Abstract
Ty1 retrotransposons in Saccharomyces cerevisiae are maintained in a state of transpositional dormancy. We isolated a mutation, rtt100-1, that increases the transposition of genomic Ty1 elements 18- to 56-fold but has little effect on the transposition of related Ty2 elements. rtt100-1 was shown to be a null allele of the FUS3 gene, which encodes a haploid-specific mitogen-activated protein kinase. In fus3 mutants, the levels of Ty1 RNA, protein synthesis, and proteolytic processing were not altered relative to those in FUS3 strains but steady-state levels of TyA, integrase, and reverse transcriptase proteins and Ty1 cDNA were all increased. These findings suggest that Fus3 suppresses Ty1 transposition by destabilizing viruslike particle-associated proteins. The Fus3 kinase is activated through the mating-pheromone response pathway by phosphorylation at basal levels in naive cells and at enhanced levels in pheromone-treated cells. We demonstrate that suppression of Ty1 transposition in naive cells requires basal levels of Fus3 activation. Substitution of conserved amino acids required for activation of Fus3 derepressed Ty1 transposition. Moreover, epistasis analyses revealed that components of the pheromone response pathway that act upstream of Fus3, including Ste4, Ste5, Ste7, and Ste11, are required for the posttranslational suppression of Ty1 transposition by Fus3. The regulation of Ty1 transposition by Fus3 provides a haploid-specific mechanism through which environmental signals can modulate the levels of retrotransposition.
Retroviruses and endogenous retrovirus-like elements are ubiquitous in the eucaryotic kingdom and have been involved in the formation of a significant portion of the typical eucaryotic genome (45). Hence, eucaryotes have evolved many types of regulatory mechanisms to control the replication and mobility of retroelements. However, only a few host genes that control retroviral replication in vertebrates have been identified. A recent example is the mouse Fv1 gene, whose product is derived from the gag domain of an endogenous retroelement and is postulated to inhibit murine leukemia virus replication by interacting with the viral capsid protein (4).
Aside from the infectivity of the retroviral particle, the steps of retrotransposition are analogous to retroviral replication. A well-characterized model system to study host regulation of retrovirus-like elements is the Ty1 retrotransposon in the yeast Saccharomyces cerevisiae (8, 50). Ty1 elements have two long terminal repeats (LTRs) surrounding a central region consisting of two overlapping open reading frames: TyA, which encodes a structural capsid protein, and TyB, which encodes protease (PR), integrase (IN), and reverse transcriptase (RT) activities. Replication of Ty1 occurs in the following sequence of events: a chromosomal Ty1 element is transcribed from LTR to LTR by RNA polymerase II into a terminally redundant RNA that is polyadenylated and transported to the cytoplasm. TyA and TyA-TyB fusion protein are synthesized from the full-length transcript, the latter requiring a translational frameshift event that occurs with about 3% efficiency (37). Subsequently, both Ty1 proteins associate with the Ty1 transcript to form viruslike particles (VLPs). The VLP is the site of proteolytic maturation of TyA protein and processing of TyB into separate PR, IN, and RT proteins. Ty1 protein processing is performed by PR and is essential for transposition (57). Reverse transcription of the Ty1 RNA into a full-length linear duplex cDNA occurs in the VLP, using tRNA-Met(i) as a primer (11, 23). The Ty1 cDNA is then transported into the nucleus of the same cell, where nonhomologous integration is catalyzed by the IN protein. In the absence of IN, Ty1 cDNA is used as a template for gene conversion of preexisting Ty1 elements or LTRs (51).
Most of the 25 to 30 copies of Ty1 in the haploid yeast genome are competent for transposition and highly transcribed; however, individual elements transpose in only 1 of 105 to 107 cells per generation (15, 17). This “transpositional dormancy” is overcome when a Ty1 element is expressed at high levels from the inducible GAL1 promoter, which increases the efficiency of Ty1 transposition at a posttranslational step, perhaps by overwhelming or evading a host-encoded inhibitor (30, 50). One type of inhibition may occur at the level of maturation of Ty1 proteins in the VLP, which is very inefficient in normal yeast cells and greatly enhanced by induction of GAL1-Ty1 expression (16). Since the formation of mature VLPs is an essential step in transposition, inhibition of VLP formation or VLP instability in normal yeast cells could account for the low levels of transposition.
The FUS3 gene encodes a haploid-specific mitogen-activated protein (MAP) kinase that is activated through a conserved signal transduction cascade in response to mating pheromone. Pheromone secreted by cells of one of the two mating types of haploid yeast, either a or α, is bound by a serpentine receptor on the surface of cells of the opposite mating type. Pheromone binding triggers the activation of a heterotrimeric G-protein consisting of Ste4, Ste18, and Gpa1, which transmits the signal to the Ste20 kinase (39). In turn, Ste20 activates a MAP kinase module consisting of Ste11 (a MAP kinase kinase kinase [MEK kinase]), Ste7 (a MAP kinase kinase [MEK]), and Fus3 (a MAP kinase) (24, 29, 44). The module is held together by interaction with the LIM domain containing protein Ste5 (12). Once activated, Fus3 phosphorylates the cyclin-dependent kinase inhibitor Far1, which mediates G1 arrest (10, 26, 46). In addition, Fus3 activates the transcription factor Ste12, which binds to pheromone response elements of mating-specific genes and induces their transcription (26, 27). The products of mating-specific genes are required for morphological changes that lead to cell fusion and formation of the a/α diploid cell.
Multiple MAP kinase cascades function in haploid yeast cells, and the components of these pathways can be used in different MAP kinase modules to respond to different extracellular signals. For example, the signalling pathway that triggers invasive growth in haploid cells utilizes Ste20, Ste7, Ste11, and Ste12 (48). Upon activation of the invasive growth pathway, the MAP kinase Kss1 is phosphorylated by Ste7, which switches Kss1 from an inhibitor to an activator of invasive growth (13, 41). Activation of Kss1 leads to the binding of Ste12 in combination with Tec1 to genes containing a Ste12/Tec1 composite binding site, referred to as a filamentous and invasive growth response element (FRE) (3, 40, 43). Subsequent expression of genes that contain FREs is required for invasive growth. Interestingly, the promoter of Ty1 contains an FRE, and Ty1 RNA levels are decreased by mutations in components of the invasive growth pathway (3, 22, 28, 40). Recently, Madhani et al. (41) have shown that in the absence of Fus3, the invasive growth pathway MAP kinase Kss1 is inappropriately activated by the mating signalling pathway, leading to transcription of both mating-specific genes and invasive growth genes. Hence, Fus3 has a kinase-independent function as a negative regulator of invasive growth.
Here we describe the isolation of a mutation, rtt100-1, that results in high levels of retrotransposition of genomic Ty1 elements. We identify the rtt100-1 mutation as a null allele of the FUS3 gene and show that Fus3 acts at a posttranslational step in the retrotransposition cycle after processing of Ty1 proteins in the maturing VLP. Our results indicate that the stability of Ty1 proteins in VLPs is increased in fus3 mutants and that this increase leads to elevated levels of Ty1 cDNA and increased transpositional integration of Ty1 cDNA. Furthermore, we demonstrate that activation of Fus3 by the basal activity of the mating pathway in vegetative cells is required to fully suppress Ty1 transposition. Hence, the modulation of Ty1 transposition by Fus3 links retrotransposition to environmental signals through evolutionarily conserved signal transduction cascades. Since the mating pathway is specific to haploid cells, we propose that this regulatory mechanism is one way in which haploid yeast cells limit insertional mutagenesis by Ty1 transposition.
MATERIALS AND METHODS
Plasmids.
Plasmid YIpFUS3 was generated by subcloning the 3.7-kb BamHI-HindIII fragment from plasmid pYEE81 (provided by G. Fink) into the yeast integrating vector, pRS406. Plasmid pFUS3ΔHIS3, containing the HIS3 gene fused to nucleotide 584 of FUS3 (25), was created by inserting the SmaI-EcoRV fragment of pGEM-HIS3 (16) in place of the HpaI fragment in YIpFUS3. Plasmid pFUS3Δ carries a fus3Δ::hisG-URA3-hisG allele, which has a deletion of FUS3 nucleotides 500 to 849. The oligomer AX032 (TAGCTAGCTAGATCTGC) containing a BglII site was inserted into the SacII site (nucleotide 500) of FUS3 in YIpFUS3. The resulting plasmid was digested with BglII, and the 3.8-kb BamHI-BglII fragment of pNKY51 (1) was inserted. The 6.0-kb BamHI-EcoRI fragment of this plasmid, containing fus3Δ::hisG-URA3-hisG, was subcloned into pBST-KS(+) (Stratagene) to create pFUS3Δ.
Yeast strains.
