Abstract
Duck Tembusu virus (DTMUV) belongs to the Flaviviridae family and mainly infects ducks. Duck Tembusu virus genome encodes one polyprotein that undergoes cleavage to produce 10 proteins. Among these, NS4B, the largest transmembrane protein, plays a crucial role in the viral life cycle. In this study, we investigated the localization of NS4B and found that it is located in the endoplasmic reticulum, where it co-localizes with DTMUV dsRNA. Subsequently, we confirmed 5 different transmembrane domains of NS4B and discovered that only its transmembrane domain 3 (TMD3) can traverse ER membrane. Then mutations were introduced in the conserved amino acids of NS4B TMD3 of DTMUV replicon and infectious clone. The results showed that V111G, V117G, and I118G mutations enhanced viral RNA replication, while Q104A, T106A, A113L, M116A, H120A, Y121A, and A122G mutations reduced viral replication. Recombinant viruses with these mutations were rescued and studied in BHK21 cells. The findings demonstrated that A113L and H120A mutations led to higher viral titers than the wild-type strain, while Q104A, T106A, V111G, V117G, and Y121A mutations attenuated viral proliferation. Additionally, H120A, M116A, and A122G mutations enhanced viral proliferation. Furthermore, Q104A, T106A, V111G, M116A, V117G, Y121A, and A122G mutants showed reduced viral virulence to 10-d duck embryos. Animal experiments further indicated that all mutation viruses resulted in lower genome copy numbers in the spleen compared to the WT group 5 days postinfection. Our data provide insights into the topological model of DTMUV NS4B, highlighting the essential role of NS4B TMD3 in viral replication and proliferation.
Key words: TMUV, NS4B, transmembrane domain, mutation, proliferation
INTRODUCTION
Duck Tembusu virus (DTMUV) belongs to the flavivirus family, which includes important human pathogens such as yellow fever virus (YFV), dengue virus (DENV), and zika virus (ZIKV). Duck Tembusu virus gained public attention in 2010 when a large outbreak occurred in Jiangxi Province and rapidly spread to other regions (Su et al., 2011; Yan et al., 2011), additionally, DTMUV has caused significant economic losses in other Southeast Asian countries, such as Thailand and Malaysia (He et al., 2017; Lei et al., 2017). Similar to other flaviviruses, DTMUV genome is a single-strand positive-sense RNA with a 10990 bp length. It is capped at the 5′ end but lacks a 3′ poly(A) tail. Upon infection, the genome is released into the cytoplasm and translated into a unique polyprotein precursor. This polyprotein is subsequently cleaved into 3 structural proteins (core, membrane, and envelope) and 7 nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, 2KNS4B, and NS5). Among these proteins, NS2A, NS2B, NS4A, and NS4B are transmembrane proteins.
NS4B is the largest transmembrane protein, with a molecular weight of approximately 27 kDa, and it possesses 5 integral transmembrane segments (Miller et al., 2006). It has been reported that the C-terminal contains a signal sequence, known as 2K due to its size of 2 kDa, which facilitates the translocation of NS4B into the lumen of the endoplasmic reticulum (ER). Subsequently, the 2K fragment is cleaved by a host protease at the 2K/NS4B cleavage site in the ER lumen, with this proteolytic process requiring prior NS2B3 proteinase-mediated cleavage at the 4A/2K site (Lin et al., 1993). Previous studies have indicated that 2K can direct NS4B to the ER lumen from the C-terminal of NS4A; however, 2K is not necessary for the membrane integration of NS4B (Gopala Reddy, et al., 2018). The N and C-terminal of NS4B are located in the ER lumen and cytoplasm, respectively (Miller et al., 2006). The structural and conformational integrity of NS4B, as a transmembrane protein, plays a crucial role in genome replication. Moreover, flavivirus NS4B contains 2 N-linked glycosylation sites, both of which are essential for replication and virus production (Chatel-Chaix et al., 2015; Naik and Wu, 2015).
In previous reports, NS4B has been identified as a critical component of the viral replication complex, showing co-localization with NS3 and viral double-stranded RNA on the ER membrane. The interaction between NS4B and NS3 is crucial for the formation of the replication complex. In this process, the structural and conformational integrity of NS4B is vital for a successful interaction with NS3 helicase (Westaway et al., 1997; Roosendaal et al., 2006; Umareddy et al., 2006; Paul and Bartenschlager, 2013; Zou et al., 2014). Additionally, NS4B can interact with NS1 and NS4A, which are essential for virus replication (Youn et al., 2012; Zou et al., 2015). Moreover, when expressed in cells, Flavivirus NS4B forms oligomers (Zou et al., 2014) and the expression of ZIKV NS4B enriches host sphingolipids (Leier et al., 2020). Furthermore, Flavivirus NS4B has been implicated in affecting the host's immune response. For instance, DENV NS4B has been reported to act as an antagonist against host type I interferon by influencing the phosphorylation of STAF1 (Munoz-Jordan et al., 2003; Munoz-Jordan et al., 2005). Furthermore, NS4B inhibits RIG-I/MADA5/MAVS/TBK1/IKKε-mediated production of IFNβ in a dose-dependent manner (Chen et al., 2017).
