Abstract
The most common form of facioscapulohumeral dystrophy (FSHD1) is caused by a partial loss of the D4Z4 macrosatellite repeat array in the subtelomeric region of chromosome 4. Patients with FSHD1 typically carry 1 to 10 D4Z4 repeats, whereas nonaffected individuals have 11 to 150 repeats. The ~150-kilobyte subtelomeric region of the chromosome 10q exhibits a ~99% sequence identity to the 4q, including the D4Z4 array. Nevertheless, contractions of the chr10 array do not cause FSHD or any known disease, as in most people D4Z4 array on chr10 is flanked by the nonfunctional polyadenylation signal, not permitting the DUX4 expression. Here, we attempted to correct the FSHD genotype by a CRISPR-Cas9–induced exchange of the chr4 and chr10 subtelomeric regions. We demonstrated that the induced t(4;10) translocation can generate recombinant genotypes translated into improved FSHD phenotype. FSHD myoblasts with the t(4;10) exhibited reduced expression of the DUX4 targets, restored PAX7 target expression, reduced sensitivity to oxidative stress, and improved differentiation capacity.
Induced chromosomal translocations are able to correct facioscapulohumeral dystrophy phenotype.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is a common myopathy (1:8000 to 1:20,000) characterized by progressive weakness and atrophy of the face, shoulder blade, and upper arm muscles (1). It is associated with the abnormal expression of the embryonic transcription factor DUX4, which is cytotoxic to postnatal muscle tissue, causing transcriptional deregulation (2, 3), myogenesis impairment (4–7), oxidative stress (8–10), and immune responses (3, 11). Multiple copies of the DUX4 gene reside in the D4Z4 macrosatellite repeat array present in the subtelomeric regions of chromosomes 4 and 10; nevertheless, in physiological conditions, DUX4 expression from the D4Z4 repeats is heavily repressed due to multiple genetic and epigenetic mechanisms [reviewed in (12)]. The D4Z4 array generally comprises 11 to 120 D4Z4 units in a healthy population. The contraction of the D4Z4 array below 11 units weakens the epigenetic repression of the array potentially allowing for the DUX4 expression.
The major form of FSHD (FSHD1) representing >90% of the cases results from the combination of two genetic events: the contraction of the D4Z4 repeat array in the 4q35 subtelomeric region of chromosome 4 below 11 units and the presence of a specific 4q haplotype downstream of the array (13). D4Z4 repeat contraction likely compromises the epigenetic repression of the array, which allows for the DUX4 expression. Notably, functional expression is only possible from the most distal D4Z4 repeat, as the typical D4Z4 unit does not contain the polyadenylation signal (PAS) stabilizing the DUX4 transcript, and the PAS can only be provided by the sequence flanking the array. The distal D4Z4 repeat and its immediate flanking sequence exist in two major variants, 4qA and 4qB, which differ in a number of insertions and deletions and are almost equally present in the healthy European population (14, 15). The major difference between the 4qA and 4qB chr4 allele is the presence of the 260–base pair (bp) sequence (pLAM) followed by a 6.2-kb β-satellite repeat directly distal to D4Z4 on 4qA (15). The pLAM sequence contains the PAS, and FSHD is associated uniquely with the 4qA variants (15). The 4qA allele is further divided into several haplotypes differing by a number of single-nucleotide polymorphisms (SNPs). One of these haplotypes, 4A166, seems to be nonpermissive for FSHD for yet unknown reasons (16). All the other 4qA haplotypes are FSHD permissive, mainly for the fact that they provide a functional PAS. Only the simultaneous presence of the D4Z4 contraction on chromosome 4 and the permissive 4qA haplotype result in the FSHD manifestation.
The 10q26 subtelomeric region of chromosome 10 exhibits an almost complete sequence identity (~98.5%) to the 4q35, including the D4Z4 array and the distal flanking sequence (15). A >90% 10q chromosomes carry the 4qA type alleles with the pLAM-10qA (15, 17). Nevertheless, the contractions of the D4Z4 array on chromosome 10 are not associated with FSHD or any known disease (15), which can be explained by the fact that all typical 10qA haplotypes carry a T/C or (very rarely) an A/T SNP within the PAS rending it dysfunctional (16–18). Recently, a hybrid 10A166H haplotype, which resulted from the de novo D4Z4 repeat exchange between chromosomes 4 and 10, was found in one family with manifested FSHD (18). In this family, chromosome 10 acquired a functional PAS from 4qA, resulting in the exceptional DUX4 expression from the hybrid chromosome 10. This suggested that the opposite exchange aimed at the restoring of the normal phenotype was possible.
In this study, we used the CRISPR-Cas9 technique to induce the targeted 4q35;10q26 translocation in FSHD myoblasts. We were able to demonstrate that t(4;10)(q35;q26) could generate recombinant genotypes, which reverted the pathological phenotype of the FSHD cells. This is the first report of the use of targeted chromosomal translocations to treat a genetic disease.
RESULTS
Induction of the t(4;10)(q35;q26) chromosomal translocation
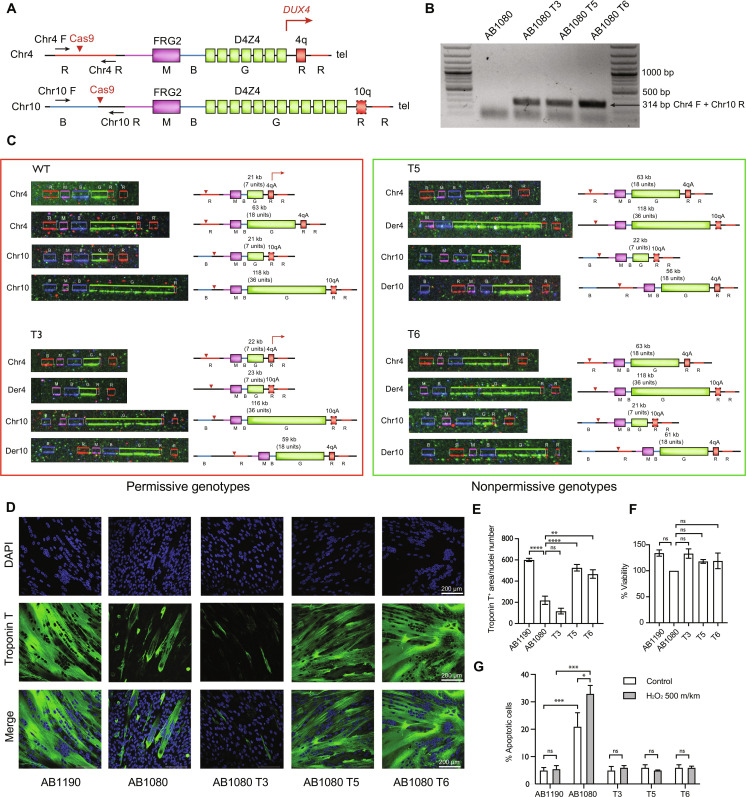
In an attempt to revert the FSHD pathological genotype, we exchanged the subtelomeric 4q35 and 10q26 regions of chromosomes 4 and 10 using the CRISPR-Cas9 system. One challenge of this strategy was to find single guide RNA (sgRNA) and primers highly specific for each chromosome. The 4q35 and 10q26 regions exhibit 98.5% sequence identity, whereas the specificity of the primers is crucial to determine the chromosomal origin of the amplified product when verifying the translocation presence. We have analyzed ~25-kb genomic regions upstream of the FRG2 gene (common for both chromosomes) on chromosomes 4 and 10 to lastly select two primer pairs capable of amplifying amplicons specific for chromosome 4 or 10 uniquely and the sgRNAs recognizing specifically the chromosome 4 or 10 upstream of the FRG2 gene (Fig. 1A).
