Abstract
DNA double-strand breaks (DSBs) are one of the most genotoxic DNA lesions, driving a range of pathological defects from cancers to immunodeficiencies. To combat genomic instability caused by DSBs, evolution has outfitted cells with an intricate protein network dedicated to the rapid and accurate repair of these lesions. Pioneering studies have identified and characterized many crucial repair factors in this network, while the advent of genome manipulation tools like CRISPR-Cas9 has reinvigorated interest in DSB repair mechanisms. This review surveys the latest methodological advances and biological insights gained by utilizing Cas9 as a precise ‘damage inducer’ for the study of DSB repair. We highlight rapidly inducible Cas9 systems that enable synchronized and efficient break induction. When combined with sequencing and genome-specific imaging approaches, inducible Cas9 systems greatly expand our capability to spatiotemporally characterize cellular responses to DSB at specific genomic coordinates, providing mechanistic insights that were previously unobtainable.
Keywords: DNA double-strand break, DSB repair, chromatin, CRISPR-Cas9, very fast CRISPR, genomic imaging
In human cells, genomic DNA spontaneously accumulates lesions as a result of replication stress, endogenous metabolites, and environmental carcinogens [1,2]. Failure to quickly repair DNA lesions can lead to permanent genomic mutation, disrupted cellular function, and ultimately cause physiological dysregulation and disease. To prevent the accumulation of DNA damage and mutations, cells have evolved a dynamic network of proteins to detect, signal, and repair various genomic lesions, collectively termed the DNA damage response (DDR). This is a truly formidable task as each cell contains over six billion DNA base pairs and experience tens of thousands of DNA lesions that must be faithfully repaired on a daily basis.
Of the DNA damage types, double-strand breaks (DSBs) are the most potent. DSBs occur when both DNA backbones are ruptured to expose two free DNA ends. Unprotected ends are at high risk of degradation and must be quickly repaired to avoid mutation and other forms of chromosome instability. Pioneering biochemical and genetic studies have identified a network of DDR factors that sense broken DNA ends, initiate a cascade of biochemical signaling, and recruit DNA end processing and ligation machinery (a summary of DSB repair has been provided in Textbox 1). The main DSB repair pathways include nonhomologous end joining (NHEJ), microhomology-mediated end joining (MMEJ), single-strand annealing (SSA), and homologous recombination (HR). While these pathways are often intertwined, each pathway comprises unique repair factors that are recruited to distinct DNA end structures and drive characteristic repair outcomes [3].
Text Box 1 – DNA Double Strand Break Repair Biology.
DNA double strand breaks (DSBs) occur when both DNA backbones are physically ruptured, exposing free DNA ends. DSBs are particularly dangerous due to the potential for genetic information loss in the form of insertions and deletions (indels) or chromosomal translocations and fusions. Efficient DSB repair paramount, as cells with impaired DSB repair networks often show increased mutation rate and genome instability which drive cancer and disease. Several pathways are responsible DSB repair, including the conventionally “error-prone” classical nonhomologous end-joining (cNHEJ), conventionally “high-fidelity” homology directed repair (HDR), alternative NHEJ (alt-NHEJ), microhomology-mediated end joining (MMEJ), and single-strand annealing (SSA) [2]. Upon DSB induction, DDR proteins rapidly translocate to the break, process the break ends, fill in single strand regions, and ligate. The main DSB sensors are PARP1, MRN, and Ku70/80 which recognize free DNA ends and trigger a signal cascade upon binding. Epigenetic spreading of phosphorylation, ubiquitin, sumo, methyl, and acetyl modifications are propagated by a variety of kinases (ATM, ATR, DNA-PKcs), ubiquitin ligases (RNF8/168), and histone remodelers [75]. cNHEJ directly ligates double strand break ends with minimal end processing using a combination of XLF, PAXX, MRI, TDP-43, FOXL2, IFFO1, RNasH2, Artemis, DNA pol λ/μ, and the LigIV-XRCC4 complex [76]. cNHEJ often produces faithful repair or small insertions. HDR is a resection dependent repair pathway that recruits MRN, MDC1, RPA, Rad51, Exo1-BLM, CtIP, BRCA1, PALB2, and BRCA2 to facilitate resection of damaged DNA and invasion of the newly formed single strand DNA into a template sequence, often the sister chromatid present during DNA replication. The decision between resection-dependent HDR and resection-independent NHEJ is a topic of intense study and is cell cycle dependent with resection potentially being spatially controlled around the break [60]. Alt-NHEJ, which often overlaps with MMEJ, relies on 2–20 nucleotide microhomologies which facilitate pairing of two broken ends which are then filled in by polӨ and ligated by XRCC1 and Ligase I/III [77]. Cellular processes like replication, meiosis, V(D)J recombination, topoisomerase relaxation, and nucleotide excision repair have the potential to generate DSBs making DSBs less rare than may be anticipated [78]. Finally, in the event of prolonged damage signaling, incomplete repair, or numerous breaks, signaling kinases p21, p53, and CDK2 are activated to drive cell cycle arrest or apoptotic cell death.
Several factors like local sequence context, epigenetic modifications, and chromatin structure play critical roles in modulating DSB repair [4]. For example, accessible euchromatin regions recruit DDR kinases within seconds that rapidly propagate histone variants and post-translational modifications across megabase pairs to create a productive repair environment [5]. Conversely, efficient repair of inaccessible heterochromatic DSBs requires serial unpacking and translocation steps and occurs more slowly [6,7]. A comprehensive understanding of the dynamic interplay between these structural, biochemical, and sequence features during repair requires technologies that can induce precise DSBs at defined loci and chromatin regions in living cells.
CRISPR-Cas9 as a programmable DSB inducer
The use of physical irradiation, chemical mutagens, and chimeric nucleases as exogenous DSB inducers have contributed significantly to our knowledge of DSB repair, and each technique has strengths and weaknesses (details provided in Textbox 2). For example, chemical mutagens are simple to implement and induce DSB with high efficiency but produce damage at genetically undefined locations, making it impossible to study DDR on predetermined genomic targets without causing excessive damage to the cell. Until recently, existing methods were unable to induce exclusively DSBs at specific targets with high synchronicity and efficiency. Originally discovered as a component of bacterial adaptive immunity, CRISPR systems have driven a broader interest in DNA repair by providing a convenient platform to induce genomic DSBs[8].
Text Box 2 – Existing Systems for Targeted DSB Induction.