Flavivirus NS4B is an essential protein for viral replication and also plays a crucial role in innate immune evasion. In recent research, some drugs have been developed to target NS4B and inhibit flaviviruses, these studies involve WNV, YFV, and DENV). (Zmurko et al., 2015; Kaptein et al., 2021), Therefore, understanding the structure and function of NS4B is essential for further research. In this study, we investigated the topological model of DTMUV NS4B and found evidence of 5 transmembrane domains, with only TMD3 being able to cross the ER membrane. Additionally, we conducted studies where we mutated the conserved amino acids in TMD3. The results indicated that TMD3 is crucial for viral replication and proliferation. Moreover, mutations in TMD3 were found to alter the distribution of the virus in DTMUV-infected ducklings.
MATERIAL AND METHODS
Ethics Statement
The animal studies were approved by the Institutional Animal Care and use Committee of Sichuan Agricultural University (No. SYXK(川)2019-187) and followed the National Institutes of Health guidelines for the performance of animal experiments.
Cells, Viruses and Antibodies
BHK-21 cells were grown supplemented with 10% FBS and maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Shanghai, China), 100 U/mL penicillin and 100 μg/mL streptomycin at 37°C in 5% CO2. Tembusu virus strain CQW1 (GenBank: KM233707.1) was isolated from Cherry Valley ducks in Southwest China by our laboratory (Zhu et al., 2015). Antibodies against EGFP, His were purchased from TransGen Biotech, rabbit anti-NS4B and mouse anti-DTMUV polyclonal antibodies were prepared by our laboratory. Duck embryos and ducklings were obtained from the poultry farm at Sichuan Agricultural University.
Plasmid Constructs
Standard molecular biology techniques were employed to construct all plasmids. The pCAGGS-2KNS4B-His was constructed, sequence of the 2KNS4B gene was amplified from the duck DTMUV CQW1 strain genome and then inserted into the pCAGGS expression vector, with a His tag added to the C terminus. To investigate the transmembrane domain of NS4B, an eGFP tag and an N-linked glycosylation sequence (-Asn-Ser-Thr-Ser-Ala, Glyc) were linked to the C-terminal of NS4B. Simultaneously, SPG-NS4AC16 was linked to the N-terminal of NS4B to ensure that the N-terminal could be located in the ER (truncated NS4B).
Indirect Immunofluorescence Assay
BHK21 cells with cover slips transfected with virus genome collected daily. After washed with PBST, cells were fixed with 1% paraformaldehyde for 1h, permeabilized with 0.22% Triton X-100 for 20 min in 4°C. and blocked with 5% BSA, then mouse anti-DTMUV polyclone was used as the primary antibody at a dilution of 1:200 with 1% BSA, cells incubated in 37°C for 2h, after washed with PBS, the cells were incubated with fluorescein isothiocyanate (FITC)-labelled goat anti-mouse IgG at a dilution of 1:200 for 1 h and then incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. Finally, the cells were analyzed by a fluorescence microscope (Nikon, Tokyo, Japan).
Real-Time Quantitative PCR
Total RNA was isolated from selected tissues using RNAiso Plus reagent. The quantity of the RNA in each sample was determined using a NanoDrop 2000 (Thermo, Waltham, MA), and RT-PCR was performed on each sample using HiScript II QRT SuperMix (Vazyme, Nanjing, China). Finally, the cDNAs were stored at -80°C until use. qPCR was used to detect the genome copy number of the virus and performed using the Bio-Rad CFX96 Real Time Detection System (Bio-Rad, CA). The qRT-PCR reaction mixture containing 5μL of 2 × SYBR Green PCR master mixtures, 0.4μL each of primers, 0.4 μL of cDNA and 3.8μL of RNAase free ddH2O was incubated for 3 min at 95°C, followed by 40 cycles of 95°C for 10s and 59°C for 30s. After the amplification phase, a melting curve program (65°C to 95°C with a heating rate of 0.5°C per second and a continuous fluorescence measurement) was routinely performed to confirm the presence of single PCR product.
Western Blot Analysis
All the cell samples were analyzed by western blot. The samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane. Followed by incubation in blocking buffer (comprising 5% skim milk), after that the membrane was wash 3 times with TBST and incubated in blocking buffer containing a mouse monoclonal antibody against His tag (1:4,000). Then, the membrane was washed 3 times with TBST again and incubated with a goat anti-mouse antibody conjugated to horseradish peroxidase (1:4,000). Finally, the membrane was wash 3 times with TBST buffer and the antibody-protein complexes were detected using the Clarity Western ECL Substrate (Bio-Rad).
PNGase F assay
Based on the predictions, we constructed 8 truncated versions of DTMUV NS4B to validate the transmembrane domains (Figure 2B). all the truncated NS4B variants were harvested and subjected to PNGase F treatment, PNGase F is an amidase enzyme that cleaves N-linked oligosaccharides from proteins by breaking the asparagine (N)-linked glycosidic bond. This enzymatic reaction occurs within the endoplasmic reticulum. If a protein undergoes truncation and, after treatment with PNGase F, exhibits a decrease in molecular weight, it suggests that its C-terminal is located within the lumen of the endoplasmic reticulum and has undergone N-glycosylation. then all the plasmud was transfected to BHK-21cells, and pCA-eGFP-Glyc was used as contral. After 24 h of transfection, wash the cells 3 times with pre cooled PBS and add 500 to each cell well μ L's solubilizing buffer was placed at 4 °C for 2 h of reaction. Subsequently, centrifuge at 12,000 r/min and 4 °C for 30 min to collect the supernatant. Divide the supernatant solution obtained from each well into 2 groups and add 50,000 U/mL PNGase F or an equal amount of PBS as a control. After reacting at 37 °C for 2 h, the sample were analyst by WB.
