Abstract
Yeast cells are keenly sensitive to the availability and quality of nutrients. Addition of glucose to cells growing on a poorer carbon source elicits a cell cycle delay during G1 phase and a concomitant increase in the cell size. The signal is transduced through the RAS-cyclic AMP pathway. Using synchronized populations of G1 cells, we show that the increase in cell size required for budding depends upon CLN1 but not other G1 cyclins. This delay in cell cycle initiation is associated specifically with transcriptional repression of CLN1. CLN2 is not repressed. Repression of CLN1 is not limited to the first cycle following glucose addition but occurs in each cell cycle during growth on glucose. A 106-bp fragment of the CLN1 promoter containing the three MluI cell cycle box (MCB) core elements responsible for the majority of CLN1-associated upstream activation sequence activity is sufficient to confer glucose-induced repression on a heterologous reporter. A mutant CLN2 promoter that is rendered dependent upon its three MCB core elements due to inactivation of its Swi4-dependent cell cycle box (SCB) elements is also repressed by glucose. The response to glucose is partially suppressed by inactivation of SWI4, but not MBP1, which is consistent with the dependence of MCB core elements upon the SCB-binding transcription factor (SBF). We suggest that differential regulation of CLN1 and CLN2 by glucose results from differences in the capacity of SBF to activate transcription driven by SCB and MCB core elements. Finally, we show that transcriptional repression is sufficient to explain the cell cycle delay that occurs in response to glucose.
The ability to mount adaptive responses to environmental changes is a fundamental property of living systems. This capacity is apparent at both the organismal and the cellular levels and reflects the existence of signaling pathways via which environmental conditions are translated into cellular responses. The stimuli provoking such responses are diverse, as are the mechanisms through which those signals are transduced. Although our understanding of machinery generating and transducing these signals is in many cases limited, it is clear that many of these signals ultimately result in alteration of the pattern of gene expression.
The availability and quality of nutrients comprise one of the environmental conditions which cells must monitor and to which they must respond. This is especially important to free-living microorganisms, such as the budding yeast Saccharomyces cerevisiae, which experience frequent dramatic changes in nutrient conditions and as a result require reliable mechanisms for coordinating growth with cell cycle progression. In those cells the decision of whether and when to initiate a new cell cycle occurs during the G1 interval. When glucose, the favored fermentable carbon source, becomes available to cells growing on poorer carbon sources, they not only change the pattern of expression of many genes involved in carbohydrate metabolism (reviewed in reference 28) but also undergo an increase in cell size (17, 19). That is, upon introduction of glucose the cells transiently delay cell cycle progression during the G1 phase and so attain a larger size than cells growing on poorer carbon sources. This increase has been attributed to a resetting of the minimal cell size required for budding (19).
Upon addition of glucose to the growth medium cells experience a transient spike in intracellular cyclic AMP (cAMP) levels followed by a resetting of basal cAMP to a higher level (4, 29, 35, 36). This increase in cAMP requires activation of adenylate cyclase, which in yeast results from activation by Ras-GTP (reviewed in reference 34). It is not clear whether the spike, the increased basal level, or both are important for the physiological responses to glucose. The only known target for cAMP in yeast cells is cAMP-dependent protein kinase. A number of observations support the view that the adjustment of cell size in response to glucose is induced via this pathway. First, mutants with reduced Ras pathway activity or with reduced but unregulated cAMP-dependent protein kinase activity bud at a smaller cell volume on poor carbon sources and fail to increase their size in response to glucose (1, 3, 36). Next, cells expressing constitutively activated Ras2, which in yeast is an activator of adenylate cyclase, display a large cell size when growing on nonglucose carbon sources and fail to further increase that size in response to glucose (1). Finally, artificially elevating the level of intracellular cAMP (by applying high levels of cAMP exogenously to cells compromised for cAMP turnover) results in an increase in the minimal budding size of cells growing on glucose (3, 36). It has not been possible to evaluate whether cAMP can mimic the effect of glucose on the size of cells growing on other carbon sources because exogenous cAMP is lethal to cells under those conditions (29). Nevertheless, these observations support a role for cAMP and, as a consequence, for cAMP-dependent protein kinase as mediators of the glucose-induced increase in the minimum budding size.
Genetic analysis suggests that the G1 cyclins are critical targets for the glucose–cAMP-induced increase in cell size (3, 36). This is consistent with the observation that cell volume at budding is largely a reflection of the activity of G1 forms of the cyclin-dependent protein kinase Cdc28 and, consequently, of the abundance of the G1 cyclins (8, 15, 21). Of the three G1 cyclins, Cln1 and Cln2 appear to be largely responsible for promoting cell cycle progression, whereas Cln3 acts primarily to promote transcription of CLN1 and CLN2 and other G1-specific genes (12, 33). Although all three G1 cyclins are important in establishing the size at which cells initiate a new cell cycle, CLN1 and, to a lesser extent, CLN2 appear to be targets of the cAMP signal (3, 36). Consistent with the results of that genetic analysis, exogenously added cAMP strongly represses expression of both CLN1 and CLN2.
Glucose is the physiologically relevant inducer of this response which resets the cell volume required for budding. Using synchronized populations of G1 daughter cells, we confirm that CLN1 is the primary target for the glucose-induced size increase. We show that the glucose-induced increase in cell size is exerted, largely, through repression of CLN1 expression, that this repression occurs during each cell cycle when cells are growing on glucose, and that repression occurs at the level of the promoter. Swi4-dependent cell cycle box (SCB)-binding transcription factor (SBF)-dependent upstream activation sequence (UAS) elements of CLN1 are probable targets for this repression. In contrast, neither the SBF-dependent gene CLN2 nor the MluI cell cycle box (MCB)-binding transcription factor (MBF)-dependent gene RNR1 is transcriptionally repressed in response to glucose. Both are strongly repressed by exogenously added cAMP. Our data support the view that differential regulation of CLN1 and CLN2 results from differences in their cis-acting binding sites for SBF. Finally, we show that regulation of the CLN1 promoter is sufficient and, at least in part, necessary for modulation of cell size in response to glucose. Thus, although CLN1 and CLN2 are coordinately regulated under many conditions (40), the present study provides one condition in which differential transcriptional regulation of these genes leads to a physiological response.
MATERIALS AND METHODS
Strains and plasmids.
The yeast strains used in this study were all congenic with 15DaubΔ, a bar1Δ ura3Δns derivative of BF264-15D (MATa ade1 his2 leu2-3,112 trp1-1a) (27). The relevant genotypes of individual strains used in this study are provided in Table 1.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotypea | Source or reference |
|---|---|---|
| 15Daub | MATa ade1 his2 leu2 trp1 ura3Δns bar1Δ | 38 |
| CWY228 | MATa cln1::TRP1 | 38 |
| CWY229 | MATa cln2::LEU2 | 38 |
| DSY560 | MATa cln3Δ ade1 his2 leu2 trp1 ura3Δns | This study |
| CWY227 | MATa cln1::TRP1 cln3::URA3 | This study |
| DSY790 | MATa cln2x/s cln3Δ LEU2::GAL1-CLN3 | This study |
| DSY12 | MATa swi4::LEU2 | 32 |
| DSY910 | MATa mbp1::URA3 | 33 |
| CWY762 | MATa cln2::LEU2 TRP1::CLN2p-CLN1 | This study |
| CWY765 | MATa cln2x/s cln3Δ LEU2::GAL1-CLN3 TRP1::CLN2p-CLN1 | This study |
| CWY780 | MATa cln1::TRP1 URA3::CLN1p-CLN2 | This study |
| CWY785 | MATa cln1Δ cln3Δ LEU2::GAL1-CLN3 URA3::CLN1p-CLN2 | This study |
All strains are derivatives of 15Daub and have the same genotype except as indicated.
