Abstract
The essential yet toxic nature of copper demands tight regulation of the copper homeostatic machinery to ensure that sufficient copper is present in the cell to drive essential biochemical processes yet prevent the accumulation to toxic levels. In Saccharomyces cerevisiae, the nutritional copper sensor Mac1p regulates the copper-dependent expression of the high affinity Cu(I) uptake genes CTR1, CTR3, and FRE1, while the toxic copper sensor Ace1p regulates the transcriptional activation of the detoxification genes CUP1, CRS5, and SOD1 in response to copper. In this study, we characterized the tandem regulation of the copper uptake and detoxification pathways in response to the chronic presence of elevated concentrations of copper ions in the growth medium. Upon addition of CuSO4, mRNA levels of CTR3 were rapidly reduced to eightfold the original basal level whereas the Ace1p-mediated transcriptional activation of CUP1 was rapid and potent but transient. CUP1 expression driven by an Ace1p DNA binding domain-herpes simplex virus VP16 transactivation domain fusion was also transient, demonstrating that this mode of regulation occurs via modulation of the Ace1p copper-activated DNA binding domain. In vivo dimethyl sulfate footprinting analysis of the CUP1 promoter demonstrated transient occupation of the metal response elements by Ace1p which paralleled CUP1 mRNA expression. Analysis of a Mac1p mutant, refractile for copper-dependent repression of the Cu(I) transport genes, showed an aberrant pattern of CUP1 expression and copper sensitivity. These studies (i) demonstrate that the nutritional and toxic copper metalloregulatory transcription factors Mac1p and Ace1p must sense and respond to copper ions in a dynamic fashion to appropriately regulate copper ion homeostasis and (ii) establish the requirement for a wild-type Mac1p for survival in the presence of toxic copper levels.
Copper (Cu) is an essential element required by all living organisms; however, it is also highly toxic. Copper functions as an important cofactor for a variety of enzymes that are required for essential biochemical processes such as cytochrome c oxidase, Cu,Zn superoxide dismutase, lysyl oxidase, and dopamine-β-hydroxylase (32). On the other hand, Cu can participate in Fenton-like reactions that can generate extremely reactive hydroxyl radicals which cause cellular damage such as the oxidation of proteins, cleavage of DNA and RNA molecules, and membrane damage due to lipid peroxidation (18). Furthermore, Cu toxicity can result from its improper incorporation into proteins. For example, Cu has been shown in vitro to replace Zn in the zinc finger DNA binding domain of the human estrogen receptor, rendering the protein unable to bind to its target DNA sequences (36). It is therefore important that organisms elaborate appropriate mechanisms for uptake and detoxification, as well as possess cellular sensors to ensure that sufficient Cu is present in the cell to drive the essential biochemical processes while preventing its accumulation to toxic levels. The importance of maintaining appropriate intracellular Cu levels is underscored by the existence of two human genetic disorders of Cu homeostasis, Menkes syndrome and Wilson’s disease (2, 3, 43, 47). Proper regulation of the Cu homeostatic machinery requires the ability of the Cu ion sensors to detect Cu and respond by appropriately regulating the expression of Cu homeostasis genes in order to maintain the delicate balance between essential and toxic levels.
The baker’s yeast Saccharomyces cerevisiae has been a powerful model organism for elucidating the components of the Cu homeostatic machinery and their mechanisms of action. Elegant genetic screens have identified genes that are responsible for Cu uptake under nutritional conditions when essential levels of Cu ions are present in the cell (10, 25), distribution to appropriate subcellular compartments (8, 15, 31, 49), and detoxification under toxic conditions when Cu ions are present in excess (7, 40, 41, 45). At the nutritional level, Cu uptake into yeast cells is mediated by two membrane-associated high-affinity Cu(I) transporters, encoded by CTR1 and CTR3 (10, 25), and a cell surface Cu(II)/Fe(III) reductase, encoded by the FRE1 gene, which reduces Cu(II) to Cu(I) prior to uptake (14, 21). In the presence of excess Cu ions, when expression of the high-affinity Cu(I) transporters is abolished, Cu ion uptake can proceed through a putative low-affinity Cu ion uptake system. The high-affinity Cu(I) transport genes CTR1, CTR3, and FRE1 are transcriptionally downregulated by Cu ions and induced by Cu ion starvation. This Cu ion-dependent regulation requires a wild-type Mac1p Cu metalloregulatory transcription factor (CuMRTF) (15, 24, 29, 48). Regulation of these genes by Mac1p is highly specific for and exquisitely sensitive to Cu ions (29). Deletion of the MAC1 gene results in Cu ion starvation phenotypes similar to those associated with deletions in the CTR1 and CTR3 genes, which can be corrected by added Cu ions. In mac1Δ strains, transcription of CTR1 and CTR3 is undetectable and the FRE1 gene is transcribed at low levels. In addition, yeast strains which possess a dominant MAC1up1 allele exhibit high basal levels of CTR1, CTR3, and FRE1 mRNAs, a lack of Cu-dependent repression of CTR1 and CTR3 (29), and hypersensitivity to Cu ions (24). The mutations in Mac1p that lead to a dominant MAC1up1 allele all map to the first of two cysteine clusters found in the carboxyl-terminal half of Mac1p (16, 24, 48, 56). Regulation by Mac1p requires the Cu-responsive cis-acting elements (CuREs) 5′-TTTTGCTC-3′ which are arranged either in tandem or inverted repeats in the promoters of the CTR1, CTR3, and FRE1 genes (29, 48). In vivo dimethyl sulfate (DMS) footprinting revealed that the CuREs are occupied under Cu ion starvation conditions in which the Cu(I) transport genes are expressed and unoccupied in the presence of sufficient Cu ion concentrations when transcription of these genes is inactivated. The CuREs are unoccupied in a mac1 deletion strain, are constitutively occupied in a MAC1up1 strain (29), and have been demonstrated by electrophoretic mobility shift assays to bind Mac1p in vitro (48).
Excess levels of Cu ions are directly sensed by the S. cerevisiae CuMRTF Ace1p. Ace1p cooperatively binds Cu(I) to form a tetra-Cu cluster through specific cysteine residues within the amino-terminal DNA binding domain (13, 40). Copper binding leads to a conformational change in this domain that results in specific binding of monomeric Ace1p to the metal response elements (MREs) 5′-TCY(4–6)GCTG-3′ (Y = pyrimidine) on the promoters of genes that are involved in Cu ion detoxification and protection against oxidative damage (53). These include CUP1 (19) and CRS5 (7), which encode small cysteine-rich metallothioneins that sequester Cu ions and protect the cell from its toxic effects, and SOD1, which encodes Cu,Zn superoxide dismutase (17). Both in vitro and in vivo footprinting analyses have demonstrated that Cu-activated Ace1p binds to four MREs on the CUP1 promoter (12, 23). In addition, Ace1p binds to single MREs on the SOD1 and CRS5 promoters to modestly activate transcription. The importance of Ace1p in Cu detoxification is underscored by the observation that deletion of the ACE1 gene renders yeast strains extremely sensitive to Cu ion toxicity (22). Interestingly, the Cu ion sensors Mac1p and Ace1p share 50% identity only within the 40 amino-terminal amino acids which contains 3 of the 11 conserved cysteine residues found in Ace1p and Amt1p, the homologous CuMRTF in the opportunistic pathogenic yeast Candida glabrata (55). This region binds Zn and may be involved in minor groove binding to the MREs (27, 42).
In this study, we characterized the dynamic regulation of the Cu ion uptake and detoxification pathways in the presence of elevated Cu ion concentrations in the growth medium. Our results show that tandem regulation by both the nutritional copper sensor, Mac1p, and the toxic copper sensor, Ace1p, of the expression of their respective target genes is required for survival of S. cerevisiae cells in the presence of toxic levels of Cu ions. In addition to mediating the Cu ion-dependent regulation of the high affinity Cu(I) uptake genes, a wild-type Mac1p is also important for proper detoxification in the absence of these genes.
