Abstract
Stat3 activation has been associated with cytokine-induced proliferation, anti-apoptosis, and transformation. Constitutively activated Stat3 has been found in many human tumors as well as v-abl- and v-src-transformed cell lines. Because of these correlations, we examined directly the relationship of activated Stat3 to cellular transformation and found that wild-type Stat3 enhances the transforming potential of v-src while three dominant negative Stat3 mutants inhibit v-src transformation. Stat3 wild-type or mutant proteins did not affect v-ras transformation. We conclude that Stat3 has a necessary role in v-src transformation.
Cytokines—extracellular signaling polypeptides—are capable of inducing cellular programs including proliferation or growth arrest and differentiation. Many cytokines activate STAT (signal transducers and activators of transcription) molecules (10, 26); therefore, the role of STATs in growth control is a crucial, incompletely explored issue. It has been known for many years that interferons (IFNs) slow the growth of a wide variety of cell types (2, 19). Both IFN-α and IFN-γ activate Stat1, and this protein is required for growth restraint in response to both IFNs (4, 8, 27).
In contrast, the results of several studies suggest that Stat3 may be involved in promoting cell growth: interleukin 6 (IL-6), which activates Stat3, is required in liver regeneration (9), and overexpression of IL-6 causes plasmacytomas (29); dominant negative Stat3 inhibits gp130-mediated anti-apoptosis (13); Stat3−/− mouse embryos implant but fail to grow (30); Stat3 has been reported to be activated constitutively in human tumors (14, 17, 25, 32, 36); and v-abl- and v-src-transformed cells have constitutively activated Stat3 (6, 21, 35).
We evaluated the role of Stat3 in cellular transformation by determining its contribution to v-src transformation. Colony formation in soft agar induced by newly introduced v-src was increased by wild-type Stat3 but not by mutant proteins that failed to be phosphorylated or to bind DNA. Also, in cells already transformed by v-src, the introduction of extra Stat3 led to an increase in the number of colonies in soft agar. In contrast, the presence of Stat3 dominant negative mutant protein had an inhibitory effect on the colony-forming capacity of v-src-transformed cells. The enhancement of v-src transformation by Stat3 was associated with increased Stat3 activity (assayed by DNA binding and transcriptional activation), while suppression of v-src transformation by the dominant negative Stat3 mutants correlated with decreased Stat3 activity. Thus, activated wild-type Stat3 has oncogenic potential and v-src transformation requires Stat3 activation.
MATERIALS AND METHODS
Plasmids.
Stat3 was cloned into RcCMV-Neo (Invitrogen) tagged at the 3′ end with a FLAG epitope (7, 33). Stat3Y705-F was generated by PCR and cloned into RcCMV-Neo and tagged at the 3′ end with a FLAG epitope. Stat3DB contains changes within its DNA-binding domain (VVV461–463AAA and EE434–435AA) and is also tagged at its 3′ end with a FLAG epitope. Individual clones having mutations at positions 461 to 463 and 434 to 435 (16) were joined by PCR and recloned. The resulting double mutant protein becomes phosphorylated in response to epidermal growth factor but does not bind DNA (15). Stat3S, which contains an alanine instead of a serine at position 727, was cloned into RcCMV (34). pBabe/v-src was a gift from H. Hanafusa, and v-src-transformed cells were selected with puromycin (24). pRSV Ha-rasLeu61 contains a constitutively active ras oncogene (11). The luciferase reporter plasmid used contains three copies of the Ly6E Stat1 and Stat3 binding site (34). A β-galactosidase expression plasmid (Invitrogen) was used in luciferase assays for normalization.
Tissue culture and soft agar assay.
NIH 3T3 cells were obtained from the American Type Culture Collection. v-src-transformed NIH 3T3 cells were obtained by transfecting NIH 3T3 cells with pBabe/v-src and selecting for transformants with 2μg of puromycin (Sigma) (24) per ml. All cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% Cosmic calf serum (HyClone). Lipofectamine reagent (Life Technologies) was utilized for all transfections. Typically, for a 35-mm-diameter dish containing 5 × 105 cells, 6 μl of Lipofectamine and 1 μg of each plasmid to be transfected was utilized. We increased the ratio of Stat3 or Stat3DN to v-src or ras by introducing 1.8 μg of RcCMV, Stat3, Stat3Y, Stat3DB, or Stat3S to 200 ng of v-src or ras. Selection and maintenance of plasmids in NIH 3T3-based cells required 800 μg of G418 sulfate (Geneticin; Life Technologies) per ml and/or 2μg of puromycin (Sigma) per ml. Soft agar assays were performed essentially as described previously (20). Twenty hours after transfection, cells were fed with DMEM plus 10% bovine calf serum (BCS). Twenty-four hours later, the cells were trypsinized and 5 × 105 cells were plated into 3 ml of soft agar (BiTek; Difco) containing 1× DMEM, 10% BCS, and the appropriate selective antibiotics. Two to 3 weeks later, colonies were counted. Typically, these assays were carried out in parallel with 4 to 10 replications. Averages of the replications with standard deviations are presented in the figures and in Table 1. For viability assays, 5 × 105 cells were plated into 3-cm-diameter dishes containing DMEM, 10% BCS, 800 μg of G418 sulfate per ml, and 2μg of puromycin per ml. Cells were counted with a Coulter counter and by a hemacytometer 5 and 7 days later. This assay was performed in triplicate. Averages with standard deviations are presented in Table 2.
