Abstract
The general transcription factor IIA (TFIIA) interacts with the TATA binding protein (TBP) and promoter DNA to mediate transcription activation in vitro. To determine if this interaction is generally required for activation of all class II genes in vivo, we have constructed substitution mutations in yeast TFIIA which compromise its ability to bind TBP. Substitution mutations in the small subunit of TFIIA (Toa2) at residue Y69 or W76 significantly impaired the ability of TFIIA to stimulate TBP-promoter binding in vitro. Gene replacement of wild-type TOA2 with a W76E or Y69A/W76A mutant was lethal in Saccharomyces cerevisiae, while the Y69F/W76F mutant exhibited extremely slow growth at 30°C. Both the Y69A and W76A mutants were conditionally lethal at higher temperatures. Light microscopy indicated that viable toa2 mutant strains accumulate as equal-size dumbbells and multibudded clumps. Transcription of the cell cycle-regulatory genes CLB1, CLB2, CLN1, and CTS1 was significantly reduced in the toa2 mutant strains, while the noncycling genes PMA1 and ENO2 were only modestly affected, suggesting that these toa2 mutant alleles disrupt cell cycle progression. The differential effect of these toa2 mutants on gene transcription was examined for a number of other genes. toa2 mutant strains supported high levels of CUP1, PHO5, TRP3, and GAL1 gene activation, but the constitutive expression of DED1 was significantly reduced. Activator-induced start site expression for HIS3, GAL80, URA1, and URA3 promoters was defective in toa2 mutant strains, suggesting that the TFIIA-TBP complex is important for promoters which require an activator-dependent start site selection from constitutive to regulated expression. We present evidence to indicate that transcription defects in toa2 mutants can be both activator and promoter dependent. These results suggest that the association of TFIIA with TBP regulates activator-induced start site selection and cell cycle progression in S. cerevisiae.
The RNA polymerase II general transcription factors are an evolutionarily conserved set of proteins required for the regulation and recognition of specific promoter start sites (reviewed in references 56 and 60). In higher eukaryotes, the general transcription factor IID (TFIID) binds to core promoter elements and can nucleate the assembly of an active preinitiation complex in vitro (reviewed in reference 10). TFIID consists of the TATA binding protein (TBP) and TBP-associated factors (TAFIIs), which modify the promoter recognition and transcriptional activities of TBP (reviewed in reference 75). In addition to the TAFIIs, multiple other factors can associate with TBP and regulate transcription initiation by modulating the binding of TBP to the core promoter (5, 19, 27, 35, 48, 52). The general transcription factor IIA (TFIIA) is a positive modulator of TBP binding to TATA box elements and is essential for regulated transcription in vitro. However, the precise function and general requirement for a TFIIA-TBP association in vivo have not been completely elucidated.
TFIIA stimulates and stabilizes the interaction of TBP with a variety of TATA elements and may make direct contact with promoter DNA upstream of the TATA box (26, 40, 55). TFIIA is required for activator-mediated transcriptional stimulation in reactions reconstituted with human or Drosophila TFIID but appears dispensable for basal-level transcription in reactions reconstituted with TBP (20, 58, 72, 74, 78). Human TFIIA binds directly to at least three viral transcriptional activators (16, 37, 58, 72, 78) and mediates an activator-induced conformational change in TFIID that allows TAFIIs to interact with promoter sequences downstream of the transcriptional initiation site (42, 58). TFIIA can also induce changes in the interaction of TAFIIs with promoter sequences in the absence of a transcriptional activator (40, 55). The TFIIA-mediated conformational change in TFIID facilitates the assembly of TFIIB, indicating that TFIIA binding stimulates productive preinitiation complex assembly (13, 14).
TFIIA activity can be reconstituted in vitro by the expression of two evolutionarily conserved genes, referred to as TOA1 and TOA2 in yeast or αβ and γ in humans (58, 59, 72, 78). The crystal structure of the yeast TFIIA-TBP-DNA ternary complex revealed that the two subunits of yeast TFIIA fold into a complex heterodimer consisting of a four-helix-bundle domain (FHB) and a β-sheet domain (22, 73). Contact with TBP is directed through a series of aromatic residues in the β-sheet domain contributed primarily from the small subunit of TFIIA (Toa2) (22, 73). Mutagenesis of the human TFIIA small subunit (γ) further corroborated the importance of these aromatic residues in forming a stable TFIIA-TBP-DNA complex in vitro (57). Mutations in these residues of the human TFIIA γ subunit were generally defective for transcriptional activation in vitro, indicating that the TFIIA-TBP interaction is absolutely required for transcription function in vitro. Interestingly, conservative mutations in these residues did not disrupt the ternary TFIIA-TBP-DNA complex in gel electrophoretic mobility shift assays (EMSA), but transcriptional activation for these mutants was still defective in vitro (57). Subsequent biochemical analysis indicated that these mutations in TFIIA increase the dissociation rate or protease sensitivity of the TFIIA-TBP-DNA complex, revealing subtle defects in the stability or conformation of the ternary complex not revealed by EMSA, yet correlating with loss of transcription activation function (58a).
TFIIA also functions to derepress transcriptional repression. The stable interaction between TFIIA and TBP precludes the inhibitory association of a variety of transcriptional repressors of TBP-promoter binding, including DR1, NC2, MOT1, DSP1, and HMG1 (5, 21, 27, 35, 53). The derepression function of TFIIA was found to be distinct from its transcriptional coactivation function. Isolation of a smaller form of human TFIIA which lacks the α subunit (the Toa1 amino-terminal homolog) was capable of binding to TBP and derepressing transcriptional inhibitors (47). However, this smaller TFIIA form was incapable of supporting transcription activation in vitro. These results are consistent with mutagenesis studies that implicate the FHB domain as being essential for coactivation function and the β-sheet domain as essential for coactivation, derepression of TBP inhibition, and formation of the TFIIA-TBP-DNA complex (33, 57).
Despite indications that TFIIA is generally important for regulated transcription of all class II promoters in vitro, relatively little is known about how TFIIA functions in vivo. The genes encoding yeast TFIIA, TOA1 and TOA2, are both essential for viability in Saccharomyces cerevisiae (59). Depletion of TFIIA in vivo results in a decrease of several RNA polymerase II-dependent gene transcripts, with no apparent effect on RNA polymerase I- or III-dependent transcripts in yeast (33). Mutations in TBP which disrupt TFIIA binding cause defective transcription activation by acidic activators in yeast and by multiple activators in human cells (8, 68). Mutations in the large subunit of TFIIA which disrupt TBP-DNA binding were found to cause temperature-sensitive phenotypes in yeast (33). However, it is not clear from these previous studies whether a stable TFIIA-TBP interaction was generally required for all class II promoters and activators, or only for a specific subclass of promoters or activators in vivo. To further investigate the general requirements for TFIIA in vivo, we have constructed mutations in TOA2 which compromise the ability of TFIIA to interact with TBP and form a stable TBP-TFIIA-DNA (T-A) complex. These S. cerevisiae mutants were examined for growth phenotypes and specific gene transcription defects in vivo.
MATERIALS AND METHODS
Plasmid constructs and yeast strains.
Wild-type (wt) genomic TOA2 in pSH343 (pRS315 ARS CEN LEU2) and pSH342 (pRS316 ARS CEN URA3) and wt TOA1 in pSH363 (pRS315 ARS CEN LEU2) were kindly provided by S. Hahn (33). toa2 mutants under the control of the wt TOA2 promoter were generated by overlap extension PCR (24, 57). The 2.0-kb PCR fragments containing site-directed mutations in TOA2 were subcloned into the PstI site of pRS315. Escherichia coli expression constructs for the wt and toa2 mutants were generated by PCR (Vent polymerase; New England Biotechnology) and subcloned into pRSETA (Invitrogen) with a BamHI restriction site immediately preceding the initiation codon and a HindIII restriction site immediately following the termination codon. pRSETA-Toa1 was constructed by the same cloning strategy. All the wt and toa2 mutant constructs were confirmed by DNA sequencing in both orientations with an ABI automated 373A DNA sequencer. The resulting pRSETA wt or mutant toa2 open reading frames were expressed in E. coli BL21 and purified as previously described (57). S. cerevisiae SHY94 MATα ade1− ura3 his4 leu2 ΔTOA2::HIS4/pSH342 (ARS CEN URA3 TOA2) was kindly provided by S. Hahn (33). The parent strain of SHY94 was BWG1-7a (MATα leu2 his4 ade1 ura3). The toa2 mutant strains used in this study were produced by transforming SHY94 with wt or mutant toa2 (pRS315) and shuttling out the wt TOA2 copy (pRS316) from SHY94 by streaking the yeast on 5-fluoro-orotic acid (5-FOA) plates. The resulting strains were assayed for a petite phenotype by streaking on several nonfermentable carbon sources (see Table 1). The GAL4(1-147)-VP16 and GAL4(1-147)-HAP4 expression constructs contain the DNA binding domain of GAL4 (amino acids [aa] 1 to 147) fused to the activation domain of herpes simplex virus VP16 (aa 413 to 490) or the yeast HAP4, as described previously (6). Both open reading frames were driven by the yeast ADH1 promoter, which was isolated as a 2-kb BamHI fragment from pDB20L (a gift of S. Berger) and was subcloned into the BamHI site of pRS416 (URA3 CEN) vector (66).
