Abstract
Pancreatic adenocarcinoma (PDAC) is one of the most deadly cancers, characterized by extremely limited therapeutic options and a poor prognosis, as it is often diagnosed during late disease stages. Innovative and selective treatments are urgently needed, since current therapies have limited efficacy and significant side effects. Through proteomics analysis of extracellular vesicles, we discovered an imbalanced distribution of amino acids secreted by PDAC tumor cells. Our findings revealed that PDAC cells preferentially excrete proteins with certain preferential amino acids, including isoleucine and histidine, via extracellular vesicles. These amino acids are associated with disease progression and can be targeted to elicit selective toxicity to PDAC tumor cells. Both in vitro and in vivo experiments demonstrated that supplementation with these specific amino acids effectively eradicated PDAC cells. Mechanistically, we also identified XRN1 as a potential target for these amino acids. The high selectivity of this treatment method allows for specific targeting of tumor metabolism with very low toxicity to normal tissues. Furthermore, we found this treatment approach is easy-to-administer and with sustained tumor-killing effects. Together, our findings reveal that exocytosed amino acids may serve as therapeutic targets for designing treatments of intractable PDAC and potentially offer alternative treatments for other types of cancers.
Graphical abstract
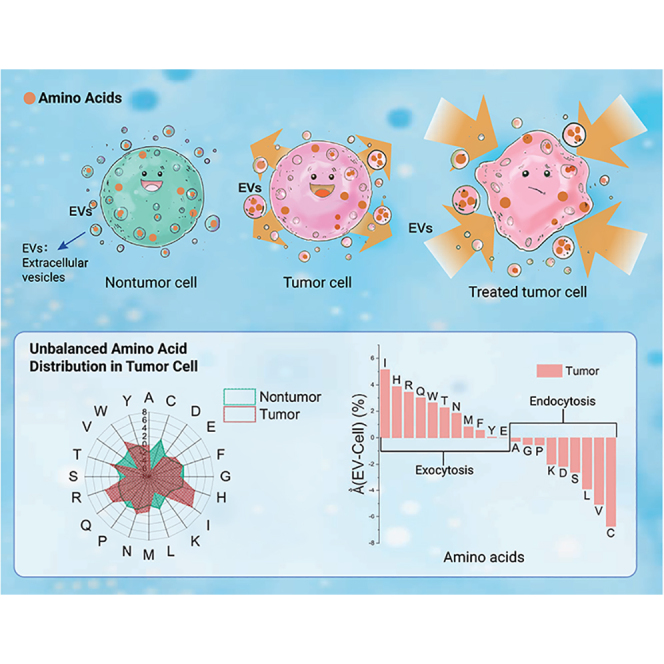
Public summary
-
•
Tumor cells exocytose amino acids via extracellular vesicles, which act as stressors to tumor cells.
-
•
Novel non-starvation approach using amino acids for treating pancreatic cancer.
-
•
Amino acid treatment is selective and effective against PDAC cells with no side effects.
-
•
The new treatment can be used as an adjuvant with chemotherapy to enhance efficacy.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death in the United States and the eighth leading cause worldwide, with an increasing incidence.1,2,3 Treatment options for pancreatic cancer are limited to surgery, chemotherapy, radiation therapy, and palliative care.4 Although surgical resection can potentially ameliorate early-stage disease, more than 80% of patients who present with locally advanced or metastatic disease at the time of diagnosis are ineligible for resection.4 Chemoradiation and systemic chemotherapy are the mainstays of treatment to slow disease progression for PDAC disease at advanced stages.5 However, these therapies attack all rapidly dividing cells, including healthy cells, and thus cause significant side effects and toxicity.6 The lack of selective protection for healthy cells has limited the effectiveness of current therapies. These challenges underscore the need for innovative and more effective treatments to achieve breakthroughs in pancreatic cancer treatment.
Recent research has identified the crucial role of extracellular vesicles (EVs) in cell-cell signaling and selective transfer of cellular information for tumor development.7,8,9,10,11 These membrane vesicles of endocytic origin, with sizes ranging from 30 to 150 nm, contain biomolecules, such as DNA, RNA, and proteins,7 which migrate between cells. EVs have been found to play a critical role in promoting the cascade of cell signaling events that lead to tumor growth, invasion, and metastasis. Studies have shown that the cargoes carried by EVs, especially proteins, and DNA (e.g., secondary protein structures, excessive mtDNA), have an uneven distribution in tumor-derived EVs, highlighting their potential as novel biomarkers for noninvasive cancer detection.12,13,14,15 Collectively, these findings suggest that EVs could be a promising information source for developing markedly improved therapies for PDAC. By utilizing EV-based approaches, we may overcome the current limitations associated with finding potential therapies for PDAC, which have been linked to the high aggressiveness of the disease and related cascade of cell signaling events that promote tumor growth, invasion, and metastasis.16
In this study, we conducted proteomics analysis of EVs to quantify the individual amino acid abundance. We linked the proteins with their primary sequences17 for statistical analysis of the relative abundance of the individual amino acids that make the protein. We compared the individual amino acid abundance between proteins of PDAC tumors and nontumor cells at both EV and cellular levels. Using this novel approach, we found a significant imbalance in the amino acid distribution between PDAC tumor cells and EVs, but not in nonmalignant counterparts. This suggests that the tumor cells selectively exocytose certain amino acids through EVs. Inspired by this EV information, we developed a new treatment that stresses PDAC tumors with amino acids of high exocytosis selectivity. We demonstrate for the first time that combined treatment of amino acids identified from selective exocytosis of PDAC cells via EVs, mediates necrosis of PDAC cells in vitro and in vivo. Moreover, we identified the potential therapeutic target of the combined amino acid treatment to be the XRN1. XRN1 is a 3′-5′ exoribonuclease that is involved in the cytoplasmic mRNA decay and stability in cellular processes critical to some disease development, such as osteosarcoma and Wilms’ tumor.18 Previously, it was reported that XRN1 silencing in melanoma cells suppressed RNA decay and stimulated antitumor immunity.19 Our studies suggest that targeting the XRN1 gene with amino acids may be crucial for the selective treatment of PDAC tumors by affecting the RNA degradation pathway.
