Abstract
Ral is a ubiquitously expressed Ras-like small GTPase which is abundantly present in human platelets. The biological function of Ral and the signaling pathway in which Ral is involved are largely unknown. Here we describe a novel method to measure Ral activation utilizing the Ral binding domain of the putative Ral effector RLIP76 as an activation-specific probe. With this assay we investigated the signaling pathway that leads to Ral activation in human platelets. We found that Ral is rapidly activated after stimulation with various platelet agonists, including α-thrombin. In contrast, the platelet antagonist prostaglandin I2 inhibited α-thrombin-induced Ral activation. Activation of Ral by α-thrombin could be inhibited by depletion of intracellular Ca2+, whereas the induction of intracellular Ca2+ resulted in the activation of Ral. Our results show that Ral can be activated by extracellular stimuli. Furthermore, we show that increased levels of intracellular Ca2+ are sufficient for Ral activation in platelets. This activation mechanism correlates with the activation mechanism of the small GTPase Rap1, a putative upstream regulator of Ral guanine nucleotide exchange factors.
RalA and RalB are very similar small GTPases that have 55% sequence identity with Ras (6, 7, 41). Ral and Ras have comparable nucleotide binding characteristics and low intrinsic GTPase activity (19). The Ral proteins are ubiquitously expressed but are particularly abundant in brain, testes, and platelets (2, 21, 39). Ral becomes posttranslationally processed and is found in the plasma membrane (16) as well as in endocytotic vesicles (41) or synaptic vesicles (50). In platelets, Ral is a major GTP binding protein that is present in the plasma membrane and specifically in dense granules, a class of secretory organelles (29, 41).
Recently, a putative effector protein of RalA and RalB has been identified; it has been designated RLIP76 but is also termed RalBP1 or RIP1 (5, 22, 40). RLIP76 interacts with the active, GTP-bound form of Ral, both in the yeast two-hybrid system and in vitro. Interaction studies using the yeast two-hybrid system and deletion mutants of RLIP76 demonstrate that the Ral binding region is located in the C-terminal region, between amino acids 403 and 499 (22). Interestingly, RLIP76 exhibits GTPase-activating protein (GAP) activity for the Rho-like GTPase Cdc42, suggesting that Ral may be involved in the regulation of Cdc42 (5, 22, 40). Cdc42 plays a role in the organization of the actin cytoskeleton and the regulation of cytoskeletal polarity.
Ral also interacts with phospholipase D1 (PLD1) (21, 28). The interaction between Ral and PLD1 is independent of the nucleotide binding of Ral and occurs via the N-terminal region of Ral (21, 28). In platelets and other cells, PLD has been implicated in vesicle transport, regulation of the actin cytoskeleton, and generation of lysophosphatidic acid, which is secreted by platelets upon activation (14, 31).
Different proteins that regulate the activity of Ral have been identified. A RalGAP with a high molecular mass was purified from brain and testes, whereas a 34-kDa RalGAP was found in human platelets (3, 13). Furthermore, several Ral guanine nucleotide exchange factors (RalGEFs) have been cloned (1, 10, 34, 52). Three of these RalGEFs, RalGDS, Rgl, and Rlf, can bind to and become activated by Ras via their C-terminally located Ras binding domains both in vitro and in vivo, indicating that RalGEFs function as Ras effector proteins (34, 48, 53). Apart from Ras, the activated versions of other members of the Ras family of proteins, Rap1, Rap2, R-Ras, and TC21, can interact with RalGEFs (27, 44, 48, 52). These might thus be involved in the activation of Ral as well, although cotransfection experiments with Cos7 cells suggested that Rap1 and R-Ras cannot activate RalGDS (48). However, the lack of information about the physiological activation of Ral by extracellular stimuli has been an obstacle to further understanding the regulation and function of Ral. The unavailability of antibodies suitable for efficient immunoprecipitation and subsequent analysis of bound nucleotides has so far precluded the analysis of growth factor-induced Ral activation.
In this report we present an alternative assay for the detection of endogenously activated Ral. We used the Ral binding domain (RalBD) of RLIP76, which interacts specifically with the GTP-bound form of Ral, to monitor the activity of Ral in human platelets, and we show that α-thrombin and other platelet agonists stimulate a rapid and strong activation of Ral. We demonstrate that the elevation of Ca2+ levels is involved in the activation of Ral. Finally, we discuss the stimulus-induced activations of Ras, Rap1, and Ral.
MATERIALS AND METHODS
Isolation of GST-RalBD.
Glutathione S-transferase (GST)-RalBD containing amino acids 397 to 518 of human RLIP76 (22) was cloned into pGEX4T3 by PCR with specific primers introducing a BamHI and a XhoI site. AD202 protease-negative bacteria were transformed with the pGEX4T3-GST-RalBD construct. GST-RalBD was isolated from isopropyl-β-d-thiogalactopyranoside (IPTG)-induced bacteria as described for GST-Rlf (53). Purified protein was concentrated and was stored in a solution containing 10% glycerol, 50 mM Tris (pH 7.4), 150 mM NaCl, 5 mM MgCl2, and 5 mM dithioerythritol (DTE) at −70°C.
Determination of Ral-GTP levels.
Cos7 cells were cultured in 100-mm dishes in Dulbecco’s modified Eagle medium–10% fetal calf serum–0.05% glutamine and tranfected with 3 μg of pMT2 expression vectors encoding hemagglutinin (HA)-Ral, HA-RalV23, or HA-RalN28, as described previously (53). For determination of levels of GTP bound to Ral, transfected Cos7 cells were put in 1.5% serum overnight and subsequently metabolically labeled with 150 μCi of 32Pi for 5 h in phosphate-free medium. Next, cells were lysed, transfected Ral proteins were recovered in 12CA5 immunoprecipitations, bound nucleotides were eluted and separated, and GTP/GDP ratios were determined with a PhosphorImager, as described previously (53).