Fig. 1. Genotype and phenotype characterization of the recombinant clones.
(A) Schematic representation of the 4q35 and 10q26 loci and the location of the primers to detect the t(4;10)(q35;q26). Chromosomal regions are colored according to the colors of the fluorescent hybridization probes from the FiberVision assay (R, red; B, blue; G, green; M, magenta). The dashed line outlining the 10qA allele represents the impaired polyadenylate signal. The red errors point to the guide RNA target sequences. (B) PCR products obtained with the Chr4 F/Chr10 R primer pair in the maternal AB1080 cells and the recombinant clones. The presence of the 314-bp hybrid chr4-chr10 product indicates successful t(4;10)(q35;q26) induction. (C) FiberVision FSHD assay characterizing the structure of the FSHD locus in the maternal AB1080 cells and the recombinant clones T3, T5, and T6. The green fluorescent probe (G) hybridizes to the D4Z4 units to determine the length of the D4Z4 array; the most proximal probe distinguishes between chr4 (red, R) and chr10 (blue, B); the most distal probe distinguishes between the A (red, R) and B (blue, B) chr4/chr10 alleles. In the AB1080 cell line, both copies of chr4 and chr10 carry the A variant. (D) Myotubes formed by control, FSHD, and the recombinant AB1080 T3, T5, and T6 myoblasts, fourth day of differentiation; staining with the monoclonal antibodies against skeletal troponin T (green) and 4′,6-diamidino-2-phenylindole (DAPI) (blue). Representative fields. (E) Differentiation efficiency assessment; troponin T. positive area normalized to the nuclei number, means ± SEM. N = 6. (F) Cell viability assessment in the control, FSHD, and the recombinant T3, T5, and T6 myoblasts; means ± SEM, N = 3. (G) Apoptosis assessment in the control, FSHD, and the recombinant T3, T5, and T6 myoblasts following the oxidative stress induction by 500 mkm of H2O2; means ± SEM, N = 3.
We next induced simultaneous double-strand breaks on chromosomes 4 and 10 by transfecting the AB1080 FSHD myoblasts with plasmids containing Cas9–green fluorescent protein (GFP) and two sgRNAs targeting specific sites in the 4q35 and 10q26 loci. This led to the induction of the t(4;10)(q35;q26) in some cells, which was detectable by polymerase chain reaction (PCR) with the primers specific to two different chromosomes (Fig. 1B and fig. S1, A and C).
Multiple clones containing the t(4;10)(q35;q26) were selected and expanded. Given that the translocation frequency is relatively low and that myoblasts require survival signaling from cell-cell contact to grow, we used a two-step strategy to select the single-cell clones. Initially, transfected cells were sorted by fluorescence-activated cell sorting (FACS), and 20 GFP-positive cells were seed into each well of a 96-well plate. A total of 400 wells were analyzed by PCR with the Chr4F and Chr10R primers, and six t(4;10)(q35;q26)-positive wells were detected. The positive wells were subjected to the second round of sorting, the sorted cells were seeded one cell per well, and the surviving cells were expanded. We lastly identified 36 clones with the t(4;10)(q35;q26) from 20,000 cells analyzed initially after 800 independent PCR reactions. The Chr4F/Chr10R and Chr10F/Chr4R PCR products from nine of these clones (named T1, T2, T3, T4, T5, T6, T7, T8, and T9) were sequenced, and the expected reciprocal t(4;10)(q35;q26) translocation was confirmed in these clones (fig. S1, B and D). Three of the clones (T3, T5, and T6) were selected for further analysis.
Characterization of the 4q35;10q26 recombinant genotypes
As CRISPR-Cas9 may introduce additional chromosomal aberrations, we first performed a karyotype analysis of chromosomal spreads for the maternal cell line and the selected clones. All of the cell lines contained the normal chromosome number and structure (46, XX), and no visible aberrations were revealed. We have also confirmed that all the translocation clones were derived from the same maternal AB1080 cell line using PCR single-locus technology analysis (file S1).
To characterize the structure of the FSHD locus in the t(4;10)(q35;q26) clones, we performed the FiberVision FSHD assay, a method to visualize the subtelomeric regions of chromosomes 4 and 10 by fluorescent hybridization on combed DNA. This assay is used in clinics to diagnose FSHD1; it effectively distinguishes between chromosomes 4 and 10 and the A and B variants of the D4Z4 flanking region (19). The maternal AB1080 cell line contained a short D4Z4 array with seven units and the permissive 4qA allele on one of the chromosomes 4, consistent with its FSHD phenotype (Fig. 1C). The other chromosome 4 had a normal number of D4Z4 units (18), and the two copies of chromosomes 10 contained 7 and 36 units. All of the four chromosomes contained the A variant of the D4Z4 flanking sequence.
Most of the 4qA haplotypes, except for the 4A166, are FSHD permissive, while most of the 10qA carry a dysfunctional PAS and are thus nonpermissive for FSHD (16–18). Nevertheless, to rule out the possibility of a hybrid permissive 10qA haplotype with the functional PAS, similar to that described in (18), we performed long read sequencing of the maternal AB1080 cell line using the Oxford Nanopore Technology. Five >1-kb reads mapped uniquely to the PAS sequence at 4q (chr4:193543619-193543623, T2T v2 genome assembly). All five bore the functional PAS (ATTAA) and the T at the T/G SNP (AF117653.3: 8035), excluding the nonpermissive 4A166 haplotype at any of the two chr4 copies (fig. S2A). Three reads mapped uniquely to the PAS distal to the last D4Z4 on chr10 (chr10:134726542-134726546, T2T v2 genome assembly). In all of them, the PAS was dysfunctional (ATCAA) and the SNP combinations suggested that both copies of chr10 had the frequent nonpathological 10A166 haplotype (fig. S2B). Therefore, the intact chromosomes 4 in the maternal and the recombinant clones could only bear the permissive 4qA haplotype, and the intact chromosomes 10 could only bear the nonpermissive 10qA, while the derivative chromosomes should have exchanged the variants and bear the nonpermissive 10qA on derivative 4 and the permissive 4qA on derivative 10.
The recombinant genotypes T3 inherited the intact chr4 with the short D4Z4 repeat and the permissive 4qA; thus, it should have preserved the DUX4 expression and the pathological phenotype. On the contrary, the genotypes T5 and T6 have lost the permissive D4Z4 length/haplotype combinations: They had the normal number of D4Z4 units on chr4, der4, and der10, while the only short D4Z4 array on chr10 was flanked by the nonpermissive 10qA. Therefore, the T5 and T6 clones had a non-FSHD genotype and a potentially normal phenotype.
It is noteworthy that the total number of D4Z4 repeats in the maternal cell line and the obtained clones with the potentially restored phenotypes is not the same. This is due to recombination that occurred in the G2 phase when the cell ploidy was 4n. The recombination in 2n cells could only result in combinations where the short D4Z4 repeat arrays are left with their initial distal PAS sequence, resulting in the unchanged overall phenotype (fig. S3A); the repeat numbers observed by us in the cells with the restored phenotype could not be explained by this recombination (Fig. 1C). On the other hand, the recombination in 4n cells resulted in a broader number of possible karyotypes, including those observed in this study (fig. S3B).
