Abstract
GAGA factor is known to remodel the chromatin structure in concert with nucleosome-remodeling factor NURF in a Drosophila embryonic S150 extract. The promoter region of the Drosophila fushi tarazu (ftz) gene carries several binding sites for GAGA factor. Both the GAGA factor-binding sites and GAGA factor per se are necessary for the proper expression of ftz in vivo. We observed transcriptional activation of the ftz gene when a preassembled chromatin template was incubated with GAGA factor and the S150 extract. The chromatin structure within the ftz promoter was specifically disrupted by incubation of the preassembled chromatin with GAGA factor and the S150 extract. Both transcriptional activation and chromatin disruption were blocked by an antiserum raised against ISWI or by base substitutions in the GAGA factor-binding sites in the ftz promoter region. These results demonstrate that GAGA factor- and ISWI-mediated disruption of the chromatin structure within the promoter region of ftz activates transcription on the chromatin template.
Chromatin structure appears to play a key role in the regulation of gene expression in eukaryotes (12, 13, 34, 44). Since chromatin structure represses transcription by preventing the binding of transcriptional regulators and transcription machinery to their target sequences, some mechanisms such as chromatin remodeling likely counteract this repressive effect (7, 17, 21). Indeed, transcriptional activation usually accompanies remodeling of chromatin. For example, the promoter region of the yeast PHO5 gene is covered by nucleosomes, but the ordered array of nucleosomes disappears from the promoter upon induction of the gene by phosphate starvation (1). Transactivation by certain regulators requires recruitment of histone acetyltransferase through coactivators (4, 25, 26, 46).
The relationship between chromatin structure and transcription was also studied in vitro. The affinity of TATA-binding protein for the TATA element is significantly reduced when the TATA element is covered by a nucleosome (17). It was shown that the binding of TFIID to the TATA element prior to chromatin assembly alleviates the nucleosome-mediated repression of transcription (45). These observations suggested that remodeling of the chromatin structure around the promoter is crucial for transcriptional activation on the chromatin template. Pazin et al. reported the NF-κB-mediated chromatin remodeling and transcriptional activation of the human immunodeficiency virus type 1 enhancer on a preassembled chromatin template (28). Their study highlighted the importance of analyses on a natural promoter to understand the regulation observed in vivo.
The Drosophila fushi tarazu (ftz) gene is expressed in a seven-striped manner during early embryogenesis (6, 14). The promoter region of ftz carries several binding sites for GAGA factor, which triggers chromatin remodeling (39, 40). Topol et al. constructed a series of transgenic flies carrying the ftz promoter-lacZ fusion gene and found that deletion of the GAGA factor-binding sites in the ftz promoter markedly reduced the reporter gene expression (37). Furthermore, the striped expression of ftz was abolished by a mutation of the Trithorax-like gene, which encodes GAGA factor (3, 11). These observations suggested that GAGA factor-mediated chromatin remodeling is required for the proper expression of ftz in vivo.
To investigate the role of chromatin remodeling in transcriptional activation directly, we have studied GAGA factor-mediated changes in the transcriptional and structural properties of the ftz chromatin template in vitro. Here we report that GAGA factor-mediated disruption of the chromatin structure within the promoter region of ftz leads to transcriptional activation from the preassembled chromatin template. This finding helps explain how the inhibitory effect of chromatin is overcome upon induction on a natural promoter.
MATERIALS AND METHODS
Preparation of proteins.
GAGA factor was expressed in Escherichia coli by using expression vector pAR-GAGA (32) and purified as previously described (41). Core histones were purified from HeLa cell nuclei by the procedure of Simon and Felsenfeld (31). Glutathione S-transferase (GST) or GST-ISWI peptide was bacterially expressed and purified with a glutathione Sepharose column (Pharmacia). GST-ISWI peptide contains the N-terminal 100 amino acids of ISWI which lack the ATPase domain.
Template DNA.
Plasmid pE(Hu)5-N carrying the ftz promoter region from −617 to +124 (23) and plasmid pFb205 harboring the promoter region from −860 to +728 of the fibroin gene of Bombyx mori (23) were used as templates. To construct the templates carrying mutations in the GAGA factor-binding sites, one, two, three, or all of the GAGAG sequences in the ftz promoter region (GAGAG at −360 and CTCTC at −348, −158, and −46) were replaced by site-directed mutagenesis (20) with the sequences GCGCG, CGCGC, GGATC, and AATTC, respectively.
Reconstitution of chromatin on a ftz plasmid.