Platelets.
Platelets were prepared as described previously (18) and resuspended in HEPES/Tyrode buffer at 3 × 108 platelets/ml. Platelets were left at room temperature for 30 min. Samples of 1.0 ml were used for each point. Stimulation with agonists at 37°C was performed without stirring. Prior to ionomycin treatment, 1 mM CaCl2 was added to the platelet suspension.
Use of RalBD as an activation-specific probe.
Platelets were lysed (2:1, vol/vol) in 3× Ral buffer (final concentrations: 10% glycerol, 1% Nonidet P-40, 50 mM Tris-HCl [pH 7.4], 200 mM NaCl, 2.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 1 μM leupeptin, 10 μg of soybean trypsin inhibitor per ml, and 0.1 μM aprotinin). Lysates were clarified by centrifugation and the supernatants of each sample were incubated with 15 μg of GST-RalBD precoupled to glutathione beads. Samples were incubated for 1 h on a tumbler at 4°C. Beads were washed four times in Ral buffer. Remaining fluid was removed with an insulin syringe, and beads were collected in Laemmli sample buffer. As a control, Ral levels in whole lysates were determined. Samples were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (SDS-PAGE) (12.5% polyacrylamide) and Western blotting with a monoclonal anti-RalA antibody (Transduction Laboratories).
For in vitro analysis, 800 μg of GST-RalBD was bound to 700 μl of glutathione-Sepharose beads (Pharmacia) in 8 ml of a solution of 15% glycerol, 1% Nonidet P-40, 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 2 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, and 25 nM Pefablock. Beads were collected by centrifugation and washed in the same buffer. A 4 μM concentration of a C-terminally truncated version of simian RalA (sRalA, representing amino acids 1 to 178) isolated from Escherichia coli was loaded with GDP or Gpp(NH)p and incubated with 3 μM GST-RalBD bound to glutathione beads for 1 h at 4°C. Beads were collected and washed three times with the same buffer and resuspended in 80 μl of Laemmli sample buffer. Ten microliters of the sample was separated by SDS–12.5% PAGE and protein was detected by Coomassie brilliant blue staining.
Ras and Rap1 activation assays.
The GTP-bound forms of Ras and Rap1 were specifically pulled down from clarified platelet lysates by incubation with the GST-tagged forms of the Raf1-Ras binding domain (RBD) or RalGDS-RBD, respectively, precoupled to glutathione beads, as described previously (12, 18). However, we used the Ral buffer indicated above for cellular lysis and washing of the beads since this resulted in a higher sensitivity without affecting the specificity (data not shown). In the experiments in which the different small GTPases were compared, the lysates were split and used for the determination of either Ras, Rap1, or Ral activity.
RESULTS
The RalBD of RLIP76 interacts specifically with RalGTP.
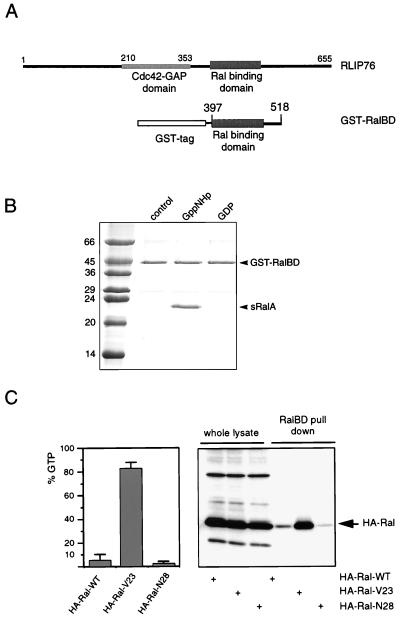
Based on structural predictions of the coiled-coil region encompassing the Ral binding region of RLIP76, we used a GST fusion protein containing amino acids 397 to 518 of RLIP76 that was predicted to be sufficient for direct interaction with Ral in vitro (GST-RalBD) (Fig. 1A). To investigate its interaction with Ral, GST-RalBD was incubated with purified sRalA (C-terminally truncated) that was bound to either GDP or a nonhydrolyzable GTP analog, Gpp(NH)p, at 4°C. Subsequently, GST-RalBD was pulled down on glutathione beads, the beads were washed, and bound proteins were analyzed by SDS-PAGE and gel staining. Figure 1B shows that GST-RalBD interacts with sRalA-Gpp(NH)p and not with sRalA-GDP.
FIG. 1.
A GST fusion protein of the RalBD of RLIP76 binds specifically to the activated form of Ral. (A) Schematic representation of RLIP76, indicating the position of the RalBD of RLIP76 that was used to generate the GST-RalBD. (B) In vitro analysis of the interaction between GST-RalBD and sRalA. A 3μM concentration of GST-RalBD was precoupled to glutathione-Sepharose beads and washed. A 4 μM concentration of a C-terminally truncated sRalA loaded with GDP or Gpp(NH)p (a nonhydrolyzable GTP analog) was incubated with the precoupled beads. After 1 h of incubation at 4°C, beads were washed three times and resuspended in 80 μl of sample buffer, of which 10 μl was loaded onto an SDS-PAGE gel. After separation, the proteins were stained by Coomassie brilliant blue. The numbers are molecular masses of the marker proteins, in kilodaltons. (C) Analysis of GTP levels of HA-Ral mutants by different methods. The left panel shows the percentage of GTP bound to ectopically expressed wild-type HA-Ral (HA-Ral-WT), activated HA-Ral (HA-RalV23), and inactive HA-Ral (HA-RalN28) in serum-starved Cos7 cells. Error bars indicate the variations between two independent experiments. The right panel shows the detection of HA-RalGTP in a GST-RalBD pull-down experiment. Therefore, the transfected Cos7 cells were serum starved, lysed, and incubated with 15 μg of GST-RalBD precoupled to glutathione beads to recover GTP-bound Ral. Beads were washed four times, and collected Ral was identified by Western analysis with an anti-HA (12CA5) monoclonal antibody (lanes 4 to 6). The first three lanes show the levels of the transfected HA-Ral mutants in the whole lysates. In these lanes, 5% of the lysate used for the pull-down experiment was loaded onto the gel.