TABLE 1.
Colony forming by NIH 3T3 cells transfected with various plasmid constructsa
| Plasmid construct | No. of NIH 3T3 colonies at ratio ofb:
|
No. of v-src-transformed NIH 3T3 colonies | |
|---|---|---|---|
| 1:2 | 1:10 | ||
| RcCMV | 0 | 0 | 51 ± 7 |
| Stat3 | 0 | 0 | 183 ± 15 |
| Stat3Y | 0 | 0 | 22 ± 5 |
| Stat3DB | 0 | 0 | 27 ± 4 |
| Stat3S | 0 | 0 | 20 ± 3 |
| v-src/RcCMV | 37 ± 5 | 32 ± 3 | NDc |
| v-src/Stat3 | 171 ± 16 | 105 ± 5 | ND |
| v-src/Stat3Y | 18 ± 5 | 7 ± 2 | ND |
| v-src/Stat3DB | 21 ± 7 | 2 ± 1 | ND |
| v-src/Stat3S | 16 ± 3 | 1 ± 1 | ND |
| v-Ha-rasLeu61 | 30 ± 5 | 16 ± 3 | ND |
| v-ras/Stat3 | 33 ± 6 | 17 ± 4 | ND |
| v-ras/Stat3Y | 36 ± 4 | 15 ± 4 | ND |
| v-ras/Stat3DB | 32 ± 3 | 15 ± 2 | ND |
| v-ras/Stat3S | 38 ± 6 | 16 ± 4 | ND |
NIH 3T3 and v-src-transformed NIH 3T3 cells were transfected with various combinations of RcCMV (control vector), Stat3, Stat3Y, Stat3DB, Stat3S, pBabe/v-src, and v-Ha-rasLeu61 and plated into soft agar as described in the legends to Fig. 2, 3, and 5.
Ratios are of v-src or v-ras to RcCMV Stat3-based constructs.
ND, not done.
TABLE 2.
Viability of transfected NIH 3T3 cellsa
| Plasmid construct | 104 Cells at:
|
|
|---|---|---|
| 5 Days | 7 Days | |
| RcCMV/pBabe | 5 ± 1 | 17 ± 3 |
| Stat3/pBabe | 6 ± 1 | 19 ± 4 |
| Stat3Y/pBabe | 7 ± 2 | 18 ± 3 |
| Stat3DB/pBabe | 7 ± 1 | 23 ± 5 |
| RcCMV/pBabe/v-src | 3 ± 1 | 18 ± 3 |
| Stat3/v-src | 3 ± 1 | 22 ± 4 |
| Stat3Y/v-src | 4 ± 2 | 23 ± 3 |
| Stat3DB/v-src | 3 ± 2 | 21 ± 3 |
| Stat3S/v-src | 4 ± 1 | 22 ± 3 |
| Stat3Y/v-src | 3 ± 1 | 23 ± 3 |
Cells were transfected with a 10:1 ratio of Stat3-based constructs to v-src, as described in the legend to Fig. 3A. Cells (5 × 105) were plated onto selective media and counted 5 and 7 days later with a Coulter counter; counts were confirmed by Trypan blue exclusion and counting with a hemacytometer. Values are averages of three independent transfections ± standard deviations.
In vitro assays.
