Abstract
Several efforts are currently directed at the creation and cellular implementation of alternative genetic systems composed of pairing components that are orthogonal to the natural dA:dT and dG:dC base pairs. In an alternative approach, Watson-Crick type pairing is conserved, but one or all of the four letters of the A, C, G and T alphabet are substituted by modified components. Thus, all four nucleobases were altered to create halogenated deazanucleic acid (DZA): dA was replaced by 7-deaza-2′-deoxyadenosine (dzA), dG by 7-deaza-2′-deoxyguanosine (dzG), dC by 5-fluoro-2′-deoxycytidine (FdC) and dT by 5-chloro-2′-deoxyuridine (CldU). This base-pairing system was previously shown to retain function in E. coli. Here we analyze, stability, hydration, structure and dynamics of a DZA Dickerson-Drew Dodecamer (DDD) of sequence 5′-FdC-dzG-FdC-dzG-dzA-dzA-CldU-CldU-FdC-dzG-FdC-dzG-3′. Contrary to similar stabilities of DDD and DZA-DDD, osmotic stressing revealed a dramatic loss of hydration for the DZA-DDD relative to the DDD. The parent DDD 5′-d(CGCGAATTCGCG)-3′ features an A-tract, a run of adenosines uninterrupted by a TpA step, and exhibits a hallmark narrow minor groove. Crystal structures – in the presence of RNase H – and MD simulations show increased conformational plasticity (‘morphing’) of DZA-DDD relative to the DDD. The narrow dzA-tract minor groove in one structure widens to resemble that in canonical B-DNA in a second structure. These changes reflect an indirect consequence of altered DZA major groove electrostatics (less negatively polarized) and hydration (reduced) compared to DNA. Therefore, chemical modifications outside the minor groove that lead to a collapse of major groove electrostatics and hydration can modulate A-tract geometry.
Graphical Abstract

INTRODUCTION
Chemically modified antisense, siRNA and aptamer oligonucleotides are being widely evaluated in the discovery and development of potential therapeutics against a host of targets.1–5 Modifications can affect RNA affinity, metabolic stability, uptake, interactions with proteins and cellular distribution. Artificial pairing systems, so-called xeno nucleic acids (XNAs), that may or may not pair with native nucleic acids potentially offer a path to semi-synthetic or synthetic organisms with non-DNA genomes.6 Besides their role as tools in synthetic biology and genetics, such artificial polymers can benefit diagnostics and the selection of new catalysts, and they may populate new regions of nucleic acid fold space. Examples of XNAs that were investigated in the context of in vitro evolution experiments and in vivo information transfer, often requiring reengineering of polymerases, include hexitol nucleic acid (HNA),7–11 threose nucleic acid (TNA),10,12,13 2′-fluoroarabino nucleic acid (FANA),14 4′-thio-modified nucleic acids,15,16 and size expanded DNA analogs (xDNA and yDNA).17–19
In addition to the creation of pairing systems with alternative backbones relative to natural DNA and RNA, efforts to design XNAs have focused on base pairs orthogonal to dA:dT and dG:dC, such that the new bases only interact with the complementary partner during replication. Examples of components of so-called Artificially Expanded Genetic Information Systems (AEGIS)20 include the Ds:Px,21,22 and S:B; Z:P; K:X and V:J base pairs.23,24 In an E. coli semi-synthetic organism unnatural hydrophobic 5SICS:NaM and TPT3:NaM base pairs were replicated correctly over several generations, although their retention remains limited compared to the natural pairs.25–27 In a further example of a building block with a modified nucleobase, albeit one that conserves Watson-Crick type pairing, 2′-deoxythymidine (dT) was replaced by 5-chloro-2′-deoxyuridine (CldU) to generate an E. coli strain that relies on ClU, A, C and G instead of the standard T, A, C and G alphabet.28 Evolving the strain that lacks thymidylate synthase and requires exogenous dT (CldU) over many generations eventually resulted in an adapted bacterium that contained less than 2% dT in its genome. Crystal structures of Dickerson-Drew Dodecamer (DDD) B-form DNA duplexes with CldU in place of dT with sequence, e.g. d(CGCGAAClUClUCGCG), or CldU in place of dC with sequence, e.g. d(CGCGAATTClUGCG), confirmed the nearly identical geometries of T:A/ClU:A pairs and T:G/ClU:G mismatch pairs, respectively.29 Furthermore, the thermodynamic stabilities of native and the corresponding CldU-modified duplexes were very similar, with melting temperatures Tm of within 1 to 2°C. The conclusion that base pair geometry and duplex conformation remain virtually unchanged as a consequence of replacement of the 5-methyl group of thymine by a chlorine atom was corroborated by the observation that CldU-modified DDDs were still recognized and cleaved by EcoRI endonuclease.29 Interestingly, there is precedence for a genome with one letter altered in nature. Thus, it was reported decades ago that 2,6-diaminopurine (DAP) completely replaces A in S-2L cyanophage DNA,30 and it was recently determined that the pathway for substituting A by DAP is widespread in phage genomes.31
In an extension of the above work on an E. coli genome chemically modified with CldU and its evolution, 7-deaza-2′-deoxyadenosine (7-deaza-dA or dzA) was identified as a suitable replacement of dA, along with several other analogs. Thus, duplexes with CldU:dzA pairs are stable and the synthesis of long >2kb constructs with the CldU/dzA combination in place of dT/dA was successfully demonstrated with Taq DNA polymersase.32 Morphing of 2′-deoxycytidine (dC) and 2′-deoxyguanosine (dG) into 5-fluoro-2′-deoxycytidine (FdC) and 7-deaza-2′-deoxyguanosine (7-deaza-dG or dzG), respectively, and combining them with CldU and dzA was used to create an alternative coding system with four noncanonical bases, so-called halogenated deazanucleic acid (DZA, Figure 1).33 The triphosphates of the four modified nucleotides were used for polymerase chain reaction (PCR) amplification with the Taq or Vent (exo-) DNA polymerases and various templates of up to 525 bases length. Remarkably, a completely morphed DZA gene encoding dihydrofolate reductase was successfully transformed into E. coli cells and shown to confer trimethoprim resistance. Moreover, it was recently demonstrated that DZA can be replicated, transcribed into ribo-deazanucleic acid (RZA) as well as reverse transcribed.34 Efficient and accurate replication using standard polymerases opens a path to library generation and in vitro evolution for selection of DZAptamers and potentially DZAzymes. An even more exciting prospect entails engineered cells capable of selectively taking up DZA triphosphates or perhaps even biosynthesizing them.33
Figure 1.

The four building blocks of halogenated deazanucleic acid (DZA).
Considered in the context of a DNA double helix, the chemical modifications of bases in DZA exclusively affect the major groove. The arrangements of H-bond acceptors and donors engaged in Watson-Crick pairing in DZA and DNA are identical. However, the base pairing strengths in the two systems most likely differ as a result of the introduction of halogen atoms at the C5 position of pyrimidines and the lack of the N7 function in purines in DZA. To gain a better understanding of the potential consequences of these chemical changes for duplex conformation, stability, hydration and dynamics, we used single crystal X-ray crystallography, UV melting, osmotic stressing and molecular dynamics (MD) simulations to analyze a pair of DZA and DNA self-complementary 12mer duplexes. The so-called Dickerson-Drew Dodecamer (DDD) of sequence d(CGCGAATTCGCG) has been studied in great detail for over 40 years.35–41 Two of its hallmarks are a short central A-tract that exhibits a contracted minor groove and an intricate double spine of hydration that dissects that groove.37–39 A-tracts are runs of A:T base pairs uninterrupted by a TpA step that are associated with characteristic sugar puckers, roll angles and propeller twists, resulting in narrow minor grooves and particular helix curvatures.42 These sequence-dependent conformational features play an important role in DNA recognition and indirect readout, whereby bending toward the minor groove and minor groove narrowing are interrelated, as exemplified by the distribution of short A-tracts that facilitate wrapping of DNA around the histone core in the nucleosome.43
Two crystal structures of DZA-DDD at resolutions of 1.5 Å and 2.3 Å reveal drastically different conformations. In one structure the shape of the DZA duplex closely resembles that of the native DNA-DDD (DDD) with a narrow (dz)A-tract minor groove. However, the other structure lacks the narrow minor groove in the central portion and is more reminiscent of the shape of canonical B-form DNA. The stability of DZA-DDD is reduced relative to DDD as established by UV melting experiments. Osmotic stressing assays using ethylene glycol and acetamide as cosolutes in melting experiments demonstrate significantly diminished hydration of the DZA-DDD compared to the parent DDD duplex, consistent with the loss of the N7 acceptor at the major groove edges of dzA and dzG. The observation of a minor groove without the characteristic contraction in the A-tract in one of the DZA-DDD structures is surprising. To probe the relative conformational plasticities of DZA- and DNA-DDD, we conducted 10 μsec MD simulations for both systems. For DZA-DDD, we generated two trajectories, one each starting from the “narrow-groove” and “wide-groove” crystal structures. We observe that the ‘wide-groove’ DZA-DDD structure is stable and maintains the crystal structure conformation during the full simulation, although we do observe some narrowing of the A-tract region. In marked contrast, the ‘narrow-groove’ simulation rapidly transitions to a structure that resembles closely the wide-groove crystal structure, The rapid structural transition we observe in these equilibrium MD simulations implies that there is a negligible energy barrier separating the two conformations, which is consistent with the suggestion that DZA-DDD possesses considerable conformational plasticity. These investigations offer insight into the effects of relatively minor chemical modifications of DNA bases in the major groove on sequence-dependent changes in duplex geometry. Moreover, our work sheds new light on the precise origins of a well-established conformational hallmark of DNA A-tracts.
EXPERIMENTAL PROCEDURES
Synthesis of DZA nucleoside phosphoramidites.
The dzA and dzG phosphoramidites were purchased from Glen Research (Sterling, VA). The CldU and FdC phosphoramidites were synthesized following previously reported protocols.29,44,45
Oligonucleotide synthesis and purification.