MATERIALS AND METHODS
Growth conditions.
Yeast cells were maintained in YPD medium (1% yeast extract, 2% Bacto Peptone, 2% dextrose) (38) with or without the addition of CuSO4 or in the corresponding dropout media for maintenance of yeast strains transformed with plasmids. Liquid cultures were seeded to an optical density of 0.4 and grown to exponential phase (optical density at 650 nm of 1.2 to 1.5) at 30°C and 400 rpm and then treated with the indicated Cu concentrations for up to 2 h. Five-milliliter samples were withdrawn from the Cu-treated cultures at the indicated time points for analysis. Plasmids were constructed and maintained in Escherichia coli DH5αF′ cells, using standard techniques (1).
Yeast strains and plasmids.
The yeast strains used in this study are listed in Table 1. Strain DTY205 is isogenic to DTY1 but contains the dominant MAC1up1 allele and was a generous gift from D. J. Kosman. To determine the role of the Ace1p carboxyl-terminal activation domain in CUP1 expression, strains KKY1, MPY2, and MPY3 were constructed as follows. Strain KKY1 was derived from SLY1 (29) by deletion of the chromosomal copy of the ACE1 gene by using plasmid pace1Δ::hisG-URA3 (4). Disruption of ACE1 was verified by testing the sensitivity of KKY1 on Cu plates and by Southern blot analysis. Plasmid p316:ACE1 contains a 1.8-kb genomic fragment encompassing the complete ACE1 open reading frame, 541 bp of the 5′ flanking region, and 612 bp of the 3′ flanking region cloned into the HindIII site of pRS316. The 1.8-kb ACE1 genomic fragment was released from p316:ACE1 by digestion with EcoRI and SalI and mobilized into the same restriction sites on the pRS303 integrating vector to generate plasmid p303:ACE1. To generate a fusion gene between the Ace1p DNA binding domain and the activation domain of the herpes simplex virus transcriptional activator VP16 (5, 6), p316:ACE1 was digested with BamHI and BglII, releasing a 937-bp fragment containing the coding region for the amino-terminal DNA binding domain of Ace1p and the 5′ flanking region. An in-frame fusion with the VP16 activation domain was constructed by cloning this fragment into the BglII site of plasmid CRF3 which contains the coding region for amino acids 402 to 479 of VP16, 119 bp of 3′ flanking region, and a 400-bp fragment containing the thymidine kinase termination signal and polyadenylation site (a generous gift from Steven Triezenberg). The ACE1-VP16 fusion gene was then released by digesting the resulting plasmid with EcoRI and SalI and mobilized into pRS303 to generate p303:ACVP. Plasmids p303:ACE1 and p303:ACVP were digested at a unique BsmI site on the HIS3 marker and transformed into strain KKY1 for integration into the chromosome at the his3 locus to generate strains MPY2 and MPY3, respectively. Proper integration of these plasmids into the genome was verified by Southern blot analysis and by testing the resistance of the resulting strains to Cu on synthetic complete medium (SC)-His plates. To evaluate the role of MAC1 in Cu detoxification, strains MPY17 and MPY18 were constructed. MPY17 was derived from SKY46, which contains a double deletion of the high-affinity transport genes CTR1 and CTR3 (25), by PCR mutagenesis of the URA3 marker in which 391 bp of the URA3 open reading frame was replaced by the kanamycin resistance cassette (44). MPY18 was derived from MPY17 by disruption of the MAC1 allele, using the plasmid pmac1::URA3 (29). Disruption of the MAC1 gene was verified by diagnostic PCR analysis. Plasmids pRSMAC1(HA) and pRSMAC1up1(HA) contain a 2.5-kb genomic fragment of the MAC1 gene and a 2.5-kb genomic fragment of MAC1up1, both cloned into the SalI and BamHI sites of the pRS313 vector (56). Both genes were tagged with a single copy of the Haemophilus influenzae hemagglutinin protein epitope at the carboxyl terminus, giving rise to proteins functionally indistinguishable from the parental proteins. For RNase protection analyses, three plasmids were constructed for making antisense RNA probes. Plasmid pSKCUP1 was constructed by inserting a 149-bp EcoRI-BamHI fragment of the CUP1 gene into the same sites of pBlueScript SK. The antisense RNA hybridizes to the region between +31 and +179 downstream from the translational start codon of CUP1. To generate pSKCTR3, a 181-bp fragment of the CTR3 gene was amplified from strain DTY1 and cloned into the EcoRI and BamHI sites of pBlueScript SK. This fragment hybridizes to the region between +86 and +267 downstream from the translational start codon of CTR3. The riboprobe derived from the plasmid pKSACT1 (29) was used to probe ACT1 mRNA as an internal control for normalization during quantitation of the RNase protection products. For in vivo DMS footprinting, plasmid pRSCUP1/CYC1-lacZ, containing the CUP1 promoter from −390 up to the first base of the translation start codon fused to a minimal CYC1 promoter and a reporter lacZ gene (a generous gift from Nicholas Santoro), was used as a template for sequencing.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| DTY1 | MATa gal1 trp1-1 his3 ade8 CUP1R | 37 |
| DTY205 | MATa gal1 trp1-1 his3 ade8 CUP1R MAC1up1 | 24 |
| SKY46 | MATa ctr1::URA3 ctr3::TRP1 his3 CUP1R lys2-801 | 25 |
| SLY2 | MATa gal1 trp1-1 his3 ade8 CUP1R ura3::Kanr mac1::URA3 | 29 |
| KKY1 | MATa gal1 trp1-1 his3 ade8 CUP1R ura3::Kanr ace1::URA3 | This study |
| MPY2 | MATa gal1 trp1-1 his3 ade8 CUP1R ura3::Kanr ace1::URA3 HIS3::ACE1 | This study |
| MPY3 | MATa gal1 trp1-1 his3 ade8 CUP1R ura3::Kanr ace1::URA3 HIS3::ACE1/VP16 | This study |
| MPY17 | MATa ctr1::ura3::Kanr ctr3::TRP1 his3 lys2-801 CUP1R | This study |
| MPY18 | MATa ctr1::ura3::Kanr ctr3::TRP1 his3 lys2-801 CUP1R mac1::URA3 | This study |
[35S]cysteine labeling.
Exponential-phase cultures of DTY1 cells were grown at 30°C in 100 ml of YPD medium to exponential phase. The cells were incubated with 5 μCi of [35S]cysteine (800 Ci/mmol; ICN Radiochemicals) per ml for 30 min prior to the addition of 0.1 M CuSO4 to a final concentration of 100 μM. Five-milliliter samples were withdrawn from the cultures after Cu treatment. Proteins were extracted and analyzed on 20% nondenaturing polyacrylamide gels and visualized by fluorography as described previously (54).
In vivo DMS footprinting.
Log-phase cultures of DTY1 cells were grown in 1.5 liters of YPD liquid medium and treated with 100 μM CuSO4. After 0, 15, 30, and 60 min, 250-ml samples were treated with 2% DMS for 5 min. In vivo DMS footprinting was performed as previously described (57). Isolated genomic DNA samples from cells that were treated or untreated with Cu or DMS were digested with BspHI prior to G,A-specific cleavage with 1.0 N NaOH. An oligonucleotide (5′-CCTCATATATGTGTATAGGTTTATACGG-3′) which hybridizes to the CUP1 promoter at positions −310 to −282 with respect to the start site of transcription was used for primer extension analysis and for dideoxy sequencing of the pRSCUP1/CYC1-lacZ template.
Standard methods.