Yeast strains are described in Table 1. Strain JC297 is a trp1::hisG derivative of a strain containing the genomic Ty1his3AI-270 element, which was isolated following induction of transposition from plasmid pGTy1-H3his3AI (17) in strain DG733 (20). The trp1::hisG allele was introduced as described by Alani et al. (1). Strain JC358, which has a URA3 gene integrated between the MATa and cry1 loci, is an ascospore derived by crossing strain JC297 to strain GRY340 and then backcrossing a selected ascospore to strain JC297 twice. Strains JC297 and JC358 are congenic.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| JC297 | MATα ura3-167 his3Δ200 trp1::hisG Ty1his3AI-270 | This work |
| JC358 | MATa-ura3-cry1 ura3 his3Δ200 ade2 Ty1his3AI-270 | This work |
| JC801 | MATα fus3-187 ura3 his3Δ200 trp1::hisG Ty1his3AI-270 | This work |
| JC935 | MATα fus3-187 ura3 his3Δ200 trp1::hisG Ty1his3AI-270 | This work |
| JC953 | MATα fus3-187 ura3 his3Δ200 Ty1his3AI-270 | This work |
| JC980 | MATα fus3-187, rad52::hisG-URA3-hisG, ura3, his3Δ200, Ty1his3AI-270 | This work |
| JC984 | MATα fus3-187 ura3 his3Δ200 trp1::hisG ade2-101 Ty1his3AI-270 | This work |
| JC1065 | MATα rad52::hisG-URA3-hisG ura3-167 his3Δ200 trp1::hisG Ty1his3AI-270 | This work |
| JC515 | MATα ura3-167 his3Δ200 trp1::hisG ade2Δ::hisG Ty1ade2AI-515 | 21 |
| JC1038 | MATα fus3Δ::HIS3 ura3-167 his3Δ200 trp1::hisG ade2Δ::hisG Ty1ade2AI-515 | This work |
| JC560 | MATα ura3-167 his3Δ200 trp1::hisG Ty2his3AI-1 | This work |
| JC1510 | MATα fus3Δ::hisG ura3-167 his3Δ200 trp1::hisG Ty2his3AI-1 | This work |
| JC563 | MATα ura3-167 his3Δ200 trp1::hisG Ty2his3AI-19 | This work |
| JC1515 | MATα fus3Δ::hisG ura3-167 his3Δ200 trp1::hisG Ty2his3AI-19 | This work |
| L1451 | MATa ade1 leu2 trp5 lys9 met10 pet9 | G. Fink |
| GRY340 | MATa-URA3-cry1 ade2 his3Δ200 ura3-52 trp1Δ1 leu2Δ1, lys2-8Δ1 tyr7-1 | 20 |
| JC242 | MATα ura3-167 his3Δ200 Ty1his3AI-242 | 17 |
| JC1516 | MATα fus3Δ::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
| JC1430 | MATα leu2::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
| JC1566 | MATα fus3Δ::hisG leu2::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
| JC1634 | MATα ste4Δ::LEU2 leu2::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
| JC1635 | MATα ste4Δ::LEU2 fus3Δ::hisG leu2::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
| JC1602 | MATα ste5Δ102::URA3 leu2::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
| JC1581 | MATα ste5Δ102::URA3 fus3Δ::hisG leu2::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
| JC1651 | MATα ste7Δ3::URA3 leu2::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
| JC1653 | MATα ste7Δ3::URA3 fus3Δ::hisG leu2::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
| JC1600 | MATα ste11Δ6::URA3 leu2::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
| JC1579 | MATα ste11Δ6::URA3 fus3Δ::hisG ura3-167 his3Δ200 Ty1his3AI-242 | This work |
The rtt100-1 strain 18-6A was isolated following ethyl methanesulfonate (EMS) mutagenesis of strain JC358 (see the next section for details). It was crossed to strain JC297 to obtain the rtt100-1 ascospore JC801. Strain JC801 was backcrossed to JC358 to obtain the rtt100-1 ascospore JC935, which was backcrossed to JC358 again to obtain rtt100-1 ascospores JC953 and JC984.
Strains JC560 and JC563, each containing a single Ty2his3AI element, were isolated following induction of Ty2 transposition from a his3AI-marked derivative of plasmid pGTy2-917 (20) in strain JC299 (MATα ura3-167 his3Δ200 trp1::hisG), as described previously (17).
The fus3Δ::hisG allele in strains JC1510, JC1515, and JC1516 was introduced by single-step gene disruption with the 6.0-kb BamHI-EcoRI fragment of plasmid pFUS3Δ, followed by selection for FOAr derivatives of Ura+ transformants. The fus3Δ::HIS3 allele in strain JC1038 was introduced by gene disruption of strain JC515 with a 1.5-kb fragment containing the 5′ end of the FUS3 open reading frame (nucleotides 1 to 584), the HIS3 gene, and the last 55 nucleotides of the FUS3 open reading frame (nucleotides 1008 to 1062). The fus3Δ::HIS3 fragment was generated by PCR from the template DNA pFUS3ΔHIS3 with oligomers LFUS3INT (ATGCCAAAGAGAATTGTATAC) and R2FUSHIS (CTAACTAAATATTTCGTTCCAAATGAGTTTCTTGAGGTCTTTCGT CGTTAGTGCTCTTGGCCTCCTCTAGTA). rad52::hisG-URA3-hisG strains JC980 and JC1065 were constructed by single-step gene disruption with plasmid pRAD52-GB (15).
Strains JC1430 and JC1566 are leu2::hisG derivatives of JC242 and JC1516, respectively, constructed with plasmid pNKY85 (provided by N. Kleckner). The ste11Δ6::URA3 allele was introduced into JC1430 and JC1566 by using plasmid pNC202, the ste5Δ102::URA3 allele was introduced by using plasmid pJB221, and the ste4Δ::LEU2 allele was introduced by using plasmid pDJ154. (All three plasmids were provided by H. Madhani and G. Fink.) The ste7Δ3::URA3 allele was introduced into strains JC1430 and JC1566 by using plasmid pNC149 (a gift of B. Errede). All transformants were verified by Southern analysis.
Media were prepared as described by Rose et al. (49). Synthetic dropout mixtures were obtained from Bio 101, Inc. Glucose (2%) was used as the carbon source except where stated.
Isolation of the rtt100-1 mutant.
Strain JC358 was mutagenized with EMS (49). EMS-treated cells, diluted and plated onto yeast extract-peptone-dextrose (YPD) medium, were grown for 5 days at 20°C. Subsequently, colonies were replicated to synthetic complete medium lacking histidine (SC-His) plates and incubated at 30°C for 3 days. Colonies with more than three His+ papillae (the parental strain yields zero or one papilla per colony) were clonally purified and retested. The rtt100-1 strain 18-6A had a significant increase in the number of His+ papillae when pregrown at 20°C but no significant increase when grown at 30°C.
Strain 18-6A was crossed to the unmutagenized strain JC297. Following tetrad dissection, an α-factor-resistant spore with a hypertransposition phenotype, JC801, was backcrossed to strain JC358 twice. The hypertransposition phenotype segregated 2:2 in 39 of 39 tetrads from the third backcross, as indicated by growing each ascospore as a patch, approximately 2 cm2, on YPD plates at 20°C for 3 to 5 days, replicating the patch to SC-His plates, and scoring His+ papillae after 3 days of growth at 30°C. Cosegregation of a bilateral mating defect with the Rtt− phenotype was demonstrated by genetic selection for diploid formation. Strain 18-6A and all a ascospores were tested for resistance to α-factor-induced growth inhibition in a halo assay (31).
Because Ty1 and RTT100 (FUS3 [see below]) are haploid-specific genes whose expression is down-regulated in a/α diploids, dominance of the rtt100-1 mutation was tested in α/α diploid strains. The MATa-URA3 allele in the rtt100-1 strain 18-6A and the RTT100 strain JC358 was used to make the MAT loci homozygous in diploids following mating to JC297 (RTT100). MATa-URA3/α diploids were grown on 5-fluoroorotic acid medium to select derivatives that had lost the URA3 marker. Derivatives that became α/α at the MAT locus were identified by their ability to mate to a MATa tester strain. His+ prototroph formation in the rtt100-1/RTT100 diploids was tested and compared to the RTT100/RTT100 control strain by the patch test described above.
The genetic distance between rtt100-1 and pet9 was determined by tetrad analysis of a cross between strains JC801 and L1451 (provided by G. Fink). The rtt100-1 mutation was scored in tetrads by a failure to form diploids when ascospores were mated to a known rtt100-1 mutant of the opposite mating type. Genetic distance was calculated from the results of 26 tetrads (19 parental ditypes, 0 nonparental ditypes, and 7 tetratypes) (49). Complementation of the rtt100-1 mutation by FUS3 was performed by integrative transformation of strains JC953 and JC984 with plasmid YIpFUS3 linearized with EcoRI.
DNA sequencing.
A 1.2-kb PCR fragment including the FUS3 transcription unit from positions −114 to +1062 was generated from genomic DNA of strain JC953 and cloned into plasmid pBST-KS(+). Two independent clones were sequenced with Sequenase version 2.0 (U.S. Biochemicals).
Tyhis3AI and Tyade2AI element transposition assays.
The rate of transposition in strains harboring a chromosomal Ty1his3AI, Ty2his3AI, or Ty1ade2AI element was determined by the maximum-likelihood method (38) as described previously (17, 21).
The frequency of Ty1his3AI-242 transposition in the fus3Δ::hisG strain JC1566 transformed with the LEU2-CEN vector pRS415 or with the LEU2-CEN plasmid pRS315 carrying FUS3, fus3-T180D, or fus3-Y182E alleles (plasmids pJB236, pJB290 and pJB291 respectively; provided by H. Madhani and G. Fink) was determined as follows. Four independent transformants of each plasmid were grown overnight in SC-Leu medium at 30°C and then diluted 1:100 into YPD medium and grown for 2 days at 20°C. Aliquots of each culture were plated on SC-Leu to determine the frequency of Leu+ cells and on SC-Leu-His to determine the frequency of His+ prototrophs.
Ty1 integration in the CAN1 locus.
The rate of canavanine resistance in strains JC297 and JC801 was determined by the maximum-likelihood method (38). Strains were grown overnight in YPD medium at 30°C. Approximately 500 cells were used to start 2-ml cultures of YPD, which were grown to saturation at 20°C. The titers of four cultures were determined by plating on YPD medium, and the number of canavanine-resistant (Canr) cells was determined by plating each culture on SC-Arg+canavanine medium (49).
One Canr colony was picked from each of 40 SC-Arg+canavanine plates, and genomic DNA was prepared. PCR analysis was performed on 250 ng of genomic DNA. Each reaction mixture also contained 10 mM Tris (pH 8.3), 50 mM KCl, 2.5 mM MgCl2 250 μM each deoxynucleoside triphosphate, 2 U of Taq polymerase (Boehringer Mannheim), and 250 ng of each of two DNA oligomers. The reaction conditions were 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min in a Perkin-Elmer 9600 thermocycler. CAN1-specific oligomers AX016 (GAAAATTTCGAGGAAGACGATAAGG) and AX019 (CAAATGCTTCTACTCCGTCTGC) were used to amplify a 2,265-bp sequence of the CAN1 gene. DNA samples that failed to amplify the CAN1 gene in this PCR were subjected to two additional PCR amplifications with the CAN1-specific oligomer AX016 or AX019 and a Ty1 LTR-specific oligomer, AX015 (GCCTTTATCAACAATGGAATCCC) to amplify a Ty1:can1 junction fragment, if present.
Northern analysis.
Northern hybridization of approximately 20 μg of total cellular RNA from each strain was performed as described previously (19). [32P]RNA probes were synthesized by in vitro transcription with SP6 or T7 polymerase, using the following DNA templates: plasmid pGEM-PYK1 for synthesis of antisense PYK1 RNA; plasmid pGEM-TyA1 for synthesis of the antisense Ty1 RNA probe; pGEM-HIS3 for synthesis of a sense strand HIS3 RNA to detect the Ty1his3AI transcript (16); and pSPADE2 for synthesis of a sense strand ADE2 RNA to detect the Ty1ade2AI transcript (21). Quantitation of hybridization bands was performed either by scanning densitometry with a Howtek Scanmaster 3+ and Scanalytics software or by phosphorimage analysis with a Molecular Dynamics PhosphorImager and ImageQuant Software.
Immunoprecipitation of 35S-labeled TyA protein.
