Extract
Cannabis use has been controversial, largely having been designated a controlled substance over the past century. While certain studies have linked cannabis smoking with harmful effects such as increased respiratory symptoms and faster lung function decline in older adults [1, 2], these findings have not been fully replicated by others [3]. The link between cannabis and disease pathogenesis may best be explored through DNA methylation. This mechanism consists of the addition or removal of a methyl group at a cytosine–guanine residue (CpG), can be influenced by exposures, and can modify transcription. Methylation changes can accumulate over time in patterns that are highly associated with age, leading to the development of epigenetic clocks that can estimate biological age [4].
Shareable abstract
Current cannabis smoking is significantly associated with faster peripheral blood epigenetic age acceleration; interestingly, cannabis smoking cessation is shown to normalise this age acceleration signal. https://bit.ly/3x7s2CU
To the Editor:
Cannabis use has been controversial, largely having been designated a controlled substance over the past century. While certain studies have linked cannabis smoking with harmful effects such as increased respiratory symptoms and faster lung function decline in older adults [1, 2], these findings have not been fully replicated by others [3]. The link between cannabis and disease pathogenesis may best be explored through DNA methylation. This mechanism consists of the addition or removal of a methyl group at a cytosine–guanine residue (CpG), can be influenced by exposures, and can modify transcription. Methylation changes can accumulate over time in patterns that are highly associated with age, leading to the development of epigenetic clocks that can estimate biological age [4].
We investigated the relationship between epigenetic age and cannabis smoking in participants within the Canadian Cohort of Obstructive Lung Disease (CanCOLD) cohort (n=93) (NCT00920348) [5]. A full description of the assessment for cannabis smoking status has been previously reported [2]. Blood samples were collected at the baseline study visit. These samples were profiled for DNA methylation using the Illumina MethylationEPIC BeadChipv1 at two separate laboratories (subset 1: n=34; subset 2: n=59), and randomised based on sex and age (subset 1), and age and cigarette smoking status (subset 2). Raw data were processed separately using filtering, quality control and normalisation steps, according to previously described methods [6]. The XY CpGs were kept as recommended by the Clock Foundation guidelines (https://dnamage.clockfoundation.org) and background correction and normalisation were applied to both subsets. No batch effects based on a singular value decomposition analysis were detected for subset 1, while subset 2 demonstrated slide and array batch effects, therefore batch correction was applied to subset 2.
The epigenetic age of each sample was calculated using the Clock Foundation tool (https://dnamage.clockfoundation.org), which estimates the following clocks: DNAmAge [4], DNAmSkinBlood [7], DNAmAgeHannum [8], DNAmGrimAge [9] and DNAmPhenoAge [10]. Briefly, DNAmAge, DNAmAgeHannum and DNAmAgeSkinBlood were trained solely on chronological age. DNAmGrimAge was derived from CpG sites correlated with chronological age, inflammatory proteins previously linked with mortality, and cigarette smoking [9]. DNAmPhenoAge [10] was derived from CpG sites correlated with clinical biomarkers and chronological age. A positive epigenetic age residual resulting from the regression of epigenetic age on chronological age indicates faster age acceleration. ANOVA was used to identify differences in the age acceleration residuals associated with cannabis smoking status (never, former and current), adjusted for chronological age, sex, body mass index (BMI), batch, cigarette smoking status, and the first two principal components of blood cell proportions (CD8 T-cells, CD4 T-cells, natural killer cells, B-cells, monocytes and granulocytes). Since predictors of DNAmPhenoAge include white blood cells, the model used for this clock did not include the principal components of blood cell proportions. We further tested the effect of lung function on the associations by repeating these analyses with adjustment for forced expiratory volume in 1 s (FEV1) to forced vital capacity (FVC) ratio. We also repeated the analyses to test the effect of years of cannabis smoking on the association between epigenetic age and former and current smoking status. Significance was set at a Tukey honestly significant difference-adjusted p<0.05.