Total RNA was extracted by the hot phenol method as previously described (28). RNase protection assays were performed as described by Koch and Thiele (27). Quantitation of the radioactive bands were performed with a PhosphorImager SP and ImageQuant 3.3 software (Molecular Dynamics). Quantitation from the PhosphorImager were plotted and analyzed by using Kaleidagraph 3.02 (Synergy Software, Reading, Pa.). Proteins were extracted as previously described (54) and quantitated by using a Bio-Rad protein assay kit with bovine serum albumin as a protein standard. Spectrophotometric measurements were performed on a Beckman DU64 spectrophotometer. DNA isolation and PCRs were performed by standard protocols (1). DNA sequencing was carried out with a Sequenase kit as specified by the manufacturer (U.S. Biochemical). Western blot analysis was performed by standard protocols (1) and visualized with horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G (Bethesda Research Laboratories) and a Renaissance chemiluminescence kit (Dupont NEN).
RESULTS
Transcriptional regulation of CUP1 and CTR3 in the chronic presence of Cu ions.
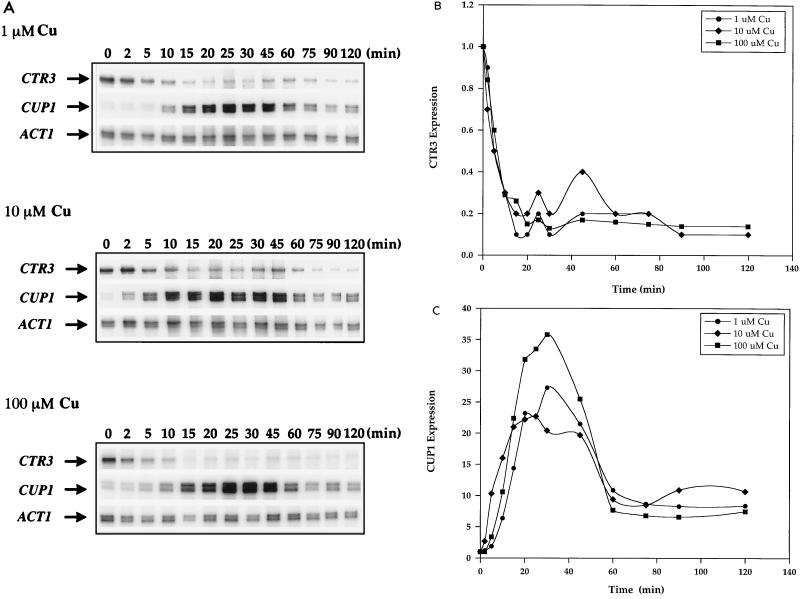
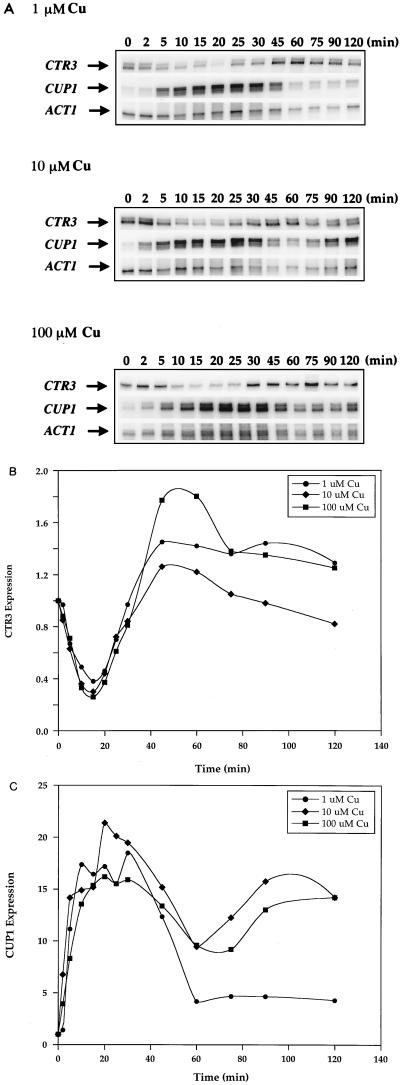
To study the interplay between the Cu ion sensors Mac1p and Ace1p, which regulate the expression of genes encoding components of the high-affinity Cu(I) uptake pathway and the Cu ion detoxification pathway, respectively, yeast cells were incubated with low and high Cu ion concentrations in the media. Expression of the high-affinity Cu(I) transport gene CTR3 and the metallothionein gene CUP1 was analyzed over time by RNase protection. In response to 1, 10, and 100 μM added Cu, CTR3 mRNA levels were quickly reduced approximately fivefold within 15 min of Cu treatment (Fig. 1A). This loss of expression was followed by a slight and transient but reproducible derepression and then subsequent repression to eightfold less than the original basal level (Fig. 1B). Simultaneously, the Ace1p-mediated transcriptional activation of CUP1 mRNA expression was rapid and robust but transient. In response to 1, 10, and 100 μM Cu, maximum levels of CUP1 mRNA induction of 16-, 27-, and 36-fold, respectively, occurred within 30 min of Cu treatment. This was followed by a precipitous reduction in CUP1 mRNA levels within 60 min to approximately eightfold the original basal level and remained at these levels throughout the time course of the experiment (Fig. 1A and C). Therefore, in response to the continued presence of Cu ions, the expression of CTR3 was extinguished whereas the CUP1 gene was strongly but transiently activated.
FIG. 1.
Expression of CTR3 and CUP1 in response to 1, 10, and 100 μM CuSO4. (A) Exponential-phase cultures of S. cerevisiae DTY1 were treated with 1, 10, and 100 μM CuSO4. Five-milliliter samples were taken after 0, 2, 5, 10, 15, 20, 25, 30, 45, 60, 75, 90, and 120 min of Cu treatment. RNA extracted from each sample was analyzed by RNase protection assays. Each reaction contained 30 μg of total RNA. CTR3, CUP1, and ACT1 RNAs are indicated by arrows. (B) Quantitation of CTR3 mRNA expression in response to Cu. (C) Quantitation of CUP1 mRNA expression in response to Cu. All values in panels B and C were normalized against those for ACT1 mRNA as an internal control.
Steady-state Cup1p levels during Cu ion treatment.
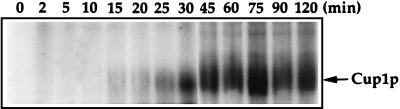
The yeast metallothionein encoded by CUP1 detoxifies Cu ions by tightly sequestering seven atoms of Cu(I) through coordination with the thiolate ligands of the abundant cysteine residues in the protein. Since CUP1 mRNA levels were transiently elevated when cells were grown in the continued presence of Cu ions, the levels of Cup1 protein were analyzed from cells treated with 100 μM Cu by metabolically labeling yeast cells with [35S]cysteine. Total soluble proteins were extracted from culture aliquots, analyzed on a 20% nondenaturing polyacrylamide gel, and visualized by fluorography (54). Figure 2 shows that although the levels of Cup1 protein increased with increasing times of Cu treatment, these levels remained high throughout the time course of the experiment. Therefore, while CUP1 mRNA levels were transiently induced, Cup1p remained at high steady-state levels.
FIG. 2.
Induction of Cup1p in response to 100 μM CuSO4. Exponential-phase cultures of S. cerevisiae DTY1 were labeled with 5 μCi of [35S]cysteine (800 Ci/mmol) per ml for 30 min and then treated with 100 μM CuSO4. Five-milliliter samples were taken at 0, 2, 5, 10, 15, 20, 25, 30, 45, 60, 75, 90, and 120 min of Cu treatment. Total protein extracts were analyzed on a 20% nondenaturing polyacrylamide gel. Each lane contains 30 μg of total protein. The gel was fixed with 10% acetic acid and 30% methanol for 1 h, fluorographed with En3Hance (Dupont), dried under vacuum, and exposed to Kodak BioMax Film with an intensifying screen at −80°C.
Ace1p occupation of MREs parallels transient CUP1 expression.
