12:664738. 10.3389/fpls.2021.664738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami AG, Inatsugi R, Jiao J, Kotake T, Kuwata K, Ootani K, Okuda S, Sankaranarayanan S, Sato Y, Maruyama D, et al. The AMOR arabinogalactan sugar chain induces pollen-tube competency to respond to ovular guidance. Curr Biol. 2016:26(8):1091–1097. 10.1016/j.cub.2016.02.040 [DOI] [PubMed] [Google Scholar]
- Mohnen D. A signaling peptide locks pollen tube walls. Science. 2023:382(6671):648–649. 10.1126/science.adl1198 [DOI] [PubMed] [Google Scholar]
- Mohnen D, Atmodjo MA, Jayawardhane P. Reconsidering pectin structure: A historical perspective informed by identity and function of the biosynthetic enzymes. In: Geitmann A, editor. The plant cell wall - research milestones and conceptual insights. Boca Raton (FL): CRC Press, Taylor & Francis Group; 2024. p. 94–126. 10.1201/9781003178309. [DOI] [Google Scholar]
- Molina-Hidalgo FJ, Franco AR, Villatoro C, Medina-Puche L, Mercado JA, Hidalgo MA, Monfort A, Caballero JL, Muñoz-Blanco J, Blanco-Portales R. The strawberry (Fragaria×ananassa) fruit-specific rhamnogalacturonate lyase 1 (FaRGLyase1) gene encodes an enzyme involved in the degradation of cell-wall middle lamellae. J Exp Bot. 2013:64(6):1471–1483. 10.1093/jxb/ers386 [DOI] [PubMed] [Google Scholar]
- Molina A, Miedes E, Bacete L, Rodríguez T, Mélida H, Denancé N, Sánchez-Vallet A, Rivière MP, López G, Freydier A, et al. Arabidopsis cell wall composition determines disease resistance specificity and fitness. Proc Natl Acad Sci U S A. 2021:118(5):e2010243118. 10.1073/pnas.2010243118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet JC, Leroux C, Dardelle F, Lehner A. Cell wall composition, biosynthesis and remodeling during pollen tube growth. Plants (Basel). 2013:2(1):107–147. 10.3390/plants2010107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Messerli MA, Shi C, Gilroy S. Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc Natl Acad Sci U S A. 2007:104(52):20996–21001. 10.1073/pnas.0708586104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montezinos D, Brown M Jr. Surface architecture of the plant cell: biogenesis of the cell wall, with special emphasis on the role of the plasma membrane in cellulose biosynthesis. J Supramol Struct. 1976:5(3):277–290. 10.1002/jss.400050303 [DOI] [PubMed] [Google Scholar]
- Moor H, Muhlethaler K, Waldner H, Frey-Wyssling A. A new freezing-ultramicrotome. J Biophys Biochem Cytol. 1961:10(1):1–13. 10.1083/jcb.10.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PJ, Staehelin LA. Immunogold localization of the cell-wall-matrix polysaccharides rhamnogalacturonan I and xyloglucan during cell expansion and cytokinesis in Trifolium pratense L.; implication for secretory pathways. Planta. 1988:174(4):433–445. 10.1007/BF00634471 [DOI] [PubMed] [Google Scholar]
- Moreira D, Kaur D, Pereira AM, Held MA, Showalter AM, Coimbra S. Type II arabinogalactans initiated by hydroxyproline-O-galactosyltransferases play important roles in pollen-pistil interactions. Plant J. 2023:114(2):371–389. 10.1111/tpj.16141 [DOI] [PubMed] [Google Scholar]
- Morgan JL, McNamara JT, Zimmer J. Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol. 2014:21(5):489–496. 10.1038/nsmb.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013:493(7431):181–186. 10.1038/nature11744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrall P, Briggs DE. Changes in cell wall polysaccharides of germinating barley grains. Phytochemistry. 1978:17(9):1495–1502. 10.1016/S0031-9422(00)94628-4 [DOI] [Google Scholar]
- Morris ER, Powell DA, Gidley MJ, Rees DA. Conformation and interactions of pectins. I. Polymorphism between gel and solid stated of calcium polygalacturonate. J Mol Biol. 1982:155(4):507–516. 10.1016/0022-2836(82)90484-3 [DOI] [PubMed] [Google Scholar]
- Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, et al. Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci U S A. 2010:107(40):17409–17414. 10.1073/pnas.1005456107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y, Karlen SD, Goacher RE, Ralph J, Mansfield SD. Metabolic engineering of p-hydroxybenzoate in poplar lignin. Plant Biotechnol J. 2023:21(1):176–188. 10.1111/pbi.13935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille G, Ralet M-C, Cavelier C, Eland C, Effroy D, Hematy K, McCartney L, Truong H-N, Gaudon V, Tibault J-F, et al. Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 2007:50(4):605–614. 10.1111/j.1365-313X.2007.03086.x [DOI] [PubMed] [Google Scholar]
- Moussu S, Ingram G. The EXTENSIN enigma. Cell Surf. 2023:9:100094. 10.1016/j.tcsw.2023.100094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussu S, Lee H-K, Haas KT, Broyart C, Rathgeb U, De Bellis D, Levasseur T, Schoenaers S, Fernandez GS, Grossniklasu U, et al. Plant cell wall patterning and expansion mediated by protein-peptide-polysaccharide interaction. Science. 2023:382(6671):719–725. 10.1126/science.adi4720 [DOI] [PubMed] [Google Scholar]
- Mueller SC, Brown RM Jr. Evidence for an intra-membrane component associated with a cellulose-microfibril-synthesizing complex in higher plants. J Cell Biol. 1980:84(2):315–326. 10.1083/jcb.84.2.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlethaler K. Die feinstruktur der zellulosemikrofibrillen. Beih Z Schweiz Forstver. 1960:30:55–64. [Google Scholar]
- Muschitz A, Riou C, Mollet J-C, Gloaguen V, Faugeron C. Modifications of cell wall pectin in tomato cell suspension in response to cadmium and zinc. Acta Physiol Plant. 2015:37:1–11. 10.1007/s11738-015-2000-y [DOI] [Google Scholar]
- Nakamura A, Furuta H, Maeda H, Takao T, Nagamatsu Y. Structural studies by stepwise enzymatic degradation of the main backbone of soybean soluble polysaccharides consisting of galacturonan and rhamnogalacturonan. Biosci Biotechnol Biochem. 2002:66(6):1301–1313. 10.1271/bbb.66.1301 [DOI] [PubMed] [Google Scholar]
- Narváez-Barragán DA, Tovar-Herrera OE, Guevara-García A, Serrano M, Martinez-Anaya C. Mechanisms of plant cell wall surveillance in response to pathogens, cell wall-derived ligands and the effect of expansins to infection resistance or susceptibility. Front Plant Sci. 2022:13:969343. 10.3389/fpls.2022.969343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature. 2017:544(7648):65–70. 10.1038/nature21725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RH, Hill SJ, Harris PJ. Wide-angle x-ray scattering and solid-state nuclear magnetic resonance data combined to test models for cellulose microfibrils in mung bean cell walls. Plant Physiol. 2013:163(4):1558–1567. 10.1104/pp.113.228262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibbering P, Petersen BL, Motawia MS, Jørgensen B, Ulvskov P, Niittylä T. Golgi-localized exo-(1,3-galactosidases involved in cell expansion and root growth in Arabidopsis. J Biol Chem. 2020:295(31):10581–10592. 10.1074/jbc.RA120.013878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998:17(19):5563–5576. 10.1093/emboj/17.19.5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Langan P, Chanzy H. Crystal structure and hydrogen-bonding system in cellulose iβ from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc. 2002:124(31):9074–9082. 10.1021/ja0257319 [DOI] [PubMed] [Google Scholar]