In summary, our study presents an amino acid-based treatment of PDAC tumors, which was developed using innovative proteomics analysis that accurately quantifies individual amino acids in PDAC cell-derived EVs compared with those from normal cells. The combined amino acid treatment used in this study demonstrates that a non-starvation treatment approach is selective for killing PDAC tumors with little or no side effects on nontumor cells. These novel findings may establish a paradigm for using EV information in drug discovery, contribute to an improved understanding of the molecular mechanism underlying PDAC development, provide the basis for developing selective and effective therapeutic interventions against PDAC, and offer translational advantages for clinical applications.
Results
Selective exocytosis of amino acids in PDAC cell-derived EVs is discovered via quantitative proteomics profiling
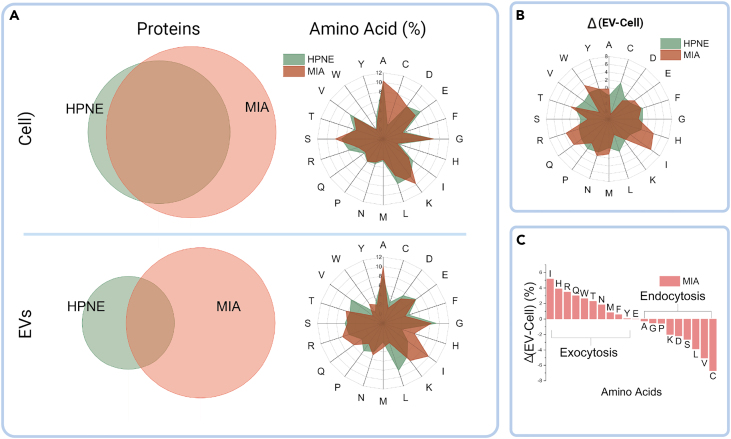
Modern mass spectrometry-based protein profiling provides not only individual identification but also quantitative information on each protein in a given sample. This has facilitated comparative proteomics to identify and quantify proteins in cells from tumor and nontumor tissues at both the cellular and EV levels. We performed comparative proteomics studies of both cells and EVs of PDAC and immortalized nontumor human ductal epithelial cells, respectively.20 We evaluated the proteins by determining the relative abundances of each identified protein within tumor and nontumor cells and EVs. We observed that the percentage of the proteins shared by tumor and nontumor tissue cells was significantly higher at the cellular level than at the EV level (Figure 1A), indicating possible selective exocytosis in EV shedding. Previous studies have shown that protein transport complexes are involved in sorting proteins and other biomolecules in EVs before shedding occurs.21 However, the subsequent sorted protein distributions specific to PDAC cells have not been evaluated comprehensively.21,22 Therefore, we investigated the differential protein distribution down to the single amino acid level17 for the first time in PDAC cells.
Figure 1.
PDAC cells selectively exocytose certain amino acids through EVs
(A) Proteomic analysis of cellular and EV level amino acid distribution.
(B) Percentage difference (Δ) of amino acids between cells and EVs.
(C) Exocytosis selectivity ranking of amino acids (Δ) in tumor cells. Single letter representation of the amino acids. EVs, extracellular vesicles; HPNE, hTert-HPNE (nonmalignant pancreas epithelial); MIA, MIA PaCa-2 (malignant pancreas epithelial).
To further evaluate our proteomics results, we developed a method to study the statistical distribution of the amino acids. We utilized the accession IDs of identified proteins to retrieve their primary amino acid sequences from the Uniprot database (https://www.uniprot.org/). By doing so, we were able to perform a detailed analysis of the amino acid content of each protein identified in our study. We defined the resulting percentage of ith amino acid in the sample as: , where is the amino acid percentage calculated by the primary sequence in the protein; is the relative abundance of the protein in the sample; is the number of the proteins identified. Evaluating the amino acid distribution at the cellular and EV level, we found that, at the cellular level, the distributions in the tumor and nontumor tissue cells are analogous except for cysteine (Figure 1A). However, the distribution varied from tumor to nontumor cells at the EV level, which implied selective exocytosis of amino acids through tumor EVs. To elucidate the selectivity, the ith amino acid percentage difference between EV and cellular level is defined as , where and are the amino acid percentage at EV and cellular levels, respectively. We summarized the selectivity (Δ) of amino acids in tumor and nontumor cells (Figure 1B). The nontumor cells featured homogeneous selectivity over all amino acids in a round profile in Figure 1B, but tumor cells present imbalanced selectivity, which implicates selective exocytosis of specific amino acids in tumor cells through EVs. We ranked the amino acids by their value in the tumor cells revealing that amino acids with positive values (PSA) may undergo selective exocytosis processes (Figure 1C), indicating the metabolic disparity of the amino acids in tumor cells. Figure 1C demonstrates that isoleucine, histidine, and arginine, are the top 3 amino acids in terms of exocytosis, while cysteine, valine, and leucine are the top in terms of endocytosis, as ranked by our analysis.