Labeling of cells with [35S]Met, chasing with excess unlabeled Met, and immunoprecipitation of 35S-TyA from denatured cell lysates were performed as described previously (16), except that TyA1 antiserum was used in place of VLP antiserum. TyA1 antiserum to a TrpE-TyA fusion protein, which was synthesized from a pATH-TrpE expression vector carrying nucleotides 1032 to 1317 of Ty1-H3, was raised in rabbits as described by Garfinkel et al. (33).
Western analysis of Ty1 proteins.
Strains were grown overnight at 30°C in YPD broth and then diluted 20-fold in fresh YPD broth and grown for 4 h at 20°C. Total-cell proteins were extracted from a 1-ml sample of each culture by trichloroacetic acid precipitation (55). Equal amounts of protein (as determined by Coomassie blue staining) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene difluoride membrane (Bio-Rad) with a Semi-dry Horizontal Transfer Cell (Bio-Rad), and probed with TyA1 antibody.
VLP-enriched cell fractions were obtained as follows. Yeast strains were grown in YPD broth overnight at 30°C. The cultures were diluted 50-fold in YPD broth and grown overnight at 20°C and then diluted 5-fold in YPD broth again and grown at 20°C to an optical density at 600 nm of 1.25. VLPs from a 1-liter culture were fractionated on a sucrose step gradient as described by Eichinger and Boeke (23) with the following modifications. Cells were resuspended and disrupted in buffer A (10 mM HEPES-KOH [pH 7.8], 15 mM KCl, 5 mM EDTA) containing 30 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, and 5 μg of leupeptin per ml. The lysate was centrifuged, and the resulting supernatant was loaded onto a sucrose step gradient composed of 5 ml of 75% sucrose in buffer A, 3 ml of 45% sucrose in buffer A, 5 ml of 30% sucrose in buffer A, and 12 ml of 20% sucrose in buffer A. The gradients were centrifuged for 3 h at 25,000 rpm in a Beckman SW28 rotor at 4°C, and the 45% layer containing VLPs was removed (∼5 ml) and diluted twofold in buffer A. VLP-enriched fractions were concentrated by ultracentrifugation in a Beckman NVt90 rotor at 55,000 rpm for 30 min at 4°C. Pellets were resuspended in 500 μl of VLP storage buffer (20 mM HEPES-KOH [pH 7.6], 140 mM KCl, 1 mM EDTA, 10% glycerol). The protein concentration of VLP-enriched fractions was determined with the Bio-Rad Dc protein assay system. Protein samples (25 μg) were subject to SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and probed with TyA1, TyB2 (57), and TyB8 (33) antisera. Immune complexes were detected with the enhanced chemiluminescence Western blot detection system (Amersham). Scanning densitometry was performed with a Howtek Scanmaster 3+ and Scanalytics software.
Southern blot analysis of total cellular Ty1 DNA.
For each strain analyzed, a single colony grown on YPD medium at 20°C was used to inoculate a 10-ml culture of YPD broth and the cultures were grown to saturation at 20°C. Spheroplasts were prepared from each culture by incubating yeast cells for 30 min at 37°C in 0.9 M sorbitol–0.1 M EDTA containing 1 μl of β-mercaptoethanol per ml and 0.6 mg of Zymolyase 100T (Seikagu) per ml, and then total cellular DNA was prepared with the G NOME kit (Bio 101). DNA samples were digested with PvuII, subjected to electrophoresis on a 1% SeaKem Gold (FMC) agarose gel, and transferred to a Hybond N+ nylon membrane (Amersham) in 0.4 N NaOH. The filter was probed with a 934-bp HindIII-BglII fragment of Ty1-H3 (nucleotides 4627 to 5561 [6]), radiolabeled with High Prime DNA labeling mix (Boehringer Mannheim) as specified by the manufacturer. Hybridization bands were quantitated by scanning densitometry with a Howtek Scanmaster 3+ and Scanalytics software.
RESULTS
Isolation of hypertransposition mutants.
Transposition of a single genomic Ty1 element under the control of its native promoter can be detected when the element is marked with the retrotransposition indicator gene his3AI (17) or ade2AI (21). These indicator genes contain an artificial intron (AI) cloned in an antisense orientation into the coding sequences of HIS3 or ADE2, respectively. The marker genes are contained in the 3′ untranslated region of a genomic Ty1 element, such that the transcriptional orientations of Ty1 and his3AI (or ade2AI) are opposed. In this orientation, the antisense strand of the marker gene is transcribed as part of Ty1 RNA and the AI can be spliced from the Ty1 transcript. Use of the spliced Ty1 RNA as a template for cDNA synthesis and genomic integration results in the formation of a Ty1 element containing a functional HIS3 (or ADE2) gene. Hence, the rate of transposition of a Ty1his3AI or Ty1ade2AI element is proportional to the rate of His+ or Ade+ prototroph formation, respectively.
To identify genes that are involved in the maintenance of Ty1 transpositional dormancy, we performed a genetic screen for mutations that increased the transposition of the Ty1his3AI-270 element in strain JC358. Yeast cells were mutagenized with EMS, and colonies were screened to identify rtt (regulator of Ty1 transposition) mutants with an elevated level of His+ papillation following nonselective growth on YPD at 20°C, a permissive temperature for retrotransposition. Chromosomal Ty1 element transposition is inhibited at 30°C, but His+ prototrophs can arise by homologous recombination of a Ty1HIS3 cDNA with Ty1 elements in the genome. To eliminate mutants with increased homologous recombination of Ty1HIS3 cDNA, only those that increased His+ papillation at 20°C but not at 30°C were selected.
One mutation identified, rtt100-1, segregated 2:2 through three backcrosses to a congenic wild-type strain containing Ty1his3AI-270. The Rtt− phenotype is recessive in α/α diploids that are heterozygous for rtt100-1. The transposition rate of Ty1his3AI-270 was determined in the wild-type strain JC297 and compared to that in JC953, a congenic rtt100-1 ascospore from the third backcross of the 18-6A mutant. The rtt100-1 mutant had a 39-fold increase in the rate of His+ prototroph formation per generation (Table 2).
TABLE 2.
Effect of fus3 mutations on Ty1 transposition
| Strain | Relevant genotype | Marked Ty element | Transposition rate | Fold effect of fus3 mu- tation |
|---|---|---|---|---|
| JC297 | WTc | Ty1his3AI-270 | (3.1 ± 1.4) × 10−7a | |
| JC953 | rtt100-1 (fus3-187) | Ty1his3AI-270 | (1.2 ± 0.5) × 10−5a | 39 |
| JC242 | WT | Ty1his3AI-242 | (2.1 ± 0.8) × 10−7a | |
| JC1516 | fus3Δ::hisG | Ty1his3AI-242 | (3.7 ± 1.4) × 10−6a | 18 |
| JC515 | WT | Ty1ade2AI-515 | (3.2 ± 1.2) × 10−8b | |
| JC1038 | fus3Δ::HIS3 | Ty1ade2AI-515 | (1.8 ± 0.6) × 10−6b | 56 |
| JC1065 | rad52 | Ty1his3AI-270 | (4.8 ± 1.7) × 10−6a | |
| JC980 | fus3-187 rad52 | Ty1his3AI-270 | (1.7 ± 0.6) × 10−5a | 4 |
| JC560 | WT | Ty2his3AI-1 | (7.1 ± 4.2) × 10−8a | |
| JC1515 | fus3Δ::hisG | Ty2his3AI-1 | (3.7 ± 2.6) × 10−7a | 5 |
| JC563 | WT | Ty2his3AI-19 | (3.3 ± 1.6) × 10−8a | |
| JC1510 | fus3Δ::hisG | Ty2his3AI-19 | (9.2 ± 6.0) × 10−8a | 3 |
The rate of His+ prototroph formation per cell per generation.
The rate of Ade+ prototroph formation per cell per generation.
WT, wild type.
Identification of RTT100 as FUS3.
A second phenotype was found to cosegregate with the hypertransposition phenotype in tetrad analysis of rtt100-1 mutants: failure to form diploids in bilateral mutant crosses. Microscopic analysis of rtt100-1 × rtt100-1 mating mixtures showed defects in conjugation typical of cell fusion mutants, which were not observed in rtt100-1 × RTT100 mixtures. In addition, haploid MATa rtt100-1 strains were found to be resistant to α-factor-induced arrest of growth. The bilateral mating defect and the failure to undergo pheromone-induced arrest in rtt100-1 mutants were reminiscent of phenotypes observed in fus3 and far1 mutants (10, 25). The rtt100-1 mutation was mapped and shown to be located 13.5 centimorgans (cM) from pet9 on chromosome II, which is the same distance reported for FUS3 (25).
To test whether FUS3 could complement the rtt100-1 allele, the FUS3 gene on a URA3-based integrating vector was introduced into rtt100-1 strains JC953 and JC984. Ura+ transformants of each strain showed complementation of the bilateral mating defect and reversion of the hypertransposition defect. Therefore, the fus3 coding region in strain JC953 was sequenced to identify the rtt100-1 mutation. A transition from G to A was found at position 561, which introduces a nonsense codon in place of the codon for Trp187, resulting in truncation of nearly half of the 353-amino-acid Fus3 protein. Herein, the rtt100-1 mutation is referred to as fus3-187. To confirm that hypertransposition of Ty1 is a null phenotype of fus3 mutants, a fus3Δ::HIS3 allele, in which sequences coding for the carboxy-terminal 158 amino acids of Fus3 are deleted, was introduced into a strain that harbors the chromosomal element, Ty1ade2AI-515. The rate of Ade+ prototroph formation in the fus3Δ::HIS3 strain JC1038 was 56-fold higher than that in the isogenic FUS3 strain JC515 (Table 2). We also tested a fus3Δ::hisG allele in strain JC242, which contains a different Ty1his3AI element from the one in the mutagenesis strains JC297 and JC358. The fus3Δ::hisG allele has a deletion of sequences encoding amino acids 167 to 284 of Fus3. The rate of His+ prototroph formation was increased 18-fold in the fus3Δ::hisG strain JC1516 relative to the isogenic wild-type strain, JC242 (Table 2). Both the fus3Δ::HIS3 and fus3Δ::hisG strains were mating defective in bilateral mutant crosses, and fus3Δ::hisG has been shown to be defective in blocking Kss1-dependent mating activity relative to the catalytic-site mutation, fus3-K42R (41). In summary, fus3 null mutants have an 18- to 56-fold increase in the rate of transposition of a single chromosomal Ty1 element.