The episomal CEN4 CYC1-lacZ reporter gene plasmids described in this report are all derivatives of pCZΔ (6). Constructions of plasmids pCLN2-lacZ (pΔ5′728) and pCLN2Δscb-lacZ (pΔscb) were as described previously (32). For the plasmid pCLN1p-lacZ, the XhoI/AflII fragment (−683 to −267) containing the CLN1 promoter was ligated to XhoI and XbaI linkers, respectively, digested with the appropriate enzyme, and cloned into a XhoI/XbaI-digested pCZΔ plasmid. The plasmid pCLN1(UAS)-lacZ contains a 106-bp fragment of the CLN1 promoter (−633 to −527) driving CYC1-lacZ. This fragment was obtained by annealing four oligonucleotides containing this sequence and EcoRI and SalI sites at the 3′ and 5′ ends, respectively. The fragment was cloned into the EcoRI and SalI site of pCZΔ.
To generate pCLN2p-CLN1, the SalI/BamHI fragment of pΔ5′728 was cloned into YIplac204. The resulting plasmid, YIplac204-CLN2p, was then digested with XhoI, blunted with T4 polymerase, ligated to a KpnI linker, and digested with SacI and KpnI. The KpnI/SacI fragment of CLN1 (−333 to 456) was ligated into SacI/KpnI-digested YIplac204-CLN2p. The resulting plasmid, YIplac204-CLN2p-CLN1, was linearized with SpeI and transformed into yeast. Transformants having the appropriate genomic structure were identified by PCR.
To generate pCLN1p-CLN2, the FspI/SpeI fragment of CLN2 (−341 to 489) was ligated to KpnI linkers, digested with KpnI, and ligated into KpnI/XbaI-digested YIplac211, resulting in YIplac211-CLN2. A KpnI fragment of CLN1 (−673 to −334) was ligated into the KpnI-digested plasmid YIplac211-CLN2. The resulting plasmid, YIplac211-CLN1p-CLN2, was linearized with XhoI and transformed into yeast. Transformants with the appropriate genomic structure were identified by PCR.
Culture conditions, cell synchronization, and cell size analysis.
All strains were grown in standard culture media: yeast extract-peptone-dextrose, minimal complete medium, or minimal medium lacking uracil. The carbon source was were either 2% glucose or 2% glycerol–1% ethanol.
Synchronous populations of small G1 cells were obtained by centrifugal elutriation. Cells were inoculated into 2 liters of minimal medium containing glycerol-ethanol as a carbon source and grown overnight at 30°C to an optical density at 600 nm of 0.5. Centrifugal elutriation was performed as described by Stuart and Wittenberg (33). The elutriated population of G1 cells was concentrated by centrifugation and divided into two aliquots which were resuspended in 200 ml of prewarmed minimal medium containing either glucose or glycerol-ethanol. Samples were taken for cell size analysis, determination of budding index, and RNA analysis at 20-min intervals for a period of 3 to 9 h.
Cell size analysis was performed with a Coulter Channelyzer. Statistical analysis was performed with software developed by Stefan Lanker and C.W. Approximately 1.5 × 105 cells were diluted into 20 ml of physiological saline, and the mean cell volume in femtoliters was determined.
For determination of budding index, 200 formaldehyde-fixed cells were counted and the proportion of budded cells was determined for each time point.
For RNA analysis from asynchronous cultures, cells were grown at 30°C to an optical density at 600 nm of 0.2 on minimal complete medium containing either 2% glucose or 2% glycerol–1% ethanol. Cells were harvested, and RNA was isolated as described below.
RNA analysis.
Total RNA was obtained from frozen cell pellets as described previously (32). RNase protection analysis was performed on 10 μg of total RNA with CLN2, ACT1, and lacZ riboprobe as described previously (32). The template for the CLN1 riboprobe consisted of a 190-bp fragment from the 5′ end of the CLN1 gene starting with the start codon. This fragment was cloned into pSP72 (Promega), and antisense riboprobes were synthesized from the RNA polymerase T7 promoter.
Northern blot analysis of 10 μg of total RNA was performed according to standard protocols (30). Probes were prepared from 1.6-kb CLN1 and CLN2 open-reading-frame, 1.6-kb ACT1 (BamHI/HindIII) and 2.6-kb RNR1 (EcoRI) fragments by random priming (Random Primed DNA Labeling Kit; Boehringer Mannheim).
Quantitation of the RNase protection and Northern blot data was performed with a PhosphorImager (Molecular Dynamics).
RESULTS
Glucose induces an increase in the minimum budding size of cells growing on poorer carbon sources.
Yeast cells undergo an increase in the minimal volume required for budding when shifted to glucose from a poorer carbon source (17, 19). This is best observed by evaluating the mean cell volume at budding in synchronous populations of small G1 daughter cells obtained by centrifugal elutriation. A population of small G1 cells was prepared from cells growing on a nonfermentable carbon source (2% glycerol–1% ethanol) and reinoculated either into the same medium or into 2% glucose; samples were then collected at 20-min intervals for analysis of cell volume and of the proportion of budded cells as an indicator of cell cycle initiation. An increase of ∼40% in the cell volume required for budding was observed in the glucose-grown cultures relative to those maintained on glycerol-ethanol medium (Fig. 1 and Fig. 2A, top panel). A similar adaptation has been shown to occur upon shift into glucose from a number of other carbon sources (19, 31). It should be noted that although the growth rate of the cells inoculated into glucose greatly exceeds that of cells reinoculated into glycerol-ethanol medium, the glucose-grown cells initiate budding at a later time than the glycerol-ethanol- grown cells. Thus, the size increase is not simply due to the increase in growth rate but results, at least in part, from a delay in the capacity of cells to initiate a new cell cycle. The terms cell size increase and cell cycle delay are used interchangeably throughout this study.
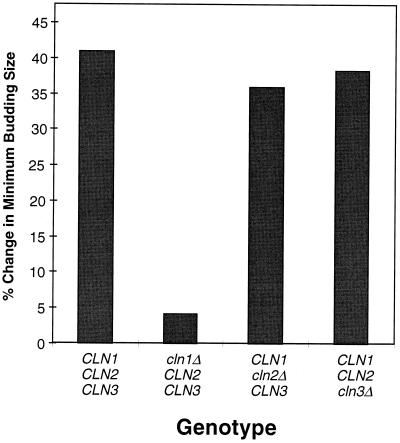
FIG. 1.