The Ace1p metalloregulatory transcription factor rapidly activates CUP1 transcription in response to Cu ions (40). The cooperative formation of a tetra-Cu cluster in the amino-terminal Ace1p DNA binding domain, via cysteine thiolate coordination, induces Ace1p to bind to two potent and two modestly active MREs in the CUP1 promoter (23). We postulated that inactivation of CUP1 expression, in the chronic presence of Cu ions, could occur via several mechanisms. First, it is possible that the half-life of CUP1 mRNA decreases at the later time points. Second, it is possible that at the later time points, Ace1p is degraded. Third, Ace1p could remain bound to the CUP1 MREs but the carboxyl-terminal transactivation domain might be rendered inactive. Fourth, the activation and inactivation of CUP1 transcription by Ace1p may simply reflect fluctuations in the amount of available intracellular Cu ions sufficient to maintain Ace1p in an active configuration for DNA binding.
Since the estimated half-life of CUP1 mRNA is approximately 12 to 16 min (35a), the reduced levels of CUP1 mRNA between 30 and 60 min may reflect the combined result of its normal decay and a reduced rate of transcription at these time points. To test the possibility that Ace1p is degraded at the later time points, DTY1 cells were incubated with 1, 10, and 100 μM Cu ions, and total cellular proteins were extracted and analyzed by immunoblotting with polyclonal antibodies raised against Ace1p expressed in and purified from E. coli. The results in Fig. 3 show that the levels of Ace1p remained constant throughout the time course of the experiment at all Cu ion concentrations. Therefore, the reduction in CUP1 mRNA levels at later time points was not the result of a Cu-dependent degradation of Ace1p. This finding is consistent with previous observations that ACE1 expression, at the level of steady-state mRNA, is constitutive and not affected by the presence or absence of Cu ions (39).
FIG. 3.
Western blot analysis of Ace1p levels during Cu ion treatment. Exponential-phase cultures of S. cerevisiae DTY1 were treated with 1, 10, or 100 μM CuSO4. Five-milliliter samples were taken after 0, 2, 5, 10, 15, 20, 25, 30, 45, 60, 75, and 90 min of Cu treatment. Thirty micrograms of total protein extract was analyzed by immunoblotting with polyclonal antibodies raised against purified recombinant Ace1p from E. coli (rACE1p).
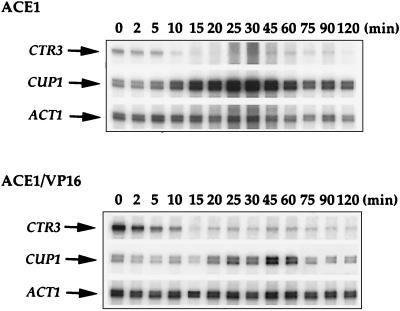
To test the role of the Ace1p transactivation domain in the inactivation of CUP1 expression, we constructed a chimeric gene in which the transactivation domain of the herpes simplex virus VP16 protein was fused to the Ace1p amino-terminal DNA binding domain. This chimeric protein contains the amino-terminal 122 amino acids of Ace1p, encompassing the minimal Cu-activated DNA binding domain (22) fused to 78 amino acids (residues 401 to 479) from the carboxyl-terminal activation domain of VP16 (5, 6). The ACE1-VP16 gene and, as a control, the wild-type ACE1 gene were integrated in single copy at the chromosomal his3 locus in an ace1Δ strain. Copper resistance tests demonstrated that the strain harboring the integrated ACE1 gene was resistant to 2 mM CuSO4, which was indistinguishable from the result for parental strain containing a genomic copy of wild-type ACE1. On the other hand, the strain harboring the integrated ACE1-VP16 fusion gene was resistant only to 200 μM CuSO4 (data not shown). Since a strain harboring only the Ace1p Cu-activated DNA binding domain, with no transactivation domain, is resistant to only approximately 25 μM CuSO4 (22), the VP16 activation domain significantly activates CUP1 expression, though not as strongly as the natural Ace1p activation domain. It is possible that Ace1p-VP16 is present at lower levels than Ace1p; however, it is also possible that since VP16 is a heterologous transactivator, it requires other factors not present in yeast or other cellular components that may not function well with the CUP1 promoter for strong activation of CUP1 transcription, and this might be responsible for the lower level of activation exhibited by this fusion protein. Cells harboring the integrated ACE1 and ACE1-VP16 genes were incubated with 100 μM CuSO4, and CTR3, CUP1, and ACT1 mRNA levels were analyzed by RNase protection assays. The results shown in Fig. 4 demonstrate that, consistent with the Cu ion resistance data, both Ace1p and Ace1p-VP16 mediate the induction with subsequent inactivation of CUP1 expression in response to Cu, although at different magnitudes. Ace1p activates CUP1 expression approximately 19-fold within 45 min, followed by reduction to approximately 6-fold above the original basal level. The Ace1p-VP16 fusion protein modestly (fourfold) activates the expression of CUP1, followed by reduction to less than twofold above the original basal level. These results suggest that the Ace1p carboxyl-terminal activation domain is required for maximum CUP1 induction in response to Cu. Furthermore, the reduction in CUP1 expression upon prolonged exposure to Cu ions does not occur specifically through the Ace1p carboxyl-terminal transactivation domain.
FIG. 4.
Transcription of CTR3 and CUP1 by ACE1-VP16 in response to 100 μM CuSO4. Exponential-phase cultures of strains MPY2 and MPY3 harboring a wild-type ACE1 gene and a gene fusion between the Ace1p DNA binding domain and the VP16 activation domain integrated at the his3 locus, respectively, were treated with 100 μM CuSO4. Five-milliliter samples were taken after 0, 2, 5, 10, 15, 20, 25, 30, 45, 60, 75, 90, and 120 min of Cu treatment. Thirty micrograms of total RNA was used in RNase protection experiments for each sample, using ACT1 mRNA as an internal control. CTR3, CUP1, and ACT1 RNase protection products are indicated by arrows.
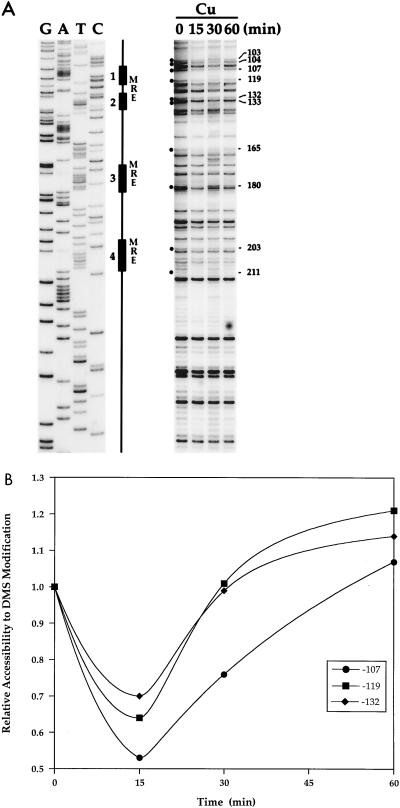
The CUP1 promoter harbors four MREs which have previously been shown to be bound by Ace1p both in vitro and in vivo (12, 23). The promoter proximal MREs (MREs 1 and 2) potently activate CUP1 transcription in response to Cu ions, while the distal MREs (MREs 3 and 4) modestly contribute to the magnitude of Cu ion-inducible CUP1 activation (23). To test the hypothesis that the inactivation of CUP1 transcription in response to prolonged treatment of cells with Cu ions is mediated through the regulation of Cu-dependent Ace1p binding to MREs, the occupancy of the CUP1 MREs was monitored over a time course of Cu ion exposure by in vivo footprinting with DMS. Log-phase cultures of DTY1 cells were treated with 100 μM Cu; samples were taken after 0, 15, 30, and 60 min of Cu ion treatment and incubated with 2% DMS to assess the occupancy of the MREs by Ace1p over time. The data in Fig. 5 show that before treatment of cells with Cu ions, the guanosine (G) residues within the MREs that are engaged in major groove contacts with Ace1p (indicated by dots in Fig. 5A) were accessible to modification by DMS. After 15 min of Cu ion treatment, when CUP1 expression was strongly induced, the G residues at positions −180, −165, −133, −132, −119, −107, −104, and −103 were relatively inaccessible to modification by DMS compared to the zero time point. This is consistent with occupation of the MREs corresponding to these positions (MREs 1 to 3) by Ace1p. Occupancy of the MREs coincides with the onset of CUP1 mRNA synthesis as shown in Fig. 1A. The intensities of these bands after 15 and 30 min of Cu treatment suggest a 50% occupancy by Ace1p of the MREs at this Cu concentration. After 60 min, when CUP1 mRNA levels were strongly reduced, the G residues were more accessible to modification by DMS. The quantitation shown in Fig. 5B shows the relative accessibility of the G residues at positions −107, −119, and −132 with respect to the start site of CUP1 transcription. These residues fall within or immediately adjacent to MREs 1 and 2, previously shown to be the most potent MREs in the CUP1 promoter (23). On the other hand, the G residues at positions −104, −165, −180, −203, and −211 were more accessible than after 15 and 30 min of Cu treatment but were not restored to basal levels (data not shown). This finding is consistent with the observation that at this time point, CUP1 mRNA levels were approximately eightfold above the basal level; thus, transcription by Ace1p is eightfold higher than in untreated cells. Based on these observations, the induction and subsequent reduction of CUP1 mRNA levels in response to Cu ions are mediated in large part through the Cu-dependent DNA binding of Ace1p to the MREs. Furthermore, the observation of reduced Ace1p occupation of the MREs at the later time points suggests that the inactivation of CUP1 expression is likely due to inactivation of the Ace1p Cu ion-dependent DNA binding domain.