We further determined whether the frequency of transposition of a Ty1ade2AI element expressed from the GAL1 promoter was affected by a fus3 null mutation. After galactose induction of plasmid pGTy1ade2AI in strain JC418 for 24 h (21), the frequency of Ade+ prototrophs was 3.1 × 10−3, whereas two independent fus3Δ::HIS3 derivatives had frequencies of 1.4 × 10−3 and 5.2 × 10−3 Ade+ prototrophs. Perhaps these data, which demonstrate that pGTy1 transposition is not increased in fus3Δ mutants, indicate that the high levels of Ty1 protein processing and VLP accumulation in cells expressing pGTy1 (16, 32) are able to overcome the negative effect of Fus3 on transposition.
The effect of fus3 is specific for the Ty1 retrotransposon.
Regulatory mechanisms for one type of retrotransposon may be shared by other classes of transposons. In Drosophila virilis, a host-mediated hybrid dysgenesis that causes mobility of the Ulysses retrotransposon also mobilizes four other classes of transposons (47). To determine if FUS3 regulates other retrotransposons in yeast, we examined the effect of a fus3 mutation on the transposition of Ty2 elements, which are highly related to Ty1 elements. The fus3Δ::hisG allele was introduced into two strains containing independent chromosomal Ty2his3AI elements, and the rate of His+ prototroph formation was determined (Table 2). Compared to isogenic wild-type strains, transposition of the Ty2his3AI-1 element was increased approximately fivefold and that of the Ty2his3AI-19 element was increased threefold in fus3Δ derivatives. Hence, fus3Δ causes a small increase in Ty2 retrotransposition, but the effect is significantly lower than the 18- to 56-fold effect of fus3 null mutations on Ty1 transposition. In contrast, expression of a pGTy1 element causes trans activation of both genomic Ty1his3AI and Ty2his3AI elements to equivalent levels (18). Our findings indicate that expression of a pGTy1 element and expression of the fus3 mutation overcome transpositional dormancy by different mechanisms, the latter of which is more specific for Ty1 retrotransposition.
Transpositional integration of Ty1 cDNA is increased in fus3-187 mutants.
Ty1 cDNA can enter the genome by two distinct pathways. The transposition pathway, mediated by IN, results in integration of Ty1 at nonhomologous sites and generates a 5-bp target duplication. A second pathway is the RAD52-dependent gene conversion of Ty1 elements or LTRs in the genome. Although cDNA-mediated gene conversion is normally rare, it is stimulated by conditions that negatively affect Ty1 IN-mediated integration (51). Either pathway could be stimulated in fus3 mutants, giving rise to increased His+ prototroph formation. To determine if the increase in His+ prototroph formation in fus3 mutants is dependent on RAD52, a rad52 mutation was introduced into the fus3-187 strain JC953 and the FUS3 strain JC297. The 39-fold increase in transposition in a fus3-187 mutant was not abolished by introduction of the rad52 mutation but instead was slightly enhanced, resulting in a 55-fold increase in transposition relative to the wild-type strain JC297 (Table 2). The data demonstrate that His+ prototroph formation in a fus3-187 strain is RAD52 independent. The rad52 mutation by itself increases Ty1his3AI-270 transposition about 15-fold, in agreement with previous results (15), and transposition in the fus3-187 rad52 double mutant is about 4-fold higher than in the rad52 mutant. The findings clearly argue that the RAD52-independent process of transposition is responsible for the increase in His+ prototrophs in fus3 mutants.
To confirm our hypothesis that transposition into de novo targets is increased, we quantitated transposition events that inactivate a selectable target gene, CAN1, in the FUS3 strain, JC297 and a congenic fus3-187 strain, JC801 (Table 3). Loss-of-function mutations in the CAN1 gene result in resistance of the strain to growth inhibition in the presence of canavanine. The rate of canavanine resistance was similar in the FUS3 and fus3-187 strains, but the fraction of independent Canr colonies that sustained a Ty1 insertion into the CAN1 gene was increased eightfold in the fus3-187 strain relative to the FUS3 strain (Table 3), yielding an overall increase of sixfold in the rate of Ty1 insertion into CAN1 in the fus3-187 mutant. The relatively smaller increase in Ty1 transposition into CAN1 compared to the increase in Ty1his3AI and Ty1ade2AI transposition into the whole genome is probably because open reading frames of RNA polymerase II genes are relatively poor targets for Ty1 transposition (36). Taken together, the data show that the fus3-187 mutation significantly increases de novo integration of Ty1.
TABLE 3.
De novo transposition into the CAN1 locus
| Strain | Relevant genotype | No. of Ty1 insertions/ no. of Canr colonies analyzed | Rate of Canr/ generation | Rate of Ty1 insertion into CAN1/generation |
|---|---|---|---|---|
| JC297 | FUS3 | 1/40 | (3.9 ± 1.1) × 10−7 | (9.8 ± 2.8) × 10−9 |
| JC801 | fus3-187 | 8/40 | (2.8 ± 0.9) × 10−7 | (5.6 ± 2.0) × 10−8 |
The Ty1 RNA level is not increased in fus3 mutants.
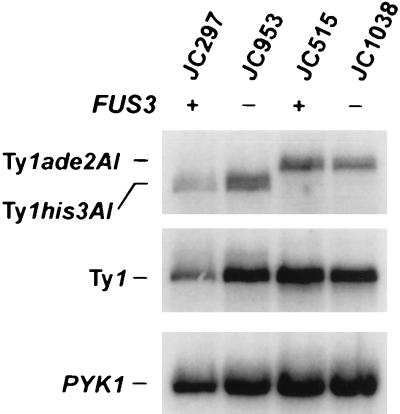
To understand how FUS3 regulates Ty1 transposition in normal cells, we analyzed different steps in the retrotransposition process in fus3 mutants. Hypertransposition could be caused by an increase in the amount of Ty1 RNA; therefore, RNA prepared from cells in the log phase of growth was analyzed by Northern blotting to quantitate Ty1his3AI-270 and Ty1ade2AI-515 transcripts from individual elements and from Ty1 elements collectively, relative to a control transcript, PYK1 RNA (Fig. 1). The level of Ty1his3AI RNA in the fus3-187 strain JC953 was not significantly increased relative to that in the congenic FUS3 strain JC297. Similarly, the level of Ty1ade2AI RNA in the fus3Δ strain JC1038 was not increased relative to that in the isogenic FUS3 strain JC515. Total Ty1 RNA levels were also not significantly altered as a result of either the fus3-187 or fus3Δ::HIS3 mutation. Although both minor increases and decreases in the level of RNA from individual Ty1 elements occur in fus3 mutants relative to isogenic or congenic FUS3 strains, a consistent increase in the Ty1 RNA level that could explain the 18- to 56-fold increase in Ty1 transposition has not been observed. In addition, Northern analysis of RNA from cells in the stationary phase showed no change in the levels of Ty1, Ty1his3AI or Ty1ade2AI RNA (normalized to PYK1 RNA) in fus3 mutants relative to FUS3 strains (data not shown). The data indicate that derepression of Ty1 transposition in fus3 mutants occurs at the posttranscriptional level.
FIG. 1.
Northern analysis of total RNA from strains JC297 (FUS3) and JC953 (fus3-187), containing the Ty1his3AI-270 element, and strains JC515 (FUS3) and JC1038 (fus3Δ), containing the Ty1ade2AI-515 element. The riboprobes used to detect Ty1his3AI and Ty1ade2AI RNA (top), total Ty1 RNA (center), and PYK1 RNA (bottom) are described in Materials and Methods.
Another possibility to explain the increased His+ prototroph formation in fus3 mutants is that the efficiency of splicing of the AI from Ty1his3AI RNA is increased. Therefore, relative levels of spliced Ty1HIS3 RNA and unspliced Ty1his3AI were measured in the fus3-187 strain 18-6A and the unmutagenized strain JC358 by quantitative reverse transcription-PCR with primers in HIS3 that flank the AI (51). No difference in the ratio of Ty1HIS3 RNA to Ty1his3AI RNA was observed (data not shown). Hence, the increased rate of Ty1his3AI transposition in fus3-187 strains cannot be accounted for by an increase in the efficiency of splicing of the AI.
Posttranslational regulation of Ty1 transposition by FUS3.
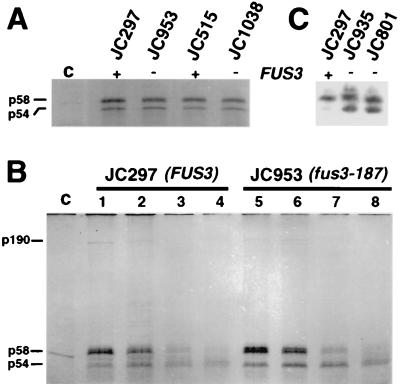
To determine if the efficiency of Ty1 protein synthesis is increased in fus3 mutants, cells were labeled for 1 h with [35S]Met and TyA protein was immunoprecipitated from denatured cell lysates (Fig. 2A). The levels of unprocessed p58-TyA and processed p54-TyA were equivalent in the fus3-187 strain JC953 and the wild-type strain JC297. Similarly, the levels of pulse-labeled TyA proteins in the fus3Δ strain JC1038 showed no difference from those in the isogenic FUS3 strain, JC515. Therefore, the higher levels of retrotransposition in fus3 mutants cannot be attributed to an increase in the synthesis of TyA protein.
FIG. 2.
Posttranslational regulation of Ty1 by FUS3. (A) JC297 (FUS3), JC953 (fus3-187), JC515 (FUS3), and JC1038 (fus3Δ) cells were labeled with [35S]Met for 1 h and immunoprecipitated with TyA1 antiserum. The control (c) contains no antiserum. The products were analyzed by SDS-PAGE (10% polyacrylamide). (B) Cells were pulse-labeled with [35S]Met for 15 min and then chased with excess unlabeled methionine. Samples were harvested 15 min (lanes c, 1, and 5), 1 h (lanes 2 and 6), 8 h (lanes 3 and 7), and 20 h (lanes 4 and 8) after the addition of [35S]Met, and denatured cell lysates were incubated with preimmune serum (lane c) or TyA1 antiserum (lanes 1 to 8). The products (p190-TyA/ TyB, p58-TyA, and p54-TyA) were analyzed by SDS-PAGE (10% polyacrylamide). (C) Total cellular proteins isolated from JC297 (FUS3), JC935 (fus3-187), and JC801 (fus3-187) were separated by SDS-PAGE (10% polyacrylamide) and subjected to Western analysis with TyA1 antiserum.