Chromatin was assembled by a salt dialysis method (33) with a slight modification. Plasmids pE(Hu)5-N (2.5 μg) and pFb205 (2.5 μg) and purified core histones (4.5 μg) were mixed in a 50-μl reaction mixture containing 10 mM Tris-HCl (pH 7.2), 0.1 mM EDTA, 2 M NaCl, and 10 mM 2-mercaptoethanol. The mixture was dialyzed for 90 min each at room temperature against buffer A (10 mM Tris-HCl [pH 7.2], 0.1 mM EDTA, 0.8 M NaCl, 10 mM 2-mercaptoethanol), buffer B (10 mM Tris-HCl [pH 7.2], 0.1 mM EDTA, 0.15 M NaCl), and then buffer C (20 mM HEPES-KOH [pH 7.9], 0.1 mM EDTA).
In vitro transcription.
Preassembled chromatin or naked template DNA (100 ng each of ftz and fibroin plasmid DNA) was incubated at 26°C for 2 h with various amounts of GAGA factor and 1.5 μl of the S150 extract in 10 μl of a solution containing 10 mM HEPES-KOH (pH 7.6), 0.5 mM EGTA, 60 mM KCl, 4 mM MgCl2, 10% glycerol, 30 mM creatine phosphate, 3 mM ATP, 1 mM dithiothreitol, 1-μg/ml creatine phosphokinase, and 100 ng of φX174 replicative-form I DNA. Half of the mixture or 100 ng of untreated naked template DNA was then subjected to a transcription reaction containing the B. mori posterior silk gland extract as described previously (23). Accurately initiated transcripts from the ftz and fibroin promoters were detected by a modified S1 nuclease assay (16).
Micrococcal nuclease assay.
Chromatin containing 20 ng of ftz plasmid DNA and 180 ng of φX174 replicative-form I DNA was incubated with GAGA factor and the S150 extract as described for transcription reactions. The mixture (10 μl) was then treated with 2.5 U of micrococcal nuclease in 60 μl of a solution containing 50 mM Tris-HCl (pH 8.6), 50 mM KCl, 3 mM CaCl2, and 10% glycerol. After 2, 5, and 20 min at 26°C, a 20-μl aliquot was removed and the reaction was terminated by addition of 1.2 μl of 2.5% Sarkosyl–0.25 M EDTA. RNase digestion was followed by proteinase K treatment. Digested DNA was electrophoresed on a 2% agarose gel and stained with SYBR green I. DNA was transferred to a nylon membrane and hybridized with an end-labeled oligonucleotide probe carrying the ftz sequence from −362 to −328 (GAWT or GAmut), from −169 to −138 (site IV), or from −56 to −26 (TATA) or the vector sequence (5′-cttcccttcctttctcgccacgttctccggc-3′) 1.2 kb downstream of the ftz transcription start site (distal). The substituted bases in GAmut were as described above.
Restriction enzyme assay.
Chromatin was incubated with GAGA factor and S150 extract as described for the micrococcal nuclease assay. The mixture (10 μl) was then treated with 0.5 U of AvaII, 2 U of FspI, or 1 U of PstI at 26°C for 1 h. After successive treatments with RNase, proteinase K, and phenol, DNA was recovered by ethanol precipitation and digested to completion with KpnI or HindIII. Resulting DNA fragments were resolved on a 2% agarose gel and detected on a Southern blot with a mixture of radioactive oligonucleotide probes GAWT and site IV or the probe distal.
RESULTS
GAGA factor activates transcription on a ftz chromatin template.
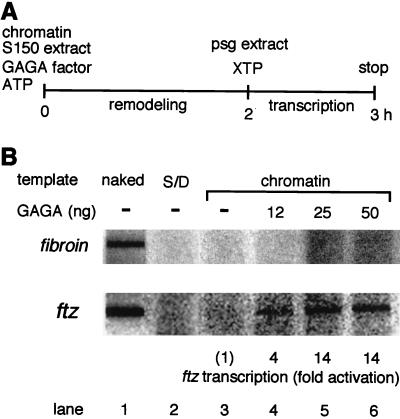
To test whether GAGA factor can activate transcription from the preassembled ftz chromatin template, we performed an in vitro transcription assay (Fig. 1A). We used a plasmid template carrying the ftz promoter sequence from −617 to +124 (nucleotides are numbered relative to the transcription initiation site). This promoter region is known as the zebra element that is sufficient for the seven-striped expression of ftz (15). As a control, a plasmid harboring the promoter region from −860 to +728 of the fibroin gene of B. mori that does not contain the GAGAG sequence was included in the reaction mixtures. Chromatin was reconstituted from core histones, ftz, and fibroin plasmid DNA by a salt dialysis method (33). When the reconstituted chromatin was subjected to transcription reactions without preincubation with GAGA factor and the S150 extract, transcription of both ftz and fibroin was repressed completely (Fig. 1B, S/D).