The main methods to induce DSB in living cells include irradiation, chemotherapy, restriction endonucleases, and chimeric nucleases. Irradiation (IR) uses high energy particles to create DNA lesions within milliseconds of illumination. IR forms a range of damage including base oxidation, single-strand breaks, inter-strand crosslinks, and double strand breaks at undefined genomic locations [79]. Micro-irradiation refers to the use of focused laser light (337 – 405 nm) in conjunction with a sensitizing agent to induce damage in a subnuclear region. The combination of micro-irradiation and fluorescence microscopy allows for visualizing DDR protein dynamics with millisecond time resolution in live cells. Micro-irradiation generates many breaks within a confined region therefore amplifying the DNA damage signal, making it a compelling method to study proteins that are recruited in low copy numbers like Ku70/80 [80]. Chemotherapeutics like topoisomerase inhibitors and platinum crosslinkers create DSB in the form of DNA-protein or DNA-DNA covalent crosslinks. Adducts cause replication fork stalling and DSB formation during DNA replication. The Haber and Jasin labs pioneered the use of meganucleases, HO and I-SceI, to generate sequence specific DSBs [80]. These restriction enzyme-like meganucleases recognize 12–40 bp DNA motifs and create double-stranded DNA breaks (DSBs) upon binding [69,81]. The meganuclease DNA recognition motif is orthogonal to the mammalian genome which necessitates the insertion of an exogenous target for use in cells and precludes the study of endogenous sites. Importantly, these and other meganucleases (I-PpoI, and I-CreI)[82] have made it possible to incorporate ChIP-qPCR and next generation sequencing (NGS) in DSB studies to measure protein recruitment at specific genomic locations with high sequence resolution [69,81]. As an alternative to meganucleases, Type II restriction enzymes like AsiSI have been adopted to cleave tens to hundreds of endogenous DNA sites for multiplexed experiments compatible with NGS [79]. Chimeric nucleases - zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) - were engineered as programable nucleases for DSB induction. These chimeric nucleases consist of promiscuous DNA cleavage domains (Fok I) and DNA binding motifs with sequence specificity achieved through major/minor groove interactions. ZNF and TALEN are the most widely used programmable nucleases before the advent of CRISPR-Cas9 system and have enabled many critical studies of DNA repair. However, it is challenging to apply these methods genome wide as non-trivial protein engineering is required to reprogram ZNF and TALEN against new genomic targets. The FokI/Lac system is also used to target engineered sites in the genome, augmenting the signal of recruited DSB repair factors for fluorescence visualization [83].
Streptococcus pyogenes CRISPR associated protein 9 (Cas9) is an endonuclease evolved to recognize and cleave foreign viral DNA elements [8] (details provided in Textbox 3). Cas9 in complex with a guide RNA (gRNA) can hybridize with specific genome targets and cleave double-stranded DNA (figure 1A). Reprogramming the Cas9 gRNA is simpler and less expensive than reengineering the sequence recognition domains of chimeric nucleases like zinc finger nucleases (ZFNs) and TAL effector nucleases (TALENs). As a result, Cas9 can be easily programmed against predetermined targets at nearly any site in the genome for highly specific and efficient break induction [9–11]. Programming gRNA is also scalable, leading to the invention of high-throughput CRISPR screens that interrogate the function of tens of thousands of genes in one experiment [12–14].
Textbox 3 – Cas9 Discovery and Mechanism of Action.
CRISPR associated protein 9 (Cas9) was first demonstrated to be a powerful tool for creating targeted DNA double strand breaks in vitro and in cells [8,9]. The bacterial Cas9 nuclease is associated with a clustered regularly interspersed short palindromic repeats (CRISPR) DNA array, a CRISPR associated RNA (crRNA), and trans-activating crRNA (tracrRNA). The crRNA/tracrRNA have also been redesigned as a single guide RNA (sgRNA) for genome editing and other molecular biology applications [8]. Cas9 contains two nuclease domains, the HNH and RuvC domains, which cleave the target and nontarget sequences respectively. This cleavage activity requires 20 nucleotides of complementarity at the 5’ end of the crRNA, called the protospacer, to hybridize with the DNA target. Cas9 unwinds its target sequentially, first binding a protospacer adjacent motif (PAM) then engaging a 10-nucleotide seed region in the PAM proximal end of the protospacer ultimately followed by unwinding of the PAM distal region. When 17 – 20 nucleotides have base paired, the HNH and RuvC domains are positioned at the third nucleotide position on the target DNA on both the target and nontarget strands. Cas9 mutants have also been developed to improve the specificity of Cas9 binding and cleavage [84,85]. To deliver Cas9 for gene editing and repair studies, early work employed plasmid or viral transfection to express Cas9 and gRNA [9]. These strategies are well suited for measuring terminal insertion/deletion (indel) products but provide limited control over the timing of Cas9 damage induction. Another important consideration when using Cas9 for DSB repair studies is that Cas9 may intrinsically biases repair pathway choice in ways that do not mirror endogenous DSBs. Like other enzymatic nucleases, Cas9 primarily creates clean DNA duplex breaks whereas cellular DSBs often result from DNA replication and produces ‘dirty’ breaks containing DNA-protein adducts or other incompatible end structures. The residency time of Cas9 may additionally influence repair pathway choice, since a 3’ flap is exposed prior to Cas9 dissociation [86]. Cas9 slowly dissociates from cleaved DNA, a process that may be facilitated in cells, at least in part, by histone chaperone complex FACT [87]. Finally, there is also evidence that Cas9 can trim the nontarget strand due to conformational flexibility in the RuvC domain leading to predominantly single strand overhangs for a subset of targets in a sequence dependent manner [88].
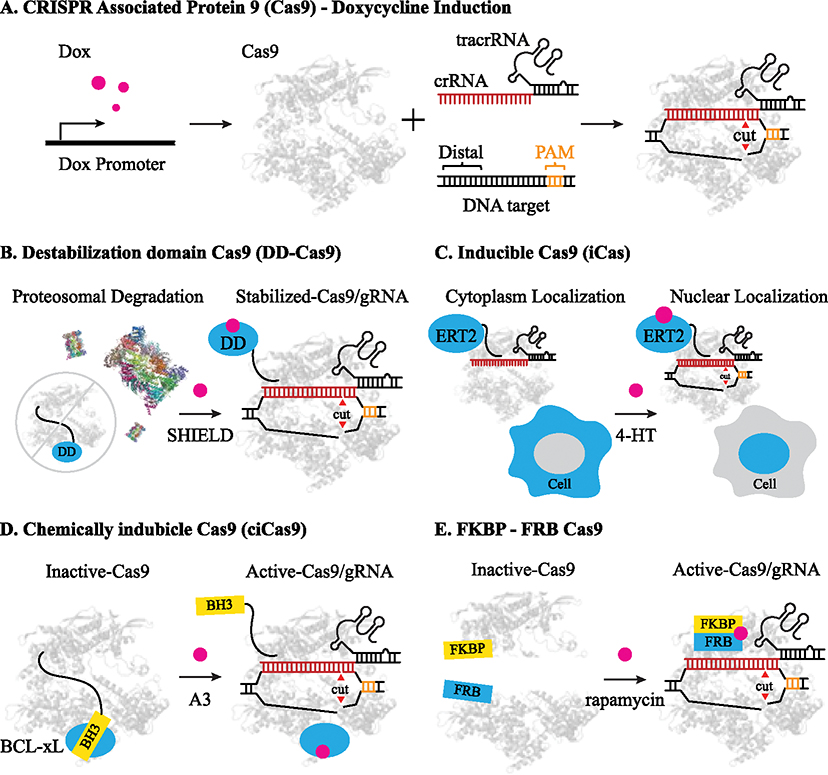
Figure 1. Chemically Inducible Cas9 Cleavage Systems.
A) CRISPR-Cas9 expression depicted driven by a doxycycline promoter. Cas9 forms a cleavage competent complex with its crRNA and tracrRNA (or sgRNA). B) DD-Cas9: proteosome depicted in multicolor. C) iCas9: cellular localization of iCas9 shown in cyan in response to 4-hydroxytamoxiphen (4-HT). D) ciCas9 depicted with BH3 domain in yellow blocking gRNA loading until the small molecule A3 is introduced for activation. E) FKBP-FRB Cas9: dimerization domains shown in yellow and cyan are independent until the introduction of the small molecule rapamycin which promotes dimerization and activates the complex.