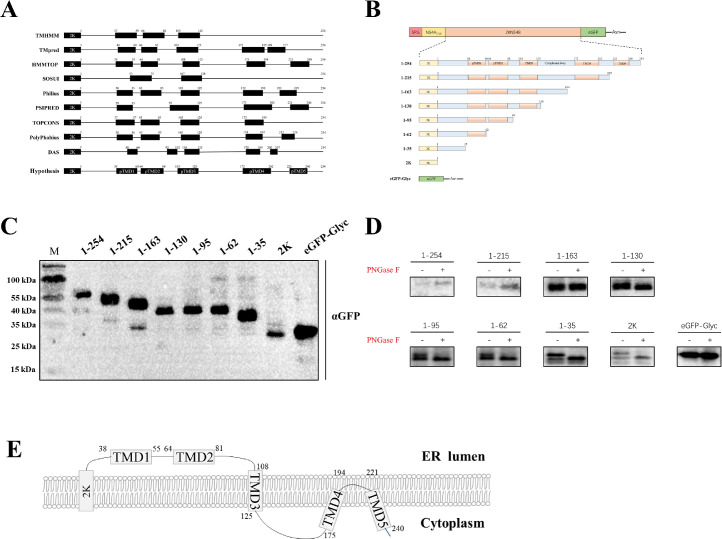
Figure 2.
DTMUV transmembrane domain in endoplasmic reticulum. (A) Predicted topological model of DTMUV NS4B using different software. (B) Different constructs of truncated DTMUV NS4B used in the study to investigate its transmembrane domains. (C) Expression of truncated DTMUV NS4B in BHK21 cells post-transfection. (D) Proteinase K protection analysis of NS4B truncations. (E) DTMUV NS4B topological model.
Transfection
Briefly, BHK-21 cells were seeded in 6-well plates containing 1 mL of DMEM and incubated overnight. Then, the BHK-21 cells were transfected with 4 μg plasmids for wild-type (pAR-DTvIc-WT and pAC-DTvRlucRep-WT) or mutant infectious clones (pAR-DTvIc-NS4B-Q104A/T106A/V111G/A113L/M116A/V117G/I118G/H120A/Y121A/A122G) and replicons (pAC-DTvRlucRep- Q104A/T106A/V111G/A113L/M116A/V117G/I118G/H120A/Y121A/A122G). At the given time points, the cells were harvested for further study. The luciferase activity of the replicon was determined according to the manufacturer's protocol.
Plaque Assay
BHK21 cells were cultured with DMEM in a 6-well plate for 16 h, and then the medium was removed and washed with PBS 3 times. Viral samples were diluted and added to cells with the 10 TCID50, 100 TCID50 and 1000 TCID50 respectively, and then the samples were incubated for 1.5 h and swirled every 15 min. After incubation, viral samples were removed, and the cells were washed with PBS 3 times. Finally, 2 ml of 0.75% methyl cellulose overlay containing 2% FBS and 1% penicillin/streptomycin was added to each well. Four days postinfection, the medium was removed, and the cells were washed 3 times with PBS and fixed with 4% paraformaldehyde for 20 min. After the fixative was removed and the cells were washed with PBS, the plate was stained with 1% crystal violet and washed carefully, and then the plaque was observed in the plate.
Animal Experiment
In the experiment, ducklings at 3-day-old were segregated into 3 separate groups. Each group received an injection of 1 mL varying virus solutions. The ducklings' body weight was monitored on a daily basis. Subsequent to infection, on d 3, 5, and 7, we selected and euthanized 3 ducks from each group. We collected tissue samples from the spleen and blood to investigate the presence of DTMUV. These samples were homogenized using a phosphate-buffered solution (PBS), then clarified through centrifugation at 12,000 g and 4°C over a 15-min period. We extracted the total RNA and preserved it at -80°C for later analysis.
Data Statistics
Statistical analyses were performed with GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA). The differences between values were evaluated by Student's t test. P < 0.05 indicated statistical significance, and all values are expressed as the mean ± SEM.