Transcriptional assays were performed by transiently transfecting NIH 3T3 or v-src-transformed cells with RcCMV-Neo-based plasmids and pBabe/v-src and Ly6E luciferase plasmids. Twenty hours after transfection, the cells were fed with DMEM plus 10% BCS. Twenty-four hours later, the cells were lysed and luciferase (Promega) assays were performed according to the Promega protocol. Each transfection was normalized to concomitant β-galactosidase expression from a control-transfected plasmid (Invitrogen) (1). Nuclear extracts were made from transiently transfected cells, as described previously (34), and electrophoretic mobility shift assays (EMSA) were performed as described previously (12) with 32P-labeled Stat binding site M67 SIE (31) oligonucleotide. Western blotting was carried out by standard methods (1). Anti-Stat3-C serum was diluted 1:1,000 for Western blotting and 1:100 for supershifting of DNA-protein complexes in EMSA gels. Anti-M2 (FLAG) monoclonal antiserum (Kodak/IBI) was used at a 1:500 dilution for Western blotting and at a 1:100 dilution for supershifting of DNA-protein complexes. Antiphosphotyrosine PY20 monoclonal antiserum (Transduction Laboratories) was used at a 1:1,000 dilution for Western blotting.
RESULTS
Expression constructs used to establish the role of Stat3 in v-src transformation.
Oncogenic transformation of cells in vitro often requires cooperation between two different types of proteins with oncogenic potential (18), and v-src transformation is known to lead to activation of endogenous Stat3 (6, 35). To test whether Stat3 cooperates with v-src in cellular transformation, we transfected NIH 3T3 cells both with v-src in a vector containing the gene for puromycin resistance (24) and with an expression vector encoding resistance to G418 that controls expression of wild-type Stat3, Stat3Y, Stat3DB, and Stat3S or the expression vector alone (Fig. 1). The three mutant STATs act as dominant negative variants of wild-type protein during IL-6 or epidermal growth factor induction of Stat3 activity through various mechanisms (15, 22, 23, 34) and could be expected to inhibit endogenous Stat3 in v-src-transformed cells by similar mechanisms.
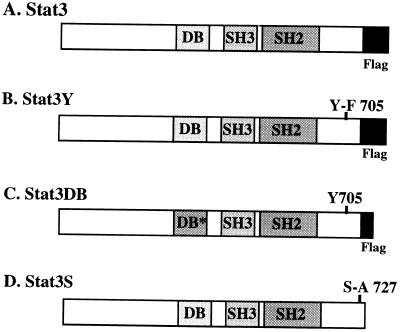
FIG. 1.
Wild-type Stat3 and dominant negative Stat3 constructs. (A) Wild-type Stat3 was cloned into RcCMV-Neo and tagged at the 3′ end with a FLAG epitope (7, 33). (B) Stat3Y705-F was generated by PCR and cloned into RcCMV-Neo and tagged at the 3′ end with a FLAG epitope. The expressed protein can be detected with FLAG (M2) monoclonal antiserum or Stat3-C antiserum. It cannot be phosphorylated on Y705 and can presumably compete with wild-type Stat3 for v-src or other kinases (5, 6). (C) Stat3DB was tagged at the 3′ end with a FLAG epitope which contains slightly fewer amino acids than those of the above constructs. Within the Stat3 DNA-binding domain, multiple changes (VVV461–463AAA and EE434–435AA), which abolish the ability of this protein to bind DNA while it retains the ability to be phosphorylated, form dimers, and translocate to the nucleus (15, 16), were made. (D) Stat3S contains an alanine instead of a serine at position 727 (34). This protein can be phosphorylated on tyrosine, form dimers, and bind DNA, but it cannot fully activate transcription (34). Stat3S was cloned into RcCMV and does not contain a FLAG epitope (34).
The Stat3Y mutant, which cannot be phosphorylated on tyrosine 705 (34), may compete with the wild-type protein for the activating kinase. It has been reported that Stat3 and v-src can be coprecipitated (6); therefore, it is possible that Stat3 is directly activated by v-src. Alternatively, some other kinase activated by v-src may be responsible for the constitutive activation of Stat3 (5).
The Stat3DB mutant combines both of the mutations (VVV to AAA and EE to AA) that were earlier shown to allow phosphorylation on tyrosine, dimerization, and nuclear localization but no DNA binding (15, 16). Stat3DB may function in a dominant negative manner by forming heterodimers with wild-type Stat3, rendering this complex inactive due to its inability to bind DNA (15).
The Stat3S mutant contains an alanine in place of a serine at position 727. This mutant protein can be phosphorylated on tyrosine 705 and can bind DNA but showed reduced transcriptional activation potential in acute transfection assays (34). Stat3S produced in excess will, upon activation, form homo- or heterodimers with the majority of the wild-type protein, resulting in poor transcriptional activation.
Potentiation of v-src transformation by Stat3.