FIG. 5.
In vivo DMS footprinting analysis of the CUP1 promoter. (A) Exponential-phase cultures of strain DTY1 were treated with 100 μM Cu. After 0, 15, 30, and 60 min of Cu treatment, 250-ml samples were taken and analyzed by in vivo DMS footprinting to assess the occupancy of the MREs by Ace1p. The positions of MREs 1 through 4 are indicated schematically, and G residues within MREs are indicated by dots. (B) Quantitation of the relative accessibility to DMS modification of representative G residues from the proximal MRE (MRE 1) closest to the CUP1 transcriptional start site. These values were normalized against the bands at positions −212 and −243 with respect to the transcriptional start site, whose intensities remained constant throughout the time course of Cu ion treatment.
Effect of increasing Cu ion concentrations on CUP1 regulation.
Inactivation of the DNA binding activity of Ace1p requires the removal of Cu ions from its DNA binding domain. To test the possibility that inactivation of the Ace1p DNA binding occurs through sequestration by Cup1p of excess Cu ions, DTY1 cells were challenged with 1 and 5 mM Cu ions and the effect on CUP1 downregulation was examined by RNase protection analyses. Since these Cu concentrations are at the threshold level of Cu resistance for this strain, expression of CUP1 is required for its survival and the excess Cu ions may surpass the chelation capacity of Cup1p. In response to 1 mM Cu, the levels of CUP1 mRNA were quickly induced 15-fold within 15 min, followed by downregulation to 4-fold above the original basal level within 60 min and then a further 14-fold induction within 2 h (Fig. 6). In response to 5 mM Cu, the CUP1 mRNA levels were quickly induced 15-fold within 15 min; however, the mRNA levels were not significantly downregulated but were maintained at approximately 15- to 20-fold above the basal level for 2 h. Therefore, at toxic Cu ion concentrations approaching the threshold for cell survival, transcription of the high-affinity Cu(I) transporters is extinguished, Cu uptake proceeds via the low-affinity uptake pathway, and CUP1 transcription is no longer downregulated at the later time points.
FIG. 6.
Effect of increasing Cu ion concentration on CUP1 regulation. Exponential-phase cultures of strain DTY1 were treated with 1 or 5 mM CuSO4. Five-milliliter samples were taken after 0, 15, 30, 45, 60, 90, and 120 min of Cu treatment. RNA was extracted from each sample and analyzed by RNase protection assays. Each reaction contained 20 μg of total RNA. CUP1 and ACT1 mRNAs are indicated by arrows. Quantitation of CUP1 mRNA is shown in the graph.
Role of the Cu(I) transporters in CUP1 regulation.
S. cerevisiae utilizes the products of the FRE1, CTR1, and CTR3 genes to carry out high-affinity Cu(I) transport. In contrast to the CUP1 gene, each of these genes is transcriptionally activated by Cu ion starvation and inactivated by Cu ion repletion (26). Both the transcriptional activation and inactivation of these genes require Mac1p (14, 24, 29, 48). Furthermore, the ability to properly activate, via metallation, the Ace1p DNA binding domain depends on the presence of functional yeast Cu(I) ion transport machinery (9, 25). The observations that CUP1 expression and Cu ion-dependent Ace1p binding to the CUP1 MREs is transient and that expression of the high-affinity Cu(I) transport machinery is inactivated under these conditions suggest a link between the proper expression of the Cu ion transport and detoxification pathways. To test the potential role of the high-affinity Cu(I) transport system in the inactivation of CUP1 expression in response to Cu ions, a strain carrying a dominant mutation in the MAC1 gene, MAC1up1, was used. MAC1up1 strains are unable to properly sense even high Cu ion concentrations and therefore express high constitutive levels of mRNA from the FRE1, CTR1, and CTR3 genes (14, 29, 48, 56). A MAC1up1 strain (DTY205) was incubated in the presence of 1, 10, or 100 μM CuSO4, and the levels of CTR3, CUP1, and ACT1 mRNAs were analyzed by RNase protection. The results shown in Fig. 7A, and quantitated in Fig. 7B and C, show that although CTR3 transcription was transiently inactivated by Cu ions in the MAC1up1 strain at all concentrations used, CTR3 mRNA levels were eventually elevated to approximately 1.3-fold the original basal level upon prolonged exposure to Cu ions. In response to 1 μM Cu, transcription of the CUP1 gene was transiently induced as in the wild-type parental strain. However, at a Cu ion concentration of 10 or 100 μM, CUP1 transcription was sustained at approximately 15-fold above the original basal level after the initial induction (Fig. 7). These results demonstrate that sustained expression of the high-affinity Cu(I) ion transport machinery also results in sustained transcription of CUP1.
FIG. 7.
Expression of CTR3 and CUP1 in a MAC1up1 strain. (A) Exponential-phase cultures of strain DTY205, which harbors the dominant MAC1up1 allele, were treated with 1, 10, and 100 μM Cu. Five-milliliter samples were taken after 0, 2, 5, 10, 15, 20, 25, 30, 45, 60, 75, 90, and 120 min of Cu ion treatment and analyzed by RNase protection assays. Thirty micrograms of total RNA was analyzed in each reaction. The CTR3, CUP1, and ACT1 RNase protection products are indicated by arrows. (B) Quantitation of CTR3 expression in response to Cu. (C) Quantitation of CUP1 expression in response to Cu. The values shown in panels B and C were normalized against values for ACT1 as an internal control.