RESULTS
NS4B Co-Located With Viral Double dsRNA in Endoplasmic Reticulum
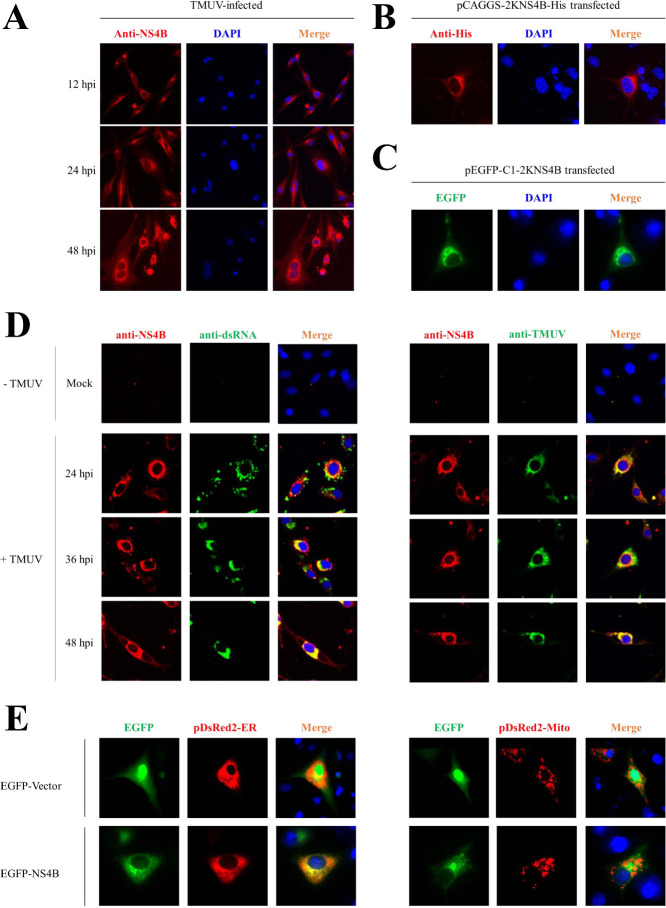
To detect the localization of NS4B in viral infection, we used mouse anti-NS4B polyclonal antibodies, as shown in Figure 1A, where NS4B is located in the cytoplasm. At the same time, we used eukaryotic expression to detect the localization of NS4B, and concurrently employed the anti-NS4B polyclonal antibody, along with the EGFP-NS4B fusion expression, to observe the location of NS4B. We found that NS4B is dispersed in a dot-like cluster distribution within the cytoplasm. (Figures 1B and 1C). Subsequently, since NS4B is involved in the construction of the replication complex, we used mouse anti-dsRNA and mouse anti-TMUV antibodies to detect the localization of NS4B with virus dsRNA and structural proteins during infection. In DTMUV-infected cells, dsRNA was found in the cytoplasm at 24 h postinfection, and it continued to distribute in the cytoplasm. However, 36 and 48 h postinfection, dsRNA aggregates were colocalized in the cytoplasm, and dsRNA also co-located with NS4B in the cytoplasm (Figure 1D). Furthermore, we used a mouse anti-DTMUV polyclonal antibody to detect the location of DTMUV. The results indicated that NS4B co-localized with DTMUV. As previously reported, flavivirus NS4B is located in the endoplasmic reticulum (ER). ER and Mito located plasmid pDsRed2-ER and pDsRed2-Mito were used. As shown in Figure 1E, NS4B was found to be located in the ER but not in the mitochondria.
Figure 1.
NS4B co-located with viral double dsRNA in endoplasmic reticulum. (A) Localization of NS4B in cells at 12, 24, and 48 h postinfection. (B) and (C) Localization of NS4B in BHK21 cells 24 h post-transfection. (D) Localization of DTMUV, DTMUV dsRNA and NS4B in DTMUV infected cells. (E) Localization of DTMUV NS4B with mitochondria and endoplasmic reticulum in BHK21 cells.
DTMUV NS4B Possesses 5 Transmembrane Domains
In the previous study, it was established that Flavivirus NS4B is a multiple transmembrane protein (Miller et al., 2006). In our present study, we employed diverse software and web tools (HMMTOP, SOSUI, Philius, PSIPRED, TOPCONS, PolyPhobius, and DAS) to predict the transmembrane domains of DTMUV NS4B. Based on these predictions, a hypothesis regarding the transmembrane domain was formulated (Figure 2A). Based on these predictions, we constructed 8 truncated of DTMUV NS4B to validate the transmembrane domains (Figure 2B). The expression of the truncated NS4B in cells was confirmed (Figure 2C), and all the truncated NS4B variants were harvested and subjected to PNGase F assay. The resulting products were then analyzed using WB (Figure 2D). Notably, NS4B 1-95aa, 1-62aa, 1-35aa, and 2K were found to react with PNGase F, indicating that their C-terminal regions are located in the ER lumen. On the other hand, NS4B 1-135aa, 1-163aa, 1-215aa, and 1-254aa did not react with PNGase F, suggesting that their C-terminal regions are not located in the ER lumen. Based on these results, we drew the topological model of DTMUV NS4B protein Figure 2E.
The Conserved Amino Acids in NS4B TMD3 are Essential for TMUV RNA Replication
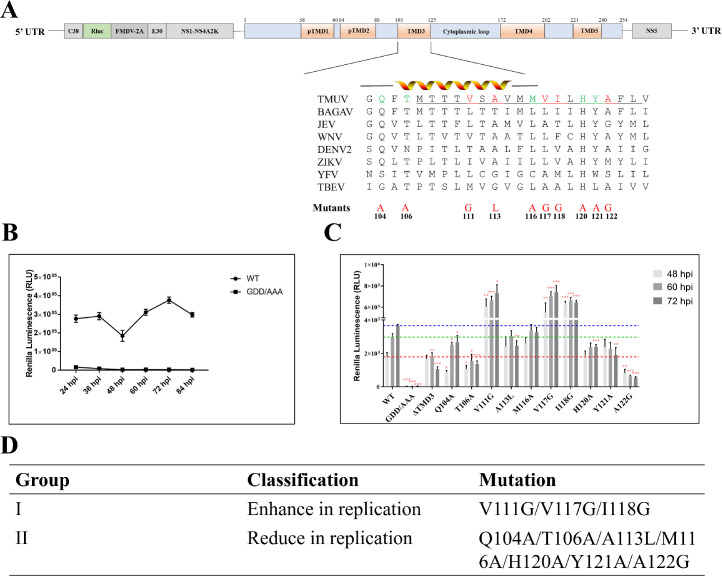
To investigate the role of NS4B TMD3 conserved amino acids in the viral life cycle, we mutated 10 conserved sites. To validate the mutational effect on viral RNA replication, we used the nano luciferase reporter replicon of duck DTMUV (Figure 3A). The luciferase activity kinetics were measured to assess the replication of the replicon.