For these reasons, CRISPR-Cas9 based genome editors have spearheaded a gene editing revolution that has driven innovation in areas from basic biology to biotechnology and medicine [15]. Many investigations have since been conducted to study the biology of Cas9-mediated DSB repair, several of which are discussed in the following sections. In this review, we summarize technical advances in CRISPR-Cas9 systems that have allowed for improved spatial and temporal control of DSB induction. We then highlight the novel insights revealed by implementing Cas9 and activatable or deactivatable Cas9 variants to study DSB repair. Looking forward, inducible CRISPR-Cas9 technologies are beginning to show strong synergy with high-resolution imaging and sequencing methods which we anticipate will further refine our understanding of DSB repair dynamics in the context of DSB biology and gene editing applications.
Advances in Inducible CRISPR-Cas9 systems
Inducible Cas9s allow for spatiotemporal control of Cas9 cleavage activity either by transcriptional control or post-transcriptional activation. Current inducible systems mainly include chemically and optically inducible CRISPR-Cas9s [16], although local genome editing has also been demonstrated using magnetic particles [17]. Doxycycline was one of the first chemically inducible systems used to transcriptionally induce CRISPR-Cas9 expression and was successfully used in early CRISPR screen experiments to study molecular components of DSB repair [13]. However, since transcriptional activation with doxycycline or similar strategies can take several hours to reach full expression, it is difficult to induce DSB rapidly and capture early DSB repair events in a synchronized way. In this review, we focus on tools to study early DSB repair dynamics and only address post-translational CRISPR activation. From the standpoint of studying the dynamics of DSB repair, we argue that the cleavage kinetics of an inducible system should ideally match or exceed the kinetics of DSB repair-on the order of minutes-which has not been possible using transcriptionally controlled CRISPR-Cas9 systems.
Chemically inducible Cas9 systems
Chemically inducible CRISPR-Cas9 systems rely broadly on controlled localization (DD-Cas9, iCas), blocked gRNA loading (ciCas9), or chemical dimerization (FKBP-FRB, Fig. 1B–E). Destabilization domain Cas9 (DD-Cas9) uses a Cas9-FKBP12 fusion to localize Cas9 to the proteasome for degradation, preventing DNA cleavage in the absence of treatment. A small molecule, SHIELD-1, is introduced to restrain the DD domain from localizing to the proteasome, which prevents degradation of the Cas9 complex and promotes cleavage [18]. A similar design, iCas, relies on a Cas9 variant fused with the hormone-binding domain of the estrogen receptor 2 (ERT2) [19]. Without 4-hydroxytamoxifen (4-HT), iCas localizes to the cytoplasm; with 4-HT, iCas translocates to the nucleus to promote genomic DNA cleavage. Both DD-Cas9 and iCas allow for controllable knockdown and editing of essential genes, functioning on a time scale of several hours [20]. An additional inducible system driven by 4-HT was reported using a self-cleaving intein domain with similar induction kinetics of 4–8 hours [21].
In 2017, a chemically inducible (ci)Cas9 was reported for rapid DNA cleavage and genome editing (Figure 1D) [22]. By replacing the Cas9 REC2 domain with a BCL domain and adding a BH3 domain to the C-terminus, the BCL/BH3 domains dimerize to sterically block gRNA association. A small molecule, A-3, disrupts the BCL/BH3 interaction to allow gRNA loading and activate the ciCas9. Compared with other chemically inducible Cas9s [16], ciCas9 demonstrated much faster cleavage kinetics, forming detectable DSBs within 10 minutes and indels within 2 hours. However, ciCas9 showed a 50% reduction in editing activity compared to wild-type Cas9 and relatively high basal activity, likely due to spontaneous dissociation of the BCL and BH3 domains. Although engineering higher affinity BH3 variants may improve this system, replacing large protein domains will likely require substantial optimization to reach the cleavage efficiency of the wild-type Cas9 nuclease.
Chemically induced dimerization has also been used to control Cas9 activation. FK506 binding protein 12 (FKBP) and FKBP rapamycin binding domain (FRB) are well-studied domains that associate in the presence of the small molecule rapamycin (Figure 1E). Using a split Cas9 FKBP/FRB design, researchers showed rapamycin-dependent indel formation after 12 days of rapamycin treatment but did not report the activation kinetics at earlier time points [23]. Catalytically dead Cas9 (dCas9) dimerization strategies have been proposed using orthogonal chemically inducible domains to modulate gene expression, with no cleavage-competent versions proposed [24].
Light inducible Cas9 systems for controlled cleavage
Light-inducible CRISPR-Cas9 systems have also been developed, mainly relying on light-dependent conformational changes, dimerization control, or chemical uncaging (Figure 2). In 2015, Hemphill et al reported the first light inducible Cas9 design by introducing a photocaged lysine into Cas9 during translation [25]. Placement of the caged lysine residue was chosen based on proximity to the gRNA binding pocket and was assumed to prevent Cas9-gRNA interaction in the absence of light. After UV irradiation, the photo-uncaged Cas9 produced a 60% reduction in mRNA and a 50% reduction in protein expression showing light controlled Cas9 gene editing for the first time. While genetically encodable, this design required unnatural amino acids to incorporate the photocaged lysine and showed reduced overall cleavage efficiency compared to the wild-type Cas9 nuclease.
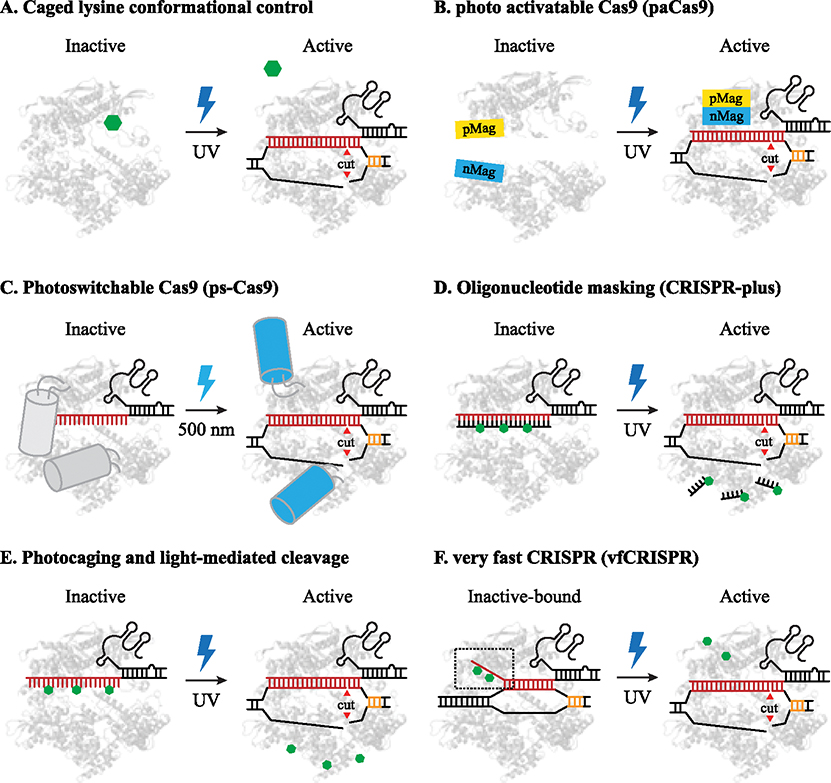
Figure 2. Light Inducible Cas9 Cleavage Systems.