FIG. 1.
Transcription on the preassembled ftz chromatin template is activated by preincubation with GAGA factor. (A) Schematic diagram of a transcription experiment. The preassembled chromatin template containing 200 ng of DNA was preincubated with or without GAGA factor in the presence of S150 extract and ATP. Subsequently, transcription was carried out by addition of the posterior silk gland (psg) extract and the remaining three nucleotides (XTP) to one-half of each preincubation mixture. (B) Transcription on the ftz chromatin template. Chromatin was preincubated with the indicated amounts of GAGA factor and then transcribed as described above. The signal intensity of each transcript relative to that obtained without GAGA factor is indicated at the bottom. Transcription on the same amount of naked template DNA without preincubation with GAGA factor is shown as a control. S/D shows transcription on the chromatin template (100 ng of DNA) without preincubation with GAGA factor and the S150 extract.
To test the effect of GAGA factor on transcription of the chromatin template, the reconstituted chromatin was first incubated with GAGA factor and a small amount of S150 extract from Drosophila early embryos (2) to allow remodeling and then tested for transcription activity. The S150 extract contains a nucleosome-remodeling factor (NURF) that acts with GAGA factor to disrupt the ordered array of nucleosomes near the GAGA factor-binding sites (41). Since the S150 extract was highly inhibitory to transcription in vitro (data not shown), the amount of the extract was adjusted to 1/10 of that required for reconstitution of the chromatin structure on the template DNA to minimize the carryover of the extract to transcription reactions. The transcripts from the ftz and fibroin promoters were analyzed by a modified S1 nuclease assay (16). Transcription on the ftz chromatin template was activated up to 14-fold by preincubation with GAGA factor and the S150 extract, but it was not activated in the absence of GAGA factor (Fig. 1B). The transcriptional activation reached its maximum level when the chromatin template was preincubated with 25 ng of GAGA factor. Under these conditions, GAGA factor was present in 13-fold molar excess over the chromatin template that carries four GAGAG sequences within the ftz promoter region. No activation occurred in the absence of the S150 extract or ATP in the preincubation mixture (data not shown). By contrast, transcription on the fibroin chromatin template was not activated by the preincubation with GAGA factor and the S150 extract (Fig. 1B).
GAGA factor does not activate transcription on a naked DNA template.
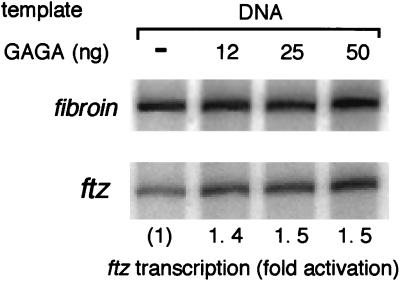
GAGA factor has been reported to activate transcription on naked DNA templates in crude extracts but not in transcription systems reconstituted from purified components (8, 19). GAGA factor-mediated transcriptional activation on a naked DNA template required the presence of a nonspecific DNA-binding protein, suggesting that GAGA factor functions as an antirepressor by preventing nonspecific inhibitory proteins such as histone H1 from binding to DNA (8, 19, 29). It is therefore possible that GAGA factor may activate transcription by a similar mechanism in our transcription assay. To test this possibility, we carried out experiments similar to those in Fig. 1, starting from the naked DNA template. Since the amount of S150 extract in the preincubation reactions was 1 order of magnitude lower than that required for full assembly of the chromatin structure on the template, nucleosomes were barely detectable by supercoiling assay after preincubation (data not shown). In contrast to the preassembled chromatin, we observed little activation (up to 1.5-fold) of ftz transcription after preincubation of the naked template DNA with GAGA factor and the S150 extract (Fig. 2). This indicates that only trace levels of activation may be caused by elimination of nonspecific DNA-binding proteins in the presence of GAGA factor. These results also suggest that the GAGA factor-mediated transcriptional activation occurs specifically on the chromatin template.
FIG. 2.
Transcription on the DNA template is not activated by preincubation with GAGA factor. Naked template DNA (100 ng each of the ftz and fibroin plasmids) was preincubated with the indicated amounts of GAGA factor in the presence of S150 extract and ATP. Half of each mixture was then used for transcription.
GAGA factor disrupts the chromatin structure in the ftz promoter region.