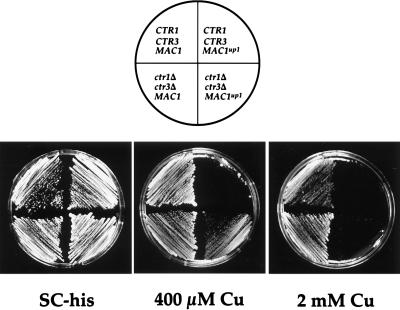
One phenotype associated with MAC1up1 strains is hypersensitivity to Cu ion toxicity, even though, as demonstrated here, CUP1 expression after prolonged exposure to at least 10 μM Cu ions (90 to 120 min) is much higher than in an isogenic MAC1 wild-type strain. This hypersensitivity may be a consequence of the high levels of Cu ion transport previously observed (24) due to the constitutive expression of the Cu ion transport genes. To assess the effect of continued Cu uptake by the Cu ion transporters on the ability of a MAC1up1 strain to detoxify Cu, strains harboring the MAC1 or MAC1up1 allele, with or without functional CTR1 and CTR3 genes, were challenged with increasing concentrations of Cu ions. Figure 8 shows that a strain harboring a wild-type MAC1 gene survived in the presence of 2 mM CuSO4. Deletion of the CTR1 and CTR3 high-affinity Cu(I) transport genes allowed these cells to grow slowly in the presence of a slightly higher Cu concentration of 2.5 mM (data not shown), thus providing the cell with a slight added advantage over strains that have both CTR1 and CTR3. On the other hand, a strain harboring the dominant MAC1up1 allele was sensitive to 400 μM Cu whereas deletion of CTR1 and CTR3 allowed these strains to grow in the presence of 400 μM Cu, but not at higher concentrations, providing only a limited advantage over strains that possessed both transporters. Thus, while continued Cu ion uptake by CTR1 and CTR3 contribute to the increased Cu sensitivity of a MAC1up1 strain, the presence of this allele may directly or indirectly influence other cellular events that can contribute to the Cu sensitivity phenotype. These results clearly show that although deletion of the Cu ion transporters Ctr1p and Ctr3p contribute modestly to Cu resistance, the nutritional Cu sensor Mac1p plays a critical role in cell survival under toxic Cu conditions.
FIG. 8.
Copper sensitivity of MAC1 and MAC1up1 strains. Yeast strains of the indicated relevant genotypes were plated on SC-His medium containing increasing concentrations of CuSO4. Strains DTY1 and DTY205 were transformed with the pRS313 vector to allow growth on SC-His plates. Both strains contain the CTR1 and CTR3 genes and harbor a wild-type MAC1 and MAC1up1 alleles, respectively. MPY18 strains lack both high-affinity Cu transport genes and harbor a chromosomal deletion of the mac1Δ allele. These strains were transformed with a plasmid expressing either the MAC1 or MAC1up1 allele on a pRS313-based centromeric vector. The MAC1 genes were both tagged with a single copy of the hemagglutinin epitope at the 3′ end of the open reading frame.
DISCUSSION
The essential yet toxic nature of Cu ions in biological systems demands tight regulation of the expression of genes involved in Cu homeostasis. Proper regulation is critical to dictate that appropriate levels of Cu ions are present in cells at all times and under all growth conditions. Consistent with this delicate regulation, the proteins involved in Cu uptake, distribution, and detoxification are regulated in response to Cu ions at the level of gene transcription, posttranscriptional events, and protein trafficking (26). The CTR3 gene, encoding a protein involved in high-affinity Cu(I) transport, and the CUP1 gene, encoding the Cu ion binding and detoxification metallothionein protein, are transcriptionally regulated in opposite directions in response to elevated Cu ion concentrations. Furthermore, although CTR3 is transcriptionally inactivated at very low Cu ion concentrations and CUP1 is activated at high Cu ion concentrations, our results establish that there is a window of overlap between the nutritional and toxic Cu ion sensing systems. Since at least two Cu-responsive regulatory circuits function in yeast, there must be a dynamic interplay between the Cu ion uptake and detoxification pathways and their regulation by the Cu-responsive transcription factors Mac1p and Ace1p.
Interestingly, we demonstrate that in the chronic presence of elevated Cu ion concentrations, CUP1 mRNA levels are rapidly but transiently activated, while expression of the CTR3 gene is inactivated. The analysis of Ace1p steady-state levels, the activity of an Ace1p DNA binding domain-VP16 activation domain fusion protein, and in vivo footprinting studies have clearly demonstrated that the transient activation of CUP1 occurs via the modulation of Cu ion-dependent Ace1p binding to the CUP1 promoter MREs. Occupation of the CUP1 promoter MREs closely paralleled the abundance of CUP1 mRNA levels. This correlation might reflect fluctuations in the availability of intracellular Cu ions for the activation of Ace1p DNA binding during the time course of the experiment. Previous studies in mice showed that administration of cadmium (Cd) transiently induced metallothionein-I gene (MT-I) transcription in the liver and kidney, with maximum transcriptional rates observed within 1 h and maximum mRNA levels observed within 4 h of Cd injection. MT-I mRNA levels are then reduced within 9 h; however, these levels never return to the constitutive basal levels (11). In these studies, however, MT-I induction was measured in response to a single injection of Cd rather than constant exposure of the mice to extracellular Cd. In other studies using Neurospora crassa, Cu treatment strongly induced Cu-MT mRNA levels within 1 h followed by repression to basal levels within 8 h, while Cu-MT protein levels reach maximum levels within 3 h and remained at the same elevated levels over the time period examined (17 h) (33). These results in other eukaryotic systems correlate well with our direct analysis of the activity of a yeast CuMRTF.
What might be the mechanisms by which CUP1 expression is turned down, through inactivation of the Ace1p DNA binding function even in the continued presence of elevated Cu ion concentrations? One mechanism might be through Cu ion efflux. Studies using BHK cell lines showed transient expression of a β-galactosidase reporter gene fused to five MREs (MRE-β-Geo) in response to zinc, and this transient expression was attributed to zinc efflux by ZnT-1 (33a, 35) and sequestration of zinc into an endosomal/lysosomal compartment by ZnT-2 (34), to prevent the accumulation of intracellular zinc to toxic levels. However, it is unlikely that the transient expression of CUP1 mRNA shown in our results is due to extrusion of Cu ions from yeast cells. Previous studies have clearly shown that over the same time course as our experiments, cells treated with elevated Cu levels in the media continue to accumulate Cu ions to a level of saturation with increasing time of incubation, without any evidence for efflux (30, 35b). In these studies, however, Cu levels were measured as total cell-associated Cu; thus, although the results preclude Cu efflux, they do not address the possibility of Cu transport into the vacuole. Since our experiments demonstrate that Cup1 protein stably accumulates over the time course of these experiments, one mechanism may be that Cup1p itself outcompetes Ace1p for available Cu ions, either through competition for available free intracellular copper or through the disassembly of the tetra-copper cluster in the Ace1p DNA binding domain that is essential for an active DNA binding configuration. It is interesting that proteins called Cu chaperones which deliver Cu ions to specific intracellular targets that include the secretory compartment, Cu,Zn superoxide dismutase, and the mitochondria have been identified in yeast and human cells (8, 15, 31). Therefore, it is possible that Cu ion chaperones exist to disassemble preformed Cu clusters or that the assembly reaction carried out by a Cu-specific chaperone is also reversible. To date, no Cu ion chaperones specific for assembly of the Cu cluster in Ace1p have been reported. On the other hand, our results show that when cells were treated with toxic levels of Cu (5 mM), CUP1 expression was no longer downregulated, suggesting that as the intracellular Cu levels increase, the chelation capacity of Cup1p is saturated and excess Cu becomes available to activate the DNA binding activity of Ace1p, resulting in the continued expression of CUP1 mRNA. Previous studies have shown that apometallothioneins can sequester Zn from the zinc finger domains on transcription factors such as Sp1 (50) and TFIIIA (51), rendering them unable to bind to target DNA sequences. In addition, it has been shown that autoregulation of CUP1 transcription by Cup1p depends on its ability to bind and detoxify Cu. Mutations in CUP1 that prevented Cu ion binding also diminished its ability to autoregulate its own transcription and detoxify Cu (46). Thus, increased synthesis of Cup1p metallothionein in response to Cu ions in yeast might sequester Cu from Ace1p, rendering it unable to bind to the MREs on the CUP1 promoter, providing a mechanism for autoregulating its own transcription. This mechanism for Cu detoxification by Cup1p and autoregulation of its own transcription in the continued presence of Cu in the media clearly requires that additional Cu uptake by the high-affinity transporters be prevented.