The percent change in minimum budding size of wild-type and CLN-deficient strains following a shift from glycerol-ethanol to glucose-containing medium. Synchronous populations of unbudded G1 daughter cells prepared by centrifugal elutriation from cells growing on glycerol-ethanol-containing medium were either reinoculated into fresh glycerol-ethanol-containing medium or inoculated into medium containing glucose. Samples were then taken at 20-min intervals and analyzed for cell size and budding index. Minimum budding size (MBS) is defined for the purpose of this study as the cell volume at which the budding index of a synchronous population of G1 daughter cells achieves 50% of its maximal level. A representative experiment is presented for each strain. The percent change is defined as follows: % change in MBS = [(MBS on glucose − MBS on glycerol-ethanol)/MBS on glycerol-ethanol] × 100.
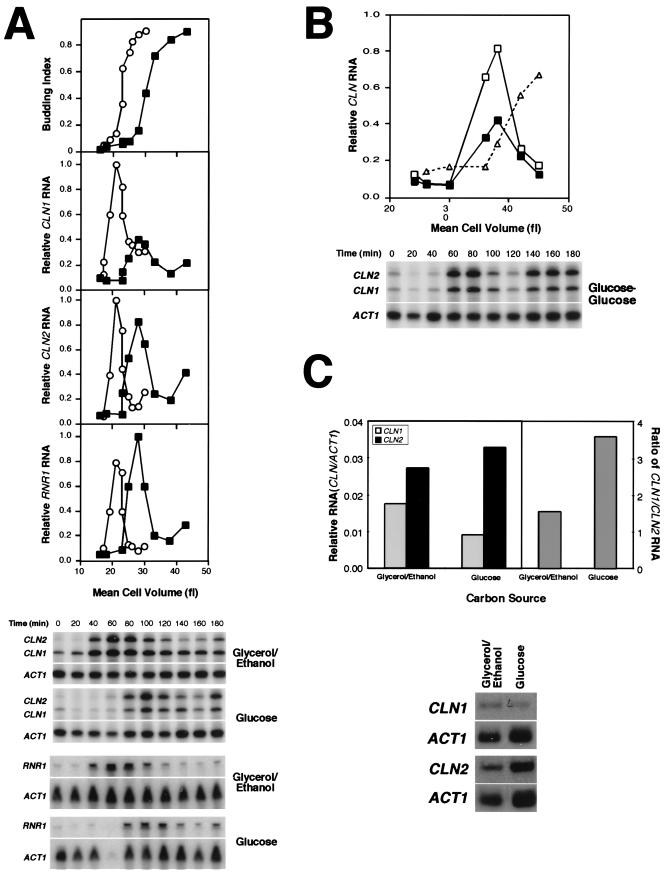
FIG. 2.
(A) Glucose differentially regulates the accumulation of CLN1 and CLN2 transcripts. A synchronous culture of small G1 cells (15Daub) pregrown on minimal complete medium containing glycerol-ethanol was prepared by centrifugal elutriation and inoculated into fresh medium containing either glycerol-ethanol (○) or glucose (▪) as a carbon source. Samples were taken at 20-min intervals over a period of 3 h and evaluated for cell volume, budding index, and CLN1, CLN2, and RNR1 transcript levels. The budding index is plotted relative to cell volume (top graph). CLN1, CLN2, and RNR1 transcript levels (normalized to ACT1 transcript level and then presented as the proportion of the maximal level of that transcript achieved on glycerol-ethanol-containing medium) were plotted as a function of cell volume (lower three graphs). Primary data from RNase protection assays for CLN1, CLN2, and ACT1 and Northern blots for RNR1 and ACT1 are presented in the lower part of panel A. (B) CLN1 transcription is repressed in cells maintained on glucose during each cell cycle. A synchronous culture of small G1 cells (15Daub) was pregrown on glucose, elutriated, and then reinoculated into glucose. CLN1 (▪) and CLN2 (□) transcripts were analyzed as described for panel A with the same preparations of probe. The transcript levels are presented as values relative to the maximal level achieved on glycerol-ethanol medium. The budding index is plotted relative to cell volume (▵) in the same graph. Primary data from RNase protection assays for CLN1, CLN2, and ACT1 are presented in the lower part of panel B. (C) Comparison of CLN1 and CLN2 levels in asynchronous cultures during continuous growth on glycerol-ethanol or glucose. Cells were grown to log phase on either glycerol-ethanol- or glucose-containing medium, and the level of CLN1 (gray bars) and CLN2 (black bars) transcript was evaluated relative to that of ACT1 by Northern blot analysis (left panel). The ratio of CLN1 to CLN2 transcript level under each condition is also presented (right panel). Primary data from the Northern blot analysis are presented in the lower part of panel C.
The glucose-induced G1 delay is mediated primarily through CLN1.
To investigate the importance of specific G1 cyclins in effecting the delay we have carried out the same analysis shown in Fig. 2A using a series of isogenic yeast strains carrying single and double mutations in the CLN1, CLN2, and CLN3 genes. The change in minimum budding size of each of these strains following a shift from glycerol-ethanol to glucose-containing media (Fig. 1) demonstrates that CLN1 is of primary importance in effecting the cell cycle delay. Cells lacking functional CLN1 did not undergo an increase in minimum budding size. Furthermore, cells with CLN1 as their only functional CLN gene (see Fig. 5B) reached a cell volume prior to budding that was almost twice that of cells growing in glycerol-ethanol-containing medium. This finding is in agreement with previous studies with glucose or exogenous cAMP (2, 36). This more extensive analysis establishes that the capacity of glucose to induce a G1-phase delay is unaffected by the characteristic cell size of the strain. That is, the extent to which cells are affected by glucose appears not to be related to the relative capacity of their functional cyclins to promote cell cycle progression but instead depends upon whether the specific cyclin that remains functional is a target for that regulation. For example, CLN3-deficient cells, which are substantially larger than wild-type cells when growing on either carbon source, undergo a comparable delay upon shift into glucose. In contrast, cells deficient in CLN1 are comparable in size to wild-type cells but fail to undergo a significant size increase in response to glucose. Finally, cells ectopically expressing CLN2 during early G1 phase (under control of the CLN3 promoter) bud at an exceedingly small cell size (33, 38) but still undergo a substantial delay in response to glucose (7). The delay in those cells remains dependent upon a functional CLN1 gene.
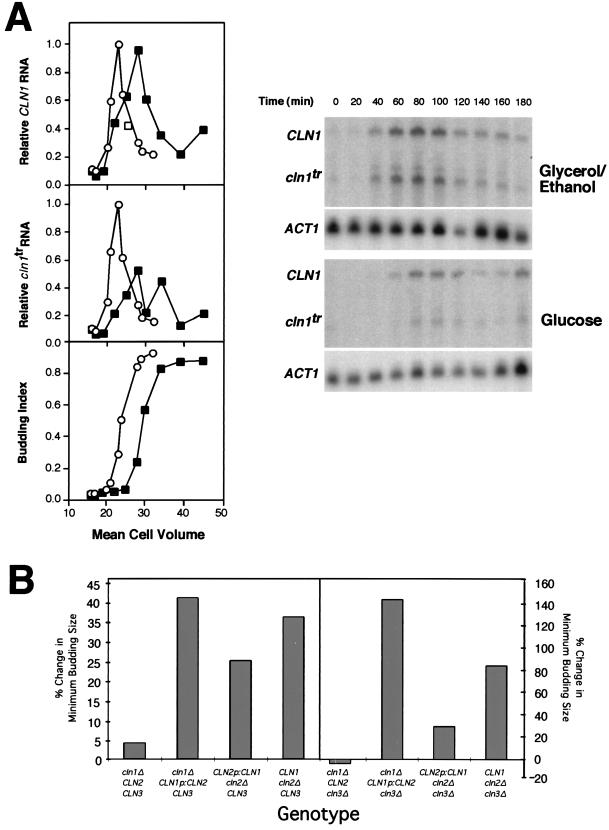
FIG. 5.