In order to further evaluate the use of this method, we transfected Cos7 cells with epitope-tagged versions of wild-type Ral or active (RalV23) or inactive (RalN28) mutants. First, the GTP levels of the different mutants were determined after labeling of the cells with [32P]orthophosphate. The left panel of Fig. 1C shows that after serum starvation, wild-type Ral contained approximately 8% GTP, while more than 85% of the HA-RalV23 molecules were GTP bound, in contrast to HA-RalN28, which was almost exclusively bound to GDP. In a parallel experiment, we used GST-RalBD to pull down HA-RalGTP from the lysates of the transfected cells. The right panel of Fig. 1C shows that the relative amounts of HA-Ral isolated from Cos7 cells as detected by immunoblotting correlated well with the GTP levels of the expressed Ral proteins. Together with the in vitro data, this shows that GST-RalBD specifically interacts with RalGTP and can be used to monitor Ral activation in cells.
α-Thrombin and platelet agonists induce rapid activation of Ral.
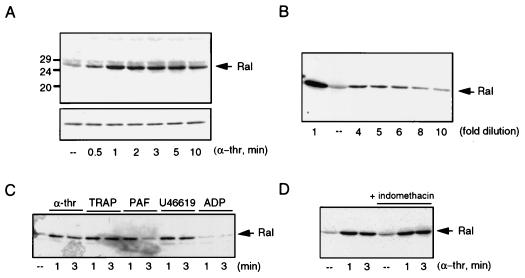
Human platelets were treated with 0.1 U of α-thrombin per ml, which induces complete aggregation of stirred platelets within 5 to 10 min (49), for different periods of time. RalGTP was pulled down from the cell lysates by GST-RalBD precoupled to glutathione beads. Subsequently, the amount of RalGTP was analyzed by Western blotting with a monoclonal anti-RalA antibody, which recognizes RalA as a single 27-kDa protein and which does not cross-react with Ras or Rap1 (data not shown). Due to the very high homology between RalA and RalB, RalB may be recognized as well.
Figure 2A shows that, whereas in resting platelets hardly any RalGTP was found, stimulation with 0.1 U of α-thrombin per ml induced a rapid increase in the amount of RalGTP collected by GST-RalBD. The activation of Ral was maximal after 1 min of α-thrombin stimulation but remained elevated for at least 10 min. In order to estimate the increase of Ral activation by α-thrombin, a range of dilutions of the RalGTP precipitated by GST-RalBD from stimulated platelets was made and compared with RalGTP isolated from resting platelets. The signal obtained from a 6- to 10-fold dilution approximated that from RalGTP isolated from resting platelets (Fig. 2B), indicating that treatment of the platelets with 0.1 U of α-thrombin per ml stimulates Ral activation at least 6-fold.
FIG. 2.
Stimulation of platelets with α-thrombin (α-thr) and other platelet agonists leads to rapid activation of Ral. (A) Activation of Ral by α-thrombin. Human platelets were isolated as described previously (18) and stimulated with 0.1 U of α-thrombin per ml for different times. The platelets were lysed and incubated with 15 μg of GST-RalBD precoupled to glutathione beads to recover GTP-bound Ral. Beads were washed four times, and collected Ral was identified by Western analysis with a monoclonal anti-RalA antibody (upper panel). The lower panel shows the levels of Ral in the whole lysates. (B) Determination of RalGTP induction. Platelets were stimulated with α-thrombin for 1 min and Ral activation was determined as described for panel A. In order to estimate the increase in RalGTP induced by α-thrombin, the α-thrombin-induced sample (far left) was diluted severalfold so it could be compared to the resting platelets (––). The gel shows that the signal obtained after a 6- to 10-fold dilution of the RalGTP obtained from α-thrombin-stimulated platelets matches the amount of Ral detected in resting platelets. (C) Ral activation by different platelet agonists. Platelets were stimulated for 1 and 3 min with 0.1 U of α-thrombin per ml, thrombin receptor-activating peptide (TRAP, 10 μM), PAF (200 nM), TxA2 analog U46619 (1 μM), or ADP (10 μM). Ral activation was analyzed as described for panel A. (D) Ral activation occurs independently of TxA2 formation. Platelets were treated with 30 μM indomethacin for 10 min to inhibit TxA2 formation, prior to stimulation with 0.1 U of α-thrombin per ml for the indicated times, or were stimulated with 0.1 U of α-thrombin per ml alone. At the concentration used, indomethacin inhibits TxA2 formation (not shown). Activation of Ral was monitored as described for panel A.
The thrombin receptor-activating peptide, which mimics α-thrombin by binding to and activating the thrombin receptor, also activates Ral (Fig. 2C). We investigated the effects of other platelet agonists on Ral activation. We observed that platelet-activating factor (PAF) was able to induce a rapid activation of Ral, although this activation was more transient than α-thrombin stimulation (Fig. 2C). As a result of α-thrombin treatment, thromboxane A2 (TxA2), a product of the cyclooxygenase-dependent arachidonic acid pathway, is formed and released and ADP is secreted from vesicles. These agents act in positive-feedback loops to platelet activation (43). ADP did not activate Ral significantly, if at all (Fig. 2C), but treatment of platelets with the TxA2 mimetic U46619 did induce Ral activation (Fig. 2C). Pretreatment with the cyclooxygenase inhibitor indomethacin did not significantly inhibit α-thrombin-induced Ral activation, demonstrating that α-thrombin-induced Ral activation is not dependent on the release of TxA2 (Fig. 2D). Taken together, these findings demonstrate that different platelet agonists stimulate a rapid activation of Ral.