- Nissen KS, Willats WGT, Malinovsky FG. Understanding CrRLK1L function: cell walls and growth control. Trends Plant Sci. 2016:21(6):516–527. 10.1016/j.tplants.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Nixon BT, Mansouri K, Singh A, Du J, Davis JK, Lee JG, Slabaugh E, Vandavasi VG, O'Neill H, Roberts EM, et al. Comparative structural and computational analysis supports eighteen cellulose synthases in the plant cellulose synthesis complex. Sci Rep. 2016:6(1):28696. 10.1038/srep28696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notley SM, Pettersson B, Wågberg L. Direct measurement of attractive van der Waals' forces between regenerated cellulose surfaces in an aqueous environment. J Am Chem Soc. 2004:126(43):13930–13931. 10.1021/ja045992d [DOI] [PubMed] [Google Scholar]
- Nuñez A, Fishman ML, Fortis LL, Cooke PH, Hotchkiss AT Jr. Identification of extensin protein associated with sugar beet pectin. J Agric Food Chem. 2009:57(22):10951–10958. 10.1021/jf902162t [DOI] [PubMed] [Google Scholar]
- Oehme DP, Shafee T, Downton MT, Bacic A, Doblin MS. Differences in protein structural regions that impact functional specificity in GT2 family beta-glucan synthases. PLoS One. 2019:14(10):e0224442. 10.1371/journal.pone.0224442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani M, Demura T. The quest for transcriptional hubs of lignin biosynthesis: beyond the NAC-MYB-gene regulatory network model. Curr Opin Biotechnol. 2019:56:82–87. 10.1016/j.copbio.2018.10.002 [DOI] [PubMed] [Google Scholar]
- Oikawa A, Joshi HJ, Rennie EA, Ebert B, Manisseri C, Heazlewood JL, Scheller HV. An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. PLoS One. 2010:5(11):e15481. 10.1371/journal.pone.0015481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olek AT, Rayon C, Makowski L, Kim HR, Ciesielski P, Badger J, Paul LN, Ghosh S, Kihara D, Crowley M, et al. The structure of the catalytic domain of a plant cellulose synthase and its assembly into dimers. Plant Cell. 2014:26(7):2996–3009. 10.1105/tpc.114.126862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olek AT, Rushton PS, Kihara D, Ciesielski P, Aryal UK, Zhang Z, Stauffacher CV, McCann MC, Carpita NC. Essential amino acids in the plant-conserved and class-specific regions of cellulose synthases. Plant Physiol. 2023:191(1):142–160. 10.1093/plphys/kiac479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MA, Eberhard S, Albersheim P, Darvill AG. Requirement of borate cross-linking of cell wall rhamnogalactaruonan II for Arabidopsis growth. Science. 2001:294(5543):846–849. 10.1126/science.1062319 [DOI] [PubMed] [Google Scholar]
- O'Neill MA, Warrenfeltz D, Kates K, Pellerin P, Doco T, Darvill AG, Albersheim P. Rhamnogalacturonan-II, a pectic polysaccharide in the walls of growing plant cells, forms a dimer that is covalently cross-linked by a borate ester. In vitro conditions for the formation and hydrolysis of the dimer. J Biol Chem. 1996:271(37):22923–22930. 10.1074/jbc.271.37.22923 [DOI] [PubMed] [Google Scholar]
- Oyarce P, De Meester B, Fonseca F, de Vries L, Goeminne G, Pallidis A, De Rycke R, Tsuji Y, Li Y, Van den Bosch S, et al. Introducing curcumin biosynthesis in Arabidopsis enhances lignocellulosic biomass processing. Nat Plants. 2019:5(2):225–237. 10.1038/s41477-018-0350-3 [DOI] [PubMed] [Google Scholar]
- Paniagua C, Bilkova A, Jackson P, Dabravolski S, Riber W, Didi V, Houser J, Gigli-Bisceglia N, Wimmerova M, Budínská E, et al. Dirigent proteins in plants: modulating cell wall metabolism during abiotic and biotic stress exposure. J Exp Bot. 2017:68(13):3287–3301. 10.1093/jxb/erx141 [DOI] [PubMed] [Google Scholar]
- Paniagua C, Posé S, Morris VJ, Kirby AR, Quesada MA, Mercado JA. Fruit softening and pectin disassembly: an overview of nanostructural pectin modifications assessed by atomic force microscopy. Ann Bot. 2014:114(6):1375–1383. 10.1093/aob/mcu149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006:312(5779):1491–1495. 10.1126/science.1126551 [DOI] [PubMed] [Google Scholar]
- Park AR, Cho SK, Yun UJ, Jin MY, Lee SH, Sachetto-Martins G, Park OK. Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J Biol Chem. 2001:276(28):26688–26693. 10.1074/jbc.M101283200 [DOI] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ. A revised architecture of primary cell walls based on biochemical changes induced by substrate-specific endoglucanases. Plant Physiol. 2012a:158(4):1933–1943. 10.1104/pp.111.192880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Cosgrove DJ. Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol. 2012b:158(1):465–475. 10.1104/pp.111.189779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Diaz-Moreno SM, Davis DJ, Wilkop TE, Bulone V, Drakakaki G. Endosidin 7 specifically arrests late cytokinesis and inhibits callose biosynthesis, revealing distinct trafficking events during cell plate maturation. Plant Physiol. 2014:165(3):1019–1034. 10.1104/pp.114.241497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Szumlanski AL, Gu F, Guo F, Nielsen E. A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat Cell Biol. 2011:13(8):973–980. 10.1038/ncb2294 [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu Rev Plant Biol. 2016:67(1):235–259. 10.1146/annurev-arplant-043015-112222 [DOI] [PubMed] [Google Scholar]
- Pauly M, Ramirez V. New insights into wall polysaccharide O-acetylation. Front Plant Sci. 2018:9:1210. 10.3389/fpls.2018.01210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen A. Memoire sur le composition du tissu propre des plantes et du ligneux. Compt Rend. 1838:7:1052–1056. [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc Natl Acad Sci U S A. 1996:93(22):12637–12642. 10.1073/pnas.93.22.12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA Jr. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci U S A. 2001:98(22):12843–12847. 10.1073/pnas.201416998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Braybrook S, Höfte H. Cell wall mechanics and growth control in plants: the role of pectins revisited. Front Plant Sci. 2012:3:121. 10.3389/fpls.2012.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Wightman R, Hofte H. The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr Biol. 2015:25(13):1746–1752. 10.1016/j.cub.2015.05.022 [DOI] [PubMed] [Google Scholar]
- Pelletier S, Van Orden J, Wolf S, Vissenberg K, Delacourt J, Ndong YA, Pelloux J, Bischoff V, Urbain A, Mouille G, et al. A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol. 2010:188(3):726–739. 10.1111/j.1469-8137.2010.03409.x [DOI] [PubMed] [Google Scholar]
- Pelloux J, Rustérucci C, Mellerowicz EJ. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007:12(6):267–277. 10.1016/j.tplants.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Pena MJ, Kong Y, York WS, O'Neill MA. A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell. 2012:24(11):4511–4524. 10.1105/tpc.112.103390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Kawagoe Y, Hogan P, Delmer D. Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science. 2002:295(5552):147–150. 10.1126/science.1064281 [DOI] [PubMed] [Google Scholar]
- Pérez S, Rodrίguez-Caravajal MA, Doco T. A complex plant cell wall polysaccharide: rhamnogalacturonan II. A structure in quest of a function. Biochimie. 2003:85(1-2):109–121. 10.1016/S0300-9084(03)00053-1 [DOI] [PubMed] [Google Scholar]
- Pérez García M, Zhang Y, Hayes J, Salazar A, Zabotina OA, Hong M. Structure and interactions of plant cell-wall polysaccharides by two and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry. 2011:50(6):989–1000. 10.1021/bi101795q [DOI] [PubMed] [Google Scholar]