4q35;10q26 translocation restores the phenotype of FSHD cells
We next sought to determine whether any of the t(4;10)(q35;q26) clones acquired the normal phenotype. FSHD cells are generally incapable to differentiate properly and form either atrophic or disorganized myotubes upon differentiation induction (5, 7). In the T5 and T6 clones with the nonpermissive recombinant genotype, we observed a marked restoration of the myotube morphology to the level of control AB1190 cells, while the T3 clone with the permissive recombinant genotype did not exhibit any notable morphology changes (Fig. 1, D and E).
FSHD myoblasts are also known to be more sensitive to oxidative stress and associated DNA damage (8–10). We thus assessed the sensitivity of the t(4;10)(q35;q26)-positive cells to hydrogen peroxide, which is a common oxidative stress inductor. Consistent with the literature data, AB1080 FSHD myoblasts were more prone to apoptosis and cell death upon H2O2 treatment, while the T3, T5, and T6 clones exhibited apoptosis levels similar to that in the control AB1190 cells after treatment (Fig. 1G). At the same time, the cell viability assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) test was similar for both the maternal cell line and the translocation clones (Fig. 1F).
All in all, the t(4;10)(q35;q26) clones with the nonpermissive recombinant genotype restored their differentiation capacity and increased their tolerance toward oxidative stress; their FSHD phenotype was thus largely reverted.
4q35;10q26 translocation partially restores the transcriptome profile of FSHD cells
To assess the effect of the t(4;10)(q35;q26) on the transcriptome of the FSHD cells, we have performed RNA sequencing (RNA-seq) in the control FSHD and the translocation cell lines. As DUX4 expression is thought to be triggered at the moment of myoblast differentiation, and many DUX4 target genes are expressed at higher levels in myotubes rather than myoblasts (20–23), the experiment was performed both in the proliferating myoblasts and the myotubes obtained after 4 days of differentiation.
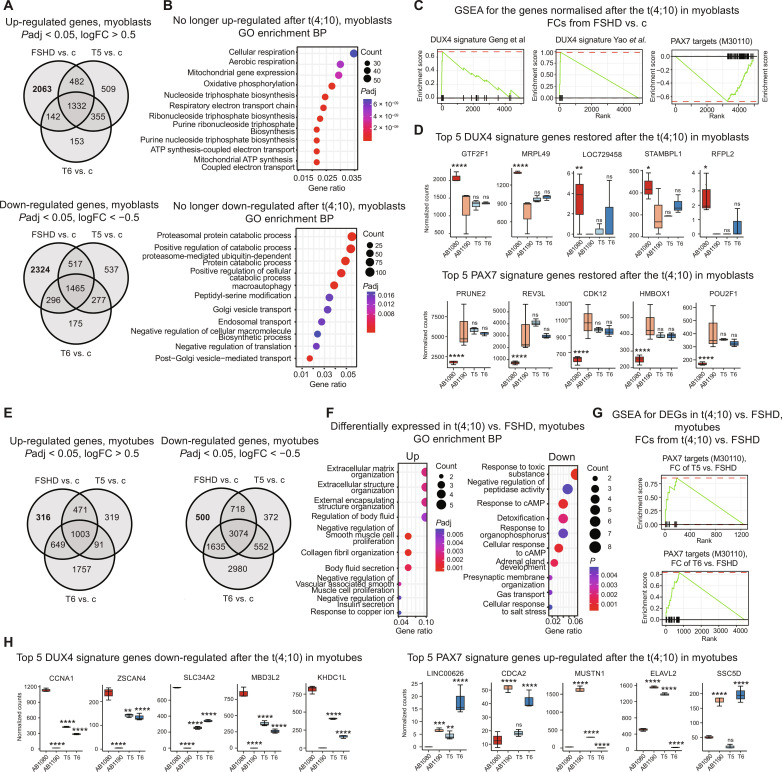
Differential expression analysis revealed more than 4000 genes that restored their normal expression levels after the t(4;10) in both translocation clones at the myoblast stage (Fig. 2A). A total of 2063 genes were no longer up-regulated in the T5 and T6 myoblasts; those genes were highly enriched for the Gene Ontology (GO) terms related to aerobic respiration and adenosine 5′-triphosphate synthesis (Fig. 2B). Twenty-eight of those genes encoded for the mitochondrial proteins and the proteins of the electron transport chain in particular (e.g., NDUFS8, NDUFA2, UQCRC1, UQCC2, COX4I1, and COX6B1), with Complex I components being most represented. These results are in line with the recent data on impaired mitochondrial function in FSHD (10, 24); notably, deregulated mitochondrial metabolism was shown to contribute significantly to the oxidative stress typical for the FSHD muscles, and Complex I was identified as the primary target for DUX4-induced mitochondrial dysfunction (24). Restored expression of Complex I and other mitochondrial proteins may explain the decreased sensitivity to oxidative stress observed in the translocation clones (Fig. 1G). A total of 2324 genes that are no longer down-regulated in the translocation clones were enriched for the catabolic processes–related genes and the genes involved in vesicle-mediated transport. This change may reflect the myoblasts readiness for differentiation, as membrane vesicle transport system was shown to mediate the actinin proteins transfer to the actin filaments (25), which is necessary for the actin-filament remodeling upon myoblast fusion.
Fig. 2. Transcriptome profiling of the recombinant clones.
(A) Up-regulated and down-regulated genes in FSHD, T5, or T6 myoblasts versus control myoblasts. (B) GO Biological Process enrichment for genes no longer deregulated in both translocation clones at the myoblast stage. (C) Gene Set Enrichment Analysis (GSEA) against the DUX4 signature gene sets from Geng et al. (3) and Yao et al. (2) and PAX7 targets gene set (M30110) (44) for the genes no longer deregulated in both translocation clones at the myoblast stage. The genes were thresholded by Padj < 0.05 and ranked by the log2FC from the FSHD versus control myoblasts comparison. (D) DUX4 and PAX7 signature genes restored in both translocation clones. Top 5 genes by Padj in the FSHD versus control myoblasts; median + range of the DESeq2 normalized counts; Padj values from the DESeq2 comparisons with the control myoblasts. (E) Up-regulated and down-regulated genes in FSHD, T5, or T6 myotubes versus control myotubes. (F) GO Biological Process enrichment for the genes commonly up-regulated or down-regulated in the T5 versus FSHD myotubes and T6 versus FSHD myotubes. (G) GSEA against the PAX7 targets gene set (M30110) (44) for the differentially expressed genes (Padj < 0.05, |log2fold| > 0.5) in the T5 versus FSHD myotubes and the T6 versus FSHD myotubes. The genes were thresholded by Padj < 0.05 and ranked by log2FC from the corresponding comparisons. (H) DUX4 and PAX7 signature genes restored in both (DUX4) or at least one of the translocation clones (PAX7). Top 5 genes by the average Padj in the T5 versus FSHD myotubes and the T6 versus FSHD myotubes; median + range of the DESeq2 normalized counts; Padj values from the DESeq2 comparisons with the FSHD myotubes. *Padj < 0.01, **Padj < 0.001, ***Padj < 0.0001, and ***Padj < 0.00001. ns, not significant; cAMP, cyclic adenosine monophosphate; ATP, adenosine triphosphate.