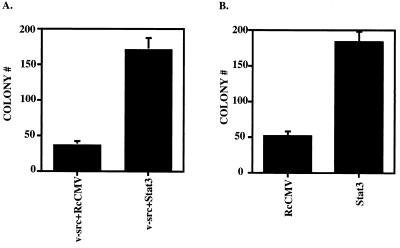
NIH 3T3 cells were transfected with v-src- and Stat3-containing plasmids and plated in soft agar containing puromycin and G418 to assay for transformed cells expressing proteins from both vectors. As expected, the cells expressing v-src and control vector (RcCMV) gave transformants, but when wild-type Stat3 supplemented v-src, three to five times as many colonies were observed (Fig. 2A and Table 1). A second assay, for the effect of additional Stat3 protein in cells already transformed by v-src, was carried out. The vectors carrying G418 resistance were transfected into cells already stably transformed by v-src, and the cells were plated in soft agar containing puromycin and G418. The v-src-transformed cells that had acquired G418 resistance formed colonies in this assay. Those that expressed additional wild-type Stat3 formed three to four times as many colonies as the v-src-transformed cells (Fig. 2B and Table 1).
FIG. 2.
Stat3 potentiates v-src transformation in soft agar colony-forming assays. (A) NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) were transfected with RcCMV plus v-src and Stat3 plus v-src. One microgram of RcCMV-based vectors was used per transfection; 500 ng of pBabe/v-src (24) was used per transfection. Twenty hours after transfection, the cells were fed with DMEM plus 10% BCS. Twenty-four hours later, the cells were trypsinized and plated (5 × 105 cells in 3ml) into soft agar with 10% BCS (20) containing 2 μg of puromycin per ml and 800 μg of G418 per ml. Seventeen days later, colonies were counted. Each bar represents the average of 10 independent transfections performed simultaneously; each error bar indicates the standard deviation. (B) v-src-transformed NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) were transfected (as above) with RcCMV and Stat3. One microgram of the RcCMV-based vectors was used per transfection. Twenty hours after transfection, the cells were fed with DMEM plus 10% BCS. Twenty-four hours later, the cells were trypsinized and plated (105 cells in 3 ml) into soft agar containing 2 μg of puromycin per ml and 800 μg of G418 per ml. Fourteen days later, colonies were counted. Each bar corresponds to the average of six independent transfections performed simultaneously; each error bar indicates the standard deviation.
Suppression of v-src transformation by Stat3 dominant negative mutants.
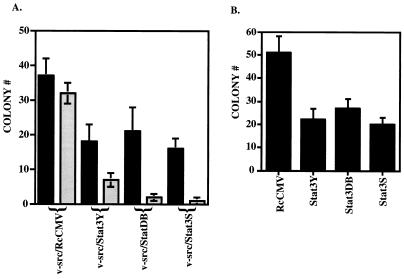
In contrast with the above findings, cotransfection of 500 ng of v-src with 1 μg of Stat3Y, Stat3DB, or Stat3S resulted in a depression of the transformation caused by v-src (Fig. 3A and Table 1). Similarly, expression of Stat3Y, Stat3DB, or Stat3S lowered the number of colonies in cells stably transformed by v-src, compared to RcCMV alone (Fig. 3B and Table 1). A more profound reduction in colony numbers relative to RcCMV/v-src was observed when the ratio of cotransfected Stat3 dominant negative mutants to v-src was increased to about 10:1 (1.8 to 0.2 μg) (Fig. 3A and Table 1). Transfection of NIH 3T3 cells with RcCMV, Stat3, Stat3Y, Stat3DB, or Stat3S alone yielded no transformants or colonies in soft agar (Table 1).
FIG. 3.
Stat3 dominant negative mutants abrogate v-src transformation. (A) The black columns indicate NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) transfected with a 1:2 ratio (500 ng to 1 μg) of v-src plus RcCMV, v-src plus Stat3Y, v-src plus Stat3DB, or v-src plus Stat3S and plated in soft agar, as described in the legend to Fig. 2A. Seventeen days later, colonies were counted. Each black bar represents the average of 10 independent transfections performed simultaneously; each error bar indicates the standard deviation. The grey columns indicate NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) transfected with a 1:10 ratio (0.2 to 1.8 μg) of v-src plus RcCMV, v-src plus Stat3Y, v-src plus Stat3DB, or v-src plus Stat3S and plated in soft agar, as described in the legend to Fig. 2A. Twenty-five days later, colonies were counted. Each grey bar represents the average of three independent transfections performed simultaneously; each error bar indicates the standard deviation. (B) v-src-transformed NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) were transfected with RcCMV, Stat3Y, Stat3DB, or Stat3S and plated in soft agar, as described in the legend to Fig. 2B. Fourteen days later, colonies were counted. Each bar corresponds to the average of six independent transfections performed simultaneously; each error bar indicates the standard deviation.