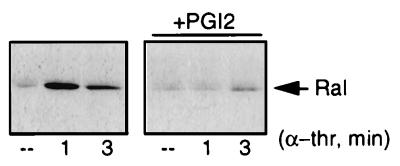
α-Thrombin-induced Ral activation is inhibited by PGI2.
Platelet activation is antagonized by agents that stimulate the cyclic AMP/protein kinase A pathway (26), such as prostaglandin I2 (PGI2). To further investigate the importance of Ral activation in platelets, we tested the effect of PGI2 on α-thrombin-induced Ral activation. Figure 3 demonstrates that the addition of 20 ng of PGI2 per ml prior to α-thrombin treatment was sufficient to almost completely block α-thrombin-induced Ral activation. PGI2 treatment did not affect the total protein levels of Ral in the lysate (data not shown). Since Ral is activated by agents that lead to the activation of platelets and blocked by a platelet antagonist, our data suggest a stimulatory role for Ral in platelet activation.
FIG. 3.
Platelet antagonist PGI2 inhibits α-thrombin-induced activation of Ral. Platelets were treated with 20 ng of PGI2 per ml for 2 min prior to stimulation with 0.1 U of α-thrombin (α-thr) per ml or were stimulated with 0.1 U of α-thrombin per ml alone. Ral activation was analyzed as described in the legend to Fig. 2. ––, resting platelets.
α-Thrombin-induced Ral activation involves Ca2+ but not PKC.
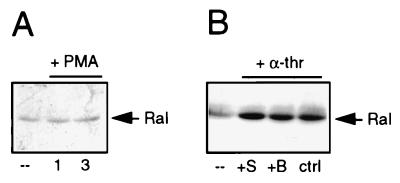
The thrombin receptor in platelets may couple to Gi, which inhibits adenylate cyclase, and to Gq, which induces phosphoinositide hydrolysis leading to diacylglycerol (DAG) generation and inositoltrisphosphate (InsP3) formation. DAG stimulates the protein kinase C (PKC) pathway, whereas InsP3 releases Ca2+ from the intracellular stores (43). In order to dissect the mechanism of α-thrombin-induced Ral activation, we examined whether PKC or increases in intracellular Ca2+ were involved in the activation of Ral. First, platelets were treated with phorbol myristate acetate (PMA) to activate PKC. PMA treatment did not result in the rapid activation of Ral (Fig. 4A). To test whether inhibition of PKC affects α-thrombin-induced Ral activation, we used the PKC inhibitor bisindolylmaleimide (47). At concentrations at which either bisindolylmaleimide or staurosporine inhibited α-thrombin-induced aggregation of the platelets completely (data not shown), neither inhibited α-thrombin-induced Ral activation (Fig. 4B). These observations indicate that α-thrombin-induced Ral activation is not mediated by PKC.
FIG. 4.
PKC is not involved in α-thrombin-induced Ral activation. (A) PKC activation does not lead to rapid Ral activation. Platelets were stimulated with PMA (10 nM) for 1 and 3 min. (B) Inhibition of PKC does not block α-thrombin-induced Ral activation. Platelets were treated with the PKC inhibitor bisindolylmaleimide (B, 5 μM, 1 min) or the kinase inhibitor staurosporine (S, 1 μM, 5 min) prior to stimulation with 0.1 U of α-thrombin (α-thr) per ml for 1 min or were stimulated with α-thrombin alone. Ral activation was analyzed as described in the legend to Fig. 2. ––, resting platelets. ctrl, control.
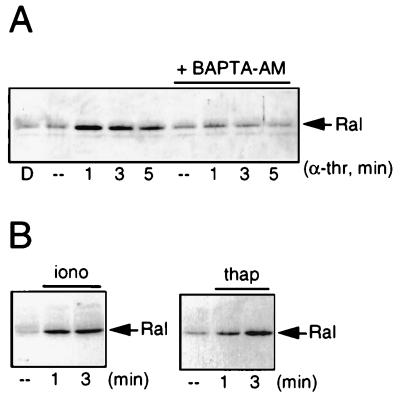
Next we examined whether an increase in the intracellular concentration of Ca2+ is involved in the activation of Ral. Pretreatment with the intracellular Ca2+ chelator BAPTA-AM [1,2-bis(o-aminophenoxy)ethane-N-N-N′-N′-tetraacetate] (11) completely blocked α-thrombin-induced Ral activation, suggesting that the elevation of intracellular Ca2+ levels is the signal by which α-thrombin stimulates Ral activation (Fig. 5A). In addition, treatment with agents that induce an elevation of Ca2+, by stimulating either a Ca2+ influx (ionomycin) or a release from intracellular Ca2+ stores (thapsigargin), induced a rapid activation of Ral (Fig. 5B). Indomethacin pretreatment (30 μM, 10 min) did not block the effect of ionomycin or thapsigargin on Ral, demonstrating that the formation of TxA2 is not necessary for Ca2+-induced Ral activation (data not shown). Therefore, we conclude that α-thrombin-induced activation of Ral is mediated by an increase in intracellular Ca2+ and that elevation of intracellular Ca2+ levels is sufficient to induce Ral activation.
FIG. 5.
Ca2+ is necessary and sufficient for activation of Ral. (A) Ca2+ is necessary for Ral activation by α-thrombin. Platelets preincubated or not for 30 min with 30 μM BAPTA-AM, a chelator of intracellular Ca2+, were stimulated with 0.1 U of α-thrombin (α-thr) per ml for the indicated times. Dimethyl sulfoxide (D) alone did not have an effect on Ral activation. Analysis of Ral in total lysates demonstrated that BAPTA-AM did not induce degradation of Ral (data not shown). (B) Ca2+ is sufficient to induce Ral activation. Platelets were treated for the indicated times with 100 nM ionomycin (iono) (in the presence of 1 mM CaCl2 in the buffer) to promote Ca2+ influx or with 100 nM thapsigargin (thap) to induce the release of Ca2+ from intracellular stores. Ral activation was analyzed as described in the legend to Fig. 2. ––, resting platelets.