Participants were divided into never (n=51), former (n=32) and current (n=10) cannabis smoking groups that were aged 55±7 years, 56±7 years and 51±6 years, respectively (Kruskal–Wallis p=0.255). 82%, 78% and 40% of the never, former and current cannabis smoking groups were females, respectively (Fisher p=0.022). There were no significant differences in BMI between the groups (Kruskal–Wallis p=0.364). 25%, 59% and 40% of the never, former and current cannabis smokers, respectively, were also current cigarette smokers (Kruskal–Wallis p<0.001). In the never and former smoking groups, ∼22% of participants were physician-diagnosed with COPD, while 30% of the current smoking group had COPD (Fisher p=0.822). COPD severity as classified by Global Initiative for Chronic Obstructive Lung Disease (GOLD) stage was as follows: seven GOLD stage 1 (FEV1 ≥80% predicted), 11 GOLD stage 2 (FEV1 50–79% predicted) and three GOLD stage 3 (FEV1 30–49% predicted). ∼40% of the participants in each group reported a history of asthma (Fisher p≈1). Out of the total cohort, 50% of the participants had either COPD (n=10), asthma (n=26) or both (n=11). Overall, there were no significant differences in FEV1/FVC (66.08±9.77%) between the three groups (Kruskal–Wallis p=0.676). The former cannabis smoking groups reported mean 6.42±13.38 joint-years, while the current cannabis smoking group reported mean 39.57±56.68 joint-years (Kruskal–Wallis p=0.10). Overall, our study cohort had a similar proportion of participants with COPD (Fisher p=0.210) and similar BMI (Kruskal–Wallis p=0.730) compared to the full CanCOLD cohort. However, our study cohort was characterised by lower post-bronchodilator FEV1 % predicted (Kruskal–Wallis p=3.44×10−05), younger chronological age (Kruskal–Wallis p<1.76×10−28), higher proportion of females (Fisher p=7.30×10−10), and higher proportion of current smoking (Fisher p=1.51×10−12) compared to the full CanCOLD cohort (n=1561).
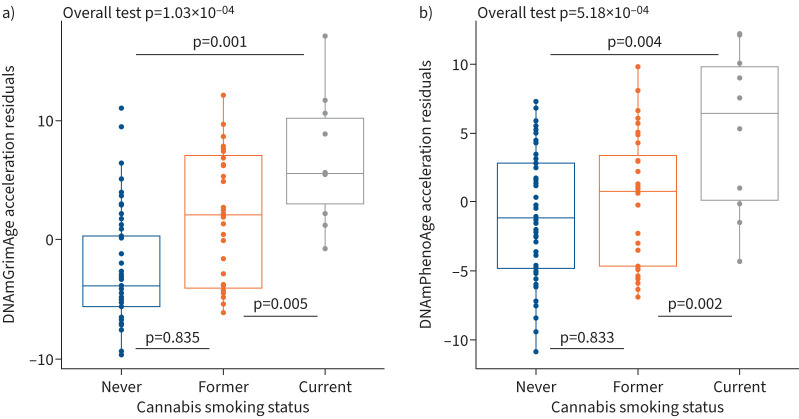
We found that current cannabis smoking was significantly associated with higher DNAmGrimAge and DNAmPhenoAge acceleration (figure 1) when compared to former smoking (Tukey adjusted p=0.005 for DNAmGrimAge and Tukey adjusted p=0.002 for DNAmPhenoAge) and never smoking (Tukey adjusted p=0.001 for DNAmGrimAge and Tukey adjusted p=0.004 for DNAmPhenoAge). There were no significant differences in either DNAmGrimAge or DNAmPhenoAge acceleration between the former and never cannabis smoking groups. No significant associations were identified between cannabis smoking status and DNAmAge, DNAmAgeHannum and DNAmAgeSkinBlood.
FIGURE 1.
Epigenetic age and cannabis smoking status. Current cannabis smoking status was associated with greater epigenetic age residuals compared to former and never cannabis smoking status. There were no significant differences in either a) DNAmGrimAge or b) DNAmPhenoAge between the former and never smoking groups. Boxes represent the interquartile range; the horizontal line represents the median value of DNAmGrimAge and DNAmPhenoAge residuals. Tukey adjusted p-values correspond to ANOVA tests that were adjusted for age, sex, body mass index, cigarette smoking status, laboratory (batch effect) and cell proportions, and are provided at the top of each panel.