In vitro demonstration of the expression and activity of Stat3 constructs.
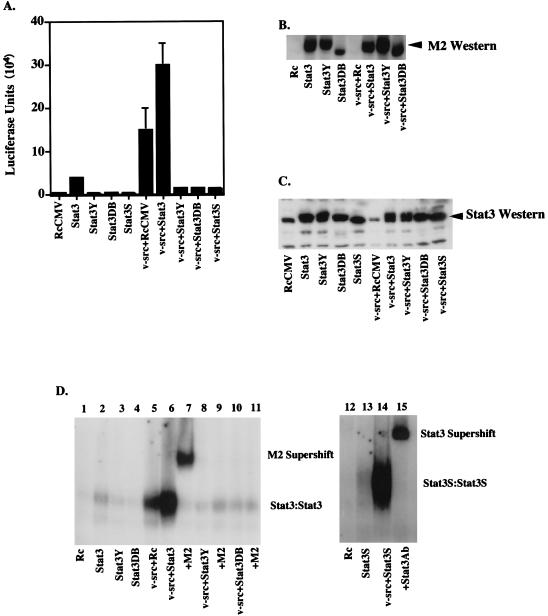
To demonstrate that activated Stat3 was present under conditions in which heightened or reduced cell transformation was observed, several tests were conducted with NIH 3T3 cells. A Stat3-driven luciferase promoter construct (34) was introduced by transfection along with various other expression constructs. First, it was clear that v-src alone was capable of driving increased transcription of the target gene in NIH 3T3 cells (Fig. 4A) in association with the activation of endogenous Stat3 (Fig. 4D, lane 5). The addition of extra Stat3 alone led to a small amount of activation, potentially due to a background activation of Stat3 in serum-grown cells (Fig. 4A). There was, however, a substantial (2.2-fold) transcriptional increase over v-src alone with the addition of extra Stat3 (Fig. 4A). In addition, the dominant negative Stat3 mutants greatly decreased (approximately fivefold) the transcriptional activation caused by v-src, indicating competition between the mutant proteins and the endogenous wild-type protein (Fig. 4A).
FIG. 4.
In vitro assays. (A) Luciferase assay. NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) were transiently transfected (as described in the legend to Fig. 3A) with RcCMV, Stat3, Stat3Y, Stat3DB, Stat3S, v-src plus RcCMV, v-src plus Stat3, v-src plus Stat3Y, v-src plus Stat3DB, or v-src plus Stat3S. All were transfected with 1 μg of Ly6E (three copies) Luc (34) and a β-galactosidase expression vector. One microgram of RcCMV-based vectors was used per transfection; 500 ng of pBabe/v-src was used in appropriate transfections. Twenty hours after transfection, the cells were fed with DMEM plus 10% BCS. Twenty-four hours later, the cells were lysed and luciferase assays were performed. Each bar represents the average of eight individual transfections with standard deviations, each performed in duplicate and normalized to β-galactosidase activity. (B) FLAG Western blotting. Thirty micrograms of protein per lane from whole-cell extracts from transiently transfected NIH 3T3 cells or v-src-transformed cells (as in Fig. 5A) were analyzed by Western blot analyses using chemiluminescence. The Stat3DB-FLAG construct is slightly smaller (see Fig. 1C) and therefore migrates faster. Stat3S expression was not determined due to its lack of a FLAG epitope. (C) Stat3 Western blotting. As in panel B, 30 μg of protein per lane was analyzed for the relative levels of Stat3 protein. Lanes 1 and 6 reveal endogenous levels of Stat3 protein, while the other lanes demonstrate that the transiently transfected cells are producing additional Stat3 protein. (D) Gel shift. NIH 3T3 cells were transiently transfected as described in the legend to Fig. 5A. Nuclear extracts from these cells were used in an EMSA using a 32P-labeled Stat3 binding site (M67) (31). Stat3-Stat3 homodimers form when Stat3 is phosphorylated on tyrosine 705. M2 (FLAG) monoclonal antiserum was used at a 1:100 dilution for supershifting of Stat3-FLAG complexes. A 1:100 dilution of Stat3-C antiserum was used for supershifting of Stat3S complexes.