Green hexagons represent photosensitive groups for light activation (Photocaged lysine: 2A, Photocleavable groups: 2C, 6-nitropiperonyloxymethyl modification (NPOM): 2E-F). A) Caged lysine (K866) prevents Cas9 activity until photouncaging. B) Photo activatable Cas9: dimerization domains shown in yellow and cyan are independent until UV stimulation which promotes dimerization and activates the complex. C) Photoswitchable-Cas9: Grey/cyan cylinders represent Dronpa domains in response to 500 nm light. D) CRISPR-plus: oligomask ‘protector’ photolyzed after UV stimulation to activate RNP binding and cleavage. E) Photocaged gRNAs block Cas9 binding before UV stimulation. F) vfCRISPR is prebound before UV stimulation and cleaves within seconds of UV photolysis of caged groups.
In the same year, Nihongaki et al reported a genetically encoded photoactivatable Cas9 (paCas9). paCas9 consists of a pair of split Cas9 domains fused to either pMag or nMag domains (Figure 1E) that dimerize under 470 nm light stimulation. paCas9 produces minimal basal indel formation in the absence of light and induces detectable indel after 9 hours of light exposure as measured by the T7E1 assay [26]. Optical control has also been achieved using a photo-dissociable dimeric protein domain, termed photoswitchable Cas9 (ps-Cas9) [27]. Although this method showed minimal basal Cas9 activity, the overall indel frequency and gene-editing efficiency of ps-Cas9 was 38% compared to wild-type Cas9, even after 24h light exposure. These results speak to the challenges of maintaining wild-type cleavage efficiency when introducing or modifying large domains of the Cas9 nuclease.
Several light-inducible Cas9 systems were recently reported using photolabile DNA or RNA oligonucleotides. One system, CRISPR-plus, uses a photocleavable ssDNA to mask gRNA and thus Cas9 activity (Figure 1F). Light stimulation triggered highly efficient cleavage of ssDNA, unleashing Cas9’s DNA cleavage in vitro and in situ [28]. Three different systems were reported in quick succession utilizing NPOM-masked gRNAs to control gene editing. The first two systems evenly distributed NPOM-modified bases along the gRNA protospacer using caged uridine and guanosine nucleotides [29,30]. These methods showed spatially controlled activation in mammalian cells and zebrafish embryos with as high as 26.2% indel formation frequency after 365 or 405 nm light activation and minimal (0.9%) editing in the absence of light [29].
In contrast, very fast CRISPR (vfCRISPR) uses NPOM modifications specifically in the PAM distal region of the gRNA protospacer, allowing the complex to bind but not cleave its DNA target before light stimulation [31–33]. Upon UV stimulation, the caging groups are rapidly removed, and the prebound vfCRISPR complex is efficiently converted to a cleavage-competent state and cleaves target DNA. vfCRISPR showed negligible basal activity and high light-dependent editing efficiency of over 90%, comparable to wild-type Cas9. vfCRISPR can also create DSBs in a subnuclear volume, like micro-irradiation. For the first time, Liu et al activated a subpopulation of vfCRISPR molecules for single allele DNA cleavage using focused laser light. Despite high efficiency and compatibility with the wild-type Cas9 nuclease, all photoactivatable oligos demand chemical synthesis and delivery into cells via lipofectamine-based transfection or electroporation.
Inducible Cas9 systems for controlled deactivation
Another critical axis for Cas9 control is the ability to deactivate its catalytic activity. Cas9 remains catalytically active in gene editing applications long after its nuclear delivery or conditional stimulation. After an accurate repair event, Cas9 is expected to cut its target repeatedly until mutations are generated (Figure 3A). Indeed, cyclic cleavage and repair have been observed using Cas9 and similar type II restriction enzymes in living cells [33–35]. Cas9 can even re-cleave single-base insertion products at some endogenous loci in living mammalian cells [33], which is supported by in vitro work showing that Cas9 can cleave near-perfect target DNA with small bulges [36]. Cyclic Cas9 cleavage poses a problem for kinetic analysis as only the first repair cycle is temporally defined, and subsequent cycles are asynchronous with respect to the onset of DNA repair. Therefore, devising a fast and complete mechanism to inactivate Cas9 is highly desirable.
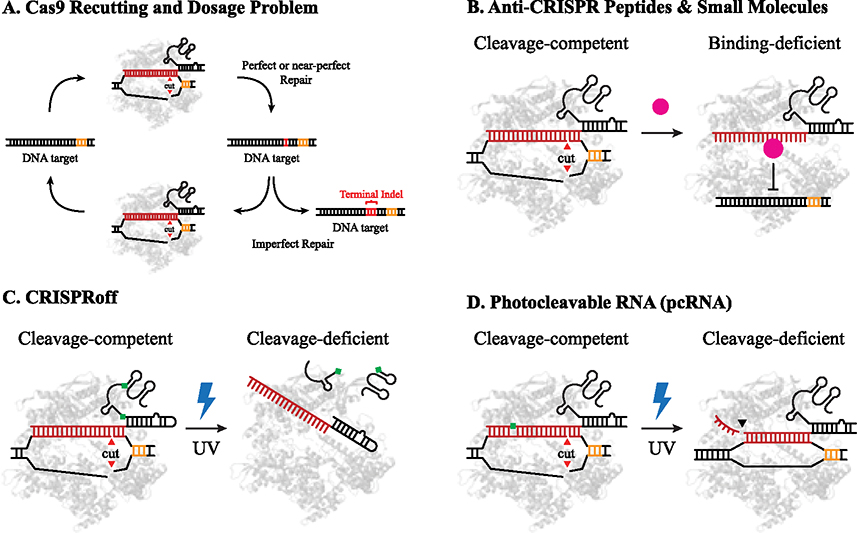
Figure 3. Inducible Cas9 deactivation strategies.
A) Enzymatic recutting and dosage problem schematic: faithfully repaired DNA and common repair products can be targeted repeatedly by sequence-specific nucleases until a terminal indel prevents the nuclease from recognizing the target. When exposed to active Cas9 for extended periods, off-targets that closely resemble the on-target are also more likely to be unintentionally cut. B) Anti-CRISPR (Acr) proteins (AcrIIA4, AcrII4, AcrII2, AcrIF9), small molecules (BRD0539), or small nucleic acid-based inhibitors (SNuBs) are used to prevent Cas9 cleavage activity by a variety of binding modes. C) CRISPR-off: o-nitrobenzyl photocleavable linkers are introduced into synthesized sgRNA which are cleaved after UV stimulation to deactivate the RNP. D) Photocleavable RNA (pcRNA): o-nitrobenzyl photocleavable groups are introduced into the protospacer to retain cleavage activity and are cleaved in the presence of UV stimulation to deactivate the RNP.
Several strategies have been developed to inhibit CRISPR-Cas9 activity, including anti-CRISPR (Acr) proteins [37], small-molecule inhibitors [38], and small nucleic acid-based inhibitors (SNuBs, Figure 3B) [39]. Generally, these inhibitors disrupt Cas9 binding to target DNA by occluding the binding pocket or impeding R-loop formation. By timing the delivery of anti-CRISPR AcrIIA4, investigators were able to preferentially reduce off-target editing while largely retaining on target editing [37]. Anti-CRISPR SNuBs were also able to slow Cas9 cleavage kinetics in cell culture, but the kinetics of deactivation were not directly measured [39]. While these methods remain promising for genome editing applications to reduce off target effects, their incomplete inhibition with residual editing between 20–30% complicates kinetic analyses.