Metabolic reprogramming is a hallmark of cancer, and PDAC tumors display distinct metabolic alterations due to the activation of KRAS mutation to meet the growth demands in relatively hypoxic and nutrient-poor niches.23,24,25 Approximately 90% of PDAC tumors exhibit KRAS mutations, and KRAS-induced metabolic alterations rely on amino acids to provide the carbon and nitrogen sources to meet the energy requirements for PDAC growth and survival.26 For instance, KRAS-mediated alteration in protein expressions, such as repression of glutamate dehydrogenase (GDH) and upregulation of cytoplasmic aspartate transaminase (GOT1), impacts glutamine metabolism and inhibits tumor suppression while favoring tumor growth and proliferation, respectively.16,27 Moreover, PDAC can uptake extracellular proteins through micropinocytosis and lysosomal degradation to provide the necessary amino acids as an alternative nutrient source.16 These dynamic changes in the regulation of specific proteins for the facilitation of essential amino acids that contribute to PDAC survival substantiate our results for the uneven distribution of amino acid contents in tumor and nontumor cells. Consequently, we raised whether the amino acids undergoing selective exocytosis could be stressors to tumor cells when reintroduced. Based on our results, we utilized these selective exocytosis amino acids (positive Δ values, PSA) as stressors in tumor cells in a non-starvation manner, as illustrated schematically in the graphics abstract.
Selective exocytosed amino acids are stressors to PDAC cells
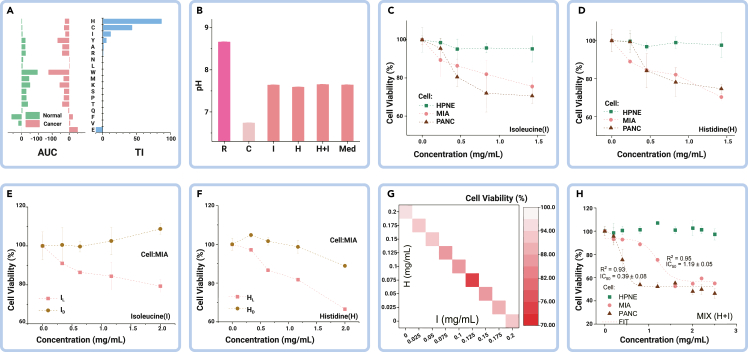
To address the question raised earlier, we initially identified the individual amino acids exocytosed through tumor EVs (as shown in Figure 1C) and evaluated their cytotoxic effects (Figure S1). The area under the curve (AUC) of each amino acid for cytotoxicity was measured, and the therapeutic index was calculated as the cytotoxicity (AUC) difference between nontumor and tumor cells (Figure 2A). The amino acids were then ranked by therapeutic index, and among the top-ranked amino acids, cysteine and arginine introduce significant pH change (Figure 2B), and cysteine exhibited selective endocytosis (Figure 1C), making them unsuitable for non-starvation treatment. Based on the results, histidine and isoleucine were chosen as the finalists for amino acid treatment. The tumor cells demonstrated a monotonic dose response to both amino acids. The effect of histidine and isoleucine treatment on nontumor cells showed no significant difference in cell viability, whereas treated tumor cells exhibited selective cytotoxic efficacy (Figures 2C and 2D). This highlights the potential of amino acid treatment with selective efficacy in tumor cells, which may result in fewer adverse drug reactions (ADRs) or side effects in nontumor cells.
Figure 2.
Histidine and isoleucine combination treatment is cytotoxic to PDAC cells with low side effects
(A) Area under the curve (AUC) and therapeutic index (TI) from amino acid cytotoxicity tests.
(B) pH change caused by amino acid addition with histidine (H) and isoleucine (I) exhibiting trivial impact to the medium.
(C and D) Viability dose response of individual H and I in the same cell lineage.
(E and F) Comparison of cell viability dose response between L and D form amino acids.
(G) Synergetic efficacy of combining H and I treatment on tumor cells.
(H) Cytotoxicity of H and I combined treatment. R, arginine; C, cysteine; Med, medium only; HPNE, hTert-HPNE (nonmalignant pancreas epithelial); MIA, MIA PaCa-2; and PANC, PANC-1 (malignant pancreas epithelial).
In addition, our proteomics results revealed an imbalance in the L form amino acids (Figures 1B and 1C), which prompted us to examine the functional efficacy of these amino acids by testing their different isomeric forms. It should be noted that amino acids can occur in L and D forms, but only the L forms are absorbable by cells. Our results demonstrated that only the L form of the amino acids had significant effects on tumor cells, indicating their potential therapeutic value (Figures 2E and 2F).
Modern medicine has always taken advantage of the combined use of several active agents to treat diseases, and it has become a standard in cancer treatment. Thus, we studied the synergetic efficacy of combining histidine and isoleucine for cancer treatment and found a remarkable efficacy burst when combining these two amino acids, than each amino acid individually to treat tumor cells (Figure 2G). We thus validated the optimized combination of the two amino acids (histidine and isoleucine), in a mass ratio of 2:1, for treatment to achieve maximal efficacy in vitro, and confirmed their highly selective cytotoxicity to the tumor cells (Figure 2H).
Histidine and isoleucine are essential amino acids that cannot be synthesized by mammalian cells but can be supplied through dietary intake and extracellular protein scavenging.16,27 Genomic alterations by oncogenic transcription factors—c-MYC, HIF2α, and NOTCH—mediate high expression of LAT1 (SLC7A5) protein, which is involved in the intracellular influx and efflux movement of histidine and isoleucine that facilitate cancer development.16,27 This suggests that the combined use of histidine and isoleucine (hereafter referred to as “AA”) may impact the expression of protein transporters and alter the metabolic milieu of tumor cells.