Although CTR1 and CTR3 play critical roles in high-affinity Cu ion transport, our data suggest that the proper regulation of these transporter genes by Mac1p, even in the presence of high Cu ion concentrations, is crucial for the normal transient expression of CUP1. In a strain expressing a mutant allele of the MAC1 gene which gives strong constitutive activation of CTR1 and CTR3, CUP1 mRNA was transcriptionally activated by Cu ions, but the mRNA levels persisted, in contrast to the transient expression under the same conditions in wild-type cells. These observations suggest that the disappearance of the Ctr1p and Ctr3p transporters from the cell surface plays an important role in limiting intracellular Cu ion levels that are available to Ace1p.
The results presented here also clearly demonstrate that the Mac1p, previously thought to function predominantly in sensing nutritional levels of Cu ions, must function properly to allow cells to mount a normal Cu detoxification response. The inability of Mac1p to properly sense Cu ions, as exhibited by the Mac1up1 protein, resulted in sustained expression of the CTR1 and CTR3 genes and a concomitant hypersensitivity to Cu ions. This background also gives rise to sustained expression of the CUP1 gene, suggesting that Cu toxicity results from continued Cu uptake that surpasses the chelation or compartmentalization capacity of the cell. Consistent with this possibility, deletion of the CTR1 and CTR3 genes in the MAC1up1 strain partially restored Cu resistance. Our results show, however, that continued Cu uptake by the high-affinity Cu ion transporters contribute minimally to the Cu hypersensitivity of a MAC1up1 strain since removal of CTR1 and CTR3 in this strain allowed cells to survive in the presence of only 400 μM, while removal of both genes in a wild-type MAC1 strain allowed this strain to grow in the presence of 2.5 mM Cu. This finding suggests that additional Mac1p target genes must be properly regulated for normal Cu ion resistance. It is possible that, as in the case of the Zn metalloregulatory factor Zap1p, which regulates the transcription of both the high- and low-affinity zinc uptake genes in yeast (52), Mac1p regulates the expression of both the high- and low-affinity Cu ion uptake genes and that a Mac1up1 protein would result in dramatic overexpression of the low-affinity Cu ion transport machinery. Additionally, Cu sensitivity in the MAC1up1 background could result from an inability to properly regulate other genes encoding proteins that may directly or indirectly protect cells from the toxic effects of Cu ions. To date, these genes and genes encoding the low-affinity Cu ion uptake machinery remain to be identified. The results presented here underscore the importance of dynamic regulation of the Cu ion transport pathway by the nutritional Cu sensor Mac1p, and the Cu detoxification pathway by the toxic Cu sensor Ace1p, to maintain appropriate intracellular Cu levels and for survival in the continued presence of elevated Cu levels in the environment.
ACKNOWLEDGMENTS
We thank the members of the Thiele laboratory for stimulating discussions and critical comments. We are grateful to Simon Labbé for providing plasmids pSKCUP1, pSKCTR3, ura3::KanMX2, and pKSACT1 and to Zhiwu Zhu for providing plasmids pRSMAC1(HA) and pRSMAC1up1(HA).
This work was supported by National Institutes of Health (NIH) grant RO1 GM41840 to D.J.T., postdoctoral fellowship-National Research Service Award F32 GM18089 from NIH to M.M.O.P., and Cellular Biotechnology Training Program NIH grant GM08353 to K.A.K. D.J.T. is a Burroughs Wellcome Toxicology Scholar.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 2.Bull P C, Cox D W. Wilson disease and Menkes disease: new handles on heavy metal transport. Trends Genet Sci. 1994;10:248–252. doi: 10.1016/0168-9525(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 3.Bull P C, Thomas G R, Rommens J M, Forbe J R, Cox D W. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet. 1993;5:327–337. doi: 10.1038/ng1293-327. [DOI] [PubMed] [Google Scholar]
- 4.Butler G, Thiele D J. ACE2, an activator of yeast metallothionein expression which is homologous to SWI5. Mol Cell Biol. 1991;11:476–485. doi: 10.1128/mcb.11.1.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cousens D J, Greaves R, Goding C R, O’Hare P. The C-terminal 79 amino acids of the herpes simplex virus regulatory protein, Vmw65, efficiently activate transcription in yeast and mammalian cells in chimeric DNA-binding proteins. EMBO J. 1989;8:2337–2342. doi: 10.1002/j.1460-2075.1989.tb08361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cress W D, Triezenberg S J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 7.Culotta V C, Howard W R, Liu X F. CRS5 encodes a metallothionein-like protein in Saccharomyces cerevisiae. J Biol Chem. 1994;269:25295–25302. [PubMed] [Google Scholar]
- 8.Culotta V C, Klomp L W J, Strain J, Casareno R L B, Krems B, Gitlin J D. The copper chaperone for superoxide dismutase. J Biol Chem. 1997;272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 9.Dancis A, Haile D, Yuan D S, Klausner R D. The Saccharomyces cerevisiae copper transport protein (Ctr1p) J Biol Chem. 1994;269:25660–25667. [PubMed] [Google Scholar]
- 10.Dancis A, Yuan D S, Haile D, Askwith C, Eide D, Moehle C, Kaplan J, Klausner R D. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell. 1994;76:393–402. doi: 10.1016/0092-8674(94)90345-x. [DOI] [PubMed] [Google Scholar]
- 11.Durnam D M, Palmiter R D. Transcriptional regulation of the mouse metallothionein-I gene by heavy metals. J Biol Chem. 1981;256:5712–5716. [PubMed] [Google Scholar]
- 12.Evans C F, Engelke D R, Thiele D J. ACE1 transcription factor produced in Escherichia coli binds multiple regions within yeast metallothionein upstream activation sequences. Mol Cell Biol. 1990;10:426–429. doi: 10.1128/mcb.10.1.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furst P, Hu S, Hackett R, Hamer D. Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell. 1988;55:705–717. doi: 10.1016/0092-8674(88)90229-2. [DOI] [PubMed] [Google Scholar]
- 14.Georgatsou E, Mavrogiannis L A, Fragiadakis G S, Alexandraki D. The yeast Fre1p/Fre2p cupric reductases facilitate copper uptake and are regulated by the copper-modulated Mac1p activator. J Biol Chem. 1997;272:13786–13792. doi: 10.1074/jbc.272.21.13786. [DOI] [PubMed] [Google Scholar]
- 15.Glerum D M, Shtanko A, Tzagoloff A. Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J Biol Chem. 1996;271:14504–14509. doi: 10.1074/jbc.271.24.14504. [DOI] [PubMed] [Google Scholar]
- 16.Graden J A, Winge D R. Copper-mediated repression of the activation domain in the yeast Mac1p transcription factor. Proc Natl Acad Sci USA. 1997;94:5550–5555. doi: 10.1073/pnas.94.11.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gralla E B, Thiele D J, Silar P, Valentine J S. ACE1, a copper-dependent transcription factor, activates expression of the yeast copper,zinc superoxide dismutase gene. Proc Natl Acad Sci USA. 1991;88:8558–8562. doi: 10.1073/pnas.88.19.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halliwell B, Gutteridge J M C. Oxygen toxicity, oxygen radicals, transition metals and diseases. Biochem J. 1984;219:1–4. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamer D H. Metallothioneins. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- 20.Hamer D H, Thiele D J, Lemontt J E. Function and autoregulation of yeast copperthionein. Science. 1985;228:685–690. doi: 10.1126/science.3887570. [DOI] [PubMed] [Google Scholar]