After adjusting for FEV1/FVC ratio, the current cannabis smoking group still had significantly increased DNAmGrimAge (current versus former, p=0.017, and current versus never, p=0.001) and DNAmPhenoAge (current versus former, p=0.006, and current versus never, p=0.005). Results remained similar after adjusting for number of years of cannabis smoking when comparing current to former smoking (DNAmGrimAge, p=0.037, and DNAmPhenoAge, p=0.002). The number of years of cannabis smoking had a significant effect on the epigenetic age of former smokers (for every year of cannabis smoking, there was an estimated increase of 0.12 years in DNAmGrimAge, p=0.010). The sample size within the current smoking group did not allow us to test this effect in these participants. The severity of smoking habit as quantified by joint-years was also associated with increased DNAmGrimAge, where for every unit of joint-year smoked there was a 0.092±0.034 year increase in DNAmGrimAge (p=0.018) after adjusting for sex, age, BMI, cigarette smoking status, batch, cannabis smoking status and cell proportions.
In comparison to cannabis smoking, cigarette smoking had a more significant impact on epigenetic ageing. Current and former cigarette smoking were both associated with increased DNAmGrimAge (current versus never, Tukey adjusted p=6.54×10−11, former versus never, Tukey adjusted p=6.24×10−09). The effect size of these increases was greater than that found in cannabis smoking. The mean differences in age acceleration based on DNAmGrimAge between the current and never cigarette smoking group was 8.86 years, while the difference between current and never cannabis smoking was 4.19 years. We further found that only current cigarette smoking was associated with increased DNAmPhenoAge compared to never smoking (Tukey adjusted p=2.96×10−05).
Our observations indicate that current cannabis smoking and higher joint-years exposure are associated with epigenetic age acceleration; cessation, however, may help to normalise in part this age acceleration. DNAmGrimAge and DNAmPhenoAge were able to capture peripheral blood age acceleration in cannabis smoking in a manner that was not reflected in the other available clocks. Previously, we have shown that blood DNAmGrimAge is associated with risk of mortality [11] and demonstrated strong performance in reflecting the epigenetic age of the airway, with a correlation of 0.93 (p=9.86×10−19) between blood and airway epithelium, likely due to the inclusion of smoking pack-years in its training algorithm [12]. We speculate that the insult that cannabis smoking may cause in the airway was more easily exhibited in peripheral blood DNAmGrimAge. We also note that the inclusion of inflammatory biomarkers in the training algorithms of both DNAmGrimAge and DNAmPhenoAge could also account for their significant associations with cannabis smoking. The links between inflammation and cannabis exposure remain somewhat conflicting. On the one hand, cannabinoids have been linked to reductions in tumour necrosis factor-α, interleukin (IL)-6, and IL-1β after exposure [13]. Users of cannabis (not restricted to smoking as method of use), on the other hand, had 54% higher levels of IL-1β compared to non-users [14]. Our finding that DNAmGrimAge is associated with cumulative exposure to cannabis is consistent with previous studies in younger populations [15]. Here, we have expanded upon their work by showing that cessation could reverse the effects of cannabis on systemic ageing as measured by the epigenetic clock. Nonetheless, the time between cessation and blood collection as well as differences in the overall cumulative exposure to cannabis could be important variables influencing our results.
Our study had several limitations. First, our small sample may not reflect all individuals who smoke cannabis. Questions remain regarding how the relationship between cannabis smoking and ageing is altered in patients with underlying lung diseases and whether sex/gender also modulate this relationship. Second, concurrent drug and alcohol use could contribute to age acceleration, and therefore future work is needed to understand the interaction between these habits. Third, we were not able to test epigenetic age longitudinally in individuals prior to and after cannabis smoking cessation, therefore our findings remain associative. However, our findings should give the growing population of people who smoke cannabis some degree of pause.