Combined AA treatment induces therapeutic effects and mediates cell death in PDAC
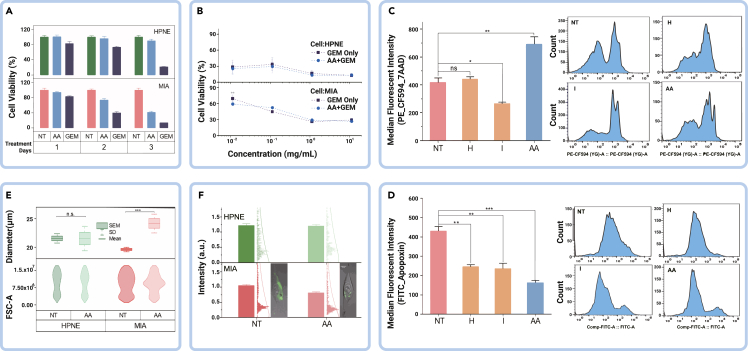
We introduced the non-starvation treatment using the combined amino acids (AA treatment) with selective exocytosis, delivering these amino acids as stressors to the tumor cells. The next question is, how effective is the AA treatment compared with the standard chemotherapy drug—gemcitabine (GEM)?28,29 To compare cytotoxicity efficacy and explore possible mechanisms, we conducted cellular-based assays. Figure 3A shows the time-dependent cytotoxicity of AA treatment compared with GEM. While GEM showed faster efficacy and is more tumoricidally effective, it exhibited a similar time-dependent cytotoxicity feature in nontumor cells, affirming the disadvantage of the standard chemotherapy regimen. On the other hand, the AA treatment demonstrated selective time-dependent cytotoxicity to tumor cells without harm to normal cells, suggesting that it could be used safely for long-term treatment. These findings suggest the potential of AA treatment as an effective and safer therapeutic option for PDAC treatment.
Figure 3.
Synergistic combination of amino acids and gemcitabine enhances cytotoxicity to PDAC cells
(A) Treatment efficacy comparison of AA and GEM. AA, histidine and isoleucine; GEM, gemcitabine.
(B) Adjuvant efficacy of combined AA + GEM treatment on tumor cells.
(C and D) Flow cytometric analyses with 7AAD/Apopxin/CytoCalcein-FITC assay for apoptosis and necrosis on tumor cells in different treatment conditions.
(E) Cell morphology changes upon AA treatment.
(F) Mitochondrial activity shifts due to AA treatment. HPNE, hTert-HPNE (nonmalignant pancreas epithelial); MIA, MIA PaCa-2 (malignant pancreas epithelial); NT, no treatment control; AA, histidine and isoleucine; GEM, gemcitabine. Error bars represent mean ± SEM; n.s., not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Next, we evaluated the synergistic effect of AA and GEM treatment in Figure 3B. No significant synergistic efficacy was observed in nontumor cells, whereas, in tumor cells, low-dose GEM in combination with AA treatment showed enhanced efficacy, suggesting the potential of amino acid supplementation as an adjuvant regimen. This also suggests that amino acid treatment may have a different tumoricidal mechanism from GEM. Histidine plays a crucial role in nucleotide synthesis, which is essential for tumor development. Alterations in the metabolic flux of histidine availability affect the cellular pool of tetrahydrofolate, which influences methotrexate sensitivity to tumor cells.30 Similarly, isoleucine plays a vital role in the modulation of the immune system against cancers by inducing β-defensin.31,32 However, GEM elicits its anticancer effect as a nucleotide analog interfering with DNA synthesis and ribonucleotide reductase.33 The observed enhanced efficacy of the combined AA + GEM treatment may be attributed to its impact on nucleotide (DNA or RNA) metabolic regulation and immune system modulation. These factors prompted us to further determine whether the treatment exerts its effects on tumor cells through apoptosis or necrosis.
To this end, we performed Apopoxin/7-AAD staining and flow cytometry analysis. The results showed that the AA treatment had the greatest therapeutic effect on tumor cells through necrosis, as demonstrated by the cell count and fluorescent intensity (Figures 3C and 3D). Necrosis is characterized by morphological changes leading to plasma membrane rupture in an inflammatory-dependent manner.34 Therefore, we examined the effect of the combined AA treatment on tumor cell morphology compared with nontumor cells. We found significant changes in tumor cell morphology, including increased cell diameter with inflammation, which further supports the occurrence of necrosis (Figure 3E).
Drug-induced necrosis has been associated with mitochondrial dysfunction in many cancers.35 Amino acids play a crucial role as nutrient sources and have a metabolic effect on PDAC. The abnormal increase in mitochondrial activity via the Warburg effect is one of the characteristics responsible for PDAC tumor development and progression.36 Thus, we evaluated the effect of AA treatment on mitochondrial activity in PDAC tumor cells compared with nontumor cells using fluorescent staining (Figure 3F). The results showed a significant decrease in mitochondrial activity in tumor cells due to the treatment, indicating an impact on metabolic activity that is critical to tumor cells. These findings suggest that the AA treatment mediates PDAC cell death through necrosis and implies its effect on metabolic activities crucial to PDAC development. Identifying the metabolic pathway and therapeutic target of the combined AA treatment will provide further insight into its therapeutic effect.