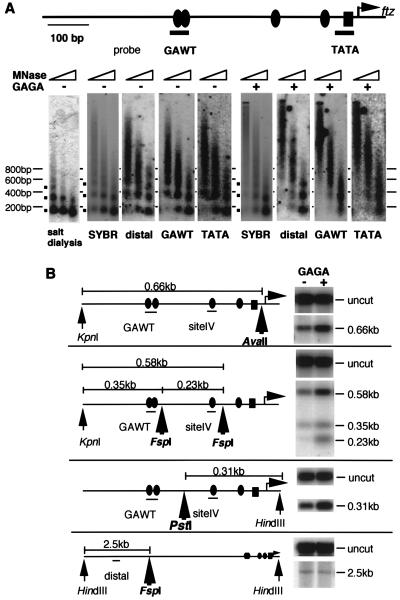
Next, we analyzed changes in the chromatin structure of the ftz template triggered by GAGA factor. After incubation of the chromatin template with GAGA factor in the presence of a small amount of S150 extract, chromatin was subjected to a micrococcal nuclease assay. After digestion with micrococcal nuclease, DNA was purified and electrophoresed on an agarose gel and then stained with SYBR green I dye. No change in the pattern of nucleosome ladders was observed upon incubation with GAGA factor, indicating that GAGA factor did not affect the bulk chromatin structure (Fig. 3A, SYBR). The DNA in the gel was then transferred to a nylon membrane, and successive Southern hybridizations were carried out. We observed GAGA factor-dependent smearing of nucleosome ladders when the Southern blot was probed with an oligonucleotide carrying the sequence in the ftz promoter (Fig. 3A, GAWT and TATA). The smearing of nucleosome ladders was not detectable when we omitted the S150 extract (data not shown). It also appeared that GAGA factor had little effect on the chromatin structure 1.2 kb downstream of the transcription start site (Fig. 3A, distal). These data indicate that the chromatin structure in the ftz promoter was specifically disrupted by incubation with GAGA factor and the S150 extract.
FIG. 3.
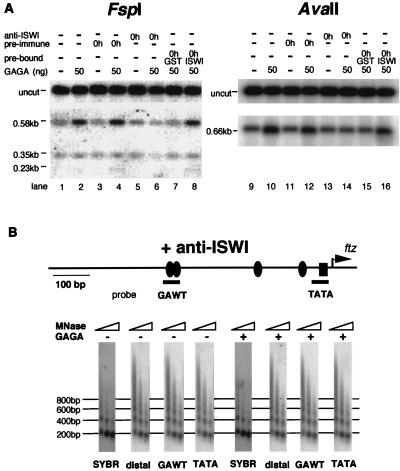
GAGA factor-mediated disruption of the chromatin structure in the ftz promoter. Preassembled chromatin (200 ng of DNA) was incubated with 0 or 50 ng of GAGA factor in the presence of S150 extract and ATP, and the chromatin structure was subsequently analyzed. (A) Micrococcal nuclease (MNase) assay. After incubation with GAGA factor, chromatin was digested with micrococcal nuclease for 2, 5, and 20 min. Deproteinized digestion products were resolved on a 2% agarose gel and stained with SYBR green I fluorescent dye. Salt dialysis showed the digestion pattern without incubation with GAGA factor and S150 extract. DNA was transferred to a nylon membrane and detected by successive Southern hybridizations using the oligonucleotide probes indicated by the thick bars. Probe distal corresponds to the vector sequence 1.2 kb downstream of the transcription start site (position not shown). The filled ovals and the rectangle represent the GAGAG sequences and TATA element, respectively. The arrow shows the start site of ftz transcription. (B) Restriction enzyme assay. After incubation with GAGA factor, chromatin was treated with AvaII, FspI, or PstI. The DNA was purified and digested to completion with KpnI or HindIII. The probes used for Southern hybridizations are indicated. Uncut represents DNA that was not digested with AvaII, FspI, or PstI. The middle regions of the gel lacking relevant hybridization signals are not shown. Thick arrows represent the restriction sites. The other symbols are the same as in panel A.
To get independent evidence for GAGA factor-mediated disruption of chromatin, we analyzed the accessibility of the ftz promoter region to restriction enzymes. After incubation of the chromatin with GAGA factor in the presence of a small amount of S150 extract, AvaII, FspI, or PstI was added to the reaction mixture and incubation was continued. DNA was purified and then cut to completion with KpnI or HindIII. The digested products were detected on a Southern blot. The recognition sites for AvaII (−9), FspI (−90 and −317), and PstI (−267) in the ftz promoter region were more susceptible to these enzymes when the chromatin template was incubated in the presence of GAGA factor than in its absence (Fig. 3B). However, GAGA factor did not affect digestion at the FspI site 2.5 kb downstream from the ftz promoter region (Fig. 3B, bottom). These results confirm that specific disruption of the chromatin structure in the ftz promoter region occurs in a GAGA factor-dependent manner.
Transcription is not activated by alteration of nucleosome spacing.