Although DUX4 expression is a key characteristic of the FSHD myoblasts (13), it was shown to be hardly detectable in multiple studies (3, 20, 21), probably due to its stochastic expression pattern (20). Similarly, we did not detect the DUX4 mRNA in our RNA-seq data. Instead, the expression of DUX4 signature genes (2, 3, 26) was assessed. DUX4 signatures from Geng et al. (3) and Yao et al. (2) were indeed enriched in the genes deregulated in AB1080 FSHD myoblasts compared to the control; however, this was not the case for the Choi et al. (26) signature in our cellular model (fig. S4A). Similarly, the genes that restored their expression to the control level in both translocation clones were enriched for the DUX4 signature genes from Geng et al. (3) and Yao et al. (2) (Fig. 2C), although not all of the genes up-regulated in the maternal cell line were normalized. A strong negative enrichment of PAX7 targets was detected both in the genes deregulated in FSHD myoblasts versus the control (fig. S4B) and the genes restored in the translocation clones (Fig. 2C). Global repression of PAX7 targets has recently been shown to be at least as major a signature of FSHD skeletal muscle as DUX4 target gene expression, outperforming the DUX4 signature as a biomarker in certain cases (27). Consistent with these data, the repression of PAX7 target genes was more pronounced than the DUX4 targets overexpression in our dataset, in both the overall genes deregulated in the AB1080 FSHD myoblasts (fig. S4B) and the genes restored in the translocation clones (Fig. 2C).
In the differentiated myotubes, 816 genes restoring their expression in the translocation clones were detected; 316 genes were no longer up-regulated, and 500 genes were no longer down-regulated following the translocation (Fig. 2E). No significant enrichment of specific ontology terms was detected in the genes completely normalizing their expression, likely due to the low overall number of genes expressed in differentiated myotubes; thus, the enrichment analysis was performed for all the genes significantly up-regulated [log2 fold change (log2FC) > 0.5, P adjusted (Padj) < 0.05] or down-regulated (log2FC < −0.5, Padj < 0.05) in the differentiated myotubes derived from the translocation clones compared to the myotubes from the FSHD cells. The genes up-regulated in the t(4;10) myotubes were related to extracellular matrix organization and negative regulation of muscle cell proliferation, reflecting the switch from proliferating myoblasts to the differentiated myotube stage, more pronounced in the translocation clones (Fig. 1, D and E). The most enriched term among the down-regulated genes was response to toxic substances, which probably reflected the decreased toxic burden following reduced DUX4 expression, as DUX4 protein is highly toxic for muscle cells (4). In line with the previous observations (20–23), certain DUX4 target genes were expressed at much higher levels in the differentiated myotubes. Although the enrichment of the overall DUX4 signature was not significant among the genes normalized in the t(4;10) myotubes, many DUX4 target genes—including ZSCAN4, CCNA1, and SLC34A2—were significantly down-regulated (Fig. 2G). Consistent with the result obtained in myoblasts, the up-regulation of the PAX7 target genes was also observed in the myotubes from the T5 and T6 cells (Fig. 2, G and H). Nevertheless, at the myotube stage, the up-regulated PAX7 targets were not the same for the two translocation clones, suggesting only a partial restoration of the PAX7 signature genes expression (Fig. 2H).
As DUX4 expression is known to be affected by the chromatin state of the D4Z4 locus (28), we have also checked the methylation status of the D4Z4 units in the control, FSHD, and t(4;10) myoblasts. Consistent with previous reports (28–30), the D4Z4 array of the control AB1190 myoblasts was more heavily methylated than that of the FSHD AB1080 myoblasts. Nevertheless, we did not observe any significant changes in D4Z4 methylation in the maternal versus translocation-bearing clones (fig. S5), as expected. In physiological conditions, DUX4 is expressed during zygotic genome activation and is afterward heavily silenced in most tissues, including muscle [reviewed in (12)]. Thus, the methylation status of the D4Z4 array should be established at a much earlier stage of the cell development and is unlikely to be modified by the translocation at the myoblast stage.
Together, the 4q35;10q26 translocation significantly affected the gene expression profile of the FSHD cells, bringing it closer to that of the normal cells. This restoration of expression was independent of the methylation status and was more prominent at the myoblast stage.
DISCUSSION
FSHD1 is caused by the contraction of a highly polymorphic D4Z4 macrosatellite repeat array in the 4q35 region of chromosome 4 combined with the permissive 4qA haplotype flanking the D4Z4 array. Soon after the discovery of the 4q35 D4Z4, sequences homologous to D4Z4 have been identified on many chromosomes (31, 32). In particular, an almost identical D4Z4 repeat array (~98.5% sequence identity) is found in the 10q26 region of chromosome 10 (14, 33), and the spontaneous exchanges between the subtelomeric regions of chromosomes 4 and 10 have been described (17). Despite strong homology in the D4Z4 array between chromosomes 4 and 10, D4Z4 contractions on chromosome 10q are not associated with FSHD (16), likely because the overwhelming majority of the 10q chromosomes contain the dysfunctional PAS flanking the most distal D4Z4 unit (16–18). This does not allow for the DUX4 mRNA stabilization and productive DUX4 expression from the typical 10q D4Z4.
Recently, a family with the 10q-linked FSHD was reported (18); in this family, a de novo D4Z4 repeat exchange between chromosomes 4 and 10 led to the replacement of the nonpermissive 10qA distal sequence on an FSHD-sized chromosome 10 with the permissive 4qA, leading to the atypical DUX4 expression from chromosome 10. This underlined the dominant role of DUX4, and not the other chromosome 4 genes, in FSHD pathogenesis. We thus questioned whether the opposite exchange of the 4qA/10qA sequences would be possible, and whether it would reduce the DUX4 expression and revert the FSHD phenotype.
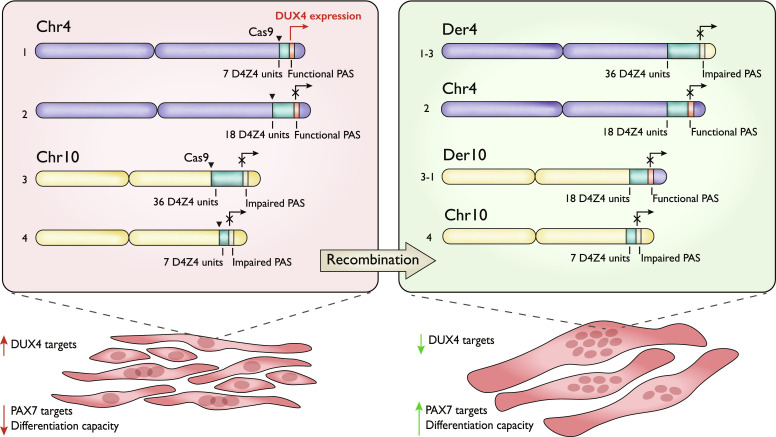
Here, we experimentally induced a reciprocal t(4;10) (q35;q26) chromosomal translocation in human FSHD myoblast using the CRISPR-Cas9 system and created a number of recombinant t(4;10) myoblasts clones with the varying combinations of the D4Z4 repeat size and the 4qA/10qA flanking sequence (Fig. 3). We demonstrated that the recombination between chromosomes 4 and 10 could produce a number of nonpathological genotypes manifested by the FSHD phenotype improvement. The nonpathological recombinant genotypes were characterized by the lack of the 4qA and a short D4Z4 array combination. Myoblasts with these genotypes greatly reduced the expression of the DUX4 targets and increased the expression of PAX7 signature genes, became less sensitive to oxidative stress, and improved their differentiation capacity. Their methylation profile at the D4Z4 unit was not changed, suggesting that it is established earlier in development.
Fig. 3. Schematic representation of the FSHD genotype correction approach.