Ca2+-induced Ral activation is inhibited by PGI2 and correlates with Rap1 activation.
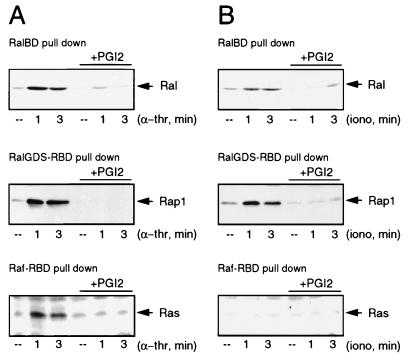
Recently, we and others have demonstrated that the formation of RalGTP can be triggered by oncogenic Ras via the activation of RalGEFs (38, 48, 53). Apart from Ras, the RalGEFs can interact with the active form of Rap1 both in vitro and in vivo (references 23, 44, and 52 and unpublished observations). Rap1, whose function is still elusive, is highly abundant in platelets (46) and becomes rapidly activated after α-thrombin stimulation. Interestingly, for the activation of both Rap1 and Ral, elevated levels of Ca2+ are both necessary and sufficient (18). In addition, the activation of Rap1 by either α-thrombin or ionomycin is sensitive to PGI2 treatment. Therefore, we compared the pattern of Ral activation with those of Ras and Rap1 in platelets, using the previously described activation-specific probes GST-Raf1-RBD and GST-RalGDS-RBD to monitor the formation of RasGTP and Rap1GTP, respectively (12, 18). α-Thrombin stimulation led to the activation of Ral, Rap1, and Ras with similar kinetics and this activation was blocked by PGI2 pretreatment (Fig. 6A). In contrast, when we measured ionomycin-induced activation of the three GTPases, we found that both Ral and Rap1, but not Ras, were activated in a PGI2-sensitive manner (Fig. 6B). This result shows that in platelets, Rap1 and Ral, but not Ras, are coregulated by a Ca2+-induced, PGI2-sensitive signaling pathway.
FIG. 6.
Ral activation by Ca2+ correlates with the activation of Rap1 but not Ras. (A) α-Thrombin-induced Ral activation correlates with Rap1 and Ras activation. Platelets preincubated or not for 2 min with 20 ng of PGI2 per ml were stimulated with 0.1 U of α-thrombin (α-thr) per ml for the indicated times. Cell lysates were split and analyzed for the presence of RalGTP, RasGTP, and Rap1GTP. Ral activation (upper panel) was analyzed as described in the legend to Fig. 2. Rap1GTP was isolated with GST-RalGDS-RBD precoupled to glutathione beads and Western blotted with polyclonal anti-Rap1 (middle panel). The lower panel shows Ras activation. RasGTP was isolated with GST-Raf1-RBD, precoupled to glutathione beads, and analyzed by Western blotting with an anti-Ras monoclonal antibody (Transduction Laboratories). These methods for the detection of Ras and Rap1 activation have been described previously (12, 18). (B) Platelets preincubated or not for 2 min with 20 ng of PGI2 per ml were stimulated with 100 nM ionomycin in the presence of 1 mM CaCl2 for the indicated times. Cell lysates were split and analyzed for the presence of RalGTP, Rap1GTP, and RasGTP, as described for panel A. ––, resting platelets.
DISCUSSION
Use of GST-RalBD as an activation-specific probe for Ral.
In this report we demonstrate that GST-RalBD, a tagged version of the Ral binding domain of RLIP76, binds to purified Ral loaded with a GTP analog but not to RalGDP. We used GST-RalBD to pull down RalGTP from cellular lysates of cells transfected with Ral mutants and showed that the amount of Ral precipitated correlates with the levels of bound GTP. The same method was used to monitor Ral activation in platelets, using a monoclonal anti-RalA antibody. Although this antibody was raised to a peptide that contains RalA-specific sequences, it may cross-react with RalB, which is 85% identical to RalA. Due to the unavailability of antibodies that can precipitate Ral, it is difficult to compare the Ral activation that we observed in platelet cell lysates by this method with the actual ratio of GTP and GDP bound to Ral. However, for Ras we found that a similar method using GST-Raf-RBD revealed increases in RasGTP that correlated very nicely with the increased ratio of GTP versus GDP bound to Ras (12). In conclusion, the procedure we describe here is a fast and reliable way to qualitatively analyze increases in RalGTP.
Rapid Ca2+-mediated activation of Ral in human platelets.
To test the assay in vivo we used human platelets, since in these cell fragments Ral is a major GTP binding protein (2). Indeed, when platelets are stimulated with α-thrombin, Ral is very rapidly activated. This provides the first example of ligand-induced activation of Ral. Also, the platelet agonists PAF and TxA2 activate Ral, suggesting that Ral activation is mediated by a common signaling event. As we show, this pathway involves an increase in the level of cytosolic Ca2+, either by the mobilization from internal stores or by the influx of Ca2+. This evidence is based on the observation that the calcium chelator BAPTA-AM completely inhibits α-thrombin-induced activation of Ral, whereas artificial Ca2+ mobilization with either thapsigargin or ionomycin induces Ral activation. Recently, it has been shown that activation of platelets by α-thrombin and other platelet agonists requires activation of the heterotrimeric G protein Gq (37). Gq mediates the activation of phospholipase C-β in platelets (35, 37). This enzyme hydrolyzes phosphatidylinositol-4,5-diphosphate, resulting in the second messengers DAG and InsP3. InsP3 in turn releases Ca2+ from internal stores, which may trigger the influx of Ca2+ (26). Consistent with this finding, ADP, which induces only a small increase in InsP3 formation and Ca2+ release (37), is a weak activator of Ral.