Transcriptional repression of CLN1 is required for full induction of the glucose-induced increase in minimum budding size. (A) The effect of glucose upon the CLN1 transcripts expressed from a CLN2p-CLN1 fusion (CLN1) and from a truncated CLN1 gene (cln1tr) under control of its own promoter was analyzed in synchronized G1 cells and plotted relative to cell volume as described in the legend to Fig. 3. Both forms of the CLN1 transcript were detected by Northern blot analysis with the CLN1 open reading frame as a probe. The budding index is plotted versus cell volume (bottom graph). Cells were grown in medium containing either glycerol-ethanol (○) or glucose (▪) as a carbon source. Primary data from Northern blot analysis of CLN1, cln1tr, and ACT1 are presented at right. (B) Exchange of the CLN1 and CLN2 promoters reveals that transcriptional regulation is sufficient and, at least in part, necessary for the glucose-induced increase in budding size. Cell volume at budding was determined in strains carrying a single integrated copy of CLN2p-CLN1 or CLN1p-CLN2 at the CLN1 or CLN2 locus, respectively. Cells were either grown continuously on glycerol-ethanol or shifted to glucose. A representative experiment is presented for each strain. Data for the relevant CLN-deficient strains were either reproduced from Fig. 1 or determined as described for Fig. 1 and in Materials and Methods. The percent change in minimum budding size is presented.
Glucose differentially regulates CLN1 and CLN2 transcription.
The relatively greater importance of CLN1 over CLN2 in mediating the cell cycle delay in response to glucose is consistent with the effect of glucose on accumulation of the CLN1 and CLN2 transcripts. The abundance of those transcripts in RNA prepared from cells responding to a shift from glycerol-ethanol into glucose was analyzed by an RNase protection assay (Fig. 2A). As suggested by previous analysis with asynchronous populations (36), the accumulation of CLN1 transcripts in small G1 daughter cells is strongly repressed in response to glucose relative to its level in cells maintained on glycerol-ethanol-containing medium. In contrast, the accumulation of a number of G1-specific transcripts, including CLN2, RNR1 (Fig. 2A), and SWI4 (14), is not repressed. Despite the differences in their expression level, all of these G1-specific transcripts, including CLN1, are delayed in response to glucose. The differential effect of glucose contrasts with the strong repression of all of these genes that is observed in response to exogenously applied cAMP (2, 36).
Since cells growing continuously on glucose are larger and have a larger minimum budding size than cells grown on poorer carbon sources, it seemed possible that repression of CLN1 expression occurred during each cell cycle in glucose-grown cells. We have examined that hypothesis in two ways. First, small G1 cells were prepared by centrifugal elutriation from an asynchronous population growing on glucose. The G1 population was then reinoculated into glucose and subjected to the same analysis as performed previously. To assess the relative level of expression, the CLN1 and CLN2 transcripts were analyzed with the same preparation of RNA probe as used for the analysis presented in Fig. 2A. The level of the CLN1 and CLN2 transcripts was then normalized to the maximal level accumulated on glycerol-ethanol medium (Fig. 2B). This experiment clearly shows that the level of the CLN1 transcript is reduced in each cell cycle during growth on glucose relative to the level in glycerol-ethanol-grown cells, whereas the level of the CLN2 transcript is unaffected. That observation predicts that CLN1 expression should be reduced in asynchronous populations of cells growing continuously on glucose compared to those growing on glycerol-ethanol. To test that prediction we analyzed the level of CLN1 transcript accumulating in unperturbed asynchronous populations growing on each carbon source (Fig. 2C). Both the absolute level of the CLN1 and the CLN2 transcripts and the ratio of the CLN1 transcript to the CLN2 transcript under each condition are presented. The ratio of CLN1 to CLN2 is utilized to eliminate the potential contribution of differences in cell cycle distribution between the two populations, which are doubling at different rates. Again, even in populations containing both mother and daughter cells, the repression of CLN1 by glucose is easily detectable. Thus, it is likely that repression of CLN1 occurs during the G1 phase of each cell cycle and contributes to the larger size of glucose-grown cells.
Glucose-induced repression of CLN1 transcription is exerted through the major CLN1 UAS.
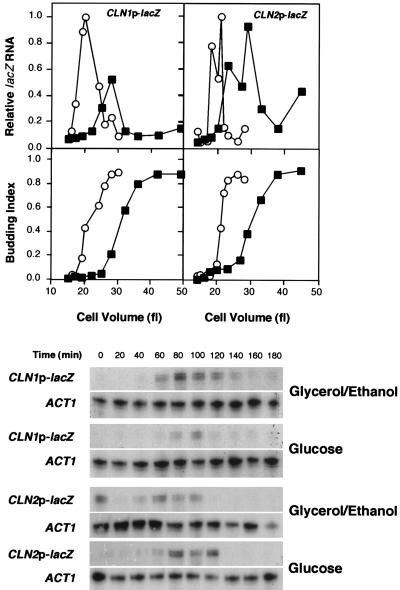
Experiments were next undertaken to establish whether the specific repression of CLN1 observed in the previous experiments occurred at the level of transcription initiation. To address this issue a UAS-deficient CYC1 promoter fused to the lacZ open reading frame (6) was placed under control of either the CLN1 promoter or the CLN2 promoter and introduced into a wild-type yeast strain. Expression of lacZ from that construct was analyzed by RNase protection in a synchronous culture of G1 daughter cells undergoing a shift from glycerol-ethanol to glucose according to the same experimental regimen described previously (Fig. 3). The behavior of the lacZ transcript expressed from these promoters was found to be identical to the behavior of the endogenous CLN1 and CLN2 transcripts. The CLN1 promoter was repressed in response to glucose, whereas the maximal activity of the CLN2 promoter was essentially unchanged. Thus, the CLN1 promoter is sufficient to confer glucose-induced repression upon the CYC1-lacZ reporter. We conclude that regulation of CLN1 by glucose is exerted at the level of transcription initiation as opposed to transcript stability.
FIG. 3.
Glucose-induced repression of CLN1 occurs at the level of the promoter. Wild-type cells (15Daub) carrying either CLN1p-CYC1-lacZ or CLN2p-CYC1-lacZ in the centromeric plasmid pCZΔ (6) were subjected to the same regimen as described for Fig. 2A. The lacZ and ACT1 transcripts were quantitated by RNase protection assay, and the values from cells growing in glycerol-ethanol (○)- or glucose (▪)-containing medium were plotted relative to the maximal level achieved on glycerol-ethanol-containing medium (top graph). The budding index is plotted versus the cell volume (bottom graph). Primary data from the RNase protection assays for lacZ and ACT1 are presented below. Low points in CLN2p-lacZ samples at 80 min on glycerol-ethanol and at 100 min on glucose resulted from degradation of high-molecular-weight RNA in those samples.