The DZA dodecamer was synthesized on an Expedite DNA synthesizer (Applied Biosystems) by the phosphoramidite approach. After deprotection and cleavage from the solid support with methylamine (40% in water), the oligomer was concentrated in aqueous ammonia (1:1, 30°C). Following gel filtration using an NAP-10® column (Sephadex G25-DNA grade, Pharmacia) and water as eluent, the crude mixture was analyzed on a Mono-Q® HR 5/5 anion exchange column. Further purification involved a Mono-Q® HR 10/10 column (Pharmacia) and the following gradient system: A=10 mM NaOH, pH 12.0, 0.1M NaCl, and B=10 mM NaOH, pH 12.0, 0.9M NaCl. After desalting on a NAP-10® column and lyophilization, the oligo was purified by RP-HPLC on a C-18 column, using a linear gradient of A, ammonium bicarbonate (25 mM in H2O, pH 7.0), and B, acetonitrile (80% in H2O). The purity was checked by capillary electrophoresis and the correct mass confirmed by mass spectrometric analysis. The native DDD was purchased from Integrated DNA Technologies, Inc. (Coralville, Iowa).
Tm measurements.
The dodecamers were dissolved in a buffer solution containing 20 mM KH2PO4, pH 7.5, 100 mM NaCl, and 0.1 mM EDTA. Concentrations were established by measuring absorbance in MilliQ water at 260–270 nm at 80°C and assuming that modified nucleosides have the same extinction coefficients per base moiety in the denatured state as the natural nucleosides. The concentration for each strand was 4 μM. Melting curves were determined with a Varian Cary 300 BIO spectrophotometer. The cuvettes were kept at constant a temperature using a Peltier element. Annealing of strands was achieved with a quick heating and cooling cycle. Samples were then heated from 10°C to 90°C at a rate of 0.2°C per minute and cooled again at the same speed. The melting temperatures Tm were determined by plotting the first derivative of the absorbance as a function of temperature and data are the average of two runs (Table 1, Table S1, supp. inform.).
Table 1.
Comparison between UV melting data for native and modified DDDs
| Oligonucleotide sequence | Tm. [°C] |
ΔTm [°C] |
strand conc. [μM] |
Na+ conc. [mM] |
ref. |
|---|---|---|---|---|---|
| d(CGCGAATTCGCG) = DDD | 59.1. | – | 4.0 | 100 | this work |
| FdC-dzG-FdC-dzG-dzA-dzA- | 53.2 | −5.9 | 4.0 | 100 | this work |
| CldU-CldU-FdC-dzG-FdC-dzG | |||||
| DDD | 57.7. | – | 10.0 | 100 | [76] |
| d(CGCGAATTCzGCG) | 50.0. | −7.7 | 10.0 | 100 | [76] |
| d(CGCGAzATTCGCG) | 49.5 | −8.2 | 10.0 | 100 | [77] |
| DDD | 62.6. | – | 7.5 | 150 | [29] |
| d(CGCGAAClUClUCGCG) | 63.8 | +1.2 | 7.5 | 150 | [29] |
Van’t Hoff analysis and osmotic stressing studies.
UV melting experiments were done on two instruments in parallel: Shimadzu UV-1800 and UV-260 UV-Vis spectrometers, both equipped with eight-position TMSPC-8 Peltier temperature controllers. The data were processed using Shimadzu LabSolutions Tm Analysis software, version 1.31. The number of repetitions varied among samples but was at least six (Table 2, Table S2).
Table 2.
Thermodynamic stability and osmotic stressa analysis (Δnw) for DNA and DZA DDDs
| Oligo | Tm [°C] b (UV melting) |
ΔG [kcalmol−1] |
ΔH [kcalmol−1] |
ΔS [eu] |
Δnw (ethylene glycol) |
Δnw (acetamide) |
|---|---|---|---|---|---|---|
| DNA-DDD | 66.7 ± 0.6 | −13.8 ± 0.4 | −51.0 ± 3.8 | −125 ± 12 | 29 ± 7 | 43 ± 5 |
| DZA-DDD | 62.1 ± 0.5 | −11.2 ± 0.2 | −34.7 ± 1.6 | −79 ± 5 | 13 ± 4 | 27 ± 4 |
In order to determine Δnw, the change in the number of water molecules associated with the melting process, we followed the approach initially described by Spink and Chaires for duplex and triplex DNA46 and subsequently used for native and chemically modified DNA and RNA duplexes47–51: Δnw = (−ΔH/R)[d(Tm−1)/d(lnaw)], where −ΔH is the enthalpy determined in the UV melting experiment and R is the universal gas constant (1.986 calmol−1K−1). Professors Spink and Chaires provided the experimentally determined values of water activity, lnaw, at given co-solute concentrations (ethylene glycol, acetamide). The slope of the plot of the reciprocal temperature of melting (in K) versus the logarithm of water activity (lnaw) at different concentrations (0, 5, 10, 15 and 20%) of the small co-solutes gave the value of d(Tm−1)/d(lnaw). Finally, Δnw values were extracted by linear fitting the data using KaleidaGraph software (Version 3.51), whereby experimental uncertainties were calculated as reported earlier.46,48
Crystallization.
The Asp132→Asn mutant of Bacillus halodurans RNase H (BhRNase H; Met58 to Lys196) was expressed in E. coli and purified as described previously.52,53 The protein solution was concentrated to 35 mg/mL. Protein and DNA solutions were mixed in a 1:1 molar ratio in the presence of 5 mM MgCl2 and crystallization experiments were performed by the sitting drop vapor diffusion technique using the crystal screen (Hampton Research, Aliso Viejo, CA).54 Briefly, 200 nL complex solution was mixed with 200 nL of reservoir solution and equilibrated against 70 μL reservoir wells using a Mosquito crystal automated liquid handler (SPT Labtech Inc., Boston, MA). Well diffracting crystals (P21) were obtained from droplets that were mixed and equilibrated with 2 M (NH4)2SO4 in about 3 days. Similarly, crystals (P62) appeared from droplets that were mixed and equilibrated with 0.2 M (NH4)2SO4, 0.1 M Sodium acetate trihydrate (pH 4.6) and 25% (w/v) PEG 4000. Crystals were mounted in nylon loops, cryo-protected in 20% glycerol containing reservoir solution and frozen in liquid nitrogen.
Data collection, phasing and refinement.
Diffraction data were collected on the 21-ID-G beam line of the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Photon Source (APS), Argonne National Laboratory (Argonne, IL). Data sets were collected for multiple RNase H:DZA-DDD crystals. Crystals were kept at 100 K during data collection, and the images were captured on a MARCCD 300 detector using a wavelength of 0.97856 Å. Diffraction data were integrated, scaled and merged with HKL2000.55 A summary of selected crystal data and data collection parameters is provided in Table 3. The structures were solved by molecular replacement with the program MOLREP,56,57 using only the protein as the search model (PDB code: 3EY1). In the case of P21 model, there were three RNase H molecules and one DZA-DDD duplex in the asymmetric unit. The initial refinement of the protein alone in Refmac558 resulted in an initial Rwork/Rfree of 0.35/0.39, respectively, by keeping aside 5% of the reflections to compute the Rfree. the Fourier 2Fo-Fc sum and Fo-Fc difference electron density maps. These maps clearly revealed the DZA duplex which was then added to the three protein molecules. Map visualization and model rebuilding were performed with the program Coot.59 All 2′-deoxynucleotides were replaced with the corresponding modified nucleotides, i.e., dA by dzA, dC by FdC, dG by dzG, and dT by CldU. Further isotropic refinements were conducted in Refmac558 after adapting the dictionary files using the program PRODRG.60 In subsequent refinement steps, water molecules were added (about 20–30 molecules in each refinement step) on the basis of superimposed positive Fourier 2Fo-Fc sum and Fo-Fc difference electron density peaks and distance and B-factor criteria. As the refinement progressed, a glycerol molecule become clear in the electron density and was added. Selected final refinement parameters and deviations from ideal geometries are summarized in Table 3.
Table 3.
Selected crystal, data collection and refinement parameters.
| Parameter | BhRNase-H: DZA-DDD Complex | |
|---|---|---|
| Data collection | ||
| DZA-DDD sequence | 5′-FdC-dzG-FdC-dzG-dzA-dzA-CldU-CldU-FdC-dzG-FdC-dzG-3′ | |
| Space group | P21 | P62 |
| Unit cell constants: a, b, c [Å] | 36.98, 89.00, 72.57 | 92.54, 92.54, 78.90 |
| Unit cell constants: α, β, γ [°] | 90.0, 100.4, 90.0 | 90.00, 90.00, 120.00 |
| Resolution [Å]a | 44.54–1.51 (1.56–1.51) | 27.60–2.30 (2.38–2.30) |
| No. of unique reflections | 72,096 | 17,002 |
| Completeness (outer shell) [%] | 99.8 (100.0) | 98.9 (90.8) |
| R-merge | 0.053 (0.370) | 0.160 (1.471) |
| R-pim | 0.028 (0.214) | 0.044 (0.485) |
| I/σ(I) | 35.3 (2.9) | 14.2 (0.6) |
| Redundancy | 4.5 (3.9) | 13.8 (7.1) |
| Refinement | ||
| Number of reflections | 68,594 (3,475) | 16,084 (1,050) |
| R-work | 0.177 (0.236) | 0.214 (0.416) |
| R-free | 0.210 (0.226) | 0.254 (0.359) |
| No. of protein/nucleotide atomsb | 3,232/596 | 1,080/495 |
| No. of waters/ions or ligands | 495/1 | 85/5 |
| R.m.s. deviations bonds [Å] | 0.009 | 0.018 |
| R.m.s. deviations angles [°] | 1.9 | 1.6 |
| Avg. B-factor, Protein/DZA atoms [Å2] | 21.2/17.1 | 48.7/55.1 |
| Avg. B-factor, H2O/ions or ligands [Å2] | 29.1/24.6 | 48.9/78.6 |
| PDB entry code | 8SV3 | 8SV4 |
Outermost shell in parentheses
Dual occupancy atoms included
In the case of P62 crystals, several data sets were collected from different crystals and processed using HKL2000.55 Following molecular replacement and initial refinement, electron density maps were visualized with the program Coot.59 In several cases, DZA duplexes exhibited positional disorder, with duplexes of partial occupancy shifted relative to one another along the direction of the helical axis. We had previously encountered this phenomenon with crystals of BhRNase in complex with a hybrid between DNA and 2′-F RNA.61 Instead, we selected a data set with a resolution limit of 2.3 Å for which phasing revealed an asymmetric unit that consists of a single DZA duplex bound to an RNase molecule. Subsequent refinements were continued in a similar fashion as described above for the P21 complex model. Selected final refinement parameters and deviations from ideal geometries are summarized in Table 3 and electron densities around the final complex models are shown in Figure 2. Nucleotides in one strand are numbered 1 to 12, and 13 to 24 in the complementary strand. All structural illustrations were prepared with the program UCSF Chimera.62
Figure 2.