Recently, two independent groups have engineered photocleavable gRNAs, CRISPRoff and pcRNA, to inactivate the Cas9 gRNA [40,41]. CRISPRoff incorporates two photocleavable groups in the tracrRNA stem loops where 345 nm light triggers a photochemical reaction that splits and inactivates the sgRNA (Figure 3C). CRISPRoff significantly reduced editing for some but not all genomic targets [40]. pcRNA incorporates photocleavable groups in the crRNA where 365 nm light photochemically cleaves the crRNA to disable Cas9 cleavage (Figure 3D). Strikingly, gene edits by pcRNA Cas9 nuclease and pcRNA base editors were reduced by over 99% after a single exposure to light for 30 seconds, which enabled the synchronized measurement of DSB repair progression and repair.
Understanding DNA Double Strand Break Repair using Cas9 Systems
The ease of reprogramming Cas9 against genomic targets has empowered researchers to precisely probe the molecular mechanisms of DSB repair at various DNA targets, flanking chromatin sequences/modifications, and genetic backgrounds. This section highlights new biological insights gained by implementing Cas9 systems to study templated and non-templated repair, followed by a discussion on the utility of high throughput Cas9 screens to characterize DSB repair networks. Finally, we showcase that combining dynamic genome imaging with targeted Cas9 damage provides a powerful strategy to reveal ultrafine structural features at DSB repair foci.
Repair Insights using wild-type Cas9
Leveraging programmable gRNAs, CRISPR-Cas9 has been deployed at various exogenous and endogenous target sites to interrogate how the surrounding nucleotide composition influences repair outcomes. In yeast, Cas9-mediated NHEJ repair showed frequent templated +1 insertions. This phenotype was attributed to polymerase fill-in of asymmetric 1-nt 5’ overhangs left by Cas9 asymmetric cleavage [42]. In human cells, Cas9-induced repair products were more diverse. Using a large-scale library containing thousands of gRNA-target pairs, researchers observed the frequent formation of short insertions and deletions. Cas9 gene editing outcomes were shown to follow either a predictable or unpredictable set of rules based on the sequence composition surrounding the cut site. At predictable edits, single base insertions or deletions were highly dependent on the fourth nucleotide in the protospacer upstream of the PAM, with insertions preferentially for A/T and deletions for C/G [43,44]. Additionally, flanking microhomology promoted short deletions (< 20nt), a result positively correlated with GC content, homology length, and the proximity of microhomology to broken DNA ends. Deletions over several kilobases and crossover events have also been reported, suggesting that repair may favor large deletions at resection-prone loci or in mitotically active cells [45]. Finally, the epigenetic state of endogenous targets contributes to the repair profile. By expressing gRNAs targeting many genomic targets, Chakrabarti et al found that histone hyperacetylation, an epigenetic mark that promotes open chromatin, increased indel formation efficiency and was inversely correlated with the endogenous levels of histone acetylation, suggesting that higher accessibility promotes indel formation [43].
Cas9-mediated DSB induction has also been widely utilized to facilitate the insertion of exogenous DNA fragments via homology-directed repair (HDR). In HDR, a repair template is supplied to the cell in addition to the Cas9 nuclease, containing homology arms ranging from 35 nucleotides to 900 base pairs [46–49]. The efficiency of HDR is generally low, and it depends heavily on the cell cycle due to the cell-cycle-dependent expression profiles of major HDR-associated proteins [50,51]. By introducing Cas9 during the S phase or mitosis, HDR efficiency was increased from ~5% to ~33% [50,51]. Interestingly, when endogenous 53BP1 is blocked using a Cas9-dominant negative 53BP1 fusion, NHEJ machinery recruitment is repressed leading to a boost in HDR frequency as high as 86% [52]. It was found that HDR can also be accomplished using a Cas9 nickase fused to Rad51, which suggests that homology search can be initiated even in the absence of a DSB as long as Rad51 is present to initiate the formation of the HDR target search complex on ssDNA [53].
The ability to genetically encode pooled Cas9 targets has enabled systematic screening of DSB repair outcomes across various genetic backgrounds. Hussmann et al conducted a CRISPR screen against 476 DNA repair-associated genes where Cas9 cleaves an exogenously introduced reporter while simultaneously knocking down a library of target genes [54]. Their screening method, termed Repair-seq, was able to deconvolute the many repair outcomes as a function of specific genetic dependencies independent of sequence or genomic context. Short insertions of different lengths showed a strong dependence on repair polymerases and suggested that the insertion profile in human cells is decidedly more complex than the yeast model proposed previously [42,54]. Similar studies have been conducted using exogenous fluorescence reporters inserted at a single genomic locus called Color Assay Tracing-Repair (CAT-R) [55,56]. Using this method, Roidos et al observed that small indels were mainly driven by NHEJ, while large deletions (2 – 8kb) were primarily a result of the resection-dependent repair pathways such as single-strand annealing and homologous recombination [55]. They also identified a potential role for nucleotide excision repair (NER) in DSB repair, showing that knockout of NER factors increased error-free repair while reducing both small indels and large deletions.
There is growing interest in determining how DSB repair proteins behave in specific genomic regions and conversely how repair changes the spatial organization of genomic regions. Cas9 has enabled repair studies at specific DNA sequences and subnuclear compartments when combined with fluorescence microscopy. For example, transient expression of Cas9 targeting ribosomal DNA induced a distinct semi-circular structure known as the ‘nucleolar cap’ [57]. This structure formed on the periphery of the nucleolus in an ATM-dependent manner. At telomeric DNA, DSBs triggered increased telomere diffusion and caused the clustering of damage foci [58]. RAD51 depletion significantly suppressed clustering suggesting that clustering is associated with homologous recombination. At centromeric and pericentromeric DNA, damage foci organization varied depending on the cell cycle and repair pathway choice[59]. In pericentromeric DNA, NHEJ was activated in G1 within the pericentromeric center, while homologous recombination was activated in G2 with foci relocated to the periphery. Both pathways were present throughout the cell cycle in the centromere, with NHEJ at the center and homologous recombination at the periphery of the centromeric DNA structures showing a cell-cycle dependent active relocation of repair features.
Additionally, Cas9-induced DNA damage foci have been visualized in relation to local TAD boundaries using super-resolution structured illumination microscopy (SIM) [60]. Ochs et al demonstrated that nanodomains of repair foci labeled with NHEJ marker 53BP1 excluded HR markers and colocalized with underlying topologically associated domains. This suggests that repair protein spreading and pathway choice may be linked to TAD maintenance. Microscopy-based studies like those above highlight the unique structural information gained by combining targeted damage with imaging techniques. We envision the application of other Cas9 imaging techniques like GOLDFISH [61,62], Live-FISH [63], and other Cas9 labeling technologies (reviewed in detail[64]) as attractive alternatives.
Understanding DSB Repair Dynamics using Inducible Cas9 Systems
While wild-type Cas9 has proven a powerful tool to profile DSB repair outcomes like endpoint mutations, chemically and optically inducible Cas9 systems provide the fine temporal control needed to quantitatively study the dynamics of early repair events within the first minutes of repair. This section highlights biological insights gained by implementing inducible Cas9 systems to study cellular responses after DSB, early repair kinetics, and rapid chromatin remodeling after DSB. Finally, we showcase the utility of combining inducible methods to measure physical accessibility changes during and after DSB repair.