Figure 3.
The key amino acids in NS4B TMD3 are essential for viral replicon. (A) conserved amino acids in NS4B TMD3. (B) Replication of DTMUV in BHK21 cells, the replicon replication 48, 60 and 72h post-transfection. (C) and (D) Different mutation in NS4B TMD3 effected the replication of replicon in BHK21 cells.
The replication curve of the WT replicon was measured, and 3 time points were selected for the experiment. Equal amounts of WT and mutated replicon plasmids were transfected into BHK21 cells, and luciferase activity was assayed at 48, 60, and 72 hours post-transfection. As shown in Figures 3B and 3C, based on the influence of the luciferase signals of DTMUV replicons, the NS4B mutations can be classified into 2 groups: Group I mutants (V111G/V117G/I118G) highly enhance viral replication, while Group II mutants (Q104A/T106A/A113L/M116A/H120A/Y121A/A122G) obviously reduce viral replication (Figure 3D). In summary, our primary data indicate that the conserved amino acids in NS4B TMD3 play important roles in viral RNA replication, although the specific mechanisms remain unknown.
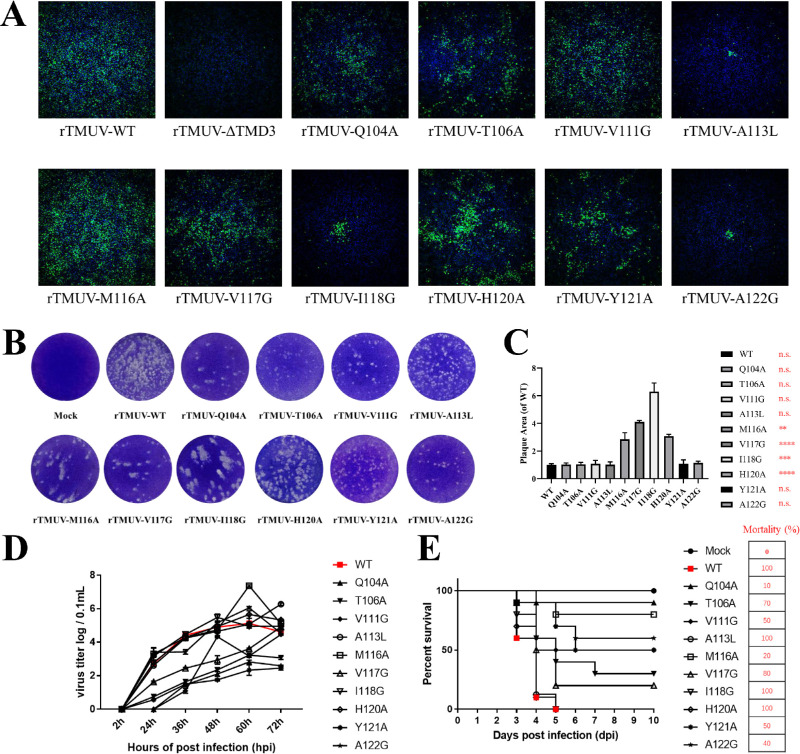
Characterization and Phenotypes of Duck DTMUV Infectious cDNA Clones With NS4B TMD3 Conserved Amino Acids Mutation
To confirm the findings obtained from the duck DTMUV replicon, we introduced the NS4B TMD3 mutants into a full-length duck DTMUV infectious cDNA clone (pACNR-rDTMUV-WT). Equal amounts of WT and NS4B mutant genome length RNAs were transiently transfected into BHK-21 cells. After 4 days post-transfection, the transfected cells were examined using an immunofluorescence assay (IFA) with an anti-duck DTMUV ployclonal antibody (Figure 4A). We observed that the ΔTMUD3 mutants were not viable and could not be rescued. Additionally, the other mutants resulted in fewer IFA-positive cells compared to the WT. As expected, the plaque morphology also exhibited similar results as the IFA (Figure 4B), plaque assay showed that the M116A, V117G, I118G and Y120A mutation virus produced significantly larger plaques than the WT (Figure 4C). The growth kinetics of the WT and NS4B TMD3 mutant recombinant viruses in BHK-21 cells were compared. The virus titers were detected at specific time points, and the results showed that the A113L and H120A can’t effect the viral proliferation. On the other hand, the Q104A, T106A, V111G, V117G, and Y121A mutations attenuated DTMUV viral proliferation. Furthermore, the H120A, M116A, and A122G mutations enhanced viral proliferation in cells (Figure 4D). The virulence of the WT and NS4B mutants was evaluated in duck embryos (Figure 4E). Each group of ten 9-day-old duck embryos was injected with 200 μL of 102 TCID50 of P1 WT and NS4B TMD3 mutant viruses. The duck embryos inoculated with the A113L, I118G, and H120A mutant viruses exhibited similar mortality rates as the WT. In contrast, the Q104A, T106A, V111G, M116A, V117G, Y121A, and A122G mutants showed attenuation in duck embryos, especially the Q104A mutants, which exhibited a lower mortality rate (10%) than WT. Combining these results with the replicon data, it can be inferred that the Q104A, T106A, V111G, M116A, V117G, Y121A, and A122G mutants have reduced viral virulence in duck embryos.