XRN1 is a potential therapeutic target of the AA treatment
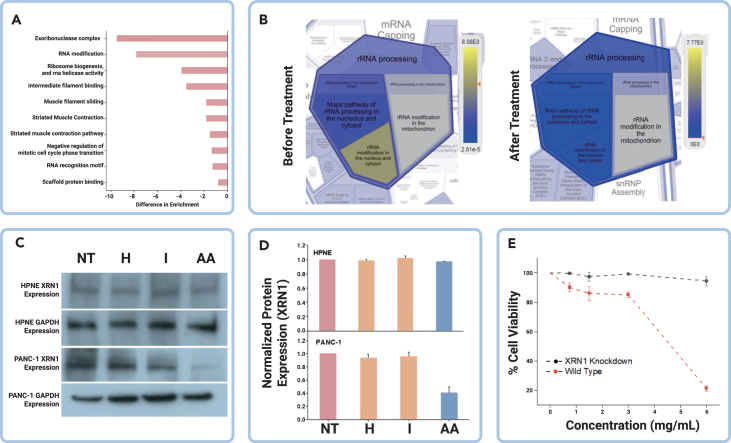
To explore the mechanism of action (MOA) of the AA treatment, we conducted a comparative proteomics analysis before and after treatment application, followed by pathway analysis using the Reactome Knowledgebase tool.37 We utilized four replicates each of treated and untreated cells, and calculated the abundance ratio of untreated and treated cells proteins. The intensity ratio and p value filters was set at fold change >2 for upregulation and <0.2 for downregulation, and significant p < 0.05 respectively, for significantly differentially expressed proteins. Enrichment scores were compared before and after AA treatment in PDAC cells (Figure S2). The pathways were ranked based on the enrichment difference before and after treatment, and the top-ranked amino acids are shown in Figure 4A. We found that the exoribonuclease complex and RNA modification were the least enriched in the treatment group. In addition, a visual comparison (Figure 4B) using Reactome revealed that the RNA metabolic pathway was the most affected, and the XRN1 gene was ranked the highest of all. These results suggest that the treatment may affect the RNA metabolic pathway and perturb downstream post-transcriptional activity that is critical to protein expression in PDAC tumor cells. The XRN1 gene encodes the XRN1 exonuclease protein, which degrades mRNA. Notably, our results may indicate that genetic alterations of the tumor cells and the exocytosis of histidine and isoleucine influence the degradation of mRNA transcripts, modulating the inflammatory response and providing a nutrient-rich milieu in the tumor cells. Therefore, reintroducing histidine and isoleucine through the AA treatment may repress the availability of the XRN1 protein, leading to the accumulation of mRNA transcripts that are toxic to the tumor cells.
Figure 4.
XRN1 is a target of the histidine and isoleucine metabolic pathway
(A) Difference in enrichment analysis before and after AA treatment.
(B) Reactome analysis before and after AA treatment.
(C and D) Western blot expression analysis of XRN1 protein in tumor and nontumor cells with combined AA treatment.
(E) Cytotoxicity effects of AA treatment on XRN1 knockdown cells and wild-type. AA: histidine and isoleucine; HPNE, hTert-HPNE (nonmalignant pancreas epithelial); XRN1, XRN1 gene knockdown in PANC-1 tumor cells; PANC-1, malignant pancreas epithelial and wild-type for the XRN1. Error bars represent mean ± SEM; n.s., not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Furthermore, the repression of the XRN1 gene by the AA treatment would lead to a reduction in the availability of the XRN1 protein. This was confirmed by western blot analysis, which showed reduced expression of the XRN1 protein in the AA-treated group compared with other groups (Figures 4C and 4D). These results nominate XRN1 as a possible MOA for AA treatment. To confirm AA treatment targeting XRN1 for tumoricidal effect, we constructed XRN1 knockdown tumor cells (Figure S3) and compared the cytotoxicity of AA with the wild-type cells. We observed that there were no significant changes in cell viability of the knockdown tumor cells compared with the wild-type cells, which experienced a significant decrease in cell viability (Figure 4E). This evidence supports the idea that XRN1 is the potential therapeutic target of the AA treatment and affects metabolic post-transcriptional activity in the tumor cells.
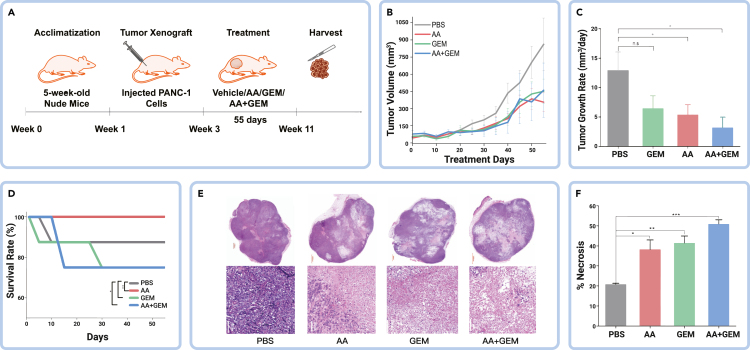
AA treatment effectively inhibits PDAC tumor growth and extends mice survival
To evaluate the therapeutic effect of AA treatment preclinically, we conducted an in vivo study using subcutaneous heterotopic mouse PDAC cancer modeling (Figure 5A) and determined treatment efficacy using statistical assessment throughout the treatment. The treatment was administered orally. As shown in Figure 5B, all treatment groups (GEM, AA, and AA + GEM) showed antitumor efficacy, consistent with the in vitro study (Figure 3A). Although AA + GEM displayed the highest tumor volume change from about 20 days through the course of 40 days of treatment, the antitumor efficacy of the three treatment groups (Figures 5B and 5C) are comparable. Upon comparing the tumor growth rates affected by the treatment, AA + GEM demonstrated the highest antitumor growth effect compared with AA and GEM individually, which is in line with the in vitro study (Figure 3B) and suggests a synergistic efficacy of the AA and GEM treatment. We also assessed the survival rate of the mice and, interestingly, the AA treatment group showed a 100% survival rate (Figure 5D). While the GEM and AA + GEM treatment groups displayed varying survival rates at the beginning of the treatment, they eventually converged at 30 days. This finding further supports the selective toxicity of AA treatment on PDAC tumor cells in vivo. The high survival rate observed in the AA treatment group suggests that it may be a promising first-line therapy, while the synergistic antitumor effect of AA + GEM treatment highlights the potential of AA treatment as adjuvant therapy.
Figure 5.
Amino acid treatment effectively impairs PDAC tumor growth and extends mice survival
(A) Scheme of the in vivo experimental design. GEM, gemcitabine; AA, histidine and isoleucine.
(B and C) Comparison of tumor volume changes among the AA treatment and other treatment conditions.
(D) Comparison of survival rate among the AA treatment and other treatment conditions.
(E) Representative images of H&E staining on xenograft tumor sections for different treatment conditions.