- Perkins ML, Schuetz M, Unda F, Chen KT, Bally MB, Kulkarni JA, Yan Y, Pico J, Castellarin SD, Mansfield SD, et al. Monolignol export by diffusion down a polymerization-induced concentration gradient. Plant Cell. 2022:34(5):2080–2095. 10.1093/plcell/koac051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins M, Smith RA, Samuels L. The transport of monomers during lignification in plants: anything goes but how? Curr Opin Biotechnol. 2019:56:69–74. 10.1016/j.copbio.2018.09.011 [DOI] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science. 1999:284(5422):1976–1979. 10.1126/science.284.5422.1976 [DOI] [PubMed] [Google Scholar]
- Petersen BL, Macalister CA, Ulvskov P. Plant Protein O-Arabinosylation. Front Plant Sci. 2021:12:645219. 10.3389/fpls.2021.645219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe F, Pelloux J, Rayon C. Plant pectin acetylesterase structure and function: new insights from bioinformatics analysis. BMC Genomics. 2017:18(1):456. 10.1186/s12864-017-3833-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phyo P, Gu Y, Hong M. Impact of acidic pH on plant cell wall polysaccharide structure and dynamics: insights into the mechanism of acid growth in plants from solid-state NMR. Cellulose. 2019:26(1):291–304. 10.1007/s10570-018-2094-7 [DOI] [Google Scholar]
- Pogorelko G, Lionetti V, Bellincampi D, Zabotina O. Cell wall integrity: targeted post-synthetic modifications to reveal its role in plant growth and defense against pathogens. Plant Signal Behav. 2013:8(9):e25435. 10.4161/psb.25435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polko JK, Kieber JJ. The regulation of cellulose biosynthesis in plants. Plant Cell. 2019:31(2):282–296. 10.1105/tpc.18.00760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potikha TS, Collins CC, Johnson DI, Delmer DP, Levine A. The involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 1999:119(3):849–858. 10.1104/pp.119.3.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier D, Roitsch T, Persson S. Cell wall regulation by carbon allocation and sugar signaling. Cell Surf. 2023:9:100096. 10.1016/j.tcsw.2023.100096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar PK, Pereira JH, Taujale R, Shao W, Bharadwaj VS, Chapla D, Yang JY, Bomble YJ, Moremen KW, Kannan N, et al. Structural and biochemical insight into a modular β-1,4-galactan synthase in plants. Nat Plants. 2023:9(3):486–500. 10.1038/s41477-023-01358-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston R. The formation of wood in forest trees. NY, USA: Academic Press; 1964, p. 169–187. [Google Scholar]
- Proseus TE, Boyer JS. Turgor pressure moves polysaccharides into growing cell walls of Chara corallina. Ann Bot. 2005:95(6):967–979. 10.1093/aob/mci113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proseus TE, Boyer JS. Tension required for pectate chemistry to control growth in Chara corallina. J Exp Bot. 2007:58(15-16):4283–4292. 10.1093/jxb/erm318 [DOI] [PubMed] [Google Scholar]
- Proseus TE, Boyer JS. Calcium deprivation disrupts enlargement of Chara corallina cells: further evidence for the calcium pectate cycle. J Exp Bot. 2012a:63(10):3953–3958. 10.1093/jxb/ers089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proseus TE, Boyer JS. Pectate chemistry links cell expansion to wall deposition in Chara corallina. Plant Signal Behav. 2012b:7(11):1490–1492. 10.4161/psb.21777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proseus TE, Ortega JKE, Boyer JS. Separating growth from elastic deformation during cell enlargement. Plant Physiol. 1999:119(2):775–784. 10.1104/pp.119.2.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proseus TE, Zhu G-L, Boyer JS. Turgor, temperature and the growth of plant cells: using Chara corallina as a model system. J Exp Bot. 2000:51(350):1481–1494. 10.1093/jexbot/51.350.1481 [DOI] [PubMed] [Google Scholar]
- Purushotham P, Cho SH, Diaz-Moreno SM, Kumar M, Nixon BT, Bulone V, Zimmer J. A single heterologously expressed plant cellulose synthase isoform is sufficient for cellulose microfibril formation in vitro. Proc Natl Acad Sci U S A. 2016:113(40):11360–11365. 10.1073/pnas.1606210113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham P, Ho R, Yu L, Fincher GB, Bulone V, Zimmer J. Mechanism of mixed-linkage glucan biosynthesis by barley cellulose synthase-like CslF6 (1,3;1,4)-β-glucan synthase. Sci Adv. 2022:8(45):eadd1596. 10.1126/sciadv.add1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham P, Ho R, Zimmer J. Architecture of a catalytically active homotrimeric plant cellulose synthase complex. Science. 2020:369(6507):1089–1094. 10.1126/science.abb2978 [DOI] [PubMed] [Google Scholar]
- Qi X, Behrens BX, West PR, Mort AJ. Solubilization and partial characterization of extensin fragments from cell walls of cotton suspension cultures. Evidence for a covalent cross-link between extensin and pectin. Plant Physiol. 1995:108(4):1691–1701. 10.1104/pp.108.4.1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Shi Y, Li C, Liu J, Chong S-L, Lim K-J, Si J, Han Z, Chen D. Glucomannan in Dendrobium catenatum: bioactivities, biosynthesis and perspective. Genes (Basel). 2022:13(11):1957. 10.3390/genes13111957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y-M, Zhu Y-X. How cotton fibers elongate: a tale of linear cell-growth mode. Curr Opin Plant Biol. 2011:14(1):106–111. 10.1016/j.pbi.2010.09.010 [DOI] [PubMed] [Google Scholar]
- Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, et al. Lignin valorization: improving lignin processing in the biorefinery. Science. 2014:344(6185):1246843. 10.1126/science.1246843 [DOI] [PubMed] [Google Scholar]
- Rajangam AS, Kumar M, Aspeborg H, Guerriero G, Arvestad L, Pansri P, Brown CJL, Hober S, Blomqvist K, Divne C, et al. MAP20, a microtubule-associated protein in the secondary cell walls of hybrid aspen, is a target of the cellulose synthesis inhibitor 2,6-dichlorobenzonitrile. Plant Physiol. 2008:148(3):1283–1294. 10.1104/pp.108.121913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Brunow G, Harris P, Dixon RA, Boerjan W. Lignification: are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication? Rec Adv Polyphenols Res. 2008:1:36–66. 10.1002/9781444302400.ch2 [DOI] [Google Scholar]
- Ralph J, Lapierre C, Boerjan W. Lignin structure and its engineering. Curr Opin Biotechnol. 2019:56:240–249. 10.1016/j.copbio.2019.02.019 [DOI] [PubMed] [Google Scholar]
- Rao X, Dixon RA. Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front Plant Sci. 2018:9:399. 10.3389/fpls.2018.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle DL, Cleland R. Enhancement of wall loosening and elongation by acid solutions. Plant Physiol. 1970:46(2):250–253. 10.1104/pp.46.2.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JSG. Cell wall storage carbohydrates in seeds-biochemistry of the seed “gums” and “hemicelluloses”. Adv Bot Res. 1985:11:125–155. 10.1016/S0065-2296(08)60170-6 [DOI] [Google Scholar]
- Reiter WD, Chapple C, Somerville CR. Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 1997:12(2):335–345. 10.1046/j.1365-313X.1997.12020335.x [DOI] [PubMed] [Google Scholar]
- Reuhs BL, Glenn J, Stephens SB, Kim JS, Christie DB, Glushka JG, Zablackis E, Albersheim P, Darvill AG. O'Neill MA. l-galactose replaces l-fucose in the pectic polysaccharide rhamnogalacturonan II synthesized by the l-fucose-deficient mur1 Arabidopsis mutant. Planta. 2004:219(1):147–157. 10.1007/s00425-004-1205-x [DOI] [PubMed] [Google Scholar]
- Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000:124(2):495–498. 10.1104/pp.124.2.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J, Ploderer M, Mongelard G, Gutierrez L, Hauser MT. Role of CrRLK1L cell wall sensors HERCULES1 and 2, THESEUS1, and FERONIA in growth adaptation triggered by heavy metals and trace elements. Front Plant Sci. 2017:8:1554. 10.3389/fpls.2017.01554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C, Keller B, Ryser U. Glycine-rich proteins as structural components of plant cell walls. Cell Mol Life Sci. 2001:58(10):1430–1441. 10.1007/PL00000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AW, Frost AO, Roberts EM, Haigler CH. Roles of microtubules and cellulose microfibril assembly in the localization of secondary-cell-wall deposition in developing tracheary elements. Protoplasma. 2004:224(3-4):217–229. 10.1007/s00709-004-0064-4 [DOI] [PubMed] [Google Scholar]
- Roberts AW, Haigler CH. Rise in chlorotetracycline fluorescence accompanies tracheary element differentiation in suspension cultures of Zinnia. Protoplasma. 1989:152(1):37–45. 10.1007/BF01354238 [DOI] [Google Scholar]
- Robinson DG, Quader H. Towards cellulose synthesis in vitro from plant extracts. J Theor Biol. 1981:92(4):483–495. 10.1016/0022-5193(81)90261-7 [DOI] [Google Scholar]
- Roelofsen PA, Houwink AL. Architecture and growth of the primary cell wall in some plant hairs and in the Phycomyces sprangiophore. Acta Bot Neerl. 1953:2(2):218–225. 10.1111/j.1438-8677.1953.tb00272.x [DOI] [Google Scholar]
- Roelofsen PA, Kreger DR. The submicroscopic structure of pectin in collenchyma cell-walls. J Exp Bot. 1951:2(3):332–343. 10.1093/jxb/2.3.332 [DOI] [Google Scholar]
- Römling U, Galperin MY. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 2015:23(9):545–557. 10.1016/j.tim.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002:43(12):1421–1435. 10.1093/pcp/pcf171 [DOI] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroomt E, van der Mareit J, van Boomt H, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987:327(6101):279–281. 10.1038/325279a0 [DOI] [PubMed] [Google Scholar]
- Roudier F, Fernandez AG, Fujita M, Himmelspach R, Borner GHH, Schindelman G, Song S, Baskin TI, Dupree P, Wasteneys GO, et al. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell. 2005:17(6):1749–1763. 10.1105/tpc.105.031732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounds CM, Bezanilla M. Growth mechanisms in tip-growing plant cells. Annu Rev Plant Biol. 2013:64(1):243–265. 10.1146/annurev-arplant-050312-120150 [DOI] [PubMed] [Google Scholar]
- Rui Y, Dinneny JR. A wall with integrity: surveillance and maintenance of the plant cell wall under stress. New Phytol. 2019:225(4):1428–1439. 10.1111/nph.16166 [DOI] [PubMed] [Google Scholar]
- Rushton PS, Olek AT, Makowski L, Badger J, Steussy CN, Carpita NC, Stauffacher CV. Rice cellulose synthaseA8 plant-conserved region is a coiled-coil at the catalytic core entrance. Plant Physiol. 2017:173(1):482–494. 10.1104/pp.16.00739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser U. Protoxylem: the deposition of a network containing glycine-rich cell wall proteins starts in the cell corners in close association with the pectins of the middle lamella. Planta. 2003:216(5):854–864. 10.1007/s00425-002-0938-7 [DOI] [PubMed] [Google Scholar]
- Ryser U, Schorderet M, Guyot R, Keller B. A new structural element containing glycine-rich proteins and rhamnogalacturonan I in the protoxylem of seed plants. J Cell Sci. 2003:117(7):1179–1190. 10.1242/jcs.00966 [DOI] [PubMed] [Google Scholar]
- Sadava D, Walker F, Chrispeels MJ. Hydroxyproline-rich cell wall protein (extension): biosynthesis and accumulation in growing pea epicotyls. Dev Biol. 1973:30(1):42–48. 10.1016/0012-1606(73)90046-8 [DOI] [PubMed] [Google Scholar]
- Salnikov VV, Grimson MJ, Delmer DP, Haigler CH. Sucrose synthase localizes to cellulose synthesis sites in tracheary elements. Phytochemistry. 2001:57(6):823–833. 10.1016/S0031-9422(01)00045-0 [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. The expansin superfamily. Genome Biol. 2005:6(12):242. 10.1186/gb-2005-6-12-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels AL, Giddings TH Jr, Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J Cell Biol. 1995:130(6):1345–1357. 10.1083/jcb.130.6.1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarko A, Muggli R. Packing analysis of carbohydrates and polysaccharides. III. Valonia cellulose and cellulose II. Macromolecules. 1974:7(4):486–494. 10.1021/ma60040a016 [DOI] [Google Scholar]
- Saxena IM, Kudlicka K, Okuda K, Brown RM Jr. Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol. 1994:176(18):5735–5752. 10.1128/jb.176.18.5735-5752.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena IM, Lin FC, Brown RM. Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol Biol. 1990:15(5):673–683. 10.1007/BF00016118 [DOI] [PubMed] [Google Scholar]
- Scanlan EM, Mackeen MM, Wormald MR, Davis BG. Synthesis and solution-phase conformation of the RG-I fragment of the plant polysaccharide pectin reveals a modification-modulated assembly mechanism. J Am Chem Soc. 2022:132(21):7238–7239. 10.1021/ja9090963 [DOI] [PubMed] [Google Scholar]
- Scavuzzo-Duggan TR, Chaves AM, Singh A, Sethaphong L, Slabaugh E, Yingling YG, Haigler CH, Roberts AW. Cellulose synthase ‘class specific regions' are intrinsically disordered and functionally undifferentiated. J Integr Plant Biol. 2018:60(6):481–497. 10.1111/jipb.12637 [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010:61(1):263–289. 10.1146/annurev-arplant-042809-112315 [DOI] [PubMed] [Google Scholar]
- Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001:15(9):1115–1127. 10.1101/gad.879101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Ehrhardt DW, Meyerowitz EM, Sampathkumar A. Tethering of cellulose synthase to microtubules dampens mechano-induced cytoskeletal organization in Arabidopsis pavement cells. Nat Plants. 2022:8(9):1064–1073. 10.1038/s41477-022-01218-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Hanak T, Persson S, Voigt CA. Cellulose and callose synthesis and organization in focus, what's new? Curr Opin Plant Biol. 2016:34:9–16. 10.1016/j.pbi.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Schneider R, Tang L, Lampugnani ER, Barkwill S, Lathe R, Zhang Y, McFarlane HE, Pesquet E, Niittyla T, Mansfield SD, et al. Two complementary mechanisms underpin cell wall patterning during xylem vessel development. Plant Cell. 2017:29(10):2433–2449. 10.1105/tpc.17.00309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenaers S, Balcerowicz D, Breen G, Hill K, Zdanio M, Mouille G, Holman TJ, Oh J, Wilson MH, Nikonorova N, et al. The auxin-regulated CrRLK1L kinase ERULUS controls cell wall composition during root hair tip growth. Curr Biol. 2018:28(5):722–732. 10.1016/j.cub.2018.01.050 [DOI] [PubMed] [Google Scholar]
- Schoenaers S, Lee HK, Gonneau M, Faucher E, Levasseur T, Akary E, Claeijs N, Moussu S, Broyart C, Balcerowicz D, et al. A pectin-binding peptide with a structural and signaling role in the assembly of the plant cell wall. BioRxiv. 2023. 10.1101/2023.08.23.554416, 23 August 2023, preprint: not peer reviewed. [DOI]
- Schrick K, Debolt S, Bulone V. Deciphering the molecular functions of sterols in cellulose biosynthesis. Front Plant Sci. 2012:3:84. 10.3389/fpls.2012.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultink A, Liu L, Zhu L, Pauly M. Structural diversity and function of xyloglucan sidechain substituents. Plants. 2014:3(4):526–542. 10.3390/plants3040526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. The biology of arabinogalactan proteins. Annu Rev Plant Biol. 2006:58(1):137–161. 10.1146/annurev.arplant.58.032806.103801 [DOI] [PubMed] [Google Scholar]