To test directly for activated Stat3 under the conditions described above, we examined cell extracts from the transfected cells for Stat3 binding activity. This requires tyrosine phosphorylation and dimerization of the Stat3 protein (34). In these transfection experiments, all of the Stat3 constructs, with the exception of Stat3S, had a short protein marker, the FLAG epitope (7, 34), inserted at the end of the molecule so that newly added protein could be recognized by a commercial FLAG antiserum, M2 (Fig. 4B). Furthermore, the relative increase of introduced Stat3 or Stat3 dominant negative mutants over endogenous Stat3 is demonstrated by a Stat3-probed Western blot (Fig. 4C; compare lane 1 or 6 to lanes 2 to 5 or 7 to 10, respectively). The DNA-binding complexes from various transfected cells were examined in native gels with a deoxyoligonucleotide to which Stat3 binds. The most important comparison in this experiment is between extracts from cells transfected with v-src alone or v-src plus wild-type Stat3. v-src induces activation and DNA binding of endogenous Stat3 (Fig. 4D, lane 5). When extra Stat3 was added along with v-src, the cell extracts gave a substantial increase in DNA-binding complexes (Fig. 4D, lane 6), in agreement with the increased transcriptional activation (Fig. 4A). All of the DNA-binding complexes could be supershifted by the M2 antiserum, indicating that in the cells receiving the transfecting vectors there was enough FLAG-tagged Stat3 produced so that almost every Stat3 dimer contained one FLAG-tagged molecule (Fig. 4D, lane 7). Likewise, extracts from Stat3S- and v-src-transfected cells gave DNA-binding complexes which could be supershifted by Stat3-C antiserum (Fig. 4D, lanes 14 and 15).
Unphosphorylated Stat3 does not cooperate with the ras oncogene.
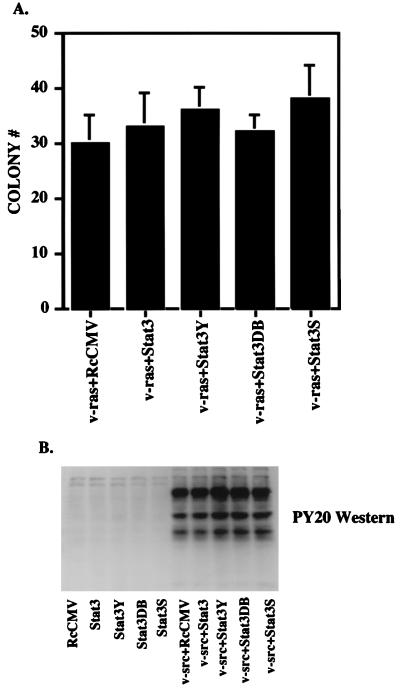
To test whether Stat3 could enhance the colony-forming capacity of cells transformed by any oncogene, we transfected NIH 3T3 cells with the series of Stat3 proteins together with v-Ha-ras (11) into NIH 3T3 cells. v-ras transformation in NIH 3T3 cells does not lead to constitutive phosphorylation of Stat3 (3). Neither wild-type Stat3 nor the dominant negative forms of the protein affected the number of v-ras-transformed colonies (Fig. 5A and Table 1). As described above for v-src, we increased the ratio of wild-type Stat3 or Stat3 dominant negative mutants to v-ras and performed colony assays, again with no effect on v-ras-induced colony formation (Table 1).
FIG. 5.
Stat3 and Stat3 dominant negative mutants do not affect v-ras transformation or overall tyrosine phosphorylation by v-src. (A) NIH 3T3 cells (5 × 105 cells in a 35-mm-diameter dish) were transfected (as in the legend to Fig. 2A) with 500 ng of v-ras (pRSV Ha-rasLeu61) (11) in conjunction with 1 μg of RcCMV, Stat3, Stat3Y, Stat3DB, or Stat3S. Twenty hours after transfection, the cells were fed with DMEM plus 10% BCS. Twenty-four hours later, the cells were trypsinized and plated (5 × 105 cells) into soft agar. Seventeen days later, colonies were counted. Each bar represents the average of four independent transfections. (B) NIH 3T3 cells were transfected (as in the legend to Fig. 2A) with 500 ng of v-src and 1 μg of RcCMV-based Stat3 constructs. Whole-cell extracts were isolated, and 100 μg per lane was analyzed by Western blotting with an antiserum to phosphotyrosine (PY20).
Introduced wild-type or dominant negative Stat3 proteins do not effect overall tyrosine phosphorylation by v-src.