S150 extract contains an activity that adjusts the nucleosome spacing from the tightly packed state to the regularly spaced one observed in biological samples (2). By incubation with S150 extract, the distance between the nucleosomes of the chromatin template assembled by the salt dialysis method was changed from 160 bp (Fig. 3A, salt dialysis) to 180 bp (Fig. 3A, SYBR) in the presence or absence of GAGA factor. However, the transcription of ftz was not activated by incubation with S150 extract when GAGA factor was absent (Fig. 1B, lanes 2 and 3). These results showed that the alteration of the nucleosome spacing was not sufficient for transcriptional activation of the ftz chromatin template. The increase in nucleosome repeat length did not occur in the absence of ATP (data not shown).
Anti-ISWI serum prevents both transcriptional activation and chromatin remodeling.
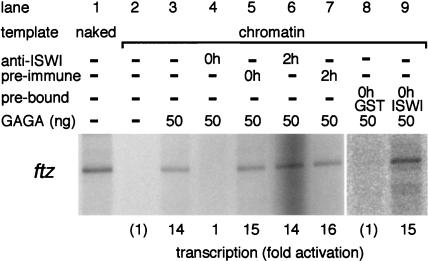
Three chromatin-remodeling complexes termed NURF, ACF, and CHRAC, all of which contain ISWI as a component, were isolated from Drosophila embryonic extracts (18, 40, 41, 43). ISWI shares similarity with yeast SWI2/SNF2 and Drosophila Brahma in its DNA-dependent ATPase domain but not in the N-terminal region (10, 22, 36, 40). To test the correlation between GAGA factor-mediated transcriptional activation and chromatin remodeling, we added a polyclonal antiserum raised against the N-terminal region of ISWI to the reaction mixtures. The antiserum, but not the preimmune serum, abolished the transcriptional activation when added at the beginning of preincubation (Fig. 4, lanes 4 and 5). Prebinding of a GST-ISWI peptide to the antibody restored the transcriptional activation (Fig. 4, lane 9). The antiserum had no effect on the GAGA factor-mediated transcriptional activation when it was added just before transcription (Fig. 4, lanes 6 and 7). Transcription on the naked template DNA was not influenced by addition of the antiserum (data not shown). This finding indicates that ISWI is involved in the GAGA factor-mediated transcriptional activation on the chromatin template. The results also showed that the chromatin template becomes competent for transcription during preincubation with GAGA factor and the S150 extract and not after transcription has been initiated.
FIG. 4.
Anti-ISWI serum prevents GAGA factor-mediated transcriptional activation on the ftz chromatin template. A polyclonal antiserum raised against the N-terminal region of ISWI or preimmune serum (0.5 μl) was added at the beginning of (lanes 4 and 5) or after (lanes 6 and 7) preincubation, and then transcription was carried out. The antiserum (0.5 μl) was mixed with 1.5 μg of bacterially expressed GST-ISWI (lane 9) or GST (lane 8) and incubated for 2 h on ice prior to addition to the preincubation mixture. Samples in lanes 1 to 7 were electrophoresed on the same gel, while those in lanes 8 and 9 were run on a different gel.
Both restriction enzyme susceptibility and micrococcal nuclease assay results showed that addition of ISWI antiserum (Fig. 5A, lanes 5, 6, 13, and 14, and B), but not preimmune serum (Fig. 5A, lanes 3, 4, 11, and 12), to the preincubation mixture abolished the GAGA factor-mediated chromatin remodeling. Addition of the antiserum that had been incubated with the GST-ISWI peptide had no effect on the chromatin remodeling (Fig. 5A, lanes 8 and 16). These data indicate that the chromatin remodeling mediated by GAGA factor and ISWI is necessary for transcriptional activation on the ftz chromatin template. Interestingly, regularly spaced nucleosomes were formed in the presence of ISWI antiserum (Fig. 5B). This suggests that the ISWI antiserum blocked transcriptional activation on the chromatin template by inhibiting the GAGA factor-mediated chromatin remodeling but not by affecting the nucleosome-spacing activity.
FIG. 5.
Anti-ISWI serum blocks GAGA factor-mediated chromatin remodeling. (A) Restriction enzyme assay. Preassembled chromatin (200 ng of DNA) was incubated with 0 or 50 ng of GAGA factor, the S150 extract, and ATP in the presence (lanes 5 to 8 and 13 to 16) or absence (lanes 1, 2, 9, and 10) of antiserum against ISWI (0.5 μl) or in the presence of preimmune serum (lanes 3, 4, 11, and 12), and the chromatin structure was subsequently analyzed by restriction enzyme assay as described in the legend to Fig. 3B. The anti-ISWI serum was prebound with GST-ISWI (lanes 8 and 16) or GST (lanes 7 and 15) as described in the legend to Fig. 4. (B) Micrococcal nuclease (MNase) assay. Preassembled chromatin (200 ng of DNA) incubated with 0 or 50 ng of GAGA factor, the S150 extract, and ATP in the presence of anti-ISWI serum (0.5 μl) was subjected to a micrococcal nuclease assay as described in the legend to Fig. 3A.