This study serves as a proof of principle for the existing genetic model of FSHD, which considers the D4Z4 repeat contraction on the 4qA-type chromosome 4 as the major FSHD predictor. This work also proves the possibility of the FSHD phenotype correction by targeted genome editing as our method has the potential to achieve a permanent correction of FSHD1. For potential clinical applications in the future, the genetic correction can be carried out in Epstein-Barr virus (EBV)–immortalized B cells from patients with FSHD; cells bearing the t(4;10) can be then expanded and converted into induced pluripotent stem cells (iPSCs). During conversion, EBV that is present as an episome in B cells will be lost and cells will recover the normal genotype (34). These iPSCs will be converted into myoblasts or mesoangioblasts using one of the existing approaches (35) and introduced into the patients’ muscles. We have previously shown that fusion between healthy and FSHD myoblasts corrects differentiation defects (36).
MATERIALS AND METHODS
Cell culture
Human immortalized myoblasts were derived from a healthy individual (AB1190) and from a patient with the FSHD (AB1080) (37) and were a gift of V. Mouly (Institute of Myology, Paris, France). AB1080 and AB1190 were cultured in a proliferation medium composed of four parts of high-glucose Dulbecco’s modified Eagle’s medium (DMEM) and one part of Medium 199 (Sigma-Aldrich, #M4530) supplemented with 20% fetal bovine serum (FBS) (Life Technologies, #10270), fetuin (25 μg/ml; Life Technologies, #10344026), human epidermal growth factor (hEGF) (5 ng/ml; Life Technologies, #PHG0311), basic fibroblast growth factor (0.5 ng/ml; Life Technologies, #PHG0026), insulin (5 μg/ml; Sigma-Aldrich, #91077C-1G), dexamethasone (0.2 μg/ml; Sigma-Aldrich, #D4902), and 1% penicillin-streptomycin (Gibco, #15140-122).
PCR primers and CRISPR sgRNA design
The subtelomeric regions of chr4 and chr10 exhibit >90% sequence homology, making it difficult to design chromosome-specific guide RNAs (gRNAs) and primers. To ensure the chromosome specificity, a double-step procedure was applied to design the primers and the gRNAs. First, several sets of primers were designed using the Primer3 platform (http://bioinfo.ut.ee/primer3-0.4.0/) (table S1). The ~25-kb region from chromosome 10 and ~16-kb region from chromosome 4 located on the centromeric side of the FRG2 gene common to both chromosomes (upstream of the D4Z4 array) (Fig. 1A) were screened for the primers satisfying the following conditions: (i) The amplicon sequence is specific for chromosome 4 or chromosome 10, and (ii) the amplicon sequence does not contain any coding genes. To ensure the selected primers amplified exclusively Chr4 or Chr10, all possible primer combinations were assessed for the nonspecific amplicons using the UCSC In-Silico PCR analysis (https://genome.ucsc.edu/cgi-bin/hgPcr), which provides the sequences of all possible amplicons and the percentage of their homology with different chromosome(s). Only the amplicons exhibiting 100% homology to chr4 or chr10 with the least possible homology to the other chromosome were retained. The selected primer pairs were lastly checked experimentally by PCR with the myoblast gDNA to amplify a single band.
Second, the amplicon from the selected primer pair was screened for the possible gRNA target sequences using the CRISPOR program (http://crispor.tefor.net/). Top X gRNAs with the best predicted specificity and efficiency were selected (table S2). The sgRNAs were cloned into the phU6 plasmid (Addgene, #53188).
PCR detection of the t(4;10)
Chr4F/Chr10R and Chr10F/Chr4R primers combinations were used to detect the intended translocation (Fig. 1A and fig. S1). Genomic DNA of the screened cells was isolated with the NucleoSpinTissue Kit (Macherey Nagel, #740952). The PCR was performed using the PowerUp SYBR Green Master Mix (Life Technologies, #A25778) and the appropriate PCR primers. PCR products were loaded on the 1% agarose gel, and the size of amplicons was determined by comparison with a 1-kb DNA ladder (Thermo Fisher Scientific, #SM0311). PCR products were isolated from the gel using the NucleoSpin gel and PCR clean-up kit (Macherey-Nagel) and subjected to Sanger sequencing (Eurofins Genomics, Cochin Sequencing Platform). Sequence data were aligned using Seqman version II (DNASTAR). Three to 4 independent experiments were performed for each clone.
Cell transfection and establishment of clones
Briefly, 2.5 × 105 of AB1080 FSHD cells were seeded into six-well plates. Following adhesion at 37°C with 5% CO2 for 24 hours, the cells were transfected with 3 μg of Cas9-GFP (Addgene, #57818), 1 μg of phU6 containing the sgRNA, and 9 μl of ViaFect (Promega, #E4981). After incubation for 48 hours, cells were trypsinized and resuspended in 1 ml of FBS. Establishment of clones was based on a two-step strategy due to myoblasts cell culture characteristics. Transfected cells were sorted into wells of 96-well plates containing 200 μl of the growth medium using the BD FACSAria III cell sorter. Twenty GFP-positive cells were seeded per well. At 80% confluence, half of the cells in each well were collected for DNA extraction and PCR analysis to detect the t(4;10) translocation. The wells positive for the t(4;10) were sorted again into a 96-well plate one cell per well. Single cells were grown for 4 weeks, and then the surviving clones were expanded and screened. The origin of the clones from the same maternal cell line AB1080 was confirmed by the Cell Line Authentication service provided by Eurofins using PCR single-locus technology. The cell line authentication report generated by Eurofin is available in file S1.
Karyotype analysis
Chromosome spreads were prepared from the input cells and the cells with the t(4;10) and analyzed at the Hôpital Saint Antoine-Paris, France. Experiments were performed in duplicate.
Fiber FISH
To characterize the composition of the 3.3 kb-D4Z4 repeat-containing loci on chromosomes 4 and 10 in the obtained clones, the Fibervision FISH FSHD assay with the FiberProbes FSHD (FSHD-HYB-002-RUO, Genomic vision) was performed. Genomic DNA from the maternal cell line and the cell lines containing the t(4;10) translocations was extracted, and the plugs were prepared using the FiberPrep DNA Extraction Kit (Genomic vision). The material was transferred to the Genomic vision facilities to perform molecular combing, hybridization, and image acquisition. Image analysis was performed using the FiberStudio FSHD CE-IVD software. Experiments were performed in duplicates.
Nanopore library preparation and sequencing
To genotype the subtelomeric PAS sequence in the maternal AB1080 cell line, Nanopore sequencing was performed. The total genomic DNA was extracted from the fresh cell pellets using the NucleoSpinTissue Kit (Macherey Nagel, #740952). Total DNA (5 μg) was sonicated in a Covaris S220 sonicator in the microTUBE AFA Fiber Pre-Slit Snap-Cap 6 × 16 mm columns with the following program: 5 s, 1000 cycles per burst. The median band size obtained after the sonication was 1500 to 4000 bp as determined by the gel electrophoresis. The DNA library was then prepared as described in (38) with some modifications. Briefly, the sheared DNA was centrifuged at 15,000g for 5 min, and the supernatant was transferred to the 30-kDa Amicon column (Millipore, UFC5030) and centrifuged again at the same conditions. The flow-through was discarded, 450 μl of 10 mM Tris-HCL pH 8.0 was applied to the column, and the column was centrifuged for 5 min at 15,000g. The concentrated DNA was transferred to a new tube and purified/size-selected using the KAPA Pure Beads (Roche, 7983271001) following the manufacturer’s instruction. The purified DNA was subjected to end repair as described in (38) and purified again using KAPA Pure Beads (Roche, 7983271001). Next, the A-tailing was performed as described in (38), followed by another round of purification of the KAPA Pure Beads.