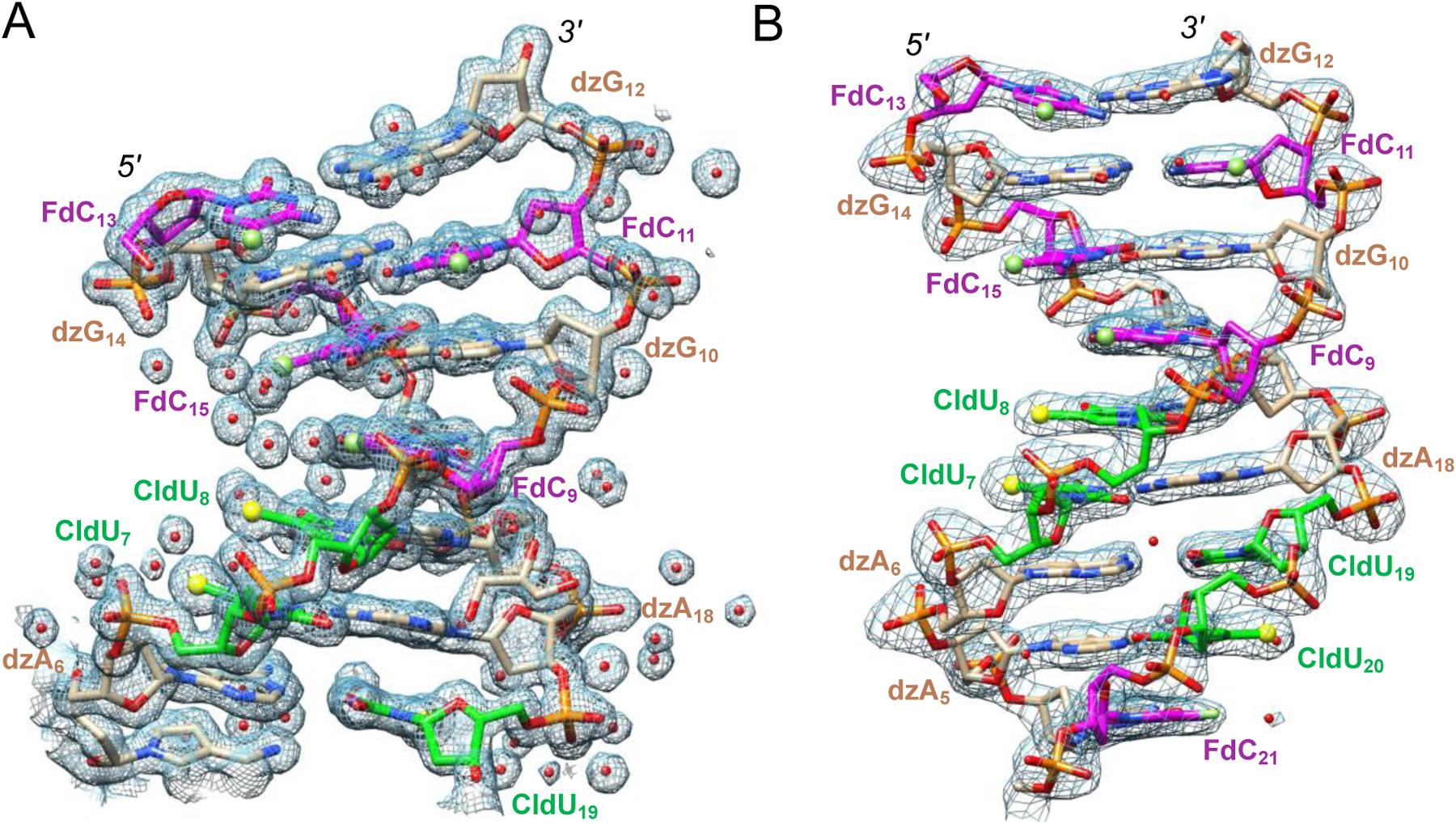
Quality of the final Fourier 2Fo-Fc sum electron density (1σ threshold). The two dodecamers are viewed across the grooves, revealing (A) a widened minor groove for DZA-DDD1 and (B) a narrow ‘A-tract’ minor groove for DZA-DDD2. Nucleotides in one strand are numbered 1 to 12, and 13 to 24 in the complementary strand. Carbon atoms of FdC, dzG/dzA and CldU are colored magenta, beige and green, respectively. Oxygen, nitrogen, phosphorus, fluorine and chlorine atoms are colored red, blue, orange, green and yellow, respectively, and water (red), Cl and F are highlighted as spheres.
Data deposition.
Coordinates and structure factors for both complexes have been deposited in the Protein Data Bank (PDB; http://www.rcsb.org). The PDB ID codes for the RNase-H-oligonucleotide complexes in space groups P21 and P62 are 8SV3 and 8SV4, respectively.
MD simulations and parameter development.
Force field parameters for the modified nucleotides were needed to perform the MD simulations described. Development of the modified pyrimidine parameters was straightforward. 5-chlorouracil (CldU) is simply the chloro homolog of thymine, so we used the OL15 force field63 template for thymine to construct CldU. Additional bond and bond angle parameters missing from the standard OL15 dataset were taken from the Generalized Amber Force Field, GAFF64. Torsion angle parameters were adapted directly from the thymine OL15 force field values. Likewise, 5-fluorocytosine (FdC) is the fluoro homolog of cytosine, so we utilized the OL15 cytosine template to constructure FdC, again utilizing the GAFF force field for missing bond and bond angle parameters, while adapting the standard OL15 cytosine torsion angle parameters.
Development of 7-deazapurine parameters is slightly more involved. However, 7-deazapurines are quite common in RNA from many species, and Aduri et al.65 have previously developed force field parameters for substituted 7-deazapurines suitable for use with the AMBER OL3 force field (i.e., the standard AMBER RNA force field). Thus, it was trivial to build upon this earlier work to generate appropriate templates for 7-deazaadenine (DzA) and 7-deazaguanine (DzG), utilizing the Aduri force field parameters and GAFF parameters to supplement the existing OL15 parameter dataset.
Partial charges for all four modified nucleotides were computed using the standard RESP protocol66 for AMBER charge development. During the charge fitting procedure, partial charge values for all atoms in the 2′-deoxyribose sugar ring and phosphate group were restrained to match the values for the standard A, C, G and T nucleotides in the OL15 force field templates. These restraints are necessary for consistency with the OL15 force field and to ensure that our modified nucleotides can be integrated seamlessly into a polynucleotide chain which also contains standard residues. The resultant partial charge assignments for CldU, FdC, DzA, and DzG are quite similar with partial charge values for the standard OL15 nucleotides, with only modest atom charge deviations in the immediate vicinity of the residue modifications, e.g., immediately at and around atom C7 of the deazapurines and atom C5 of CldU and FdC. Files for the modified nucleotide templates containing atom types, bond connectivity, geometry and partial charges, as well as a file containing new OL15 force field parameters for the modified nucleotides are available in Supplemental Information.
Equilibrium MD simulations were performed with the PMEMD AMBER module67 utilizing our extended OL15 parameter set together with the TIP3P water model68 and Joung/Cheatham parameters for monovalent sodium ions69 for all calculations. The DZA-DDD (both wide-groove and narrow-groove conformations) and standard DDD molecules were each solvated in a truncated octahedron box with a 14Å buffer zone between any nucleotide atom and the closest box wall. Sodium cations were added to each system to impose system charge neutrality, yielding solutions of ~5 mM DDD or DZA-DDD and ~130 mM sodium cations. Each starting complex was subjected to a three-step minimization procedure. First, DNA atoms were relaxed for 10,000 steps of conjugate gradient energy minimization while water molecules and counterions were restrained at starting positions. Next, all solvent and counterions were relaxed for 10,000 steps while DNA atoms were restrained. Finally, all restraints were removed, and the entire system was minimized for 10,000 additional steps. Each minimized nucleotide dodecamer complex was then heated gradually from 0 to 300 K during a 200 ns canonical ensemble (NVT) MD simulation, followed by a 300 ns NPT ensemble simulation. Production MD simulations were run with a 1.5 fs timestep for 10 μs. Energy and force calculations were performed using minimal image periodic boundary conditions, a 12Å nonbonded cutoff for real space interactions, a homogeneity assumption to approximate the contributions of long-range Lennard-Jones forces to the virial tensor, and staggered particle-mesh Ewald for long-range electrostatics correction.70 A Langevin thermostat with collision frequency 3 ps−1 was used to maintain the system temperature.71 All bonds containing hydrogen were constrained using the SHAKE algorithm72 and the SETTLE method was used to maintain rigid water geometry.73 Final analysis of all MD trajectories was performed using the cpptraj package.74 DNA structural parameters were calculated using the 3DNA75 algorithms as implemented in the cpptraj package.