Both GAGA factor-mediated transcriptional activation and chromatin remodeling require GAGA factor-binding sites.
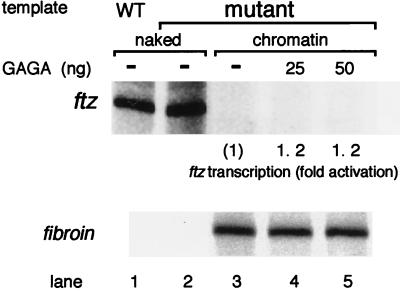
To test whether transcriptional activation and chromatin remodeling are dependent on GAGA factor-binding sites, we designed a template in which all four of the GAGAG sequences in the ftz promoter were replaced with other sequences. Preassembled chromatin on the mutant template was preincubated with GAGA factor and the S150 extract, and then transcription was carried out. The mutations abolished the GAGA factor-mediated transcriptional activation on the chromatin template. In the same reactions, naked fibroin template DNA was transcribed efficiently (Fig. 6, lanes 3 to 5). The mutations had no effect on transcription from the naked template DNA (Fig. 6, lanes 1 and 2).
FIG. 6.
Mutations in the GAGA factor-binding sites abolish transcriptional activation on the chromatin template. The preassembled chromatin template (200 ng of DNA) carrying base substitutions in four GAGAG sequences was preincubated with the indicated amounts of GAGA factor in the presence of S150 extract and ATP; subsequently, half of each preincubation mixture was subjected to a transcription reaction. As a control, naked fibroin plasmid DNA (50 ng) was added to the transcription reaction mixtures (lanes 3 to 5). The wild-type (WT; lane 1) or mutant (lane 2) naked template DNAs were also tested for transcriptional activity without preincubation with GAGA factor.
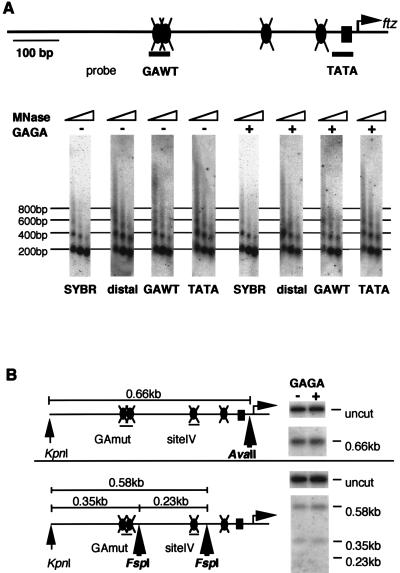
GAGA factor-mediated disruption of the chromatin structure in the ftz promoter region was not observed on the mutant template when either a micrococcal nuclease assay (Fig. 7A) or a restriction enzyme assay (Fig. 7B) was used. When we compared transcription and restriction enzyme sensitivity among chromatin templates carrying a mutation(s) in one to four GAGAG sequences, the levels of both transcriptional activation and nuclease sensitivity decreased in proportion to the number of altered GAGAG sequences (data not shown). These results showed that the GAGA factor-binding sites in the ftz promoter are necessary for both chromatin remodeling and subsequent transcriptional activation. The alteration of nucleosome spacing occurred on the mutant template lacking GAGAG sequences (Fig. 7A). However, it did not lead to transcriptional activation, supporting our conclusion that the formation of regularly spaced nucleosomes is not sufficient for transcriptional activation.
FIG. 7.
Mutations in GAGA factor-binding sites prevent chromatin remodeling. (A) Micrococcal nuclease (MNase) assay. Preassembled chromatin (200 ng of DNA) carrying mutations in four GAGAG sequences was incubated with 0 or 50 ng of GAGA factor in the presence of S150 extract and ATP and then subjected to a micrococcal nuclease assay as described in the legend to Fig. 3A. (B) Restriction enzyme assay. Preassembled chromatin carrying mutations was incubated with GAGA factor and the S150 extract under conditions identical to those used for panel A and subjected to a restriction enzyme assay.
DISCUSSION
In this study, we showed chromatin remodeling and transcriptional activation on the preassembled ftz chromatin template. Both of these processes require GAGA factor, S150 extract, and intact GAGA factor-binding sites on the template, and both can be inhibited by an antiserum against ISWI. From these results, we conclude that the GAGA factor- and ISWI-mediated chromatin remodeling activates transcription on the preassembled ftz chromatin template.