TABLE 1.
Growth phenotypes of toa2 mutants in different mediaa
| toa2 genotype | 5-FOA viability | OD600 doubling time (min) at 30°C in YPD | Growthb on:
|
||||
|---|---|---|---|---|---|---|---|
| SC medium
|
YPGly
|
Lactate, EtOH, KAc (30°C) | |||||
| 30°C | 37°C | 30°C | 37°C | ||||
| wt | Yes | 111 | ++++ | ++++ | ++++ | ++++ | ++++ |
| Y69A | Yes | 191 | ++ | + | + | − | +++ |
| Y69F | Yes | 142 | +++ | ++ | ++ | − | ++++ |
| F71A | Yes | ND | ++++ | ++++ | ++++ | +++ | ++++ |
| F71E | Yes | 128 | ++++ | ++++ | ++++ | +++ | ++++ |
| F71R | Yes | 141 | ++++ | ++++ | ++++ | +++ | ++++ |
| W76A | Yes | 170 | ++ | + | + | − | +++ |
| W76F | Yes | ND | ++++ | +++ | +++ | ++ | ++++ |
| Y69F/W76F | Yes | 244 | + | − | ND | ND | + |
| W76E | No | ||||||
| Y69A/W76A | No | ||||||
YPD, YP dextrose medium; YPGly, YP medium with glycerol as a nonfermentable carbon source; KAc, potassium acetate.
−, no growth; +, pinpoint colonies; ++, very slow growth; +++, slow growth; ++++, wt growth; ND, not determined.
Protein preparations.
The pRSET wt or toa2 mutant constructs were purified under denaturing conditions on Ni-nitrilotriacetic acid agarose columns (Qiagen). The recombinant wt or Toa2 mutant proteins were isolated by column fractionation with elution denaturant (8 M urea–0.1 M NaH2PO4–0.01 M Tris [pH 8.0]–7 mM β-mercaptoethanol [β-ME]–1 mM phenylmethylsulfonyl fluoride [PMSF]) of decreasing pH. wt Toa1 was similarly expressed and purified. Purified Toa2 proteins were renatured with equal molar amounts of wt Toa1 by stepwise dialysis into D100 buffer (20 mM HEPES [pH 7.9] [KOH]–20% glycerol–0.2 mM EDTA Na2+–100 mM KCl–7 mM β-ME–1 mM PMSF) as described previously (57, 58). Recombinant γ TFIIA was more than 85% pure based on Coomassie blue staining in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (data not shown). The stepwise dialysis protocol yielded approximately 50% higher soluble concentrations of γ TFIIA mutant compared to wt γ TFIIA (data not shown). γ TBP was purified as described elsewhere (44).
DNA binding reactions.
EMSA of TBP-TFIIA DNA binding reactions with the adenovirus E1B TATA 30-bp oligonucleotide have been described previously (57). TBP (5 ng) was incubated with 50 fmol of 32P-labeled TATA oligonucleotide in a 12.5-μl reaction volume in the absence or presence of 250, 50, or 20 ng of wt TFIIA or mutant TFIIA, as indicated in Fig. 1B. Complexes were resolved by native 6% polyacrylamide–45 mM Tris-base–45 mM boric acid–1.25 mM EDTA gels at 22°C for 2.5 h.
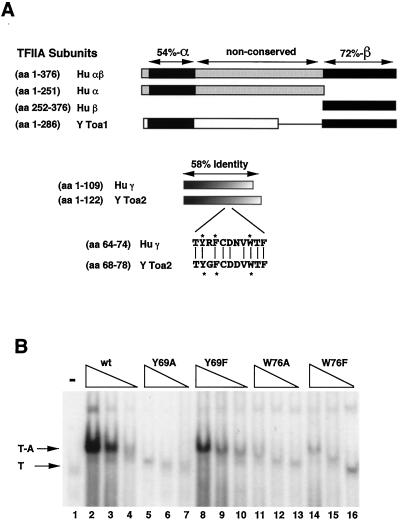
FIG. 1.
TFIIA is a highly conserved heterodimer. (A) The TFIIA subunits from humans (Hu) and yeast (Y) are aligned. The human TFIIA large subunit (αβ) (aa 1 to 376) is aligned to yeast Toa1 (aa 1 to 286). Hu αβ is proteolyzed in vivo to produce individual α and β subunits, which are indicated. The human α subunit is 54% conserved with the yeast homolog (Toa1; aa 7 to 58), while the β subunit is 72% conserved (Toa1; aa 226 to 286). The γ subunit is 58% conserved throughout its length. The conserved Toa2 residues that are mutated in this study are indicated by stars. (B) Yeast TBP-TFIIA complex formation in EMSA. Recombinant yeast TBP and TFIIA proteins were expressed in E. coli, purified, and used in EMSA. The 32-P labeled 30-bp adenoviral E1B TATA box was used as a probe. Decreasing amounts (250, 50, and 20 ng) of TFIIA proteins were added to the EMSA reaction for wt and Toa2 mutant proteins, as indicated above the gel. Arrows point to the TBP-DNA (T) and TBP-TFIIA-DNA (T-A) complexes.
β-Gal assays.
The β-galactosidase (β-Gal) assays were performed as described elsewhere (61). For the CUP1 expression experiment, the yeast cultures were grown to mid-log phase and were transferred to a 37°C water bath for 10 min prior to being placed on a shaker at 37°C for 4 h. CuSO4 was added to a final concentration of 50 μM, and the cultures were placed for an additional 2 h at 37°C on a shaker prior to making the extracts for the β-Gal analysis (30). The CUP1 experiments were performed twice; the averages of these values are shown in Fig. 5, and the error was less than 20% for each sample (wt or mutant). Similar CUP1 expression experiments were performed in duplicate at 30 and at 37°C, with shorter preincubation times (20 min and 2.5 h at 37°C) prior to CuSO4 addition (2 h). The results of all these variations were very similar to those shown in Fig. 5 (data not shown). pCLUC (CUP1 LacZ URA3 CEN4) was a gift of S. Berger and D. Thiele. For the PHO5 expression experiments, yeast cultures were grown as described elsewhere (1). For the PHO5 β-Gal assays, induction was performed for 5 h at 30°C in medium without phosphorus prior to extract production. The PHO5 experiments were performed twice; the averages of these values are shown in Fig. 5, and the error was less than 10% for each sample (wt or mutant). β-Gal activity is expressed as units per milligram of protein and was calculated as described elsewhere (12). The PHO5-LacZ construct (pMH313) was a gift of M. Grunstein.
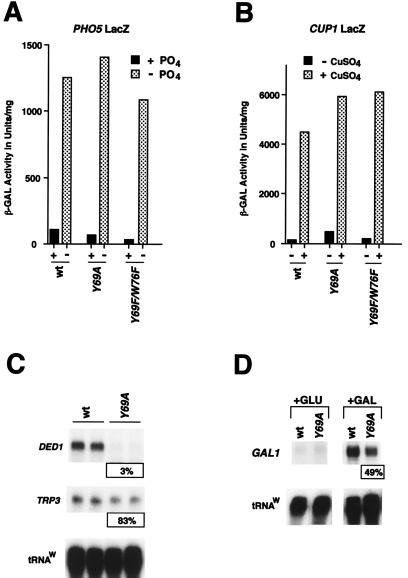
FIG. 5.
A stable T-A complex is not generally required for transcription in vivo. (A) PHO5-driven LacZ expression at 30°C in SC medium was assayed for wt, toa2 Y69A, and Y69F/W76F strains by β-Gal assays. In high-PO4 medium, PHO5 expression was repressed. In synthetic medium in the absence of PO4, PHO5 induction is shown. β-Gal activity is expressed as units per milligram of protein. (B) CUP1-driven β-Gal activity was assayed under conditions similar to those for the experiment for which results are shown in panel A, except that after reaching mid-log phase, samples were grown for an additional 4 h at the nonpermissive temperature (37°C) prior to Cu ion addition. (C) The toa2 Y69A mutant shows defective expression of endogenous DED1, but not of TRP3, in vivo. The wt and toa2 Y69A mutant strains were grown to mid-log phase in SC medium at 30°C and were shifted to the nonpermissive temperature (37°C) for 3 h prior to RNA isolation. Endogenous DED1 and TRP3 expression was assayed by S1 nuclease protection. (D) GAL1 expression is induced in the toa2 Y69A mutant. The wt and toa2 Y69A strains were used to assay GAL1 expression in vivo. Expression levels were measured under conditions of glucose repression (+GLU) and under galactose induction (+GAL). PhosphorImager quantitation indicates that GAL1 expression was induced about 600-fold in the wt and 300-fold in the toa2 Y69A strain. For the GAL1 samples, 4 μg of total RNA was used per reaction, while 40 μg of total RNA was used for all other S1 reactions. PhosphorImager quantitation (expression in the mutant as a percentage of that in the wt) is shown in panels C and D.