Figure 4.
The proliferation and virulence of recombination TMUV in BHK21 cells and duck embryo. (A) Identification of the effect of NS4B-TMD3 mutation on recombinant virus phenotypes by IFA. (B) Plaque morphologies of NS4B-TMD3 mutation recombinant virus in BHK21 cells. (C) Measurement of plaque size in BHK-21 cells infected with mutataed viruses. (D) Growth curves of NS4B-TMD3 mutation recombinant on BHK21 cells. (E) Virulence of NS4B-TMD3 mutation recombinant viruses in duck embryos.
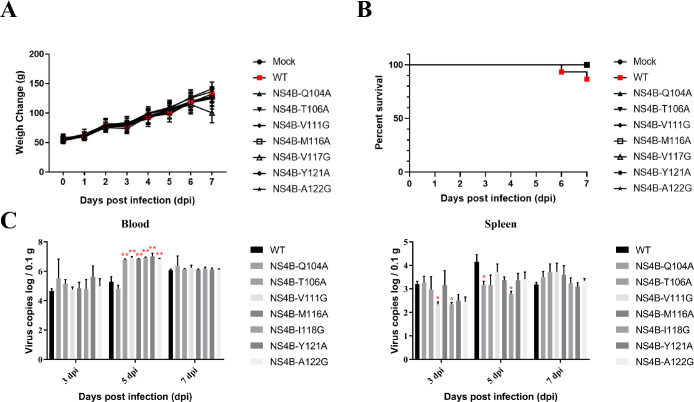
Mutation on NS4B Affect Viral Virulence and Distribution in Duckling
To compare the virulence of the WT and NS4B mutant recombinant viruses in ducklings, 3-day-old ducklings were infected with WT and NS4B mutant viruses. The body weight and mortality were recorded each day postinfection. Seven days postinfection, the ducklings infected with the V117G mutant virus showed a decrease in body weight, while the other mutation groups had body weights similar to the WT group, but lower than the mock-infected group (Figure 5A). Additionally, ducklings infected with the WT virus exhibited a 13.3% mortality rate, while no ducklings died after being infected with the mutated virus (Figure 5B). Five days post-infection, the Q104A mutant showed an equal genome copy number in blood compared to the WT (Figure 5C). Regarding the spleen, 3 days postinfection, the viral load of the Q104A, T106A, and M116A mutation groups was equal to the WT group, while the V111G, I118G, Y121A, and A122G groups showed lower viral loads than the WT group. Five days postinfection, all mutation viruses resulted in lower genome copy numbers in the spleen compared to the WT group. Seven days postinfection, the Q104A, T106A, V111G, and M116A mutations enhanced the genome copy in the spleen, while the I118G, Y121A, and A122G mutations decreased the genome copy in the spleen (Figure 5D). These results indicate that mutations in NS4B-TMD3 can affect the distribution of the virus in different tissues.
Figure 5.
Animal experimental infection of NS4B-TMD3 mutation recombinant viruses. (A) Changes in body weight gain of rTMUV infected ducks. (B) Mortality rate of rTMUV infected ducks. (C) and (D) Detection of viral genome copies in blood and spleen of the infected ducklings.
DISCUSSION
As a nonstructural protein, NS4B is not a component of the viral particle; however, it plays a crucial role in other processes in the virus life cycle. In our study, we detected the localization of NS4B using immunofluorescence assay (IFA) and found that NS4B is located in the endoplasmic reticulum (ER) and co-localizes with viral double-stranded RNA (dsRNA), which aligns with its known function. During flavivirus replication in cells, NS4B is translated and anchored to the ER, where it participates in the formation of the replication complex and facilitates virus RNA replication. RNA replication is a pivotal step in the virus life cycle, occurring within the replication complex. The flavivirus replication complex comprises several nonstructural proteins, including NS4B. Despite not possessing an enzymatic activity domain, NS4B's function in the replication complex involves binding with other non-structural proteins and anchoring to the ER, which is crucial for the stability of the replication complex within the cell.
To further understand NS4B's role in anchoring to the ER, we investigated its transmembrane domains. Through experimentation and the use of different software, we confirmed the presence of 5 distinct transmembrane domains that are associated with NS4B's localization in the endoplasmic reticulum. Our deglycosylation experiment revealed that TMD1 and TMD2 of DTMUV NS4B are located in the lumen of the ER, TMD3 can cross the ER membrane, while TMD4 and TMD5 are embedded in the ER membrane. Interestingly, the structures of TMD4 and TMD5 in DTMUV NS4B differed from those observed in DENV-2 NS4B, possibly due to the difference in experimental conditions, where NS4B was translated separately without being connected to NS5 protein. We speculate that when NS4B is expressed alongside NS5 and cleaved by NS2B3, TMD4 and TMD5 may display different characteristics (Miller et al., 2006; Zou et al., 2014).