To determine if the addition of supplementary wild-type Stat3 or dominant negative Stat3 altered the tyrosine kinase activity of v-src, we transfected NIH 3T3 cells with the series of STAT constructs without and with v-src at a 2:1 ratio. Phosphotyrosine (PY20) Western blot analysis was performed on whole-cell extracts from transfected cells (Fig. 5B), demonstrating no obvious differences in overall phosphotyrosine pattern or intensity with or without the addition of wild-type Stat3 or dominant negative Stat3 (Fig. 5B, lane 5 versus lanes 6 to 10). The same analysis was performed on extracts from cells transfected with a 10:1 ratio of Stat3 constructs to v-src. No differences in relative phosphotyrosine levels were observed (data not shown). Furthermore, no differences in v-src levels, as determined by Western blot analysis, were seen (data not shown).
Cell viability is not affected by the addition of Stat3 constructs.
We have demonstrated that the addition of Stat3 dominant negative protein in conjunction with v-src in NIH 3T3 cells leads to fewer colonies in soft agar. To differentiate between a decrease in transformation efficiency versus a decrease in cell viability, we plated an aliquot of transfected cells onto plates containing selective media. Thus, only cells expressing both pBabe (puromycin) constructs and RcCMV (neomycin) constructs would grow. The presence of Stat3 dominant negative mutants had no effect on cell viability (Table 2).
DISCUSSION
The experiments described herein demonstrate that activated Stat3 has oncogenic potential. Colony growth in soft agar of v-src-transformed cells is potentiated by Stat3 and inhibited by dominant negative Stat3. This enhancement of transformation is accompanied by Stat3 activation (assayed both as DNA binding and transcriptional activation) either directly or indirectly by v-src. Three different Stat3 dominant negative mutants were utilized, each with a different potential mechanism of action. Stat3Y cannot be phosphorylated and can presumably compete with wild-type Stat3 for the kinase. Stat3DB can heterodimerize with wild-type Stat3, leading to a defective DNA-binding complex (15, 16). Stat3S as a homodimer, and presumably as a heterodimer, cannot activate transcription efficiently (34) and when overproduced would greatly reduce any wild-type Stat3 dimers. This is the first demonstration that phosphorylation of Stat3 on serine 727 is required for biological activity. The decrease of v-src transformation by the three Stat3 dominant negative constructs correlated with inhibition of endogenous Stat3 DNA binding and transcriptional activation. These results allow the conclusion that v-src transformation is at least abetted by, if not dependent upon, Stat3.
The addition of Stat3 or Stat3 dominant negative proteins did not affect v-ras-mediated transformation, indicating that the introduced Stat3 molecules do not disrupt normal cellular functions on their own. Furthermore, no effect on cell viability was observed when wild-type or dominant negative Stat3 was cotransfected with v-src. Phosphotyrosine blot analysis of extracts derived from Stat3 constructs and v-src and cotransfected cells suggests that the presence of the various supplemental Stat3 molecules does not markedly alter the tyrosine kinase activity of v-src.
Among the interesting points these experiments raise is the possible effect on growth of simultaneous Stat1 and Stat3 activation, which occurs in response to many ligands (10, 26, 37). Stat1, as mentioned in the introduction, is required for the growth restraint imposed by IFN-α and IFN-γ (4, 8, 28). Since, as we demonstrate here, Stat3 has a growth-promoting capacity, it may be that Stat1 and Stat3 are often stimulated together to keep growth at a balanced level. The present findings and those with IFN activation of Stat1 establish conditions under which searches for specific Stat3- or Stat1-activated genes that contribute to growth regulation might be made. Finally, these findings and similar future experiments will be useful in clinical oncology because constitutively activated Stat3 has been reported in cells and tissues from a number of human tumors (14, 17, 25, 32, 36).
ACKNOWLEDGMENTS
We thank Lois Cousseau for preparing the manuscript.
J.F.B. was supported by a Howard Hughes Postdoctoral Fellowship for Physicians and NIH K08 grant CA67950. C.M.H. is a special fellow at the Leukemia Society of America. D. B. was supported by the Swiss National Science Foundation. This work was supported by NIH grants AI32489 and AI34420 to J.E.D.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1994. [Google Scholar]
- 2.Balkwill F, Taylor-Papadimitriou J. Interferon affects both G1 and S+G2 in cells stimulated from quiescence to growth. Nature. 1978;274:798–801. doi: 10.1038/274798a0. [DOI] [PubMed] [Google Scholar]
- 3.Bromberg, J., and J. E. Darnell, Jr. Unpublished observations.