Yeast phenotype analysis and RNA isolation.
Yeast manipulations and growth protocols are described in detail elsewhere (62). Yeast cultures were grown to an optical density at 600 nm (OD600) range of 0.8 to 1.1 prior to RNA isolation. One hundred-milliliter cultures were pelleted, washed with 0.5 volume of sterile H2O, resuspended in 2.5 ml of sterile filtered TES buffer (10 mM Tris [pH 7.5]–10 mM EDTA–0.5% SDS), frozen on dry ice, and placed at −80°C prior to total RNA purification. The total yeast RNA was isolated as described previously (29). The isolated total RNA was aliquoted into 40-μg quantities, reprecipitated, and stored at −20°C until used for S1 nuclease analysis. For URA1 induction, 10 μg of 6-azauracil/ml (final concentration) was added for 2.5 h prior to harvesting. For galactose induction, samples were grown in synthetic complete (SC) lactate medium overnight and switched to SC galactose medium for 3 h at 30°C. Control samples were grown in SC glucose medium. Doubling times were calculated as described previously (34).
S1 nuclease analysis.
Oligonucleotides complementary to the genes assayed by S1 nuclease analysis are as follows: CLB1, 5′-TCATTACTATTAATGGTTCTACTATTCTCTACCAAAAGGGATCGTGACATGGGTGT-3′; CLB2, 5′-TTCTATTGGGTTGGACATCTATAAGATCAATGAAGAGAGAGAGGCCC GGG-3′; CLN1, 5′-CAGTGACAATTAACCCAGTTTTCACTTCTGAGTGGTTCATCGGGGG-3′; CTS1, 5′-TCCTAGGGACTGGCAAGTTTCAATAT CTTCAGCAATCTGGGTGCAGTGAAGTAAGCCATCGGGGG-3′; DED1, 5′-CCGCCACGGCCACCGTTGTAGCCGCCGTTGTTGTTATTGTAGCGATGGAGA-3′; ENO2, 5′-CGGGAGTCGTAGACGGATCTAGCGTAAACTTTAGAGACAGCCTAATAA-3′; GAL1, 5′-CTAGAATTGAACTCAGGTACAATCACTTCTTCTGAATGAGATTTAGTCATGCGCGCGC-3′; GAL80, 5′-AGATCTCTTGTTGTAGTCCATGACGGGAGTGGAAAGAACGGGAAA CCAACTATCGAGATTGTAGCTATA-3′; HIS3, 5′-GGTTTCATTTGTAATACGCTTTACTAGGGCTTTCTGCTCTGTCATCTTTGCCTTCGTTTATCT TGCCTGCTCATTTT-3′; PMA1, 5′-GCAGGCTTTTCTTGAGTTGGCTGATGAGCTGAAACAGAAGATGCACTTCT-3′; SEC72, 5′-AACAACAGCATCACTCGCAGTGATCAGTTTACTGTTTGCATTGTATTCAAGGGTAACCATCCGGCC-3′; TRP3, 5′-GGTAAAGGAATCGTAGTTGTCAATTAGAACCACATGCTTACCTTAG-3′; tRNAW, 5′-GGAATTTCCAAGATTTAATTGGAGTCGAAAGCTCGCCTTA-3′; URA3, 5′-GATTTATCTTCGTTTCCTGCAGGTTTTTGTTCTGTGCAGTTGGGTTAAGAATACTGGGCAGGGGGG -3′; and URA1, 5′-GTTTGGTACGGAAGTTCAATTTTTTTTTGAGTAATTGTGTATATCTATTTGAAACGTCTACGGCGG-3′.
S1 probes were end labeled in a 10-μl reaction mixture (30 pM oligonucleotide–50 mM Tris-HCl [pH 8.2]–10 mM MgCl2–0.1 mM EDTA–5 mM dithiothreitol–0.1 mM spermidine–15 U of T4 polynucleotide kinase [Boehringer Mannheim]–333 μCi of [γ-32P]ATP [7,000 Ci/mmol; ICN]) at 37°C for 30 min. The reaction was stopped with a phenol-chloroform extraction, and the unincorporated label was separated from the end-labeled oligonucleotide by using a G25 spin column (5 Prime-3 Prime, Inc.). The probe was precipitated by adding 300 mM Na acetate and 2.5 volumes of 100% ethanol (EtOH), resuspended in 50 mM Tris (pH 8.3)–5 mM EDTA, and stored at −20°C until it was used in S1 assays. The probes were analyzed on 10% denaturing polyacrylamide gels to confirm the extent of incorporation and efficiency of oligonucleotide synthesis. The oligonucleotides were synthesized by Integrated DNA Technologies Corp. Approximately 0.5 pM of oligonucleotide probe was resuspended with 40 μg of total yeast RNA in 10 μl of H2O, unless noted otherwise in the figure legend. The annealing reaction and S1 digestion reaction have been described previously (29). For the cyclins, 80 μg of total RNA was used. For GAL1 experiments, 4 μg of total RNA was used. The samples were analyzed on 10% denaturing polyacrylamide gels. Audioradiographs were made with X-Omat AR film (Kodak), and quantitation was performed on a PhosphorImager (Molecular Dynamics). S1 assays were performed in duplicate at least three times, and PhosphorImager quantitation showed less than 15% error. Representative experiments are shown. tRNAW levels were used as a control for intact RNA and are indicated for all samples. tRNAW autoradiographs were exposed for equal times.
FACS and light microscopy analysis.
Yeast cultures were grown in SC media and harvested in mid-logarithmic phase, and nuclei were stained with acriflavine by a modification of a method previously described (7). Cells were fixed in 70% EtOH for 20 min at 4°C, followed by a 20-min incubation at 22°C in 4 N HCl. Cellular DNA was then stained with 1 ml of staining solution (0.02% acriflavine HCl [Sigma]–20 mM K2O5S2 [Sigma]–0.05 N HCl) for 20 min at 22°C. To remove nonspecific cellular staining, three sets of washes followed, each consisting of two steps: (i) a 2-min incubation at 22°C in 0.12 N HCl in 70% EtOH and (ii) brief resuspension of the pellet in 4 N HCl. Stained cells were stored in H2O at 4°C for no more than 2 days prior to fluorescence-activated cell sorter (FACS) analysis. Haploid (1N), diploid (2N), or clumpy peaks were determined by acriflavine fluorescence intensity by using an EPICS XL flow cytometer (Coulter Corporation, Hialeah, Fla.). Each wt and toa2 mutant cell haploid, diploid, and multiploid peak was FACS sorted, viewed by light microscopy on a Nikon AFX-IIA light microscope (magnification, ×40), and photographed with black-and-white TMAX film. These experiments were performed in duplicate at both the permissive (30°C) and nonpermissive growth temperatures at least three independent times. The FACS and light microscopy results were quite similar at 30, 34, and 37°C, with greater clumping seen with Y69F/W76F mutants at higher temperatures (data not shown). Sonication of yeast cells was performed at 4°C three times for 20-s pulses (on setting 3) on a Misonix ultrasonic processor XL sonicator (see Table 2).
TABLE 2.
Cell cycle phenotypes of toa2 mutantsa
| Mutant | % of cellsb
|
% of cells with the following bud size after sonicationc:
|
||||
|---|---|---|---|---|---|---|
| Single | Double | Clumpy | Small | Medium | Large | |
| wt | 22 | 59 | 19 | 32 | 38 | 29 |
| Y69A | 9 | 51 | 40 | 5 | 17 | 78 |
| Y69F/W76F | 5 | 41 | 54 | |||
| W76A | 7 | 32 | 61 | |||
At least 400 cells were counted for each mutant in each experiment.
Determined by light microscopy from unsonicated cell cultures grown at 37°C in SC medium.
Clumpy cells were disrupted by sonication.
RESULTS
TFIIA-TBP interaction is essential for growth and viability in yeast.
Previous work showed that aromatic residues Y65 and W72 in the small subunit of human TFIIA (γ) were important for forming the TFIIA-TBP-DNA ternary complex (57). Human TFIIA γ is 58% conserved with the yeast TFIIA small subunit (Toa2) (Fig. 1A). Crystal structure revealed that the homologous aromatic residues in Toa2 (Y69 and W76) make the primary stabilizing contact with TBP in the ternary complex with DNA (22, 73). Recombinant yeast TFIIA reconstituted with single-substitution mutants of Toa2 were analyzed for ternary complex formation by EMSA. Substitution of alanine for Y69 and W76 in Toa2 significantly reduced complex formation ∼65-fold (Fig. 1B; compare lane 2 with lanes 5 and 11). Phenylalanine substitution of W76 reduced T-A complex formation 45-fold, and the Y69F mutation reduced it 4.5-fold relative to that in wt TFIIA (Fig. 1B; compare lane 2 with lanes 8 and 14). As mentioned previously, the human homologs of Toa2 Y69F and W76F (Hu Y65F and W76F) stimulate normal levels of T-A complex but fail to stimulate transcription in vitro with most activators (57). Recent biochemical studies indicate that the Y65F mutant has an increased T-A dissociation rate in EMSA and the W76F mutant forms an altered T-A complex that is highly sensitive to proteolytic digestion (data not shown). These results further indicate that mutations in Toa2 at residues Y69 and W76 affect the stability and/or the conformation of the T-A complex.