NEXTFLEX DNA-seq barcode adapters (NEXTFLEX Rapid DNA-Seq Kit 2.0, NOVA-5188-01) were ligated to the A-tailed DNA with the T4 DNA ligase (5 U/μl; Thermo Fisher Scientific, EL0012) overnight at room temperature. Next day, the barcoded pre-PCR library was purified on the KAPA Pure Beads and amplified with the KAPA HiFi HotStart PCR Kit (Roche, catalog no. 07958897001) and the NEXTFLEX PCR primers (NEXTFLEX Rapid DNA-Seq Kit 2.0, NOVA-5188-01). Six amplification cycles were used. The amplified libraries were purified on the KAPA Pure Beads, quantified with the Qubit dsDNA High Sensitivity Kit (Invitrogen, Q32854), and stored at 4°C until sequencing.
Long-read sequencing was conducted using Oxford Nanopore Technology using the FLO-PRO002 flow cell and Promethion. The library was prepared according to the ligation sequencing genomic DNA (SQK-LSK109) protocol, which involves end repair and A-tailing of the DNA and then adapter ligation. The raw sequencing data are available at the European Nucleotide Archive (ENA) database (www.ebi.ac.uk/ena/browser) with the accession number PRJEB61799.
Nanopore sequencing analysis and haplotype identification
The Nanopore reads were aligned to the T2T CHM13v2.0 Telomere-to-Telomere genome assembly using the QAlign software. The aligned reads were visualized using the IGV-2.16.0 genome browser. The sequences of the reads of interest were extracted from the fastq files by read name using samtools 1.3.1 (www.htslib.org/doc/1.3.1/samtools.html) and seqtk 1.3-r106 (https://docs.csc.fi/apps/seqtk/). The reads spanning the PAS sequences were aligned to the reference sequences containing the distal D4Z4 unit and the regions distal to it including PAS (GenBank: AF117653.3 for chr4 and GenBank: HM190188.1 for chr10) using SnapGene v3.2.1. The sequences of the pathological and nonpathological 4q and 10q subtelomere haplotypes were taken from (17) and (18) for the hybrid 10A166H1 genotype.
RNA-seq sample preparation and processing
RNA-seq was performed for the control (AB1190) and FSHD (AB1080) cell lines, as well as the translocation clones with the improved phenotype (AB1080-T5 and AB1080-T6). The cells were collected at the myoblasts stage and at the stage of myotubes after four full days of differentiation. For the nondifferentiated samples, 5 × 105 cells were collected and subjected to the RNA extraction right away. For the differentiated samples, 5 × 105 cells were seeded to six-well plates to obtain ~90% confluency on the next day, when the differentiation was induced by changing the proliferation medium to the differentiation medium with low serum content [98% DMEM (Sigma-Aldrich, #D0822), 2% FBS (Life Technologies, #10270), and 1% penicillin-streptomycin (Gibco, #15140-122)]. On the fifth day of differentiation, the myotubes were detached with Trypsin-EDTA (Sigma-Aldrich, #T2601) and subjected to the RNA extraction. Total RNA was isolated using the Zymo Quick-RNA Miniprep Kit (R1055). All samples were prepared in triplicates. mRNA libraries were prepared using Novogene NGS RNA Library Prep Set (PT042) and sequenced on the Illumina NovaSeq 6000 system.
Raw sequencing data were processed by the nfcore RNA-seq pipeline (v3.10.1) with default parameters (39). Briefly, the reads were trimmed with trimgalore (0.6.7) and cutadapt (3.4) to remove the Illumina adapter sequences from the 3′-ends. The trimmed reads were mapped to the GRCh38 genome from the Illumina iGenomes using STAR (2.6.1d) and quantified with Salmon (1.9.0) (40). The obtained gene counts were then used for differential expression analysis with DESeq2 (...) (41). The genes with the |log2FC| > 0.5 and Padj < 0.05 were considered differentially expressed.
The genes significantly up-regulated/down-regulated in the FSHD versus c comparison and no longer up-regulated/down-regulated in both the T5 versus c and T6 versus c comparisons were considered restored after the translocation and were used for the GO enrichment analysis and Gene Set Enrichment Analysis (GSEA). GO enrichment was performed against the Biological Processes domain of the GO database with the P value of 0.05 and q value cutoffs of 0.2 using the clusterProfile (4.8.2) R package (42). The formerly up-regulated and down-regulated gene sets were analyzed separately. All the genes with nonzero count detected in the RNA-seq experiment were used as a universe. GSEA was performed with the fgsea (1.26.0) R package (43). The genes restored in both T5 and T6 clones were ranked by log2Fold changes from the FSHD versus c comparison and tested against the PAX7 targets gene set (M30110) derived from the Molecular Signature Database (44) and the DUX4 signature gene sets derived manually from the previously published works (2, 3, 26) and a recent meta-analysis (27). Raw and processed sequencing data have been submitted to Gene Expression Omnibus (GEO) under the accession number GSE254310.
Methylation analysis using Oxford Nanopore Technology
Genomic DNA samples [AB1190, AB1080, AB1080 (T3), AB1080 (T5), and AB1080 (T6)] were prepared for sequencing using the NBD 114.96 kit library from Oxford Nanopore Technologies. Briefly, the samples were subjected to end prepping via end repair/dA-tailing, followed by purification using AMPure XP Beads at a concentration of 40% v/v. The next step involved native barcode ligation using Blunt/TA Ligase Master Mix (NEB), followed by termination with EDTA before stoichiometrically pooling and cleaning with AMPure XP Beads (40% v/v). Last, sequencing adapter ligation was performed at the DNA ends using NEBNext Quick Ligation (NEB), followed by purification of the library with AMPure XP Beads (40% v/v).
This library was next sequenced four times during 72 hours using R10.4.1 flow cells (FLO-MIN114) and a PromethION 2 Solo. Analysis of DNA methylation has been obtained by base calling of Pod5 files with dorado-0.5.2 and the basecalling model dna_r10.4.1_e8.2_400bps_sup@v4.3.0 with the option “--modified-bases 5mCG_5hmCG”. Alignment was conducted using minimap2 with T2T CHM13v2.0-HS1 as the reference genome. Subsequently, BAM files were merged using samtools, and plots of methylation data were generated using modBAMtools.
MTT assay for cell viability
For analysis of cell viability, 1.5 × 104 myoblast cells were seeded in 96-well plates. 10mkl of the MTT reagent (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; Sigma-Aldrich #M2128) was added to each well, and the cells were incubated at 37°C for 3 hours and lysed in the MTT lysis buffer [30% SDS, 30% N,N′-dimethylformamide, 2% glacial acetic acid, and 25 mM HCl (pH 4.7)] overnight. The next day, the absorbance at 570 nm was measured using the Victor 1420 multilabel counter. Three independent experiments were performed.
Apoptosis assessment
Сells were stained for annexin V with the eBioscience Annexin V-FITC Apoptosis Detection Kit (Life Technologies, #BMS500FI-20) according to the manufacturer’s instructions and analyzed by flow cytometry on the BD/Accuri C6 PLus instrument. Three independent experiments were performed.