To obtain the correct structure of NS4B, we constructed 2K with NS4B, which helps direct NS4B to the ER lumen from the C-terminal of NS4A and assists in proper folding of NS4B. The correct structure of flavivirus membrane proteins is crucial for virus replication, as unfolded proteins are subject to degradation by the unfolded protein response (Hwang and Qi, 2018; Lin et al., 2019; Ngo, et al., 2019). Studies have reported that mutation of YFV NS2B transmembrane domain is significant for virus replication and assembly (Li, et al., 2016). Through our replicon experiments, we identified Q104, T106, A113, M116, H120, Y121, and A122 as essential for viral replication. Subsequently, we utilized infection clones to rescue various recombinant viruses and observed that some mutations in TMD3 (Q104A, T106A, V111G, M116A, V117G, Y121A, A122G) reduced viral replication in cells, leading to attenuated virulence. As TMD3 can cross the ER membrane, it may play a critical role in forming the correct structure of NS4B, which serves as the basis for the replication complex. Mutation in TMD3 could potentially disrupt the structure of NS4B, leading to an incorrect replication complex. Proper structure of transmembrane proteins is vital for viral replication, and unfolded transmembrane proteins can be degraded by proteases (Lin et al., 2019; Ngo et al., 2019; Welsch et al., 2009). Moreover, mutations in TMD3 reduced the genome copy of the virus in the spleen, indicating that replication of the virus in the spleen was inhibited by TMD3 mutation. The spleen is integral to the innate immune system, and NS4NB can participate in immune evasion (Chen et al., 2017), Thus, TMD3 mutations may impair the functions of NS4B in immune evasion, leading to inhibited viral replication and proliferation in the spleen.
In summary, our data indicate that NS4B possesses 5 different transmembrane domains, with only TMD3 capable of crossing the ER membrane. Mutations in TMD3 can influence viral replication, Q104A, T106A, V111G, V117G, and Y121A mutations attenuated viral proliferation, H120A, M116A, and A122G mutations enhanced viral proliferation. Additionally, mutations in TMD3 affect the distribution of the virus in different tissues. These findings shed light on the importance of NS4B's structure and function in viral replication and provide valuable insights for potential drug development targeting NS4B to inhibit flavivirus replication.
ACKNOWLEDGMENTS
This work was funded by the National Key Research and Development Program of China (2022YFD1801900), the National Natural Science Foundation of China (32272976), the Sichuan Provincial Department of Science and Technology International scientific and technological innovation cooperation (2022YFH0026), the China Agriculture Research System of Ministry of Finance (MOF) and Ministry of Agriculture and Rural Affairs (MARA), and grant SCCXTD-2021-18 from the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System.
Ethical Statement: All animal experimental procedures were approved by the Institutional Animal Care and Use Committee of Sichuan Agriculture University in Sichuan, China (Protocol Permit Number: SYXK(川)2019-187).
Author Contributions: Conceptualization: Bowen Jiang, Wei Zhang, Jiaqi Guo, Tao Hu, Mingshu Wang, Renyong Jia, Dekang Zhu, Mafeng Liu, Xinxin Zhao, Qiao Yang, Ying Wu, Shaqiu Zhang, Juan Huang, Sai Mao, Xumin Ou, Qun Gao, Di Sun, Yunya Liu, Ling Zhang, Yanling Yu Xin pan, Jingyi Zhong and Anchun Cheng; Data curation, Bowen Jiang and Shun Chen; Formal analysis, Wei Zhang; Funding acquisition, Shun Chen and Anchun Cheng; Project administration, Shun Chen and Anchun Cheng; Resources, Shun Chen; Supervision, Shun Chen; Writing – original draft, Bowen Jiang; Writing – review & editing, Bowen Jiang, Wei Zhang, Jiaqi Guo, Tao Hu, Mingshu Wang, Renyong Jia, Dekang Zhu, Mafeng Liu, Xinxin Zhao, Qiao Yang, Ying Wu, Shaqiu Zhang, Juan Huang, Sai Mao, Xumin Ou, Qun Gao, Di Sun, Yunya Liu, Ling Zhang, Yanling Yu Xin pan, Jingyi Zhong and Anchun Cheng
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- Chatel-Chaix L., Fischl W., Scaturro P., Cortese M., Kallis S., Bartenschlager M., Fischer B., Bartenschlager R. A combined genetic-proteomic approach identifies residues within dengue virus NS4B critical for interaction with NS3 and viral replication. J. Virol. 2015;89:7170–7186. doi: 10.1128/JVI.00867-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Wu Z., Wang M., Cheng A. Innate immune evasion mediated by flaviviridae non-structural proteins. Viruses. 2017;9:291. doi: 10.3390/v9100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopala Reddy S.B., Chin W.X., Shivananju N.S. Dengue virus NS2 and NS4: minor proteins, mammoth roles. Biochem. Pharmacol. 2018;154:54–63. doi: 10.1016/j.bcp.2018.04.008. [DOI] [PubMed] [Google Scholar]
- He Y., Wang A., Chen S., Wu Z., Zhang J., Wang M., Jia R., Zhu D., Liu M., Yang Q., Wu Y., Sun K., Chen X., Cheng A. Differential immune-related gene expression in the spleens of duck Tembusu virus-infected goslings. Vet. Microbiol. 2017;212:39–47. doi: 10.1016/j.vetmic.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Hwang J., Qi L. Quality control in the endoplasmic reticulum: crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 2018;43:593–605. doi: 10.1016/j.tibs.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptein S.J.F., Goethals O., Kiemel D., Marchand A., Kesteleyn B., Bonfanti J.F., Bardiot D., Stoops B., Jonckers T.H.M., Dallmeier K., Geluykens P., Thys K., Crabbe M., Chatel-Chaix L., Munster M., Querat G., Touret F., de Lamballerie X., Raboisson P., Simmen K., Chaltin P., Bartenschlager R., Van Loock M., Neyts J. A pan-serotype dengue virus inhibitor targeting the NS3-NS4B interaction. Nature. 2021;598:504–509. doi: 10.1038/s41586-021-03990-6. [DOI] [PubMed] [Google Scholar]