Recent studies have suggested the existence of distinct multiprotein complexes that can act by chromatin remodeling (5, 18, 41, 43). Four such complexes have been identified in Drosophila. The first contains brm, a Drosophila homolog of yeast SWI2/SNF2 (9, 36). The second is NURF, which consists of four proteins that include ISWI (40, 41). The third and fourth are ACF and CHRAC, which also contain ISWI (18, 43). While NURF perturbs the regularly spaced nucleosome, ACF and CHRAC serve as an ATP-dependent nucleosome assembly and spacing factor, respectively (18, 43). The present study shows that formation of regularly spaced nucleosomes is not sufficient but GAGA factor-mediated chromatin remodeling is required for transcriptional activation of the ftz chromatin template. Moreover, CHRAC is less effective in GAGA factor-dependent chromatin remodeling than NURF (43). These findings suggest that NURF plays a key role in the GAGA factor-dependent transcriptional activation in our system. The transcription extract may also contain some chromatin-remodeling activities, possibly containing ISWI homologs. But these activities, if any, do not seem to contribute so much to transcriptional activation in our system, because addition of the ISWI antiserum just before transcription had no effect on transcription. However, we cannot exclude the possibility that these activities might reduce the extent of the activation observed.
Although GAGA factor activated transcription on the chromatin template in the presence of S150 extract, it barely activated transcription on the naked template DNA. This supports the notion that GAGA factor is not an ordinary transcriptional regulator which controls the efficiency of transcription by interacting with general transcription factors (13). Instead, the factor may serve as a landmark to recruit a remodeling factor close to its binding site. However, there is no evidence for direct interaction between GAGA factor and chromatin-remodeling factors, and mechanisms of the recruitment of these factors remain unknown. Genetic analyses have suggested that products of the trithorax group genes, including Trithorax-like, form a multimeric protein complex to execute their function (27). This hypothetical complex may be responsible for the recruitment of remodeling factor. The in vitro system employed in this study enabled us to remodel a chromatin assembled from purified histones and DNA. The system will be useful for functional characterization of the putative protein complex.
Micrococcal nuclease assays showed that the nucleosome structure around −350 and the TATA element of ftz was disrupted by incubation with GAGA factor and the S150 extract. A restriction enzyme assay demonstrated that the AvaII site at −9, the FspI sites at −90 and −317, and the PstI site at −267 on the ftz chromatin template were more susceptible to digestion after incubation with GAGA factor and the S150 extract. Furthermore, base substitutions of all four GAGAG sequences in the ftz promoter at −360, −348, −158, and −46 were required to completely suppress the GAGA factor-mediated chromatin remodeling. These results indicate that chromatin is remodeled throughout the proximal region of the ftz promoter.
Analyses using transgenic fly lines have identified two major groups of cis elements within the ftz promoter that are required for the striped expression of ftz (37, 38, 42). One group comprises the GAGA factor-binding sites described above. The other is a binding site for the nuclear receptor FTZ-F1 located 280 bp upstream of the transcription initiation site. Our in vitro transcription studies revealed that activation of ftz by FTZ-F1 requires two coactivators, termed MBF1 and MBF2 (23). MBF1 is a bridging molecule that interconnects FTZ-F1 and TATA-binding protein and recruits positive cofactor MBF2 to a promoter carrying the FTZ-F1-binding site. MBF2 activates transcription through its contact with TFIIA. This allows the selective activation of ftz in a FTZ-F1 binding site-dependent manner (24, 35). It is most likely that the GAGA factor-mediated chromatin remodeling in the proximal region of the ftz promoter is a prerequisite for the formation of active complexes containing FTZ-F1, MBF1, MBF2, TFIIA, and TBP. Shopland et al. have reported that a mutation in the GAGA site on the hsp70 promoter affects the accessibility of the heat shock transcription factor (30). This suggests that a mechanism similar to that described here may operate in the activation of the hsp70 promoter.
ACKNOWLEDGMENTS
We thank T. Tsukiyama and C. Wu for pAR-GAGA and important suggestions on preparation of the S150 extract, T. Ohta for ISWI antiserum, H. Ueda for helpful discussions during the course of this work, and J. Tomizawa, M. Muramatsu, and M. Jindra for critical reading of the manuscript.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (S.H.) and the Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists (M.O.).