Myogenic differentiation and analysis of myotubes
Myoblasts were seeded at a density of 5 × 105 cells per well in a six-well plate and cultured in 2 ml of the growth medium. When the cells reached 90% confluence, myogenic differentiation was induced by replacing the proliferation medium with the differentiation medium (DMEM containing 2% FBS). The cells were incubated for 4 days under these conditions. On day 4, the cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, #30525-89-4) for 5 min, permeabilized with 0.25% Triton X-100 (Sigma-Aldrich, #T8787) for 5 min, and blocked with 0.5% bovine serum albumin (Euromedex, #04-100-812-C) for 1 hour. Myotubes were then stained with anti–troponin T antibodies (Sigma-Aldrich, #T6277) diluted at 1:50 for 2 hours at 37°C, revealed using anti-mouse Alexa-488 IgG conjugated antibodies (1:100; Life Technologies, #A-21200) for 1 hour at room temperature, mounted with a medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, #H-1200), and observed on a fluorescent microscope (Microvision instruments). The images from the adjacent fields of view were stitched together using the Cartograph software (Microvision) to create the large images of a big field. Three large images were generated per sample, and six independent experiments were performed.
Differentiation assessment
Analysis of differentiation was performed using the ImageJ software (National Institutes of Health, USA). For each large image of the specimen, the normalized troponin area (NTA) was calculated by dividing the overall troponin-positive area by the overall number of nuclei. Then, the mean NTA was obtained for each specimen. The procedure was repeated in six independent experiments. Statistical analysis was performed using Kruskal-Wallis test with Dunn’s correction for multiple comparison (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001).
Acknowledgments
We thank V. Mouly for a gift of immortalized myoblasts and T. Manoliu and Imaging and Cytometry Core Facility (PFIC) (Unit AMMICa, Gustave Roussy) for the expertise and advice in using instruments and methodological developments.
Funding: This work was supported by grants from the ANRS to Y.S.V. and E.A., AFM, FSHD Society, and the IDB RAS government basic research program 0088-2024-0010 to Y.S.V.
Author contributions: Y.S.V. and A.S. conceptualized the study. Y.S.V. and E.A. acquired funding. Y.M., A.S., C.D., and C.G. performed experiments. Y.S.V., Y.M., A.S., C.D., C.G., O.H., and E.A. performed formal data analysis. A.S. and C.G. performed bioinformatics analysis. E.A. and O.H. provided material and equipment. Y.S.V. and A.S wrote the manuscript and prepared figures. Y.S.V., A.S, E.A., and C.D. edited the manuscript and performed article revision. All authors read and approved the final version of the manuscript.
Competing interests: All authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw sequencing data are available in the open access databases GEO (GSE254310) and ENA (PRJEB61799).
Supplementary Materials
This PDF file includes:
Tables S1 and S2
Figs. S1 to S4
REFERENCES AND NOTES
- 1.Tawil R., van der Maarel S. M., Tapscott S. J., Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet. Muscle 4, 12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao Z., Snider L., Balog J., Lemmers R. J. L. F., Van Der Maarel S. M., Tawil R., Tapscott S. J., DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum. Mol. Genet. 23, 5342–5352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geng L. N., Yao Z., Snider L., Fong A. P., Cech J. N., Young J. M., van der Maarel S. M., Ruzzo W. L., Gentleman R. C., Tawil R., Tapscott S. J., DUX4 activates germline genes, retroelements, and immune mediators: Implications for facioscapulohumeral dystrophy. Dev. Cell 22, 38–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosnakovski D., Xu Z., Gang E. J., Galindo C. L., Liu M., Simsek T., Garner H. R., Agha-Mohammadi S., Tassin A., Coppée F., Belayew A., Perlingeiro R. R., Kyba M., An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 27, 2766–2779 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barro M., Carnac G., Flavier S., Mercier J., Vassetzky Y., Laoudj-Chenivesse D., Myoblasts from affected and non-affected FSHD muscles exhibit morphological differentiation defects. J. Cell. Mol. Med. 14, 275–289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosnakovski D., Gearhart M. D., Toso E. A., Ener E. T., Choi S. H., Kyba M., Low level DUX4 expression disrupts myogenesis through deregulation of myogenic gene expression. Sci. Rep. 8, 16957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanderplanck C., Ansseau E., Charron S., Stricwant N., Tassin A., Laoudj-Chenivesse D., Wilton S. D., Coppée F., Belayew A., The FSHD atrophic myotube phenotype is caused by DUX4 expression. PLOS ONE 6, e26820 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dmitriev P., Bou Saada Y., Dib C., Ansseau E., Barat A., Hamade A., Dessen P., Robert T., Lazar V., Louzada R. A. N., Dupuy C., Zakharova V., Carnac G., Lipinski M., Vassetzky Y. S., DUX4-induced constitutive DNA damage and oxidative stress contribute to aberrant differentiation of myoblasts from FSHD patients. Free Radic. Biol. Med. 99, 244–258 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Bou Saada Y., Dib C., Dmitriev P., Hamade A., Carnac G., Laoudj-Chenivesse D., Lipinski M., Vassetzky Y. S., Facioscapulohumeral dystrophy myoblasts efficiently repair moderate levels of oxidative DNA damage. Histochem. Cell Biol. 145, 475–483 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Karpukhina A., Galkin I., Ma Y., Dib C., Zinovkin R., Pletjushkina O., Chernyak B., Popova E., Vassetzky Y., Analysis of genes regulated by DUX4 via oxidative stress reveals potential therapeutic targets for treatment of facioscapulohumeral dystrophy. Redox Biol. 43, 102008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerji C. R. S., Panamarova M., Zammit P. S., DUX4 expressing immortalized FSHD lymphoblastoid cells express genes elevated in FSHD muscle biopsies, correlating with the early stages of inflammation. Hum. Mol. Genet. 29, 2285–2299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karpukhina A., Tiukacheva E., Dib C., Vassetzky Y. S., Control of DUX4 expression in facioscapulohumeral muscular dystrophy and cancer. Trends Mol. Med. 27, 588–601 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Lemmers R. J. L. F., van der Vliet P. J., Klooster R., Sacconi S., Camaño P., Dauwerse J. G., Snider L., Straasheijm K. R., van Ommen G. J., Padberg G. W., Miller D. G., Tapscott S. J., Tawil R., Frants R. R., van der Maarel S. M., A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329, 1650–1653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Geel M., Dickson M. C., Beck A. F., Bolland D. J., Frants R. R., van der Maarel S. M., de Jong P. J., Hewitt J. E., Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics 79, 210–217 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Lemmers R. J. L. F., de Kievit P., Sandkuijl L., Padberg G. W., van Ommen G.-J. B., Frants R. R., van der Maarel S. M., Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 32, 235–236 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Lemmers R. J. L. F., Wohlgemuth M., van der Gaag K. J., van der Vliet P. J., van Teijlingen C. M. M., de Knijff P., Padberg G. W., Frants R. R., van der Maarel S. M., Specific sequence variations within the 4q35 region are associated with Facioscapulohumeral Muscular Dystrophy. Am. J. Hum. Genet. 