(F) Comparison of percentage necrosis of the AA treatment and other treatment conditions evaluated using histology image analysis. Xenografts: n = 5 for each treatment group. Tumor section image sizes −1,000 and 100 μm. Error bars represent mean ± SEM; n.s., not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To further investigate the pathological efficacy of the treatment, we performed H&E staining of tissue sections from the different treatment groups at the end of the in vivo study. The resulting tumor tissue images were analyzed using the QuPath software.38 Tissue sections from the untreated group exhibited a cellular solid nested pattern with hypercellularity and nuclear pleomorphism (Figure 5E). In contrast, the treated groups displayed varying degrees of necrosis as suggested by cytoplasmic vacuolation, reduced staining intensity, and infiltration of inflammatory cells with lower cellular density. Among the treatment groups, the AA + GEM-treated group showed the most extensive ablation areas characterized by a reduction in tumor cellularity. Thus, the AA + GEM treatment group exhibited the highest level of necrosis (Figures 5E and 5F). These results are consistent with the in vitro study (Figures 3C–3F). Collectively, our studies provide strong evidence of the therapeutic efficacy of the AA treatment for killing PDAC tumor cells, which may serve as alternative or combination therapy for combating pancreatic cancers.
Discussion
Our study uncovered distinct differences in the proteome composition of PDAC tumor-derived EVs compared with nontumor cells.15 Although mass spectrometry is a standard method for evaluating cellular proteins, most analyses end at only protein identification and quantification of unique peptides. In our study, we went one step further to characterize the individual amino acid distribution involved in the proteome of EVs and cells derived from PDAC. Our findings revealed that tumor cells have selective exocytosis of certain amino acids compared with nontumor cells. Despite striking, this is not unexpected, as amino acids play essential roles in providing nutrients, redox balance, energetic regulation, biosynthetic support, and homeostatic maintenance in various cells.39 Moreover, amino acids have demonstrated metabolic reprogramming features to meet the nutritional demands of tumor cells to facilitate its progression dynamics.16 Thus, tumor cells may exocytose specific amino acids to meet their metabolic requirements for growth. This observation inspired us to formulate an idea using amino acid-based treatment, and our results proved that the exocytosed amino acids are stressors for PDAC tumor cells, which can inhibit their growth and progression. This novel approach may open a new avenue for the development of precision medicine based on tracking information from tumor-derived EVs.
Histidine and isoleucine are essential amino acids involved in nucleotide (DNA and RNA) metabolism and immune system modulation, respectively.30,31 Some studies have utilized a starvation approach in amino acid treatment, limiting essential amino acids for disease progression.40,41,42,43 However, the starvation approach is problematic as it alters mitochondria function, which may be detrimental to diseased patients43 and subsequently contributes to the cachexia-anorexia syndrome experienced by most patients with advanced cancer.44 Other studies have attempted the use of histidine supplementation in different cancers,30,45 and isoleucine supplementation in inhibiting tumor growth and angiogenesis in various cancers.32,46,47 However, our study, to the best of our knowledge, is the first non-starvation amino acid treatment approach for pancreatic cancer, combining the use of histidine and isoleucine. This approach has several translational advantages, including selective toxicity against tumor cells, and eliminating severe side effects exhibited by standard chemotherapeutic drugs, such as GEM. In addition, amino acids are among the most accessible nutrients, with no side effects, making them best-suited for counteracting malnutrition and cachexia in advanced-stage cancer patients, thus predisposing them to more effective treatment. Furthermore, amino acids can be applied as adjuvant therapy for various diseases. Our study demonstrated that the amino acid treatment can be used as an adjuvant therapy in combination with GEM, providing better efficacy in treating PDAC.
XRN1 protein, implicated as the potential therapeutic target, is a highly conserved 5′-3′ exoribonuclease that regulates gene expression in eukaryotes by coupling nuclear DNA transcription to cytosolic mRNA decay.48 XRN1 is involved in transcription, mRNA translation, and mRNA decay, and its modification plays a crucial role in post-transcriptional and translational activities that promote tumor development and chemoresistance.18,48,49 Therefore, changes in the availability of the XRN1 protein and alterations in mRNA metabolism can lead to anomalous mRNA accumulation in PDAC tumor cells, which may trigger inflammatory responses and/or necrosis.19,48 Thus, we consider XRN1 protein as the potential therapeutic target of AA treatment, complementary to the targets for GEM. This discovery highlights a novel approach for non-ADR treatment targeting mRNA translation and decay.
Although our study has yielded promising results, it is important to acknowledge certain limitations. In proteomics analysis, some genes may produce various forms of proteins (proteoforms) that may affect the overall amino acid estimation. We preliminarily looked on the necrosis-based tumoricidal effect for the treatment in vitro, but further studies are necessary to confirm the detailed MOA and evaluate the pharmacodynamics/pharmacokinetics of the AA treatment in vivo. In addition, even though we observed promising results of the treatment effect on tumor cell invasion and migration in vitro (Figure S4), we noted that the use of heterotopic subcutaneous mouse models for preclinical validation may not fully recapitulate the complexity of human tumorigenesis, particularly the interactions between tumor cells and the surrounding microenvironment. Therefore, we will need to conduct engineered orthotopic mouse models that better mimic human pancreatic tumorigenesis for a more comprehensive evaluation of the treatment, including its impact on metastasis. Moreover, we opted for oral administration in our study due to the likelihood that intratumoral administration to the subcutaneous mouse model would behave similarly to our in vitro study, and the potential risk of unexpected death associated with intravenous and intraperitoneal injections. Oral intake also highlights the potential for diet modification as a preventive intervention, but further studies are required to explore its potential as a therapeutic method. Despite being preliminary, our study serves as the first indication of the potential of AA treatment and the use of EVs as a source of information for guiding personalized precision medicine.
In summary, this study demonstrates that PDAC tumor cells exocytose a unique group of amino acids via EVs, which act as stressors to the tumor cells. Our non-starvation isoleucine and histidine diet feeding represents a promising strategy for direct therapy and/or combination therapies with GEM for the deadly pancreatic duct carcinoma. Mechanistically, XRN1 is characterized as the target of this amino acid-mediated RNA regulation pathways. Through different functional biochemistry, this AA treatment may have the potential for clinical application as direct or alternative therapy.