- Selvendran RR. Developments in the chemistry and biochemistry of pectic and hemicellulosic polymers. J Cell Sci Suppl. 1985:2(Suppl 2):51–88. 10.1242/jcs.1985.Supplement_2.4 [DOI] [PubMed] [Google Scholar]
- Sénéchal F, Wattier C, Rustérucci C, Pelloux J. Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J Exp Bot. 2014:65(18):5125–5160. 10.1093/jxb/eru272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafee T, Bacic A, Johnson K, Wilke C. Evolution of sequence-diverse disordered regions in a protein family: order within the chaos. Mol Biol Evol. 2020:37(8):2155–2172. 10.1093/molbev/msaa096 [DOI] [PubMed] [Google Scholar]
- Sharma P, Sharma S, Ramakrishna G, Srivastava H, Gaikwad K. A comprehensive review on leguminous galactomannans: structural analysis, functional properties, biosynthesis process and industrial applications. Critical Rev Food Sci Nutr. 2022:62(2):443–465. 10.1080/10408398.2020.1819196 [DOI] [PubMed] [Google Scholar]
- Shedletzky E, Shmuel M, Delmer DP, Lamport DTA. Adaptation and growth of tomato cells on the herbicide 2,6-dichlorobenzonitrile leads to production of unique cell walls virtually lacking a cellulose-xyloglucan network. Plant Physiol. 1990:94(3):980–987. 10.1104/pp.94.3.980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedletzky E, Shmuel M, Trainin T, Kalman S, Delmer D. Cell wall structure in cells adapted to growth on the cellulose-synthesis inhibitor 2, 6-dichlorobenzonitrile: a comparison between two dicotyledonous plants and a graminaceous monocot. Plant Physiol. 1992:100(1):120–130. 10.1104/pp.100.1.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Liu J, Li JF. Type-II metacaspases mediate the processing of plant elicitor peptides in Arabidopsis. Mol Plant. 2019:12(11):1524–1533. 10.1016/j.molp.2019.08.003 [DOI] [PubMed] [Google Scholar]
- Shi D-C, Wang J, Hu R-B, Zhou G-K, O'Neill MA, Kong Y-Z. Boron-bridged RG-II and calcium are required to maintain the pectin network of the Arabidopsis seed mucilage ultrastructure. Plant Mol Biol. 2017:94(3):267–280. 10.1007/s11103-017-0606-8 [DOI] [PubMed] [Google Scholar]
- Shigenaga AM, Berens ML, Tsuda K, Argueso CT. Towards engineering of hormonal crosstalk in plant immunity. Curr Opin Plant Biol. 2017:38:164–172. 10.1016/j.pbi.2017.04.021 [DOI] [PubMed] [Google Scholar]
- Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993:5(1):9–23. 10.1105/tpc.5.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010:153(2):485–513. 10.1104/pp.110.156554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Ferraz R, Dupree P, Showalter AM, Coimbra S. Three decades of advances in arabinogalactan-protein biosynthesis. Front Plant Sci. 2020:11:610377. 10.3389/fpls.2020.610377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões MS, Ferreira SS, Grandis A, Rencoret J, Persson S, Floh EIS, Ferraz A, del Río JC, Buckeridge MS, Cesarino I. Differentiation of Tracheary Elements in Sugarcane Suspension Cells Involves Changes in Secondary Wall Deposition and Extensive Transcriptional Reprogramming. Front. Plant Sci. 2020:11:617020. 10.3389/fpls.2020.617020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Avci U, Eichler Inwood SE, Grimson MJ, Landgraf J, Mohnen D, Sorensen I, Wilkerson CG, Willats WG, Haigler CH. A specialized outer layer of the primary cell wall joins elongating cotton fibers into tissue-like bundles. Plant Physiol. 2009:150(2):684–699. 10.1104/pp.109.135459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Kwansa AL, Kim HS, Williams JT, Yang H, Li NK, Kubicki JD, Roberts AW, Haigler CH, Yingling YG. In silico structure prediction of full-length cotton cellulose synthase protein (GhCESA1) and its hierarchical complexes. Cellulose. 2020:27(10):5597–5616. 10.1007/s10570-020-03194-7 [DOI] [Google Scholar]
- Sloep AC. Onderzoekingen over pektinestoffen en hare enzymatische ontleding [PhD Diss.] University of Delft; 1928. [Google Scholar]
- Smith HC. The plasma membrane with notes on the history of botany. Circulation. 1962:26:987–1012. 10.1161/01.CIR.26.5.987 [DOI] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Roach M, Mansfield SD, Ellis B, Samuels L. Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell. 2013:25(10):3988–3999. 10.1105/tpc.113.117176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM, Stone BA. β-Glucan synthesis by cell-free extracts from Lolium multiforme endosperm. Biochim Biophys Acta. 1973:313(1):72–94. 10.1016/0304-4165(73)90189-X [DOI] [PubMed] [Google Scholar]
- Smith PJ, Wang H-T, York WS, Peña MJ, Urbanowicz BR. Designer biomass for next-generation biorefineries: leveraging recent insights into xylan structure and biosynthesis. Biotechnol Biofuels. 2017:10:286. 10.1186/s13068-017-0973-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobry S, Havelange A, Van Cutsem P. Immunocytochemistry of pectins in shoot apical meristems: consequences for intercellular adhesion. Protoplasma. 2005:225(1-2):15–22. 10.1007/s00709-005-0089-3 [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brinistool G, Facette M, Hamann T, Milne J, Osborne E, Paradez A, Persson S, Raab T, et al. Toward a systems approach to understanding plant cell walls. Science. 2004:306(5705):2206–2211. 10.1126/science.1102765 [DOI] [PubMed] [Google Scholar]
- Song B, Zhao S, Shen W, Collings C, Ding S-Y. Direct measurement of plant cellulose microfibril and bundles in native cell walls. Front Plant Sci. 2020:11:479. 10.3389/fpls.2020.00479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinski EE, Westman BM, Redmond CR, Kong Y, Olek AT, Olek J, McCann MC, Carpita NC. Lack of xyloglucan in the cell walls of the Arabidopsis xxt1/xxt2 mutant results in specific increases in homogalacturonan and glucomannan. Plant J. 2022:110(1):212–227. 10.1111/tpj.15666 [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Hepler PK. Cytokinesis in higher plants. Cell. 1996:94(6):821–824. 10.1016/S0092-8674(00)81060-0 [DOI] [PubMed] [Google Scholar]
- Staehelin LA, Pickett-Heaps JD. The ultrastructure of Scenedesmus (Chlorophyceae). I. Species with the “reticulate” or “warty” type ornamental layer. J Phycol. 1975:11:163–185. 10.1111/j.1529-8817.1975.tb02765.x [DOI] [Google Scholar]
- Staudinger H. Der Aufbau der Hochmolecularen Organischen Verbindungen. die Naturwissenschaften. In Naturwissenschaften 1932: Berlin: Springer, pp. 84–89. [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science. 2017:355(6322):287–289. 10.1126/science.aal2541 [DOI] [PubMed] [Google Scholar]
- Sterjiades R, Dean JFD, Eriksson K-EL. Laccase from sycamore maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol. 1992:99(3):1162–1168. 10.1104/pp.99.3.1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kolli VSK, Quigley HF, Hahn MG, Mohnen D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci U S A. 2006:103(13):5236–5241. 10.1073/pnas.0600120103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suslov D, Verbelen JP, Vissenberg K. Onion epidermis as a new model to study the control of growth anisotropy in higher plants. J Exp Bot. 2009:60(14):4175–4187. 10.1093/jxb/erp251 [DOI] [PubMed] [Google Scholar]
- Swords KMM, Staehelin LA. Analysis of extensin structure in plant cell walls. In: Linskens HF, Jackson JF, editors. Plant fibers, modern methods of plant analysis. Vol. 10. Germany: Springer-Verlag; 1989. p. 219–233. [Google Scholar]