How Ca2+ subsequently activates Ral is unclear, although several possibilities can be envisioned. First, Ca2+ may have a direct effect on Ral activity: RalA from erythrocytes can bind to calmodulin in a Ca2+-dependent fashion, which might influence Ral activation (51). Alternatively, phosphorylation by Ca2+-dependent kinases may regulate the activity of RalGEFs or RalGAPs, or there might exist Ral-specific homologs of the Ca2+-sensitive Ras guanine nucleotide-releasing factor (RasGRF) (15).
Second, Ral activation may occur via a mechanism involving Ras family members acting as upstream activators of RalGEFs (17). Interestingly, the pattern of Ral activation in platelets reveals a good correlation with the activation of Rap1 but a less clear correlation with the activation of Ras, since (i) Ca2+ is necessary and sufficient for rapid activation of both Rap1 and Ral in platelets, but not for Ras activation, and (ii) α-thrombin-induced activation of Rap1 and Ral is not inhibited by PKC inhibition in platelets (Fig. 5 and reference 18), in contrast to α-thrombin-induced activation of Ras (reference 42 and data not shown). Furthermore, Rap1 activation and Ral activation exhibit similar kinetics in response to the different platelet agonists: α-thrombin-stimulated activation of both Rap1 and Ral is maximal after 1 min and remains elevated for several minutes, and the activation of both Rap1 and Ral by PAF is transient (Fig. 1C and reference 18). Contradicting the idea that Rap1 may act as an upstream activator of Ral is the observation that a putative activated form of Rap1, Rap1-E63, interacted with but did not activate RalGDS in a transient-transfection experiment with Cos7 cells (48). However, Rap1 signaling in Cos7 cells and that in platelets could be different, for instance, because the localization of Rap1 and Ral in Cos7 cells may differ from that in platelets. Recently, it was shown that upon incorporation in liposomes, Rap can stimulate GDP release from Ral through RalGDS, providing evidence that, in principle, Rap1 can activate RalGEFs (24). It is also possible that a second, Ca2+-induced signal cooperates with Rap1 in the activation of Ral. The possible connection between Rap1 and Ral does not exclude the possibility that one of the other Ras-like GTPases which can interact with RalGEFs is involved in the activation of Ral as well (27, 34, 48, 52).
Downstream targets of Ral signal transduction.
The only putative target of active Ral that has been described so far is RLIP76, which exhibits GAP activity for Cdc42. This indicates that Ral activation may function either to downregulate the activity of Cdc42 or to inhibit Cdc42 GAP activity, resulting in activation of Cdc42. In turn, Cdc42 regulates cytoskeletal rearrangements, which might involve the Cdc42 effector molecule Wiskott-Aldrich syndrome protein (45). Mutations in this molecule lead to severe abnormalities in the cytoskeletons of platelets (23).
The intracellular localization of the Ral proteins has led to the speculation that Ral might be involved in exo- or endocytosis: Ral has been detected in endocytotic vesicles (16) and in synaptic vesicles (4, 16, 36, 50) and was found to be associated with specialized secretory organelles, the dense granules, in platelets (29). Furthermore, our results demonstrate that Ral can become activated by agents which can also stimulate secretion in platelets. For example, treatment of platelets with 0.1 to 0.5 U of α-thrombin per ml induces the fusion of dense granules with the plasma membrane and the open canalicular system (33), which may lead to the release of ADP functioning in positive feedback. Interestingly, there is evidence that Ca2+ can rapidly and strongly stimulate exocytosis in platelets and other cells (8, 25). PLD1, which binds to RalA, has also been implicated in secretion in platelets (8, 9, 20, 25, 32). It is not clear how PLD1 activity is regulated by Ral: although dominant negative Ral can inhibit Src-induced PLD activation, the interaction between Ral and PLD1 is independent of the activation state of Ral (21, 28). Stimulation of PLD activity in platelets has been reported to be mainly the result of PKC activation (30) and therefore does not correlate with Ral activation. It might be that Ral is involved in specifically targeting PLD1 to certain membranes, where it can be activated by other factors, such as PKC or the small GTPases Arf and Rho (14).
ACKNOWLEDGMENTS
We thank our colleagues for assistance and support and Boudewijn Burgering for critically reading the manuscript.
This work was supported by the Dutch Organization for Scientific Research (GB-MW; NWO), The Netherlands Heart Foundation (grant 94.136), the EC (grant BIO-4-CT96-1110), and the Dutch Cancer Society (KWF).