The coordinate expression of CLN1 and CLN2 during the cell cycle has been presumed to be a consequence of the presence of SCB elements (9, 22, 23, 32) in both promoters. SCB elements are target sites for SBF (reviewed in reference 5), one of the primary mediators of G1-specific transcription. However, recent findings of Partridge et al. (24) demonstrate that the major UAS activity associated with the CLN1 promoter is confined to a 106-bp fragment which lacks recognizable SCB elements and that the SCB elements which can be recognized have no associated UAS activity. Instead, the major CLN1 UAS has multiple sequences related to MCB elements, the target site for MBF (reviewed in reference 5) (Fig. 4A), a second G1-specific transcriptional activator. That study further showed that these MCB core elements are responsible for approximately 75% of G1-specific expression of CLN1 and that they act as binding sites for SBF in vitro. We therefore considered the possibility that these sequences were the targets for negative regulation of CLN1 transcription by glucose.
FIG. 4.
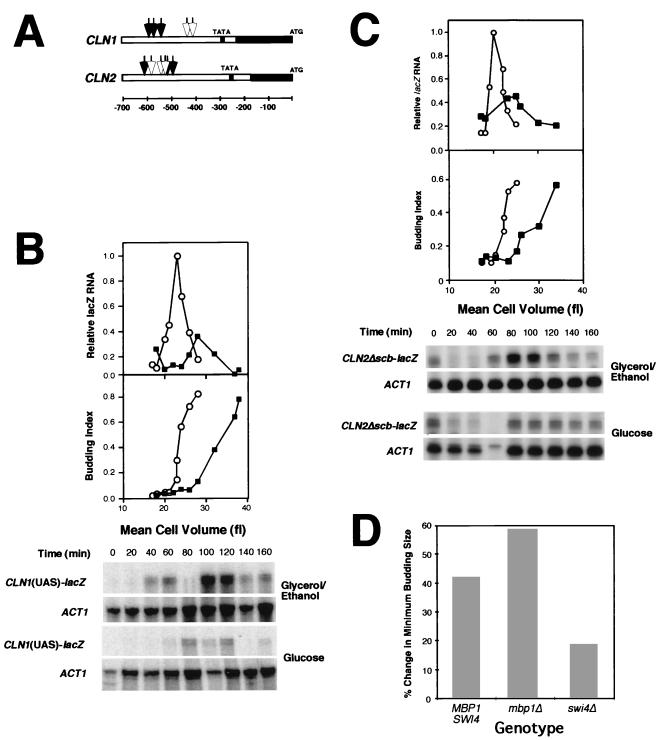
(A) Diagram of relevant features of the CLN1 and CLN2 promoters. Open arrows indicate the position of SCB elements. Closed arrows indicate the position of MCB core elements. The solid black bars represent the transcribed regions, and the open bars represent the nontranscribed promoter sequences. The gray regions represent the sequences surrounding the TATA element that are conserved between the CLN1 and CLN2 promoters. The scale is designated below in base pairs. (See references 9, 23, 24, and 32.) (B) Glucose-induced transcriptional repression of CLN1 is conferred by the major CLN1 UAS. The effect of glucose upon the expression of the CYC1-lacZ reporter of pCZΔ driven by a 106-bp fragment containing the major CLN1 UAS (24) was analyzed in synchronized G1 cells and plotted relative to cell volume as described in the legend to Fig. 3 (top graph). The budding index is plotted as a function of cell volume (bottom graph). Cells were grown in medium containing either glycerol-ethanol (○) or glucose (▪) as a carbon source. Primary data from RNase protection assays for lacZ and ACT1 are presented in the lower part of panel B. The 80-min sample from the glycerol-ethanol-grown culture is not plotted because in that sample there was significant degradation of high-molecular-weight RNA (see lower panel). (C) The effect of glucose upon the expression the CYC1-lacZ reporter in pCZΔ driven by a CLN2 promoter deficient in SCB elements, pCLN2Δscb (32), was analyzed in synchronized G1 cells and plotted relative to cell volume as described in the legend to Fig. 3 (top graph). The budding index is plotted versus cell volume (bottom graph). Cells were grown in medium containing either glycerol-ethanol (○) or glucose (▪) as a carbon source. Primary data from RNase protection assays for lacZ and ACT1 are presented below. Data from 20 through 160 min are represented on the graph. (D) Effect of swi4Δ and mbp1Δ mutations on the glucose-induced increase in minimum budding size. Cell volume at budding was determined in synchronous cultures of swi4Δ and mbp1Δ cells either growing on glycerol-ethanol medium or shifted to glucose medium. The percent change in minimum budding size is presented. A representative experiment is presented for each strain. All values were determined as described for Fig. 1 and in Materials and Methods.
To evaluate the involvement of the major CLN1 UAS in glucose-induced repression of CLN1 transcription, a 106-bp fragment of the CLN1 promoter containing the MCB core sequences was placed upstream of the CYC1-lacZ fusion in pCZΔ and introduced into a wild-type yeast strain. This strain was then subjected to the same experimental regimen as that utilized in the previous experiments, and the accumulation of lacZ transcripts was analyzed (Fig. 4B). As in the previous experiment in which the intact CLN1 promoter was used, the accumulation of lacZ transcripts was repressed when cells were transferred from glycerol-ethanol to glucose. This repression correlated with the G1 delay induced by glucose. Thus, the 106-bp fragment carrying the major CLN1 UAS was sufficient to confer G1-specific repression on CYC1-lacZ.
Since this small fragment of the CLN1 promoter, which contains three MCB core elements, was sufficient to explain glucose repression of CLN1 expression, we considered the possibility that repression was exerted through those elements. The presence of active MCB core elements in the CLN2 promoter allowed us to test this hypothesis (22, 23, 32) (Fig. 4A). In contrast to their primary role in the CLN1 promoter, the MCB core elements of the CLN2 promoter contribute less than half of its UAS activity (32). Thus, mutational inactivation of all three SCB elements yielded a promoter (CLN2Δscb) that drives cell cycle-dependent transcription to approximately 30% of the wild-type level, the majority of which can be attributed to the MCB core elements (32). Cells carrying the CLN2Δscb promoter fused to CYC1-lacZ in pCZΔ were subjected to the same synchrony and carbon source shift regimen as in the previous experiments, and the accumulation of the lacZ reporter transcript was then analyzed (Fig. 4C). In striking contrast to the wild-type CLN2 promoter, the CLN2Δscb promoter is strongly repressed in response to glucose. The extent of repression is similar to that observed with the CLN1 promoter driving the same reporter. Although this experiment does not directly assign that repression to the MCB core elements, these results are consistent with regulation exerted either directly through those elements or through elements that depend upon MCB core elements for their repressive activity. The failure of the wild-type promoter to be significantly repressed under the same conditions (Fig. 3) demonstrates that SCB elements are not targets for this regulation.
Since MCB core elements, as well as SCB elements, are targets for SBF (reviewed in reference 5), we would predict that the inactivation of SBF but not MBF would suppress the effect of glucose on cell size. To test this hypothesis we evaluated the effect of mutational inactivation of SWI4 and MBP1, the DNA binding components of SBF and MBF, respectively, on the ability of glucose to induce an increase in the minimal cell volume at budding. Strains containing disrupted alleles of either SWI4 or MBP1 were subjected to the carbon source shift regimen previously described and the cell volume at budding was determined (Fig. 4D). As predicted, whereas mbp1Δ mutants exhibited a substantial increase in cell size in response to glucose, that response was largely eliminated in swi4Δ mutants. Thus, SBF is required to achieve a glucose-induced delay in cell cycle progression equal to that of the wild type.