Materials and methods
See supplemental information for details.
Data and code availability
All data are available in the main text or supplemental information. Proteomics data are available via ProteomeXchange with ID PXD047958.
Acknowledgments
The authors acknowledge North Dakota State University Center for Computationally Assisted Science and Technology for computing resources. This work was financially supported by grants from the National Cancer Institute (R21CA270748, R03CA252783) and the National Institute of General Medical Sciences (U54GM128729) of National Institutes of Health to D.S., NDSU EPSCoR STEM Research and Education fund (FAR0032086) to D.S., ND EPSCoR: Advancing Science Excellence in ND (FAR0030554) to D.S., and National Science Foundation (NSF) under NSF EPSCoR Track-1 Cooperative Agreement (OIA no. 1355466) to D.S., the National Institue of General Medical Sciences (P20GM109036) to J.F., NSF under NSF OIA ND-ACES (award no. 1946202) to W.X., and NDSU Foundation and Alumni Association to D.S.
Author contributions
D.S. conceived the idea, obtained financial support, and supervised the project. A.A. (Akinlalu), Z.F., and D.S. wrote the manuscript. K.R., S.M.M., A.B., S.E., and N.E. assisted with in vitro and in vivo experiments. S.M. and J.F. conducted quantitative proteomics, and Z.F. and A.A. (Arshad) helped with the analysis. D.S., W.X., J.F., A.G., M.W., and S.M. designed the experiments, validated the data, and edited the manuscript. D.S. approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published Online: April 9, 2024
Footnotes
It can be found online at https://doi.org/10.1016/j.xinn.2024.100626.
Supplemental information
References
- 1.Lockhart A.C., Rothenberg M.L., Berlin J.D. Treatment for pancreatic cancer: Current therapy and continued progress. Gastroenterology. 2005;128(6):1642–1654. doi: 10.1053/j.gastro.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 2.McGuigan A., Kelly P., Turkington R.C., et al. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018;24(43):4846–4861. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pancreatic Cancer: Statistics | Cancer.Net . American Society of Clinical Oncology; 2019. https://www.cancer.net/cancer-types/pancreatic-cancer/statistics [Google Scholar]
- 4.Kamisawa T., Wood L.D., Itoi T., et al. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 5.Saung M.T., Zheng L. Current standards of chemotherapy for pancreatic cancer. Clin. Therapeut. 2017;39(11):2125–2134. doi: 10.1016/j.clinthera.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallinson C.N., Rake M.O., Cocking J.B., et al. Chemotherapy in pancreatic cancer: Results of a controlled, prospective, randomised, multicentre trial. Br. Med. J. 1980;281(6255):1589–1591. doi: 10.1136/bmj.281.6255.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa-Silva B., Aiello N.M., Ocean A.J., et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17(6):816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu C., Li L., Liu L., et al. Research of exosome in bone metastasis through dual aptamer recognition based entropy-driven amplification. Anal. Biochem. 2022;636 doi: 10.1016/j.ab.2021.114433. [DOI] [PubMed] [Google Scholar]
- 9.Casari I., Emmanouilidi A., Domenichini A., et al. Extracellular vesicles derived from pancreatic cancer cells are enriched in the growth factor Midkine. Adv. Biol. Regul. 2022;83 doi: 10.1016/j.jbior.2021.100857. [DOI] [PubMed] [Google Scholar]
- 10.Arance E., Ramírez V., Rubio-Roldan A., et al. Determination of exosome mitochondrial dna as a biomarker of renal cancer aggressiveness. Cancers. 2021;14(1):199. doi: 10.3390/cancers14010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei X.C., Liu L.J., Zhu F. Exosomes as potential diagnosis and treatment for liver cancer. World J. Gastrointest. Oncol. 2022;14(1):334–347. doi: 10.4251/wjgo.v14.i1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshioka Y., Kosaka N., Konishi Y., et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat. Commun. 2014;5:3591. doi: 10.1038/ncomms4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow A., Zhou W., Liu L., et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-κB. Sci. Rep. 2014;4:5750. doi: 10.1038/srep05750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Melo S.A., Luecke L.B., Kahlert C., et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasuleva K., Elamurugan S., Bauer A., et al. β-sheet richness of the circulating tumor-derived extracellular vesicles for noninvasive pancreatic cancer screening. ACS Sens. 2021;6(12):4489–4498. doi: 10.1021/acssensors.1c02022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu R., Yang J., Ren B., et al. Reprogramming of amino acid metabolism in pancreatic cancer: Recent advances and therapeutic strategies. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.572722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fauchon M., Lagniel G., Aude J.C., et al. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell. 2002;9(4):713–723. doi: 10.1016/s1097-2765(02)00500-2. [DOI] [PubMed] [Google Scholar]
- 18.Pashler A.L., Towler B.P., Jones C.I., et al. The roles of the exoribonucleases DIS3L2 and XRN1 in disease. Biochem. Soc. Trans. 2016;44(5):1377–1384. doi: 10.1042/BST20160107. [DOI] [PubMed] [Google Scholar]
- 19.Ran X.B., Ding L.W., Sun Q.Y., et al. Targeting RNA exonuclease XRN1 Potentiates Efficacy of Cancer Immunotherapy. Cancer Res. 2023;83(6):922–938. doi: 10.1158/0008-5472.CAN-21-3052. [DOI] [PubMed] [Google Scholar]