REFERENCES
- 1.Albright C F, Giddings B W, Liu J, Vito M, Weinberg R A. Characterization of a guanine nucleotide dissociation stimulator for a ras-related GTPase. EMBO J. 1993;12:339–347. doi: 10.1002/j.1460-2075.1993.tb05662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhullar R P, Chardin P, Haslam R J. Identification of multiple ral gene products in human platelets that account for some but not all of the platelet Gn-proteins. FEBS Lett. 1990;260:48–52. doi: 10.1016/0014-5793(90)80063-o. [DOI] [PubMed] [Google Scholar]
- 3.Bhullar R P, Seneviratne H D. Characterization of human platelet GTPase activating protein for the Ral GTP-binding protein. Biochim Biophys Acta. 1996;1311:181–188. doi: 10.1016/0167-4889(96)00002-x. [DOI] [PubMed] [Google Scholar]
- 4.Bielinski D F, Pyun H Y, Linko-Stentz K, Macara I G, Fine R E. Ral and Rab3a are major GTP-binding proteins of axonal rapid transport and synaptic vesicles and do not redistribute following depolarization stimulated synaptosomal exocytosis. Biochim Biophys Acta. 1993;1151:246–256. doi: 10.1016/0005-2736(93)90109-d. [DOI] [PubMed] [Google Scholar]
- 5.Cantor S B, Urano T, Feig L A. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol Cell Biol. 1995;15:4578–4584. doi: 10.1128/mcb.15.8.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chardin P, Tavitian A. Coding sequences of human ralA and ralB cDNAs. Nucleic Acids Res. 1989;17:4380. doi: 10.1093/nar/17.11.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chardin P, Tavitian A. The Ral gene: a new Ras-related gene isolated by the use of a synthetic probe. EMBO J. 1986;5:2203–2208. doi: 10.1002/j.1460-2075.1986.tb04485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coorssen J R. Phospholipase activation and secretion: evidence that PLA2, PLC, and PLD are not essential to exocytosis. Am J Physiol. 1996;270:C1153–C1163. doi: 10.1152/ajpcell.1996.270.4.C1153. [DOI] [PubMed] [Google Scholar]
- 9.Coorssen J R, Schmitt H, Almers W. Ca2+ triggers massive exocytosis in Chinese hamster ovary cells. EMBO J. 1996;15:3787–3791. [PMC free article] [PubMed] [Google Scholar]
- 10.D’Adamo D R, Novick S, Kahn J M, Leonardi P, Pellicer A. Rsc: a novel oncogene with structural and functional homology with the gene family of exchange factors for Ral. Oncogene. 1997;14:1295–1305. doi: 10.1038/sj.onc.1200950. [DOI] [PubMed] [Google Scholar]
- 11.Dash D, Aepfelbacher M, Siess W. The association of pp125FAK, pp60Src, CDC42Hs and Rap1B with the cytoskeleton of aggregated platelets is a reversible process regulated by calcium. FEBS Lett. 1995;363:231–234. doi: 10.1016/0014-5793(95)00320-9. [DOI] [PubMed] [Google Scholar]
- 12.de Rooij J, Bos J L. Minimal Ras-binding domain of Raf1 can be used as an activation-specific probe for Ras. Oncogene. 1997;14:623–625. doi: 10.1038/sj.onc.1201005. [DOI] [PubMed] [Google Scholar]
- 13.Emkey R, Freedman S, Feig L A. Characterization of a GTPase-activating protein for the Ras-related Ral protein. J Biol Chem. 1991;266:9703–9706. [PubMed] [Google Scholar]
- 14.Exton J H. New developments in phospholipase D. J Biol Chem. 1997;272:15579–15582. doi: 10.1074/jbc.272.25.15579. [DOI] [PubMed] [Google Scholar]
- 15.Farnsworth C L, Freshney N W, Rosen L B, Ghosh A, Greenberg M E, Feig L A. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- 16.Feig L A, Emkey R. Ral gene products and their regulation. In: Lacal J C, McCormick F, editors. The ras superfamily of GTPases. London, England: CRC Press; 1993. pp. 247–258. [Google Scholar]
- 17.Feig L A, Urano T, Cantor S. Evidence for a Ras/Ral signaling cascade. Trends Biochem Sci. 1996;21:438–441. doi: 10.1016/s0968-0004(96)10058-x. [DOI] [PubMed] [Google Scholar]
- 18.Franke B, Akkerman J W N, Bos J L. Rapid Ca2+-mediated activation of Rap1 in human platelets. EMBO J. 1997;16:252–259. doi: 10.1093/emboj/16.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frech M, Schlichting I, Wittinghofer A, Chardin P. Guanine nucleotide binding properties of the mammalian RalA protein produced in Escherichia coli. J Biol Chem. 1990;265:6353–6359. [PubMed] [Google Scholar]
- 20.Haslam R J, Coorssen J R. Evidence that activation of phospholipase D can mediate secretion from permeabilized platelets. Adv Exp Med Biol. 1993;344:149–164. doi: 10.1007/978-1-4615-2994-1_11. [DOI] [PubMed] [Google Scholar]
- 21.Jiang H, Luo J Q, Urano T, Frankel P, Lu Z, Foster D A, Feig L A. Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature. 1995;378:409–412. doi: 10.1038/378409a0. [DOI] [PubMed] [Google Scholar]
- 22.Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A, Gacon G, Camonis J H. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J Biol Chem. 1995;270:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 23.Kikuchi A, Demo S D, Ye Z-H, Chen Y-W, Williams L T. ralGDS family members interact with the effector loop of ras p21. Mol Cell Biol. 1994;14:7483–7491. doi: 10.1128/mcb.14.11.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kishida S, Koyama S, Matsubara K, Kishida M, Matsuura Y, Kikuchi A. Colocalization of Ras and Ral on the membrane is required for Ras-dependent Ral activation through Ral GDP dissociation stimulator. Oncogene. 1997;15:2899–2907. doi: 10.1038/sj.onc.1201473. [DOI] [PubMed] [Google Scholar]
- 25.Knight D E, Niggli V, Scrutton M C. Thrombin and activators of protein kinase C modulate secretory responses of permeabilised human platelets induced by Ca2+ Eur J Biochem. 1984;143:437–446. doi: 10.1111/j.1432-1033.1984.tb08391.x. [DOI] [PubMed] [Google Scholar]
- 26.Kroll M H, Schafer A I. Biochemical mechanisms of platelet activation. Blood. 1989;74:1181–1195. [PubMed] [Google Scholar]
- 27.Lopez-Barahona M, Bustelo X R, Barbacid M. The TC21 oncoprotein interacts with the Ral guanosine nucleotide dissociation factor. Oncogene. 1996;12:463–470. [PubMed] [Google Scholar]