- Tai H-C, Chang C-H, Cai W, Lin J-H, Huang S-J, Lin Q-Y, Yuan EC-Y, Li S-L, Lin Y-CJ, Chan JCC, et al. Wood cellulose microfibrils have a 24-chain core-shell nanostructure in seed plants. Nat Plants. 2023:9(7):1154–1168. 10.1038/s41477-023-01430-z [DOI] [PubMed] [Google Scholar]
- Takenaka Y, Kato K, Ogawa-Ohnishi M, Tsuruhama K, Kajiura H, Yagyu K, Takeda A, Takeda Y, Kunieda T, Hara-Nishimura I, et al. Pectin RG-I rhamnosyltransferases represent a novel plant-specific glycosyltransferase family. Nat Plants. 2018:4(9):669–676. 10.1038/s41477-018-0217-7 [DOI] [PubMed] [Google Scholar]
- Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, et al. An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell. 2013:25(1):270–287. 10.1105/tpc.112.107334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Xu J, Held M, Lamport DTA, Kieliszewski M. Structures of repetitive serinehydroxyproline glycomodule expressed by Arabidopsis cell suspension cultures. Plants (Basel). 2023a:12:1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Zhang L, Black I, Glushka J, Urbanowicz B, Heiss C, Azadi P. Most of the rhamnogalacturonan-I from cultured Arabidopsis cell walls is covalently linked to arabinogalactan-protein. Carbohydr Polym. 2023b:301(Part B):120340. 10.1016/j.carbpol.2022.120340 [DOI] [PubMed] [Google Scholar]
- Tang W, Lin W, Zhou X, Guo J, Dang X, Li B, Li D, Yang Z. Mechano-transduction via the pectin-FERONIA complex activates ROP6 GTPase signaling in Arabidopsis pavement cell morphogenesis. Curr Biol. 2022:32(3):508–517.e3. 10.1016/j.cub.2021.11.031 [DOI] [PubMed] [Google Scholar]
- Temple H, Phyo P, Yang W, Lyczakowski JJ, Echevarria-Poza A, Yakunin I, Parra-Rojas JP, Terrett OM, Saez-Aguayo S, Dupree R, et al. Golgi-localized putative S-adenosyl methionine transporters required for cell wall polysaccharide methylation. Nat Plants. 2022:8(6):656–669. 10.1038/s41477-022-01156-4 [DOI] [PubMed] [Google Scholar]
- Tenhaken R. Cell wall remodeling under abiotic stress. Front Plant Sci. 2014:5:771. 10.3389/fpls.2014.00771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima N, Fukushima K. Heterogeneity in formation of lignin. XI. An autoradiographic study of the heterogeneous formation and structure of pine lignin. Wood Sci Technol. 1988:22(3):259–270. 10.1007/BF00386021 [DOI] [Google Scholar]
- Tierney ML, Varner JE. The extensins. Plant Physiol. 1987:84(1):1–2. 10.1104/pp.84.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryfona T, Bourdon M, Marques RD, Busse-Wicher M, Vilaplana F, Stott K, Dupree P. Grass xylan structural variation functional specialization and distinctive interaction with cellulose and lignin. Plant J. 2023:113(5):1004.–. https://doi.org/10.1111tpj.16096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuyama T, Kawai R, Shitan N, Matoh T, Sugiyama J, Yoshinaga A, Takabe K, Fujita M, Yazaki K. Proton-dependent coniferin transport, a common major transport event in differentiating xylem tissue of woody plants. Plant Physiol. 2013:162(2):918–926. 10.1104/pp.113.214957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuyama T, Takabe K. Coniferin β-glucosidase is ionically bound to cell wall in differentiating xylem of poplar. J Wood Sci. 2015:61(4):438–444. 10.1007/s10086-015-1486-7 [DOI] [Google Scholar]
- Turlapati PV, Kim K-W, Davin LB, Lewis NG. The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta. 2011:233(3):439–470. 10.1007/s00425-010-1298-3 [DOI] [PubMed] [Google Scholar]
- Turner S, Gallois P, Brown D. Tracheary element differentiation. Annu Rev Plant Biol. 2007:58(1):407–433. 10.1146/annurev.arplant.57.032905.105236 [DOI] [PubMed] [Google Scholar]
- Turner S, Kumar M. Cellulose synthase complex organization and cellulose microfibril structure. Philos Trans A Math Phys Eng Sci. 2018:376(2112):20170048. 10.1098/rsta.2017.0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio SP, Teichmann T. Poplar genomics is getting popular: the impact of the poplar genome project on tree research. Plant Biol (Stuttg). 2004:6(1):2–4. 10.1055/s-2003-44715 [DOI] [PubMed] [Google Scholar]
- Tuttle JR, Nah G, Duke MV, Alexander DC, Guan X, Song Q, Chen ZJ, Scheffler BE, Haigler CH. Metabolomic and transcriptomic insights into how cotton fiber transitions to secondary wall synthesis, represses lignification, and prolongs elongation. BMC Genomics. 2015:16(1):477. 10.1186/s12864-015-1708-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah M, Liu P, Xie S, Sun S. Recent advancements and challenges in lignin valorization: green routes towards sustainable bioproducts. Molecules. 2022:27(18):6055. 10.3390/molecules27186055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS. Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J. 2014:80(2):197–206. 10.1111/tpj.12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustun S, Hafren A, Hofius D. Autophagy as a mediator of life and death in plants. Curr Opin Plant Biol. 2017:40:122–130. 10.1016/j.pbi.2017.08.011 [DOI] [PubMed] [Google Scholar]
- Vaahtera L, Schulz J, Hamann T. Cell wall integrity maintenance during plant development and interaction with the environment. Nat Plants. 2019:5(9):924–932. 10.1038/s41477-019-0502-0 [DOI] [PubMed] [Google Scholar]
- Vain T, Crowell EF, Timpano H, Biot E, Desprez T, Mansoori N, Trindade LM, Pagant S, Robert S, Hofte H, et al. The cellulase KORRIGAN is part of the cellulose synthase complex. Plant Physiol. 2014:165(4):1521–1532. 10.1104/pp.114.241216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandavasi VG, Putnam DK, Zhang Q, Petridis L, Heller WT, Nixon BT, Haigler CH, Kalluri U, Coates L, Langan P, et al. A structural study of CESA1 catalytic domain of Arabidopsis cellulose synthesis complex: evidence for CESA trimers. Plant Physiol. 2016:170(1):123–135. 10.1104/pp.15.01356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science. 2013:341(6150):1103–1106. 10.1126/science.1241602 [DOI] [PubMed] [Google Scholar]
- Vanholme R, De Meester B, Ralph J, Boerjan W. Lignin biosynthesis and its integration into metabolism. Curr Opin Biotechnol. 2019:56:230–239. 10.1016/j.copbio.2019.02.018 [DOI] [PubMed] [Google Scholar]
- van Parijs FRD, Morreel K, Ralph J, Boerjan W, Merks RMH. Modeling lignin polymerization. I. Simulation model of dehydrogenation polymers. Plant Physiol. 2010:53(3):1332–1344. 10.1104/pp.110.154468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter W-D. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in the fucosyltransferase AtFUT1. Proc Natl Acad Sci U S A. 2002:99(5):3340–3345. 10.1073/pnas.052450699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D, Kinoshita T, Packer NH, Prestegard JH, et al. Essentials of glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2022. p. 1–859. [PubMed] [Google Scholar]
- Varner JE, Lin L-S. Plant cell wall architecture. Cell. 1989:56(2):231–239. 10.1016/0092-8674(89)90896-9 [DOI] [PubMed] [Google Scholar]
- Verbančič J, Lunn JE, Stitt M, Persson S. Carbon supply and the regulation of cell wall synthesis. Mol Plant. 2018:11(1):75–94. 10.1016/j.molp.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Verma P, Kwansa AL, Ho R, Yingling YG, Zimmer J. Insights into substrate coordination and glycosyl transfer of poplar cellulose synthase-8. bioRxiv. 2023. 10.1101/2023.02.07.527505, 7 February 2023, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaas JV, Dixon RA, Chen F, Mansfield SD, Boerjan W, Ralph J, Crowley MF, Beckham GT. Passive membrane transport of lignin-related compounds. Proc Natl Acad Sci U S A. 2019:116(46):23117–23123. 10.1073/pnas.1904643116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DMA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009:47(1):177–206. 10.1146/annurev.phyto.050908.135202 [DOI] [PubMed] [Google Scholar]