RESULTS
Thermal Stability of DDD and DZA-DDD
The UV melting temperatures Tm of the parent DDD and the corresponding DZA duplex were measured in a solution composed of 20 mM KH2PO4, pH 7.5, 100 mM NaCl, and 0.1 mM EDTA, and using an oligo concentration of 4 μM. The melting temperature Tm of the DDD is 59.1°C and that of the DZA-DDD duplex is 53.2°C (Table 1). Given the chemical changes at the edge of the major groove in the DZA duplex that affect all 24 nucleobases and eliminate an H-bond acceptor as well as potential Na+ binding site in purines and replace a methyl group by chlorine and hydrogen by fluorine in T and C, respectively, this difference in Tm is quite small. The loss in stability for the DZA duplex amounts to just −0.5°C per base pair. Replacement of a single G by dzG in the DDD, d(CGCGAATTCzGCG), was reported to reduce the Tm by −7.7°C (from 57.7 to 50.0°C) in 100 mM NaCl using a 10 μM strand concentration.76 Similarly, replacement of a single A by dzA in the DDD, d(CGCGAzATTCGCG), was reported to reduce the Tm by −8.2°C (from 57.7 to 49.5°C) in 100 mM NaCl using a 10 μM strand concentration.77 Conversely, replacement of both Ts by CldU in the DDD, d(CGCGAAClUClUCGCG), in 150 mM NaCl and using a 7.5 μM strand concentration increased the Tm by 1.2°C (from 62.6 to 63.8°C; 0.6°C per residue).29 The van’t Hoff analysis of thermodynamic parameters based on concentration-dependent UV melts at 300 mM salt concentration confirmed the rather small difference in Tm values (66.7°C and 62.1°C for native and DZA-DDD, respectively; Table 2, Table S1, supp. inform.). The difference in free energy between the two duplexes is just 2.6 kcalmol−1. The consequence of FdC incorporation alone into the DDD is not available. However, the stability of a DNA hairpin 5′-d(CGGAAGG[UCCG]CFCUUCC)-3′ with a single FdC opposite G (underlined, loop residues in brackets) was reduced by 1.1°C relative to the native hairpin (75.6°C vs. 76.7°C, respectively).78
Osmotic Stressing
To measure the effects of base chemical modification on the hydration of the DZA duplex compared to DNA we used osmotic stressing.47 UV melting experiments in the presence of cosolutes that do not stably bind to the macromolecule but displace water molecules from its surface afford a qualitative estimate of the number of waters that are released upon strand separation. Both in the presence of either ethylene glycol or acetamide, the DZA-DDD sheds fewer water molecules than the parent duplex during melting (Table 2, Table S2). With ethylene glycol, ΔΔnw amounts to 16 and the DDD released more than twice as many waters compared to the DZA-DDD. With acetamide the assay gave an identical value for ΔΔnw, but the overall numbers for DDD and DZA-DDD indicate a less dramatic reduction in hydration as a result of modifying all four bases (Δnw=43 and 26 for the two duplexes, respectively; Table 2). However, the data indicate clearly that the loss of the N7 acceptor in all 12 purines of the DDD leads to a significantly diminished hydration in the B-DNA major groove.
Crystal Structures of DZA-DDD
In order to examine potential effects of 7-deaza- and 5-halogen-modified purines and pyrimidines, respectively, in DZA-DNA on its conformation relative to DNA, we turned to the Dickerson-Drew Dodecamer (DDD) B-form duplex. DDD-based structures account for a significant portion (some 15%) of the ca. 1,100 DNA-only crystal structures determined to date.40 Initially we directed our efforts at growing crystals and determining the structure of DZA-DDD alone. However, these attempts ultimately remained unsuccessful because of the insufficient quality of crystals. Instead we turned to a strategy that we had previously used with chemically modified DDDs that resisted crystallization.79 The complex between DDD and B. halodurans RNase H (inhibitor complex) produced well diffracting crystals, whereby the interactions with the enzyme do not alter the conformation of the DDD relative to that of the duplex alone (i.e. the narrow minor groove in the central A-tract section is maintained).53 Using this approach, we determined two crystal structures of DZA-DDD:RNase H complexes at resolutions of 1.5 Å and 2.3 Å (named DZA-DDD1 and DZA-DDD2, respectively). Examples of the quality of the final electron density for the two complexes are depicted in Figure 2 and selected crystal data, data collection and refinement parameters are listed in Table 3.
The two crystal structures in the space groups monoclinic P21 and hexagonal P62 feature different contents of their respective asymmetric unit (a.u.). The a.u. of the former contains one duplex (DZA-DDD1) and three RNase H molecules; the a.u. of the latter contains one duplex (DZA-DDD2) and a single RNase H molecule. A look at the two duplexes reveals a striking difference between them: Compared to the native DDD, the minor groove of DZA-DDD1 is clearly wider, whereas the DZA-DDD2 minor groove exhibits the same characteristic contraction in the central A-tract portion (Figure 3). The figure depicts the crystal structure of the parent DDD in the free form (leftmost duplex).80 In all structures of the DDD alone and independent of resolution (e.g., at ca. 1.5 Å80 or at atomic resolution up to 0.95 Å37–40) the A-tract minor groove is always extremely narrow. This narrow groove is preserved in the crystal structure of the DDD forming a non-specific complex with RNase H from Bacillus halodurans.53 RNase H cleaves the RNA strand of RNA:DNA hybrids and the enzyme bound to the DDD thus constitutes an inhibitor complex. A comparison between the minor groove widths of the DDD alone and in complex with RNase H is shown in the publication reporting the first structure of the complex.53
Figure 3.

Conformational properties of DNA-DDD (PDB ID 355D)80 and DZA-DDD duplexes. The three dodecamers are viewed into the central minor groove, revealing a wider groove typical for canonical B-form DNA in the case of DZA-DDD1 (center) and a narrow ‘A-tract’ groove in DZA-DDD 2 (right) that is a hallmark of the native DDD (left).
We used the programs 3DNA81 and Curves82 to calculate helical parameters for the various duplexes. Minor groove widths were taken from the latter outputs and are graphically displayed in Figure 4. This analysis demonstrates that the A-tract minor groove in the DZA-DDD1 duplex is about 4 Å wider than the corresponding groove in the DZA-DDD2 duplex. The groove in the latter is unchanged relative to the native duplex (both bound to RNase H; Figure 4), and also relative to the native DDD in the free form (as shown before in ref. 53). However, in the flanking G-tracts (or zG-tracts in DZA), the minor groove widths in all duplexes remain more-or-less the same. Inspection of the helical parameters offers insight into some of the changes that contribute to the difference in minor groove width between DZA-DDD1 and DZA-DDD2/native DDD. One of these parameters is inclination (base pairs relative to helix axis); the average numbers from 3DNA are 14.2° and 7.2° for DZA-DDD1 and DZA-DDD2, respectively, and the numbers from Curves are 9.4° and 3.5° for DZA-DDD1 and DZA-DDD2, respectively. Therefore, groove widening is accompanied by an increase in inclination. Two further parameters that exhibit clear differences between the two DZA duplexes are helical twist and x-displacement. For twist, the average values are 33° and 35° for DZA-DDD1 and DZA-DDD2, respectively (3DNA and Curves provide similar numbers). For x-displacement, the values are −1.8 Å (−1.7 Å) and −0.9 Å (−1.0 Å) for DZA-DDD1 and DZA-DDD2, respectively (Curves numbers in parentheses).
Figure 4.

Minor groove widths of native DDD and DZA-DDD duplexes in complex with RNase H. Values were calculated with the program Curves.82 The DDD:RNase H crystal structure contains two independent complexes, DDD:RNaseH1 and DDD:RNaseH2, that exhibit very similar conformations and sit on a crystallographic dyad (note the symmetrical shape of the corresponding blue and orange groove width curves, respectively).79
Besides the above helical parameters, the average values of which differ between the DZA-DDD1 and DZA-DDD2 duplexes, the analysis of their helix geometries identified further distinct features. Sugar puckers in the native DDD and its complex with RNase H are with a few exceptions limited to the C2′-endo and C1′-exo ranges (Figure 5). There are some excursions into the Eastern range (O4′-endo) and an occasional C3′-endo (North) pucker for residues at or near the 5′- or 3′-terminal ends of strands. The same applies to the DZA-DDD2 duplex that exhibits a narrow minor groove. Conversely, there are six nucleotides in the DZA-DDD1 duplex that adopt a C3′-endo pucker and another two with a C4′-exo pucker. Despite this trend toward an RNA-like sugar conformation for a third of the nucleotides, the DZA-DDD1 duplex is not of the A-form. The diameters of B-form and A-form duplexes differ considerably, but notwithstanding the observed geometric differences between the DZA-DDD1 duplex on the one hand and the DZA-DDD2 and native DDD duplexes on the other, they all exhibit similar diameters.
Figure 5.

Sugar puckers in the native DDD and DZA-DDDs. The graph covers the Eastern half of the pseudorotation phase cycle and shows phase angles for individual nucleotides in the 12mer duplexes. DZA1 and DZA2 correspond to the DZA-DDD1 and DZA-DDD2 duplexes, respectively.