To determine the effects of these and related toa2 mutations on cell viability and growth, mutant toa2 alleles were introduced into yeast by the plasmid shuffle technique (62). toa2 mutants with radical substitution of W76 with glutamic acid (W76E) and those with the double alanine substitution mutation (Y69A/W76A) were inviable (Fig. 2A). The toa2 mutant with the conservative double phenylalanine substitution (Y69F/W76F) was viable but extremely slow in growth at 30°C (Fig. 2A and C). Growth rates show that even single alanine substitution mutations at residues Y69 and W76 are more deleterious than radical mutations in a neighboring conserved aromatic residue, F71, underscoring the general importance of the Y69 and W76 residues in yeast growth and viability (Table 1). Maintaining either the Y69A or the W76A allele on high-copy-number plasmids (2μm) failed to rescue the toa2 strains’ growth defect in SC media at 30°C, indicating that increased expression could not rescue the transcription defects (data not shown).
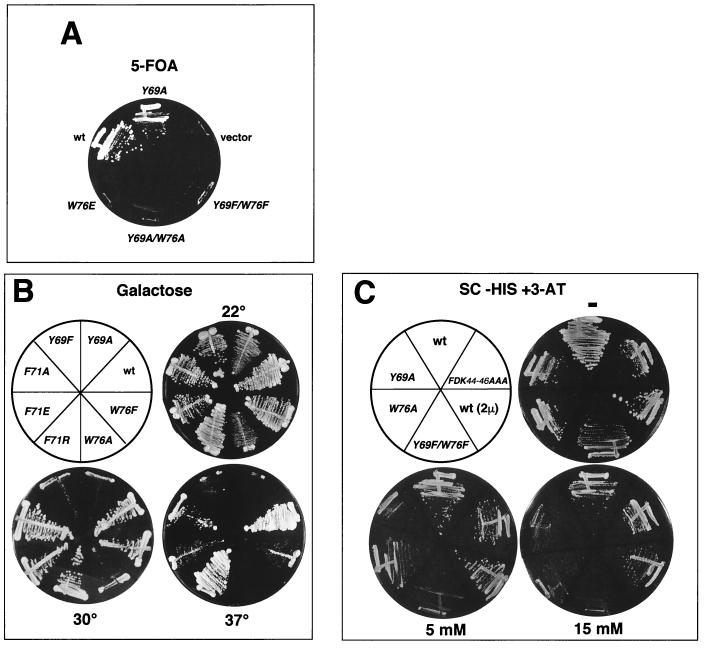
FIG. 2.
toa2 mutants defective in TBP-TFIIA complex formation are conditionally lethal in vivo. (A) toa2 aromatic mutants exhibit lethal and conditionally lethal growth phenotypes in vivo on 5-FOA. Haploid S. cerevisiae cells with the indicated wt or toa2 mutant genotype were grown on SC plates at 30°C with 5-FOA to select for cells that lost wt TOA2. (B) toa2 mutants are conditionally lethal on YP galactose plates. Haploid S. cerevisiae cells with the indicated wt or toa2 mutant genotype were grown at the growth temperatures shown. (C) toa2 mutants are conditionally lethal on SC-His plates without (−) or with 3-AT. Haploid S. cerevisiae cells with the indicated wt or toa2 mutant genotype were grown at 30°C with the indicated concentrations of 3-AT.
The panel of TFIIA substitution mutants was further analyzed for growth defects and conditional lethality (summarized in Table 1). Among the most dramatic growth defects were the failure of Y69F, Y69A, and W76A mutants to grow on galactose-containing media at 30°C, while the F71A, F71E, and W76F mutants grew slowly at 37°C on galactose (Fig. 2B). The Y69 and W76 mutants grew extremely slowly on glycerol at 30°C and were lethal at 37°C on yeast extract-peptone (YP) glycerol plates. In contrast, these mutants grew significantly better on several other nonfermentable carbon sources (Table 1; data not shown). The inability of toa2 alleles to grow on various carbon sources suggests that they are incapable of expressing genes essential for either galactose or glycerol utilization. Furthermore, we examined the ability of toa2 mutants to grow on increasing concentrations of 3-aminotriazole (3-AT), a HIS3 competitor which requires high-level HIS3 gene expression for cell viability. toa2 Y69A and W76A strains showed significantly impaired growth on 15 mM 3-AT, while the Y69F/W76F strain was incapable of growth on 5 mM 3-AT at 30°C (Fig. 2C). We have also generated a mutation in the FHB domain of TFIIA (Toa2 FDK44-46AAA) that causes a temperature-sensitive phenotype in SC media at 37°C (data not shown). In the human system, similar mutants preclude normal interaction of the FHB α and γ helices in glutathione S-transferase assays (57), and TFIIA derivatives lacking the FHB domain stimulate T-A formation in EMSA (47, 58). The TFIIA FHB mutant strain had no effect on 3-AT-dependent growth, indicating that a 3-AT growth defect correlates with mutations in the β-sheet domain of TFIIA which interfere with TBP binding (Fig. 2C). These results suggest that TFIIA must interact efficiently with TBP to support high-level activation of the HIS3 gene.
TFIIA mutants disrupt cell cycle progression.
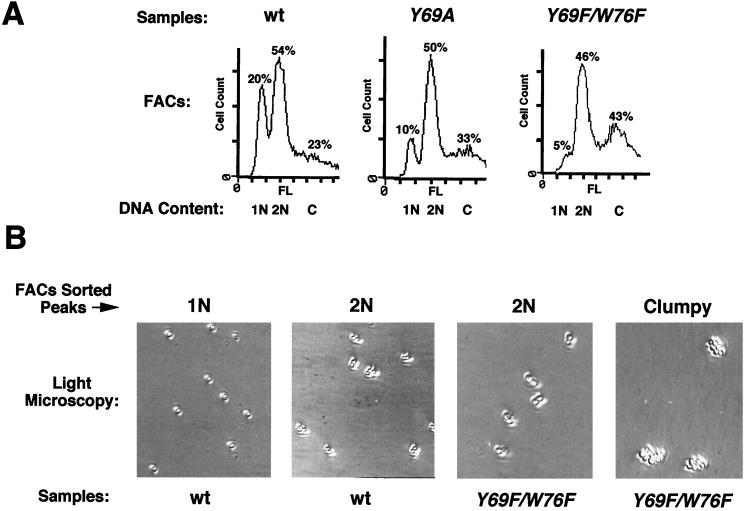
Since several yeast TAFII mutations were found to cause cell cycle arrest phenotypes (3, 77), we further inspected toa2 mutant alleles for effects on cell cycle progression. Light microscopy revealed that toa2 mutants accumulated as fused cell pairs and multipaired clumps with equal-sized buds under permissive conditions, and more so under nonpermissive conditions. We visually counted wt, Y69A, and Y69F/W76F cells (>400 for each experiment) and found a significant decrease in the number of single or small-budded cells in the mutants relative to the wt, with an accumulation of multibudded clumps (Table 2). FACS analysis confirmed that these TFIIA mutants had decreased numbers of cells with a 1N copy of DNA and an increase in multibudded complexes, or clumps (Fig. 3A). The various major FACS peaks of the wt and Y69F/W76F strains were subjected to cell sorting and then analyzed by light microscopy to confirm that the peaks were indeed fused and unbudded cell twins (2N) and aggregated multibudded complexes (Fig. 3B). Sonication was capable of disrupting the clumps into unbudded single and fused cell pairs, with a noticeable loss of small-budded cells (Table 2). The clumpy phenotype, accumulation of fused cell pairs, and the loss of small-budded cells after sonication suggest that toa2 mutant strains may be arresting in G2/M or cytokinesis (3, 31, 32, 38, 39, 63).
FIG. 3.
toa2 mutants accumulate in the G2/M phase of the cell division cycle. (A) FACS analysis of the toa2 Y69A and Y69F/W76F mutants. For each sample, haploid (1N), diploid (2N), and clumpy cells (C) are indicated beneath each peak. Cell count is plotted as a function of fluorescence (FL). For cell count, each dash represents 25 cells. The percentage of sorted cells in each peak is shown above the peak. (B) Light microscopy of FACS-sorted peaks. For the wt and the Y69F/W76F mutant, haploid, diploid, or clumpy peaks were individually FACS sorted and viewed by light microscopy (magnification, ×32) and photographed. For each sample, representative pictures of major peaks are shown (either 1N, 2N, or clumpy).