- Voiniciuc C. Modern mannan: a hemicellulose's journey. New Phytol. 2022:234(4):1175–1184. 10.1111/nph.18091 [DOI] [PubMed] [Google Scholar]
- Voxeur A, Habrylo O, Guénin S, Miart F, Soulié MC, Rihouey C, Pau-Roblot C, Domon JM, Gutierrez L, Pelloux J, et al. Oligogalacturonide production upon Arabidopsis thaliana–Botrytis cinerea interaction. Proc Natl Acad Sci U S A. 2019:116(39):19743–19752. 10.1073/pnas.1900317116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkinshaw MD, Arnott S. Conformations and interactions of pectins. I. X-ray diffraction analyses of sodium pectate in neutral and acidified forms. J Mol Biol. 1981a:53(4):1055–1073. 10.1016/0022-2836(81)90467-8 [DOI] [PubMed] [Google Scholar]
- Walkinshaw MD, Arnott S. Conformations and interactions of pectins. II. Models for junction zones in pectinic acid and calcium pectate gels. J Mol Biol. 1981b:153(4):1075–1085. 10.1016/0022-2836(81)90468-X [DOI] [PubMed] [Google Scholar]
- Wallace IS, Somerville CR. Plant cell wall patterning and cell shape. Wiley Online; 2014. p. 65–95. 10.1002/9781118647363.ch3. [DOI] [Google Scholar]
- Wang Y, Alonso AP, Wilkerson CG, Keegstra K. Deep EST profiling of developing fenugreek endosperm to investigate galactomannan biosynthesis and its regulation. Plant Mol Biol. 2012:79(3):243–258. 10.1007/s11103-012-9909-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Avci U, Nakashima J, Hahn MG, Chen F, Dixon RA. Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc Natl Acad Sci U S A. 2010:107(51):22338–22343. 10.1073/pnas.1016436107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cao Y, Liang X, Zhuang J, Wang X, Qin F, Jiang C. A dirigent family protein confers variation of Casparian strip thickness and salt tolerance in maize. Nat Commun. 2022b:13(1):2222. 10.1038/s41467-022-29809-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Hong M. Solid-state NMR investigations of cellulose structure and interactions with matrix polysaccharides in plant primary cell walls. J Exp Bot. 2016:67(2):503–514. 10.1093/jxb/erv416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, Matthews ML, Williams CM, Shi R, Yang C, Tunlaya-Anukit S, Chen H-C, Li Q, Liu J, Lin C-Y, et al. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat Commun. 2018:9(1):1579. 10.1038/s41467-018-03863-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, McFarlane H E, Persson S. The impact of abiotic factors on cellulose synthesis. J Exp Bot. 2016a:67(2):543–552. 10.1093/jxb/erv488 [DOI] [PubMed] [Google Scholar]
- Wang T, Park YB, Cosgrove DJ, Hong M. Cellulose-pectin spatial contacts are inherent to never-dried Arabidopsis primary cell walls: evidence from solid state nuclear magnetic resonance. Plant Physiol. 2015:168(3):871–884. 10.1104/pp.15.00665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Viljamaa S, Hodek O, Moritz T, Niittyla T. Sucrose synthase activity is not required for cellulose biosynthesis in Arabidopsis. Plant J. 2022a:110(5):1493–1497. 10.1111/tpj.15752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Xiao S, Shi S, Cai L. Mechanical strength, thermal stability, and hydrophobicity of fiber materials after removal of residual lignin. BioRes. 2017:13(1):71–85. 10.15376/biores.13.1.71-85 [DOI] [Google Scholar]
- Wang T, Yang H, Kubicki JD, Hong M. Cellulose structural polymorphism in plant primary cell walls investigated by high-field 2D solid-state NMR spectroscopy and density functional theory calculations. BioMacromolecules. 2016b:17(6):2010–2022. 10.1021/acs.biomac.6b00441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Meents MJ, McDonnell M, Barkwill S, Sampathkumar A, Cartwright HN, Demura T, Ehrhardt DW, Samuels L, Mansfield SD. Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science. 2015:350(6257):198–203. 10.1126/science.aac7446 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Schneider R, Barkwill S, Gonzales-Vigil E, Hill JL Jr, Samuels AL, Persson S, Mansfield SD. Cellulose synthase complexes display distinct dynamic behaviors during xylem transdifferentiation. Proc Natl Acad Sci U S A. 2018:115(27):E6366–E6374. 10.1073/pnas.1802113115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells B, McCann MC, Shedletzky E, Delmer D, Roberts K. Structural features of cell walls from tomato cells adapted to grow on the herbicide 2,6-dichlorobenzonitrile. J Microscopy. 1994:173(2):155–164. 10.1111/j.1365-2818.1994.tb03438.x [DOI] [Google Scholar]
- Wen X, Zhai Y, Zhang L, Chen Y, Zhu Z, Chen G, Wang K, Zhu Y. Molecular studies of cellulose synthase supercomplex from cotton fiber reveal its unique biochemical properties. Sci China Life Sci. 2022:65(9):1776–1793. 10.1007/s11427-022-2083-9 [DOI] [PubMed] [Google Scholar]
- Wilkerson CG, Mansfield SD, Lu F, Withers S, Park J-Y, Karlen SD, Gonzales-Vigil E, Padmakshan D, Unda F, Rencoret J, et al. Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science. 2014:344(6179):90–93. 10.1126/science.1250161 [DOI] [PubMed] [Google Scholar]
- Willats WGT, Knox JP, Mikkelsen JD. Pectin: new insights into an old polymer are starting to gel. Trends Food Sci Technol. 2006:17(3):97–104. 10.1016/j.tifs.2005.10.008 [DOI] [Google Scholar]
- Willats WGT, McCartney L, Steele-King CG, Marcus SE, Mort A, Huisman M, van Alebeek GJ, Schols HA, Voragen AGJ, Le Goff A, et al. A xylogalacturonan epitope is specifically associated with plant cell detachment. Planta. 2004:218(4):673–681. 10.1007/s00425-003-1147-8 [DOI] [PubMed] [Google Scholar]
- Wilson TH, Kumar M, Turner SR. The molecular basis of plant cellulose synthase complex organisation and assembly. Biochem Soc Trans. 2021:49(1):379–391. 10.1042/BST20200697 [DOI] [PubMed] [Google Scholar]
- Winter H and Huber SC. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Plant Sci. 2010:19:31–67. [DOI] [PubMed] [Google Scholar]
- Wisniak J. Henri Braconnot. Revista CENIC Ciencias Químicas. 2007:38(2):345–355. [Google Scholar]
- Wolf S. Cell wall signaling in plant development and defense. Annu Rev Plant Biol. 2022:73(1):323–353. 10.1146/annurev-arplant-102820-095312 [DOI] [PubMed] [Google Scholar]
- Wolf S, Hofte H. Growth control: a saga of cell walls, ROS, and peptide receptors. Plant Cell. 2014:26(5):1848–1856. 10.1105/tpc.114.125518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Mouille G, Pelloux J. Homogalacturonan methyl-esterification and plant development. Mol Plant. 2009:2(5):851–860. 10.1093/mp/ssp066 [DOI] [PubMed] [Google Scholar]
- Wolf S, van der Does D, Ladwig F, Sticht C, Kolbeck A, Schurholz AK, Augustin S, Keinath N, Rausch T, Greiner S, et al. A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc Natl Acad Sci U S A. 2014:111(42):15261–15266. 10.1073/pnas.1322979111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock C, Sarko A. Packing analysis of carbohydrates and polysaccharides. 11. Molecular and crystal structure of native ramie cellulose. Macromolecules. 1980:13(5):1183–1187. 10.1021/ma60077a030 [DOI] [Google Scholar]
- Wu S-Z, Chaves AM, Li R, Roberts AW, Bezanilla M. Cellulose synthase-like D movement in the plasma membrane requires enzymatic activity. J Cell Biol. 2023:222(6):e202212117. 10.1083/jcb.202212117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, Chai J. Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature. 2019:572(7768):270–274. 10.1038/s41586-019-1409-7 [DOI] [PubMed] [Google Scholar]