The wider minor groove of DZA-DDD1 compared to the DZA-DDD2 and parent DDD duplexes has consequences for the interaction with RNase H in the crystal of the complex. The enzyme probes the minor width of duplexes and uses it to discriminate between RNA:DNA hybrid substrates and B-form DNA and A-form RNA inhibitors, in addition to scanning for the presence of 2′-hydroxyl groups on one strand. The DDD A-tract minor groove is extremely narrow (as tight as 3 Å; Figure 4) compared to that of an RNA:DNA hybrid duplex (8 Å).52,83 By comparison, the minor groove width of B- and A-form duplexes are 5.7 Å and 11 Å, respectively.83 At the ends of the DDD, in the G-tract region, the minor groove opens up compared to the A-tract (Figure 4) and B. halodurans RNase H clamps onto both G-tract regions in the crystal of the complex with the native DDD.53
In the crystals of the complexes between RNase H and DZA-DDD1 and DZA-DDD2, RNase H only binds to one end of the duplex and the opposite end stacks onto an adjacent duplex. In the crystal of the complex with DZA-DDD1 that contains three RNase H molecules per a.u., the protein bound to the dzG-tract is RNase H-a (Figure 6A). Because the central minor groove in the DZA-DDD1 duplex is wider than in the DDD (expanded to ca. 7 Å on average, Figure 4, and thus similar to that in an RNA:DNA hybrid, a second RNase H molecule termed RNase H-c (Figure 6A) binds to its dzA-tract. The binding modes of the two RNase H molecules are virtually identical (Figure 6B,C). Most importantly, protein binding is not at the origin of the wider minor groove in the DZA-DDD1 duplex. Rather, unlike the tight DZA-DDD2 or DDD minor grooves, the expanded DZA-DDD1 groove is suitable for docking by the protein, consistent with minor groove width being the major determinant of RNase H substrate recognition.
Figure 6.

RNase H-DZA interactions. (A) Two RNase H molecules are lodged in the relatively wide minor groove of the DZA-DDD1 duplex, and the non-specific binding modes of (B) RNase H-a (dark-gray ribbon) in the dzG-tract and (C) RNase H-c (light-gray ribbon) in the dzA-tract are very similar. The DZA-DDD1 duplex is shown as a surface model with coloring identical to that in Figure 3. In panels B and C, the so-called phosphate binding site is in the foreground, the ‘active site’ phosphate is in the background and selected H-bonds are drawn with thin solid lines.
MD simulations with DDD and DZA-DDD
The MD simulations reveal a (largely) converged conformation for DZA-DDD that corresponds closely with the “wide-groove” crystal structure. In the simulation that originated from the wide-groove structure, ensemble-averaged values for base inclination (9.3°), twist (32.1°) and x-displacement (−1.9Å) are all in excellent agreement with values observed in the reference crystal structure as noted above. We do observe that the minor groove A-tract region gradually narrows during the simulation: the mean narrow groove width in the A-tract region is ~14.0 ± 0.7 Å initially but diminishes to ~11.3 ± 1.0 Å during the simulation. This is nonetheless still much wider than the narrow groove width we observe for the A-tract region in the native DDD structure (groove width ~9.5 ± 0.3 Å).
The MD simulation that originated from the narrow-groove crystal structure exhibited markedly different behavior. We observed relatively rapid transitions from the crystal structure values for inclination (initial: 4.3°, ensemble average: 9.3°), twist (initial: 34.8°, ensemble average: 32.1°) and x-displacement (initial: −0.6 Å, ensemble average: −1.9 Å) to values nearly identical to those observed for the wide-groove crystal structure and the MD simulation propagated from that structure. These transitions occurred over the first 500–800 ns and the ensemble average values were stable for the duration of the simulation. Concurrent with the transitions in base inclination, twist and x-displacement, we also observed a gradual widening of the minor groove A-tract region (albeit on a somewhat slower time scale of several microseconds). Thus, the initial minor groove width in the A-tract region for this simulation was ~9.9 ± 0.7 Å and the final groove width was ~11.3 ± 1.0 Å. We also observe rare, but occasional, Watson-Crick base pair disruptions in this simulation, particularly in the earlier stages of the trajectory when we also observe the structural transitions and minor groove widening. However, the base pair disruptions are quite rare: excluding the terminal base pairs, Watson-Crick base pairing is 99.97% maintained in this narrow groove simulation, compared to 99.99% Watson-Crick base pairing maintenance in both the wide-groove DZA and native DDD simulations. Thus, base pairing is well maintained for all simulations, and it seems unlikely that these transient base pair disruptions are strongly correlated with the observed structural changes in the narrow-groove DZA-DDD trajectory. In essence, we observed that the simulation initiated from the narrow-groove crystal structure converged to a nearly identical structure as that observed in the “wide-groove” simulation, and the groove widening appeared to be coupled to changes in base inclination, twist, and displacement.
An analysis of the sugar pucker behavior for DZA-DDD reveals no noticeable differences between the two simulations, at least in an ensemble average sense. This is not surprising given that the simulations “converge” to very similar structures. We did note that sugar puckers for the bases in and immediately adjacent to the A-tract region do tend to transition between C2’-endo and O4’-endo conformations much more frequently than the distal base pairs.
As noted above, the crystal structures suggest that hydration is reduced in the immediate vicinity of the C7 position in deazapurine residues (compared to the N7 atom in the native adenine and guanine residues), as would be expected. Both our DZA-DDD simulations confirm this trend, as we observe ~70% fewer water molecules in the first hydration shell around the C7 purine atoms and ~20% fewer water molecules total in the combined first and second hydration shells as compared to the hydration profile near N7 atoms in the DDD simulation (3.4Å and 5.0Å radius values were used to compute first and second hydration shell statistics).
The general sodium cation distribution in a 4.0Å shell around each polynucleotide complex is quite similar. However, there are some subtle, and interesting, differences in the fine details of sodium ion locations. We focused our analysis on sodium cations that form specific interactions with the polynucleotides, defined as those cations with a contact distance shorter than 3.0Å. The ensemble-averaged number of sodium ions present in the major groove is nearly identical for all three simulations, but there is a notable difference in the specific interactions observed. In the native DDD complex, sodium cations form specific interactions primarily with the N7 atom of the purine bases (~65% of all sodium interactions observed in the major groove), while specific interactions between sodium cation and the O4 atom of thymine or the O6 atom of guanine are less prevalent. In the DZA-DNA complexes, which lack the N7 purine atom, specific sodium cation interactions are distributed evenly between the O4 atom of 5-chlorouracil and the O6 atom of 7-deazaguanine.
The minor groove exhibits more notable differences between native DDD and DZA-DNA complexes, as we observe approximately twice as many specific minor groove sodium interactions in the native DDD complex. The DZA-DNA complexes exhibit fewer specific minor groove sodium cation interactions but a larger number of strong cation interactions with background phosphates (i.e., sodium-phosphate contact distance less than 3.0Å) as compared to the native DNA complex. In the native DDD structure, ~80% of the specific sodium cation interactions are with the O2 atom in cytosine and thymine, and the remaining interactions are with the purine N3 atoms. In the DZA-DNA minor groove, specific sodium interactions are distributed evenly between the O2 atoms of 5-chlorouracil and 5-fluorocytosine, and the N3 atom of the deaza-purines. Our simulations reveal an intriguing apparent correlation between minor groove width and the extent of specific cation interactions in the minor groove. The DZA-DNA simulation initiated from the crystal structure that displays a narrow minor groove exhibits a gradual transition to a wider groove geometry, as noted above. During the first 1–2 microseconds of the simulation, the minor groove still retains the narrow geometry characteristic of the native DDD complex, and we observe a larger number of specific sodium cation interactions, comparable to the native DDD simulation. As the minor groove gradually widens during this simulation, the number of specific minor groove cation interactions diminishes, and a few additional strong interactions are formed instead with the backbone phosphate groups. During the last 2–3 microseconds of this simulation, sodium cation distributions and behavior are indistinguishable from those observed for the DZA-DNA simulation initiated from the “wide-groove” crystal structure.
DISCUSSION
Stability.
DZA represents an alternative pairing system with a natural 2′-deoxyribose sugar-phosphate backbone and chemically modified nucleobases dzA, dzG, CldU and FdC (in place of A, G, T and C, respectively) capable of forming Watson-Crick H-bonds. The stability of a DZA dodecamer, the DZA-DDD studied here, is only moderately reduced compared to the parent DNA dodecamer (DDD). At 100 mM NaCl and a 4 μM strand concentration, the melting temperature Tm of the DZA-DDD is 53.2°C and just 5.9°C lower than the Tm of the DDD. At higher ionic strength (300 mM NaCl) and using a 2 μM strand concentration, the Tm values of DZA-DDD and DDD differ by less than 5°C (66.7°C and 62.1°C, respectively). These relatively modest differences stand in sharp contrast to substantially reduced Tm values observed for DDDs with a single dzA in place of dA or a single dzG in place of dG per strand relative to the parent duplex. In both cases the loss amounted to ca. 8°C (100 mM NaCl, 10 μM strand concentration),76,77 a difference that is 2°C higher than that seen between the Tms of the DDD and a completely modified DZA-DDD.
The steep loss in duplex stability due to incorporation of a single, chemically modified nucleotide into a DNA strand compared to stable pairing without additional reduction in stability seen for the all-modified oligonucleotide is surprising. More often, changes in the melting temperature of a DNA or RNA duplex caused by chemically modified nucleotides, either a loss or a gain, are additive.84 It appears that replacing a single dA by dzA or a dG by dzG in the DDD results in a conformational or electrostatic heterogeneity that is more-or-less ironed out in the all-modified DZA-DDD. There is certainly precedence for poor tolerance of a nucleotide analogue inside DNA or RNA as indicated by a drastic loss in stability that contrasts with stable self-pairing of the nucleic acid analogue. For example, inserting a single glycol (or glycerol) nucleic acid (GNA) residue into DNA prompts a loss of more than 10°C in the thermal stability of the duplex.85 GNA strands do not pair with DNA strands, but pair weakly with RNA (S-GNA isomer),86 and GNA modifications inside RNA are very destabilizing.87,88 However, GNA duplexes are more stable than RNA duplexes.86 Similarly, a destabilizing, conformational (and/or electrostatic) heterogeneity introduced by a single dzA/G inside DNA gives way to homogeneous and stable pairing in the all-modified DZA-DNA system. The influenc of the 7-deazapurine modification must be dominant in the reversal of the stability trend between single modification and all-modified strand as a single FdC or CldU does not significantly affect the stability of the modified DNA relative to the parent duplex. FdC is slightly destabilizing at the level of a single modification inside DNA78 and CldU is slightly stabilizing.29
Hydration and Role of Metal Ions.