Transcriptional repression is sufficient and, at least in part, necessary for the glucose-induced increase in the size at budding.
The previous experiments demonstrate that the MCB core elements of the CLN1 promoter are sufficient to explain the behavior of the CLN1 transcript in response to glucose. To address the importance of transcriptional repression by glucose in delaying cell cycle initiation, the CLN1 gene was placed under control of the CLN2 promoter, which is not repressed in response to glucose. This was accomplished by fusing the CLN2 promoter to the CLN1 gene just upstream of their highly conserved TATA elements (Fig. 4A). The CLN1 gene in this construct was truncated within the open reading frame. When integrated at the CLN1 locus by homologous recombination, this construct results in a hybrid gene (CLN2p-CLN1) that has all of the known UAS activities of the CLN2 promoter yet retains the transcriptional start site and the transcribed sequences of the CLN1 gene. The integrated fusion gene was shown to be functional based upon its ability to complement a deficiency of all three CLN genes (data not shown). Another useful consequence of this integration was that the native CLN1 promoter was left driving a nonfunctional truncated gene, producing an RNA transcript (cln1tr) that was detectable with the CLN1 probe.
The CLN2p-CLN1 construct was introduced into both a cln2Δ strain and a cln2Δ cln3Δ strain to evaluate its effect on the extent of the glucose-induced delay in cell cycle initiation. The cln2Δ strain carrying this construct was also analyzed to evaluate the effect of glucose on the expression of CLN1 transcripts. Northern blot analysis with the full-length CLN1 open reading frame as a probe enabled us to detect transcripts expressed from both the native CLN1 promoter (cln1tr) and the CLN2 promoter (CLN2p-CLN1) (Fig. 5A). Two important observations can be made from this experiment. First, CLN2p-CLN1 is not repressed in response to glucose, whereas the truncated CLN1 transcript continues to be repressed. This demonstrates that regulation at the level of the promoter is not only sufficient but also necessary for glucose repression of CLN1 transcript accumulation. Second, failure to repress the CLN1 transcript results in partial suppression of the glucose-induced increase in budding size. This is most easily observed by comparing the strains carrying the CLN2p-CLN1 construct to equivalent strains having either a wild-type CLN1 or a wild-type CLN2 gene (Fig. 5B). That comparison reveals that expression of CLN1 under control of the CLN2 promoter eliminates a portion of the increase in cell size that is observed in the comparable strains having wild-type CLN1. Thus, transcriptional regulation of CLN1 accounts for approximately one-half of the increase in budding size observed in response to glucose. Posttranscriptional mechanisms are likely to account for the remainder of the delay.
The experiment described above shows that posttranscriptional mechanisms can contribute to the repression of CLN1 in response to glucose. However, a similar analysis of CLN2 expressed under control of the CLN1 promoter (CLN1p-CLN2) suggests that such mechanisms are not necessary. The CLN1p-CLN2 fusion was constructed essentially as described above for CLN2p-CLN1 and integrated into either cln1Δ or cln1Δ cln3Δ strains. These strains, in which the CLN1p-CLN2 construct replaces CLN2, were analyzed by the same regimen described previously, and the cell volume at budding was established (Fig. 5B). Unlike the CLN2p-CLN1 strains, these strains exhibited an increase in budding size in response to glucose that was either similar to or more dramatic than the equivalent strain expressing CLN1 under control of its own promoter. In contrast, when CLN2 is expressed under control of its own promoter, little or no size increase is observed. We conclude that transcriptional repression of the CLN1 promoter is sufficient to account for the cell cycle delay observed in response to glucose.
DISCUSSION
The G1 cyclins Cln1 and Cln2 in conjunction with the Cdc28 cyclin-dependent protein kinase act to promote cell cycle initiation (reviewed in reference 26). Their activity during the cell cycle is regulated primarily at the level of protein abundance, which is a consequence of periodic transcriptional activity (reviewed in reference 5) coupled with regulated protein instability (11, 18, 39). The expression of these critical cell cycle regulators has been shown to be coordinately regulated both during the cell cycle and in response to environmental regulators of cell proliferation, including mating pheromone and nutrient limitation (40). Coordinate expression is predicted by their common mode of transcriptional regulation. Both genes are targets of SBF and are dependent upon CLN3 and CDC28 for activation (reviewed in reference 5).
However, genetic analysis of CLN mutants has suggested that these genes are, in some circumstances, independently regulated. Inactivation of CLN1, but not of other G1 cyclin genes, suppresses the glucose-induced increase in minimum size required for entry into a new cell cycle, suggesting that CLN1 is a specific target for negative regulation by glucose (36). In this study we have used synchronized populations of G1 daughter cells to confirm that CLN1 is required for glucose to effect an increase in the minimum size for cell cycle initiation. We extend previous studies by establishing here that CLN1 transcription is specifically repressed in glucose-grown cells. Neither CLN2 nor other G1-specific transcripts are repressed in response to glucose. CLN1 repression is not a transient response to a shift to glucose from poorer carbon sources, since it occurs during each cell cycle when cells are growing continuously on glucose. This suggests that repression of CLN1 transcription is a consequence of the higher basal level of cAMP observed in cells maintained on glucose (4, 29) and is not solely a response to the glucose-induced spike of cAMP.
In contrast to the rather specific effect of glucose on CLN1 expression, treatment of cells with exogenous cAMP, the presumed mediator of the glucose signal, results in dramatic repression of CLN2, RNR1, and a number of other G1-specific genes. Thus, although the effect of added cAMP is in many ways similar to the effect of glucose, there are significant differences. Whether this is simply due to a distinction between the intracellular level of cAMP attained in response to glucose and that attained by treatment of cells with cAMP or whether it is instead a reflection of a qualitative difference between the signals generated by these two stimuli is unclear. It is known that elements of the RAS-cAMP pathway are required for the glucose-induced increase in minimum budding size, and cAMP treatment appears to mimic effects of hyperactivation of that pathway.