Repair insights using chemically inducible Cas9 systems
Chemically inducible Cas9 systems have been used to study cellular responses and chromatin context during DSB repair. Using DD-Cas9, it was observed that a single DSB was sufficient to delay cell cycle progression and that additional breaks had a compounding effect to further delay cell cycle [65]. DD-Cas9 was also combined with a barcoded reporter library to induce DSBs and insertions across thousands of genomic sites, showing that differentially modified chromatin contexts impacted the relative frequency of NHEJ, MMEJ, and SSTR [66]. The reporter library adopted the chromatin marks of genomic location surrounding the insertion, and the authors attributed increased MMEJ at heterochromatin to slower chromatin reorganization around the break. In general, heterochromatin regions showed lower indel frequencies than euchromatic regions. Significant variation existed between heterochromatin regions with lamin-associated domains demonstrating the highest propensity towards MMEJ over NHEJ [66]. It is worth noting that DD-Cas9 expression accumulates gradually at the onset of the experiment with repair products detected on the hour time scale.
Prior studies predominantly infer in vivo Cas9 cleavage activity by measuring indel formation. Rose et al developed a digital droplet PCR assay to directly measure DSBs (DSB-ddPCR) after ciCas9 activation, whereby simultaneously tracking the kinetics of the DSB and indel frequencies becomes feasible. For example, rapid DNA cleavage was observed within 10 minutes at several loci, and their frequencies peaked around 1–2 hours, whereas the earliest indel formation occurred 30 minutes after induction [22]. In a separate study, Brinkman et al measured Cas9 repair kinetics after DD-Cas9 activation and created an analytical model to account for the gradual increase in Cas9 activity [20]. Using this method, they estimated a half-life of DNA cleavage to be approximately 6 hours, matching experimental results. A predominant indel profile of +1 insertion and −7 deletion was observed and were attributed to cNHEJ and MMEJ, respectively. Interestingly, the rate of cNHEJ was much faster than that of MMEJ, and inhibiting DNA-PKcs increased the rate of MMEJ by three-fold, suggesting a kinetic competition between the two pathways.
Kinetic insights into DSB repair using optically inducible Cas9
Light inducible strategies have further improved the temporal control over Cas9 cleavage for detailed kinetic studies of DNA repair. Particularly, vfCRISPR and pcRNA have been used to make high-resolution measurements of the DSB repair process, including break sensing and repair fidelity, recruitment and restoration rates of repair proteins, and structural changes to the chromatin environment.
As the first step following Cas9 cleavage, DNA break sensing is fast, on the order of minutes, but overall repair rates are often reported on the scale of hours [20,22]. It remains unclear if the repair is slow or if broken DNAs are faithfully repaired so that it does not contribute to indel formation. In fact, it is technically challenging to directly measure the rate and extent of faithful DSB repair. Using vfCRISPR and ChIP-seq, the recruitment of MRE11 was detected as early as 2 minutes after a single Cas9-induced DSB, matching closely with irradiation studies [33,67]. The rate and fidelity of DSB repair were measured by examining the Mre11 ChIP-seq reads that span the cut site, where spanning reads represented damaged then ligated DNA. Interestingly, ligated repair products were observed just 15 minutes after damage induction with 10–15% of reads showing NHEJ-mediated insertions [33]. This suggested a relatively high repair accuracy with fast kinetics on the minute timescale. Therefore, in genome editing applications, Cas9 nucleases must likely cleave the target DNA multiple times to produce NHEJ-mediated indels.
After damage sensing, the break site is epigenetically modified to promote the recruitment of additional repair proteins. Damage sensing is initiated by a handful of protein kinases that sense DSB ends and propagate phosphorylated histone variant H2AX (γH2AX), a well-studied DSB signal, over megabases of chromatin around the site of damage [68]. The mechanism of γH2AX spreading is a topic of ongoing study, with yeast studies suggesting that ATM, the kinases responsible for H2AX modification, undergoes directed one-dimensional diffusion along DNA [69]. By inducing synchronized DSBs using vfCRISPR, γH2AX propagation was first measured in mammalian cells with a rate of 2 kb/sec bidirectionally. This rate is similar to that of cohesin-mediated DNA loop extrusion measured in vitro [70,71] and was consistent with rates reported by Arnould et al showing that cohesin reorganizes damaged chromatin in an ATM-dependent manner after enzymatic DSB induction [5].
Restoring the chromatin environment after DSB repair is critical to maintaining epigenetic integrity and resuming normal transcriptional activity. Irradiation has been used to monitor repair restoration but creates complex DNA lesions [67]. For specific DSB lesions, residual activity and slow kinetics of other induction methods have made kinetic studies of DSBs difficult. pcRNA is a valuable tool for monitoring the removal of repair factors after light induced Cas9 deactivation because of its high deactivation efficiency and fast kinetics [41]. For example, Mre11 departure was quantified with a half-life of 42–50 min, comparable with the ~33 min half-life observed for Rad50, another component alongside Mre11 in the MRN complex [41,67]. Across hundreds of sites, pcRNA showed a 75% reduction in MRE11 signal within the first 15 min, likely corresponding to completion of DNA repair [72].
Finally, the physical accessibility of DNA surrounding a DSB is modified after damage. Micro-irradiation assays show that damaged chromatin de-compacts within minutes at the micron scale [73,74]. To observe accessibility changes at individual damage sites, multitargeting vfCRISPR and pcRNA were used in conjunction with ATAC-seq to measure chromatin accessibility across hundreds of endogenous DSB sites [72]. Mre11 recruitment was quickly detected within 2 min while ATAC-seq measurements showed increased accessibility within 500bp surrounding the cleavage site after 30 min, suggesting that DSB break sensing preceded local chromatin accessibility changes. Because Cas9-generated DSB are likely repaired within as early as 15 minutes, the slower ATAC signal may suggest that chromatin accessibility increases occur only when the DSB is not quickly repaired. On average, chromatin accessibility is restored within 1h after terminating Cas9 cleavage activity, as measured using pcRNA and ATAC-seq. These findings represent a disconnect between the reported micron-scale chromatin decompaction seen after micro-irradiation [73,74] and the nanoscale accessibility changes within hundreds of base pairs at single cuts. This result also suggests that local epigenetic integrity is quickly restored after DSB repair via an unknown mechanism that is likely distinct from larger-scale changes in chromatin structure [72].
Concluding Remarks and Future Directions
DNA repair is critical for maintaining genomic integrity. The emerging picture of a dynamic 3D genome provides important context for the DSB repair process and enforces the need for DSB induction tools with high spatiotemporal control. Cas9 and newly designed inducible Cas9 systems provide both the temporal and spatial control to enable quantitative studies of DSB repair dynamics. By combining the kinetic control of inducible Cas9 systems with advanced microscopy and sequencing methods, it is now possible to measure single DNA repair events on a spatiotemporal scale that was previously unachievable. The combination of advanced light controllable multi-target gRNAs has already begun to unravel the temporal complexity of repair while simultaneously deconvoluting the role of epigenetic and chromatin reorganization events that drive DSB repair.