REFERENCES
- 1.Almer A, Horz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker P B, Wu C. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol Cell Biol. 1992;12:2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhat K M, Farkas G, Karch F, Gyurkovics H, Gausz J. The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development. 1996;122:1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- 4.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 5.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 6.Carroll S B, Scott M P. Localization of the fushi tarazu protein during Drosophila embryogenesis. Cell. 1985;43:47–57. doi: 10.1016/0092-8674(85)90011-x. [DOI] [PubMed] [Google Scholar]
- 7.Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 8.Croston G E, Kerrigan L A, Lira L M, Marshak D R, Kadonaga J T. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 9.Dingwall A K, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elfring L K, Deuring R, McCallum C M, Peterson C L, Tamkun J W. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 12.Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;355:219–223. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 13.Granok H, Leibovitch B A, Shaffer C D, Elgin S C R. Ga-ga over GAGA factor. Curr Biol. 1995;5:238–241. doi: 10.1016/s0960-9822(95)00048-0. [DOI] [PubMed] [Google Scholar]
- 14.Hafen E, Kuroiwa A, Gehring W J. Spatial distribution of transcripts from the segmentation gene fushi tarazu during Drosophila embryonic development. Cell. 1984;37:833–841. doi: 10.1016/0092-8674(84)90418-5. [DOI] [PubMed] [Google Scholar]
- 15.Hiromi Y, Kuroiwa A, Gehring W J. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985;43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- 16.Hirose S, Tsuda M, Suzuki Y. Enhanced transcription of fibroin gene in vitro on covalently closed circular templates. J Biol Chem. 1985;260:10557–10562. [PubMed] [Google Scholar]
- 17.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 18.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 19.Kerrigan L A, Croston G E, Lira L M, Kadonaga J T. Sequence-specific transcriptional antirepression of the Drosophila Kruppel gene by the GAGA factor. J Biol Chem. 1991;266:574–582. [PubMed] [Google Scholar]
- 20.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 22.Laurent B C, Treitel M A, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F-Q, Ueda H, Hirose S. Mediators of activation of fushi tarazu gene transcription by BmFTZ-F1. Mol Cell Biol. 1994;14:3013–3021. doi: 10.1128/mcb.14.5.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li F-Q, Takemaru K, Goto M, Ueda H, Handa H, Hirose S. Transcriptional activation through interaction of MBF2 with TFIIA. Genes Cells. 1997;2:143–153. doi: 10.1046/j.1365-2443.1997.1090306.x. [DOI] [PubMed] [Google Scholar]
- 25.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Berger S L, Kouzavides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 26.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 27.Orland V, Paro R. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr Opin Genet Dev. 1995;5:174–179. doi: 10.1016/0959-437x(95)80005-0. [DOI] [PubMed] [Google Scholar]
- 28.Pazin M J, Sheridan P L, Cannon K, Cao Z, Keck J G, Kadonaga J T, Jones K A. NF-kappa B-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- 29.Sandaltzopoulos R, Mitchelmore C, Bonte E, Wall G, Becker P B. Dual regulation of the Drosophila hsp26 promoter in vitro. Nucleic Acids Res. 1995;23:2479–2487. doi: 10.1093/nar/23.13.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shopland L S, Hirayoshi K, Fernandes M, Lis J T. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- 31.Simon R H, Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979;6:689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soeller W C, Oh C E, Kornberg T B. Isolation of cDNAs encoding the Drosophila GAGA transcription factor. Mol Cell Biol. 1993;13:7961–7970. doi: 10.1128/mcb.13.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein A. Reconstitution of chromatin from purified components. Methods Enzymol. 1989;170:585–603. doi: 10.1016/0076-6879(89)70066-5. [DOI] [PubMed] [Google Scholar]
- 34.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 35.Takemaru K, Li F-Q, Ueda H, Hirose S. Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein. Proc Natl Acad Sci USA. 1997;94:7251–7256. doi: 10.1073/pnas.94.14.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamkun J T, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. Brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 37.Topol J, Dearolf C R, Prakash K, Parker C S. Synthetic oligonucleotides recreate Drosophila fushi tarazu zebra-stripe expression. Genes Dev. 1991;5:855–867. doi: 10.1101/gad.5.5.855. [DOI] [PubMed] [Google Scholar]
- 38.Tsai C, Gergen J P. Gap gene properties of the pair-rule gene runt during Drosophila segmentation. Development. 1994;120:1671–1683. doi: 10.1242/dev.120.6.1671. [DOI] [PubMed] [Google Scholar]
- 39.Tsukiyama T, Becker P B, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–531. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 40.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1028. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 41.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 42.Ueda H, Sonoda S, Brown J L, Scott M P, Wu C. A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes Dev. 1990;4:624–635. doi: 10.1101/gad.4.4.624. [DOI] [PubMed] [Google Scholar]
- 43.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 44.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 45.Workman J L, Roeder R G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 46.Yang X-J, Ogryzko V V, Nishikawa J-I, Howard B, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral E1A oncoprotein. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]