To determine if the rate of Ty1 protein processing is increased in fus3 mutants, TyA proteins were immunoprecipitated from lysates of cells pulse-labeled with [35S]Met for 15 min and chased with excess unlabeled Met for up to 20 h (Fig. 2B). Processing of 35S-labeled p58-TyA to 35S-labeled p54-TyA was relatively inefficient in both wild-type and mutant strains, which has been shown previously for wild-type strains (16). An increase in TyA processing was not detected in the fus3-187 strain JC953 relative to the FUS3 strain JC297. Furthermore, both 35S-labeled p58-TyA and 35S-labeled p190-TyA/TyB were detected after a 15-min pulse-labeling step in this analysis (Fig. 2B, lanes 1 and 5), and there was no difference in their level of synthesis in fus3-187 and FUS3 strains. In summary, these results indicate that in fus3 null mutants, the 18- to 56-fold increase in Ty1 transposition is not a result of a detectable increase in the level of Ty1 RNA, Ty1 protein synthesis, or proteolytic processing of TyA.
A small increase in the ratio of p54-TyA to p58-TyA was noticed in fus3 mutants after 8 h (Fig. 2B, compare lanes 3 and 7), suggesting that the p54-TyA protein might be stabilized. Therefore, Western blot analysis of total-cell extracts was performed with TyA1 antibody (Fig. 2C). Steady-state levels of TyA proteins were increased in fus3-187 mutants JC935 and JC801 relative to the congenic FUS3 strain JC297, and this difference was attributable mostly to an increase in the level of the processed form of TyA, p54. The increase in the level of p54 was also observed in the fus3Δ mutants JC1038 and JC1516 relative to the isogenic FUS3 strains (data not shown). Since neither an increase in TyA protein synthesis nor an increase in TyA processing was detected, the data argue that the stability of p54-TyA is increased in fus3 mutants.
The levels of VLP-associated Ty1 proteins and total Ty1 cDNA are increased in fus3 mutants.
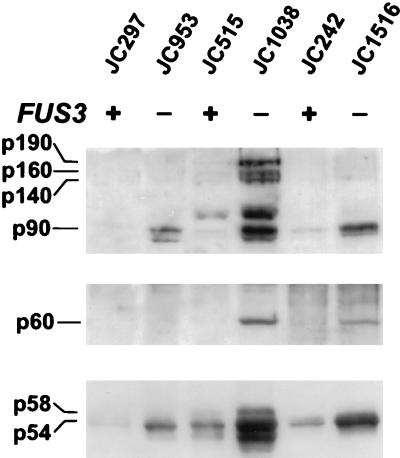
The preferential accumulation of the processed form of TyA in fus3 mutants suggested that Ty1 proteins were stabilized after the formation of VLPs. Therefore, a cell fractionation procedure developed to isolate VLPs from cells expressing a GAL1:Ty1 element (transposition-induced cells) was used to obtain a cytoplasmic fraction that was enriched for VLPs from uninduced cells. Although Ty1 proteins are not a major component of this sucrose gradient fraction prepared from uninduced cells as they are when prepared from transposition-induced cells (33), the VLPs were significantly concentrated to allow both TyA and TyB proteins to be detected by Western analysis (Fig. 3). Equal amounts (25 μg) of protein from VLP-enriched cell fractions were analyzed by immunoblotting with TyA1, TyB2, and TyB8 antibodies against the TyA, IN, and RT domains, respectively (Fig. 3). TyA proteins p58 and p54 were five- to sevenfold more abundant in VLP fractions from the fus3 strains JC953, JC1038, and JC1516 than in those from the FUS3 control strains JC297, JC515, and JC242, respectively (Fig. 3, bottom panel). A more significant increase in the level of processed TyB proteins from VLP-enriched fractions of fus3 strains was observed. The level of p90-IN was 21- to 43-fold higher (Fig. 3, top panel) and the level of p60-RT was >14- to 200-fold higher (Fig. 3, center panel) in fus3 mutants than in FUS3 strains. In addition, processing intermediates p140, p160, and p190 are detected in strain JC515 by the B2 antibody, and the level of these proteins in VLP fractions is also increased in the isogenic fus3Δ strain JC1038 (Fig. 3, top panel). In summary, TyA and TyB proteins that cosediment with VLPs are significantly more abundant in fus3 mutants. Taken together, the data argue that the stability of VLP-associated proteins is enhanced in fus3 mutants. Alternatively, the assembly of Ty1 proteins into VLPs could be increased in fus3 mutants, which might enhance their stability relative to Ty1 proteins not associated with VLPs.
FIG. 3.
Ty1-VLP synthesis is increased in fus3 mutants. Western blot analysis of Ty1-VLPs from FUS3 (+) and fus3 (−) strains. VLPs were separated by SDS-PAGE (7.5% polyacrylamide). Western blots were probed with B2 antiserum to detect p90-IN (top panel). Size labels on the left indicate the position of p90-IN, which appears as a doublet (9), p190-TyA/TyB, and processing intermediates (p140 and p160). The band at 110 kDa is attributable to nonspecific proteolysis of TyB (33). The relative levels of p90 doublet in fus3 strains compared to that in the isogenic or congenic FUS3 strains, determined by scanning densitometry, are as follows: JC953/JC297, 43; JC1038/JC515, 38; JC1516/JC242, 21. B8 antiserum (center panel) was used to detect p60-RT. The p60 band from JC953 is visible in darker exposures of the Western blot. The relative levels of p60 in fus3 strains compared to the isogenic or congenic FUS3 strain are as follows: JC953/JC297, >14; JC1038/JC515, 200; JC1516/JC242, 66. TyA1 antiserum (bottom panel) detected p58 and p54 TyA. Lower-molecular-weight species recognized by this antiserum are probably a result of nonspecific proteolysis. The relative levels of p58 and p54 in fus3 strains compared to the isogenic or congenic FUS3 strain are as follows: JC953/JC297, 7; JC1038/JC515, 5; JC1516/JC242, 5.
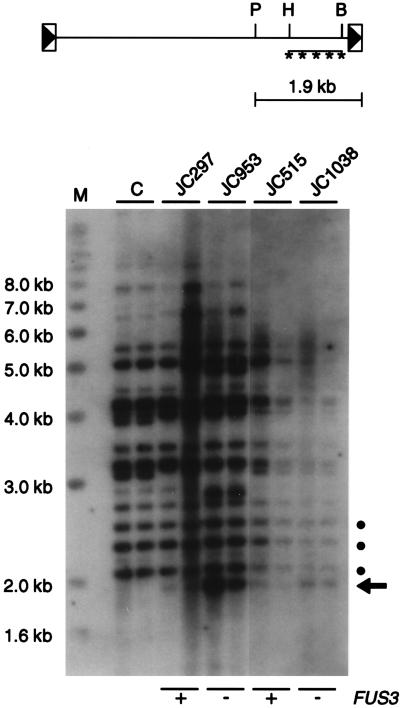
If the increase in TyA and TyB levels leads to an increase in activity that is responsible for the elevated rates of transposition in fus3 strains, the amount of Ty1 cDNA might be increased in fus3 mutants. The Ty1 cDNA present in VLP-enriched fractions was difficult to detect and varied between different preparations of the same strain (data not shown). Therefore, we compared the amount of unintegrated, linear Ty1 cDNA present in total cellular DNA in FUS3 strains to that in total cellular DNA in fus3 strains. DNA was prepared from each strain grown at 20°C, digested with PvuII, and subjected to Southern hybridization with a probe derived from the 3′ end of Ty1 (Fig. 4). The probe detected a 1.9-kb fragment of unintegrated Ty1 cDNA from the PvuII site at nucleotide 3944 of Ty1 to the 3′ end. In addition, larger PvuII fragments representing the junction between the 3′ end of integrated Ty1 elements and genomic DNA were detected.
FIG. 4.
The level of Ty1 cDNA is increased in fus3 mutants. (Top) Structure of the unintegrated linear Ty1 cDNA and location of relevant PvuII (P), HindIII (H), and BglII (B) sites. The HindIII-BglII Ty1 DNA fragment that was used as a probe in Southern analysis is indicated by the horizontal line marked with asterisks. The 1.9-kb Ty1 cDNA band generated by PvuII digestion is represented as the line with crossbars. (Bottom) Total cellular DNA from two independent cultures of FUS3 (+) and fus3 (−) strains or an spt3-101 control strain (lane C) grown at 20°C was digested with PvuII, subjected to electrophoresis on a 1% agarose gel, and hybridized to the Ty1 DNA probe described above. Random-primer labeled 1-kb marker DNA (lane M) was included on the agarose gel as a size control. The 1.9-kb Ty1 cDNA band, indicated by the arrow, and three Ty1-genomic DNA junction fragments, indicated by solid circles, were quantitated by scanning densitometry. The ratio of the 1.9-kb Ty1 cDNA band to each of the three Ty1-genomic DNA junction bands was determined, and the three ratios from each of two independent DNA samples were averaged for each strain analyzed. The average ratio of Ty1 cDNA fragment to Ty1-genomic DNA fragment was 0.4 for strain JC297, 2.7 for strain JC953, 0.3 for strain JC515, and 3.1 for strain JC1038.
In an spt3-101 strain in which Ty1 elements are not expressed (54), the 1.9-kb Ty1 cDNA fragment was not detected, even when the autoradiogram was overexposed, indicating that its appearance is dependent on Ty1 expression (Fig. 4). The level of the Ty1 cDNA band was quantitated relative to the levels of three different Ty1-genomic DNA junction fragments in FUS3 and fus3 strains (Fig. 4). The fus3-187 strain JC953 had a 7-fold increase in Ty1 cDNA relative to the congenic FUS3 strain JC297, and the fus3Δ strain JC1038 had a 10-fold increase relative to the isogenic FUS3 strain JC515. The data demonstrate that higher levels of Ty1 transposition are correlated with an increase in total cellular Ty1 cDNA levels in fus3 mutants.
Fus3 is activated by the mating-pheromone response pathway in vegetative cells.
In response to mating pheromone, Fus3 is rapidly phosphorylated on threonine-180 and tyrosine-182 residues, and this phosphorylation is required for activation of Fus3 to mediate the mating response (34). During vegetative growth, Fus3 kinase is partially activated by the basal activity of the mating-pheromone response pathway (52). We investigated the role of activation of Fus3 in suppression of Ty1 transposition in vegetative cells by examining the ability of phosphorylation site substitution alleles fus3-T180D and fus3-Y182E to suppress transposition in a fus3Δ strain. The fus3-T180D and fus3-Y182E alleles are completely unable to rescue the mating defect of a fus3Δ kss1Δ double mutant (data not shown), indicating that these alleles cannot be activated. The frequency of His+ prototroph formation from the genomic Ty1his3AI-242 element was determined for each allele (Table 4). FUS3 lowered transposition 74-fold compared to the LEU2-CEN vector pRS415 alone, while the fus3-T180D and fus3-Y182E alleles decreased transposition 15- and 9-fold, respectively. Hence, the activity of the fus3-T180D and fus3-Y182E alleles in suppressing Ty1 transposition is significantly lower than that of FUS3. These findings argue that basal levels of Fus3 phosphorylation are involved in maintaining normal levels of suppression of Ty1 transposition.