We envision the use of light controllable Cas9 systems in conjunction with advanced sequencing methods like high throughout Chromatin Conformation Capture (Hi-C), ChIP, or single-cell sequencing and imaging methods like STED or STORM to characterize the nanostructures that scaffold DNA repair. It is also possible to optically deconvolute chromatin context information in single cells using DNA FISH, GOLDFISH, barcoded sequential FISH techniques like multiplexed error-robust FISH (MERFISH), or single particle tracking. Together, these methods have the exciting potential to enable spatiotemporal measurements within living cells at near-single molecule resolution and to characterize physical processes like phase separated compartment formation or chromatin diffusion after DNA damage. Combining inducible Cas9 systems with genetic, sequencing, and imaging technologies will continue to inform on the structural and epigenetic dynamics of DSB repair to refine our understanding of physiological DNA maintenance and gene editing from the molecular to the organismal scale.
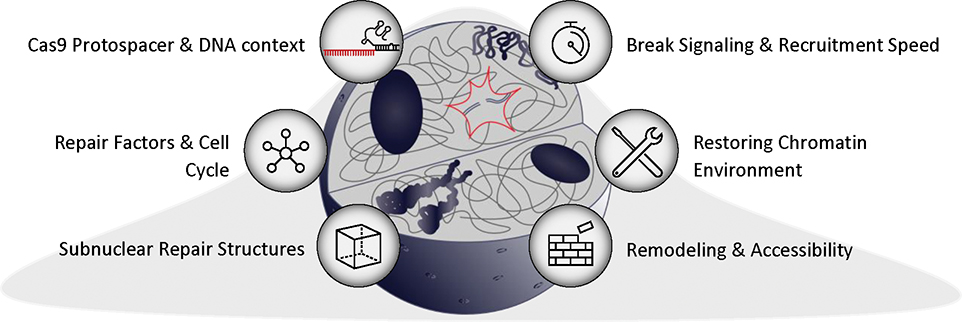
Figure 4.
Insights into the biological factors driving repair of Cas9-mediated DSBs.
Table 1.
Methods for establishing DNA double-stranded breaks (DSB)
| DNA Damage | Target(s) | Synchronization | Efficiency | |
|---|---|---|---|---|
| Physical Irradiation | Mixed DNA Lesions | Non-specific | High (seconds) | High |
| Chemical Mutagen | Mixed DNA Lesions | Non-specific | Mid (min – hr) | Med - High |
| Meganuclease | Double-strand break | Specific but limited | Low (>0.5 – 1h) | Med - High |
| Chimeric Nucleases | Double-strand break | Specific and numerous | Low (>0.5 – 1hr) | Med – High |
| CRISPR Cas9 | Double-strand break | Specific and numerous | Mid (min – hr) | High |
Table 2.
Comparison of Inducible Cas9 Strategies
| Strategy | Stimulator | Induction Speed | Activation Area | Efficiency | Ref. | |
|---|---|---|---|---|---|---|
| Chemically Inducible Systems | Dox-Inducible | Doxycycline | Hours to days | N/A | High | 2015 Nat Biotech |
| FKBP-FRB Dimerization | Rapamycin | Hours | N/A | Moderate | 2015 Nat Biotech | |
| Estrogen Receptor-fused Cas9 | 4-hydroxytamoxifen | Minutes to hours | N/A | Low | 2016 Nat Chem Biol | |
| Autoinhibitory Cas9 | A-385358 | Minutes to hours | N/A | Low | 2017 Nat Methods | |
| Degron-fused Cas9 | SHIELD1 | Hours | N/A | High | 2017 Nat Comm | |
| Optically Inducible Systems | Photo-inducible dimerization | Light | Hours | 2 mm | Low | 2015 Nat Biotech |
| Caged Cas9 | Light | Hours | 5 mm | High | 2015 JACS | |
| Photo-inducible RNA | Light | Seconds to hours | not shown 1 mm 0.2 mm 1 um |
High | 2016 Angew Chem 2020 Angew Chem 2020 ACS Cent. Sci. 2020 Science |
Acknowledgements
This work was supported by grants from the National Institutes of Health (R35 GM 122569 and U01 DK 127432 to T.H.), and the National Science Foundation (PHY 1430124 to T.H.). Y.L. was supported by the Cottrell Postdoctoral Fellowship from Research Corporation for Science Advancement. W.T.C. was supported by the NIH training grant T32 GM007445 and an NSF GRFP Fellowship Award DGE2139757. T.H. is an investigator of the Howard Hughes Medical Institute. We thank all the scientists whose results were discussed in this review.
References
- 1.Jackson SP and Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polo SE and Jackson SP Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. DOI: 10.1101/gad.2021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serrano-Benítez A et al. (2020) “An End to a Means”: How DNA-End Structure Shapes the Double-Strand Break Repair Process. Front Mol Biosci 6, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.House NCM et al. (2014) Chromatin modifications and DNA repair: Beyond double-strand breaks. Front Genet 5, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnould C et al. (2021) Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature 590, 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryu T et al. (2015) Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol 17, 1401–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caridi CP et al. (2018) Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (1979) 337, 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M et al. (2013) RNA-programmed genome editing in human cells. Elife 2013, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P et al. (2013) RNA-guided human genome engineering via Cas9. Science 339, 823–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalem O et al. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science (1979) 343, 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T et al. (2014) Genetic screens in human cells using the CRISPR-Cas9 system. Science (1979) 343, 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koike-Yusa H et al. (2013) Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature Biotechnology 2013 32:3 32, 267–273 [DOI] [PubMed] [Google Scholar]
- 15.Anzalone A. v. et al. (2020) Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38, 824–844 [DOI] [PubMed] [Google Scholar]
- 16.Gangopadhyay SA et al. (2019) Precision Control of CRISPR-Cas9 Using Small Molecules and Light. Biochemistry 58, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu H et al. (2019) Spatial control of in vivo CRISPR–Cas9 genome editing via nanomagnets. Nat Biomed Eng 3, 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senturk S et al. (2017) Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat Commun 8, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu KI et al. (2016) A chemical-inducible CRISPR-Cas9 system for rapid control of genome editing. Nat Chem Biol 12, 980–987 [DOI] [PubMed] [Google Scholar]
- 20.Brinkman EK et al. (2018) Kinetics and Fidelity of the Repair of Cas9-Induced Double-Strand DNA Breaks. Mol Cell 70, 801–813.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis KM et al. (2015) Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol 11, 316–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rose JC et al. (2017) Rapidly inducible Cas9 and DSB-ddPCR to probe editing kinetics. Nat Methods 14, 891–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zetsche B et al. (2015) A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol 33, 139–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y et al. (2016) Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat Methods 13, 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemphill J et al. (2015) Optical control of CRISPR/Cas9 gene editing. J Am Chem Soc 137, 5642–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nihongaki Y et al. (2015) Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol 33, 755–760 [DOI] [PubMed] [Google Scholar]
- 27.Zhou XX et al. (2018) A Single-Chain Photoswitchable CRISPR-Cas9 Architecture for Light-Inducible Gene Editing and Transcription. ACS Chem Biol 13, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain PK et al. (2016) Development of Light-Activated CRISPR Using Guide RNAs with Photocleavable Protectors. Angewandte Chemie - International Edition 55, 12440–12444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou W et al. (2020) Spatiotemporal Control of CRISPR/Cas9 Function in Cells and Zebrafish using Light-Activated Guide RNA. Angewandte Chemie - International Edition 59, 8998–9003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moroz-Omori E. v. et al. (2020) Photoswitchable gRNAs for Spatiotemporally Controlled CRISPR-Cas-Based Genomic Regulation. ACS Cent Sci 6, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sternberg SH et al. (2015) Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527, 110–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh D et al. (2018) Mechanisms of improved specificity of engineered Cas9s revealed by single-molecule FRET analysis. Nat Struct Mol Biol 25, 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y et al. (2020) Very fast CRISPR on demand. Science (1979) 368, 1203C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H et al. (2019) CRISPR-mediated live imaging of genome editing and transcription. Biotechnology 365, 2–6 [DOI] [PubMed] [Google Scholar]