- Lei W., Guo X., Fu S., Feng Y., Tao X., Gao X., Song J., Yang Z., Zhou H., Liang G. The genetic characteristics and evolution of Tembusu virus. Vet. Microbiol. 2017;201:32–41. doi: 10.1016/j.vetmic.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Leier H.C., Weinstein J.B., Kyle J.E., Lee J.Y., Bramer L.M., Stratton K.G., Kempthorne D., Navratil A.R., Tafesse E.G., Hornemann T., Messer W.B., Dennis E.A., Metz T.O., Barklis E., Tafesse F.G. A global lipid map defines a network essential for Zika virus replication. Nat. Commun. 2020;11:3652. doi: 10.1038/s41467-020-17433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.D., Deng C.L., Ye H.Q., Zhang H.L., Zhang Q.Y., Chen D.D., Zhang P.T., Shi P.Y., Yuan Z.M., Zhang B. Transmembrane domains of NS2B contribute to both viral RNA replication and particle formation in Japanese encephalitis virus. J. Virol. 2016;90:5735–5749. doi: 10.1128/JVI.00340-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Amberg S.M., Chambers T.J., Rice C.M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 1993;67:2327–2335. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.L., Inoue T., Chen Y.J., Chang A., Tsai B., Tai A.W. The ER membrane protein complex promotes biogenesis of dengue and zika virus non-structural multi-pass transmembrane proteins to support infection. Cell Rep. 2019;27:1666–1674. doi: 10.1016/j.celrep.2019.04.051. e1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S., Sparacio S., Bartenschlager R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J. Biol. Chem. 2006;281:8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- Munoz-Jordan J.L., Laurent-Rolle M., Ashour J., Martinez-Sobrido L., Ashok M., Lipkin W.I., Garcia-Sastre A. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 2005;79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Jordan J.L., Sanchez-Burgos G.G., Laurent-Rolle M., Garcia-Sastre A. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. U S A. 2003;100:14333–14338. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik N.G., Wu H.N. Mutation of putative n-glycosylation sites on dengue virus NS4B decreases RNA replication. J Virol. 2015;89:6746–6760. doi: 10.1128/JVI.00423-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo A.M., Shurtleff M.J., Popova K.D., Kulsuptrakul J., Weissman J.S., Puschnik A.S. The ER membrane protein complex is required to ensure correct topology and stable expression of flavivirus polyproteins. Elife. 2019;8:e48469. doi: 10.7554/eLife.48469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2013;2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosendaal J., Westaway E.G., Khromykh A., Mackenzie J.M. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 2006;80:4623–4632. doi: 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Li S., Hu X., Yu X., Wang Y., Liu P., Lu X., Zhang G., Hu X., Liu D., Li X., Su W., Lu H., Mok N.S., Wang P., Wang M., Tian K., Gao G.F. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related flavivirus. PLoS One. 2011;6:e18106. doi: 10.1371/journal.pone.0018106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umareddy I., Chao A., Sampath A., Gu F., Vasudevan S.G. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J. Gen. Virol. 2006;87:2605–2614. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway E.G., Mackenzie J.M., Kenney M.T., Jones M.K., Khromykh A.A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Yan P., Zhou J., Teng Q., Li Z. Establishing a TaqMan-based real-time PCR assay for the rapid detection and quantification of the newly emerged duck Tembusu virus. Virol. J. 2011;8:464. doi: 10.1186/1743-422X-8-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn S., Li T., McCune B.T., Edeling M.A., Fremont D.H., Cristea I.M., Diamond M.S. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J. Virol. 2012;86:7360–7371. doi: 10.1128/JVI.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K., Huang J., Jia R., Zhang B., Wang M., Zhu D., Chen S., Liu M., Yin Z., Cheng A. Identification and molecular characterization of a novel duck Tembusu virus isolate from Southwest China. Arch. Virol. 2015;160:2781–2790. doi: 10.1007/s00705-015-2513-0. [DOI] [PubMed] [Google Scholar]
- Zmurko J., Neyts J., Dallmeier K. Flaviviral NS4b, chameleon and jack-in-the-box roles in viral replication and pathogenesis, and a molecular target for antiviral intervention. Rev. Med. Virol. 2015;25:205–223. doi: 10.1002/rmv.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Xie X., Lee le T., Chandrasekaran R., Reynaud A., Yap L., Wang Q.Y., Dong H., Kang C., Yuan Z., Lescar J., Shi P.Y. Dimerization of flavivirus NS4B protein. J. Virol. 2014;88:3379–3391. doi: 10.1128/JVI.02782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Xie X., Wang Q.Y., Dong H., Lee M.Y., Kang C., Yuan Z., Shi P.Y. Characterization of dengue virus NS4A and NS4B protein interaction. J. Virol. 2015;89:3455–3470. doi: 10.1128/JVI.03453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]