TABLE 4.
Complementation of Ty1his3AI-242 hypertransposition in a fus3Δ strain
| Allelea | Transposition frequencyb | Relative decrease in transposition frequencyc |
|---|---|---|
| Vector only | 2.0 × 10−4 | 1 |
| FUS3 | 2.7 × 10−6 | 74 |
| fus3-T180D | 1.3 × 10−5 | 15 |
| fus3-Y182E | 2.2 × 10−5 | 9 |
Contained on a LEU2-CEN vector.
Number of His+ Leu+ prototrophs as a fraction of the total number of Leu+ colonies.
Relative to the transposition frequency in the strain containing the vector only.
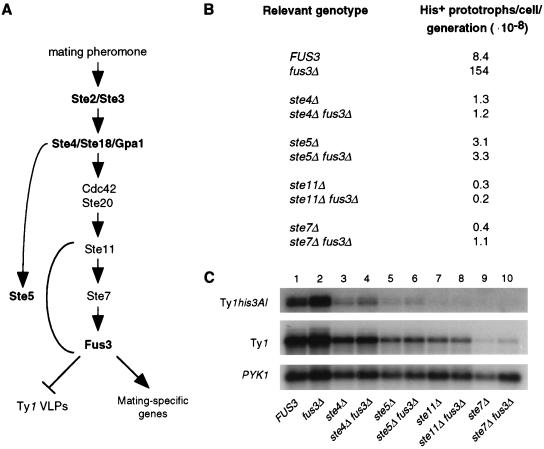
To determine if an intact pheromone response pathway is required for Fus3 to repress Ty1 transposition, we examined the effect of deleting genes that act upstream of Fus3 in the pheromone response pathway (Fig. 5A). ste11Δ, ste7Δ, ste4Δ, and ste5Δ mutations were introduced into the FUS3 strain JC1430 or the isogenic fus3Δ strain JC1566 to allow a comparison of ste fus3Δ double mutants to single ste mutants (Fig. 5B). The rate of Ty1his3AI-242 transposition in a ste11Δ fus3Δ mutant was equivalent to that in a ste11Δ mutant. Similarly, Ty1his3AI-242 transposition was increased less than threefold in the ste7Δ fus3Δ mutant relative to the ste7Δ mutant. This small increase may be because Fus3 is present in a complex with Ste7 in naive cells (2) and hence may have increased access to its targets in the absence of Ste7. In addition, transposition was not increased in ste4Δ fus3Δ or ste5Δ fus3Δ double mutants relative to ste4Δ or ste5Δ mutants, respectively. The level of total Ty1 RNA or Ty1his3AI-242 RNA was the same in each ste fus3Δ mutant relative to the corresponding single ste mutant (Fig. 5C), indicating that the absence of an increase in transposition in ste fus3Δ double mutants is not a consequence of lower Ty1 RNA levels. Hence, deletion of FUS3 is insufficient to derepress transposition in the absence of upstream components of the mating pathway.
FIG. 5.
An intact mating-pheromone response pathway is required for Fus3-mediated repression of Ty1 retrotransposition. (A) Major components of the mating-pheromone response pathway. The receptor Ste2 (or Ste3), the components of the heterotrimeric G protein (Ste4, Ste18, and Gpa1), the scaffolding protein Ste5, and the MAP kinase Fus3 are shown in boldface type to indicate that they are specific to the pheromone response signal cascade. (B) The rate of His+ prototroph formation was determined for each strain as described in Materials and Methods. (C) Northern analysis of steady-state RNA from pheromone response pathway mutants. Each lane is labeled below with the relevant genotype of the strain analyzed. Riboprobes to detect Ty1his3AI-242 RNA (top), total Ty1 RNA (center) and PYK1 RNA (bottom) are described in Materials and Methods. The level of Ty1his3AI-242 RNA was normalized to PYK1 RNA. The ratios of Ty1his3AI-242 RNA to PYK1 RNA in each strain, relative to the FUS3 strain, were as follows: FUS3, 1.0; fus3Δ, 1.3; ste4Δ, 0.5; ste4Δ fus3Δ, 0.5; ste5Δ, 0.3; ste5Δ fus3Δ, 0.3; ste11Δ, 0.1; ste11Δ fus3Δ, 0.1; ste7Δ, 0.1; ste7Δ fus3Δ, 0.1. The ratios of total Ty1 RNA to PYK1 RNA relative to the FUS3 strain were as follows: FUS3, 1.0; fus3Δ, 0.9; ste4Δ, 1.1; ste4Δ fus3Δ, 1.0; ste5Δ, 0.7; ste5Δ fus3Δ, 0.7; ste11Δ, 0.5; ste11Δ fus3Δ, 0.4; ste7Δ, 0.1; ste7Δ fus3Δ, 0.1.
Because activation of Fus3 is required for suppression of transposition, mutations that prevent Fus3 activation might be expected to cause the same phenotype as fus3 null mutations do. However, strains with mutations in upstream components of the mating pathway showed decreased rather than increased transposition. Compared to the fus3Δ strain, the rate of Ty1his3AI-242 transposition was reduced 513-fold in a ste11Δ mutant, 385-fold in a ste7Δ mutant, 118-fold in a ste4Δ mutant, and 50-fold in a ste5Δ mutant (Fig. 5B). One explanation is that the production of Ty1 RNA is dependent on components of the mating pathway. We found a 2-fold reduction in the level of Ty1 RNA in ste11Δ mutants and a 10-fold reduction in ste7Δ mutants (Fig. 5C, center panel, lanes 7 and 9). However, RNA from the Ty1his3AI-242 element was reduced ten-fold in both ste11Δ and ste7Δ mutants (Fig. 5C, top panel, lanes 7 and 9). The levels of Ty1 RNA were equivalent to the wild-type level in ste4Δ mutants and 70% of the wild-type level in ste5Δ mutants (Fig. 5C, center panel, lanes 3 and 5). The ste4Δ and ste5Δ mutations had a more significant effect on Ty1his3AI-242 RNA, reducing it to 50 and 30% of the level in wild-type strains, respectively (Fig. 5C, top panel, lanes 3 and 5). The pattern of regulation for total Ty1 RNA is consistent with previous results (22, 28) and with the idea that transcription of many Ty1 elements is at least partially under the control of the invasive growth pathway. The invasive growth pathway uses Ste11 and Ste7 proteins but does not use Ste4 and Ste5 proteins, which are unique to the mating pathway. Hence, the finding that Ty1his3AI-242 RNA shows some degree of dependence on all four components of the mating pathway indicates that individual Ty1 elements may also be regulated as mating-specific genes.
The absence of Ste4 or Ste5 has a small effect on Ty1 RNA levels but significantly reduces Ty1his3AI-242 transposition. These findings argue that fus3Δ is not epistatic to these ste mutations for the regulation of transposition. (Likewise, fus3Δ is not epistatic to ste mutations in the mating response.) In ste mutants, neither Fus3 nor Kss1 can activate mating-specific gene expression but Kss1 can substitute for the activation of mating-specific genes in the absence of Fus3 (24, 41). Therefore, ste4Δ and ste5Δ mutants may not have the same hypertransposition phenotype as fus3Δ mutants because they lose expression of a mating-specific gene whose product activates Ty1 transposition at the posttranscriptional level.
Kss1 is not redundant with Fus3 in regulating Ty1 transposition.
To determine if Kss1 is redundant with Fus3 in regulating transposition, we examined the effect of a kss1Δ mutation on Ty1 transposition. The rate of transposition of the Ty1his3AI-242 element was 2.0 × 10−8 in a kss1Δ mutant, which is fourfold lower than in the isogenic wild-type strain (Fig. 5B). Ty1 RNA levels are also reduced about twofold in a kss1Δ mutant relative to the isogenic wild-type strain (data not shown). These data demonstrate that Kss1 is not redundant with Fus3 in repressing Ty1 transposition. The rate of transposition in a kss1Δ fus3Δ double mutant was 1.0 × 10−9, which is similar to that in the ste7Δ and ste11Δ mutants. There is a concomitant decrease in the level of Ty1 RNA in the kss1Δ fus3Δ double mutant to a level similar to that in a ste7Δ mutant (data not shown). The regulation of transpositional dormancy by Fus3 but not by Kss1 is consistent with two models: (i) Ty1 transposition in fus3Δ mutants is increased because the invasive growth response is activated, resulting in increased expression of an invasive growth gene whose product is an activator of Ty1 transposition; and (ii) transposition is increased in fus3Δ mutants by the loss of a negative regulatory interaction between the Fus3 kinase and a Ty1-encoded target protein.
DISCUSSION
The yeast MAP kinase Fus3 is a specific negative regulator of Ty1 transposition.
Transpositional dormancy of Ty1 elements in yeast results from multiple levels of regulation that affect Ty1 transcription, protein synthesis, VLP formation and maturation, cDNA synthesis, and integration (5, 8, 16, 30, 50, 51). We have used a his3AI-based assay for transposition of individual genomic Ty1 elements to screen for host regulators of transpositional dormancy. The hypertransposition mutant rtt100-1 was isolated in this screen, and RTT100 was shown to be allelic with FUS3 (25). Null mutations in FUS3 increase the transposition rates of individual Ty1his3AI and Ty1ade2AI elements 18- to 56-fold, depending on the individual chromosomal Ty1 element that is assayed (Table 2). The increase in His+ prototroph formation in fus3 mutants occurs independently of the RAD52 gene and therefore cannot be attributed to elevated levels of homologous recombination of Ty1HIS3 cDNA (Table 2). Moreover, Ty1 integration into the CAN1 gene is elevated in fus3 mutants (Table 3). These results demonstrate that integration of Ty1 into de novo targets is increased by null mutations in the FUS3 gene.