- 21.Hassett R, Kosman D J. Evidence for Cu(II) reduction as a component of Cu uptake by Saccharomyces cerevisiae. J Biol Chem. 1996;270:128–134. doi: 10.1074/jbc.270.1.128. [DOI] [PubMed] [Google Scholar]
- 22.Hu S, Furst P, Hamer D. The DNA and Cu binding functions of ACE1 are interdigitated within a single domain. New Biol. 1990;2:544–555. [PubMed] [Google Scholar]
- 23.Huibregtse J M, Engelke D R, Thiele D J. Copper-induced binding of cellular factors to yeast metallothionein upstream activation sequences. Proc Natl Acad Sci USA. 1989;86:65–69. doi: 10.1073/pnas.86.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jungmann J, Reins H A, Lee J, Romeo A, Hassett R, Kosman D, Jentsch S. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 1993;12:5051–5056. doi: 10.1002/j.1460-2075.1993.tb06198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight S A B, Labbé S, Kwon L F, Kosman D J, Thiele D J. A widespread transposable element masks expression of a yeast copper transport gene. Genes Dev. 1996;10:1917–1929. doi: 10.1101/gad.10.15.1917. [DOI] [PubMed] [Google Scholar]
- 26.Koch K A, Peña M M O, Thiele D J. Copper-binding motifs in catalysis, transport, detoxification and signaling. Chem Biol. 1997;4:549–560. doi: 10.1016/s1074-5521(97)90241-6. [DOI] [PubMed] [Google Scholar]
- 27.Koch K A, Thiele D J. Autoactivation by a Candida glabrata copper metalloregulatory transcription factor requires critical minor groove interactions. Mol Cell Biol. 1996;16:724–734. doi: 10.1128/mcb.16.2.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 29.Labbé S, Zhu Z, Thiele D J. Copper-specific transcriptional repression of yeast genes encoding critical components in the copper transport pathway. J Biol Chem. 1997;272:15951–15958. doi: 10.1074/jbc.272.25.15951. [DOI] [PubMed] [Google Scholar]
- 30.Lin C, Kosman D J. Copper uptake in wild type and copper metallothionein-deficient Saccharomyces cerevisiae. J Biol Chem. 1990;265:9194–9200. [PubMed] [Google Scholar]
- 31.Lin S-J, Pufahl R A, Dancis A, O’Halloran T V, Culotta V C. A role for the Saccharomyces cerevisiae ATX1 gene in copper trafficking and iron transport. J Biol Chem. 1997;272:9215–9220. [PubMed] [Google Scholar]
- 32.Linder M C. Biochemistry of copper. New York, N.Y: Plenum Press; 1991. [Google Scholar]
- 33.Münger K, Germann U A, Lerch K. Isolation and regulation of expression of the Neurospora crassa copper metallothionein gene. Basel, Germany: Birkhauser Verlag; 1987. [DOI] [PubMed] [Google Scholar]
- 33a.Palmiter, R. Personal communication.
- 34.Palmiter R D, Cole T B, Findley S D. ZnT-2, a mammalian protein that confers resistance to zinc by facilitating vesicular sequestration. EMBO J. 1996;15:1784–1791. [PMC free article] [PubMed] [Google Scholar]
- 35.Palmiter R D, Findley S D. Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 1995;14:639–649. doi: 10.1002/j.1460-2075.1995.tb07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Parker, R. Personal communication.
- 35b.Peña, M. M. O., and D. J. Thiele. Unpublished data.
- 36.Predki P F, Sarkar B. Effect of replacement of “zinc finger” zinc on estrogen receptor DNA interactions. J Biol Chem. 1992;267:5842–5846. [PubMed] [Google Scholar]
- 37.Rymond B C, Zitomer R S, Schumperli D, Rosenberg M. The expression in yeast of the Escherichia coli galk gene on CYC1::galk fusion plasmids. Gene. 1983;25:249–262. doi: 10.1016/0378-1119(83)90229-9. [DOI] [PubMed] [Google Scholar]
- 38.Sherman F, Fink G R, Hicks J. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 39.Szczypka M S, Thiele D J. A cysteine-rich nuclear protein activates yeast metallothionein gene transcription. Mol Cell Biol. 1989;9:421–429. doi: 10.1128/mcb.9.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiele D J. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein gene. Mol Cell Biol. 1988;8:2745–2752. doi: 10.1128/mcb.8.7.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiele D J. Metal-regulated transcription in eukaryotes. Nucleic Acids Res. 1992;20:1183–1191. doi: 10.1093/nar/20.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorvaldsen J L, Sewell A K, Tanner A M, Peltier J M, Pickering I J, George G N, Winge D R. Mixed Cu+ and Zn2+ coordination in the DNA-binding domain of the AMT1 transcription factor from Candida glabrata. Biochemistry. 1994;33:9566–9577. doi: 10.1021/bi00198a024. [DOI] [PubMed] [Google Scholar]
- 43.Vulpe C, Levinson B, Whitney S, Packman S, Gitschier J. Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper transporting ATPase. Nat Genet. 1993;3:7–13. doi: 10.1038/ng0193-7. [DOI] [PubMed] [Google Scholar]
- 44.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 45.Welch J, Fogel S, Buchman C, Karin M. The CUP2 gene product regulates the expression of the CUP1 gene coding for yeast metallothionein. EMBO J. 1989;8:255–260. doi: 10.1002/j.1460-2075.1989.tb03371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright C F, Hamer D H, McKenney K. Autoregulation of the yeast copper metallothionein gene depends on metal binding. J Biol Chem. 1988;263:1570–1574. [PubMed] [Google Scholar]
- 47.Yamaguchi Y, Heiny M E, Gitlin J D. Isolation and characterization of a human liver cDNA as a candidate gene for Wilson disease. Biochem Biophys Res Commun. 1993;197:271–277. doi: 10.1006/bbrc.1993.2471. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi-Iwai Y, Serpe M, Haile D, Yang W, Kosman D J, Klausner R D, Dancis A. Homeostatic regulation of copper uptake in yeast via direct binding of MAC1 protein to upstream regulatory sequences of FRE1 and CTR1. J Biol Chem. 1997;272:17711–17718. doi: 10.1074/jbc.272.28.17711. [DOI] [PubMed] [Google Scholar]
- 49.Yuan D S, Stearman R, Dancis A, Dunn T, Beeler T, Klausner R D. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc Natl Acad Sci USA. 1995;92:2632–2636. doi: 10.1073/pnas.92.7.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng J, Heuchel R, Schaffner W, Kagi J H R. Thionein (apometallothionein) can modulate DNA binding and transcription by zinc finger containing factor Sp1. FEBS Lett. 1991;279:310–312. doi: 10.1016/0014-5793(91)80175-3. [DOI] [PubMed] [Google Scholar]
- 51.Zeng J, Vallee B, Kagi J H R. Zinc transfer from transcription factor IIIA fingers to thionein clusters. Proc Natl Acad Sci USA. 1991;88:9984–9988. doi: 10.1073/pnas.88.22.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao H, Eide D J. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:5044–5052. doi: 10.1128/mcb.17.9.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou P, Thiele D J. Copper and gene regulation in yeast. BioFactors. 1993;4:105–115. [PubMed] [Google Scholar]
- 54.Zhou P, Thiele D J. Rapid transcriptional autoregulation of a yeast metalloregulatory factor is essential for high-level copper detoxification. Genes Dev. 1993;7:1824–1835. doi: 10.1101/gad.7.9.1824. [DOI] [PubMed] [Google Scholar]
- 55.Zhou P, Thiele D J. Isolation of a metal-activated transcription factor gene from Candida glabrata by complementation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:6112–6116. doi: 10.1073/pnas.88.14.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Z, Labbé S, Peña M M O, Thiele D J. Copper differentially regulates the activity and degradation of yeast Mac1 transcription factor. J Biol Chem. 1998;273:1277–1288. doi: 10.1074/jbc.273.3.1277. [DOI] [PubMed] [Google Scholar]
- 57.Zhu Z, Thiele D J. A specialized nucleosome modulates transcription factor access to a C. glabrata metal responsive promoter. Cell. 1996;87:459–470. doi: 10.1016/s0092-8674(00)81366-5. [DOI] [PubMed] [Google Scholar]