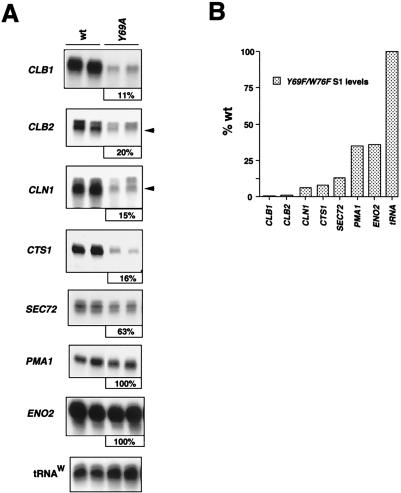
To further characterize a potential cell cycle defect, we determined if transcription levels of several cell cycle-regulatory genes were reduced in these toa2 mutant strains. We analyzed the CLB1, CLB2, CLN1, SEC72/SIM2, and CTS1 genes by S1 nuclease protection (Fig. 4A). CLB1 and CLB2 are specifically expressed in G2, and their protein products are required for progression into mitosis (2). SEC72/SIM2 is expressed in late G2 and prevents rereplication of the genome prior to mitosis or start (18). CLN1 is expressed in G1 and is required for progression through G1/S (2). CTS1 encodes chitinase and is required for completion of cytokinesis (39). In the toa2 Y69A strain, CLB1, CLB2, CLN1, and CTS1 expression were reduced significantly (to less than 20% of that of the wt), while SEC72/SIM2 expression was reduced to 63% of that of the wt (Fig. 4A). In contrast, expression of ENO2 and PMA1, which are not cell cycle dependent (3), was unaffected by the toa2 Y69A allele (Fig. 4A). In the toa2 Y69F/W76F double mutant, which has a more severe growth arrest phenotype, the cell cycle-specific CLB1, CLB2, and CLN1 transcripts were 0.6, 4, and 6% of wt levels, respectively, while the cell cycle-independent ENO2 and PMA1 transcripts were reduced to only ∼35% of wt levels (Fig. 4B). These results indicate that some cell cycle-specific promoters and/or activators are preferentially sensitive to TFIIA mutations and that a stable association of TFIIA with TBP is required for efficient transcription of genes required for cell cycle progression.
FIG. 4.
Reduced expression of cell cycle-regulatory genes in toa2 mutant strains. (A) wt and toa2 Y69A samples were grown at the nonpermissive temperature (37°C). The cell cycle-regulatory genes CLB1, CLB2, CLN1, and CTS1, and the noncycling genes PMA1 and ENO2, were assayed by S1 nuclease protection. Eighty micrograms of total RNA was used for CLB1, CLB2, and SEC72, expression, and 40 μg of total RNA was used for the other S1 reactions. S1 assays were performed in duplicate at least three times, and PhosphorImager quantitation showed less than 20% error. PhosphorImager quantitation (expression in the mutant shown as a percentage of SEC72 wt expression) is given below each panel. Results of representative experiments are shown. tRNAW levels were used as a control for intact RNA and are indicated for all samples. (B) Expression of cell cycle-regulatory genes is severely reduced in the toa2 Y69F/W76F mutant. S1 assays were quantitated by PhosphorImager, and the averages from at least six experiments are plotted in graph format. PhosphorImager quantitation showed less than 10% error.
A stable TFIIA-TBP interaction is not generally required for transcription of all genes in vivo.
S1 analysis of cell cycle-regulatory genes suggested that transcription of some genes is preferentially affected by toa2 mutations (Fig. 4). To better characterize the class of genes affected by toa2 mutations, we assayed the ability of the PHO5 and CUP1 promoters to respond to inducing agents. Using β-Gal assays, we found that viable mutants with substitutions at residues Y69, F71, and W76 had no significant transcriptional defect with PHO5 induction on medium without phosphorus, or with CUP1 induction in the presence of copper ions (Fig. 5A and B and data not shown). Figures 5A and B show that the most severely growth-defective toa2 mutants, Y69A and Y69F/W76F, were indistinguishable from wt strains with regard to PHO5 and CUP1 induction, even at the nonpermissive temperature (Fig. 5B). This suggests that a compromised TFIIA-TBP interaction is not generally required for all class II transcription. To determine if a subset of genes were affected by these TFIIA mutants, we examined two constitutively active promoters and another inducible promoter by S1 analysis. Steady-state RNA levels of the TRP3 gene were only slightly reduced (83%) in the Y69A mutant relative to the wt (Fig. 5C). In contrast, DED1 RNA levels were dramatically reduced in the Y69A mutant to just 3% of wt RNA levels (Fig. 5C). wt GAL1 mRNA could be induced almost 600-fold by switching to galactose- from lactate-containing medium, while in the Y69A mutant, GAL1 induction was impaired ∼2-fold, to 49% of wt levels (Fig. 5D). In the S1 assays, RNA levels were normalized for tRNAW expression, which appears to be unaffected by mutations in TFIIA (33). Together, these results indicate that a subset of promoters are highly sensitive to defects in the TFIIA-TBP interaction (e.g., DED1), while other promoters are seemingly unaffected in vivo (e.g., CUP1).
TFIIA-TBP interaction is required for promoter selection in vivo.
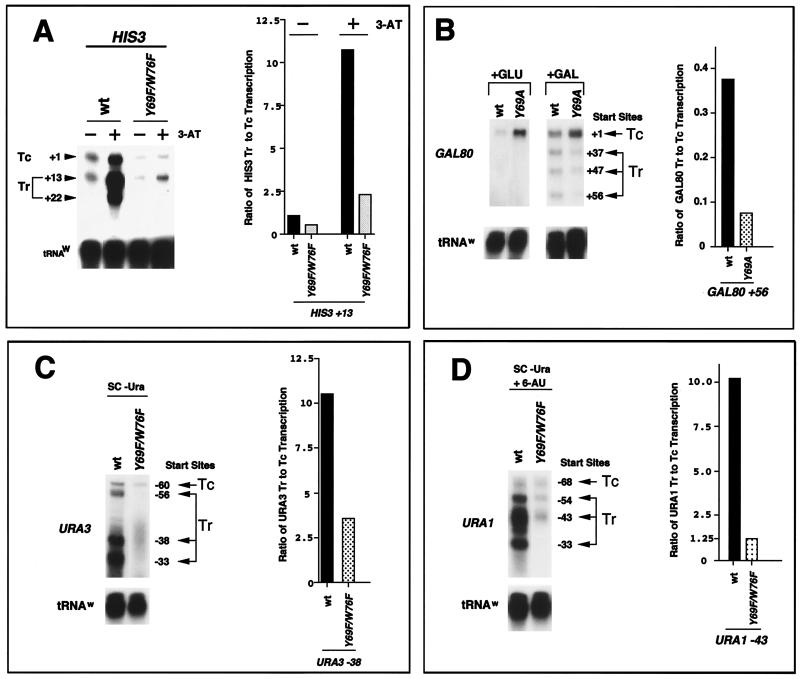
For a subset of promoters in yeast, inducible expression is characterized by the utilization of several transcriptional initiation sites (71). Constitutive transcription (TC) initiates from the furthest upstream start site, while inducible transcription initiates from one or multiple downstream initiation sites (TR). Utilization of TR start sites is important for high-level activator-dependent gene expression of the HIS3, GAL80, URA3, and URA1 promoters (46, 64, 70). These promoters were all examined for activator-induced start site selection in mutant versus wt toa2 strains by S1 analysis (Fig. 6). The HIS3 gene has been extensively studied for selection from the constitutive +1 initiation site (TC) to the activator-induced +13 and +22 initiation site (TR) (28, 49, 70). In wt strains, addition of 3-AT induced transcription by ∼16-fold at +13 and 9-fold at +22. In contrast, transcription was induced only 1.5-fold at +1 (Fig. 6A). In wt strains, the utilization of TR (+13) after induction with 3-AT was ∼11-fold greater than transcription from TC. In contrast, the utilization of TR (+13) was only 2.5-fold greater than that of TC for the toa2 Y69F/W76F strain (Fig. 6A). While utilization of TC was modestly reduced, the ability to activate transcription at TR was most significantly affected by the toa2 Y69F/W76F allele. Similar HIS3 results were seen for the toa2 Y69A single mutant allele (data not shown). Thus, TFIIA mutants in which TBP binding is compromised disrupt high-level transcription initiating from the HIS3 TR start sites.
FIG. 6.
toa2 Y69A and Y69F/W76F mutants show defects in activator-induced start site switching. (A) HIS3 TC (+1) and TR (+13 and +22) start site expression for wt and toa2 Y69A. 3-AT (45 mM) was added to the medium for 2.5 h at 30°C prior to RNA isolation. PhosphorImager quantitation of the S1 assays is presented as the ratio of TR (+13) to TC (+1). (B) The wt and toa2 Y69A strains were used to assay GAL80 TR expression in vivo. Expression levels were measured under glucose repression (+GLU) and under galactose induction (+GAL). GAL80 TC (+1) and TR (+37, +47, and +56) start site expression is shown (64). (C) The wt and toa2 Y69F/W76F strains, containing a wt URA3 CEN plasmid, were grown in SC medium at 30°C, and total RNA was isolated. S1 analyses were performed to determine transcription efficiency from the multiple URA3 start sites. The TC (−60) and TR (−56, −38, and −33) start sites relative to the translation start site (AUG) are shown (46). (D) S1 analyses were similar to those described for panel C, except that endogenous URA1 expression was induced by 6-azauracil (10 μg/ml) for 2.5 h at 30°C. The URA1 TC (−68) and TR (−54, −43, and −33) start sites relative to AUG are shown (46). The ratio of TR to TC transcription is indicated on the right for each experiment. Forty micrograms total RNA was used per S1 reaction.