81, 884–894 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemmers R. J. L. F., van der Vliet P. J., van der Gaag K. J., Zuniga S., Frants R. R., de Knijff P., van der Maarel S. M., Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete interchromosomal sequence transfers in human evolution. Am. J. Hum. Genet. 86, 364–377 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemmers R. J. L. F., van der Vliet P. J., Blatnik A., Balog J., Zidar J., Henderson D., Goselink R., Tapscott S. J., Voermans N. C., Tawil R., Padberg G. W. A. M., van Engelen B. G., van der Maarel S. M., Chromosome 10q-linked FSHD identifies DUX4 as principal disease gene. J. Med. Genet. 59, 180–188 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen K., Broucqsault N., Chaix C., Roche S., Robin J. D., Vovan C., Gerard L., Mégarbané A., Urtizberea J. A., Bellance R., Barnérias C., David A., Eymard B., Fradin M., Manel V., Sacconi S., Tiffreau V., Zagnoli F., Cuisset J.-M., Salort-Campana E., Attarian S., Bernard R., Lévy N., Magdinier F., Deciphering the complexity of the 4q and 10q subtelomeres by molecular combing in healthy individuals and patients with facioscapulohumeral dystrophy. J. Med. Genet. 56, 590–601 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Snider L., Geng L. N., Lemmers R. J. L. F., Kyba M., Ware C. B., Nelson A. M., Tawil R., Filippova G. N., van der Maarel S. M., Tapscott S. J., Miller D. G., Facioscapulohumeral dystrophy: Incomplete suppression of a retrotransposed gene. PLOS Genet. 6, e1001181 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones T. I., Chen J. C. J., Rahimov F., Homma S., Arashiro P., Beermann M. L., King O. D., Miller J. B., Kunkel L. M., Emerson C. P., Wagner K. R., Jones P. L., Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 21, 4419–4430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krom Y. D., Dumonceaux J., Mamchaoui K., den Hamer B., Mariot V., Negroni E., Geng L. N., Martin N., Tawil R., Tapscott S. J., van Engelen B. G. M., Mouly V., Butler-Browne G. S., van der Maarel S. M., Generation of isogenic D4Z4 contracted and noncontracted immortal muscle cell clones from a mosaic patient: A cellular model for FSHD. Am. J. Pathol. 181, 1387–1401 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tassin A., Laoudj-Chenivesse D., Vanderplanck C., Barro M., Charron S., Ansseau E., Chen Y.-W., Mercier J., Coppée F., Belayew A., DUX4 expression in FSHD muscle cells: How could such a rare protein cause a myopathy? J. Cell. Mol. Med. 17, 76–89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heher P., Ganassi M., Weidinger A., Engquist E. N., Pruller J., Nguyen T. H., Tassin A., Declèves A.-E., Mamchaoui K., Banerji C. R. S., Grillari J., Kozlov A. V., Zammit P. S., Interplay between mitochondrial reactive oxygen species, oxidative stress and hypoxic adaptation in facioscapulohumeral muscular dystrophy: Metabolic stress as potential therapeutic target. Redox Biol. 51, 102251 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y., Jin Y., Zhao P., Wu J., Ren Z., Lipid droplets contribute myogenic differentiation in C2C12 by promoting the remodeling of the acstin-filament. Cell Death Dis. 12, 1102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi S. H., Gearhart M. D., Cui Z., Bosnakovski D., Kim M., Schennum N., Kyba M., DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res. 44, 5161–5173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerji C. R. S., Panamarova M., Hebaishi H., White R. B., Relaix F., Severini S., Zammit P. S., PAX7 target genes are globally repressed in facioscapulohumeral muscular dystrophy skeletal muscle. Nat. Commun. 8, 2152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butterfield R. J., Dunn D. M., Duval B., Moldt S., Weiss R. B., Deciphering D4Z4 CpG methylation gradients in fascioscapulohumeral muscular dystrophy using nanopore sequencing. Genome Res. 33, 1439–1454 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaillard M., Roche S., Dion C., Tasmadjian A., Bouget G., Salort-Campana E., Vovan C., Chaix C., Broucqsault N., Morere J., Puppo F., Bartoli M., Levy N., Bernard R., Attarian S., Nguyen K., Magdinier F., Differential DNA methylation of the D4Z4 repeat in patients with FSHD and asymptomatic carriers. Neurology 83, 733–742 (2014). [DOI] [PubMed] [Google Scholar]
- 30.van Overveld P. G. M., Lemmers R. J. F. L., Sandkuijl L. A., Enthoven L., Winokur S. T., Bakels F., Padberg G. W., van Ommen G.-J. B., Frants R. R., van der Maarel S. M., Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 35, 315–317 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Hewitt J. E., Lyle R., Clark L. N., Valleley E. M., Wright T. J., Wijmenga C., van Deutekom J. C., Francis F., Sharpe P. T., Hofker M., Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum. Mol. Genet. 3, 1287–1295 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Lyle R., Wright T. J., Clark L. N., Hewitt J. E., The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics 28, 389–397 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Bakker E., Wijmenga C., Vossen R. H., Padberg G. W., Hewitt J., van der Wielen M., Rasmussen K., Frants R. R., The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve Suppl. 2, S39–S44 (1995). [PubMed] [Google Scholar]
- 34.Rajesh D., Dickerson S. J., Yu J., Brown M. E., Thomson J. A., Seay N. J., Human lymphoblastoid B-cell lines reprogrammed to EBV-free induced pluripotent stem cells. Blood 118, 1797–1800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poulin H., Martineau L., Racine V., Puymirat J., Chahine M., Differentiation of lymphoblastoid-derived iPSCs into functional cardiomyocytes, neurons and myoblasts. Biochem. Biophys. Res. Commun. 516, 222–228 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Dib C., Bou Saada Y., Dmitriev P., Richon C., Dessen P., Laoudj-Chenivesse D., Carnac G., Lipinski M., Vassetzky Y. S., Correction of the FSHD myoblast differentiation defect by fusion with healthy myoblasts. J. Cell. Physiol. 231, 62–71 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Mamchaoui K., Trollet C., Bigot A., Negroni E., Chaouch S., Wolff A., Kandalla P. K., Marie S., Di Santo J., St Guily J. L., Muntoni F., Kim J., Philippi S., Spuler S., Levy N., Blumen S. C., Voit T., Wright W. E., Aamiri A., Butler-Browne G., Mouly V., Immortalized pathological human myoblasts: Towards a universal tool for the study of neuromuscular disorders. Skelet. Muscle 1, 34 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golov A. K., Ulianov S. V., Luzhin A. V., Kalabusheva E. P., Kantidze O. L., Flyamer I. M., Razin S. V., Gavrilov A. A., C-TALE, a new cost-effective method for targeted enrichment of Hi-C/3C-seq libraries. Methods 170, 48–60 (2020). [DOI] [PubMed] [Google Scholar]
- 39.H. Patel, P. Ewels, A. Peltzer, J. Manning, O. Botvinnik, G. Sturm, M. U. Garcia, D. Moreno, P. Vemuri, nf-core bot, M. Binzer-Panchal, silviamorins, L. Pantano, M. Zepper, R. Syme, A. Talbot, G. Kelly, F. Hanssen, J. A. F. Yates, J. Espinosa-Carrasco, rfenouil, C. Cheshire, marchoeppner, E. Miller, P. Zhou, S. Guinchard, G. Gabernet, C. Mertes, D. Straub, P. D. Tommaso, nf-core/rnaseq: nf-core/rnaseq v3.14.0 - Hassium Honey Badger, version 3.14.0 (Zenodo, 2024); 10.5281/zenodo.10471647. [DOI]
- 40.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.G. Korotkevich, V. Sukhov, N. Budin, B. Shpak, M. N. Artyomov, A. Sergushichev, Fast gene set enrichment analysis. bioRxiv 060012 [Preprint]. 01 February 2021. 10.1101/060012. [DOI]
- 44.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J. P., Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2
Figs. S1 to S4