- 28.Luo J Q, Liu X, Hammond S M, Colley W C, Feig L A, Frohman M A, Morris A J, Foster D A. RalA interacts directly with the Arf-responsive, PIP2-dependent phospholipase D1. Biochem Biophys Res Commun. 1997;235:854–859. doi: 10.1006/bbrc.1997.6793. [DOI] [PubMed] [Google Scholar]
- 29.Mark B L, Jilkina O, Bhullar R P. Association of Ral GTP-binding protein with human platelet dense granules. Biochem Biophys Res Commun. 1996;225:40–46. doi: 10.1006/bbrc.1996.1128. [DOI] [PubMed] [Google Scholar]
- 30.Martinson E A, Scheible S, Greinacher A, Presek P. Platelet phospholipase D is activated by protein kinase C via an integrin alpha IIb beta 3-independent mechanism. Biochem J. 1995;310:623–628. doi: 10.1042/bj3100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moolenaar W H, Kranenburg O, Postma F R, Zondag G C. Lysophosphatidic acid: G-protein signalling and cellular responses. Curr Opin Cell Biol. 1997;9:168–173. doi: 10.1016/s0955-0674(97)80059-2. [DOI] [PubMed] [Google Scholar]
- 32.Morgan C P, Sengelov H, Whatmore J, Borregaard N, Cockcroft S. ADP-ribosylation-factor-regulated phospholipase D activity localizes to secretory vesicles and mobilizes to the plasma membrane following N-formymethionyl-leucyl-phenylalanine stimulation of human neutrophils. Biochem J. 1997;325:581–585. doi: 10.1042/bj3250581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgenstern E. The formation of compound granules from different types of secretory organelles in human platelets (dense granules and alpha-granules). A cryofixation/-substitution study using serial sections. Eur J Cell Biol. 1995;68:183–190. [PubMed] [Google Scholar]
- 34.Murai H, Ikeda M, Kishida S, Ishida O, Okazaki-Kishida M, Matsuura Y, Kikuchi A. Characterization of Ral GDP dissociation stimulator-like (RGL) activities to regulate c-fos promoter and the GDP/GTP exchange of Ral. J Biol Chem. 1997;272:10483–10490. doi: 10.1074/jbc.272.16.10483. [DOI] [PubMed] [Google Scholar]
- 35.Neer E J. Heterotrimeric G proteins: organizers of transmembrane signals. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 36.Ngsee J K, Elferink L A, Scheller R H. A family of ras-like GTP-binding proteins expressed in electromotor neurons. J Biol Chem. 1991;266:2675–2680. [PubMed] [Google Scholar]
- 37.Offermans S, Toombs C F, Hu Y-H, Simon M I. Defective platelet activation in Gαq-deficient mice. Nature. 1997;389:183–186. doi: 10.1038/38284. [DOI] [PubMed] [Google Scholar]
- 38.Okazaki M, Kishida S, Hinoi T, Hasegawa T, Tamada M, Kataoka T, Kikuchi A. Synergistic activation of c-fos promoter activity by Raf and Ral GDP dissociation stimulator. Oncogene. 1997;14:515–521. doi: 10.1038/sj.onc.1200860. [DOI] [PubMed] [Google Scholar]
- 39.Olofsson B, Chardin P, Touchot N, Zahraoui A, Tavitian A. Expression of the ras-related ralA, rho12 and rab genes in adult mouse tissues. Oncogene. 1988;3:231–234. [PubMed] [Google Scholar]
- 40.Park S H, Weinberg R A. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene. 1995;11:2349–2355. [PubMed] [Google Scholar]
- 41.Polakis P G, Weber R F, Nevins B, Didsbury J R, Evans T, Snyderman R. Identification of the ral and rac1 gene products, low molecular mass GTP-binding proteins from human platelets. J Biol Chem. 1989;264:16383–16389. [PubMed] [Google Scholar]
- 42.Shock D D, He K, Wencel D J, Parise L V. Ras activation in platelets after stimulation of the thrombin receptor, thromboxane A2 receptor or protein kinase C. Biochem J. 1997;321:525–530. doi: 10.1042/bj3210525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siess W. Molecular mechanisms of platelet activation. Physiol Rev. 1989;69:58–178. doi: 10.1152/physrev.1989.69.1.58. [DOI] [PubMed] [Google Scholar]
- 44.Spaargaren M, Bischoff J R. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras and Rap. Proc Natl Acad Sci USA. 1994;91:12609–12613. doi: 10.1073/pnas.91.26.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Symons M, Derry J M, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 46.Torti M, Lapetina E G. Structure and function of rap proteins in human platelets. Thromb Haemostasis. 1994;71:533–543. [PubMed] [Google Scholar]
- 47.Toullec D, Pianetti P, Coste H, Bellevergue P, Grand-Perret T, Ajakane M, Baudet V, Boissin P, Boursier E, Loriolle F, et al. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J Biol Chem. 1991;266:15771–15781. [PubMed] [Google Scholar]
- 48.Urano T, Emkey R, Feig L A. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- 49.van Willigen G, Hers I, Gorter G G, Akkerman J W N. Exposure of ligand-binding sites on platelet integrin alphaII-beta3 by phosphorylation of the beta3 subunit. Biochem J. 1996;314:769–779. [PMC free article] [PubMed] [Google Scholar]
- 50.Volknandt W, Pevsner J, Elferink L A, Scheller R H. Association of three small GTP-binding proteins with cholinergic synaptic vesicles. FEBS Lett. 1993;317:53–56. doi: 10.1016/0014-5793(93)81490-q. [DOI] [PubMed] [Google Scholar]
- 51.Wang K L, Khan M T, Roufogalis B D. Identification and characterization of a calmodulin-binding domain in Ral-A, a Ras-related GTP-binding protein purified from human erythrocyte membrane. J Biol Chem. 1997;272:16002–16009. doi: 10.1074/jbc.272.25.16002. [DOI] [PubMed] [Google Scholar]
- 52.Wolthuis R M F, Bauer B, van’t Veer L J, de Vries-Smits A M M, Cool R H, Spaargaren M, Wittinghofer A, Burgering B M T, Bos J L. RalGDS-like factor (Rlf) is a novel Ras and Rap 1A-associating protein. Oncogene. 1996;13:353–362. [PubMed] [Google Scholar]
- 53.Wolthuis R M F, de Ruiter N D, Cool R H, Bos J L. Stimulation of gene induction and cell growth by the Ras effector Rlf. EMBO J. 1997;22:6748–6761. doi: 10.1093/emboj/16.22.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]