Galactose induction of the GAL80 gene results in the stimulation of multiple TR start sites (+37, +47, +56, and +67) which are downstream of the constitutive TC (+1) start site (64). Only GAL80 TC expression is observed under glucose repression (64). Surprisingly, in SC medium with glucose, GAL80 TC levels are severalfold higher for the toa2 Y69A mutant compared to the wt (Fig. 6B and data not shown). In the presence of galactose, GAL80 TR expression was significantly reduced in the toa2 Y69A strain relative to the wt (Fig. 6B). For the GAL80 +56 start site, the ratio of the TR to the TC level was fourfold lower in the mutant toa2 Y69A strain relative to the wt (Fig. 6B).
The URA3 TC start site at −60 (relative to AUG) is weakly expressed in the absence of the PPR1 activator, while the TR start sites at the −56, −38, and −33 positions are induced to high levels by the PPR1 activator (46). We found that yeast strains carrying the toa2 Y69F/W76F mutant allele were able to express the URA3 TC transcript but were severely defective in mediating activator-dependent expression from multiple TR sites for this gene (Fig. 6C).
Similarly, URA1 gene TC expression (−68) was barely affected by the toa2 Y69F/W76F allele, but TR expression at the −54, −43, and −33 start sites was significantly defective in the mutant strain (Fig. 6D). URA1 expression at the −43 start site (TR/TC ratio) was more than eightfold lower in the toa2 Y69F/W76F mutant compared to the wt, while there is barely detectable expression from the −33 start site in the mutant background (Fig. 6D). The RNA samples used for both URA1 and URA3 S1 analyses had wt levels of both PMA1 and ENO2 expression (data not shown). These results clearly show that yeast strains carrying a TFIIA mutation, with compromised TBP binding, exhibited defective high-level transcriptional activation of the inducible TR start sites, with modest TC defects, for multiple genes in vivo.
Activator- and promoter-dependent defects with mutant TFIIA.
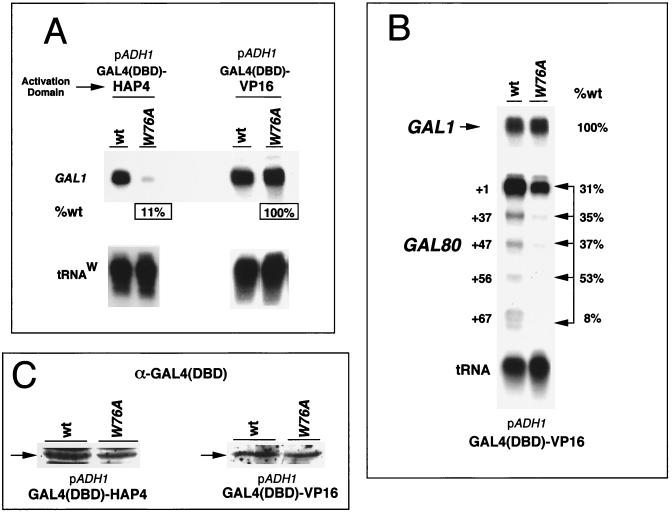
Activation of GAL1 and GAL80 genes is largely dependent upon the interaction of GAL4 with the UASG of each promoter (64). The toa2 Y69A mutation affected the steady-state transcription level of GAL1 ∼2-fold (Fig. 5) and the start site selection of GAL80 TR sites up to ∼4-fold (Fig. 6B). To determine if these defects were partly a result of the transcriptional activator, we compared the ability of two distinct transcriptional-activation domains to activate the same promoter in a wt or a toa2 mutant strain. The activation domains of the herpesvirus VP16 and yeast HAP4 transcriptional activators were fused to the GAL4 DNA binding domain and expressed at high levels by the ADH1 promoter under conditions of glucose repression (Fig. 7A) (6). Both the HAP4 and VP16 activation domains stimulated GAL1 expression to similar levels in the wt strain (Fig. 7A). However, in the toa2 W76A mutant strain, stimulation of GAL1 by the HAP4 activation domain was reduced to 11% of that of the wt, while stimulation by the VP16 activation domain was identical to that observed in the wt strain (Fig. 7A). Western blotting confirmed that the GAL4-fusion proteins were expressed at similar levels in the wt and mutant toa2 strains (Fig. 7C). These results indicate that different activation domains have different requirements for a stable TFIIA-TBP interaction.
FIG. 7.
Activator- and promoter-specific defects of toa2 mutant alleles. (A) The GAL4-VP16 and GAL4-HAP4 activators were compared for their ability to activate the endogenous GAL1 promoter in wt or toa2 W76A strains. The activators were expressed by the ADH1 promoter under conditions of glucose repression. RNA levels were determined by S1 analysis and quantitated by PhosphorImager. Percentage of wt activity is indicated for the W76A mutant-derived mRNA. DBD, DNA binding domain. (B) The endogenous GAL1 and GAL80 promoters were compared for activation by the GAL4-VP16 activator in the wt and toa2 W76A strains. RNA levels were assayed by S1 analysis, and PhosphorImager quantitation is presented as the percentage of wt levels. Multiple TR start sites are indicated for GAL80. TR induction by GAL4-VP16 in SC medium with 2% glucose was compared for the wt and toa2 Y69A strains. The percentage of wt transcription for each GAL80 start site is indicated. Experiments were performed at least twice in duplicate, and the error was less than 15% (data not shown). (C) Western blot analysis of GAL4-VP16 and GAL4-HAP4 expression levels in the wt and toa2 W76A strains. Cell extracts were derived from cultures grown under identical conditions to those used for panels A and B. GAL4-VP16 and GAL4-HAP4 were expressed from the ADH1 promoter.
Promoter structure is also likely to contribute to the differential requirement for TFIIA in transcription activation. We have already observed that GAL4-mediated activation of GAL80 was more sensitive to toa2 mutation than was activation of GAL1 (compare Fig. 5D and 6B). We now show that toa2 mutations affect activation of GAL80 but not of GAL1 when the two genes are activated by GAL4-VP16 under identical conditions (Fig. 7B). The GAL4-VP16 fusion protein was expressed by the ADH1 promoter under conditions of glucose repression, and levels of transcription of GAL80 and GAL1 were directly compared. Activation of the GAL1 promoter was similar in the wt and toa2 W76A mutant strains (Fig. 7B). In contrast, GAL80 transcription was highly sensitive to toa2 mutation (Fig. 7B). RNA levels at all of the GAL80 start sites were significantly reduced in the W76A strain relative to the wt, with the most dramatic defects occurring at the most distal start site, +67, which was reduced to 8% of wt activation levels. These results indicate that different promoters regulated by the same activator can have differential requirements for TFIIA.
DISCUSSION
TFIIA-TBP complex formation is essential for a subset of promoters in vivo.
The interaction of TFIIA with TBP has been shown to be important for transcriptional activation in vivo in both human and yeast systems (8, 67). Selection of random mutations in TBP which specifically affect response to acidic activators in yeast predominantly affect the DNA binding surface of TBP or disrupt the association of TBP with TFIIA (4, 68, 69). Facilitated recruitment of TBP to the promoter can bypass the need for an activator in yeast, indicating that some promoters require enhancement of TBP binding in vivo (11, 36). TFIIA has been shown to augment TBP binding to TATA sequences and to function as a coactivator for several human and viral activators in vitro (25, 37, 58). Consistent with this, we found that TFIIA mutants in which binding to TBP was compromised were defective for transcription at a subset of promoters in vivo. TFIIA mutations had dramatic effects on the expression of DED1, and the induced expression of HIS3, GAL80, URA1, and URA3 (Fig. 5 and 6). Cell cycle-regulated CLB1, CLB2, CLN1, and CTS1 expression was significantly reduced in TFIIA mutant strains, while SEC72 levels were modestly reduced (Fig. 4). The interaction of TFIIA with TBP may regulate the activity of these promoters by mediating an association between activators and TBP, or by directly enhancing TBP binding to specific core promoters. For these promoters, the interaction of TFIIA with TBP is likely to be rate limiting in vivo.