CLN1 is the only gene among those examined in this study which undergoes substantial transcriptional repression in response to glucose. In contrast, glucose induces a delay in the accumulation of all G1-specific transcripts, including CLN1. The source of this delay is not understood. One hypothesis to explain this pattern of gene expression is that all G1-specific transcription is initially repressed in response to glucose and that cells are initially delayed during the G1 phase. However, under these conditions the CLN2 transcript continues to accumulate until it reaches a level equivalent to that observed in cells growing on a poorer carbon source, thereby generating sufficient cyclin-dependent kinase (CDK) activity to promote cell cycle progression. This scenario holds that CLN1 is more effectively repressed than other G1-specific genes and, unlike those genes, never achieves the levels observed in cells maintained in medium without glucose. Several observations appear to be at odds with this hypothesis. First, this hypothesis predicts that, like the CLN2 transcript in wild-type cells, the CLN1 transcript in CLN2-deficient cells would be delayed but would ultimately reach the same level as that achieved in medium lacking glucose. Yet, the kinetics of CLN1 transcript accumulation in cln2Δ mutants is similar to that observed in wild-type cells in being both delayed and repressed (7). Next, where it has been examined the duration of the delay in G1-specific transcripts appears to be equivalent to the duration of the delay in budding (7). Since the timing of budding is a reflection of the timing of expression of G1 cyclins, the lack of a cell cycle delay in CLN1-deficient cells suggests that in those cells there is no delay in the program of G1-specific transcription. This leaves us with the hypothesis that under the conditions of the glucose-induced delay, CLN1 not only influences the timing of cell cycle initiation but also affects the timing of activation of G1-specific transcription. Although that proposal appears inconsistent with evidence that the activation of G1-specific transcription is independent of the activity of CLN1 and CLN2 (12, 33), these genes have been shown to be capable of promoting their own expression under some conditions (10, 13, 33). Resolution of this issue awaits further analysis.
To understand the basis for the differential regulation of the CLN1 and CLN2 promoters by glucose, we have attempted to localize the cis-acting targets required for repression. We have shown that a 106-bp fragment encompassing the major CLN1 UAS is sufficient to confer glucose-induced repression upon a UAS-deficient reporter (Fig. 4C). The UAS activity associated with this fragment is derived from three MCB core elements (24). Related MCB core elements in the CLN2 promoter also appear to be repressed by growth on glucose-containing medium. However, this is only observed when the SCB elements, the primary cis-acting elements of the CLN2 promoter, are eliminated (32). Unexpectedly, the MCB core elements of both the CLN1 and CLN2 promoters depend primarily upon SWI4, the DNA binding component of SBF, and not upon MBP1, the DNA binding component of MBF, for maximal expression (24, 31, 32). Partridge and coworkers have shown that SBF binds effectively to the MCB core elements from CLN1 in vitro (24). Consistent with the importance of SBF for activation of those elements, inactivation of SWI4 but not MBP1 partially suppresses the extent of the glucose-induced increase in budding size.
We speculate that the differential effect of glucose upon these two genes is explained by the difference in their cis-acting targets for SBF. For instance, it is possible that cells respond to glucose by altering the capacity of SBF to activate expression of MCB core elements without affecting its activity toward SCB elements. Alternatively, a lower affinity for MCB core elements than for SCB elements may be an inherent property of SBF, and glucose may simply act to lower its overall activity. It must also be true that the activity of MBF is not similarly affected since RNR1 and other MBF-dependent genes are not repressed by glucose. At present we cannot exclude the possibility that a cis-acting element distinct from the MCB core elements is responsible for the modulation of activity we observed in response to glucose since the minimal CLN1 promoter fragment used in this study includes other sequences. However, if such elements are present in the CLN1 promoter, then similar elements must also exist in the CLN2 promoter since the SCB-deficient promoter is repressed by glucose. The recently described Xbp1 transcriptional repressor (20) is clearly not responsible for CLN1 repression since the Xbp1 binding site is absent both from the glucose-responsive 106-bp fragment of the CLN1 promoter and from the CLN2 promoter.
Analysis of cells carrying CLN1 expressed under control of the wild-type CLN2 promoter, which is not repressed by glucose, shows that posttranscriptional mechanisms can contribute to the cell cycle delay caused by glucose. Nevertheless, cells carrying a complementary construct with CLN2 under the control of the CLN1 promoter are very sensitive to glucose, suggesting that transcriptional repression alone is sufficient. Since the CLN1 and CLN2 promoters drive transcription at comparable levels and with similar cell cycle timing, this experiment eliminates several caveats associated with previous experiments that used constitutive hyperexpression of CLN1 (2, 36). The high level of expression used in those experiments resulted in full suppression of the cAMP-induced delay.
Our results suggest that posttranscriptional mechanisms can contribute to this response. Although a number of regulatory mechanisms are possible, the involvement of cAMP-dependent protein kinase in this response (2, 3, 36) suggests a role for protein phosphorylation. Phosphorylation of G1 cyclins, including Cln1, has been well documented (18, 37, 40). At least some of those modifications are involved in inducing protein turnover (18, 39, 41). However, analysis of the Cln1 protein expressed from the CLN2 promoter failed to reveal a decrease in abundance in response to glucose (14). Furthermore, there is still no evidence for direct phosphorylation of G1 cyclins by cAMP-dependent protein kinase. Alternatively, protein kinase A may affect the activity of the Cln1-CDK complex. This issue has yet to be investigated.
In contrast to the regulation of CLN1 expression described in this study, other cyclins appear to be induced on glucose. CLN3 is one example. The addition of glucose to a culture growing on a carbon-poor source increases the level of CLN3 expression as much as 10-fold (16, 31). Conversely, the number of CLN3 transcripts decreases as cells deplete glucose and undergo the diauxic shift (16). Surprisingly, this dramatic change in CLN3 expression which occurs upon addition of glucose fails to have a significant effect on the expression of CLN1 and CLN2. It is unclear whether this results from a failure of the increase in CLN3 transcripts to result in a commensurate increase in Cln3 protein due to translational regulation (25) and/or Cln3-associated CDK activity or whether it instead results from a greater requirement for Cln3 to activate CLN1 and CLN2 transcription on glucose. Yet, it is clear that a deficiency in CLN3 has a dramatic effect on both the timing and the extent of CLN1 and CLN2 gene expression on both glucose and poorer carbon sources (12, 33, 37). Therefore, glucose and CLN3 must act independently to affect CLN1 gene expression and thereby the timing of cell cycle initiation.
The physiological relevance for the glucose-induced delay in cell cycle initiation remains an enigma. It is likely to reflect an important adaptation to the change in growth rate. For example, it seems reasonable to suggest that cells growing on nonfermentable carbon sources benefit from an increase in the surface/volume ratio that is not necessary when cells are growing on fermentable carbon sources. However, this interpretation appears to be incorrect since the effect on cell size is restricted to glucose and a few other carbon sources that support rapid growth. Other fermentable sugars, such as sucrose and raffinose, fail to induce a cell size increase.
An explanation that is perhaps more trivial is that the increase in cell size results solely from the inability of generation time to keep pace with rapid growth. This hypothesis is supported by the observation that cells growing continuously in a chemostat with glucose as a carbon source initiate budding at a cell size characteristic of the age of the mother (number of bud scars) when glucose is limiting (17). However, as the concentration of glucose increases and cells approach the minimum generation time their size at budding increases. The magnitude of that increase is comparable to the increase observed when cells were shifted to glucose in the present study. This effect on cell size is related to growth rate and not glucose per se since nitrogen limitation results in an increase in cell cycle time and an accompanying decrease in cell volume at budding (19). This explanation would be consistent with the failure of the fermentable carbon sources that are incapable of supporting rapid growth to cause a size increase.