In contrast, the fus3Δ mutation increases the transposition of Ty2his3AI elements only three- to fivefold (Table 2). One interpretation of this data is that Fus3 interacts directly or indirectly with a Ty gene product that is divergent in Ty1 and Ty2 elements. These elements have two regions of sequence divergence, one encompassing TYA and the PR domain of TYB and the other including the carboxy-terminal domain of IN encoded within TYB. There is 50% identity and 66% similarity in the primary amino acid sequence of the TYA product between Ty1 and Ty2 elements, whereas the corresponding TYB gene products have 77% identity and 86% similarity in the primary amino acid sequence. Hence, the specificity of Fus3 for the regulation of Ty1 transposition may suggest that TyA1 is the target of the Fus3 kinase.
Null mutations in FUS3 increase stability of VLP-associated Ty1 proteins.
Our analysis of Ty1 RNA levels suggests that hypertransposition in fus3 mutants does not result from increases in Ty1 RNA synthesis or stability. Steady-state levels of Ty1 RNA were not detectably increased in fus3-187 or fus3Δ strains relative to those in congenic or isogenic control strains (Fig. 1 and 5). Similarly, the RNA level from marked Ty1his3AI and Ty1ade2AI elements was not consistently increased in fus3 mutants. The simplest explanation of this data is that Ty1 transposition is regulated at a posttranscriptional step by Fus3. However, we cannot rule out the possibility of small increases in the Ty1 RNA level that were not detected in our Northern analyses and that lead to significant increases in the rate of genomic Ty1 element transposition. However, this possibility seems quite unlikely, given that increasing the copy number of Ty1 does not derepress the transposition of Ty1 elements (7, 17). Moreover, a 25% increase in the total Ty1 RNA level created when a Ty1ade2AI element on a high-copy-number plasmid is maintained in the cell actually decreases the transposition of the genomic Ty1his3AI-242 element (14).
Immunoprecipitation of pulse-labeled Ty1 proteins demonstrated that p58-TyA and p190-TyA/TyB were synthesized at equivalent levels in fus3 and wild-type strains (Fig. 2A and B). These results are consistent with the observation that Ty1 RNA levels are not altered and indicate that higher levels of Ty1 retrotransposition in fus3 mutants are not a result of an increase in the synthesis of Ty1 proteins. The rate of maturation of p58-TyA to p54-TyA, a step in retrotransposition that is very inefficient in normal cells (16), was also not detectably altered in fus3 strains. However, there was a higher level of processed p54-TyA in fus3 total-cell extracts (Fig. 2C). Furthermore, Western analysis of cytoplasmic fractions enriched for VLPs revealed that the levels of TyA, p90-IN, and p60-RT proteins were all increased in fus3 mutants. We were unable to detect the highly unstable p23-PR protein (33), but if its levels are increased in fus3 mutants, it does not significantly enhance the rate of TyA protein processing (Fig. 2B). Taken together, these findings suggest that transposition is inhibited by Fus3 at a posttranslational step after Ty1 protein processing in VLPs and that Fus3 contributes to the instability of Ty1 proteins. Similar to the regulation of retrotransposition in fus3 mutants, induction of expression of a pGTy1 element does not change the efficiency of protein synthesis per Ty1 transcript. However, the rate of TyA protein processing is increased in transposition-induced cells, and this gives rise to dramatically higher steady-state levels of TyA and TyB proteins, Ty1 VLPs, and transposition per Ty1 RNA transcript. (16, 32). Our analysis of fus3 mutants suggests that increasing the stability of Ty1 proteins can also stimulate Ty1 retrotransposition. A posttranslational increase in VLP-associated proteins in fus3 mutants is likely to be the direct cause of the increase in transposition, because fus3 mutants also have 7- to 10-fold-higher levels of total cellular Ty1 cDNA. Although we do not know if this cDNA is VLP associated, the simplest explanation of this finding is that higher levels of VLPs result in an increase in the synthesis of the substrate for integration, which in turn leads to higher levels of Ty1 transposition in fus3 mutants.
The pheromone response pathway activates Fus3 to mediate the suppression of Ty1 transposition.
Our results indicate that basal activation of Fus3 is required to suppress Ty1 transposition at the posttranslational level. Alleles of FUS3 with amino acid substitutions in the activating residues, fus3-T180D and fus3-Y182E, were significantly less active than wild-type FUS3 in complementing Ty1 hypertransposition in a fus3Δ strain, suggesting that phosphorylation of Fus3 is involved in mediating the repression of Ty1 transposition in vegetative cells (Table 4). The fus3-T180D and fus3-Y182E alleles did exhibit a low level of activity in suppressing transposition. This activity could result from these alleles having retained the kinase-independent inhibitory function of Fus3 whereas null fus3Δ alleles lack this function (41).
The requirement for activation of Fus3 was confirmed by demonstrating that in the absence of Ste4, Ste5, Ste11, and Ste7, Ty1 transposition is not suppressed by Fus3 (Fig. 5B). Both Ste4 and Ste5 are specific to the mating-pheromone pathway. Hence, the data suggest that for Fus3 to suppress Ty1 transposition, it must be activated by upstream components of the pheromone response pathway. This modulation of Fus3 activity in regulating Ty1 transposition at the posttranslational level is therefore entirely consistent with its modulation in response to mating pheromone. In contrast to the increase in Ty1 transposition in fus3 mutants, Ty1 transposition is reduced in the four ste mutants analyzed and in a fus3Δ kss1Δ double mutant. Since ste4Δ and ste5Δ mutants had nearly wild-type levels of Ty1 RNA, this finding suggests that an activator of Ty1 transposition is encoded by a mating-specific gene, whose expression would be blocked in ste and kss1Δ fus3Δ mutants but not in a fus3Δ mutant. Despite this possibility, the rate of transposition remains sufficiently high in ste4Δ and ste5Δ mutants (6.5- and 2.7-fold lower than in an isogenic wild-type strain, respectively [Fig. 5B]) that the postulated activator must not be absolutely required for Ty1 transposition and the effect of deleting FUS3 could be easily detected if it occurred in the absence of activation by upstream components of the pheromone response pathway.
Complex regulation of Ty1 RNA levels by components of the pheromone response and invasive growth pathways.
The promoter of Ty1 contains an FRE that is bound by Ste12 and Tec1 and is responsive to conditions that stimulate invasive growth genes (3, 40, 43). Mutations in components of the invasive growth pathway, including Ste11, Ste7, Ste12, and Tec1, abolish the expression of an FRE(Ty1)::lacZ reporter. However, components specific to the mating pathway, including Ste4 or Ste5, do not affect the expression of FRE(Ty1)::lacZ (40, 43). In general agreement with these findings, we have found that Ty1 RNA levels are reduced in ste7Δ and ste11Δ mutants, slightly lowered in a ste5Δ mutant, and unaltered in a ste4Δ mutant (Fig. 5B). However, our findings suggest that Ty1 promoters in their native context may not all be regulated identically to FRE(Ty1)::lacZ. For example, FRE(Ty1)::lacZ expression was abolished in a ste11Δ mutant (40), but we found that the Ty1 RNA level was reduced only twofold in ste11Δ mutants. Second, while FRE(Ty1)::lacZ expression in a fus3Δ kss1Δ double mutant is similar to that in a wild-type strain, Ty1 RNA levels decrease dramatically in a fus3Δ kss1Δ double mutant (data not shown), which is more characteristic of mating-specific genes than of invasive growth genes. These differences might be attributable to a posttranscriptional effect on Ty1 RNA levels, since we are measuring steady-state levels of Ty1 RNA rather than promoter activity. Alternatively, there might be heterogeneous regulation of individual Ty1 promoters. The effect of ste mutations on RNA from the marked Ty1his3AI-242 element is consistent with the possibility that at least some Ty1 elements are regulated with characteristics of mating-specific genes. The Ty1his3AI-242 RNA level was lowered three- and twofold in ste5Δ and ste4Δ mutants, respectively, which is a more significant decrease than that of the total Ty1 RNA level in these strains (Fig. 5B). Hence, the expression of individual Ty1 elements may be affected to different extents by the mating and invasive growth pathways.
Direct or indirect interaction of Fus3 with Ty1?
TyA is a phosphoprotein (42, 56) whose steady-state levels are elevated in fus3 mutants (Fig. 2C and 3). Therefore, a simple hypothesis to explain the repression of transposition by Fus3 is that TyA is phosphorylated by the Fus3 kinase and that this modification targets TyA for degradation. Fus3 has multiple targets (26), and TyA has several MAP kinase consensus sites (S/T-P), but the phosphorylation sites in TyA that are used in vivo have not been identified. Xu and Boeke (56) found that when cells were exposed to mating pheromone, thereby activating Fus3, transposition of a pGTy1 element was inhibited. Moreover, TyA was hyperphosphorylated and VLP-associated p90-IN and Ty1 cDNA levels were reduced. Taken together with the results presented here, these findings are consistent with the hypothesis that Fus3 phosphorylates TyA and that this modification decreases the stability of VLPs and reduces transposition levels. An intermediate protein that is phosphorylated by Fus3 and in turn affects the phosphorylation state of TyA and the stability of VLP-associated proteins would also explain these observations.
Another mechanism by which Fus3 might mediate the posttranslational regulation of Ty1 is through its role in activating Ste12, which in turn promotes the expression of mating-specific genes. In this scenario, Fus3 would suppress transposition by stimulating the expression of a mating-specific gene that encodes an inhibitor of Ty1 transposition. Several observations argue against this interpretation. First, mating-specific genes are still expressed in a fus3 mutant, because Kss1 can substitute for Fus3 in the activation of Ste12 (24, 41). Second, the large decrease in Ty1his3AI-242 transposition in ste4Δ and ste5Δ mutants relative to that in a fus3Δ mutant (Fig. 5B) suggests that there is an activator of transposition encoded by a mating-specific gene, rather than an inhibitor. Third, the fact that Kss1 is not redundant with Fus3 for the suppression of Ty1his3AI-242 transposition suggests that transpositional dormancy is not regulated via an inhibitor encoded by a mating-specific gene.
A third scenario by which Fus3 might regulate transposition is via its role as a negative regulator of invasive growth and FRE-dependent gene expression (40, 48). Fus3 has a kinase-independent inhibitory activity that prevents the inappropriate activation of invasion by the mating cascade; therefore, Kss1 can be erroneously activated by the mating pathway in fus3Δ mutants, resulting in stimulation of both mating-specific and invasive-growth gene expression (41). Hence, Fus3 may inhibit the expression of an invasive growth gene that encodes an activator of Ty1 transposition. Several of our observations are consistent with this hypothesis. Unactivable alleles of Fus3, fus3-T180D and fus3-Y182E, retain some ability to suppress transposition (Table 4), consistent with the expectation that they retain the kinase-independent function of blocking the invasive-growth pathway. A second relevant observation is that Ty1 transposition is dramatically reduced in ste11Δ mutants even though the total Ty1 RNA level is reduced by only twofold (Fig. 5). Similarly, invasive growth and FRE-dependent gene expression are completely blocked in ste11Δ mutants (40, 48). In contrast, transposition is only slightly reduced in kss1Δ mutants, even though Ty1 RNA levels are comparable to those in ste11Δ mutants. This is consistent with the moderate reduction in FRE-dependent gene expression and weak invasion defect in kss1Δ mutants that results from loss of both positive and negative regulation of invasion by Kss1 (13, 40, 41). Further experiments will be needed to determine whether the Fus3 kinase interacts directly with Ty1 proteins or whether it acts through the invasive growth pathway to mediate the posttranslational suppression of Ty1 transposition.