TFIIA mutants in which TBP binding was compromised did not generally defective in transcription activation of all class II promoters. Activation of the CUP1 and PHO5 promoters was unaffected by TFIIA mutations, as was expression of the constitutively expressed TRP3, ENO2, and PMA1 genes (Fig. 4 and 5). Previously, we had found that human TFIIA mutants in which TBP binding was compromised exhibited defective transcription from all promoters and most activators tested in vitro (57). The finding that homologous yeast TFIIA mutants have more complex phenotypes in vivo is not unprecedented. TFIIB has been reported to be a rate-limiting target of several eukaryotic activators in vitro, yet mutations in TBP which compromise TFIIB binding had no detectable effect on transcription activation in vivo in yeast (15, 41, 45). Similarly, TAFIIs are essential for activated transcription in vitro but may be dispensable for regulation of many genes in vivo in yeast (54, 76). More recent examination of the TAFIIs in yeast indicate that core promoter differences contribute to the requirement for particular TAFIIs (65, 77). Thus, promoter structure may dictate which general factors and coactivators are rate limiting for transcriptional regulation. Our results indicate that a subset of promoters, but not all, require a stable interaction between TFIIA and TBP for efficient expression in vivo in yeast.
Role of TFIIA in the regulation of cell cycle progression.
TFIIA mutants with compromise TBP binding accumulated as aggregated clumps which, when sonicated, were reduced to single or twin buds of equal size. S1 analysis of cell cycle-regulated genes revealed a significant reduction in RNA levels of cyclin genes required for cell cycle progression, with little or no effect on several genes not involved in the cell cycle. The chitinase-encoding CTS1 RNA was also significantly reduced in toa2 mutant strains. Reduction in CTS1 expression may account for the clumpy phenotype, since chitinase is required for progression through cytokinesis (39). Similar clumpy phenotypes with reductions in CTS1 transcription levels have been observed for yeast strains in which the SIN4 and RGR1 transcriptional regulators were deleted (31, 32, 63). The large accumulation of clumpy cells suggests that cytokinesis is blocked in toa2 mutant strains, and the lack of small- and medium-budded cells after sonication supports this conclusion (Table 2). However, we cannot exclude the possibility that toa2 mutants may be arresting at additional points in the cell cycle. Interestingly, mutations causing temperature-sensitive phenotypes in yTAFII90 cause a G2/M arrest, while depletion of TAFII145 causes a G1/S cell cycle arrest, indicating that different TAFIIs are required for transcription of distinct subclasses of cell cycle-regulated genes (3, 77). TFIIA, like TAFIIs, appears to also be required for the transcriptional regulation of multiple genes controlling cell cycle progression.
TFIIA and TAFIIs have different effects on start site switching.
TFIIA mutants with compromised TBP binding showed defective activation of genes with inducible start sites. Several extensively characterized yeast core promoters have two control elements referred to as TR and TC (28, 64, 70). TR resembles a consensus TATA element and is important for regulated transcriptional initiation in vivo. TC does not have a clear consensus sequence but is important for directing constitutive transcription from the proximal initiation site in vivo. The TC element has been hypothesized to consist of a collection of weak TATA elements, but it is also conceivable that TBP does not directly bind to this sequence (28). Our results indicate that a stable TFIIA-TBP interaction is important for the efficient utilization of the consensus TATA element in TR. TFIIA has also been shown to be important for the selection of the proximal promoter start site found in the Drosophila ADH promoter, which appears to possess a consensus TATA element relative to the nonconsensus TATA element at the distal promoter (23). Our results further suggest that TFIIA is required for the efficient utilization of consensus TATA elements found in many eukaryotic promoters.
Genetic and biochemical evidence clearly indicate that TAFIIs play a regulatory role in transcriptional activation and promoter selection and that this function may be largely dependent upon the presence of TFIIA (23, 54, 76). TAFIIs allow TFIID to utilize the initiator element found in many higher eukaryotic TATA-less promoters (51, 79). The GAL80 core promoter consists of two control elements, a consensus TATA at −20 and an Inr-like element at +1 (64). Mutagenesis of the TATA element results in an abrogation of activator-inducible transcription from the downstream (TR) start sites, while the Inr controls the constitutive expression of the +1 start site (TC) (64). Mutations in TFIIA resulted in reduced TR transcription and a slight but reproducible increase in GAL80 TC expression (Fig. 6 and 7). This is consistent with the findings of Sakurai et al. (64), who describe a competition between the TC and TR initiation sites. For GAL80 expression, TFIIA may function to mediate an isomerization of TFIID from a TC/Inr recognition complex to a TR/TATA binding complex that allows high-level transcription.
TFIIA interacts biochemically and genetically with other TBP-associated polypeptides. Mot1/ADI is an ATP-dependent inhibitor of TBP-DNA binding in vitro. The inhibition of TBP binding by Mot1p could be prevented by TFIIA in vitro (5). The MOT1 gene was originally identified as a global negative regulator of a class of genes in yeast (19). Interestingly, Mot1 mutants showed defects in +1 (TC) start site expression of the HIS3 gene (17). In contrast, our data show that TFIIA mutants were defective primarily for +13 (TR) start site expression of HIS3. Thus, Mot1 and TFIIA appear to affect the two distinct promoter start sites of the HIS3 gene. Similarly, TAFIIs may also compete with TFIIA for directing TBP activation function. Depletion of yeast TAFII145 and TAFII19 in vivo caused phenotypes similar to those of Mot1 mutants, resulting in a decrease in TC expression, but had no effect on TR expression of the HIS3 gene (54). Collart has proposed that Mot1p dissociates TBP from consensus TATA elements and that this release may be important for the activation of genes with nonconsensus TATA elements (17). This interpretation is consistent with our findings and suggests that TFIIA promotes TBP interactions at a class of activator-dependent consensus TATA elements (TR) in vivo.
Our results also reveal differences between constitutively expressed promoters in their sensitivity to TFIIA mutants. We found that TRP3 expression was relatively insensitive, while DED1 expression was dramatically reduced by the Y69A allele. TRP3 has been shown to contain a nonconsensus TATA element similar to TC (50). In contrast, strong constitutive expression of DED1 is synergistically activated by a T-rich element and a UAS which binds to ABF1 (9). Interestingly, DED1 expression is also significantly reduced in a TBP mutant (P109A) which binds poorly to the TATA box in EMSA and may affect TFIIA binding, since the mutated residue is in close proximity to the TFIIA recognition site (4, 22, 73). Thus, even constitutively expressed genes may show differential sensitivity to mutations which compromise TFIIA-TBP complex formation.
Activator and promoter dependence of TFIIA defects.
The differential requirements for TFIIA-TBP interaction may depend on either the activator or the promoter structure. Our results suggest that both activator structure and promoter structure contribute to the requirement for a stable TFIIA-TBP interaction. Activation of the GAL1 gene by the VP16 activation domain was unaffected by the toa2 W76A mutation, while the HAP4 activation domain was extremely sensitive to it (Fig. 7A). Thus, different activation domains may require more stable interactions between TFIIA and TBP to execute their function. We found that the GAL80 promoter was significantly more sensitive to the toa2 W76A mutation than was the GAL1 promoter when both were activated by the same activator, GAL4-VP16 (Fig. 7B). This indicates that different promoters can have differential requirement for a stable interaction between TFIIA and TBP. A similar observation has been made for the Zta transcriptional activator in vitro. Zta stimulates TFIIA-TFIID-promoter complex formation and can partially overcome a transcriptional defect resulting from similar human TFIIA mutants in which TBP interaction is compromised in vitro (43, 57). Other activation domains fail to stimulate this interaction and cannot overcome TFIIA mutant transcriptional defects. This suggests that some activators can overcome defects in TFIIA-TBP interaction by introducing compensatory and stabilizing interactions. TFIIA recruitment by Zta was also found to be important for a subset of promoters, further indicating that promoter structure contributes to a requirement for TFIIA recruitment by an activator (43). Together, these results demonstrate that TFIIA can be used variantly by different activators and promoters to regulate transcription initiation.
Conclusion.
The interaction of TFIIA with TBP is highly conserved between humans and yeast and is likely to be important for multiple levels of gene regulation. Using site-directed mutagenesis of TFIIA amino acid residues critical for stable interaction with TBP, we were able to characterize the importance of this interaction for the growth phenotypes and RNA expression of several class II genes in S. cerevisiae. In this study, the stable interaction of TFIIA with TBP was found to be particularly important for activator-induced expression of promoters with consensus TATA elements that direct multiple downstream initiation sites (TR) and for a subset of cell cycle-specific genes. These results confirm biochemical studies which suggest that TFIIA is a core promoter-dependent coactivator and further suggest that the TFIIA-TBP interaction is rate limiting for the transcriptional regulation of a subset of genes in vivo.
ACKNOWLEDGMENTS
We thank S. Berger, S. Hahn, M. Grunstein, K. Struhl, and F. Winston for the generous gifts of plasmids and yeast strains. We thank D. Gursel, F. Arroyo, and D. Lee for excellent technical support, J. S. Faust for running the flow cytometry samples, Allison Borenstein for assistance in figure preparation, and the Wistar core facilities for automated DNA sequencing and oligonucleotide synthesis. We thank S. Dalton, R. Candau, N. Barlev, C.-J. Chen, and S. Triezenberg for helpful comments during this study.
J.O. was supported by an NIH NRSA postdoctoral fellowship and a VFW postdoctoral cancer fellowship during this study. This work was supported by NIH grant GM 12345-01 to P.M.L., who is also a Leukemia Society of America Scholar.