Although the latter explanation is consistent with most experimental observations it is not without its inconsistencies. Notably, Baroni et al. (3) have shown that in glucose-grown cells a G1 delay and an associated increase in cell size at budding occur in response to exogenously added cAMP. Yet cAMP fails to increase the biosynthetic capacity of those cells. Furthermore, the apparent increase in the minimum size required for budding resulting from cAMP treatment can occur in cells which are restricted from growth by starvation for nitrogen (2). Thus, it appears that cAMP treatment can effectively separate the effect of glucose on the growth rate from its effect on the minimum size required for budding, suggesting that the explanation for this finding is more complex. Furthermore, the observation that the response is associated with alterations in gene expression that are sufficient to explain the behavior indicates that it is unlikely to simply reflect a passive effect of the relationship between growth rate and cell cycle progression. We conclude that the ability of cells to increase in size in response to glucose is either physiologically important or a manifestation of a regulatory pathway that plays some other physiologically important role.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank Steve Haase and Peter Kaiser for careful reading of the manuscript and helpful suggestions. We also thank Linda Breeden, Warren Heideman, and M. David Baroni for communication of results prior to publication and for stimulating discussion. We are grateful to Linda Breeden, Steve Elledge, and Mike Tyers for plasmids and Stefan Lanker for development and maintenance of software for analysis of Coulter Channelyzer data.
We acknowledge support for D.C.-S. by a fellowship from the U.S. Public Health Service (NIH F32 GM16857). D. Stuart is the recipient of a Special Fellowship from the Leukemia Society of America. This work was supported by U.S. Public Health Service grants GM43487 and GM46006 from the National Institutes of Health.
REFERENCES
- 1.Baroni M D, Martegani E, Monti P, Alberghina L. Cell size modulation by CDC25 and RAS2 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:2715–2723. doi: 10.1128/mcb.9.6.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baroni M D, Monti P, Alberghina L. Repression of growth-regulated G1 cyclin expression by cyclic AMP in budding yeast. Nature. 1994;371:339–342. doi: 10.1038/371339a0. [DOI] [PubMed] [Google Scholar]
- 3.Baroni M D, Monti P, Marconi G, Alberghina L. cAMP-mediated increase in the critical cell size required for the G1 to S transition in Saccharomyces cerevisiae. Exp Cell Res. 1992;201:299–306. doi: 10.1016/0014-4827(92)90277-f. [DOI] [PubMed] [Google Scholar]
- 4.Beullens M, Mbonyi K, Geerts L, Gladines D, Detremerie K, Jans A W H, Thevelein J M. Studies on the mechanism of the glucose-induced cAMP signal in glycolysis and glucose repression mutants of the yeast Saccharomyces cerevisiae. Eur J Biochem. 1988;172:227–231. doi: 10.1111/j.1432-1033.1988.tb13877.x. [DOI] [PubMed] [Google Scholar]
- 5.Breeden L. Start specific transcription in yeast. Curr Top Microbiol Immunol. 1996;208:95–127. doi: 10.1007/978-3-642-79910-5_5. [DOI] [PubMed] [Google Scholar]
- 6.Buchman A R, Kornberg R D. A yeast ars-binding protein activates transcription synergistically in combination with other weak activating factors. Mol Cell Biol. 1990;10:887–897. doi: 10.1128/mcb.10.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman-Shimshoni, D. Unpublished data.
- 8.Cross F. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cross F R, Hoek M, McKinney J D, Tinkelenberg A H. Role of Swi4 in cell cycle regulation of CLN2 expression. Mol Cell Biol. 1994;14:4779–4787. doi: 10.1128/mcb.14.7.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cross F R, Tinkelenberg A H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 11.Deshaies R J, Chau V, Kirschner M. Ubiquitination of the G1 cyclin Cln2p by a Cdc34p-dependent pathway. EMBO J. 1995;14:303–312. doi: 10.1002/j.1460-2075.1995.tb07004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirick L, Böhm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- 14.Flick, K., and C. Wittenberg. Unpublished data.
- 15.Hadwiger J A, Wittenberg C, de Barros Lopes M A, Richardson H E, Reed S I. A family of cyclin homologs that control G1 phase in yeast. Proc Natl Acad Sci USA. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heideman, W. Personal communication.
- 17.Johnston G C, Ehrhardt C W, Lörincz A, Carter B L A. Regulation of cell size in the yeast Saccharomyces cerevisiae. J Bacteriol. 1979;137:1–5. doi: 10.1128/jb.137.1.1-5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin, Cln2, induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 19.Lörincz A T, Carter B L A. Control of cell size at bud initiation in Saccharomyces cerevisiae. J Gen Microbiol. 1979;113:287–295. [Google Scholar]
- 20.Mai B, Breeden L. Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol Cell Biol. 1997;17:6491–6501. doi: 10.1128/mcb.17.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nash R, Tokiwa G, Anand S, Erickson K, Futcher A B. The WHI1+ gene of S. cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of the G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- 23.Ogas J, Andrews B, Herskowitz I. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- 24.Partridge J F, Mikesell G E, Breeden L L. Cell cycle-dependent transcription of CLN1 involves Swi4 binding to MCB-like elements. J Biol Chem. 1997;272:9071–9077. doi: 10.1074/jbc.272.14.9071. [DOI] [PubMed] [Google Scholar]
- 25.Polymenis M, Schmidt E V. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–2531. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed S. START and the G1-S phase transition in budding yeast. In: Barnes B D, Glover D M, editors. Cell cycle control. Oxford, England: IRL Press; 1995. pp. 40–62. [Google Scholar]
- 27.Richardson H E, Wittenberg C, Cross F R, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 28.Ronne H. Glucose repression in fungi. Trends Genet. 1995;11:12–17. doi: 10.1016/s0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 29.Russell M, Bradshaw-Rouse J, Markwardt D, Heideman W. Changes in gene expression in the Ras/adenylate cyclase system of Saccharomyces cerevisiae: correlation with cAMP levels and growth arrest. Mol Biol Cell. 1993;4:757–765. doi: 10.1091/mbc.4.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Stuart, D. Unpublished data.
- 32.Stuart D, Wittenberg C. Cell cycle-dependent transcription of CLN2 is conferred by multiple distinct cis-acting regulatory elements. Mol Cell Biol. 1994;14:4788–4801. doi: 10.1128/mcb.14.7.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 gene expression in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 34.Thevelein J M. The RAS-adenylate cyclase pathway and cell cycle control in Saccharomyces cerevisiae. Antonie Leeuwenhoek. 1992;62:109–130. doi: 10.1007/BF00584466. [DOI] [PubMed] [Google Scholar]
- 35.Thevelein J M, Beullens M. Cyclic AMP and the stimulation of trehalase activity in the yeast Saccharomyces cerevisiae by carbon sources, nitrogen sources and inhibitors of protein synthesis. J Gen Microbiol. 1985;131:3199–3209. doi: 10.1099/00221287-131-12-3199. [DOI] [PubMed] [Google Scholar]
- 36.Tokiwa G, Tyers M, Volpe T, Futcher B. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature. 1994;371:342–345. doi: 10.1038/371342a0. [DOI] [PubMed] [Google Scholar]
- 37.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdivieso M H, Sugimoto K, Jahng K-Y, Fernandes P M B, Wittenberg C. FAR1 is required for posttranscriptional regulation of CLN2 gene expression in response to mating pheromone. Mol Cell Biol. 1993;13:1013–1022. doi: 10.1128/mcb.13.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willems A R, Lanker S, Patton E E, Craig K L, Nasson T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 40.Wittenberg C, Sugimoto K, Reed S I. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- 41.Yaglom J, Linskens M H, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]