Shareable PDF
Footnotes
Ethics statement: The research ethics board for each participating institution approved the study protocol.
Conflicts of interest: D.D. Sin has received a stipend for giving talks on COPD from AstraZeneca, GlaxoSmithKline and Boehringer Ingelheim, and is deputy chief editor of the European Respiratory Journal. The remaining authors have no potential conflicts of interest to disclose.
Support statement: The Canadian Cohort Obstructive Lung Disease (CanCOLD; NCT00920348) study is currently funded by the Canadian Respiratory Research Network and industry partners AstraZeneca Canada Ltd, Boehringer Ingelheim Canada Ltd, GlaxoSmithKline Canada Ltd and Novartis. This work was supported by funding from the Canadian Institutes of Health Research. A.I. Hernandez Cordero is supported by the Michael Smith Health Research BC Trainee Award. M.S. Kobor, D.D. Sin and J.M. Leung are supported by the Canada Research Chairs programme. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Winhusen T, Theobald J, Kaelber DC, et al. . Regular cannabis use, with and without tobacco co-use, is associated with respiratory disease. Drug Alcohol Depend 2019; 204: 107557. doi: 10.1016/j.drugalcdep.2019.107557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan WC, Bourbeau J, Aaron SD, et al. . The effects of marijuana smoking on lung function in older people. Eur Respir J 2019; 54: 1900826. doi: 10.1183/13993003.00826-2019 [DOI] [PubMed] [Google Scholar]
- 3.Barjaktarevic I, Cooper CB, Shing T, et al. . Effect of marijuana smoking on lung function change in older ever tobacco smokers. Eur Respir J 2022; 60: 2201133. doi: 10.1183/13993003.01133-2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013; 14: R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourbeau J, Tan WC, Benedetti A, et al. . Canadian Cohort Obstructive Lung Disease (CanCOLD): Fulfilling the need for longitudinal observational studies in COPD. COPD 2014; 11: 125–132. doi: 10.3109/15412555.2012.665520 [DOI] [PubMed] [Google Scholar]
- 6.Hernandez Cordero AI, Yang CX, Obeidat M, et al. . DNA methylation is associated with airflow obstruction in patients living with HIV. Thorax 2021; 76: 448–455. doi: 10.1136/thoraxjnl-2020-215866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horvath S, Oshima J, Martin GM, et al. . Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging (Albany NY) 2018; 10: 1758–1775. doi: 10.18632/aging.101508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannum G, Guinney J, Zhao L, et al. . Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013; 49: 359–367. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu AT, Quach A, Wilson JG, et al. . DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019; 11: 303–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine ME, Lu AT, Quach A, et al. . An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018; 10: 573–591. doi: 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez Cordero AI, Yang CX, Milne S, et al. . Epigenetic blood biomarkers of ageing and mortality in COPD. Eur Respir J 2021; 58: 2101890. doi: 10.1183/13993003.01890-2021 [DOI] [PubMed] [Google Scholar]
- 12.Hernandez Cordero AI, Yang CX, Li X, et al. . The blood DNA methylation clock GrimAge is a robust surrogate for airway epithelia aging. Biomedicines 2022; 10: 3094. doi: 10.3390/biomedicines10123094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henshaw FR, Dewsbury LS, Lim CK, et al. . The effects of cannabinoids on pro- and anti-inflammatory cytokines: a systematic review of in vivo studies. Cannabis Cannabinoid Res 2021; 6: 177–195. doi: 10.1089/can.2020.0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krsak M, Wada NI, Plankey MW, et al. . Self-reported cannabis use and markers of inflammation in men who have sex with men with and without HIV. Cannabis Cannabinoid Res 2021; 6: 165–173. doi: 10.1089/can.2019.0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nannini DR, Zheng Y, Joyce BT, et al. . Marijuana use and DNA methylation-based biological age in young adults. Clin Epigenetics 2022; 14: 134. doi: 10.1186/s13148-022-01359-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00458-2024.Shareable (430.9KB, pdf)