The modifications of purine and pyrimidine nucleobases in DZA all map to the duplex major groove. Removing the N7 H-bond acceptor and potential cation binding site in all purines significantly alters the electrostatics in that groove, whereas replacement of the C5 methyl group in dT by chlorine in CldU and the C5 hydrogen in dC by fluorine in FdC affects sterics at the edge of the major groove only to a limited degree. Indeed, our osmotic stressing data demonstrate that the loss of all N7 nitrogen atoms in the DZA-DDD decreases the hydration of the duplex by as much as 50%. Considering the location of N7, the reduced number of water molecules in the DZA-DDD is mainly associated with a change in the major groove water structure. A previous crystal structure of a DDD with a single dzG at G22 offers an interesting perspective in this context as removal of N7 led to loss of a Mg2+ ion that links G2 and G22 across strands37,39,40 and a rearrangement of water molecules in the vicinity.89 Structures of the DDD at atomic resolution revealed a major groove water ribbon that consists of two central water hexagons and flanking pentagons.39 Four sides of the hexagons are formed by water tandems that are H-bonded to N6 and N7 of adenines in the A-tract. The majority of guanine bases are also contacted by two water molecules that bridge the O6 and N7 atoms. Thus, removal of N7 in all purines in the DZA-DDD is expected to fundamentally alter major groove hydration and diminish the negative electrostatic surface potential (ESP) inside the groove. These changes will likely affect stacking interactions between bases as well as the conformation of the sugar-phosphate backbone and therefore the width of the major groove.
The crystal structures of DZA-DDD1 and DZA-DDD2 in complex with RNase H did not reveal cations bound inside the grooves or to sugar-phosphate backbones. Previous investigations of DDDs with a single dzG [d(CGCGAATTCzGCG)]76 or dzA [d(CGCGAzATTCGCG)]77 residue per strand offer some insight into the changes in monovalent cation coordination (Na+) as a function of ionic strength. The values of ΔΔnNa+ at 10, 100 and 200 mM NaCl for the native DDD and the analogue with a single dzG per strand were determined to be 0.6, 0.5 and 0.3, respectively (whereby cation coordination is higher for the DDD). Thus, the net decrease of 0.4 mol Na+/mol duplex in counterion binding for the modified duplex translated to 0.25 mol Na+/mol duplex per modification.76 The introduction of the dzA modification into the DDD decreased the association with Na+, ΔΔnNa+, by 0.9 and 0.7 (10 and 100 mM NaCl, respectively) relative to the parent DDD duplex.77 Unlike for Mg2+ in the major groove of the G-tract, the reduced uptake in Na+ cannot be attributed to the loss of high-affinity sites near dzA base edges in the major groove. This conclusion is supported by the lack of such sites near As in the major groove in high-resolution structures of DDD crystals obtained in the presence of Tl+ 90 or either Rb+ or Cs+.38
As noted above, we do observe some differences in both the extent and exact nature of sodium cation binding in the minor groove for native DDD versus DZA-DDD, even though there are no base alterations present in the minor groove (all modifications involve atoms/substituents that project into the major groove). These differences in sodium cation minor groove binding appear to be correlated with minor groove width; as the minor groove becomes wider, more sodium cations interact strongly with the phosphate backbone rather than bases directly in the minor groove. By contrast, the changes observed in sodium cation major groove interactions for native DDD versus DZA-DNA simulations are minimal and rather subtle. Frankly, this is somewhat surprising given the significant changes in major groove electrostatics and the resultant significant changes in major groove hydration observed in the crystal structures and in the simulations. Thus far, we cannot deduce from the current simulation data any simple mechanistic explanation for the groove widening, but it does appear that changes in the hydration patterns must be a major factor. More extensive MD simulations will be needed to probe details of DZA-DNA plasticity in greater detail.
Groove Widths.
The observed difference in the minor groove widths between the DZA-DDD1 and DZA-DDD2 duplexes is the most intriguing aspect of the analysis of DZA structure and stability relative to the parent DNA dodecamer. This is because the geometry of A-tracts is largely invariant in crystal structures of DNA and protein-DNA complexes. The extremely narrow minor groove width and conformational rigidity for ≥ four base pairs are hallmarks of this so-called B’- or B*-DNA along with bending.43,91 Indeed, the narrow groove width is preserved in the DZA-DDD2 duplex that resembles the native DDD. But the minor groove in the DZA-DDD1 duplex is significantly wider and falls between the widths of canonical B-form DNA and an RNA:DNA hybrid. Interestingly, the groove is wide enough to prompt binding by RNase H in the dzA-tract, whereas in the native DDD and in DZA-DDD2 the protein can only clamp onto the backbones in the more open G-tract and dzG-tract, respectively, minor groove regions. The conformational plasticity exhibited by DZA was not seen in the structure of the DDD with all four dTs replaced by CldU.29 Clearly, eliminating all N7 positions in purine bases is of more importance for conformation than replacing dT by CldU and most likely also replacing dC by FdC. Opening of the DZA-DDD minor groove is accompanied by enhanced nucleobase x-displacement in the dzA-tract, increased helical twist and inclination between bases and helix axis as well as excursions of multiple sugars into the Northern pucker region. Since none of the chemical changes in DZA concern the minor groove, its widening in DZA-DDD1 is an indirect effect as a result of altered major groove electrostatics and hydration that trigger a collapse of the central portion of the major groove. That the major groove undergoes a significant transformation in DZA compared to parent DNA or an analog with dT completely replaced by CldU is demonstrated by the altered activity of restriction enzymes. Thus, we showed earlier that EcoRI still cleaves the DDD with all dTs substituted by CldU.29 However, DZA was found to be protected against cleavage by eight different endonucleases.92
Interestingly, the two MD simulations initiated from the ‘wide-groove’ and ‘narrow-groove’ crystal structures both converge to nearly identical structures during the 10 μs trajectories, and both ensemble average structures are in good agreement with the wide-groove crystal structure, although they exhibit a somewhat narrower minor groove in the A-tract region. The relatively rapid transition of the narrow-groove simulation to a wide-groove structure in this 298K equilibrium simulation suggests that the energy barrier for interconversion must be rather small, i.e., there is significant structural plasticity for this DZA-DDD sequence. While it is possible that the narrow-groove crystal structure could represent a ‘trapped’ meta-stable conformation, there is no experimental evidence to support this conjecture. As noted above in the discussion of the crystal structure details, there are no observable protein-nucleic acid interactions that could potentially induce or stabilize this alternate conformation, so it seems more plausible that the conformational interconversion we observe is a function of DZA-DDD plasticity. However, it does seem likely that the final ensemble average structure observed in both simulations is a lower energy conformation.
It appears that transitions in base inclination, twist, and x-displacement, as well as sugar pucker fluctuations between the C2′-endo and O4′-endo conformations, may be coupled to the narrowing or widening of the minor groove A-tract region, although the precise mechanism or pathway for this conformational transition cannot be deciphered easily from the current simulations. Undoubtedly, the notable difference in major groove hydration around the purine N7/C7 positions observed in both the crystal structures and the MD simulations must have a significant impact on the increased plasticity of DZA-DDD versus native DDD.
CONCLUSIONS
We have determined the first crystal structures for DZA, a DNA analog with a 2′-deoxyribose phosphate backbone and A, C, G and T replaced by dzA, FdC, dzG and CldU, respectively. Although the difference in thermal stability between the DZA-DDD and the parent DNA dodecamer is surprisingly small (−5.9°C, 100 mM NaCl, and −4.6°C, 300 mM NaCl), 7-deaza purine modifications in the DZA major groove drastically alter electrostatics and hydration as demonstrated by osmotic stressing experiments that attest to a loss of solvation caused by removing all N7 H-bond acceptors in DZA. These changes in major groove ESP and water structure are accompanied by an altered A- (dzA-) tract geometry and enhanced conformational plasticity of DZA compared to DNA. Thus, one of the crystal structures determined for the DZA-DDD exhibits the familiar, narrow central minor groove. However, the other structure exhibits a widened minor groove that resembles that in canonical B-form DNA. MD simulations for DNA-DDD and DZA-DDD support softer conformational constraints in the latter and transitions between narrow and wide dzA-tract minor grooves. These differences are essentially caused by 7-deaza purine modification and not the introduction of FdC and CldU in place of dC and dT, respectively. Therefore, it appears that eliminating adenine N7 acceptors that face the major groove can ultimately widen the narrow A-tract minor groove and soften its conformational rigidity. Studies of alternative genetic systems at the structural, stability and dynamic levels can yield insights into long-known features of native DNA that remain incompletely understood.
Supplementary Material
ACKNOWLEDGMENTS
Financial support by Vanderbilt University and NIGMS MIRA GM130207 (E.R.) is gratefully acknowledged. Some calculations were performed using resources at the Vanderbilt University Advanced Computing Center for Research and Education (supported in part by NIH grants S10 OD020154 and S10 RR031634).
Footnotes
Associated Content
The coordinates and reflection files for both crystal structures have been deposited in the Protein Data Bank under PDB ID codes 8SV3 and 8SV4.
Supplemental Information
Results of osmotic stressing experiments (melting temperatures), thermodynamic analysis of melting experiments and Δnw calculations. Files with modified nucleotide templates and additional force field parameters.
The authors declare no competing interests.