REFERENCES
- 1.Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 3.Apone L M, Virbasius C-M A, Reece J C, Green M R. Yeast TAFII90 is required for cell cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 4.Arndt K M, Ricupero-Hovasse S, Winston F. TBP mutants defective in activated transcription in vivo. EMBO J. 1995;14:1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 6.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:252–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 7.Block D E, Eitzman P D, Wangensteen J D, Srienc F. Slit scanning of Saccharomyces cerevisiae cells: quantification of asymmetric cell division and cell cycle progression in asynchronous culture. Biotechnol Prog. 1990;6:512–514. doi: 10.1021/bp00006a015. [DOI] [PubMed] [Google Scholar]
- 8.Bryant G O, Martel L S, Burley S K, Berk A J. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 9.Buchman A R, Kornberg R D. A yeast ARS-binding protein activates transcription synergistically in combination with other weak activating factors. Mol Cell Biol. 1990;10:887–897. doi: 10.1128/mcb.10.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 11.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Barlev N A, Westergaard O, Jakobsen B K. Identification of the C-terminal activator domain in yeast heat shock factor: independent control of transient and sustained transcriptional activity. EMBO J. 1993;12:5007–5018. doi: 10.1002/j.1460-2075.1993.tb06194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 14.Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 15.Choy B, Green M R. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature. 1993;366:531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- 16.Clemens K E, Piras G, Radonovich M F, Choi K S, Duvall J F, DeJong J, Roeder R, Brady J N. Interaction of the human T-cell lymphotropic virus type 1 Tax transactivator with transcription factor IIA. Mol Cell Biol. 1996;16:4656–4664. doi: 10.1128/mcb.16.9.4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collart M A. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahmann C, Diffley J F, Nasmyth K A. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 19.Davis J L, Kunisawa R, Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeJong J, Bernstein R, Roeder R G. Human general transcription factor TFIIA: characterization of a cDNA encoding the small subunit and requirement for basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:3313–3317. doi: 10.1073/pnas.92.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge H, Roeder R G. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J Biol Chem. 1994;269:17136–17140. [PubMed] [Google Scholar]
- 22.Geiger J H, Hahn S, Lee S, Sigler P B. The crystal structure of the yeast TFIIA/TBP/DNA complex. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 23.Hansen S K, Tjian R. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 24.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 25.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 26.Imbalzano A N, Zaret K S, Kingston R E. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J Biol Chem. 1994;269:8280–8286. [PubMed] [Google Scholar]
- 27.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 28.Iyer V, Struhl K. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol Cell Biol. 1995;15:7059–7066. doi: 10.1128/mcb.15.12.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeyaprakash A, Welch J W, Fogel S. Multicopy CUP1 plasmids enhance cadmium and copper resistance levels in yeast. Mol Gen Genet. 1991;225:363–368. doi: 10.1007/BF00261675. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y W, Dohrmann P R, Stillman D J. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcription regulation. Genetics. 1995;140:47–54. doi: 10.1093/genetics/140.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Y W, Stillman D J. Regulation of HIS4 expression by the Saccharomyces cerevisiae SIN4 transcriptional regulator. Genetics. 1995;140:103–114. doi: 10.1093/genetics/140.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang J J, Auble D T, Ranish J A, Hahn S. Analysis of the yeast transcription factor TFIIA: distinct functional regions and a polymerase II-specific role in basal and activated transcription. Mol Cell Biol. 1995;15:1234–1243. doi: 10.1128/mcb.15.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kayne P S, Kim U-J, Han M, Mullen J R, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 35.Kirov N, Lieberman P, Rushlow C. The mechanism of repression by DSP1 (dorsal switch protein) involves interference with preinitiation complex formation. EMBO J. 1995;15:7079–7087. [PMC free article] [PubMed] [Google Scholar]
- 36.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi N, Boyer T G, Berk A J. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 39.Kuranda M J, Robbins P W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991;266:19758–19767. [PubMed] [Google Scholar]
- 40.Lagrange T, Kim T-K, Orphanides G, Ebright Y W, Ebright R H, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photo-crosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee M, Struhl K. A severely defective TATA-binding protein-TFIIB interaction does not preclude transcriptional activation in vivo. Mol Cell Biol. 1997;17:1336–1345. doi: 10.1128/mcb.17.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 43.Lieberman P M, Ozer J, Gursel D B. Requirement for TFIIA-TFIID recruitment by an activator depends on promoter structure and template competition. Mol Cell Biol. 1997;17:6624–6632. doi: 10.1128/mcb.17.11.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lieberman P M, Schmidt M C, Kao C C, Berk A J. Two distinct domains in the yeast transcription factor IID and evidence for a TATA box-induced conformational change. Mol Cell Biol. 1991;11:63–74. doi: 10.1128/mcb.11.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y S, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 46.Losson R, Fuchs R P, Lacroute F. Yeast promoters URA1 and URA3. Examples of positive control. J Mol Biol. 1985;185:65–81. doi: 10.1016/0022-2836(85)90183-4. [DOI] [PubMed] [Google Scholar]
- 47.Ma D, Olave I, Merino A, Reinberg D. Separation of the transcriptional coactivator and antirepression functions of transcription factor IIA. Proc Natl Acad Sci USA. 1996;93:6583–6588. doi: 10.1073/pnas.93.13.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madison J M, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahadevan S, Struhl K. TC, an unusual promoter element required for constitutive transcription of the yeast HIS3 gene. Mol Cell Biol. 1990;10:4447–4455. doi: 10.1128/mcb.10.9.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martens J A, Brand C J. GCN4p activation of the yeast TRP3 gene is enhanced by ABF1p and uses a suboptimal TATA element. J Biol Chem. 1994;269:15661–15667. [PubMed] [Google Scholar]
- 51.Martinez E, Zhou Q, L’Etoile N D, Oelgeschlager T, Berk A J, Roeder R G. Core promoter-specific function of a mutant transcription factor TFIID defective in TATA-box binding. Proc Natl Acad Sci USA. 1995;92:11864–11868. doi: 10.1073/pnas.92.25.11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meisterernst M, Roeder R G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 53.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 54.Moqdateri Z, Poon D, Bai Y, Weil P A, Struhl K. TBP-associated factors are not generally required for transcription activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 55.Oelgeschlager T, Chiang C M, Roeder R G. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- 56.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 57.Ozer J, Bolden A H, Lieberman P M. Transcription factor IIA mutations show activator-specific defects and reveal a IIA function distinct from stimulation of TBP-DNA binding. J Biol Chem. 1996;271:11182–11190. doi: 10.1074/jbc.271.19.11182. [DOI] [PubMed] [Google Scholar]
- 58.Ozer J, Moore P A, Bolden A H, Lee A, Rosen C A, Lieberman P M. Molecular cloning of the small (gamma) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 58a.Ozer, J., and P. M. Lieberman. Unpublished data.
- 59.Ranish J A, Lane W S, Hahn S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- 60.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 61.Rose M, Botstein D. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- 62.Rose M, Winston R, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 63.Sakai A, Shimizu Y, Kondou S, Chibazakura T, Hishinuma F. Structure and molecular analysis of RGR1, a gene required for glucose repression of Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:4130–4138. doi: 10.1128/mcb.10.8.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakurai H, Ohishi T, Fukasawa T. Two alternative pathways of transcription initiation in the yeast negative regulatory gene GAL80. Mol Cell Biol. 1994;14:6819–6828. doi: 10.1128/mcb.14.10.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen W-C, Green M C. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 66.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stargell L, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 68.Stargell L A, Struhl K. The TBP-TFIIA interaction in response to acidic activators in vivo. Science. 1995;269:75–78. doi: 10.1126/science.7604282. [DOI] [PubMed] [Google Scholar]
- 69.Stargell L A, Struhl K. A new class of activation-defective TATA binding protein mutants: evidence for two steps of transcriptional activation in vivo. Mol Cell Biol. 1996;16:4456–4464. doi: 10.1128/mcb.16.8.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986;6:3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Struhl K. Yeast transcriptional regulatory mechanisms. Annu Rev Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 72.Sun X, Ma D, Sheldon M, Yeung K, Reinberg D. Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev. 1994;8:2336–2348. doi: 10.1101/gad.8.19.2336. [DOI] [PubMed] [Google Scholar]
- 73.Tan S, Hunziker Y, Sargent D F, Richmond T J. Crystal structure of a yeast TFIIA/TBP/DNA complex. Nature. 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 74.Tyree C M, George C P, Lira-DeVito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J T. Identification of a minimal set of proteins that is sufficient for accurate transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 75.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 76.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 77.Walker S S, Shen W-C, Reese J C, Apone L M, Green M R. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 78.Yokomori K, Zeidler M P, Chen J L, Verrijzer C P, Mlodzik M, Tjian R. Drosophila TFIIA directs cooperative DNA binding with TBP and mediates transcriptional activation. Genes Dev. 1994;8:2313–2323. doi: 10.1101/gad.8.19.2313. [DOI] [PubMed] [Google Scholar]
- 79.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]