- Xie Y, Sun P, Li Z, Zhang F, You C, Zhang Z. FERONIA receptor kinase integrates with hormone signaling to regulate plant growth, development, and responses to environmental stimuli. Int J Mol Sci. 2022:23(7):3730. 10.3390/ijms23073730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang M, Shi D, Zhou G, Niu T, Hahn MG, O’Neill MA, Kong Y. DGE-seq analysis of MUR3-related Arabidopsis mutants provide insight into how dysfunctional xyloglucan affects cell elongation. Plant Sci. 2017:258:156–159. 10.1016/j.plantsci.2017.01.005 [DOI] [PubMed] [Google Scholar]
- Yang J, Bak G, Burgin T, Barnes WJ, Mayes HB, Pen MJ, Urbanowicz BR, Nielsen E. Biochemical and genetic analysis identify CSLD3 as a β-1,4-glucan synthase that functions during plant cell wall synthesis. Plant Cell. 2020b:32(5):1749–1767. 10.1105/tpc.19.00637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Benatti MR, Karve RA, Fox A, Meilan R, Carpita NC, McCann MC. Rhamnogalacturonan-I is a determinant of cell-cell adhesion in poplar wood. Plant Biotechnol J. 2020a:18(4):1027–1040. 10.1111/pbi.13271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Liu R, Pang J, Ren B, Zhou H, Wang G, Wang E, Liu J. Poaceae-specific cell wall-derived oligosaccharides activate plant immunity via OsCERK1 during Magnaporthe oryzae infection in rice. Nat Commun. 2021:12(1):2178. 10.1038/s41467-021-22456-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yu Y, Liang Y, Anderson CT, Cao J. A profusion of molecular scissors for pectins: classification, expression, and functions of plant polygalacturonases. Front Plant Sci. 2018:9:1208. 10.3389/fpls.2018.01208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D, Kiemle SN, Rongpipi S, Wang X, Wang C, Cosgrove DJ, Gomez EW, Gomez ED. Resonant soft X-ray scattering reveals cellulose microfibril spacing in plant primary cell walls. Sci Rep. 2018:8(1):12449. 10.1038/s41598-018-31024-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z-H, Zhong R. Outstanding questions on xylan bioxynthesis. Plant Sci. 2022:325:111476. 10.1016/j.plantsci.2022.111476 [DOI] [PubMed] [Google Scholar]
- Yin Y, Johns MA, Cao H, Rupani M. A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the cellulose synthase superfamily. BMC Genomics. 2014:15(1):260. 10.1186/1471-2164-15-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A, Ohtani M, Katagiri D, Hosokawa Y, Demura T. Hechtian strands transmit cell wall integrity signals in plant cells. Plants. 2020:9(5):604. 10.3390/plants9050604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Yoshimi Y, Cresswell R, Wightman R, Lyczakowski JJ, Wilson LFL, Ishida K, Stott K, Yu X, Charalambous S, et al. Eudicot primary cell wall glucomannan is related in synthesis, structure, and function to xyloglucan. Plant Cell. 2022:34(11):4600–4622. 10.1093/plcell/koac238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou Y-H, Eberhard S, Danhof L, Keegstra K, Hahn MG. Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol. 2012:159(4):1367–1384. 10.1104/pp.112.198119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, Zhang N, Weerts R. Polysaccharide biosynthesis: glycosyltransferases and their complexes. Front Plant Sci. 2021:12:625307. 10.3389/fpls.2021.625307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakzeski J, Bruijnincx PC, Jongerius AL, Weckhuysen BM. The catalytic valorization of lignin for the production of renewable chemicals. Chem Rev. 2010:110(6):3552–3599. 10.1021/cr900354u [DOI] [PubMed] [Google Scholar]
- Zandleven J, Sørensen SO, Harholt J, Beldman G, Schols HA, Scheller HV, Voragen AGJ. XGA exists in cell walls from various tissues of Arabidopsis thaliana. Phytochemistry. 2007:68(8):1219–1226. 10.1016/j.phytochem.2007.01.016 [DOI] [PubMed] [Google Scholar]
- Zarattini M, Corso M, Kadowaki MA, Monclaro A, Magri S, Milanese I, Jolivet S, de Godoy MO, Hermans C, Fagard M, et al. LPMO-oxidized cellulose oligosaccharides evoke immunity in Arabidopsis conferring resistance towards necrotrophic fungus B. cinerea. Commun Biol. 2021:4(1):727. 10.1038/s42003-021-02226-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdunek A, Pieczywek PM, Cybulska J. The primary, secondary, and structures of higher levels of pectin polysaccharides. Compr Rev Food Sci Food Safety. 2021:20(1):1101–1117. 10.1111/1541-4337.12689 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Saar BG, Friedrich MG, Chen F, Liu Y-S, Dixon RA, Himmel ME, Xie XS, Ding S-Y. Imaging lignin-down-regulated alfalfa using Coherent Anti-Stoke Raman Scattering Microscopy. BioEnerg Res. 2010:3(3):272–277. 10.1007/s12155-010-9079-1 [DOI] [Google Scholar]
- Zhang X, Bian A, Li T, Ren L, Li L, Su Y, Zhang Q. ROS and calcium oscillations are required for polarized root hair growth. Plant Signal Behav. 2022b:17(1):2106410. 10.1080/15592324.2022.2106410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Guo Z, Zhuang Y, Suo Y, Du J, Gao Z, Pan J, Li L, Wang T, Xiao L, et al. MicroRNA775 regulates intrinsic leaf size and reduces cell wall pectin levels by targeting a galactosyltransferase gene in Arabidopsis. Plant Cell. 2021a:33(3):581–602. 10.1093/plcell/koaa049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu Y, Li C, Yin B, Liu X, Guo X, Zhang C, Liu D, Hwang I, Li H, et al. PtomtAPX is an autonomous lignification peroxidase during the earliest stage of secondary wall formation in Populus tomentosa carr. Nat Plants. 2022a:8(7):828–839. 10.1038/s41477-022-01181-3 [DOI] [PubMed] [Google Scholar]
- Zhang X, Xue Y, Guan Z, Zhou C, Nie Y, Men S, Wang Q, Shen C, Zhang D, Jin S, et al. Structural insights into homotrimeric assembly of cellulose synthase CesA7 from Gossypium hirsutum. Plant Biotechnol J. 2021b:19(8):1579–1587. 10.1111/pbi.13571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yu J, Wang X, Durachko DM, Zhang S, Cosgrove DJ. Molecular insights into the complex mechanics of plant epidermal cell walls. Science. 2021c:372(6543):706–711. 10.1126/science.abf2824 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang Z-Y, Dixon RA. LACCASE is necessary and non-redundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis thaliana. Plant Cell. 2013:25(10):3976–3987. 10.1105/tpc.113.117770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Fu Q, Xu F, Zheng H, Yu, F. New paradigms in cell adaptation: decades of discoveries on the CrRLK1L receptor kinase signalling network. New Phytol. 2021:232:1168–1183. doi: 10.1111/nph.17683 [DOI] [PubMed] [Google Scholar]
- Zhuo C, Wang X, Docampo-Palacios M, Xiao X, Sanders BC, Engle NL, Tschaplinski TJ, Hendry J, Maranas C, Chen F, et al. Developmental changes in lignin composition are driven by both monolignol supply and laccase specificity. Sci Adv. 2022:8(10):eabm8145. 10.1126/sciadv.abm8145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zugenmaier P. Order in cellulosics: historical review of crystal structure research on cellulose. Carbohydr Polym. 2021:254:117417. 10.1016/j.carbpol.2020.117417 [DOI] [PubMed] [Google Scholar]
- Zykwinska AW, Ralet MC, Garnier CD, Thibault JF. Evidence for in vitro binding of pectin side chains to cellulose. Plant Physiol. 2005:139(1):397–407. 10.1104/pp.105.065912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykwinska A, Thibault J-F, Ralet M-C. Organization of pectic arabinan and galactan side chains in association with cellulose microfibrils in primary cell walls and related models envisaged. J Exp Bot. 2007:58(7):1795–1802. 10.1093/jxb/erm037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This historical review contains no new data.