REFERENCES
- (1).Egli M; Manoharan M Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Res. 2023, 51, 2529–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Crooke ST; Witztum JL; Bennett CF; Baker BF RNA-targeted therapeutics. Cell Metab. 2018, 27, 714–739. [DOI] [PubMed] [Google Scholar]
- (3).Zhou J; Rossi J Aptamers as targeted therapeutics: current potential and challenges. Nature Reviews Drug Discovery 2017, 16, 181–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Egli M; Herdewijn P (Edts.) Chemistry and Biology of Artificial Nucleic Acids, Wiley-VCH Publishers, Weinheim, Germany, 2012. [Google Scholar]
- (5).Deleavey GF; Damha MJ Deleavey GF; Damha MJ Chem. Biol 2012, 19, 937–954. Chem. Biol. 2012, 19, 937–954. [DOI] [PubMed] [Google Scholar]
- (6).Anosova I; Kowal EA; Dunn MR; Chaput JC; van Horn WD; Egli M The structural diversity of artificial genetic polymers. Nucleic Acids Res. 2016, 44, 1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Herdewijn P; Marlière P Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem. Biodiv 2009, 6, 791–808. [DOI] [PubMed] [Google Scholar]
- (8).Pezo V; Liu FW; Abramov M; Froeyen M; Herdewijn P; Marlière P Binary genetic cassettes for selecting XNA-templated DNA synthesis in vivo. Angew. Chem. Int. Ed 2013, 52, 8139–8143. [DOI] [PubMed] [Google Scholar]
- (9).Pezo V; Schepers G; Lambertucci C; Marlière P; Herdewijn P Probing ambiguous base-pairs by genetic transformation with XNA templates. ChemBioChem 2014, 15, 2255–2258. [DOI] [PubMed] [Google Scholar]
- (10).Pinheiro VB; Taylor AI; Cozens C; Abramov M; Renders M; Zhang S; Chaput JC; Wengel J; Peak-Chew S-Y; McLaughlin SH; Herdewijn P; Holliger P Synthetic genetic polymers capable of heredity and evolution. Science 2012, 336, 341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Taylor AI; Pinheiro VB; Smola MJ; Morgunov AS; Peak-Chew S; Cozens C; Weeks KM; Herdewijn P; Holliger P Catalysts from synthetic genetic polymers. Nature 2014, 518, 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yu H; Zhang S; Chaput JC Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem 2012, 4, 183–187. [DOI] [PubMed] [Google Scholar]
- (13).Yu H; Zhang S; Dunn MR; Chaput JC An efficient and faithful in vitro replication system for threose nucleic acid. J. Am. Chem. Soc 2013, 135, 3583–3591. [DOI] [PubMed] [Google Scholar]
- (14).Alves Ferreira-Bravo I; Cozens C; Holliger P; DeStefano JJ Selection of 2’-deoxy-2’-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015, 43, 9587–9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Maruyama H; Furukawa K; Kamiya H; Minakawa N; Matsuda A Transcription of 4′-thioDNA templates to natural RNA in vitro and in mammalian cells. Chem. Comm 2015, 51, 7887–7890. [DOI] [PubMed] [Google Scholar]
- (16).Tarashima N; Ando H; Kojima T; Kinjo N; Hashimoto Y; Furukawa K; Ishida T; Minakawa N Gene silencing using 4′-thioDNA as an artificial template to synthesize short hairpin RNA without inducing a detectable innate immune response. Mol. Ther. Nucleic Acids 2016, 5, e274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Delaney JC; Gao J; Liu H; Shrivastav N; Essigmann JM; Kool ET Efficient replication bypass of size-expanded DNA base pairs in bacterial cells. Angew. Chem. Int. Ed 2009, 48, 4524–4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Krueger AT; Peterson LW; Chelliserry J; Kleinbaum DJ; Kool ET Encoding phenotype in bacteria with an alternative genetic set. J. Am. Chem. Soc 2011, 133, 18447–18451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Chelliserrykattil J; Lu H; Lee AHF; Kool ET Polymerase amplification, cloning, and gene expression of benzo-homologous “yDNA” base pairs. ChemBioChem 2008, 9, 2976–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Biondi E; Benner SA Artificially expanded genetic information systems for new aptamer technologies. Biomedicines 2018, 6, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kimoto M; Yamashige R; Matsunaga K; Yokoyama S; Hirao I Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol 2013, 31, 453–457. [DOI] [PubMed] [Google Scholar]
- (22).Matsunaga K; Kimoto M; Hirao I High-affinity DNA aptamer generation targeting von Willebrand Factor A1-Domain by genetic alphabet expansion for systematic evolution of ligands by exponential enrichment using two types of libraries composed of five different bases. J. Am. Chem. Soc 2017, 139, 324–334. [DOI] [PubMed] [Google Scholar]
- (23).Zhang L; Yang Z; Sefah K; Bradley KM; Hoshika S; Kim M-J; Kim H-J; Zhu G; Jimenez E; Cansiz S; Teng I-T; Champanhac C; McLendon C; Liu C; Zhang W; Gerloff DL; Huang Z; Tan W; Benner SA Evolution of functional six-nucleotide DNA. J. Am. Chem. Soc 2015, 137, 6734–6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hoshika S; Leal NA; Kim M-J; Kim M-S; Karalkar NB; Kim H-J; Bates AM; Watkins NE Jr.; SantaLucia HA; Meyer AJ; DasGupta S; Piccirilli JA; Ellington AD; SantaLucia J Jr.; Georgiadis MM; Benner SA Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 2019, 363, 884–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Malyshev DA; Dhami K; Lavergne T; Chen T; Dai N; Foster JM; Corrêa IR; Romesberg FE A semi-synthetic organism with an expanded genetic alphabet. Nature 2014, 509, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Malyshev DA; Romesberg FE The expanded genetic alphabet. Angew. Chem. Int. Ed 2015, 54, 11930–11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Zhang Y; Lamb BM; Feldman AW; Zhou AX; Lavergne T; Li L; Romesberg FE A semisynthetic organism engineered for the stable expansion of the genetic alphabet. Proc. Natl. Acad. Sci. USA 2017, 114, 1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Marlière P; Patrouix J; Döring V; Herdewijn P; Tricot S; Cruveiller S; Bouzon M; Mutzel R Chemical evolution of a bacterium’s genome. Angew. Chem. Int. Ed 2011, 50, 7109–7114. [DOI] [PubMed] [Google Scholar]
- (29).Patra A; Harp J; Pallan PS; Zhao L; Abramov M; Herdewijn P; Egli M Structure, stability and function of 5-chlorouracil modified A:U and G:U base pairs. Nucleic Acids Res. 2013, 41, 2689–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kirnos MD; Khudyakov IY; Alexandrushkina NI; Vanyushin BF 2-Aminoadenine is an adenine substituting for a base in S-2L cyanophage DNA. Nature 1977, 270, 369–370. [DOI] [PubMed] [Google Scholar]
- (31).Zhou Y; Xu X; Wei Y; Cheng Y; Guo Y; Khudyakov I; Liu F; He P; Song Z; Li Z; Gao Y; Lui Ang E; Zhao H; Zhang Y; Zhao S A widespread pathway for substitution of adenine by diaminopurine in phage genomes. Science 2021, 372, 512–516. [DOI] [PubMed] [Google Scholar]
- (32).Eremeeva E; Abramov M; Marlière P; Herdewijn P The 5-chlorouracil:7-deazaadenine base pair as an alternative to the dT:dA base pair. Org. Biomol. Chem 2017, 15, 168–176. [DOI] [PubMed] [Google Scholar]
- (33).Eremeeva E; Abramov M; Margamuljana L; Rozenski J; Pezo V; Marlière P; Herdewijn P Chemical morphing of DNA containing four noncanonical bases. Angew. Chem. Int. Ed 2016, 55, 7515–7519. [DOI] [PubMed] [Google Scholar]
- (34).Yang H; Eremeeva E; Abramov M; Herdewijn P The network of replication, transcription, and reverse transcription of a synthetic genetic cassette. Angew. Chem. Int. Ed 2021, 60, 4175–4182. [DOI] [PubMed] [Google Scholar]
- (35).Wing R; Drew H; Takano T; Broka C; Takano S; Itakura K; Dickerson RE Crystal structure analysis of a complete turn of B-DNA. Nature 1980, 287, 755–758. [DOI] [PubMed] [Google Scholar]
- (36).Drew H; Wing R; Takano T; Broka C; Tanaka S; Itakura K; Dickerson RE Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. U.S.A 1981, 78, 2179–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Tereshko V; Minasov G; Egli M The Dickerson-Drew B-DNA dodecamer revisited at atomic resolution. J. Am. Chem. Soc 1999, 121, 470–471. [Google Scholar]
- (38).Tereshko V; Minasov G; Egli MA “hydrat-ion” spine in a B-DNA minor groove. J. Am. Chem. Soc 1999, 121, 3590–3595. [Google Scholar]
- (39).Minasov G; Tereshko V; Egli M Atomic-resolution crystal structures of B-DNA reveal specific influences of divalent metal ions on conformation and packing. J. Mol. Biol 1999, 291, 83–99. [DOI] [PubMed] [Google Scholar]
- (40).Egli M; Tereshko V Lattice- and sequence-dependent binding of Mg2+ in the crystal structure of a B-DNA dodecamer. In: Curvature and deformation of nucleic acids: recent advances, new paradigms (Stellwagen N; Mohanty U; Eds), ACS Symp. Ser 2004, 884, 87–109. [Google Scholar]
- (41).Egli M; Pallan PS The many twists and turns of DNA: template, telomere, tool and target. Curr. Opin. Struct. Biol 2010, 20, 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Strahs D; Schlick T A-tract bending: insights into experimental structures by computational models. J. Mol. Biol 2000, 301, 643–663. [DOI] [PubMed] [Google Scholar]
- (43).Rohs R; Jin X; West SM; Joshi R; Honig B; Mann RS Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem 2010, 79, 233–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Schmidt S; Pein C-D; Fritz H-J; Cech D Chemical synthesis of 2’-deoxyoligonucleotides containing 5-fluoro-2’-deoxycytidine. Nucleic Acids Res. 1992, 20, 2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Marasco CJ Jr.; Sufrin JR A convenient method for the direct incorporation of 5-fluoro-2’-deoxycytidine into oligodeoxynucleotides. J. Org. Chem 1992, 57, 6363–6365. [Google Scholar]
- (46).Spink CH; Chaires JB Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry 1999, 38, 496–508. [DOI] [PubMed] [Google Scholar]