- 35.Aymard F et al. (2014) Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 21, 366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y et al. (2014) CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res 42, 7473–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin J et al. (2017) Disabling Cas9 by an anti-CRISPR DNA mimic. Sci Adv DOI: 10.1126/sciadv.1701620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maji B et al. (2019) A High-Throughput Platform to Identify Small-Molecule Inhibitors of CRISPR-Cas9. Cell 177, 1067–1079.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barkau CL et al. (2019) Rationally Designed Anti-CRISPR Nucleic Acid Inhibitors of CRISPR-Cas9. Nucleic Acid Ther 29, 136–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carlson-Stevermer J et al. (2020) CRISPRoff enables spatio-temporal control of CRISPR editing. Nat Commun 11, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou RS et al. (2021) Cas9 deactivation with photocleavable guide RNAs. Mol Cell 81, 1553–1565.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lemos BR et al. (2018) CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc Natl Acad Sci U S A 115, E2010–E2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakrabarti AM et al. (2019) Target-Specific Precision of CRISPR-Mediated Genome Editing. Mol Cell 73, 699–713.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Overbeek M et al. (2016) DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol Cell 63, 633–646 [DOI] [PubMed] [Google Scholar]
- 45.Kosicki M et al. (2018) Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36, 765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paix A et al. (2016) Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs. Nucleic Acids Res 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paix A et al. (2017) Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proceedings of the National Academy of Sciences DOI: 10.1073/pnas.1711979114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu VT et al. (2015) Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33, 543–548 [DOI] [PubMed] [Google Scholar]
- 49.Zhang JP et al. (2017) Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol 18, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutschner T et al. (2016) Post-translational Regulation of Cas9 during G1 Enhances Homology-Directed Repair. Cell Rep 14, 1555–1566 [DOI] [PubMed] [Google Scholar]
- 51.Lin S et al. (2014) Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3, e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jayavaradhan R et al. (2019) CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat Commun 10, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rees HA et al. (2019) Development of hRad51–Cas9 nickase fusions that mediate HDR without double-stranded breaks. Nat Commun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hussmann JA et al. (2021) Mapping the genetic landscape of DNA double-strand break repair. Cell 184, 5653–5669.e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roidos P et al. (2020) A scalable CRISPR/Cas9-based fluorescent reporter assay to study DNA double-strand break repair choice. Nat Commun 11, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eki R et al. (2020) A robust CRISPR-Cas9-based fluorescent reporter assay for the detection and quantification of DNA double-strand break repair. Nucleic Acids Res 48, E126–E126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Sluis M and McStay B (2015) A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev 29, 1151–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mao P et al. (2016) Homologous recombination-dependent repair of telomeric DSBs in proliferating human cells. Nat Commun 7, 12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsouroula K et al. (2016) Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol Cell 63, 293–305 [DOI] [PubMed] [Google Scholar]
- 60.Ochs F et al. (2019) Stabilization of chromatin topology safeguards genome integrity. Nature 574, 571–574 [DOI] [PubMed] [Google Scholar]
- 61.Wang Y et al. (2021) Genome oligopaint via local denaturation fluorescence in situ hybridization. Mol Cell 81, 1566–1577.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y et al. (2022) Achieving single nucleotide sensitivity in direct hybridization genome imaging Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Bloomberg School of Public Health, Johns Hopkins University School of Medicine, Baltim [Google Scholar]
- 63.Wang H et al. (2019) CRISPR-mediated live imaging of genome editing and transcription. Science (1979) 365, 1301–1305 [DOI] [PubMed] [Google Scholar]
- 64.Wu X et al. (2019) Progress and Challenges for Live-cell Imaging of Genomic Loci Using CRISPR-based Platforms. Genomics Proteomics Bioinformatics 17, 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van den Berg J et al. (2018) A limited number of double-strand DNA breaks is sufficient to delay cell cycle progression. Nucleic Acids Res 46, 10132–10144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schep R et al. (2021) Impact of chromatin context on Cas9-induced DNA double-strand break repair pathway balance. Mol Cell 81, 2216–2230.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aleksandrov R et al. (2018) Protein Dynamics in Complex DNA Lesions. Mol Cell 69, 1046–1061.e5 [DOI] [PubMed] [Google Scholar]
- 68.Rogakou EP et al. (1999) Megabase chromatin domains involved in DNA double-strand breaks in vivo. Journal of Cell Biology 146, 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li K et al. (2020) Yeast ATM and ATR kinases use different mechanisms to spread histone H2A phosphorylation around a DNA double-strand break. Proceedings of the National Academy of Sciences 117, 21354 LP – 21363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davidson IF et al. (2019) DNA loop extrusion by human cohesin. Science (1979) 366, 1338 LP – 1345 [DOI] [PubMed] [Google Scholar]
- 71.Kim Y et al. (2019) Human cohesin compacts DNA by loop extrusion. Science (1979) 366, 1345 LP – 1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zou RS et al. (2022) Massively parallel genomic perturbations with multi-target CRISPR interrogates Cas9 activity and DNA repair at endogenous sites. Nat Cell Biol DOI: 10.1038/s41556-022-00975-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kruhlak MJ et al. (2006) Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. Journal of Cell Biology 172, 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strickfaden H et al. (2016) Poly(ADP-ribosyl)ation-dependent transient chromatin decondensation and histone displacement following laser microirradiation. Journal of Biological Chemistry 291, 1789–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fernandez A et al. (2021) Epigenetic Mechanisms in DNA Double Strand Break Repair: A Clinical Review. Front Mol Biosci 8, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sishc BJ and Davis AJ (2017) The role of the core non-homologous end joining factors in carcinogenesis and cancer. Cancers (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang H et al. (2020) Methods favoring homology-directed repair choice in response to crispr/cas9 induced-double strand breaks. Int J Mol Sci 21, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pfeiffer P et al. (2000) Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 15, 289–302 [DOI] [PubMed] [Google Scholar]
- 79.Clouaire T and Legube G (2019) A Snapshot on the Cis Chromatin Response to DNA Double-Strand Breaks. Trends in Genetics 35, 330–345 [DOI] [PubMed] [Google Scholar]
- 80.Rouet P et al. (1994) Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol 14, 8096–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clouaire T et al. (2018) Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures. Mol Cell 72, 250–262.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vítor AC et al. (2020) Studying DNA Double-Strand Break Repair: An Ever-Growing Toolbox. Front Mol Biosci 7, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shanbhag NM et al. (2010) ATM-Dependent chromatin changes silence transcription in cis to dna double-strand breaks. Cell 141, 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Slaymaker IM et al. (2016) Rationally engineered Cas9 nucleases with improved specificity. Science (1979) 351, 84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kleinstiver BP et al. (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richardson CD et al. (2016) Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34, 339–344 [DOI] [PubMed] [Google Scholar]
- 87.Wang AS et al. (2020) The Histone Chaperone FACT Induces Cas9 Multi-turnover Behavior and Modifies Genome Manipulation in Human Cells. Mol Cell 79, 221–233.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jr SKJ et al. (2021) Massively parallel kinetic profiling of natural and engineered CRISPR nucleases. Nat Biotechnol 39, 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]