- 20.Sun D., Zhao Z., Spiegel S., et al. Dye-free spectrophotometric measurement of nucleic acid-to-protein ratio for cell-selective extracellular vesicle discrimination. Biosens. Bioelectron. 2021;179 doi: 10.1016/j.bios.2021.113058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colombo M., Moita C., van Niel G., et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013;126(Pt 24):5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 22.Jones S., Zhang X., Parsons D.W., et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin C., Yang G., Yang J., et al. Metabolism of pancreatic cancer: Paving the way to better anticancer strategies. Mol. Cancer. 2020;19(1):50. doi: 10.1186/s12943-020-01169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biankin A.V., Waddell N., Kassahn K.S., et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamphorst J.J., Nofal M., Commisso C., et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75(3):544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neinast M.D., Jang C., Hui S., et al. Quantitative analysis of the whole-body metabolic fate of branched-chain amino acids. Cell Metabol. 2019;29(2):417–429.e4. doi: 10.1016/j.cmet.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei Z., Liu X., Cheng C., et al. Metabolism of amino acids in cancer. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.603837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conroy T., Bachet J.B., Ayav A., et al. Current standards and new innovative approaches for treatment of pancreatic cancer. Eur. J. Cancer. 2016;57:10–22. doi: 10.1016/j.ejca.2015.12.026. [DOI] [PubMed] [Google Scholar]
- 29.Manji G.A., Olive K.P., Saenger Y.M., et al. Current and emerging therapies in metastatic pancreatic cancer. Clin. Cancer Res. 2017;23(7):1670–1678. doi: 10.1158/1078-0432.CCR-16-2319. [DOI] [PubMed] [Google Scholar]
- 30.Kanarek N., Keys H.R., Cantor J.R., et al. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature. 2018;559(7715):632–636. doi: 10.1038/s41586-018-0316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu C., Mao X., Chen D., et al. Isoleucine plays an important role for maintaining immune function. Curr. Protein Pept. Sci. 2019;20(7):644–651. doi: 10.2174/1389203720666190305163135. [DOI] [PubMed] [Google Scholar]
- 32.Murata K., Moriyama M. Isoleucine, an essential amino acid, prevents liver metastases of colon cancer by antiangiogenesis. Cancer Res. 2007;67(7):3263–3268. doi: 10.1158/0008-5472.CAN-06-3739. [DOI] [PubMed] [Google Scholar]
- 33.Ciccolini J., Serdjebi C., Peters G.J., et al. Pharmacokinetics and pharmacogenetics of Gemcitabine as a mainstay in adult and pediatric oncology: An EORTC-PAMM perspective. Cancer Chemother. Pharmacol. 2016;78(1):1–12. doi: 10.1007/s00280-016-3003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alberts B., Johnson A., Lewis J., et al. Molecular Biology of the Cell. 4th edition. Garland Science; 2002. Programmed Cell Death (Apoptosis) [Google Scholar]
- 35.Bastian A., Thorpe J.E., Disch B.C., et al. A small molecule with anticancer and antimetastatic activities induces rapid mitochondrial-associated necrosis in breast cancer. J. Pharmacol. Exp. Therapeut. 2015;353(2):392–404. doi: 10.1124/jpet.114.220335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vyas S., Zaganjor E., Haigis M.C. Mitochondria and cancer. Cell. 2016;166(3):555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillespie M., Jassal B., Stephan R., et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50(D1):D687–D692. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bankhead P., Loughrey M.B., Fernández J.A., et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieu E.L., Nguyen T., Rhyne S., et al. Amino acids in cancer. Exp. Mol. Med. 2020;52(1):15–30. doi: 10.1038/s12276-020-0375-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fung M.K.L., Chan G.C.F. Drug-induced amino acid deprivation as strategy for cancer therapy. J. Hematol. Oncol. 2017;10(1):144. doi: 10.1186/s13045-017-0509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lind D.S. Arginine and cancer. J. Nutr. 2004;134(10):2837S–2853S. doi: 10.1093/jn/134.10.2837S. [DOI] [PubMed] [Google Scholar]
- 42.Choi Y.K., Park K.G. Targeting glutamine metabolism for cancer treatment. Biomol. Ther. 2018;26(1):19–28. doi: 10.4062/biomolther.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson M.A., Vidoni S., Durigon R., et al. Amino acid starvation has opposite effects on mitochondrial and cytosolic protein synthesis. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0093597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ragni M., Fornelli C., Nisoli E., et al. Amino acids in cancer and cachexia: An integrated view. Cancers. 2022;14(22):5691. doi: 10.3390/cancers14225691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng X., Zhang X., Cheng W., et al. Tumor-specific delivery of histidine-rich glycoprotein suppresses tumor growth and metastasis by anti-angiogenesis and vessel normalization. Curr. Gene Ther. 2014;14(2):75–85. doi: 10.2174/1566523214666140305223912. [DOI] [PubMed] [Google Scholar]
- 46.Wang H., Chen S., Kang W., et al. High dose isoleucine stabilizes nuclear PTEN to suppress the proliferation of lung cancer. Discov. Oncol. 2023;14(1):25. doi: 10.1007/s12672-023-00634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preciado D., Moreno G., Cardona W., et al. Discovery of novel trihybrids based on salicylic acid/isoleucine/N-acylhydrazone: A promising therapeutic opportunity in colorectal cancer. J. Appl. Pharmaceut. Sci. 2022;12(11):10–20. [Google Scholar]
- 48.Blasco-Moreno B., de Campos-Mata L., Böttcher R., et al. The exonuclease Xrn1 activates transcription and translation of mRNAs encoding membrane proteins. Nat. Commun. 2019;10(1):1298. doi: 10.1038/s41467-019-09199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akinlalu A.O., Njoku P.C., Nzekwe C.V., et al. Recent developments in the significant effect of mRNA modification (M6A) in glioblastoma and esophageal cancer. Sci. Afr. 2022;17 doi: 10.1016/j.sciaf.2022.e01347. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or supplemental information. Proteomics data are available via ProteomeXchange with ID PXD047958.