- (47).Rozners E Determination of nucleic acid hydration using osmotic stress. Curr. Protoc. Nucleic Acid Chem 2010, 43, 7.14.1–7.4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Rozners E; Moulder J Hydration of short DNA, RNA, and 2′-OMe oligonucleotides determined by osmotic stressing. Nucleic Acids Res. 2004, 32, 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kolarovic A; Schweizer E; Greene E; Gironda M; Pallan PS; Egli M; Rozners E Interplay of structure, hydration and thermal stability in formacetal modified oligonucleotides: RNA may tolerate nonionic modifications better than DNA. J. Am. Chem. Soc 2009, 131, 14932–14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Li F; Pallan PS; Maier MA; Rajeev KG; Mathieu SL; Kreutz C; Fan Y; Sanghvi J; Micura R; Rozners E; Manoharan M; Egli M Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribo-difluorotoluyl nucleotide: insights into substrate requirements by the human RISC Ago2 enzyme. Nucleic Acids Res. 2007, 35, 6424–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Rozners E; Smicius R; Uchiyama C Expanding functionality of RNA: synthesis and properties of RNA containing imidazole modified tandem G-U wobble base pairs. Chem. Commun 2005, 41, 5778–5780. [DOI] [PubMed] [Google Scholar]
- (52).Nowotny M; Gaidamakov SA; Crouch RJ; Yang W Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 2005, 121, 1005–1016. [DOI] [PubMed] [Google Scholar]
- (53).Pallan PS; Egli M Insights into RNA/DNA hybrid recognition and processing by RNase H from the crystal structure of a non-specific enzyme-dsDNA complex. Cell Cycle 2008, 7, 2562–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Berger I; Kang CH; Sinha N; Wolters M; Rich A A highly efficient 24-condition matrix for the crystallization of nucleic acid fragments. Acta Cryst. D 1996, 52, 465–468. [DOI] [PubMed] [Google Scholar]
- (55).Otwinowski Z; Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276, 307–326. [DOI] [PubMed] [Google Scholar]
- (56).Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Cryst. D 1994, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- (57).Vagin A; Teplyakov A MOLREP: an Automated program for molecular replacement. J. Appl. Cryst 1997, 30, 1022–1025. [Google Scholar]
- (58).Murshudov GN; Skubák P; Lebedev AA; Pannu NS; Steiner RA; Nicholls RA; Winn MD; Long F; Vagin AA REFMAC5 for the refinement of macromolecular crystal structures. Acta Cryst. D, 2011, 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Emsley P; Cowtan K Coot: model-building tools for molecular graphics. Acta Cryst. D 2004, 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- (60).Schüttelkopf AW; van Aalten DMF PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Cryst. 2004, 60, 1355–1363. [DOI] [PubMed] [Google Scholar]
- (61).Pallan PS; Prakash TP; de Leon AR; Egli M Limits of RNA 2’-OH mimicry by fluorine: crystal structure of Bacillus halodurans RNase H bound to a 2’F-RNA:DNA hybrid. Biochemistry 2016, 55, 5321–5325. [DOI] [PubMed] [Google Scholar]
- (62).Pettersen EF; Goddard TD; Huang CC; Couch GS; Greenblatt DM; Meng EC; Ferrin TE UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (63).Galindo-Murillo R; Robertson JC; Zgarbovic M; Sponer J; Otyepka M; Jureska P; Cheatham TE Assessing the current state of AMBER force field modifications for DNA. J. Chem. Theory Comput 2016, 12, 4114–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Wang J; Wolf RM; Caldwell JW; Kollman PA; Case DA Development and testing of a general AMBER force field. J. Comput. Chem 2004, 25, 1157–1174. [DOI] [PubMed] [Google Scholar]
- (65).Aduri R; Psciuk BT; Saro P; Taniga H; Schlegel HB; SantaLucia J Jr. AMBER force field parameters for the naturally occurring modified nucleosides in RNA. J. Chem. Theory Comput 2007, 3, 1465–1475. [DOI] [PubMed] [Google Scholar]
- (66).Bayly CI; Cieplak P; Cornell WD; Kollman PA A well-behaved electrostatic potential based method using charge restraints for determining atom-centered charges: The RESP model. J. Phys. Chem 1993, 97, 10269–10280. [Google Scholar]
- (67).Case DA; Babin V; Berryman JT,;Betz RM; Cai Q; Cerutti DS; Cheatham TE III; Darden TA; Duke RE; Gohlke H; Goetz AW; Gusarov S; Homeyer N; Janowski P; Kaus J; Kolossváry I; Kovalenko A; Lee TS; LeGrand S; Luchko T; Luo R; Madej B; Merz KM; Paesani F; Roe DR; Roitberg A; Sagui C; Salomon-Ferrer R; Seabra G; Simmerling CL; Smith W; Swails J; Walker RC; Wang J; Wolf RM; Wu X and Kollman PA 2014, AMBER14, University of California, San Francisco. [Google Scholar]
- (68).Jorgensen WL; Chandrasekhar J; Madura J; Klein ML Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 1983, 79, 926–935. [Google Scholar]
- (69).Joung IS; Cheatham TE III. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 2008, 112, 9020–9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Cerutti DS; Duke RE; Darden TA; Lybrand TP Staggered Mesh Ewald: an extension of the Smooth Particle-Mesh Ewald method adding great versatility. J. Chem. Theory Comp 2009, 5, 2322–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Izaguirre JA; Catarello DP; Wozniak JM; Skeel RD Langevin stabilization of molecular dynamics. J. Phys. Chem 2001, 114, 2090–2098. [Google Scholar]
- (72).Ryckaert JP; Ciccotti G; Berendsen HJC; Hirasawa K Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys 1977, 23, 327–341. [Google Scholar]
- (73).Miyamoto S; Kollman PA Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem 1992, 13, 952–962. [Google Scholar]
- (74).Roe DR; Cheatham TE III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput 2013, 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- (75).Lu XJ; Olson WK 3dna: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003, 31, 5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Ganguly M; Wang F; Kaushik M; Stone MP; Marky LA; Gold B A study of 7-deaza-2′-deoxyguanosine-2′-deoxycytidine base pairing in DNA. Nucleic Acids Res. 2007, 35, 6181–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Kowal EA; Ganguly M; Pallan PS; Marky LA; Gold B; Egli M; Stone MP Altering the electrostatic potential in the major groove: Thermodynamic and structural characterization of 7-deaza-2′-deoxyadenosine:dT base pairing in DNA. J. Phys. Chem. B 2011, 115, 13925–13934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Puffer B; Kreutz C; Rieder U; Ebert M-O; Konrat R; Micura R 5-Fluoro pyrimidines: labels to probe DNA and RNA secondary structures by 1D 19F NMR spectroscopy. Nucleic Acids Res. 2009, 37, 7728–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Egli M; Pallan PS Generating crystallographic models of DNA dodecamers from structures of RNase H:DNA complexes. In: Nucleic Acid Crystallography: Methods and Protocols, Methods in Molecular Biology (Ennifar E, Ed.), Chapter 8, Humana Press, Springer Science and Business Media, New York, NY, 2015, 1320, 111–126. [DOI] [PubMed] [Google Scholar]
- (80).Shui X; McFail-Isom L; Hu GG; Williams LD The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry 1998, 37, 8341–8355. [DOI] [PubMed] [Google Scholar]
- (81).Zheng G; Lu XJ; Olson WK Web 3DNA - a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009, 37, W240–W246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Lavery R; Sklenar H Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn 1989, 6, 655–667. [DOI] [PubMed] [Google Scholar]
- (83).Egli M In: Nucleic Acids in Chemistry and Biology (Blackburn GM; Egli M; Gait MJ; Watts JK, Eds.), 4th Ed., Chapter 2, R. Soc. Chem., Cambridge, UK, 2022, 20–95. [Google Scholar]
- (84).Freier SM; Altmann KH The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997. 25, 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Schneider KC; Benner SA Oligonucleotides containing flexible nucleoside analogs. J. Am. Chem. Soc 1990, 112, 453–455. [Google Scholar]
- (86).Zhang L; Peritz A; Meggers E 2005. A simple glycol nucleic acid. J. Am. Chem. Soc 2005, 127, 4174–4175. [DOI] [PubMed] [Google Scholar]
- (87).Schlegel MK; Foster DJ; Kel’in AV; Zlatev I; Bisbe A; Jayaraman M; Lackey JG; Rajeev KG; Charisse K; Harp J; Pallan PS; Maier MA; Egli M; Manoharan M Chirality dependent potency enhancement and structural impact of glycol nucleic acid modification on siRNA. J. Am. Chem. Soc 2017, 139, 8537–8546. [DOI] [PubMed] [Google Scholar]
- (88).Egli M; Schlegel MK; Manoharan M Acyclic (S)-glycol nucleic acid (S-GNA) modification of siRNAs improves the safety of RNAi therapeutics while maintaining potency. RNA 2023, 29, 402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Wang F; Li F; Ganguly M; Marky LA; Gold B; Egli M; Stone MP A bridging water anchors the tethered 5-(3-aminopropyl)-2′-deoxyuridine amine in the DNA major groove proximate to the N+2 C•G base pair: implications for formation of interstrand 5′-GNC-3′ cross-links by nitrogen mustards. Biochemistry 2008, 47, 7147–7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Howerton SB; Sines CC; VanDerveer D; Williams LD Locating monovalent cations in the grooves of B-DNA. Biochemistry 2001, 40, 10023–10031. [DOI] [PubMed] [Google Scholar]
- (91).Hud NV; Plavec J A unified model for the origin of DNA sequence-directed curvature. Biopolymers 2003, 69, 144–159. [DOI] [PubMed] [Google Scholar]
- (92).Eremeeva E; Abramov M; Margamuljana L; Herdewijn P Base-modified nucleic acids as a powerful tool for synthetic biology and biotechnology. Chem. Europ. J 2017, 23, 9560–9567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