- 4.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell G S, Yu C L, Jove R, Carter-Su C. Constitutive activation of JAK1 in Src-transformed cells. J Biol Chem. 1997;272:2591–2594. doi: 10.1074/jbc.272.5.2591. [DOI] [PubMed] [Google Scholar]
- 6.Cao X, Tay A, Guy G R, Tan Y H. Activation and association of Stat3 with Src in v-Src transformed cell lines. Mol Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang C M, Roeder R G. Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Pept Res. 1993;6:62–64. [PubMed] [Google Scholar]
- 8.Chin Y E, Kitagawa M, Su W C, You Z H, Iwamoto Y, Fu X Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 9.Cressman D E, Greenbaum L E, DeAngelis R A, Ciliberto G, Furth E E, Poli V, Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 10.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 11.de Vries-Smits A M, Burgering B M, Leevers S J, Marshall C J, Bos J L. Involvement of p21ras in activation of extracellular signal-regulated kinase 2. Nature. 1992;357:602–604. doi: 10.1038/357602a0. [DOI] [PubMed] [Google Scholar]
- 12.Fried M, Crothers D M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of Stat3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 14.Gouilleux-Gruart V, Gouilleux F, Desaint C, Claisse J F, Capiod J C, Delobel J, Weber-Nordt R, Dusanter-Fourt I, Dreyfus F, Groner B, Prin L. STAT-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood. 1996;87:1692–1697. [PubMed] [Google Scholar]
- 15.Horvath, C. M., J. Bromberg, and J. E. Darnell, Jr. Unpublished observations.
- 16.Horvath C M, Wen Z, Darnell J E., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 17.Karras J G, Wang Z H, Huo L, Howard R G, Frank D A, Rothstein T L. Signal transducer and activator of transcription-3 (Stat3) is constitutively activated in normal, self renewing B-1 cells but only inducibly expressed in conventional B lymphocytes. J Exp Med. 1997;185:1035–1042. doi: 10.1084/jem.185.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Land H, Parada L F, Weinberg R A. Tumorigenic conversion of primary fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 19.Lin S, Kikuchi T, Pledger W J, Tamm I. Interferon inhibits the establishment of competence in G0/S-phase transition. Science. 1986;233:356–359. doi: 10.1126/science.3726533. [DOI] [PubMed] [Google Scholar]
- 20.MacAuley A, Pawson T. Cooperative transforming activities of ras, myc, and src viral oncogenes in nonestablished rat adrenocortical cells. J Virol. 1988;62:4712–4721. doi: 10.1128/jvi.62.12.4712-4721.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Migone T S, Lin J X, Cereseto J C, Mulloy J C, O’Shea J J, Franchini G, Leonard W J. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-1. Science. 1995;269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 22.Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira S. Stat3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 24.Sabe H, Okada M, Nakagawa H, Hanafusa H. Activation of c-Src in cells bearing v-Crk and its suppression by Csk. Mol Cell Biol. 1992;12:4706–4713. doi: 10.1128/mcb.12.10.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sartor C I, Dziubinski M L, Yu C L, Jove R, Ethier S P. Role of epidermal growth factor receptor and STAT-3 activation in autonomous proliferation of SUM-102PT huma breast cancer cells. Cancer Res. 1997;57:978–987. [PubMed] [Google Scholar]
- 26.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 27.Shuai K, Liao J, Song M M. Enhancement of antiproliferative activity of gamma interferon by the specific inhibition of tyrosine dephosphorylation of Stat1. Mol Cell Biol. 1996;16:4932–4941. doi: 10.1128/mcb.16.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuai K, Schindler C, Prezioso V R, Darnell J E., Jr Activation of transcription by IFN-γ: tyrosine phosphorylation of a 91 kD DNA binding protein. Science. 1992;259:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 29.Suematsu S, Matsusaka T, Matsuda T, Ohno S, Miyazaki J, Yamamura K, Hirano T, Kishimoto T. Generation of plasmacytomas with the chromosomal translocation t(12;15) in interleukin 6 transgenic mice. Proc Natl Acad Sci USA. 1992;89:232–235. doi: 10.1073/pnas.89.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner B J, Hayes T E, Hoban C J, Cochran B H. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber-Nordt R M, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, Finke J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–816. [PubMed] [Google Scholar]
- 33.Wen, Z., and J. E. Darnell, Jr. Unpublished data.
- 34.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 35.Yu C L, Meyer D, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q, Nowak I, Vonderheid E C, Rook A H, Kadin M E, Nowell P C, Shaw L M, Wasik M A. Activation of Jak/STAT proteins involved in signal transduction pathway mediated by receptor for interleukin 2 in malignant T lymphocytes derived from cutaneous anaplastic large T-cell lymphoma and Sezary syndrome. Proc Natl Acad Sci USA. 1996;93:9148–9153. doi: 10.1073/pnas.93.17.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong Z, Wen Z, Darnell J E., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]