Significance of regulating Ty1 transposition through conserved signal transduction cascades.
Because insertional mutations caused by Ty1 transposition can be deleterious in haploids, the haploid-specific Fus3 kinase may have been adapted to specifically repress transposition during the haploid stage of the yeast life cycle. This mechanism of regulation also provides flexibility for the host cell to modulate transposition in response to environmental signals. We have demonstrated that Ty1 transposition is regulated not only by Fus3 but also by Ste4, Ste5, Ste11, Ste7, and Kss1. Therefore, transposition is potentially responsive to environmental signals in both haploid and diploid yeasts. The inhibition of transposition upon exposure of cells to mating pheromone is likely to be a direct effect of activating the Fus3 kinase. The fact that the Ty1 promoter is regulated as a filamentous or invasive growth gene predicts that environmental conditions that promote invasive growth will also activate Ty1 transposition, potentially in both haploids and diploids. Furthermore, the activity of Fus3 can be repressed by activation of other signal transduction cascades, such as the osmoregulatory pathway (35). Therefore, growth in high-osmolarity medium could increase Ty1 transposition in haploids. Although transposition is potentially deleterious, yeast cells might encounter severe conditions in which it is advantageous to activate transposition because of the ability of Ty1 to cause adaptive mutations by altering the regulation of target genes (53). Hence, under conditions of stress (such as conditions that induce invasive growth) where survival is threatened, Ty1 could be activated to induce genetic change, thereby providing a means for the cell to adapt to its environment.
ACKNOWLEDGMENTS
We are grateful to B. Faiola and W.-Y. Hu for their assistance at an early stage in this project, to H. Madhani for helpful suggestions and for sharing results prior to publication, to M. Bryk and R. Zitomer for helpful comments on the manuscript, and to the Molecular Genetics Core Facility for oligomer synthesis.
This work was funded by National Institutes of Health grant GM52072 to M.J.C. The research of D.J.G. was sponsored by the National Cancer Institute, DHHS, under contract with ABL.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell L, Cook J G, Chang E C, Cairns B R, Thorner J. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baur M, Esch R K, Errede B. Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol Cell Biol. 1997;17:4330–4337. doi: 10.1128/mcb.17.8.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Best S, Letissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 5.Boeke J D, Corces V G. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- 6.Boeke J D, Eichinger D, Castrillon D, Fink G R. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol Cell Biol. 1988;8:1432–1442. doi: 10.1128/mcb.8.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke J D, Eichinger D J, Natsoulis G. Doubling Ty1 element copy number in Saccharomyces cerevisiae: host genome stability and phenotypic effects. Genetics. 1991;129:1043–1052. doi: 10.1093/genetics/129.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke J D, Sandmeyer S B. Yeast transposable elements. In: Broach J R, Pringle J, Jones E, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 193–262. [Google Scholar]
- 9.Braiterman L T, Monokian G M, Eichinger D J, Merbs S L, Gabriel A, Boeke J D. In-frame linker insertion mutagenesis of yeast transposon Ty1: phenotypic analysis. Gene. 1994;139:19–26. doi: 10.1016/0378-1119(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 10.Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63:999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- 11.Chapman K B, Bystrom A S, Boeke J D. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc Natl Acad Sci USA. 1992;89:3236–3240. doi: 10.1073/pnas.89.8.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi K Y, Satterberg B, Lyons D M, Elion E A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 13.Cook J G, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;360:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 14.Curcio, M. J. Unpublished results.
- 15.Curcio M J, Garfinkel D J. Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics. 1994;136:1245–1259. doi: 10.1093/genetics/136.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curcio M J, Garfinkel D J. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol Cell Biol. 1992;12:2813–2825. doi: 10.1128/mcb.12.6.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curcio M J, Garfinkel D J. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curcio, M. J., and D. J. Garfinkel. Unpublished results.
- 19.Curcio M J, Hedge A M, Boeke J D, Garfinkel D J. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol Gen Genet. 1990;220:213–221. doi: 10.1007/BF00260484. [DOI] [PubMed] [Google Scholar]
- 20.Curcio M J, Sanders N J, Garfinkel D J. Transpositional competence and transcription of endogenous Ty elements in Saccharomyces cerevisiae: implications for regulation of transposition. Mol Cell Biol. 1988;8:3571–3581. doi: 10.1128/mcb.8.9.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalgaard J Z, Banerjee M, Curcio M J. A novel Ty1-mediated fragmentation method for native and artificial yeast chromosomes reveals that the mouse Steel gene is a hotspot for Ty1 integration. Genetics. 1996;143:673–683. doi: 10.1093/genetics/143.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubois E, Jacobs E, Jauniaux J-C. Expression of the ROAM mutations in Saccharomyces cerevisiae: involvement of trans-acting regulatory elements and relation with the Ty1 transcription. EMBO J. 1982;1:1133–1139. doi: 10.1002/j.1460-2075.1982.tb01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eichinger D J, Boeke J D. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988;54:955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 24.Elion E A, Brill J A, Fink G R. Functional redundancy in the yeast cell cycle: FUS3 and KSS1 have both overlapping and unique functions. Cold Spring Harbor Symp Quant Biol. 1991;56:41–49. doi: 10.1101/sqb.1991.056.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Elion E A, Grisafi P L, Fink G R. FUS3 encodes a cdc2+/CDC28− related kinase required for the transition from mitosis into conjugation. Cell. 1990;60:649–664. doi: 10.1016/0092-8674(90)90668-5. [DOI] [PubMed] [Google Scholar]
- 26.Elion E A, Satterberg B, Kranz J E. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Errede B, Ammerer G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 1989;3:1349–1361. doi: 10.1101/gad.3.9.1349. [DOI] [PubMed] [Google Scholar]
- 28.Errede B, Cardillo T S, Sherman F, Dubois E, Deschamps J, Wiame J M. Mating signals control expression of mutations resulting from insertion of a transposable repetitive element adjacent to diverse yeast genes. Cell. 1980;22:427–436. doi: 10.1016/0092-8674(80)90353-0. [DOI] [PubMed] [Google Scholar]
- 29.Errede B, Gartner A, Zhou Z, Nasmyth K, Ammerer G. MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature. 1993;362:261–264. doi: 10.1038/362261a0. [DOI] [PubMed] [Google Scholar]
- 30.Farabaugh P J. Post-transcriptional regulation of transposition by Ty retrotransposons of Saccharomyces cerevisiae. J Biol Chem. 1995;270:10361–10364. doi: 10.1074/jbc.270.18.10361. [DOI] [PubMed] [Google Scholar]
- 31.Fink G R, Styles C A. Curing of a killer factor in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1972;69:2846–2849. doi: 10.1073/pnas.69.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garfinkel D J, Boeke J D, Fink G R. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985;42:507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 33.Garfinkel D J, Hedge A M, Youngren S D, Copeland T D. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991;65:4573–4581. doi: 10.1128/jvi.65.9.4573-4581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gartner A, Nasmyth K, Ammerer G. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev. 1992;6:1280–1292. doi: 10.1101/gad.6.7.1280. [DOI] [PubMed] [Google Scholar]
- 35.Hall J P, Cherkasova V, Elion E, Gustin M C, Winter E. The osmoregulatory pathway represses mating pathway activity in Saccharomyces cerevisiae: isolation of a FUS3 mutant that is insensitive to the repression mechanism. Mol Cell Biol. 1996;16:6715–6723. doi: 10.1128/mcb.16.12.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji H, Moore D P, Blomberg M A, Braiterman L T, Voytas D F, Natsoulis G, Boeke J D. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-x. [DOI] [PubMed] [Google Scholar]
- 37.Kawakami K, Pande S, Faiola B, Moore D P, Boeke J D, Farabaugh P J, Strathern J N, Nakamura Y, Garfinkel D J. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics. 1993;135:309–320. doi: 10.1093/genetics/135.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 39.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madhani H D, Fink G R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 41.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 42.Mellor J, Fulton A M, Dobson M J, Roberts N A, Wilson W, Kingsman A J, Kingsman S M. The Ty transposon of Saccharomyces cerevisiae determines the synthesis of at least three proteins. Nucleic Acids Res. 1985;13:6249–6263. doi: 10.1093/nar/13.17.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosch H U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neiman A M, Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc Natl Acad Sci USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nouvel P. The mammalian genome shaping activity of reverse transcriptase. Genetica. 1994;93:191–201. doi: 10.1007/BF01435251. [DOI] [PubMed] [Google Scholar]
- 46.Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 47.Petrov D A, Schutzman J L, Hartl D L, Lozovskaya E R. Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci USA. 1995;92:8050–8054. doi: 10.1073/pnas.92.17.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts R L, Fink G R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 49.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 50.Sandmeyer S B. Yeast retrotransposons. Curr Opin Genet Dev. 1992;2:705–711. doi: 10.1016/s0959-437x(05)80130-3. [DOI] [PubMed] [Google Scholar]
- 51.Sharon G, Burkett T J, Garfinkel D J. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14:6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sprague G F, Thorner J W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- 53.Wilke C M, Adams J. Fitness effects of Ty transposition in Saccharomyces cerevisiae. Genetics. 1992;131:31–42. doi: 10.1093/genetics/131.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winston F, Durbin K J, Fink G R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984;39:675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- 55.Wright A P, Bruns M, Hartley B S. Extraction and rapid inactivation of proteins from Saccharomyces cerevisiae by trichloroacetic acid precipitation. Yeast. 1989;5:51–53. doi: 10.1002/yea.320050107. [DOI] [PubMed] [Google Scholar]
- 56.Xu H, Boeke J D. Inhibition of Ty1 transposition by mating pheromones in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2736–2743. doi: 10.1128/mcb.11.5.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Youngren S D, Boeke J D, Sanders N J, Garfinkel D J. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol Cell Biol. 1988;8:1421–1431. doi: 10.1128/mcb.8.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]